Abstract
Determination of cell fate within the prosensory domain of the developing cochlear duct relies on the temporal and spatial regulation of the bHLH transcription factor Atoh1. Auditory hair cells and supporting cells arise in a wave of differentiation that patterns them into discrete rows mediated by Notch-dependent lateral inhibition. However, the mechanism responsible for selecting sensory cells from within the prosensory competence domain remains poorly understood. We show in mice that rather than being upregulated in rows of cells, Atoh1 is subject to transcriptional activation in groups of prosensory cells, and that highly conserved sites for Hes/Hey repressor binding in the Atoh1 promoter are needed to select the hair cell and supporting cell fate. During perinatal supporting cell transdifferentiation, which is a model of hair cell regeneration, we show that derepression is sufficient to induce Atoh1 expression, suggesting a mechanism for priming the 3′ Atoh1 autoregulatory enhancer needed for hair cell expression.
KEY WORDS: Cochlear development, Organ of Corti, Hair cell regeneration, Transdifferentiation, Atoh1, Hes5, Mouse
Summary: Defining and patterning cell fate in the mouse cochlea depends on integration of developmental signals at highly conserved regions of the Atoh1 promoter.
INTRODUCTION
The auditory sensory epithelium in mammals, the organ of Corti, is composed of a linear array of inner and outer sensory hair cells surrounded by a variety of supporting cells that lie in discrete rows on a basilar membrane within the mature cochlear duct. These cells first differentiate from a postmitotic prosensory domain that forms within the cochlear duct between approximately embryonic day (E) 12.5 and E14.5 in the mouse (Ruben, 1967; Lee et al., 2006). Cells within the prosensory region are competent to form either sensory hair cells or the supporting cells that surround them (Woods et al., 2004; Driver et al., 2013). Patterning of the prosensory domain occurs under the influence of the basic helix-loop-helix (bHLH) transcription factor ATOH1, which is first observed in nascent hair cells near the base of the cochlea and drives differentiation in a wave towards the apex over a period of ∼4 days from E13.5 to E17.5 or later (Bermingham et al., 1999; Lanford et al., 2000; Chen et al., 2002; Woods et al., 2004). In the current model, and by analogy with Drosophila sensory organ precursor selection, once Atoh1 expression is established in nascent hair cells, Notch-mediated lateral inhibition stimulates the activity of the Hairy/Enhancer of Split families of transcriptional repressors (Hes/Hey) in the adjacent prosensory cells (reviewed by Jarman and Groves, 2013). This inhibits further differentiation of prosensory cells to hair cells (Lanford et al., 1999; Kiernan et al., 2005), leading them to differentiate as supporting cells (Woods et al., 2004). Although a number of factors are known to influence Atoh1 expression (reviewed by Jarman and Groves, 2013), primarily through interaction with a previously characterized 3′ enhancer (Helms et al., 2000), the mechanism by which prosensory cells are selected for Atoh1 upregulation has not been identified.
The Hes and Hey families of bHLH transcriptional repressors function throughout development to mediate Notch signaling (Jarriault et al., 1995; Nishimura et al., 1998; Ohtsuka et al., 1999). In the developing nervous system, HES1 and HES5 are important for maintaining the neural progenitor status, and their loss leads to premature cell cycle exit and neuronal differentiation (Kageyama et al., 2000; Nakamura et al., 2000; Ohtsuka et al., 2001). Previous reports suggest that repression is accomplished by the recruitment of co-repressors, such as those encoded by Groucho-related genes (Grg1-4; also known as Tle1-4) in the case of the Hes family, which in turn are able to recruit histone deacetylases (HDACs) to mediate gene inactivation (Fisher et al., 1996; Grbavec and Stifani, 1996; Winkler et al., 2010). In the organ of Corti, Hes/Hey genes are expressed in a complex combinatorial pattern (Doetzlhofer et al., 2009; Tateya et al., 2011) and are needed to maintain supporting cell fate (Zine et al., 2001; Zine and de Ribaupierre, 2002; Hayashi et al., 2008; Li et al., 2008), but the molecular mechanism by which this is accomplished has not been investigated.
The model in which Atoh1 is selectively upregulated in nascent hair cells has recently come into question following reports that Atoh1 may be initially upregulated in progenitors that go on to form not only all the hair cells but also some of the supporting cells (Matei et al., 2005; Yang et al., 2010; Cai et al., 2013; Driver et al., 2013). This suggests a second model in which the final configuration of hair cells does not rely on the positive selection by Atoh1 of specific cells within the prosensory domain, but rather on the complex regulation of repressive mechanisms that shut down Atoh1 expression in those cells that are to become the different supporting cell types. In this paper, we show that this second model is likely to be correct. We identify a set of highly evolutionarily conserved Hes/Hey binding sites in the promoter of the Atoh1 gene that are needed for timely repression of Atoh1 within the cells of the prosensory domain destined to be supporting cells. In functional studies in neural progenitors we show that Hes/Hey factors bind directly to these sites and, in the case of HES5, actively repress Atoh1 expression by recruiting the Groucho co-repressor in a Notch-dependent manner. We show that in FACS-purified supporting cells, inhibition of Notch leads to an increase in histone H3K9 acetylation (H3K9ac) at the Atoh1 promoter, commensurate with increased expression, and that inhibition of acetylation limits Notch-mediated Atoh1 expression during transdifferentiation. We also show that the transcriptional silence of Atoh1 in supporting cells at postnatal day (P) 1, and thus the maintenance of the supporting cell fate, although dependent on active repression by Hes/Hey factors, is reinforced by activator insufficiency and a switch-like behavior of the Atoh1 3′ autoregulatory enhancer. We suggest that derepression of the Atoh1 promoter is sufficient to stimulate Atoh1 expression and thus prime the autoregulatory function of this enhancer during transdifferentiation of supporting cells.
RESULTS
Derepression of Atoh1 is sufficient to induce Atoh1 transcription
In non-mammalian vertebrates, hair cell loss leads to Atoh1 upregulation in surrounding supporting cells and their direct transdifferentiation, followed later by a wave of proliferation and additional transdifferentiation (Roberson et al., 2004; Cafaro et al., 2007; Stone and Cotanche, 2007). By contrast, in mammals, hair cells in the cochlea fail to regenerate after damage (Chardin and Romand, 1995; Brigande and Heller, 2009). However, we and others have shown that blocking Notch signaling in the perinatal mouse organ of Corti leads to transdifferentiation of supporting cells to a sensory hair cell-like state (Yamamoto et al., 2006; Doetzlhofer et al., 2009). To confirm this result, we treated cochlear cultures with and without DAPT (a γ-secretase inhibitor) for 72 h. Supporting cell transdifferentiation was visualized using a well-characterized transgenic mouse line that expresses GFP in hair cells under the control of the Atoh1 3′ autoregulatory enhancer (Atoh1 enhancer/β-globin promoter/GFP), which is sufficient to direct accurate expression during organ of Corti development (although a weak expression in inner phalangeal cells is occasionally noted) (Lumpkin et al., 2003; see below). Inhibition of Notch signaling by DAPT increased the number of GFP-positive hair cell-like cells and reduced the number of supporting cells labeled with PROX1, an early marker of supporting cell differentiation, suggesting their direct transdifferentiation (Fig. 1A). Using a different reporter line carrying a p27/GFP BAC transgene that is expressed in supporting cells (Lee et al., 2006; White et al., 2006), we FACS purified supporting cells from DAPT-treated and DMSO-treated organ cultures. qPCR analysis showed that, 24 h after inhibition of Notch signaling, Atoh1 expression increased more than 10-fold, whereas the expression of Hey1 and Hes5 decreased ∼70% and 90%, respectively (P<0.005; Fig. 1B).
Fig. 1.
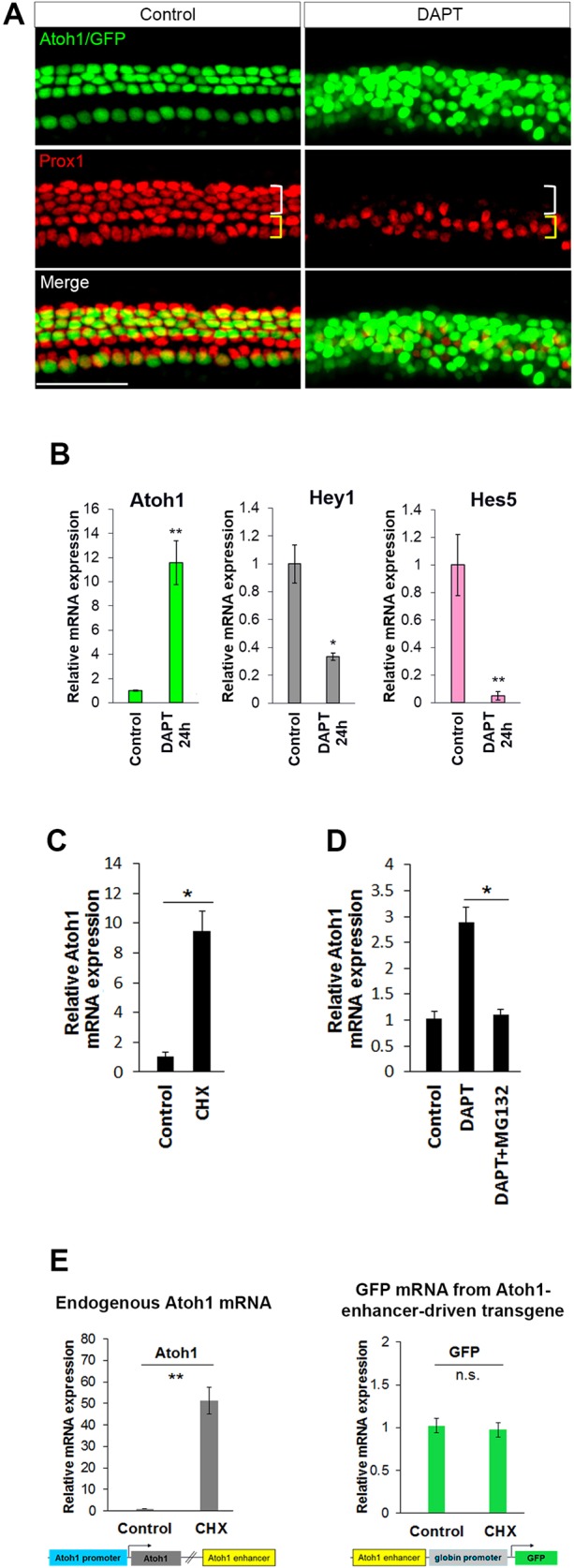
Derepression of Atoh1 in supporting cells is sufficient to drive expression and occurs through promoter elements, not through the 3′ autoregulatory enhancer. (A) P1 Atoh1 enhancer/β-globin promoter/GFP transgenic organ of Corti was cultured for 72 h in DMSO (control) or DAPT (γ-secretase inhibitor) and immunostained with anti-PROX1 antibody to label supporting cells (Deiters' and pillar cells). The appearance of ectopic GFP+ hair cell-like cells in DAPT is accompanied by loss of PROX1+ supporting cells (white bracket). (B-E) Changes in mRNA expression level were examined by real-time quantitative PCR (qPCR). (B) The observed transdifferentiation of supporting cells in A correlates with upregulation of Atoh1 and downregulation of Hes/Hey factors mRNA in FACS-purified supporting cells (p27/GFP+) after 24 h treatment with DAPT. n=4. (C) Inhibition of protein synthesis with cycloheximide (CHX) for 6 h induces Atoh1 expression in FACS-purified supporting cells (p27/GFP+). n=3. (D) Inhibition of protein degradation by MG132 prevents DAPT-induced upregulation of Atoh1 in FACS-purified supporting cells (p27/GFP+) treated for 12 h. n=3. (E) CHX induces endogenous Atoh1, but not the Atoh1 enhancer/β-globin promoter/GFP transgene. P1 cochlear organ cultures from Atoh1 enhancer/β-globin promoter/GFP mice were incubated without and with CHX for 6 h, after which organ cultures were dissociated and FACS sorted to eliminate hair cells. The GFP− population from these transgenic organs includes supporting cells, cells from the greater epithelial ridge and lesser epithelial ridge. Atoh1 and GFP levels were measured in GFP− cells (non-hair cells of the cochlear epithelia). n=4. All values are mean±s.e.m. *P<0.05, **P<0.005; n.s., not statistically significant. Scale bar: 100 µm. See also Fig. S1.
In other systems, the Hes/Hey family members HES1 and HES7 have been shown to have short protein half-lives (Hirata et al., 2002, 2004). Transfecting flag-tagged HES5 into HEK 293 cells indicated that its protein half-life is ∼2.5 h, and that selective inhibition of the proteasome pathway significantly increased HES5 stability (Fig. S1). Within the Hes/Hey family, Hes5 is expressed strongly in Deiters' cells, a subpopulation of supporting cells that readily transdifferentiate into hair cells when Notch signaling is blocked. Hes5 appears to be the most highly expressed Hes/Hey family member in supporting cells (Tateya et al., 2011), and appears to be the most sensitive to loss of Notch signaling (Fig. 1B) (Doetzlhofer et al., 2009). We hypothesized that if HES5 is needed to directly silence constitutive Atoh1 transcriptional activity, inhibition of protein synthesis might derepress Atoh1 expression as a consequence of HES5 protein turnover in supporting cells and, similarly, stabilizing HES5 protein would prevent Atoh1 upregulation in supporting cells in response to blocking Notch signaling. To test this hypothesis in supporting cells, we treated P1 organ cultures for 6 h with cycloheximide (CHX) to block protein synthesis, and then FACS purified the supporting cells. qPCR analysis indicated a more than 9-fold increase in Atoh1 expression, showing that derepression is sufficient to drive endogenous Atoh1 expression (Fig. 1C). If upregulation requires Atoh1 derepression through HES5 turnover, then blocking HES5 degradation should inhibit Atoh1 upregulation. To test this we simultaneously blocked Notch signaling to induce Atoh1, and inhibited HES5 turnover by blocking the activity of the proteasome pathway with MG132 (Hirata et al., 2002). Analysis of FACS-purified supporting cells indicated that Atoh1 failed to be upregulated following inhibition of Notch when proteasome activity was simultaneously inhibited (Fig. 1D).
The Atoh1 3′ enhancer contains a conserved E-box, which has been shown to bind ATOH1 (Helms et al., 2000), upon which the enhancer's autoregulatory function and Atoh1 expression depend (Helms et al., 2000). In perinatal supporting cells, the Atoh1 transgene containing the 3′ autoregulatory enhancer (Atoh1 enhancer/β-globin promoter/GFP) is silent (Chen et al., 2002; Lumpkin et al., 2003) (Fig. 1A), suggesting that lateral inhibition might repress Atoh1 through sites in the enhancer. Indeed, the E-box site is overlapped by a consensus N-box, a potential site of Hes/Hey binding (Helms et al., 2000). Interestingly, although blocking protein synthesis with CHX stimulated endogenous Atoh1 expression, it failed to stimulate expression from the Atoh1 enhancer transgene (Fig. 1E), suggesting that the repressive effects of HES5 are mediated through elements that are not in the enhancer but elsewhere at the Atoh1 locus. The reason for enhancer inactivity might be activator insufficiency (see below). These results also show that the enhancer sequence alone is not sufficient to initiate expression, even when repression is relieved.
HES5 targets the Atoh1 promoter region for repression
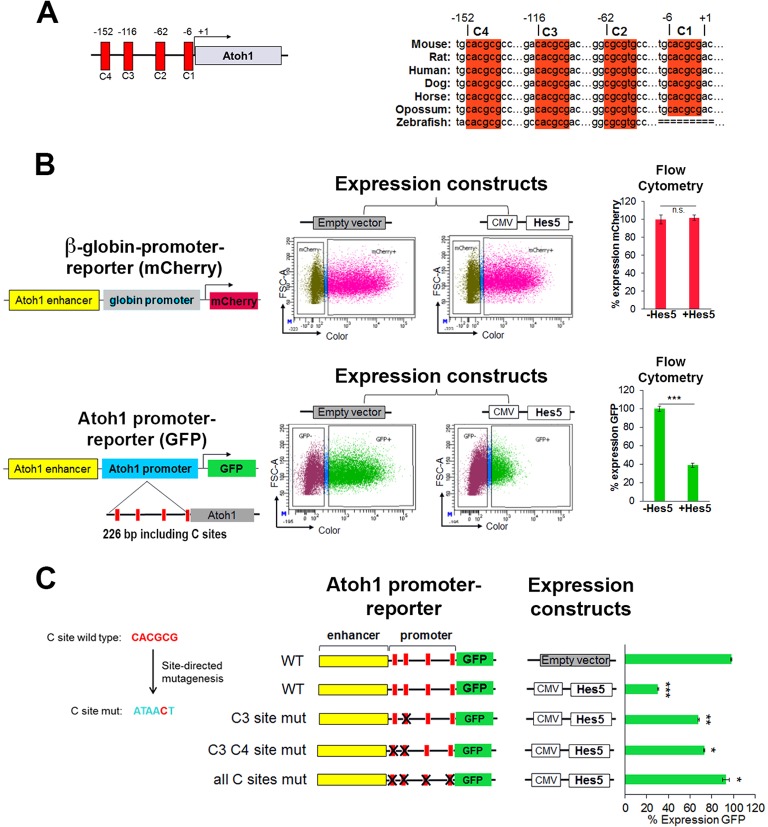
bHLH proteins of the Hes/Hey family have been shown to bind a canonical N-box (CACNAG) or C-site (CACGNG) in their target genes (Liu et al., 2006; Grogan et al., 2008; Zheng et al., 2011). In addition to the N-box and a C-site in the 3′ enhancer, the Atoh1 promoter contains four highly evolutionarily conserved sites within 226 bp of the transcription start site (Fig. 2A), suggesting that HES5 might directly regulate Atoh1 expression through the promoter. To test the functional significance of the HES5 binding sites in the Atoh1 promoter and enhancer region, we compared the ability of HES5, transfected into HEK 293 cells, to repress a co-transfected Atoh1 promoter-reporter construct driving GFP, relative to the heterologous β-globin promoter, which does not contain C-sites, driving mCherry. The percentage of cells expressing GFP and mCherry was determined by flow cytometry (Fig. 2B). The reporter with the β-globin minimal promoter (mCherry) was unaffected by the expression of HES5, whereas the Atoh1 promoter containing four C-sites (GFP) was strongly inhibited (>60%).
Fig. 2.
HES5 functions to repress Atoh1 through highly conserved binding sites in the promoter region. (A) Schematic of the murine Atoh1 promoter indicating the C-sites and their conservation among vertebrates. Numbers are relative to the transcription start site (+1) in mouse. (B) Ectopic Hes5 expression requires the Atoh1 promoter to repress Atoh1 expression in heterologous HEK 293 cells [compare β-globin promoter (top) with Atoh1 promoter (bottom)]. Reporters and expression constructs were transiently transfected and expression was quantified by flow cytometry. (Top) Reporter contains the Atoh1 enhancer and β-globin basal promoter driving the expression of mCherry. (Bottom) Reporter contains the Atoh1 enhancer and 226 bp of the Atoh1 promoter (containing the predicted C-sites) driving the expression of GFP. Reporters were cotransfected with a control (empty vector) or a plasmid expressing Hes5. The number of cells expressing GFP or mCherry was determined by flow cytometry after 48 h. Values are mean±s.e.m.; n=6. ***P<1×10−7. (C) Mutational analysis of C-site function. Five out of six nucleotides of each C-site were mutated. Quantification was performed as in B with the indicated plasmids reported as percentage of cells expressing GFP relative to the CMV-RFP co-transfected control (not shown) set to 100% in the absence of Hes5 expression (empty vector). Values are mean±s.e.m.; n=4. *P<0.0003, **P<2×10−5, ***P<4×10−7. See also Fig. S2.
To determine if the predicted Hes/Hey binding sites in the Atoh1 promoter region are required for HES5-mediated repression of Atoh1, we mutated the C-sites in the Atoh1 reporter construct (Fig. 2C) and again tested them in HEK 293 cells. In the absence of HES5 expression, the mutation of C-sites had no effect on the expression of GFP (data not shown). However, mutation of the C-sites in the Atoh1 promoter had an additive effect: the more sites that were mutated, the greater the loss of HES5 inhibitory activity on the Atoh1 promoter (Fig. 2C).
HEY2, which is also expressed in the organ of Corti with other Hes/Hey family members (Li et al., 2008; Doetzlhofer et al., 2009), was tested using the same reporter constructs co-transfected into HEK 293 cells. Similar to HES5, HEY2 represses expression in an Atoh1 promoter-dependent manner, although site usage varied from that seen with HES5 (Fig. S2).
Atoh1 is upregulated in prosensory progenitors, not just in nascent hair cells
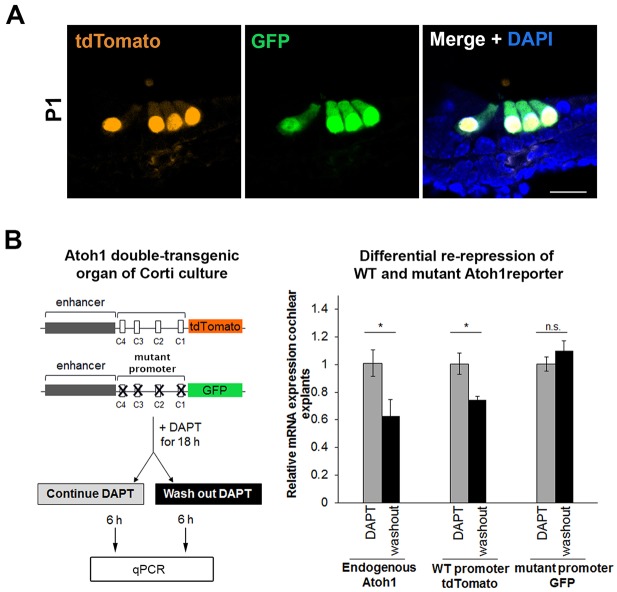
The mutational analysis of the Atoh1 promoter-reporter in vitro demonstrated the importance of the C-sites for Atoh1 repression in HEK 293 cells. To better understand the mechanism of HES-mediated lateral inhibition in vivo, we generated a double-transgenic mouse line carrying both the wild-type and mutated Atoh1 promoter-reporter constructs. During organ of Corti development, Atoh1 is upregulated in nascent hair cells in a basal-to-apical wave (Chen et al., 2002). Evidence suggests that lateral inhibition is responsible for repressing Atoh1 in the prosensory progenitors surrounding the nascent hair cells, allowing their differentiation as supporting cells (Kiernan et al., 2005; Yamamoto et al., 2006; Doetzlhofer et al., 2009). We hypothesized that if Atoh1 is initially upregulated in groups of prosensory progenitors, followed by rapid repression in those selected progenitors that will become supporting cells, the mutant promoter-reporter (GFP) would be deficient in this process and would report the presence of GFP in the nascent progenitors destined to be supporting cells, as well as the nascent hair cells. By contrast, the wild-type promoter-reporter (tdTomato) would accurately identify only the nascent hair cells. As hypothesized, at E16.5 in the middle region of the cochlear duct, the expression of the wild-type promoter-tdTomato was observed only in inner and outer hair cells, whereas GFP was strongly expressed in the hair cell region as well as being misexpressed in the differentiating supporting cell layer (Fig. 3B, arrowheads). In a slightly more apical and thus less differentiated position along the cochlear duct, only nascent inner hair cells expressing the wild-type tdTomato transgene were observed, with a commensurate increase in the number of progenitors expressing GFP from the mutant promoter. The appearance of only the inner hair cells is consistent with the well documented earlier differentiation of inner, relative to outer, hair cells (Sher, 1971; Lim and Anniko, 1985; Chen et al., 2002). Together, these results indicate that Atoh1 is upregulated in groups of prosensory progenitors, not just in nascent hair cells, and that the promoter-proximal Hes/Hey binding motifs are crucial for the silencing of Atoh1 in these prosensory progenitors.
Fig. 3.
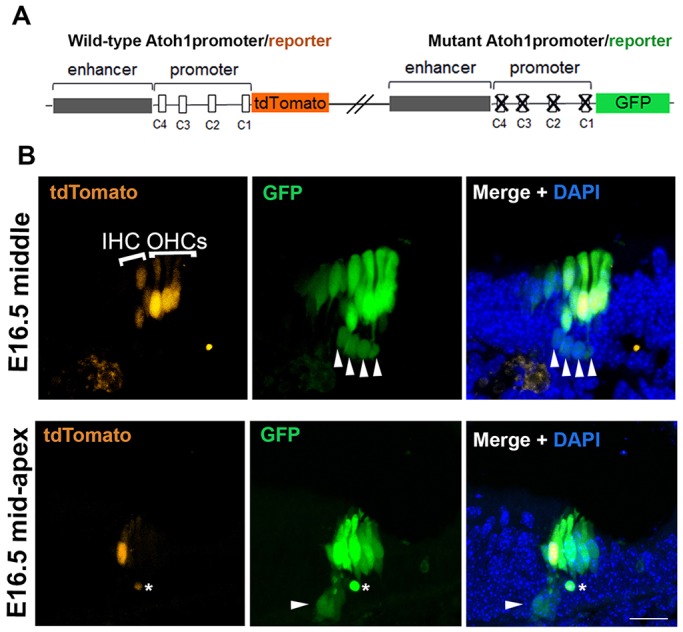
The misexpression of a mutant promoter transgene in vivo shows that Hes/Hey binding sites in the Atoh1 promoter are required for the proper silencing of Atoh1 in supporting cells. (A) Schematics showing wild-type and mutant constructs used to generate the double-transgenic mouse (see Materials and Methods). (B) The expression pattern of the wild-type and mutant transgenes at E16.5 is shown in transverse section at two positions (middle and mid-apex) of the cochlear duct. The GFP from the mutant promoter transgene is misexpressed in the supporting cell layer (white arrowheads; asterisk indicates debris), in addition to the expected expression in the hair cells, indicating the need for active repression for supporting cell silencing. tdTomato from the wild-type transgene is absent from the supporting cell layer. IHC, inner hair cell; OHCs, outer hair cells. Scale bar: 20 µm.
At P1, Atoh1 mRNA expression is localized to the inner and outer hair cells (Lanford et al., 1999; Woods et al., 2004), as is expression of both the wild-type and mutant promoter transgenic constructs (Fig. 4A). The lack of expression of the mutant promoter-reporter, which is unable to undergo Hes/Hey-mediated repression, indicates that active repression is unlikely to be responsible for the silence of this construct in supporting cells. Rather, it suggests that Hes/Hey repression of endogenous Atoh1 mRNA is sufficient to keep the expression of endogenous ATOH1 protein below the threshold needed to trigger the Atoh1 enhancer in vivo – an example of activator insufficiency (Barolo and Posakony, 2002; see below).
Fig. 4.
Hes/Hey binding sites are required to rerepress Atoh1 in supporting cells after Notch signaling is first inhibited, allowing Atoh1 levels to rise, and then restored. (A) At P1, the expression of both wild-type and mutant promoters is limited to hair cells, indicating that low levels of ATOH1 (activator insufficiency) are sufficient to maintain silencing from the mutant transgene. Scale bar: 20 µm. (B) Schematic shows experimental time course: P1 cochlear cultures from the double-transgenic mouse line were treated with DAPT for 18 h, after which DAPT was washed out from half of the explants, and the explants were then collected after an additional 6 h in culture. Expression analysis of endogenous Atoh1, GFP and tdTomato by qPCR shows that endogenous Atoh1 and the wild-type (WT) transgene are actively repressed by returning Notch signaling, but the mutant transgene fails to be rerepressed in the same time period (6 h). Values are mean±s.e.m.; n=3. *P<0.05.
To test the importance of C-site repression in the maintenance of supporting cell fate at P1, we treated cochlear explants with DAPT for 18 h to inhibit Notch signaling and induce the transdifferentiation of supporting cells (Fig. 1) and, by doing so, also induce expression of the two transgenic reporters, which contain identical copies of the 3′ autoregulatory enhancer (Fig. 4B). We hypothesized that, upon washing out DAPT after 18 h, lateral inhibition would return and repression of Atoh1 expression would be re-established at the endogenous promoter and the wild-type transgene, but that the mutant promoter would be unable to re-establish repression. As expected, expression of the wild-type promoter-reporter (tdTomato) transgene and the endogenous Atoh1 mRNA were partially re-repressed at 6 h following DAPT washout, but expression from the promoter-reporter mutant (GFP) transgene lagged significantly behind.
HES5 recruits the co-repressor GRG to the Atoh1 promoter in a Notch-dependent fashion
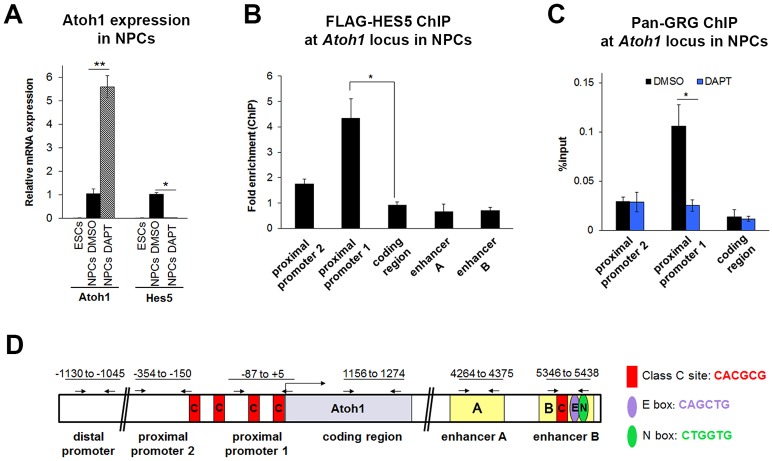
Hes proteins have been reported to recruit GRG co-repressors (GRG1-4) through their conserved C-terminal WRPW motif (Grbavec and Stifani, 1996; Grbavec et al., 1998). Our in vitro results suggested that loss of the HES5 WRPW motif severely reduces its ability to inhibit the Atoh1 reporter, and that HES5 and GRG interact through the WRPW motif of HES5 (Fig. S3). To test whether the interaction of HES5 and GRG co-repressor could be responsible for mediating the repression of Atoh1 we used neural progenitor cells (NPCs) as a model (Fig. S4), since the paucity of supporting cells in the organ of Corti makes reliable transcription factor ChIP currently impractical. NPCs, like supporting cells, rely on active Notch signaling for maintenance of their progenitor state (Ohtsuka et al., 2001; Hatakeyama et al., 2004; Crawford and Roelink, 2007; Borghese et al., 2010). Consistent with this, we observed more than 5-fold (P<0.005) upregulation of Atoh1 and a complete loss of Hes5 mRNA in NPCs following DAPT treatment (Fig. 5A). Also like supporting cells, NPCs express GRG4 at higher levels than the other GRG factors (Fig. S5).
Fig. 5.
GRG localizes to the Atoh1 locus in a Notch-dependent manner in neural progenitor cells. Neural progenitor cells (NPCs) were differentiated from ESCs and FACS purified (Sox1-GFP+). (A) Atoh1 and Hes5 mRNA expression levels in 46C ESCs and NPCs that were treated with control (DMSO) or γ-secretase inhibitor (DAPT) for 48 h. Similar to supporting cells, inhibition of Notch signaling by DAPT induces the expression of Atoh1 and represses the expression of Hes5 in NPCs. n=4. (B) HES5 localizes to the proximal promoter and not the enhancer by ChIP-qPCR of NPCs transfected with CMV-FLAG-Hes5 plasmid (or CMV-RFP control). Results are reported as fold enrichment (FLAG-Hes5 transfected percentage input/RFP transfected percentage input). n=3. (C) GRG localizes to the Atoh1 promoter in a Notch-dependent manner. ChIP-qPCR with anti-pan-GRG antibody and primers scanning the Atoh1 locus in NPCs treated with control (DMSO) or γ-secretase inhibitor (DAPT) for 24 h after 7 days of differentiation. n=3. (D) Schematic of the Atoh1 locus in mouse showing the promoter and the autoregulatory enhancer regions. Numbers refer to the regions amplified in ChIP-qPCR. The primers used to measure the enrichment after ChIP are shown by arrows. All values are mean±s.e.m. *P<0.05, **P<0.005. See also Figs S3-S5.
To test whether HES5 directly interacts with the Atoh1 promoter region, NPCs were transfected with a flag-tagged HES5 expression plasmid, and ChIP-qPCR indicated that HES5 interacts with the Atoh1 promoter region, but not the enhancer, confirming the reporter assay findings (Fig. 5B). Similarly, ChIP-qPCR in untransfected NPCs with a pan-GRG antibody showed that endogenous GRG is also normally enriched at the Atoh1 promoter region (Fig. 5C). Interestingly, the association of endogenous GRG with the Atoh1 promoter region was Notch dependent (Fig. 5C), suggesting that it is the interaction of HES5 with this co-repressor in NPCs that maintains them in an undifferentiated state.
Inhibition of Notch induces histone acetylation at the Atoh1 promoter in supporting cells
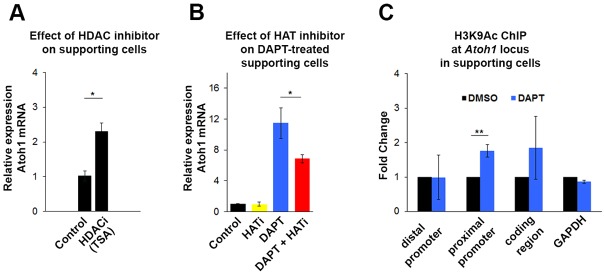
GRG-mediated repression has been shown to be partially dependent on HDAC recruitment in Drosophila (Winkler et al., 2010). In neonatal supporting cells purified from cochlear cultures treated with the HDAC inhibitor trichostatin A (TSA) for 6 h, Atoh1 was upregulated (P<0.05; Fig. 6A), suggesting that the repression of Atoh1 in supporting cells is dependent on maintenance of the deacetylated state of histones at the Atoh1 locus. Similar experiments in Hes5-transfected HEK 293 cells suggested a similar conclusion (Fig. S6). Furthermore, inhibition of histone acetyltransferase (HAT) activity by curcumin partially blocks the DAPT-induced accumulation of Atoh1 mRNA, further indicating a need for ongoing histone acetylation to induce Atoh1 (Fig. 6B). Curcumin, either on its own or in conjunction with DAPT, had no effect on the expression level of the Notch effectors Hes5 and Hey1 (data not shown). This prompted us to ask if blocking Notch signaling in supporting cells brings about changes in the acetylation status of histones at the Atoh1 locus. We performed ChIP-qPCR with FACS-purified DMSO-treated and DAPT-treated supporting cells for H3K9ac, a histone modification highly correlated with actively transcribed genes (Wang et al., 2008). Inhibition of Notch signaling for 24 h increased the H3K9ac level at the promoter region in transdifferentiating supporting cells (Fig. 6C). This result strongly suggests that the upregulation of Atoh1 in supporting cells is accompanied by epigenetic changes that render the chromatin more permissive at the Atoh1 locus.
Fig. 6.
Inhibition of Notch leads to increased histone acetylation at the Atoh1 promoter in supporting cells. (A) Inhibition of HDAC activity by trichostatin A (TSA) for 6 h induces Atoh1 expression in FACS-purified supporting cells (p27/GFP+). Values are mean±s.e.m.; n=3. (B) Inhibition of HAT activity by curcumin (Balasubramanyam et al., 2004) for 24 h blocks DAPT-induced Atoh1 activation in FACS-purified supporting cells (Lfng/GFP+). n=3. (C) Inhibition of Notch signaling (DAPT for 24 h) increases H3K9ac, as analyzed by ChIP-qPCR at the Atoh1 promoter in FACS-purified supporting cells (p27/GFP+). Values are mean±s.e.m.; n=3. *P<0.05, **P<0.005. See also Fig. S6.
DISCUSSION
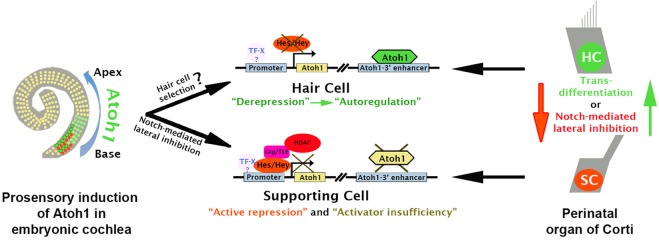
Our data help explain several important aspects of Atoh1 regulation that underlie embryonic sensory epithelia differentiation and patterning, including the identification of the sites of active Hes/Hey-mediated repression of Atoh1, the sufficiency of derepression to stimulate Atoh1 expression, and the role of activator insufficiency in reinforcing the transcriptional silence of Atoh1 in prosensory and supporting cells. These results suggest a model of Atoh1 regulation during embryonic development that emphasizes the importance of the repressive activity of Hes/Hey factors to select the rows of hair cells and supporting cells, as opposed to the selective upregulation of Atoh1 in prosensory populations destined to be hair cells. These issues are discussed below and the results presented in a model of Atoh1 regulation (Fig. 7).
Fig. 7.
Model of Atoh1 regulation during organ of Corti development. Upregulation of Atoh1 in groups of cells within the embryonic prosensory domain occurs in a basal-to-apical wave. Once the Atoh1 autoregulatory threshold is achieved in selected nascent hair cells, Notch-mediated active repression is triggered in the surrounding prosensory cells to stimulate the silencing of Atoh1 through Hes/Hey binding, leading to the onset of supporting cell differentiation and the patterning of the cellular mosaic of the organ of Corti. At P1, cell fate is maintained by continuing Notch-mediated repression through recruitment of GRG/HDACs. Loss of Notch activity leads to derepression of the Atoh1 promoter and increased Atoh1 expression through hypothetical positive transcriptional activity (TF-X), leading to autoregulation and transdifferentiation of supporting cells (SC) to a hair cell-like state (HC).
Atoh1 repression is mediated through highly conserved Hes/Hey binding motifs in the Atoh1 promoter
Our results show that HES5 represses Atoh1 expression by directly binding to the promoter region of Atoh1 (Fig. 2B, Fig. 5B) and that this binding is dependent on four highly conserved C-sites located within the proximal 226 bp of the transcription start site (Figs 2–4). Mutation in all four C-sites is required to abolish the inhibitory effect of ectopically expressed HES5 on promoter function (Fig. 2C), indicating a synergistic role in this context. However, a comparison of the effects of HES5 and HEY2 on the repression of Atoh1 revealed differences in strength of inhibition and binding (Fig. S2), suggesting the possibility that different members of the Hes/Hey families could play independent roles in Atoh1 regulation, as previously shown (Zine et al., 2001; Li et al., 2008; Doetzlhofer et al., 2009). While Hes/Hey repressor sequences are present in regulatory elements of atonal homologs separated by hundreds of millions of years of evolution, there are fewer recognizable Hes/Hey motifs associated with proneural genes in non-vertebrate lineages, just as there are fewer homologs of Hes/Hey genes (Rebeiz et al., 2005), suggesting that duplication of the binding sites in the Atoh1 promoter might have paralleled the expansion of the Hes/Hey family of genes, leading to specialization rather than redundancy in mammals. The complex combinatorial pattern of Hes/Hey gene expression in the organ of Corti (Doetzlhofer et al., 2009) and the differing patterning phenotypes of the Hes/Hey mutant mice suggest a more subtle role for these sites in sculpting organ of Corti structure.
Atoh1 is upregulated in groups of prosensory progenitors
Earlier observations using a transgenic reporter (Atoh1 enhancer/β-globin promoter/GFP), and by antibody staining, suggested that Atoh1 is upregulated only in nascent hair cells during the embryonic patterning of the organ of Corti (Chen et al., 2002). However, recent evidence suggests an alternate model in which Atoh1 transcriptional activity may begin in groups of prosensory cells and then be rapidly refined to just the nascent hair cells. First, loss of Notch signaling through the conditional mutation of Rbpj leads to the broader expression of Atoh1 within the prosensory domain (Basch et al., 2011; Yamamoto et al., 2011). Second, lineage-tracing experiments indicated that significant numbers of supporting cells derive from Atoh1-cre-expressing prosensory cells (Yang et al., 2010; Driver et al., 2013). Third, Cai et al. (2013) report that, in an Atoh1-GFP knock-in mouse, low levels of GFP are detectable in multiple rows of cells at the leading edge of hair cell differentiation (Cai et al., 2013). Our current transgene data support these observations by showing that the mutant Atoh1 promoter allows expression in both nascent hair cell types, as well as in additional prosensory cells, which are likely destined to become supporting cells (Fig. 3B).
This upregulation of Atoh1 in groups of cells within the prosensory domain is reminiscent of recent observations regarding the selection of the sensory organ precursors (SOP) that go on to form the micro- and macrochaete, the sensory bristles in the Drosophila notum (Troost et al., 2015). Here, the standard theory holds that small stochastic differences in proneural gene expression in single cells within a proneural cluster are amplified by strong Notch-mediated feedback to select them as SOPs (reviewed by Fortini, 2009). However, in their re-evaluation of this system, Troost et al. (2015) discovered that, similar to the situation in the cochlea, the proneural genes achaete and scute, which are involved in SOP selection, are first upregulated in a band of cells that crosses the proneural domain and are subsequently repressed in all but the SOP. Thus, although the mechanism by which SOPs are selected remains unknown, this re-evaluation recognizes that it is unlikely to be strictly stochastic.
Atoh1 silencing in supporting cells is dependent on active repression and reinforced by ‘activator insufficiency’
Our early observation that the transgenic Atoh1 3′ enhancer linked to the β-globin minimal promoter (Chen et al., 2002; Lumpkin et al., 2003) is not expressed in supporting cells led us to the initial hypothesis that the site of Notch-mediated repression of Atoh1 is likely to be in the enhancer. However, our current results indicate that this is not the case, as shown by the failure to repress Atoh1 enhancer-mediated expression in our co-transfection assay with HES5 (Fig. 2B) and HEY2 (Fig. S2B), and by the role of the Atoh1 promoter in active repression (Fig. 2). This, in turn, raises the question of why the Atoh1 3′ enhancer transgene, linked to the β-globin minimal promoter, is silent in supporting cells. We hypothesize that this silencing is an example of ‘activator insufficiency’ (Barolo and Posakony, 2002), in which active repression of endogenous Atoh1 through the promoter maintains the ATOH1 protein below a threshold needed to stimulate the autoregulatory enhancer activity. In this context, the Atoh1 enhancer acts in a switch-like manner, so that once the threshold is crossed Atoh1 is robustly stimulated (Fig. S7). Activator insufficiency is further confirmed by the absence of expression of this transgene in the neural tube or the ear of the Atoh1 knockout mouse (Helms et al., 2000; Raft et al., 2007), and demonstrated by the failure of the Atoh1 enhancer alone (Atoh1 enhancer/β-globin promoter/GFP) to initiate transcriptional upregulation when protein synthesis is blocked, whereas the endogenous Atoh1 gene is upregulated (Fig. 1E). This later observation strongly indicates the presence of positively acting transcriptional mechanisms that are engaged at the endogenous Atoh1 promoter in the perinatal supporting cells, but held in check by active repression and activator insufficiency.
Our data also suggest that the reported low level of Atoh1 transcription in the prosensory domain prior to E13.5 (Lanford et al., 2000; Woods et al., 2004), before the overt upregulation of Atoh1 in the nascent hair cells (Chen et al., 2002; Lumpkin et al., 2003), does not rise to the threshold needed to trigger enhancer autoregulation (Fig. 7, activator insufficiency). This begs the question of what is responsible for the onset of Atoh1 expression. This could occur either through a derepression signal, such as the downregulation of Hey factors, as recently suggested (Benito-Gonzalez and Doetzlhofer, 2014), or by the active stimulation of Atoh1 transcription through other binding motifs in the 226 bp promoter.
Repression and derepression through the Atoh1 promoter regulates Atoh1 expression
Notch signaling has been shown to prevent the recruitment of the HAT p300 and the acetylation of histones (Krishnamoorthy et al., 2015), and transcriptional repression by Hes/Hey proteins has been linked to interaction with HDACs (Iso et al., 2001; Takata and Ishikawa, 2003; Weber et al., 2015). Our results indicate that HES5-dependent repression of the Atoh1 promoter is mediated by the recruitment of GRG co-repressors, and that the presence of HDAC activity is Notch dependent (Figs 5, 6, S3 and S5). Further studies are required to identify which HDAC(s) are specifically involved in Atoh1 repression in supporting cells.
We show that blocking de novo protein synthesis with CHX is sufficient to rapidly stimulate Atoh1 expression, and leads to the rapid loss of HES5, not unlike blocking Notch signaling activity with DAPT in supporting cells (Fig. 1C, Fig. S1). Similarly, HES1 protein has previously been shown to have a short half-life (Hirata et al., 2002; Kobayashi et al., 2009) and loss of HES1 expression leads to selective derepression and transcriptional stimulation of target genes (Riccio et al., 2008).
Conclusion
Together, these data suggest that Atoh1 levels transiently rise in groups of cells in the prosensory domain at the onset of differentiation, followed by a rapid repression in those cells not destined to be hair cells. It seems likely that this initial upregulation is sufficient to stimulate the production of Notch ligands, but is likely to just skirt the threshold needed to stimulate the autoregulatory function of the Atoh1 enhancer. Active repression integrated at the Atoh1 promoter is required to inhibit this process of transcriptional activation in nascent supporting cells. Our results argue for a model in which the complex pattern of Hes/Hey expression, interacting with multiple binding motifs in the Atoh1 promoter, sculpts the precise hair cell and supporting cell mosaic required for organ of Corti function.
MATERIALS AND METHODS
Experimental animals, generation of transgenic mouse lines and genotyping
The experimental procedures were approved by the Animal Care and Use Committees of the University of Southern California and the House Research Institute. p27/GFP BAC transgenic (Lee et al., 2006; White et al., 2006), Atoh1 enhancer/β-globin promoter/GFP transgenic (Lumpkin et al., 2003) and Lfng/GFP BAC transgenic (Korrapati et al., 2013) mice were described previously. For generating the Atoh1 reporter double-transgenic line, Atoh1 enhancer/Atoh1 promoterwt/tdTomato and Atoh1 enhancer/Atoh1 promotermut/GFP plasmids were linearized, and founder animals were generated by pronuclear co-injection (UCI transgenic mouse facility; FVB background). Positive founders were identified by PCR genotyping using tail DNA and the primers listed in Table S3. Postnatal transgenic mice were identified by direct observation of GFP or tdTomato fluorescence in the cerebellum. Animals were put together in the evening, and the next morning was designated as E0.5.
Cell culture
HEK 293 cells (ATCC) and OC1 (organ of Corti 1) cells (generous gift from Federico Kalinec, UCLA) were cultured in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (FBS; GIBCO), 2 mM L-glutamine, and 10 mM HEPES maintained in a 5% CO2 incubator at 37°C and 33°C, respectively. Mouse embryonic stem cell (ESC) line 46C, a Sox1-GFP knock-in line, was a generous gift from Qi-Long Ying (University of Southern California, USA). ESCs were cultured and differentiated to NPCs as reported (Ying and Smith, 2003); for details see the supplementary Materials and Methods.
Cochlear explant culture
Cochlea of P1 pups were dissected and cochlear explants were cultured on SPI black membranes as previously described (Doetzlhofer et al., 2009), except that B27 supplement was not added to the media. All cultures were incubated in a 5% CO2, 5% O2 humidified incubator (Forma Scientific).
Immunohistochemistry and imaging
Cochlear explants, HEK 293 cells and NPCs were fixed in 4% paraformaldehyde in PBS and kept in PBS at 4°C until the staining was performed. E16.5 whole heads or P1 inner ears were prepared for immunohistological analysis as previously reported (White et al., 2006). Anti-nestin (1:500; Developmental Studies Hybridoma Bank) and anti-PROX1 (1:500; Novus Biologicals, NBP1-18605) primary antibodies with species-specific Alexa Fluor secondary antibodies (1:1000; Life Technologies) were used. Confocal images were taken on a Zeiss LSM 780 scanning confocal microscope. Fluorescent imaging of NPCs was performed with a Zeiss Axio Observer.A1.
Plasmid construction and site-directed mutagenesis
A description of the plasmid constructs used is provided in the supplementary Materials and Methods. Detailed cloning and mutagenesis procedures are available upon request. Point mutations in the Atoh1 reporter plasmids and deletion in the Hes5 expression plasmid were introduced with the aid of the Finnzyme Phusion site-directed mutagenesis kit following the manufacturer's instructions.
Small-molecule inhibitors
The inhibitors/chemicals used in this study and their final concentrations were as follows: 10 µM γ-secretase inhibitor DAPT (Calbiochem, EMD Millipore); 100 µM proteasome inhibitor MG132 (Sigma); 100 µg/ml protein synthesis inhibitor cycloheximide (Sigma); 1 mM protease inhibitor PMSF (Sigma); 10 µM proteasome inhibitor lactacystin (Sigma); 30 µM HAT inhibitor curcumin (Enzo Life Sciences); and 200 nM HDAC inhibitor trichostatin A (Sigma).
FACS purification
All cells were sorted on a BD FACSAria II cell sorter with lasers at 488 nm and 561 nm wavelength and 100 µm nozzle. For RNA extraction, cells were sorted directly into RNA lysis buffer (Zymo Research) and kept at −80°C until the RNA isolation was performed. Preparation of cochlear samples for FACS was undertaken as described (White et al., 2006). FACS-purified supporting cell samples (GFP+) were analyzed by qPCR and compared with the GFP− population. Only samples that showed more than 40-fold enrichment for Hes5 mRNA and less than 0.8-fold enrichment for Atoh1 mRNA were used for further analysis. NPCs transfected with pCS2-CMV-FLAG-Hes5 and/or pCS2-CMV-H2BmRFP (control) were collected 48 h post transfection, and RFP+ cells were FACS purified.
RNA extraction and real-time quantitative PCR (qPCR)
Total RNA was isolated using the Zymo Research Quick-RNA MicroPrep Kit, with DNase digestion (Qiagen). RNA from supporting cells (8000 cells), NPCs (10,000 cells) and non-hair cells (100,000 cells) was reverse transcribed using the Quanta qScript cDNA Synthesis Kit. qPCR was performed with Power SYBR Master Mix (Applied Biosystems) on a ViiA 7 real-time PCR system with fast 96-well block (Applied Biosystems). Relative quantification of gene expression was performed by the comparative ΔΔCt method (Livak and Schmittgen, 2001) with Gapdh as the endogenous reference. Two-tailed Student's t-test was used to determine statistical significance. P<0.05 was considered statistically significant. The primers used in qPCR are listed in Table S1.
Western blotting
Western blot analysis of proteins extracted from transfected HEK 293 cells is described in the supplementary Materials and Methods.
Proximity ligation assay
To visualize GRG/TLE:FLAG-HES5 interaction, the proximity ligation assay was performed as described in the supplementary Materials and Methods.
Chromatin immunoprecipitation (ChIP)-qPCR
For HES5 ChIP, NPCs in each well of a 6-well culture dish were transfected with 15 µg pCS2-CMV-FLAG-Hes5 and/or pCS2-CMV-H2BmRFP (control) using Lipofectamine LTX (Invitrogen). NPCs were FACS purified into DMEM containing 2% FBS, 1 mM PMSF (Sigma) and Protease Inhibitor Cocktail (Calbiochem). ChIP from NPCs was performed as described (Dahl and Collas, 2008), using 2×105 (FLAG-Hes5) or 1×106 (pan-GRG) cells. For H3K9ac ChIP, supporting cells were FACS purified from the p27/GFP transgenic mouse line (∼25,000 supporting cells per sample). A detailed ChIP protocol and primers are provided in the supplementary Materials and Methods and Table S2.
Reporter assay
Reporter assays were conducted using HEK 293 cells (2×105) seeded in each well of a 24-well culture dish and transfected 14-16 h later with 150 ng Atoh1 reporter plasmid (GFP or mCherry, Fig. 2) and 600 ng pCS2-CMV-FLAG-Hes5 plasmid, or empty plasmid control using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. For the mutational analysis of the C-sites in the Atoh1 promoter region, 150 ng pCS2-CMV-H2BmRFP plasmid was also cotransfected as an internal control for transfection efficiency. 48 h post-transfection, the number of positive cells was determined by flow cytometry using 1×105 cells and analyzed by BD FACSDiva software. Two-tailed Student's t-test was used to determine statistical significance. Further details of FACS analysis of reporter assays are provided in the supplementary Materials and Methods.
Acknowledgements
We thank Juan Llamas and Welly Makmura for expert technical assistance; Qi-Long Ying for the mouse ESC line; Jane Johnson for the Atoh1 enhancer-reporter plasmid; Greta Segil for the Atoh1-mCherry reporter; Manfred Gessler for the Hey2 expression plasmid; and Verdon Taylor for the Hes5 plasmid. We acknowledge the University of California, Irvine, transgenic mouse facility, and the USC Eli and Edythe Broad Center Flow Cytometry Core for their expert work. We also thank Andrew Groves and Cristy Lytal for comments on the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Y.A. and N.S. developed the concepts, analyzed the data and wrote the manuscript; Y.A. designed and performed the experiments; Z.S. performed histone ChIP, contributed intellectual advice and edited the manuscript.
Funding
This work was supported by the National Institutes of Health [grant # DC04189], the Sidgmore Family Foundation, and the Hearing Health Foundation. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.129320/-/DC1
References
- Balasubramanyam K., Varier R. A., Altaf M., Swaminathan V., Siddappa N. B., Ranga U. and Kundu T. K. (2004). Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J. Biol. Chem. 279, 51163-51171. 10.1074/jbc.M409024200 [DOI] [PubMed] [Google Scholar]
- Barolo S. and Posakony J. W. (2002). Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev. 16, 1167-1181. 10.1101/gad.976502 [DOI] [PubMed] [Google Scholar]
- Basch M. L., Ohyama T., Segil N. and Groves A. K. (2011). Canonical Notch signaling is not necessary for prosensory induction in the mouse cochlea: insights from a conditional mutant of RBPjkappa. J. Neurosci. 31, 8046-8058. 10.1523/JNEUROSCI.6671-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito-Gonzalez A. and Doetzlhofer A. (2014). Hey1 and Hey2 control the spatial and temporal pattern of mammalian auditory hair cell differentiation downstream of Hedgehog signaling. J. Neurosci. 34, 12865-12876. 10.1523/JNEUROSCI.1494-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham N. A., Hassan B. A., Price S. D., Vollrath M. A., Ben-Arie N., Eatock R. A., Bellen H. J., Lysakowski A. and Zoghbi H. Y. (1999). Math1: an essential gene for the generation of inner ear hair cells. Science 284, 1837-1841. 10.1126/science.284.5421.1837 [DOI] [PubMed] [Google Scholar]
- Borghese L., Dolezalova D., Opitz T., Haupt S., Leinhaas A., Steinfarz B., Koch P., Edenhofer F., Hampl A. and Brüstle O. (2010). Inhibition of notch signaling in human embryonic stem cell-derived neural stem cells delays G1/S phase transition and accelerates neuronal differentiation in vitro and in vivo. Stem Cells 28, 955-964. 10.1002/stem.408 [DOI] [PubMed] [Google Scholar]
- Brigande J. V. and Heller S. (2009). Quo vadis, hair cell regeneration? Nat. Neurosci. 12, 679-685. 10.1038/nn.2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafaro J., Lee G. S. and Stone J. S. (2007). Atoh1 expression defines activated progenitors and differentiating hair cells during avian hair cell regeneration. Dev. Dyn. 236, 156-170. 10.1002/dvdy.21023 [DOI] [PubMed] [Google Scholar]
- Cai T., Seymour M. L., Zhang H., Pereira F. A. and Groves A. K. (2013). Conditional deletion of Atoh1 reveals distinct critical periods for survival and function of hair cells in the organ of Corti. J. Neurosci. 33, 10110-10122. 10.1523/JNEUROSCI.5606-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chardin S. and Romand R. (1995). Regeneration and mammalian auditory hair cells. Science 267, 707-711. 10.1126/science.7839151 [DOI] [PubMed] [Google Scholar]
- Chen P., Johnson J. E., Zoghbi H. Y. and Segil N. (2002). The role of Math1 in inner ear development: uncoupling the establishment of the sensory primordium from hair cell fate determination. Development 129, 2495-2505. [DOI] [PubMed] [Google Scholar]
- Crawford T. Q. and Roelink H. (2007). The notch response inhibitor DAPT enhances neuronal differentiation in embryonic stem cell-derived embryoid bodies independently of sonic hedgehog signaling. Dev. Dyn. 236, 886-892. 10.1002/dvdy.21083 [DOI] [PubMed] [Google Scholar]
- Dahl J. A. and Collas P. (2008). A rapid micro chromatin immunoprecipitation assay (microChIP). Nat. Protoc. 3, 1032-1045. 10.1038/nprot.2008.68 [DOI] [PubMed] [Google Scholar]
- Doetzlhofer A., Basch M. L., Ohyama T., Gessler M., Groves A. K. and Segil N. (2009). Hey2 regulation by FGF provides a Notch-independent mechanism for maintaining pillar cell fate in the organ of Corti. Dev. Cell 16, 58-69. 10.1016/j.devcel.2008.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driver E. C., Sillers L., Coate T. M., Rose M. F. and Kelley M. W. (2013). The Atoh1-lineage gives rise to hair cells and supporting cells within the mammalian cochlea. Dev. Biol. 376, 86-98. 10.1016/j.ydbio.2013.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher A. L., Ohsako S. and Caudy M. (1996). The WRPW motif of the hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein-protein interaction domain. Mol. Cell. Biol. 16, 2670-2677. 10.1128/MCB.16.6.2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortini M. E. (2009). Notch signaling: the core pathway and its posttranslational regulation. Dev. Cell 16, 633-647. 10.1016/j.devcel.2009.03.010 [DOI] [PubMed] [Google Scholar]
- Grbavec D. and Stifani S. (1996). Molecular interaction between TLE1 and the carboxyl-terminal domain of HES-1 containing the WRPW motif. Biochem. Biophys. Res. Commun. 223, 701-705. 10.1006/bbrc.1996.0959 [DOI] [PubMed] [Google Scholar]
- Grbavec D., Lo R., Liu Y. and Stifani S. (1998). Transducin-like Enhancer of split 2, a mammalian homologue of Drosophila Groucho, acts as a transcriptional repressor, interacts with Hairy/Enhancer of split proteins, and is expressed during neuronal development. Eur. J. Biochem. 258, 339-349. 10.1046/j.1432-1327.1998.2580339.x [DOI] [PubMed] [Google Scholar]
- Grogan S. P., Olee T., Hiraoka K. and Lotz M. K. (2008). Repression of chondrogenesis through binding of notch signaling proteins HES-1 and HEY-1 to N-box domains in the COL2A1 enhancer site. Arthritis Rheum. 58, 2754-2763. 10.1002/art.23730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama J., Bessho Y., Katoh K., Ookawara S., Fujioka M., Guillemot F. and Kageyama R. (2004). Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development 131, 5539-5550. 10.1242/dev.01436 [DOI] [PubMed] [Google Scholar]
- Hayashi T., Kokubo H., Hartman B. H., Ray C. A., Reh T. A. and Bermingham-McDonogh O. (2008). Hesr1 and Hesr2 may act as early effectors of Notch signaling in the developing cochlea. Dev. Biol. 316, 87-99. 10.1016/j.ydbio.2008.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms A. W., Abney A. L., Ben-Arie N., Zoghbi H. Y. and Johnson J. E. (2000). Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development 127, 1185-1196. [DOI] [PubMed] [Google Scholar]
- Hirata H., Yoshiura S., Ohtsuka T., Bessho Y., Harada T., Yoshikawa K. and Kageyama R. (2002). Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science 298, 840-843. 10.1126/science.1074560 [DOI] [PubMed] [Google Scholar]
- Hirata H., Bessho Y., Kokubu H., Masamizu Y., Yamada S., Lewis J. and Kageyama R. (2004). Instability of Hes7 protein is crucial for the somite segmentation clock. Nat. Genet. 36, 750-754. 10.1038/ng1372 [DOI] [PubMed] [Google Scholar]
- Iso T., Sartorelli V., Poizat C., Iezzi S., Wu H.-Y., Chung G., Kedes L. and Hamamori Y. (2001). HERP, a novel heterodimer partner of HES/E(spl) in Notch signaling. Mol. Cell. Biol. 21, 6080-6089. 10.1128/MCB.21.17.6080-6089.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarman A. P. and Groves A. K. (2013). The role of Atonal transcription factors in the development of mechanosensitive cells. Semin. Cell Dev. Biol. 24, 438-447. 10.1016/j.semcdb.2013.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarriault S., Brou C., Logeat F., Schroeter E. H., Kopan R. and Israel A. (1995). Signalling downstream of activated mammalian Notch. Nature 377, 355-358. 10.1038/377355a0 [DOI] [PubMed] [Google Scholar]
- Kageyama R., Ohtsuka T. and Tomita K. (2000). The bHLH gene Hes1 regulates differentiation of multiple cell types. Mol. Cells 10, 1-7. 10.1007/s10059-000-0001-0 [DOI] [PubMed] [Google Scholar]
- Kiernan A. E., Cordes R., Kopan R., Gossler A. and Gridley T. (2005). The Notch ligands DLL1 and JAG2 act synergistically to regulate hair cell development in the mammalian inner ear. Development 132, 4353-4362. 10.1242/dev.02002 [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Mizuno H., Imayoshi I., Furusawa C., Shirahige K. and Kageyama R. (2009). The cyclic gene Hes1 contributes to diverse differentiation responses of embryonic stem cells. Genes Dev. 23, 1870-1875. 10.1101/gad.1823109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korrapati S., Roux I., Glowatzki E. and Doetzlhofer A. (2013). Notch signaling limits supporting cell plasticity in the hair cell-damaged early postnatal murine cochlea. PLoS ONE 8, e73276 10.1371/journal.pone.0073276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamoorthy V., Carr T., de Pooter R. F., Akinola E. O., Gounari F. and Kee B. L. (2015). Repression of ccr9 transcription in mouse T lymphocyte progenitors by the notch signaling pathway. J. Immunol. 194, 3191-3200. 10.4049/jimmunol.1402443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanford P. J., Lan Y., Jiang R., Lindsell C., Weinmaster G., Gridley T. and Kelley M. W. (1999). Notch signalling pathway mediates hair cell development in mammalian cochlea. Nat. Genet. 21, 289-292. 10.1038/6804 [DOI] [PubMed] [Google Scholar]
- Lanford P. J., Shailam R., Norton C. R., Ridley T. and Kelley M. W. (2000). Expression of Math1 and HES5 in the cochleae of wildtype and Jag2 mutant mice. J. Assoc. Res. Otolaryngol. 1, 161-171. 10.1007/s101620010023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.-S., Liu F. and Segil N. (2006). A morphogenetic wave of p27Kip1 transcription directs cell cycle exit during organ of Corti development. Development 133, 2817-2826. 10.1242/dev.02453 [DOI] [PubMed] [Google Scholar]
- Li S., Mark S., Radde-Gallwitz K., Schlisner R., Chin M. T. and Chen P. (2008). Hey2 functions in parallel with Hes1 and Hes5 for mammalian auditory sensory organ development. BMC Dev. Biol. 8, 20 10.1186/1471-213X-8-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim D. J. and Anniko M. (1985). Developmental morphology of the mouse inner ear: a scanning electron microscopic observation. Acta Otolaryngol. 99 Suppl. 422, 5-69. 10.3109/00016488509121766 [DOI] [PubMed] [Google Scholar]
- Liu A., Li J., Marin-Husstege M., Kageyama R., Fan Y., Gelinas C. and Casaccia-Bonnefil P. (2006). A molecular insight of Hes5-dependent inhibition of myelin gene expression: old partners and new players. EMBO J. 25, 4833-4842. 10.1038/sj.emboj.7601352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. and Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402-408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lumpkin E. A., Collisson T., Parab P., Omer-Abdalla A., Haeberle H., Chen P., Doetzlhofer A., White P., Groves A., Segil N. et al. (2003). Math1-driven GFP expression in the developing nervous system of transgenic mice. Gene Expr. Patterns 3, 389-395. 10.1016/S1567-133X(03)00089-9 [DOI] [PubMed] [Google Scholar]
- Matei V., Pauley S., Kaing S., Rowitch D., Beisel K. W., Morris K., Feng F., Jones K., Lee J. and Fritzsch B. (2005). Smaller inner ear sensory epithelia in Neurog1 null mice are related to earlier hair cell cycle exit. Dev. Dyn. 234, 633-650. 10.1002/dvdy.20551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., Sakakibara S., Miyata T., Ogawa M., Shimazaki T., Weiss S., Kageyama R. and Okano H. (2000). The bHLH gene hes1 as a repressor of the neuronal commitment of CNS stem cells. J. Neurosci. 20, 283-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura M., Isaka F., Ishibashi M., Tomita K., Tsuda H., Nakanishi S. and Kageyama R. (1998). Structure, chromosomal locus, and promoter of mouse Hes2 gene, a homologue of Drosophila hairy and Enhancer of split. Genomics 49, 69-75. 10.1006/geno.1998.5213 [DOI] [PubMed] [Google Scholar]
- Ohtsuka T., Ishibashi M., Gradwohl G., Nakanishi S., Guillemot F. and Kageyama R. (1999). Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 18, 2196-2207. 10.1093/emboj/18.8.2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka T., Sakamoto M., Guillemot F. and Kageyama R. (2001). Roles of the basic helix-loop-helix genes Hes1 and Hes5 in expansion of neural stem cells of the developing brain. J. Biol. Chem. 276, 30467-30474. 10.1074/jbc.M102420200 [DOI] [PubMed] [Google Scholar]
- Raft S., Koundakjian E. J., Quinones H., Jayasena C. S., Goodrich L. V., Johnson J. E., Segil N. and Groves A. K. (2007). Cross-regulation of Ngn1 and Math1 coordinates the production of neurons and sensory hair cells during inner ear development. Development 134, 4405-4415. 10.1242/dev.009118 [DOI] [PubMed] [Google Scholar]
- Rebeiz M., Stone T. and Posakony J. W. (2005). An ancient transcriptional regulatory linkage. Dev. Biol. 281, 299-308. 10.1016/j.ydbio.2005.03.004 [DOI] [PubMed] [Google Scholar]
- Riccio O., van Gijn M. E., Bezdek A. C., Pellegrinet L., van Es J. H., Zimber-Strobl U., Strobl L. J., Honjo T., Clevers H. and Radtke F. (2008). Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO Rep. 9, 377-383. 10.1038/embor.2008.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson D. W., Alosi J. A. and Cotanche D. A. (2004). Direct transdifferentiation gives rise to the earliest new hair cells in regenerating avian auditory epithelium. J. Neurosci. Res. 78, 461-471. 10.1002/jnr.20271 [DOI] [PubMed] [Google Scholar]
- Ruben R. J. (1967). Development of the inner ear of the mouse: a radioautographic study of terminal mitoses. Acta Otolaryngol. Suppl. 220, 221-244. [PubMed] [Google Scholar]
- Sher A. E. (1971). The embryonic and postnatal development of the inner ear of the mouse. Acta Otolaryngol. Suppl. 285, 1-77. [PubMed] [Google Scholar]
- Stone J. S. and Cotanche D. A. (2007). Hair cell regeneration in the avian auditory epithelium. Int. J. Dev. Biol. 51, 633-647. 10.1387/ijdb.072408js [DOI] [PubMed] [Google Scholar]
- Takata T. and Ishikawa F. (2003). Human Sir2-related protein SIRT1 associates with the bHLH repressors HES1 and HEY2 and is involved in HES1- and HEY2-mediated transcriptional repression. Biochem. Biophys. Res. Commun. 301, 250-257. 10.1016/S0006-291X(02)03020-6 [DOI] [PubMed] [Google Scholar]
- Tateya T., Imayoshi I., Tateya I., Ito J. and Kageyama R. (2011). Cooperative functions of Hes/Hey genes in auditory hair cell and supporting cell development. Dev. Biol. 352, 329-340. 10.1016/j.ydbio.2011.01.038 [DOI] [PubMed] [Google Scholar]
- Troost T., Schneider M. and Klein T. (2015). A re-examination of the selection of the sensory organ precursor of the bristle sensilla of Drosophila melanogaster. PLoS Genet. 11, e1004911 10.1371/journal.pgen.1004911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Zang C., Rosenfeld J. A., Schones D. E., Barski A., Cuddapah S., Cui K., Roh T.-Y., Peng W., Zhang M. Q. et al. (2008). Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 40, 897-903. 10.1038/ng.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber D., Heisig J., Kneitz S., Wolf E., Eilers M. and Gessler M. (2015). Mechanisms of epigenetic and cell-type specific regulation of Hey target genes in ES cells and cardiomyocytes. J. Mol. Cell. Cardiol. 79, 79-88. 10.1016/j.yjmcc.2014.11.004 [DOI] [PubMed] [Google Scholar]
- White P. M., Doetzlhofer A., Lee Y. S., Groves A. K. and Segil N. (2006). Mammalian cochlear supporting cells can divide and trans-differentiate into hair cells. Nature 441, 984-987. 10.1038/nature04849 [DOI] [PubMed] [Google Scholar]
- Winkler C. J., Ponce A. and Courey A. J. (2010). Groucho-mediated repression may result from a histone deacetylase-dependent increase in nucleosome density. PLoS ONE 5, e10166 10.1371/journal.pone.0010166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods C., Montcouquiol M. and Kelley M. W. (2004). Math1 regulates development of the sensory epithelium in the mammalian cochlea. Nat. Neurosci. 7, 1310-1318. 10.1038/nn1349 [DOI] [PubMed] [Google Scholar]
- Yamamoto N., Tanigaki K., Tsuji M., Yabe D., Ito J. and Honjo T. (2006). Inhibition of Notch/RBP-J signaling induces hair cell formation in neonate mouse cochleas. J. Mol. Med. 84, 37-45. 10.1007/s00109-005-0706-9 [DOI] [PubMed] [Google Scholar]
- Yamamoto N., Chang W. and Kelley M. W. (2011). Rbpj regulates development of prosensory cells in the mammalian inner ear. Dev. Biol. 353, 367-379. 10.1016/j.ydbio.2011.03.016 [DOI] [PubMed] [Google Scholar]
- Yang H., Xie X., Deng M., Chen X. and Gan L. (2010). Generation and characterization of Atoh1-Cre knock-in mouse line. Genesis 48, 407-413. 10.1002/dvg.20633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q.-L. and Smith A. G. (2003). Defined conditions for neural commitment and differentiation. Methods Enzymol. 365, 327-341. 10.1016/S0076-6879(03)65023-8 [DOI] [PubMed] [Google Scholar]
- Zheng X., Tsuchiya K., Okamoto R., Iwasaki M., Kano Y., Sakamoto N., Nakamura T. and Watanabe M. (2011). Suppression of hath1 gene expression directly regulated by hes1 via notch signaling is associated with goblet cell depletion in ulcerative colitis. Inflamm. Bowel Dis. 17, 2251-2260. 10.1002/ibd.21611 [DOI] [PubMed] [Google Scholar]
- Zine A. and de Ribaupierre F. (2002). Notch/Notch ligands and Math1 expression patterns in the organ of Corti of wild-type and Hes1 and Hes5 mutant mice. Hear. Res. 170, 22-31. 10.1016/S0378-5955(02)00449-5 [DOI] [PubMed] [Google Scholar]
- Zine A., Aubert A., Qiu J., Therianos S., Guillemot F., Kageyama R. and de Ribaupierre F. (2001). Hes1 and Hes5 activities are required for the normal development of the hair cells in the mammalian inner ear. J. Neurosci. 21, 4712-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]