Abstract
The heterochronic genes lin-28, let-7 and lin-41 regulate fundamental developmental transitions in animals, such as stemness versus differentiation and juvenile versus adult states. We identify a new heterochronic gene, lep-2, in Caenorhabditis elegans. Mutations in lep-2 cause a delay in the juvenile-to-adult transition, with adult males retaining pointed, juvenile tail tips, and displaying defective sexual behaviors. In both sexes, lep-2 mutants fail to cease molting or produce an adult cuticle. We find that LEP-2 post-translationally regulates LIN-28 by promoting LIN-28 protein degradation. lep-2 encodes the sole C. elegans ortholog of the Makorin (Mkrn) family of proteins. Like lin-28 and other heterochronic pathway members, vertebrate Mkrns are involved in developmental switches, including the timing of pubertal onset in humans. Based on shared roles, conservation and the interaction between lep-2 and lin-28 shown here, we propose that Mkrns, together with other heterochronic genes, constitute an evolutionarily ancient conserved module regulating switches in development.
KEY WORDS: Mkrn, Heterochronic pathway, LIN-28/let-7 axis, Developmental timing
Highlighted article: The C. elegans Makorin homolog lep-2 is a heterochronic gene required for the proper timing of the juvenile-to-adult transition. It acts in the LIN-28/let-7 pathway.
INTRODUCTION
LIN-28 is a well-conserved RNA-binding protein that acts at the core of a molecular switch regulating fundamental developmental transitions. For example, LIN-28 is involved in the transition between stemness and differentiation in normal development, wound healing, induced pluripotent stem cells and cancer (Shyh-Chang et al., 2013; Thornton and Gregory, 2012; Tsialikas and Romer-Seibert, 2015). Another transition regulated by LIN-28 is the juvenile-to-adult (J/A) transition in mammals, flies and nematodes (Thornton and Gregory, 2012; Tsialikas and Romer-Seibert, 2015).
In the nematode Caenorhabditis elegans, the J/A and larval transitions are regulated by the ‘heterochronic pathway’, of which LIN-28 is a founding member (Ambros and Horvitz, 1984). During the transition from the fourth larval stage to the adult, C. elegans worms synthesize an adult-specific cuticle and undergo morphogenesis to form a vulva in hermaphrodites and the copulatory structure called the ‘male tail’ in males (Cox et al., 1980; Sulston et al., 1980; Sulston and Horvitz, 1977). Also, the seam cells, a row of stem-like cells along each side of the animal, exit the cell cycle, fuse and form a ridged cuticular structure called alae (Ambros and Horvitz, 1984). These cells and their stage-specific division pattern have been the focus of most studies elucidating the heterochronic pathway in C. elegans (Rougvie and Moss, 2013). Mutations in heterochronic genes cause alae to appear earlier or later than normal. Loss-of-function lin-28 mutations result in the precocious formation of alae prior to the adult stage (Ambros and Horvitz, 1984; Moss et al., 1997). By contrast, constitutive expression of LIN-28 causes reiteration of early seam cell states, thus preventing adult alae from forming (Moss et al., 1997).
That LIN-28 regulates the J/A transition in mammals as well as invertebrates has been demonstrated by several studies. In humans and mice, genetic variation in lin-28 homologs is associated with heterochronic transformations at the age when the J/A transition (pubertal onset) occurs (Gajdos et al., 2010; Ong et al., 2009; Perry et al., 2009; Zhu et al., 2010). Transgenic mice overexpressing Lin28a have delayed puberty; Lin28a knockout mice exhibit precocious puberty phenotypes similar to those associated with LIN28B genetic variants in humans (Gajdos et al., 2010; Hannon et al., 2006; Ong et al., 2009; Perry et al., 2009; Zhu et al., 2010, 2011).
It is increasingly clear from studies in several systems that lin-28 usually works with two other genes, let-7 and lin-41/Trim71, in a conserved ‘axis’ of negative interactions (Thornton and Gregory, 2012; Tsialikas and Romer-Seibert, 2015). Besides being involved in the J/A transition, this axis controls many stemness-differentiation transitions. LIN-28 is highly expressed in undifferentiated embryonic stem cells (ESCs) and declines during subsequent development (Thornton and Gregory, 2012; Tsialikas and Romer-Seibert, 2015). LIN-28 inhibits the biogenesis of the mature form of the let-7 microRNA (miRNA) (Newman et al., 2008; Piskounova et al., 2008; Van Wynsberghe et al., 2011; Viswanathan et al., 2008). When LIN-28 is downregulated, mature let-7 miRNA is produced, directing cells toward differentiated/adult fates (Copley et al., 2013; Hayes and Ruvkun, 2006; Rybak et al., 2008; Thornton and Gregory, 2012; Tsialikas and Romer-Seibert, 2015; Yu et al., 2007) by inhibiting the translation of primarily lin-41 and other target mRNAs (Ecsedi and Grosshans, 2013; Long et al., 2007; Slack et al., 2000; Vella et al., 2004a,b). LIN-41 homologs are proposed to silence the translation of target mRNAs (Ecsedi and Grosshans, 2013; Ecsedi et al., 2015; Loedige et al., 2013; Sonoda and Wharton, 2001; Vella et al., 2004a,b) that otherwise promote an undifferentiated/juvenile state (Chang et al., 2012; Ecsedi and Grosshans, 2013; Rybak et al., 2009).
Because of the conservation of the lin-28/let-7/lin-41 axis and its role in developmental transitions, the C. elegans J/A transition provides a good model for studying how this axis is regulated. In addition to studying the J/A transition in seam cells, we focus on male tail tip morphogenesis, which has allowed us to uncover novel heterochronic regulators. Here, we report the discovery that another conserved protein, Makorin (Mkrn), regulates LIN-28 at the post-translational level. Evidence accumulated from vertebrate systems (Abreu et al., 2013; Kim et al., 2005; Mai et al., 2011; Salvatico et al., 2010) points to roles for Mkrns in developmental processes that overlap with those regulated by LIN-28. Thus, the regulatory interaction between Mkrn and LIN-28 that we find in C. elegans might be of general significance in animal development.
RESULTS
Four alleles of lep-2 identified in screens for male defects
The C. elegans male tail tip provides a simple model with which to study the regulation of the J/A transition (Nguyen et al., 1999). This structure comprises four epithelial cells that originate during embryogenesis and form a conical, pointed tail tip. The tail tip remains pointed throughout the lifespan of the hermaphrodite. In males, however, it undergoes a morphogenetic change during the J/A transition resulting in the rounded tail of the adult. During this tail tip morphogenesis (TTM), the four tail tip cells fuse to form a syncytium, which changes shape and moves anteriorly (Fig. 1A). TTM is known to be under the control of the heterochronic genes lin-41 and let-7 (Del Rio-Albrechtsen et al., 2006). In lin-41 loss-of-function (lf) mutants, TTM occurs precociously in the L3 stage, whereas gain-of-function (gf) mutations cause a delay (Del Rio-Albrechtsen et al., 2006; Nguyen et al., 1999). let-7(lf) males also fail to undergo TTM in L4, and adults retain the juvenile tail tip (Del Rio-Albrechtsen et al., 2006; Vadla et al., 2012). This phenotype is called Lep (Nguyen et al., 1999).
Fig. 1.
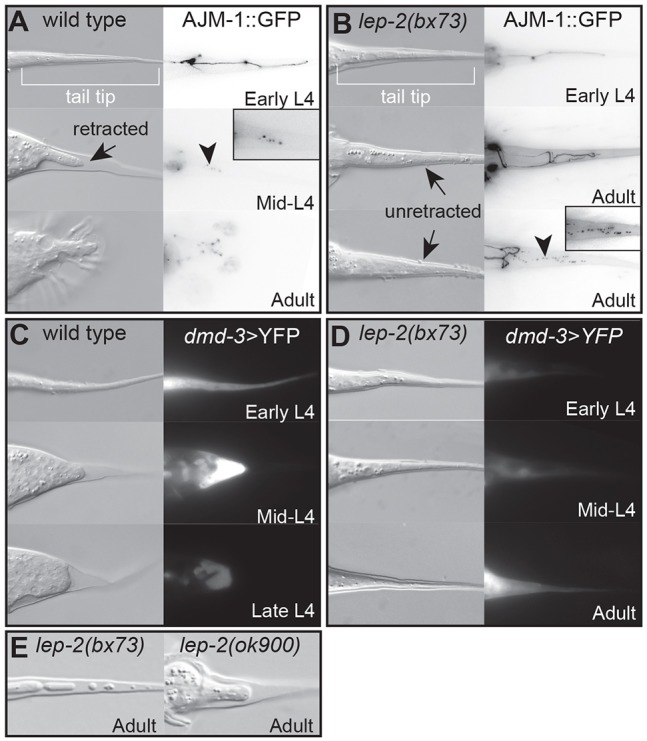
Tail tip morphogenesis in wild-type and lep-2(lf) mutant C. elegans. (A,B) Developmental profiles of tail tip morphogenesis (TTM) in wild-type (A) and lep-2(lf) mutant (B) males. Left panels show DIC images; right panels shows adherens junctions at the cell boundaries visualized by AJM-1::GFP. The tail tip is conical in early L4. In wild type, tail tip retraction (arrow) and cell fusion (visible as breakdown of the adherens junctions, arrowhead and inset) begins in mid L4. The adult tail tip is fully retracted. lep-2(bx73) male tail tips do not undergo morphogenesis during L4. The adult tail tips are long, unretracted (arrows) and the tail tip cells are unfused. Punctate AJM-1::GFP staining indicates tail tip cell fusion in adult males (arrowhead and inset). (C,D) Expression of a dmd-3 transcriptional reporter (right) during TTM in wild-type males (C) and lep-2(lf) mutants (D). In wild type, dmd-3>YFP is expressed in the tail tip from early to mid-L4. Expression diminishes in late L4. In lep-2(bx73) mutants, the reporter is not expressed in early or mid-L4, but turns on in the tail tips of some late L4 and adult males. (E) DIC images of tail tips of an adult lep-2(bx73) and lep-2(ok900) mutant, showing adult tail tip retraction.
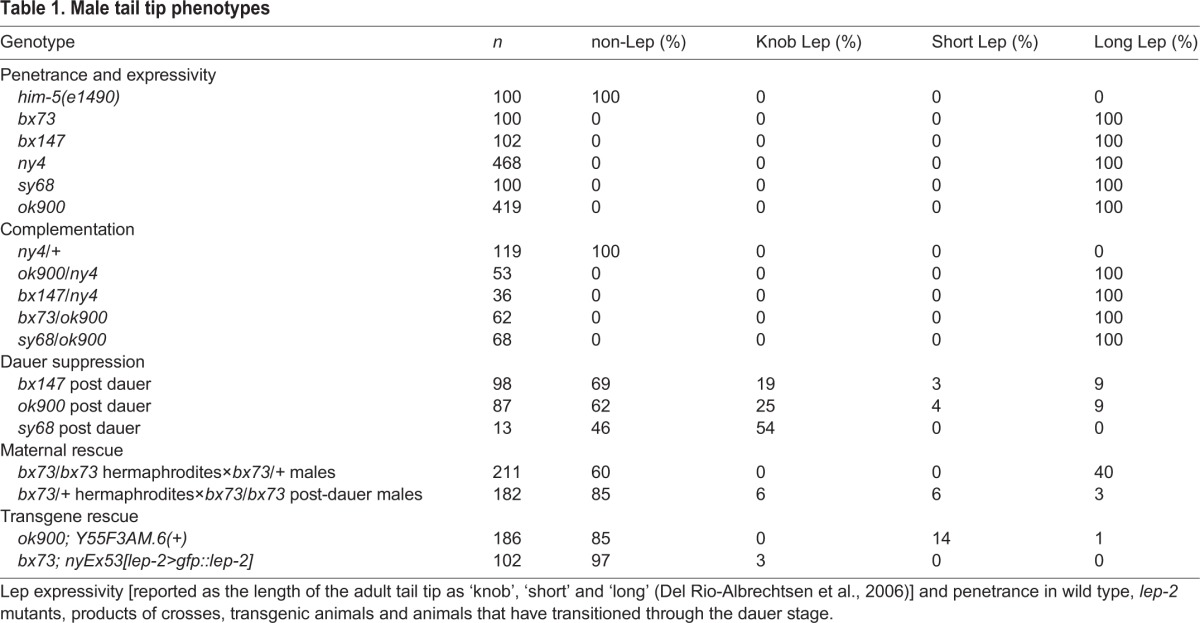
To identify genes involved in TTM, we performed a TMP/UV mutagenesis screen for Lep male tails. One mutation identified in this screen, ny4, failed to complement three other recessive Lep mutants (bx73, bx147 and sy68) identified in independent forward genetic screens (S. Baird, S. Emmons and P. Sternberg, personal communication) (Table 1). A further mutation, ok900, with the same phenotype was obtained from the C. elegans Deletion Mutant Consortium (C. elegans Deletion Mutant Consortium, 2012). In adult homozygous lep-2(lf) males, the tail tip extends far beyond the fan, and L4 males show no evidence of retraction or fusion of tail tip cells (Fig. 1A,B). This phenotype is 100% penetrant (Table 1). lep-2(lf) males exhibit otherwise normal male organs (i.e. gonads, spicules, fans and rays), yet are unable to sire progeny. Homozygous lep-2(lf) males born to a heterozygous mother have wild-type tails (Table 1), indicating that lep-2 may be deposited by the mother.
Table 1.
Male tail tip phenotypes
lep-2 is required for the proper timing of TTM
TTM is controlled by the master regulatory gene dmd-3, which integrates temporal, spatial and sexual cues and controls downstream effectors of tail tip cell fusion and retraction (Mason et al., 2008; Nelson et al., 2011). dmd-3 is expressed in tail tip cells of wild-type males from early to mid-L4 (Fig. 1C) (Mason et al., 2008). By investigating the expression of a dmd-3>YFP transcriptional reporter we found that, in lep-2 mutant males, dmd-3 transcription does not occur during L4 but is delayed to adulthood (Fig. 1D).
The Lep phenotype can result from a general failure of TTM, as in dmd-3 mutants (Mason et al., 2008), or from a failure to initiate TTM at the correct time, as in lin-41(gf) and let-7(lf) mutants in which TTM occurs in adults (Del Rio-Albrechtsen et al., 2006). To test if lep-2 is involved in the timing of TTM, we studied tail tips of young adult lep-2(lf) males. We observed that the tail tip cells had detached from the cuticle and began to round up and retract (Fig. 1E). We also found evidence of tail tip cell fusion in older lep-2(lf) males (Fig. 1B, inset). Delayed, adult TTM was observed in all lep-2 alleles.
Taken together, these observations suggest that lep-2 is required for the proper timing of TTM. We thus hypothesized that lep-2 acts in the heterochronic pathway.
lep-2 affects tissues other than the tail tip
Several mutants for lep-2 were isolated in screens for male phenotypes, but never in screens that focused on the development of seam cells. We therefore tested if lep-2 mutations affect the male tail tip specifically or also other tissues.
Observation of lep-2 mutants on culture plates revealed that some males and hermaphrodites had undergone an additional molt. Such males usually failed to fully shed the molted cuticle and died by rupturing through the cloaca. Occasional survivors had no spicules or fan and the rays appeared as finger-like processes. A systematic study of males 1 day after they had reached adulthood revealed that the majority had initiated an additional molt (Table S1). Thus, mutations in lep-2 affect hypodermal tissues other than the tail tip and the animals fail to exit the molting cycle. To test whether lep-2(lf) adults have an adult cuticle, we examined the adult collagen ROL-1, which is absent from the larval cuticle (Cox et al., 1980). Mutation of rol-1 causes a highly penetrant (95.2%, n=289) Rol phenotype in adults only (Table S1). However, in rol-1(e91); lep-2(ok900) mutants, the Rol phenotype is rare (8.5%, n=258) (Table S1). Thus, lep-2 is required for formation of the adult cuticle.
Interestingly, although lep-2(lf) adults lack the adult collagen ROL-1, they display normal alae. To test whether seam cell fusion or seam cell exit from the cell cycle was affected in lep-2 mutants, we investigated young adults in which adherens junctions and seam cell nuclei were labeled with GFP. All had a fully fused seam and the normal number of seam cells (Table S2). We conclude that lep-2 is required for the terminal differentiation of most of the hypodermis, but that in the seam cells it is either not needed or acts redundantly with other factors.
Two alleles of lep-2 were isolated in screens for defects in male mating: lep-2(sy68) was found in a screen for males that do not sire progeny (P. Sternberg, personal communication); and lep-2(bx147) was found in a screen for males that do not show the typical leaving behavior in the absence of hermaphrodites (Lipton et al., 2004). Indeed, all mutant alleles of lep-2 cause male infertility, although gonads, spicules and fans appear normal, and it is known that a Lep tail alone does not affect male fertility (Del Rio-Albrechtsen et al., 2006). To test if lep-2 affects mating behavior, we investigated the performance of lep-2 mutant males in a standard mating assay. Wild-type C. elegans males mate with a stereotypical pattern of behaviors (Barr and Garcia, 2006) and, in our assays, all wild-type males quickly located a vulva and copulated (Fig. 2, Fig. S1, Movie 1). By contrast, many of the lep-2 mutant males failed to contact a hermaphrodite during the 60 min assay time and only two out of 32 copulated (Fig. 2). Of the lep-2(lf) males that initiated mating behavior, several lost contact with the hermaphrodite (Movie 2). These observations indicate that lep-2 males are infertile because they do not perform proper mating behavior. Considering the other developmentally delayed phenotypes of lep-2 mutants, it is possible that the behavioral defects are due to retarded differentiation of the nervous system.
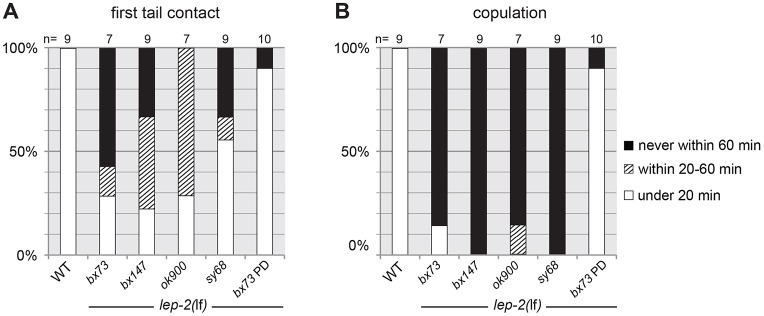
Fig. 2.
Male mating behavior is impaired in lep-2(lf). Mating behavior of individual wild-type (WT) and lep-2 mutant males was recorded over a period of 60 min. Wild-type males showed the known suite of mating behaviors (Barr and Garcia, 2006), which include the initial contact of the male tail with the hermaphrodite, scanning the hermaphrodite for the vulva, precise ventral turns when the male reaches the end of its mate, and a successful copulation. lep-2(lf) males were unable to perform normal mating behavior. Shown are data for (A) the time it took a male to make first contact with a hermaphrodite and (B) the time it took for the male to copulate. Wild-type males contacted a hermaphrodite and copulated in less than 20 min. Most mutant males took much longer to contact a mate and all but two males never copulated. The defect is rescued in males that had developed as dauer larvae (PD).
The mutant phenotypes of the heterochronic genes lin-4, lin-14, lin-28 and hbl-1 are suppressed when the animals pass through the dauer stage (Abrahante et al., 2003; Euling and Ambros, 1996; Liu and Ambros, 1991). We found that the same is true for lep-2. After passage through the dauer stage, lep-2 mutant males had fully retracted tails (Table 1), there was no evidence of supernumerary molts (Table S1) and the Rol phenotype was restored in rol-1(e91); lep-2(lf) animals, indicating that they synthesized an adult cuticle (Table S1). We also found that nine out of ten adult males that had developed as dauers performed normal mating behavior and sired progeny (Fig. 2, Fig. S1).
Taken together, our observations of the different mutant phenotypes confirm that lep-2 is a bona fide heterochronic gene affecting the J/A transition.
LEP-2 is a Makorin
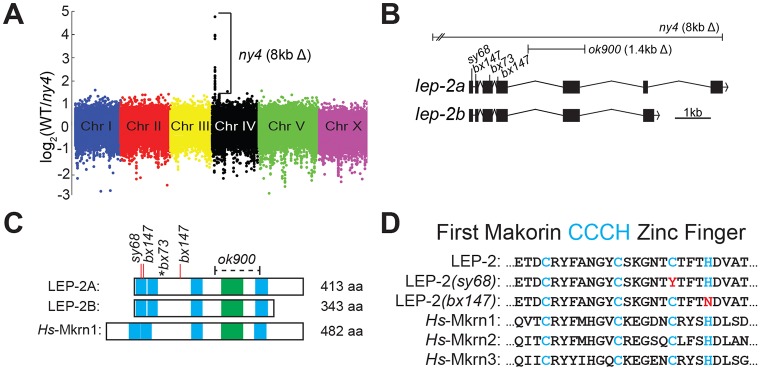
The lep-2(ny4) allele was generated with TMP/UV, which results in genomic deletions (Yandell et al., 1994). Therefore, we chose to map this allele by array comparative genomic hybridization (array CGH) (Flibotte and Moerman, 2008). Array CGH identified a region of ∼8 kb on the left arm of chromosome IV that is absent in ny4 (Fig. 3A, Fig. S2). This deletion removes coding sequences from Y55F3AM.6 and Y55F3AM.7 (Fig. S2B). A mutant allele, ok900, in which 1.4 kb of the Y55F3AM.6 coding sequence is deleted, produces males with a Lep phenotype and fails to complement the other lep-2 alleles (Table 1). Transformation of ok900 animals with wild-type Y55F3AM.6 DNA rescues the Lep phenotype to 95% (Table 1). Thus, lep-2 corresponds to Y55F3AM.6.
Fig. 3.
Location and mapping of lep-2. (A) Array comparative genomic hybridization (array CGH) graph of the C. elegans whole-genome tiling array, color-coded by chromosome. The dots represent the log2 ratio of wild-type to mutant [lep-2(ny4)] signal for each probe. Chromosome IV shows a ∼8 kb long region with a higher DNA content in the wild-type sample relative to the mutant, indicating a deletion. (B) lep-2 gene structure, depicting two isoforms. The lesions affecting this locus are labeled: ny4 and ok900 are deletions; bx73, bx147 and sy68 harbor point mutations within the coding sequence of lep-2. (C) Schematic of the predicted LEP-2 protein: two predicted isoforms (LEP-2A and LEP-2B), both of which include the full complement of Mkrn motifs (zinc fingers in blue, RING domain in green) with the lesions in lep-2 alleles indicated by a bracket (deletion in ok900), red bars (amino acid substitutions in bx147 and sy68) and an asterisk (premature stop in bx73). Human MKRN1 is shown for comparison. (D) An alignment of the first Mkrn zinc finger from LEP-2, two mutant forms, and three human Mkrns. Blue letters indicate conserved amino acids; red letters indicate amino acid substitutions in lep-2 mutants.
lep-2 encodes a protein with two predicted isoforms (Fig. 3B) and is the sole C. elegans Makorin (Mkrn). Mkrns are putative developmental regulators that can bind nucleic acid and act as E3 ubiquitin ligases and are conserved in eukaryotes from humans to rice (Arumugam et al., 2007; Gray et al., 2000). Mkrns, including LEP-2, possess a conserved array of protein motifs: C3H-type zinc fingers (ZFs) flanking a RING domain (Fig. 3C). The deletion ny4 removes most of Y55F3AM.6, suggesting that it is a molecular null mutation. We found that the lep-2 alleles sy68 and bx147 are point mutations that cause amino acid changes in conserved cysteine or histidine residues within the first C3H-ZF (Fig. 3C,D). The bx73 mutation introduces a premature stop codon after the first two C3H-ZFs (Fig. 3C).
lep-2 is widely expressed throughout development
To examine where and when lep-2 is expressed, we generated a GFP::LEP-2 translational reporter that includes the coding region of the shorter isoform plus 235 bp downstream, and is driven by a 2 kb region upstream of the start codon (Fig. S3A). Transformation of lep-2(bx73) with this construct rescued the Lep phenotype (eight lines, n>100, Table 1). GFP::LEP-2 is expressed throughout development from embryo to adult. It is seen in many tissues, including seam, tail tip and other hypodermal cells, head and tail neurons, pharynx, intestine and the developing hermaphroditic somatic gonad (Fig. S3). Expression seems to be excluded from body muscle cells. The reporter is diffusely expressed in the cytoplasm but absent from nuclei. Therefore, the cytoplasm might be the primary site of action for LEP-2.
lep-2 acts upstream of lin-28 in the heterochronic pathway
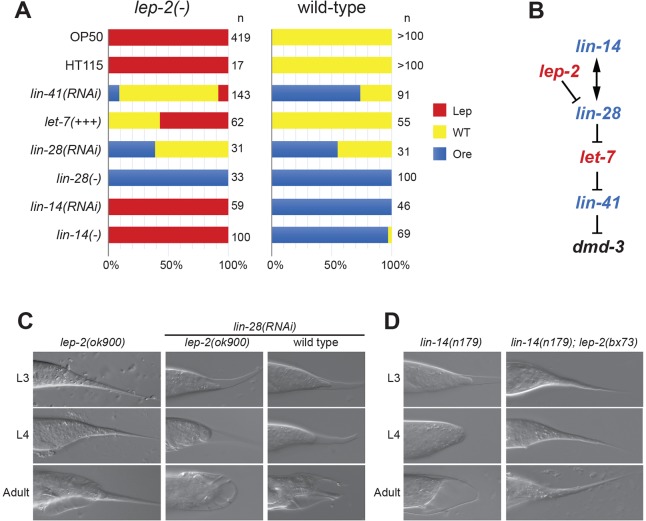
Two heterochronic genes known to be involved in TTM are lin-41 and let-7 (Del Rio-Albrechtsen et al., 2006; Vadla et al., 2012). To establish the genetic position of lep-2 in the heterochronic pathway relative to lin-41 and let-7, we performed epistasis experiments with RNAi and an overexpression construct. Knockdown of lin-41 with RNAi in wild-type animals causes TTM to begin one stage too early, leading to over-retracted (Ore) male tails (Del Rio-Albrechtsen et al., 2006). lin-41 RNAi mostly abolished the Lep phenotype of lep-2 mutants (Fig. 4A). Thus, lep-2 acts upstream of, or in parallel to, lin-41 in TTM. lin-41 mRNA is negatively regulated by let-7, the loss-of-function alleles of which have a strong Lep phenotype (Del Rio-Albrechtsen et al., 2006; Vadla et al., 2012). To test the genetic interaction between lep-2 and let-7, we used the let-7 overexpression construct zaIs3[let-7(+); myo-3::GFP], which increases the level of mature let-7 ∼5-fold (Büssing et al., 2008). In the wild-type background, this transgene causes 15% of males to precociously retract their tail tips during the L3 stage (indicated by a rounded L4 tail tip) (Table 2). Adult male tails appear wild type (Fig. 4A), suggesting that TTM is robust to moderate changes in let-7 levels. However, addition of let-7 via the zaIs3 transgene in the lep-2(lf) genetic background was sufficient to suppress the Lep phenotype in 43% of males (Fig. 4A). We conclude that lep-2 acts upstream of, or in parallel to, let-7.
Fig. 4.
Genetic interaction of lep-2 with other heterochronic genes. (A) Penetrance and expressivity of the adult tail tip phenotypes in lep-2(lf) (left) and wild-type (right) males alone and in combination with RNAi knockdown, gain-of-function or loss-of-function of other heterochronic genes. Data are for lep-2(ok900), except in lin-28(−) and lin-14(−) double mutants, where lep-2(ny4) was used. The penetrance of the Ore phenotype in RNAi-treated wild-type males indicates that lin-41(RNAi) had an efficiency of ∼75%, which is comparable to the findings of Del Rio-Albrechtsen et al. (2006). lin-28(RNAi) was 50% efficient, since loss of lin-28 by mutation led to 100% Ore tails. OP50 indicates that the animals were raised under standard culture conditions. HT115 is the bacterial strain used as control for RNAi experiments. let-7(+++) represents the transgene zaIs3, which leads to overexpression of let-7. (B) Genetic interactions between heterochronic genes evaluated in the epistasis experiments. Red letters indicate retarded phenotypes, blue letters precocious phenotypes in loss-of-function mutants. (C,D) DIC micrographs of male tail tips in single-mutant and double-mutant or RNAi-treated animals. (C) Wild-type and lep-2(lf) mutant males missing LIN-28 either by mutation or RNAi show precocious TTM as early as L3, have rounded L4 tails and display the Ore phenotype in adults. The lep-2(lf) phenotype is shown for comparison on the left. (D) TTM occurs precociously in lin-14(n179) males, but not in lin-14(n179); lep-2(bx73) males.
Table 2.
Timing of precocious TTM in heterochronic gene mutants, RNAi knockdowns or transgene insertion
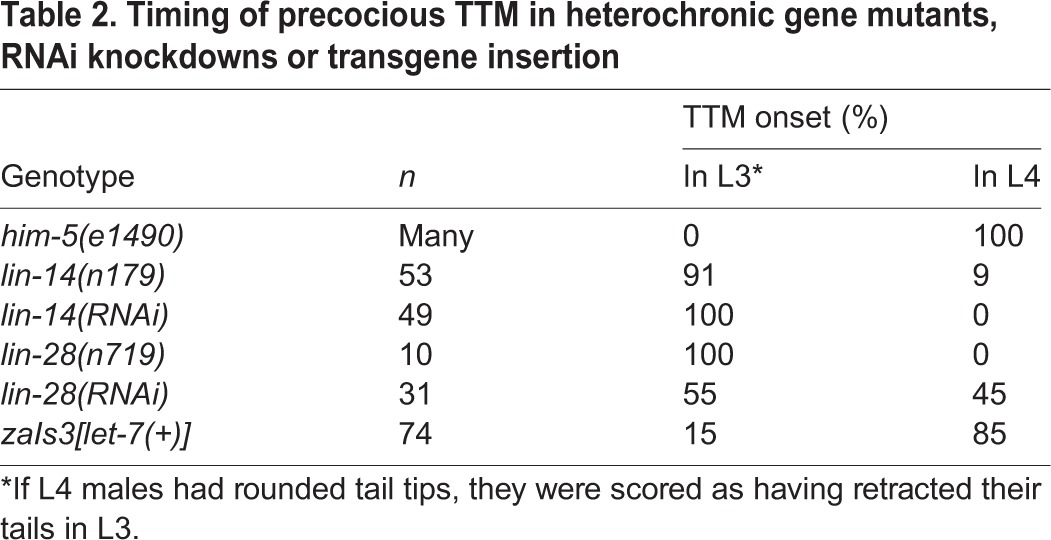
From work on seam cell development it is known that LIN-28 inhibits the biogenesis of mature let-7 miRNA, thereby preventing early L3-to-L4 transition and precocious adult fates (Vadla et al., 2012; Van Wynsberghe et al., 2011). However, it was not known if lin-28 plays a role in TTM. We therefore investigated L3, L4 and adult males in which lin-28 was mutated or knocked down by RNAi. In both situations, TTM began precociously during the L3 stage and continued during L4, leading to a severe Ore phenotype in adults (Fig. 4A,C, Table 2). Knowing that lin-28 regulates the timing of TTM, we next tested whether it is epistatic to lep-2 by knocking down lin-28 by RNAi in a lep-2 mutant and by creating a lep-2(lf); lin-28(lf) double mutant. In both experiments, males initiated TTM in the L3 stage, leading to adults with wild-type-like or Ore tails, an almost complete copy of the lin-28(lf) phenotype (Fig. 4C). Therefore, lep-2 acts upstream of, or in parallel to, lin-28.
Another heterochronic gene with a precocious phenotype is lin-14. In the context of seam development, lin-14 and lin-28 promote each other's expression via an indirect feedback circuit (Pepper et al., 2004). We found that loss of LIN-14 causes a strong precocious phenotype, with TTM beginning in L3 and Ore tails in adult males (Fig. 4A,D, Table 2). Thus, lin-14 is also involved in the timing of TTM. Males that were mutant for both lin-14 and lep-2 showed no change in penetrance or expressivity of the Lep phenotype typical for lep-2.
We conclude that lep-2 acts downstream of lin-14 and upstream of lin-28, and that lep-2 is a negative regulator of lin-28.
LEP-2 regulates LIN-28 at the protein level by affecting its stability
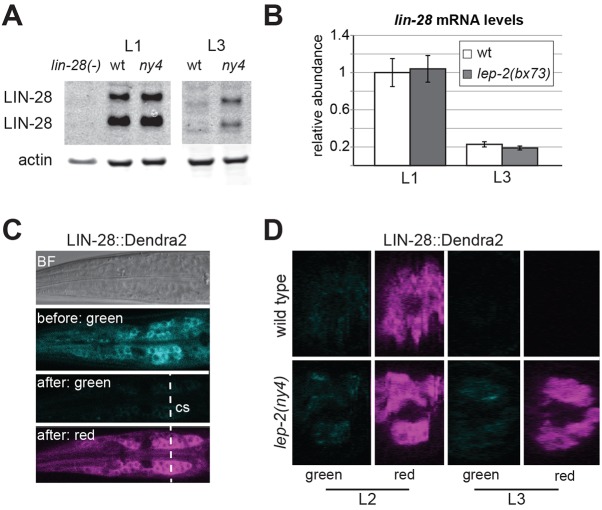
Our epistasis experiments indicated that lep-2 represses lin-28. We thus hypothesized that the level of LIN-28 is elevated in lep-2(lf) animals. To test this, we examined the expression of a lin-28>LIN-28::GFP::lin-28_3′UTR reporter in lep-2(lf) mutants. In wild-type L1 and L2 larvae, this reporter is cytoplasmically expressed in the epidermis, including the seam and the tail tip, in the pharynx and in the nervous system. Fluorescence is particularly bright in ganglia in the pharynx and anal regions. Reporter expression diminishes during the L3 stage and is largely absent by L4. In lep-2(lf), expression of the reporter in L1 and L2 was similar to that observed in wild-type animals. However, fluorescence remained bright until adulthood in all tissues including the tail tip (Fig. S4). Thus, in lep-2(lf) mutants, LIN-28 protein is not properly downregulated. To determine if endogenous LIN-28 protein levels are affected by lep-2(lf), we performed western blot analysis on proteins isolated from L1 and L3 wild-type and lep-2(ny4) animals (Fig. 5A) using polyclonal anti-LIN-28 antibodies that recognize two LIN-28 isoforms (Seggerson et al., 2002; Weaver et al., 2014). We found that in wild-type animals LIN-28 levels between L1 and L3 decreased by a factor of 6, consistent with the literature (Seggerson et al., 2002; Weaver et al., 2014). However, in lep-2 mutants (bx73, ny4, ok900 were tested), LIN-28 levels decreased by only a factor of 3, resulting in L3 levels twice as high as in wild type (Fig. 5A, Fig. S4B).
Fig. 5.
LIN-28 is regulated by LEP-2. (A) Western blot analysis of LIN-28 levels in wild-type (wt) and lep-2(ny4) L1 and L3 larvae. (B) lin-28 mRNA levels at L1 and L3 stages in wild-type and lep-2(bx73) mutants normalized to wild-type L1 levels. Error bars indicate s.d. (C,D) Expression of LIN-28::Dendra2 in the pharynx region of wild-type and lep-2(ny4) animals. (C) Images of the same focal plane in the pharynx region showing signal in (from top to bottom), bright field (BF), green channel before photoconversion, green channel after photoconversion, red channel after photoconversion. The dashed line indicates the position of the cross-section view through the z-plane shown in D. (D) Optical cross-section through the pharynx in one wild-type (top row) and one lep-2(ny4) (bottom row) animal at the L2 and L3 stage showing LIN-28::Dendra2 expression after photoconversion. The absence of green fluorescence in L2 animals shows that photoconversion was near complete. In the wild-type L3 animal, LIN-28::Dendra2 could not be detected in either channel, showing that LIN-28::Dendra2 is degraded normally. In the lep-2(ny4) animal, fluorescence was detected in the red channel only, indicating that LIN-28::Dendra2 persisted and no new protein was produced. 22 wild-type and 23 lep-2 mutant animals were examined, with results similar to the examples shown in D.
To assess whether the downregulation of lin-28 by LEP-2 occurs at the level of lin-28 mRNA or at the protein level, we performed quantitative PCR with mRNA isolated from synchronized L1 and L3 larvae. In wild-type animals, lin-28 mRNA levels decreased by a factor of 5 between L1 and L3, similar to previous findings (Bagga et al., 2005; Morita and Han, 2006; Stadler et al., 2012). In lep-2(lf) mutants, wild-type levels of lin-28 mRNA were observed for both stages (Fig. 5B), showing that lin-28 mRNA is downregulated normally. Thus, lep-2 does not act on the amount of lin-28 mRNA but is required to reduce the level of LIN-28 protein after the L2 stage.
LEP-2 could act to downregulate LIN-28 protein in two ways: by slowing or blocking translation of lin-28 mRNA; or by promoting protein degradation. To distinguish between these possibilities, we used Dendra2, a fluorescent protein with an emission wavelength that can be irreversibly changed from green (507 nm) to red (573 nm) by exposure to short-wavelength light (Chudakov et al., 2007) (Fig. 5C). This strategy allows one to test how long a protein endures after photoconversion (indicated by persistence of red). We created a lin-28>LIN-28::Dendra2::lin-28_3′UTR reporter, evaluated its expression in wild-type and lep-2(lf) animals and found it to be consistent with that of the LIN-28::GFP reporter described above. For photoconversion experiments, we focused on the bright fluorescence of neurons in the pharynx region because immobilization was required for photoconversion and image acquisition, and tail tips could not be fully immobilized. Exposure to 405 nm light photoconverted almost all of the Dendra2, leading to a bright red signal with very little green. When wild-type larvae had reached the L3 stage, both green and red signals were almost completely absent (Fig. 5D). This suggests that LIN-28::Dendra2 was efficiently degraded and that no new translation of LIN-28::Dendra2 occurred. lep-2(lf) animals showed a bright fluorescent signal 24 h after photoconversion (Fig. 5D). In areas where LIN-28::Dendra2 protein was photoconverted in L2, fluorescence at the L3 stage was red. Thus, the excess LIN-28::Dendra2 seen in lep-2(lf) L3 larvae was already present at L2 and not nascent. We conclude that LEP-2 is required, directly or indirectly, for the degradation of LIN-28 after the L2 stage.
DISCUSSION
By studying male TTM in C. elegans, an event that occurs during the J/A transition, we discovered a new heterochronic gene, lep-2. In lep-2(lf) mutants, TTM is delayed into the adult stage. This retarded heterochronic effect is not limited to the tail tip; lep-2 is also required for proper male mating behavior, as well as for adult cuticle formation and exit from the molting cycle in both sexes. Surprisingly, lep-2 is not required in the seam cells. This general requirement for lep-2 in many tissues suggests that it is likely to be an important component of the heterochronic pathway. We find that LEP-2 negatively regulates LIN-28 post-translationally by promoting LIN-28 degradation. Without LEP-2, LIN-28 is not properly degraded, delaying let-7-dependent adult differentiation.
Although several mechanisms have been identified that downregulate lin-28 mRNA stability or translation, little is known about the mechanism that eliminates LIN-28 protein. Translation of LIN-28 is downregulated by the lin-4 miRNA, possibly in concert with SEA-2 (Huang et al., 2011; Moss et al., 1997; Seggerson et al., 2002). LIN-66 and DAF-12 regulate translation from the lin-28 mRNA independently of lin-4 (Huang et al., 2011; Morita and Han, 2006). Most recently, it has been found that the caspase CED-3 cleaves the LIN-28 protein, presumably leading to subsequent degradation, but the heterochronic effect of ced-3(−) is subtle (Weaver et al., 2014). Here, we show that lep-2 also affects LIN-28 stability but that loss of lep-2 results in strong heterochronic phenotypes. How LEP-2 regulates LIN-28 is not yet clear, but one simple model is that LEP-2 functions as an E3 ubiquitin ligase to tag LIN-28 for proteasomal degradation. Like other Mkrns, LEP-2 has a RING domain that is typical of many E3 ligases (Bohne et al., 2010; Gray et al., 2000). Indeed, Mkrn1 in mammals functions as an E3 ligase, targeting several proteins and even itself, although LIN-28 has not yet been identified as one of its targets (Kim et al., 2005; Salvatico et al., 2010; Lee et al., 2009, 2012; Ko et al., 2012; Shimada et al., 2009; Ko et al., 2010; Kim et al., 2014; Cassar et al., 2015). In other contexts, Mkrns are also RNA-binding proteins, are localized to ribonucleoprotein granules, and have been found to sequester some mRNAs or promote their translation (Cano et al., 2010; Cassar et al., 2015; Gajdos et al., 2010; Kwon et al., 2013; Miroci et al., 2012; Cheung et al., 2010; Yang et al., 2008). Thus, an alternative mechanism for LEP-2/Mkrn function could be to sequester, protect from degradation and/or promote the translation of an mRNA encoding another protein that promotes LIN-28 destruction. Testing these hypotheses is the focus of future studies in our lab.
Mkrns and LIN28 homologs are highly conserved; it is therefore possible that a regulatory connection between them is also conserved, such that they act together in similar developmental processes. Synthesizing data from the literature, we find two areas of overlap between LIN-28 and Mkrn function: both proteins are involved in transitions between undifferentiated and differentiated states and in the J/A transition.
Just as LIN-28 governs transitions between undifferentiated and differentiated states (Shyh-Chang et al., 2013; Rougvie and Moss, 2013; Thornton and Gregory, 2012; Tsialikas and Romer-Seibert, 2015), upregulation of Mkrns in vertebrates is often linked with differentiation. For example, mouse and human Mkrn3 are expressed in differentiated cells derived from ESCs and are not expressed in proliferating ESCs (Jong et al., 1999a; Mai et al., 2011). In fibroblasts that transition out of the cell cycle, Mkrn1 is upregulated 6-fold; in several cancer cell lines that are induced to differentiate using retinoic acid, Mkrn1 is upregulated as well (Salvatico et al., 2010). MKRN1 ubiquitylates the human telomerase TERT, which is downregulated in switches from stemness toward differentiation and in tumor suppression (Kim et al., 2005). During the reprogramming of mouse fibroblasts into induced pluripotent stem cells, Mkrn1 is co-regulated with a set of known differentiation markers (O'Malley et al., 2013). Also, ectopic Mkrn2 in Xenopus results in a smaller head and shorter tail than normal (Yang et al., 2008), a phenotype consistent with a precocious switch from proliferation to differentiation. These phenomena are consistent with a role for Mkrns in promoting differentiation by switching off LIN-28, which otherwise maintains an undifferentiated state. This role for Mkrn and LIN-28 might be context dependent, as there are instances in which LIN-28 promotes differentiation and where Mkrn is associated with undifferentiated states (Cassar et al., 2015; O'Malley et al., 2013; Polesskaya et al., 2007; Walker et al., 2007).
Also like LIN-28, MKRN3 is involved in timing the onset of the human J/A transition (Abreu et al., 2013; Gajdos et al., 2010; Ong et al., 2009; Perry et al., 2009; Zhu et al., 2010). Specifically, alleles expected to reduce or abolish MKRN3 function were implicated as causal factors of precocious pubertal onset as early as 5 years of age (Abreu et al., 2013). MKRN3 resides in a paternally imprinted region called the Prader-Willi syndrome (PWS) critical region, and individuals with PWS have defects in pubertal timing (Crino et al., 2003; Jong et al., 1999b). Interestingly, both MKRN3 and LIN28B variants are associated with precocious puberty (Abreu et al., 2013). This is different from C. elegans, in which lin-28(lf) mutants have precocious and lep-2(lf) mutants have retarded J/A transitions. Therefore, the mode of the regulatory interaction in the J/A transition must have changed during evolution.
In summary, Mkrns and LIN-28/let-7 are highly conserved and are involved in juvenile-to-adult and stemness-to-differentiation transitions in animals as diverse as nematodes and humans. Thus, both are also likely to be part of a well-conserved axis that governs several fundamental developmental switches.
MATERIALS AND METHODS
Strains
Strains (Table 3) were cultured according to standard protocols (Brenner, 1974) and fed with Escherichia coli OP50-1. Unless otherwise mentioned, non-DF strains were obtained from the Caenorhabditis Genetics Center.
Table 3.
C. elegans strains

Microscopy
Worms were placed in a drop of 20 mM sodium azide or 1 mM levamisole on 5% (w/v) agar pads and studied at 400× or 1000× magnification with a Zeiss Axioskop with Nomarski (DIC) and epifluorescence optics. Images were recorded with a C4742-95 ‘Orca' Hamamatsu digital camera and Openlab 3.0.9 (Improvision). Confocal images were obtained on a Leica TCS SP8 X microscope. The 63× objective was used for image acquisition and z-stacks were obtained with 0.5 or 1 µm steps. Image editing was performed with ImageJ (http://rsb.info.nih.gov/ij/) and Photoshop (Adobe).
Mutagenesis screen for TTM defects
CB4088 hermaphrodites were mutagenized with trimethylpsoralen (TMP) and exposure to UV light. F1 hermaphrodites were isolated to produce the F2 generation, which were segregated. F3 male progeny of individual F2 were scored at 400× magnification for adult male tails that exhibited Lep tail tips.
Mapping lep-2
Genomic DNA was purified from wild-type (reference) and lep-2(ny4) (mutant) animals for comparative hybridization onto C. elegans whole-genome tiling arrays by Roche NimbleGen. The tiling arrays contained 385,179 randomly distributed (50-85mer) probes covering the entire C. elegans genome. The DNA samples were labeled with Cy3 or Cy5 and hybridized to the array. The ratio of fluorescence intensities was log2 normalized using MATLAB (MathWorks). An 8.4 kb region on chromosome IV including sequences 2 kb upstream and 1.3 kb downstream of Y55F3AM.6 was PCR amplified from the wild-type and lep-2 mutant strains using primers #9 and #10 (Table S3), then sequenced by GENEWIZ. The sequences were checked against the CB4088 genome sequence provided by K. Gunsalus (New York University) and S. Emmons (Albert Einstein College of Medicine).
Complementation tests
Although lep-2 males are normally sterile, those that have developed through dauer diapause can sire progeny. Genetic non-complementation was determined by crossing post-dauer lep2 mutant males with dpy-17-marked lep-2 hermaphrodites and examining the F1 non-Dpy cross progeny for the Lep phenotype at 400× magnification.
DNA transgene construction
Wild-type hermaphrodites were microinjected with DNA transgenes and pRF4=rol-6(su1006), a transformation marker producing a dominant Rol phenotype. To generate recombinant DNA constructs, we performed overlap-extension PCR as described previously (Nelson et al., 2011). lep-2>GFP::lep-2 was made by amplifying lep-2 sequences from N2 DNA and GFP from pPD95.75 (Addgene) with primers #1-10 (Table S3). lin-28>lin-28::Dendra2::lin-28_3′UTR was made by amplifying lin-28 from N2 DNA and Dendra2 from pEG345 (Addgene) with primers #11-18 (Table S3).
RNAi knockdown
RNAi was performed by feeding E. coli transformed with inducible RNAi vectors to mothers or larvae as previously described (Nelson et al., 2011). The dsRNA-expressing bacterial strains for lin-14 and lin-41 were from the J. Ahringer library, and those for lin-28 from the M. Vidal library (Source Bioscience). For lin-14 and lin-28 RNAi, L4 hermaphrodites were treated and their male progeny scored as L3, L4 and adults for defects in TTM. For lin-41 RNAi, L1 larvae were fed and scored as L4 and adults. The L4440 plasmid was used as a negative control.
Mating assays
Mating assays were performed as previously described (Barr and Garcia, 2006) between unc-119(e2498) hermaphrodites and lep-2 mutant males. The day before the assay, 6-cm NGM plates were seeded with 18 µl OP50-1 at room temperature. L4 males and hermaphrodites from healthy, non-starved cultures were picked to same-sex plates and kept for 12 h to mature. On the assay day, three hermaphrodites and one test male were picked to the food patch of the mating plates and recovered for 5 min. Videos of the male behavior were recorded and evaluated for aspects of mating behavior, i.e. the time it took for the male tail to make contact with the hermaphrodite and copulate. Spicule insertion was used to identify copulation; sperm transfer was not assayed.
Quantitative PCR
lin-28 mRNA levels were measured in him-5(e1490) and lep-2(bx73) backgrounds at the L1 and L3 stages. Animals were collected using the hatch-off method with a 4-h window (Pepper et al., 2004). For the L3 sample, L1 were fed for 27 h at 20°C. Total RNA was extracted using a modification of the tissue protocol for the RNeasy Micro Kit (Qiagen). DNase treatment was performed on-column. cDNA was generated from 250 ng total RNA using the double-primed RNA to cDNA EcoDry Premix system (Clontech). Quantitative PCR was performed with intron-spanning primers #19 and #20 (Table S3) using the iQ SYBR Green Supermix (Bio-Rad) with a two-step PCR protocol (3 min 95°C; 40 cycles of 10 s 95°C, 60 s 60°C; followed by melt curve analysis) on the MyiQ single-color real-time PCR detection system (Bio-Rad). Y45F10D.4 served as reference gene (Hoogewijs et al., 2008). Unknowns were run in triplicates, standards in duplicates.
Western blot
Western blot analysis by SDS-PAGE was performed according to standard procedure. For the L1 sample, arrested L1 were placed on food and collected a few hours later. L3 samples were collected 24 h after plating arrested L1 at 20°C. Animals were washed twice with PBS, frozen in liquid nitrogen and ground in the presence of a protease inhibitor (Halt, Pierce), then SDS-PAGE loading buffer was added. Samples were heated to 95°C for 10 min and centrifuged to pellet the insoluble fraction. An aliquot was kept for Bradford assay. 10-20 µg total protein was loaded onto a 10% bis-Tris Bolt gel (Invitrogen). After electrophoresis and transfer to a nitrocellulose membrane, the blot was incubated overnight at 4°C with rabbit anti-LIN-28 polyclonal antibody (gifts from E. Moss, Rowan University, and M. Han, University of Colorado Boulder; 1:1000) and a mouse anti-actin monoclonal antibody (Sigma, A3853; 1:5000) to provide a loading control. The blot was incubated 45-60 min in the dark with fluorescent secondary antibodies (Licor, 827-11081 and 827-08364; 1:30,000) and scanned on a Licor infrared fluorescence scanner.
Photoconversion of Dendra2
To observe LIN-28 dynamics in lep-2 mutants, we transformed wild-type worms with a lin-28>lin-28::Dendra2::lin-28_3′UTR transgene. Fluorescence of the transgene was visualized by confocal microscopy. L2 stage animals were examined and fluorescence in the pharynx region was captured for the green and red channels by sequential scans. Dendra2 photoconversion was performed by exposing a section of the pharynx to brief flashes of 405 nm light with short rest periods between in 10-20 slices with ten successive scans of the z-plane. Sequential z-stacks of the exposed region were taken for the red and green channel to record the post-photoconversion fluorescent signal. The animal was recovered onto a plate with food and kept at 20°C for 24 h. The worms (now L3) were remounted, and another sequential z-stack was recorded. The image stacks were analyzed with ImageJ.
Acknowledgements
We thank E. Moss, B. Weaver and M. Han for anti-LIN-28 antibodies; V. Ambros, S. Emmons, A. Mason, D. Portman, A. Rougvie, F. Slack and P. Sternberg for strains; D. Moerman for help designing the array CGH; and S. Ahn for contributions to the post-dauer analysis. Some strains were provided by the Caenorhabditis Genetics Center (University of Minnesota), which is funded by National Institutes of Health Office of Research Infrastructure Programs [P40 OD010440]. Some deletion mutations were provided by the International C. elegans Deletion Mutant Consortium (C. elegans Gene Knockout Facility at the Oklahoma Medical Research Foundation, funded by the National Institutes of Health; and the C. elegans Reverse Genetics Core Facility at the University of British Columbia, funded by the Canadian Institute for Health Research, Genome Canada, Genome BC, the Michael Smith Foundation, and the National Institutes of Health). Several analyses in this work depended on data provided by WormBase (www.wormbase.org).
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization, methodology, investigation and writing (review and editing), R.A.H., K.K. and D.H.A.F.; writing (original draft), R.A.H.; visualization, R.A.H. and K.K.; funding acquisition, D.H.A.F.
Funding
This work was supported by a grant from the National Institutes of Health National Institute of General Medical Sciences [R01-GM100140 to D.H.A.F.]; and research funds provided by New York University-Shanghai. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.132738/-/DC1
References
- Abrahante J. E., Daul A. L., Li M., Volk M. L., Tennessen J. M., Miller E. A. and Rougvie A. E. (2003). The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev. Cell 4, 625-637. 10.1016/S1534-5807(03)00127-8 [DOI] [PubMed] [Google Scholar]
- Abreu A. P., Dauber A., Macedo D. B., Noel S. D., Brito V. N., Gill J. C., Cukier P., Thompson I. R., Navarro V. M., Gagliardi P. C. et al. (2013). Central precocious puberty caused by mutations in the imprinted gene MKRN3. N. Engl. J. Med. 368, 2467-2475. 10.1056/NEJMoa1302160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. and Horvitz H. R. (1984). Heterochronic mutants of the nematode Caenorhabditis elegans. Science 226, 409-416. 10.1126/science.6494891 [DOI] [PubMed] [Google Scholar]
- Arumugam T. U., Davies E., Morita E. H. and Abe S. (2007). Sequence, expression and tissue localization of a gene encoding a makorin RING zinc-finger protein in germinating rice (Oryza sativa L. ssp. Japonica) seeds. Plant Physiol. Biochem. 45, 767-780. 10.1016/j.plaphy.2007.07.006 [DOI] [PubMed] [Google Scholar]
- Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R. and Pasquinelli A. E. (2005). Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122, 553-563. 10.1016/j.cell.2005.07.031 [DOI] [PubMed] [Google Scholar]
- Barr M. M. and Garcia L. R. (2006). Male mating behavior. WormBook, 1-11. 10.1895/wormbook.1.78.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohne A., Darras A., D'cotta H., Baroiller J.-F., Galiana-Arnoux D. and Volff J.-N. (2010). The vertebrate makorin ubiquitin ligase gene family has been shaped by large-scale duplication and retroposition from an ancestral gonad-specific, maternal-effect gene. BMC Genomics 11, 721 10.1186/1471-2164-11-721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büssing I., Slack F. J. and Grosshans H. (2008). let-7 microRNAs in development, stem cells and cancer. Trends Mol. Med. 14, 400-409. 10.1016/j.molmed.2008.07.001 [DOI] [PubMed] [Google Scholar]
- C. elegans Deletion Mutant Consortium. (2012). Large-scale screening for targeted knockouts in the Caenorhabditis elegans genome. G3 (Bethesda). 2, 1415-1425. 10.1534/g3.112.003830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano F., Miranda-Saavedra D. and Lehner P. J. (2010). RNA-binding E3 ubiquitin ligases: novel players in nucleic acid regulation. Biochem. Soc. Trans. 38, 1621-1626. 10.1042/BST0381621 [DOI] [PubMed] [Google Scholar]
- Cassar P. A., Carpenedo R. L., Samavarchi-Tehrani P., Olsen J. B., Park C. J., Chang W. Y., Chen Z., Choey C., Delaney S., Guo H. et al. (2015). Integrative genomics positions MKRN1 as a novel ribonucleoprotein within the embryonic stem cell gene regulatory network. EMBO Rep. 16, 1334-1357. 10.15252/embr.201540974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H.-M., Martinez N. J., Thornton J. E., Hagan J. P., Nguyen K. D. and Gregory R. I. (2012). Trim71 cooperates with microRNAs to repress Cdkn1a expression and promote embryonic stem cell proliferation. Nat. Commun. 3, 923 10.1038/ncomms1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung W. K. C., Yang P.-H., Huang Q.-H., Chen Z., Chen S.-J., Lin M. C. M. and Kung H.-F. (2010). Identification of protein domains required for makorin-2-mediated neurogenesis inhibition in Xenopus embryos. Biochem. Biophys. Res. Commun. 394, 18-23. 10.1016/j.bbrc.2010.02.041 [DOI] [PubMed] [Google Scholar]
- Chudakov D. M., Lukyanov S. and Lukyanov K. A. (2007). Tracking intracellular protein movements using photoswitchable fluorescent proteins PS-CFP2 and Dendra2. Nat. Protoc. 2, 2024-2032. 10.1038/nprot.2007.291 [DOI] [PubMed] [Google Scholar]
- Copley M. R., Babovic S., Benz C., Knapp D. J. H. F., Beer P. A., Kent D. G., Wohrer S., Treloar D. Q., Day C., Rowe K. et al. (2013). The Lin28b-let-7-Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells. Nat. Cell Biol. 15, 916-925. 10.1038/ncb2783 [DOI] [PubMed] [Google Scholar]
- Cox G. N., Laufer J. S., Kusch M. and Edgar R. S. (1980). Genetic and phenotypic characterization of Roller mutants of Caenorhabditis elegans. Genetics 95, 317-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crino A., Schiaffini R., Ciampalini P., Spera S., Beccaria L., Benzi F., Bosio L., Corrias A., Gargantini L., Salvatoni A. et al. (2003). Hypogonadism and pubertal development in Prader-Willi syndrome. Eur. J. Pediatr. 162, 327-333. [DOI] [PubMed] [Google Scholar]
- Del Rio-Albrechtsen T., Kiontke K., Chiou S.-Y. and Fitch D. H. A. (2006). Novel gain-of-function alleles demonstrate a role for the heterochronic gene lin-41 in C. elegans male tail tip morphogenesis. Dev. Biol. 297, 74-86. 10.1016/j.ydbio.2006.04.472 [DOI] [PubMed] [Google Scholar]
- Ecsedi M. and Grosshans H. (2013). LIN-41/TRIM71: emancipation of a miRNA target. Genes Dev. 27, 581-589. 10.1101/gad.207266.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecsedi M., Rausch M. and Grosshans H. (2015). The let-7 microRNA directs vulval development through a single target. Dev. Cell 32, 335-344. 10.1016/j.devcel.2014.12.018 [DOI] [PubMed] [Google Scholar]
- Euling S. and Ambros V. (1996). Reversal of cell fate determination in Caenorhabditis elegans vulval development. Development 122, 2507-2515. [DOI] [PubMed] [Google Scholar]
- Flibotte S. and Moerman D. G. (2008). Experimental analysis of oligonucleotide microarray design criteria to detect deletions by comparative genomic hybridization. BMC Genomics 9, 497 10.1186/1471-2164-9-497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajdos Z. K. Z., Henderson K. D., Hirschhorn J. N. and Palmert M. R. (2010). Genetic determinants of pubertal timing in the general population. Mol. Cell. Endocrinol. 324, 21-29. 10.1016/j.mce.2010.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray T. A., Hernandez L., Carey A. H., Schaldach M. A., Smithwick M. J., Rus K., Marshall Graves J. A., Stewart C. L. and Nicholls R. D. (2000). The ancient source of a distinct gene family encoding proteins featuring RING and C(3)H zinc-finger motifs with abundant expression in developing brain and nervous system. Genomics 66, 76-86. 10.1006/geno.2000.6199 [DOI] [PubMed] [Google Scholar]
- Hannon T. S., Janosky J. and Arslanian S. A. (2006). Longitudinal study of physiologic insulin resistance and metabolic changes of puberty. Pediatr. Res. 60, 759-763. 10.1203/01.pdr.0000246097.73031.27 [DOI] [PubMed] [Google Scholar]
- Hayes G. D. and Ruvkun G. (2006). Misexpression of the Caenorhabditis elegans miRNA let-7 is sufficient to drive developmental programs. Cold Spring Harb. Symp. Quant. Biol. 71, 21-27. 10.1101/sqb.2006.71.018 [DOI] [PubMed] [Google Scholar]
- Hoogewijs D., Houthoofd K., Matthijssens F., Vandesompele J. and Vanfleteren J. R. (2008). Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol. Biol. 9, 9 10.1186/1471-2199-9-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Zhang H. and Zhang H. (2011). The zinc-finger protein SEA-2 regulates larval developmental timing and adult lifespan in C. elegans. Development 138, 2059-2068. 10.1242/dev.057109 [DOI] [PubMed] [Google Scholar]
- Jong M. T. C., Carey A. H., Caldwell K. A., Lau M. H., Handel M. A., Driscoll D. J., Stewart C. L., Rinchik E. M. and Nicholls R. D. (1999a). Imprinting of a RING zinc-finger encoding gene in the mouse chromosome region homologous to the Prader-Willi syndrome genetic region. Hum. Mol. Genet. 8, 795-803. 10.1093/hmg/8.5.795 [DOI] [PubMed] [Google Scholar]
- Jong M. T. C., Gray T. A., Ji Y., Glenn C. C., Saitoh S., Driscoll D. J. and Nicholls R. D. (1999b). A novel imprinted gene, encoding a RING zinc-finger protein, and overlapping antisense transcript in the Prader-Willi syndrome critical region. Hum. Mol. Genet. 8, 783-793. 10.1093/hmg/8.5.783 [DOI] [PubMed] [Google Scholar]
- Kim J. H., Park S.-M., Kang M. R., Oh S.-Y., Lee T. H., Muller M. T. and Chung I. K. (2005). Ubiquitin ligase MKRN1 modulates telomere length homeostasis through a proteolysis of hTERT. Genes Dev. 19, 776-781. 10.1101/gad.1289405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.-H., Park K. W., Lee E.-W., Jang W.-S., Seo J., Shin S., Hwang K.-A. and Song J. (2014). Suppression of PPARgamma through MKRN1-mediated ubiquitination and degradation prevents adipocyte differentiation. Cell Death Differ. 21, 594-603. 10.1038/cdd.2013.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko A., Lee E.-W., Yeh J.-Y., Yang M.-R., Oh W., Moon J.-S. and Song J. (2010). MKRN1 induces degradation of West Nile virus capsid protein by functioning as an E3 ligase. J. Virol. 84, 426-436. 10.1128/JVI.00725-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko A., Shin J.-Y., Seo J., Lee K.-D., Lee E.-W., Lee M.-S., Lee H.-W., Choi I.-J., Jeong J. S., Chun K.-H. et al. (2012). Acceleration of gastric tumorigenesis through MKRN1-mediated posttranslational regulation of p14ARF. J. Natl. Cancer Inst. 104, 1660-1672. 10.1093/jnci/djs424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S. C., Yi H., Eichelbaum K., Fohr S., Fischer B., You K. T., Castello A., Krijgsveld J., Hentze M. W. and Kim V. N. (2013). The RNA-binding protein repertoire of embryonic stem cells. Nat. Struct. Mol. Biol. 20, 1122-1130. 10.1038/nsmb.2638 [DOI] [PubMed] [Google Scholar]
- Lee E.-W., Lee M.-S., Camus S., Ghim J., Yang M.-R., Oh W., Ha N.-C., Lane D. P. and Song J. (2009). Differential regulation of p53 and p21 by MKRN1 E3 ligase controls cell cycle arrest and apoptosis. EMBO J. 28, 2100-2113. 10.1038/emboj.2009.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.-W., Kim J.-H., Ahn Y.-H., Seo J., Ko A., Jeong M., Kim S.-J., Ro J. Y., Park K.-M., Lee H.-W. et al. (2012). Ubiquitination and degradation of the FADD adaptor protein regulate death receptor-mediated apoptosis and necroptosis. Nat. Commun. 3, 978 10.1038/ncomms1981 [DOI] [PubMed] [Google Scholar]
- Lipton J., Kleemann G., Ghosh R., Lints R. and Emmons S. W. (2004). Mate searching in Caenorhabditis elegans: a genetic model for sex drive in a simple invertebrate. J. Neurosci. 24, 7427-7434. 10.1523/JNEUROSCI.1746-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. and Ambros V. (1991). Alternative temporal control systems for hypodermal cell differentiation in Caenorhabditis elegans. Nature 350, 162-165. 10.1038/350162a0 [DOI] [PubMed] [Google Scholar]
- Loedige I., Gaidatzis D., Sack R., Meister G. and Filipowicz W. (2013). The mammalian TRIM-NHL protein TRIM71/LIN-41 is a repressor of mRNA function. Nucleic Acids Res. 41, 518-532. 10.1093/nar/gks1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long D., Lee R., Williams P., Chan C. Y., Ambros V. and Ding Y. (2007). Potent effect of target structure on microRNA function. Nat. Struct. Mol. Biol. 14, 287-294. 10.1038/nsmb1226 [DOI] [PubMed] [Google Scholar]
- Mai X., Mai Q., Li T. and Zhou C. (2011). Dynamic expression patterns of imprinted genes in human embryonic stem cells following prolonged passaging and differentiation. J. Assist Reprod. Genet. 28, 315-323. 10.1007/s10815-010-9524-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason D. A., Rabinowitz J. S. and Portman D. S. (2008). dmd-3, a doublesex-related gene regulated by tra-1, governs sex-specific morphogenesis in C. elegans. Development 135, 2373-2382. 10.1242/dev.017046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miroci H., Schob C., Kindler S., Olschlager-Schutt J., Fehr S., Jungenitz T., Schwarzacher S. W., Bagni C. and Mohr E. (2012). Makorin ring zinc finger protein 1 (MKRN1), a novel poly(A)-binding protein-interacting protein, stimulates translation in nerve cells. J. Biol. Chem. 287, 1322-1334. 10.1074/jbc.M111.315291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K. and Han M. (2006). Multiple mechanisms are involved in regulating the expression of the developmental timing regulator lin-28 in Caenorhabditis elegans. EMBO J. 25, 5794-5804. 10.1038/sj.emboj.7601451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss E. G., Lee R. C. and Ambros V. (1997). The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 88, 637-646. 10.1016/S0092-8674(00)81906-6 [DOI] [PubMed] [Google Scholar]
- Nelson M. D., Zhou E., Kiontke K., Fradin H., Maldonado G., Martin D., Shah K. and Fitch D. H. (2011). A bow-tie genetic architecture for morphogenesis suggested by a genome-wide RNAi screen in Caenorhabditis elegans. PLoS Genet 7, e1002010 10.1371/journal.pgen.1002010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman M. A., Thomson J. M. and Hammond S. M. (2008). Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 14, 1539-1549. 10.1261/rna.1155108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen C. Q., Hall D. H., Yang Y. and Fitch D. H. A. (1999). Morphogenesis of the Caenorhabditis elegans male tail tip. Dev. Biol. 207, 86-106. 10.1006/dbio.1998.9173 [DOI] [PubMed] [Google Scholar]
- O'Malley J., Skylaki S., Iwabuchi K. A., Chantzoura E., Ruetz T., Johnsson A., Tomlinson S. R., Linnarsson S. and Kaji K. (2013). High-resolution analysis with novel cell-surface markers identifies routes to iPS cells. Nature 499, 88-91. 10.1038/nature12243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong K. K., Elks C. E., Li S., Zhao J. H., Luan J., Andersen L. B., Bingham S. A., Brage S., Smith G. D., Ekelund U. et al. (2009). Genetic variation in LIN28B is associated with the timing of puberty. Nat. Genet. 41, 729-733. 10.1038/ng.382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper A. S.-R., McCane J. E., Kemper K., Yeung D. A., Lee R. C., Ambros V. and Moss E. G. (2004). The C. elegans heterochronic gene lin-46 affects developmental timing at two larval stages and encodes a relative of the scaffolding protein gephyrin. Development 131, 2049-2059. 10.1242/dev.01098 [DOI] [PubMed] [Google Scholar]
- Perry J. R. B., Stolk L., Franceschini N., Lunetta K. L., Zhai G., McArdle P. F., Smith A. V., Aspelund T., Bandinelli S., Boerwinkle E. et al. (2009). Meta-analysis of genome-wide association data identifies two loci influencing age at menarche. Nat. Genet. 41, 648-650. 10.1038/ng.386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskounova E., Viswanathan S. R., Janas M., LaPierre R. J., Daley G. Q., Sliz P. and Gregory R. I. (2008). Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J. Biol. Chem. 283, 21310-21314. 10.1074/jbc.C800108200 [DOI] [PubMed] [Google Scholar]
- Polesskaya A., Cuvellier S., Naguibneva I., Duquet A., Moss E. G. and Harel-Bellan A. (2007). Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 21, 1125-1138. 10.1101/gad.415007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie A. E. and Moss E. G. (2013). Developmental transitions in C. elegans larval stages. Curr. Top. Dev. Biol. 105, 153-180. 10.1016/B978-0-12-396968-2.00006-3 [DOI] [PubMed] [Google Scholar]
- Rybak A., Fuchs H., Smirnova L., Brandt C., Pohl E. E., Nitsch R. and Wulczyn F. G. (2008). A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 10, 987-993. 10.1038/ncb1759 [DOI] [PubMed] [Google Scholar]
- Rybak A., Fuchs H., Hadian K., Smirnova L., Wulczyn E. A., Michel G., Nitsch R., Krappmann D. and Wulczyn F. G. (2009). The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat. Cell Biol. 11, 1411-1420. 10.1038/ncb1987 [DOI] [PubMed] [Google Scholar]
- Salvatico J., Kim J. H., Chung I. K. and Muller M. T. (2010). Differentiation linked regulation of telomerase activity by Makorin-1. Mol. Cell. Biochem. 342, 241-250. 10.1007/s11010-010-0490-x [DOI] [PubMed] [Google Scholar]
- Seggerson K., Tang L. and Moss E. G. (2002). Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev. Biol. 243, 215-225. 10.1006/dbio.2001.0563 [DOI] [PubMed] [Google Scholar]
- Shimada H., Shiratori T., Yasuraoka M., Kagaya A., Kuboshima M., Nomura F., Takiguchi M., Ochiai T., Matsubara H. and Hiwasa T. (2009). Identification of Makorin 1 as a novel SEREX antigen of esophageal squamous cell carcinoma. BMC Cancer 9, 232 10.1186/1471-2407-9-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N., Zhu H., Yvanka de Soysa T., Shinoda G., Seligson M. T., Tsanov K. M., Nguyen L., Asara J. M., Cantley L. C. and Daley G. Q. (2013). Lin28 enhances tissue repair by reprogramming cellular metabolism. Cell 155, 778-792. 10.1016/j.cell.2013.09.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack F. J., Basson M., Liu Z., Ambros V., Horvitz H. R. and Ruvkun G. (2000). The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell 5, 659-669. 10.1016/S1097-2765(00)80245-2 [DOI] [PubMed] [Google Scholar]
- Sonoda J. and Wharton R. P. (2001). Drosophila Brain Tumor is a translational repressor. Genes Dev. 15, 762-773. 10.1101/gad.870801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler M., Artiles K., Pak J. and Fire A. (2012). Contributions of mRNA abundance, ribosome loading, and post- or peri-translational effects to temporal repression of C. elegans heterochronic miRNA targets. Genome Res. 22, 2418-2426. 10.1101/gr.136515.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J. E. and Horvitz H. R. (1977). Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56, 110-156. 10.1016/0012-1606(77)90158-0 [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Albertson D. G. and Thomson J. N. (1980). The Caenorhabditis elegans male: postembryonic development of nongonadal structures. Dev. Biol. 78, 542-576. 10.1016/0012-1606(80)90352-8 [DOI] [PubMed] [Google Scholar]
- Thornton J. E. and Gregory R. I. (2012). How does Lin28 let-7 control development and disease? Trends Cell Biol. 22, 474-482. 10.1016/j.tcb.2012.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsialikas J. and Romer-Seibert J. (2015). LIN28: roles and regulation in development and beyond. Development 142, 2397-2404. 10.1242/dev.117580 [DOI] [PubMed] [Google Scholar]
- Vadla B., Kemper K., Alaimo J., Heine C. and Moss E. G. (2012). lin-28 controls the succession of cell fate choices via two distinct activities. PLoS Genet. 8, e1002588 10.1371/journal.pgen.1002588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wynsberghe P. M., Kai Z. S., Massirer K. B., Burton V. H., Yeo G. W. and Pasquinelli A. E. (2011). LIN-28 co-transcriptionally binds primary let-7 to regulate miRNA maturation in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 18, 302-308. 10.1038/nsmb.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella M. C., Choi E.-Y., Lin S.-Y., Reinert K. and Slack F. J. (2004a). The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev. 18, 132-137. 10.1101/gad.1165404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella M. C., Reinert K. and Slack F. J. (2004b). Architecture of a validated microRNA::target interaction. Chem. Biol. 11, 1619-1623. 10.1016/j.chembiol.2004.09.010 [DOI] [PubMed] [Google Scholar]
- Viswanathan S. R., Daley G. Q. and Gregory R. I. (2008). Selective blockade of microRNA processing by Lin28. Science 320, 97-100. 10.1126/science.1154040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker E., Ohishi M., Davey R. E., Zhang W., Cassar P. A., Tanaka T. S., Der S. D., Morris Q., Hughes T. R., Zandstra P. W. et al. (2007). Prediction and testing of novel transcriptional networks regulating embryonic stem cell self-renewal and commitment. Cell Stem Cell 1, 71-86. 10.1016/j.stem.2007.04.002 [DOI] [PubMed] [Google Scholar]
- Weaver B. P., Zabinsky R., Weaver Y. M., Lee E. S., Xue D. and Han M. (2014). CED-3 caspase acts with miRNAs to regulate non-apoptotic gene expression dynamics for robust development in C. elegans. Elife 3, e04265 10.7554/eLife.04265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yandell M. D., Edgar L. G. and Wood W. B. (1994). Trimethylpsoralen induces small deletion mutations in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 91, 1381-1385. 10.1073/pnas.91.4.1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P.-H., Cheung W. K. C., Peng Y., He M.-L., Wu G.-Q., Xie D., Jiang B.-H., Huang Q.-H., Chen Z., Lin M. C. M. et al. (2008). Makorin-2 is a neurogenesis inhibitor downstream of Phosphatidylinositol 3-Kinase/Akt (PI3K/Akt) signal. J. Biol. Chem. 283, 8486-8495. 10.1074/jbc.M704768200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Yao H., Zhu P., Zhang X., Pan Q., Gong C., Huang Y., Hu X., Su F., Lieberman J. et al. (2007). let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 131, 1109-1123. 10.1016/j.cell.2007.10.054 [DOI] [PubMed] [Google Scholar]
- Zhu H., Shah S., Shyh-Chang N., Shinoda G., Einhorn W. S., Viswanathan S. R., Takeuchi A., Grasemann C., Rinn J. L., Lopez M. F. et al. (2010). Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nat. Genet. 42, 626-630. 10.1038/ng.593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Shyh-Chang N., Segre A. V., Shinoda G., Shah S. P., Einhorn W. S., Takeuchi A., Engreitz J. M., Hagan J. P., Kharas M. G. et al. (2011). The Lin28/let-7 axis regulates glucose metabolism. Cell 147, 81-94. 10.1016/j.cell.2011.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]