Abstract
Lipid droplets (LDs) are dynamic subcellular organelles whose growth is closely linked to obesity and hepatic steatosis. Cell death-inducing DNA fragmentation factor-α-like effector (CIDE) proteins, including Cidea, Cideb, and Cidec (also called Fsp27), play important roles in lipid metabolism. Cidea and Cidec are LD-associated proteins that promote atypical LD fusion in adipocytes. Here, we find that CIDE proteins are all localized to LD-LD contact sites (LDCSs) and promote lipid transfer, LD fusion, and growth in hepatocytes. We have identified two types of hepatocytes, one with small LDs (small LD-containing hepatocytes, SLHs) and one with large LDs (large LD-containing hepatocytes, LLHs) in the liver. Cideb is localized to LDCSs and promotes lipid exchange and LD fusion in both SLHs and LLHs, whereas Cidea and Cidec are specifically localized to the LDCSs and promote lipid exchange and LD fusion in LLHs. Cideb-deficient SLHs have reduced LD sizes and lower lipid exchange activities. Fasting dramatically induces the expression of Cidea/Cidec and increases the percentage of LLHs in the liver. The majority of the hepatocytes from the liver of obese mice are Cidea/Cidec-positive LLHs. Knocking down Cidea or Cidec significantly reduced lipid storage in the livers of obese animals. Our data reveal that CIDE proteins play differential roles in promoting LD fusion and lipid storage; Cideb promotes lipid storage under normal diet conditions, whereas Cidea and Cidec are responsible for liver steatosis under fasting and obese conditions.
Keywords: cell biology, lipid, lipid droplet, lipid metabolism, liver, metabolism, CIDE proteins, hepatic steatosis, lipid droplet fusion, lipid storage
Introduction
Obesity and its associated diseases, including type II diabetes, cardiovascular disease, and hepatic steatosis, have become alarmingly common diseases (1). Liver plays a major regulatory role in whole-body lipid metabolism. Disruption of the hepatic lipid metabolism could lead to the initiation and progression of several metabolic disorders (2, 3). The accumulation of fat in the form of lipid droplets (LDs)4 is an early pathophysiological feature of altered liver metabolism that is linked to insulin resistance and the potential progression of severe liver diseases, such as liver steatosis, liver cirrhosis, and hepatocellular carcinoma (4). LDs are subcellular organelles composed of a neutral lipid core surrounded by a phospholipid monolayer that is coated with various types of proteins (5). LDs are involved in several biologically significant processes, including neutral lipid storage, protein storage and degradation, as well as viral packaging (6, 7). LDs can grow in size via a tightly regulated mechanism (8, 9). The sizes of LDs reflect different biological processes. Several models projecting the growth of LDs have been proposed, including targeted lipid delivery from the endoplasmic reticulum to LDs mediated by fat storage-inducing transmembrane proteins 1 and 2 (FITM1/2) (10), local lipid synthesis on LDs mediated by CTP:phosphocholine cytidylyltransferase and diacylglycerol acyltransferase 2 (DGAT2) (11, 12), and the fusion of smaller LDs into larger LDs mediated by CIDEs in adipocytes (13).
Many proteins are localized on the surface of LDs. These proteins play important roles in regulating the size and function of LDs (14). Perilipin1/2/3/4/5 (Plin1/2/3/4/5) are LD-associated proteins in mammalian cells (15). The expression levels of Plin2, Plin3, and Plin5 are up-regulated in fatty livers (16). CIDE proteins, including Cidea, Cideb, and Cidec (also called Fsp27), are novel LD-associated proteins (17, 18). CIDEs play important roles in LD morphology and function. Cidea and Cidec are predominantly expressed in adipocytes, although Cideb is specifically expressed in the liver of wild-type mice. Cidea and Cidec are up-regulated in the steatotic liver (19, 20). Fasting can induce the expression of Cidec in the liver of wild-type mice (21–24). Both Cidea and Cidec localize on the surface of LDs and are particularly enriched at LD-LD contact sites (LDCSs) to promote atypical LD fusion and growth by lipid exchange and transfer in adipocytes (25–28). Several factors are involved in Cidea/Cidec-mediated LD fusion, including Plin1, Rab8a, MSS4, and AS160 (29–31). However, the role of Cidea/Cidec in LD fusion in the liver has not been investigated. Cideb is localized on the endoplasmic reticulum and LDs (32), and its deficiency results in the accumulation of smaller LDs in the liver (33). However, whether Cideb has the ability to localize on LDCSs and promote lipid exchange and LD fusion is still unknown.
Here, we systematically analyzed the role of CIDEs in LD fusion and lipid storage in hepatocytes under normal, fasting, and obese conditions. We found that Cideb is localized to LDCSs and promotes LD fusion and growth in hepatocytes. In the livers of normal diet-fed mice, the majority of hepatocytes express Cideb alone and contain small LDs. A small proportion of hepatocytes express Cidea and Cidec and contain large LDs. Under fasting and obese conditions, the percentage of hepatocytes expressing Cidea and Cidec increases dramatically. Finally, using Cideb−/−, ob/ob hepatocytes, we found that CIDEs play important roles in LD size and lipid storage in hepatocytes.
Materials and Methods
Mice
Wild-type, Cideb−/−, ob/ob mice were maintained as described previously (33–35). All mice used were on a C57BL/6J background. Four-month-old male mice were used for hepatocyte isolation. In vivo delivery of siRNAs was performed using Invivofectamine 2.0 (Invitrogen, 1377501). One week after the injection of siRNAs, the mice were fasted for 16 h. Liver tissues were harvested for further analysis. Liver TAG was measured as described previously (20). All animals were maintained in the animal facility of the Center of Biomedical Analysis, Tsinghua University (Beijing, China). The laboratory animal facility has been accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC). The Institutional Animal Care and Use Committee (IACUC) of Tsinghua University approved all animal protocols.
Cell Culture and Transfection
293T cells and HepG2 cells were cultured in DMEM (Invitrogen) containing 10% FBS (Invitrogen). Mouse primary hepatocytes were isolated as described previously (35). The isolated hepatocytes were seeded at a density of 106 cells per dish in glass bottom microwell dishes (P35G-1.5-14-C, MatTek Corp.) in DMEM (Invitrogen) containing 10% FBS (Invitrogen). Plasmid DNAs were transfected into 293T cells, HepG2 cells, and primary hepatocytes using Lipofectamine 2000 according to the manufacturer's instruction (Invitrogen, 11668019). For the siRNA experiment, 24 h after seeding, hepatocytes were transfected with siRNA using Lipofectamine 2000 (Invitrogen). The sequences used to target the CIDEs and Plin2 are as follows: Cidea, ACACGCATTTCATGATCTT; Cideb, CCTCTGCATGGAGTACCTT; Cidec, AATCGTGGAGACAGAAGAATA; Plin2, GAATATGCACAGTGCCAAC. Cells were visualized using an Axiovert 200 M microscope (Carl Zeiss) or an LSM710 confocal microscope (Carl Zeiss) 48 h after transfection.
Fluorescent Microscopic Imaging
Twelve hours after seeding, hepatocytes were fixed with 4% paraformaldehyde for 1 h at room temperature. Cells were then treated with 0.4% Triton X-100 for 20 min and then blocked with 10% goat serum for 1 h at room temperature. The primary antibodies for Cidea, Cideb and Cidec were added and the reaction was incubated for 1 h at room temperature. Anti-rabbit IgG antibodies conjugated with Alexa Fluor 568 (Molecular Probes, A11011) were used as secondary antibodies. Bodipy 493/503 (Molecular Probes, D3922) was used for neutral lipid staining. For the Cideb and Cidea co-staining and the Cideb and Cidec co-staining (Figs. 2E and 6, C and D), a Cideb antibody (from goat) obtained from Santa Cruz Biotechnology (sc-8733, 1:50) was used. Cidea and Cidec antibodies were generated from rabbit (20). Donkey anti-rabbit 488 (Molecular Probes, A21206) and donkey anti-goat 568 (Molecular Probes, A11057) were used as secondary antibodies. LDs were stained with Lipidtox (Molecular Probes, H34477). For the staining in Fig. 3C, a Cideb antibody (generated from rabbit) was used. Anti-rabbit IgG antibodies conjugated with Alexa Fluor 405 (Molecular Probes, A31556) were used as secondary antibodies. The sections were observed using a Zeiss 200 M inverted microscope, and the images were collected using an AxioCam MRm camera and Axio Vision software.
FIGURE 2.
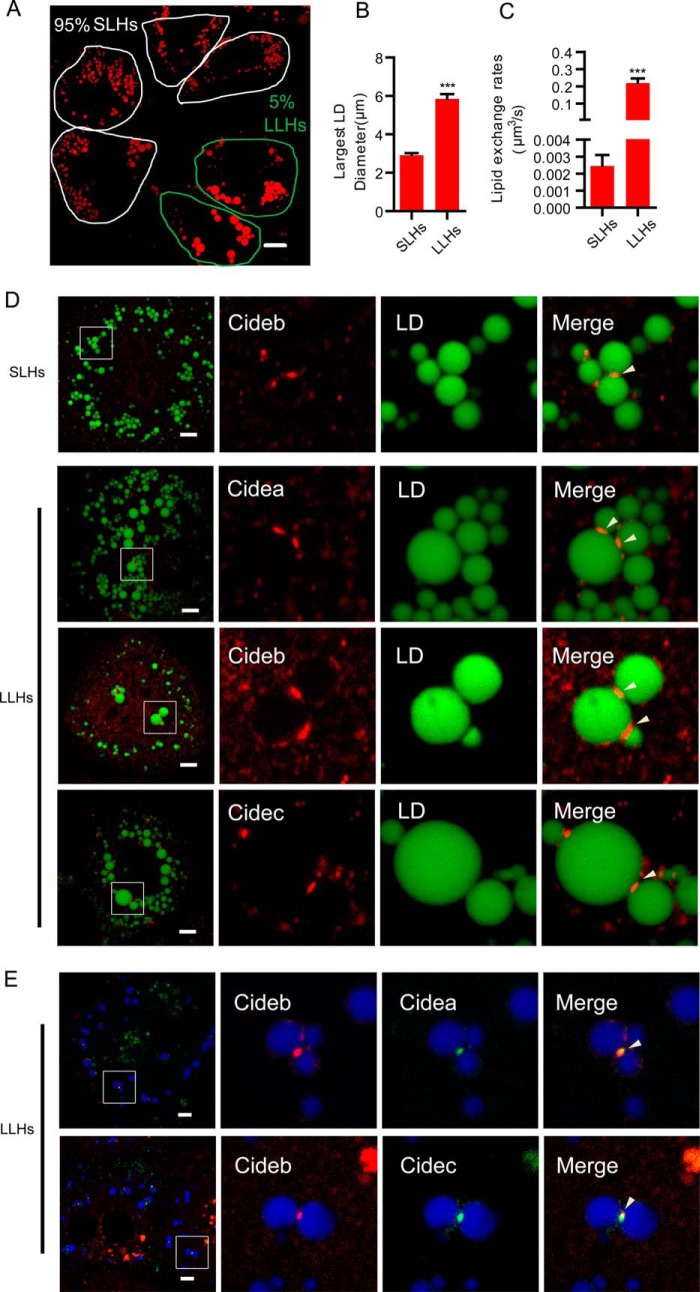
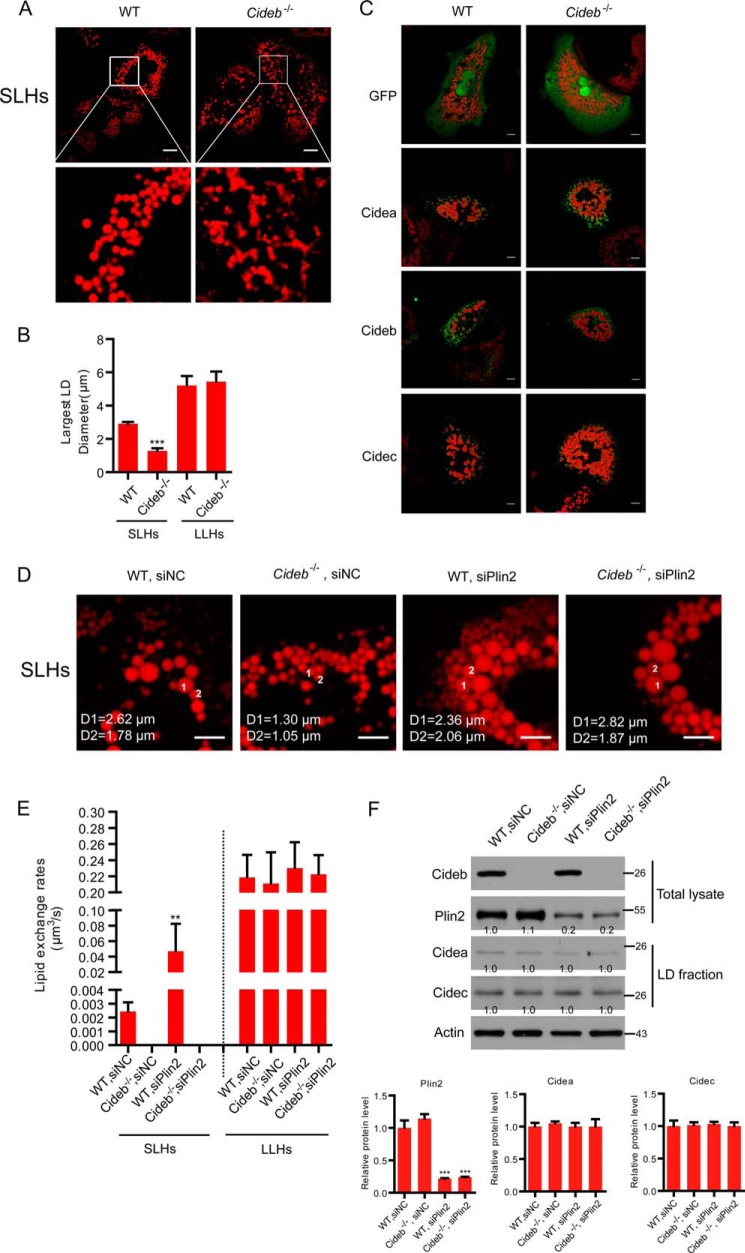
Endogenous CIDE proteins are localized at LDCSs in wild-type hepatocytes. A, representative image showing the LD morphology in wild-type hepatocytes. A total of 95% of the hepatocytes have small LDs with a diameter below 4 μm (SLHs). A total of 5% of the hepatocytes have large LDs with a diameter above 4 μm (LLHs). LDs were labeled with Bodipy 556/568 (C12, red). Scale bars, 10 μm. B, largest LD diameter in SLHs and LLHs. n = 20 for each group. C, lipid exchange rates in SLHs and LLHs. n = 8 for each group. D, wild-type hepatocytes were stained with antibodies against Cidea, Cideb, and Cidec. LDs were labeled with Bodipy 493/503 (green). Scale bars, 5 μm. E, LLHs were stained with antibodies against Cideb (red) and Cidea/Cidec (green). Scale bars, 5 μm. D and E, arrowheads point to LDCSs. Quantitative data are presented as the mean ± S.E. Differences were considered significant at p < 0.05. ***, p < 0.001.
FIGURE 6.
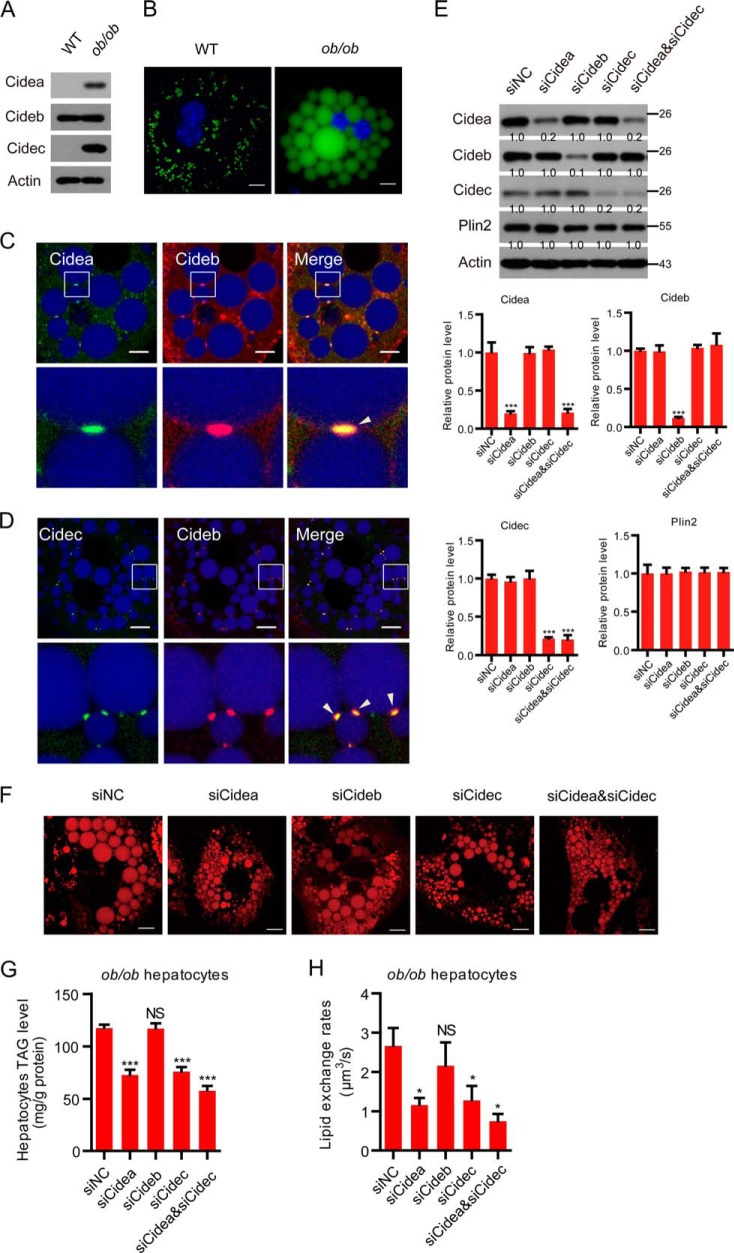
CIDE proteins promote lipid storage in ob/ob hepatocytes. A, expression levels of CIDEs in the livers of wild-type (WT) and ob/ob mice. B, representative image showing the lipid droplet morphology in wild-type and ob/ob hepatocytes. Scale bar, 10 μm. C, Cidea and Cideb localized on LDCSs. ob/ob hepatocytes were stained with antibodies against Cidea (green) and Cideb (red). Scale bar, 10 μm. D, Cidec and Cideb localized on LDCSs. ob/ob hepatocytes were stained with antibodies against Cidec (green) and Cideb (red). Scale bar, 10 μm. Arrowheads point to LDCSs. E, expression levels of the indicated proteins. Quantitation of the bands was performed using Quantity One software and are expressed as the fold change, after correction for actin levels. Values are averages obtained from three experiments. F, representative image showing the LD morphology after the depletion of different proteins. Scale bar, 10 μm. G, hepatocytes TAG level (n = 4). H, lipid exchange rates (n = 4). Quantitative data are presented as the mean ± S.E. Differences were considered significant at p < 0.05. *, p < 0.05; ***, p < 0.001; NS, no significant difference.
FIGURE 3.
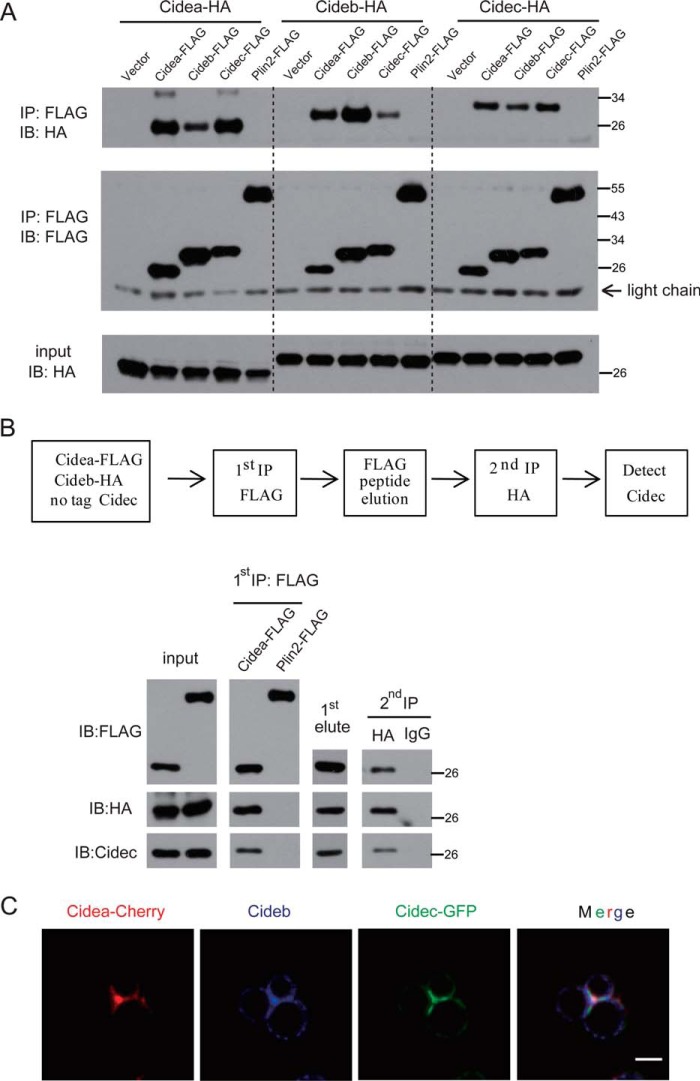
CIDE proteins interact with each other and form a complex at LDCSs. A, FLAG-tagged CIDEs and Plin2 together with HA-tagged CIDEs were coexpressed in 293T cells. Anti-FLAG M2 beads were used for immunoprecipitation. The immunoprecipitated products were detected by antibodies against FLAG or HA. IP, immunoprecipitation; IB, immunoblot. B, two-step coimmunoprecipitation of complex containing Cidea, Cideb, and Cidec. Top, schematic showing procedures for two-step coimmunoprecipitation assay. Cidea-FLAG or Plin2-FLAG (as a control) was transfected into 293T cells with Cideb-HA and untagged Cidec. C, Cidea, Cideb, and Cidec were colocalized at the LDCS. HepG2 cells were cotransfected with Cidea-Cherry (red), Cidec-GFP (green), and Cideb. Cells were stained with antibodies against Cideb (blue). Scale bar, 2 μm.
Antibodies and Western Blot Analysis
Methods for LD isolation, tissue homogenization, immunoprecipitation, two-step co-immunoprecipitation, and Western blot sample preparation were previously described (30, 35). The proteins were subjected to Western blot analysis with the desired antibodies. The antibodies against Cidea, Cideb, and Cidec were used as described previously (35). Antibodies against β-actin (Sigma, A5441, 1:2000), FLAG (Sigma, F1804, 1:1000), HA (Santa Cruz Biotechnology, sc-7392, 1:1000), and Plin2 (Fitzgerald Industries, 20R-Ap002, 1:8000) were used for Western blot analysis. The blots were detected using HRP-conjugated secondary antibodies (GE Healthcare, UK) and the ECL-Plus system.
Measurement of LD Sizes
Quantitative analysis of LD size in hepatocytes has been previously described (26, 29, 30). The diameter of the largest LD in each hepatocyte was measured. At least 50 hepatocytes were analyzed for each condition.
Calculation of Neutral Lipid Exchange Rate
The calculation of the lipid exchange rate was essentially the same as described previously (30). In brief, hepatocytes transfected with siRNAs were incubated with 200 μm BSA-bound oleic acid (Sigma) and 1 μg/ml Bodipy 558/568 C12 fatty acids (Molecular Probes, D3835) for 15 h and then transferred to fresh medium 1 h before the experiment. Live cells were viewed under a confocal microscope (LSM710) using a ×63 oil immersion objective. LD pairs were selected for bleaching. Selected regions were bleached by 500 interactions at 100% laser power (543 diode laser), followed by time-lapse scanning with 12.5-s intervals for 2 min for Cidea/Cidec-mediated exchange and a total recovery of 6 min for Cideb-mediated exchange.
Statistics
The statistical data reported include results from at least three biological replicates. All results are expressed as the mean ± S.E. Quantitation of the Western blot bands was performed using Quantity One software (Bio-Rad) and was expressed as the fold change after correction for relative control levels. All statistical analyses were performed in GraphPad Prism Version 5 (GraphPad Software). Significance was established using a two-tailed Student's t test. Differences were considered significant at p < 0.05. p values are indicated in each figure as follows: *, p < 0.05; **, p < 0.01, or ***, p < 0.001.
Results
CIDE Proteins Promote LD Fusion and Growth When Overexpressed in HepG2 Cells
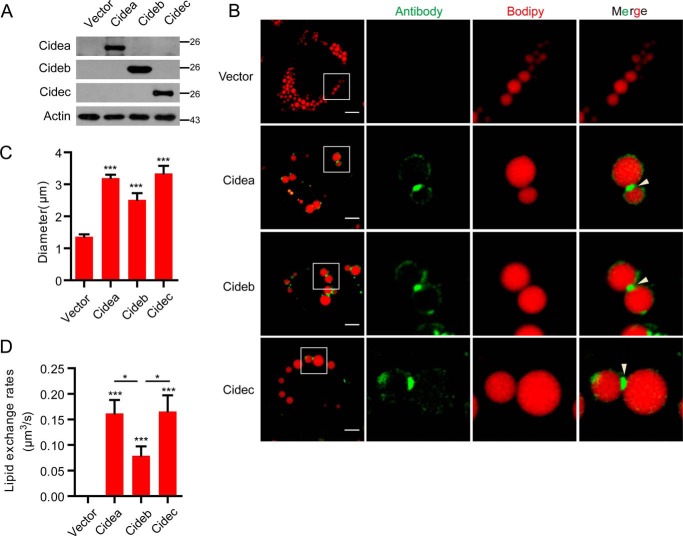
To investigate the role of CIDE proteins in LD fusion and growth in hepatocytes, we transfected HepG2 cells with no-tagged CIDE cDNAs. The expression levels of these proteins were detected by their corresponding antibodies (Fig. 1A). Similar to our observation in adipocytes (26), Cidea and Cidec were enriched at the LDCS in HepG2 cells (Fig. 1B). Interestingly, Cideb proteins were also enriched at LDCSs (Fig. 1B). We then measured the LD sizes and observed that cells expressing Cidea and Cidec accumulated larger LDs compared with control cells (Fig. 1C). Cideb also showed high activity in promoting large LD accumulation, albeit lower than that of Cidea and Cidec (Fig. 1C). Next, we measured lipid exchange activity, a hallmark of Cidea/Cidec-mediated LD fusion, between LD pairs that were positive for CIDE proteins. Lipid exchange activity for LD pairs positive for Cidea or Cidec was high (all are 0.16 μm3/s) and was similar to that in adipocytes (0.13 μm3/s) (26). Lipid exchange was also observed in Cideb-positive LD pairs and appeared to be lower (0.078 μm3/s) than that of Cidea and Cidec (Fig. 1D), consistent with the accumulation of smaller LDs in Cideb-expressing HepG2 cells. Overall, these data indicate that all CIDE proteins have the ability to promote LD fusion and growth in hepatocytes.
FIGURE 1.
CIDE proteins promote LD fusion and large LD formation in HepG2 cells. A, Vector, Cidea, Cideb, and Cidec plasmids were transfected into HepG2 cells. Expression levels of CIDEs were detected by Western blot. B, protein localization of CIDEs in HepG2 cells as in A. Oleic acid was added to promote the formation of LDs for 15 h. LDs were labeled with Bodipy 556/568 (C12, red). Scale bars, 10 μm. Arrowheads point to LDCSs. C, largest lipid droplet size per cell was measured in A. Ten cells were analyzed in each group. D, lipid exchange rates were measured in A. Five pairs of LDs were measured. Quantitative data are presented as the mean ± S.E. Differences were considered significant at p < 0.05. *, p < 0.05; ***, p < 0.001.
Identification of Two Types of Hepatocytes with Differential Expression of CIDE Proteins and Lipid Storage Capacity
We then investigated the precise subcellular distribution and function of endogenous CIDEs in isolated wild-type hepatocytes. We found that ∼95% of hepatocytes had small LDs, and the size (in diameter) of the largest LD in these cells was below 4 μm. We defined these hepatocytes as small LD-containing hepatocytes (SLHs). The remaining ∼5% of hepatocytes had large LDs, and the size (diameter) of the largest LD in these cells was above 4 μm. We defined those hepatocytes as large LD-containing hepatocytes (LLHs) (Fig. 2, A and B). In LLHs, the average size of the largest LD in each cell was ∼6 μm in diameter, and the average size of the largest LD in SLHs was 3 μm in diameter (Fig. 2B). Lipid exchange activity between contacted LD pairs in SLHs was 0.0025 μm3/s. Lipid exchange activity was 0.22 μm3/s in LLHs, nearly 90-fold higher than that in the SLHs (Fig. 2C). The heterogeneity in LD size and lipid exchange activity in the hepatocytes prompted us to check the levels of CIDE proteins in SLHs and LLHs using their corresponding antibodies. Cideb proteins were detected in both SLHs and LLHs and were found to be enriched at the LDCSs (Fig. 2D). However, Cidea and Cidec were only detected in LLHs and were enriched at LDCSs of contacted LDs (Fig. 2D). Co-staining of Cidea- or Cidec-positive hepatocytes with the Cideb antibody showed that Cideb was also present in LLHs and was enriched at the LDCSs (Fig. 2E). These data indicate that hepatocytes can be separated into two types according to their lipid storage capacity. Hepatocytes with high lipid storage capacity express all three CIDE proteins, whereas hepatocytes with low lipid storage capacity express only Cideb.
CIDE Proteins Form a Complex at LDCSs
We then investigated whether CIDEs interact with each other at LDCSs. We co-expressed Cidea-FLAG, Cideb-FLAG, Cidec-FLAG, or Plin2-FLAG with Cidea-HA in 293T cells. Then we used anti-FLAG M2 beads to immunoprecipitate FLAG-tagged proteins. The immunoprecipitated products were detected by using antibody against HA. Cidea was pulled down by Cidea, Cideb, and Cidec but not Plin2 (Fig. 3A). The interaction between Cidea and Cideb was slightly weaker than that of the Cidea-Cidea or Cidea-Cidec interaction. We also found that Cidea-FLAG, Cideb-FLAG, and Cidec-FLAG, but not Plin2-FLAG, were able to immunoprecipitate Cideb-HA or Cidec-HA (Fig. 3A). These data clearly showed that CIDE proteins can interact with each other. We then did a two-step coimmunoprecipitation analysis and to check whether Cidea, Cideb, and Cidec could form a ternary complex. Cidea-FLAG, Cideb-HA, and non-tagged Cidec were coexpressed in 293T cells. 1st step immunoprecipitation was carried out using anti-FLAG antibody. The coimmunoprecipitate components were then eluted with the FLAG peptide, and immunoprecipitation was carried out for the second round using HA antibody. Cideb and Cidec were observed in the final products (Fig. 3B). Contrary to that, Plin2 was not pulled down by either Cideb or Cidec. Consistent with their complex formation, Cidea, Cideb, and Cidec were colocalized and enriched at the LDCSs (Fig. 3C).
Reduced LD Size and Lipid Exchange Activity in Cideb−/− SLHs
To investigate the physiological role of Cideb in controlling LD fusion and lipid storage in hepatocytes, we isolated wild-type and Cideb−/− hepatocytes. The ratio of SLHs and LLHs was similar between the wild-type and Cideb−/− hepatocytes (data not shown). However, the sizes of the LDs were dramatically reduced in the Cideb−/− SLHs (Fig. 4, A and B), although the sizes of LDs in the LLHs were not affected by Cideb depletion (Fig. 4B). Introduction of Cidea, Cideb, or Cidec into Cideb−/− hepatocytes was able to increase the LD sizes (Fig. 4C). Therefore, Cidea and Cidec could functionally substitute for Cideb in enlargement of LDs. No lipid exchange activity was detected in Cideb−/− SLHs (Fig. 4, D and E), consistent with their reduced LD sizes and lower lipid storage capacity. Our previous studies have shown that knockdown of Plin2 enhanced LD size in WT and Cideb−/− hepatocytes (36). We then tested the lipid exchange rate in hepatocytes with Plin2 knockdown (with a knockdown efficiency of ∼80%, Fig. 4F). Plin2 knockdown did not affect the expression level of Cidea and Cidec (Fig. 4F). The lipid exchange rate was dramatically increased in wild-type SLHs with the depletion of Plin2 (18-fold higher, Fig. 4, D–F). However, the knockdown of Plin2 did not affect the lipid exchange activity of Cideb−/− SLHs and LLHs (Fig. 4E). Therefore, Cideb is responsible for LD fusion and lipid storage in SLHs.
FIGURE 4.
Lower LD fusion activity and reduced LD size in Cideb−/− hepatocytes. A, reduced LD sizes in Cideb−/− SLHs. LDs were labeled with Bodipy 556/568 (C12, red). Scale bar, 5 μm. B, largest LD sizes in SLHs and LLHs were measured (n = 20). C, representative image showing the LD size of wild-type (WT) and Cideb−/− hepatocytes transfected with CIDEs. Non-tagged CIDE proteins were transfected to hepatocytes. These proteins were stained by their corresponding antibodies. LDs were labeled with Bodipy 556/568 (C12, red). Scale bar, 5 μm. D, representative image showing the lipid droplet size of SLHs from WT, Cideb−/−, WT with siPlin2, and Cideb−/− with siPlin2. Scale bar, 5 μm. E, lipid exchange rates in SLHs as in D and LLHs (n = 6). F, protein expression levels in the total lysate and LD fraction of the indicated liver tissues. Quantitation of the bands was performed using Quantity One software and are expressed as the fold change, after correction for actin levels. Values are averages obtained from three independent experiments. Quantitative data are presented as the mean ± S.E. Differences were considered significant at p < 0.05. **, p < 0.01; ***, p < 0.001.
Fasting-induced LD Growth and Lipid Storage in the Liver Is Controlled by Cidea and Cidec
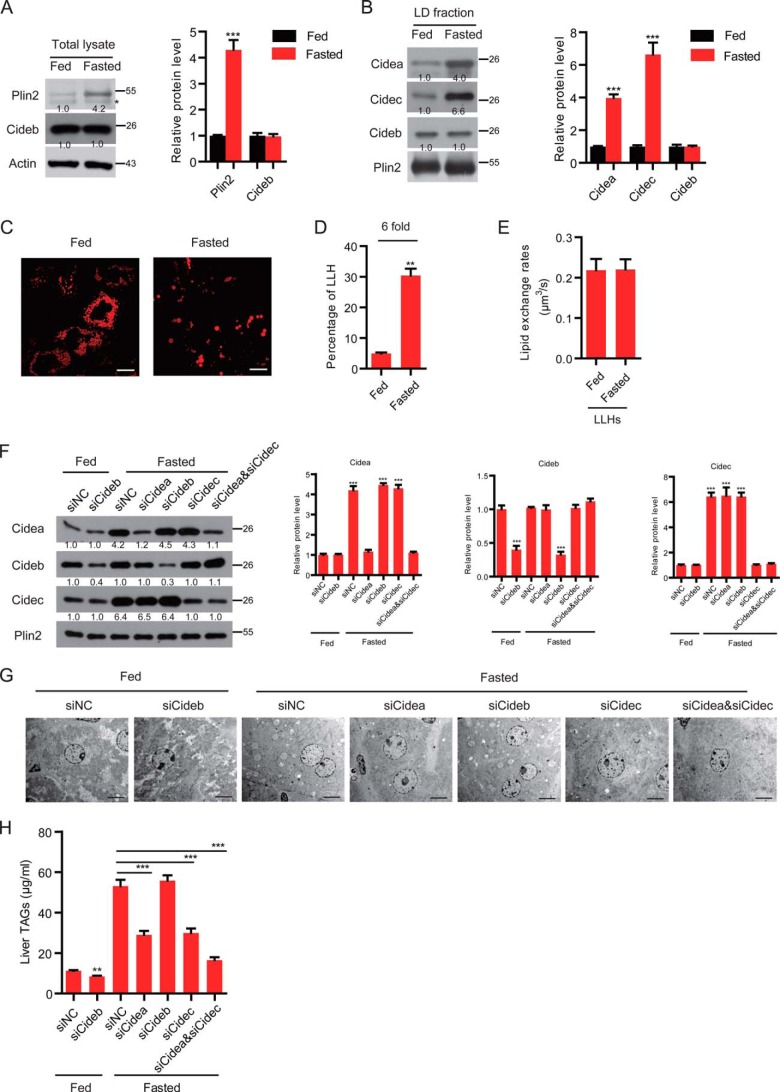
Many studies have shown that fasting induces the accumulation of large LDs in hepatocytes and induces higher hepatic lipid storage in mice (21–23). To investigate the role of CIDE proteins in fasting-induced liver steatosis, we analyzed expression levels of CIDE proteins, LD sizes, and lipid exchange and fusion activity in the livers and in isolated hepatocytes of animals fasted up to 16 h. The expression levels of Cideb in the liver were similar before and after fasting (Fig. 5A). Levels of Plin2 in the fasted liver were increased compared with that in the fed liver (4-fold, Fig. 5A). Interestingly, the levels of Cidea and Cidec were both increased in the LD fraction of liver tissue under the fasting condition (Fig. 5B). The percentage of LLHs isolated from the livers of fasted mice was dramatically increased compared with that of fed animals (30% versus 5%, 6-fold higher in fasted livers, Fig. 5, C and D). The lipid exchange rate in LLHs was similar between the fed and fasted hepatocytes (Fig. 5E). To better understand the relevance of Cidea, Cideb, and Cidec on the metabolic adaptation of the liver to fasting, we knocked down Cidea, Cideb, or Cidec using an Invivofectamine-mediated siRNA delivery system. As shown in Fig. 5F, the expression levels of Cidea, Cideb, and Cidec in the liver were significantly reduced by targeted delivery of siRNAs against Cidea, Cideb, or Cidec. Cideb knockdown under fed conditions indeed led to 25% reduction in TAG storage in liver (Fig. 5, G and H). However, in fasting wild-type mice, Cideb knockdown alone did not affect hepatic TAG levels (Fig. 5H). Lipid accumulation was dramatically reduced in the livers of Cidea- or Cidec-depleted animals that were fasted for 16 h (Fig. 5, G and H). The knockdown of Cidea and Cidec led to a further reduction in hepatic TAG levels and to the accumulation of smaller LDs under fasting conditions (Fig. 5, G and H). These data indicate that Cidea and Cidec mediate the elevated hepatic lipid storage under the fasting condition by increasing the LD fusion in LLHs and switching SLHs to LLHs.
FIGURE 5.
Fasting induced the expression of Cidea and Cidec. A, left, representative Western blot showing the protein expression profiles of the liver total lysate. The asterisk designates a nonspecific band. Right, quantitative analysis of the relative level of Plin2 and Cideb. n = 3. Actin was used as a loading control. B, left, representative Western blot showing the protein expression profiles of the LD fraction. Right, quantitative analysis of the relative level of CIDE proteins. n = 3. Plin2 was used as a loading control. C, representative image showing the lipid droplet morphology in fed and fasted wild-type (WT) hepatocytes. Scale bar, 10 μm. D, ratio of LLHs was increased in fasted conditions (n = 4). E, lipid exchange rates in LLHs (n = 6). F, protein expression levels in the LD fraction of the liver. Wild-type mice were injected with different siRNAs. n = 3. Quantitation of the bands was performed using Quantity One software and are expressed as the fold change, after correction for Plin2 levels. Values are averages obtained from three independent experiments. G, electron microscopy (EM) showing the morphology of the liver. H, liver TAG level. n = 3. Quantitative data are presented as the mean ± S.E. Differences were considered significant at p < 0.05. **, p < 0.01; ***, p < 0.001.
Cidea and Cidec Promote Hepatic Lipid Storage in ob/ob Mice
Cidea and Cidec are both shown to be dramatically unregulated in the livers of ob/ob mice (Fig. 6A). Hepatocytes isolated from ob/ob mice contained very large LDs (Fig. 6B). Expression levels of Cidea and Cidec were detected in nearly all hepatocytes and were enriched at LDCSs (Fig. 6, C and D). Cideb was also detected and enriched at LDCSs of all ob/ob hepatocytes (Fig. 6, C and D). We knocked down individual CIDE protein alone or in combination in ob/ob hepatocytes using a Lipofectamine-mediated siRNA delivery system. This strategy resulted in a substantial knockdown of Cidea (80% knockdown efficiency), Cideb (90% knockdown efficiency), and Cidec (80% knockdown efficiency), respectively (Fig. 6E). The knockdown of Cidea and Cidec alone or in combination in the ob/ob hepatocytes resulted in reduced lipid storage and accumulation of smaller LDs (Fig. 6, F and G). However, the knockdown of Cideb did not affect the LD size of ob/ob hepatocytes (Fig. 6, F and G). Consistently, the depletion of Cidea or Cidec in ob/ob hepatocytes led to reduced lipid exchange activity (Fig. 6H). The knockdown of Cideb did not affect the lipid exchange activity of contacted LD pairs (Fig. 6H). The knockdown of both Cidea and Cidec reduced the lipid exchange rate further compared with Cidea or Cidec single depletion (Fig. 6H). These data indicate that Cidea and Cidec play important roles in hepatic lipid storage in ob/ob animals.
Discussion
CIDE family proteins (Cidea and Cidec) act as important regulators of lipid storage and lipid metabolism in adipocytes by promoting LD fusion and growth (17). Here, we examined the precise function of individual CIDEs in controlling LD fusion and growth in the liver. We found that overexpression of CIDE proteins dramatically induced the formation of large LDs and promoted the fusion and growth of LDs in hepatocytes. In addition, all CIDE proteins are enriched at LDCSs in hepatocytes or when they were overexpressed in liver cell lines.
Interestingly, we identified two populations of hepatocytes based on the size of LDs (SLHs and LLHs). We further observed that ∼5% of wild-type hepatocytes are LLHs that express all three CIDE proteins and have higher LD fusion activity. SLHs express Cideb alone and have lower LD fusion activity (Fig. 7). Cideb−/− SLHs have smaller LDs and lower lipid exchange activity. In contrast, depletion of Cideb did not affect the LD sizes and LD fusion activity of LLHs. The lipid storage capacity in SLHs is primarily determined by Cideb, whereas Cidea and Cidec regulate lipid storage in LLHs. Under fasting conditions, the percentage of LLHs and the expression levels of both Cidea and Cidec increased dramatically. However, the lipid exchange activity in individual LLHs was similar to that of control hepatocytes. Therefore, fasting induced the expression of Cidea/Cidec in SLHs, promoting the conversion of SLHs to LLHs, but did not affect the Cidea/Cidec expression in LLHs. Moreover, the percentage of Cidea/Cidec-expressing cells reached nearly 100% in ob/ob hepatocytes. All hepatocytes from ob/ob livers exhibited high expression levels of Cidea/Cidec and had higher LD fusion activity (Fig. 7). The increased expression of Cidea and Cidec in LLHs could be due to their transcriptional regulation by various factors (22–24). Alternatively, increased Cidea and Cidec protein levels in fasted or obese hepatocytes are most likely due to the enhanced stability resulting from exposure to high fatty acid levels and TAG synthesis (20–23, 37). We think the expansion of LLHs that express Cidea and Cidec contributes primarily to the development of hepatic steatosis. In addition, the percentage of LLHs may reflect the nutrition supply to the liver and may play an important role in the development of liver steatosis. Further characterization of gene expression profiles and physiological function (insulin sensitivity) of SLHs and LLHs will be useful in the investigation of the origin and regulation of SLHs and LLHs.
FIGURE 7.
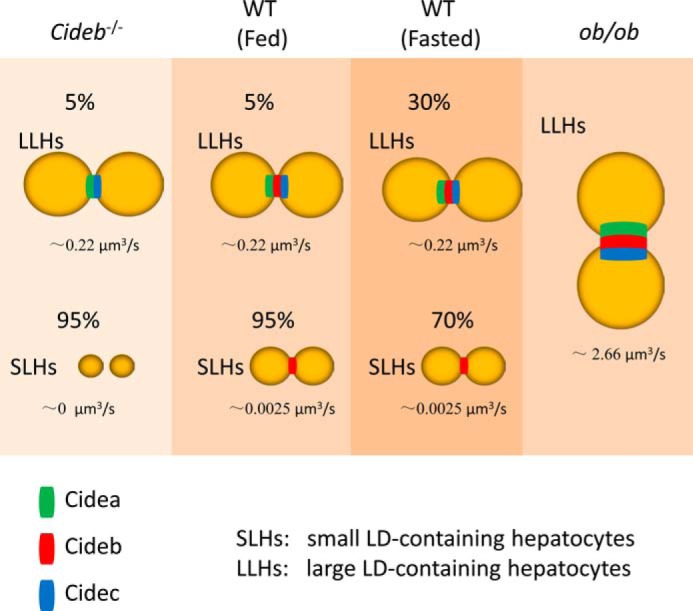
Proposed model of CIDE-mediated LD fusion and growth in hepatocytes. In wild-type (WT) hepatocytes, two types (SLHs and LLHs) are observed. Cideb is expressed in SLHs and LLHs. Cidea and Cidec are expressed in LLHs. Fasting enhances the percentage of LLHs. Cideb deficiency leads to reduced LD size in SLHs. ob/ob hepatocytes have higher Cidea and Cidec expression levels and higher lipid exchange rates.
Previous data showed that the expressions levels of Cidea and Cidec were significantly increased in human steatotic liver (20, 38, 39). A single nucleotide polymorphism of a G to T transversion in CIDEA exon 4, which is equivalent to a V115F substitution, is associated with body mass index in Swedish male and female obese patients (40). In Japanese and Chinese patients, the CIDEA V115F polymorphism is associated with obesity and metabolic syndrome (41, 42). A single nucleotide polymorphism involving a G to T transversion in CIDEC exon 6, which causes a E186X nonsense mutation was also observed in a female patient who showed partial lipodystrophy (43). Controlling the correct amount of CIDE proteins was important in maintaining lipid homeostasis in the liver. If there was insufficient CIDE proteins in the liver, this would lead to reduced LD sizes and lower lipid storage capacity. Free fatty acid levels may increase in hepatocytes, resulting in lipotoxicity and increased oxidative stress and inflammatory response (44). Previous data showed that overexpression of CIDE proteins in many cell types resulted in a caspase-independent cell death (45). However, the rate of cell death was significantly reduced in CIDE-overexpressing cells when exogenous oleic acid was introduced (46). In the presence of lipid-rich medium, CIDE proteins localized to LDs promote LD fusion and lipid storage, resulting in reduced lipotoxicity and oxidative stress.
In conclusion, we have shown that CIDE proteins are localized to LDCSs and promote LD fusion in hepatocytes. According to the differential expression of CIDE proteins and the sizes of LDs, hepatocytes can be characterized into two populations (SLHs and LLHs). Cideb plays an important role in promoting lipid storage and maintaining lipid homeostasis in the liver under normal conditions. Cidea and Cidec are crucial regulators of hepatic lipid storage under fasting and obese conditions.
Author Contributions
L. Z., W. X., L. W., and P. L. designed the research; W. X., L. Z., L. W., M. Y., H. R., F. C., and J. Y. performed the experiments; L. Z., L. W., L. X., D. X., J. Z. L., X. X., and M. A. analyzed the data; and P. L. and L. Z. wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank the members of the P. Li Laboratory at Tsinghua University for their helpful discussions.
This work was supported by National Basic Research Program Grants 2013CB530602 (to P. L.), National Natural Science Foundation of China Grants 31430040 and 31321003 (to P. L.) and Grant 31501089 (to J. Y.), and Grants 31271268 and 81471079 (to J. Z. L.), and China Postdoctoral Science Foundation Grants 2012M520249 and 2013T60103 (to L. Z.). The authors declare that they have no conflicts of interest regarding the contents of this article.
This article was selected as a Paper of the Week.
- LD
- lipid droplet
- CIDE
- cell death-inducing DNA fragmentation factor-α-like effector
- LDCS
- LD-LD contact site
- LLHs
- large lipid droplet-containing hepatocytes
- and SLHs
- small lipid droplet-containing hepatocytes
- TAG
- triacylglycerol.
References
- 1. Cornier M. A., Dabelea D., Hernandez T. L., Lindstrom R. C., Steig A. J., Stob N. R., Van Pelt R. E., Wang H., and Eckel R. H. (2008) The metabolic syndrome. Endocr. Rev. 29, 777–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tessari P., Coracina A., Cosma A., and Tiengo A. (2009) Hepatic lipid metabolism and non-alcoholic fatty liver disease. Nutr. Metab. Cardiovasc. Dis. 19, 291–302 [DOI] [PubMed] [Google Scholar]
- 3. Musso G., Gambino R., and Cassader M. (2009) Recent insights into hepatic lipid metabolism in non-alcoholic fatty liver disease (NAFLD). Prog. Lipid Res. 48, 1–26 [DOI] [PubMed] [Google Scholar]
- 4. Dietrich P., and Hellerbrand C. (2014) Non-alcoholic fatty liver disease, obesity and the metabolic syndrome. Best Pract. Res. Clin. Gastroenterol. 28, 637–653 [DOI] [PubMed] [Google Scholar]
- 5. Wilfling F., Haas J. T., Walther T. C., and Farese R. V. Jr. (2014) Lipid droplet biogenesis. Curr. Opin. Cell Biol. 29, 39–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Farese R. V. Jr., and Walther T. C. (2009) Lipid droplets finally get a little R-E-S-P-E-C-T. Cell 139, 855–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Thiam A. R., Farese R. V. Jr., and Walther T. C. (2013) The biophysics and cell biology of lipid droplets. Nat. Rev. Mol. Cell Biol. 14, 775–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang H., Galea A., Sytnyk V., and Crossley M. (2012) Controlling the size of lipid droplets: lipid and protein factors. Curr. Opin. Cell Biol. 24, 509–516 [DOI] [PubMed] [Google Scholar]
- 9. Wilfling F., Thiam A. R., Olarte M. J., Wang J., Beck R., Gould T. J., Allgeyer E. S., Pincet F., Bewersdorf J., Farese R. V. Jr., and Walther T. C. (2014) Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting. Elife 3, e01607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gross D. A., Zhan C., and Silver D. L. (2011) Direct binding of triglyceride to fat storage-inducing transmembrane proteins 1 and 2 is important for lipid droplet formation. Proc. Natl. Acad. Sci. U.S.A. 108, 19581–19586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Krahmer N., Guo Y., Wilfling F., Hilger M., Lingrell S., Heger K., Newman H. W., Schmidt-Supprian M., Vance D. E., Mann M., Farese R. V. Jr., and Walther T. C. (2011) Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab. 14, 504–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McFie P. J., Banman S. L., Kary S., and Stone S. J. (2011) Murine diacylglycerol acyltransferase-2 (DGAT2) can catalyze triacylglycerol synthesis and promote lipid droplet formation independent of its localization to the endoplasmic reticulum. J. Biol. Chem. 286, 28235–28246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xu L., Zhou L., and Li P. (2012) CIDE proteins and lipid metabolism. Arterioscler. Thromb. Vasc. Biol. 32, 1094–1098 [DOI] [PubMed] [Google Scholar]
- 14. Crunk A. E., Monks J., Murakami A., Jackman M., Maclean P. S., Ladinsky M., Bales E. S., Cain S., Orlicky D. J., and McManaman J. L. (2013) Dynamic regulation of hepatic lipid droplet properties by diet. PLoS One 8, e67631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bickel P. E., Tansey J. T., and Welte M. A. (2009) PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim. Biophys. Acta 1791, 419–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Okumura T. (2011) Role of lipid droplet proteins in liver steatosis. J. Physiol. Biochem. 67, 629–636 [DOI] [PubMed] [Google Scholar]
- 17. Gong J., Sun Z., and Li P. (2009) CIDE proteins and metabolic disorders. Curr. Opin. Lipidol. 20, 121–126 [DOI] [PubMed] [Google Scholar]
- 18. Yonezawa T., Kurata R., Kimura M., and Inoko H. (2011) Which CIDE are you on? Apoptosis and energy metabolism. Mol. Biosyst. 7, 91–100 [DOI] [PubMed] [Google Scholar]
- 19. Matsusue K., Kusakabe T., Noguchi T., Takiguchi S., Suzuki T., Yamano S., and Gonzalez F. J. (2008) Hepatic steatosis in leptin-deficient mice is promoted by the PPARγ target gene Fsp27. Cell Metab. 7, 302–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou L., Xu L., Ye J., Li D., Wang W., Li X., Wu L., Wang H., Guan F., and Li P. (2012) Cidea promotes hepatic steatosis by sensing dietary fatty acids. Hepatology 56, 95–107 [DOI] [PubMed] [Google Scholar]
- 21. Vilà-Brau A., De Sousa-Coelho A. L., Gonçalves J. F., Haro D., and Marrero P. F. (2013) Fsp27/CIDEC is a CREB target gene induced during early fasting in liver and regulated by FA oxidation rate. J. Lipid Res. 54, 592–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Langhi C., and Baldán Á. (2015) CIDEC/FSP27 is regulated by peroxisome proliferator-activated receptor α and plays a critical role in fasting- and diet-induced hepatosteatosis. Hepatology 61, 1227–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xu X., Park J. G., So J. S., and Lee A. H. (2015) Transcriptional activation of Fsp27 by the liver-enriched transcription factor CREBH promotes lipid droplet growth and hepatic steatosis. Hepatology 61, 857–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Puri V. (2013) Fasting regulates FSP27 expression in the liver. J. Lipid Res. 54, 569–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jambunathan S., Yin J., Khan W., Tamori Y., and Puri V. (2011) FSP27 promotes lipid droplet clustering and then fusion to regulate triglyceride accumulation. PLoS One 6, e28614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gong J., Sun Z., Wu L., Xu W., Schieber N., Xu D., Shui G., Yang H., Parton R. G., and Li P. (2011) Fsp27 promotes lipid droplet growth by lipid exchange and transfer at lipid droplet contact sites. J. Cell Biol. 195, 953–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu L., Zhou L., Chen C., Gong J., Xu L., Ye J., Li D., and Li P. (2014) Cidea controls lipid droplet fusion and lipid storage in brown and white adipose tissue. Sci. China Life Sci. 57, 107–116 [DOI] [PubMed] [Google Scholar]
- 28. Barneda D., Planas-Iglesias J., Gaspar M. L., Mohammadyani D., Prasannan S., Dormann D., Han G. S., Jesch S. A., Carman G. M., Kagan V., Parker M. G., Ktistakis N. T., Dixon A. M., Klein-Seetharaman J., Henry S., and Christian M. (2015) The brown adipocyte protein CIDEA promotes lipid droplet fusion via a phosphatidic acid-binding amphipathic helix. Elife 4, e07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sun Z., Gong J., Wu H., Xu W., Wu L., Xu D., Gao J., Wu J. W., Yang H., Yang M., and Li P. (2013) Perilipin1 promotes unilocular lipid droplet formation through the activation of Fsp27 in adipocytes. Nat. Commun. 4, 1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu L., Xu D., Zhou L., Xie B., Yu L., Yang H., Huang L., Ye J., Deng H., Yuan Y. A., Chen S., and Li P. (2014) Rab8a-AS160-MSS4 regulatory circuit controls lipid droplet fusion and growth. Dev. Cell 30, 378–393 [DOI] [PubMed] [Google Scholar]
- 31. Grahn T. H., Zhang Y., Lee M. J., Sommer A. G., Mostoslavsky G., Fried S. K., Greenberg A. S., and Puri V. (2013) FSP27 and PLIN1 interaction promotes the formation of large lipid droplets in human adipocytes. Biochem. Biophys. Res. Commun. 432, 296–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ye J., Li J. Z., Liu Y., Li X., Yang T., Ma X., Li Q., Yao Z., and Li P. (2009) Cideb, an ER- and lipid droplet-associated protein, mediates VLDL lipidation and maturation by interacting with apolipoprotein B. Cell Metab. 9, 177–190 [DOI] [PubMed] [Google Scholar]
- 33. Li J. Z., Ye J., Xue B., Qi J., Zhang J., Zhou Z., Li Q., Wen Z., and Li P. (2007) Cideb regulates diet-induced obesity, liver steatosis, and insulin sensitivity by controlling lipogenesis and fatty acid oxidation. Diabetes 56, 2523–2532 [DOI] [PubMed] [Google Scholar]
- 34. Toh S. Y., Gong J., Du G., Li J. Z., Yang S., Ye J., Yao H., Zhang Y., Xue B., Li Q., Yang H., Wen Z., and Li P. (2008) Up-regulation of mitochondrial activity and acquirement of brown adipose tissue-like property in the white adipose tissue of fsp27 deficient mice. PLoS One 3, e2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhou L., Park S. Y., Xu L., Xia X., Ye J., Su L., Jeong K. H., Hur J. H., Oh H., Tamori Y., Zingaretti C. M., Cinti S., Argente J., Yu M., Wu L., et al. (2015) Insulin resistance and white adipose tissue inflammation are uncoupled in energetically challenged Fsp27-deficient mice. Nat. Commun. 6, 5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li X., Ye J., Zhou L., Gu W., Fisher E. A., and Li P. (2012) Opposing roles of cell death-inducing DFF45-like effector B and perilipin 2 in controlling hepatic VLDL lipidation. J. Lipid Res. 53, 1877–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nian Z., Sun Z., Yu L., Toh S. Y., Sang J., and Li P. (2010) Fat-specific protein 27 undergoes ubiquitin-dependent degradation regulated by triacylglycerol synthesis and lipid droplet formation. J. Biol. Chem. 285, 9604–9615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hall A. M., Brunt E. M., Klein S., and Finck B. N. (2010) Hepatic expression of cell death-inducing DFFA-like effector C in obese subjects is reduced by marked weight loss. Obesity 18, 417–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xu M. J., Cai Y., Wang H., Altamirano J., Chang B., Bertola A., Odena G., Lu J., Tanaka N., Matsusue K., Matsubara T., Mukhopadhyay P., Kimura S., Pacher P., Gonzalez F. J., Bataller R., and Gao B. (2015) Fat-specific protein 27/CIDEC promotes development of alcoholic steatohepatitis in mice and humans. Gastroenterology 149, 1030–1041.e1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dahlman I., Kaaman M., Jiao H., Kere J., Laakso M., and Arner P. (2005) The CIDEA gene V115F polymorphism is associated with obesity in Swedish subjects. Diabetes 54, 3032–3034 [DOI] [PubMed] [Google Scholar]
- 41. Zhang L., Miyaki K., Nakayama T., and Muramatsu M. (2008) Cell death-inducing DNA fragmentation factor α-like effector A (CIDEA) gene V115F (G→T) polymorphism is associated with phenotypes of metabolic syndrome in Japanese men. Metabolism 57, 502–505 [DOI] [PubMed] [Google Scholar]
- 42. Wu J., Zhang L., Zhang J., Dai Y., Bian L., Song M., Russell A., and Wang W. (2013) The genetic contribution of CIDEA polymorphisms, haplotypes and loci interaction to obesity in a Han Chinese population. Mol. Biol. Rep. 40, 5691–5699 [DOI] [PubMed] [Google Scholar]
- 43. Rubio-Cabezas O., Puri V., Murano I., Saudek V., Semple R. K., Dash S., Hyden C. S., Bottomley W., Vigouroux C., Magré J., Raymond-Barker P., Murgatroyd P. R., Chawla A., Skepper J. N., Chatterjee V. K., et al. (2009) Partial lipodystrophy and insulin resistant diabetes in a patient with a homozygous nonsense mutation in CIDEC. EMBO Mol. Med. 1, 280–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Anstee Q. M., and Goldin R. D. (2006) Mouse models in non-alcoholic fatty liver disease and steatohepatitis research. Int. J. Exp. Pathol. 87, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen Z., Guo K., Toh S. Y., Zhou Z., and Li P. (2000) Mitochondria localization and dimerization are required for CIDE-B to induce apoptosis. J. Biol. Chem. 275, 22619–22622 [DOI] [PubMed] [Google Scholar]
- 46. Liu K., Zhou S., Kim J. Y., Tillison K., Majors D., Rearick D., Lee J. H., Fernandez-Boyanapalli R. F., Barricklow K., Houston M. S., and Smas C. M. (2009) Functional analysis of FSP27 protein regions for lipid droplet localization, caspase-dependent apoptosis, and dimerization with CIDEA. Am. J. Physiol. Endocrinol. Metab. 297, E1395–E1413 [DOI] [PubMed] [Google Scholar]