Abstract
Glycosaminoglycans (GAGs) are polysaccharides that play vital functional roles in numerous biological processes, and compounds belonging to this class have been implicated in a wide variety of diseases. Chondroitin AC lyase (ChnAC) (EC 4.2.2.5) catalyzes the degradation of various GAGs, including chondroitin sulfate and hyaluronic acid, to give the corresponding disaccharides containing an Δ4-unsaturated uronic acid at their non-reducing terminus. ChnAC has been isolated from various bacteria and utilized as an enzymatic tool for study and evaluating the sequencing of GAGs. Despite its substrate specificity and the fact that its crystal structure has been determined to a high resolution, the direction in which ChnAC catalyzes the cleavage of oligosaccharides remain unclear. Herein, we have determined the structural cues of substrate depolymerization and the cleavage direction of ChnAC using model substrates and recombinant ChnAC protein. Several structurally defined oligosaccharides were synthesized using a chemoenzymatic approach and subsequently cleaved using ChnAC. The degradation products resulting from this process were determined by mass spectrometry. The results revealed that ChnAC cleaved the β1,4-glycosidic linkages between glucuronic acid and glucosamine units when these bonds were located on the reducing end of the oligosaccharide. In contrast, the presence of a GlcNAc-α-1,4-GlcA unit at the reducing end of the oligosaccharide prevented ChnAC from cleaving the GalNAc-β1,4-GlcA moiety located in the middle or at the non-reducing end of the chain. These interesting results therefore provide direct proof that ChnAC cleaves oligosaccharide substrates from their reducing end toward their non-reducing end. This conclusion will therefore enhance our collective understanding of the mode of action of ChnAC.
Keywords: chondroitin sulfate, glycosaminoglycan, mass spectrometry (MS), oligosaccharide, substrate specificity, Arthrobacter species, Catalytic Direction, Chondroitin exolyase, elimination reaction, structurally defined substrate
Introduction
Glycosaminoglycans (GAGs)3 are long unbranched polysaccharides consisting of repeating disaccharide units that can be found in the connective tissues of animals (1, 2). In terms of their structure, GAGs are composed of repeating disaccharide units containing a glucosamine (e.g. galactosamine or glucosamine) and uronic acid, such as glucuronic acid (GlcA) or iduronic acid (IdoA) (2). Heparan sulfate (HS), chondroitin sulfate (CS), and hyaluronic acid (HA) are three of most extensively studied classes of GAGs (2). The glucosamine and uronic acid residues in HS and CS are modified by the sulfation process, whereas HA is not naturally sulfated. These polysaccharides are thought to be involved in the regulation of various cellular processes such as adhesion, differentiation, migration, and proliferation (1, 3–6). GAGs and their low molecular weight derivates, which can be prepared by various methods of depolymerization, have therefore received considerable interested in terms of their potential clinical applications as anticoagulant, antitumor, anti-inflammatory, and anti-complement agents (7–11). Several different degradation methods, including chemical and enzymatic approaches, can be used to provide access to structurally distinct GAG chains, which can display a wide variety of different pharmacological and pharmacokinetic properties (12–14). Among the GAGs, CS consists of repeating disaccharide units of GlcA and N-acetyl-galactosamine (GalNAc) with β-1,3 and β-1,4 linkages; each of which is capable of carrying sulfonate groups. CS can be classified into several different types depending on the position of the sulfate groups. For example, CS-A, CS-C, and CS-E contain GalNAc-4-O-sulfate, GalNAc-6-O-sulfate, and GalNAc-4,6-O-sulfate groups, respectively. CS-B (dermatan sulfate, DS) differs from CS-A in that the GlcA unit has been replaced by an IdoA unit. Compared with CS-E, CS-D contains an additional sulfated modification on carbon 2 of the GlcA unit.
A variety of bacterial CS lyases have been prepared with the ability to affect the enzymatic degradation of CS polysaccharides (15, 16). Although these CS lyases followed the same general mechanism, in that they catalyzed a β-elimination reaction at the C4 position of GlcA to yield Δ4,5-unsaturated GlcA at the non-reducing end of the oligosaccharide products, these enzymes exhibited different substrate specificities (17–19). Briefly, chondroitin AC lyase (ChnAC, EC 4.2.2.5) cleaves HA, CS-A, and CS-C. Chondroitin-sulfate-ABC exolyase (exChnABC, EC 4.2.2.21) and chondroitin-sulfate-ABC endolyase (enChnABC, EC 4.2.2.20) can both degrade CS-A, CS-C, and DS, although the latter of these enzymes shows endolytic activity. Notably, chondroitin B lyase (ChnB, EC 4.2.2.19) is specific for DS. To date, a number of ChnAC enzymes have been obtained from various bacteria, including Arthrobacter sp (16, 20), Flavobacterium heparinum (21, 22), Serratia marcescens (23) and Bacteroides stercoris (24), and these enzymes have been subjected to extensive biochemical and x-ray crystallographic analyses. Interestingly, the enzymatic characterization of the ChnAC from F. heparinum showed that it acted as an endolyase toward a polysaccharide substrate, producing a mixture of unsaturated oligosaccharides of different sizes (25). This behavior was in contrast to that of the other bacterial ChnAC lyases listed above, which acted as exolyases. Crystal structures of the ChnAC isolated from Arthrobacter sp. bound to various oligosaccharide substrates revealed that the lytic mechanism of this enzyme was dependent of the (α/α)5 toroidal fold. These data also showed that the Tyr-242 residue acted as a base to abstract a proton from the C5 position of GlcA, while the Asn-183 and His-233 residues neutralized the charge on the acidic group of GlcA (16). ChnAC and several other CS lyases have been widely used in conjunction with modern separation and analytical methods for disaccharide analysis, polysaccharide sequencing, and biological evaluations (19). However, very little is known about the direction of the ChnAC-catalyzed degradation of polysaccharides. This lack of understanding regarding the direction of the degradation is mainly due to the lack of structurally suitable oligosaccharides. An advanced chemoenzymatic method was recently developed for the efficient synthesis of GAG oligosaccharides with high purity (26–28). Furthermore, the bacterial enzymes involved in the synthesis of the GAG backbone, including N-acetyl glucosaminyl transferase from Escherichia coli K5 strain (KfiA), heparosan synthase 2 from Pasteurella multocida (PmHS2), and E. coli K4 chondroitin polymerase (KfoC), show broad donor substrate specificities and acceptor promiscuity (29).4 These developments have allowed us to prepare structurally defined chimeric oligosaccharide substrates to probe the directionality of this process in greater detail.
The primary goal of this study was to uncover the direction of the ChnAC-catalyzed polysaccharide degradation using a library of structurally defined oligosaccharides. A similar strategy to this was successfully applied to the bacterial heparosan lyase and mammalian heparanase (30, 31). However, the oligosaccharides involved in these previous studies were synthesized based on the natural structure of the GAG polysaccharides. In this study, we decided to synthesize chimeric oligosaccharides concurrently containing repeating HS and CS disaccharide units. This selection was based on the fact that the CS backbone structure (-GalNAc-β-1,4-GlcA-) can be cleaved by ChnAC, whereas the HS backbone structure (-GlcNAc-α-1,4-GlcA-) remains unchanged (16, 19). The site resistant to being cleaved by the ChnAC enzyme was positioned on the reducing or the non-reducing terminus, and the products resulting from the treatment of these substrates with ChnAC were identified by electrospray ionization mass spectrometry (ESI-MS). The results of these experiments therefore provided an opportunity to determine the preference of this enzyme for endolytic cleavage at the reducing or non-reducing ends of the different substrates. The ChnAC isolated from Arthrobacter sp.5 displayed cleavage activity toward the oligosaccharide substrate when the GalNAc-β1,4-GlcA residue was present at the reducing end. In contrast, this enzyme displayed no activity toward the degradation of the oligosaccharide substrate were the GalNAc-β-1,4-GlcA residues at the reducing end had been replaced with GlcNAc-α-1,4-GlcA residues. These data demonstrate, for the first time, that ChnAC cleaves polysaccharides from their reducing end toward their non-reducing end. The results of this study will therefore advance our understanding of the mode of action of ChnAC enzymes.
Experimental Procedures
Materials
1-O-(para-nitrophenyl)-glucuronide (GlcA-pNP),uridine 5′-diphosphate (UDP)-N-acetylglucosamine (GlcNAc),UDP-N-acetylgalactosamine (GalNAc), and UDP-GlcA were purchased from Sigma Aldrich.
Enzymes
Arthrobacter species chondroitin AC lyase genes, AsChnAC (GenBankTM accession number KT985049), were cloned into a pET28a(+) expression plasmid2, followed by a transformation step into E. coli BL21 (DE3). The resulting recombinant strain was used as a source of recombinant ChnAC. The N-His-tagged AsChnAC were purified over a Ni Sepharose 6 Fast Flow column (GE Healthcare Bio-Sciences AB, Uppsala, Sweden) following the protocol provided by the manufacturer. KfiA and PmHS2 were expressed in E. coli and purified using an appropriate affinity chromatography system as described previously (27, 32–34). KfoC (GenBankTM accession number AB079602) was recombinantly expressed in E. coli BL21 (DE3) cells as a soluble N-His6-tagged fusion protein and purified using an appropriate affinity chromatography system.4
Preparation of Model Oligosaccharide Substrates
Four structurally defined oligosaccharides were prepared using enzymatic synthesis following previously published procedures from the literature (32, 33, 35). All four of the oligosaccharides prepared in the current study were synthesized from the commercially available monosaccharide of GlcA-pNP. Notably, the α-(1,4) linkages between the GlcNAc and GlcA residues and the β-(1,4) linkages between the GalNAc and GlcA residues were built with PmHS2 and KfoC, respectively. Briefly, the CS-trisaccharide (GlcA-β1,3-GalNAc-β1,4-GlcA-pNP) was synthesized using KfoC, and the subsequent elongation of this trisaccharide to the HP-CS-hexasaccharide was completed by KfiA and PmHS2. In contrast, the HP-trisaccharide (GlcA-β1,4-GlcNAc-α1,4-GlcA-pNP) was initially built by KfiA and pmHS2, and its subsequent elongation to the desired size was achieved using a KfoC-catalyzed glycosyl transferase reaction. For a typical elongation reaction, 20 mg of acceptor was incubated with 1.5 equivalents of UDP-sugar and 2 mg of enzyme in 40 ml of buffer containing 25 mm Tris-HCl (pH 7.2) and 10 mm MnCl2 at 37 °C for 12 h. To confirm that the acceptor had been completely converted to the desired elongation product, the reactions were monitored by polyamine-based anion exchange (PAMN)-HPLC. For HPLC, the column (4.6 × 250 mm, Shimogyoku, Kyoto, Japan) was eluted with a linear gradient of KH2PO4 from 0 to 0.6 m over 40 min at a flow rate of 0.5 ml/min. The para-nitrophenyl (pNP) group was detected at 310 nm. The product was then purified over a Bio-Gel P-2 column, with the desired fractions being detected based on their UV absorbance at 310 nm. In this way, we synthesized milligram quantities of these model oligosaccharide substrates and confirmed their structures by ESI-MS and NMR analyses.
Cleavage of Synthetic Oligosaccharides
To uncover the catalytic direction of ChnAC, we evaluated the elimination products of this enzyme toward several structurally defined oligosaccharides, including CS-trisaccharide, HP-trisaccharide, HP-CS-hexasaccharide, and CS-HP-hexasaccharide. A typical elimination reaction was conducted in a total volume of 1.5 ml (0.4 mm HP-CS-hexasaccharide, pH 7.0) containing 1 μg of purified ChnAC for 0, 1, 10 min, or 16 h at 37 °C, followed by the subsequent heating of the mixture at 100 °C for 5 min and separation by centrifugation. The reaction mixture was monitored by (PAMN)-HPLC, followed ESI-MS analysis.
ESI-MS Analysis
ESI-MS analyses were performed on a Thermo LCQ-Deca system (Thermo Fisher Scientific Inc.). The samples were dissolved in 50% methanol. Experiments were performed in the negative ionization mode with a spray voltage of 3.5 kV and a capillary temperature of 275 °C.
Results and Discussion
Our efforts for determining the direction of the chondroitin AC lyase-catalyzed degradation of the chondroitin sulfate (CS) chains required the expression of recombinant chondroitin AC lyase and the preparation of structurally defined oligosaccharide substrates. A novel eliminase gene from an Arthrobacter species capable of degrading CS and hyaluronan (HA) was cloned and successfully expressed in E. coli.5 Studies pertaining to the substrate specificity of this enzyme clearly demonstrated that AsChnAC was a chondroitin AC exolyase (EC 4.2.2.5). Briefly, purified recombinant AsChnAC affected the degradation of polysaccharides containing 1,4-β-d-hexosaminyl and 1,3-β-d-glucuronosyl linkages to give the corresponding unsaturated disaccharides containing 4-deoxy-β-d-gluc-4-enuronosyl groups.
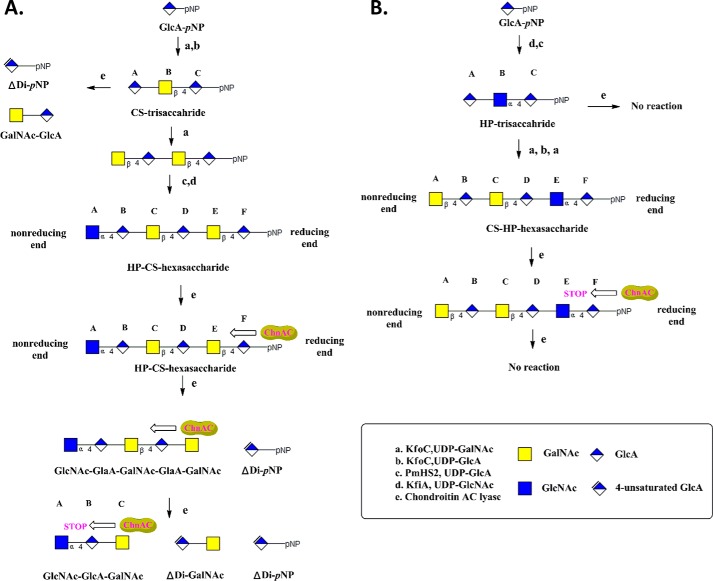
Two trisaccharides (HP- and CS-trisaccharides) and two hexasaccharides (CS-HP- and HP-CS-hexasaccharides) were synthesized as model substrates according to a similar procedure to that described in our previous work (32, 33, 35) (Fig. 1). The structures of these homogenous oligosaccharides differed from each other in terms of the arrangement of their repeating units (i.e. -GlcNAc-α-1,4-GlcA- or -GalNAc-β-1,4-GlcA-) and the fact that they contain different glycosidic bonds between their N-acetylhexosamine and GlcA residues. The HP-CS-hexasaccharide had two repeating -GalNAc-β-1,4-GlcA- units at its reducing end. In contrast, the repeating units in the CS-HP-hexasaccharide were configured with the opposite arrangement to those of the HP-CS-hexasaccharide, in that it has one α-1,4 glycosidic bond at its reducing end. All four of the oligosaccharides in this study were synthesized from the commercially available monosaccharide GlcA-pNP. Notably, the progress of each reaction was readily monitored by exploiting the UV absorbance of the pNP tag at 310 nm. The elongation of GlcA-pNP to give the HP-trisaccharide was accomplished using two bacterial glycosyltransferases: KfiA and pmHS2. The synthesis of the CS-HP-hexasaccharide was initiated from the HP-trisaccharide, and the subsequent elongation steps were accomplished using KfoC. Although the HP-trisaccharide behaved as a suitable acceptor for the β-1,4-N-acetylgalactosaminyltransferase activity of KfoC,4 the synthesis of the first β-1,4 glycosidic bond remained a significant synthetic challenge because of the low reaction rate. A carefully designed ratio of acceptor to donor was therefore employed to deliver the desired product with high purity and yield from this step. Similar procedures and approaches were also used to synthesize the CS-trisaccharide and the HP-CS-hexasaccharide. All of these oligosaccharides were purified using conventional techniques and their purities and structures were confirmed by (PAMN)-HPLC and ESI-MS analyses, respectively (Figs. 2, A, B, D, E; 3, A, B; 4, A, B).
FIGURE 1.
Scheme for the synthesis of homogenous oligosaccharides and the schematic representation of AsChnAc cleavage of these substrates. The synthesis was initiated from GlcUA-pNP using a chemoenzymatic approach. The starting monosaccharide was elongated to the desired size using glycosyltransferases according to the methods described in “Experimental Procedures.” Four oligosaccharides were synthesized in this way, and their purities were assessed by (PAMN)-HPLC (Figs. 2, A, D; 3A; and 4A). The structures of these oligosaccharides were evaluated by ESI-MS analysis (Figs. 2, B and E; 3B; and 4B). KfiA is an N-acetyl glucosaminyl transferase from E. coli strain K5. pmHS2 was obtained from P. multocida heparosan synthase. KfoC is a chondroitin polymerase from E. coli K4. A, scheme for the synthesis of the CS-trisaccharide and HP-CS-hexasaccharide, and the cleavage of these oligosaccharide substrates by AsChnAC. The degradation products resulting from the treatment of CS-trisaccharide with AsChnAC were determined by (PAMN)-HPLC and ESI-MS analyses, which revealed the presence of two new compounds (GalNAc-GlcA and ΔDi-pNP). When the β-1,4 linkages between the N-acetylhexosamine and GlcA residues (i.e. residues Cys and Asp or residues Glu and Phe) were present at the reducing end of the HP-CS-hexasaccharide, AsChnAC displayed a consecutive cleavage pattern. The products of this degradation process were determined to contain GlcNAc-GlcA-GalNAc, ΔDi-GalNAc, and ΔDi-pNP by ESI-MS analysis. The pentasaccharide GlcNAc-GlaA-GalNAc-GlaA-GalNAc was identified following the partial digestion of the HP-CS-hexasaccharide. B, scheme for the synthesis of the HP-trisaccharide and CS-HP-hexasaccharide, and the cleavage of these oligosaccharide substrates by AsChnAC. When there was one α-1,4 linkage between the N-acetylhexosamine and GlcA residues at the non-reducing end of the substrate, as in the HP-trisaccharide and the CS-HP-hexasaccharide, ChnAC did not exhibit any cleavage activity. By setting the ChnAC-uncleavable site on the reducing or non-reducing terminus, we have shown that ChnAC cleaves oligosaccharide chains from the reducing end toward the non-reducing end.
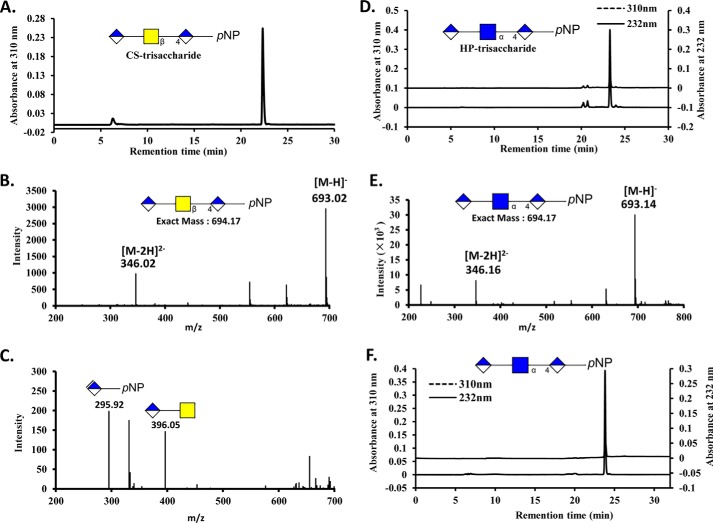
FIGURE 2.
HPLC and MS analysis of CS-trisaccharide and HP- trisaccharide treated with AsChnAC. The CS-trisaccharide and HP-trisaccharide were prepared using a chemoenzymatic process, and their purities were analyzed by (PAMN)-HPLC (panels A and D). The structures of these two trisaccharides were also analyzed by ESI-MS according to the methods described in “Experimental Procedures.” B, ESI-MS spectrum of the CS-trisaccharide. E, ESI-MS spectrum of the HP-trisaccharide. C, ESI-MS spectrum of the CS-trisaccharide was after it had been treated with AsChnAC for 16 h at 37 °C. This treatment process resulted in the formation of two newly compounds with m/z values of 396.05 and 295.92, which were attributed to the [M-H]−− ions of GalNAc-GlcA and ΔDi-pNP, respectively. F, (PAMN)-HPLC analysis of the HP- trisaccharide was after it had been treated with AsChnAC for 16 h at 37 °C. The result of this analysis revealed that there were no peaks with measurable absorption at 232 nm, suggesting that the HP-trisaccharide was a poor substrate for ChnAC.
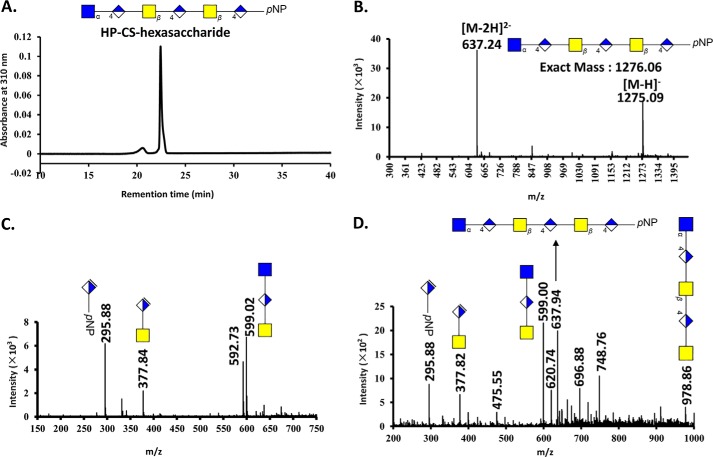
FIGURE 3.
HPLC and MS analysis of HP-CS-hexasaccharide treated with AsChnAC. The HP-CS-hexasaccharide was prepared using a chemoenzymatic process, and the purity and structural characteristics of the resulting material were confirmed by (PAMN)-HPLC (panel A) and ESI-MS (panel B) analyses, respectively. The HP-CS-hexasaccharide was incubated for 0, 1, or 10 min at 37 °C, followed by a subsequent period of heating at 100 °C for 5 min. The structures of the partially and completely ChnAC-digested HP-CS-hexasaccharide samples were analyzed by ESI-MS. C, ESI-MS spectrum of the fully digested products resulting from the ChnAC-catalyzed degradation of the HP-CS-hexasaccharide. This reaction was incubated for 10 min at 37 °C, followed by subsequent heating at 100 °C for 5 min. The ESI-MS spectrum of the fully digested material revealed the presence of three new peaks with m/z values of 599.02, 377.84, and 295.89, which were attributed the [M-H]−− values of GlcNAc-GlcA-GalNAc, ΔDi-GalNAc, and ΔDi-pNP, respectively. D, ESI-MS spectrum of the products resulting from the partial digestion of the HP-CS-hexasaccharide. The reaction was incubated for 1 min at 37 °C, followed by subsequent heating at 100 °C for 5 min. ESI-MS analysis of this mixture revealed a pentasaccharide with a molecular mass of 978.80 Da.
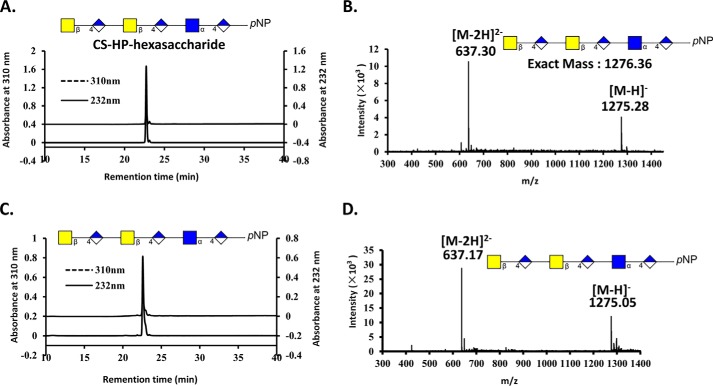
FIGURE 4.
HPLC and MS analyses of the CS-HP-hexasaccharide following its treatment with AsChnAC. The CS-HP-hexasaccharide was prepared using a chemoenzymatic method, and the purity and structural characteristics of the resulting material were confirmed by (PAMN)-HPLC (panel A) and ESI-MS (panel B) analyses, respectively. The CS-HP-hexasaccharide was incubated for 16 h at 37 °C. Analysis by (PAMN)-HPLC revealed that there were no peaks with measurable absorption at 232 nm, suggesting that the CS-HP-hexasaccharide was a poor substrate for ChnAC (panel C). D, ESI-MS spectrum of the products resulting from the complete digestion of the CS-HP-hexasaccharide. The ESI-MS spectrum contained a peak corresponding to the CS-HP-hexasaccharide with an m/z value of 1275.05 for the [M-H]−− ion. Notably, no new peaks were identified, suggesting that the CS-HP-hexasaccharide was a poor substrate for ChnAC. The presence of the GlcNAc-α-1,4-GlcA unit at the reducing terminus therefore appeared to prevent the ChnAC-mediated cleavage of the GalNAc-β-1,4-GlcA unit located in the middle or at the non-reducing end of the CS-HP-hexasaccharide.
The results of x-ray crystallography studies pertaining to GAG eliminase and its enzyme-substrate complexes have allowed for the molecular mechanisms of polysaccharide degradation to be elucidated, as well as providing information concerning the potential direction of the reaction (16, 22, 36, 37, 39). However, the lack of conclusive evidence to support the direction of this process means that the direction currently remains unclear, which is primarily due to a lack of appropriate homologous model substrates. Herein, we have prepared structurally defined oligosaccharides to determine the catalytic direction of the ChnAC directly. We decided to synthesize chimeric oligosaccharides containing repeating HS (-GlcNAc-α-1,4-GlcA-) and CS (-GalNAc-β-1,4-GlcA-) disaccharide units, and positioned the ChnAC-resistant site on the reducing or non-reducing end of the oligosaccharide, respectively. First, the degradation products resulting from the treatment of the HP- and CS-trisaccharides with purified AsChnAC were purified and analyzed by (PAMN)-HPLC and ESI-MS, respectively (Fig. 2). As expected, two new compounds (GalNAc-GlcA and ΔDi-pNP) were obtained following the depolymerization of the CS-trisaccharide (Fig. 2C). In contrast, no peaks were detected at 232 nm following the treatment of the HP-trisaccharide sample with ChnAC, which suggested that the HP-trisaccharide was a poor substrate for ChnAC (Fig. 2F). Taken together, these results once again confirmed that ChnAC can selectively recognize and cleave the β-1,4 linkages between the N-acetylhexosamine and GlcA residues, but connot act on the α-1,4 linkages (40, 41). These results also suggested that the pNP tag did not affect the substrate specificity of the ChnAC enzyme and that trisaccharides bearing a pNP tag were not too big to be degraded by ChnAC.
We then sought to determine whether the ChnAC-catalyzed cleavage occurred in a specific direction using two homologous oligosaccharides (i.e. the CS-HP- and HP-CS-hexasaccharides). It is noteworthy that these two substrates contained different linkages between the N-acetylhexosamine and GlcA residues at the ends of their chains. For example, the CS-HP- and HP-CS-hexasaccharides had α-1,4 and β-1,4 linkages at their reducing ends, respectively. The degradation products resulting from the treatment of these two oligosaccharides with ChnAC were identified by ESI-MS analysis, thereby providing an opportunity to determine whether this enzyme exhibited a preference for endolytic cleavage at the reducing or non-reducing end of these substrates. The reactions were incubated for 0, 1, and 10 min at 37 °C, followed by a subsequent period of heating at 100 °C for 5 min. The resulting mixtures of partially and completely ChnAC-digested HP-CS-hexasaccharide were analyzed by ESI-MS (Fig. 3). The complete digestion of the HP-CS-hexasaccharide substrate yielded three products. ESI-MS analysis revealed that one of these products had a molecular mass of 599.02 Da, which was very close to the molecular mass calculated for GlcNAc-GlcA-GalNAc (599.52 Da). Using a similar method, we also identified the disaccharide ΔDi-GalNAc (calculated molecular mass of 379.32 Da) and the pNP-tagged monosaccharide ΔDi-pNP (calculated molecular mass of 297.22 Da). In contrast, the treatment of the CS-HP-hexasaccharide substrate under the same conditions did not result in any degradation, even when it was incubated with the enzyme for 16 h at 37 °C (Fig. 4, C and D). This result suggested that the presence of the GlcNAc-α-1,4-GlcA unit at the reducing end of the substrate prevented it from being cleavage by ChnAC. These data therefore indicated that the saccharide chain was being degraded from the reducing end toward the non-reducing end. Interestingly, a pentasaccharide with a molecular mass of 978.86 Da, which was close to the calculated molecular mass of GlcNAc-GlaA-GalNAc-GlaA-GalNAc (979.84 Da) (Fig. 3C), was identified after the partial digestion of the HP-CS-hexasaccharide. This result therefore provided further evidence in support of our conclusion regarding the direction of the ChnAC cleavage process.
ChnAC, which belongs to the CS lyase family, can be used to achieve the eliminative degradation of HA, CS-A, and CS-C polysaccharides with the release of the corresponding Δ4-unsaturated disaccharides. Over a dozen of ChnAC enzymes have been obtained from various bacteria and their biochemical and zymological characteristics studied extensively. ArthroAC, the ChnAC lyase from A. aurescens, has been studied extensively, and the results of these studies provided a satisfactory explanation for its exolytic mode of action. Lunin et al. recently reported a high-resolution x-ray crystal structure of ArthroAC as a complex with various oligosaccharides (16). Numerous critical interactions between the enzyme and substrate were identified, including the (α/α)5 toroidal fold responsible for the lytic mechanism. This work also identified the Tyr-242 residue, which acted as a general base for the abstraction of the proton from the C5 position of the GlcA unit, as well as the Asn-183 and His-233 residues, which neutralized the charge on the glucuronic acid group. A novel ChnAC lyase from an Arthrobacter species (AsChnAC) was cloned and overexpressed in E. coli by our group,5 and the purified enzyme was employed to determine the direction of the ChnAC-catalyzed degradation of the polysaccharide in this paper. This enzyme showed the same substrate specificity and had a high sequence similarity to that of the ArthroAC lyase, thereby demonstrating that this enzyme acted as a typical chondroitin AC exolytic lyase. AsChnAC also showed 65% amino acid similarity to ArthroAC, which were relatively high compared with those of the other lyases belonging to the GAG family. The essential amino acid residues (Tyr-242, Asn-183, and His-233) required for the lytic activity of ArthroAC (16), were determined to be Tyr-275, Asn-216, and His-266, respectively, in the AsChnAC enzyme. These results therefore indicate that these two enzymes (AsChnAC and ArthroAC) share the same lytic mechanism. Taken together with the results of this study, which show that this enzyme cleaves polysaccharides from their reducing end toward their non-reducing end, these data provide important information pertaining to the exolytic mode of action of ChnAC lyases. Interestingly, it has recently been reported that the enChnABC from proteus vulgaris NCTC 4636 cleaved polysaccharides or oligosaccharides from their non-reducing end to their reducing end (38). This conclusion was based on an analysis of the oligosaccharide intermediates generated at different stages of the digestion of the enChnABC-treated CS-C. The three-dimensional structure of this enzyme (PDB code 1HN0) showed that the catalytic domain of this chondroitin ABC lyase had an (α/α)5 topology, which led to the proposal of a plausible mechanism for this enzyme based on the replacement of an arginine residue remote to the linear sequence (39). However, with the exception of the C-terminal domain, the chondroitin ABC lyase showed poor sequence similarity compared with other lyases belonging to the CS family, while the substrate-binding site also showed very little sequence conservation (16, 39).
In this study, we have designed and synthesized two homogenous hexasaccharides that differed only in terms of the stereo-configuration of the first glycosidic bond between the N-acetylhexosamine and GlcA residues at their reducing ends. These hexasaccharides were subsequently degraded in the presence of purified ChnAC and the resulting cleavage products were analyzed by (PAMN)-HPLC and ESI-MS. The results revealed that ChnAC cleaves polysaccharides from their reducing end toward their non-reducing end, and will therefore provide valuable information to enhance our collective understanding of the mode of action of ChnAC and related enzymes.
As mentioned above, ChnAC and several other CS lyases have been widely used in conjunction with modern separation and analytical methods for disaccharide analysis, polysaccharide sequencing, and biological evaluations (19). The conclusion, ChnAC cleaves substrates from their reducing end toward their non-reducing end, enhanced our collective understanding of the mode of action of ChnAC, and also would assist in the designing future strategies for directed evolution of this enzyme in order to create mutants toward user-defined goals. Additionally, an advanced chemoenzymatic method was recently developed for the efficient synthesis of homologous GAG oligosaccharides with high purity in recent years (2, 26–28). For example, a series of synthetic structurally defined heparin oligosaccharides were prepared by cost-effective chemoenzymatic methods with high efficiency (27, 34). By coupling with these synthetic systems, the specific-direction of the ChnAC-catalyzed degradation make it is possible to create desired structure of the final products through the strategy of targeted introduction and precise excision of specific glucosidic bonds.
Author Contributions
J. S. and F. W. designed and coordinated the work. F. Y. carried out the experiments. J. S. and F. Y. wrote the manuscript. All of the authors have read and approved the final manuscript.
Acknowledgments
We thank Dr. Cao Hongzhi and Dr. Liu Chunhui gratefully for their many helpful discussions and suggestions.
This work was supported in part by the National Natural Science Foundation of China (Project no. 31300673), the Specialized Research Fund for the Doctoral Program of Higher Education (Project no. 20130131120046), the Projects of Science and Technology Dept. of Shandong Province (Project no. 2015GSF121002 and 2015ZDJS04002), and the Fundamental Research Funds (Project no. 2014YQ002) of Shandong University. A patent (CN201510289566) has been published relating to this article. Despite this publication, the authors have strictly adhered to all the policies provide by the The Journal of Biological Chemistry regarding the sharing of data and materials.
J. J. Xue, L. Jin, X. K. Zhang, F. S. Wang, P. X. Ling, and J. Z. Sheng, manuscript in preparation.
F. X. Yin, F. S. Wang, and J. Z. Sheng, unpublished observations.
- GAG
- glycosaminoglycan
- ChnAC
- chondroitin AC lyase
- CS
- chondroitin sulfate
- HA
- hyaluronan
- AsChnAC
- Arthrobacter ChnAC
- PAMN-HPLC
- polyamine-based anion exchange
- ESI-MS
- electrospray ionization mass spectrometry
- HS
- heparan sulfate.
References
- 1. Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Bertozzi C. R., Hart G. W., and Etzler M. E. (eds.) (2009) Essentials of Glycobiology, 2nd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- 2. DeAngelis P. L., Liu J., and Linhardt R. J. (2013) Chemoenzymatic synthesis of glycosaminoglycans: re-creating, re-modeling and re-designing nature's longest or most complex carbohydrate chains. Glycobiology 23, 764–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Taylor K. R., and Gallo R. L. (2006) Glycosaminoglycans and their proteoglycans: host-associated molecular patterns for initiation and modulation of inflammation. FASEB J. 20, 9–22 [DOI] [PubMed] [Google Scholar]
- 4. Xu D., and Esko J. D. (2014) Demystifying Heparan Sulfate–Protein Interactions. Annu. Rev. Biochem. 83, 129–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gandhi N. S., and Mancera R. L. (2008) The Structure of Glycosaminoglycans and their Interactions with Proteins. Chem. Biol. Drug Des. 72, 455–482 [DOI] [PubMed] [Google Scholar]
- 6. Pantazaka E., and Papadimitriou E. (2014) Chondroitin sulfate-cell membrane effectors as regulators of growth factor-mediated vascular and cancer cell migration. Biochim. Biophys. Acta 1840, 2643–2650 [DOI] [PubMed] [Google Scholar]
- 7. Li P., Sheng J., Liu Y., Li J., Liu J., and Wang F. (2013) Heparosan-derived heparan sulfate/heparin-like compounds: one kind of potential therapeutic agents. Med. Res. Rev. 33, 665–692 [DOI] [PubMed] [Google Scholar]
- 8. Li L., Li Y., Ijaz M., Shahbaz M., Lian Q., and Wang F. (2015) Review on complement analysis method and the roles of glycosaminoglycans in the complement system. Carbohydr. Polym. 134, 590–597 [DOI] [PubMed] [Google Scholar]
- 9. Migliore A., and Procopio S. (2015) Effectiveness and utility of hyaluronic acid in osteoarthritis. Clin. Cases Miner. Bone Metab. Off. J. Ital. Soc. Osteoporos. Miner. Metab. Skelet. Dis. 12, 31–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu J., and Pedersen L. C. (2007) Anticoagulant heparan sulfate: structural specificity and biosynthesis. Appl. Microbiol. Biotechnol. 74, 263–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang Q., Li J., Liu C., Song C., Li P., Yin F., Xiao Y., Li J., Jiang W., Zong A., Zhang X., and Wang F. (2015) Protective effects of low molecular weight chondroitin sulfate on amyloid beta (Aβ)-induced damage in vitro and in vivo. Neuroscience 305, 169–182 [DOI] [PubMed] [Google Scholar]
- 12. Bisio A., Vecchietti D., Citterio L., Guerrini M., Raman R., Bertini S., Eisele G., Naggi A., Sasisekharan R., and Torri G. (2009) Structural features of low-molecular-weight heparins affecting their affinity to antithrombin. Thromb. Haemost. 102, 865–873 [DOI] [PubMed] [Google Scholar]
- 13. Merli G. J., and Groce J. B. (2010) Pharmacological and Clinical Differences Between Low-Molecular-Weight Heparins. Pharm. Ther. 35, 95–105 [PMC free article] [PubMed] [Google Scholar]
- 14. Pempe E. H., Xu Y., Gopalakrishnan S., Liu J., and Harris E. N. (2012) Probing structural selectivity of synthetic heparin binding to Stabilin protein receptors. J. Biol. Chem. 287, 20774–20783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ernst S., Langer R., Cooney C. L., and Sasisekharan R. (1995) Enzymatic Degradation of GlycosaminogIycans. Crit. Rev. Biochem. Mol. Biol. 30, 387–444 [DOI] [PubMed] [Google Scholar]
- 16. Lunin V. V., Li Y., Linhardt R. J., Miyazono H., Kyogashima M., Kaneko T., Bell A. W., and Cygler M. (2004) High-resolution Crystal Structure of Arthrobacter aurescens Chondroitin AC Lyase: An Enzyme–Substrate Complex Defines the Catalytic Mechanism. J. Mol. Biol. 337, 367–386 [DOI] [PubMed] [Google Scholar]
- 17. Lombard V., Bernard T., Rancurel C., Brumer H., Coutinho P. M., and Henrissat B. (2010) A hierarchical classification of polysaccharide lyases for glycogenomics. Biochem. J. 432, 437–444 [DOI] [PubMed] [Google Scholar]
- 18. Garron M.-L., and Cygler M. (2010) Structural and mechanistic classification of uronic acid-containing polysaccharide lyases. Glycobiology 20, 1547–1573 [DOI] [PubMed] [Google Scholar]
- 19. Linhardt R. J., Avci F. Y., Toida T., Kim Y. S., and Cygler M. (2006) CS Lyases: Structure, Activity, and Applications in Analysis and the Treatment of Diseases (Pharmacology B.-A. in ed), pp. 187–215, Chondroitin Sulfate: Structure, Role and Pharmacological Activity, Academic Press, 53, 187–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kale V., Friðjónsson Ó., Jónsson J. Ó., Kristinsson H. G., Ómarsdóttir S., and Hreggviðsson G. Ó. (2015) Chondroitin Lyase from a Marine Arthrobacter sp. MAT3885 for the Production of Chondroitin Sulfate Disaccharides. Mar. Biotechnol. N. Y. N. 17, 479–492 [DOI] [PubMed] [Google Scholar]
- 21. Hernáiz M. J., and Linhardt R. J. (2001) Degradation of chondroitin sulfate and dermatan sulfate with chondroitin lyases. Methods Mol. Biol. 171, 363–371 [DOI] [PubMed] [Google Scholar]
- 22. Huang W., Boju L., Tkalec L., Su H., Yang H. O., Gunay N. S., Linhardt R. J., Kim Y. S., Matte A., and Cygler M. (2001) Active site of chondroitin AC lyase revealed by the structure of enzyme-oligosaccharide complexes and mutagenesis. Biochemistry 40, 2359–2372 [DOI] [PubMed] [Google Scholar]
- 23. Ke T., Zhangfu L., Qing G., Yong T., Hong J., Hongyan R., Kun L., and Shigui L. (2005) Isolation of Serratia marcescens as a chondroitinase-producing bacterium and purification of a novel chondroitinase AC. Biotechnol. Lett. 27, 489–493 [DOI] [PubMed] [Google Scholar]
- 24. Shim K.-W., and Kim D.-H. (2008) Cloning and expression of chondroitinase AC from Bacteroides stercoris HJ-15. Protein Expr. Purif. 58, 222–228 [DOI] [PubMed] [Google Scholar]
- 25. Capila I., Wu Y., Rethwisch D. W., Matte A., Cygler M., and Linhardt R. J. (2002) Role of arginine 292 in the catalytic activity of chondroitin AC lyase from Flavobacterium heparinum. Biochim. Biophys. Acta. 1597, 260–270 [DOI] [PubMed] [Google Scholar]
- 26. Liu J., and Linhardt R. J. (2014) Chemoenzymatic synthesis of heparan sulfate and heparin. Nat. Prod. Rep. 31, 1676–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xu Y., Masuko S., Takieddin M., Xu H., Liu R., Jing J., Mousa S. A., Linhardt R. J., and Liu J. (2011) Chemoenzymatic Synthesis of Homogeneous Ultralow Molecular Weight Heparins. Science 334, 498–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen Y., Li Y., Yu H., Sugiarto G., Thon V., Hwang J., Ding L., Hie L., and Chen X. (2013) Tailored Design and Synthesis of Heparan Sulfate Oligosaccharide Analogues Using Sequential One-Pot Multienzyme Systems. Angew. Chem. Int. Ed. 52, 11852–11856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Y., Yu H., Thon V., Chen Y., Muthana M. M., Qu J., Hie L., and Chen X. (2014) Donor substrate promiscuity of the N-acetylglucosaminyltransferase activities of Pasteurella multocida heparosan synthase 2 (PmHS2) and Escherichia coli K5 KfiA. Appl. Microbiol. Biotechnol. 98, 1127–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peterson S., and Liu J. (2012) Deciphering Mode of Action of Heparanase Using Structurally Defined Oligosaccharides. J. Biol. Chem. 287, 34836–34843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. O'Leary T. R., Xu Y., and Liu J. (2013) Investigation of the substrate specificity of K5 lyase A from K5A bacteriophage. Glycobiology 23, 132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sheng J., Liu R., Xu Y., and Liu J. (2011) The Dominating Role of N-Deacetylase/N-Sulfotransferase 1 in Forming Domain Structures in Heparan Sulfate. J. Biol. Chem. 286, 19768–19776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sheng J., Xu Y., Dulaney S. B., Huang X., and Liu J. (2012) Uncovering biphasic catalytic mode of C5-epimerase in heparan sulfate biosynthesis. J. Biol. Chem. 287, 20996–21002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xu Y., Cai C., Chandarajoti K., Hsieh P.-H., Li L., Pham T. Q., Sparkenbaugh E. M., Sheng J., Key N. S., Pawlinski R., Harris E. N., Linhardt R. J., and Liu J. (2014) Homogeneous low-molecular-weight heparins with reversible anticoagulant activity. Nat. Chem. Biol. 10, 248–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu C., Sheng J., Krahn J. M., Perera L., Xu Y., Hsieh P.-H., Dou W., Liu J., and Pedersen L. C. (2014) Molecular mechanism of substrate specificity for heparan sulfate 2-O-sulfotransferase. J. Biol. Chem. 289, 13407–13418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kawaguchi Y., Sugiura N., Kimata K., Kimura M., and Kakuta Y. (2013) The crystal structure of novel chondroitin lyase ODV-E66, a baculovirus envelope protein. FEBS Lett. 587, 3943–3948 [PubMed] [Google Scholar]
- 37. Li S., and Jedrzejas M. J. (2001) Hyaluronan Binding and Degradation by Streptococcus agalactiae Hyaluronate Lyase. J. Biol. Chem. 276, 41407–41416 [DOI] [PubMed] [Google Scholar]
- 38. Hamai A., Hashimoto N., Mochizuki H., Kato F., Makiguchi Y., Horie K., and Suzuki S. (1997) Two Distinct Chondroitin Sulfate ABC Lyases An Endoeliminase Yielding Tetrasaccharides And An Exoeliminase Preferentially Acting On Oligosaccharides. J. Biol. Chem. 272, 9123–9130 [DOI] [PubMed] [Google Scholar]
- 39. Huang W., Lunin V. V., Li Y., Suzuki S., Sugiura N., Miyazono H., and Cygler M. (2003) Crystal structure of Proteus vulgaris chondroitin sulfate ABC lyase I at 1.9 Å resolution. J. Mol. Biol. 328, 623–634 [DOI] [PubMed] [Google Scholar]
- 40. Linhardt R. J., Galliher P. M., and Cooney C. L. (1986) Polysaccharide lyases. Appl. Biochem. Biotechnol. 12, 135–176 [DOI] [PubMed] [Google Scholar]
- 41. Sutherland I. W. (1995) Polysaccharide lyases. FEMS Microbiol. Rev. 16, 323–347 [DOI] [PubMed] [Google Scholar]