Abstract
Recognition and repair of double-stranded DNA breaks (DSB) involves the targeted recruitment of BRCA tumor suppressors to damage foci through binding of both ubiquitin (Ub) and the Ub-like modifier SUMO. RAP80 is a component of the BRCA1 A complex, and plays a key role in the recruitment process through the binding of Lys63-linked poly-Ub chains by tandem Ub interacting motifs (UIM). RAP80 also contains a SUMO interacting motif (SIM) just upstream of the tandem UIMs that has been shown to specifically bind the SUMO-2 isoform. The RAP80 tandem UIMs and SIM function collectively for optimal recruitment of BRCA1 to DSBs, although the molecular basis of this process is not well understood. Using NMR spectroscopy, we demonstrate that the RAP80 SIM binds SUMO-2, and that both specificity and affinity are enhanced through phosphorylation of the canonical CK2 site within the SIM. The affinity increase results from an enhancement of electrostatic interactions between the phosphoserines of RAP80 and the SIM recognition module within SUMO-2. The NMR structure of the SUMO-2·phospho-RAP80 complex reveals that the molecular basis for SUMO-2 specificity is due to isoform-specific sequence differences in electrostatic SIM recognition modules.
Keywords: DNA damage response, nuclear magnetic resonance (NMR), protein phosphorylation, protein-protein interaction, sumoylation
Introduction
The DNA repair process in eukaryotic cells is an indispensible life process responsible for maintaining the fidelity of the genome (1, 2). The genomic information encoded within the molecular structure of DNA is relentlessly compromised as a result of factors that include free radicals arising from metabolic processes, radiation, and replication errors (1). By virtue of highly regulated and efficient DNA repair mechanisms, most DNA damage does not progress to viable malignant tumors (1). Among the different kinds of damage that alter DNA structure, double strand breaks are the most deleterious (1). Depending on the nature of DNA damage, checkpoint activation and cell cycle arrest accompany a number of repair pathways, including homologous recombination, non-homologous end joining, or alternative non-homologous end joining repair, which function to combat the damage (3). Similar to many life processes, homologous recombination is governed by the hierarchical and synergistic action of various post-translational modifications, such as phosphorylation, ubiquitination, and SUMOylation (4, 5). The severed ends of damaged DNA are sensed by the MRE11/Rad50/NBS1 (MRN) protein complex, followed by recruitment of ATM kinase and its concomitant activation (3, 5, 6). This results in the phosphorylation of nearby histones, which serves as a marker for initiation of repair (3, 5). Phosphorylated histones comprise the binding site for MDC1, which is also phosphorylated by ATM kinase; subsequently, phosphorylated MDC1 recruits the ubiquitination enzyme RNF8, which acts with the Ubc13/Mms2 heterodimer to attach Lys63-linked Ub chains at damaged sites, in combination with the Ub2 ligase RNF168 (3, 7, 8). One of the biological functions for Lys63-linked poly-Ub chains is to serve as a signal for BRCA1 recruitment, a key protein that is obligatory for repair of DNA damage, and cell cycle checkpoint activation. RAP80, an 80-kDa nuclear protein, is responsible for recruitment of the BRCA1 A complex (BRCA1, BARD1, BRCC36, Abraxas, and RAP80) to sites of DNA damage by binding Lys63-linked Ub chains through tandem α-helical UIMs (9, 10). In addition to ubiquitination, SUMOylation of different DNA repair proteins by PIAS4, a SUMO-specific E3 ligase, is involved in BRCA1 recruitment (11, 12). The well established role for Lys63-linked poly-Ub recognition by RAP80 in BRCA1 recruitment was recently modified by the finding that RAP80 possesses a SIM, N-terminal to the tandem UIMs, which is partly responsible for BRCA1 A complex recruitment to DNA damage sites (13, 14). Optimal recruitment of BRCA1 to damage sites depends on the combined action of the SIM and UIM of RAP80; this implies that there are two possibilities for SUMO and poly-Ub binding: independent recognition of the individual modifier proteins, or recognition of SUMO-Ub hybrid chains (13, 14). Hybrid chain recognition is appealing in comparison to independent modifier binding, as a result of an ∼80-fold higher affinity for RAP80 as compared with SUMO and Ub binding alone (14). In addition, the requirement of RNF4, a SUMO-binding Ub ligase, for BRCA1 recruitment by RAP80, suggests that hybrid chains are the preferred candidates for RAP80 binding (14).
There are four SUMO isoforms in mammalian cells: SUMO-1, -2, -3, and 4. SUMO-1 shows 45% sequence identity to SUMO-2 and SUMO-3, whereas SUMO-2 and SUMO-3 share 95% sequence identity, and can form poly-SUMO chains. The function of SUMO-4 is currently unknown. Although their sequences and chain forming capabilities vary, all SUMO isoforms assume a Ub-like fold (15). From a structural perspective, the binding of SUMO to its cognate partners typically involves electrostatic and hydrophobic interactions, unlike Ub interactions, which typically involve a hydrophobic patch centered on Ile44 (16–18). The SIM is the most extensively studied SUMO binding motif, with a hydrophobic module (V/I)X(V/I)(V/I), bordered by N- and C-terminal acidic modules (18). The three isoforms of SUMO bind SIMs within a hydrophobic groove between the α1 helix and the β2 strand, typically forming an intermolecular β-sheet at the interface. The orientation of the β strand has been observed in both parallel and antiparallel conformations depending on the specific SIM sequence and SUMO isoform (19). This is believed to result from the distribution of negatively charged residues adjacent to the hydrophobic SUMO-interacting module from the SIM. This region also possesses serine residues that are typically phosphorylation sites, and play a role in determining SUMO isoform preference (17, 18). Phosphorylation of the SIM serine residues provides enhanced electrostatic interactions, which generally result in a substantial increase in the affinity of the SUMO-SIM interaction (20, 21). A number of structures for phosphorylated SIMs bound to SUMO-1 have been reported (19, 21–25). However, the molecular basis for the interaction between SUMO-2 and its cognate phosphorylated SIM is unknown. The DNA repair protein RAP80 has been shown to preferentially interact with SUMO-2 (14), and possesses a canonical CK2 phosphorylation site within its SIM. In this study, the structure of the N-terminal UIM and SIM domains from RAP80, as well as the molecular basis for binding of SUMO-2 to the SIM were investigated using NMR spectroscopy. We also determined the first structure of SUMO-2 bound to a phosphorylated SIM, which in conjunction with measurement of the thermodynamics and kinetics of SUMO-2 binding for the phosphorylated and non-phosphorylated states of RAP80, provide insight into the molecular determinants underlying the SUMO-2 specificity of this critical DNA repair protein.
Experimental Procedures
Cloning, Protein Expression, and Purification of RAP80
RAP80 is a 719-residue, multidomain protein consisting of N-terminal nuclear localization signals (∼residues 3–35), two N-terminal tandem UIMs (∼residues 80–120), an N-terminal SIM (∼residues 35–50), and in the C-terminal half, an Abraxas interacting region and two putative zinc fingers (26). The central Abraxas interacting region binds phosphorylated Abraxas within the BRCA1 complex (9, 10, 27). This function, combined with the independent SUMO and Ub binding properties of the N-terminal region (∼residues 30- 120) (13, 14, 28, 29), facilitates DNA damage recognition and repair by the BRCA1 complex. To study the SUMO binding properties of human RAP80, residues 33–131 were cloned into the EcoRI and BamHI sites of pHis-P1, and the insert sequence was verified by sequencing. Expression of the His6-tagged fusion construct results in an N-terminal GAMDP cloning artifact following cleavage with tobacco etch virus protease. For expression of unlabeled proteins, 100 μl of electrocompetent Escherichia coli strain BL21(DE3)-RIPL cells were transformed with 300 ng of plasmid, and allowed to grow overnight on agar plates containing ampicillin and chloramphenicol at 37 °C. A single colony was picked and used to inoculate 50 ml of LB starter culture, which was incubated at 37 °C overnight. LB containing ampicillin and chloramphenicol (500 ml) was inoculated with 5 ml of starter culture and incubated at 37 °C with shaking at 250 rpm. Upon optimal growth to A600 ∼0.6–0.8, cells were induced with 0.4 mm isopropyl 1-thio-β-d-galactopyranoside. Post-induction, cells were grown overnight at 25 °C and subsequently harvested. Cells were suspended in 100 ml of lysis buffer containing 20 mm imidazole, 500 mm NaCl, 20 mm sodium phosphate, 2 mm DTT, 10 mm MgSO4, 5 μg/ml of DNase I, and 0.5% protease inhibitor mixture II (Calbiochem catalogue number 538132), pH 7.3, and subjected to sonication. Following cell rupture, lysate was centrifuged at 25,000 rpm in a Beckman JA-25.5 rotor for 30 min at 4 °C. The supernatant was filtered using a Millipore steriflip 0.45-μm vacuum filtration unit. The filtrate was affinity-purified using a His-prep FF 16/10 column equilibrated with buffer containing 20 mm imidazole, 500 mm NaCl, 20 mm sodium phosphate, and 2 mm DTT, pH 7.3. Bound protein was eluted using a gradient of increasing imidazole concentration ranging from 20 to 500 mm. Fractions containing protein, as detected by UV absorbance, were pooled and the His6 tag was cleaved by addition of 100 μl of 210 μm tobacco etch virus protease with incubation at 4 °C overnight. The cleaved affinity tag was removed by passing the sample over a His-prep FF 16/10 column; unbound protein was collected and exchanged using a dialysis membrane with a 3.5-kDa cutoff, into buffer containing 50 mm Tris, 150 mm NaCl, and 2 mm DTT, pH 7.3. Final purification was carried out by size exclusion with a HiLoad 26/60 Superdex 75 column equilibrated with 50 mm Tris, 150 mm NaCl, and 2 mm DTT, pH 7.3. Fractions containing protein were pooled and concentrated using an Amicon Ultra 15 centrifugal membrane filtration device with a cutoff of 3 kDa. For expression of [U-15N]- and [U-13C,15N]-labeled protein for NMR studies, cells were grown to A600 ∼0.6–0.8 in 2 liters of LB media, and pelleted by centrifugation using a Beckman JA-10.5 rotor for 30 min at 5,000 rpm. Cells were washed in M9 medium, and suspended in 250 ml of M9 media containing 15N-labeled ammonium sulfate as the sole nitrogen source or both 15N-labeled ammonium sulfate and 13C-labeled glucose as the sole nitrogen and carbon sources, respectively. Cells were acclimatized to the change in media conditions for ∼2 h through incubation at 25 °C with shaking at 250 rpm. Protein expression was induced using 0.4 mm isopropyl 1-thio-β-d-galactopyranoside. Following induction, the incubation temperature was reduced to 18 °C for overnight growth. Purification was achieved as described for unlabeled proteins.
Protein Expression and Purification of SUMO-2
pET28a plasmid harboring residues 1–93 of human SUMO-2 was a gift from Dr. Lawrence McIntosh, University of British Columbia. Expressed SUMO-2 contained an N-terminal His6 tag, which results in a GSH cloning artifact following cleavage by thrombin. Protein expression and purification strategies for unlabeled, [U-15N]- and [U-13C,15N]-labeled SUMO-2 were similar to those for RAP80-(33–131), except where noted. Thrombin cleavage of the SUMO-2 His6 tag was carried out using a Thrombin CleanCleave kit (Sigma). Cleaved SUMO-2 was dialyzed against cleavage buffer containing 50 mm Tris-HCl and 2.5 mm CaCl2, pH 7.9, followed by incubation with thrombin-immobilized agarose beads at room temperature for 48 h. Post-cleavage, thrombin beads were removed by centrifugation and the protein was dialyzed in nickel column binding buffer (20 mm imidazole, 20 mm sodium phosphate, 500 mm NaCl, 2 mm DTT, pH 7.3). Purification by size exclusion chromatography was similar to that for RAP80-(33–131). For all protein samples, purity and molecular weight were confirmed using SDS-PAGE and MALDI-TOF mass spectrometry.
RAP80 SIM Peptide Synthesis and Phosphorylation
Peptide containing residues 35–50 (RLEDAFIVISDSDGEE) from RAP80, with an acetylated N terminus and amidated C terminus, was chemically synthesized (Biomatik, 99% pure). Doubly phosphorylated peptide was synthesized through incubation of 0.18 mm peptide with 10000 units of CK2 (New England Biolabs), in reaction buffer containing 0.63 mm ATP, 50 mm Tris, 10 mm MgCl2 0.1 mm EDTA, 2 mm DTT, 0.01% Brij35, pH 7.5. The reaction was carried out at 30 °C for 4 h. Complete phosphorylation at residues Ser44 and Ser46 was confirmed by MALDI-TOF mass spectrometry. For NMR-monitored binding studies and structure determination of SUMO-2 with phosphorylated RAP80 peptide complex, phosphopeptide containing RAP80 residues 37–49 (EDAFIVIpSDpSDGE) was chemically synthesized (Biomatik, 95.96% pure).
NMR Chemical Shift Assignment
All NMR experiments were carried out on a Varian Unity INOVA 600-MHz spectrometer, except where noted. All samples were prepared in SHIGEMI microcell NMR tubes. Main chain resonance assignments for RAP80-(33–131) were accomplished at 25 °C using a [U-13C,15N] NMR sample in 50 mm Tris, 150 mm NaCl, 45 mm Tris(2-carboxyethyl)phosphine, pH 6.1, with a protein concentration of 0.3 mm, 0.1 mm DSS as an internal reference, and 10% D2O, and the three-dimensional HNCACB (30, 31), CBCA(CO)NNH (31, 32), HNN(CA,CO) (33, 34), HNCO (35), and HN(CA)CO (36) experiments. Backbone 1HN, 15N, 13Cα, 13CO, and 13Cβ chemical shifts were assigned manually using the SPARKY program (37) and verified using the automatic assignment feature of CARA (38). Main chain resonance assignments for SUMO-2 were obtained in a similar fashion using a [U-13C/15N] NMR sample containing 0.4 mm SUMO-2 in 50 mm Tris, 150 mm NaCl, 2 mm DTT, 10% D2O, and 0.1 mm DSS as an internal standard at pH 7.3. Chemical shifts were verified against those deposited in the BMRB (accession number 6801).
Main chain 1HN, 1Hα, and side chain proton chemical shifts for chemically synthesized RAP80-(35–50) were assigned using two-dimensional TOCSY and NOESY experiments (39) at 5 and 25 °C, for a sample containing 0.7 mm peptide in 50 mm Tris, 150 mm NaCl, 2 mm DTT, and 0.1 mm DSS, and 10% D2O, pH 7.3. Chemical shifts for synthesized, doubly phosphorylated pRAP80-(37–49) were assigned similarly using a sample containing 0.5 mm peptide in 50 mm Tris, 150 mm NaCl, 2 mm DTT, 0.1 mm DSS, and 10% D2O, pH 7.3.
Chemical Shift Perturbation Mapping for [U-15N]-RAP80-(33–131) upon SUMO-2 Addition
We determined RAP80-(33–131) main chain 1HN and 15N chemical shift changes upon interaction with SUMO-2, by recording two-dimensional 1H-15N HSQC NMR spectra for 0.3 mm [U-15N]-RAP80-(33–131) in 50 mm Tris, 150 mm NaCl, 2 mm DTT, and 10% D2O at pH 6.1 until there were no significant changes in RAP80 resonances upon SUMO-2 addition (>10-fold excess of SUMO-2).
NMR Monitored Titrations for [U-15N]SUMO-2 with RAP80-(33–131), RAP80-(35–49), and pRAP80-(37–49)
Generally, changes in main chain amide chemical shifts for SUMO-2 upon addition of various RAP80 peptides were followed at different titration points using two-dimensional 1H-15N HSQC NMR spectra (40, 41). Titration of 225 μm [U-15N]-SUMO-2 in 50 mm Tris, 150 mm NaCl, 10 mm DTT, and 10% D2O, pH 7.3, was carried out by adding increasing amounts of stock solution containing unlabeled RAP80-(33–131) in the same buffer. During titration, the increase in RAP80-(33–131) is accompanied by dilution of SUMO-2 to concentrations of 225, 211, 198, 184, 171, 157, 144, 117, 91, 64, and 11 μm, with RAP80-(33–131) concentrations of 0, 35, 70, 104, 139, 174, 208, 278, 347, 416, and 555 μm. For these titration points, the RAP80-(33–131)/SUMO-2 concentration ratios were 0, 0.16, 0.35, 0.56, 0.81, 1.12, 1.44, 2.36, 3.82, 6.49, and 51.98. For this, and all subsequent titrations, the combined 15N and 1HN chemical shift change for each residue was calculated according to Δδ = [(Δδ15N/5)2 + (Δδ1HN)2]1/2, where Δδ15N and Δδ1HN are the respective 15N and 1HN chemical shift changes in ppm. These combined chemical shift changes were numerically fit to a 1:1 binding isotherm to determine the dissociation constant (KD), as previously described (41). For all titrations, protein concentrations were measured using the BCA assay and verified with amino acid analyses. For NMR-monitored titrations of SUMO-2 with RAP80-(35–50), a stock solution of unlabeled RAP80-(35–50) was titrated into a 380 μm [U-15N]-SUMO-2 sample in 50 mm Tris, 150 mm NaCl, 2 mm DTT, and 10% D2O, pH 7.3. The concentrations of SUMO-2 and RAP80-(35–50) at the various titration points were 376, 373, 368, 359, 342, 324, 303, 288, 253, 217, 181, 145, 108, 72, and 36 μm, and 0, 5, 15, 31, 64, 98, 136, 163, 229, 296, 364, 431, 498, 565, and 633 μm, respectively, giving concentration ratios for RAP80-(35–50)/SUMO-2 of 0, 0.013, 0.040, 0.08, 0.18, 0.30, 0.44, 0.56, 0.90, 1.36, 2.01, 2.97, 4.6, 7.84, and 17.5.
Titration of SUMO-2 with synthetic pRAP80-(37–49) peptide was carried out by adding increasing concentrations of peptide to a 190 μm [U-15N]-SUMO-2 sample in 50 mm Tris, 150 mm NaCl, 2 mm DTT, and 10% D2O, pH 7.3. Concentrations of SUMO-2 and pRAP80-(37–49) for the different titration points were 190, 188, 185, 179, 172, 159, 147, 136, 124, 113, 102, 91, 80, 69, 68, 61, 51, and 46 μm, and 0, 1, 4, 7, 12, 22, 30, 38, 46, 54, 62, 70, 78, 86, 88, 100, 120, and 135 μm, respectively, with corresponding concentration ratios for pRAP80-(37–49)/SUMO-2 of 0, 0.006, 0.02, 0.04, 0.07, 0.14, 0.20, 0.28, 0.37, 0.47, 0.60, 0.77, 0.97, 1.24, 1.29, 1.63, 2.35, and 2.93.
Chemical shift differences were calculated for the first and last points from the titrations of SUMO-2 with various RAP80 peptides, and mapped onto the structure of free SUMO-2 (PDB code 1WM2). For these chemical shift maps, common missing residues included N- and C-terminal residues and prolines: Met1, Ala2, Asp3, Asn15, Asp16, Gln90-Gly93, Pro6, Pro39, Pro66, Pro73, and broadened residues: Lys33, Lys35. For the titration with RAP80-(33–131), missing residues in the 1H-15N NMR spectra due to low SUMO-2 concentration include Glu4, Gln25, Arg36, Arg50, Met55, and Glu81.
NMR Structure Determination for RAP80-(33–131) and the SUMO-2·pRAP80-(37–49) Complex
The secondary structure for RAP80-(33–131) was assigned based on protein backbone (φ, ψ) and side chain (χ1) torsion angles, calculated by the TALOS-N program (42), using backbone 1HN, 1Hα, 13CO, 13Cα, and side chain 13Cβ chemical shifts.
To determine a structure for pRAP80-(37–49) in complex with SUMO-2, a sample containing 0.4 mm [U-13C/15N]-SUMO-2 and 1.2 mm pRAP80-(37–49) was prepared in buffer containing 25 mm Tris, 2 mm DTT, and 10% D2O, pH 7.3. For assignment of bound SUMO-2 backbone 1HN, 15N, 13Cα, 13CO, and side chain 13Cβ chemical shifts, three-dimensional HNCACB, CBCA(CO)NNH, HNCO, HN(CA)CO, and two-dimensional 1H-15N HSQC NMR experiments were collected. Side chain carbon and proton chemical shifts were obtained using the three-dimensional (H)CCTOCSY(CO)NNH and H(CC)TOCSY(CO)NNH NMR experiments (43–46). Chemical shift assignment was accomplished manually using the SPARKY NMR software, and automatically using CARA. The six aromatic side chains of SUMO-2 His17, Phe32, His37, Tyr47, Phe60, Phe62, and Phe87 were assigned using an in-house modified two-dimensional 1H-13C aromatic TROSY HSQC (47), an in-house two-dimensional 1H-13C aromatic NOESY-TROSY-HSQC pulse sequence, as well as the two-dimensional (HB)CB(CGCD)HD and (HB)CB(CGCDCE)HE aromatic side chain correlation experiments (48). To assign pRAP80-(37–49) side chain and backbone proton chemical shifts for peptide bound to SUMO-2, two-dimensional TOCSY and NOESY experiments (49) with suppression of SUMO-2 signals from protons bound to 13C and 15N were carried out on a Unity INOVA 800 MHz NMR spectrometer using the same sample. To determine interacting residues between [U-13C/15N]-SUMO-2 and unlabeled pRAP80-(37–49), two-dimensional 13C/15N F1-filtered, F3-edited NOESY experiments for aliphatic and aromatic protons were carried out (49). Intermolecular NOEs were assigned using side chain proton assignments for bound SUMO-2, and side chain proton assignments for bound pRAP80-(37–49). A total of 15 intermolecular NOEs between SUMO-2 and pRAP80-(37–49) were assigned, as well as 28 intramolecular pRAP80-(37–49) NOEs. Restraints for NOEs were determined on the basis of peak intensities, and assigned to distance ranges using the auxiliary programs from the Amber 15 biomolecular simulation suite of programs (50). Main chain torsion angle restraints for bound SUMO-2 were determined with the TALOS-N program using main chain 1HN, 1Hα, 13CO, 13Cα, and side chain 13Cβ chemical shifts. Torsion angles for bound peptide were determined from 1HN and 1Hα chemical shifts using the PREDITOR program (51). Torsion angle restraints were used in structure calculations for residues with a PREDITOR confidence score higher than 0.7.
For structure calculations, the starting model for SUMO-2 was derived from the high-resolution crystal structure (PDB code 1WM2) (52). N terminally acetylated and C terminally amidated pRAP80-(37–49) peptide was manually docked to strand β2 from SUMO-2, in the parallel β-strand conformation, to generate a starting model for the complex. The choice for this initial model was based on the following experimental observations, which suggested a parallel β-strand conformation for peptide: intermolecular NOEs between SUMO-2 and RAP80 residues Ile41/Ile43, combined with a lack of intermolecular NOEs between Phe40 and Val42 from RAP80 and SUMO-2, significant main chain amide chemical shift perturbations for a network of positively charged residues including His17, His37, and Lys42 for SUMO-2, and corresponding large 1Hα and 1HN chemical shift changes for Ser(P)44, Asp45, and Ser(P)46 for SIM peptide, upon SUMO-SIM interaction. This initial model was solvated in a truncated octahedral TIP3P water box, with a distance of 24 Å between protein atoms from images in adjacent unit cells. The starting model was energy minimized using the ff14SB force field and the sander program within the Amber 15 suite of biomolecular simulation programs, with pairwise long range electrostatics and van der Waals interactions cut off at 8 Å. In addition, default parameters for phosphoserine bearing a −2 charge (S2P residue) were employed (53). The simulation system was heated for 50 ps to a temperature of 298 K, solute atoms were subjected to 2 kcal/mol restraints, and allowed to equilibrate to 1 atmosphere pressure for a further 50 ps. The system was then subjected to production dynamics for 40 ps with the inclusion of NMR-derived distance and dihedral restraints. Structural statistics for 20 snapshots from the last 40 ps of the simulations with NMR restraints are given in Table 1.
TABLE 1.
Structural statistics for 20 NMR-derived structures for SUMO-2/pRAP80-(37–49)
| pRAP80-(37–49) | SUMO-2 | |
|---|---|---|
| Distance restraints | ||
| Total | 30 | |
| Intraresidue | 3 | |
| Sequential (|i–j| = 1) | 21 | |
| Medium (2 ≤ |i–j| ≤ 4) | 6 | |
| Long (|i–j| ≥ 5) | 0 | |
| Intermolecular | 15 | 15 |
| Dihedral restraints | 11 φ, 11 ψ, 0 χ1 | 68 φ, 68 ψ, 30 χ1 |
| Restraint violations | ||
| Distance >0.5 Å | 0 | 0 |
| Dihedral >5° | 4 | 8 |
| φ/ψ in the most favored region (%) | 88.8 | |
NMR Lineshape Analyses
For titrations of SUMO-2 with RAP80-(35–50) and pRAP80-(37–49), lineshape analyses were carried out using the Bloch-McConnell equations for two-site chemical exchange, as previously described (54), to yield the kinetics (kon and koff) of the RAP80 SIM/SUMO-2 interaction, as well as changes in kinetics upon phosphorylation.
Results
Secondary Structure for RAP80-(33–131) from Quantitative Chemical Shift Analysis
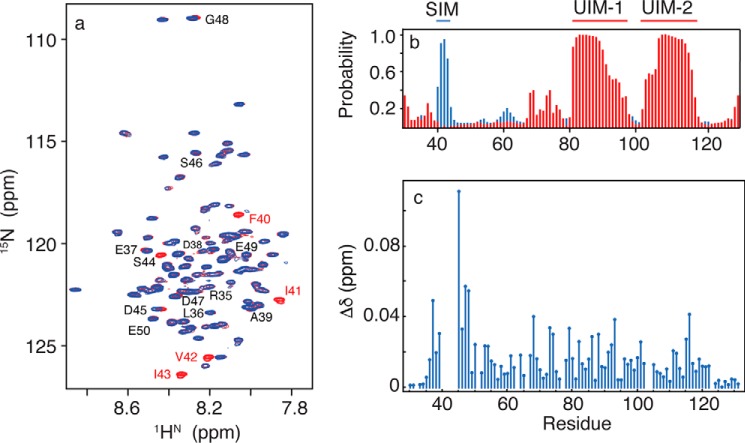
The Ub and SUMO binding properties of RAP80 are confined approximately to residues 30–120 (13, 14, 28, 29), with the tandem UIMs (residues 80–120) forming α-helical structure (55). To determine the molecular basis of SUMO-2 recognition by residues 30–50 from RAP80, we explored the structure of free RAP80-(33–131) using NMR spectroscopy. Two-dimensional 1H-15N HSQC NMR spectra for RAP80-(33–131) are shown in Fig. 1a. The 1HN chemical shifts range from ∼7.7 to 8.9 ppm; this relatively narrow dispersion suggests that for RAP80-(33–131), other than the tandem α-helical UIM domains, a specific global-fold is not adopted. Quantitative chemical shift analysis of main chain torsion angles using the TALOS-N program indicates that residues 33–39, N-terminal to the SIM, adopt a random coil conformation (Fig. 1b), and are flexible, as indicated by random coil index derived S2 values of ∼0.50 (56). TALOS-N chemical shift analysis indicates that residues 40–47 that form the SIM, adopt a β strand conformation with a high probability of ∼0.95 (Fig. 1b). Residues 48–78 that connect the SIM to the tandem UIMs also adopt a primarily random coil conformation, with higher flexibility, as indicated by an average random coil-derived S2 value of ∼0.3. Residues belonging to the tandem UIMs adopt α-helical conformations, as previously described (28, 29). The LR motif (residues 60–78), believed to assist in recruitment of BRCA1 to damage sites (57), does not adopt a specific secondary structure and appears flexible.
FIGURE 1.
a, two-dimensional 1H-15N HSQC NMR spectra for free RAP80-(33–131) (red) and bound to SUMO-2 (blue). Residues 40–44 are broadened beyond detection upon SUMO-2 binding. b, secondary structure for free RAP80-(33–131) derived from quantitative chemical shift analysis, with α-helix shown in red, and β-strand shown in blue. c, chemical shift perturbations for RAP80-(33–131) bound to an 11-fold excess of SUMO-2.
Chemical Shift Perturbation Mapping for [U-15N]-RAP80-(33–131) upon Interaction with SUMO-2
To delineate the N-terminal residues of RAP80 that interact with SUMO-2, we followed changes in main chain amide resonances for RAP80-(33–131) upon SUMO-2 binding using two-dimensional 1H-15N HSQC NMR spectroscopy (Fig. 1a). In general, a significant chemical shift change accompanying a protein-protein interaction can be expected to have a threshold value of ∼0.1 ppm, up to a maximum of ∼0.5 ppm (55). There are few residue-specific, significant chemical shift changes for RAP80-(33–131) within the typical range (Fig. 1c). However, the main chain 1HN and 15N resonances for residues expected to be directly involved in the interaction with SUMO-2, Phe40, Ile41, Val42, Ile43, and Ser44, could not be detected upon interaction with SUMO-2 due to extensive line broadening. The resonances for residues adjacent to the hydrophobic SIM residues, on the other hand, Asp45, Ser46, and Asp47, shifted linearly with increasing SUMO-2, without severe line broadening, consistent with fast chemical exchange. These results suggest that RAP80 interacts with SUMO-2 exclusively via the SIM, similar to the interaction of RAP80 with Ub, wherein only UIM residues are directly involved in binding (29).
NMR-monitored Titrations for the [U-15N]-SUMO-2/RAP80-(33–131) Interaction
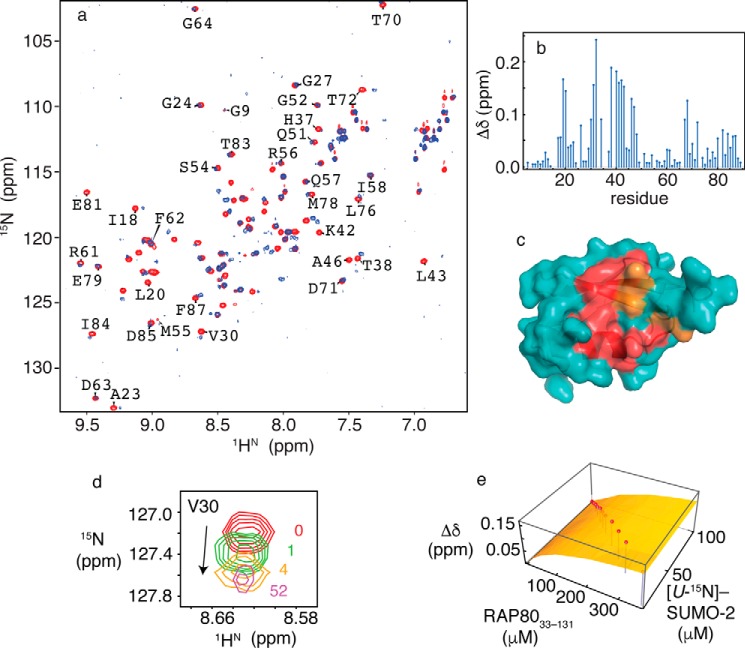
To gain insight into the molecular basis underlying specificity of the SUMO-2/RAP80 interaction, we employed NMR spectroscopy to determine the RAP80 binding site on SUMO-2 using chemical shift mapping, and to determine a quantitative dissociation constant for the interaction. Two-dimensional 1H-15N HSQC NMR spectra of SUMO-2 in the absence and presence of RAP80-(33–131) are shown in Fig. 2a with the per residue combined chemical shift changes shown in Fig. 2b. SUMO-2 main chain amide resonances showing significant changes upon interaction with RAP80-(33–131) are located within strand β2, helix α1, the loop connecting them, as well as loop residues near the 310 helix in free SUMO-2 (Fig. 2c). These regions form the typical SIM interaction site for the various SUMO isoforms (18). Residues Lys33 and Lys35 are undetectable as a result of line broadening, and remained unobservable upon saturation of the binding site. Other residues in the binding cleft, including Val30 (Fig. 2d), Gln31, Phe32, Ile34, Arg36, Thr38, Leu40, Ser41, Lys42, and Leu43, showed linear chemical shift changes that could be followed throughout the titration. The main chain amide resonances for Phe32 and Leu40 were broadened at high RAP80-(33–131)/SUMO-2 ratios. Fitting chemical shift changes for 20 SUMO-2 residues that could be followed during the titration to a 1:1 binding isotherm gives an average dissociation constant (KD) of 195 ± 33 μm. Representative chemical shift changes for Val30, and the fit to a 1:1 binding isotherm are shown in Fig. 2, d and e.
FIGURE 2.
a, two-dimensional 1H-15N HSQC NMR spectra for free SUMO-2 (red) and bound to RAP80-(33–131) (blue). b, chemical shift perturbations for SUMO-2 bound to a 52-fold excess of RAP80-(33–131). c, residues experiencing chemical shift changes greater than 1 S.D. from the mean (red) and those broadened beyond detection (orange) are mapped on the surface of SUMO-2 (PDB code 1WM2). d, expanded region from the two-dimensional 1H-15N HSQC NMR spectra taken during titration of SUMO-2 with RAP80-(33–131), showing chemical shift changes for Val30, with ligand/protein ratios indicated. e, fits of chemical shift perturbation data to 1:1 binding isotherms for the SUMO-2 interaction with RAP80-(33–131). Chemical shift changes for Val30 are indicated on the vertical axis, and concentrations for SUMO-2 and unlabeled RAP80-(33–131) are indicated on the horizontal axes. Experimentally determined chemical shift changes are shown as points, and the best fit to a 1:1 binding isotherm is shown as a surface.
Chemical Shift Perturbation Mapping, NMR Monitored Titrations, and Lineshape Analysis for Interaction of [U-15N]-SUMO-2 with RAP80-(35–50)
To facilitate NMR and structural studies, we synthesized peptides encompassing the minimal binding motif from RAP80 (residues 35–50 or 37–49), given the lack of structure adjacent to these regions, and that no significant chemical shift changes are observed beyond these regions in NMR binding studies employing the longer RAP80-(33–131) construct. The synthetic peptides were N terminally amidated and C terminally acetylated to maintain neutrality for the terminal residues, thereby avoiding introduction of non-physiologically relevant electrostatic interactions. To ensure that the peptides retained the SUMO interactions of the longer RAP80-(33–131) construct, we conducted chemical shift mapping and NMR-monitored titrations to determine the binding site of the RAP80-(35–50) peptide on SUMO-2, as well as the KD and kinetic constants for the interaction, as described in detail below.
Main chain amide 1HN chemical shifts from SUMO-2 residues showing significant changes upon interaction with RAP80-(35–50) are evident in two-dimensional 1H-15N HSQC NMR spectra (Fig. 3, a and b), and the results recapitulate those for titrations with the longer RAP80-(33–131) construct. Similar to the RAP80-(33–131)/SUMO-2 interaction, residues Lys33 and Lys35 could not be detected due to extensive line broadening. Residues in the vicinity of Lys33 and Lys35, such as Phe32, Ile34, Arg36, His37, and Thr38 showed linear main chain amide chemical shift changes upon interaction with RAP80-(35–50), with significant chemical shift changes indicating that the binding site is similar to that for the longer RAP80-(33–131) construct (Fig. 3c). Fitting chemical shift changes for 17 residues to a 1:1 binding isotherm yields an average KD of 239 ± 54 μm. Linear chemical shift changes for Val30, and the corresponding fit to a 1:1 binding isotherm are shown in Fig. 3, d and e. The KD for the SUMO-2 interaction with RAP80-(35–50) is the same within error for interaction of the longer RAP80-(33–131) construct with SUMO-2 (195 ± 33 μm). We conducted lineshape analysis for the chemical shift changes of Val30 during the titration to determine the kinetics of interaction (Fig. 3f). Fits of the lineshape changes to two site chemical exchange using the Bloch-McConnell equations yields a kon value of 5.9 × 106 m−1 s−1, and a koff value of 1511 s−1.
FIGURE 3.
a, two-dimensional 1H-15N HSQC NMR spectra for free SUMO-2 (red) and bound to RAP80-(35–50) (blue). b, chemical shift perturbations for SUMO-2 bound to a 17-fold excess of RAP80-(35–50). c, residues experiencing chemical shift changes greater than 1 S.D. from the mean (red) and those broadened beyond detection (orange) are mapped on the surface of SUMO-2 (PDB code 1WM2). d, fits of chemical shift changes to 1:1 binding isotherms for the SUMO-2 interaction with RAP80-(35–50). Chemical shift changes for Val30 are indicated on the vertical axis, and concentrations for SUMO-2 and unlabeled RAP80-(35–50) are indicated on the horizontal axes. Experimentally determined chemical shift changes are shown as points, and the best fit to a 1:1 binding isotherm is shown as a surface. e, expanded region from two-dimensional 1H-15N HSQC NMR spectra taken during titration of SUMO-2 with RAP80-(35–50), with ligand/protein ratios indicated. f, lineshape analysis for Val30 15N chemical shift changes taken from two-dimensional 1H-15N HSQC NMR spectra taken during titration of SUMO-2 with RAP80-(35–50), experimental data are shown as blue dots connected by lines, and the best fits are shown as green lines.
Chemical Shift Perturbation Mapping for RAP80-(35–50) upon Phosphorylation
RAP80 contains a canonical CK2 recognition motif (S/T)XX(D/E) (58) adjacent to the hydrophobic module of the SIM, that possesses the sequence: Ser44-Asp45-Ser46-Asp47. We assessed the phosphorylation of RAP80 by CK2 using 1H NMR spectroscopy. N terminally acetylated, C terminally amidated RAP80-(35–50) was doubly phosphorylated at Ser44 and Ser46 with CK2. Phosphorylation of the two serines was confirmed by mass spectrometry, and the observation of significant chemical shift changes for the backbone and side chain protons from Ser44/Ser46 (Fig. 4). In addition, a significant change for the 1HN chemical shift for Asp47 is also observed. The ∼0.3 ppm downfield shift for the 1HN of serine upon phosphorylation is suggestive of a hydrogen bond between the amide proton and phosphoryl group, as previously observed (59).
FIGURE 4.
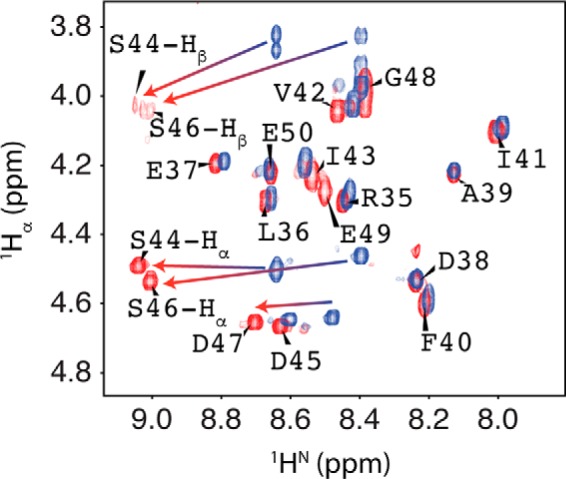
1Hα-1HN fingerprint region from the NMR spectra for RAP80-(35–50) (blue) and phosphorylated RAP80-(35–50) (red).
Chemical Shift Perturbation Mapping, NMR Monitored Titrations, and Lineshape Analysis for the SUMO-2 Interaction with pRAP80-(37–49)
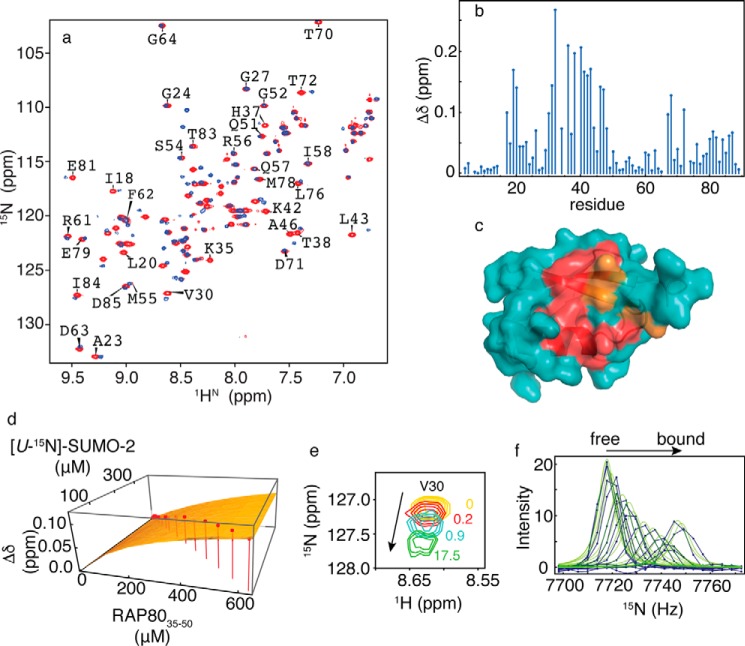
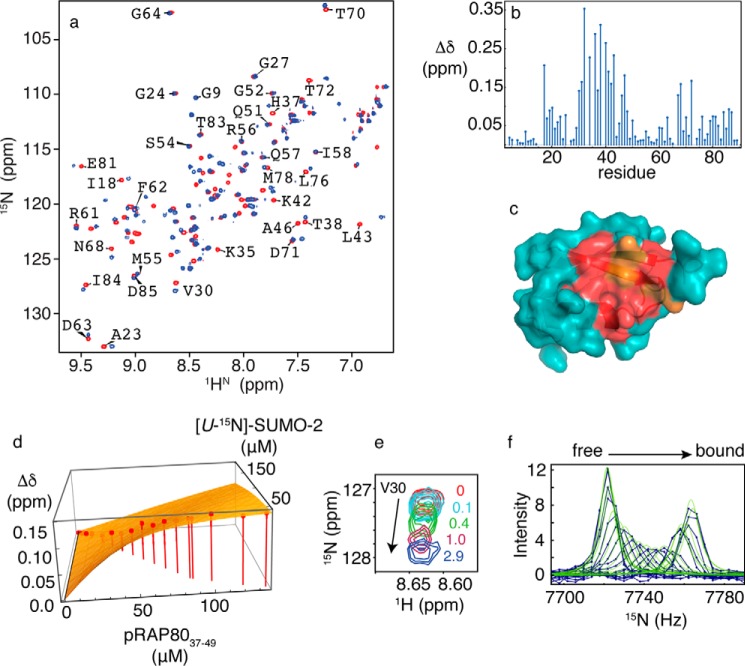
To assess the impact of RAP80 phosphorylation on the specificity of the RAP80/SUMO-2 interaction, we determined the RAP80 binding site on SUMO-2, as well as the dissociation and kinetic rate constants using NMR spectroscopy. The interaction of [U-15N]-SUMO-2 with SIM peptide phosphorylated at Ser44 and Ser46 (pRAP80-(37–49)) was studied using chemical shift titrations. The two-dimensional 1H-15N HSQC NMR spectra, and associated chemical shift maps for SUMO-2 in the absence and presence of pRAP80-(37–49) are shown in Fig. 5, a and b. For the interaction between pRAP80-(37–49) and SUMO-2, the binding site determined by chemical shift mapping is similar to that for unphosphorylated RAP80-(35–50) (Fig. 5c). However, the NMR resonances for residues Phe32, Ile34, Arg36, Thr38, Leu40, and Leu43 showed extensive line broadening and concomitant signal loss for [pRAP80-(37–49)]:[SUMO-2] ratios <1. For peptide/protein ratios >1, the resonances appear less broad, and upon saturation, become more intense in comparison to titrations with unphosphorylated peptide. This behavior indicates that SUMO-2 interaction with the RAP80 SIM peptide becomes more specific upon phosphorylation. However, the main chain amide chemical shifts for residues Lys33 and Lys35 remain broad, even at saturating concentrations of phosphorylated SIM peptide. For 10 residues that could be followed during the titration, the average KD value was determined to be 9 ± 3 μm, a ∼25-fold increase in binding affinity upon phosphorylation. Representive chemical shift changes and the fit to a 1:1 binding isotherm, for Val30, are shown in Fig. 5, d and e. From lineshape analysis (Fig. 5f), the values of kon and koff were determined to be 1.1 × 108 m−1 s−1 and 1020 s−1, respectively. Compared with the kinetics of binding for unphosphorylated RAP80 SIM, kon increases by ∼100-fold, whereas the decrease in koff is 1.5-fold.
FIGURE 5.
a, two-dimensional 1H-15N HSQC NMR spectra for free SUMO-2 (red) and bound to doubly phosphorylated RAP80-(37–49) (blue). b, chemical shift perturbations for SUMO-2 bound to a 3-fold excess of pRAP80-(37–49). c, residues experiencing chemical shift changes greater than 1 S.D. from the mean (red) and those broadened beyond detection (orange) are mapped on the surface of SUMO-2 (PDB code 1WM2). d, fits of chemical shift changes to 1:1 binding isotherms for the SUMO-2 interaction with pRAP80-(37–49). Chemical shift changes for Val30 are indicated on the vertical axis, and concentrations for SUMO-2 and unlabeled pRAP80-(37–49) are indicated on the horizontal axes. Experimentally determined chemical shift changes are shown as points, and the best fit to a 1:1 binding isotherm is shown as a surface. e, expanded region from two-dimensional 1H-15N HSQC NMR spectra taken during titration of SUMO-2 with pRAP80-(37–49), with the ligand/protein ratios indicated. f, lineshape analysis for Val30 15N chemical shift changes taken from two-dimensional 1H-15N HSQC NMR spectra taken during titration of SUMO-2 with pRAP80-(37–49), experimental data are shown as blue dots connected by lines, and the best fits are shown as green lines.
NMR Structure for the SUMO-2·pRAP80-(37–49) Complex
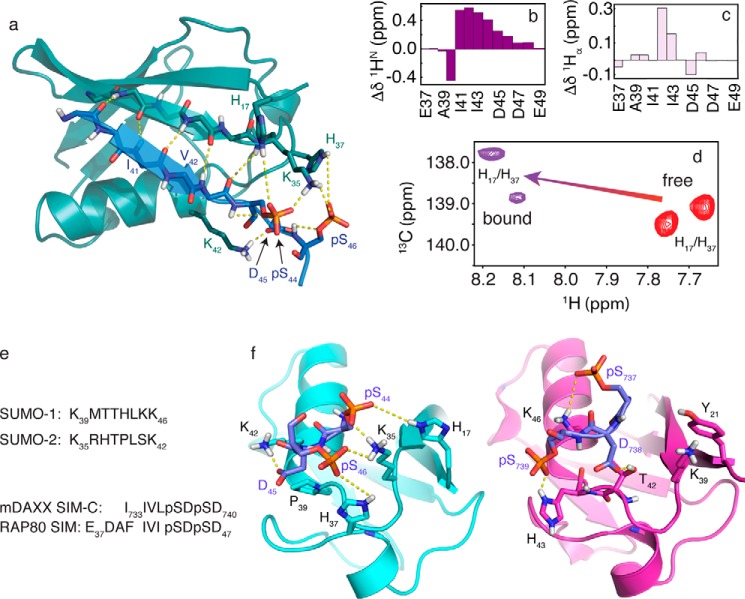
In general, the time scale for exchange between the free and bound forms of SUMO·SIM complexes results in significant NMR resonance broadening, rendering structural studies of the complex difficult (24). For the SUMO-2·pRAP80-(37–49) complex, intermolecular NOEs between the hydrophobic region of the SIM (Phe40-Ile41-Val42-Ile43), and NMR experiments selective for SUMO-2 aromatic residues, particularly Phe32 at the interface, help define the structure of the complex for this region. In contrast, for the electrostatic binding region of the interface, extensive line broadening for SUMO-2 residues, and the inability to obtain NOE-based NMR distance restraints between SIM phosphate groups and SUMO-2, present a significant challenge to NMR structure determination. To overcome these difficulties, we adopted an NMR-guided structural approach wherein sparse NMR data are combined with molecular dynamics simulations using a modern force field, in explicit solvent, to determine a model for the structure. To that end, for both SUMO-2 and doubly phosphorylated RAP80 SIM peptide, main chain φ and ψ dihedral angles derived from chemical shifts, intramolecular peptide NOEs, and intermolecular SIM-SUMO-2 NOEs were used as constraints in molecular dynamics calculations with the state of the art AMBER ff14SB force field (50), to obtain structural models for the complex. A representative SUMO-2-pRAP80-(37–49) structure (Fig. 6a) highlights the parallel SIM orientation with phosphorylated serine residues interacting with a basic region within the SIM binding cleft from SUMO-2. The parallel β-strand from pRAP80-(37–49) completes a β sheet in the intermolecular complex with strand β2 from SUMO-2, consistent with large, positive chemical shift changes (Δδ = bound − free) in the 1HN-1Hα fingerprint region of the bound peptide (Fig. 6, b and c) (60). The fingerprint region also indicates that the N and C termini of the peptide are flexible and not involved in binding, as supported by a lack of significant changes for the main chain proton chemical shifts of Glu37, Asp38, Ala39, Gly48, and Glu49 upon binding SUMO-2 (Fig. 6, b and c). At the N terminus, Ala39 and Phe40 side chain atoms do not directly contact SUMO-2. The side chain from SIM residue Ile41 forms multiple intermolecular contacts with the side chains of Val30, Phe32, Ala46, and Arg50, from SUMO-2. In addition, SIM residue Ile43 contacts the side chains from SUMO-2 residues Lys42 and Leu43. The phosphate group from Ser44 is involved in electrostatic interactions with the side chains from Lys35 and His17 from SUMO-2. This close interaction is supported by an ∼0.5 ppm downfield shift for the histidine side chain Hδ2 protons, as shown in Fig. 6d. Asp45 from pRAP80-(37–49) is involved in favorable electrostatic interactions with Lys42 and the Thr38 hydroxyl group from SUMO-2. The Ser(P)46 phosphate group from the SIM peptide shows favorable electrostatic interactions with the side chains of His37 and Lys35 from SUMO-2. The involvement of His37 in the interaction is supported by a downfield shift for the histidine Hδ2 protons by ∼0.5 ppm (Fig. 6d). At the C terminus of the SIM peptide, residues Asp47, Gly48, and Glu49 appear flexible, consistent with narrower 1HN resonances in 13C/15N-filtered experiments for the protein-peptide complex, in comparison to directly interacting residues.
FIGURE 6.
a, NMR structure for the SUMO-2·pRAP80-(37–49) complex. The main chain atoms are shown in the schematic representation (RAP80 SIM, blue; SUMO-2, teal). Key electrostatic interactions are indicated by yellow dashed lines, and include hydrogen bonds between the main chain atoms of the SIM hydrophobic module, and interactions between the SUMO-2 specificity module and the negatively charged side chains of the SIM. b, 1HN main chain amide chemical shift changes (bound-free) for pRAP80-(37–49) upon binding of SUMO-2. c, 1Hα main chain chemical shift changes (bound-free) for pRAP80-(37–49) upon binding of SUMO-2. d, two-dimensional 1H-13C HSQC NMR spectra showing side chain chemical shifts for the SUMO-2 Hδ2 atoms from residues His17 and His37 in the free state (red) and upon interaction with pRAP80-(37–49) (purple). e, sequence alignment for the SIM binding loop from SUMO-1 and -2, and the alignment for the SIM from RAP80 and that from “modified” DAXX, which contains residues from the PML SIM N-terminal to Ile733 to facilitate crystallization. f, comparison of the structure of the SUMO-2·pRAP80-(37–49) complex with the SUMO-1·pDAXX-(733–740) complex (PDB code 4WJP). SUMO-1 and -2 are shown in purple and cyan, respectively, the electrostatic SUMO recognition modules from the pSIMs are shown in blue. Dashed yellow lines highlight key intermolecular electrostatic interactions. The hydrophobic SUMO recognition modules from the pSIMs are not shown for clarity.
Discussion
For RAP80-(33–131), two-dimensional 1H-15N NMR spectra, and quantitative chemical shift analyses were used to derive per residue flexibility and main chain secondary structure, and indicate that the N-terminal region of RAP80 consists of three independent domains. These domains include a partially structured SIM, and two partially structured, tandem UIMs, surrounded by flexible regions. From chemical shift perturbation mapping and NMR-monitored titrations for various RAP80 constructs, we observe that the minimal region sufficient for interaction with SUMO-2 encompasses residues 40–47 from the RAP80 SIM. Similar to Ub binding to the tandem UIMs (29), the RAP80 SIM binds SUMO-2 independently. Furthermore, we determined that residues 50–78 between the SIM and tandem UIMs are not involved in Ub binding. The affinity of the RAP80/SUMO-2 interaction is weak, with a KD of ∼200 μm, and interestingly, double phosphorylation of the SIM at Ser44 and Ser46 gives rise to a ∼25-fold increase in the affinity of the interaction. NMR lineshape analysis indicates that this is due to a substantial increase in kon for SUMO-2/RAP80 SIM association; a result of enhanced electrostatic interactions between the phosphate groups of Ser44/Ser46, and a positively charged SUMO-2 region at one end of the SIM interaction site. The key role for electrostatics in the modulation of SUMO/SIM interactions is underscored by a prominent dependence on salt concentration (24). The binding affinity of unphosphorylated RAP80 SIM for SUMO-2 is comparable with the affinities of SIM-C from DAXX for both SUMO-1 and SUMO-2 with KD values of ∼140 μm at 200 mm KCl, but weaker in comparison to the ∼40 μm KD for SIM-N binding to both SUMO-1 and SUMO-2 at 200 mm KCl (24). It should be noted that for these affinity comparisons, SIM-C from DAXX shares the SDSD electrostatic module sequence with RAP80, whereas SIM-N from DAXX does not (DDDD). Upon double phosphorylation of RAP80, the SUMO-2-SIM interaction affinity increases, with a KD ∼9 μm; this is comparable with a KD of ∼1 μm for the tetraphosphorylated PML SIM/SUMO-1 interaction, although the greater affinity for the phospho-PML/SUMO-1 interaction, in comparison to that for RAP80/SUMO-2, is likely a result of lower salt concentration for the former interaction (21, 23).
The NMR-derived solution state structure of SUMO-2 in complex with pRAP80-(37–49) is the first structure of SUMO-2 bound to a phosphorylated SIM. The structure of the complex shows a rich variety of electrostatic interactions, as well as key hydrophobic interactions, generally separated into two distinct, but adjacent regions, or modules, of the binding interface, as generally observed for SUMO/SIM interactions. The negatively charged region of RAP80 (Ser(P)44-Asp45-Ser(P)46-Asp47), adjacent to hydrophobic SIM module (Phe40-Ile41-Val42-Ile43), is involved in extensive electrostatic interactions with the positively charged residues of the SIM binding interface on SUMO-2. Specifically, the observed chemical shift changes for the SUMO-2 His17 and His37 side chain Hδ2 protons upon phospho-SIM binding, suggest electrostatic interactions with a distance range of 3–5 Å between the histidine Nϵ2 and SIM phosphate oxygen atoms for the histidine-phosphoserine pairs. This is consistent with the known relationship between crystallographic structures and chemical shift changes for histidine side chain Hδ2 protons from RNase A upon interaction with nucleotide phosphate groups (61, 62). Other key electrostatic interactions include the intermolecular hydrogen bonds across the peptide planes from RAP80 and SUMO-2 that form the intermolecular β-sheet, as well as intramolecular SIM hydrogen bonds between the phosphoserine side chain γ-oxygens and their respective main chain amide protons.
The RAP80 SIM-SUMO-2 structure is similar to the mDAXX C-terminal SIM/SUMO-1 interaction (PDB code 4WJP) (21), although there are important differences in the electrostatic module of the binding interface that form the basis of specificity determinants (Fig. 6, e and f). The residues from the SIM hydrophobic modules, FIVI for RAP80 and IIVL for DAXX, insert into the hydrophobic SUMO binding cleft similarly, with the side chain of the second and fourth residues buried, and in direct contact with SUMO. As with other SUMO/SIM interactions, key structural differences are found in the electrostatic binding module adjacent to the hydrophobic module. Specifically, there are key sequence differences for positively charged residues in the electrostatic SIM binding module of SUMO, which includes a SIM binding loop flanking the interaction site (Fig. 6e, residues Met40 to Lys46, and Arg36 to Lys42 in SUMO-1 and SUMO-2, respectively). In the case of the electrostatic phosphorylated SDSD modules from both RAP80 and the C-SIM from DAXX (PDB code 4WJP), the first phosphoserine from RAP80, Ser(P)44, forms favorable electrostatic interactions with Lys35 from strand β2 adjacent to the N terminus of the binding loop, as well as His17 from strand β1. In contrast, the first phosphoserine from DAXX, interacts with Lys46 from helix α1 of SUMO-1, which lies on the opposite, or C-terminal side of the SIM binding loop. In addition, SUMO-1 lacks an equivalent to the positively charged His17 SUMO-2 residue at the N-terminal side of the binding loop, and possesses Tyr21 instead. Furthermore, a key SIM binding loop histidine, His37 from SUMO-2 and His43 from SUMO-1, occurs at opposite ends of the SIM binding loop. These differences result in the second phosphate from DAXX interacting with the C-terminal portion of the SIM binding loop, whereas the second phosphate from the RAP80 SIM interacts with the N-terminal portion of the SIM binding loop (Fig. 6f). Thus, the change in orientation for various SIM phosphoserine side chains upon interacting with cognate SUMO isoforms can be attributed to different charge distributions in the electrostatic binding modules from SUMO.
From a broader biological perspective, the N-terminal region from RAP80 connects numerous signaling pathways, including SUMOylation, ubiquitination, phosphorylation, and lysine and acetylation, with DNA damage repair (63, 64). For example, it has recently been suggested that phosphorylated SIMs function as a node in a network that connects CK2 signaling with SUMOylation (20). CK2 has also been shown to be involved in double strand break repair through homologous recombination by assisting the association of the DNA repair protein Rad51 to the MRN complex through phosphorylation (63). Double phosphorylation of the canonical CK2 site in RAP80 substantially elevates the binding affinity for SUMO-2, and suggests a deeper role for CK2 in the repair of double-stranded DNA breaks, at least within the context of RAP80-mediated recruitment of BRCA1 to DNA damage sites. Interestingly, the phosphorylation of SIMs may be cross-regulated through acetylation of Lys33 in SUMO-2 and Lys37 in SUMO-1, although additional lysines near the SIM binding cleft may be involved (65–67). From a structural perspective, Lys33 and Lys35 are critical for electrostatic interactions with SIM or phosphorylated SIM; their acetylation abolishes SUMO-SIM binding, although not in all cases. In the case of RAP80, it will be of interest to determine whether the interaction with SUMO-2 is regulated by acetylation of Lys33 and Lys35, linking regulation of SUMO acetylation with DNA repair by homologous recombination.
The molecular basis underlying the role of ubiquitination and SUMOylation in the RAP80-mediated function of the DNA damage response is not yet fully understood. It is unclear if these post-translational modifications function independently, or if hybrid SUMO-Ub chain recognition is necessary for RAP80 recruitment to DNA damage sites. The latter model is alluring, given that the poly-SUMO binding Ub ligase RNF4 has been shown to be necessary for BRCA1 recruitment (14), and can catalyze the covalent attachment of Ub to SUMO chains (68). This study provides the first molecular details underlying the SUMO/RAP80 interaction, and its regulation by phosphorylation. It will be of interest to gain a deeper understanding of the interplay between various regulatory and signaling processes in RAP80-mediated BRCA1 recruitment to the DNA damage sites.
Author Contributions
L. S. and Anamika designed the study and wrote the paper. Anamika labeled, expressed, and purified RAP80 and SUMO-2, performed NMR experiments, and analyzed NMR data with assistance from L. S. MD simulations with NMR restraints, and NMR lineshape analyses were conducted and analyzed by L. S. and Anamika. Aromatic HSQC NMR pulse sequences were written by L. S., set up and analyzed by Anamika. All authors analyzed the results and approved the final version of the manuscript.
Acknowledgments
We thank Drs. Manoj K. Rout, Brian L. Lee, and Craig J. Markin for assistance with experimental design, data analyses, and helpful discussions.
This work was supported by Canadian Institutes of Health Research Grant MOP 110964 (to L. S.). The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (code 2N9E) have been deposited in the Protein Data Bank (http://wwpdb.org/).
NMR chemical shifts have been deposited in the BioMagRes Bank under accession number 104587.
- Ub
- ubiquitin
- UIM
- ubiquitin interacting motif
- SUMO
- small ubiquitin like modifier
- SIM
- SUMO interacting motif
- CK2
- casein kinase 2
- DSS
- 4,4-dimethyl-4-silapentane-1-sulfonic acid
- PDB
- Protein Data Bank
- HSQC
- heteronuclear single quantum coherence.
References
- 1. Jackson S. P., and Bartek J. (2009) The DNA-damage response in human biology and disease. Nature 461, 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Giglia-Mari G., Zotter A., and Vermeulen W. (2011) DNA damage response. Cold Spring Harbor Perspect. Biol. 3, 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bekker-Jensen S., and Mailand N. (2010) Assembly and function of DNA double-strand break repair foci in mammalian cells. DNA Repair 9, 1219–1228 [DOI] [PubMed] [Google Scholar]
- 4. Bologna S., and Ferrari S. (2013) It takes two to tango: ubiquitin and SUMO in the DNA damage response. Front. Genet. 4, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huen M. S., and Chen J. (2008) The DNA damage response pathways: at the crossroad of protein modifications. Cell Res. 18, 8–16 [DOI] [PubMed] [Google Scholar]
- 6. Matsuoka S., Ballif B. A., Smogorzewska A., McDonald E. R. 3rd, Hurov K. E., Luo J., Bakalarski C. E., Zhao Z., Solimini N., Lerenthal Y., Shiloh Y., Gygi S. P., and Elledge S. J. (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316, 1160–1166 [DOI] [PubMed] [Google Scholar]
- 7. Kolas N. K., Chapman J. R., Nakada S., Ylanko J., Chahwan R., Sweeney F. D., Panier S., Mendez M., Wildenhain J., Thomson T. M., Pelletier L., Jackson S. P., and Durocher D. (2007) Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318, 1637–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., and Lukas J. (2007) RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131, 887–900 [DOI] [PubMed] [Google Scholar]
- 9. Kim H., Chen J., and Yu X. (2007) Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 316, 1202–1205 [DOI] [PubMed] [Google Scholar]
- 10. Sobhian B., Shao G., Lilli D. R., Culhane A. C., Moreau L. A., Xia B., Livingston D. M., and Greenberg R. A. (2007) RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 316, 1198–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Galanty Y., Belotserkovskaya R., Coates J., Polo S., Miller K. M., and Jackson S. P. (2009) Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 462, 935–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Morris J. R., Boutell C., Keppler M., Densham R., Weekes D., Alamshah A., Butler L., Galanty Y., Pangon L., Kiuchi T., Ng T., and Solomon E. (2009) The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature 462, 886–890 [DOI] [PubMed] [Google Scholar]
- 13. Hu X., Paul A., and Wang B. (2012) RAP80 protein recruitment to DNA double-strand breaks requires binding to both small ubiquitin-like modifier (SUMO) and ubiquitin conjugates. J. Biol. Chem. 287, 25510–25519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guzzo C. M., Berndsen C. E., Zhu J., Gupta V., Datta A., Greenberg R. A., Wolberger C., and Matunis M. J. (2012) RNF4-dependent hybrid SUMO-ubiquitin chains are signals for RAP80 and thereby mediate the recruitment of BRCA1 to sites of DNA damage. Sci. Signal 5, ra88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kerscher O., and William C. (2007) SUMO junction: what's your function? New insights through SUMO-interacting motifs. EMBO Rep. 8, 550–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Müller S., Hoege C., Pyrowolakis G., and Jentsch S. (2001) SUMO, ubiquitin's mysterious cousin. Nat. Rev. Mol. Cell Biol. 2, 202–210 [DOI] [PubMed] [Google Scholar]
- 17. Hecker C.-M., Rabiller M., Haglund K., Bayer P., and Dikic I. (2006) Specification of SUMO1- and SUMO2-interacting motifs. J. Biol. Chem. 281, 16117–16127 [DOI] [PubMed] [Google Scholar]
- 18. Gareau J. R., and Lima C. D. (2010) The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11, 861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Song J., Zhang Z., Hu W., and Chen Y. (2005) Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J. Biol. Chem. 280, 40122–40129 [DOI] [PubMed] [Google Scholar]
- 20. Stehmeier P., and Muller S. (2009) Phospho-regulated SUMO interaction modules connect the SUMO system to CK2 signaling. Mol. Cell 33, 400–409 [DOI] [PubMed] [Google Scholar]
- 21. Cappadocia L., Mascle X. H., Bourdeau V., Tremblay-Belzile S., Chaker-Margot M., Lussier-Price M., Wada J., Sakaguchi K., Aubry M., Ferbeyre G., and Omichinski J. G. (2015) Structural and functional characterization of the phosphorylation-dependent interaction between PML and SUMO1. Structure 23, 126–138 [DOI] [PubMed] [Google Scholar]
- 22. Sekiyama N., Ikegami T., Yamane T., Ikeguchi M., Uchimura Y., Baba D., Ariyoshi M., Tochio H., Saitoh H., and Shirakawa M. (2008) Structure of the small ubiquitin-like modifier (SUMO)-interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3. J. Biol. Chem. 283, 35966–35975 [DOI] [PubMed] [Google Scholar]
- 23. Chang C. C., Naik M. T., Huang Y. S., Jeng J. C., Liao P. H., Kuo H. Y., Ho C. C., Hsieh Y. L., Lin C. H., Huang N. J., Naik N. M., Kung C. C., Lin S. Y., Chen R. H., Chang K. S., Huang T. H., and Shih H. M. (2011) Structural and functional roles of Daxx SIM phosphorylation in SUMO paralog-selective binding and apoptosis modulation. Mol. Cell 42, 62–74 [DOI] [PubMed] [Google Scholar]
- 24. Escobar-Cabrera E., Okon M., Lau D. K., Dart C. F., Bonvin A. M., and McIntosh L. P. (2011) Characterizing the N- and C-terminal small ubiquitin-like modifier (SUMO)-interacting motifs of the scaffold protein DAXX. J. Biol. Chem. 286, 19816–19829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Reverter D., and Lima C. D. (2005) Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature 435, 687–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yan Z., Kim Y. S., and Jetten A. M. (2002) RAP80, a novel nuclear protein that interacts with the retinoid-related testis-associated receptor. J. Biol. Chem. 277, 32379–32388 [DOI] [PubMed] [Google Scholar]
- 27. Wang B., Matsuoka S., Ballif B. A., Zhang D., Smogorzewska A., Gygi S. P., and Elledge S. J. (2007) Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science 316, 1194–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sims J. J., and Cohen R. E. (2009) Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of RAP80. Mol. Cell 33, 775–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Markin C. J., Xiao W., and Spyracopoulos L. (2010) Mechanism for recognition of polyubiquitin chains: balancing affinity through interplay between multivalent binding and dynamics. J. Am. Chem. Soc. 132, 11247–11258 [DOI] [PubMed] [Google Scholar]
- 30. Wittekind M., and Mueller L. (1993) HNCACB, a high-sensitivity 3D NMR experiment to correlate amide proton and nitrogen resonances with the alpha- and beta-carbon resonances in proteins. J. Magn. Reson. B 101, 201–205 [Google Scholar]
- 31. Muhandiram D. R., and Kay L. E. (1994) Gradient-enhanced triple-resonance three-dimensional NMR experiments with improved sensitivity. J. Magn. Reson. B 103, 203–216 [Google Scholar]
- 32. Grzesiek S., and Bax A. (1992) Correlating backbone amide and side chain resonances in larger proteins by multiple relayed triple resonance NMR. J. Am. Chem. Soc. 114, 6291–6293 [Google Scholar]
- 33. Szyperski T., Braun D., Fernandez C., Bartels C., and Wüthrich K. (1995) A novel reduced-dimensionality triple-resonance experiment for efficient polypeptide backbone assignment, 3D CO HN N CA. J. Magn. Reson. B 108, 197–203 [Google Scholar]
- 34. Panchal S. C., Bhavesh N. S., and Hosur R. V. (2001) Improved 3D triple resonance experiments, HNN and HN(C)N, for HN and 15N sequential correlations in (13C, 15N) labeled proteins: application to unfolded proteins. J. Biomol. NMR 20, 135–147 [DOI] [PubMed] [Google Scholar]
- 35. Kay L. E., Xu G. Y., and Yamazaki T. (1994) Enhanced-sensitivity triple-resonance spectroscopy with minimal H2O saturation. J. Magn. Reson. A 109, 129–133 [Google Scholar]
- 36. Yamazaki T., Lee W., Arrowsmith C. H., Muhandiram D. R., and Kay L. E. (1994) A suite of triple resonance experiments for the backbone assignment of 15N,13C,2H labeled proteins with high sensitivity. J. Am. Chem. Soc. 116, 11655–11666 [Google Scholar]
- 37. Goddard T. D., and Kneller D. G. (2008) SPARKY 3. University of California, San Francisco [Google Scholar]
- 38. Keller R. L. (2004) The Computer-aided Resonance Assignment Tutorial. Cantina Verlag, Zürich [Google Scholar]
- 39. Wüthrich K. (1986) NMR of Proteins and Nucleic Acids. John Wiley & Sons, New York [Google Scholar]
- 40. Kay L. E., Keifer P., and Saarinen T. (1992) Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc. 114, 10663–10665 [Google Scholar]
- 41. Markin C. J., and Spyracopoulos L. (2012) Increased precision for analysis of protein-ligand dissociation constants determined from chemical shift titrations. J. Biomol. NMR 53, 125–138 [DOI] [PubMed] [Google Scholar]
- 42. Shen Y., and Bax A. (2013) Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR 56, 227–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Grzesiek S., Anglister J., and Bax A. (1993) Correlation of backbone amide and aliphatic side chain resonances in 13C/15N-enriched proteins by isotropic mixing of 13C magnetization. J. Magn. Reson. B 101, 114–119 [Google Scholar]
- 44. Logan T. M., Olejniczak E. T., Xu R. X., and Fesik S. W. (1993) A general method for assigning NMR spectra of denatured proteins using 3D HC(CO)NH-TOCSY triple resonance experiments. J. Biomol. NMR 3, 225–231 [DOI] [PubMed] [Google Scholar]
- 45. Lyons B. A., and Montelione G. T. (1993) An HCCNH triple-resonance experiment using C-13 isotropic mixing for correlating backbone amide and side-chain aliphatic resonances in isotopically enriched proteins. J. Magn. Reson. B 101, 206–209 [Google Scholar]
- 46. Gardner K. H., Konrat R., Rosen M. K., and Kay L. E. (1996) An (H)C(CO)NH-TOCSY pulse scheme for sequential assignment of protonated methyl groups in otherwise deuterated 15N,13C-labeled proteins. J. Biomol. NMR 8, 351–356 [DOI] [PubMed] [Google Scholar]
- 47. Pervushin K., Riek R., Wider G., and Wüthrich K. (1998) Transverse relaxation-optimized spectroscopy (TROSY) for NMR studies of aromatic spin systems in 13C-labeled proteins. J. Am. Chem. Soc. 120, 6394–6400 [Google Scholar]
- 48. Yamazaki T., Forman-Kay J. D., and Kay L. E. (1993) Two-dimensional NMR experiments for correlating 13Cβ and 1Hδ/ϵ chemical shifts of aromatic residues in 13C-labeled proteins via scalar couplings. J. Am. Chem. Soc. 115, 11054–11055 [Google Scholar]
- 49. Otting G., and Wüthrich K. (1989) Extended heteronuclear editing of 2D 1H NMR spectra of isotope-labeled proteins, using the X(ω1, ω2) double half filter. J. Magn. Reson. 85, 586–594 [Google Scholar]
- 50. Case D. A., Berryman J. T., Betz R. M., Cerutti D. S., Cheatham T. E., Darden T. A., Duke R. E., Giese T. J., Gohlke H., Goetz A. W., Homeyer N., Izadi S., Janowski P., Kaus J., Kovalenko A., Lee T. S., Le Grand S., Li P., Luchko T., Luo R., Madej B., Merz K. M., Monard G., Needham P., Nguyen H., Nguyen H. T., Omelyan I., Onufriev A., Roe D. R., Roitberg A., Salomon-Ferrer R., Simmerling C., Smith W., Swails J., Walker R. C., Wang J., Wolf R. M., Wu X., York D. M., and Kollman P. A. (2015) AMBER 2015, University of California, San Francisco, CA [Google Scholar]
- 51. Berjanskii M. V., Neal S., and Wishart D. S. (2006) PREDITOR: A web server for predicting protein torsion angle restraints. Nucleic Acids Res. 34, W63–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Huang W. C., Ko T. P., Li S. S., and Wang A. H. (2004) Crystal structures of the human SUMO-2 protein at 1.6 Å and 1.2 Å resolution: implication on the functional differences of SUMO proteins. Eur. J. Biochem. 271, 4114–4122 [DOI] [PubMed] [Google Scholar]
- 53. Homeyer N., Horn A. H., Lanig H., and Sticht H. (2006) AMBER force-field parameters for phosphorylated amino acids in different protonation states: phosphoserine, phosphothreonine, phosphotyrosine, and phosphohistidine. J. Mol. Model 12, 281–289 [DOI] [PubMed] [Google Scholar]
- 54. Markin C. J., and Spyracopoulos L. (2012) Accuracy and precision of protein-ligand interaction kinetics determined from chemical shift titrations. J. Biomol. NMR 54, 355–376 [DOI] [PubMed] [Google Scholar]
- 55. Anamika, Markin C. J., Rout M. K., and Spyracopoulos L. (2014) Molecular basis for impaired DNA damage response function associated with the RAP80 ΔE81 defect. J. Biol. Chem. 289, 12852–12862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Berjanskii M. V., and Wishart D. S. (2005) A simple method to predict protein flexibility using secondary chemical shifts. J. Am. Chem. Soc. 127, 14970–14971 [DOI] [PubMed] [Google Scholar]
- 57. Panier S., Ichijima Y., Fradet-Turcotte A., Leung C. C., Kaustov L., Arrowsmith C. H., and Durocher D. (2012) Tandem protein interaction modules organize the ubiquitin-dependent response to DNA double-strand breaks. Mol. Cell 47, 383–395 [DOI] [PubMed] [Google Scholar]
- 58. Olsten M. E., and Litchfield D. W. (2004) Order or chaos? An evaluation of the regulation of protein kinase CK2. Biochem. Cell Biol. 82, 681–693 [DOI] [PubMed] [Google Scholar]
- 59. Tholey A., Lindemann A., Kinzel V., and Reed J. (1999) Direct effects of phosphorylation on the preferred backbone conformation of peptides: a nuclear magnetic resonance study. Biophys. J. 76, 76–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wishart D. S., Sykes B. D., and Richards F. M. (1991) Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J. Mol. Biol. 222, 311–333 [DOI] [PubMed] [Google Scholar]
- 61. Rüterjans H., and Witzel H. (1969) NMR studies on the structure of the active site of pancreatic ribonuclease A. Eur. J. Biochem. 9, 118–127 [DOI] [PubMed] [Google Scholar]
- 62. Leonidas D. D., Chavali G. B., Oikonomakos N. G., Chrysina E. D., Kosmopoulou M. N., Vlassi M., Frankling C., and Acharya K. R. (2003) High-resolution crystal structures of ribonuclease A complexed with adenylic and uridylic nucleotide inhibitors. Implications for structure-based design of ribonucleolytic inhibitors. Protein Sci. 12, 2559–2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yata K., Lloyd J., Maslen S., Bleuyard J. Y., Skehel M., Smerdon S. J., and Esashi F. (2012) Plk1 and CK2 act in concert to regulate Rad51 during DNA double strand break repair. Mol. Cell 45, 371–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Krejci L., Altmannova V., Spirek M., and Zhao X. (2012) Homologous recombination and its regulation. Nucleic Acids Res. 40, 5795–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., and Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 66. Cheema A., Knights C. D., Rao M., Catania J., Perez R., Simons B., Dakshanamurthy S., Kolukula V. K., Tilli M., Furth P. A., Albanese C., and Avantaggiati M. L. (2010) Functional mimicry of the acetylated C-terminal tail of p53 by a SUMO-1 acetylated domain, SAD. J. Cell. Physiol. 225, 371–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ullmann R., Chien C. D., Avantaggiati M. L., and Muller S. (2012) An acetylation switch regulates SUMO-dependent protein interaction networks. Mol. Cell 46, 759–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tatham M. H., Plechanovová A., Jaffray E. G., Salmen H., and Hay R. T. (2013) Ube2W conjugates ubiquitin to α-amino groups of protein N-termini. Biochem. J. 453, 137–145 [DOI] [PMC free article] [PubMed] [Google Scholar]