Abstract
The expansion of cells for regenerative therapy will require the genetic dissection of complex regulatory mechanisms governing the proliferation of non-transformed human cells. Here, we report the development of a high-throughput RNAi screening strategy specifically for use in primary cells and demonstrate that silencing the cell cycle-dependent kinase inhibitors CDKN2C/p18 or CDKN1A/p21 facilitates cell cycle entry of quiescent adult human pancreatic beta cells. This work identifies p18 and p21 as novel targets for promoting proliferation of human beta cells and demonstrates the promise of functional genetic screens for dissecting therapeutically relevant state changes in primary human cells.
Keywords: beta cell (β-cell), cell cycle, high throughput screening (HTS), pancreatic islet, RNA interference (RNAi), primary human cells
Introduction
Discovery-based research using cultured cells, including chemical and functional genomic high-throughput screens (HTS),6 are recognized as powerful, comprehensive tools that identify novel components of signaling pathways (1–3). However, as cell lines often display genomic instability and loss of biochemical regulatory mechanisms, screens using primary human cells are now necessary to gain physiological insight in areas most relevant to disease. The use of primary human cells for HTS introduces several new challenges, including limited source material, cellular quiescence, heterogeneity in cell type, and variability from donor-to-donor. Phenotypic HTS of collections of small molecules can identify lead compounds for cell biological research and hypothesis generation, yet a lack of understanding of the target and the mechanism of action remains a major impediment to their utility due to off target effects.
Pancreatic islets are composed of five endocrine cell types, including beta cells that respond to elevations in blood glucose and nutrients by secreting insulin. Insulin maintains euglycemia in part by promoting glucose uptake and storage in metabolically sensitive tissues, including liver, muscle, and fat. Type 1 diabetes is characterized by insulin deficiency and hyperglycemia due to an autoimmune-mediated destruction of beta cells. Transplantation of donor islets as a treatment for type 1 diabetes is limited by several factors as follows: donor shortages, side effects associated with immunosuppression, loss of insulin secretion, and even cell death, problems common to treatments for other degenerative diseases (4–15). Alternative replacement strategies are currently being developed in which stem cell-derived progenitors are expanded ex vivo prior to transplantation. An additional potential source is adult beta cells themselves; thus acquiring a working knowledge of the molecular mechanisms governing the proliferative behavior of adult beta cells and their progenitors is central to the success of all replacement strategies. However, as adult beta cells proliferate infrequently, and the precise molecular events involved in deriving mature beta cells from precursors is complex, generating an alternative adult source of human pancreatic beta cells for replacement strategies has not yet been achieved (16–20). An additional impediment has been the lack of glucose-responsive human beta cell lines, limiting our understanding of the signaling mechanisms involved in beta cell proliferation in cell lines derived from rodent beta cell tumors (insulinomas) and animals (16). However, recent work has indicated that species differences in the cell cycle proteome may invalidate the use of rodents to understand human beta cell biology (21–25).
Taken together, there is now a critical need for approaches that elicit proliferative behavior of mature human beta cells and their progenitors and to establish a working understanding of the underlying mechanisms for precise intervention so as to avoid the risk of unrestricted growth. Here, we address these issues and describe the development and implementation of an HTS approach using RNA interference to analyze proliferation following gene silencing in a mixed population of primary human pancreatic islet cells, and we report that the cell cycle-dependent kinase (CDK) inhibitors CDKN2C/p18 or CDKN1A/p21 are critical negative regulators of human beta cell proliferation.
Experimental Procedures
Antibodies/Reagents
Antibodies for Western blotting were as follows: β-actin (Sigma, 1:20,000); p53 (Santa Cruz Biotechnology, 1:1000); pRb (Pharmingen, 1:500) and p21 (Pharmingen 1:500); p27 (Cell Signaling, 1:500); PTEN (Santa Cruz Biotechnology, 1:200); pAKT (Cell Signaling, 1:1000) and AKT (Cell Signaling, 1:1000); LKB1 (Santa Cruz Biotechnology, 1:1000), p18 (Cell Signaling 1:200), and HSP90 (Santa Cruz Biotechnology, 1:1000). Antibodies for immunofluorescence were as follows: insulin (Dako, 1:1000), somatostatin (Dako, 1:1000), and glucagon (Dako, 1:1000). Click-it EdU imaging kit (Life Technologies, Inc.) was used according to the manufacturer's recommendations. The following vectors were from Addgene: pLKO.1-TRC (catalog no. 10878) (26) and pHCMV-LCMV-WE (catalog no. 15793) (27).
Islet Culture
We used two sources of human pancreatic islets, the NIDDK (National Institutes of Health)-funded Integrated Islet Distribution Program (IIDP islets) at City of Hope and the J. Shapiro laboratory, University of Alberta, Edmonton, Canada (Edmonton islets). Male and female deceased donors were used, ranging in age from 16 to 79 years and body mass index from 20.1–35.7 kg/m2, none of which had a prior diagnosis of diabetes. The average purity of the Integrated Islet Distribution Program and Edmonton islets were 91.3 and 42.7%, viability of 94 and 80% respectively. When islet purity was under 85%, islets were picked manually and cultured for up to 30 days at 37 °C in a 5% CO2 atmosphere in non-tissue culture-treated Petri dishes in PIM(S) media supplemented with 5% human AB serum, glutamine/glutathione mixture, and penicillin/streptomycin (all reagents from Prodo Laboratories Inc.). Medium was changed every 2–3 days.
Islet Dissociation and Seeding
Islets were washed in PBS and dissociated with Accutase (1 ml/1000 islets) for 20–30 min at 37 °C, triturating every 5 min for 10 s. Dissociated islets were counted and seeded at a density of 15,000–20,000 cells/well in a 384-well plate for fluorescence or 60,000 cells/well in a 96-well plate to generate protein extracts. With the exception of the initial plate coating experiment, islets were always seeded on a PDL-coated plate (described below) to facilitate attachment of dissociated cells.
Coated Plate Assay
Six different matrices/surfaces were compared to determine the optimal surface to promote dissociated islet adherence. 384-Well plates were left untreated (tissue culture polystyrene) as a control surface. Matrigel (BD Biosciences), collagen type 1 (Sigma), applied cell extracellular matrix (Applied Biological Materials Inc.), and poly-d-lysine (Sigma) were coated onto the wells of 384-well plates according to the manufacturer's recommendations. HTB-9 extracellular matrix was coated onto 384-well plates as described previously (28, 29). Dissociated islets were then seeded onto the coated/uncoated plates, infected with non-targeting shRNA for 96 h, and fixed for immunofluorescence as described below.
GSIS Assay
Human islets were dissociated and seeded at a density of 20,000 cells per well into a 384-well plate in triplicate 96 h before treatment. Dissociated islet cells or whole islets (10 islets per replicate) were equilibrated in 50 μl of Krebs-Ringer Buffer (KRB) containing 2.8 mm glucose for 15 min and then incubated in 2.8 mm glucose in KRB (low glucose) for 30 min (cells) or 1 h (islets) prior to stimulation with 16.7 mm glucose in KRB for 30 min (cells) or 1 h (islets). KRB supernatants from 2.8 and 16.7 mm glucose treatments were collected, and insulin amount was determined using an HTRF insulin assay (Cisbio). All conditions were done in triplicate.
Lentiviral Constructs
The pLKO.1eGFP vector was constructed by replacing the puromycin gene in pLKO.1 with a BamHI-KpnI enhanced GFP PCR product. Three independent dsDNA sequences per target were designed using the Sigma Mission shRNA tool and inserted into AgeI-EcoRII-digested pLKO.1eGFP vector.
Lentiviral Infection Optimization
Pilot experiments as outlined in Table 1 showed that the GP-VSVG envelope protein coding vector (pMD2G) provided greater infection efficiency than GP-LCMV (pHCMV) as evaluated by GFP and titer (supplemental Fig. 1 a and Table 1). The optimized ratio selected for all experiments is highlighted in Table 1. We found the best ratio of pMD2G vector to use by measuring LKB1 protein level (supplemental Fig. 1b). Virus was packaged in HEK293T for 72 h with pLKO1.1-eGFP plasmid encoding GFP and shRNA (supplemental Fig. 1c) and pMD2G and pCMV8.74 encoding coat and structural genes, respectively. 10 ml of culture supernatant was concentrated 100× by ultracentrifugation at 28,000 rpm for 2 h on a 20% sucrose layer (supplemental Fig. 1d) as described (30). The shRNA library sequences for the screen are listed in Table 2. Table 3 lists the additional shRNA constructs made against p18 and p21. The virus was then resuspended overnight at 4 °C in serum-free PIMS medium without antibiotics, arrayed into 384-well screening plates, and stored at −80 °C until use. The titer was determined using quantitative PCR lentivirus titration (titer) kit (Applied Biological Materials Inc.) according to the manufacturer's recommendations. In a direct comparison of crude versus concentrated virus (supplemental Fig. 1 e), we saw that concentrated virus resulted in greater knockdown efficiency and less toxicity than crude virus-treated cells using LKB1 shRNA and LKB1 protein levels as a readout (Fig. 1e). Large T antigen was cloned into pLenti6/V5 destination (DEST) vector (Invitrogen) and packaged as described above. The following primers were used: SV40 T antigen forward, GGGGACAAGTTTGTACAAAAAAGCAGGCTTCAGAACCATGGATAAAGTTTTAAACAGAG, and SV40 T antigen reverse, GGGGACCACTTTGTACAAGAAAGCTGGGTCTGTTTCAGGTTCAGGGGGAGGTGTGGGAG.
TABLE 1.
Conditions to package virus
| Envelope coding vector pMD2.G (GP-VSVG) | Envelope coding vector pHCMV (GP-LCMV) | Lentiviral vector pLKO1.1 enhanced GFP | Packaging vector pCMV8.74 | Envelope coding vector/total transfected DNA (final %) | Titer of lentivirus (108 IU/ml) |
|---|---|---|---|---|---|
| μg | μg | μg | μg | ||
| 5 | 10 | 10 | 20 | 8.9 | |
| 1 | 10 | 10 | 5 | 12.6 | |
| 0.2 | 10 | 10 | 1 | 9.9 | |
| 5 | 10 | 10 | 20 | 7.9 | |
| 1 | 10 | 10 | 5 | 5.9 | |
| 0.2 | 10 | 10 | 1 | 2.8 |
TABLE 2.
shRNA library used for the screens
| Targeted protein | Gene name | shRNA |
|---|---|---|
| CDK6 | CDK6 | sh1-CCGGCTTCTGAAGTGTTTGACATTTCTCGAGAAATGTCAAACACTTCAGAAGTTTTTG |
| CDK6 | CDK6 | sh2-CCGGCGTGGAAGTTCAGATGTTGATCTCGAGATCAACATCTGAACTTCCACGTTTTTG |
| CDK6 | CDK6 | sh3-CCGGTCTGGAGTGTTGGCTGCATATCTCGAGATATGCAGCCAACACTCCAGATTTTTG |
| CDKN3 | CDKN3 | sh1-CCGGTGTTCTCAGTTTCTCGGTTTACTCGAGTAAACCGAGAAACTGAGAACATTTTTG |
| CDKN3 | CDKN3 | sh2-CCGGCAGACCATCAAGCAATACAATCTCGAGATTGTATTGCTTGATGGTCTGTTTTTG |
| CDKN3 | CDKN3 | sh3-CCGGAGAACTAAAGAGCTGTGGTATCTCGAGATACCACAGCTCTTTAGTTCTTTTTTG |
| CyclinD1 | CCND1 | sh1-CCGGATTGGAATAGCTTCTGGAATCTCGAGATTCCAGAAGCTATTCCAATCTTTTTG |
| CyclinD1 | CCND1 | sh2-CCGGGAACAAACAGATCATCCGCAACTCGAGTTGCGGATGATCTGTTTGTTCTTTTTG |
| CyclinD1 | CCND1 | sh3-CCGGCCACAGATGTGAAGTTCATTTCTCGAGAAATGAACTTCACATCTGTGGTTTTTG |
| GSK3 beta | GSK3B | sh1-CCGGGCTGAGCTGTTACTAGGACAACTCGAGTTGTCCTAGTAACAGCTCAGCTTTTTG |
| GSK3 beta | GSK3B | sh2-CCGGCCCAAACTACACAGAATTTAACTCGAGTTAAATTCTGTGTAGTTTGGGTTTTTG |
| GSK3 beta | GSK3B | sh3-CCGGCATGAAAGTTAGCAGAGACAACTCGAGTTGTCTCTGCTAACTTTCATGTTTTTG |
| p15 | CDKN2B | sh1-CCGGACGGAGTCAACCGTTTCGGGACTCGAGTCCCGAAACGGTTGACTCCGTTTTTTG |
| p15 | CDKN2B | sh2-CCGGACTAGTGGAGAAGGTGCGACACTCGAGTGTCGCACCTTCTCCACTAGTTTTTTG |
| p15 | CDKN2B | sh3-CCGGGCGCGGATCCCAACGGAGTCACTCGAGTGACTCCGTTGGGATCCGCGCTTTTTG |
| p16 | CDKN2A | sh1-CCGGGCTCTGAGAAACCTCGGGAAACTCGAGTTTCCCGAGGTTTCTCAGAGCTTTTTG |
| p16 | CDKN2A | sh2-CCGGATCAGTCACCGAAGGTCCTACCTCGAGGTAGGACCTTCGGTGACTGATTTTTTG |
| p16 | CDKN2A | sh3-CCGGCACTACCGTAAATGTCCATTTCTCGAGAAATGGACATTTACGGTAGTGTTTTTG |
| p18 | CDKN2C | sh1-CCGGCTATGGGAGGAATGAGGTTGTCTCGAGACAACCTCATTCCTCCCATAGTTTTTG |
| p18 | CDKN2C | sh2-CCGGACTGGTTTCGCTGTCATTCATCTCGAGATGAATGACAGCGAAACCAGTTTTTTG |
| p18 | CDKN2C | sh3-CCGGTGGATTTGGAAGGACTGCGCTCTCGAGAGCGCAGTCCTTCCAAATCCATTTTTG |
| p19 | CDKN2D | sh1-CCGGCCAATCCATCTGGCAGTTCAACTCGAGTTGAACTGCCAGATGGATTGGTTTTTG |
| p19 | CDKN2D | sh2-CCGGGCGCTGCAGGTCATGATGTTTCTCGAGAAACATCATGACCTGCAGCGCTTTTTG |
| p19 | CDKN2D | sh3-CCGGGCAGCTGAATCTGATCTCCATCTCGAGATGGAGATCAGATTCAGCTGCTTTTTG |
| p21 | CDKN1A | sh1-CCGGCTGATCTTCTCCAAGAGGAACTCGAGTTCCTCTTGGAGAAGATCAGCTTTTTG |
| p21 | CDKN1A | sh2-CCGGCGCTCTACATCTTCTGCCTTACTCGAGTAAGGCAGAAGATGTAGAGCGTTTTTG |
| p21 | CDKN1A | sh3-CCGGGAGCGATGGAACTTCGACTTTCTCGAGAAAGTCGAAGTTCCATCGCTCTTTTTG |
| p27 | CDKN1B | sh1-CCGGAGCAATGCGCAGGAATAAGGCTCGAGCCTTATTCCTGCGCATTGCTCTTTTTG |
| p27 | CDKN1B | sh2-CCGGAATGGTGATCACTCCAGGTACTCGAGTACCTGGAGTGATCACCATTCTTTTTG |
| p27 | CDKN1B | sh3-CCGGGTAGGATAAGTGAAATGGATACTCGAGTATCCATTTCACTTATCCTACTTTTTG |
| p53 | TP53 | sh1-CCGGGTCCAGATGAAGCTCCCAGAACTCGAGTTCTGGGAGCTTCATCTGGACTTTTTG |
| p53 | TP53 | sh2-CCGGGACTCCAGTGGTAATCTACCTCGAGGTAGATTACCACTGGAGTCTTTTTG |
| p53 | TP53 | sh3-CCGGGCTCCCAGAATGCCAGAGGCTCGAGCCTCTGGCATTCTGGGAGCTTTTTG |
| p57 | CDKN1C | sh1-CCGGACATCCACGATGGAGCGTCTTCTCGAGAAGACGCTCCATCGTGGATGTTTTTTG |
| p57 | CDKN1C | sh2-CCGGCCAGAACCGCTGGGATTACGACTCGAGTCGTAATCCCAGCGGTTCTGGTTTTTG |
| p57 | CDKN1C | sh3-CCGGTATTCTGCACGAGAAGGTACACTCGAGTGTACCTTCTCGTGCAGAATATTTTTG |
| p107 | RBL1 | sh1-CCGGCATCGATAGTGATGCAGAATCCTCGAGGATTCTGCATCACTATCGATGTTTTTG |
| p107 | RBL1 | sh2-CCGGCCCACTGTGGTAATTCCACATCTCGAGATGTGGAATTACCACAGTGGGTTTTTG |
| p107 | RBL1 | sh3-CCGGCCAAGCTAATAGTCACGTATACTCGAGTATACGTGACTATTAGCTTGGTTTTTG |
| p130 | RBL2 | sh1-CCGGATCTTCATTGGTTAGCATGTCTCGAGACATGCTAACCAATGAAGATCTTTTTG |
| p130 | RBL2 | sh2-CCGGTACTTCAGCAACAGTCCTTCACTCGAGTGAAGGACTGTTGCTGAAGTATTTTTG |
| p130 | RBL2 | sh3-CCGGCAAGATACTGCTACGTGTAATCTCGAGATTACACGTAGCAGTATCTTGTTTTTG |
| pRB | RB1 | sh1-CCGGCCACATTATTTCTAGTCCAAACTCGAGTTTGGACTAGAAATAATGTGGTTTTTG |
| pRB | RB1 | sh2-CCGGCGAAATTGGATCACAGCGATACTCGAGTATCGCTGTGATCCAATTTCGTTTTTG |
| pRB | RB1 | sh3-CCGGGTGCGCTCTTGAGGTTGTAATCTCGAGATTACAACCTCAAGAGCGCACTTTTTG |
| PTEN | PTEN | sh1-CCGGAGGCGCTATGTGTATTATTATCTCGAGATAATAATACACATAGCGCCTTTTTTG |
| PTEN | PTEN | sh2-CCGGCCACAGCTAGAACTTATCAAACTCGAGTTTGATAAGTTCTAGCTGTGGTTTTTG |
| PTEN | PTEN | sh3-CCGGCTAGAACTTATCAAACCCTTTCTCGAGAAAGGGTTTGATAAGTTCTAGTTTTTG |
TABLE 3.
Screen donor information
| Screen no. | No. of donors | Age | Sex | Body mass index |
|---|---|---|---|---|
| years | ||||
| 1 | 2 (50%/50%) | 46 | Female | 20.1 |
| 21 | Male | NA | ||
| 2 | 1 | 36 | Male | 33.8 |
| 3 | 1 | 53 | Male | 27.2 |
FIGURE 1.
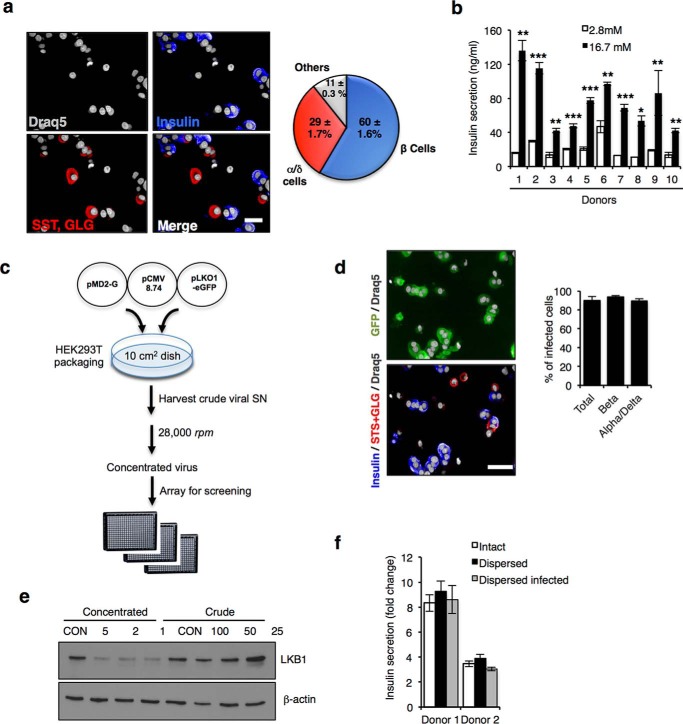
Assay development to screen in human dissociated islets. a, left, representative images of dissociated human islets stained for nuclei (Draq5, gray), insulin (blue), glucagon (GLG, alpha cell marker), and somatostatin (SST, delta cell marker). Scale bar, 10 μm. Right, pie chart showing percentages of dissociated human islet cells belonging to given cell groups shown below. b, GSIS assay using dissociated human islets from 10 different donors. Islets were dissociated into single cells and seeded for 90 h on a 384-well poly-d-lysine (PDL)-coated plate. Islet cells were treated with low (2.8 mm) and high (16.7 mm) glucose and shown as a fold change. Bars indicate ± S.E. c, flow chart of lentivirus preparation for screen. d, representative images of GFP+ cells 4 days after infection of dissociated islet cultures with control shRNA-expressing lentivirus stained for insulin (blue-beta cells), glucagon, and somatostatin (GLG + SST; red-alpha and delta cells, respectively) and nuclei (gray). Scale bar, 20 μm. Histogram showing infection rate is shown at right. e, Western blot showing LKB1 protein level following infection with crude and concentrated virus preparations. β-Actin is shown as loading and toxicity control. f, GSIS assay showing fold change in insulin secretion (16.7/2.8 mm glucose) using intact, dissociated, and dissociated and infected human islets from two independent donors. Error is mean ± S.E.
The human p21 and p18 open reading frames were both cloned into pLenti6/V5 DEST vector (Invitrogen). shRNA-resistant constructs were generated by the QuikChange Mutagenesis kit (Stratagene). The cDNA sequences used are listed in Table 5.
TABLE 5.
shRNA/resistant constructs for p18 and p21 validation experiments
| Construct | Vector | Sequence |
|---|---|---|
| p18 shRNA 4 | pLKO1.1 eGFP | CCGGCTGCGCTGCAGGTTATGAAACCTCGAGGTTTCATAACCTGCAGCGCAGTTTTTG |
| p18 cDNA 5′ | pLKO1.1 eGFP | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCAGAACCATGGCCGAGCCTTGGGGGAACGAG |
| p18 cDNA 3′ | pLenti6-V5 DEST | GGGGACCACTTTGTACAAGAAAGCTGGGTCTTGAAGATTTGTGGCTCCCCCAGCC |
| p21 shRNA 4 | pLenti6-V5 DEST | CCGGGACACCACTGGAGGGTGACTTCTCGAGAAGTCACCCTCCAGTGGTGTCTTTTTG |
| p21 cDNA 5′ | pLenti6-V5 DEST | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCAGAACCATGTCAGAACCGGCTGGGGATGTCC |
| p21 cDNA 3′ | pLenti6-V5 DEST | GGGGACCACTTTGTACAAGAAAGCTGGGTCGGGCTTCCTCTTGGAGAAGATCAGC |
Islet Cell Infection
Dissociated islet cells were infected at the time of seeding by adding the cell suspension to multiwell plates (96- or 384-well) in which purified adenovirus or lentivirus had been arrayed. After incubation for the indicated times, cells were fixed and processed for imaging or Western blot analysis. Lentiviral islet infection was performed with 1 μl per well for TAg encoding virus and 2 μl per well of shRNA concentrated virus in 384-well plate and 5 μl per well in a 96-well plate.
Immunofluorescence
After seeding/infection (72 h), EdU was added to the medium (20 μm) for the remaining time of the experiment, and medium was changed every 48–72 h. At the appropriate time point, dissociated islet cells were fixed by adding 3.7% paraformaldehyde for 15 min at 37 °C using a Multidrop Combi dispenser (Fisher), quenched with 0.75% glycine in PBS at room temperature, and gently rinsed twice with PBS using an automated plate washer (Biotek). All subsequent steps were performed at room temperature. Cells were permeabilized with 0.1% Triton X-100 in PBS for 20 min, rinsed with PBS and blocked with 3% BSA in PBS for 2 h. Primary antibodies were incubated for 2 h at room temperature or overnight at 4 °C. Secondary antibodies in 3% BSA in PBS were incubated for 1 h. Nuclei were stained with Draq5 (2.5 μm) or Hoechst (5 μg/ml) for 15 min. EdU detection was performed according to the manufacturer's protocol for 30 min in the dark, before the blocking step. Each step was followed by three washes in 1× PBS performed using a BioTek 405CW multiwell plate washer. Western blotting was performed as described previously (30).
Statistical Analysis consisted of two-tail unpaired t tests. NS or no asterisk = not significant; p ≥ 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Imaging
Fluorescent images were captured using a PerkinElmer Life Sciences Opera® automated confocal multiwell plate microscope fitted with a ×40 high NA water lens. Forty fields per well were acquired per fluorescent channel: blue 405 (Hoechst); green 488 (GFP); red 594 (insulin); and far-red 647 (EdU). For HTS, images were analyzed using an algorithm that calculates the number of infected cells (GFP+) that were also insulin+ (beta cells) and scored the presence of EdU in the nucleus. Each condition was done in triplicate. All cells identified as positive by the algorithm for both EdU and insulin were confirmed manually.
Results
To enable HTS in primary human cells, we developed an imaging assay in 384-well microplates using freshly isolated human islets from non-diabetic male and female subjects. Donors were 16–68 years and had body mass index ranging from 20 to 36 kg/m2. Fluorescence imaging using markers of the major islet endocrine lineages in dissociated islet cell preparations confirmed the expected ratio between beta cell (insulin-positive cells, 60% of total) to alpha and delta cell populations (glucagon and somatostatin-positive cells, 30% of total; Fig. 1a). GSIS assays demonstrated a glucose-induced increase (2–8-fold) in insulin secretion over basal in islets from all donors, after dispersion and seeding as single cells in 384-well imaging plates even after long term culture (up to 32 days; Fig. 1b).
The least time-consuming approach to performing HTS withRNAi is to use commercially available libraries of arrayed siRNA duplexes. However, lipid-mediated transfection of siRNA is both inefficient and interferes with central primary beta cell functions, including GSIS (data not shown) (31, 32). In our hands, lentivirus-mediated delivery of shRNA confers knockdown in both rodent and human beta cells without adverse effects on beta cell function (30, 33, 34). Furthermore, lentivirus infects target cells independent of the cell cycle stage, and the integration of the lentiviral genome delivery enables stable and long term silencing, a feature that is particularly important for slower growing or quiescent cells. To determine whether lentiviral shRNA delivery was feasible in human islet cells, we generated lentivirus using a modified pLKO1.1 vector that encodes shRNA and expresses enhanced green fluorescent protein (eGFP) to achieve maximal titer and infection efficiency (Fig. 1c and Table 1). Lentivirus preparation was optimized to achieve >90% beta cell infection (Fig. 1d) and to increase knockdown reproducibility and avoid toxicity triggered by exogenous factors present in the supernatant from HEK293T viral packaging cells (Fig. 1e). Importantly, GSIS observed with intact islets and dispersed cells was also unaffected by viral infection, confirming their integrity under screening conditions (Fig. 1f).
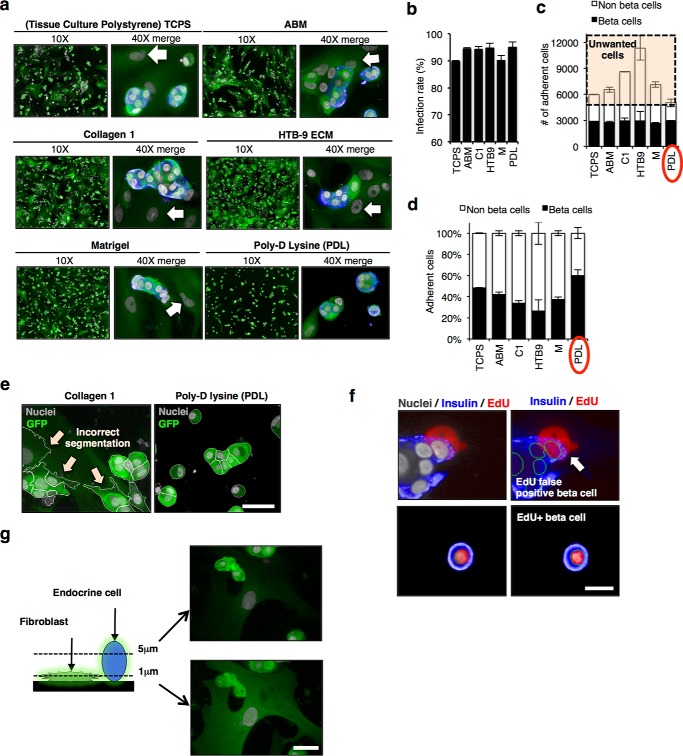
To optimize cell attachment for automated processing during screening, we compared retention of dissociated human islet cells infected with lentivirus on surfaces previously used for culturing human islets (Fig. 2a). Infection rates of endocrine cells were ≥90%, and the absolute number of beta cells retained was the same on all surfaces (Fig. 2, b and c). However, the percentage of beta cells remaining attached following processing for imaging was highest on poly-d-lysine (PDL; Fig. 2d). Moreover, a distinct non-islet fibroblast-like cell accumulated on all surfaces except PDL, which interfered with segmentation analysis (Fig. 2e) and contributed to a false-positive signal (Fig. 2f). To further improve segmentation efficiency, we imaged in a single plane 5 μm from the bottom of the plate to avoid capturing these flatter non-endocrine cells (Fig. 2g).
FIGURE 2.
Imaging human dispersed islets. a, confocal images of dispersed islets cultured on different coating matrices. Islets were dispersed and infected with GFP+ lentivirus encoding NT shRNA for 96 h. Image on left (×10) of paired images shows GFP (green) and Draq5 nuclear stain (gray). Higher magnification (×40) images merged with insulin (blue) are shown at right. White arrows indicate non-endocrine cells (fibroblast-like). b, representative bar graph showing the percent infection rate for dissociated beta cells (insulin+) seeded on the six different surface coatings. c, bar graph showing total number of adherent cells split into two populations beta cells (insulin positive, black bars) and non-beta cells (insulin negative, white bars) seen with the different surface coatings. The highlighted portion shows non-beta “unwanted” cells that accumulate on surfaces other than PDL (red circle). d, representative bar graph showing the percentage of beta and non-beta cells (%) remaining on the different coatings following automated staining. Each condition was done in triplicate, in two different donors. Bars indicate means ± S.E. of one donor. e, representative confocal images showing automated segmentation of cells on collagen (left) and PDL (right). White arrows show inaccurate segmentation when non-beta fibroblast-like cells are present. Scale bar, 20 μm. f, representative confocal images of EdU+ beta cells, arrow showing an example of false EdU+ beta cell picked up by the algorithm, EdU signal coming from fibroblast at the bottom. Scale bar, 10 μm. g, schematic and representative confocal images of dispersed human islets taken at two scanning plane heights (5 and 1 μm above the culture surface). NT, non-transfected. Scale bar, 20 μm.
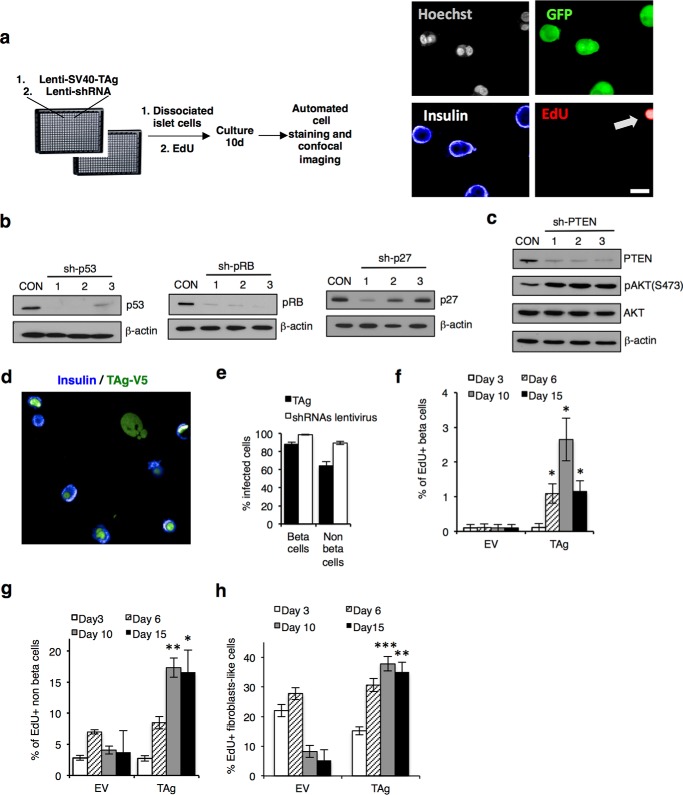
To perform a proof of principal screen, we generated a lentiviral library of shRNA sequences targeting known tumor suppressors and negative regulators of the cell cycle machinery (Table 2) and infected primary human islets cells according to the scheme in Fig. 3a. After 4 days of gene silencing, we performed Western blots on protein extracts from lentivirus-infected cells and confirmed efficient knockdown of several targets, including pRb, p53, CDKN1B/p27, and PTEN with one or more shRNAs (Fig. 3b). Moreover, when PTEN was silenced, we observed an increase in phosphorylation of the PTEN target AKT at Ser-473 (35), confirming functionality of this approach (Fig. 3c).
FIGURE 3.
HTS proliferation assay. a, HTS workflow. Images of dissociated islets 10 days after seeding/infection, following fixation and staining. Scale bar, 10 μm. b, representative Western blots showing knockdown efficiency of various genes targeted in the screen. Dissociated islets were infected with lentiviral shRNAs against p53, pRB, and p27. c, Western blot analysis of PTEN downstream effector phospho-AKT, with total AKT and β-actin control. d, representative confocal images showing SV40 TAg-V5 (green) and insulin (blue) staining. e, bar graph showing the percentage of beta and non-beta cells stained positive for TAg-V5 (black bars) or GFP+ shRNA plasmid marker (white bars) from the three screens. Error bars indicate ± S.E. of the three screens in triplicate for islet cells. f, bar graphs showing the percentage of EdU+ beta cells following infection with control lentivirus (empty vector, EV) or lentivirus encoding TAg at 3, 6, 10, and 15 days post-infection. g, bar graph showing the percentage of EdU+, insulin-negative cells at 3, 6, 10, and 15 days post-infection. h, bar graph showing the percentage of EdU+ fibroblast-like cells at 3, 6, 10, and 15 days post-infection. For data in c–e, n.s. not significant (p ≥ 0.05); *, p < 0.05; **, p < 0.01; ***, p < 0.001, two-tailed unpaired t test.
To monitor cellular proliferation in dissociated islet cultures, we cultured islet cells with EdU, a thymidine analog that incorporates into DNA during S-phase, as its high signal/noise is optimal for HTS imaging (36, 37). Interestingly, dissociated human beta cells did not incorporate EdU or express Ki67 antigen following 10 days in culture indicative of stable quiescence (data not shown), confirming that beta cells are a useful model of post-mitotic cells. As preliminary screens using library shRNAs alone failed to reveal genes that block proliferation after 10 days of gene silencing, we next primed human islet cells to enter the cell cycle by co-infecting human islet cells with lentivirus that expresses SV40 large T antigen (TAg), similarly achieving >90% infection rate in beta cells (Fig. 3, d and e). Surprisingly, but consistent with the proliferative intransigence of primary human cells, only 1–3% of insulin+ cells infected with TAg-V5 incorporated EdU after 2 weeks in culture (Fig. 3f), compared with 18% of non-beta cells (Fig. 3g) or fibroblasts (Fig. 3h).
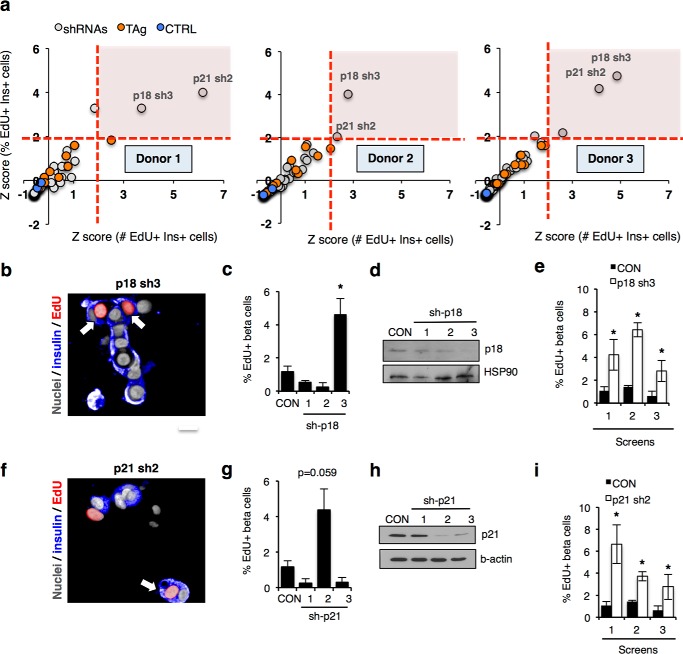
Given these observations, we next repeated the screen to determine whether silencing cell cycle genes in cells expressing TAg would result in an increase in EdU incorporation. We performed three independent screens using islets from different donors (Fig. 4a; see Table 4 for donor information), all of which exhibited Z′-factors between 0.46 and 0.80 indicative of a robust assay with a high degree of fidelity (38). To eliminate potential false positives due to a low number of beta cells, Z scores (representing the number of standard deviations a candidate gene scored above the mean of the total population in the library) for both the absolute number of proliferating beta cells and the percentage of proliferative beta cells were determined and presented for each screen. Z scores and % EdU+ cells for each screen are provided in Table 5. In each screen, silencing the CDK inhibitors CDKN2C/p18 or CDKN1A/p21 resulted in a statistically significant 2.6–6.1-fold increase in EdU-positive cells, with Z scores >2. Fluorescence micrographs taken from the screen, histograms showing the percentage of insulin and EdU double-positive cells, together with Western blots for cells silenced for p18 (Fig. 4, b–e) and p21 (Fig. 4, f–i) are shown.
FIGURE 4.
HTS in primary human cells identifies p18 and p21 as inhibitors of beta cell cycle entry. a, Z-score plots of screen results from three independent donors of islets. Control cells not expressing SV40 large T antigen (TAg) (blue circles), TAg+ cells infected with control shRNA (orange circles), and TAg+ cells infected with library shRNAs (gray circles) are shown. Dotted red lines mark the region of both Z-scores greater than 2. The x axis represents Z-scores of double-positive (EdU, insulin) cells based on the absolute number; the y axis represents Z-scores for the percentage of total beta cells. b, screen images showing EdU-positive cells (red) in beta cells silenced for p18. Scale bar, 10 μm. c, histogram showing percentage of EdU+ insulin+ cells following p18 silencing over a 10-day assay period compared with non-targeting control (CON). d, Western blot showing knockdown of p18 following p18 silencing. e, histogram showing percentage of EdU+ insulin+ cells in each screen replicate following p18 silencing over a 10-day assay period compared with non-targeting control (CON). f, screen images showing EdU-positive cells (red) in beta cells silenced for p21 using lenti-shRNA constructs used in the screen. Scale bar, 10 μm. g, histogram showing percentage of EdU+ insulin+ cells following p21 silencing over a 10-day assay period compared with non-targeting control (CON). h, Western blot showing knockdown of p21 following silencing using lenti-shRNA constructs used in the screen. i, histogram showing percentage of EdU+ insulin+ cells in each screen replicate following p21 silencing over a 10-day assay period compared with non-targeting control (CON). For data in c, e, and i, * indicates p < 0.05, two-tailed unpaired t test.
TABLE 4.
Z scores and % EdU+ cells from screens
NA means not applicable.
| shRNA | Islet screen 1 |
Islet screen 2 |
Islet screen 3 |
|||
|---|---|---|---|---|---|---|
| % EdU | Z score | % EdU | Z score | % EdU | Z score | |
| CDK6 sh1 | 0.1 | −0.447 | 0.2 | −0.536 | 0.1 | −0.377 |
| CDK6 sh2 | 0.8 | 0.209 | 0.0 | −0.682 | 0.0 | −0.553 |
| CDK6 sh3 | 0.0 | −0.577 | 0.0 | −0.682 | 0.2 | −0.274 |
| CDKN3 sh1 | 0.6 | −0.006 | 1.5 | 0.389 | 0.0 | −0.553 |
| CDKN3 sh2 | 0.0 | −0.577 | 0.1 | −0.595 | 0.0 | −0.553 |
| CDKN3 sh3 | 1.1 | 0.532 | 0.3 | −0.488 | 0.0 | −0.553 |
| CyclinD1 sh1 | NA | NA | 0.8 | −0.125 | 0.1 | −0.436 |
| CyclinD1 sh2 | 0.0 | −0.577 | 0.4 | −0.389 | 0.0 | −0.553 |
| CyclinD1 sh3 | 1.3 | 0.700 | 2.7 | 1.271 | 0.2 | −0.236 |
| Gsk3b sh1 | 1.6 | NA | 0.5 | NA | 0.7 | NA |
| Gsk3b sh2 | 0.1 | NA | 0.7 | NA | 1.1 | NA |
| Gsk3b sh3 | 0.0 | NA | 0.6 | NA | 1.7 | NA |
| p15 sh1 | 1.3 | 0.745 | 1.9 | 0.680 | 0.4 | 0.141 |
| p15 sh2 | 0.3 | −0.319 | 0.3 | −0.431 | 0.1 | −0.417 |
| p15 sh3 | 0.0 | −0.577 | 0.2 | −0.511 | 0.2 | −0.261 |
| p16 sh1 | 0.4 | −0.170 | 0.5 | −0.324 | 0.2 | −0.170 |
| p16 sh2 | 0.7 | 0.106 | 1.6 | 0.510 | 0.6 | 0.420 |
| p16 sh3 | 0.0 | −0.577 | 0.0 | −0.682 | 0.1 | −0.429 |
| p18 sh1 | 0.7 | 0.152 | 0.5 | −0.302 | 0.3 | 0.016 |
| p18 sh2 | 0.0 | −0.577 | 0.0 | −0.682 | 0.0 | −0.553 |
| p18 sh3 | 4.2 | 3.709 | 6.4 | 4.024 | 3.2 | 4.855 |
| p19 sh1 | 0.0 | −0.577 | 0.5 | −0.308 | 0.3 | −0.039 |
| p19 sh2 | 1.6 | 1.045 | 1.4 | 0.371 | 1.0 | 1.071 |
| p19 sh3 | 1.5 | 0.914 | 2.3 | 1.031 | 0.3 | 0.028 |
| p21 sh1 | 0.0 | −0.577 | 0.0 | −0.682 | 0.2 | −0.244 |
| p21 sh2 | 6.6 | 6.165 | 3.7 | 2.048 | 2.7 | 4.106 |
| p21 sh3 | 0.0 | −0.577 | 0.1 | −0.608 | 0.8 | 0.843 |
| p27 sh1 | 1.1 | 0.527 | 1.0 | 0.080 | 0.1 | −0.305 |
| p27 sh2 | 0.7 | 0.113 | 0.5 | −0.279 | 0.4 | 0.092 |
| p27 sh3 | 0.2 | −0.418 | 0.6 | −0.271 | 0.5 | 0.286 |
| p53 sh1 | 0.0 | −0.577 | 0.1 | −0.590 | 0.3 | −0.123 |
| p53 sh2 | 0.0 | −0.577 | 0.2 | −0.555 | 0.0 | −0.553 |
| p53 sh3 | 0.0 | −0.577 | 0.6 | −0.263 | 0.4 | 0.091 |
| p57 sh1 | 0.0 | −0.577 | 0.0 | −0.682 | 0.0 | −0.553 |
| p57 sh2 | 0.4 | −0.168 | 0.4 | −0.379 | 0.2 | −0.161 |
| p57 sh3 | 0.8 | 0.233 | 0.5 | −0.311 | 0.3 | 0.024 |
| p107 sh1 | 0.5 | −0.073 | 2.2 | 0.926 | 0.0 | −0.553 |
| p107 sh2 | 0.2 | −0.370 | 0.1 | −0.610 | 1.0 | 1.143 |
| p107 sh3 | 0.0 | −0.577 | 0.0 | −0.682 | 0.1 | −0.392 |
| p130 sh1 | 0.3 | −0.323 | 1.8 | 0.612 | 0.4 | 0.092 |
| p130 sh2 | 1.0 | 0.479 | 1.0 | 0.072 | 0.0 | −0.553 |
| p130 sh3 | 0.0 | −0.577 | 0.5 | −0.331 | 0.0 | −0.553 |
| pRB sh1 | 0.0 | −0.577 | 0.0 | −0.682 | 0.0 | −0.553 |
| pRB sh2 | 0.2 | −0.376 | 3.2 | 1.673 | 0.0 | −0.553 |
| pRB sh3 | 0.0 | −0.577 | 0.6 | −0.270 | 0.1 | −0.426 |
| PTEN sh1 | 0.5 | −0.059 | 0.6 | −0.237 | 0.1 | −0.410 |
| PTEN sh2 | 2.4 | 1.850 | 1.0 | 0.050 | 1.2 | 1.451 |
| PTEN sh3 | 0.8 | 0.211 | 0.9 | −0.013 | 1.3 | 1.716 |
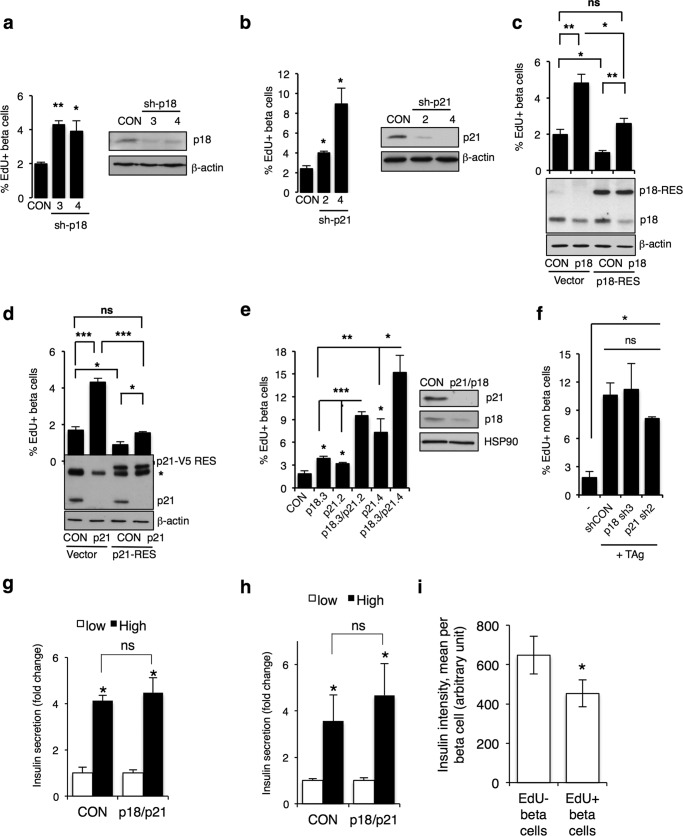
To validate p18 and p21 as regulators of beta cell quiescence, first we silenced p18 or p21 with an additional shRNA for each gene and observed increased EdU incorporation, confirming the screen results (Fig. 5, a and b). Importantly, reintroduction of cDNAs encoding shRNA-resistant p18 or p21 proteins in the presence of the corresponding shRNAs reduced EdU incorporation to levels seen in control cells (Fig. 5, c and d). These data confirm that the observed proliferative enhancement results from an on-target effect of the shRNAs targeting p18 and p21. Furthermore, silencing both p18 and p21 together elicited a synergistic increase in EdU incorporation in beta cells (Fig. 5e) but no other cells in the human islet cell cultures (Fig. 5f), indicating that p18 and p21 function in independent pathways to maintain beta cell quiescence. Importantly, knockdown of these two candidates did not affect insulin secretion capacity in dispersed and intact islets, showing functional viability of beta cells (Fig. 5, g and h). However, when the intensity of insulin staining in EdU+ versus EdU− dispersed beta cells is compared in cells where we observed the greatest percentage of proliferating cells (i.e. in cells expressing TAg and silenced for p18 + p21), EdU+ beta cells had 30% less insulin than EdU cells (Fig. 5i), consistent with dedifferentiation of proliferative beta cells.
FIGURE 5.
p18 and p21 cooperate to prevent human beta cell proliferation. a, histogram showing percentage of EdU+ beta cells following silencing with additional shRNA targeting p18 with Western blots showing p18 knockdown. b, histogram showing percentage of EdU+ beta cells following silencing with additional shRNA targeting p21 with Western blots showing p21 knockdown. c and d, histogram showing percentage of EdU+ beta cells following silencing p18 (c) or p21 (d), in the presence of the corresponding shRNA-resistant cDNA tagged with V5 (p18-RES, p21-RES). Empty vector control (vector) is shown. Western blots showing expression of endogenous and exogenous p18 and p21 are shown below. Note that the V5 tag shifts the shRNA-resistant protein higher on the blot. e, histogram showing percentage of EdU+ beta cells following silencing p18 with one shRNA (p18.3) or p21 with two shRNAs (p21.2 and p21.4) alone or together. Western blot at right shows knockdown of p18 and p21 in p18.3 + p21.4 condition. * indicates a nonspecific band detected with p21 antibody. n = 3 independent biological replicates done in triplicate for c–e. n.s., p > 0.05; *, ≤ 0.05; **, ≤ 0.01; ***, ≤ 0.001, two-tailed unpaired t test. f, histogram showing the percentage of EdU+, TAg+, insulin-negative cells infected with control non-silencing shRNA (CON) or shRNA targeting p18 (p18 sh3) or p21 (p21 sh2). ns, not significant, p ≥ 0.05, two-tailed unpaired t test. g, islets were dissociated and plated on poly-d-lysine-coated plates and infected with TAg in combination with empty vector control (CON) virus, or p18 and p21 silencing. GSIS was performed following 3 days. h, intact islets were infected with TAg in combination with control vector (CON) or p18 and p21 silencing. GSIS was performed 10 days after infection. Bars indicate the mean of three independent donors ± S.E. i, insulin staining intensity in EdU+ and EdU-beta cells, calculated with Columbus software. Bars represent the mean of insulin intensity per cell, based on all insulin+ cells in the experiments shown in e (TAg + sh-p18.3 + sh-p21.4). ns, not significant, p ≥ 0.05; *, p ≤ 0.05, two-tailed unpaired t test.
Discussion
Here, we demonstrate the feasibility of genetic HTS using limited freshly isolated primary human tissue and identify the cell cycle inhibitors p18 and p21 as required for maintenance of adult human beta cell quiescence. Furthermore, we adapt this strategy to use with primary human neurons, expanding beyond cell lines the systems that can be used to identify disease-relevant genes.
Silencing p18, p21 alone or together increased S-phase entry of human beta cells only in the presence of SV40 TAg. TAg inhibits pRb and p53 activity and thus short circuits entry into the G1 and S stages of the cell cycle (39). p18 inhibits cyclin-CDK4/CDK6 complexes that are required for G0/G1 progression (40), and p21 prevents G1 progression and entry into S phase by blocking cyclin-CDK2/CDK4 activity (41, 42). Taken together, our data indicate that mature beta cells reside in G0/G1 and that silencing p18 and p21 lowers the threshold for G1/S phase entry. Furthermore, genetic rescue with RNAi-resistant p18 and p21 constructs establishes these CDK inhibitors as bona fide targets to facilitate human beta cell cycling behavior. Of note, mild overexpression of p18 or p21 (Fig. 5, b and c) reduces proliferation in control cells, indicating that proliferative potential in human beta cells is finely tuned to the levels of these two CDK inhibitors. Parenthetically, we did not observe an effect of silencing the CDK inhibitor CDKN1C/p57 (43) perhaps due to insufficient silencing in this context.
Recent work indicates that the small molecule harmine, which targets serine/threonine kinases in the CMGC branch of the kinome, can induce 1% of human beta cells to proliferate (44), but its precise target(s) remains unclear. Interestingly, under the same assay conditions >8% of rodent beta cells proliferate following harmine treatment, supporting the observations that adult human beta cells have evolved additional safeguards against inappropriate proliferation (17–20). Consistent with this, we achieve only ∼15% EdU incorporation by silencing p18 and p21 together in the presence of TAg, suggesting that multiple redundant mechanisms likely evolved to limit inappropriate S-phase entry and the development of fatal beta cell-derived tumors.
As a consequence, intervening to elicit cell proliferation raises concerns about the possibility of neoplastic growth. Thus, it stands to reason that strategies designed to restore lost or damaged tissue must be subject to reversal. Interestingly, these molecular impediments can be overcome in physiological scenarios such as pregnancy in both rodents and humans (45–48). We contend that identification and validation of these targets will require a genetic approach. Both p18 and p21 were identified as hits using islets from independent donors, supporting the feasibility of pooling islets from multiple donors for larger scale RNAi screens (28, 49). We anticipate that such studies will provide a comprehensive functional framework to permit precise control over the proliferative behavior of primary cells, with potentially broad general application to regenerative approaches in multiple settings.
Author Contributions
K. R., J. R., and J. E. M. researched data and wrote the manuscript. A. F. researched data and edited the manuscript. S. B. and Q. D. researched data. T. K. and A. M. J. S. provided human islets and edited the manuscript. R. A. S. designed the project and wrote the manuscript.
Supplementary Material
Acknowledgment
We acknowledge the Integrated Islet Distribution Program for additional human pancreatic islets.
This work was supported in part by the Juvenile Diabetes Research Foundation and Canadian Institutes of Health Research Grant MOP 111186. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Fig. 1.
- HTS
- high throughput screen
- GSIS
- glucose-stimulated insulin secretion
- PDL
- poly-d-lysine
- TAg
- T antigen
- CDK
- cyclin-dependent kinase
- EdU
- 5-ethynyl-2′-deoxyuridine.
References
- 1. Mayr L. M., and Bojanic D. (2009) Novel trends in high-throughput screening. Curr. Opin. Pharmacol. 9, 580–588 [DOI] [PubMed] [Google Scholar]
- 2. Bernards R. (2014) Finding effective cancer therapies through loss of function genetic screens. Curr. Opin. Genet. Dev. 24, 23–29 [DOI] [PubMed] [Google Scholar]
- 3. Mohr S., Bakal C., and Perrimon N. (2010) Genomic screening with RNAi: results and challenges. Annu. Rev. Biochem. 79, 37–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cooper D. K., Bottino R., Satyananda V., Wijkstrom M., and Trucco M. (2013) Toward clinical islet xenotransplantation-are revisions to the IXA guidelines warranted? Xenotransplantation 20, 68–74 [DOI] [PubMed] [Google Scholar]
- 5. Jeong J. H., Yook S., Lee H., Ahn C. H., Lee D. Y., and Byun Y. (2013) Effects of surface camouflaged islet transplantation on pathophysiological progression in a db/db type 2 diabetic mouse model. Biochem. Biophys. Res. Commun. 433, 513–518 [DOI] [PubMed] [Google Scholar]
- 6. Ryan E. A., Lakey J. R., Paty B. W., Imes S., Korbutt G. S., Kneteman N. M., Bigam D., Rajotte R. V., and Shapiro A. M. (2002) Successful islet transplantation: continued insulin reserve provides long-term glycemic control. Diabetes 51, 2148–2157 [DOI] [PubMed] [Google Scholar]
- 7. Ryan E. A., Lakey J. R., and Shapiro A. M. (2001) Clinical results after islet transplantation. J. Invest. Med. Res. 49, 559–562 [DOI] [PubMed] [Google Scholar]
- 8. Ryan E. A., Paty B. W., Senior P. A., Bigam D., Alfadhli E., Kneteman N. M., Lakey J. R., and Shapiro A. M. (2005) Five-year follow-up after clinical islet transplantation. Diabetes 54, 2060–2069 [DOI] [PubMed] [Google Scholar]
- 9. Jansson D., Ng A. C., Fu A., Depatie C., Al Azzabi M., and Screaton R. A. (2008) Glucose controls CREB activity in islet cells via regulated phosphorylation of TORC2. Proc. Natl. Acad. Sci. U.S.A. 105, 10161–10166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ali F., Dua A., and Cronin D. C. (2015) Changing paradigms in organ preservation and resuscitation. Curr. Opin. Organ Transplant. 20, 152–158 [DOI] [PubMed] [Google Scholar]
- 11. Bruni A., Gala-Lopez B., Pepper A. R., Abualhassan N. S., and Shapiro A. J. (2014) Islet cell transplantation for the treatment of type 1 diabetes: recent advances and future challenges. Diabetes Metab. Syndr. Obes. 7, 211–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lucidi V., Gustot T., Moreno C., and Donckier V. (2015) Liver transplantation in the context of organ shortage: toward extension and restriction of indications considering recent clinical data and ethical framework. Curr. Opin. Crit. Care 21, 163–170 [DOI] [PubMed] [Google Scholar]
- 13. McDonald-Hyman C., Turka L. A., and Blazar B. R. (2015) Advances and challenges in immunotherapy for solid organ and hematopoietic stem cell transplantation. Sci. Transl. Med. 7, 280rv2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ruttens D., Vandermeulen E., Verleden S. E., Bellon H., Vos R., Van Raemdonck D. E., Dupont L. J., Vanaudenaerde B. M., and Verleden G. M. (2015) Role of genetics in lung transplant complications. Ann. Med. 47, 106–115 [DOI] [PubMed] [Google Scholar]
- 15. Thomas K. A., Valenzuela N. M., and Reed E. F. (2015) The perfect storm: HLA antibodies, complement, FcγRs, and endothelium in transplant rejection. Trends Mol. Med. 10.1016/j.molmed.2015.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kulkarni R. N., Mizrachi E. B., Ocana A. G., and Stewart A. F. (2012) Human beta-cell proliferation and intracellular signaling: driving in the dark without a road map. Diabetes 61, 2205–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Butler A. E., Janson J., Bonner-Weir S., Ritzel R., Rizza R. A., and Butler P. C. (2003) Beta-cell deficit and increased beta-cell apoptosis in humans with type 2 diabetes. Diabetes 52, 102–110 [DOI] [PubMed] [Google Scholar]
- 18. Tyrberg B., Ustinov J., Otonkoski T., and Andersson A. (2001) Stimulated endocrine cell proliferation and differentiation in transplanted human pancreatic islets: effects of the ob gene and compensatory growth of the implantation organ. Diabetes 50, 301–307 [DOI] [PubMed] [Google Scholar]
- 19. Kassem S. A., Ariel I., Thornton P. S., Scheimberg I., and Glaser B. (2000) Beta-cell proliferation and apoptosis in the developing normal human pancreas and in hyperinsulinism of infancy. Diabetes 49, 1325–1333 [DOI] [PubMed] [Google Scholar]
- 20. Meier J. J., Butler A. E., Saisho Y., Monchamp T., Galasso R., Bhushan A., Rizza R. A., and Butler P. C. (2008) Beta-cell replication is the primary mechanism subserving the postnatal expansion of beta-cell mass in humans. Diabetes 57, 1584–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cozar-Castellano I., Harb G., Selk K., Takane K., Vasavada R., Sicari B., Law B., Zhang P., Scott D. K., Fiaschi-Taesch N., and Stewart A. F. (2008) Lessons from the first comprehensive molecular characterization of cell cycle control in rodent insulinoma cell lines. Diabetes 57, 3056–3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Martín J., Hunt S. L., Dubus P., Sotillo R., Néhmé-Pélluard F., Magnuson M. A., Parlow A. F., Malumbres M., Ortega S., and Barbacid M. (2003) Genetic rescue of Cdk4 null mice restores pancreatic beta-cell proliferation but not homeostatic cell number. Oncogene 22, 5261–5269 [DOI] [PubMed] [Google Scholar]
- 23. Fiaschi-Taesch N. M., Salim F., Kleinberger J., Troxell R., Cozar-Castellano I., Selk K., Cherok E., Takane K. K., Scott D. K., and Stewart A. F. (2010) Induction of human beta-cell proliferation and engraftment using a single G1/S regulatory molecule, cdk6. Diabetes 59, 1926–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fiaschi-Taesch N. M., Kleinberger J. W., Salim F. G., Troxell R., Wills R., Tanwir M., Casinelli G., Cox A. E., Takane K. K., Scott D. K., and Stewart A. F. (2013) Human pancreatic beta-cell G1/S molecule cell cycle atlas. Diabetes 62, 2450–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fiaschi-Taesch N., Bigatel T. A., Sicari B., Takane K. K., Salim F., Velazquez-Garcia S., Harb G., Selk K., Cozar-Castellano I., and Stewart A. F. (2009) Survey of the human pancreatic beta-cell G1/S proteome reveals a potential therapeutic role for cdk-6 and cyclin D1 in enhancing human beta-cell replication and function in vivo. Diabetes 58, 882–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moffat J., Grueneberg D. A., Yang X., Kim S. Y., Kloepfer A. M., Hinkle G., Piqani B., Eisenhaure T. M., Luo B., Grenier J. K., Carpenter A. E., Foo S. Y., Stewart S. A., Stockwell B. R., Hacohen N., et al. (2006) A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124, 1283–1298 [DOI] [PubMed] [Google Scholar]
- 27. Sena-Esteves M., Tebbets J. C., Steffens S., Crombleholme T., and Flake A. W. (2004) Optimized large-scale production of high titer lentivirus vector pseudotypes. J. Virol. Methods 122, 131–139 [DOI] [PubMed] [Google Scholar]
- 28. Walpita D., Hasaka T., Spoonamore J., Vetere A., Takane K. K., Fomina-Yadlin D., Fiaschi-Taesch N., Shamji A., Clemons P. A., Stewart A. F., Schreiber S. L., and Wagner B. K. (2012) A human islet cell culture system for high-throughput screening. J. Biomol. Screen. 17, 509–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Beattie G. M., Cirulli V., Lopez A. D., and Hayek A. (1997) Ex vivo expansion of human pancreatic endocrine cells. J. Clin. Endocrinol. Metab. 82, 1852–1856 [DOI] [PubMed] [Google Scholar]
- 30. Eberhard C. E., Fu A., Reeks C., and Screaton R. A. (2013) CRTC2 is required for beta-cell function and proliferation. Endocrinology 154, 2308–2317 [DOI] [PubMed] [Google Scholar]
- 31. Moore F., Cunha D. A., Mulder H., and Eizirik D. L. (2012) Use of RNA interference to investigate cytokine signal transduction in pancreatic beta cells. Methods Mol. Biol. 820, 179–194 [DOI] [PubMed] [Google Scholar]
- 32. Beck A., Vinik Y., Shatz-Azoulay H., Isaac R., Streim S., Jona G., Boura-Halfon S., and Zick Y. (2013) Otubain 2 is a novel promoter of beta cell survival as revealed by siRNA high-throughput screens of human pancreatic islets. Diabetologia 56, 1317–1326 [DOI] [PubMed] [Google Scholar]
- 33. Fu A., Robitaille K., Faubert B., Reeks C., Dai X. Q., Hardy A. B., Sankar K. S., Ogrel S., Al-Dirbashi O. Y., Rocheleau J. V., Wheeler M. B., MacDonald P. E., Jones R., and Screaton R. A. (2015) LKB1 couples glucose metabolism to insulin secretion in mice. Diabetologia 58, 1513–1522 [DOI] [PubMed] [Google Scholar]
- 34. Sakamaki J., Fu A., Reeks C., Baird S., Depatie C., Al Azzabi M., Bardeesy N., Gingras A. C., Yee S. P., and Screaton R. A. (2014) Role of the SIK2-p35-PJA2 complex in pancreatic beta-cell functional compensation. Nat. Cell Biol. 16, 234–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stambolic V., Suzuki A., de la Pompa J. L., Brothers G. M., Mirtsos C., Sasaki T., Ruland J., Penninger J. M., Siderovski D. P., and Mak T. W. (1998) Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell 95, 29–39 [DOI] [PubMed] [Google Scholar]
- 36. Walpita D., and Wagner B. K. (2014) Evaluation of compounds in primary human islet cell culture. Curr. Protoc. Chem. Biol. 6, 157–168 [DOI] [PubMed] [Google Scholar]
- 37. Shen W., Tremblay M. S., Deshmukh V. A., Wang W., Filippi C. M., Harb G., Zhang Y. Q., Kamireddy A., Baaten J. E., Jin Q., Wu T., Swoboda J. G., Cho C. Y., Li J., Laffitte B. A., et al. (2013) Small-molecule inducer of beta cell proliferation identified by high-throughput screening. J. Am. Chem. Soc. 135, 1669–1672 [DOI] [PubMed] [Google Scholar]
- 38. Zhang J. H., Chung T. D., and Oldenburg K. R. (1999) A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4, 67–73 [DOI] [PubMed] [Google Scholar]
- 39. Ahuja D., Sáenz-Robles M. T., and Pipas J. M. (2005) SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene 24, 7729–7745 [DOI] [PubMed] [Google Scholar]
- 40. Stein J., Milewski W. M., and Dey A. (2013) The negative cell cycle regulators, p27(Kip1), p18(Ink4c), and GSK-3, play critical role in maintaining quiescence of adult human pancreatic beta-cells and restrict their ability to proliferate. Islets 5, 156–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. LaBaer J., Garrett M. D., Stevenson L. F., Slingerland J. M., Sandhu C., Chou H. S., Fattaey A., and Harlow E. (1997) New functional activities for the p21 family of CDK inhibitors. Genes Dev. 11, 847–862 [DOI] [PubMed] [Google Scholar]
- 42. Herrup K., and Yang Y. (2007) Cell cycle regulation in the postmitotic neuron: oxymoron or new biology? Nat. Rev. Neurosci. 8, 368–378 [DOI] [PubMed] [Google Scholar]
- 43. Avrahami D., Li C., Yu M., Jiao Y., Zhang J., Naji A., Ziaie S., Glaser B., and Kaestner K. H. (2014) Targeting the cell cycle inhibitor p57Kip2 promotes adult human beta cell replication. J. Clin. Invest. 124, 670–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang P., Alvarez-Perez J. C., Felsenfeld D. P., Liu H., Sivendran S., Bender A., Kumar A., Sanchez R., Scott D. K., Garcia-Ocaña A., and Stewart A. F. (2015) A high-throughput chemical screen reveals that harmine-mediated inhibition of DYRK1A increases human pancreatic beta cell replication. Nat. Med. 21, 383–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nir T., Melton D. A., and Dor Y. (2007) Recovery from diabetes in mice by beta cell regeneration. J. Clin. Invest. 117, 2553–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gupta R. K., Gao N., Gorski R. K., White P., Hardy O. T., Rafiq K., Brestelli J. E., Chen G., Stoeckert C. J. Jr., and Kaestner K. H. (2007) Expansion of adult beta-cell mass in response to increased metabolic demand is dependent on HNF-4α. Genes Dev. 21, 756–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Parsons J. A., Brelje T. C., and Sorenson R. L. (1992) Adaptation of islets of Langerhans to pregnancy: increased islet cell proliferation and insulin secretion correlates with the onset of placental lactogen secretion. Endocrinology 130, 1459–1466 [DOI] [PubMed] [Google Scholar]
- 48. Cano D. A., Rulifson I. C., Heiser P. W., Swigart L. B., Pelengaris S., German M., Evan G. I., Bluestone J. A., and Hebrok M. (2008) Regulated beta-cell regeneration in the adult mouse pancreas. Diabetes 57, 958–966 [DOI] [PubMed] [Google Scholar]
- 49. Lakey J. R., Warnock G. L., Rajotte R. V., Suarez-Alamazor M. E., Ao Z., Shapiro A. M., and Kneteman N. M. (1996) Variables in organ donors that affect the recovery of human islets of Langerhans. Transplantation 61, 1047–1053 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.