Abstract
1,1,1-Trichloro-2,2-bis(p-chlorophenyl)ethane (DDT), the first organochlorine insecticide, and pyrethroid insecticides are sodium channel agonists. Although the use of DDT is banned in most of the world due to its detrimental impact on the ecosystem, indoor residual spraying of DDT is still recommended for malaria control in Africa. Development of resistance to DDT and pyrethroids is a serious global obstacle for managing disease vectors. Mapping DDT binding sites is necessary for understanding mechanisms of resistance and modulation of sodium channels by structurally different ligands. The pioneering model of the housefly sodium channel visualized the first receptor for pyrethroids, PyR1, in the II/III domain interface and suggested that DDT binds within PyR1. Previously, we proposed the second pyrethroid receptor, PyR2, at the I/II domain interface. However, whether DDT binds to both pyrethroid receptor sites remains unknown. Here, using computational docking of DDT into the Kv1.2-based mosquito sodium channel model, we predict that two DDT molecules can bind simultaneously within PyR1 and PyR2. The bulky trichloromethyl group of each DDT molecule fits snugly between four helices in the bent domain interface, whereas two p-chlorophenyl rings extend into two wings of the interface. Model-driven mutagenesis and electrophysiological analysis confirmed these propositions and revealed 10 previously unknown DDT-sensing residues within PyR1 and PyR2. Our study proposes a dual DDT-receptor model and provides a structural background for rational development of new insecticides.
Keywords: molecular modeling, molecular pharmacology, receptor, sodium channel, toxicology, toxin, DDT, insect sodium channel, pyrethroids
Introduction
Voltage-gated sodium channels are transmembrane proteins that are critical for the initiation and propagation of action potentials in neurons and other excitable cells. In response to membrane depolarization, sodium channels open (activate) and allow sodium ions to flow into the cell, causing depolarization of the membrane potential. Activation of sodium channels is responsible for the rapidly rising phase of action potential. A few milliseconds after channel opening, the channel pore is occluded by an inactivation particle in the process known as fast inactivation that plays an important role in the termination of action potentials. Because of their crucial role in regulating membrane excitability, sodium channels are the primary target site of a broad range of neurotoxins, including insecticides.
The pore-forming α-subunit of the sodium channel consists of four homologous repeat domains (I–IV). Each domain has six transmembrane segments (S1–S6) that are connected by intracellular and extracellular loops (Fig. 1). The S1–S4 segments in each domain serve as the voltage-sensing modules, whereas the S5 and S6 segments from each of the four domains and the four extracellular membranes re-entering S5–S6 loops, known as P-regions, constitute the pore-forming module. In response to membrane depolarization, the S4 segments move in the extracellular direction, initiating conformational changes that lead to the pore opening. Short intracellular linkers L45, which connect segments S4 and S5 in each domain, are believed to transmit the movements of the voltage-sensing modules to the S6 segments during the channel gating.
FIGURE 1.
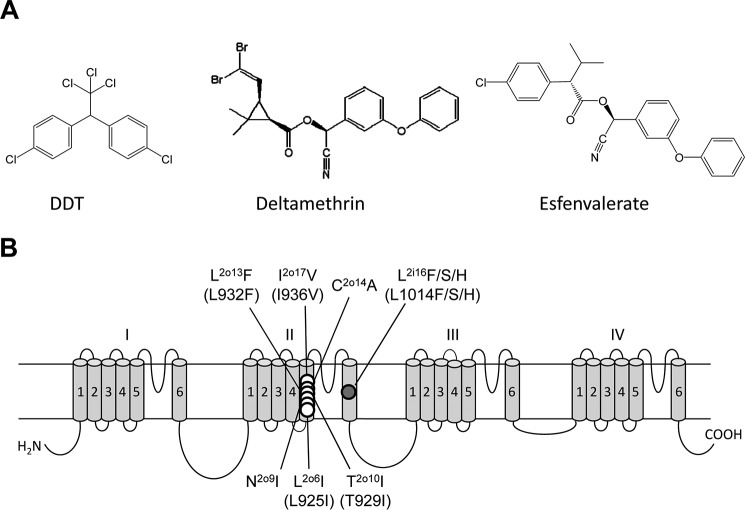
A, chemical structure of DDT, deltamethrin, and esfenvalerate. B, topology of the voltage-gated sodium channel indicating residues that have been experimentally confirmed to be critical for the action of DDT prior to this study. We use a residue-labeling scheme that is universal for P-loop channels (14, 33). A residue label includes the domain number (1–4), segment type (k, the linker-helix between S4 and S5; i, the inner helix S6; and o, the outer helix S5), and relative number of the residue in the segment (see Fig. 2). This provides the same labels to residues in the matching positions of the sequence alignment of sodium channels from different organisms whose genuine residue numbers are different. The scheme also highlights symmetric location of residues in different channel domains. Corresponding positions of naturally occurring kdr mutations (designated based on housefly numbering (GenBank accession number: AAB47604)) are indicated in parentheses.
1,1,1-Trichloro-2,2-bis(p-chlorophenyl)ethane (DDT)3 is an early generation lipid-soluble organochlorine insecticide (Fig. 1A). Because of its environmental toxicity, usage of DDT for pest control is restricted or forbidden in many countries, but DDT is permitted for control of disease vectors when safe, effective, and affordable alternatives are not available (1). DDT is produced in large amounts (mainly in India), and it is widely used in Africa. DDT is an agonist of voltage-gated sodium channels (2), and its mode of action of DDT is similar to that of pyrethroid insecticides, which are extensively used in the control of agricultural and urban arthropod pests and human disease vectors (3). DDT and pyrethroids, such as deltamethrin and esfenvalerate (Fig. 1A), stabilize the open conducting state of sodium channels by inhibiting their transition to non-conducting inactivated or deactivated states (4, 5).
Mutations in the sodium channel confer knockdown resistance (kdr) to DDT and pyrethroids and also provide valuable information for mapping the receptor sites of DDT and pyrethroids on sodium channels (6–8). The first computational study of binding of pyrethroids and DDT in the Kv1.2-based model of the house fly open sodium channel visualized the insecticides in the lipid-exposed side of the interface between domains II and III (9). We refer to this site as the first pyrethroid receptor, PyR1 (10). Superimposed three-dimensional structures of insecticide molecules, which target the sodium channel, suggest that the bulky trichloroethane moiety of DDT matches the dimethylcyclopropyl moiety of deltamethrin and the isopropyl moiety of fenvalerate (9), supporting the notion that these insecticides have overlapping binding sites. Six mutations in IIS5 that are critical for the action of pyrethroids are also confirmed to reduce the potency of DDT on insect sodium channels (Fig. 1B) (11, 12), suggesting that DDT and pyrethroids bind at the overlapping receptor site(s) within the sodium channel. These pioneering studies led to a model in which the trichloroethane moiety of DDT binds in the domain II/III interface (fenestration), between helices IIS5, IIP, and IIIS6, with one p-chlorophenyl ring oriented toward lipids and the other p-chlorophenyl ring extending along the IIS5 helix toward the kink between IIL45 and IIS5.
The classic kdr mutation L1014F (i.e. L2i16F) (Fig. 1B) in the housefly and in analogous positions in many other insects substantially decreases the action of pyrethroids and DDT (7, 8, 13), but Leu1014 does not contribute to PyR1 (9). Subsequently, we have shown that Leu1014 is located within a second pyrethroid receptor, PyR2, in the interface between domains I and II (14). In our PyR2 model, the dimethylcyclopropyl group of deltamethrin binds between helices IL45, IS5, IS6, and IIS6 below the fenestration. Our more recent study suggests a rotational symmetry of the two pyrethroid receptors, PyR1 and PyR2, and proposes a model in which two deltamethrin molecules simultaneously bind to these receptors (15). In the overlay of the two channel models (9, 15), the dimethylcyclopropyl groups of deltamethrin bound in the same domain interface are more than 12 Å away from each other. Because the trichloromethyl group of DDT and the dimethylcyclopropyl group of deltamethrin are believed to bind at the same loci, the two models imply essentially different positions of DDT. To resolve this problem, unbiased mapping of DDT binding sites is warranted.
In this study, we first employed a reduced x-ray structure of the open potassium channel, Kv1.2, to search for folding-specific regions in subunit interfaces to which DDT has the maximal shape complementarity. We further used a full-fledged Kv1.2-based model of the mosquito sodium channel AaNav1-1 and Monte Carlo energy minimizations to dock DDT from many starting positions and orientations that were randomly generated around the maximum shape-complementarity position found in the reduced Kv1.2 structure. Calculations predicted 14 sodium channel residues within the PyR1 and PyR2 sites that formed direct contacts with two DDT molecules. We mutated these as well as some other nearby residues in the mosquito sodium channel (AaNav1-1) and cockroach sodium channel (BgNav1-1a) and conducted functional analysis of the mutant channels in Xenopus oocytes. We found that 10 mutations significantly affected the DDT-induced inhibition of sodium channel inactivation, thus confirming predictions of our model. Our study elaborates the dual DDT-receptor model, confirms that DDT molecules bind to the pyrethroid receptors, PyR1 and PyR2, and provides a structural background for rational development of new insecticides.
Experimental Procedures
Computer Modeling
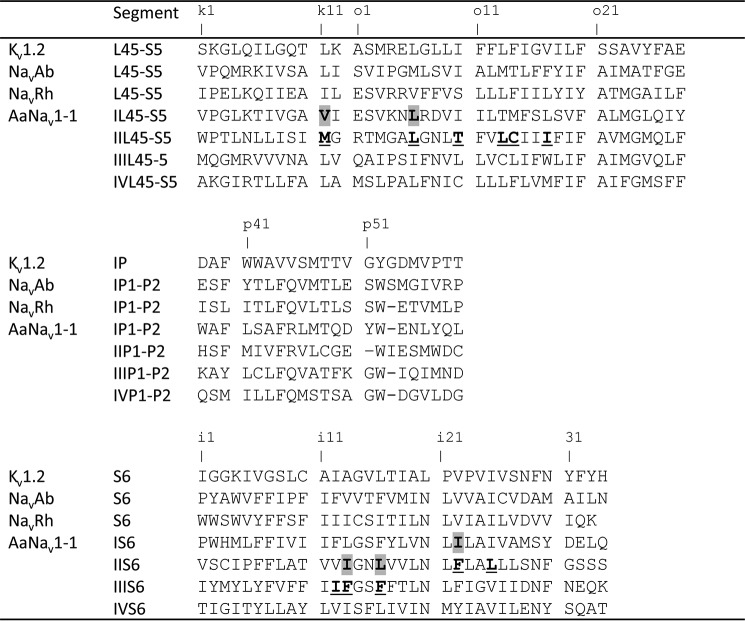
The x-ray structures of an open potassium channel, Kv1.2 (16), and a closed sodium channel, NavRh (17), were used as templates to build, respectively, the open and closed state models of the mosquito sodium channel AaNav1-1. The transmembrane topology of the sodium channel is shown in Fig. 1, and the sequence alignment of selected regions of different channels is shown in Fig. 2. A bacterial sodium channel NavMs is crystallized in a presumably open state (18), but we did not use the respective x-ray structure because it lacks the L45 linkers whose analogues in eukaryotic sodium channels contribute to pyrethroid receptors. Homology modeling and ligand docking were performed with the ZMM program (19) and the Monte Carlo minimization (MCM) protocol (20) as described in our previous study (14). Molecular images were created using the PyMOL Molecular Graphics System, Version 0.99rc6 (Schrödinger, LLC, New York).
FIGURE 2.
Sequence alignment of the pore module in Kv1.2 and sodium channels. Residues in the interfaces between domains II/II or I/II whose mutations affect DDT action are highlighted and underlined, respectively (see also Table 1).
Homology models of heterotetrameric asymmetric eukaryotic sodium channels are less precise than x-ray structures of homotetrameric symmetric ion channels. Therefore, an apparent global minimum found by hands-free docking of a ligand is unlikely to correspond to the real structure of the ligand-channel complex. In an attempt to resolve this problem, we have used a two-stage approach. At the first stage, we employed the x-ray structures of Kv1.2 and NavRh channels in which all side chains, except glycines, alanines, and prolines, were substituted by alanine (hereafter reduced x-ray structures), and we used these structures to search for folding-specific regions in the subunit interfaces with the maximal shape complementarity to DDT. In the reduced x-ray structures of both channels, these regions were found in interfaces between three helices of one subunit (L45, S5, and S6) and helix S6 of another subunit.
At the second stage, we used Monte Carlo minimizations to dock DDT in the full-fledged Kv1.2-based model of the mosquito sodium channel AaNav1-1 pore module. A thousand starting positions and orientations of DDT were randomly generated around its maximum-complementarity position found in the reduced Kv1.2 structure. We additionally focused the search by applying distance constraints between DDT and four residues (one from each of the helices IL45, IS5, IS6, and IIS6) that contribute to the maximal complementarity region in the reduced x-ray structures. Because specific atom-atom interactions between DDT and individual residues are unknown, we used ligand-side chain constraints. Each constraint specified the ligand (DDT), a channel residue, and the upper limit of the ligand side chain distance that was set at 5 Å. For each constraint, the closest pair of atoms between DDT and the side chain was selected in the beginning of each round of energy minimization so that the closest pair of atoms between DDT and a side chain may have been switched during the MCM trajectory. To preclude large deviations of the model backbones from the x-ray templates and thus preserve the channel folding during docking of the flexible ligand to the flexible protein, another set of distance constraints, pins, was imposed between matching α-carbons in the template and the model. A pin constraint is a flat-bottom parabolic energy function that allows an atom (in this study, an α-carbon) to deviate, penalty-free, up to 1 Å from the template and imposes a penalty of 10 kcal mol−1 Å−1 for larger deviations. The pin constraints are necessary because the initial relaxation of an unconstrained homology model with a bulky ligand would cause large deviations of the model backbones from the template due to steric clashes between the ligand and the channel. The MC minimization protocol optimized the system energy that included the penalty energy. For the final analysis, we considered low energy binding modes within 7 kcal/mol from the apparent global minimum. In all these binding modes, the penalty energy was equal to zero, indicating that all the distance constraints, including pins, were satisfied.
Site-directed Mutagenesis
We used a mosquito sodium channel, AaNav1-1, from Aedes aegypti or BgNav1-1a from Blattella germanica to generate all mutants used in this study. Site-directed mutagenesis was performed by polymerase chain reaction (PCR) using Phusion High-Fidelity DNA polymerase (New England Biolabs, Ipswich, MA). All mutagenesis results were confirmed by DNA sequencing.
Expression of insect sodium channels in Xenopus oocytes and electrophysiology. Procedures for preparation of oocytes and cRNA and injection were identical to those described previously (21). Methods and data analysis for two-electrode voltage clamp recording of sodium currents were identical to those previously described (21, 22). All experiments were performed at room temperature. Sodium currents were measured with an OC725C oocyte clamp (Warner Instruments, Hamden, CT) and a Digidata 1440A interface (Axon Instruments Inc., Foster City, CA); pCLAMP 10.2 software (Axon Instruments Inc.) was used for data acquisition and analysis.
The inhibitory effect of DDT on channel inactivation was assayed by measuring the remaining current at the end of a 20-ms depolarization to −10 mV from a holding potential of −120 mV and normalized to peak current. Oocytes expressing AaNav1-1 and BgNav1-1a were incubated in DDT solution for 3–4 h before the assay. DDT was kindly provided by Dr. Robert Hollingworth (Michigan State University) and was dissolved in dimethyl sulfoxide (DMSO) in a 100 mm stock. The working concentrations were prepared in ND96 recording solution immediately prior to experiments. The concentration of DMSO in the final solution was <0.5%, which had no effect on the function of expressed sodium channels.
Statistical Analysis
Results are reported as mean ± S.E. Statistical significance was determined by using one-way analysis of variance with Scheffe's post hoc analysis, and significant values were set at p < 0.05.
Results
Predicting Shape-complementarity Complexes between DDT and the Reduced X-ray Structure of Kv1.2
DDT is a bulky semi-rigid molecule. MC minimization of DDT yielded 20 conformations within 7 kcal/mol from the apparent global energy minimum. All the conformations are rather similar with some variations of the torsional angles C-Ph and C-CCl3 and bond angles (Fig. 3, A and B). Despite the rather inflexible three-dimensional structure of DDT, the relevance of the DDT-sodium channel model to the real structure is questionable due to several causes. First, a homology model of a heterotetrameric sodium channel cannot be as precise as the x-ray structures of the respective homotetrameric template. Second, DDT binding may change the side-chain conformations versus those in ligand-free homology models of the channel. Third, a millisecond molecular dynamics trajectory analysis of the Kv1.2 potassium channel (23) showed that the three-dimensional structure of the channel undergoes significant changes (breathings) during gating, and similar changes are expected to occur in sodium channels. Even if the full-fledged homology model of AaNav1-1 were precise, the straightforward search for the lowest energy complex between DDT and the flexible channel would require sampling and MC minimizations from a very large number of starting points, which is a huge computational job without a guaranteed success.
FIGURE 3.
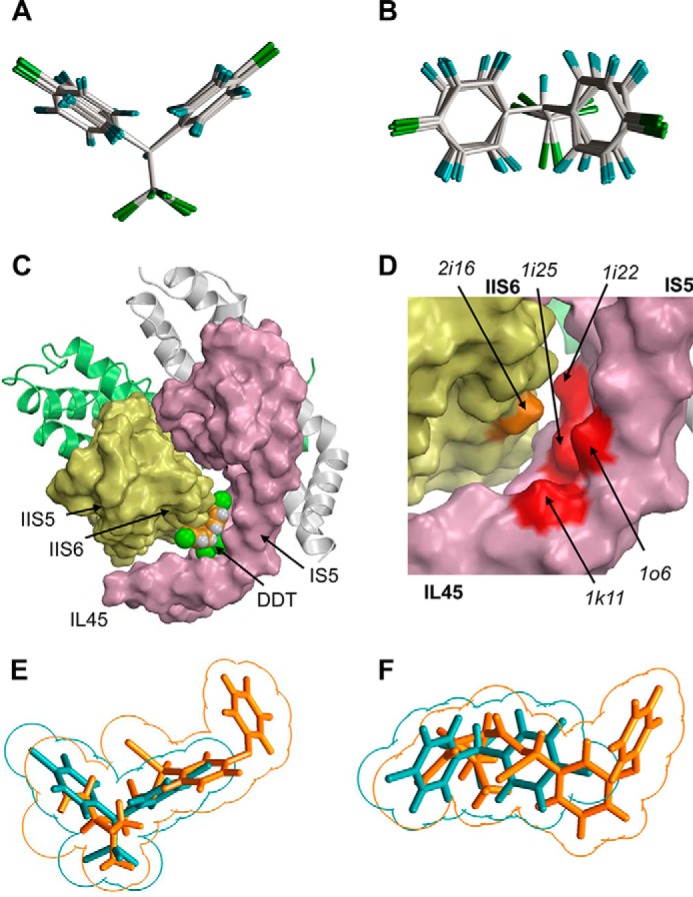
Predicting shape-complementarity complexes between DDT and the reduced x-ray structure of Kv1.2. A and B, orthogonal views of the global minimum conformation of DDT and local minimum conformations (within 7 kcal/mol from the global minimum). C, surface image of the x-ray structure of the open Kv1.2 channel in which all residues, except glycines, alanines, and prolines, are replaced with alanine to show potential ligand-binding clefts. The DDT molecule fits snugly into the interface between IL45, IS5, and S6. D, enlargement of C with the DDT molecule removed to show highlighted surfaces of some residues that contribute to the DDT-binding pocket. E and F, orthogonal views of the superimposed DDT (cyan) and deltamethrin (orange) molecules. Note an approximate size/shape similarity between the bulky hydrophobic dimethylcyclopropyl and C-CCl3 groups, leftward moieties bound to these groups, and similar angles at which two fragments of each ligand extend from of the bulky groups.
To address this problem we first searched for shape-complementarity complexes between DDT and a reduced Kv1.2 model (see “Experimental Procedures”). In this model, potential DDT binding sites are more accessible than in the full-fledged model. Therefore, the inter-subunit channel cavities and clefts, which are determined by the protein folding rather than conformations of specific side chains, provide potential ligand-binding sites where the ligand would experience minimal sterical repulsions from the channel and maximal van der Waals attractions to it. Following the strategy described elsewhere (19), we generated 10,000,000 starting points with random orientations of DDT and random positions of its mass center within a 30 × 30 × 30 Å cube. The latter was centered at the widest point of the fenestration between different Kv1.2 subunits, i.e. between two S6 helices and one P-helix. In each point, the energy was calculated but not minimized. In the vast majority of the starting points, DDT clashed with the channel, and these were filtered out. However, even the lowest energy starting point had a positive energy due to DDT-channel repulsions that are unavoidable without energy minimizations. Next, we MC-minimized 1000 lowest energy structures accumulated from the 10,000,000 starting points in 1000 independent MCM trajectories. The protein conformation was kept rigid, whereas DDT position, orientation, and conformation were allowed to vary. Each trajectory was terminated when 100 consecutive energy minimizations did not decrease the energy of the lowest energy structure found in the trajectory. The lowest energy complex found in the 1000 trajectories is shown in Fig. 3C. The bulky trichloroethane moiety of DDT fits snugly between helices L45, S5, and S6 in subunit I and helix S6 of neighboring subunit II. (Because of the 4-fold symmetry of Kv1.2, any subunit can be assigned number I, but number II would denote a neighboring subunit that at the extracellular view is clockwise to subunit I.) The two p-chlorophenyl rings bound in the interfaces between helices IS5 and IL45 and helix IIS6. Importantly, the angle between the p-chlorophenyl rings matched that between helices IL45 and IS5. The DDT-Kv1.2 energy in this structure is −13.4 kcal/mol. No alanine residue experienced repulsion from DDT, and the energetic favorability of the complex was due to the van der Waals attractions between the channel and DDT. In other words, the complex had a large number of contacts between DDT and the channel and the maximal shape complementarity between DDT and the reduced Kv1.2 structure. Because of the 4-fold symmetry of the channel, the same maximal complementarity complexes are expected in the four interfaces between Kv1.2 subunits.
Fig. 3D shows some alanine residues (positions 1k11, 1o6, 1i22, 1i25, and 2i16) in the domain I/II interface of the reduced Kv1.2 model that contribute to the DDT binding site. These positions correspond to PyR2 residues, which interact with the dimethyl-cyclopropyl group of deltamethrin (14, 15). This result supports the notion that the binding sites for DDT and pyrethroids may overlap and are consistent with the fact that at the maximum overlap superposition of the ligands, the dimethyl-cyclopropyl group of deltamethrin (Fig. 1A) coincides with the trichloromethyl group of DDT (Fig. 3, E and F).
Predicting Shape-complementarity Complexes between DDT and the Reduced X-ray Structure of NavRh
Both DDT and pyrethroids inhibit deactivation and inactivation of sodium channels and stabilize the open state, causing prolonged channel opening. Type II pyrethroids, like deltamethrin, preferably bind to open sodium channels, whereas type I pyrethroids, like permethrin, can also modify resting or inactivated channels (24). In terms of the chemical structure, DDT is more similar to type I pyrethroids than to type II pyrethroids, and DDT is believed to bind to the closed channel and stabilize the open state upon channel opening (25). However, it remains unknown whether or not DDT binds to the same region in the open and closed channel states.
To address this question, we searched for the DDT binding site in the closed channel by using the reduced x-ray structure of a bacterial sodium channel NavRh (17), which, despite certain asymmetry of the channel tetramer, appears a better template for homology modeling of eukaryotic channels than the x-ray structure of another bacterial sodium channel, NavAb (26). Indeed, in the aligned sequences of AaNav1-1 (domains I, II, and IV), NavRh, and Kv1.2, all the channels have a serine or threonine residue in position o2 at the N-end of theS5 helix (Fig. 2). It is well known that an H-bond between the serine/threonine side chain and the main-chain carbonyls in the preceding helical turn may cause a kink in the α-helix (27). These H-bonds appear to cause the kinks between the L45 and S5 helices in the x-ray structures of NavRh, NavAb, and Kv1.2.
We searched for the maximal shape complementarity for the DDT binding site in the reduced structure of NavRh by using the same approach as with Kv1.2. Calculations predicted the lowest energy complex in which DDT fits between helices IL45, IS5, IS6, and IIS6 (Fig. 4) in a way that is similar to that found in the reduced Kv1.2 structure. This result suggested that the maximal shape-complementarity binding sites for DDT in the closed and open channel states are located in the same channel regions, implying that upon opening DDT would simply readjust its position, not moving significantly, to fit better in the open-state binding site, thus stabilizing the open channel. Therefore, our subsequent efforts focused on exploring the binding site of DDT using the Kv1.2-based model of the open AaNav1-1 channel, which is available from our previous study (14).
FIGURE 4.
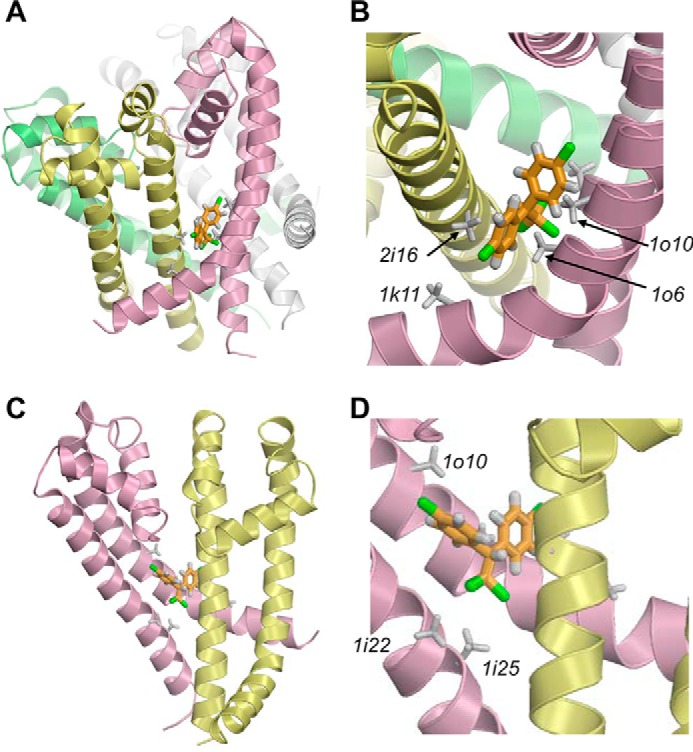
Lowest energy complex of DDT with the reduced x-ray structure of the NavRh channel pore module in which all residues, except glycine, alanine, and proline, are substituted by alanine. A and B, complex and its enlarged views from lipids. C and D, complex and its enlarged views from the pore with the two front subunits removed for clarity.
Docking of DDT in the Kv1.2-based Model of AaNav1-1
Initially, we generated 1000 starting positions and orientations of DDT within a 20 × 20 × 20-Å cube whose center corresponds to the mass center of DDT in the maximum shape-complementarity position in the I/II domain interface of the reduced Kv1.2 model. We additionally biased the search toward the interface of helices IL45, IS5, IS6, and IIS6 by applying distance constraints between DDT and side chains of residues Val1k11, Leu1o6, Ile1i25, and Leu2i16 whose alanine substitutions contribute to the DDT binding site in the reduced model of the Kv1.2 x-ray structure (Fig. 3D). Each trajectory was terminated when 100 consecutive energy minimizations did not decrease the lowest energy found in the trajectory. Then we docked the second DDT molecule in the II/III domain interface by using the same protocol with distance constraints between DDT and side chains of residues Met2k11, Leu2o6, Leu2i25, and Phe3i16. Finally, we optimized the channel complex with two DDT molecules in a long MCM trajectory without any constraints. This trajectory was terminated when 2000 consecutive energy minimizations did not decrease the energy of the apparent global minimum found.
The predicted lowest energy complex of the channel with two DDT molecules is shown in Fig. 5. Both ligands fit snugly in the respective domain interfaces and establish favorable interactions with many hydrophobic residues. Because of the sequential asymmetry of individual AaNav1-1 domains, the ligand-binding modes in the I/II and II/III domain interfaces are similar but not identical. Despite that the DDT binding sites in the full-fledged sodium channel model are obviously tighter than analogous sites in the reduced Kv1.2 structure, none of the sodium channel residues provided a positive (repulsive) contribution to the DDT-channel energy. The interaction energies of the channel with DDT molecules bound in the I/II and II/III domain interfaces were −27.3 and −25.5 kcal/mol, respectively, indicating high complementarity of the ligands to their binding sites.
FIGURE 5.
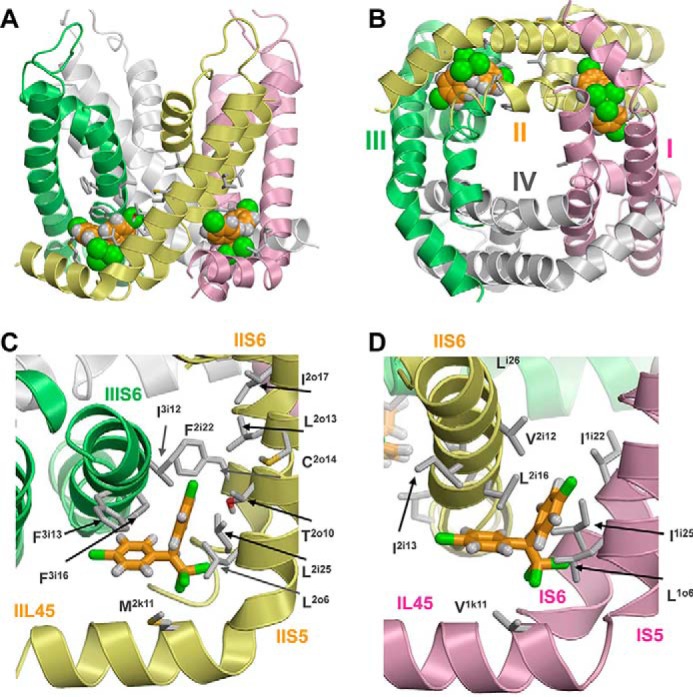
Kv1.2-based model of the open AaNav1-1 channel pore module with two DDT molecules. Domains I–IV are shown by pink, yellow, green, and white ribbons, respectively. Known DDT-sensing residues are shown as sticks. A and B, side and cytoplasmic views of the AaNav1-1 channel with two DDT molecules (space-filled) docked in the lipid-exposed domain interfaces II/III and I/II. C, side view of the domain II/III interface along helix IIIS6. D, side view of the domain I/II interface along helix IIS6.
Common features of the DDT-binding modes in the I/II and II/III domain interfaces include location of the trichloromethyl group between the helices L45, S5, and S6 in one domain and helix S6 of the “next” domain, binding of one p-chlorophenyl group between the L45 and S6 helices of two domains, and binding of the second p-chlorophenyl group between an S5 helix and two helices S6. It should be noted that the results of DDT docking in the reduced Kv1.2 model allowed us to focus the search within domain interfaces, but specific positions and orientations of the ligands within these interfaces were determined by the energetics of the interactions between DDT and the channel. In addition to the binding modes shown in Fig. 5, energetically and geometrically similar binding modes are found in the MCM protocol (data not shown). Calculations predicted a total of 14 residues within the domain I/II and II/III domain interfaces that formed direct contacts with DDT (Table 1).
TABLE 1.
Effects of mutations within and around the PyR1 and PyR2 receptors of insect sodium channels on the action of DDT
The following symbols are used: ↓, ↑, and ≈ indicate, respectively, decrease, increase, or insignificant change of the ligand potency.
| PyR1 |
PyR2 |
||||
|---|---|---|---|---|---|
| Mutant | Ref.a | Effect | Mutant | Ref.a | Effect |
| L2k7(914)F/I | 11 | = | |||
| M2k11(918)T | 11 | ↑ | V1k11A | This study | ↓ |
| L2o6(925)I | 11 | ↓ | L1o6I | This study | ↑ |
| T2o10(929)I | 11 | ↓ | I1o10C | This study | ≈ |
| L2o13(932)F | 11 | ↓ | T1o13A | This study | ≈ |
| C2o14(933)A | 11 | ↓ | |||
| I2o17(936)V | 11 | ↓ | |||
| F2i22S | This study | ↓ | I1i22A | This study | ↑ |
| L2i25A | This study | ↓ | I1i25A | This study | ≈ |
| I3i12A | This study | ↓ | V2i12A | This study | ≈ |
| F3i13C | This study | ↓ | I2i13M | This study | ↓ |
| F3i16A/S | This study | ↓ | L2i16F | This study, 9 | ↓ |
Identifying New DDT-sensing Residues within PyR1 and PyR2 of Insect Sodium Channels
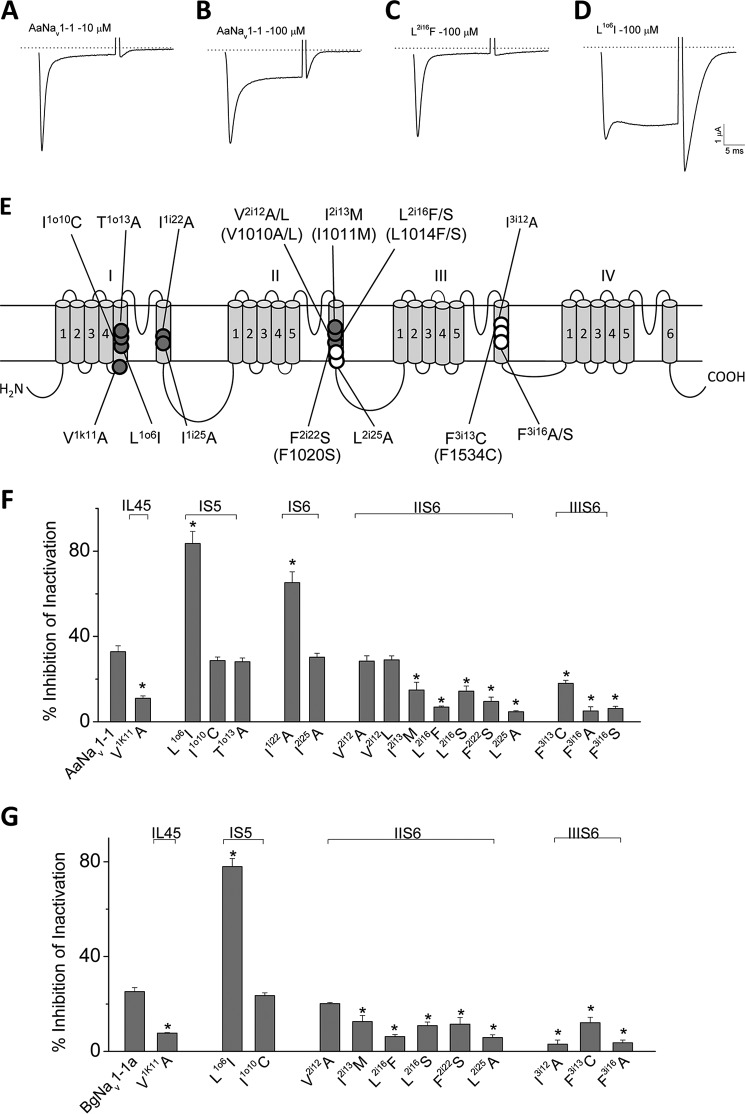
To test our model, we generated 16 mutants of the AaNav1-1 channel and functionally characterized these mutants in Xenopus oocytes (Fig. 6). Twelve mutants were examined in BgNav1-1a. Eleven of them were identical to those in the AaNav1-1 background, but one mutant, I3i12A, was examined only in BgNav1-1a channels (Fig. 6). Most of the mutations we made in this study were alanine substitutions or naturally occurring kdr mutations with a few exceptions, such as I1o10C, which has been shown to affect pyrethroid action in a previous study (Du et al. (14)). DDT inhibits inactivation of AaNav1-1 channels in a dose-dependent manner as shown in Fig. 6, A and B. Channel sensitivity to DDT was assessed by determining the percentage of inhibition of inactivation by DDT (28). Representative current traces from a DDT-resistant channel, L2i16F, and a channel with enhanced sensitivity to DDT, L1o6I, are presented in Fig. 5, C and D. Besides the L2i16F channels, an additional eight mutant channels were more resistant to DDT in both AaNav1-1 (Fig. 6F) and/or BgNav1-1a (Fig. 6G) backgrounds, whereas L1o6I and I1i22A channels were more sensitive to DDT compared with the wild-type channels. These experiments strongly support the existence of two DDT binding sites on sodium channels and the involvement of four helices in each of the DDT binding sites.
FIGURE 6.
Effects of DDT on AaNav1-1 and BgNav1-1a and mutant channels. A–D, representative sodium current traces elicited by a 20-ms depolarization from −120 to −10 mV from AaNav1-1, L2i16F, and L1o6I sodium channels after DDT treatment (10 or 100 μm). E, topology of a sodium channel indicating the position of all mutations that were tested in this study. Residues in PyR1 and PyR2 are indicated with open and solid circles, respectively. Corresponding positions of naturally occurring kdr mutations in the AaNav1-1or BgNav1-1a channel are indicated in parentheses. F and G, percentages of channel inactivation inhibited by DDT (100 μm). Eleven pairs of mutations were tested in both AaNav1-1or BgNav1-1a channels. The number of oocytes for each mutant construct was >8. Error bars indicate mean ± S.E. The asterisks indicate significant differences (p < 0.05) in sensitivity of mutants versus wild type to DDT as determined using one-way analysis of variance with Scheffé's post hoc analysis.
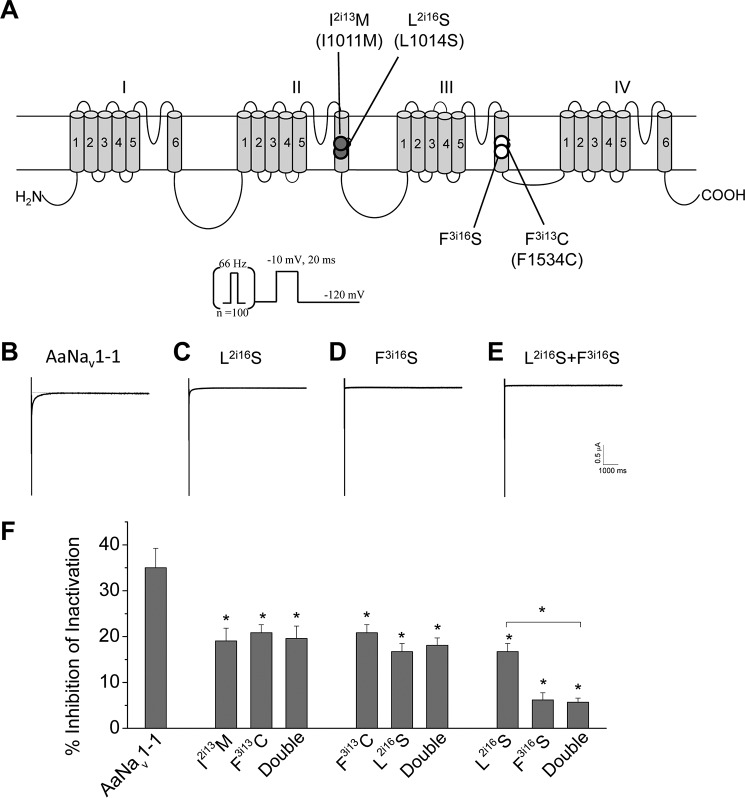
Furthermore, we made three double mutants I2i13M/F3i13C, L2i16S/F3i13C, and L2i16S/F3i16S, in which one residue in each binding site was mutated, and we compared the DDT sensitivity of these channels with respective single mutation channels (Fig. 7). The DDT-induced tail current in the wild-type channels was very small (less than 1 μA even at the concentration of 100 μm DDT) and also decayed extremely rapidly (Fig. 7B). The double mutations almost completely abolished the tail currents (Fig. 7B), indicating that these mutations also affected the modification of sodium channel deactivation by DDT. For the quantitative analysis of the mutations shown in Figs. 6 and 7, we focused on the effect of DDT on channel inactivation. The levels of DDT resistance of the I2i13M/F3i13C and F3i13C/L2i16S double mutation channels were similar to their respective single mutation channels (Fig. 7C). The third double mutant, L2i16S/F3i16S, was more resistant to DDT than one of the single mutants, L2i16S, but was as resistant as the other single mutant F3i16S (Fig. 7C). These results indicate that the effects of these double mutations on DDT action are not additive.
FIGURE 7.
Effects of double mutations (one from each receptor site) on DDT inhibition of AaNav1-1 channels. A, topology of a sodium channel indicating the position of four mutations that were made into double mutations in three combinations. Residues in PyR1 and PyR2 are indicated with open and solid circles, respectively. Corresponding positions of naturally occurring kdr mutations in the AaNav1-1or BgNav1-1a channel are indicated in parentheses. B–E, representative tail currents induced by DDT (100 μm) from AaNav1-1 and mutant channels. The protocol that was used to elicit tail currents is presented above the tail current traces. F, percentages of channel inactivation inhibited by DDT (100 μm). Error bars indicate mean ± S.E. The asterisks indicate significant differences in DDT sensitivity of mutants in comparison with the wild type, and a significant difference in DDT sensitivity of the L2i16S/F3i16S channel in comparison with the L2i16S channel was determined using one-way analysis of variance with Scheffé's post hoc analysis (p < 0.05).
Docking Esfenvalerate in the II/III Domain Interface
A common feature between insecticides, which stabilize the sodium channel open state, is the presence of a large hydrophobic group, e.g. trichloromethyl in DDT and dimethylcyclopropyl in pyrethroids. Another pyrethroid insecticide, esfenvalerate (Fig. 1A), includes isopropyl and p-chlorophenyl moieties that at the superposition with DDT match its trichloromethyl and p-chlorophenyl moieties (9). Here, we systematically searched for the lowest energy esfenvalerate binding mode in the domain II/III interface using the same approach as for DDT. Calculations predicted the lowest energy complex in which the ligand interacts with several residues, including Met2k11, Leu2o6, Phe3i16, and Phe3i17, and its isopropyl group fits between helices IIL45, IIS5, IIS6, and IIIS6 (Fig. 8). Super-kdr mutation M2k11T, which increases the resistance of housefly sodium channels to fenvalerate by 170-fold (29), is within the predicted binding site. Recently, several kdr mutations, including F3i17L, were detected in Meligethes aeneus, a pollen beetle population resistant to esfenvalerate (30). Residue Phe3i17 forms close contact with esfenvalerate in our model (Fig. 8). Results of these calculations provide additional evidence that the region between helices L45, S5, and S6 of one domain and S6 in the next domain is attractive for a bulky hydrophobic moiety, which is a fingerprint of different sodium channel agonists that are used as insecticides.
FIGURE 8.
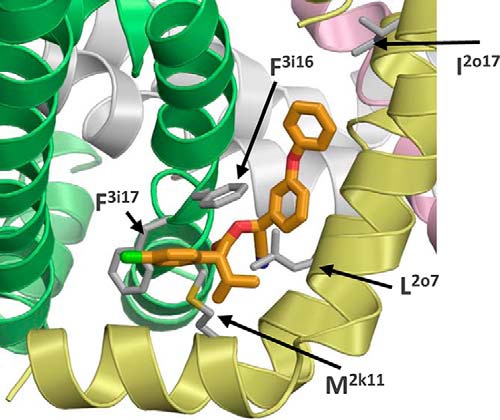
Docking of esfenvalerate in the PyR1 site of the Kv1.2-based models of AaNav1-1. The isopropyl moiety binds in the IIL45-IIS5 kink region that accommodates the dimethyl-cyclopropyl group of pyrethroids. The 4-chlorphenyl moiety binds in the interface between IIL45/IIIS6 similarly to the same group of DDT, where it interacts with the esfenvalerate-sensing residue Phe3i17 (30) as well as with Met2k11 whose substitution is identified as the super-kdr mutation that increases resistance of the housefly sodium channel to esfenvalerate by 170-fold (29).
Discussion
Dual Receptor Sites for DDT in Insect Sodium Channels
DDT shares several common structural features with pyrethroids. Thus, mapping DDT binding site(s) in sodium channels is important for a better understanding of atomic mechanisms of action of the sodium channel agonists and developing new insecticides. In this study, we used two computational approaches to predict DDT-channel complexes. The first approach included a systematic search for maximal shape-complementarity complexes with DDT using the reduced x-ray structures of the Kv1.2 and NavRh channels. The search defined the region for intensive MCM docking of DDT in the full-fledged pore module model of the open AaNav1-1 channel. Our computations predicted that the bulkiest moiety of DDT, trichloromethyl, fits in the region between L45, S5, and two S6 helices. The angle between the planes of the two p-chlorophenyl rings approximately matches that between the L45 and S5 helices of one domain, and the opposite faces of the p-chlorophenyl moieties embrace the S6 helix of the next domain. To some extent, this binding mode resembles that of pyrethroid insecticides whose dimethylcyclopropyl groups bind in the same region where the trichloromethyl moiety of DDT binds. Our calculations predicted that two deltamethrin molecules can bind within two pyrethroid receptors, PyR1 and PyR2, that we recently described (14, 15).
Functional characterization of 16 single mutation channels of the AaNav1-1 channel and 12 single mutation channels of the cockroach channel BgNav1-1 showed that analogous point mutants of both channels displayed similar changes in DDT sensitivity versus the wild-type channels (Fig. 6). Our experiments revealed five new DDT-sensing residues in the domain II/III interface and five new DDT-sensing residues in the domain I/II interface. All these residues also contribute to either PyR1 or PyR2 (14, 15). The DDT-binding modes suggested by our calculations are consistent with the maximal shape-complementarity complex of DDT with the reduced x-ray structure of Kv1.2. Several hydrophobic residues form direct contacts with DDT in the domain I/II interface. In the domain II/III interface, all DDT-interacting residues, except Thr2o10, are also hydrophobic. One cannot completely rule out that the tested mutations affect DDT action allosterically, but in our view this is a low probability scenario.
Our analysis of double-mutation channels carrying one mutation in each receptor site revealed that the effects of these double mutations were not additive (Fig. 7). If DDT binding to each receptor independently contributed to the inhibition of sodium channel inactivation, we would have observed the effects of point mutations being summed in the double mutants. The double mutation results suggest that the observed inhibitory effects on sodium channel inactivation is due to simultaneous binding of two DDT molecules to the two receptor sites.
Common and Different Features of DDT Binding to Initial and Revised Models of the Pyrethroid Receptor PyR1
The initial pioneering PyR1 model predicts that the trichloromethyl group of DDT binds in the widest part of the fenestration between domains II and III; one chlorophenyl group interacts with IIS5; another chlorophenyl group interacts with IIIS6; and unlike pyrethroids, DDT is far from the linker helix IIL45 (9). A subsequent mutational study confirmed the presence of DDT-sensing residues in IIS5 (11), and the contribution of IIIS6 to the DDT is also a correct prediction of the initial PyR1 model.
There are several features of DDT binding that are different between the revised PyR1 model and the initial PyR1 model (9, 11). First, in the initial DDT-binding model, the trichloromethyl group of the ligand binds in the widest part of the fenestration between domains II and III, whereas in our model this group binds between helices IIL45, IIS5, IIS6, and IIIS6. Second, in the initial PyR1 model DDT interacts with Cys2o14 and Ile2o17 in the middle part of IIS5, whereas in our model Cys2o14 and Ile2o17 are rather far from the ligand (see details below). Third, in the initial PyR1 model, DDT binds at the protein surface and interacts with only two helices, IIS5 and IIIS6, whereas in our model DDT binds deeply in the domain interface and interacts with four helices (IIL45, IIS5, IIS6, and IIIS6).
Mutation I2o17V deteriorates DDT potency in the housefly. The same mutation was recently described as a kdr mutation in pollen beetle populations that are resistant to esfenvalerate (30). According to our model, residue Ile2o17 does not contribute to the DDT binding site (Fig. 5C) and is located rather far from the ligand in esfenvalerate-channel complexes (Fig. 8). Isoleucine Ile2o17 forms close hydrophobic contacts with four residues in helix IIP1, namely Phe2p40, Val2p43, Phe2p44, and Leu2p47 (Fig. 9B). We suggest that substitution of Ile2o17 by a smaller valine would shift helix IIS5 toward helices IIP and IIIS6, thus tightening the domain II/III interface and deforming the PyR1 geometry where DDT and esfenvalerate bind.
FIGURE 9.
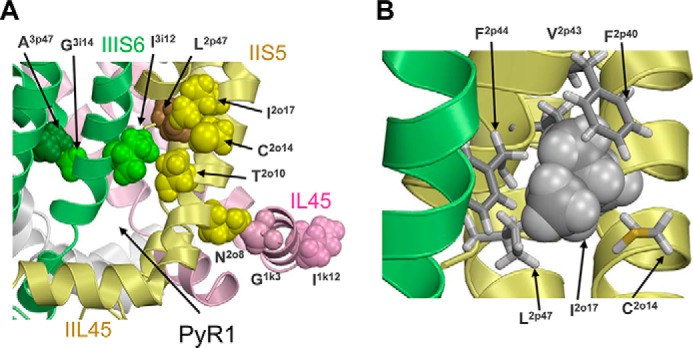
DDT-sensing residues beyond the DDT receptors that are involved in inter-segment contacts with residues whose mutations may allosterically affect the action of DDT. A, in the Kv1.2-based model of the open AaNav1-1 channel, the DDT-sensing residues Ala3p47, Asn2o8, and Ile2o17 make tight contacts with residues Gly3i14, Gly1k3, and Leu2p47, respectively. Substitution of the DDT-sensing residues may change the contacts and thus cause shifts in the position of helices IIS5 and IIIS6 that contribute to PyR1. DDT-sensing residues Ile3i12 and Thr2o10 at the top of the DDT-binding pocket contact each other, and substitution of these residues may affect DDT binding directly and/or allosterically, through changing the distance between the IIS5 and IIIS6 helices. B, enlarged view of isoleucine Ile2o17 (space-filled), which is involved in intersegment contacts with Phe2p40, Val2p43, Phe2p44, and Leu2p47 and intrasegment contact with Cys2o14.
Mutation C2o14A decreased the sensitivity of channels to DDT. In our model, Cys2o14 is located not far from DDT bound to PyR-1, but it does not form direct contacts with DDT (Fig. 5). We suggest that alanine substitution of the large cysteine (Cys2o14) would provide more freedom for the Ile2o17 side chain (Fig. 9B), permitting it to change conformation, and release a tight contact with Leu2p47 and other hydrophobic residues in the IIP1 helix. Thus, the consequence of the C2o14A mutation would be allosteric and similar to that of the I2o17V mutation.
Mutation N2o8I (11) is located on the IIS5 face that is opposite to the face, which accommodates the DDT-sensing residues Leu2o6 and Thr2o10. There is a close interdomain contact of Asn2o8 with the backbone of Gly1k1 in IL45 (Fig. 9A). Substitution of Asn2o8 by a bigger isoleucine may shift IIS5 toward IIIS6, thus shifting the DDT-sensing residues within the PyR1 site. Another possible consequence of mutation N2o8I is that a shift of Gly1k1 would affect IL45 that contains a DDT-sensing valine within the PyR2 site (Table 1).
Rotational Quasi-symmetry of the DDT-binding Sites in Insect Sodium Channels
Fig. 5B illustrates rotational quasi-symmetry of DDT binding sites within PyR1 and PyR2. Clockwise rotation of the cytoplasmic view by 90° would overlay DDT that is bound in the domain II/III interface (PyR1) directly over DDT bound to the domain I/II interface (PyR2). The geometric quasi-symmetry of the DDT binding sites is consistent with some, but not all, changes in potency of DDT following mutations in symmetric positions. Thus, mutations in positions 2i16/3i16 decrease the channel sensitivity to DDT, and the same is true for positions 2i13/3i13 (Table 1). Mutations in several other symmetric positions (2k11/1k11, 1o6/2o6, and 2i22/1i22) also affect the activity of DDT, suggesting that respective residues do interact with DDT. However, mutations at these symmetric positions have opposite effects on the activity of DDT (Table 1). This may be due to the different nature of the wild-type residues and their substitutions at the symmetric positions, as well as significant sequential asymmetry of the four channel domains that likely underline the structural asymmetry of the heterotetrameric channels. These factors may also explain why mutations T2o10I, L2o13F, F2i22S, and L2i25A within PyR1, but not mutations I1o10C, T1o13A, I1i25A, and V2i12A in analogous positions within PyR2, decreased the sensitivity of channels to DDT (Table 1). Given the limited precision of the homology model, we did not attempt to suggest structural interpretation of these data. However, taken together our experimental and modeling data strongly support the existence of two distinct DDT binding sites in the interfaces between domains I/II and II/III and certain symmetry of the DDT-binding modes at these sites.
DDT Binding within the PyR2 Site May Contribute to DDT-selective Toxicity
There are several differences in the PyR1 or PyR2 sites between insect and mammalian sodium channels that may contribute to the selective toxicity of DDT between insects and mammals. One of them is at position 2k11 in the IIL45 linker helix, which contributes to the PyR1 site in insects. Mutation I2k11M significantly enhances sensitivity of rat sodium channels, rNav1.2 and rNav1.4, to pyrethroids (31, 32), whereas the kdr mutation M2k11T decreases the sensitivity of insect sodium channels to pyrethroids (29). Intriguingly, M2k11T increases rather than decreases the sensitivity of insect sodium channels to DDT (11, 12). In our models both DDT and esfenvalerate (the active isomer of fenvalerate) approach Met2k11 with their bulky hydrophobic moieties (cf. Figs. 5C and 8). We suggest that the electrostatic repulsion between the sulfur atom of Met2k11 and chlorine atoms in the trichloromethyl group of DDT may bring destabilizing contributions to the DDT-channel energy, whereas the Thr2k11 hydroxy group of the mutant can attract the trichloromethyl moiety of DDT. In contrast to DDT, esfenvalerate forms a close hydrophobic contact with Met2k11 (Fig. 8). Thus, our model is consistent with these observations and explains the opposite effects of the M2k11T mutation on the action of DDT and fenvalerate, despite their occupying the same receptor site. Furthermore, the model supports the experimental data (11, 12) according to which the interaction of DDT with Met2k11 is unlikely to contribute to the selective toxicity of DDT.
Another position where insect and mammalian sodium channels have different residues is 1k11 within PyR2, Val1k11 in insect, and Leu1k11 in rat channels (Table S6 in Ref. 14). In our model both methyl groups of V1k11 interact with a chlorine atom of DDT bound in PyR2 (Fig. 5D), and we demonstrated that the mutation V1k11A significantly decreased the sensitivity of insect sodium channels to DDT (Fig. 6). Thus, our data suggest that DDT binding within PyR2 of the insect sodium channel may contribute to the observed selective toxicity of DDT between insects and mammals.
Conclusions
In this study, we discovered a new DDT binding site in the interface between domains I and II by elaborating the atomic model of the mosquito sodium channel with two DDT molecules bound within the pyrethroid receptors, PyR1 and PyR2, and performing model-driven mutagenesis. Together with findings from previous studies, our results suggest that simultaneous binding of two sodium channel agonists in the lipid-exposed domain interfaces of the insect sodium channel is necessary to exert the potent insecticidal action.
Author Contributions
B. S. Z. and K. D. participated in research design. Y. D., Y. N., and B. S. Z. conducted experiments and performed data analysis. Y. D., B. S. Z., and K. D. wrote or contributed to the writing of the manuscript
Acknowledgments
We thank Dr. Kris Silver for critical review of this manuscript. Computations were performed using the facilities of the Shared Hierarchical Academic Research Computing Network.
This work was supported by the National Institutes of Health Grant GM057440 from NIGMS (to K. D. and B. S. Z.) and the Natural Sciences and Engineering Research Council of Canada Grant RGPIN-2014-04894 (to B. S. Z.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- DDT
- 1,1,1-trichloro-2,2-bis(p-chlorophenyl) ethane
- AaNav1-1
- mosquito sodium channel from A. aegypti
- BgNav1-1a
- cockroach sodium channel from B. germanica
- L45
- linker connecting S4 and S5 of sodium channels
- MC
- Monte Carlo
- MCM
- Monte Carlo minimization
- PyR1
- pyrethroid receptor site 1
- PyR2
- pyrethroid receptor site 2
- kdr
- knockdown resistance.
References
- 1. van den Berg H. (2009) Global status of DDT and its alternatives for use in vector control to prevent disease. Environ. Health Perspect. 117, 1656–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hille B. (1968) Pharmacological modifications of the sodium channels of frog nerve. J. Gen. Physiol. 51, 199–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Soderlund D. M., and Bloomquist J. R. (1989) Neurotoxic actions of pyrethroid insecticides. Annu. Rev. Entomol. 34, 77–96 [DOI] [PubMed] [Google Scholar]
- 4. Vijverberg H. P., and van den Bercken J. (1990) Neurotoxicological effects and the mode of action of pyrethroid insecticides. Crit. Rev. Toxicol. 21, 105–126 [DOI] [PubMed] [Google Scholar]
- 5. Bloomquist J. R. (1996) Ion channels as targets for insecticides. Annu. Rev. Entomol. 41, 163–190 [DOI] [PubMed] [Google Scholar]
- 6. Davies T. G., Field L. M., Usherwood P. N., and Williamson M. S. (2007) DDT, pyrethrins, pyrethroids, and insect sodium channels. IUBMB Life 59, 151–162 [DOI] [PubMed] [Google Scholar]
- 7. Dong K., Du Y., Rinkevich F., Nomura Y., Xu P., Wang L., Silver K., and Zhorov B. S. (2014) Molecular biology of insect sodium channels and pyrethroid resistance. Insect Biochem. Mol. Biol. 50, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Soderlund D. M. (2005) in Comprehensive Molecular Insect Science (Gilbert L. I., Iatrou K., and Gill S. S., eds) pp. 1–24, Elsevier, Amsterdam [Google Scholar]
- 9. O'Reilly A. O., Khambay B. P., Williamson M. S., Field L. M., Wallace B. A., and Davies T. G. (2006) Modelling insecticide-binding sites in the voltage-gated sodium channel. Biochem. J. 396, 255–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang L., Nomura Y., Du Y., Liu N., Zhorov B. S., and Dong K. (2015) A mutation in the intracellular loop III/IV of mosquito sodium channel synergizes the effect of mutations in helix IIS6 on pyrethroid resistance. Mol. Pharmacol. 87, 421–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Usherwood P. N., Davies T. G., Mellor I. R., O'Reilly A. O., Peng F., Vais H., Khambay B. P., Field L. M., and Williamson M. S. (2007) Mutations in DIIS5 and the DIIS4-S5 linker of Drosophila melanogaster sodium channel define binding domains for pyrethroids and DDT. FEBS Lett. 581, 5485–5492 [DOI] [PubMed] [Google Scholar]
- 12. Usherwood P. N., Vais H., Khambay B. P., Davies T. G., and Williamson M. S. (2005) Sensitivity of the Drosophila para sodium channel to DDT is not lowered by the super-kdr mutation M918T on the IIS4-S5 linker that profoundly reduces sensitivity to permethrin and deltamethrin. FEBS Lett. 579, 6317–6325 [DOI] [PubMed] [Google Scholar]
- 13. Burton M. J., Mellor I. R., Duce I. R., Davies T. G., Field L. M., and Williamson M. S. (2011) Differential resistance of insect sodium channels with kdr mutations to deltamethrin, permethrin, and DDT. Insect. Biochem. Mol. Biol. 41, 723–732 [DOI] [PubMed] [Google Scholar]
- 14. Du Y., Nomura Y., Satar G., Hu Z., Nauen R., He S. Y., Zhorov B. S., and Dong K. (2013) Molecular evidence for dual pyrethroid-receptor sites on a mosquito sodium channel. Proc. Natl. Acad. Sci. U.S.A. 110, 11785–11790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Du Y., Nomura Y., Dong K., and Zhorov B. S. (2015) Rotational symmetry of two pyrethroid receptor sites in the mosquito sodium channel. Mol. Pharmacol. 88, 273–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Long S. B., Campbell E. B., and Mackinnon R. (2005) Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science 309, 897–903 [DOI] [PubMed] [Google Scholar]
- 17. Zhang X., Ren W., DeCaen P., Yan C., Tao X., Tang L., Wang J., Hasegawa K., Kumasaka T., He J., Wang J., Clapham D. E., and Yan N. (2012) Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature 486, 130–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McCusker E. C., Bagnéris C., Naylor C. E., Cole A. R., D'Avanzo N., Nichols C. G., and Wallace B. A. (2012) Structure of a bacterial voltage-gated sodium channel pore reveals mechanisms of opening and closing. Nat. Commun. 3, 1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Garden D. P., and Zhorov B. S. (2010) Docking flexible ligands in proteins with a solvent exposure- and distance-dependent dielectric function. J. Comput. Aided Mol. Des. 24, 91–105 [DOI] [PubMed] [Google Scholar]
- 20. Li Z., and Scheraga H. A. (1987) Monte Carlo-minimization approach to the multiple-minima problem in protein folding. Proc. Natl. Acad. Sci. U.S.A. 84, 6611–6615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tan J., Liu Z., Wang R., Huang Z. Y., Chen A. C., Gurevitz M., and Dong K. (2005) Identification of amino acid residues in the insect sodium channel critical for pyrethroid binding. Mol. Pharmacol. 67, 513–522 [DOI] [PubMed] [Google Scholar]
- 22. Tatebayashi H., and Narahashi T. (1994) Differential mechanism of action of the pyrethroid tetramethrin on tetrodotoxin-sensitive and tetrodotoxin-resistant sodium channels. J Pharmacol. Exp. Ther. 270, 595–603 [PubMed] [Google Scholar]
- 23. Jensen M. Ø., Jogini V., Borhani D. W., Leffler A. E., Dror R. O., and Shaw D. E. (2012) Mechanism of voltage gating in potassium channels. Science 336, 229–233 [DOI] [PubMed] [Google Scholar]
- 24. Silver K. S., Du Y., Nomura Y., Oliveira E. E., Salgado V. L., Zhorov B. S., and Dong K. (2014) in Advances in Insect Physiology (Cohen E., ed) pp. 389–434, Academic Press, Oxford, UK: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Narahashi T. (2000) Neuroreceptors and ion channels as the basis for drug action: past, present, and future. J. Pharmacol. Exp. Ther. 294, 1–26 [PubMed] [Google Scholar]
- 26. Payandeh J., Scheuer T., Zheng N., and Catterall W. A. (2011) The crystal structure of a voltage-gated sodium channel. Nature 475, 353–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Levitt M. (1978) Conformational preferences of amino acids in globular proteins. Biochemistry 17, 4277–4285 [DOI] [PubMed] [Google Scholar]
- 28. Rinkevich F. D., Du Y., Tolinski J., Ueda A., Wu C. F., Zhorov B. S., and Dong K. (2015) Distinct roles of the DmNav and DSC1 channels in the action of DDT and pyrethroids. Neurotoxicology 47, 99–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Davies T. G. E., and Williamson M. S. (2009) Interactions of pyrethroids with the voltage-gated sodium channel. Bayer CropScience J. 62, 159–178 [Google Scholar]
- 30. Wrzesińska B., Czerwoniec A., Wieczorek P., Węgorek P., Zamojska J., and Obrępalska-Stęplowska A. (2014) A survey of pyrethroid-resistant populations of Meligethes aeneus F. in Poland indicates the incidence of numerous substitutions in the pyrethroid target site of voltage-sensitive sodium channels in individual beetles. Insect Mol. Biol. 23, 682–693 [DOI] [PubMed] [Google Scholar]
- 31. Vais H., Atkinson S., Eldursi N., Devonshire A. L., Williamson M. S., and Usherwood P. N. (2000) A single amino acid change makes a rat neuronal sodium channel highly sensitive to pyrethroid insecticides. FEBS Lett. 470, 135–138 [DOI] [PubMed] [Google Scholar]
- 32. Wang S. Y., Barile M., and Wang G. K. (2001) A phenylalanine residue at segment D3-S6 in Nav1.4 voltage-gated Na+ channels is critical for pyrethroid action. Mol. Pharmacol. 60, 620–628 [PubMed] [Google Scholar]
- 33. Zhorov B. S., and Tikhonov D. B. (2004) Potassium, sodium, calcium and glutamate-gated channels: pore architecture and ligand action. J. Neurochem. 88, 782–799 [DOI] [PubMed] [Google Scholar]