Abstract
Inter-α-inhibitor is a proteoglycan of unique structure. The protein consists of three subunits, heavy chain 1, heavy chain 2, and bikunin covalently joined by a chondroitin sulfate chain originating at Ser-10 of bikunin. Inter-α-inhibitor interacts with an inflammation-associated protein, tumor necrosis factor-inducible gene 6 protein, in the extracellular matrix. This interaction leads to transfer of the heavy chains from the chondroitin sulfate of inter-α-inhibitor to hyaluronan and consequently to matrix stabilization. Divalent cations and heavy chain 2 are essential co-factors in this transfer reaction. In the present study, we have investigated how divalent cations in concert with the chondroitin sulfate chain influence the structure and stability of inter-α-inhibitor. The results showed that Mg2+ or Mn2+, but not Ca2+, induced a conformational change in inter-α-inhibitor as evidenced by a decrease in the Stokes radius and a bikunin chondroitin sulfate-dependent increase of the thermodynamic stability. This structure was shown to be essential for the ability of inter-α-inhibitor to participate in extracellular matrix stabilization. In addition, the data revealed that bikunin was positioned adjacent to both heavy chains and that the two heavy chains also were in close proximity. The chondroitin sulfate chain interacted with all protein components and inter-α-inhibitor dissociated when it was degraded. Conventional purification protocols result in the removal of the Mg2+ found in plasma and because divalent cations influence the conformation and affect function it is important to consider this when characterizing the biological activity of inter-α-inhibitor.
Keywords: chondroitin sulfate, glycosaminoglycan, metal ion-protein interaction, protease inhibitor, protein cross-linking, proteoglycan structure
Introduction
Inter-α-inhibitor (IαI)3 is a proteoglycan composed of three separately expressed polypeptide chains called heavy chain 1 (HC1), heavy chain 2 (HC2), and bikunin (1). The protein is unique by being held together in a covalent, intracellularly assembled complex (2–5) by a chondroitin sulfate chain (6, 7). The GAG is a low-sulfated chondroitin 4-sulfate (CS) chain that consists of 15 ± 3 disaccharide units (5, 8) attached to Ser-10 of bikunin by an O-glycosidic bond that attaches the CS chain to Ser-10 of bikunin (6). The HCs, meanwhile, are linked to the CS chain, and consequently to bikunin, through an ester bond between their C-terminal Asp residue and C-6 of an N-acetylgalactosamine in the CS chain (4, 6). This type of GAG-mediated linkage is called a protein-glycosaminoglycan-protein (PGP) cross-link (6).
The physiological functions of IαI are still mostly undefined; however, an involvement of IαI in extracellular matrix (ECM) remodeling and stabilization and in inflammation has been recurring themes in the literature (9). IαI is produced primarily in the liver and is considered to be a plasma protein. In the ECM, IαI interacts with a protein called tumor necrosis factor-stimulated gene-6 protein (TSG-6) (10, 11). TSG-6 catalyzes the transfer of the HCs from the CS in IαI to hyaluronan (HA), an abundant extracellular matrix GAG. The transfer reaction involves two sequential trans-esterifications and a covalent HC·TSG-6 intermediate (10, 11). The presence of HC2 and Ca2+, Mg2+, or Mn2+ is essential for the reaction with TSG-6 (12–14). The transfer reaction is believed to take place during inflammation and inflammation-like processes (e.g. ovulation) in which TSG-6 expression is induced (15).
The transfer of HC to HA alters both structural and functional characteristics of HA and can lead to changes in cell adhesion and migration in HA-rich ECMs (16–18). Patients with rheumatoid arthritis and osteoarthritis generate high amounts of the HC·HA complex in inflamed synovial fluid (19, 20). This has been related to an increased infiltration of leukocytes into the inflamed joints (18). The formation of the complex is, furthermore, essential for fertility in female mice. Both bikunin- and TSG-6-deficient mice are unable to form a stable ECM around oocytes and ovulation consequently fails, thereby leading to infertility (21–23). Other functions of IαI may relate to bikunin, which accounts for the protease inhibitory activity of IαI (9).
HC1 and HC2 contain a von Willebrand factor type A (vWA) domain in which one metal ion-dependent adhesion site (MIDAS) motif is present (24). However, the tertiary structures of the HCs have never been described in details, and to this date, the only IαI-related high-resolution structure available is the crystal structure of bikunin (25). Additionally, the structure of the heterotrimeric IαI complex, including the CS, is not well described. An electron microscopy study has previously indicated that the overall structure of IαI is an extended and dumbbell-like shape (26). In these electron microscopy-based analyses, the N-terminals of the HCs are observed as globular domains, with the C-terminals extending as flexible tails, and bikunin is observed as a small spherical structure (26).
The aim of the present study was to gain further insight into the structure and stability of IαI and investigate how the CS and divalent cations influenced both. We used a combination of biochemical and biophysical methods to show that IαI adopted a more compact conformation in the presence of Mg2+ or Mn2+, but not in the presence of Ca2+. This was evident by a faster migration during native electrophoresis, a decrease in the Stokes radius and an increase in the thermodynamic stability. Furthermore, cross-linking mass spectrometry revealed that the protein components of IαI interact directly and that these interactions depended on the presence of both the CS and/or Mg2+ or Mn2+ ions. It was further shown that the loose IαI structure was unable to form the HC·TSG-6 complex. Without the formation of the HC·TSG-6 complex, the ECM stabilizing HC·HA complex cannot be generated. The plasma concentration of Mg2+ is around 1 mm and it is thus likely that IαI in vivo is in the compact conformation. Conventional purification protocols results in slow IαI. Because this might not be the physiologically relevant conformation, it is important to consider this during the investigations of IαI biology and function.
Experimental Procedures
Materials
Human plasma was obtained from Aarhus University Hospital, Skejby, Denmark. Chondroitinase ABC from Proteus vulgaris was purchased from AMSBIO. Bis(sulfosuccinimidyl) suberate (BS3) and sulfo-N-hydroxysulfosuccinimide (NHS) acetate were from Pierce. Polyclonal rabbit antibodies against HC1, HC2 (cross-reactivity with HC1), bikunin, and TSG-6 were produced as previously described (3, 27). As described shortly, the proteins were run in SDS-PAGE and electroeluted. Antisera to the purified proteins were raised commercially in rabbits. The IgG fraction of the serum was recovered by affinity chromatography on a protein G Fast Flow column. Sequence grade trypsin was from Sigma.
SDS/Native-PAGE, Western Blotting, and Trypsin Inhibitor Counterstain
Samples were boiled in SDS sample buffer containing 30 mm DTT. SDS-PAGE was performed in 5–15% gradient gels (10 × 10 × 0.15 cm) using the glycine/2-amino-2-methyl-1,3-propandiol HCl system as previously described (28). The same system was used for native-PAGE, but the sample was not boiled and SDS and DTT were omitted from the sample buffer and the buffer system. Bikunin was visualized using trypsin inhibitor counterstaining following non-reducing SDS-PAGE when indicated (1, 29). For Western blotting, proteins were transferred to PVDF membranes as previously described (30). The membranes were blocked in 20 mm Tris-HCl, 137 mm NaCl, 0.1% Tween, pH 7.4 (TBS-T), containing 5% dry milk overnight at 4 °C. Rabbit antibodies raised against HC1, HC2, or bikunin were added and the blot was incubated for 2 h at 23 °C. After washing for 3 × 15 min in TBS-T, the membrane was incubated in TBS-T containing the secondary anti-rabbit Cy3-labeled antibody in TBS-T and 5% dry milk. After 2 h of incubation and additional washing an image of the blot was acquired on a FluorChem Q system (Cell Biosciences).
Purification of Inter-α-inhibitor
IαI was purified from human plasma as previously described but with some modifications (1). Human plasma was made 5% in polyethylene glycol 8000, incubated for 1 h at 4 °C, and pelleted by centrifugation at 3800 × g for 15 min. The supernatant was made 16% in polyethylene glycol 8000, incubated for 1 h at 4 °C, and centrifuged to collect the precipitated protein that subsequently was dissolved in 25 mm Tris-HCl, 150 mm NaCl, 10 mm EDTA, pH 7.4 (buffer A1). The sample was applied to a 5-ml Hitrap Q HP (GE Healthcare) anion exchange column connected to an ÄKTA purifier HPLC system (GE Healthcare) equilibrated in buffer A1. The column was eluted with a linear gradient of buffer B1 (buffer A1 containing 1 m NaCl) to 75% at 2% B/min with a flow rate of 3 ml/min. Fractions containing IαI according to SDS-PAGE were pooled and dialyzed into 150 mm NaCl, 25 mm Tris-HCl, pH 7.4 (Buffer A2). The sample was applied to a 5-ml Hitrap Blue HP (GE Healthcare) equilibrated in Buffer A2. The column was eluted using a linear gradient of buffer B2 (buffer A2 containing 2 m NaCl) to 90% at 0.5% B/min with a flow rate of 5 ml/min. Fractions containing IαI were pooled and dialyzed into 150 mm NaCl, 25 mm Tris-HCl, pH 7.4 (Buffer A3). The sample was applied to a 1-ml Hitrap Q HP (GE healthcare). The column was eluted with a linear gradient of buffer B3 (buffer A3 containing 1 m NaCl) to 75% at 2% B/min with a flow rate of 1 ml/min. The purified protein was dialyzed into 20 mm Hepes, 137 mm NaCl, pH 7.4, and frozen in aliquots.
Chondroitinase ABC Treatment of IαI
Purified IαI was treated with chondroitinase ABC (ChonABC) to partially or completely digest the CS chain. In the case of limited ChonABC digestion, IαI was incubated with 1 milliunit of ChonABC/10 μg of IαI in 20 mm Hepes, 137 mm NaCl, pH 7.4, at 37 °C for 2.5 h. In the case of extensive ChonABC digestion, IαI was incubated with 1 milliunit of ChonABC/0.15 μg of IαI at 37 °C for 18 h. Aliquots of the digests were analyzed by SDS-PAGE and the digests were kept at −20 °C.
Chemical Cross-linking and Alkaline Hydrolysis
IαI was titrated with BS3 at protein:cross-linker molar ratios of 1:50, 1:100, 1:500, 1:1000, 1:2500, and 1:5000. The titration was performed to find a low, but efficient cross-linker concentration. A ratio of 500 was used throughout the study. IαI was incubated with 1 mm BS3 in 20 mm Hepes, 137 mm NaCl, pH 7.4, for 30 min at 20 °C in the presence of 5 mm EDTA, 1 mm CaCl2, 1 mm MgCl2, or 1 mm MnCl2, as indicated. IαI was incubated with EDTA or the specified cations for 30 min prior to cross-linking. BS3 solutions were prepared immediately before use to avoid hydrolysis. The reaction was quenched with 150 mm NH4HCO3 and left for 30 min. NaOH was added to 300 mm to dissociate the protein components and the samples were left on ice for 30 min (6). Tris-HCl, pH 7.4, was added to a final concentration of 450 mm to lower the pH. The samples were separated by both SDS-PAGE and native-PAGE and visualized by either Coomassie Brilliant Blue staining or Western blotting.
Titration with Sulfo-NHS-acetate
IαI was titrated with increasing amounts of sulfo-NHS-acetate at molar ratios from 1:65 to 1:2100 in 20 mm Hepes, 137 mm NaCl, pH 7.4, to block Lys residues. The samples were incubated for 60 min at 20 °C before the reaction was quenched with 150 mm Tris-HCl, pH 7.4. Sample buffer for native-PAGE was added after 30 min and the samples were analyzed by native-PAGE.
Size Exclusion Chromatography
IαI and ChonABC-treated IαI were applied to a Superdex 200 increase 10/300 GL column (GE healthcare) equilibrated in 20 mm Hepes, 137 mm NaCl, pH 7.4, containing 5 mm EDTA, 1 mm CaCl2, 1 mm MgCl2, or 1 mm MnCl2. The column was calibrated with a size exclusion chromatography markers kit (Sigma) containing carbonic anhydrase (29 kDa and Rs 2.01 nm,), albumin (66 kDa and Rs 3.6 nm), alcohol dehydrogenase (150 kDa and Rs 4.6 nm), β-amylase (200 kDa and Rs 4.8 nm), apoferritin (443 kDa and Rs 6.1 nm), and thyroglobulin (669 kDa and Rs 8.5 nm). Blue dextran was used to obtain V0 (2000 kDa). The Stokes' radius of IαI and the protein components were calculated using the linear relationship between Ve/V0 and Stokes radius in a semi-logarithmic plot. Elution position of protein standards (repeated for each buffer condition) were used to generate a standard curve of Stokes radius versus (−log Kav)1/2 that was used to calculate the Stokes radii of the IαI species (31).
Far UV Circular Dichroism Spectroscopy Thermal Scans
IαI and ChonABC-treated IαI were diluted to a protein concentration of 0.2 mg/ml using 20 mm Hepes, 137 mm NaCl, pH 7.4, with 5 mm EDTA, 1 mm CaCl2, 1 mm MgCl2, or 1 mm MnCl2 and stored at 5 °C. Circular dichroism (CD) was performed on a J-810 CD-spectrometer (Jasco) using a 1-mm quartz cuvette with lid (Hellma Analytics). Thermo scans were performed from 20 to 95 °C at a wavelength of 222 nm with a step size of 0.2 °C, bandwidth of 8 nm, response time of 8 s, and scan speed of 60 °C/h. The thermal scan data were fitted, as described previously, using the software KaleidaGraph (version 4.0 Synergy Software) (32). The fitted parameters were used to calculate the fraction of folded protein at different temperatures. All samples were analyzed in triplicates.
Identification of Sulfo-NHS-acetate Differentially Modified Residues
IαI with 1 mm MgCl2, IαI with 5 mm EDTA, or ChonABC-treated IαI with 1 mm MgCl2 were titrated with increasing amounts of sulfo-NHS-acetate at molar ratios from 0 to 1:1000 in 20 mm Hepes, 137 mm NaCl, pH 7.4. The reaction was quenched after 1 h by addition of 200 mm Tris-HCl, pH 8. The sample was denatured and reduced in 6 m urea containing 5 mm DTT for 1 h, alkylated with 15 mm iodoacetamide for 1 h, and diluted to 0.8 m urea with 50 mm NH4HCO3. Samples were treated with either trypsin alone or endoproteinase GluC and subsequently trypsin. The proteases were added at a (w/w) ratio of 1:20 and the sample was incubated for 18 h at 37 °C. The sample was desalted using self-packed reverse phase microcolumns containing POROS R2 (33).
Cross-linked Peptides for Mass Spectrometry Analyses
IαI was cross-linked with BS3 in the presence of 1 mm MgCl2, 1 mm MnCl2, or 5 mm EDTA. Following quenching of the cross-linking reaction, the sample was concentrated using a 30-kDa cutoff centrifugal filter (Amicon Ultra) and the buffer was exchanged to 20 mm Hepes, 137 mm NaCl, pH 7.4, to remove the cross-linker. The sample was reduced, alkylated, treated with trypsin, and desalted as described above. The desalted peptides were dissolved in 10 mm KH2PO4, 20% acetonitrile, pH 2.8 (Buffer A), and applied to a PolySULFOETHYL A column (PolyLC) equilibrated in buffer A. The peptides were eluted using a flow rate at 0.15 ml/min and a gradient of 0–60% buffer B (1 m KCl in buffer A) over 30 min and 60% Buffer B for 10 min. The tryptic peptides were collected and micro-purified using self-pack microcolumns containing POROS R2 and subsequently lyophilized. Samples enriched for cross-linked peptides were analyzed by mass spectrometry in 3 replicate runs.
Mass Spectrometry
Nano LC-MS/MS was performed using an EASY-nLC II system (Thermo Scientific) connected to a TripleTOF 5600+ mass spectrometer (AB Sciex) equipped with a NanoSpray III source (AB Sciex) operated under Analyst TF 1.5.1 control. Peptides were dissolved in 0.1% formic acid, injected, trapped, and desalted on a ReproSil-Pur C18-AQ trap column (2 cm × 100-μm inner diameter packed in-house with 3 μm resin; Dr. Marisch GmbH, Ammerbuch-Entringen, Germany). The peptides were eluted from the trap column and separated on a 15-cm analytical column (75 μm inner diameter) packed in-house in a pulled emitter with ReproSil-Pur C18-AQ 3 μm resin (Dr. Marisch GmbH, Ammerbuch-Entringen, Germany). Peptides were eluted using a flow rate of 250 nl/min and a 50-min gradient from 5 to 35% phase B (0.1% formic acid and 90% acetonitrile or 0.1% formic acid, 90% acetonitrile and 5% DMSO). The collected MS files were converted to Mascot generic format (MGF) using the AB SCIEX MS Data Converter β 1.1 (AB SCIEX) and the “proteinpilot MGF” parameters.
Analysis of MS Data
For cross-linked samples data analyses were performed using the MassAI software package (MassAI Bioinformatics) (34). The MGF files were preprocessed with the MGF-filter tool using default values and keeping the 125 most intense MS/MS peaks. The files were searched against the IαI sequences using the following settings: MS error tolerance of 0.02 Da for cross-linked peptides, MS/MS error tolerance of 0.1 Da, three missed cleavages, iodoacetamide as fixed modification, oxidized methionine variable modification (M-ox), one modification per peptide, and BS3 cross-linking between lysine residues. In the case of multiple assignments to the same MS/MS scan, the highest scoring match was selected. For every unique cross-link, the MS/MS scan of the highest scoring match was manually inspected. A list of the identified cross-links can be found in the supplemental material. Cross-linked peptides were only accepted if the precision on MS was below 10 ppm and the score above 25. For the sulfo-NHS-acetate titration the generated peak-list (MGF) was searched against an in-house database containing mature HC1, HC2, and Bikunin using the Mascot search engine (Matrix Science) with the following search parameters: MS tolerance of 10 ppm, MS/MS tolerance of 0.1 Da, trypsin with 2 miscleavages, carbamidomethyl as fixed modifications, and oxidized methionine and acetyl as variable modification. The search result from Mascot was used in Skyline for MS1 filtering to extract the ion intensity of all identified peptides (35, 36). The individual peptide intensities were normalized to the total peptide intensity. The average normalized intensity and standard deviation were calculated based on three LC-MS/MS analysis.
HC·TSG-6 Complex Formation after Sulfo-NHS-acetate Treatment of IαI
IαI was titrated with increasing amounts of sulfo-NHS-acetate at molar ratios from 0 to 1:1000 as described above in the presence of either 1 mm MgCl2 or 5 mm EDTA. After acetylation, MgCl2 was added to a final concentration of 10 mm. The acetylated IαI (0.8 μg) was incubated with a fixed amount of TSG-6 (0.8 μg) for 1 h at 37 °C in a total volume of 15 μl. The total samples were separated by SDS-PAGE and visualized by Western blotting.
Results
IαI Adopts a Compact Structure in the Presence of Mg2+ and Mn2+
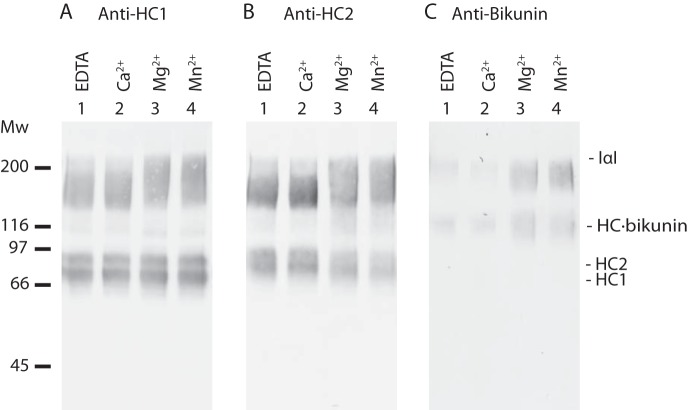
BS3 is a homobifunctional chemical cross-linker that reacts with the ϵ-amino group of lysine residues and the N termini. The maximal length of the BS3 spacer is 11.4 Å and may thus be used to cross-link primary amines that are within spatial proximity (37). IαI was incubated with BS3 in the presence of Ca2+, Mg2+, and Mn2+. These divalent cations are essential for the TSG-6-HC2 mediated transfer of HCs to HA (12). The cross-linked samples were analyzed by SDS-PAGE (Fig. 1A) and native-PAGE (Fig. 1B). Prior to SDS-PAGE, the PGP cross-link was cleaved by alkaline hydrolysis, thereby releasing the BS3 cross-linked proteins from the CS. Incubation of IαI with BS3 results in the formation of an alkaline hydrolysis insensitive protein complex, which migrated approximately as a 200-kDa protein during reduced SDS-PAGE (Fig. 1A). Hence, the protein components were cross-linked and indicate that they interacted in the native complex.
FIGURE 1.
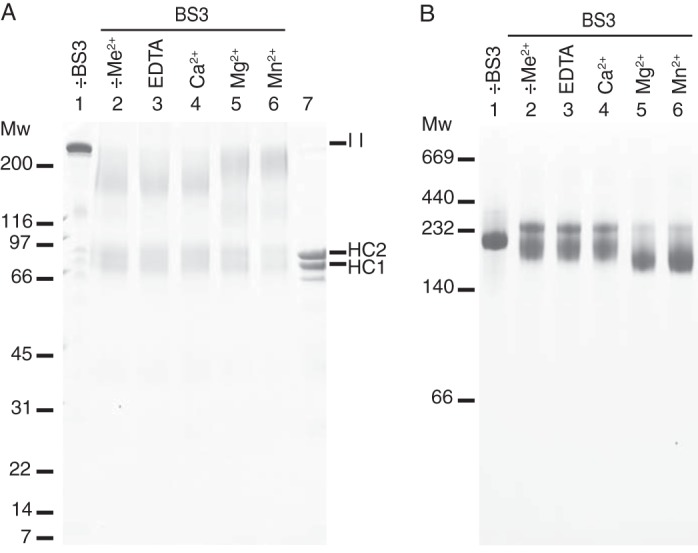
Chemical cross-linking of IαI reveals cation-induced conformational changes. IαI was incubated with the chemical cross-linker, BS3, in the presence of EDTA, Ca2+, Mg2+, or Mn2+. The cross-linked proteins were analyzed by native PAGE (panel B) or dissociated by NaOH and analyzed by SDS-PAGE (panel A). The gels were stained with Coomassie Brilliant Blue. In the presence of Mg2+ or Mn2+, IαI adopts a more compact structure as demonstrated by an increased migration during native-PAGE. Individual protein components are cross-linked into a product of the same size as IαI as demonstrated by SDS-PAGE. However, cross-linking is more efficiently achieved in the presence of Mg2+ and Mn2+. Non-cross-linked IαI (lane 1, −BS3, panels A and B), IαI “as purified” cross-linked with BS3 (lane 2, −Me2+, panels A and B), and dissociated IαI (lane 7, panel A) controls were included. The data represent more than three independent experiments.
There were subtle qualitative differences in the outcome of the cross-linking reaction when it was performed in the presence of Mg2+ or Mn2+ compared with EDTA or Ca2+. In the presence of Mg2+ or Mn2+, cross-linking resulted in the formation of a band of higher molecular weight and more homogenous migration (Fig. 1A, lanes 5 and 6). The molecular weight of this cross-linked product was similar to that of IαI (Fig. 1A, IαI in lane 1 compared with lanes 5 and 6). This suggested that the presence of Mg2+ or Mn2+ is essential for the efficient cross-linking of the three protein components and that the protein-protein interactions were divalent cation-dependent. Protein bands of lower molecular weight could, as judged by their size, arise from cross-linking of HC1 and HC2, HC1 and bikunin, or HC2 and bikunin. Chemical cross-linking analysis indicated that the protein components of IαI interacted as opposed to functioning as separate entities bound to the CS chain.
The analysis of cross-linked IαI by native-PAGE revealed further divalent cation-dependent differences (Fig. 1B). Addition of Mg2+ or Mn2+, instead of EDTA or Ca2+, prior to the cross-linking reaction resulted in a faster migrating IαI molecule. Apparently, Mg2+ or Mn2+ ions induce conformational changes in the molecule that cause the protein to obtain a more compact structure, which is maintained by the covalent cross-linking and thus remain evident after native-PAGE. Without prior cross-linking, divalent cation-dependent differences in the migration of IαI could not be observed, indicating that the metal ion-dependent interactions are disrupted during electrophoresis (data not shown). The cross-linking of IαI gave rise to two bands during native-PAGE (Fig. 1B, lanes 2-6). The upper band was present in all cross-linking conditions and was likely a less compact IαI species. We suspect that the upper band represents an IαI species with disrupted protein or protein-CS interactions. This could be caused by the presence of BS3-modified Lys residues. Hydrolysis of one of the reactive groups in BS3 prior to the formation of a cross-link will result in a modified Lys residue rather than cross-linking. Titration of IαI with sulfo-NHS-acetate, which reacted with primary amines, gave rise to a band of similar size, thus supporting this hypothesis (Fig. 2). Moreover, the reduction in the amount of the upper band species in the presence of Mg2+ and Mn2+ indicated that these two ions induce a more compact IαI structure in which cross-linking of protein components is efficiently achieved.
FIGURE 2.
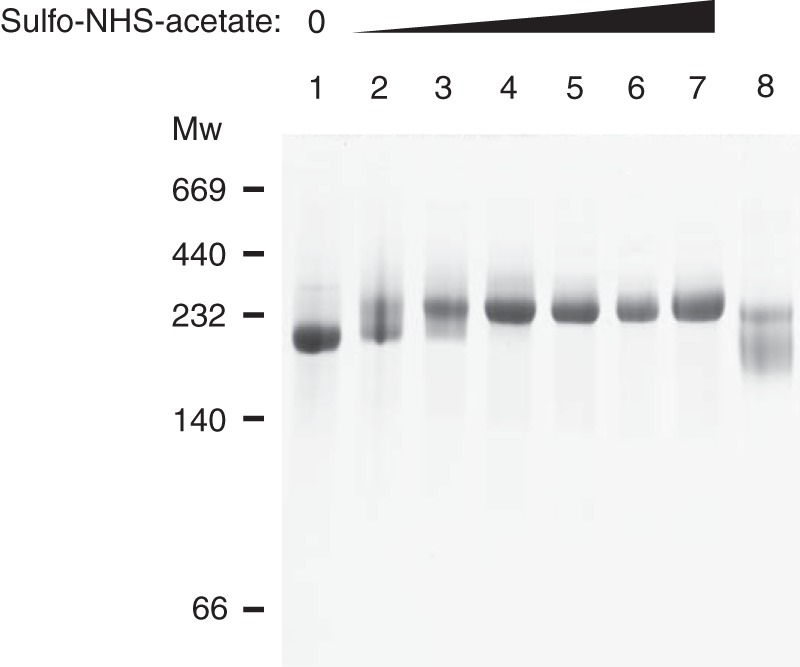
Titration of IαI with sulfo-NHS-acetate. IαI was titrated with increasing amounts of the primary amine blocking agent sulfo-NHS-acetate (molar ratios from 1:65 to 1:2100). The reaction was quenched with 150 mm Tris-HCl after 60 min and the samples were analyzed by native-PAGE. When Lys residues are blocked with sulfo-NHS-acetate, the migration of IαI into the gel is reduced and IαI appears as a less compact protein. IαI (lane 1) and IαI cross-linked with BS3 (lane 8) controls were included. This migration is similar to the migration seen when IαI is cross-linked with BS3. The data represent 3 independent experiments.
Mg2+ and Mn2+ Decrease the Stokes Radius of IαI
To ensure that the compact structure, produced in the presence of Mg2+ and Mn2+, was not an artifact of the cross-linking reaction, the IαI structural changes induced by divalent cations was also investigated using size exclusion chromatography (Fig. 3A). The retention time of IαI was determined in buffers containing a physiological salt concentration and EDTA, Ca2+, Mg2+, or Mn2+. From these experiments, it was evident that the retention of IαI changed in the presence of Mg2+ and Mn2+, even without prior cross-linking. The size of IαI was estimated using a size exclusion chromatography standard calibration kit (Fig. 3 and Table 1). In the presence of EDTA or Ca2+, the Stokes radius of IαI was calculated to be 60 Å while in the presence of Mg2+ or Mn2+, the Stokes radius of IαI changed to 55 and 56 Å, respectively. Hence, IαI seems to change its conformation in the presence of Mg2+ or Mn2+. This conformational change resulted in a reduction of the Stokes radius, which is consistent with the altered migration of cross-linked IαI during native-PAGE.
FIGURE 3.
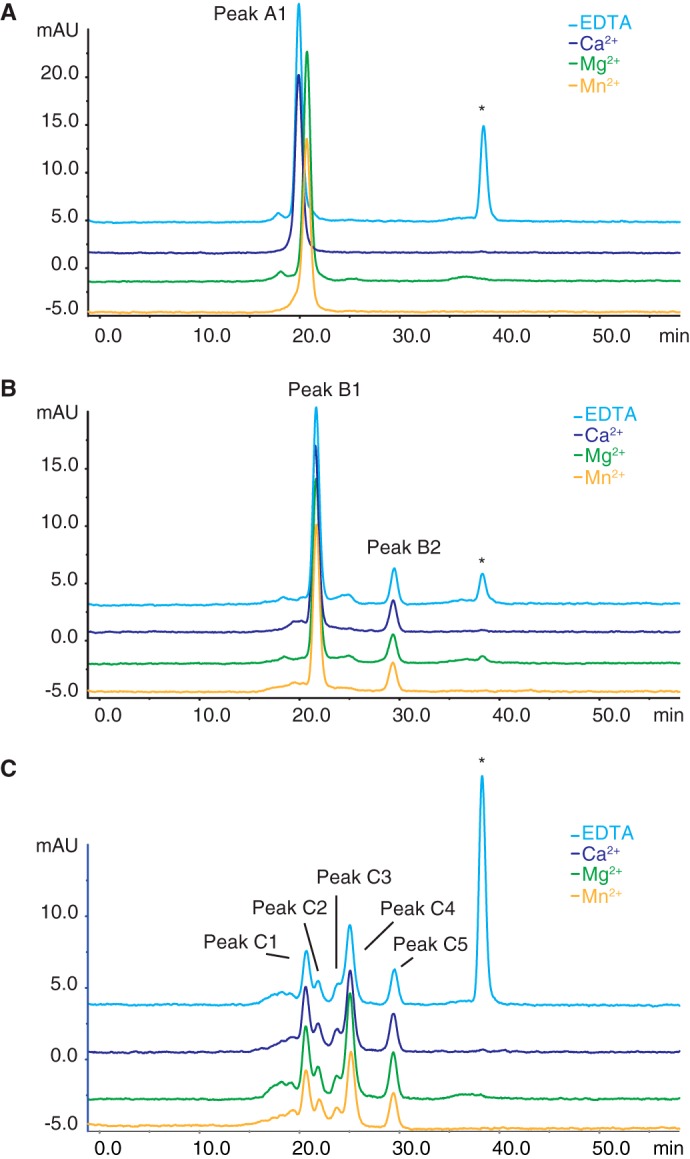
The Stokes radius of IαI is decreased in the presence of Mg2+ and Mn2+ compared with EDTA and Ca2+. Calibrated size exclusion chromatography was used to compare retention times of IαI under physiological buffer conditions with added EDTA (blue), Ca2+ (purple), Mg2+ (green), or Mn2+ (yellow) (panel A). The shift in the retention of IαI in the presence of Mg2+ or Mn2+ corresponds to a decrease in the calculated size of IαI from 350 to 290 kDa. Size exclusion chromatography of IαI following limited ChonABC treatment (panel B) and extensive ChonABC treatment (panel C) revealed no divalent cation-dependent differences in the size of the individual components. Thus, Mg2+ and Mn2+ cause IαI to adopt a compact conformation. However, the cations only affect the Stokes radius of the proteins in the presence of the CS. The content of the labeled peaks are A1, IαI; B1, HC1·HC2; B2, Bikunin; C1 to C3, HC2; C4, HC1 and C5, Bikunin (see also Table 1). *, peak eluting at Vc (total column volume) and containing EDTA added to the sample prior to loading on the column.
TABLE 1.
The calculated Stokes radii of IαI and ChonABC-treated IαI
| EDTA | Ca2+ | Mg2+ | Mn2+ | |
|---|---|---|---|---|
| Stokes radius (Å) | ||||
| Native IαI | ||||
| Peak A1 (IαI) | 60 | 60 | 55 | 56 |
| Limited ChonABC digest | ||||
| Peak B1 (HC1·HC2) | 51 | 51 | 51 | 51 |
| Peak B2 (bikunin) | 26 | 26 | 26 | 26 |
| Extensive ChonABC digest | ||||
| Peak C1 (HC2) | 56 | 56 | 56 | 56 |
| Peak C2 (HC2) | 50 | 50 | 50 | 50 |
| Peak C3 (HC2) | 43 | 43 | 43 | 43 |
| Peak C4 (HC1) | 38 | 38 | 38 | 38 |
| Peak C5 (bikunin) | 26 | 26 | 26 | 26 |
Because the CS chain is highly negatively charged, it may interact with residues in IαI and with the added divalent cations. We wanted to investigate if the removal of CS affected the protein structure. IαI was therefore treated with ChonABC prior to size exclusion chromatography, to produce either incomplete or complete degradation of the CS chain. Partial degradation released bikunin, whereas a short stretch of the CS chain still connects HC1 and HC2. This covalent complex had been named HC1·HC2 (5). There was no apparent cation-dependent difference in the retention of HC1·HC2 or bikunin when these IαI species were analyzed by size exclusion chromatography in the presence of EDTA, Ca2+, Mg2+, or Mn2+ (Fig. 3B and Table 1). This indicates that the CS is essential for the structural change observed in the presence of MG2+ or Mn2+.
Extensive ChonABC digestion results in complete digestion of the CS and causes the release of all protein components from the covalent complex (5). After extensive ChonABC digestion, IαI gives rise to multiple peaks (Fig. 3C). Western blot analysis of the eluting peaks revealed that HC2 elutes in the first three peaks, followed by HC1 and finally bikunin (Fig. 3C and Table 1). This indicated that protein interactions were lost when the CS was digested by ChonABC. Furthermore, released free HC2 appeared to polymerize. Additionally, the presence of Ca2+, Mg2+, or Mn2+ did not result in changes of the retention time during size exclusion chromatography of the released protein components (Fig. 3C and Table 1). In summary, size exclusion chromatography revealed that Mg2+ or Mn2+ and the CS chain in concert cause IαI to adopt a more compact structure.
The Thermal Stability of IαI Is Affected by Divalent Cations and the CS
To further substantiate the effects of divalent cations we employed a thermal shift assay by following the thermal unfolding of IαI using far-UV CD. CD data were recorded at 222 nm during a thermal scan from 20 to 95 °C in the presence of EDTA, Ca2+, Mg2+, or Mn2+ (Fig. 4A). The CD data (Fig. 4) were fitted to a two-state transition to obtain the midpoint of the transition (Tm) (Fig. 5A). In the presence of Mg2+ and Mn2+ the Tm was increased to 55.7 and 57.3 °C, respectively, from 53.8 °C in the presence of EDTA, whereas the addition of Ca2+ did not induce a Tm shift. The data demonstrated that both Mg2+ and Mn2+ are able to stabilize the structure of IαI during thermal unfolding, with the largest effect being observed in the presence of Mn2+.
FIGURE 4.
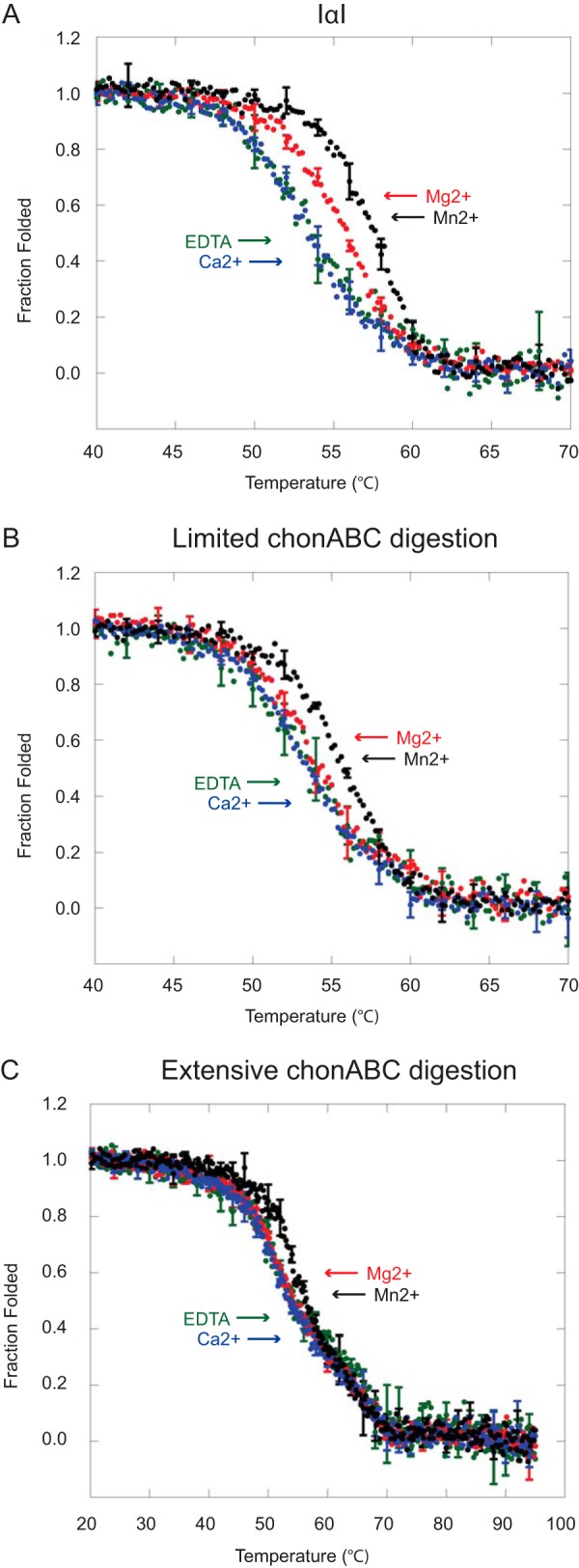
Cations and the CS chain influence the thermal unfolding of IαI. Thermal denaturation of IαI (panel A), IαI following limited ChonABC digestion (panel B), and IαI following extensive ChonABC treatment (panel C) was followed by far-UV CD at 222 nm in the presence of EDTA (green), Ca2+ (blue), Mg2+ (red), and Mn2+ (black). Mg2+ and Mn2+ cause a shift in the transition of IαI from folded to unfolded. Degradation of the CS chain by ChonABC abolishes the effect of Mg2+. It is evident that dissociated IαI deviates from a two-state transition. The data represent three technical replicates and error bars show the S.D.
FIGURE 5.
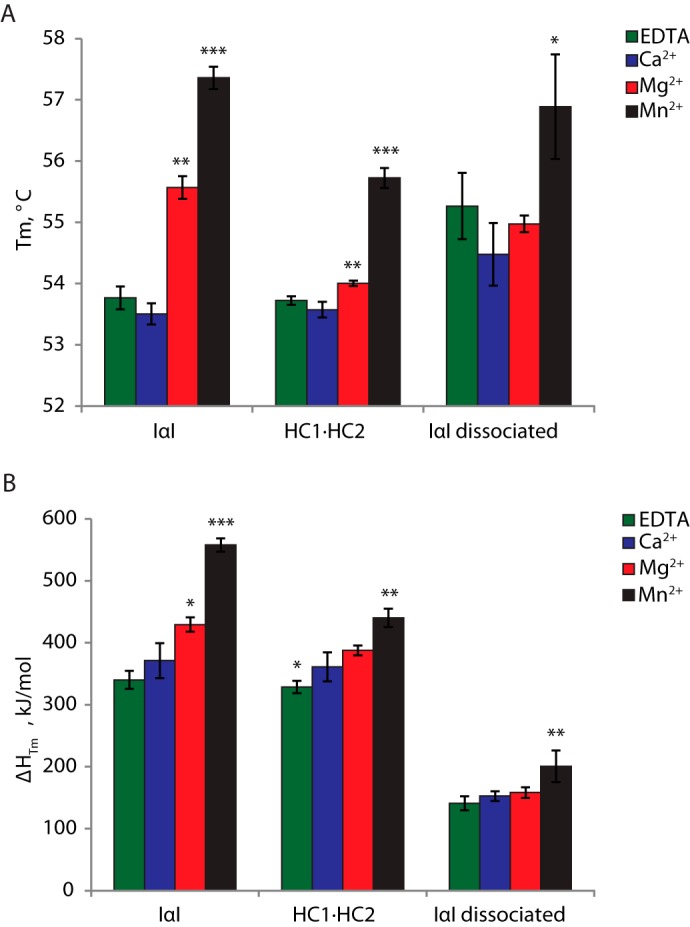
The Tm and ΔHTm of unfolding depend on the presence of cations and the CS. The midpoint of the unfolding transition (Tm) (panel A) and the enthalpy of unfolding at the midpoint of denaturation (ΔHTm) (panel B) were estimated from the thermal unfolding curves obtained in Fig. 4 (n = 3). Both the Tm and the ΔHTm of IαI unfolding are increased in the presence of Mg2+ or Mn2+. When the CS is removed (HC1·HC2 and IαI dissociated), the effect of Mg2+ is no longer observed. There still seems to be an effect of Mn2+. Mg2+ or Mn2+ thus seem to stabilize IαI and cause increased cooperatively in the unfolding. The effect depends on the presence of the CS. Significant values are indicated as: *, p < 0.05; **, p < 0.01; and ***, p < 0.001. The calculations are based on three technical replicates and error bars show the S.D.
In parallel to the size exclusion chromatography analysis, we investigated if CS was involved in the metal ion-mediated stabilization (Fig. 4, B and C). The Tm of the HC1·HC2 complex was unchanged in the presence of EDTA and Ca2+ compared with IαI (Fig. 5A). The thermal shift for Mg2+ and Mn2+ were reduced, compared with IαI, and only Mn2+ was significantly thermal shifted. A similar tendency was observed after extensive digestion of IαI with ChonABC (Fig. 4C). Only the addition of Mn2+ altered the Tm (Fig. 5A). However, the thermal scan of dissociated IαI did not fit a two-state unfolding scheme. This suggests that the process monitored during the thermal scan involves several unfolding steps. It is likely that this can be attributed to complete dissociation of IαI leading to an independent thermal unfolding of HC1, HC2, and bikunin. Although we do not observe distinct unfolding steps, the three proteins may unfold close to each other in temperature so that the process becomes a composite of different events. This is consistent with HC dissociation observed by SEC.
The enthalpy of unfolding at the midpoint of denaturation (ΔHTm) was likewise obtained by fitting the CD data (see Fig. 4) to a two-state transition (Fig. 5B). ΔHTm is a measure of the cooperativity of protein unfolding. A highly cooperative unfolding indicated that IαI exists as a compact structure in which unfolding of one part of the structure affects the unfolding of other parts (38). Low cooperativity, on the other hand, indicates that the complex consists of individually folded structures that unfold independently (39). Addition of Mg2+ or Mn2+ led to an increase in ΔHTm, thus indicating that these cations stabilize protein interactions in IαI or induce higher cooperativity of unfolding. Similarly to the thermal stability, the effect of Mg2+ and Mn2+ on ΔHTm also depends on the presence of the CS.
The Divalent Cation-dependent Interactions within IαI Depend on the Presence of the CS
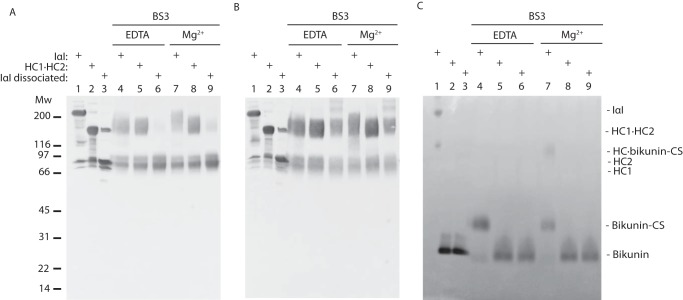
Chemical cross-linking, size exclusion chromatography, and thermal denaturation showed that Mg2+ or Mn2+ induced conformational changes in IαI. This resulted in the formation of a more compact protein structure. To decipher which protein interactions contributed to the formation of this structure, the outcome of the cross-linking reaction was further characterized by Western blotting. IαI was cross-linked with BS3 in the presence of EDTA, Ca2+, Mg2+, or Mn2+. The cross-linked protein components were dissociated from CS by mild hydrolysis and separated by SDS-PAGE. We used antibodies against HC1, HC2, and bikunin to visualize the products of the cross-linking reaction (Fig. 6). Hydrolysis of the PGP cross-link would usually result in the appearance of three bands representing HC2, HC1, and bikunin. However, cross-linking generated protein bands of higher molecular weight showing molecular weights and antigenicity corresponding to HC1·HC2, HC2·bikunin, HC1·HC2, and HC1·HC2·bikunin suggesting that all of the protein components interacted with each other (Fig. 6). The double band observed migrating similar to the HCs appears due to intramolecular cross-linking within a single HC generating two different versions of the same HC (Fig. 6, A and B).
FIGURE 6.
BS3 can cross-link all protein components of IαI. SDS-PAGE analysis of IαI cross-linked with BS3 in the presence of EDTA, Ca2+, Mg2+, or Mn2+. After quenching the cross-linking reaction, mild alkaline treatment dissociated the protein components from the CS. Western blotting using antibodies against HC1 (panel A), HC2 (panel B), and bikunin (panel C) visualized the protein components. The migration of IαI and HCs without BS3 treatment is indicated. Free bikunin loses reactivity with the antibody after BS3 treatment and is therefore not observed after hydrolysis of the PGP-cross-link (panel C). It is evident that all protein components can be cross-linked in all conditions. However, the presence of Mg2+ or Mn2+ produces a band containing HC1 and HC2 of higher molecular weight. Cross-linking of bikunin seems to be more efficient in the presence of Mg2+ or Mn2+. Evidently, there are qualitative and quantitative differences in the cross-linking of IαI when adding Mg2+ or Mn2+, compared with the addition of Ca2+ or EDTA. The data represent more than 3 independent technical replicates.
There was a qualitative and a quantitative difference between cross-linking that has been performed in the presence of EDTA or Ca2+ compared with Mg2+ or Mn2+ (Fig. 6, lanes 1 and 2, compared with 3 and 4). In the presence of Mg2+ or Mn2+ the cross-linked products migrating in the region of HC1·HC2 and IαI were more intense, had a more homogenous migration, and migrated at higher mass more similar to IαI. These data suggest that the inter-molecular cross-linking of bikunin is more efficient in the presence of Mg2+ or Mn2+. This consequently increases the amounts of cross-linked material containing all of the protein components: HC1·HC2·bikunin. Mg2+ and Mn2+, thus, seem to stabilize or induce interactions between the protein components in IαI.
To study the interplay between Mg2+ and CS, three variants of IαI were studied by BS3 cross-linking with and without Mg2+ (Fig. 7). The three types of IαI were: (a) IαI with no treatment (Fig. 7, A–C, lane 1); (b) limited ChonABC digestion producing free bikunin and the HC1·HC2 complex (Fig. 7, A–C, lane 2); and (c) extensive ChonABC digestion generating dissociated IαI where all three protein components are released from the CS (Fig. 7, A–C, lane 3). The cross-linked samples were separated by SDS-PAGE. Products containing HCs were visualized by Western blotting using antibodies toward HC1 (Fig. 7A) or HC2 (Fig. 7B). After BS3 treatment free bikunin lost the antibody reactivity and was alternatively visualized by trypsin inhibitor counterstain (Fig. 7C) (1). Limited CS digestion did not alter cross-linking of the two HCs (Fig. 7, A and B, lanes 5 and 8). Cross-linking of bikunin to the HCs is both dependent on Mg2+ (Fig. 7C, lanes 4 and 7) and CS (Fig. 7C, lanes 6 and 8). Extensive ChonABC digestion completely abolished BS3-mediated cross-linking between HC1, HC2, and/or bikunin (Fig. 7, A–C, lanes 6 and 9). Cross-linking still resulted in the formation of several high molecular weight HC2 species (Fig. 7B, lane 9). The high molecular weight HC2 species supported the evidence of HC2 polymerization, which was observed during size exclusion chromatography. These data shows that the interaction of bikunin and the HCs are both Mg2+ and CS dependent and that HC1 and HC2 interact in a CS-dependent manner.
FIGURE 7.
The PGP cross-link is essential for interactions between the protein components of IαI. Differential ChonABC treatment of IαI released bikunin (HC1·HC2) or completely removed the CS (IαI dissociated). The samples were cross-linked with BS3 in the presence of either Mg2+ or EDTA. After cross-linking mild alkaline treatment dissociated the protein components from the CS and the samples were analyzed by SDS-PAGE and Western blotting using antibodies against HC1 (panel A) and HC2 (panel B). SDS-PAGE under non-reducing conditions and trypsin inhibitor counterstaining was used to visualize bikunin (panel C). The Western blots reveal that BS3 cross-linking is only possible when the CS chain links the protein components. Moreover, a full-length CS chain is essential for the effect of divalent cations. Remarkably, upon complete dissociation of IαI, HC2 forms polymers (panel B, lanes 6 and 9). Apart from these polymers, it is evident that the interactions between IαI protein components depend on the presence of the CS. The data represent 3 independent experiments.
Chemical Footprint Reveals CS and Mg2+-mediated Interaction Sites in IαI
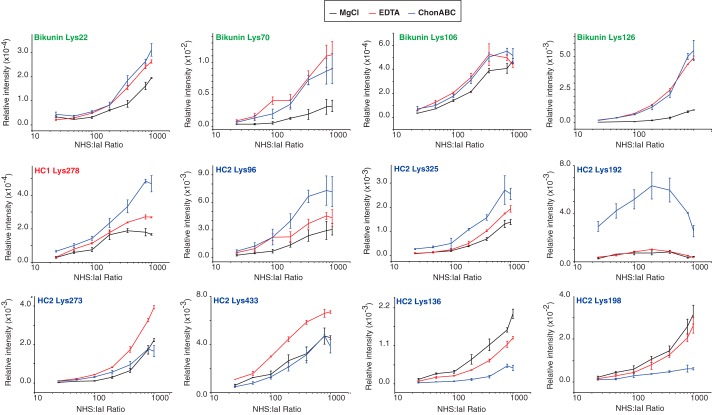
In the sulfo-NHS-acetate acetylation experiment a destabilization/unfolding of the IαI structure was observed indicating that at least one lysine residue is essential for the correct fold (Fig. 2). Chemical footprinting was applied to study the involvement of lysine residues in the CS- and Mg2+-dependent interactions as well as probing the overall structure. Basically, IαI was incubated with increasing amounts of sulfo-NHS-acetate. After acetylation, the samples were treated with either trypsin or a combination of trypsin and endoproteinase GluC. The acetylated residues were identified by MS and the amount of each peptide was quantified based on MS1 intensities (XIC) (Fig. 8). IαI was studied under three different conditions: with MgCl2, EDTA, or after enzymatic removal of the CS. The degree of acetylation can be used to measure how exposed particular Lys residues are during labeling. Bikunin had four residues (Lys-22, Lys-70, Lys-106, and Lys-126), which showed increased acetylation when either Mg2+ or the CS chain was removed. The largest effect was seen for Lys-70 and Lys-126. The increased acetylation, upon removal of Mg2+ or CS shows that Lys-70 and Lys-126 either directly binds the CS or is shielded by the CS. These two residues are located in close proximity within the bikunin structure in a region with several basic amino acid residues (25). Proteins often bind GAGs by clusters of basic residues. Our data thus indicates the presence of an Mg2+-dependent CS binding site in bikunin. A single residue in HC1 (Lys-278) showed increased acetylation upon removal of Mg2+, which further increased when the CS was removed. HC2 showed the most diverse pattern of acetylation. Lys-96 and Lys-325 showed a minor increase in acetylation by EDTA treatment and additional acetylation without CS, a pattern similar to Lys-278 in HC1. The sequential increased acetylation by EDTA and ChonABC treatment indicate the presence of an Mg2+-dependent interaction with CS in both heavy chains. HC2 Lys-192 became highly acetylated when the CS was removed, but was unaffected to the absence of Mg2+. The acetylation profile of Lys-192 indicates that the residue is either directly involved in binding to CS or highly shielded by the CS. In addition, HC2 contains two sites (Lys-273 and -433) only affected by EDTA, indicating the presence of an Mg2+-mediated interaction independent of the CS. Finally, in HC2 there are two sites (Lys-136 and Lys-198) where the acetylations are decreased by removal of CS. The lower signal from Lys-198 can be explained by the high degree of acetylation in Lys-192, however, the lower amount of acetylation can also be caused by the multimerization of HC2 seen when CS is removed.
FIGURE 8.
Chemical footprint reveals CS- and Mg2+-mediated interaction sites in IαI. IαI was incubated with increasing amounts of sulfo-NHS-acetate. After proteolytic fragmentation acetylated residues were identified by LC-MS/MS. The amount of each peptide was quantified based on MS1 intensities (XIC). The degree of acetylation was studied under three different conditions: with MgCl2 (black), EDTA (red), or with the CS removed by ChonABC (blue). The analysis revealed 12 Lys residues with differential acetylation patterns. Chemical footprinting analysis revealed Mg2+- and CS-dependent binding regions in all three subunits of IαI. The data represent three technical replicates and error bars show the S.D.
The CS and Mg2+ Mediates a Contraction of Bikunin
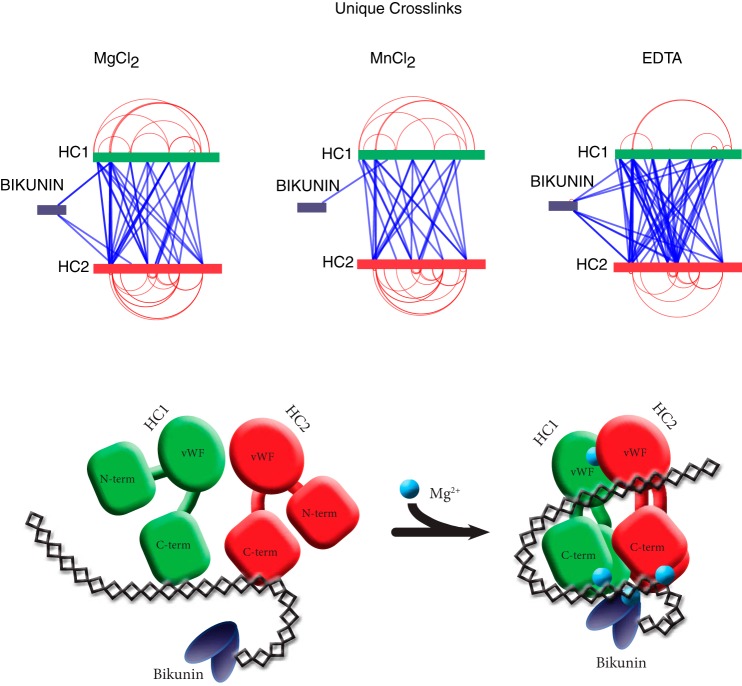
Identification of chemically cross-linked residues by mass spectrometry may provide additional insight to the structure of IαI. To identify interfacing protein regions IαI was cross-linked with BS3 in the presence of Mg2+, Mn2+, or EDTA and digested with trypsin. After enrichment of cross-linked peptides by strong cation exchange, an LC-MS/MS analysis identified the cross-linked peptides (Fig. 9 and supplemental material). The LC-MS/MS analysis resulted in the identification of both intra-protein and inter-protein cross-links (Fig. 9). In this context, “intra” refers to cross-links within the same subunit and “inter” refers to cross-links between different protein components within the same IαI molecule. Bikunin became cross-linked to the N-terminal region of both HC1 and HC2 when Mg2 was included during BS3 cross-linking. In contrast, in the presence of Mn2+ bikunin was only cross-linked to HC1. Cross-linking in the presence of either Mg2+ or Mn2+ resulted in a similar pattern of inter-protein and intra-protein cross-links of the HCs. Intra-protein cross-links that connect the N- and C-terminal were found in both HCs. This corresponds well with the known long-range disulfide bond found in HC1 (41). The identified intra-protein cross-links suggest that the N- and C-terminals of the HC are in close proximity in the native folded proteins. Inter-protein cross-links are present along the entire amino acid sequences of the HCs. Most cross-linked residues have multiple contact points at the opposing HC supporting an overall compact fold of the HCs. The combined pattern of inter-protein and intra-protein cross-links support that the N- and C-terminal are in close proximity in the native IαI complex (Fig. 9). Cross-linking in the presences of EDTA produced a more random cross-linking pattern especially evident by the multiple contact points between bikunin and the HCs, but also by the increased number of unique cross-links between the HCs. In addition, EDTA results in less intra-protein cross-links of the HCs indicating that divalent cations are involved in maintaining a more compact and rigid fold.
FIGURE 9.
Schematic representation of the identified cross-links. IαI was cross-linked with BS3 in the presence of MgCl2, MnCl2, or EDTA (upper panel). The identified cross-links are mapped to the sequences of bikunin, HC1, and HC2. Intra-protein cross-links (red) and inter-protein cross-links (blue) are shown. The cross-linking pattern of IαI indicates that the HCs have a C- to N-terminal packing where bikunin is located close to the N-terminal (lower panel). The graphic representation of IαI shows the transition from a loose and dynamic structure to a more rigid structure with interactions between the proteins subunits as well as the CS in the presence of MgCl2. In the compact conformation, bikunin is via CS- and Mg2+-dependent interactions located in vicinity of the N-terminal of the HCs.
The Compact Structure Stabilized by Mg2+ and CS Is Essential for HC·TSG-6 Complex Formation
The biological relevance of the loose and compact structures were investigated by preventing the formation of compact IαI using sulfo-NHS-acetate (see Fig. 2) and analyzing the ability to form the HC·TSG-6 complex. The results of the sulfo-NHS-acetate titration and the chemical footprinting experiments supported that Mg2+- and CS-dependent interactions exist and that these are responsible for inducing the observed compact structure. This was underscored by the ability to prevent the formation of compact IαI after altering the charge of lysine residues including those involved in the compact structure (see Figs. 2 and 8). In addition, we have previously shown that formation of a HC·TSG-6 complex is dependent on the presence of divalent cations (13). To test if TSG-6 complex formation depended on the compact IαI structure we incubated TSG-6 and sulfo-NHS-acetate pretreated IαI in the presence of either MgCl2 or EDTA (Fig. 10). The ability to form a HC·TSG-6 complex was detected by Western blotting using a TSG-6 antibody. In the presence of Mg2+ TSG-6 formed complexes with the IαI HCs (lane 1), however, increasing the sulfo-NHS-acetate concentration abolished complex formation (lanes 2-8) suggesting that the loose IαI conformation is unable to form the complex. Furthermore, when IαI was treated with sulfo-NHS-acetate in the presence of EDTA, a lower concentration of sulfo-NHS-acetate was required to prevent complex formation. These data suggest that the loose IαI conformation is unable to form the HC·6TSG-6 complex.
FIGURE 10.
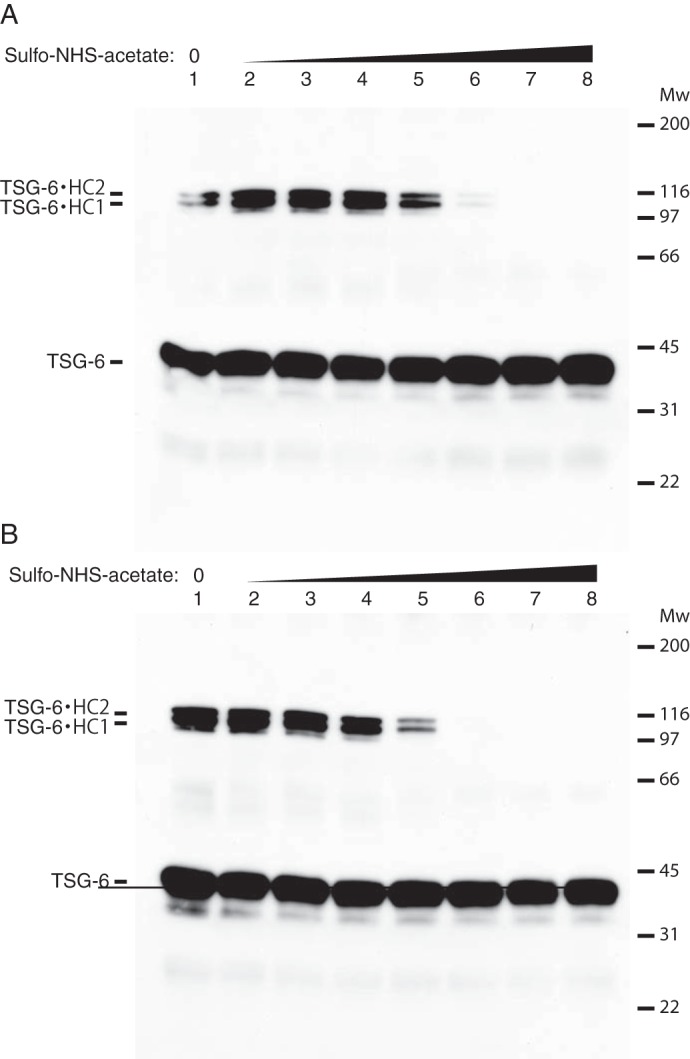
The compact IαI structure is essential for HC·TSG-6 complex formation. IαI was titrated with increasing amounts of sulfo-NHS-acetate (molar ratios from 1:10 to 1:1000, lanes 2-8) in the presence of either MgCl2 (panel A) or EDTA (panel B). After acetylation MgCl2 was added to a final concentration of 10 mm and the samples were incubated with TSG-6. The samples were analyzed by SDS-PAGE and HC·TSG-6 complexes were visualized by Western blotting using an anti-TSG-6 antibody. At a 1:400 molar ratio of IαI:Sulfo-NHS-acetate in the presence of MgCl2 the TSG-6·HC complex formation is significantly reduced (panel A, lane 6) and completely lost at a molar ratio of 1:800 (lane 7). IαI in the loose conformation (EDTA, panel B) was more susceptible to sulfo-NHS-acetate treatment and complex formation with TSG-6 is significantly reduced at a molar ratio of 1:200 (lane 5) and completely abolished at a 1:400 ratio. The data represent 3 technical replicates.
Discussion
In the present study, we have used a range of biochemical and biophysical methods to show that IαI changes its conformation and adopts a more compact structure in the presence of Mg2+ and Mn2+. Additionally, we have shown that CS is essential for the overall fold and stability of IαI. Previous studies have shown that Mg2+, Mn2+, or Ca2+ are able to act as cofactors in the TSG-6-mediated transfer of HCs from IαI to HA (13). Here we show that Mg2+ or Mn2+, but not Ca2+, cause IαI to adopt a more compact conformation and that this compact conformation is essential for the interaction with TSG-6.
The presence of either Mg2+ or Mn2+ shifts the retention of IαI during size exclusion chromatography. The altered retention time corresponds to a decrease in the calculated size of IαI, from a Stokes radius of 60 Å in the absence of cations to 55 and 56 Å in the presence of Mg2+ or Mn2+, respectively. Although IαI is compacted by Mg2+ and Mn2+, the calculated size is still larger than expected. The molecular mass of IαI including all known modifications is ∼179 kDa. If IαI were to be a fully globular protein it would result in a Stokes radius of 37 Å (42). The increased Stokes radius may be a result of a moderate elongation and/or be a result of a large hydration shell. In solution, proteoglycans typically obtain a large hydration shell due to the presence of the negatively charged GAG chains (43). Mg2+ and Mn2+ also increase the thermal stability and cooperativity of unfolding of IαI. This indicates that the conformational changes induced by Mg2+ and Mn2+ lead to a stabilization of the protein interactions in IαI. The increased cross-linking efficiency in the presence of Mg2+ and Mn2+ supports this observation. In plasma, the concentration of Ca2+ and Mg2+ is around 1 mm, whereas Mn2+ only is present in trace amounts (44). Based on the physiological concentrations, Mg2+, rather than Mn2+, is most likely the physiologically relevant ion in relationship to IαI structure and functions.
The structure of the native IαI complex is not well described. Two studies have examined the structure of IαI, however, in light of our new data showing that the structure of IαI involves both Mg2+ and CS it seems that these studies are based on either the EDTA or ChonABC-treated variant of IαI (26, 45). In the present study, we have used chemical footprinting and cross-linking to identify potential CS and Mg2+-dependent interaction sites as well as potential interfacing protein regions. The chemical footprinting revealed that bikunin has an Mg2+- and CS-dependent interaction, most likely directly to the CS. The Lys-70 and Lys-126 in bikunin are located in proximity in a region containing most of the charged residues in bikunin (25). This region is located on the opposing side of the protease inhibitor sites and could form a lysine- and Mg2+-dependent CS binding region, which would position bikunin for optimal interaction with targeted proteases. Lys-106 are found in the outskirts of this region and binds a sulfate group in the crystal structure, which could have substituted for the CS. Chemical footprinting also reveal that both HCs have Mg2+- and CS-dependent interactions. HC2 had what seems to be a direct interaction with the CS at Lys-193. Furthermore, HC2 had two lysine residues only affected by EDTA treatment, indicating an Mg2+-dependent interaction site. One of the residues, Lys-273, is located close to the MIDAS found in both HCs (residues 262–266 in HC2). An Mg2+-dependent interaction between HC1 and HC2 via their MIDAS sites could explain our observation. The biological significance of these interactions is emphasized by the observation that complex formation between TSG-6 and HCs is disrupted when the lysine-dependent interactions are prevented by acetylation.
The HC1-HC2 interaction is supported by chemical cross-linking data. Chemical cross-linkers are commonly used to characterize the protein structure and protein complexes by identifying residues that are spatially close (37). The maximal length of the BS3 spacer is 11.4 Å, thus allowing Cα-atoms of cross-linked lysine residues to be 26–30 Å apart in a proposed structure (46). We have identified both intra-protein and inter-protein cross-links in IαI. In both HCs, intra-protein cross-links that span from the N-terminal region to the C-terminal region are present, thus indicating that the N- and C-terminal regions are within close proximity in the native structure. Although the similar pattern of intra-protein cross-links in HC1 and HC2 suggests that there are similarities in the overall structure, the chemical footprinting and the fact that HC2 polymerize also reveal a clear difference between the two proteins. Inter-protein cross-links were identified between all protein components of IαI. Removal of divalent cations by EDTA resulted in a more loose IαI structure with a more dynamic distribution of bikunin. This falls in line with other results, showing that the packing of bikunin is strongly affected by divalent cations.
The HCs contain a vWA domain and within this domain the previously mentioned MIDAS motif. Our cross-linking data agrees with the common fold of the vWA domain in which the N and C termini of the domain are located at the same site of the domain and close to each other (47). The vWA domain is commonly associated with protein-protein interactions and with the formation of multiprotein complexes (24). Interactions between vWA containing proteins and their ligands often depend on the binding of divalent cations to a MIDAS motif and can be associated with conformational changes within the vWA domain. This is at least the case for vWA domain-containing integrins. However, among the vWA domain-containing proteins the mechanism of ligand binding is diverse (47). Our results show that divalent cation binding is associated with conformational changes in IαI. A possible site of cation binding is the MIDAS motif of the HCs. The coordination of Mg2+ ions at the MIDAS motifs in IαI could, similarly to Mg2+ ions in integrins, mediate interactions to other proteins or mediate interactions between the IαI protein components. Interestingly, interaction between IαI and another ECM protein, pentraxin 3, is also Mg2+ dependent (48). Pentraxin 3 is, similarly to IαI, essential for fertility in female mice, and it seems that the interactions between IαI and pentraxin 3 are important for organizing the ECM around oocytes. Interactions between IαI and complement components have similarly been suggested to depend on divalent cations and interactions with the vWA domain in IαI (40, 49).
In the present study, we have shown that interactions between the protein components exist within the native structure of IαI. These interactions are GAG-dependent and/or divalent cation-dependent. Furthermore, we have shown that divalent cations induce a more compact conformation of IαI in which the protein interactions are stabilized and the structural stability is increased. The cation-induced compact conformation is essential for the HC·TSG-6 complex formation. The compact structure is most likely the native fold of IαI. Because IαI is normally purified from citrated or EDTA-treated human plasma, this might have been overlooked in the past.
Author Contributions
C. L. N., C. S., K. W. S., and J. J. E. designed the study, analyzed the results and wrote the paper. C. L. N., I. B. T., and C. S. performed the majority of the experiments. M. S. and D. O. performed the circular dichroism spectroscopy and analyzed the data. L. W. and H. G. W. performed experiments on the TSG-6 interactions. All authors approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Peter Højrup and Morten I. Rasmussen for helpful suggestions on the use of the CrossWork/MassAI software and for implementing new features in the program.
The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental material.
- IαI
- inter-α-inhibitor
- HC
- heavy chain
- CS
- chondroitin 4-sulfate
- PGP
- protein-glycosaminoglycan-protein
- ECM
- extracellular matrix
- TSG-6
- tumor necrosis factor stimulated gene-6 protein
- HA
- hyaluronan
- wWA
- von Willebrand factor type A
- MIDAS
- metal-ion-dependent adhesion sites
- BS3
- bis(sulfosuccinimidyl) suberate
- NHS
- N-hydroxysulfosuccinimide
- ChonABC
- chondroitinase ABC
- ΔHTm
- enthalpy of unfolding at the midpoint of denaturation
- MGF
- Mascot generic format.
References
- 1. Enghild J. J., Thøgersen I. B., Pizzo S. V., and Salvesen G. (1989) Analysis of inter-α-trypsin inhibitor and a novel trypsin inhibitor, pre-α-trypsin inhibitor, from human plasma: polypeptide chain stoichiometry and assembly by glycan. J. Biol. Chem. 264, 15975–15981 [PubMed] [Google Scholar]
- 2. Thøgersen I. B., and Enghild J. J. (1995) Biosynthesis of bikunin proteins in the human carcinoma cell line HepG2 and in primary human hepatocytes: polypeptide assembly by glycosaminoglycan. J. Biol. Chem. 270, 18700–18709 [DOI] [PubMed] [Google Scholar]
- 3. Enghild J. J., Salvesen G., Thøgersen I. B., Valnickova Z., Pizzo S. V., and Hefta S. A. (1993) Presence of the protein-glycosaminoglycan-protein covalent cross-link in the inter-α-inhibitor-related proteinase inhibitor heavy chain 2/bikunin. J. Biol. Chem. 268, 8711–8716 [PubMed] [Google Scholar]
- 4. Morelle W., Capon C., Balduyck M., Sautiere P., Kouach M., Michalski C., Fournet B., and Mizon J. (1994) Chondroitin sulphate covalently cross-links the three polypeptide chains of inter-α-trypsin inhibitor. Eur. J. Biochem. 221, 881–888 [DOI] [PubMed] [Google Scholar]
- 5. Enghild J. J., Thøgersen I. B., Cheng F., Fransson L. A., Roepstorff P., and Rahbek-Nielsen H. (1999) Organization of the inter-α-inhibitor heavy chains on the chondroitin sulfate originating from Ser(10) of bikunin: posttranslational modification of IαI-derived bikunin. Biochemistry 38, 11804–11813 [DOI] [PubMed] [Google Scholar]
- 6. Enghild J. J., Salvesen G., Hefta S. A., Thøgersen I. B., Rutherfurd S., and Pizzo S. V. (1991) Chondroitin 4-sulfate covalently cross-links the chains of the human blood protein pre-α-inhibitor. J. Biol. Chem. 266, 747–751 [PubMed] [Google Scholar]
- 7. Jessen T. E., Faarvang K. L., and Ploug M. (1988) Carbohydrate as covalent crosslink in human inter-α-trypsin inhibitor: a novel plasma-protein structure. FEBS Lett. 230, 195–200 [DOI] [PubMed] [Google Scholar]
- 8. Ly M., Leach F. E. 3rd, Laremore T. N., Toida T., Amster I. J., and Linhardt R. J. (2011) The proteoglycan bikunin has a defined sequence. Nat. Chem. Biol. 7, 827–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhuo L., Hascall V. C., and Kimata K. (2004) Inter-α-trypsin inhibitor, a covalent protein-glycosaminoglycan-protein complex. J. Biol. Chem. 279, 38079–38082 [DOI] [PubMed] [Google Scholar]
- 10. Rugg M. S., Willis A. C., Mukhopadhyay D., Hascall V. C., Fries E., Fülöp C., Milner C. M., and Day A. J. (2005) Characterization of complexes formed between TSG-6 and inter-α-inhibitor that act as intermediates in the covalent transfer of heavy chains onto hyaluronan. J. Biol. Chem. 280, 25674–25686 [DOI] [PubMed] [Google Scholar]
- 11. Sanggaard K. W., Karring H., Valnickova Z., Thøgersen I. B., and Enghild J. J. (2005) The TSG-6 and IαI interaction promotes a transesterification cleaving the protein-glycosaminoglycan-protein (PGP) cross-link. J. Biol. Chem. 280, 11936–11942 [DOI] [PubMed] [Google Scholar]
- 12. Sanggaard K. W., Sonne-Schmidt C. S., Krogager T. P., Lorentzen K. A., Wisniewski H. G., Thøgersen I. B., and Enghild J. J. (2008) The transfer of heavy chains from bikunin proteins to hyaluronan requires both TSG-6 and HC2. J. Biol. Chem. 283, 18530–18537 [DOI] [PubMed] [Google Scholar]
- 13. Sanggaard K. W., Sonne-Schmidt C. S., Jacobsen C., Thøgersen I. B., Valnickova Z., Wisniewski H. G., and Enghild J. J. (2006) Evidence for a two-step mechanism involved in the formation of covalent HC x TSG-6 complexes. Biochemistry 45, 7661–7668 [DOI] [PubMed] [Google Scholar]
- 14. Sanggaard K. W., Scavenius C., Rasmussen A. J., Wisniewski H. G., Thøgersen I. B., and Enghild J. J. (2010) The TSG-6/HC2-mediated transfer is a dynamic process shuffling heavy chains between glycosaminoglycans. J. Biol. Chem. 285, 21988–21993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Milner C. M., and Day A. J. (2003) TSG-6: a multifunctional protein associated with inflammation. J. Cell Sci. 116, 1863–1873 [DOI] [PubMed] [Google Scholar]
- 16. Selbi W., Day A. J., Rugg M. S., Fülöp C., de la Motte C. A., Bowen T., Hascall V. C., and Phillips A. O. (2006) Overexpression of hyaluronan synthase 2 alters hyaluronan distribution and function in proximal tubular epithelial cells. J. Am. Soc. Nephrol. 17, 1553–1567 [DOI] [PubMed] [Google Scholar]
- 17. de la Motte C. A., Hascall V. C., Drazba J., Bandyopadhyay S. K., and Strong S. A. (2003) Mononuclear leukocytes bind to specific hyaluronan structures on colon mucosal smooth muscle cells treated with polyinosinic acid:polycytidylic acid: inter-α-trypsin inhibitor is crucial to structure and function. Am. J. Pathol. 163, 121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhuo L., Kanamori A., Kannagi R., Itano N., Wu J., Hamaguchi M., Ishiguro N., and Kimata K. (2006) SHAP potentiates the CD44-mediated leukocyte adhesion to the hyaluronan substratum. J. Biol. Chem. 281, 20303–20314 [DOI] [PubMed] [Google Scholar]
- 19. Yingsung W., Zhuo L., Morgelin M., Yoneda M., Kida D., Watanabe H., Ishiguro N., Iwata H., and Kimata K. (2003) Molecular heterogeneity of the SHAP-hyaluronan complex: isolation and characterization of the complex in synovial fluid from patients with rheumatoid arthritis. J. Biol. Chem. 278, 32710–32718 [DOI] [PubMed] [Google Scholar]
- 20. Zhao M., Yoneda M., Ohashi Y., Kurono S., Iwata H., Ohnuki Y., and Kimata K. (1995) Evidence for the covalent binding of SHAP, heavy chains of inter-α-trypsin inhibitor, to hyaluronan. J. Biol. Chem. 270, 26657–26663 [DOI] [PubMed] [Google Scholar]
- 21. Zhuo L., Yoneda M., Zhao M., Yingsung W., Yoshida N., Kitagawa Y., Kawamura K., Suzuki T., and Kimata K. (2001) Defect in SHAP-hyaluronan complex causes severe female infertility: a study by inactivation of the bikunin gene in mice. J. Biol. Chem. 276, 7693–7696 [DOI] [PubMed] [Google Scholar]
- 22. Sato H., Kajikawa S., Kuroda S., Horisawa Y., Nakamura N., Kaga N., Kakinuma C., Kato K., Morishita H., Niwa H., and Miyazaki J. (2001) Impaired fertility in female mice lacking urinary trypsin inhibitor. Biochem. Biophys. Res. Commun. 281, 1154–1160 [DOI] [PubMed] [Google Scholar]
- 23. Fülöp C., Szántó S., Mukhopadhyay D., Bárdos T., Kamath R. V., Rugg M. S., Day A. J., Salustri A., Hascall V. C., Glant T. T., and Mikecz K. (2003) Impaired cumulus mucification and female sterility in tumor necrosis factor-induced protein-6 deficient mice. Development 130, 2253–2261 [DOI] [PubMed] [Google Scholar]
- 24. Whittaker C. A., and Hynes R. O. (2002) Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol. Biol. Cell 13, 3369–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xu Y., Carr P. D., Guss J. M., and Ollis D. L. (1998) The crystal structure of bikunin from the inter-α-inhibitor complex: a serine protease inhibitor with two Kunitz domains. J. Mol. Biol. 276, 955–966 [DOI] [PubMed] [Google Scholar]
- 26. Blom A. M., Mörgelin M., Oyen M., Jarvet J., and Fries E. (1999) Structural characterization of inter-α-inhibitor: evidence for an extended shape. J. Biol. Chem. 274, 298–304 [DOI] [PubMed] [Google Scholar]
- 27. Lee T. H., Wisniewski H. G., and Vilcek J. (1992) A novel secretory tumor necrosis factor-inducible protein (TSG-6) is a member of the family of hyaluronate binding proteins, closely related to the adhesion receptor CD44. J. Cell Biol. 116, 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bury A. (1981) Analysis of protein and peptide mixtures: evaluation of three sodium dodecyl sulphate-polyacrylamide gel electrophoresis buffer systems. J. Chromatogr. A 213, 491–500 [Google Scholar]
- 29. Uriel J., and Berges J. (1968) Characterization of natural inhibitors of trypsin and chymotrypsin by electrophoresis in acrylamide-agarose gels. Nature 218, 578–580 [DOI] [PubMed] [Google Scholar]
- 30. Matsudaira P. (1987) Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J. Biol. Chem. 262, 10035–10038 [PubMed] [Google Scholar]
- 31. Laurent T. C., and Killander J. (1964) Theory of gel filtration and its experimental verification. J. Chromatogr. A 14, 317–330 [Google Scholar]
- 32. Mogensen J. E., Ipsen H., Holm J., and Otzen D. E. (2004) Elimination of a misfolded folding intermediate by a single point mutation. Biochemistry 43, 3357–3367 [DOI] [PubMed] [Google Scholar]
- 33. Kussmann M., Lässing U., Stürmer C. A., Przybylski M., and Roepstorff P. (1997) Matrix-assisted laser desorption/ionization mass spectrometric peptide mapping of the neural cell adhesion protein neurolin purified by sodium dodecyl sulfate polyacrylamide gel electrophoresis or acidic precipitation. J. Mass Spectrom. 32, 483–493 [DOI] [PubMed] [Google Scholar]
- 34. Rasmussen M. I., Refsgaard J. C., Peng L., Houen G., and Højrup P. (2011) CrossWork: software-assisted identification of cross-linked peptides. J. Proteomics 74, 1871–1883 [DOI] [PubMed] [Google Scholar]
- 35. Schilling B., Rardin M. J., MacLean B. X., Zawadzka A. M., Frewen B. E., Cusack M. P., Sorensen D. J., Bereman M. S., Jing E., Wu C. C., Verdin E., Kahn C. R., Maccoss M. J., and Gibson B. W. (2012) Platform-independent and label-free quantitation of proteomic data using MS1 extracted ion chromatograms in skyline: application to protein acetylation and phosphorylation. Mol. Cell. Proteomics 11, 202–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., and MacCoss M. J. (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rappsilber J. (2011) The beginning of a beautiful friendship: cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes. J. Struct. Biol. 173, 530–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jackson M. B. (2006) Molecular and Cellular Biophysics, Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 39. Carra J. H., Murphy E. C., and Privalov P. L. (1996) Thermodynamic effects of mutations on the denaturation of T4 lysozyme. Biophys. J. 71, 1994–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Okroj M., Holmquist E., Sjölander J., Corrales L., Saxne T., Wisniewski H. G., and Blom A. M. (2012) Heavy chains of inter α inhibitor (IαI) inhibit the human complement system at early stages of the cascade. J. Biol. Chem. 287, 20100–20110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Olsen E. H., Rahbek-Nielsen H., Thogersen I. B., Roepstorff P., and Enghild J. J. (1998) Posttranslational modifications of human inter-α-inhibitor: identification of glycans and disulfide bridges in heavy chains 1 and 2. Biochemistry 37, 408–416 [DOI] [PubMed] [Google Scholar]
- 42. Erickson H. P. (2009) Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol. Proced. Online 11, 32–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gandhi N. S., and Mancera R. L. (2008) The structure of glycosaminoglycans and their interactions with proteins. Chem. Biol. Drug Des. 72, 455–482 [DOI] [PubMed] [Google Scholar]
- 44. Zhang K., and Chen J. (2012) The regulation of integrin function by divalent cations. Cell Adh. Migr. 6, 20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Flahaut C., Mizon C., Aumercier-Maes P., Colson P., Bailly C., Sautiere P., and Mizon J. (1998) Disulphide bonds assignment in the inter-α-inhibitor heavy chains: structural and functional implications. Eur. J. Biochem. 255, 107–115 [DOI] [PubMed] [Google Scholar]
- 46. Merkley E. D., Rysavy S., Kahraman A., Hafen R. P., Daggett V., and Adkins J. N. (2014) Distance restraints from cross-linking mass spectrometry: mining a molecular dynamics simulation database to evaluate lysine-lysine distances. Protein Sci. 23, 747–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Springer T. A. (2006) Complement and the multifaceted functions of VWA and integrin I domains. Structure 14, 1611–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Scarchilli L., Camaioni A., Bottazzi B., Negri V., Doni A., Deban L., Bastone A., Salvatori G., Mantovani A., Siracusa G., and Salustri A. (2007) PTX3 interacts with inter-α-trypsin inhibitor: implications for hyaluronan organization and cumulus oophorus expansion. J. Biol. Chem. 282, 30161–30170 [DOI] [PubMed] [Google Scholar]
- 49. Garantziotis S., Hollingsworth J. W., Ghanayem R. B., Timberlake S., Zhuo L., Kimata K., and Schwartz D. A. (2007) Inter-α-trypsin inhibitor attenuates complement activation and complement-induced lung injury. J. Immunol. 179, 4187–4192 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.