Abstract
DNA replication in almost all organisms depends on the activity of DNA primase, a DNA-dependent RNA polymerase that synthesizes short RNA primers of defined size for DNA polymerases. Eukaryotic and archaeal primases are heterodimers consisting of small catalytic and large accessory subunits, both of which are necessary for the activity. The mode of interaction of primase subunits with substrates during the various steps of primer synthesis that results in the counting of primer length is not clear. Here we show that the C-terminal domain of the large subunit (p58C) plays a major role in template-primer binding and also defines the elements of the DNA template and the RNA primer that interact with p58C. The specific mode of interaction with a template-primer involving the terminal 5′-triphosphate of RNA and the 3′-overhang of DNA results in a stable complex between p58C and the DNA/RNA duplex. Our results explain how p58C participates in RNA synthesis and primer length counting and also indicate that the binding site for initiating NTP is located on p58C. These findings provide notable insight into the mechanism of primase function and are applicable for DNA primases from other species.
Keywords: DNA primase, DNA replication, gel electrophoresis, human, RNA synthesis, DNA-protein interaction, RNA primer length counting, p58 subunit, primase activity, RNA/DNA duplex, 5′-triphosphate
Introduction
The four-subunit primase-polymerase α (Prim-Polα)3 complex possessing DNA primase and DNA polymerase active sites is important for genome replication in eukaryotes (1, 2). Prim-Polα synthesizes the chimeric RNA-DNA primers for replicative DNA polymerases ϵ and δ (3, 4). In humans, the primase heterodimer contains a small catalytic subunit (p49; also known as p48, PRIM1, Pri1, and PriS) and a large regulatory subunit (p58; also known as PRIM2, Pri2, and PriL). Polα is composed of a large catalytic subunit (p180) and a small accessory subunit (p70). p58 and p70 are connected with p180 through the interaction with a small C-terminal domain (p180C) that defines the tight coordination of the RNA- and DNA-polymerizing activities (5–7).
Eukaryotic primases have a minimal specific recognition site on DNA and only require a pyrimidine to template the 5′-terminal nucleotide of the primer (8, 9). RNA primer synthesis begins with a rate-limiting initiation step that includes binding of the DNA template and two NTPs followed by dinucleotide synthesis (10). Subsequently, primase elongates the generated dinucleotide to the unit-length primer (8–10-mer) and terminates synthesis. This unique counting ability of DNA primases, which results in RNA primers that are optimal for extension by Polα, has a complex mechanism that is currently unclear. Recent structural data revealed that the primase active site located on p49 uses the common mechanism of nucleic acids synthesis, where the catalytic aspartates coordinate two divalent ions and the triphosphate moiety of the incoming NTP (11, 12).
The large subunit of human primase is composed of two separate domains connected with a long linker, which indicates a significant conformational flexibility of this subunit (13). The N-terminal domain (p58N) provides a platform for interactions with p49 and Polα (6, 12) and also plays some role in the orientation of p58C relative to p49. p58C is an all-helical domain with a buried iron-sulfur cluster coordinated by four conservative cysteines, which plays an important structural role (14, 15). Biochemical data indicate that p58C contributes to primase activity and participates in substrate binding by an unknown mechanism (16–19). Our recent report provided several pieces of evidence that p58C and p49 cooperate in a cis orientation during primer synthesis, meaning that the template-primer is handled by two functional domains located on the same two-subunit molecule (13).
In this report we defined two structural elements of the template-primer that play a critical role in primase-substrate interaction. We determined that the binding sites for the 5′-triphosphate of an RNA primer and the 3′-overhang of a DNA template are located on p58C. This finding suggests a new mechanism for participation of p58C in primase activity by taking responsibility for substrate binding and primer length counting.
Experimental Procedures
Cloning, Expression, and Purification
Cloning, expression, and purification to homogeneity of the human primase heterodimer (p49-p58; p49 contains 420 amino acids, p58 contains 509), the C-terminal domain of p58 (p58C; amino acids 266–456), primase without p58C (p49-p58N; p58N contains amino acids 1–265), and the human Polα without the catalytic core (p70-p180C; p180C contains amino acids 1265–1462) have been described elsewhere (13, 20, 21). The complexes p49-p58-p70-p180C (Prim-PolαΔcat) and p49-p58N-p70-p180C (PrimΔFeS-PolαΔcat) have been obtained in the same way as Prim-Polα (6); the pure primase and Polα samples were mixed at an equimolar ratio and incubated on ice for 30 min, and the reconstituted complexes were purified by gel-filtration on a Superose 12 column (GE Healthcare). The expression vector for T7 RNA polymerase (BH161A) was kindly provided by Dr. Dmitry Temiakov (Rowan University). T7 RNA polymerase was expressed in Escherichia coli strain Rosetta 2(DE3) at 30 °C for 4 h after induction with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside at an A600 of 1, then purified to homogeneity by 3 consecutive columns: Ni-IDA (Bio-Rad), Heparin HP HiTrap (GE Healthcare), and Superose 12 (GE Healthcare).
DNA and RNA Constructs
All oligonucleotides without 5′-triphosphate were obtained from IDT Inc. (Table 1). RNA primers containing 5′-triphosphate (P1 and P3) were synthesized by T7 RNA polymerase according to Milligan et al. (22) and purified by a Mono Q column (GE Healthcare). DNA duplex T9-P5 was used for synthesis of P1, and the duplex T10-P5 was used for synthesis of P3. The duplexes were obtained at 0.1 mm concentration by 1-min heating to 80 °C and passive chilling to room temperature for 15 min in the buffer containing 20 mm Tris-HCl (pH 7.9) and 0.1 m NaCl. Annealing temperatures for DNA/DNA and DNA/RNA duplexes were calculated using the web-based software OligoAnalyzer 3.1 (IDT Inc.).
TABLE 1.
Oligonucleotides used in the current study
| Name | Length | DNA/RNA | Sequence (template region involved in duplex formation is underlined) |
|---|---|---|---|
| T1 | 18 | DNA | 5′-AAACACCGAGCCAACATA |
| T2 | 13 | DNA | 5′-CCGAGCCAACATA |
| T3 | 13 | DNA | 5′-AAACACCGAGCCA |
| T4 | 12 | DNA | 5′-AAACACCGAGCC |
| T5 | 20 | DNA | 5′-Cy3-AACTAACCCGAGCCAATACA |
| P1 | 7 | RNA | 5′-pppGGCUCGG |
| P2 | 7 | RNA | 5′-GGCUCGG |
| T6 | 60 | DNA | 5′-gtgagagaaggagaaggagaggaagagaaggagaagaggagagaaacgccgccaaaaaaa |
| T7 | 53 | DNA | 5′-gtgagagaaggagaaggagaggaagagaaggagaagaggagagaaacgccgcc |
| T8 | 33 | DNA | 5′-GGAGTACGAATCAGTTAGACGCCGCCAAGAATA |
| P3 | 6 | RNA | 5′-pppGGCGGC |
| P4 | 6 | RNA | 5′-GGCGGC |
| T9 | 24 | DNA | 5′-CCGAGCCTATAGTGAGTCGTATTA |
| T10 | 23 | DNA | 5′-GCCGCCTATAGTGAGTCGTATTA |
| P5 | 18 | DNA | 5′-AATACGACTCACTATAGG |
| T11 | 33 | DNA | 5′-AAAAAAAAAAAAAAAAAAAAAAAGATAAAAAAA |
Electrophoretic Mobility Gel Shift Assay (EMSA)
Reactions containing 6 μm Prim-PolαΔcat or 10 μm p58C and 10% molar excess of a template-primer were incubated in 10 μl for 10 min at room temperature in the buffer containing 10 mm Tris-HCl (pH 7.7), 50 mm KCl, 1% glycerol, and 1 mm DTT; 5 μl was then loaded on 5% native PAGE. Gel was stained for 10 min in running buffer containing 0.5 μg/ml ethidium bromide and 2 mm MgCl2. After recording the image of DNA staining, the gel was stained by 1% Coomassie R-250. In experiments to measure the apparent Kd, the reaction buffer was supplemented with 0.1 mg/ml BSA and 2 μm T11 to stabilize samples at a low concentration and to prevent nonspecific interactions. Samples labeled with Cy3 dye were visualized using the Typhoon 9410 imager (emission of fluorescence at 580 nm). Bands were quantified using ImageJ software (version 1.45s, National Institutes of Health). The apparent Kd values were calculated from three independent experiments by non-linear regression using the Hill equation (one-site specific binding mode) with the Prism 6 (GraphPad Software, Inc.).
Primase Assay
Activity of primase in extension of GGCGGC ribo-primers (with or without terminal 5′-triphosphate) annealed to different DNA templates was tested in a 20-μl reaction containing 30 mm Hepes-KOH (pH 7.9), 50 mm KCl, 1 mm DTT, 2 mm MgCl2, 100 μm UTP, 100 μm CTP, 0.25 μm [α-33P]GTP (3000 Ci/mmol; PerkinElmer Life Sciences), 1 μm template-primer, and 50 nm Prim-PolαΔcat. Reactions were incubated at 35 °C in a thermal cycler (Thermolyne Amplitron I) and stopped by mixing with an equal volume of formamide loading buffer (95% v/v formamide, 5 mm EDTA 0.02% bromphenol blue, 0.02% xylene cyanol, and 0.025% SDS), heated at 65 °C for 10 min, and resolved by 20% urea-PAGE (UreaGel System (19:1 acrylamide/bisacrylamide); National Diagnostics) for 5 h at 2000 V. The gel was dried at 65 °C for 45 min using a Bio-Rad 583 gel dryer. The reaction products were visualized by phosphorimaging (Typhoon 9410, GE Healthcare). All activity gels were repeated at least two times. The intensities of bands were quantified using ImageQuant software (version 5.2, GE Healthcare).
Primase activity in extension of the rA15 primer was tested in a 20-μl reaction containing 30 mm Hepes-KOH (pH 7.9), 50 mm KCl, 1 mm DTT, 4 mm MgCl2, 100 μm ATP, 0.2 μm 5′-TYE665 fluorophore-labeled rA15 primer (obtained from IDT, Inc.) annealed to dT70 template, and 40 nm Prim-PolαΔcat. Reactions were incubated for 10 min at 30 °C and stopped by mixing with an equal volume of formamide loading buffer (95% formamide, 0.025% Orange G, 5 mm EDTA, and 0.025% SDS), heated at 65 °C for 10 min, and resolved by 16% urea-PAGE (UreaGel System (19:1 acrylamide/bisacrylamide); National Diagnostics) for 5 h at 2000 V. The products were visualized using the Typhoon 9410 imager (emission of fluorescence at 670 nm) and quantified as described previously. The IC50 values were calculated from two to three independent experiments by non-linear regression using Prism 6 (GraphPad Software, Inc.).
Results
Triphosphate and the 3′-Overhang of DNA/RNA Duplex Are Important for Interaction with Primase
RNA primers synthesized by primases or RNA polymerases contain the triphosphate moiety at the 5′-terminus (TriP) (23, 24). To examine the role of the TriP in the interaction of the human primase with the substrate, we used the DNA template T1 (oligonucleotides and their sequences are listed in Table 1), which could be annealed with a 7-mer RNA primer with or without TriP (P1 and P2, correspondingly) and has the 5′- and 3′-overhangs to mimic the natural primase substrate. In addition, we studied the role of the overhangs on the stability of the Prim-DNA/RNA complex using a series of additional templates (T2-T4, Table 1). To monitor the Prim-DNA/RNA interactions that are specific for the primase, we used the Prim-Polα complex with the Polα catalytic domain deleted (Prim-PolαΔcat) and applied EMSA, which allows visualization of protein-DNA interactions by a shift in the mobility of the protein and DNA molecules when they make a complex (25).
The addition of T1-P1 (contains a 7-bp duplex, both overhangs, and TriP; Fig. 1A) to Prim-PolαΔcat results in the formation of a stable DNA-protein complex with higher electrophoretic mobility than Prim-PolαΔcat (Fig. 1B, comparison of lanes 1 and 2). The absence of the 5′-overhang does not affect the complex of Prim-PolαΔcat with DNA/RNA (lane 4), whereas the absence of the 3′-overhang or TriP results in complete disruption of the complex (lanes 3 and 6). The weak interaction between Prim-PolαΔcat and the substrate was detected when the length of the 3′-overhang was reduced from six to one nucleotide (lane 5; only a small portion of protein was shifted on the right panel in comparison to lane 1). Altogether, these data indicate that Prim-PolαΔcat specifically binds the DNA/RNA junction at the primer 5′-terminus containing TriP.
FIGURE 1.
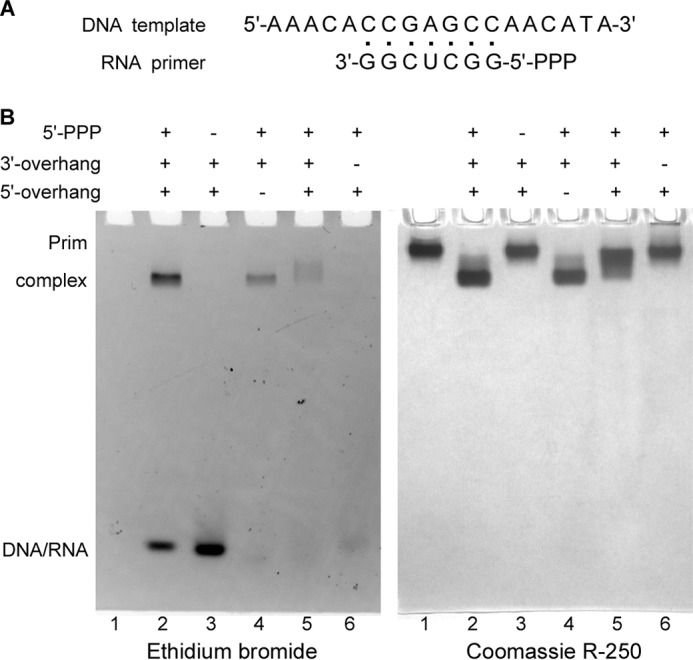
Analysis of Prim-PolαΔcat interaction with DNA/RNA duplexes by EMSA. A, structure of the T1-P1 substrate. B, 5′-triphosphate and the 3′-overhang are critical for interaction of DNA/RNA substrate with the human primase. The total amount of Prim-PolαΔcat loaded on each of all lanes was 6 μg (30 pmol); the amount of DNA/RNA substrate in each of lanes 2–6 was 35 pmol. Lane 1, no substrate; lane 2, T1-P1; lane 3, T1-P2; lane 4, T2-P1; lane 5, T3-P1 (1-nucleotide-long 3′-overhang); lane 6, T4-P1. Samples were analyzed by electrophoresis in 5% native PAGE and stained first with ethidium bromide (left panel) and then with Coomassie R-250 (right panel). The observed difference in efficiency of DNA/RNA staining is most likely caused by effect of the template overhangs on the ethidium bromide-DNA/RNA complex in the case of short duplexes. The duplexes used in lanes 4–6 are missing the 5′- or 3′-overhang, or the overhang is shortened to one nucleotide. Reduced DNA staining in lanes 5 and 6 is also due to the smearing effect because of the protein-DNA/RNA complex dissociation during electrophoresis.
TriP and the 3′-Overhang Binding Sites Are Located on p58C
Several pieces of evidence obtained earlier indicate that p58C somehow participates in the interaction with substrates because it plays an important role in primase function, especially during dinucleotide synthesis (8, 16, 17, 26–28). We decided to analyze the interaction of p58C alone with the same set of substrates as was used for Prim-PolαΔcat. p58C is a basic protein (calculated pI is 9.1) that migrates during electrophoresis in reverse direction, which precludes its visualization in the absence of complex with the negatively charged substrate (Fig. 2, lane 1 on the right panel). When p58C makes a stable complex with DNA/RNA, it migrates in the normal direction. Amazingly, the p58C-DNA/RNA complex demonstrated the same dependence on TriP and the 3′-overhang (Fig. 2, lanes 2–6) as the entire Prim-PolαΔcat. This result supports the idea that binding sites for both of these elements of the RNA-primed DNA template are located on p58C. Thus, p58C, TriP, and the 3′-overhang play key roles in the interaction of Prim-PolαΔcat and, consequently, primase with a substrate during RNA primer synthesis.
FIGURE 2.
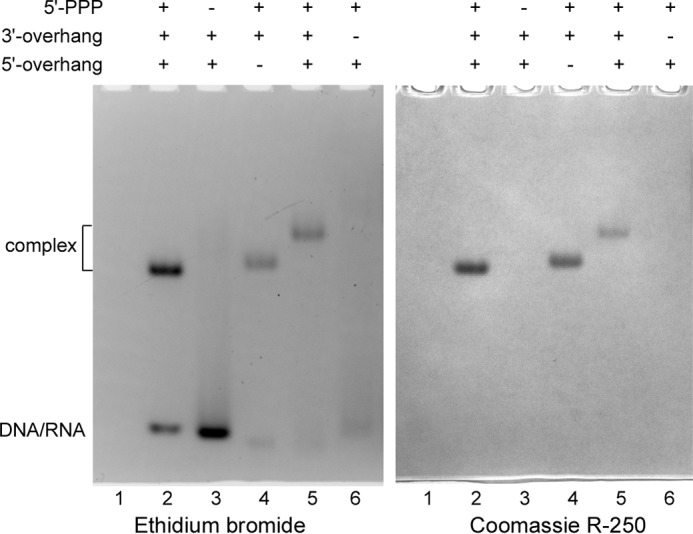
Analysis of p58C interaction with DNA/RNA duplexes by EMSA. The total amount of p58C loaded on each lane was 1.1 μg (50 pmol); the amount of DNA/RNA ligand in each one of lanes 2–6 was 55 pmol. Lane 1, no substrate; lane 2, T1-P1; lane 3, T1-P2; lane 4, T2-P1; lane 5, T3-P1 (1-nucleotide-long 3′-overhang); lane 6, T4-P1. Samples were analyzed by electrophoresis in 5% native PAGE and stained first with ethidium bromide (left panel) and then with Coomassie R-250 (right panel). p58C is a basic protein and moves in reverse direction when not in the complex with DNA/RNA.
Structural Elements of Substrates Defining Primase Affinity
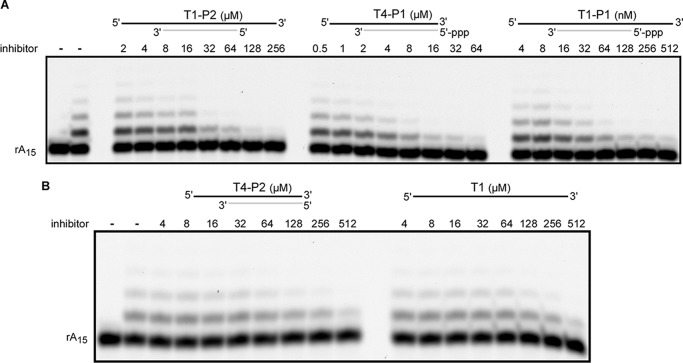
To estimate the contribution of TriP and the 3′-overhang in primase affinity for DNA/RNA, we determined the apparent Kd values for different substrates. EMSA is not suitable for measuring the dissociation constant of low affinity complexes (when Kd is >3 μm), so the competitive inhibition assay has been employed. The dependence of the primase ability to extend the rA15 primer annealed to the dT70 template on the concentration of substrates T1-P1, T1-P2, T4-P1, and T4-P2 was analyzed using urea-PAGE (Fig. 3). We considered dT70-rA15 as an analog of T1-P2 because it is missing TriP. Assuming that primase has a similar affinity for these substrates, it was considered that the level of the Prim-dT70/rA15 complex is negligibly low at the given reaction conditions, and the obtained IC50 values were directly converted to the apparent Kd values (Table 2, columns 3–7). The optimal T1-P1 substrate showed a fairly high affinity for primase (apparent Kd is 36.0 nm), whereas the absence of the 3′-overhang or TriP or both reduced the Prim-DNA/RNA complex strength ∼90-, 360-, and 4,000-fold, respectively. Moreover, primase exhibited similar affinities for ssDNA (T1) and the DNA/RNA duplex without TriP and the 3′-overhang (T4-P2). These data revealed the key role of TriP and the 3′-overhang for template-primer binding. The importance of TriP for interaction with a DNA/RNA duplex has also been shown previously for the primase of bacteriophage T7 (29).
FIGURE 3.
Inhibition of rA15 primer extension by substrates with different structures. Panels A and B correspond to two representative gels. Reactions containing 40 nm Prim-PolαΔcat and 0.2 μm 5′-TYE665 fluorophore-labeled rA15 primer annealed to the dT70 template were conducted for 10 min at 30 °C. The products were analyzed by electrophoresis in 16% urea-PAGE and visualized using the Typhoon 9410 imager (emission of fluorescence at 670 nm). The left lane is the control incubation without enzyme. On the top schematics, DNA and RNA strands are shown by black and gray lines, respectively.
TABLE 2.
Effect of the template-primer structure on the affinity for primase and p58C
| Protein | Apparent Kd values with 95% confidence intervals |
|||||
|---|---|---|---|---|---|---|
| T5-P1a | T1-P1 | T4-P1b | T1-P2c | T4-P1d | T1 | |
| nm | nm | μm | μm | μm | μm | |
| Primase | 27.6 25.3–29.9 |
36.0 28.6–45.3 |
3.3 2.1–5.1 |
12.9 10.3–16.2 |
153.1 114.2–205.1 |
195.4 139.9–272.9 |
| p58C | 32.7 28.9–36.6 |
|||||
| Assay | EMSA | Competitive inhibition | ||||
a T5-P1 substrate has a similar structure with T1-P1 and contains Cy3 dye at the 5′-terminus of T5.
b The 3′-overhang of DNA template is absent.
c The 5′-triphosphate of RNA primer is absent.
d The 5′-triphosphate and 3′-overhang are absent.
By measuring the fluorescence anisotropy of fluorescein-labeled oligonucleotides, it was shown that yeast primase binds dT40 with the apparent Kd of 1.7 μm (27). Approximately 100-fold higher ssDNA binding affinity of the yeast versus human primase is difficult to explain by the differences in ssDNA sequence and length. Moreover, similar structural organization of eukaryotic primases suggests the common mode of template binding. Potentially, ssDNA can interact with primase non-specifically as well. Our Kd determination method detects only specific interactions between primase and ssDNA, whereas the anisotropy method would detect both types of interactions. We compared the interactions of Prim and Prim-PolαΔcat with T1 and T1-P1 (Fig. 4). Both proteins efficiently bind T1-P1 (lanes 2 and 6), but their ssDNA binding properties differ: Prim-PolαΔcat does not interact with T1 at the 5 μm concentration (lane 5), whereas approximately half of T1 makes a complex with Prim (lane 1). This result means that the apparent Kd for Prim-T1 nonspecific complex is around 5 μm, which is close to the value obtained for the complex of yeast primase and dT40 (27). These data indicate that human primase has a nonspecific DNA binding site at the Prim-Polα interaction interface. Therefore, due to a very low Prim affinity for ssDNA or DNA/RNA duplexes without TriP and 3′-overhang, nonspecific interactions with these substrates can significantly affect the measurement of specific binding. Despite weak binding to ssDNA, primase loading on the template in vivo might be facilitated by Prim-Polα integration into the replisome (30, 31).
FIGURE 4.
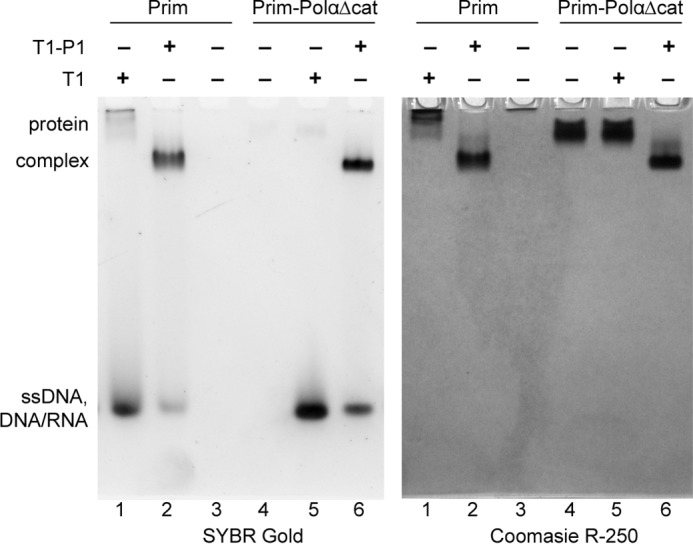
Association with PolαΔcat prevents nonspecific interaction of Prim with ssDNA. All reactions contained 5 μm protein and/or DNA. Samples were analyzed by electrophoresis in 5% native PAGE and stained first with SYBR Gold (left panel) and then with Coomassie R-250 (right panel). Prim is a basic protein and moves in reverse direction when not in the complex with ssDNA or the DNA/RNA duplex.
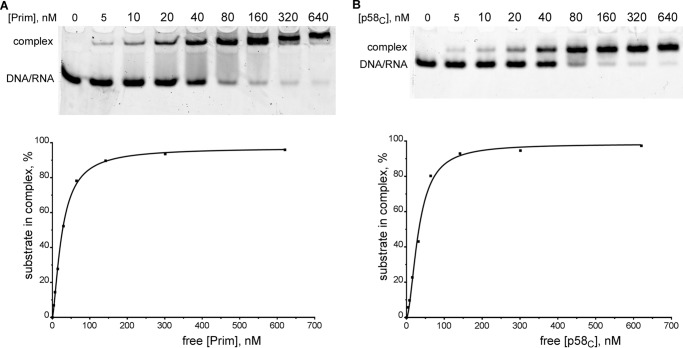
To better understand the contribution of p58C to the interaction between Prim and DNA/RNA, we have compared the affinities of Prim and p58C to T5-P1 by EMSA (Fig. 5). T5 has a similar structure to T1 and is labeled at the 5′-end with a Cy3-dye (Table 1). The obtained apparent Kd values were almost the same (Table 2; second row), which indicates that p58C domain in primase is the main element responsible for template-primer binding. This finding is consistent with the early report where only p58 reacted with photo-reactive DNA upon photolysis in experiments with photo-cross-linking of ssDNA-primase complexes before and after primer synthesis (17).
FIGURE 5.
Primase and p58C bind the template-primer with a similar affinity. Primase (A) or p58C (B) of varying concentrations were incubated with T5-P1 (20 nm; T5 is labeled with Cy3 at the 5′-end). The reaction products were separated by 5% native PAGE with subsequent visualization using the Typhoon 9410 imager (emission of fluorescence at 580 nm). The graphic analyses of the obtained data were performed using the Prism 6 software (one-site specific binding with a Hill slope). Concentration of the free protein was calculated by subtracting the concentration of the protein-DNA/RNA complex from the total protein concentration.
TriP and the 3′-Overhang Are Important for Primase Activity
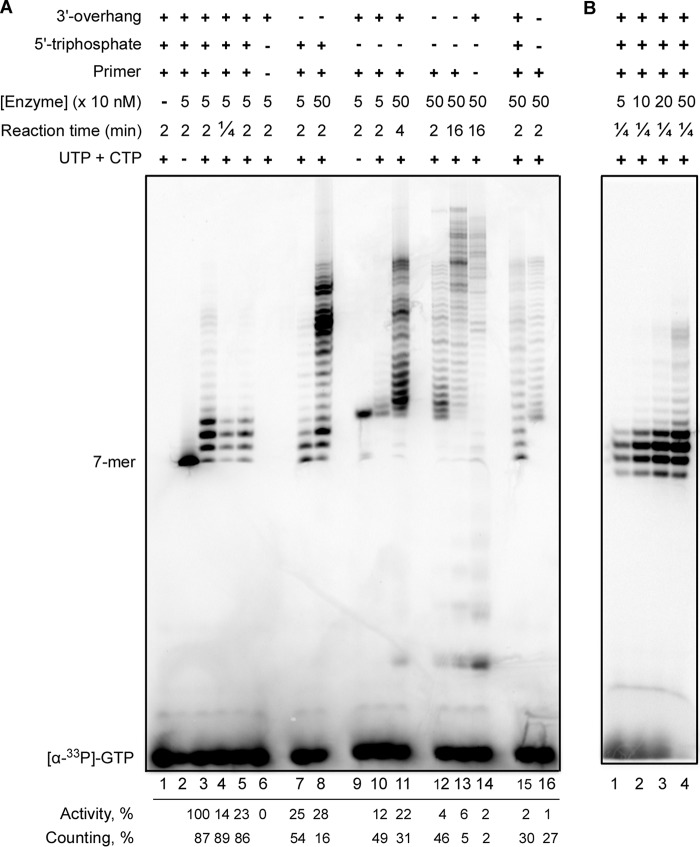
Most of the substrates used so far for the analysis of primase activity in extension of RNA primers contained the fluorescent label at the primer's 5′-end, which simplified the experiments but abrogated the usage of natural 5′-terminal triphosphate. Furthermore, other factors that make traditionally used substrates more artificial are the length of RNA primers (they were usually longer than primase products generated in vivo), absence of the 3′-overhang, and homopolymeric sequences of the templates. Here we designed a new template-primer (T6-P3, Table 1) that mimics the natural primase substrate. The duplex length was reduced to the minimum (six base pairs) sufficient to have a stable template-primer complex at the reaction conditions. The products were labeled only at one site by incorporation of [α-33P]GTP at the primer's seventh position. To avoid de novo synthesis, the pyrimidines were excluded from the single-stranded (ss) regions of the DNA template. Four different substrates including the optimal one (T6-P3) were used to analyze the effects of TriP (T6-P4), the 3′-overhang (T7-P3), and both (T7-P4) on primase activity. The reaction time and Prim-PolαΔcat concentration were varied to see the whole range of activity for each substrate. The cold GTP was typically not added to the reactions to avoid dilution of the radioactive label, and as a result, reduced band intensity further complicating the analysis of low activity samples. The addition of cold GTP does not, however, affect the distribution of product sizes (Fig. 6A, lane 5; compare with lanes 3 and 4). Reactions conducted in the absence of NTPs, except [α-33P]GTP, allow for unambiguous identification of +1 primer extension products in the presence and absence of TriP (lanes 2 and 9; note that TriP significantly affects the mobility of RNA primers).
FIGURE 6.
Effects of the template-primer structure and p58C on primase activity. Primase activity was analyzed in reactions of extension of 6-mer RNA primers (with or without TriP) annealed with 53- or 60-mer DNA templates. The products were labeled by incorporation of [α-33P]GTP at the seventh position. Note that the negatively charged TriP moiety significantly increased the mobility of RNA primers. A, analysis of the primase activity of Prim-PolαΔcat (lanes 1–14) and PrimΔFeS-PolαΔcat (lanes 15 and 16). Lanes 1–5 and 15, T6-P3; lanes 6 and 14, T6; lanes 7 and 8, T7-P3; lanes 9–11, T6-P4; lanes 12, 13, and 16, T7-P4. Reactions corresponding to lane 5 contained 10 μm GTP. Quantification of the data is provided under each lane below the gel. The intensity of all bands was determined for estimation of the relative primase activity normalized to the protein concentration (the highest activity level was taken for 100%). The primase counting ability was calculated as the ratio of the combined intensity of the bands corresponding to the 8, 9, and 10-mer primers to the total intensity of all products, multiplied by 100%. B, effect of the enzyme-template ratio on the primase ability to terminate primer synthesis at the defined position. Reactions were run at the same conditions as for lane 4 (panel A) except the concentration of Prim-PolαΔcat was varied.
Extension of the RNA primer P3 (GGCGGC) containing TriP and annealed with a 60-mer DNA template T6 resulted in the appearance of the product pattern (Fig. 6A, lanes 3 and 4) similar to de novo synthesis (32). The primase activity was estimated semi-quantitatively as described in the legend to Fig. 6. The absence of the 3′-overhang results in a 4-fold reduction in activity (comparison of lanes 3 and 7). TriP has a stronger impact on the primase activity in comparison to the 3′-overhang (comparison of lanes 7 and 10).
Primase efficiently counts the primer length on the T6-P3 substrate; the major products have the length of 8–10 nucleotides independent of reaction time (Fig. 6A, lanes 3 and 4). The absence of the 3′-overhang or TriP affects synthesis termination at these positions, which results in a significantly elevated ratio of the products with a length of >10 nucleotides (lanes 7 and 8 and lanes 10 and 11, respectively). The increase of enzyme concentration altering the enzyme to a substrate ratio from 1:20 to 1:2 has little impact on the intrinsic primase ability to terminate synthesis at the defined primer length (Fig. 6B). In the case of the substrate without both p58C binding elements, the activity is severalfold lower in comparison to the substrates where only one element is missing (Fig. 6A, comparison of lane 12 with lanes 7 and 8 or lanes 10 and 11, respectively). Moreover, the primase ability to count the primer length was almost completely disrupted on the T7-P4 substrate (lane 12), which indicates that binding of TriP and the 3′-overhang by p58C is important for counting. These results indicate that TriP and the 3′-overhang exhibit a synergistic effect on the substrate binding and RNA-polymerizing activities of the human primase.
p58C Plays an Important Role in Primer Extension and Length Counting
We analyzed the activity of PrimΔFeS-PolαΔcat (the iron-sulfur cluster binding domain p58C and Polα catalytic core are absent) on the optimal (T6-P3) and the worst (T7-P4) substrates. In the absence of p58C, primase does not sense TriP and the 3′-overhang and cannot count the primer length even on the optimal substrate (Fig. 6A, lanes 15 and 16). These data are consistent with the previous report that primase counting in de novo synthesis was affected by deletion of the Met-288–Leu-313 region in p58C (16). The deletion of p58C dramatically reduces RNA-polymerizing activity to a level close to that for the wild-type primase on the worst substrate T7-P4 (Fig. 6A, comparison of lanes 12 and 15). These data are in accordance with the results of EMSA studies that TriP and the 3′-overhang binding sites are both located on p58C (Figs. 1 and 2). These findings help to explain why in previous primer extension studies, where the substrate was missing TriP and sometimes also the 3′-overhang, the effect of p58C deletion was not so prominent, and its significance for primer elongation was underestimated (13, 18, 33). Earlier, Podust et al. (34) compared the activity of Prim-Polα purified from the human placenta on ribooligoadenylates of varying length (from 2 to 10 nucleotides) and found that deletion of the β- and γ-phosphates from the 5′-terminus of RNA primers significantly diminishes their extension. Their findings together with the data reported here indicate that the β- and γ-phosphates are important for interaction of an RNA primer with p58C, and this interaction exists over the whole cycle of primer synthesis.
Primase Prefers to Extend an Existing Primer Versus Initiating New Primer Synthesis
Primase could potentially perform two types of reactions on the T6-P3 substrate (as well as on the other three substrates analyzed in Fig. 6): extension of the existing 6-mer primer and synthesis of a new primer de novo, beginning with the generation of a dinucleotide. The template region complementary to the primer contains several cytidines as potential de novo start positions. According to the calculated annealing temperature for the T6-P3 hybrid duplex (38.5 °C), approximately half of the template is not in the complex with the primer at the specified reaction conditions and could serve as a template for de novo synthesis. No primase de novo activity was observed on the T6 template in the absence of the primer after 2 min of incubation with a 50 nm enzyme (Fig. 6A, lane 6), whereas the 8-fold increase of incubation time and the 10-fold increase of enzyme concentration (80-fold total) resulted in a significant level of products synthesized de novo (Fig. 6A, lane 14). The level of de novo synthesis observed in lane 14 (no primer) is enhanced ∼2-fold in comparison to lane 13, where approximately half of DNA molecules are primed by RNA and could not serve as templates for dinucleotide synthesis. Taking this into account, we can conclude that most of the products seen in lane 13 (Fig. 6A) are generated during the extension of a 6-mer primer. Moreover, in reactions containing PrimΔFeS-PolαΔcat, we observed only the extension of primer (Fig. 6A, lanes 15 and 16), because p58C is essential for the initiation of primer synthesis (13, 18, 33). These data indicate that the optimal DNA/RNA duplex (with TriP and the 3′-overhang) is a much more favorable substrate for primase in comparison to ssDNA, which is consistent with an ∼6000-fold difference in affinity (Table 2). In the absence of TriP and the 3′-overhang, primase activities in primer extension and in de novo synthesis become comparable. These observations will be helpful in understanding the dependence of primase counting on the type of assay discussed in the next section.
Primase Counting Depends on Many Factors
We have recently shown that primase counting depends on the template sequence (13); in the de novo assay the human primase synthesized only the unit-length primers (8–10-mer) on the oligo(dA) template and the full-length primers (16-mer) on the similar template with added guanines (pyrimidines except one thymine were excluded to keep a single-start position). Now, in an extension assay using a similar homopurine template, we observe that primase stops synthesis when the primer is 8–10 nucleotides long (Fig. 6A, lane 3). The main differences between our recent and current studies are the type of assay (de novo or extension, respectively) and the type of divalent metal used (manganese or magnesium, respectively). To study the influence of these factors on the product's length we analyzed the dependence of primase activity and counting on the concentration of divalent metals and salt using the optimal substrate (T6-P3). Primase activity was inhibited severalfold by decreasing Mg2+ concentration from 5 mm to 0.5 mm (Fig. 7, lanes 4, 6, and 8), whereas the reduction of the Mn2+ concentration enhanced activity (lanes 9, 11, and 13). Moreover, primase demonstrated a different response to salt concentration depending on the type of divalent metal in the reaction. In the presence of Mg2+, primase activity was inhibited almost 4-fold by increasing the salt concentration from 15 mm to 150 mm (lanes 5–7), whereas in the presence of Mn2+, salt stimulates RNA synthesis (lanes 10–12).
FIGURE 7.
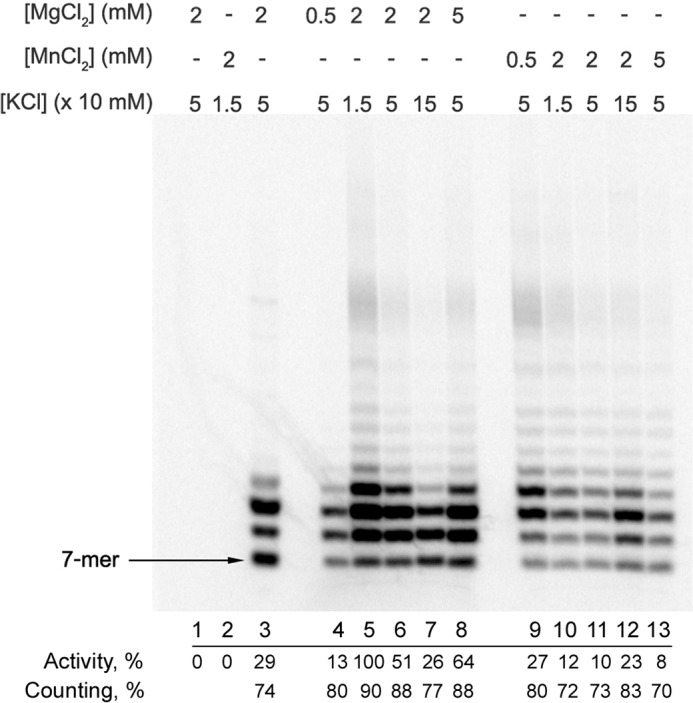
Effects of salt and divalent metals on primase activity and counting. Primase activity in the extension of a 6-mer RNA primer was analyzed by incubation at 35 °C of 50 nm Prim-PolαΔcat with 1 μm T6-P3 (lanes 4–13) or T8-P3 (lane 3) substrates in the presence of 100 μm each UTP and CTP and at varying salt and divalent metal concentrations. Reactions corresponding to lane 3 also contained 10 μm GTP and 100 μm ATP. The products were labeled by incorporation of [α-33P]GTP at the seventh position. Lane 1, control incubation of T6-P3 without enzyme; lane 2, same conditions as in lane 3 except the primer is absent (de novo synthesis conditions). Results of data quantification (performed as described in the legend to Fig. 6) are provided under each lane below the gel.
We directly compared primase counting at the previously used conditions (13) and the current ones. At 50 mm KCl, the level of the unit-length primers was lower in the presence of 2 mm Mn2+ in comparison to 2 mm Mg2+ (Fig. 7, lanes 6 and 11). The increase of salt concentration from 50 mm to 150 mm reduced the counting effect in the presence of 2 mm Mg2+ (lanes 6 and 7), whereas the opposite effect was observed in the presence of 2 mm Mn2+ (lanes 11 and 12). The pronounced ability of primase to count the primer length on the T6-P3 substrate is not due to peculiarities of the template sequence. Primase demonstrated the same behavior on the heterogeneous template T8 (Fig. 7, lane 3).
An additional important factor affecting the analysis of primase counting is the type of assay. It was mentioned previously that the DNA/RNA duplex with TriP and the 3′-overhang is the preferred substrate for primase versus ssDNA. That is why a de novo assay cannot provide a single-hit condition, which requires the comparable or higher affinity of the enzyme for the substrate versus the product. When primase dissociates from the template after synthesis of the 8–10-mer primer, there is a high probability that it will re-bind this product and continue primer extension rather than initiating new primer synthesis. In support of this conclusion, during de novo synthesis on the T6 template that has a significant number of guanines providing higher duplex stability, Prim-PolαΔcat mostly generates the primers that are >10 nucleotides long (Fig. 6A, lane 14). Actually, a de novo assay can provide a single-hit condition if the generated duplex is unstable at the reaction conditions, so the dissociation of primase from the unit-length primer will result in template-primer dissociation precluding the next round of primer synthesis. The unit-length hybrid duplexes containing only A-T pairs are not stable at temperatures higher than 25 °C (10, 32). Therefore, the counting observed on poly(dT) or poly(dA) templates is an intrinsic primase property rather than the effect of unusual structures of such templates as we proposed previously (13).
Discussion
There is no structural information for any primase in the complex with substrates during the initiation or elongation steps; thus, the mechanism of primase action remains unclear. It has been proposed that DNA primases initiate primer synthesis after the formation of the quaternary complex with a DNA template and two NTP molecules (8, 10, 23). Consequently, the primase must have two NTP binding sites, referred to as the initiation site and the elongation (active) site. The active site is located on the catalytic subunit near the catalytic aspartates 109 and 111 and has been structurally characterized (11–13). The location of the binding site for the initiating NTP has not been defined yet. It has been shown that the triphosphate group of the initiating nucleotide is important for dinucleotide synthesis, and it was proposed that it interacts with Arg-304 of p49 (35). However, recent structural data indicate that Arg-304 is buried inside the molecule and, accordingly, most likely plays a purely structural role; this explains the serious defect in primase activity when it is changed to another amino acid (12, 13). The complete abrogation of dinucleotide synthesis when p58C is absent suggests that the initiation site is located on this domain (8, 13, 27, 28). The significance of p58C for primer extension was underestimated due to the usage of unnatural primers. Here we have shown that p58C plays a critical role during the elongation step as well.
Based on the accumulated biochemical and structural data, it is possible to predict the arrangement of primase on the template-primer during all steps of RNA primer synthesis (Fig. 8). We propose that TriP of the initiating nucleotide, which forms the 5′-end of the RNA primer, interacts with p58C during both stages of primer synthesis; therefore, p58C always stays bound to the DNA/RNA junction at the 5′-end of the primer, whereas p49-p58N moved away together with the 3′-end. This mode of template-primer binding predicts the increasing distance between the two functional domains during primer elongation and offers a mechanism for how the primase may count the primer length. The “hinge” model proposed earlier was based on the general idea that the counting could be due to the decrease or increase of the distance between primase subunits during primer synthesis (24). The ability of p58C to bind DNA and NTP was predicted previously using bioinformatics tools. The region spanning residues 270–343 of p58 has a significant sequence similarity with an 8-kDa domain of the human Polβ that regulates the processivity of this polymerase (16). Moreover, the N-terminal portion of p58C comprising that region revealed a striking structural similarity with the ssDNA and FAD binding domain of the cryptochrome 3 from Arabidopsis thaliana (27).
FIGURE 8.
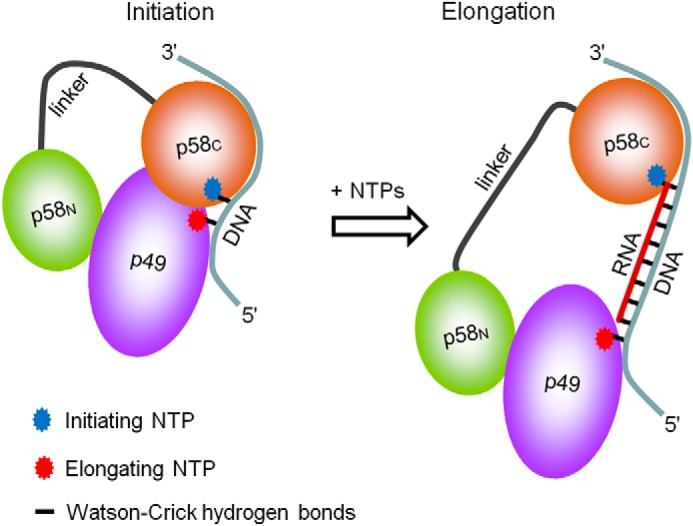
A model of interaction of the human primase with substrates at the initiation and elongation steps of primer synthesis. p58C stays bound to the initiating NTP and the 3′-overhang during the whole cycle of RNA primer synthesis, whereas p49 slides along the template toward the 5′-end.
The 18-residue linker between p58C and the rest of the primase molecule, revealed in our recent structural study, provides significant conformational freedom that allows p58C to be close to p49 during initiation and move apart during elongation (13). The current results indicate a division of work in the human DNA primase: the small catalytic subunit is responsible for catalysis, whereas the large subunit is responsible for template-primer binding. This mechanism is consistent with our recent model of the Prim-DNA/RNA-CTP complex, which revealed only a few potential contacts between the catalytic subunit and DNA (13). The model suggests a weak interaction between p49 and DNA/RNA and the strong possibility of p49 dissociation from the substrate during synthesis, which explains the distributive character of RNA primer synthesis (Fig. 6A, lane 3). The interaction between p58C and TriP might facilitate the correct positioning of the template on the primase molecule during primer extension. The comparable affinity for ssDNA and the DNA/RNA duplex without TriP and the 3′-overhang suggests only a few contacts between the human primase and the RNA primer, where most of them are mediated by the interaction of p58C with TriP. The important role of the C-terminal domain of the large primase subunit for template binding is consistent with the recent finding that this domain is required not only for primase activity but also for the loading of Prim-Polα on early replication origins (36).
According to the data in the literature and our results, we can propose that DNA primases from other species, including phages, bacteria, and archaea, bind DNA/RNA in a similar way, where the flexibly tethered accessory domain participates in binding of the template and the initiating NTP (37–42). Depending on the organism studied, the functional counterpart of p58C may be located on the catalytic or a different subunit, may contain an [FeS]-cluster or zinc or none of them, and may work in cis or trans orientation. The proposed common mode of RNA primer binding only at two distant points (the 5′ and 3′ ends) is consistent with the significant translesion synthesis activity of DNA primases because they cannot sense the modified nucleotides in the newly synthesized strand (43–46). Due to this unique mode of RNA primer binding, DNA primases are able to “write and count” but cannot “read what they wrote.” The data reported here will facilitate the design of RNA-primed DNA templates for structural studies of a broad range of DNA primases in functional complexes with their substrates, hence facilitating the design of a new class of antibiotics targeting a flexibly tethered accessory domain of primases.
Author Contributions
T. H. T. initiated and supervised the project. A. G. B. wrote the manuscript with contributions and critical comments from the other authors. A. G. B. and J. G. designed and constructed the vectors for protein expression. N. D. B. provided the technical assistance and contributed to the preparation of the figures. A. G. B., J. G., and Y. S. expressed and purified the proteins. A. G. B. performed the EMSA experiments. Y. Z. analyzed the primase activity under the supervision of Y. I. P. and A. G. B. All authors analyzed the results and approved the final version of the manuscript.
Acknowledgments
We thank K. Jordan and K. Berger for critical reading of this manuscript and D. Temiakov (Rowan University) for kindly providing the expression vector for T7 RNA polymerase.
This work was supported by National Institutes of Health Grants GM101167 (NIGMS; to T. H. T.) and, in part, by CA129925 (NCI; to Y. I. P.). DNA Sequencing Core and Structural Biology Facilities are supported by the Fred and Pamela Buffett Cancer Center Support Grant P30CA03672 (NCI; to Kenneth H. Cowan). The authors declare that they have no competing conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- Prim
- human DNA primase
- Polα
- DNA polymerase α
- p58N and p58C
- N- and C-terminal domains of the p58 subunit
- p180C
- C-terminal domain of the Polα catalytic subunit
- TriP
- 5′-triphosphate of RNA or NTP
- PrimΔFeS
- Prim without p58C
- PolαΔcat
- Polα with deleted catalytic core
- ssDNA
- single-stranded DNA.
References
- 1. Loeb L. A., and Monnat R. J. Jr. (2008) DNA polymerases and human disease. Nat. Rev. Genet. 9, 594–604 [DOI] [PubMed] [Google Scholar]
- 2. Garg P., and Burgers P. M. (2005) DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Rev. Biochem. Mol. Biol. 40, 115–128 [DOI] [PubMed] [Google Scholar]
- 3. Pellegrini L. (2012) The Polα-primase complex. Subcell. Biochem. 62, 157–169 [DOI] [PubMed] [Google Scholar]
- 4. Muzi-Falconi M., Giannattasio M., Foiani M., and Plevani P. (2003) The DNA polymerase α-primase complex: multiple functions and interactions. ScientificWorldJournal 3, 21–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kilkenny M. L., De Piccoli G., Perera R. L., Labib K., and Pellegrini L. (2012) A conserved motif in the C-terminal tail of DNA polymerase α tethers primase to the eukaryotic replisome. J. Biol. Chem. 287, 23740–23747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang Y., Baranovskiy A. G., Tahirov T. H., and Pavlov Y. I. (2014) The C-terminal domain of the DNA polymerase catalytic subunit regulates the primase and polymerase activities of the human DNA polymerase α-primase complex. J. Biol. Chem. 289, 22021–22034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Suwa Y., Gu J., Baranovskiy A. G., Babayeva N. D., Pavlov Y. I., and Tahirov T. H. (2015) Crystal structure of the human Polα B subunit in complex with the C-terminal domain of the catalytic subunit. J. Biol. Chem. 290, 14328–14337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Copeland W. C., and Wang T. S. (1993) Enzymatic characterization of the individual mammalian primase subunits reveals a biphasic mechanism for initiation of DNA replication. J. Biol. Chem. 268, 26179–26189 [PubMed] [Google Scholar]
- 9. Grosse F., and Krauss G. (1985) The primase activity of DNA polymerase α from calf thymus. J. Biol. Chem. 260, 1881–1888 [PubMed] [Google Scholar]
- 10. Sheaff R. J., and Kuchta R. D. (1993) Mechanism of calf thymus DNA primase: slow initiation, rapid polymerization, and intelligent termination. Biochemistry 32, 3027–3037 [DOI] [PubMed] [Google Scholar]
- 11. Vaithiyalingam S., Arnett D. R., Aggarwal A., Eichman B. F., Fanning E., and Chazin W. J. (2014) Insights into eukaryotic primer synthesis from structures of the p48 subunit of human DNA primase. J. Mol. Biol. 426, 558–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kilkenny M. L., Longo M. A., Perera R. L., and Pellegrini L. (2013) Structures of human primase reveal design of nucleotide elongation site and mode of Polα tethering. Proc. Natl. Acad. Sci. U.S.A. 110, 15961–15966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baranovskiy A. G., Zhang Y., Suwa Y., Babayeva N. D., Gu J., Pavlov Y. I., and Tahirov T. H. (2015) Crystal structure of the human primase. J. Biol. Chem. 290, 5635–5646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Weiner B. E., Huang H., Dattilo B. M., Nilges M. J., Fanning E., and Chazin W. J. (2007) An iron-sulfur cluster in the C-terminal domain of the p58 subunit of human DNA primase. J. Biol. Chem. 282, 33444–33451 [DOI] [PubMed] [Google Scholar]
- 15. Agarkar V. B., Babayeva N. D., Pavlov Y. I., and Tahirov T. H. (2011) Crystal structure of the C-terminal domain of human DNA primase large subunit: implications for the mechanism of the primase-polymerase α switch. Cell Cycle 10, 926–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zerbe L. K., and Kuchta R. D. (2002) The p58 subunit of human DNA primase is important for primer initiation, elongation, and counting. Biochemistry 41, 4891–4900 [DOI] [PubMed] [Google Scholar]
- 17. Arezi B., Kirk B. W., Copeland W. C., and Kuchta R. D. (1999) Interactions of DNA with human DNA primase monitored with photoactivatable cross-linking agents: implications for the role of the p58 subunit. Biochemistry 38, 12899–12907 [DOI] [PubMed] [Google Scholar]
- 18. Copeland W. C. (1997) Expression, purification, and characterization of the two human primase subunits and truncated complexes from Escherichia coli. Protein Expr. Purif. 9, 1–9 [DOI] [PubMed] [Google Scholar]
- 19. Foiani M., Lindner A. J., Hartmann G. R., Lucchini G., and Plevani P. (1989) Affinity labeling of the active center and ribonucleoside triphosphate binding site of yeast DNA primase. J. Biol. Chem. 264, 2189–2194 [PubMed] [Google Scholar]
- 20. Baranovskiy A. G., Lada A. G., Siebler H. M., Zhang Y., Pavlov Y. I., and Tahirov T. H. (2012) DNA polymerase δ and ζ switch by sharing accessory subunits of DNA polymerase δ. J. Biol. Chem. 287, 17281–17287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Baranovskiy A. G., Gu J., Babayeva N. D., Agarkar V. B., Suwa Y., and Tahirov T. H. (2014) Crystallization and preliminary x-ray diffraction analysis of human DNA primase. Acta Crystallogr. F Struct. Biol. Commun. 70, 206–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Milligan J. F., Groebe D. R., Witherell G. W., and Uhlenbeck O. C. (1987) Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 15, 8783–8798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Frick D. N., and Richardson C. C. (2001) DNA primases. Annu. Rev. Biochem. 70, 39–80 [DOI] [PubMed] [Google Scholar]
- 24. Kuchta R. D., and Stengel G. (2010) Mechanism and evolution of DNA primases. Biochim. Biophys. Acta 1804, 1180–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mitchell S. F., and Lorsch J. R. (2014) Standard in vitro assays for protein-nucleic acid interactions: gel shift assays for RNA and DNA binding. Methods Enzymol. 541, 179–196 [DOI] [PubMed] [Google Scholar]
- 26. Francesconi S., Longhese M. P., Piseri A., Santocanale C., Lucchini G., and Plevani P. (1991) Mutations in conserved yeast DNA primase domains impair DNA replication in vivo. Proc. Natl. Acad. Sci. U.S.A. 88, 3877–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sauguet L., Klinge S., Perera R. L., Maman J. D., and Pellegrini L. (2010) Shared active site architecture between the large subunit of eukaryotic primase and DNA photolyase. PLoS ONE 5, e10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vaithiyalingam S., Warren E. M., Eichman B. F., and Chazin W. J. (2010) Insights into eukaryotic DNA priming from the structure and functional interactions of the 4Fe-4S cluster domain of human DNA primase. Proc. Natl. Acad. Sci. U.S.A. 107, 13684–13689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee S. J., Zhu B., Hamdan S. M., and Richardson C. C. (2010) Mechanism of sequence-specific template binding by the DNA primase of bacteriophage T7. Nucleic Acids Res. 38, 4372–4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fien K., Cho Y. S., Lee J. K., Raychaudhuri S., Tappin I., and Hurwitz J. (2004) Primer utilization by DNA polymerase α-primase is influenced by its interaction with Mcm10p. J. Biol. Chem. 279, 16144–16153 [DOI] [PubMed] [Google Scholar]
- 31. Simon A. C., Zhou J. C., Perera R. L., van Deursen F., Evrin C., Ivanova M. E., Kilkenny M. L., Renault L., Kjaer S., Matak-Vinković D., Labib K., Costa A., and Pellegrini L. (2014) A Ctf4 trimer couples the CMG helicase to DNA polymerase α in the eukaryotic replisome. Nature 510, 293–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kuchta R. D., Reid B., and Chang L. M. (1990) DNA primase: Processivity and the primase to polymerase α activity switch. J. Biol. Chem. 265, 16158–16165 [PubMed] [Google Scholar]
- 33. Klinge S., Hirst J., Maman J. D., Krude T., and Pellegrini L. (2007) An iron-sulfur domain of the eukaryotic primase is essential for RNA primer synthesis. Nat. Struct. Mol. Biol. 14, 875–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Podust V. N., Vladimirova O. V., Manakova E. N., and Lavrik O. I. (1992) Eukaryotic DNA primase appears to act as oligomer in DNA-polymerase-α–primase complex. Eur. J. Biochem. 206, 7–13 [DOI] [PubMed] [Google Scholar]
- 35. Kirk B. W., and Kuchta R. D. (1999) Arg-304 of human DNA primase is a key contributor to catalysis and NTP binding: primase and the family X polymerases share significant sequence homology. Biochemistry 38, 7727–7736 [DOI] [PubMed] [Google Scholar]
- 36. Liu L., and Huang M. (2015) Essential role of the iron-sulfur cluster binding domain of the primase regulatory subunit Pri2 in DNA replication initiation. Protein Cell 6, 194–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Matsui E., Nishio M., Yokoyama H., Harata K., Darnis S., and Matsui I. (2003) Distinct domain functions regulating de novo DNA synthesis of thermostable DNA primase from hyperthermophile Pyrococcus horikoshii. Biochemistry 42, 14968–14976 [DOI] [PubMed] [Google Scholar]
- 38. Geibel S., Banchenko S., Engel M., Lanka E., and Saenger W. (2009) Structure and function of primase RepB' encoded by broad-host-range plasmid RSF1010 that replicates exclusively in leading-strand mode. Proc. Natl. Acad. Sci. U.S.A. 106, 7810–7815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Beck K., Vannini A., Cramer P., and Lipps G. (2010) The archaeo-eukaryotic primase of plasmid pRN1 requires a helix bundle domain for faithful primer synthesis. Nucleic Acids Res. 38, 6707–6718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kato M., Ito T., Wagner G., and Ellenberger T. (2004) A molecular handoff between bacteriophage T7 DNA primase and T7 DNA polymerase initiates DNA synthesis. J. Biol. Chem. 279, 30554–30562 [DOI] [PubMed] [Google Scholar]
- 41. Corn J. E., Pease P. J., Hura G. L., and Berger J. M. (2005) Crosstalk between primase subunits can act to regulate primer synthesis in trans. Mol. Cell 20, 391–401 [DOI] [PubMed] [Google Scholar]
- 42. Cavanaugh N. A., Ramirez-Aguilar K. A., Urban M., and Kuchta R. D. (2009) Herpes simplex virus-1 helicase-primase: roles of each subunit in DNA binding and phosphodiester bond formation. Biochemistry 48, 10199–10207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Urban M., Joubert N., Purse B. W., Hocek M., and Kuchta R. D. (2010) Mechanisms by which human DNA primase chooses to polymerize a nucleoside triphosphate. Biochemistry 49, 727–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jozwiakowski S. K., Borazjani Gholami F., and Doherty A. J. (2015) Archaeal replicative primases can perform translesion DNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 112, E633–E638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bianchi J., Rudd S. G., Jozwiakowski S. K., Bailey L. J., Soura V., Taylor E., Stevanovic I., Green A. J., Stracker T. H., Lindsay H. D., and Doherty A. J. (2013) PrimPol bypasses UV photoproducts during eukaryotic chromosomal DNA replication. Mol. Cell 52, 566–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. García-Gómez S., Reyes A., Martínez-Jiménez M. I., Chocrón E. S., Mourón S., Terrados G., Powell C., Salido E., Méndez J., Holt I. J., and Blanco L. (2013) PrimPol, an archaic primase/polymerase operating in human cells. Mol. Cell 52, 541–553 [DOI] [PMC free article] [PubMed] [Google Scholar]