Abstract
Cholesterol metabolism is subject to complex transcriptional and nontranscriptional regulation. Herein, the role of ubiquitylation is emerging as an important post-translational modification that regulates cholesterol synthesis and uptake. Similar to other post-translational modifications, ubiquitylation is reversible in a process dependent on activity of deubiquitylating enzymes (DUBs). Yet whether these play a role in cholesterol metabolism is largely unknown. As a first step to test this possibility, we used pharmacological inhibition of cellular DUB activity. Short term (2 h) inhibition of DUBs resulted in accumulation of high molecular weight ubiquitylated proteins. This was accompanied by a dramatic decrease in abundance of the LDLR and attenuated LDL uptake into hepatic cells. Importantly, this occurred in the absence of changes in the mRNA levels of the LDLR or other SREBP2-regulated genes, in line with this phenotype being a post-transcriptional event. Mechanistically, we identify transcriptional induction of the E3 ubiquitin ligase IDOL in human and rodent cells as the underlying cause for ubiquitylation-dependent lysosomal degradation of the LDLR following DUB inhibition. In contrast to the established transcriptional regulation of IDOL by the sterol-responsive liver X receptor (LXR) transcription factors, induction of IDOL by DUB inhibition is LXR-independent and occurs in Lxrαβ−/− MEFs. Consistent with the role of DUBs in transcriptional regulation, we identified a 70-bp region in the proximal promoter of IDOL, distinct from that containing the LXR-responsive element, which mediates the response to DUB inhibition. In conclusion, we identify a sterol-independent mechanism to regulate IDOL expression and IDOL-mediated lipoprotein receptor degradation.
Keywords: cholesterol metabolism, deubiquitylation (deubiquitination), E3 ubiquitin ligase, lipoprotein receptor, nuclear receptor, post-transcriptional regulation, IDOL, LDLR, LXR
Introduction
Elevated levels of plasma LDL represent a major risk factor for development of atherosclerosis and cardiovascular disease (1). Because of its ability to promote LDL uptake into cells, the LDL receptor (LDLR)2 is a major determinant of plasma LDL levels (2). The pivotal role of the LDLR in LDL metabolism is exemplified by the fact that LDLR mutations account for most incidences of familial hypercholesterolemia, a disease characterized by reduced hepatic LDL clearance, elevated plasma cholesterol levels, and accelerated cardiovascular disease (1, 3).
The LDLR is subject to tight transcriptional and post-transcriptional regulation, which is largely governed by the intracellular levels of cholesterol (4). At the level of transcription, these pathways are regulated by two nuclear transcription factor families: SREBP1 and SREBP2 (5–7), and the liver X receptor α and β (LXRs) (8, 9). When cellular sterol levels decline, SREBPs are activated to induce genes required for de novo cholesterol biosynthesis, as well as the LDLR to increase uptake of LDL cholesterol (4). In contrast, when sterol levels rise, LXRs become activated by their endogenous ligands. These ligands include oxidized cholesterol derivatives (oxysterols) and intermediates of the cholesterol synthesis pathway, the most potent being desmosterol and 24,25-epoxycholesterol (10–12). Once activated, LXRs induce the expression of a set of genes whose main function is to reduce the cellular cholesterol burden, such as the sterol efflux pumps ABCA1 and ABCG1 (9) and the E3 ubiquitin ligase (E3) IDOL (inducible degrader of the LDL receptor) (13).
As an E3, IDOL binds to the cytoplasmic tail of LDLR and promotes ubiquitylation of specific residues in this domain in conjunction with the E2 UBE2D1/E1 (13–15). Although IDOL can interact with LDLR at multiple steps in its cellular itinerary, plasma membrane-localized LDLR is particularly sensitive to IDOL-mediated ubiquitylation (16). Once ubiquitylated, LDLR is rapidly removed from the plasma membrane and sorted by the ESCRT (endosomal sorting complexes required for transport) machinery toward the lysosome for degradation (16, 17). The clinical relevance of IDOL in humans is highlighted by recent studies that found an association between common and rare genetic variance in the IDOL locus and circulating levels of LDL cholesterol. This establishes the E3 IDOL as a potential therapeutic target to treat hypercholesterolemia (18, 19).
Substrate ubiquitylation promoted by E3s can be effectively reversed through the opposing activity of deubiquitylases (DUBs) (20). The human genome encodes ∼100 DUBs, the majority of which belong to the family of ubiquitin-specific proteases (USPs) that function as cysteine proteases (21, 22). In contrast to E3s, whose role in cholesterol metabolism has gained attention in recent years, the role of DUBs in this process is largely unexplored. Recently, we (16) and Scotti et al. (17) implicated the DUB USP8, an ESCRT-associated DUB, in IDOL-mediated degradation of the LDLR. However, this likely represents nonspecific removal of ubiquitin from ubiquitylated cargo by USP8, prior to it entering MVBs, as a means to salvage ubiquitin for reuse.
In view of their diverse actions, we reasoned therefore that additional DUBs might regulate the LXR-IDOL-LDLR axis. To test this idea, we tested the effect of pharmacological DUB inhibition on the LDLR pathway. Herein, we report that pan-DUB inhibition by two established inhibitors, PR-619 and HBX41-108, results in rapid, robust, and specific transcriptional induction of IDOL that leads to subsequent degradation of the LDLR. Uniquely, this occurs in an LXR-independent manner, thereby revealing a sterol-independent mechanism to promote ubiquitylation of lipoprotein receptors by IDOL.
Experimental Procedures
Reagents
HBX41-108 was from Tocris and PR-619 from Millipore. Simvastatin and MG-132 were from Calbiochem. Lipoprotein-deficient serum was prepared as previously reported (23) and confirmed to contain no lipoproteins. GW3965, LG100268, actinomycin D, and bafilomycin A1 were from Sigma.
Cell Culture and Transient Transfections
HeLa, RAW264.7, Huh7, J774, and HepG2 cells were obtained from the ATCC. Cells were maintained in DMEM supplemented with 10% FBS at 37 °C and 5% CO2. Human umbilical vein endothelial cells were obtained from Lonza and cultured as described (24). The human immortalized hepatocyte cell line IHH was a kind gift from Geesje Dallinga-Thie (Academic Medical Center, Amsterdam, The Netherlands) and cultured in William's E medium supplemented with 2 mm glutamine, 10% FBS, 20 millunits/ml bovine insulin, and 50 nm dexamethasone, as previously reported (25). Human A431 cells were a kind gift from Dr. Elina Ikonen (University of Helsinki, Helsinki, Finland). Wild type, Lxrαβ−/−, and Idol−/− MEFs were a kind gift from Dr. Peter Tontonoz (University of California at Los Angeles) (26). HepG2 cells stably expressing a FLAG-ubiquitin were generated by transfecting cells with a FLAG-ubiquitin encoding plasmid and subsequent selection with puromycin. Where indicated, cells were sterol-depleted by culture in sterol depletion medium (DMEM supplemented with 10% lipoprotein-deficient serum, 2.5 μg/ml simvastatin, and 100 μm mevalonate). To inhibit cellular DUBs, cells were treated with DUB inhibitors for the time and at the concentration indicated in the figure legends. The cells were transfected using the JetPrime transfection reagent (Polyplus), and transfection efficiency was monitored by co-transfection of an expression plasmid for GFP.
Plasmids and Expression Constructs
Expression plasmids encoding IDOL, LDLRWT, LDLRK830R/C839A, VLDLRWT, VLDLRK811R, GFP, pCMV-hLXRα, pCMV-hRXRα, and an LXR response element (LXRE)-luciferase reporter (pTK-LXRE3x-LUC) were previously reported (13, 27). pGL3-hABCA1-LUC was kindly provided by Herbert Stangl (University of Vienna, Vienna, Austria) (28). pGL2-SV40min was a kind gift of Dr. Phil Barnett (Academic Medical Center). A 3.287-base pair (Chr6:16,126,030–16,129,524, genome version: grch37) proximal promoter region of hIDOL was PCR-amplified with Phusion high fidelity DNA polymerase (New England Biolabs) using HepG2 genomic DNA as template and cloned into pGL3-basic-LUC (Promega). Sequential deletion constructs were generated using standard cloning procedures. All proximal promoter regions contained the TATA element and stopped +207 base-pairs downstream of the IDOL transcriptional start site (TSS). Based on published LXR ChIP-Seq studies, we amplified a 490-base pair human IDOL (Chr6:16,131,159–16,131,649) and 586 base-pairs mouse IDOL (Chr13:45,486,794–45,487,380) long LXRE-containing region downstream of Exon2 of IDOL (human and mouse) from genomic DNA. The amplified fragments were cloned into pGL2-SV40min as an XhoI fragment. All plasmids used in this study were isolated by CsCl2 gradient centrifugation, and their correctness was verified by sequencing and digestion analysis.
ChIP Sequencing Analysis
Data analysis was performed using bowtie and HOMER (29) on previously published ChIP experiments: GSE50944 (30), GSE35262 (31), and GSE28319 (32). The sequencing experiment was normalized to a total of 107 uniquely mapped tags and visualized by preparing custom tracks for the UCSC Genome Browser.
Dual Luciferase Assay
HepG2 cells were transiently transfected, as described above, with luciferase reporter plasmids together with pCMV-LXRa, pCMV-RXRa, or pCMV-empty. The Renilla reporter plasmid pRL-TK (Promega) was co-transfected as a control for transfection efficiency. To induce LXR/RXR activation, cells were treated for 18 h with the LXR and RXR synthetic agonists, 1 μm GW3965 and 100 nm LG100268, or with DMSO (vehicle control). Subsequently, cells were treated with 5 μm HBX41-108 for 8 h. Luciferase activity measurements were performed using the dual luciferase reporter assay system (Promega) on a Glowmax Multi detection system (Promega) according to the manufacturer's protocol. Each experiment was repeated at least three times.
MTT Assay
Following treatments, MTT tetrazolium was added to cells at 1 mg/ml for 30 min at 37 °C. The cells were subsequently washed and lysed in isopropanol in the dark at room temperature. Absorbance was read at 550 nm and mitochondrial activity expressed as percentage of control.
Antibodies and Immunoblot Analysis
Total cell lysates were prepared in radioimmune precipitation assay buffer (150 mm NaCl, 1% Nonidet P-40, 0.1% sodium deoxycholate, 0.1% SDS, 100 mm Tris-HCl, pH 7.4) supplemented with protease inhibitors. Lysates were cleared by centrifugation at 4 °C for 10 min at 10,000 × g. Protein concentration was determined with the Bradford assay with BSA as reference. Samples (10–40 μg) were separated on NuPAGE BisTris gels and transferred to nitrocellulose. The membranes were probed with the following antibodies: LDLR (Epitomics, EP1553Y 1:4000), tubulin (Sigma, DM1A, 1:5000), β-actin (Millipore, C4, 1:5000), FLAG-HRP (Sigma, M2, 1:1000), GFP (Santa Cruz, B-2, 1:3000), HA (Covance, 16B12, 1:10000), ubiquitin (Sigma, U5379, 1:200), EGFR (affinity purified rabbit polyclonal anti-EGFR sera was from Dr. Simona Polo, 1:40,000), and TfR (Invitrogen, H68.4, 1:1000). Secondary HRP-conjugated antibodies (Zymed Laboratories Inc.) were used and visualized with chemiluminescence on a LAS4000 (GE Healthcare). All immunoblots are representative of at least three independent experiments.
Cy5-Ub-PA DUB Activity Probe Labeling
The Cy5-UB-PA DUB activity probe was used as previously described (33). Briefly, HepG2 cells were harvested, washed with PBS, and lysed in lysis buffer (50 nm Tris, 250 mm sucrose, 5 mm MgCl2, 1 mm DTT, 0.5% CHAPS, 0.1% Nonidet P-40). Samples were incubated on ice for 30 min, and debris was precleared by centrifugation. Protein concentrations were measured using the Bradford protein assay, and a total of 25 μg of lysate was incubated with HBX41-108, PR-619, or DMSO vehicle control for 15 min at room temperature. Subsequently, 1 μm Cy5-UB-PA and 50 nm NaOH were added to the samples and incubated for 15 min in the dark at room temperature. Sample buffer was added, samples were boiled and loaded onto SDS-PAGE gels as described above. Fluorescence signal was imaged in-gel using a Typhoon imager (GE Healthcare).
LDL Uptake Assay
DyLight 488-labeled LDL was produced as previously described (23). Briefly, the cells were incubated in sterol depletion medium for 16 h to increase LDLR abundance. To initiate LDL uptake, cells were washed twice with PBS and incubated with 5 μg/ml DyLight488-labeled LDL in DMEM supplemented with 0.5% BSA for the indicated time at 37 °C. Subsequently, cells were washed twice with PBS supplemented with 10% FCS, followed by an additional wash with PBS. Internalization of LDL was measured by lysing cells in radioimmune precipitation assay buffer and quantification of the fluorescence signal on a Typhoon imager. Specific LDL uptake was calculated by subtracting fluorescence in cells treated also with excess nonlabeled LDL and presented as mean ± S.D.
Determination of Cell Surface LDLR
Surface LDLR density in the different cells was determined as previously reported (16). Briefly, the cells were treated as indicated, dissociated, and incubated in FACS blocking buffer for 30 min on ice. Subsequently, 100,000 cells were stained with a R-phycoerythrin-conjugated mouse anti-human LDLR antibody for 1 h on ice. The cells were subsequently washed three times with FACS buffer and directly analyzed on a FACSCalibur flow cytometer (BD Biosciences). Relative surface LDLR was calculated from geomean values after correction for background using FlowJo and presented as mean ± S.D. For confocal imaging, HepG2-LDLR-GFP cells (13) were grown on glass slides and treated as indicated. Subsequently, cells were fixed with paraformaldehyde and stained with DAPI. A Leica TCS SP8 confocal microscope equipped with a 405-nm laser (for DAPI) and 488-nm laser (for DyLight 488-labeled LDL) and a 63× objective (Leica Microsystems, Mannheim, Germany) was used.
RNA Isolation and Quantitative PCR
Total RNA was isolated using the Direct-zol RNA kit (Zymo Research), and 1 μg of total RNA was reverse-transcribed using iScript (Bio-Rad). SYBR Green real time quantitative PCR assays were performed on a Lightcycler 480 II (Roche). Oligonucleotide sequences are available upon request.
Statistical Analysis
Data are presented as means ± S.D. The statistical analyses were performed using GraphPad Prism 5.0 software using unpaired t test and one-way analysis of variance with Bonferroni correction for grouped analyses. A p value < 0.05 was considered statistically significant.
Results
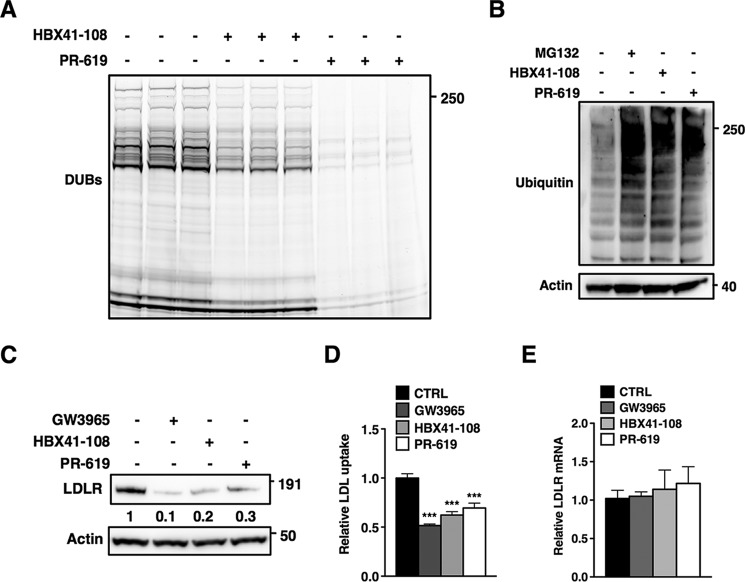
Ubiquitylation is a highly dynamic post-translational modification that is governed by the opposing actions of E3s and DUBs. Given the reversible nature of ubiquitylation, we hypothesized that DUBs could be involved in the LXR-IDOL-LDLR axis at different stages. To test this hypothesis, we decided to use two established inhibitors of cysteine protease DUBs, PR-619 and HBX41-108, that have been reported to inhibit a broad range of DUBs in in vitro reconstituted assays (34–36). As a first step, we determined that both inhibit cellular DUBs in HepG2 cells using an activity-based probe that fluorescently labels active (i.e. noninhibited) DUBs. In cell lysates from HepG2 cells HBX41-108, and even more so PR-619, effectively inhibited labeling of DUBs, consistent with their pan DUB inhibition profile (Fig. 1A). At doses reported to block cellular DUBs by these agents, in vitro inhibition by the two inhibitors was mirrored by a rapid in vivo accumulation of high molecular weight ubiquitylated proteins in HepG2 cells to a level comparable to that obtained following proteasome inhibition with MG132 (Fig. 1B). Having established that acute (2 h) inhibition of cellular DUBs by these inhibitors is effective, we tested their influence on the level of the LDLR. We observed that pan DUB inhibition resulted in a dramatic decrease in the amount of LDLR protein in HepG2 cells, which was comparable to that observed following induction of IDOL expression with the synthetic LXR agonist, GW3965 (Fig. 1C). Consistent with this, inhibition of cellular DUBs with PR-619 and HBX41-108 attenuated uptake of DyLight 488-labeled LDL into HepG2 cells, as could be anticipated from the reduced LDLR protein (Fig. 1D). Because DUBs have been implicated in transcriptional control, a simple explanation for this observation could be that acute pan-DUB inhibition represses transcription of the LDLR. However, this was not the case, because LDLR mRNA was unchanged following treatment with the DUB inhibitors (Fig. 1E). Our results thus suggest that DUB inhibition underlies the observed phenotype.
FIGURE 1.
DUB inhibition reduces LDLR protein and LDL uptake. A, a DUB activity probe was used to assess cellular DUB activity in the absence or presence of HBX41-108 or PR-619. Total cell lysates of HepG2 cells were treated as described under “Experimental Procedures.” Subsequently, lysates were separated on SDS-PAGE gels and imaged for probe-dependent fluorescence (n = 3). B, HepG2 cells were treated with 25 μm MG132, 5 μm HBX41-108, or 50 μm PR-619 for 2 h, and total cell lysates were immunoblotted as indicated. C, HepG2 cells were cultured in sterol depletion medium for 16 h prior to treatment with 1 μm GW3965 (6 h) or 5 μm HBX41-108 and 50 μm PR-619 for 2 h. Total cell lysates were immunoblotted as indicated. Immunoblots were quantified, and the mean intensity of the LDLR normalized to β-actin is indicated (n = 3). D, HepG2 cells were treated as above and incubated for 1 h with 5 μg/ml Dylight488-LDL cholesterol. Specific LDL uptake is plotted with each bar representing the mean ± S.D. (n = 4). E, expression of LDLR mRNA was determined in cells treated as described in A. Each bar represents the mean ± S.D. (n = 4). All immunoblots are representative of at least three independent experiments. ***, p < 0.001.
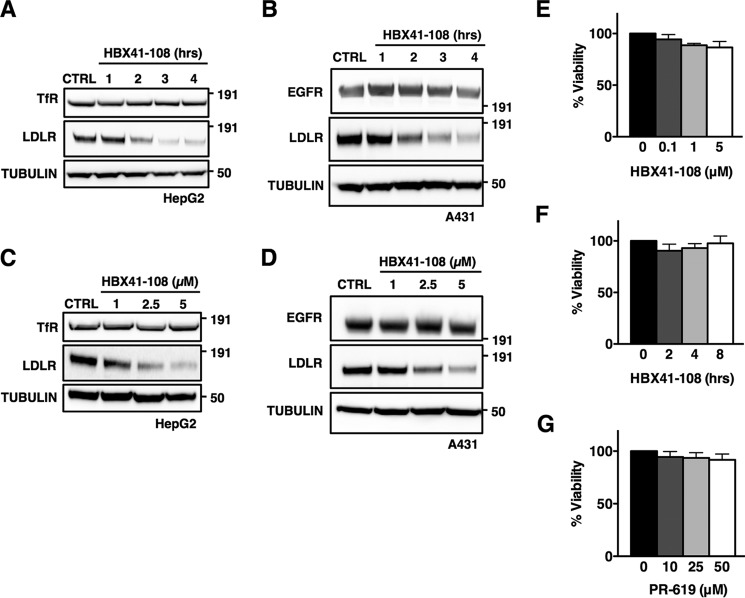
We went on to characterize the reduction of LDLR abundance by DUB inhibition in HepG2 and A431 cells, because both cell lines contain high LDLR levels upon sterol depletion. We determined that the reduction in LDLR protein induced by HBX41-108 was both time- (Fig. 2, A and B) and dose-dependent (Fig. 2, C and D) in these cells. We considered the possibility that our observations are a result of general toxicity induced by short term inhibition of cellular DUBs by these inhibitors. However, at the maximal HBX41-108 and PR-619 doses and duration tested (5 μm for 8 h and 50 μm for 2 h, respectively), these inhibitor did not induce cellular toxicity as assessed by the MTT assay (Fig. 2, E–G), even though they inhibited cellular DUBs and increased overall ubiquitylation in HepG2 cells (Fig. 1B). Upon prolonged incubation, a time-dependent decrease in cellular viability was observed, in particular with PR-619 (data not shown). Because we obtained similar results with both inhibitors, we opted therefore to do most subsequent experiments with HBX41-108. Further supporting the notion that the effect of DUB inhibition on the LDLR is specific, HBX41-108 and PR-619 treatment was not a result of overt changes in trafficking or the level of other endocytic cargo receptors, because the receptors for transferrin and EGF (TfR and EGFR, respectively) remained unchanged (Fig. 2, A–D, and data not shown).
FIGURE 2.
Degradation of the LDLR induced by DUB inhibition is time- and dose-dependent and not associated with cellular toxicity. A and B, HepG2 (A) or A431 (B) was treated with 5 μm HBX41-108 for the indicated time, and the total cell lysates were immunoblotted as indicated (n = 3). C and D, HepG2 (C) or A431 (D) were treated with the indicated concentration of HBX41-108 for 2 h, and the total cell lysates were immunoblotted as indicated (n = 3). E–G, HepG2 cells were treated with 5 μm HBX41-108 for the indicated time (E) or for 2 h (F and G) with the indicated concentration of HBX41-108 or PR-619. Subsequently, cellular viability was assessed by an MTT assay and expressed as a percentage of vehicle control. Each bar represents the mean ± S.D. (n = 3). All immunoblots are representative of at least three independent experiments. CTRL, control.
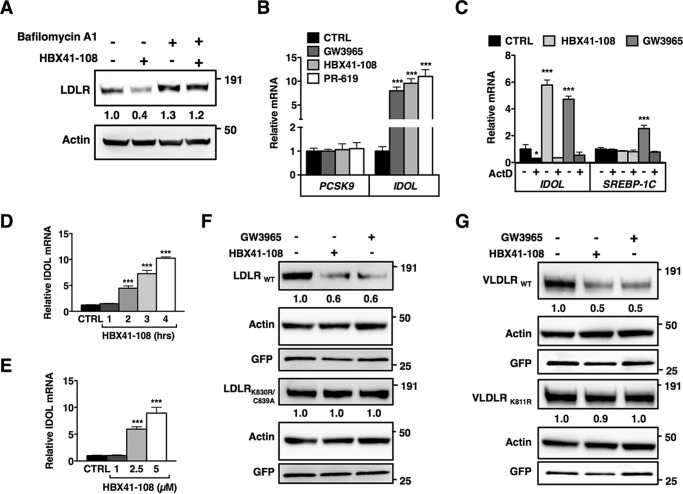
The discrepancy between reduced LDLR protein and unchanged mRNA levels in response to HBX41-108 and PR-619 may suggest that these inhibitors induce post-transcriptional degradation of the LDLR, reported to occur primarily in the lysosome. Supporting this contention, we found that pharmacological blocking of lysosomal degradation using the lysosomotropic agent bafilomycin A prevented the reduction in LDLR protein by HBX41-108 (Fig. 3A). Currently, two independent pathways have been described for directing the LDLR toward lysosomal degradation. The first depends on SREBP2-regulated PCSK9 (proprotein convertase subtilisin/kexin type 9) (37), a secreted protein that binds to the ectodomain of the LDLR and prevents it from recycling to the plasma membrane (38–40). The second pathway depends on ubiquitylation of conserved residues in the short intracellular tail of the LDLR by the LXR-regulated E3 ubiquitin ligase IDOL (13, 41). To distinguish the possible involvement of these two pathways, we initially examined whether the expression levels of PCSK9 or IDOL were sensitive to DUB inhibition in HepG2 cells (Fig. 3B). PCSK9 mRNA was unchanged by HBX41-108 and PR-619 in these cells. In contrast to PCSK9, we observed a large increase in IDOL mRNA in cells treated with both inhibitors, which was prevented by blocking transcription with actinomycin D (Fig. 3C). The increase in IDOL expression was comparable in magnitude to that seen with the synthetic LXR ligand GW3965. Furthermore, the induction of IDOL expression by HBX41-108 was both time- and dose-dependent (Fig. 3, D and E) and mirrored the levels of the LDLR protein (Fig. 2A).
FIGURE 3.
DUB inhibition promotes ubiquitylation-dependent lysosomal degradation of the LDLR. A, HepG2 cells were pretreated for 4 h with vehicle or bafilomycin A1 (100 nm) and subsequently treated for an additional 2 h treatment with 5 μm HBX41-108. Total cell lysates were immunoblotted as indicated. Immunoblots were quantified, and the mean intensity of the LDLR normalized to β-actin relative to control treated cells is indicated (n = 4). B, HepG2 cells were treated with 1 μm GW3965 (6 h), 5 μm HBX41-108, or 50 μm PR-619 (2 h). Subsequently, mRNA expression of PCSK9 and IDOL were determined by qPCR. Each bar represents the mean ± S.D. relative to vehicle-treated control cells (n = 3). C, HepG2 cells were treated with 2 μm actinomycin D (2 h) and then treated with 1 μm GW3965 or 5 μm HBX41-108 for 4 h. Expression of IDOL and SREBP-1c was determined by qPCR. Each bar represents the mean ± S.D. relative to vehicle-treated control cells (n = 3). D and E, HepG2 cells were treated with 5 μm HBX41-108 for the indicated time (D) or for 2 h (E) with the indicated concentration of HBX41-108 and mRNA expression of IDOL was determined by qPCR (n = 4). F and G, HeLa cells were co-transfected with expression plasmids for LDLRWT, VLDLRWT, the ubiquitylation mutants LDLRK830R,C839A and VLDLRK811R, and GFP (to monitor transfection efficiency). The cells were subsequently treated for 24 h with 1 μm GW3965 or for 4 h with 5 μm HBX41-108. The total cell lysates were analyzed by immunoblotting as indicated. Immunoblots are representative of at least three independent experiments. Images were quantified and intensity of the LDLR and VLDLR was normalized and displayed as mean relative to vehicle-treated control cells (n = 3). *, p < 0.05; **, p < 0.01; ***, p < 0.001. CTRL, control.
Induction of IDOL activity provides a plausible explanation for the reduced LDLR protein seen following HBX41-108 and PR-619 treatment. Unfortunately, current commercial and self-made antibodies are unable to detect endogenous levels of IDOL. We therefore used an LDLR degradation assay in HeLa cells as a proxy for IDOL activity. In this assay, treating HeLa cells overexpressing wild type LDLR with either GW3965 or HBX41-108 led to a marked decrease in LDLR protein (Fig. 3F). We have previously reported that IDOL-stimulated ubiquitylation of the LDLR takes place on conserved residues in the intracellular tail of the receptor and that mutating these residues renders LDLR insensitive to IDOL. We therefore performed the same assay in cells expressing an IDOL resistant LDLR (LDLRK830R/C839A) (13). Consistent with ubiquitylation of the LDLR being an important determinant of HBX41-108-stimulated degradation of the LDLR, this mutant receptor was resistant to the treatment (Fig. 3F). IDOL also promotes ubiquitylation-dependent degradation of the related lipoprotein receptor, the VLDLR (27). Accordingly, similar results to those obtained with the LDLR were seen with wild type VLDLR and an IDOL-resistant mutant, VLDLRK811R (Fig. 3G).
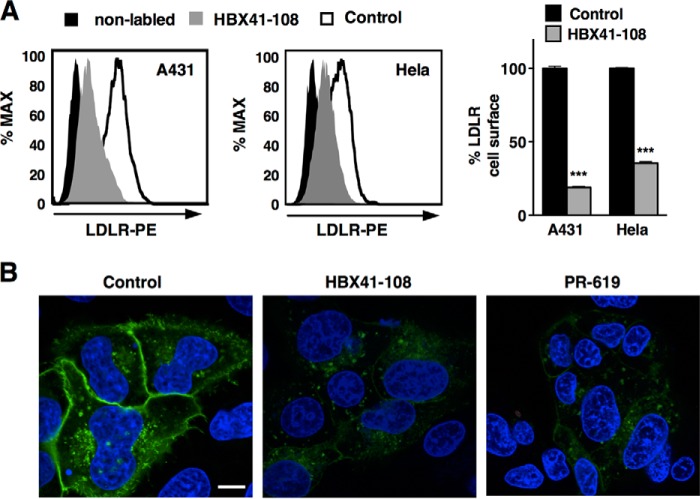
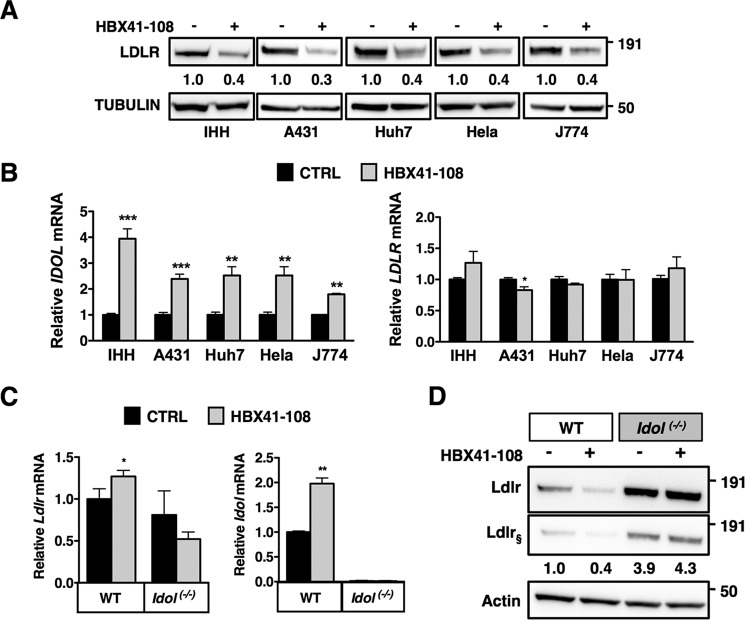
Our results indicate induction of IDOL as a key determinant of HBX41-108 action. We have previously reported that IDOL preferentially targets the LDLR pool present at the plasma membrane (16). Therefore, to substantiate the notion that the effect of DUB inhibition requires IDOL activity, we determined the cell surface levels of the LDLR in cells. By FACS analysis, we demonstrate that HBX41-108 induces a robust reduction in the level of cell surface LDLR in A431 and HeLa cells (Fig. 4A). We also evaluated this in HepG2 cells that stably express an LDLR-GFP construct. In these cells, both DUB inhibitors (2-h treatment) led to a marked removal of the LDLR from the plasma membrane as determined by confocal imaging (Fig. 4B). To further corroborate the functional DUB-IDOL link, we tested a panel of additional human (IHH, A431, HeLa, and Huh7) and mouse (J774) cell lines. Invariably, treatment with HBX41-108 resulted in a marked decrease of LDLR protein in these cells (Fig. 5A). As was seen in HepG2 cells, this was paralleled by an increase in IDOL expression, whereas LDLR mRNA remained largely unchanged (Fig. 5B). Finally, we compared the action of HBX41-108 in wild type and Idol−/− MEFs (26). Treating wild type cells with HBX41-108 increased IDOL expression (Fig. 5C) and reduced LDLR protein (Fig. 5D). The basal level of Ldlr protein in Idol−/− MEFs was elevated, as previously reported (26). However, in the absence of Idol, these cells were resistant to the LDLR-lowering effect of HBX41-108. Collectively, our experiments demonstrate that the LDLR-lowering activity of HBX41-108 is active in cells from multiple lineages and is dependent on transcriptional induction of IDOL activity.
FIGURE 4.
DUB inhibition promotes removal of cell surface LDLR. A, the human A431 and HeLa cell lines were cultured in sterol depletion medium 18 h prior to being treated with 5 μm HBX41-108 for 2 h. Cell surface LDLR was determined by FACS analysis. A representative FACS histogram and the quantification of the experiments are shown (n = 3). B, HepG2-LDLR-GFP cells were treated with vehicle (control), 5 μm HBX41-108, or 50 μm PR-619 for 2 h. Representative images are shown with LDLR in green and staining of nuclei with DAPI in blue. Size bar, 10 μm. ***, p < 0.001.
FIGURE 5.
HBX41-108 promotes LDLR degradation in human and rodent cells in an IDOL-dependent manner. A, the human cell lines IHH, Huh7, A431, and HeLa and the mouse macrophage-like J774 cell line were cultured in sterol depletion medium 18 h prior being treated with 5 μm HBX41-108 for 2 h. Total cell lysates were immunoblotted as indicated (n = 3). B, mRNA expression of IDOL and LDLR were determined by qPCR. Each bar represents the mean ± S.D. relative to vehicle-treated control cells (n = 3). C, WT and Idol−/− MEFs were cultured as above, and mRNA expression of Ldlr and Idol was determined by qPCR. Each bar represents the mean ± S.D. relative to vehicle-treated control cells (n = 6). D, WT and Idol−/− MEFs were cultured and treated as in C, and the total cell lysates were immunoblotted as indicated. Immunoblots were quantified, and the mean intensity of the Ldlr normalized to β-actin relative to control treated cells is indicated (n = 4, § represents short exposure). *, p < 0.05; **, p < 0.01; ***, p < 0.001. CTRL, control.
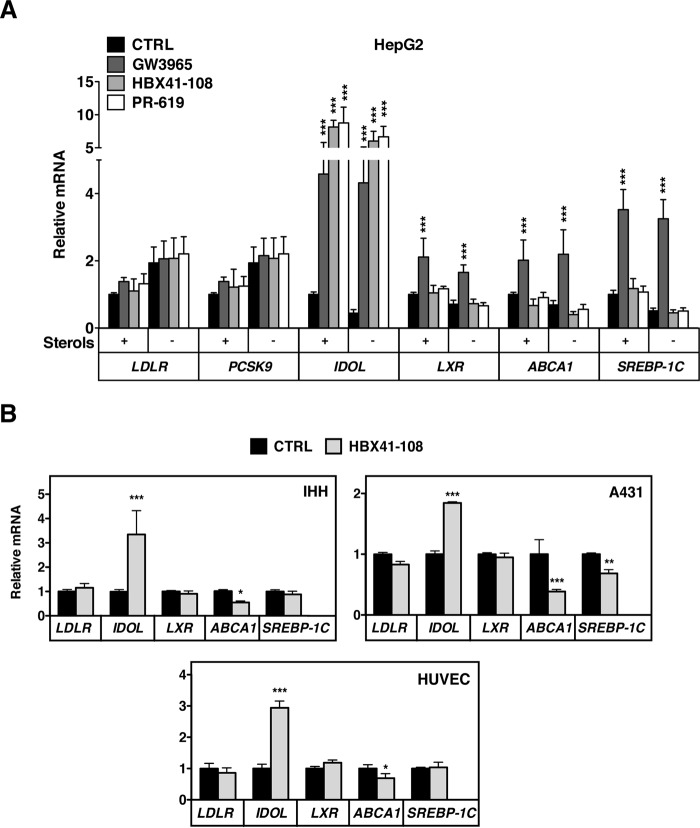
IDOL is an LXR-regulated target gene (13). The most parsimonious explanation therefore for its induction by HBX41-108 is that this inhibitor enhances the complete LXR-regulated genetic program, akin to an LXR ligand. We therefore assessed the transcriptional status of established LXR targets in HepG2 cells treated with HBX41-108 (5 μm, 2 h) or PR-619 (50 μm, 2 h) in the presence or absence of sterols in the medium (Fig. 6A). As previously shown, IDOL was induced by both inhibitors to a similar extent as by an LXR synthetic ligand, irrespective of the presence or absence of sterols in the culture medium. In response to the LXR ligand, expression of the other LXR-regulated genes we evaluated (ABCA1, SREBP-1C, and LXRα) increased. However, in contrast to our hypothesis, expression of none of these genes increased in response to DUB inhibition, and ABCA1 was even slightly decreased (Fig. 6A). Expression of the two SREBP2 targets PCSK9 and LDLR increased with sterol depletion but even under this condition remained unresponsive to HBX41-108 and PR-619. The lack of a pan-LXR response to DUB inhibition was unexpected. We therefore also analyzed IHH and A431 cells but obtained similar results (Fig. 6B). Finally, to extend the physiological relevance of our observations, we also investigated the response of primary human umbilical vein endothelial cells to HBX41-108. Using a similar experimental set up as that used above, we also observed specific induction of IDOL by HBX41-108, whereas other LXR-regulated genes remained unaffected (Fig. 6B).
FIGURE 6.
Induction of IDOL mRNA by DUB inhibition is independent of global LXR activation. A, HepG2 cells cultured in sterol depletion or sterol-rich medium for 18 h. Subsequently, cell were treated with 1 μm GW3965 (6 h), 5 μm HBX41-108, or 50 μm PR-619 (2 h) after which mRNA expression of LDLR, PCSK9, IDOL, LXR, ABCA1, and SREBP-1C were determined by qPCR. Each bar represents the mean ± S.D. relative to vehicle-treated control cells (n = 3). B, human IHH and A431 cell lines and primary human umbilical vein endothelial cells were treated with 5 μm HBX41-108 for 2 h. The mRNA expression of the indicated genes was determined by qPCR. Each bar represents the mean ± S.D. relative to vehicle-treated control cells (n = 3). *, p < 0.05; **, p < 0.01; ***, p < 0.001. CTRL, control.
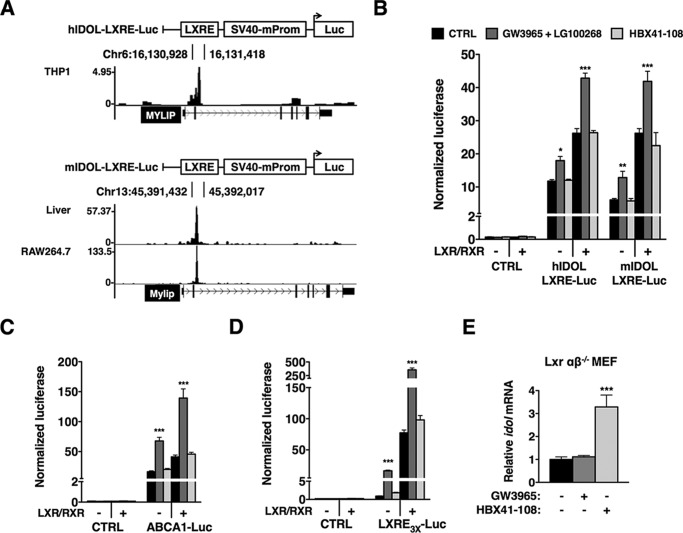
The only transcriptional pathway known thus far to induce IDOL expression is that by LXRs. A distinct requirement or competition for co-activators has been proposed to offer a potential explanation for how LXRs and other members of the nuclear receptor family can differentially regulate their target genes (42). It is therefore possible that HBX41-108 affects a unique transcriptional co-factor used by LXRs to control IDOL expression. To study this possibility, one would need to know the location of the LXR-responsive element(s) in the IDOL promoter. Because these elements have not been definitively established, we analyzed published LXR ChIP-seq experiments done in mouse macrophage-like RAW cells, mouse hepatocytes, and human THP1 macrophages (30–32). We aligned these data sets in the UCSC Genome Browser and observed a prominent LXR peak immediately following IDOL exon 2 (human chr6:16,130,500–16,131,500 and mouse chr13:45,486,900–45,487,200) (Fig. 7A). Importantly, this region is highly conserved between different species. Consistent with this genomic area harboring an active LXRE, reporter constructs containing this region responded to treatment with the LXR (GW3965) and RXR (LG100268) agonists in HepG2 cells (Fig. 7B). Additionally, the basal- and ligand-induced response of these reporters was further enhanced by co-transfection of LXRα and RXRα. Importantly, unlike its effect on IDOL mRNA, HBX41-108 did not enhance the activity of this reporter. Similarly, HBX41-108 did not increase expression of a reporter containing the LXR-responsive proximal promoter of ABCA1 (Fig. 7C) or of an artificial tandem LXRE construct (Fig. 7D), whereas both readily responded to the LXR ligand. These analyses support our findings that pan DUB inhibition does not result in global LXR activation and suggest that regulation of IDOL expression is LXR-independent. This implies that HBX41-108 should increase expression of IDOL in cells devoid of LXRs. Accordingly, in Lxrαβ−/− MEFs that lack active LXR signaling, expression of Idol was significantly increased by HBX41-108 but not by a synthetic LXR ligand (Fig. 7E).
FIGURE 7.
Induction of IDOL by HBX41-108 is LXR-independent. A, LXR ChIP-seq experiments were analyzed and used to identify an active LXRE in human THP1 cells (GSE28319), mouse hepatocytes (GSE35262), and RAW macrophage-like cells (GSE50944), as graphically illustrated. The LXRE-containing regions were cloned into a pGL2-SV40 luciferase reporter. B, HepG2 cells were co-transfected with LXR and RXR expression plasmids together with the indicated luciferase reporters. Subsequently, cells were treated with 1 μm GW3965 and 100 nm LG100268 (24 h) or 5 μm HBX41-108 (8 h). In all luciferase experiments, the transfection efficiency was normalized using Renilla luciferase, which was co-transfected. C, HepG2 cells were transfected as in B, but with control and ABCA1 proximal promoter reporters as indicated, and treated as in B. Each bar represents the mean ± S.D. relative to vehicle-treated control cells (n = 4). D, HepG2 cells were transfected as in B, but with the empty pTK-luciferase reporter or the pTK- LXRE3x luciferase reporter. E, LXRαβ−/− MEFs were stimulated with either 1 μm GW3965 (6 h) or 5 μm HBX41-108 (2 h). Subsequently, mRNA expression of Idol was determined by qPCR. In B–E, each bar represents the mean ± S.D. relative to vehicle-treated control cells (n = 4). *, p < 0.05; **, p < 0.01; ***, p < 0.001. CTRL, control.
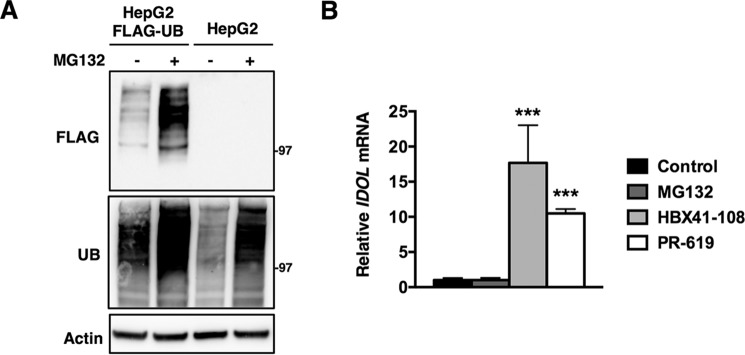
One possible outcome of treating cells with the DUB inhibitors is reduced availability of free ubiquitin, because DUBs are required to recycle ubiquitin for new rounds of conjugation (22). It is therefore possible that under conditions of limited free ubiquitin availability, cells triage E3 ligases according to physiological necessity. This is not without precedent, because, for example, yeast mutants lacking the deubiquitylase Doa4 display reduced viability because of lower free ubiquitin levels and can be fully rescued by forced expression of ubiquitin (43). To test this possibility, we generated HepG2 cells that stably overexpress FLAG-tagged ubiquitin. These cells have elevated levels of ubiquitin as assessed by immunoblotting and functionally respond to inhibition of the proteasome by MG132 by increasing ubiquitylated protein species (Fig. 8A). However, despite increased ubiquitin in these cells, both DUB inhibitors increased IDOL expression (Fig. 8B). Furthermore, effectively inhibiting the proteasome with MG132 did not increase IDOL expression. This result suggests that in these cells limited availability of ubiquitin is not the underlying mechanism for selective induction of IDOL expression.
FIGURE 8.
Forced expression of ubiquitin does not prevent IDOL induction by DUB inhibition. A, HepG2 or HepG2-FLAG ubiquitin cells were treated with control or 25 μm MG132 for 2 h. Total cell lysates were immunoblotted as indicated. B, HepG2-FLAG-ubiquitin cells were treated with vehicle (control), 25 μm MG132, 5 μm HBX41-108, or 50 μm PR-619 for 2 h. Subsequently, mRNA expression of Idol was determined by qPCR. Each bar represents the mean ± S.D. relative to vehicle-treated control cells (n = 3). ***, p < 0.001. UB, ubiquitin.
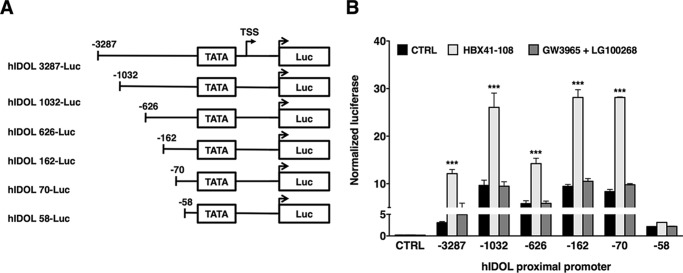
To address the transcriptional mechanism underlying LXR-independent regulation of IDOL expression, we used a promoter truncation strategy (Fig. 9A). We cloned a 3.287-kb proximal promoter fragment of human IDOL (Ch6:16,126,030–16,129,524) into a reporter construct and found that it responded to HBX41-108 (Fig. 9B). Concurrent with the absence of an LXR peak in this genomic region in the ChIP-seq analysis, this reporter did not respond to LXR/RXR ligands (Fig. 9B) or to co-transfection with LXRα and RXRα expression plasmids (data not shown). Serial truncations of the promoter region allowed us to define a minimal, 70-bp region upstream of the transcriptional start site of IDOL that remained responsive to HBX41-108. Deletion of an additional 12 bp reduced reporter activity, even though it remained substantially higher than control. This deletion also largely abolished the response to HBX41-108. We considered that these 12 bp represent the HBX41-108-responsive genomic region and generated a reporter construct with three tandem repeats of this sequence. However, this reporter was unresponsive to HBX41-108 (data not shown). Collectively, our experiments identify an LXR-independent mechanism to regulate IDOL activity that has a strong impact on the associated ubiquitylation-dependent degradation of lipoprotein receptors.
FIGURE 9.
Analysis of the proximal promoter of IDOL identifies an HBX41-108 responsive region. A, graphical representation of the human IDOL proximal promoter luciferase reporter constructs cloned into pGL3-basic. All constructs contain the hIDOL TATA element and an additional 207 base pairs downstream of the transcriptional start site (TSS; +1). B, HepG2 cells were transfected with pGL3 empty or the indicated reporter constructs and treated with 1 μm GW3965 and 100 nm LG100268 (24 h) or 5 μm HBX41-108 (8 h). Renilla luciferase was co-transfected and used to normalize for transfection efficiency. Each bar represents the mean ± S.D. relative to vehicle-treated control cells (n = 4). ***, p < 0.001. CTRL, control.
Discussion
There is growing evidence for involvement of the ubiquitin-proteasomal system in the control of lipid metabolism and in particular of E3 ubiquitin ligases (44). Ubiquitylation is a reversible process, and recent studies have highlighted the role of DUBs in a broad range of cellular processes (20). However, their particular contribution to lipid and cholesterol metabolism is largely unexplored. As such, the main findings of our paper are that pharmacological inhibition of cellular deubiquitylase activity leads to a dramatic decrease in LDLR protein and associated lipoprotein uptake and that this is a result of LXR-independent induction of the E3 ubiquitin ligase IDOL.
To date, the only pathway reported to transcriptionally regulate inducible expression of IDOL is that controlled by LXRs (13). In response to elevated cellular cholesterol levels, for example following uptake of lipoprotein-derived cholesterol, LXRs induce expression and activity of IDOL to promote degradation of the LDLR as a means to limit further accretion of lipoprotein-derived cholesterol. We map here an LXRE-containing genomic region in intron 2 of the IDOL/Idol locus in human and mouse that mediates this. Importantly, our results indicate that induction of IDOL expression by pharmacological inhibition of cellular DUBs is independent of this LXRE-containing region. Moreover, we demonstrate that IDOL induction is also not associated with global enhancement of LXR signaling and expression of LXR target genes. One could speculate that this represents differential regulation of LXR-controlled genes. Indeed, Wagner et al. (45) have previously shown that Lxr target genes display differential regulation in macrophages from Lxrαβ−/− mice. In these cells, expression of the cholesterol-related targets Abca1 and Abcg1 was increased because of derepression, whereas that of the lipogenic targets Srebp1c, Scd1, and Lpl was unchanged. This outcome was proposed to depend on gene-dependent recruitment and utilization of transcriptional co-activators and repressors by LXRs (45). Similarly, tissue- and species-dependent regulation of LXR target genes is also well established, with CYP7A1 and APOE being notable examples (8, 46, 47). The rate-limiting enzyme in bile acid synthesis Cyp7a1 is regulated by LXRs in mice, but not in humans, because of a loss of the LXRE in the human CYP7A1 locus (8, 46). Additionally, LXRs are known to control inducible expression of APOE in macrophages and in adipose tissue, but not in the liver (47). In fact, IDOL also displays tissue- and species-dependent regulation by LXRs; although highly regulated by LXRs in most tissues (13), IDOL is not regulated by LXRs in human and rodent neurons (Ref. 27 and not shown) and in livers of mice (48). Anecdotal evidence suggests that SREBP1c and ABCA1 are regulated by insulin and the co-activator p300, respectively, independent of LXRs (49, 50). However, experiments in Lxr-null mice and hepatocytes have largely demonstrated the crucial role of Lxrs in controlling basal- and inducible expression of these genes (8, 51–53). In that respect, our results showing that DUB inhibition results in specific induction of IDOL, but no other LXR target genes, in an LXR-independent manner are distinct from those previously reported.
We initiated our experiments by evaluating two pan-DUB inhibitors and found that they were able to acutely and robustly induce IDOL expression and lysosomal degradation of the LDLR. Of these, HBX41-108 was initially proposed to act as a selective USP7 inhibitor (34), a DUB implicated in histone H2B ubiquitylation, hormone signaling, PcG silencing, and control of the tumor suppressors phosphatase and tensin homology (PTEN), p16INK4a and p53 (reviewed in Ref. 54). If lack of USP7 activity underlies induction of IDOL by DUB inhibition, one would expect silencing expression of this DUB to mimic pharmacological DUB inhibition. We tested this in HepG2 cells, and despite obtaining effective silencing of USP7, both expression of IDOL and the response to DUB inhibition remained unaffected (data not shown). A potential explanation for this result is recent evidence indicating that HBX41-108 has a much broader substrate specificity and is able to inhibit a large panel of cysteine DUBs, similar to PR-619 (35, 36). The fact that multiple DUBs are sensitive to these inhibitors has precluded us from identifying a specific DUB responsible for the induction of IDOL. We point out that it is also possible that the effect of these DUB inhibitors on IDOL levels is not dependent on inhibition of one specific DUB but rather on combinatorial inhibition of cellular DUBs that converges on the IDOL proximal promoter. DUB activity has been demonstrated to regulate transcriptional responses by influencing, among others, histone modifications, transcription factor activity, and epigenetic modifiers (55). We speculate that this also underlies the ability of HBX41-108 to acutely induce IDOL expression, particularly as the identified responsive genomic region is predicted to contain multiple transcription factor-binding sequences by bioinformatic analysis. Clearly, studies to elucidate the mechanistic basis for induction of IDOL expression by DUB inhibition are required.
An important question raised by our study pertains to the physiological significance of LXR-independent regulation of IDOL expression. In the context of cholesterol metabolism, induction of IDOL in response to cellular cholesterol overload is readily explained as a homeostatic process (13). Yet the role of non-sterol (i.e. LXR-independent) regulation of IDOL expression and activity is less obvious. Next to promoting degradation of the LDLR, we previously reported that IDOL also accelerates degradation of the related lipoprotein receptor VLDLR and APOER2 (27) and show here that VLDLR degradation is also stimulated by HBX41-108. These two are receptors for the extracellular matrix protein Reelin and are particularly important in guiding neuronal migration during development of the central nervous system (56). Accordingly, mice lacking these receptors have developmental problems (57). IDOL is highly expressed in the brain (58), and unlike other LXR targets (e.g. ABCA1) is not subject to LXR-dependent regulation in neurons (27). In fact, what regulates IDOL in the brain and the significance of IDOL to development of the central nervous system is currently unknown. Potentially, our study uncovers a transcriptional process important for controlling IDOL expression in neurons that may impact the Reelin-signaling pathway. Next to this sterol-independent function, our finding may still have implications for cholesterol metabolism. Common genetic variation in the IDOL locus has been identified in genome-wide association studies as a modifier of circulating levels of LDL in humans (18), and we have recently identified the first rare IDOL loss of function variant in individuals with low levels of circulating LDL cholesterol (19). Most of the studies on IDOL, ours included, have largely focused on functional coding variants. Yet variation in a promoter element can have a large impact on expression of the studied gene, as elegantly demonstrated for SORT1 in relation to its effect on the level of circulating cholesterol (18, 59). As such, our experiments demonstrating that a limited region in the IDOL promoter can have a large effect on its expression warrants further investigation into the contribution of genetic variation in this region to variation in LDL cholesterol levels in humans.
In conclusion, we report here that inducible expression of IDOL is subject to robust and rapid regulation by a process that is sensitive to DUB inhibition in cell lines and primary human cells. We further demonstrate that in contrast to the established sterol-dependent regulation of IDOL, this process is LXR-independent. The identification of the genomic region that is subject to regulation by DUB inhibition can direct future experiments geared toward elucidating the underlying transcriptional mechanism.
Author Contributions
J. K. N., E. C., A. L., M. A. H., S. S., P. L. H., and I. K. designed, performed, and analyzed the experiments. J. K. N., H. O., and N. Z. conceived, designed, and wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank members of the Zelcer group, Ben Distel, Eric Reits, and Irith Koster for comments and suggestions.
This work was supported in part by Landsteiner Foundation for Blood Transfusion Research Fellowship 1311 (to I. K.); Netherlands Organization for Scientific Research VICI Grant 724.013.002 (to H. O.); and Dutch Heart Foundation Established Investigator Grant 2013T111, Netherlands Organization of Scientific Research VIDI Grant 17.106.355, and European Research Council Consolidator Grant 617376 (to N. Z.). The authors declare that they have no conflicts of interest with the contents of this article.
- LDLR
- LDL receptor
- LXR
- liver X receptor
- SREBP
- sterol regulatory element binding protein
- DUB
- deubiquitinating enzyme
- USP
- ubiquitin specific protease
- LXRE
- LXR response element
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- qPCR
- quantitative PCR
- IDOL
- inducible degrader of the LDLR
- ESCRT
- endosomal sorting complexes required for transport.
References
- 1. Hobbs H. H., Russell D. W., Brown M. S., and Goldstein J. L. (1990) The LDL receptor locus in familial hypercholesterolemia: mutational analysis of a membrane protein. Annu. Rev. Genet. 24, 133–170 [DOI] [PubMed] [Google Scholar]
- 2. Brown M. S., and Goldstein J. L. (1986) A receptor-mediated pathway for cholesterol homeostasis. Science 232, 34–47 [DOI] [PubMed] [Google Scholar]
- 3. Tolleshaug H., Hobgood K. K., Brown M. S., and Goldstein J. L. (1983) The LDL receptor locus in familial hypercholesterolemia: multiple mutations disrupt transport and processing of a membrane receptor. Cell 32, 941–951 [DOI] [PubMed] [Google Scholar]
- 4. Goldstein J. L., DeBose-Boyd R. A., and Brown M. S. (2006) Protein sensors for membrane sterols. Cell 124, 35–46 [DOI] [PubMed] [Google Scholar]
- 5. Yokoyama C., Wang X., Briggs M. R., Admon A., Wu J., Hua X., Goldstein J. L., and Brown M. S. (1993) SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell 75, 187–197 [PubMed] [Google Scholar]
- 6. Hua X., Yokoyama C., Wu J., Briggs M. R., Brown M. S., Goldstein J. L., and Wang X. (1993) SREBP-2, a second basic-helix-loop-helix-leucine zipper protein that stimulates transcription by binding to a sterol regulatory element. 90, 11603–11607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Horton J. D., Goldstein J. L., and Brown M. S. (2002) SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109, 1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Peet D. J., Turley S. D., Ma W., Janowski B. A., Lobaccaro J. M., Hammer R. E., and Mangelsdorf D. J. (1998) Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXRα. Cell 93, 693–704 [DOI] [PubMed] [Google Scholar]
- 9. Zelcer N., and Tontonoz P. (2006) Liver X receptors as integrators of metabolic and inflammatory signaling. J. Clin. Invest. 116, 607–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Spann N. J., Garmire L. X., McDonald J. G., Myers D. S., Milne S. B., Shibata N., Reichart D., Fox J. N., Shaked I., Heudobler D., Raetz C. R. H., Wang E. W., Kelly S. L., Sullards M. C., Murphy R. C., Merrill A. H., Brown H. A., Dennis E. A., Li A. C., Ley K., Tsimikas S., Fahy E., Subramaniam S., Quehenberger O., Russell D. W., and Glass C. K. (2012) Regulated accumulation of desmosterol integrates macrophage lipid metabolism and inflammatory responses. Cell 151, 138–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang C., McDonald J. G., Patel A., Zhang Y., Umetani M., Xu F., Westover E. J., Covey D. F., Mangelsdorf D. J., Cohen J. C., and Hobbs H. H. (2006) Sterol intermediates from cholesterol biosynthetic pathway as liver X receptor ligands. J. Biol. Chem. 281, 27816–27826 [DOI] [PubMed] [Google Scholar]
- 12. Janowski B. A., Willy P. J., Devi T. R., Falck J. R., and Mangelsdorf D. J. (1996) An oxysterol signalling pathway mediated by the nuclear receptor LXRα. Nature 383, 728–731 [DOI] [PubMed] [Google Scholar]
- 13. Zelcer N., Hong C., Boyadjian R., and Tontonoz P. (2009) LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor. Science 325, 100–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang L., Fairall L., Goult B. T., Calkin A. C., Hong C., Millard C. J., Tontonoz P., and Schwabe J. W. R. (2011) The IDOL-UBE2D complex mediates sterol-dependent degradation of the LDL receptor. Genes Dev. 25, 1262–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sorrentino V., Scheer L., Santos A., Reits E., Bleijlevens B., and Zelcer N. (2011) Distinct functional domains contribute to degradation of the low density lipoprotein receptor (LDLR) by the E3 ubiquitin ligase inducible Degrader of the LDLR (IDOL). J. Biol. Chem. 286, 30190–30199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sorrentino V., Nelson J. K., Maspero E., Marques A. R. A., Scheer L., Polo S., and Zelcer N. (2013) The LXR-IDOL axis defines a clathrin-, caveolae-, and dynamin-independent endocytic route for LDLR internalization and lysosomal degradation. J. Lipid Res. 54, 2174–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Scotti E., Calamai M., Goulbourne C. N., Zhang L., Hong C., Lin R. R., Choi J., Pilch P. F., Fong L. G., Zou P., Ting A. Y., Pavone F. S., Young S. G., and Tontonoz P. (2013) IDOL stimulates clathrin-independent endocytosis and multivesicular body-mediated lysosomal degradation of the low-density lipoprotein receptor. Mol. Cell. Biol. 33, 1503–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Teslovich T. M., Musunuru K., Smith A. V., Edmondson A. C., Stylianou I. M., Koseki M., Pirruccello J. P., Ripatti S., Chasman D. I., Willer C. J., Johansen C. T., Fouchier S. W., Isaacs A., Peloso G. M., Barbalic M., Ricketts S. L., Bis J. C., Aulchenko Y. S., Thorleifsson G., Feitosa M. F., Chambers J., Orho-Melander M., Melander O., Johnson T., Li X., Guo X., Li M., Shin Cho Y., Jin Go M., Jin Kim Y., Lee J. Y., Park T., Kim K., Sim X., Twee-Hee Ong R., Croteau-Chonka D. C., Lange L. A., Smith J. D., Song K., Hua Zhao J., Yuan X., Luan J., Lamina C., Ziegler A., Zhang W., Zee R. Y., Wright A. F., Witteman J. C., Wilson J. F., Willemsen G., Wichmann H. E., Whitfield J. B., Waterworth D. M., Wareham N. J., Waeber G., Vollenweider P., Voight B. F., Vitart V., Uitterlinden A. G., Uda M., Tuomilehto J., Thompson J. R., Tanaka T., Surakka I., Stringham H. M., Spector T. D., Soranzo N., Smit J. H., Sinisalo J., Silander K., Sijbrands E. J., Scuteri A., Scott J., Schlessinger D., Sanna S., Salomaa V., Saharinen J., Sabatti C., Ruokonen A., Rudan I., Rose L. M., Roberts R., Rieder M., Psaty B. M., Pramstaller P. P., Pichler I., Perola M., Penninx B. W., Pedersen N. L., Pattaro C., Parker A. N., Pare G., Oostra B. A., O'Donnell C. J., Nieminen M. S., Nickerson D. A., Montgomery G. W., Meitinger T., McPherson R., McCarthy M. I., McArdle W., Masson D., Martin N. G., Marroni F., Mangino M., Magnusson P. K., Lucas G., Luben R., Loos R. J., Lokki M. L., Lettre G., Langenberg C., Launer L. J., Lakatta E. G., Laaksonen R., Kyvik K. O., Kronenberg F., König I. R., Khaw K. T., Kaprio J., Kaplan L. M., Johansson A., Jarvelin M. R., Janssens A. C., Ingelsson E., Igl W., Kees Hovingh G., Hottenga J. J., Hofman A., Hicks A. A., Hengstenberg C., Heid I. M., Hayward C., Havulinna A. S., Hastie N. D., Harris T. B., Haritunians T., Hall A. S., Gyllensten U., Guiducci C., Groop L. C., Gonzalez E., Gieger C., Freimer N. B., Ferrucci L., Erdmann J., Elliott P., Ejebe K. G., Döring A., Dominiczak A. F., Demissie S., Deloukas P., de Geus E. J., de Faire U., Crawford G., Collins F. S., Chen Y. D., Caulfield M. J., Campbell H., Burtt N. P., Bonnycastle L. L., Boomsma D. I., Boekholdt S. M., Bergman R. N., Barroso I., Bandinelli S., Ballantyne C. M., Assimes T. L., Quertermous T., Altshuler D., Seielstad M., Wong T. Y., Tai E. S., Feranil A. B., Kuzawa C. W., Adair L. S., Taylor HA Jr, Borecki I. B., Gabriel S. B., Wilson J. G., Holm H., Thorsteinsdottir U., Gudnason V., Krauss R. M., Mohlke K. L., Ordovas J. M., Munroe P. B., Kooner J. S., Tall A. R., Hegele R. A., Kastelein J. J., Schadt E. E., Rotter J. I., Boerwinkle E., Strachan D. P., Mooser V., Stefansson K., Reilly M. P., Samani N. J., Schunkert H., Cupples L. A., Sandhu M. S., Ridker P. M., Rader D. J., van Duijn C. M., Peltonen L., Abecasis G. R., Boehnke M., and Kathiresan S. (2010) Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sorrentino V., Fouchier S. W., Motazacker M. M., Nelson J. K., Defesche J. C., Dallinga-Thie G. M., Kastelein J. J. P., Kees Hovingh G., and Zelcer N. (2013) Identification of a loss-of-function inducible degrader of the low-density lipoprotein receptor variant in individuals with low circulating low-density lipoprotein. Eur. Heart J. 34, 1292–1297 [DOI] [PubMed] [Google Scholar]
- 20. Reyes-Turcu F. E., Ventii K. H., and Wilkinson K. D. (2009) Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 78, 363–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sowa M. E., Bennett E. J., Gygi S. P., and Harper J. W. (2009) Defining the Human Deubiquitinating Enzyme Interaction Landscape. Cell 138, 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Komander D., Clague M. J., and Urbé S. (2009) Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563 [DOI] [PubMed] [Google Scholar]
- 23. Motazacker M. M., Pirruccello J., Huijgen R., Do R., Gabriel S., Peter J., Kuivenhoven J. A., Defesche J. C., Kastelein J. J. P., Hovingh G. K., Zelcer N., Kathiresan S., and Fouchier S. W. (2012) Advances in genetics show the need for extending screening strategies for autosomal dominant hypercholesterolaemia. Eur. Heart J. 33, 1360–1366 [DOI] [PubMed] [Google Scholar]
- 24. Schaefer A., Riet, Te J., Ritz K., Hoogenboezem M., Anthony E. C., Mul F. P. J., de Vries C. J., Daemen M. J., Figdor C. G., van Buul J. D., and Hordijk P. L. (2014) Actin-binding proteins differentially regulate endothelial cell stiffness, ICAM-1 function and neutrophil transmigration. J. Cell Sci. 127, 4470–4482 [DOI] [PubMed] [Google Scholar]
- 25. Schippers I. J., Moshage H., Roelofsen H., Müller M., Heymans H. S., Ruiters M., and Kuipers F. (1997) Immortalized human hepatocytes as a tool for the study of hepatocytic (de-)differentiation. Cell Biol. Toxicol. 13, 375–386 [DOI] [PubMed] [Google Scholar]
- 26. Scotti E., Hong C., Yoshinaga Y., Tu Y., Hu Y., Zelcer N., Boyadjian R., de Jong P. J., Young S. G., Fong L. G., and Tontonoz P. (2011) Targeted disruption of the idol gene alters cellular regulation of the low-density lipoprotein receptor by sterols and liver x receptor agonists. Mol. Cell. Biol. 31, 1885–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hong C., Duit S., Jalonen P., Out R., Scheer L., Sorrentino V., Boyadjian R., Rodenburg K. W., Foley E., Korhonen L., Lindholm D., Nimpf J., van Berkel T. J. C., Tontonoz P., and Zelcer N. (2010) The E3 ubiquitin ligase IDOL induces the degradation of the low density lipoprotein receptor family members VLDLR and ApoER2. J. Biol. Chem. 285, 19720–19726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Röhrl C., Eigner K., Winter K., Korbelius M., Obrowsky S., Kratky D., Kovacs W. J., and Stangl H. (2014) Endoplasmic reticulum stress impairs cholesterol efflux and synthesis in hepatic cells. J. Lipid Res. 55, 94–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H., and Glass C. K. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li P., Spann N. J., Kaikkonen M. U., Lu M., Oh D. Y., Fox J. N., Bandyopadhyay G., Talukdar S., Xu J., Lagakos W. S., Patsouris D., Armando A., Quehenberger O., Dennis E. A., Watkins S. M., Auwerx J., Glass C. K., and Olefsky J. M. (2013) NCoR repression of LXRs restricts macrophage biosynthesis of insulin-sensitizing omega 3 fatty acids. Cell 155, 200–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Boergesen M., Pedersen T. Å., Gross B., van Heeringen S. J., Hagenbeek D., Bindesbøll C., Caron S., Lalloyer F., Steffensen K. R., Nebb H. I., Gustafsson J.-A., Stunnenberg H. G., Staels B., and Mandrup S. (2012) Genome-wide profiling of liver X receptor, retinoid X receptor, and peroxisome proliferator-activated receptor α in mouse liver reveals extensive sharing of binding sites. Mol. Cell. Biol. 32, 852–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pehkonen P., Welter-Stahl L., Diwo J., Ryynanen J., Wienecke-Baldacchino A., Heikkinen S., Treuter E., Steffensen K. R., and Carlberg C. (2012) Genome-wide landscape of liver X receptor chromatin binding and gene regulation in human macrophages. BMC Genomics 13, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ekkebus R., van Kasteren S. I., Kulathu Y., Scholten A., Berlin I., Geurink P. P., de Jong A., Goerdayal S., Neefjes J., Heck A. J. R., Komander D., and Ovaa H. (2013) On terminal alkynes that can react with active-site cysteine nucleophiles in proteases. J. Am. Chem. Soc. 135, 2867–2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Colland F., Formstecher E., Jacq X., Reverdy C., Planquette C., Conrath S., Trouplin V., Bianchi J., Aushev V. N., Camonis J., Calabrese A., Borg-Capra C., Sippl W., Collura V., Boissy G., Rain J.-C., Guedat P., Delansorne R., and Daviet L. (2009) Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Mol. Cancer Ther. 8, 2286–2295 [DOI] [PubMed] [Google Scholar]
- 35. Ritorto M. S., Ewan R., Perez-Oliva A. B., Knebel A., Buhrlage S. J., Wightman M., Kelly S. M., Wood N. T., Virdee S., Gray N. S., Morrice N. A., Alessi D. R., and Trost M. (2014) Screening of DUB activity and specificity by MALDI-TOF mass spectrometry. Nat Commun. 5, 4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reverdy C., Conrath S., Lopez R., Planquette C., Atmanene C., Collura V., Harpon J., Battaglia V., Vivat V., Sippl W., and Colland F. (2012) Discovery of specific inhibitors of human USP7/HAUSP deubiquitinating enzyme. Chem. Biol. 19, 467–477 [DOI] [PubMed] [Google Scholar]
- 37. Jeong H. J., Lee H.-S., Kim K.-S., Kim Y.-K., Yoon D., and Park S. W. (2008) Sterol-dependent regulation of proprotein convertase subtilisin/kexin type 9 expression by sterol-regulatory element binding protein-2. J. Lipid Res. 49, 399–409 [DOI] [PubMed] [Google Scholar]
- 38. Abifadel M., Varret M., Rabès J.-P., Allard D., Ouguerram K., Devillers M., Cruaud C., Benjannet S., Wickham L., Erlich D., Derré A., Villéger L., Farnier M., Beucler I., Bruckert E., Chambaz J., Chanu B., Lecerf J.-M., Luc G., Moulin P., Weissenbach J., Prat A., Krempf M., Junien C., Seidah N. G., and Boileau C. (2003) Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 34, 154–156 [DOI] [PubMed] [Google Scholar]
- 39. Horton J. D., Cohen J. C., and Hobbs H. H. (2009) PCSK9: a convertase that coordinates LDL catabolism. J. Lipid Res. 50, (suppl.) S172–S177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cohen J., Pertsemlidis A., Kotowski I. K., Graham R., Garcia C. K., and Hobbs H. H. (2005) Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat. Genet. 37, 161–165 [DOI] [PubMed] [Google Scholar]
- 41. Sorrentino V., and Zelcer N. (2012) Post-transcriptional regulation of lipoprotein receptors by the E3-ubiquitin ligase inducible degrader of the low-density lipoprotein receptor. Curr. Opin. Lipidol. 23, 213–219 [DOI] [PubMed] [Google Scholar]
- 42. Dasgupta S., Lonard D. M., and O'Malley B. W. (2014) Nuclear receptor coactivators: master regulators of human health and disease. Annu. Rev. Med. 65, 279–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Swaminathan S., Amerik A. Y., and Hochstrasser M. (1999) The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol. Biol. Cell 10, 2583–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sharpe L. J., Cook E. C. L., Zelcer N., and Brown A. J. (2014) The UPS and downs of cholesterol homeostasis. Trends Biochem. Sci. 39, 527–535 [DOI] [PubMed] [Google Scholar]
- 45. Wagner B. L., Valledor A. F., Shao G., Daige C. L., Bischoff E. D., Petrowski M., Jepsen K., Baek S. H., Heyman R. A., Rosenfeld M. G., Schulman I. G., and Glass C. K. (2003) Promoter-specific roles for liver X receptor/corepressor complexes in the regulation of ABCA1 and SREBP1 gene expression. Mol. Cell. Biol. 23, 5780–5789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Menke J. G., Macnaul K. L., Hayes N. S., Baffic J., Chao Y.-S., Elbrecht A., Kelly L. J., Lam M.-H., Schmidt A., Sahoo S., Wang J., Wright S. D., Xin P., Zhou G., Moller D. E., and Sparrow C. P. (2002) A novel liver X receptor agonist establishes species differences in the regulation of cholesterol 7α-hydroxylase (CYP7a). Endocrinology 143, 2548–2558 [DOI] [PubMed] [Google Scholar]
- 47. Laffitte B. A., Repa J. J., Joseph S. B., Wilpitz D. C., Kast H. R., Mangelsdorf D. J., and Tontonoz P. (2001) LXRs control lipid-inducible expression of the apolipoprotein E gene in macrophages and adipocytes. Proc. Natl. Acad. Sci. U.S.A. 98, 507–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hong C., Marshall S. M., McDaniel A. L., Graham M., Layne J. D., Cai L., Scotti E., Boyadjian R., Kim J., Chamberlain B. T., Tangirala R. K., Jung M. E., Fong L., Lee R., Young S. G., Temel R. E., and Tontonoz P. (2014) The LXR-Idol axis differentially regulates plasma LDL levels in primates and mice. Cell Metab. 20, 910–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huuskonen J., Fielding P. E., and Fielding C. J. (2004) Role of p160 coactivator complex in the activation of liver X receptor. Arterioscler. Thromb. Vasc. Biol. 24, 703–708 [DOI] [PubMed] [Google Scholar]
- 50. Bobard A., Hainault I., Ferré P., Foufelle F., and Bossard P. (2005) Differential regulation of sterol regulatory element-binding protein 1c transcriptional activity by insulin and liver X receptor during liver development. J. Biol. Chem. 280, 199–206 [DOI] [PubMed] [Google Scholar]
- 51. Repa J. J., Liang G., Ou J., Bashmakov Y., Lobaccaro J. M., Shimomura I., Shan B., Brown M. S., Goldstein J. L., and Mangelsdorf D. J. (2000) Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRα and LXRβ. Genes Dev. 14, 2819–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tobin K. A. R., Ulven S. M., Schuster G. U., Steineger H. H., Andresen S. M., Gustafsson J.-A., and Nebb H. I. (2002) Liver X receptors as insulin-mediating factors in fatty acid and cholesterol biosynthesis. J. Biol. Chem. 277, 10691–10697 [DOI] [PubMed] [Google Scholar]
- 53. Tangirala R. K., Bischoff E. D., Joseph S. B., Wagner B. L., Walczak R., Laffitte B. A., Daige C. L., Thomas D., Heyman R. A., Mangelsdorf D. J., Wang X., Lusis A. J., Tontonoz P., and Schulman I. G. (2002) Identification of macrophage liver X receptors as inhibitors of atherosclerosis. 99, 11896–11901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Clague M. J., Coulson J. M., and Urbé S. (2012) Cellular functions of the DUBs. J. Cell Sci. 125, 277–286 [DOI] [PubMed] [Google Scholar]
- 55. Frappier L., and Verrijzer C. P. (2011) Gene expression control by protein deubiquitinases. Curr. Opin. Genet. Dev. 21, 207–213 [DOI] [PubMed] [Google Scholar]
- 56. Hiesberger T., Trommsdorff M., Howell B. W., Goffinet A., Mumby M. C., Cooper J. A., and Herz J. (1999) Direct binding of Reelin to VLDL receptor and ApoE receptor 2 induces tyrosine phosphorylation of disabled-1 and modulates tau phosphorylation. Neuron. 24, 481–489 [DOI] [PubMed] [Google Scholar]
- 57. Trommsdorff M., Gotthardt M., Hiesberger T., Shelton J., Stockinger W., Nimpf J., Hammer R. E., Richardson J. A., and Herz J. (1999) Reeler/disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell 97, 689–701 [DOI] [PubMed] [Google Scholar]
- 58. Olsson P. A., Korhonen L., Mercer E. A., and Lindholm D. (1999) MIR is a novel ERM-like protein that interacts with myosin regulatory light chain and inhibits neurite outgrowth. J. Biol. Chem. 274, 36288–36292 [DOI] [PubMed] [Google Scholar]
- 59. Musunuru K., Strong A., Frank-Kamenetsky M., Lee N. E., Ahfeldt T., Sachs K. V., Li X., Li H., Kuperwasser N., Ruda V. M., Pirruccello J. P., Muchmore B., Prokunina-Olsson L., Hall J. L., Schadt E. E., Morales C. R., Lund-Katz S., Phillips M. C., Wong J., Cantley W., Racie T., Ejebe K. G., Orho-Melander M., Melander O., Koteliansky V., Fitzgerald K., Krauss R. M., Cowan C. A., Kathiresan S., and Rader D. J. (2010) From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature 466, 714–719 [DOI] [PMC free article] [PubMed] [Google Scholar]