Abstract
Adenosine deaminases acting on double-stranded RNA (ADARs) catalyze the deamination of adenosine (A) to produce inosine (I) in double-stranded (ds) RNA structures, a process known as A-to-I RNA editing. dsRNA is an important trigger of innate immune responses, including interferon (IFN) production and action. We examined the role of A-to-I RNA editing by two ADARs, ADAR1 and ADAR2, in the sensing of self-RNA in the absence of pathogen infection, leading to activation of IFN-induced, RNA-mediated responses in mouse embryo fibroblasts. IFN treatment of Adar1−/− cells lacking both the p110 constitutive and p150 IFN-inducible ADAR1 proteins induced formation of stress granules, whereas neither wild-type (WT) nor Adar2−/− cells displayed a comparable stress granule response following IFN treatment. Phosphorylation of protein synthesis initiation factor eIF2α at serine 51 was increased in IFN-treated Adar1−/− cells but not in either WT or Adar2−/− cells following IFN treatment. Analysis by deep sequencing of mouse exonic loci containing A-to-I-editing sites revealed that the majority of editing in mouse embryo fibroblasts was carried out by ADAR1. IFN treatment increased editing in both WT and Adar2−/− cells but not in either Adar1−/− or Adar1−/−p150 cells or Stat1−/− or Stat2−/− cells. Hyper-edited sites found in predicted duplex structures showed strand bias of editing for some RNAs. These results implicate ADAR1 p150 as the major A-to-I editor in mouse embryo fibroblasts, acting as a feedback suppressor of innate immune responses otherwise triggered by self-RNAs possessing regions of double-stranded character.
Keywords: innate immunity, interferon, protein kinase RNA-activated (PKR), RNA editing, stress granule, adenosine deaminase acting on RNA (ADAR)
Introduction
One mechanism by which cells respond to different forms of stress is the global reduction of protein synthesis (1). Phosphorylation of protein synthesis initiation factor eIF2α on serine 51 is recognized as a universal mechanism of translation suppression in response to cellular stress (1, 2). Four different protein kinases are activated in mammalian cells in response to different stresses, and they all catalyze Ser-51 phosphorylation of eIF2α as follows: protein kinase regulated by RNA (PKR) activated by double-stranded RNA; heme-regulated inhibitor kinase activated by hemin deficiency and oxidative stress; general control non-derepressible kinase 2 (GCN2) activated by amino acid deficiency; and PKR-like endoplasmic reticulum kinase activated by protein misfolding and endoplasmic reticulum stress (2, 3). The global inhibition of translation in cultured cells mediated by eIF2α phosphorylation is linked to the formation of stress granules (SG),2 cytoplasmic aggregates of stalled 40S ribosomal subunit-containing translation initiation complexes, translation initiation factors, and RNA-binding proteins, including Ras GTPase-activating protein-Src homology 3 domain binding protein 1 G3BP1 (4). Stress granule formation is one hallmark of virus infection, serving as an index of innate immune responses, and is PKR-dependent for a variety of viruses (5–8).
A cornerstone of innate antiviral immunity is the interferon (IFN) system (9, 10). IFNs act through the transcriptional induction of IFN-stimulated genes that encode products responsible for the biological activities of IFNs, including the ability of IFNs to inhibit virus multiplication and enhance apoptosis (11–13). Among the IFN-stimulated gene products is the PKR kinase induced by IFN through canonical JAK-STAT signaling (14–16). PKR is activated by binding dsRNA that mediates PKR dimerization and autophosphorylation; activated PKR catalyzes the phosphorylation of eIF2α (17–20). In the case of several viruses, PKR displays antiviral and proapoptotic activities (12, 21, 22). Exemplified by measles virus, PKR sufficiency and kinase activation correlate with reduced virus growth (23, 24), enhanced induction of IFNβ (25, 26), increased SG responses (7), and increased cytotoxicity and apoptosis (27, 28). The central importance of PKR furthermore is illustrated both by the large number of viruses that encode gene products that antagonize PKR activation and function, and by the effects of genetic disruption of the Pkr gene (12, 13, 29).
Similar to PKR, ADAR1 (adenosine deaminase acting on RNA1) is an IFN-inducible dsRNA-binding protein with enzyme activity (30, 31). However, unlike PKR, ADAR1 uses dsRNA as a substrate (32). ADAR1 catalyzes the C6 deamination of adenosine (A) in dsRNA structures to generate inosine (I), a process known as A-to-I editing (33, 34). Adenosine deamination catalyzed by ADARs can lead to destabilization of dsRNA structures, because I base-pairs (bp) as G instead of A, and I:U mismatch bp are less stable than A:U bp (31, 33, 35). Two mammalian genes, Adar1 and Adar2, encode enzymatically active ADARs (31). The Adar1 gene encodes two quite different protein size isoforms that are both active deaminases, the large or p150 IFN-inducible protein found in both the cytoplasm and nucleus and a short or p110 constitutively expressed nuclear protein (30). Expression of ADAR1 proteins, inducible p150 and constitutive p110, occurs by a mechanism involving alternative promoters and alternative exon 1 splicing; the promoter driving p150 expression is IFN-inducible by JAK-STAT signaling (36–38). The Adar2 gene is constitutively expressed, and the ADAR2 protein, like ADAR1 p110, localizes predominantly if not exclusively to the nucleus (31).
In contrast to PKR, ADAR1 is typically proviral and antiapoptotic (31, 39). The proviral and antiapoptotic activities of ADAR1 have been demonstrated with several viruses, including measles virus, hepatitis delta virus, Venezuelan equine encephalitis virus, yellow fever virus, and HIV (27, 31, 40, 41). ADAR1 deficiency, as illustrated with measles virus, correlates with increased PKR activation (24), increased IFNβ induction (25), increased SG responses (7), and increased cytotoxicity and apoptosis (25, 42). Conversely, ADAR1 sufficiency correlates with increased virus growth, impaired SG responses, impaired IFNβ induction, and reduced cytotoxicity and apoptotsis (7, 25, 27, 42). Furthermore, in the absence of virus infection, IFNβ treatment alone induces SG formation in HeLa cells stably deficient in both p110 and p150 but not in ADAR1 sufficient cells (43). We therefore wanted to test whether IFN treatment in the absence of infection affected the SG response in cells genetically null for a single ADAR gene product and, if so, to assess the effect of IFN treatment on both the global A-to-I RNA editing profile and on PKR activation measured by eIF2α phosphorylation.
Using mouse MEF cell lines genetically null for only the inducible p150 (Adar1−/−p150), for both the p150 and p110 ADAR1 proteins (Adar1−/−), or for only the ADAR2 protein (Adar2−/−), we examined the role that these ADARs play in the formation of SG and the phosphorylation of eIF2α in response to IFN treatment, and how the IFN treatment affected the pattern of A-to-I editing observed in MEFs. Surprisingly, we found Adar1−/−p150 cells lacking only p150 readily formed SG in response to IFNα/β treatment, as did Adar1−/− cells lacking both p150 and p110. By contrast, IFN treatment did not lead to SG formation in Pkr−/− cells, Stat2−/− cells, or wild-type MEF cells and only poorly in Adar2−/− cells. When over 500 exonic loci containing about 11,000 editing sites were analyzed, the vast majority of the editing events were dependent upon ADAR1 rather than ADAR2, and many were enhanced by IFN treatment of Adar2−/− cells and wild-type MEF cells but not Adar1−/− or Adar1−/−p150 MEF cells. Stat1−/− and Stat2−/− cells showed significant editing that was not enhanced by IFN. Finally, elevated phosphorylation of eIF2α was found in the Adar1−/− cells but not in Adar2−/− or wild-type cells following IFN treatment. These results suggest that ADAR1 p150 plays an important role in destabilization of dsRNA structures present in cellular RNAs found in IFN-treated cells that otherwise, in the absence of ADAR1 p150, accumulate to a sufficiently high concentration above the threshold necessary to trigger the activation of dsRNA-dependent innate responses illustrated by eIF2α phosphorylation and SG formation.
Experimental Procedures
Cells, Maintenance, and Interferon Treatment
MEF cells homozygous null in adar1−/− (44) or adar2−/− (45) and wild-type (WT) MEF cells were provided by Dr. Kazuko Nishikura (The Wistar Institute, Philadelphia). Adar1−/−p150 MEFs homozygous null for the p150 IFN-inducible isoform of ADAR1 were as described previously (42). Stat1−/− (46) and Stat2−/− (47) MEF cells were provided by Dr. Robert Schreiber (Washington University, St. Louis) and Dr. Christopher Schindler (Columbia University, New York), respectively. Cells were maintained in Dulbecco's modification of Eagle's minimum essential medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (Hyclone), 1 mm sodium pyruvate, 100 mg/ml penicillin, and 100 units/ml streptomycin (Invitrogen). Interferon (IFN) treatment was with 1000 units/ml, unless otherwise indicated, of recombinant human IFNαA/D or mouse IFNβ (PBL Assay Science, Piscataway, NJ) for 24 h. Parallel cultures were left untreated as controls.
Immunofluorescence Analysis
Immunofluorescence was carried out essentially as described (7). Briefly, cells seeded onto 18-mm glass coverslips in 12-well plates were either left untreated or IFN-treated as indicated, and then they were fixed after 24 or 48 h with 10% (v/v) neutral buffered formalin (Sigma). Permeabilization and blocking were carried out using Tris-buffered saline (TBS) containing 2% (v/v) normal donkey serum (Jackson ImmunoResearch) and 0.2% (v/v) Triton X-100 (CBP buffer). Permeabilized cells then were incubated overnight at 4 °C with primary antibody diluted in CBP buffer. Primary antibodies used were mouse monoclonal anti-G3BP1 antibody (Sigma), rabbit anti-G3BP1 antibody (Sigma), and goat polyclonal anti-TIA-1 (Santa Cruz Biotechnology). Coverslips with cells treated with primary antibodies were washed with TBS containing 0.7% fish gelatin and 0.05% Triton X-100 (QW buffer) prior to incubation at 37 °C for 1 h with secondary antibody in CBP. Secondary antibodies used for immunofluorescence were Alexa Fluor 350 anti-rabbit and Alexa Fluor 594 anti-mouse (both from Molecular Probes) and Texas Red-conjugated anti-goat (Jackson ImmunoResearch). Coverslips were washed with QW buffer and then with TBS before mounting with ProLong Gold (Molecular Probes). Slides were analyzed using a fluorescence microscope (Olympus IX71), and images were captured with a Retiga-2000R camera and Q-Capture PRO software (version 6.0, QImaging). Images were processed using IrfanView (version 4.33), and the individual channels were overlaid using GIMP (version 2.8.2).
Quantification of Stress Granules
To quantify the number of SG-positive cells, three representative wide field ×40 images were selected per experiment, and a minimum of 500 cells were counted. Cells that exhibited punctate immunofluorescent cytoplasmic foci with the SG marker G3BP1 were scored as SG-positive. Percentages were determined as the number of SG-containing cells divided by the total number of cells ×100.
Western Immunoblot Analysis
Western blotting was carried out, as described previously (27, 43), using whole-cell extracts (48). Primary antibodies used were as follows: rabbit monoclonal anti-ADAR1 (Santa Cruz Biotechnology, catalog no. 73408); rabbit polyclonal anti-mouse PKR (49); mouse monoclonal anti-tubulin and rabbit polyclonal anti-β-actin (Sigma); rabbit polyclonal anti-eIF2α (Cell Signaling Technology); and rabbit monoclonal anti-phospho-eIF2α Ser-51 (Epitomics-Abcam). Secondary antibodies were anti-rabbit IRDye800 and anti-mouse IRDye680 (both from LI-COR Biosciences). Membranes were scanned using a LI-COR Odyssey FX imaging system at optimized intensities and quantified using the Odyssey image processing software (version 3.0, LI-COR Biosciences). Images were further processed using GIMP (version 2.8.2). Total PKR protein amounts were normalized to β-actin as the loading control. Phospho-eIF2α is relative to total eIF2α protein normalized to β-actin as the loading control.
Microfluidics-based Multiplex PCR and Deep Sequencing (mmPCR-seq)
RNA was isolated from cells treated with IFN or left untreated using TRIzol reagent following the manufacturer's protocol. cDNA was synthesized using the iScript Advanced cDNA synthesis kit (Bio-Rad). 1 μg of cDNA was used as input to mmPCR-seq. Briefly, 557 loci containing 11,103 A-to-I-editing sites3 were PCR-amplified as described previously (50). The samples were sequenced on an Illumina MiSeq instrument using 145-bp paired-end reads. Sequencing reads were mapped onto the mm9 genome and editing levels calculated as described previously (50). We enforced a minimum sequencing coverage of 50 reads for editing level quantification. Editing sites with reproducible editing level measurements in two biological replicates (within 10%) were used in the comparisons. The raw sequencing data is deposited in the Sequence Read Archive (GEO accession number GSE71834).
RNA Secondary Structure Prediction
Secondary structure predictions were generated using 200 nucleotides flanking the PCR amplicon sequence in searches for editing complementary sequences using the RNAfold program. The free energy of the predicted structure was calculated both for the unedited sequence and also for the sequence with inosine in place of adenosine for hyper-edited RNA.
Results
Interferon Treatment Leads to Stress Granule Formation in Cells Lacking ADAR1 but Not in Cells Lacking ADAR2
Formation of SG is a characteristic host response to infection observed in cell culture with a number of different viruses, including measles virus (6, 7). Unexpectedly, IFN treatment of HeLa cells deficient in ADAR1 also was found to cause formation of SG even in the absence of infection, reaching SG levels approaching that seen in measles virus-infected cells (43). ADAR1 is one of the two known catalytically active mammalian A-to-I RNA-editing enzymes. It is not known whether IFN treatment of cells deficient in ADAR2, the other active ADAR, also results in the formation of SG, nor is it known whether deficiency of the p110 or the p150 isoform of ADAR1 is responsible for the IFN-induced SG response seen in HeLa cells. Using MEF cell lines genetically null for either ADAR2 or ADAR1, or only ADAR1 p150, we tested the effect of ADAR deficiency on the IFN response in MEF cells measured by SG formation. As background, ADAR1 expression is unchanged in Adar2−/− cells from WT cells (45); ADAR2 is not detectably altered from WT in Adar1−/− cells (44, 51); and ADAR1 p110 expression is normal in Adar1−/−p150 cells (42). We monitored SG formation in untreated and IFN-treated cells by the staining pattern of G3BP1 or TIA-1, established markers of SG (7, 52, 53).
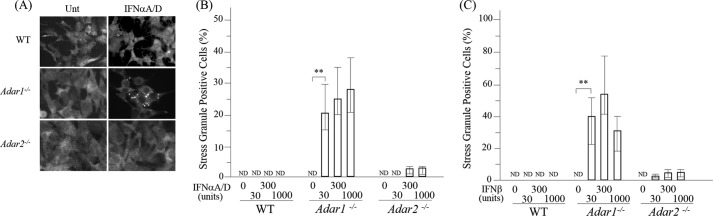
As shown in Fig. 1A, treatment of Adar1−/− cells with IFNα resulted in a G3BP1 staining pattern of punctate cytoplasmic foci, whereas in IFN-treated Adar2−/− cells and IFN-treated wild-type cells the G3BP1 signal was homogeneously dispersed in the cytoplasm. Likewise, the G3PB1 signal in untreated MEF cells, either Adar1−/−or Adar2−/− or WT, was not punctate but rather disperse. Both IFNα (Fig. 1B) and IFNβ (Fig. 1C) triggered the formation of SG in Adar1−/− cells treated with 30 units/ml, whereas doses as high as 1000 units/ml of either IFNα or IFNβ did not detectably induce SG in WT cells and only poorly induced SG in Adar2−/− cells. Quantitation revealed that IFNβ was a more potent inducer of SG than IFNα in Adar1−/− cells (Fig. 1, B and C).
FIGURE 1.
Stress granule formation is enhanced by IFN treatment of Adar1−/− cells but not Adar2−/− or wild-type cells. MEF cells, either wild-type (WT) or Adar null mutant, were left untreated (Unt) or were treated with IFN as indicated and then analyzed for stress granule formation. A, WT, Adar1−/−, and Adar2−/− mutant MEF cells analyzed by immunofluorescence microscopy as described under “Experimental Procedures” using antibody to G3BP1 as a marker for SG formation. IFN treatment was with 1000 units/ml of IFNαA/D. B, quantification of SG-positive WT, Adar1−/−, and Adar2−/− mutant cells, untreated or IFNαA/D-treated as indicated. Three representative wide field ×40 images were selected, and a minimum of 500 cells was examined for the presence or absence of SG as described under “Experimental Procedures.” Results are expressed as the percentage of cells positive for SG. Results are mean values and standard errors from three independent experiments. Statistical significance was determined by Student's t test; **, p < 0.05; ND, not detected. C, quantification of SG-positive WT, Adar1−/−, and Adar2−/− MEF cells, either untreated or IFN-treated as described under B, except treatment was with IFNβ at the indicated concentration.
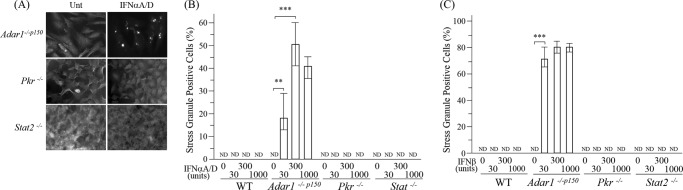
Adar1−/− cells do not express either isoform of ADAR1, p110 or p150 (44, 51). To test whether only deficiency of the inducible p150 size isoform of ADAR1 was sufficient to trigger SG formation in response to IFN treatment, Adar1−/−p150 cells that express nuclear p110 but not cytoplasmic p150 were examined. As shown in Fig. 2, formation of SG was readily detected following treatment of Adar1−/−p150 cells with either IFNα (Fig. 2, A and B) or IFNβ (Fig. 2C) when monitored by the G3BP1 staining pattern. Results for the IFN-induced SG formation in ADAR1 mutant cells seen with the G3BP1 marker (Figs. 1 and 2) were confirmed with TIA-1 (data not shown), another marker of SG (7, 52). Similar to the observations with Adar1−/− cells, IFNβ was more potent than IFNα in inducing SG formation in the Adar1−/−p150 cells (Fig. 2, B and C). A high percentage (∼40–70%) of the Adar1−/−p150 and Adar1−/− cells were SG-positive when treated with IFNβ. As controls, WT and mutant Pkr −/− and Stat2−/− MEF cells were also examined. IFN treatment did not detectably trigger SG formation in any of these ADAR1-sufficient MEF cells, either wild-type (WT) or genetic null mutant Pkr−/− or Stat2−/− cells (Fig. 2).
FIGURE 2.
Stress granule formation is enhanced by IFN treatment of Adar1−/−p150 cells but not Pkr−/− or Stat2−/− cells. MEF cells, either wild-type (WT) or null mutant as indicated, were left untreated (Unt) or were treated with IFN and then analyzed for stress granule formation. A, Adar1−/−p150, Pkr−/−, and Stat2−/− mutant MEF cells were analyzed by immunofluorescence microscopy as described under “Experimental Procedures” using antibody to G3BP1 as a marker for SG formation. IFN treatment was with 1000 units/ml of IFNαA/D. B, quantification of SG-positive Adar1−/−p150, Pkr−/−, and Stat2−/− mutant MEF cells, either untreated or IFNαA/D-treated as indicated, as described for Fig. 1B. Statistical significance was determined by Student's t test; **, p < 0.05; ***, p < 0.005; ND, not detected. C, quantification of SG-positive Adar1−/−p150, Pkr−/−, and Stat2−/− mutant MEF cells, either untreated or IFN-treated as described under Fig. 1B, except treatment was with IFNβ at the indicated concentration.
Interferon Treatment Leads to Enhanced Phosphorylation of eIF2α in Adar1−/− Cells but Not in Adar2−/− or Wild-type Cells
Stress granules are cytoplasmic aggregates of stalled translation initiation complexes (6, 54). Virus-mediated stress granule formation is PKR- and phospho-eIF2α-dependent (7, 8). The best characterized substrate of PKR is protein synthesis initiation factor eIF2α (19), which when phosphorylated on serine 51 of the α subunit leads to a global inhibition of translation (1, 12, 13). Because PKR is activated by dsRNA (19, 55) and because deficiency of the dsRNA-editing enzyme ADAR1 led to SG formation following IFN treatment (Figs. 1 and 2), we next tested the effect of IFN treatment and ADAR deficiency on eIF2α phosphorylation in WT and in mutant Adar1−/− and Adar2−/− cells (Fig. 3).
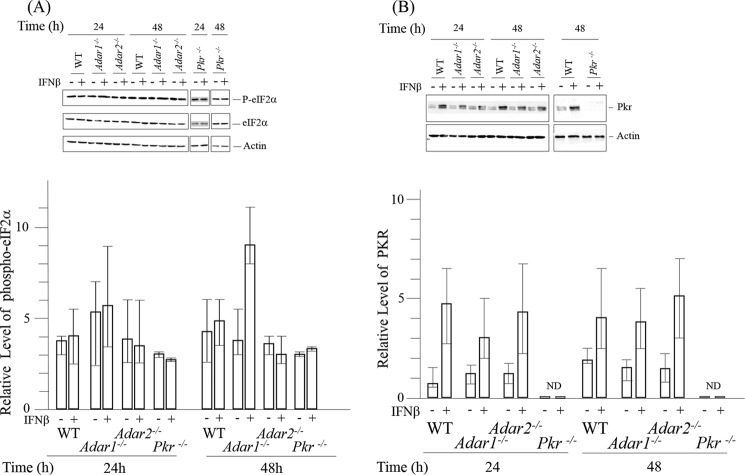
FIGURE 3.
Phosphorylation of eIF2α in ADAR1-deficient cells following interferon treatment correlates with enhanced stress granule formation. A, relative level of Ser-51 phospho-eIF2α in wild-type (WT) and Adar1−/− and Adar2−/− mutant MEF cells, either untreated or treated with IFNβ, was determined by quantification of Western blots using a LI-COR Odyssey infrared imager system. Whole-cell extracts were prepared and analyzed by immunoblot assay with antibodies against eIF2α, phospho_T446 P-eIF2α, and actin. Upper, representative blot. Lower, results shown are mean values and standard errors from three independent experiments. Phospho-eIF2α is expressed relative to total eIF2α protein, normalized to β-actin as the loading control. B, Western immunoblot analysis for PKR expression. Whole-cell extracts were prepared from WT cells or mutant Adar1−/−, Adar2−/−, Pkr−/−, or Adar1−/−p150 MEF cells, either untreated or IFN-treated, and analyzed by Western blotting using antibodies against PKR and β-actin. Upper, representative blot. Lower, results shown are mean values and standard errors from independent experiments. Total PKR protein amount is relative, normalized to β-actin as the loading control. ND, not detected.
Phosphorylation of eIF2α Ser-51 was increased following IFN treatment of Adar1−/− cells that lack both p110 and p150 but not in either WT or Adar2−/− cells (Fig. 3A). As a control, IFN treatment did not increase eIF2α phosphorylation in Pkr−/− cells. The relative basal levels of eIF2α phosphorylation in the four kinds of MEFs, WT, Adar1−/−, Adar2−/−, and Pkr−/−, were comparable (Fig. 3A). Furthermore, the amount of PKR was comparable in untreated cells and was similarly inducible by IFN in the WT, Adar1−/−, and Adar2−/− cells, but PKR was not detectable in either untreated or IFN-treated Pkr−/− cells (Fig. 3B). These results suggest that the enhanced SG formation seen in IFN-treated cells lacking ADAR1 (Fig. 1) correlates with the enhanced phosphorylation of eIF2α seen in IFN-treated cells lacking ADAR1 (Fig. 3A).
Interferon Treatment Increases A-to-I Editing at Many Sites and ADAR1 Is Responsible for Most Editing Events in MEFs, Both Constitutive and Interferon-inducible
The deamination of adenosine to inosine in duplex RNA structures is catalyzed by editing enzymes encoded by Adar1 and Adar2 (31, 33). Although A-to-I editing of mouse cellular RNAs is well established (56, 57), little is known regarding the effect of IFN on RNA editing profiles. Because the formation of SG and the phosphorylation of eIF2α both were increased by IFN treatment in ADAR1-deficient cells but not in ADAR1-sufficient cells either possessing (WT) or lacking (Adar2−/−) the ADAR2 enzyme, we next examined the effect of IFN treatment on the overall editing profile in MEFs using mmPCR-seq, a targeted deep sequencing approach (50). The mmPCR-seq method utilizes a microfluidic chip to simultaneously amplify 557 loci containing ∼11,000-editing sites from 48 different samples. Editing levels are accurately quantified by deep sequencing of the PCR products. Editing profiles were determined for RNA isolated from untreated cells and IFN-treated cells, either WT or mutant Adar1−/−, Adar2−/−, Adar1−/−p150, Stat1−/−, or Stat2−/− MEF cells (Fig. 4).
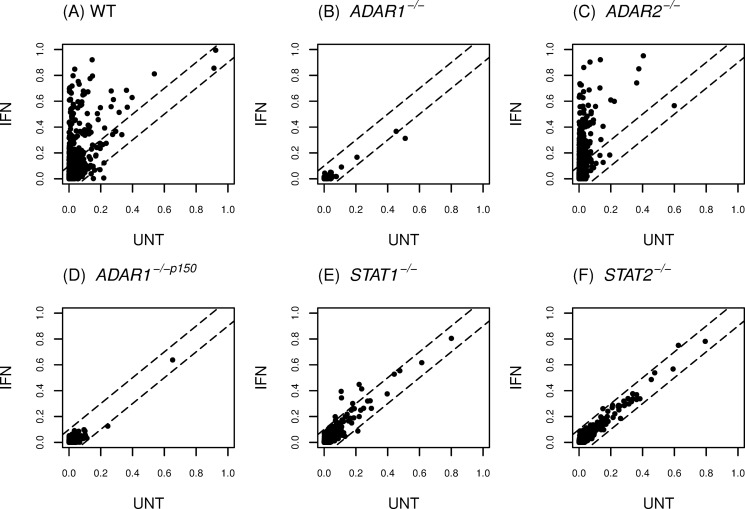
FIGURE 4.
IFN treatment enhances A-to-I editing in MEFs by canonical STAT1/2-dependent signaling, and ADAR1 is responsible for most of the observed editing, both constitutive and inducible. Editing profiles were determined for RNA isolated from untreated and IFNαA/D-treated MEF cells: A, wild-type (WT); B, Adar1−/−; C, Adar2−/−; D, Adar1−/−p150; E, Stat1−/−; and F, Stat2−/−. IFN treatment was with 1000 units/ml for 24 h. Microfluidics-based multiplex PCR and deep sequencing were carried out as described under “Experimental Procedures” for 557 mouse exonic loci that contain 11,103 editing sites. The editing levels for sites in IFN-treated cells compared with untreated cells are shown.
IFN treatment of WT MEFs greatly enhanced the level of A-to-I editing at many sites compared with the editing seen in untreated cells (Fig. 4A). By contrast, editing was greatly reduced in both IFN-treated and -untreated mutant Adar1−/− cells (Fig. 4B), indicating that either ADAR1 p110 or p150 or both ADAR1 isoforms were responsible for most A-to-I editing that occurred in cultured MEFs. The editing profile found for mutant Adar2−/− MEFs was similar to that of WT MEFs (Fig. 4C). The overall enhancement of editing in Adar2−/− MEFs by IFN treatment (Fig. 4C) was comparable with that observed for WT MEFs (Fig. 4A), consistent with the notion that ADAR2 uniquely edits very few sites in the mouse transcriptome (45). Low levels of editing were observed with Adar1−/−p150 cells (Fig. 4D), both untreated and IFN-treated, comparable with the low editing seen for Adar1−/− cells (Fig. 4B), suggesting that ADAR1 p150 was required for most A-to-I editing events in MEFs. Finally, A-to-I editing was not increased by IFN treatment in either Stat1−/− (Fig. 4E) or Stat2−/− (Fig. 4F) MEFs, indicating that the IFN-inducible enhancement of editing was by canonical STAT1/2-dependent signaling. The similar editing levels seen in untreated and IFN-treated Stat1−/− and Stat2−/− MEFs presumably are due to the activities of the constitutively expressed ADAR enzymes, ADAR1 p110 and ADAR2.
A-to-I Editing of Cellular Transcripts Occurs within Regions of Predicted Secondary Structure
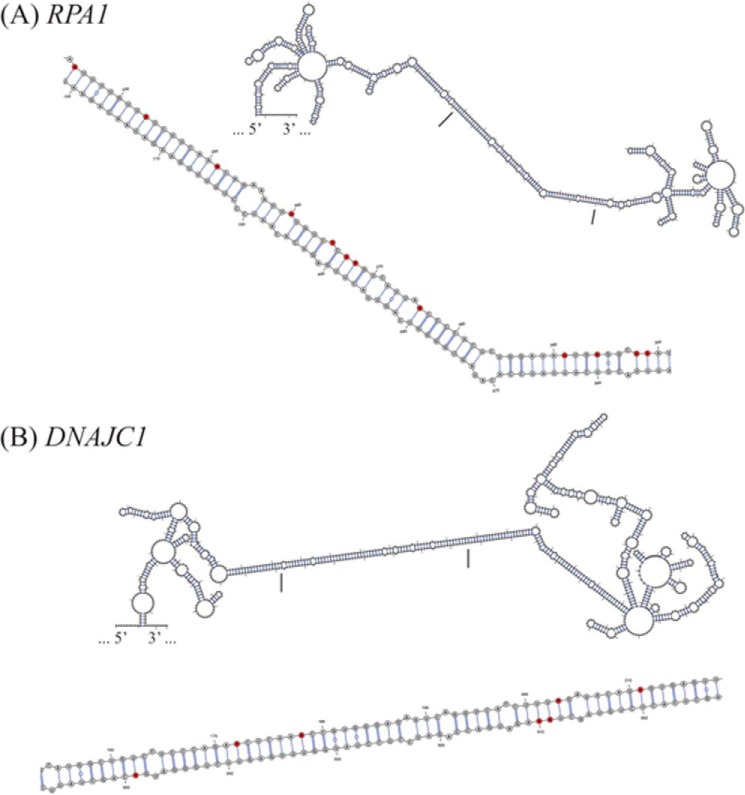
The catalytic activity of ADARs requires double-stranded RNA binding domains (58, 59) that are present in multiple copies, three in ADAR1 p110 and p150 proteins and two copies in ADAR2 (34). The double-stranded RNA binding domains bind dsRNAs greater than ∼16 bp in length in a sequence-independent manner, including dsRNAs with secondary structure defects (13, 55, 60, 61). We searched for RNA secondary structures arising from inverted complementary sequences that include the unedited adenosine form of the exonic sites that undergo A-to-I editing. RNA secondary structures were predicted for some unedited RNAs within 200 nucleotides flanking the PCR amplicon (Fig. 5). As illustrated for the RPA1 (Fig. 5A) and DNAJC1 (Fig. 5B) transcripts, both of these RNAs possessed multiple sites of A-to-I editing. The observed sites of adenosine editing within the predicted duplex structure were largely clustered on one strand of the duplex region of RPA1 but were present on both strands of the DNAJC1 duplex. When the secondary structures of the RPA1 and DNAJC1 RNAs were then modeled with inosine in place of adenosine at the experimentally observed sites of editing, the A-to-I substitution editing was predicted to give rise to RNA structures with higher free energy states, a difference of ∼25 kcal/mol for RPA1 RNA and ∼39 kcal/mol for DNAJC1 RNA, and hence destabilized structures.
FIGURE 5.
Clustered A-to-I editing occurs within regions of predicted secondary structure of cellular transcripts. RNA secondary structure predicted for unedited RNA within 200 nucleotides flanking the PCR amplicon for RPA1 (A) and DNAJC1 (B) transcripts that possess multiple A-to-I-editing sites. The region denoted by the bars corresponds to the enlarged image region and shows edited adenosines that largely cluster on one strand of the predicted duplex region of RPA1 but on both strands of DNAJC1.
Discussion
Double-stranded RNA is a well established trigger of both the production and the action of type I IFNs in virus-infected cells. When present in the cytoplasm, viral dsRNA is detected by the retinoic acid-inducible gene I (RIG-I) family of nucleic acid sensors, thereby leading to transcriptional activation of IFN gene expression via the IPS/MAVS adaptor signaling pathway (62, 63). dsRNA also activates IFN-induced gene products, including PKR, a key mediator of the actions of IFNs through phosphorylation of protein synthesis initiation factor eIF2α (13). These dsRNA-mediated cellular responses first described for virus-infected cells are suppressed by ADAR1, as illustrated initially by observations with measles virus. We earlier found that ADAR1 suppressed multiple dsRNA-dependent innate activities, including the activation of PKR, the IPS-dependent induction of IFN, and the formation of SG in cells infected with WT and V mutant measles virus (7, 25, 27). However, suppression of these dsRNA-dependent responses by ADAR1 remarkably was not observed with the measles C mutant (7, 25, 27), a mutant now understood to produce much higher amounts of viral dsRNA than either WT or the V mutant virus (23, 27, 64). Our results described herein, together with those of others (65–67), reveal that in the absence of ADAR1 at least some of the cytoplasmic sensors of dsRNA are also triggered by cellular (self) dsRNAs. Transcriptional profiling and pathway analyses carried out with mouse embryo cells revealed a strong association between ADAR1 catalytic deficiency and the gene expression signature of IFN-treated or virus-infected cells characteristic of RIG-I and MDA5 activation (65, 68). These findings taken together further establish ADAR1, and in particular the p150 isoform of ADAR1, as a key suppressor of dsRNA-mediated innate cellular responses, presumably through destabilization of duplex RNA structures.
Analysis of nearly 11,000 editing sites in untreated and IFN-treated WT and null mutant MEFs revealed that the vast majority of the A-to-I editing events were dependent upon ADAR1 and not ADAR2, and furthermore, this editing was enhanced by IFN treatment in a manner dependent upon ADAR1 p150, STAT1, and STAT2. The IFN-induced increase in editing attributed to ADAR1 p150 correlated both with the suppression of IFN-induced SG formation (Figs. 1 and 2) and the suppression of an IFN-induced increase in eIF2α phosphorylation (Fig. 3). These findings are consistent with the notion that the increase in A-to-I editing by ADAR1 caused a reduced steady-state concentration of cellular dsRNA structures, falling below the functional threshold concentration required to trigger eIF2α phosphorylation and SG formation. By contrast, in the absence of ADAR1 activity, the functional concentration of cellular dsRNA structures instead rises to a level above the threshold necessary to trigger the eIF2α phosphorylation and SG responses. The combined effects of a higher level of PKR expression due to induction by IFN, together with the accumulation of RNAs with duplex structure due to the absence of the destabilizing effect of ADAR1, provide an explanation for the enhanced eIF2α phosphorylation and SG formation that were observed. Interestingly, ADAR enzymatic activity was first described as a dsRNA unwinding activity during antisense RNA studies in Xenopus. But instead of unwinding, the deamination caused duplex RNA destabilization through hyper-editing and formation of I:U mismatch pairs that are less stable than A:U bp (35, 69).
Among the first ADAR RNA substrates identified in mammalian cells were transcripts encoding the glutamate GluRB/AMPA and serotonin 2C receptors (70–72). GluRB and 5HT2cR RNA editing is highly selective in the coding region and causes amino acid substitutions that alter the functional properties of the encoded receptors in the brain. However, such highly specific A-to-I editing events within exonic ORF sequences are the exception in the context of present understanding. Of the several thousand A-to-I-editing sites identified through next generation sequencing studies, the vast majority is found within noncoding sequences of genes that are not inducible by IFN (56, 73–78). What then is the molecular basis of the IFN-induced, dsRNA-dependent innate responses exemplified by eIF2α phosphorylation and SG formation that we now find are suppressed by A-to-I editing but are de-repressed in the absence of ADAR1 following IFN treatment?
dsRNA is the substrate of ADARs (31, 33, 58). IFN treatment induces ADAR1 p150 (38, 79), and ADARs (33, 35), including ADAR1 (herein), destabilize RNA secondary structures that otherwise accumulate to a sufficiently high concentration to trigger dsRNA-dependent innate responses. Among these responses is the activation of PKR and eIF2α phosphorylation (Fig. 3) (13, 19), and the activation of RIG-receptor signaling and enhanced IFN gene transcription (63, 65, 67, 80). These cellular responses are suppressed by ADAR1 (7, 25, 27, 81). As described earlier in virus-infected cells, in the absence of catalytically active p150, WT and V mutant measles viruses that ordinarily are poor inducers of IFNβ (26) and SG formation (7) in cultured human cells become excellent inducers, comparable with that of the C mutant virus (7, 25). However, in the presence of ADAR1 p150, the WT and V mutant viruses are poor inducers, even though the C mutant virus, a virus that produces high amounts of dsRNA, remains a robust inducer (7, 25). Likewise, in mice lacking catalytically active ADAR1, RIG receptor signaling occurs, thereby leading to a type 1 IFN signature (65, 68).
In the context of biologic responses, ADAR1 deficiency leads to enhanced apoptosis, both in cultured cells and intact animals. Knockdown of ADAR1 in HeLa cells caused increased cytotoxicity characteristic of apoptosis mediated by PKR following viral infection (24, 27). ADAR1 p150 deaminase activity is required both for suppression of PKR activation and for suppression of IFNβ induction and the type I IFN signature (7, 65). Genetic disruptions of the Adar1 gene, either knocking out both p110 and p150 or disrupting only p150 alone, or the knock-in of the editing-deficient Adar1E861A mutant, result in embryonic lethality characterized by increased cell death in the hematopoietic system and disintegration of the fetal liver (42, 44, 51, 65). Independent roles for ADAR1 isoforms have been described, with p150 affecting the MDA5 receptor response and both p150 and p110 isoforms affecting multiorgan development in the mouse (67). ADAR1 is expressed in a broad range of different tissues in adult mice and during embryonic development (37, 44, 51). The conditional Adar1 mouse knock-out and the knock-in of a catalytically deficient E861A mutant ADAR1 both give rise to an IFN signature (65, 68). Likewise, human Aicardi-Goutieres syndrome patients with mutations in ADAR1 show a type I IFN signature (66). These results, taken together, suggest that ADAR1 p150 catalytic activity and hence the generation of inosine in dsRNA lead to a suppression of the type 1 IFN response. A simple explanation is that the suppression results from the destabilization of duplex RNAs, either cellular dsRNA regions as shown herein (Figs. 4 and 5) and also by Walkley and co-workers (65) or viral dsRNA structures as seen with the measles virus (23, 64).
We found that the editing within noncoding 3′-UTRs was clustered and in instances biased on one strand of the predicted duplex RNA structure as illustrated by the cellular RPA1 transcript (Fig. 5). The strand-bias observed for RNAs edited in vivo is suggestive of a processive ADAR1 activity, consistent with earlier biochemical analyses with synthetic RNA substrates (33, 82). As illustrated by the RPA1 and DNAJC1 RNAs, the observed editing is predicted to give rise to RNA structures with higher free energy states, consistent with the notion that A-to-I editing by ADAR1 causes destabilization of the dsRNA regions generated by inverted repeat stem structures within self-cellular transcripts.
The schematic shown in Fig. 6 summarizes a model consistent with observations for uninfected as well as virus-infected cells. Because the IFN-induced changes in SG formation and eIF2α phosphorylation, like the majority of the IFN-induced A-to-I editing events, are dependent upon ADAR1 p150 (Figs. 2 and 3), then one or more properties of the ADARs unique to the p150 protein presumably are responsible for the changes. ADAR1 p150 is the only known ADAR that is cytoplasmic; ADAR1 p150 also is the only known IFN-inducible ADAR (30, 31, 83). Because the formation of cytoplasmic SG is impaired by ADAR1 both in uninfected IFN-treated mouse MEFs as shown herein (Figs. 1 and 2) and in untreated and IFN-treated HeLa cells following infection (7, 43), and because SG formation is PKR-dependent (6, 7) and the IFN-induced phosphorylation of eIF2α likewise is PKR-dependent (Fig. 3), it appears that in uninfected ADAR1 p150-deficient cells there is a population of cellular (self) RNAs that possess sufficient dsRNA structure to activate dsRNA sensors. Of the exonic loci that we analyzed for editing, very few sites were present in IFN-stimulated genes. The enhanced editing observed following IFN treatment most likely is due to the induction of the ADAR1 p150 enzyme by IFN rather than the induction of substrate RNAs, as neither Adar1−/− cells lacking both p150 and p110 (44, 51) nor Adar1−/−p150 cells lacking only p150 (42) showed the increased editing. Furthermore, neither Stat1−/− nor Stat2−/− MEFs showed IFN-induced editing. ADAR1 is induced by IFN through JAK-STAT signaling, both in mouse MEFs and human 2fTGH cells (36, 38). Likewise, PKR is induced by IFN through JAK-STAT signaling (14, 15). IFN treatment increased PKR protein levels in both ADAR1-deficient (Adar1−/−) and ADAR1-sufficient (WT, Adar2−/−) MEFs; however, the IFN treatment led to an increased eIF2α phosphorylation and SG formation only in the Adar1−/− cells (Figs. 1–3). It is conceivable that the different cytoplasmic dsRNA sensors, exemplified by PKR, and by RIG-I and MDA5, possess different thresholds for activation by the population of RNAs with double-stranded regions present within uninfected ADAR1-deficient cells (self-RNAs) as well as in virus-infected ADAR1-sufficient cells (non-self RNAs). Both mouse and human cells deficient in ADAR1 protein display selective marker induction following IFN treatment, including stress granule formation and PKR-dependent phosphorylation. This suggests a universal response to ADAR1 deficiency, illustrated phenotypically by the genetic knock-out of ADAR1 in mouse MEF cells described herein and the short hairpin RNA knockdown of ADAR1 in human HeLa cells (43).
FIGURE 6.
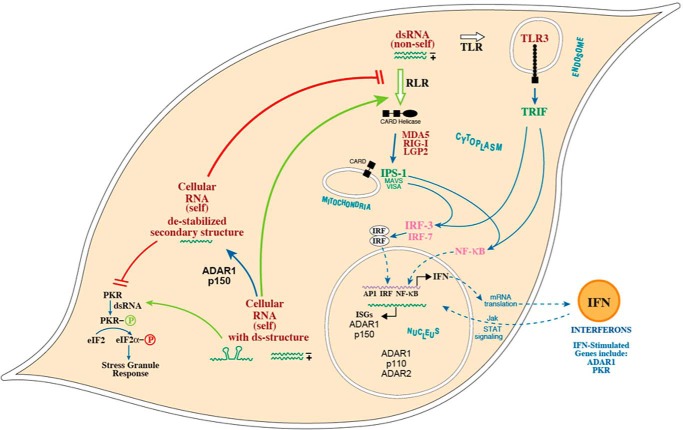
Model for the role of ADAR1 as a suppressor of cytoplasmic dsRNA sensing. dsRNA sensors mediate the production of type I interferons and proinflammatory cytokines through activation of IRF3 and IRF7 and NF-κB transcription factors. The sensors of dsRNA include the following: the cytosolic RIG-I-like receptor (RLR) family of proteins (RIG-I, Mda5, and LGP2) that signal via the MAVS (IPS-1 and VISA) mitochondrial adaptor protein and the endosomal membrane-associated Toll-like receptor 3 (TLR3) that signals via the TRIF adaptor. ADARs catalyze the deamination of A in duplex RNA structures producing I that base-pairs as G. ADAR1 p150 is cytoplasmic, whereas ADAR1 p110 and ADAR2 are nuclear enzymes. A-to-I editing leading to I:U mismatch base pairs destabilizes dsRNA structures, both in cellular (self) dsRNAs and viral (non-self) dsRNAs, thereby resulting in reduced levels of functional dsRNA and the impaired activation of dsRNA-dependent sensors. In the absence of ADAR1 p150, dsRNA accumulates and subsequently triggers dsRNA-dependent responses, including activities by RIG-I-like receptors and PKR. Viral infection leads to elevated levels of dsRNA (non-self) compared with levels of cellular (self) dsRNA present in uninfected cells.
Author Contributions
C. X. G., G. R., J. B. L., and C. E. S. designed the experiments and wrote the manuscript; C. X. G. and G. R. performed the experiments.
Acknowledgment
We thank Dr. Meng How Tan for designing and providing the primers used in the multiplex PCR amplification.
This work was supported in part by National Institutes of Health Research Grants AI-12520 and AI-20611 from the NIAID (to C. E. S.) and GM-202484 from the NIGMS (to J. B. L.), a Stanford Graduate Fellowship (to G. R.), and by the Ellison Medical Foundation (to J. B. L.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
M. H. Tan, G. Ramaswami, and J. B. Li, unpublished data.
- SG
- stress granule
- ADAR1
- adenosine deaminase acting on RNA1
- MEF
- mouse embryo fibroblast
- mmPCR-seq
- microfluidics-based multiplex PCR and deep sequencing
- RIG
- retinoic acid-inducible gene.
References
- 1. Holcik M., and Sonenberg N. (2005) Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 6, 318–327 [DOI] [PubMed] [Google Scholar]
- 2. Samuel C. E. (1993) The eIF-2α protein kinases, regulators of translation in eukaryotes from yeasts to humans. J. Biol. Chem. 268, 7603–7606 [PubMed] [Google Scholar]
- 3. Wek R. C., Jiang H. Y., and Anthony T. G. (2006) Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 34, 7–11 [DOI] [PubMed] [Google Scholar]
- 4. Anderson P., and Kedersha N. (2009) RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 10, 430–436 [DOI] [PubMed] [Google Scholar]
- 5. Beckham C. J., and Parker R. (2008) P bodies, stress granules, and viral life cycles. Cell Host Microbe 3, 206–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lloyd R. E. (2013) Regulation of stress granules and P-bodies during RNA virus infection. Wiley Interdiscip. Rev. RNA 4, 317–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Okonski K. M., and Samuel C. E. (2013) Stress granule formation induced by measles virus is protein kinase PKR-dependent and impaired by RNA adenosine deaminase ADAR1. J. Virol. 87, 756–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ruggieri A., Dazert E., Metz P., Hofmann S., Bergeest J. P., Mazur J., Bankhead P., Hiet M. S., Kallis S., Alvisi G., Samuel C. E., Lohmann V., Kaderali L., Rohr K., Frese M., et al. (2012) Dynamic oscillation of translation and stress granule formation mark the cellular response to virus infection. Cell Host Microbe 12, 71–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Borden E. C., Sen G. C., Uze G., Silverman R. H., Ransohoff R. M., Foster G. R., and Stark G. R. (2007) Interferons at age 50: past, current and future impact on biomedicine. Nat. Rev. Drug. Discov. 6, 975–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Isaacs A., and Lindenmann J. (1957) Virus interference. I. The interferon. Proc. R. Soc. Lond. B Biol. Sci. 147, 258–267 [PubMed] [Google Scholar]
- 11. Randall R. E., and Goodbourn S. (2008) Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus counter measures. J. Gen. Virol. 89, 1–47 [DOI] [PubMed] [Google Scholar]
- 12. Sadler A. J., and Williams B. R. (2007) Structure and function of the protein kinase R. Curr. Top. Microbiol. Immunol. 316, 253–292 [DOI] [PubMed] [Google Scholar]
- 13. Samuel C. E. (2001) Antiviral actions of interferons. Clin. Microbiol. Rev. 14, 778–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Das S., Ward S. V., Tacke R. S., Suske G., and Samuel C. E. (2006) Activation of the RNA-dependent protein kinase PKR promoter in the absence of interferon is dependent upon Sp proteins. J. Biol. Chem. 281, 3244–3253 [DOI] [PubMed] [Google Scholar]
- 15. Kuhen K. L., and Samuel C. E. (1997) Isolation of the interferon-inducible RNA-dependent protein kinase Pkr promoter and identification of a novel DNA element within the 5′-flanking region of human and mouse Pkr genes. Virology 227, 119–130 [DOI] [PubMed] [Google Scholar]
- 16. Tanaka H., and Samuel C. E. (1994) Mechanism of interferon action: structure of the mouse PKR gene encoding the interferon-inducible RNA-dependent protein kinase. Proc. Natl. Acad. Sci. U.S.A. 91, 7995–7999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cole J. L. (2007) Activation of PKR: an open and shut case? Trends Biochem. Sci. 32, 57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McCormack S. J., Thomis D. C., and Samuel C. E. (1992) Mechanism of interferon action: identification of a RNA binding domain within the N-terminal region of the human RNA-dependent P1/eIF-2α protein kinase. Virology 188, 47–56 [DOI] [PubMed] [Google Scholar]
- 19. Samuel C. E. (1979) Mechanism of interferon action: phosphorylation of protein synthesis initiation factor eIF-2 in interferon-treated human cells by a ribosome-associated kinase processing site specificity similar to hemin-regulated rabbit reticulocyte kinase. Proc. Natl. Acad. Sci. U.S.A. 76, 600–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thomis D. C., and Samuel C. E. (1995) Mechanism of interferon action: characterization of the intermolecular autophosphorylation of PKR, the interferon-inducible, RNA-dependent protein kinase. J. Virol. 69, 5195–5198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dalet A., Gatti E., and Pierre P. (2015) Integration of PKR-dependent translation inhibition with innate immunity is required for a coordinated anti-viral response. FEBS Lett. 589, 1539–1545 [DOI] [PubMed] [Google Scholar]
- 22. García M. A., Meurs E. F., and Esteban M. (2007) The dsRNA protein kinase PKR: virus and cell control. Biochimie 89, 799–811 [DOI] [PubMed] [Google Scholar]
- 23. Pfaller C. K., Radeke M. J., Cattaneo R., and Samuel C. E. (2014) Measles virus C protein impairs production of defective copyback double-stranded viral RNA and activation of protein kinase R. J. Virol. 88, 456–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Toth A. M., Devaux P., Cattaneo R., and Samuel C. E. (2009) Protein kinase PKR mediates the apoptosis induction and growth restriction phenotypes of C protein-deficient measles virus. J. Virol. 83, 961–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li Z., Okonski K. M., and Samuel C. E. (2012) Adenosine deaminase acting on RNA 1 (ADAR1) suppresses the induction of interferon by measles virus. J. Virol. 86, 3787–3794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McAllister C. S., Toth A. M., Zhang P., Devaux P., Cattaneo R., and Samuel C. E. (2010) Mechanisms of protein kinase PKR-mediated amplification of β interferon induction by C protein-deficient measles virus. J. Virol. 84, 380–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Toth A. M., Li Z., Cattaneo R., and Samuel C. E. (2009) RNA-specific adenosine deaminase ADAR1 suppresses measles virus-induced apoptosis and activation of protein kinase PKR. J. Biol. Chem. 284, 29350–29356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang P., and Samuel C. E. (2007) Protein kinase PKR plays a stimulus- and virus-dependent role in apoptotic death and virus multiplication in human cells. J. Virol. 81, 8192–8200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Langland J. O., Cameron J. M., Heck M. C., Jancovich J. K., and Jacobs B. L. (2006) Inhibition of PKR by RNA and DNA viruses. Virus Res. 119, 100–110 [DOI] [PubMed] [Google Scholar]
- 30. Patterson J. B., and Samuel C. E. (1995) Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol. Cell. Biol. 15, 5376–5388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Samuel C. E. (2011) Adenosine deaminases acting on RNA (ADARs) are both antiviral and proviral. Virology 411, 180–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Toth A. M., Zhang P., Das S., George C. X., and Samuel C. E. (2006) Interferon action and the double-stranded RNA-dependent enzymes ADAR1 adenosine deaminase and PKR protein kinase. Prog. Nucleic Acids Res. Mol. Biol. 81, 369–434 [DOI] [PubMed] [Google Scholar]
- 33. Bass B. L. (2002) RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 71, 817–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. George C. X., Gan Z., Liu Y., and Samuel C. E. (2011) Adenosine deaminases acting on RNA, RNA editing, and interferon action. J. Interferon Cytokine Res. 31, 99–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bass B. L., and Weintraub H. (1988) An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell 55, 1089–1098 [DOI] [PubMed] [Google Scholar]
- 36. George C. X., and Samuel C. E. (2015) STAT2-dependent induction of RNA adenosine deaminase ADAR1 by type I interferon differs between mouse and human cells in the requirement for STAT1. Virology 485, 363–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. George C. X., Wagner M. V., and Samuel C. E. (2005) Expression of interferon-inducible RNA adenosine deaminase ADAR1 during pathogen infection and mouse embryo development involves tissue-selective promoter utilization and alternative splicing. J. Biol. Chem. 280, 15020–15028 [DOI] [PubMed] [Google Scholar]
- 38. George C. X., and Samuel C. E. (1999) Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible. Proc. Natl. Acad. Sci. U.S.A. 96, 4621–4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pfaller C. K., Li Z., George C. X., and Samuel C. E. (2011) Protein kinase PKR and RNA adenosine deaminase ADAR1: new roles for old players as modulators of the interferon response. Curr Opin Immunol 23, 573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gélinas J. F., Clerzius G., Shaw E., and Gatignol A. (2011) Enhancement of replication of RNA viruses by ADAR1 via RNA editing and inhibition of RNA-activated protein kinase. J. Virol. 85, 8460–8466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schoggins J. W., Wilson S. J., Panis M., Murphy M. Y., Jones C. T., Bieniasz P., and Rice C. M. (2011) A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472, 481–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ward S. V., George C. X., Welch M. J., Liou L. Y., Hahm B., Lewicki H., de la Torre J. C., Samuel C. E., and Oldstone M. B. (2011) RNA editing enzyme adenosine deaminase is a restriction factor for controlling measles virus replication that also is required for embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 108, 331–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. John L., and Samuel C. E. (2014) Induction of stress granules by interferon and down-regulation by the cellular RNA adenosine deaminase ADAR1. Virology 454, 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang Q., Miyakoda M., Yang W., Khillan J., Stachura D. L., Weiss M. J., and Nishikura K. (2004) Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J. Biol. Chem. 279, 4952–4961 [DOI] [PubMed] [Google Scholar]
- 45. Higuchi M., Maas S., Single F. N., Hartner J., Rozov A., Burnashev N., Feldmeyer D., Sprengel R., and Seeburg P. H. (2000) Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 406, 78–81 [DOI] [PubMed] [Google Scholar]
- 46. Meraz M. A., White J. M., Sheehan K. C., Bach E. A., Rodig S. J., Dighe A. S., Kaplan D. H., Riley J. K., Greenlund A. C., Campbell D., Carver-Moore K., DuBois R. N., Clark R., Aguet M., and Schreiber R. D. (1996) Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell 84, 431–442 [DOI] [PubMed] [Google Scholar]
- 47. Park C., Li S., Cha E., and Schindler C. (2000) Immune response in Stat2 knockout mice. Immunity 13, 795–804 [DOI] [PubMed] [Google Scholar]
- 48. George C. X., and Samuel C. E. (2011) Host response to polyomavirus infection is modulated by RNA adenosine deaminase ADAR1 but not by ADAR2. J. Virol. 85, 8338–8347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rende-Fournier R., Ortega L. G., George C. X., and Samuel C. E. (1997) Interaction of the human protein kinase PKR with the mouse PKR homolog occurs via the N-terminal region of PKR and does not inactivate autophosphorylation activity of mouse PKR. Virology 238, 410–423 [DOI] [PubMed] [Google Scholar]
- 50. Zhang R., Li X., Ramaswami G., Smith K. S., Turecki G., Montgomery S. B., and Li J. B. (2014) Quantifying RNA allelic ratios by microfluidic multiplex PCR and sequencing. Nat. Methods 11, 51–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hartner J. C., Schmittwolf C., Kispert A., Müller A. M., Higuchi M., and Seeburg P. H. (2004) Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J. Biol. Chem. 279, 4894–4902 [DOI] [PubMed] [Google Scholar]
- 52. Kedersha N. L., Gupta M., Li W., Miller I., and Anderson P. (1999) RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J. Cell Biol. 147, 1431–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tourrière H., Gallouzi I. E., Chebli K., Capony J. P., Mouaikel J., van der Geer P., and Tazi J. (2001) RasGAP-associated endoribonuclease G3Bp: selective RNA degradation and phosphorylation-dependent localization. Mol. Cell. Biol. 21, 7747–7760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kedersha N., and Anderson P. (2009) Regulation of translation by stress granules and processing bodies. Prog. Mol. Biol. Transl. Sci. 90, 155–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bevilacqua P. C., George C. X., Samuel C. E., and Cech T. R. (1998) Binding of the protein kinase PKR to RNAs with secondary structure defects: role of the tandem A-G mismatch and noncontiguous helixes. Biochemistry 37, 6303–6316 [DOI] [PubMed] [Google Scholar]
- 56. Neeman Y., Levanon E. Y., Jantsch M. F., and Eisenberg E. (2006) RNA editing level in the mouse is determined by the genomic repeat repertoire. RNA 12, 1802–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Danecek P., Nellåker C., McIntyre R. E., Buendia-Buendia J. E., Bumpstead S., Ponting C. P., Flint J., Durbin R., Keane T. M., and Adams D. J. (2012) High levels of RNA-editing site conservation amongst 15 laboratory mouse strains. Genome Biol. 13, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Goodman R. A., Macbeth M. R., and Beal P. A. (2012) ADAR proteins: structure and catalytic mechanism. Curr. Top. Microbiol. Immunol. 353, 1–33 [DOI] [PubMed] [Google Scholar]
- 59. Liu Y., and Samuel C. E. (1996) Mechanism of interferon action: functionally distinct RNA-binding and catalytic domains in the interferon-inducible, double-stranded RNA-specific adenosine deaminase. J. Virol. 70, 1961–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Barraud P., and Allain F. H. (2012) ADAR proteins: double-stranded RNA and Z-DNA binding domains. Curr. Top. Microbiol. Immunol. 353, 35–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ryter J. M., and Schultz S. C. (1998) Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 17, 7505–7513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Vazquez C., and Horner S. M. (2015) MAVS Coordination of antiviral innate immunity. J. Virol. 89, 6974–6977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yoneyama M., Onomoto K., Jogi M., Akaboshi T., and Fujita T. (2015) Viral RNA detection by RIG-I-like receptors. Curr. Opin. Immunol. 32, 48–53 [DOI] [PubMed] [Google Scholar]
- 64. Pfaller C. K., Mastorakos G. M., Matchett W. E., Ma X., Samuel C. E., and Cattaneo R. (2015) Measles virus defective-interfering RNAs are generated frequently and early in the absence of C protein and can be destabilized by adenosine deaminase acting on RNA 1-like hypermutations. J. Virol. 89, 7735–7747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Liddicoat B. J., Piskol R., Chalk A. M., Ramaswami G., Higuchi M., Hartner J. C., Li J. B., Seeburg P. H., and Walkley C. R. (2015) RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 349, 1115–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rice G. I., Kasher P. R., Forte G. M., Mannion N. M., Greenwood S. M., Szynkiewicz M., Dickerson J. E., Bhaskar S. S., Zampini M., Briggs T. A., Jenkinson E. M., Bacino C. A., Battini R., Bertini E., Brogan P. A., et al. (2012) Mutations in ADAR1 cause Aicardi-Goutieres syndrome associated with a type I interferon signature. Nat. Genet. 44, 1243–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pestal K., Funk C. C., Snyder J. M., Price N. D., Treuting P. M., and Stetson D. B. (2015) Isoforms of RNA-editing enzyme ADAR1 independently control nucleic acid sensor MDA5-driven autoimmunity and multi-organ development. Immunity 43, 933–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hartner J. C., Walkley C. R., Lu J., and Orkin S. H. (2009) ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat. Immunol. 10, 109–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wagner R. W., Smith J. E., Cooperman B. S., and Nishikura K. (1989) A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc. Natl. Acad. Sci. U.S.A. 86, 2647–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hood J. L., and Emeson R. B. (2012) Editing of neurotransmitter receptor and ion channel RNAs in the nervous system. Curr. Top. Microbiol. Immunol. 353, 61–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Liu Y., Emeson R. B., and Samuel C. E. (1999) Serotonin-2C receptor pre-mRNA editing in rat brain and in vitro by splice site variants of the interferon-inducible double-stranded RNA-specific adenosine deaminase ADAR1. J. Biol. Chem. 274, 18351–18358 [DOI] [PubMed] [Google Scholar]
- 72. Liu Y., and Samuel C. E. (1999) Editing of glutamate receptor subunit B pre-mRNA by splice-site variants of interferon-inducible double-stranded RNA-specific adenosine deaminase ADAR1. J. Biol. Chem. 274, 5070–5077 [DOI] [PubMed] [Google Scholar]
- 73. Li J. B., Levanon E. Y., Yoon J. K., Aach J., Xie B., Leproust E., Zhang K., Gao Y., and Church G. M. (2009) Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science 324, 1210–1213 [DOI] [PubMed] [Google Scholar]
- 74. Pinto Y., Cohen H. Y., and Levanon E. Y. (2014) Mammalian conserved ADAR targets comprise only a small fragment of the human editosome. Genome Biol. 15, R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ramaswami G., and Li J. B. (2014) RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 42, D109–D113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ramaswami G., Lin W., Piskol R., Tan M. H., Davis C., and Li J. B. (2012) Accurate identification of human Alu and non-Alu RNA editing sites. Nat. Methods 9, 579–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bazak L., Haviv A., Barak M., Jacob-Hirsch J., Deng P., Zhang R., Isaacs F. J., Rechavi G., Li J. B., Eisenberg E., and Levanon E. Y. (2014) A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 24, 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ramaswami G., Zhang R., Piskol R., Keegan L. P., Deng P., O'Connell M. A., and Li J. B. (2013) Identifying RNA editing sites using RNA sequencing data alone. Nat. Methods 10, 128–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Patterson J. B., Thomis D. C., Hans S. L., and Samuel C. E. (1995) Mechanism of interferon action: double-stranded RNA-specific adenosine deaminase from human cells is inducible by α and γ interferons. Virology 210, 508–511 [DOI] [PubMed] [Google Scholar]
- 80. Mannion N. M., Greenwood S. M., Young R., Cox S., Brindle J., Read D., Nellåker C., Vesely C., Ponting C. P., McLaughlin P. J., Jantsch M. F., Dorin J., Adams I. R., Scadden A. D., Ohman M., et al. (2014) The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep. 9, 1482–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Li Z., Wolff K. C., and Samuel C. E. (2010) RNA adenosine deaminase ADAR1 deficiency leads to increased activation of protein kinase PKR and reduced vesicular stomatitis virus growth following interferon treatment. Virology 396, 316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Mizrahi R. A., Phelps K. J., Ching A. Y., and Beal P. A. (2012) Nucleoside analog studies indicate mechanistic differences between RNA-editing adenosine deaminases. Nucleic Acids Res. 40, 9825–9835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Nishikura K. (2010) Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 79, 321–349 [DOI] [PMC free article] [PubMed] [Google Scholar]