Abstract
Retrotransposons are eukaryotic mobile genetic elements that transpose by reverse transcription of an RNA intermediate and are derived from retroviruses. The Ty1 retrotransposon of Saccharomyces cerevisiae belongs to the Ty1/Copia superfamily, which is present in every eukaryotic genome. Insertion of Ty1 elements into the S. cerevisiae genome, which occurs upstream of genes transcribed by RNA Pol III, requires the Ty1 element-encoded integrase (IN) protein. Here, we report that Ty1-IN interacts in vivo and in vitro with RNA Pol III-specific subunits to mediate insertion of Ty1 elements upstream of Pol III-transcribed genes. Purification of Ty1-IN from yeast cells followed by mass spectrometry (MS) analysis identified an enrichment of peptides corresponding to the Rpc82/34/31 and Rpc53/37 Pol III-specific subcomplexes. GFP-Trap purification of multiple GFP-tagged RNA Pol III subunits from yeast extracts revealed that the majority of Pol III subunits co-purify with Ty1-IN but not two other complexes required for Pol III transcription, transcription initiation factors (TF) IIIB and IIIC. In vitro binding studies with bacterially purified RNA Pol III proteins demonstrate that Rpc31, Rpc34, and Rpc53 interact directly with Ty1-IN. Deletion of the N-terminal 280 amino acids of Rpc53 abrogates insertion of Ty1 elements upstream of the hot spot SUF16 tRNA locus and abolishes the interaction of Ty1-IN with Rpc37. The Rpc53/37 complex therefore has an important role in targeting Ty1-IN to insert Ty1 elements upstream of Pol III-transcribed genes.
Keywords: integrase, retrovirus, RNA polymerase III, Saccharomyces cerevisiae, transposable element (TE), Ty1 element, retrotransposon, tRNA gene
Introduction
Retrotransposons are transposable elements that share similarities to retroviruses, although they lack an infectious phase in their life cycle. The genome of the yeast Saccharomyces cerevisiae contains five families of Ty retrotransposons (Ty1–5) of which Ty1 is the most abundant (1). Ty1–5 are long terminal repeat retrotransposons because each ∼6-kilobase pair (kbp) Ty element is flanked by a ∼330-base pair (bp) direct repeat sequence. The remainder of the element comprises both TyA and TyB open reading frames (ORFs), analogous to retroviral Gag and Pol, respectively. Gag encodes the structural elements of the viral coat protein, whereas Pol is a polyprotein that is cleaved to generate a protease (PR),3 IN, and reverse transcriptase (RT) (2, 3). Ty elements propagate by reverse transcribing their RNA into cDNA in virus-like particles and incorporating the cDNA into a preintegration complex with Ty-IN, which is imported into the nucleus for targeting into the genome. Ty1, Ty2, and Ty4 elements insert within a ∼1-kbp window upstream of genes transcribed by RNA Pol III such as tRNA genes, and Ty1 insertion is blocked by mutations in the Pol III promoter, suggesting that active Pol III transcription is required for Ty1 element insertion (4, 5). Ty3 elements insert within a few bp of the RNA Pol III transcription start site, and an interaction between Ty3-IN and the TFIIIB factor Brf1 is sufficient to target Ty3 to Pol III transcription start sites in vitro (6–8). Ty5, however, interacts with Sir4 to mediate insertion into the silenced regions of the genome such as the silent mating type loci and telomeric regions (9, 10). Ty1 insertion occurs with a periodicity of ∼80 bp and is coincident with nucleosome positioning (11–14). Disruption of the chromatin structure upstream of tRNA genes disrupts the periodicity of Ty1 element insertion but not integration site selection (15, 16). Although many nuclear factors have been identified that either restrict or promote Ty1 integration, proteins that physically interact with Ty1-IN to target it upstream of Pol III-transcribed genes have remained elusive until recently (17–22). Ty1-IN interacts with separase, a protein that cleaves the cohesin complex to mediate separation of sister chromatids (23). However, despite the fact that separase mutants have reduced and cohesin mutants have increased Ty1 insertion frequency, no change in targeting specificity was demonstrated (23). Recently, a yeast two-hybrid screen using the RNA Pol III AC40 subunit as bait identified an interaction with Ty1-IN (24). Replacing the S. cerevisiae AC40 subunit with Schizosaccharomyces pombe AC40 disrupted the interaction with Ty1-IN and redirected Ty1 element insertion to telomeric and subtelomeric regions (24). This exciting new discovery provides significant insight into the mechanism of Ty1 element insertion. However, despite the lack of interaction between S. pombe AC40 and Ty1-IN, overall Ty1 mobility was not affected, suggesting that other factors could still mediate Ty1 element insertion (24).
In this study, we performed purification of Ty1-IN from yeast cells followed by MS analysis and identified an enrichment of RNA Pol III subunits in our Ty1-IN purifications. The RNA Pol III complex is a 17-subunit complex composed of a 10-subunit core with five subunits shared between all three Pols and two of the remaining five shared between Pol I and Pol III (Rpc40/AC40 and Rpc19/AC19) (25). There are two Pol III-specific subcomplexes composed of Rpc82/34/31 and Rpc53/37 and the Rpb4/7-like subcomplex containing Rpc25/17 (25). We demonstrate that multiple RNA Pol III proteins, including the Pol III-specific proteins, co-immunoprecipitate with Ty1-IN from yeast lysates. A purified RNA Pol III complex from yeast binds to Ty1-IN in vitro as well as Escherichia coli purified Rpc31, Rpc34, and Rpc53. We demonstrate that removing the N terminus of Rpc53 prevents Ty1 targeting to the SUF16 tRNA locus and abolishes the interaction of Ty1-IN with Rpc37. Our data suggest that the Rpc53/37 Pol III-specific subcomplex is required for specific targeting of Ty1-IN upstream of Pol III-transcribed genes.
Experimental Procedures
Yeast Strain Construction
S. cerevisiae strains used in this study are listed in Table 1. All C-terminal epitope tags (GFP and HA) were generated by PCR and homologous recombination as described (26).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Ref. |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 | 76 |
| LV1491 | MATα ura3-167 his3Δ200 Ty1his3AI-242 rpc40Δ::hphMX pCEN AmpR URA3 TetO7-RPC40 | 24 |
| LV1492 | MATα ura3-167 his3Δ200 Ty1his3AI-242 rpc40Δ::hphMX pCEN AmpR URA3 TetO7-Rpc40+ (S. pombe) | 24 |
| Y7092 | MATα can1Δ::STE2pr-Sp_his5 lyp1Δ his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 | 77 |
| YKH638 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ ura3Δ0 spt3::KanMX6 | 78 |
| YLM2119A | MATa his3Δ1 leu2Δ0 met15Δ ura3Δ0 trp::KanMX6 RPC82-GFP::HIS3 | This study |
| YLM2120 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ ura3Δ0 RPC34-HA::HIS3 | This study |
| YLM2124 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ ura3Δ0 RPC25-HA::HIS3 | This study |
| YM2371 | MATa his3Δ1 leu2Δ0 met15Δ ura3Δ0 RPC82-GFP::HIS3 | 42 |
| YM2346 | MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 rpo31-698::kanMX6 | 52 |
| YM2347 | MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 rpc40-V78R::kanMX6 | 52 |
| YM2348 | MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 rpc34-1::kanMX6 | 52 |
| YM2367 | MATa his3Δ1 leu2Δ0 met15Δ ura3Δ0 RPC17-GFP::HIS3 | 42 |
| YM2369 | MATa his3Δ1 leu2Δ0 met15Δ ura3Δ0 MAF1-GFP::HIS3 | 42 |
| YM2370 | MATa his3Δ1 leu2Δ0 met15Δ ura3Δ0 RPC160-GFP::HIS3 | 42 |
| YM2372 | MATa his3Δ1 leu2Δ0 met15Δ ura3Δ0 RPB2-GFP::HIS3 | 42 |
| YM2374 | MATa his3Δ1 leu2Δ0 met15Δ ura3Δ0 RPC40-GFP::HIS3 | 42 |
| YM2447 | MATα ade2::his3G his3Δ200 leu2Δ0 met15Δ0 lys2Δ0 trp1Δ63 ura3Δ0 [C53::KanMX4] pRPC53-WT/pRS315 | 36 |
| YM2448 | MATα ade2::his3G his3Δ200 leu2Δ0 met15Δ0 lys2Δ0 trp1Δ63 ura3Δ0 [C53::KanMX4] pRPC53Δ (11–40)/pRS315 | 36 |
| YM2449 | MATα ade2::his3G his3Δ200 leu2Δ0 met15Δ0 lys2Δ0 trp1Δ63 ura3Δ0 [C53::KanMX4] pRPC53Δ (316–350)/pRS315 | 36 |
| YM2450 | MATα ade2::his3G his3Δ200 leu2Δ0 met15Δ0 lys2Δ0 trp1Δ63 ura3Δ0 [C53::KanMX4] pRPC53Δ (2–280)/pRS315 | 36 |
| YSC16 | MATa his3Δ1 leu2Δ0 met15Δ ura3Δ0 RPO21-GFP::HIS3 | 42 |
| YSC17 | MATa his3Δ1 leu2Δ0 met15Δ ura3Δ0 RPC128-GFP::HIS3 | 42 |
| YSC18 | MATa his3Δ1 leu2Δ0 met15Δ ura3Δ0 RPB6-GFP::HIS3 | 42 |
| YSC19 | MATa his3Δ1 leu2Δ0 met15Δ ura3Δ0 RPC19-GFP::HIS3 | 42 |
| YSC42 | MATa his3Δ1 leu2Δ0 met15Δ ura3Δ0 BRF1-GFP::HIS3 | 42 |
| YSC43 | MATa his3Δ1 leu2Δ0 met15Δ ura3Δ0 BDP1-GFP::HIS3 | 42 |
| YSC44 | MATa his3Δ1 leu2Δ0 met15Δ ura3Δ0 TFC1-GFP::HIS3 | 42 |
| YSC45 | MATa his3Δ1 leu2Δ0 met15Δ ura3Δ0 TFC3-GFP::HIS3 | 42 |
| YSC47 | MATa his3Δ1 leu2Δ0 met15Δ ura3Δ0 TFC7-GFP::HIS3 | 42 |
| YSC62 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ ura3Δ0 RPC82-HA::HIS3 | This study |
| YSC97 | MATa ade2::his3G his3Δ1 leu2Δ0 met15Δ ura3Δ0 RPC17-GFP::HIS3 [C53::KanMX4] pRPC53-WT/pRS315 | This study |
| YSC99 | MATa ade2::his3G his3Δ1 leu2Δ0 met15Δ ura3Δ0 RPC17-GFP::HIS3 [C53::KanMX4] pRPC53 (Δ2–280)/pRS315 | This study |
| YSC103 | MATa his3Δ1 leu2Δ0 met15Δ ura3Δ0 RPC19-GFP::HIS3 [C53::KanMX4] pRPC53-WT/pRS315 | This study |
| YSC106 | MATa ade2::his3G his3Δ1 leu2Δ0 met15Δ ura3Δ0 RPC37-GFP::HIS3 [C53::KanMX4] pRPC53 (Δ2–280)/pRS315 | |
| YSC107 | MATa his3Δ1 leu2Δ0 met15Δ ura3Δ0 RPC19-GFP::HIS3 [C53::KanMX4] pRPC53 (Δ2–280)/pRS315 | This study |
| YSC109 | MATa ade2::his3G his3Δ1 leu2Δ0 met15Δ ura3Δ0 RPC37-GFP::HIS3 [C53::KanMX4] pRPC53-WT/pRS315 | This study |
| YSC111 | MATa ade2::his3G his3Δ1 leu2Δ0 met15Δ ura3Δ0 RPC82-GFP::HIS3 [C53::KanMX4] pRPC53-WT/pRS315 | This study |
| YSC113 | MATa ade2::his3G his3Δ1 leu2Δ0 met15Δ ura3Δ0 RPC82-GFP::HIS3 [C53::KanMX4] pRPC53 (Δ2–280)/pRS315 | This study |
Plasmid Construction
Ty1-IN-S (BSC3A) was constructed by amplifying Ty1-IN from yeast genomic DNA using the following primers: OSC8, 5′-CCCCATATGAATGTCCATACAAGTGAAAGTAC-3′; OSC9, 5′-CCCCTCGAGTGCAATCAGGTGAATTCGTTTCT-3′. The PCR product was digested with NdeI and XhoI and cloned into the pETDuet-1 vector (Novagen) digested with the same restriction enzymes.
Ty1-IN Purification and Mass Spectrometry
200 ml of Y7092 transformed with pJEF724 (pGAL1-Ty1-H3) or empty vector (pRS326) was grown to log phase in SC-Ura + 2% raffinose + 0.1% dextrose at 25 °C. Cells were washed, diluted, and resuspended in 1 liter of SC-Ura + 2% galactose to induce pGAL1-Ty1-H3 expression for 24 h. Cells were harvested and resuspended in lysis buffer (50 mm Hepes, pH 7.5, 0.1% Nonidet P-40, 150 mm NaCl, 5 mm EDTA, and protease inhibitors) on ice followed by addition of glass beads and lysed. Lysates were centrifuged at 12,000 rpm for 5 min at 4 °C, and supernatant was retained as whole cell protein lysate. 10 μl of anti-IN 8b11 antibody (a kind gift from Jef Boeke (27)) was incubated for 10 min with 360 μl of washed PureProteome Protein G magnetic beads and then washed three times with PBS + 0.1% Tween 20. The anti-IN-bound beads were split in half, added to 50 mg of protein lysate from each of the pGAL1-Ty1-H3 and pRS326 harvested cells, and incubated for 2.5 h at 4 °C. The magnetic beads were washed three times with PBS, supernatant was removed, and 45 μl of 1× SDS loading buffer without β-mercaptoethanol was added and placed at 65 °C for 15 min. Beads were spun down, and the supernatant was run on an SDS-polyacrylamide gel. Samples were processed using in-gel digestion (28) in which gel slices corresponding to IgG chains (25 and 55 kDa) were treated separately. Purified peptides were analyzed using an LTQ-Orbitrap Velos (Thermo Fisher Scientific) on line-coupled to an Agilent 1100 Series nanoflow HPLC using a nanospray ionization source (Thermo Fisher Scientific) using the same settings as described previously (29). A 60-min LC gradient (5–35% acetonitrile in 0.5% acetic acid) was used. The LTQ-Orbitrap was set to acquire a full-range scan at 60,000 resolution from 300 to 1,600 thomsons. 10 MS/MS collision-induced dissociation spectra were collected.
Analysis of Mass Spectrometry Data
The spectrum files were processed using MaxQuant (v1.5.1.0) (30–32), and the “label free quantification (LFQ)” and “dependent peptide search” functions were selected. The derived peak list was searched against the reference yeast proteome downloaded from UniProt on June 12, 2012 (6,674 sequences), and the false discovery rate was set to 1%.
GFP and Anti-HA Co-immunoprecipitations (IPs)
GFP and anti-HA IPs were performed as described (23) with the following modifications. For GFP IPs in Figs. 2–4, 1.5 mg of yeast lysate was used, and after Rpc-GFP protein purification, beads were washed three times in wash buffer (50 mm Tris-HCl, pH 7.5, 0.5% Nonidet P-40, and 0.5 mm EDTA) with low salt (300 mm NaCl) or high salt (500 mm NaCl). For GFP IPs in Fig. 9, 2 mg of yeast lysate was used, and beads were washed three times in low salt (300 mm NaCl) wash buffer. For anti-HA IPs (Fig. 2), 2 mg of lysate was mixed with 20 μl of a 1:1 slurry of anti-HA-agarose beads (Covance) for 1.5 h at 4 °C. After washes, the beads were resuspended in 25 μl of 2× SDS loading buffer, and 9 μl of the immunoprecipitate was loaded on an SDS-polyacrylamide gel for immunoblotting analysis.
FIGURE 2.
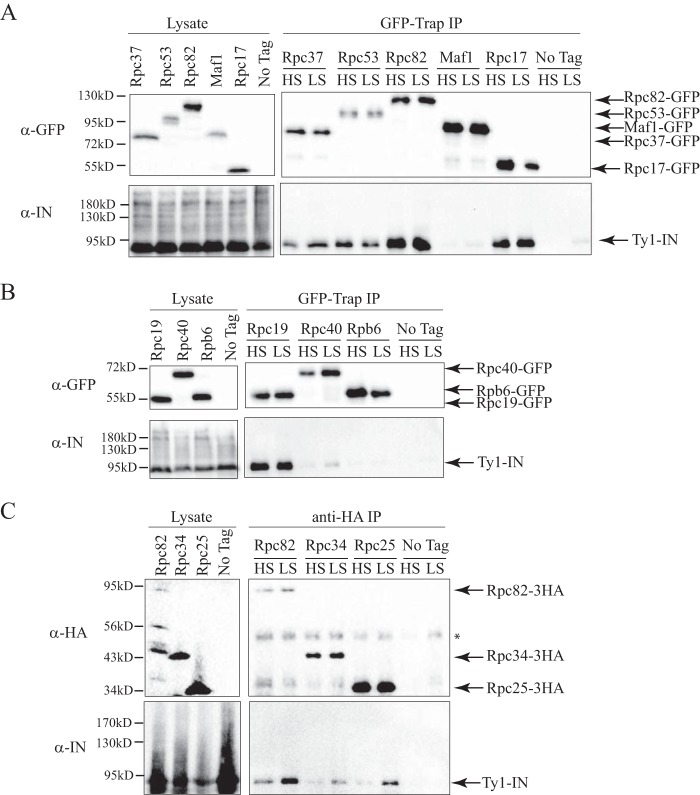
Ty1-IN interacts with RNA Pol III subunits. A, immunoblot of whole cell lysate (Lysate) and GFP-Trap IP carried out from Rpc37-GFP, Rpc53-GFP, Rpc82-GFP, Maf1-GFP, Rpc17-GFP, and untagged wild type (No Tag) cells. B, immunoblot of whole cell lysate (Lysate) and GFP-Trap IP carried out from Rpc19-GFP, Rpc40-GFP, Rpb6-GFP, and untagged wild type (No Tag) cells. C, immunoblot of whole cell lysate (Lysate) and anti-HA IP performed from Rpc82-3HA, Rpc34-3HA, Rpc25-3HA, and untagged wild type (No Tag) cells. The asterisk marks the heavy chain antibody. For A–C, expression of a Ty1 element (pGAL1-Ty1-H3, pJEF724) was induced in all strains for 24 h prior to cell lysis. Immunoprecipitates were washed with either a high salt (HS; 500 mm NaCl) or low salt (LS; 300 mm NaCl) wash. Immunoblots were probed with anti-GFP (A and B), anti-HA (C), and anti-IN (8b11) (A–C) antibodies.
FIGURE 3.
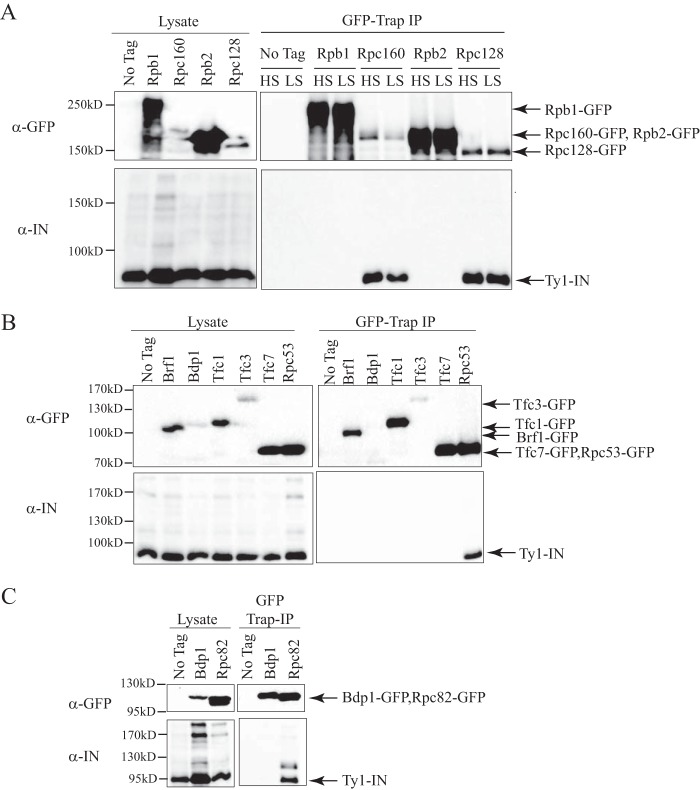
Ty1-IN interacts with Rpc160 and Rpc128 Pol III core subunits but not Pol II, TFIIIB, or TFIIIC subunits. A, immunoblot of whole cell lysate (Lysate) and GFP-Trap IP performed from untagged wild type (No Tag), Rpb1-GFP, Rpc160-GFP, Rpb2-GFP, and Rpc128-GFP cells. B, immunoblot of whole cell lysate (Lysate) and GFP-Trap IP performed from untagged wild type (No Tag), Brf1-GFP, Bdp1-GFP, Tfc1-GFP, Tfc3-GFP, Tfc7-GFP, and Rpc53-GFP cells. C, immunoblot of whole cell lysate (Lysate) and GFP-Trap IP performed from untagged wild type (No Tag), Bdp1-GFP, and Rpc82-GFP cells. For B, 1 mg of lysate was used per immunoprecipitate. For C, 3 mg of lysate was used for the GFP-Trap IP from the untagged and Bdp1-GFP strains, whereas 1.5 mg was used for the GFP-Trap IP from the Rpc82-GFP strain. For A–C, expression of a Ty1 element (pGAL1-Ty1-H3, pJEF724) was induced in all strains for 24 h prior to cell lysis. Blots were probed with anti-GFP and anti-IN (8b11) antibodies. HS, high salt (500 mm); LS, low salt (300 mm).
FIGURE 4.
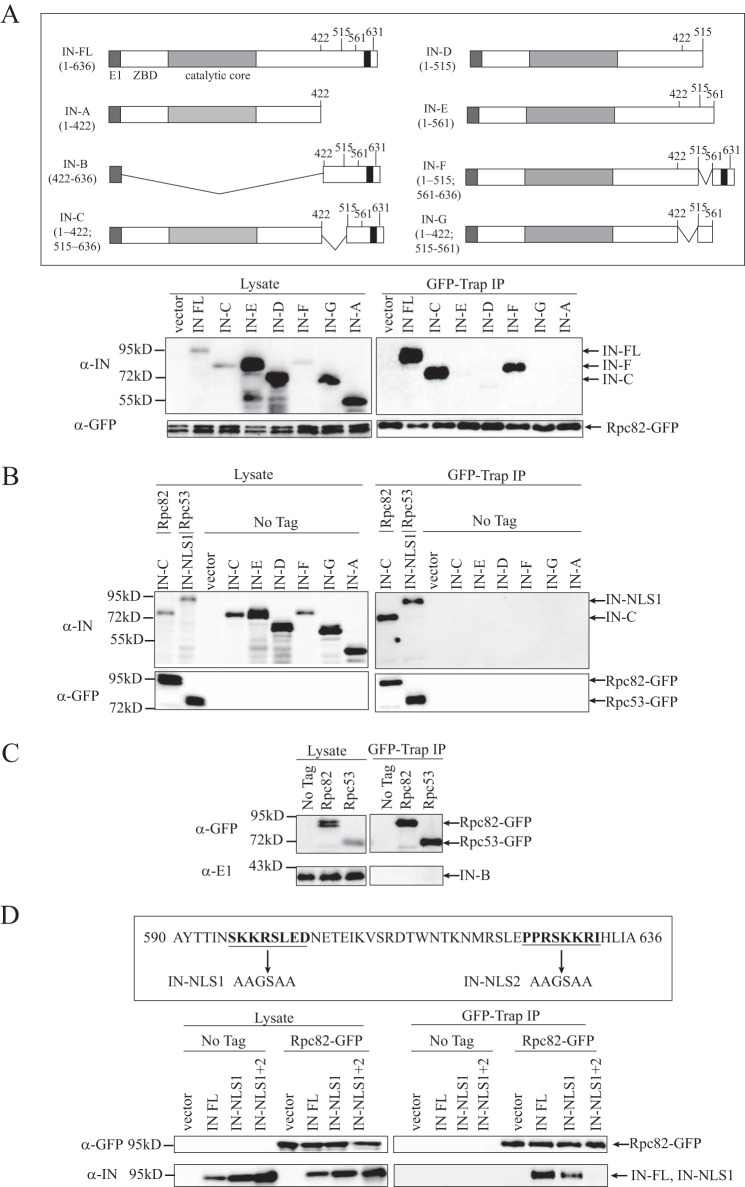
Rpc82 interacts with the C terminus of Ty1-IN. A, upper panel, schematic of E1 epitope-tagged Ty1-IN fusion proteins as described (49). E1 tag is dark gray, zinc binding domain (ZBD) is white, the IN catalytic core is light gray, and the NLS is black. Numbers shown represent amino acids. Lower panel, immunoblot of whole cell lysate (Lysate) and GFP-Trap IP performed from Rpc82-GFP-tagged cells carrying the indicated vector alone, E1-tagged full-length Ty1-IN fusion (IN-FL), or E1-tagged fusions to the indicated Ty1-IN truncations. To the right of the panel, the IN fusions that interact with Rpc82-GFP are shown. B, immunoblot of whole cell lysate (Lysate) and GFP-Trap IP performed from Rpc82-GFP cells carrying the IN-C fusion, Rpc53-GFP cells carrying the IN-NLS1 fusion, and untagged (No Tag) cells carrying vector alone and the indicated Ty1-IN fusions. C, immunoblot of whole cell lysate (Lysate) and GFP-Trap IP performed from untagged (No Tag), Rpc82-GFP-, or Rpc53-GFP-tagged cells carrying the C-terminal 422–636 amino acids of IN fused to E1 (IN-B). D, upper panel, location of the IN-NLS1 and IN-NLS2 mutations at the C terminus of Ty1-IN (amino acids 590–636 as described (49)). The mutated amino acid residues are in bold and underlined, and the arrow points to the mutated amino acid sequence. Lower panel, immunoblot of whole cell lysate (Lysate) and GFP-Trap IP performed from Rpc82-GFP-tagged cells carrying vector or the E1-tagged IN-FL, IN-NLS1 mutant, or IN-NLS1+2 mutant. For A–D, expression of Ty1-IN fusion proteins was induced for 8 h. For A, B, and D, blots were probed with anti-GFP and anti-IN (8b11) antibodies. For C, the blot was probed with anti-E1 and anti-GFP antibodies.
FIGURE 9.
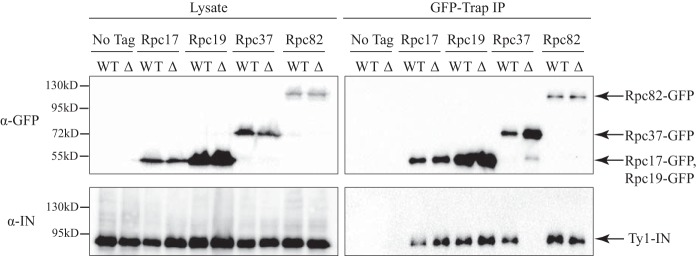
The interaction between Ty1-IN and Rpc37 is disrupted in the rpc53Δ2–280 mutant. An immunoblot of whole cell lysate (Lysate) and GFP-Trap IP carried out from untagged (No Tag), Rpc17-GFP, Rpc19-GFP, Rpc37-GFP, and Rpc82-GFP cell lysates in a wild type or rpc53Δ2–280 (Δ) strain background. Expression of a Ty1 element (pGAL1-Ty1-H3, pJEF724) was induced in all strains for 24 h prior to cell lysis. Immunoblots were probed with anti-GFP and anti-IN (8b11) antibodies.
Purification of Ty1-IN-S from E. coli
The BSC3A Ty1-IN-S expression vector was transformed into BL21(DE3) E. coli cells. Transformants were inoculated into 5 ml of LB with ampicillin, grown overnight at 37 °C, then diluted 1:100 in 200 ml, and regrown to an A600 of 0.6. Ty1-IN-S expression was induced by adding 1 mm isopropyl 1-thio-β-d-galactopyranoside and growing cells for 1 h at 37 °C. Cells were harvested, resuspended in 20 ml of ice-cold lysis buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 0.1% Triton X-100), and lysed by sonication on ice with 35% output amplitude for 3 min (QSonica; 5 s on, 10 s off). The soluble lysate was separated from the cell debris by a 20-min centrifugation at 39,000 × g at 4 °C. Ty1-IN-S was purified by adding 400 μl of a 1:1 S-protein-agarose slurry (Novagen) to the lysate and incubating for 1 h at 4 °C followed by washing three times with lysis buffer. The S-protein-agarose with captured Ty1-IN-S was stored at 4 °C and used for the in vitro binding assay within 1 week.
Purification of RNA Pol III Proteins
For RNA Pol III purification, S. cerevisiae strain NZ16 (33) (originally constructed by Nick Zecherle and Benjamin Hall) containing a hexahistidine tag followed by four tandem FLAG epitope tags at the N terminus of the Rpc128 gene, was grown to an A600 of 6 at 30 °C in 200 liters of yeast extract-peptone-dextrose medium. The 200-liter yeast culture was harvested and subsequently lysed by bead beating. The purification scheme was based on the method developed by Cramer and co-workers (34); however, the columns used for tandem chromatography were modified to heparin-Sepharose (GE Healthcare), Talon metal affinity resins (Clontech), DEAE-Sepharose (GE Healthcare), and Mono Q (GE Healthcare). The yield was ∼1 mg. Detailed procedures and the chromatography program are available upon request. Recombinant Rpc82, Rpc34, Rpc31, Rpc53, and Rpc37 protein samples were purified as described previously (35, 36). The recombinant Rpc25/17 complex protein sample was purified with a modified purification scheme based on a method described previously (37). The following columns were utilized for tandem chromatography: nickel-Sepharose (GE Healthcare), heparin-Sepharose, and Mono Q.
Ty1-IN-S in Vitro Binding Assay
5 μg of purified RNA Pol III protein was used in each in vitro binding assay. 200 μl of lysis buffer was added and incubated with 10 μl of the Ty1-IN-S-agarose slurry (∼5-μg eq of Ty1-IN-S protein) for 1 h at 4 °C. 5 μg of purified RNA Pol III protein was added to an equal amount of S-protein-agarose (without Ty1-IN-S bound) for the negative control. The agarose resin was spun down at 5,000 × g for 1 min, and the supernatant was collected (unbound fraction). The resin was washed three times with 1 ml of lysis buffer. For the Rpc34 binding assay, the resin was washed an additional three times with lysis buffer containing 300 mm NaCl. Proteins were eluted (bound fraction) by boiling in 20 μl of 2× SDS loading buffer, separated by SDS-PAGE, and detected by immunoblotting. 5 μl of the supernatant was loaded for the unbound fraction, and 5 μl of the bound fraction was loaded. 333 ng of purified protein was loaded to represent of the input RNA Pol III protein. A monoclonal antibody against the S-tag (EMD Millipore) was used to detect Ty1-IN-S. A monoclonal antibody against His (HIS.H8, AbLab, University of British Columbia) was used to detect Rpc53-His6, Rpc37-His6, Rpc82-V5-His6, and His6-MBP-Gal4. A monoclonal antibody against FLAG (Sigma) was used to detect Rpc25/17-FLAG and RNA Pol III with FLAG4His6-tagged Rpc128. A polyclonal antibody raised against RNA Pol III was used to detect Rpc31 and Rpc34 (38).
SUF16 PCR Assay
The SNR33 and Ty1 PCR primers and CPR7 primers have been previously described (39). 1 and 2 μg of yeast genomic DNA were used for the SUF16 PCR in Figs. 6 and 7, respectively. 5 ng of yeast genomic DNA was used for all CPR7 PCR assays.
FIGURE 6.
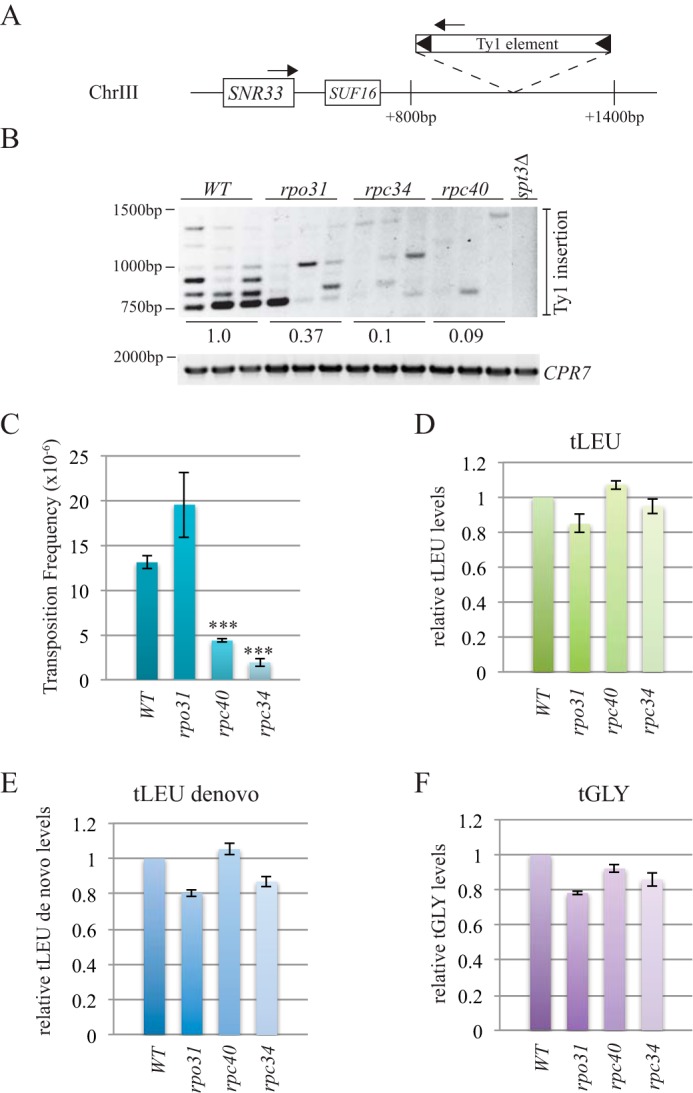
rpc34 and rpc40 mutants have decreased Ty1 element insertion at the SUF16 locus and reduced Ty1 mobility. A, schematic of the SUF16 genomic locus with PCR primers (arrows) designed to hybridize within the SNR33 gene and the newly inserted Ty1 elements. B, SUF16 PCR analysis of yeast genomic DNA extracted from wild type (WT), rpo31-698 (rpo31), rpc34-1 (rpc34), and rpc40-V78R (rpc40) strains grown in triplicate and spt3Δ (single isolate) at 20 °C for 3 days to induce transposition. The upper panel is the result of the PCR assay with primers shown in A; the lower panel is a control PCR for the CPR7 locus to demonstrate that yeast genomic DNA was present in each sample. Quantification of Ty1 insertion events is shown relative to wild type. C, quantification of Ty1 mobility in WT, rpo31-698, rpc40-V78R, and rpc34-1 mutants induced to transpose for 5 days at 20 °C. A minimum of five transformants were quantified per strain. *** represents a significant difference from wild type (p < 0.001). D–F, qPCR analysis of tLEU, tLEU de novo, and tGLY gene expression in WT, rpo31-698, rpc40-V78R, and rpc34-1 mutant strains. The RQ of wild type is set to a value of 1.0, and the mutant RQ values are compared with wild type. Error bars are S.E. and were calculated as described under “Experimental Procedures.”
FIGURE 7.
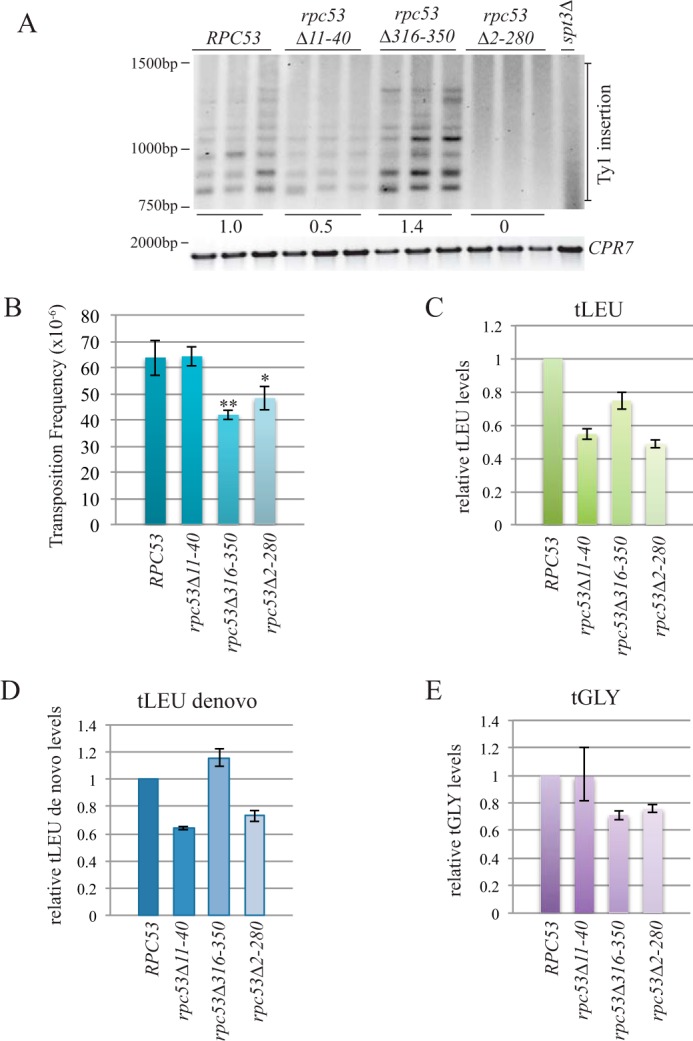
The N terminus of Rpc53 is necessary for Ty1 element insertion upstream of the SUF16 tRNA gene. A, upper panel, SUF16 PCR analysis of yeast genomic DNA extracted from the indicated strains grown in triplicate for 3 days at 20 °C to induce transposition. Lower panel, control PCR for the CPR7 locus to demonstrate that yeast genomic DNA was present in each sample. Quantification of Ty1 insertion events is shown relative to wild type. B, transposition frequency of wild type (RPC53) and rpc53 mutants (rpc53Δ11–40, rpc53Δ316–350, and rpc53Δ2–280) strains induced to transpose for 5 days at 20 °C. A minimum of eight transformants were quantified per strain. * and ** represent significant difference from wild type (p < 0.1 and p < 0.01, respectively). C–E, qPCR analysis of tLEU, tLEU de novo, and tGLY gene expression in wild type (RPC53) and rpc53 mutant (rpc53Δ11–40, rpc53Δ316–350, and rpc53Δ2–280) strains. The RQ of wild type is set to a value of 1.0, and the mutant RQ values are compared with wild type. Error bars are S.E. and were calculated as described under “Experimental Procedures.”
Subtelomere PCR Assay
The following primers were used: OSC66, 5′-CCAAGGATCTAGGTGAGGCTTTGAGAA-3′ (Chr XIV left end 13,634–13,669 bp (SNZ2) and Chr VI left end 11,739–11,765 bp (SNZ3)); OSC68, 5′-GACATGGGCCCCTGTTGCTTATATTGT-3′ (Chr IV left end 12,021–12,047 bp (HXT15), Chr X right end 733,753–733,779 bp (HXT16)); OSC70, 5′-CACAGAGCTTCAGGTTAGGAGCTTCTG-3′ (Chr XV right end 1,072,647–1,072,673 bp (FDH1), Chr XVI left end 18,198–18,224 bp (FDH2)). 1.5 μg of yeast genomic DNA was used for the PCR in Fig. 8. 5 ng of yeast genomic DNA was used for the CPR7 PCR assay in Fig. 8.
FIGURE 8.
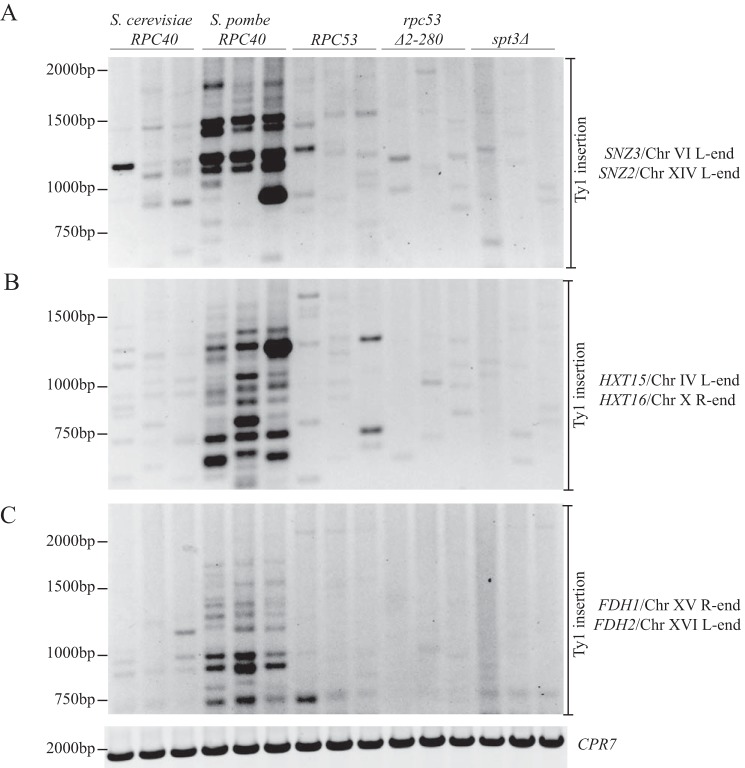
Ty1 insertion is not redirected to telomere-proximal regions in the rpc53Δ2–280 mutant. PCR analysis of yeast genomic DNA extracted from the indicated strains grown in triplicate for 3 days at 20 °C is shown. Each PCR contained a Ty1 element primer and a telomere-proximal primer. The telomere-proximal primers hybridize to two genomic regions due to identical DNA sequences at the following loci: SNZ3/SNZ2 (A), HXT15/HXT16 (B), and FDH1/FDH2 (C). CPR7 PCR is shown as a control to demonstrate that equal genomic DNA is present in each sample. R, right; L, left.
Transposition Assays
Ty1 transposition mobility assays were performed as described (23) with the following modifications. For Fig. 6C (WT and rpo31-698, rpc34-1, and rpc40-V78R mutants), 2,000 cells were inoculated into 1 ml of SC-Ura medium, and for Fig. 7C (RPC53 and rpc53Δ11–40, rpc53Δ316–350, and rpc53Δ2–280 mutants), 200,000 cells were inoculated into 1 ml of SC-Ura-Leu medium. Cells were grown for 5 days at 20 °C, 200 cells were plated for viability (SC-Ura plates for rpc mutants and SC-Ura-Leu for rpc53 mutants), and 107 cells were plated for transposition frequency (SC-Ura-His plates for rpc mutants and SC-Ura-Leu-His plates for rpc53 mutants).
Quantitative PCR (qPCR)
Cells were grown in yeast extract-peptone-dextrose medium at 20 °C to logarithmic growth, and 1 × 107 cells were spun down and stored at −80 °C. Total RNA was prepared with a Qiagen RNeasy kit, and 1 μg of RNA was used for cDNA synthesis with a Superscript VILLO cDNA synthesis kit (Invitrogen). Real time PCR was performed in triplicate with 2 μl of diluted cDNA (1:50) and Power SYBR Green Master Mix (Invitrogen). A standard 2-h comparative PCR analysis was performed using a 7500 Real Time PCR System (Applied Biosystems). The primers for the tLEU, tLEU de novo, and tGLY genes have been described (24). qPCR analysis was performed using the comparative CT method (ΔΔCT method, Applied Biosystems) with TAF10 used as an internal control. The TAF10 primers were: OVM695 5′-GGCGTGCAGCAGATTTCAC-3′ and OVM696 5′-TGAGCCCGTATTCAGCAACA-3′.
The ΔCT mean value for each sample was calculated by subtracting the CT mean value for TAF10 from the CT mean value of the target (tLEU, tLEU de novo, or tGLY). The ΔΔCT was calculated by subtracting the ΔCT mean value of the target gene from each mutant from the value of the target gene from the wild type. The relative quantity (RQ) was calculated as 2−ΔΔCT so that the wild type (20) is a value of 1.0 and every other value is a relative comparison with wild type. The ΔCT standard error (S.E.) was both added and subtracted from the ΔΔCT, and the resultant value (ΔΔCT S.E.) was calculated as 2−ΔΔCT S.E. and then subtracted from the 2−ΔΔCT value to generate the S.E. error bars for the graphs in Fig. 7, D–F.
Results
Ty1-IN Co-purifies with RNA Pol III Subunits
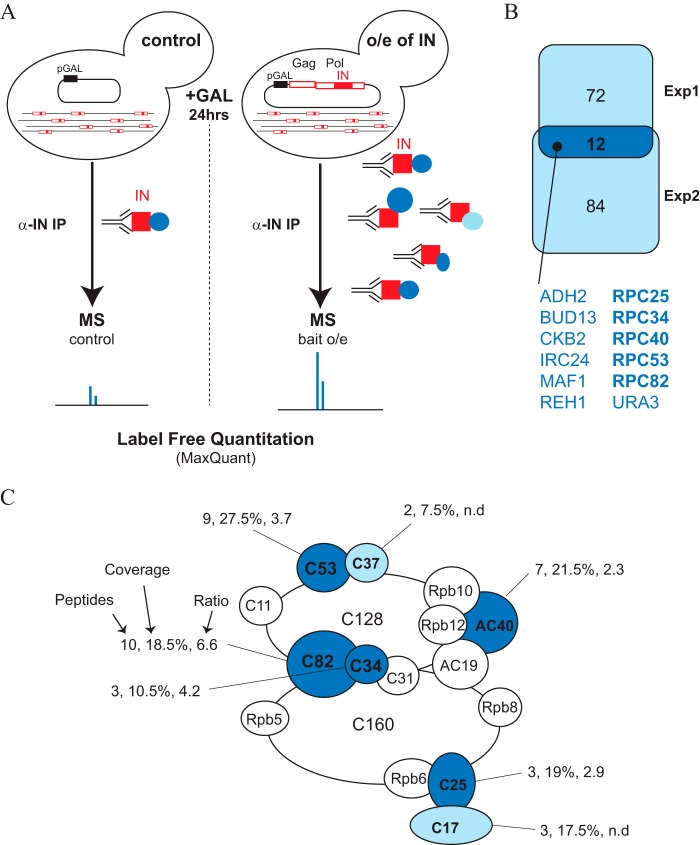
To identify proteins that interact with Ty1-IN and target Ty1 transposon insertion upstream of Pol III-transcribed genes, we purified Ty1-IN from yeast cells. Despite the fact that there are 32 full size Ty1 elements in the S288C genome, Ty1-IN is produced at very low levels. Therefore, we induced expression of Ty1-IN from a GAL1 promoter-driven Ty1 element, pGAL1-Ty1-H3, which induces overexpression of Gag and Pol polypeptides and therefore transposition (Fig. 1A and Ref. 40). Ty1-IN was purified using a monoclonal antibody (8b11) after overexpression of pGAL1-Ty1-H3 and, as a control, from cells carrying vector alone (Fig. 1A and Ref. 27). We performed two independent Ty1-IN purifications that were analyzed by label-free quantitative MS analysis. The peak intensity areas for each peptide were measured and summed for a given protein in each of the four samples analyzed (2 × Ty1-IN and control IPs). We then compared the intensities between the Ty1-IN and control purifications and only further considered proteins enriched more than 2-fold. Our MS data confirmed the enrichment of Ty1-IN peptides in the purification from cells with Ty1-IN overexpression versus control in both experiments (Table 2). In the second experiment, other Ty1 polypeptides were also enriched to a lower extent, presumably due to purification of a small portion of uncleaved or partially cleaved Ty1. In agreement, we also identified a few peptides spanning two polypeptides (Gag/PR and IN/RT; Table 2). In addition to IN peptides, we also identified 84 and 96 proteins that were enriched more than 2-fold in the Ty1-IN overexpressed purification compared with control in the first and second experiments, respectively (Fig. 1B). Of the 12 proteins that were enriched in both experiments, five are RNA Pol III subunits (Rpc25, -34, -40, -53, and -82), whereas Maf1 is a Pol III repressor protein (Fig. 1, B and C). We also identified Rpc37 and Rpc17 peptides in experiment 1 (Fig. 1C). Five Pol III subunits are specific to Pol III: the Rpc53/37 heterodimer, which is the structural counterpart of TFIIF, and the Rpc82/34/31 heterotrimer, which is related to TFIIE in Pol II-mediated transcription (25, 41). We identified four of the five Pol III-specific subunits in our Ty1-IN purifications: Rpc53/37 and Rpc82/34. We also identified the Rpc25/17 subcomplex, which is analogous to Rpb4/7 and Rpa14/43 in Pol II and Pol I, respectively, and Rpc40 (AC40), which is a subunit of both Pol I and Pol III (Fig. 1C and Ref. 25).
FIGURE 1.
Ty1-IN co-purifies with RNA Pol III subunits. A, schematic of the Ty1-IN purification and MS procedure. Control cells carry a pGAL1 plasmid vector, whereas Ty1-IN-overexpressing (o/e of IN) cells carry the pGAL1-Ty1-H3 vector (pJEF724) that induces Ty1 element expression upon galactose (+GAL) addition (40). After 24 h of galactose treatment, Ty1-IN (red squares) was bound to anti-IN antibodies (horizontal Y shape), and the co-purified proteins (blue circles) were purified using IgG-Sepharose beads (not depicted). Purified proteins were subjected to MS analysis using label-free quantitation. B, Venn diagram representing proteins enriched in each and both experiments (experiment 1 (Exp1) and experiment 2 (Exp2)). C, schematic of RNA Pol III adapted from Ref. 46 and colored subunits that were identified in the Ty1-IN MS data. Dark blue proteins were identified as Ty1-IN interactors in both experiments 1 and 2, whereas light blue proteins were identified in one experiment only. For each Pol III subunit identified by MS, the number of peptides, percent coverage of the protein, and peptide intensity ratios in the IN overexpression IP versus the control IP are shown. Only intensity ratios of peptides identified in the two replicates are shown (n.d., not determined).
TABLE 2.
Total intensities of Ty peptides in the indicated IPs
| Polypeptide | Peptides Exp1 | Total intensity (IN IP) | Total intensity (control IP) | Ratio | Peptides Exp2 | Total intensity (IN IP) | Total intensity (control IP) | Ratio |
|---|---|---|---|---|---|---|---|---|
| Gag | 9 | 4.35 × 109 | 2.42 × 109 | 1.8 | 11 | 1.45 × 1010 | 3.41 × 109 | 4.3 |
| PR | 3 | 1.15 × 108 | 8.84 × 107 | 1.3 | 1 | 3.33 × 108 | 8.78 × 107 | 3.8 |
| IN | 10 | 3.4 × 109 | 7.56 × 108 | 4.5 | 11 | 1.95 × 109 | 1.47 × 108 | 13.3 |
| RT | 3 | 8.64 × 108 | 8.55 × 108 | 1 | 4 | 2.06 × 108 | 6.87 × 107 | 3 |
We validated the physical interaction of Ty1-IN with Pol III subunits by selecting Rpc17,19,37,40,53,82-GFP tagged proteins from the GFP-tagged protein collection and performing a reciprocal IP (42). As described above, Ty1-IN was overproduced using the pGAL1-Ty1-H3 expression plasmid (40). We purified each Rpc-GFP protein using GFP-Trap beads followed by immunoblotting analysis and probed for the presence of Ty1-IN. We confirmed that Ty1-IN co-purified with Rpc17, -37, -53, and -82 but was not in the immunoprecipitate from the untagged strain after both low salt and high salt washes (Fig. 2A). We only detected processed Ty1-IN in the lower molecular weight region of the immunoblot and not in the region where unprocessed PR-IN or IN-RT polypeptides migrate as shown in the α-IN blot of the whole cell lysate (Fig. 2A, left panel). We detected a faint amount of Ty1-IN in the Rpc40-GFP purification after a low salt wash, but this was removed after a high salt wash (Fig. 2B). Next, we tested Rpc19 (AC19), which interacts with AC40 and is also a shared Pol I and Pol III subunit (43, 44). Ty1-IN was able to co-purify with Rpc19-GFP (Fig. 2B). Although the Pol III Maf1 repressor was identified in our Ty1-IN MS data, we did not detect an interaction between Ty1-IN and Maf1-GFP or between Ty1-IN and Rpb6, which is part of the central core of Pol III and also shared with Pol I and Pol III (Fig. 2, A and B).
We were not able to test Rpc25, Rpc31, or Rpc34 using the GFP-Trap system because our attempts to tag these proteins with GFP resulted in cell lethality. Similarly, 3HA and 13Myc epitope tagging was also lethal for Rpc31. However, Rpc34 and Rpc25 were successfully tagged with 3HA, and we performed a co-IP using anti-HA resin with 3HA-tagged cells carrying the pGAL1-Ty1-H3 expression plasmid (Fig. 2C). Both Rpc34-3HA and Rpc25-3HA were able to interact with Ty1-IN after a low salt wash, but the interaction was minimal after a high salt wash (Fig. 2C). As seen with the GFP system, Rpc82-3HA interacted with Ty1-IN after both a low and high salt wash (Fig. 2C).
Our co-IP interaction data indicate that Ty1-IN interacts with multiple RNA Pol III proteins, including the RNA Pol III-specific Rpc82/34/31 and Rpc53/37 subcomplexes (Fig. 2, A–C). A weaker interaction, or no interaction, is detected with some of the shared RNA Pol components such as Rpc40 (Pols I and III) and Rpb6 (Pols I, II, and III), respectively, although Rpc19 (Pols I and III) interacts well with Ty1-IN (Fig. 2B).
Ty1-IN Does Not Interact with RNA Pol II, TFIIIB, or TFIIIC
RNA Pol III is a stable complex and requires partially denaturing conditions to remove subcomplexes; therefore it is likely that when individual subunits are purified the remainder of the Pol III complex is co-purified (45, 46). To test whether Ty1-IN also interacts with the two largest core RNA Pol III subunits, we purified Rpc160-GFP and Rpc128-GFP and demonstrated that indeed Ty1-IN is present in the purification (Fig. 3A). Because Ty1 elements insert within a ∼1-kbp window upstream of tRNA genes, there remained the possibility that Ty1-IN could also interact with RNA Pol II. However, we found no evidence of interaction between Ty1-IN and the counterparts of Rpc160 and Rpc128, Rpb1 and Rpb2, respectively (Fig. 3A).
Because Ty1 element insertion requires active Pol III transcription, we were curious whether other factors required for Pol III transcription may associate with Ty1-IN. To activate tRNA transcription, TFIIIC recognizes and binds to tRNA promoters followed by TFIIIB assembly and recruitment of the RNA Pol III complex (47). An interaction between Ty3-IN and the TFIIIB factor Brf1 is sufficient to target the Ty3 element to Pol III transcription start sites in vitro (6–8). We tested whether a subset of the TFIIIB (Brf1 and Bdp1) and TFIIIC (Tfc1, -3, and -7) subunits co-purify with Ty1-IN using the GFP tag system. However, we were unable to detect any interaction between Ty1-IN and TFIIIB or TFIIIC subunits (Fig. 3, B and C). Therefore the mechanism of Ty1-IN and Ty3-IN interaction with the Pol III transcription apparatus is different.
RNA Pol III Interacts with the C Terminus of Ty1-IN
The domain structure of Ty1-IN is similar to other retroviral integrases and contains an N-terminal HHCC zinc-binding domain, a catalytic core domain, and a less conserved C-terminal domain (Fig. 4A and Ref. 48). A bipartite nuclear localization signal (NLS) resides at the end of Ty1-IN with two basic regions (NLS1 and NLS2) separated by 21 amino acids (Fig. 4D and Refs. 49 and 50). We used previously generated Ty1-IN deletion constructs to map the domain of Ty1-IN that interacts with RNA Pol III (Fig. 4A and Ref. 49). Co-IP analysis was performed by GFP-Trap purification of Rpc82-GFP in a strain expressing either full-length Ty1-IN or a Ty1-IN deletion construct. Rpc82-GFP interacted with full-length Ty1-IN, Ty1-IN with amino acids 422–515 removed (IN-C), or Ty1-IN with amino acids 515–561 removed (IN-F) but not any construct with the C-terminal amino acids 561–635 removed (IN-D, ΙN-E, and IN-G; Fig. 4A). As well, the N-terminal 422 amino acids of Ty1 were not able to interact with Rpc82-GFP (IN-A; Fig. 4A). None of the Ty1-IN fusion proteins interacted with the GFP-Trap beads in the absence of Rpc82-GFP (Fig. 4B). We also tested whether Rpc82-GFP could interact with Ty1-IN carrying mutations in basic region 1 (NLS1) or both basic regions (IN-NLS1+2) of the NLS (Fig. 4D and Ref. 49). Mutation of the Ty1-IN NLS1 and NLS2 regions completely abrogated the interaction with Rpc82 (Fig. 4D). Because the cell lysis procedure disrupts the nuclear and cytoplasmic compartments, we do not think the lack of interaction is due to IN-NLS1+2 being excluded from the nucleus, although we cannot entirely rule out this possibility. However, when combined with the Ty1-IN truncation analysis, our data suggest that the C-terminal 75 amino acids of Ty1-IN interact with RNA Pol III, which concurs with a recent studying showing a two-hybrid interaction between Rpc40 and Ty1-IN that was abrogated by removal of the C-terminal amino acids 578–635 (24). Although Bridier-Nahmias et al. (24) found that Ty1-IN amino acids 578–635 were also sufficient to interact with Rpc40 using a two-hybrid assay, we were unable to detect an interaction of a C-terminal Ty1-IN fragment (amino acids 422–636; IN-B) with Rpc82 or Rpc53 by co-IP (Fig. 4C). Possibly there is an inhibitory domain in Ty1-IN amino acids 422–578, or the requirements for Ty1-IN interaction with different RNA Pol III subunits have subtle differences.
RNA Pol III Interacts with Ty1-IN in Vitro
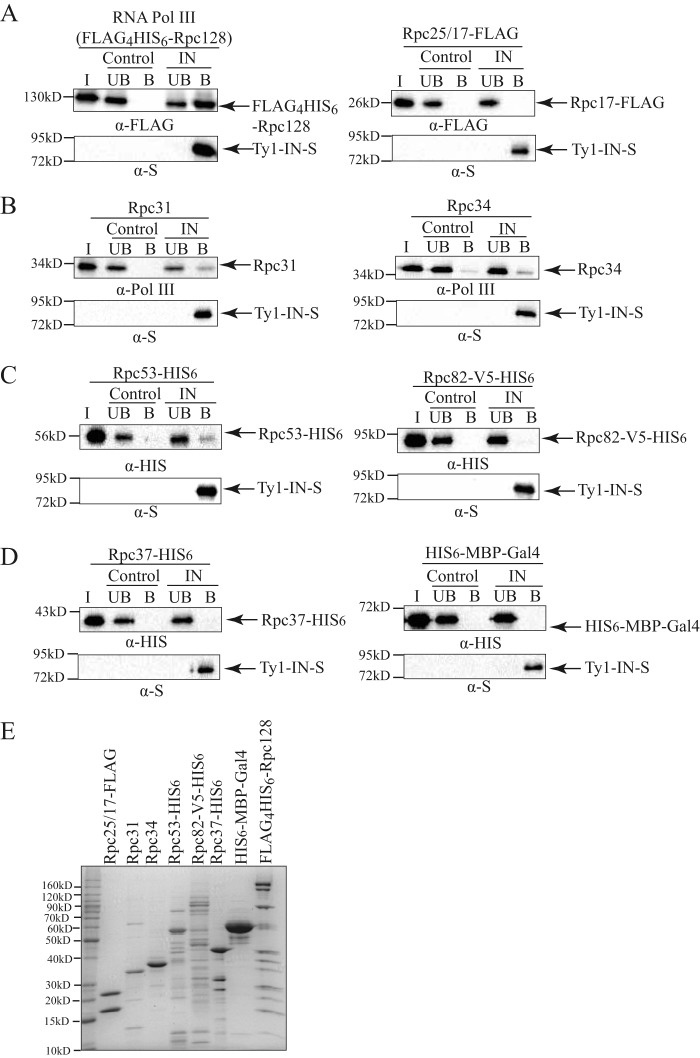
Our co-IP analysis demonstrates that Ty1-IN interacts with multiple RNA Pol III proteins when purified from yeast extracts (Fig. 2). However, due to the stability of the RNA Pol III complex, the entire complex is likely purified in each IP, and therefore the Pol III protein(s) that interacts directly with Ty1-IN cannot be determined by this method. To determine which RNA Pol III proteins Ty1-IN may interact with directly, we used an in vitro approach. Ty1-IN containing a C-terminal S-tag was expressed and purified from E. coli with S-protein-agarose and mixed with a FLAG4His6-Rpc128-tagged RNA Pol III complex purified from S. cerevisiae (Fig. 5, A (left panel) and E). S-protein-agarose with no Ty1-IN bound was used as a control. The bound and unbound fractions from the Ty1-IN-S beads and the S beads alone were analyzed by immunoblotting along with the input Pol III protein and probed for the presence of FLAG4His6-Rpc128. The RNA Pol III complex specifically interacted with the Ty1-IN-S beads but not the S beads alone (Fig. 5A). Next, RNA Pol III-specific subunits were purified from E. coli and tested individually for interaction with Ty1-IN-S. We were unable to detect an interaction between Rpc25/17-FLAG and Ty1-IN, which also served as a negative control for the FLAG tag binding to the S beads (Fig. 5, A (right panel) and E). Rpc31 and Rpc34 were both purified without epitope tags and were therefore visualized with antibody raised against the entire Pol III complex (38). Both Rpc31 and Rpc34 were identified in the bound fraction of the Ty1-IN-S beads; albeit some background binding of Rpc34 to the S beads alone was detected (Fig. 5, B and E). Rpc31 and Rpc34 interact with Rpc82 to form the Rpc82/34/31 Pol III-specific subcomplex; therefore it was of interest to test whether Rpc82 could interact directly with Ty1-IN. Rpc82 was C-terminally fused to a V5-His6 tag and expressed in E. coli as described (35); however, the yield was low as most of Rpc82-V5-His6 was insoluble. This could be because proper folding of Rpc82 requires a post-translational modification, and Rpc82 is known to be sumoylated in yeast (51). Nevertheless, we tested whether our preparation of Rpc82-V5-His6 was able to interact with Ty1-IN but failed to detect an interaction with Rpc82 and Ty1-IN-S based on immunoblotting analysis (Fig. 5, C (right panel) and E). Finally, we tested whether the Rpc53 or Rpc37 Pol III-specific proteins could interact directly with Ty1-IN. We found that purified Rpc53-His6 interacted with Ty1-IN-S beads but not S beads alone (Fig. 5, C (left panel) and E). However, neither purified Rpc37-His6 nor the His6-MBP-Gal4 protein (control for His6 tag) were able to interact with Ty1-IN-S (Fig. 5, D and E). In summary, we found that a subset of RNA Pol III proteins (Rpc31, Rpc34, and Rpc53) purified from E. coli and the purified Pol III complex from yeast are able to interact with Ty1-IN in vitro.
FIGURE 5.
Ty1-IN interacts with RNA Pol III in vitro. A–D, Ty1-IN fused to a C-terminal S-tag (Ty1-IN-S) was bound to S-protein-agarose beads and incubated with purified RNA Pol III proteins as described under “Experimental Procedures.” The purified RNA Pol III protein is indicated above each immunoblot. For all immunoblots, Ty1-IN-S was visualized with an anti-S (α-S) antibody. “Control” is S-protein-agarose alone; “IN” is Ty1-IN-S bound to S-protein-agarose. The amounts of input protein (I), unbound fraction (UB), and bound fraction (B) loaded on each gel are described under “Experimental Procedures.” For A, left panel, the entire RNA Pol III complex is co-purified with N-terminal FLAG4His6-tagged Rpc128. For A, right panel, Rpc25/17-FLAG is Rpc17-FLAG co-purified with Rpc25. FLAG4His6-Rpc128 and Rpc17-FLAG are visualized with anti-FLAG (α-FLAG) antibody. B, Rpc31 and Rpc34 are purified without epitope tags and are visualized with anti-Pol III antibody (38). C–D, Rpc53-His6, Rpc82-V5-His6, Rpc37-His6, and His6-MBP-Gal4 are all visualized with an anti-His (α-HIS) antibody. E, Coomassie-stained gel of purified proteins used for in vitro assays in A–D. 5 μg of each purified protein was loaded except for FLAG4His6-Rpc128-tagged Pol III for which 20 μg was loaded. All proteins were purified from E. coli except Pol III (A, left panel), which was purified from yeast as described under “Experimental Procedures.”
rpc34 and rpc40 Temperature-sensitive (ts) Mutants Have Defects in Ty1 Insertion Upstream of the SUF16 tRNA Gene and in Genome-wide Ty1 Mobility
Our in vivo co-IP and in vitro binding studies suggest that Ty1-IN may interact with multiple RNA Pol III subunits. RNA Pol III ts mutants that were available in a published ts collection (52) were tested for Ty1 element insertion upstream of the SUF16 tRNA gene, which is a hot spot for Ty1 transposition (53). Although the ts mutants are lethal at high temperatures, Ty1 transposition is induced by growing cells at 20 °C at which temperature the ts mutants are viable. De novo Ty1 element transposition upstream of SUF16 was detected using an established PCR assay with the indicated primers, and the spt3Δ mutant is a control that lacks Ty1 element expression (Fig. 6A and Ref. 39). We found that all three RNA Pol III mutants (rpo31-698, rpc34-1, and rpc40-V78R) displayed defects in the typical periodic Ty1 element insertion pattern typical of wild type strains (Fig. 6B and Ref. 39). The rpo31-698 RNA Pol III mutant, which carries a mutation in the largest Pol III subunit (Rpc160), had the most similar insertion pattern to wild type, although the levels of insertion were reduced to 37% of wild type (Fig. 6B). Both rpc34-1 and rpc40-V78R had highly reduced Ty1 insertion at the SUF16 locus (10 and 9%, respectively, of wild type). We used a qPCR assay to demonstrate that the ts mutants have similar levels of tRNA gene transcripts to wild type cells; therefore the lack of Ty1 element insertion at SUF16 is not due to Pol III transcription defects (Fig. 6, D–F). Because the SUF16 tRNA assay analyzes Ty1 insertions at a single locus, we also performed a quantitative assay for Ty1 mobility to examine de novo Ty1 insertions in the whole genome. We used a plasmid (pBDG922) that carries a Ty1 element containing the HIS3 gene with an artificial intron (AI) that interrupts HIS3 expression (HIS3AI). The AI intron is removed after Ty1 expression, and the cell becomes HIS3+ after Ty1 cDNA insertion into DNA, which can be quantitated (54). Wild type cells underwent Ty1 transposition at a frequency of 13.1 × 10−6 compared with 4.4 × 10−6 for rpc40-V78R and 1.9 × 10−6 for rpc34-1 (∼3- and 7-fold, respectively, less than wild type; p < 0.0001). Despite the reduction in Ty1 insertion at the SUF16 tRNA locus, rpo31-698 mutant cells did not have a statistically significant change in Ty1 mobility compared with wild type cells (Fig. 6C).
The N Terminus of Rpc53 Is Required for Ty1 Insertion Upstream of the SUF16 tRNA Gene
We analyzed a panel of viable RPC53 deletion mutants for Ty1 element insertion upstream of the SUF16 tRNA locus (36). Although an Rpc53 C-terminal mutant (rpc53Δ316–350) had slightly increased (∼1.4-fold) Ty1 insertion, an N-terminal Rpc53 deletion mutant, rpc53Δ11–40, had a 50% reduction in Ty1 insertion, and a larger N-terminal deletion mutant (rpc53Δ2–280) was completely deficient in Ty1 element insertion (Fig. 7A). We tested tRNA gene expression in rpc53Δ2–280 cells and found that tLEU and tGLY transcripts were 48 and 76%, respectively, of wild type values, whereas tLEU de novo (tLEU transcript prior to splicing) was 73% of wild type (Fig. 7, C–E). Although there is a decrease in tRNA gene expression in rpc53Δ2–280 cells, it is not sufficient to explain a complete lack of Ty1 element insertion upstream of the SUF16 tRNA gene. However the 50% reduction of SUF16 Ty1 element insertion in rpc53Δ11–40 mutant cells could be attributed to defects in tRNA gene transcription, which was reduced to 55% for tLEU and 64% for tLEU de novo transcripts but at wild type levels for tGLY transcripts (Fig. 7, C and D). Despite the fact that no Ty1 element insertion was detected upstream of the SUF16 locus in rpc53Δ2–280 cells, overall Ty1 mobility was reduced to only ∼75% of wild type with a frequency of 48.2 × 10−6 compared with 63.7 × 10−6 for wild type cells (Fig. 7B). Even though the rpc53Δ11–40 mutant had reduced Ty1 element insertion at the SUF16 locus, the overall Ty1 mobility was equivalent to wild type cells (Fig. 7B). Finally, the rpc53Δ316–350 mutant, which had an ∼1.4-fold increase in Ty1 element insertion at the SUF16 locus, had the most reduced overall Ty1 mobility with a frequency of 41.9 × 10−6, or 65% of wild type. The moderate changes we detected in genome-wide Ty1 insertion frequency in the rpc53 mutants compared with the 50% reduction in the rpc53Δ11–40 mutant, and the complete lack of insertion in the rpc53Δ2–280 mutant at the SUF16 locus could be because Pol III transcription is differentially affected at different tRNA genes or because the Ty1 element is being targeted elsewhere in the genome.
Ty1 Insertion Is Not Redirected to Chromosome Ends in the rpc53Δ2–280 Mutant
Replacing S. cerevisiae Rpc40 with S. pombe Rpc40 resulted in retargeting of Ty1 elements to telomere-proximal regions at the ends of chromosomes (24). We tested whether Ty1 elements were being similarly mistargeted in our rpc53Δ2–280 mutant by designing primers that hybridized to the chromosome ends identified in the Bridier-Nahmias et al. (24) study as preferred Ty1 integration sites. Each insertion site is within a gene that has an almost identical homologue (e.g. SNZ2/SNZ3); therefore each set of primers (one within the Ty1 element and the other within a gene) simultaneously amplified Ty1 elements at two telomere-proximal genomic locations (e.g. Chr VI and Chr XIV left end; Fig. 8A). The S. pombe RPC40 and rpc53Δ2–280 mutant strains and their wild type counterparts were grown for 3 days at 20 °C, and genomic DNA was extracted. It was evident that Ty1 elements were highly amplified from Chr IV, VI, XIV, and XVI left telomere- and Chr X and XV right telomere-proximal regions in the S. pombe RPC40 strain but not in the S. cerevisiae RPC40 wild type strain (Fig. 8). We did not find any evidence of hyperamplification of Ty1 elements from these same regions in the rpc53Δ2–280 mutant or the isogenic wild type strain (Fig. 8). Therefore Ty1 element insertion is not redirected toward chromosome ends in the rpc53Δ2–280 mutant.
Ty1-IN Interaction with Rpc37 Is Disrupted in rpc53Δ2-280
The lack of Ty1 element targeting upstream of SUF16 in the rpc53Δ2–280 mutant suggests that the interaction between Ty1-IN and either the entire Pol III complex or a specific Pol III subunit has been abrogated. We asked whether the interaction between Ty1-IN; the Rpc82/34/31, Rpc53/37, and Rpc17/25 subcomplexes; and Pol III core in the rpc53Δ2–280 mutant was affected by GFP-Trap pulldown of representative GFP-tagged proteins from each complex (Fig. 9). The Pol III core (Rpc19-GFP), Rpc82/34/31 (Rpc82-GFP), and Rpc17/25 (Rpc17-GFP) subcomplexes were all able to interact with Ty1-IN in the rpc53Δ2–280 mutant; however, the Rpc53/37 subcomplex was not (Rpc37-GFP; Fig. 9). Therefore, insertion of Ty1 elements upstream of SUF16 requires the interaction of Rpc37 and probably also Rpc53 with Ty1-IN.
Discussion
We discovered that Ty1-IN co-purifies from yeast cells with multiple RNA Pol III subunits using MS analysis and validated the interaction by performing co-IPs from yeast lysates (Figs. 1 and 2). Ty1-IN interacts with the two largest Pol III core subunits, Rpc160 and Rpc128, but not the RNA Pol II homologues, Rpb1 and Rpb2, respectively, suggesting that the interaction is specific to RNA Pol III (Fig. 3A). The interaction between Ty1-IN and RNA Pol III requires the C terminus of Ty1-IN, but we did not find that the Ty1-IN C terminus alone co-purified with Rpc82 or Rpc53 (Fig. 4). Whether or not the Ty1-IN C terminus is sufficient to interact with RNA Pol III may depend on the subunit tested and the experimental method used. Using the yeast two-hybrid system, the C-terminal 578–635 amino acids of Ty1-IN were sufficient to interact with Rpc40; however, this system cannot exclude that binding is mediated via another protein (24). We performed in vitro binding assays to demonstrate that E. coli-produced Ty1-IN interacts with RNA Pol III purified from yeast as well as the Pol III-specific proteins Rpc31, Rpc34, and Rpc53 purified from E. coli (Fig. 5). The rpc34-1 and rpc40-V78R mutant strains have defects in Ty1 element insertion upstream of the SUF16 tRNA hot spot locus and in genome-wide transposition frequency (Fig. 6). Interestingly, a strain carrying only the C-terminal third of Rpc53 (rpc53Δ2–280) is incapable of Ty1 element insertion upstream of the SUF16 locus even though Ty1 mobility is only reduced to ∼75% of wild type levels (Fig. 7). The interaction of Rpc37 with Ty1-IN is abolished in the rpc53Δ2–280 mutant, suggesting that the Rpc53/37 subcomplex is important for targeting Ty1-IN upstream of SUF16 (Fig. 9). Altogether our data suggest that multiple Pol III-specific subunits are able to interact with Ty1-IN to promote insertion of Ty1 elements upstream of Pol III-transcribed genes.
To identify host proteins that interact with Ty1-IN and target the integration of Ty1 elements into the genome, we used an unbiased approach in which we purified untagged Ty1-IN using monoclonal anti-IN (8b11) antibody followed by MS analysis (27). Remarkably, about half of the proteins identified as Ty1-IN interactors in two independent purifications were subunits of RNA Pol III. Rpc25, Rpc34, Rpc40, Rpc53, and Rpc82 peptides were identified as well as the Maf1 Pol III inhibitor protein. We also identified Rpc37 and Rpc17 as candidate interactors in the first MS experiment, but these proteins were not identified in the second MS experiment. We then validated several of these interactions using an orthogonal approach in which we assessed the ability of several RNA Pol III subunits to co-immunoprecipitate with Ty1-IN. These results indicated that Ty1-IN binds to a subset of Pol III subunits but not to TFIIIB or TFIIIC subunits.
The in vivo and in vitro RNA Pol III Ty1-IN interaction data presented in this study, combined with the recently published work demonstrating an interaction between Rpc40 and Ty1-IN, begs the question of which RNA Pol III subunit(s) interact specifically with Ty1-IN (24). Our in vitro binding data demonstrate that at least three Pol III-specific subunits, Rpc31, Rpc34, and Rpc53, are capable of interacting with Ty1-IN (Figs. 5 and 10). Because Rpc31, Rpc34, Rpc53, and Ty1-IN were purified from E. coli, a post-translational modification such as phosphorylation, acetylation, or ubiquitylation is not absolutely required for these interactions to occur. It remains possible that Rpc82 is also capable of interacting directly with Ty1-IN because we were unable to purify Rpc82 to homogeneity (Fig. 5, C and E). As well, the interaction of purified FLAG4His6-Rpc128-tagged Pol III from yeast with Ty1-IN could be mediated via any of the co-purified Pol III subunits (Fig. 5, A and E). Because many of the RNA Pol III subunits interact with each other, it is possible that Ty1-IN could interact with both of the Rpc82/34/31 and Rpc53/37 subcomplexes (Fig. 10). For example, cross-linking studies have revealed that Rpc53 interacts with the Rpc82/34/31 subcomplex, the stalk, and likely the active site of Rpc160/Rpc128 (36). Although the structure of Ty1-IN has not been determined, the crystal structure of IN from the prototype foamy virus in complex with DNA, termed the prototype foamy virus intasome, revealed that prototype foamy virus IN is a homotetramer formed from two homodimers (55). Given that Ty1-IN is a 636-amino acid protein with a predicted molecular mass of 71.5 kDa, or 286kDa for a homotetramer, it is possible that Ty1-IN could interact with multiple RNA Pol III subunits.
FIGURE 10.
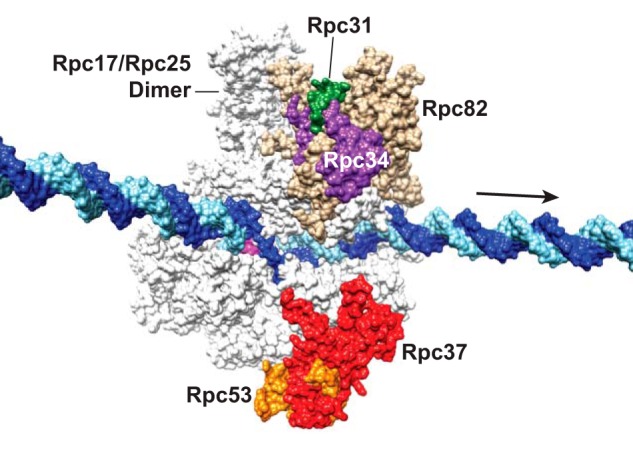
Location of Pol III-specific subunits on a surface view of elongating Pol III. The image is based on the cryoelectron microscopy structure of elongating Pol III (Protein Data Bank code 5FJ8) (56), and the DNA was extended based on the complete transcription bubble reported previously (75). The DNA template and non-template strands are colored in light blue and dark blue, respectively. The arrow points to downstream DNA, and the RNA molecule in the active site cleft is colored magenta. The highlighted Pol III subunits are Rpc31 (green), Rpc34 (purple), Rpc37 (red), Rpc53 (orange), and Rpc82 (tan). Note that the N-terminal 270 amino acids of Rpc53 are flexible and are not depicted due to the lack of structural data.
We discovered that the N-terminal 280 amino acids of Rpc53 are required for Ty1 element insertion upstream of the SUF16 tRNA gene (Fig. 7A). Deletion of a short region of the Rpc53 N terminus (rpc53Δ11–40) reduced Ty1 element insertion upstream of SUF16 to 50% of wild type levels (Fig. 7A). Interestingly, cross-linking data have shown an interaction between the N-terminal 20 amino acids of Rpc53 with Rpc25, Rpc34, and Rpc82 (36). The extended N-terminal 280 amino acids of Rpc53 cross-link to Tfc4 (TFIIIC), Rpc17, Rpc25, Rpc37, Rpc128, and Rpc160 (35, 36). A recent cryoelectron microscopy structure of RNA polymerase III suggests that the N-terminal 270 amino acids of Rpc53 are unstructured and flexible, which may explain the cross-links with multiple proteins (56). We tested whether the interaction of Ty1-IN with a selection of Pol III subunits was affected in the rpc53Δ2–280 mutant strain. We found that the interaction between Ty1-IN and Rpc37 is abolished in the rpc53Δ2–280 mutant, but the interaction with Rpc17, Rpc19, and Rpc82 remains intact (Fig. 9). The in vitro interaction of Rpc53 with Ty1-IN, lack of Ty1 element insertion upstream of the SUF16 locus, and loss of Ty1-IN interaction with Rpc37 in the absence of the Rpc53 N terminus strongly suggest that the Rpc53/37 subcomplex is important for targeting Ty1 elements upstream of Pol III-transcribed genes (Figs. 5, 7, 9, and 10).
Despite the lack of Ty1 element insertion upstream of SUF16, our Ty1 mobility assays revealed that Ty1 element mobility in the rpc53Δ2–280 mutant occurs at a frequency of 48.2 × 10−6, or 76% of wild type frequency (Fig. 7B). In the Bridier-Nahmias et al. (24) study, S. cerevisiae Rpc40 was substituted with S. pombe Rpc40, which no longer interacted with Ty1-IN but had a similar Ty1 retrotransposition frequency to wild type due to increased Ty1 element insertion at telomeric and subtelomeric regions. We tested whether Ty1 elements were also misdirected to telomeric regions in the rpc53Δ2–280 mutant; however, we did not detect enrichment of Ty1 elements at these loci (Fig. 8). It will be interesting to determine where in the genome Ty1 elements are targeted in the absence of the N terminus of Rpc53.
In addition to the rpc53Δ2–280 mutant, we identified two mutant strains, rpc34-1 and rpc40-V78R, that had both highly reduced Ty1 element SUF16 targeting and genome-wide Ty1 insertion frequency (Fig. 6, B and C). The location of the rpc34-1 mutation is unknown, but the rpc40-V78R mutant likely affects the interaction with AC19/Rpc19 because overexpression of RPC19 rescues the rpc40-V78R ts defect (44). As well, Rpc19 and Rpc40 interact by yeast two-hybrid analysis and based on homology to bacterial polymerase subunits are presumed to form a heterodimer (43, 44). Either Rpc34 or Rpc40 is a candidate for directly interacting with Ty1-IN, and we have demonstrated that Rpc34 and Rpc31 (both part of the Rpc82/34/31 subcomplex) are capable of interacting with Ty1-IN in vitro (Fig. 5B).
Until detailed structural analyses are performed with purified proteins, it is difficult to pinpoint precisely which Pol III subunit(s) interacts directly with Ty1-IN. Rpc82/34/31 interacts with the initiation factor TFIIIB to recruit RNA Pol III and functions in promoter opening and transcription initiation (57–64). Rpc53/37 also functions in promoter opening but has an additional role in transcription termination and reinitiation (36, 65, 66). Macromolecular MS studies have shown that both the Rpc82/34/31 and Rpc53/37 subcomplexes dissociate from the Pol III core upon perturbation of the Pol III complex with changes in pH or organic buffers (45, 46). The ability of the Rpc82/34/31 and Rpc53/37 subcomplexes to dissociate may aid in the mechanism of Ty1 element insertion, which occurs in a ∼1-kbp window upstream of tRNA genes. Another remaining question is whether Ty1-IN associates with the RNA Pol III preintegration complex or the RNA Pol III complex during transcriptional elongation.
The S. cerevisiae Ty3/Gypsy long terminal repeat retrotransposon is also targeted upstream of Pol III-transcribed genes; however, the Ty3 element integrates precisely within a few base pairs of the Pol III transcription start site (67, 68). Ty3-IN is targeted to Pol III-transcribed genes by an interaction with TFIIIB subunits (Brf1 and TATA-binding protein/Spt15), and Ty3 element orientation depends on an interaction of Ty3-IN with TFIIIC (Tfc1) (8, 69, 70). In contrast, we found no evidence that Ty1-IN interacts with subunits of TFIIIB (Brf1 and Bfp1) or TFIIIC (Tfc1, Tfc3, and Tfc7) (Fig. 3, B and C). Our data and those of Bridier-Nahmias et al. (24) suggest that Ty1-IN interacts with Pol III subunits, whereas purified Pol III inhibits Ty3 element integration in vitro (24, 71). Another major difference between Ty1 and Ty3 element insertion is that Ty1 elements preferentially insert into nucleosomes, in particular near the H2A/H2B interface, in a periodic manner (11–14, 73). Ty1 periodic integration, but not target site specificity, requires the ISW2 chromatin remodeling complex and the N terminus of Bdp1 (15, 16). Whether or not the interaction among Ty1-IN, Pol III, and nucleosomes requires another host factor such as a chromatin-modifying complex remains to be determined. Lentiviruses such as human immunodeficiency virus type 1 preferentially integrate into active transcription units that correlate with specific histone modifications (74). Our study may imply a general mechanism for retroviral integrases to associate with RNA polymerase complexes to mediate entry into the genome of their hosts.
Author Contributions
S. C. performed and analyzed experiments in Figs. 2 and 5–9. L. M. performed and analyzed experiments in Figs. 1, 3, 4, 6, and 7. P. H. W. C. performed and analyzed experiments for Fig. 1. H.-L. H. performed protein purification for Fig. 5. T. M. helped design and analyze experiments for Fig. 1. H.-T. C. helped design and analyze experiments for Figs. 5–7 and generated Fig. 10. V. M. conceived and coordinated the study, performed experiments for Fig. 6, and wrote the paper.
Acknowledgments
We thank Joel Acker for sending us anti-RNA Pol III antibodies, Jef Boeke for the kind gift of anti-Ty1-IN antibodies and the Ty1-IN NLS and truncation constructs, and Pascale Lesage for sending us Ty1 insertion coordinates for the S. pombe AC40 mutant.
The authors declare that they have no conflicts of interest with the contents of this article.
- PR
- protease
- Pol
- polymerase
- IN
- integrase
- TF
- transcription initiation factor
- RT
- reverse transcriptase
- IP
- immunoprecipitation
- SC
- synthetic complete medium
- MBP
- maltose-binding protein
- Chr
- chromosome(s)
- qPCR
- quantitative PCR
- RQ
- relative quantity
- NLS
- nuclear localization signal
- ts
- temperature-sensitive
- AI
- artificial intron.
References
- 1. Lesage P., and Todeschini A. L. (2005) Happy together: the life and times of Ty retrotransposons and their hosts. Cytogenet. Genome Res. 110, 70–90 [DOI] [PubMed] [Google Scholar]
- 2. Beauregard A., Curcio M. J., and Belfort M. (2008) The take and give between retrotransposable elements and their hosts. Annu. Rev. Genet. 42, 587–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Curcio M. J., Lutz S., and Lesage P. (2015) The Ty1 LTR-retrotransposon of budding yeast. Microbiol. Spectr. 3, 1–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Devine S. E., and Boeke J. D. (1996) Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev. 10, 620–633 [DOI] [PubMed] [Google Scholar]
- 5. Kim J. M., Vanguri S., Boeke J. D., Gabriel A., and Voytas D. F. (1998) Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 8, 464–478 [DOI] [PubMed] [Google Scholar]
- 6. Qi X., Daily K., Nguyen K., Wang H., Mayhew D., Rigor P., Forouzan S., Johnston M., Mitra R. D., Baldi P., and Sandmeyer S. (2012) Retrotransposon profiling of RNA polymerase III initiation sites. Genome Res. 22, 681–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Qi X., and Sandmeyer S. (2012) In vitro targeting of strand transfer by the Ty3 retroelement integrase. J. Biol. Chem. 287, 18589–18595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yieh L., Kassavetis G., Geiduschek E. P., and Sandmeyer S. B. (2000) The Brf and TATA-binding protein subunits of the RNA polymerase III transcription factor IIIB mediate position-specific integration of the gypsy-like element, Ty3. J. Biol. Chem. 275, 29800–29807 [DOI] [PubMed] [Google Scholar]
- 9. Xie W., Gai X., Zhu Y., Zappulla D. C., Sternglanz R., and Voytas D. F. (2001) Targeting of the yeast Ty5 retrotransposon to silent chromatin is mediated by interactions between integrase and Sir4p. Mol. Cell. Biol. 21, 6606–6614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zou S., and Voytas D. F. (1997) Silent chromatin determines target preference of the Saccharomyces retrotransposon Ty5. Proc. Natl. Acad. Sci. U.S.A. 94, 7412–7416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bachman N., Eby Y., and Boeke J. D. (2004) Local definition of Ty1 target preference by long terminal repeats and clustered tRNA genes. Genome Res. 14, 1232–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Baller J. A., Gao J., Stamenova R., Curcio M. J., and Voytas D. F. (2012) A nucleosomal surface defines an integration hotspot for the Saccharomyces cerevisiae Ty1 retrotransposon. Genome Res. 22, 704–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bridier-Nahmias A., and Lesage P. (2012) Two large-scale analyses of Ty1 LTR-retrotransposon de novo insertion events indicate that Ty1 targets nucleosomal DNA near the H2A/H2B interface. Mob. DNA 3, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mularoni L., Zhou Y., Bowen T., Gangadharan S., Wheelan S. J., and Boeke J. D. (2012) Retrotransposon Ty1 integration targets specifically positioned asymmetric nucleosomal DNA segments in tRNA hotspots. Genome Res. 22, 693–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bachman N., Gelbart M. E., Tsukiyama T., and Boeke J. D. (2005) TFIIIB subunit Bdp1p is required for periodic integration of the Ty1 retrotransposon and targeting of Isw2p to S. cerevisiae tDNAs. Genes Dev. 19, 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gelbart M. E., Bachman N., Delrow J., Boeke J. D., and Tsukiyama T. (2005) Genome-wide identification of Isw2 chromatin-remodeling targets by localization of a catalytically inactive mutant. Genes Dev. 19, 942–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Curcio M. J., Kenny A. E., Moore S., Garfinkel D. J., Weintraub M., Gamache E. R., and Scholes D. T. (2007) S-phase checkpoint pathways stimulate the mobility of the retrovirus-like transposon Ty1. Mol. Cell. Biol. 27, 8874–8885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dakshinamurthy A., Nyswaner K. M., Farabaugh P. J., and Garfinkel D. J. (2010) BUD22 affects Ty1 retrotransposition and ribosome biogenesis in Saccharomyces cerevisiae. Genetics 185, 1193–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Griffith J. L., Coleman L. E., Raymond A. S., Goodson S. G., Pittard W. S., Tsui C., and Devine S. E. (2003) Functional genomics reveals relationships between the retrovirus-like Ty1 element and its host Saccharomyces cerevisiae. Genetics 164, 867–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Risler J. K., Kenny A. E., Palumbo R. J., Gamache E. R., and Curcio M. J. (2012) Host co-factors of the retrovirus-like transposon Ty1. Mob. DNA 3, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Scholes D. T., Banerjee M., Bowen B., and Curcio M. J. (2001) Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics 159, 1449–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sundararajan A., Lee B. S., and Garfinkel D. J. (2003) The Rad27 (Fen-1) nuclease inhibits Ty1 mobility in Saccharomyces cerevisiae. Genetics 163, 55–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ho K. L., Ma L., Cheung S., Manhas S., Fang N., Wang K., Young B., Loewen C., Mayor T., and Measday V. (2015) A role for the budding yeast separase, Esp1, in Ty1 element retrotransposition. PLoS Genet. 11, e1005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bridier-Nahmias A., Tchalikian-Cosson A., Baller J. A., Menouni R., Fayol H., Flores A., Saib A., Werner M., Voytas D. F., and Lesage P. (2015) Retrotransposons. An RNA polymerase III subunit determines sites of retrotransposon integration. Science 348, 585–588 [DOI] [PubMed] [Google Scholar]
- 25. Cramer P., Armache K. J., Baumli S., Benkert S., Brueckner F., Buchen C., Damsma G. E., Dengl S., Geiger S. R., Jasiak A. J., Jawhari A., Jennebach S., Kamenski T., Kettenberger H., Kuhn C. D., et al. (2008) Structure of eukaryotic RNA polymerases. Annu. Rev. Biophys. 37, 337–352 [DOI] [PubMed] [Google Scholar]
- 26. Longtine M. S., McKenzie A. 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., and Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 27. Eichinger D. J., and Boeke J. D. (1988) The DNA intermediate in yeast Ty1 element transposition copurifies with virus-like particles: cell-free Ty1 transposition. Cell 54, 955–966 [DOI] [PubMed] [Google Scholar]
- 28. Shevchenko A., Tomas H., Havlis J., Olsen J. V., and Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 29. Fang N. N., Ng A. H., Measday V., and Mayor T. (2011) Hul5 HECT ubiquitin ligase plays a major role in the ubiquitylation and turnover of cytosolic misfolded proteins. Nat. Cell Biol. 13, 1344–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cox J., Hein M. Y., Luber C. A., Paron I., Nagaraj N., and Mann M. (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 32. Tyanova S., Temu T., Carlson A., Sinitcyn P., Mann M., and Cox J. (2015) Visualization of LC-MS/MS proteomics data in MaxQuant. Proteomics 15, 1453–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lannutti B. J., Persinger J., and Bartholomew B. (1996) Probing the protein-DNA contacts of a yeast RNA polymerase III transcription complex in a crude extract: solid phase synthesis of DNA photoaffinity probes containing a novel photoreactive deoxycytidine analog. Biochemistry 35, 9821–9831 [DOI] [PubMed] [Google Scholar]
- 34. Vannini A., Ringel R., Kusser A. G., Berninghausen O., Kassavetis G. A., and Cramer P. (2010) Molecular basis of RNA polymerase III transcription repression by Maf1. Cell 143, 59–70 [DOI] [PubMed] [Google Scholar]
- 35. Wu C. C., Herzog F., Jennebach S., Lin Y. C., Pai C. Y., Aebersold R., Cramer P., and Chen H. T. (2012) RNA polymerase III subunit architecture and implications for open promoter complex formation. Proc. Natl. Acad. Sci. U.S.A. 109, 19232–19237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu C. C., Lin Y. C., and Chen H. T. (2011) The TFIIF-like Rpc37/53 dimer lies at the center of a protein network to connect TFIIIC, Bdp1, and the RNA polymerase III active center. Mol. Cell. Biol. 31, 2715–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jasiak A. J., Armache K. J., Martens B., Jansen R. P., and Cramer P. (2006) Structural biology of RNA polymerase III: subcomplex C17/25 x-ray structure and 11 subunit enzyme model. Mol. Cell 23, 71–81 [DOI] [PubMed] [Google Scholar]
- 38. Huet J., Riva M., Sentenac A., and Fromageot P. (1985) Yeast RNA polymerase C and its subunits. Specific antibodies as structural and functional probes. J. Biol. Chem. 260, 15304–15310 [PubMed] [Google Scholar]
- 39. Nyswaner K. M., Checkley M. A., Yi M., Stephens R. M., and Garfinkel D. J. (2008) Chromatin-associated genes protect the yeast genome from Ty1 insertional mutagenesis. Genetics 178, 197–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Boeke J. D., Garfinkel D. J., Styles C. A., and Fink G. R. (1985) Ty elements transpose through an RNA intermediate. Cell 40, 491–500 [DOI] [PubMed] [Google Scholar]
- 41. Carter R., and Drouin G. (2010) The increase in the number of subunits in eukaryotic RNA polymerase III relative to RNA polymerase II is due to the permanent recruitment of general transcription factors. Mol. Biol. Evol. 27, 1035–1043 [DOI] [PubMed] [Google Scholar]
- 42. Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., and O'Shea E. K. (2003) Global analysis of protein localization in budding yeast. Nature 425, 686–691 [DOI] [PubMed] [Google Scholar]
- 43. Flores A., Briand J. F., Gadal O., Andrau J. C., Rubbi L., Van Mullem V., Boschiero C., Goussot M., Marck C., Carles C., Thuriaux P., Sentenac A., and Werner M. (1999) A protein-protein interaction map of yeast RNA polymerase III. Proc. Natl. Acad. Sci. U.S.A. 96, 7815–7820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lalo D., Carles C., Sentenac A., and Thuriaux P. (1993) Interactions between three common subunits of yeast RNA polymerases I and III. Proc. Natl. Acad. Sci. U.S.A. 90, 5524–5528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lane L. A., Fernández-Tornero C., Zhou M., Morgner N., Ptchelkine D., Steuerwald U., Politis A., Lindner D., Gvozdenovic J., Gavin A. C., Müller C. W., and Robinson C. V. (2011) Mass spectrometry reveals stable modules in holo and apo RNA polymerases I and III. Structure 19, 90–100 [DOI] [PubMed] [Google Scholar]
- 46. Lorenzen K., Vannini A., Cramer P., and Heck A. J. (2007) Structural biology of RNA polymerase III: mass spectrometry elucidates subcomplex architecture. Structure 15, 1237–1245 [DOI] [PubMed] [Google Scholar]
- 47. Acker J., Conesa C., and Lefebvre O. (2013) Yeast RNA polymerase III transcription factors and effectors. Biochim. Biophys. Acta 1829, 283–295 [DOI] [PubMed] [Google Scholar]
- 48. Cherepanov P., Maertens G. N., and Hare S. (2011) Structural insights into the retroviral DNA integration apparatus. Curr. Opin. Struct. Biol. 21, 249–256 [DOI] [PubMed] [Google Scholar]
- 49. Kenna M. A., Brachmann C. B., Devine S. E., and Boeke J. D. (1998) Invading the yeast nucleus: a nuclear localization signal at the C terminus of Ty1 integrase is required for transposition in vivo. Mol. Cell. Biol. 18, 1115–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Moore S. P., Rinckel L. A., and Garfinkel D. J. (1998) A Ty1 integrase nuclear localization signal required for retrotransposition. Mol. Cell. Biol. 18, 1105–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Panse V. G., Hardeland U., Werner T., Kuster B., and Hurt E. (2004) A proteome-wide approach identifies sumoylated substrate proteins in yeast. J. Biol. Chem. 279, 41346–41351 [DOI] [PubMed] [Google Scholar]
- 52. Li Z., Vizeacoumar F. J., Bahr S., Li J., Warringer J., Vizeacoumar F. S., Min R., Vandersluis B., Bellay J., Devit M., Fleming J. A., Stephens A., Haase J., Lin Z. Y., Baryshnikova A., et al. (2011) Systematic exploration of essential yeast gene function with temperature-sensitive mutants. Nat. Biotechnol. 29, 361–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ji H., Moore D. P., Blomberg M. A., Braiterman L. T., Voytas D. F., Natsoulis G., and Boeke J. D. (1993) Hotspots for unselected Ty1 transposition events on yeast chromosome III are near tRNA genes and LTR sequences. Cell 73, 1007–1018 [DOI] [PubMed] [Google Scholar]
- 54. Curcio M. J., and Garfinkel D. J. (1991) Single-step selection for Ty1 element retrotransposition. Proc. Natl. Acad. Sci. U.S.A. 88, 936–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hare S., Gupta S. S., Valkov E., Engelman A., and Cherepanov P. (2010) Retroviral intasome assembly and inhibition of DNA strand transfer. Nature 464, 232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hoffmann N. A., Jakobi A. J., Moreno-Morcillo M., Glatt S., Kosinski J., Hagen W. J., Sachse C., and Müller C. W. (2015) Molecular structures of unbound and transcribing RNA polymerase III. Nature 528, 231–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Brun I., Sentenac A., and Werner M. (1997) Dual role of the C34 subunit of RNA polymerase III in transcription initiation. EMBO J. 16, 5730–5741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Thuillier V., Stettler S., Sentenac A., Thuriaux P., and Werner M. (1995) A mutation in the C31 subunit of Saccharomyces cerevisiae RNA polymerase III affects transcription initiation. EMBO J. 14, 351–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang Z., and Roeder R. G. (1997) Three human RNA polymerase III-specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes Dev. 11, 1315–1326 [DOI] [PubMed] [Google Scholar]
- 60. Andrau J. C., Sentenac A., and Werner M. (1999) Mutagenesis of yeast TFIIIB70 reveals C-terminal residues critical for interaction with TBP and C34. J. Mol. Biol. 288, 511–520 [DOI] [PubMed] [Google Scholar]
- 61. Bartholomew B., Durkovich D., Kassavetis G. A., and Geiduschek E. P. (1993) Orientation and topography of RNA polymerase III in transcription complexes. Mol. Cell. Biol. 13, 942–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Persinger J., and Bartholomew B. (1996) Mapping the contacts of yeast TFIIIB and RNA polymerase III at various distances from the major groove of DNA by DNA photoaffinity labeling. J. Biol. Chem. 271, 33039–33046 [DOI] [PubMed] [Google Scholar]
- 63. Tate J. J., Persinger J., and Bartholomew B. (1998) Survey of four different photoreactive moieties for DNA photoaffinity labeling of yeast RNA polymerase III transcription complexes. Nucleic Acids Res. 26, 1421–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Werner M., Chaussivert N., Willis I. M., and Sentenac A. (1993) Interaction between a complex of RNA polymerase III subunits and the 70-kDa component of transcription factor IIIB. J. Biol. Chem. 268, 20721–20724 [PubMed] [Google Scholar]
- 65. Kassavetis G. A., Prakash P., and Shim E. (2010) The C53/C37 subcomplex of RNA polymerase III lies near the active site and participates in promoter opening. J. Biol. Chem. 285, 2695–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Landrieux E., Alic N., Ducrot C., Acker J., Riva M., and Carles C. (2006) A subcomplex of RNA polymerase III subunits involved in transcription termination and reinitiation. EMBO J. 25, 118–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chalker D. L., and Sandmeyer S. B. (1992) Ty3 integrates within the region of RNA polymerase III transcription initiation. Genes Dev. 6, 117–128 [DOI] [PubMed] [Google Scholar]
- 68. Sandmeyer S., Patterson K., and Bilanchone V. (2015) Ty3, a position-specific retrotransposon in budding yeast. Microbiol. Spectr. 3, MDNA3-0057-2014 [DOI] [PubMed] [Google Scholar]
- 69. Aye M., Dildine S. L., Claypool J. A., Jourdain S., and Sandmeyer S. B. (2001) A truncation mutant of the 95-kilodalton subunit of transcription factor IIIC reveals asymmetry in Ty3 integration. Mol. Cell. Biol. 21, 7839–7851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yieh L., Hatzis H., Kassavetis G., and Sandmeyer S. B. (2002) Mutational analysis of the transcription factor IIIB-DNA target of Ty3 retroelement integration. J. Biol. Chem. 277, 25920–25928 [DOI] [PubMed] [Google Scholar]
- 71. Connolly C. M., and Sandmeyer S. B. (1997) RNA polymerase III interferes with Ty3 integration. FEBS Lett. 405, 305–311 [DOI] [PubMed] [Google Scholar]
- 72. Deleted in proof
- 73. Rinckel L. A., and Garfinkel D. J. (1996) Influences of histone stoichiometry on the target site preference of retrotransposons Ty1 and Ty2 in Saccharomyces cerevisiae. Genetics 142, 761–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Craigie R., and Bushman F. D. (2014) Host factors in retroviral integration and the selection of integration target sites. Microbiol. Spectr. 2, MDNA3–0026-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Barnes C. O., Calero M., Malik I., Graham B. W., Spahr H., Lin G., Cohen A. E., Brown I. S., Zhang Q., Pullara F., Trakselis M. A., Kaplan C. D., and Calero G. (2015) Crystal structure of a transcribing RNA polymerase II complex reveals a complete transcription bubble. Mol. Cell 59, 258–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., and Boeke J. D. (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132 [DOI] [PubMed] [Google Scholar]
- 77. Tong A. H., Lesage G., Bader G. D., Ding H., Xu H., Xin X., Young J., Berriz G. F., Brost R. L., Chang M., Chen Y., Cheng X., Chua G., Friesen H., Goldberg D. S., et al. (2004) Global mapping of the yeast genetic interaction network. Science 303, 808–813 [DOI] [PubMed] [Google Scholar]
- 78. Giaever G., Chu A. M., Ni L., Connelly C., Riles L., Véronneau S., Dow S., Lucau-Danila A., Anderson K., André B., Arkin A. P., Astromoff A., El-Bakkoury M., Bangham R., Benito R., et al. (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391 [DOI] [PubMed] [Google Scholar]