Abstract
The small GTPase DiRas1 has tumor-suppressive activities, unlike the oncogenic properties more common to small GTPases such as K-Ras and RhoA. Although DiRas1 has been found to be a tumor suppressor in gliomas and esophageal squamous cell carcinomas, the mechanisms by which it inhibits malignant phenotypes have not been fully determined. In this study, we demonstrate that DiRas1 binds to SmgGDS, a protein that promotes the activation of several oncogenic GTPases. In silico docking studies predict that DiRas1 binds to SmgGDS in a manner similar to other small GTPases. SmgGDS is a guanine nucleotide exchange factor for RhoA, but we report here that SmgGDS does not mediate GDP/GTP exchange on DiRas1. Intriguingly, DiRas1 acts similarly to a dominant-negative small GTPase, binding to SmgGDS and inhibiting SmgGDS binding to other small GTPases, including K-Ras4B, RhoA, and Rap1A. DiRas1 is expressed in normal breast tissue, but its expression is decreased in most breast cancers, similar to its family member DiRas3 (ARHI). DiRas1 inhibits RhoA- and SmgGDS-mediated NF-κB transcriptional activity in HEK293T cells. We also report that DiRas1 suppresses basal NF-κB activation in breast cancer and glioblastoma cell lines. Taken together, our data support a model in which DiRas1 expression inhibits malignant features of cancers in part by nonproductively binding to SmgGDS and inhibiting the binding of other small GTPases to SmgGDS.
Keywords: breast cancer; glioblastoma; GTPase Kras (KRAS); Ras homolog gene family, member A (RhoA); Ras-related protein 1 (Rap1); Di-Ras1; NF-kappaB (NF-KB); Rap1GDS1; Rig; SmgGDS
Introduction
Small GTPases, including oncogenic Rho and Ras family members, are important in the development and progression of a number of malignancies (reviewed in Refs. 1–3). Small GTPase activation depends upon binding to GTP, which is facilitated when guanine nucleotide exchange factors (GEFs)2 promote the exchange of GDP for GTP. Inactivation of small GTPases is promoted by GTPase-activating proteins (GAPs) that increase the rate of hydrolysis of GTP to GDP. Altering the levels or availability of GEFs and GAPs for small GTPases can modulate their activation in a number of cancers. Novel ways in which to alter the balance of these proteins may prove to be important therapeutically for a variety of malignancies.
SmgGDS (Rap1GDS1) is a noncanonical GEF for RhoA and RhoC (4) and also promotes the pro-oncogenic functions of several other small GTPases with C-terminal polybasic regions (PBRs) (5). There are two predominant isoforms of SmgGDS, SmgGDS-607 and SmgGDS-558, which differ by one armadillo domain (5). Recent studies have demonstrated that SmgGDS-558 plays a more significant role than SmgGDS-607 in activating RhoA in breast cancer cells, despite lower levels of endogenous SmgGDS-558 protein (6, 7). In addition, SmgGDS promotes RhoA-mediated NF-κB transcriptional activity (6, 8, 9), which is critical to cancer cell growth and proliferation (10).
Although the unique DiRas (Distinct subgroup of the Ras family) family of small GTPases shares homology with the pro-oncogenic Ras GTPases, it has tumor-suppressive actions. DiRas1 (also known as Di-Ras1 or Rig) has been reported to be a tumor suppressor in gliomas (11, 12) and in esophageal squamous cell carcinomas (13). DiRas1 is closely related to its family members DiRas2 (Di-Ras2) and DiRas3 (ARHI, Noey2) (11). DiRas2 appears to be predominantly expressed in the brain (11), whereas DiRas3 is a tumor suppressor in breast and ovarian cancers (14). It has not been determined whether DiRas1, like DiRas3, is also expressed in normal breast tissue and lost in a proportion of breast cancers. Moreover, the mechanisms by which DiRas1 inhibits tumor growth are not fully characterized.
Overexpression of DiRas1 inhibits Ras-mediated transformation of NIH3T3 cells and inhibits the growth of glioblastoma and esophageal squamous cell cancer cell lines (12, 13). In addition, DiRas1 signaling diminished BAD serine phosphorylation, which can promote cell death, and decreased matrix metalloproteinase 2/9 expression (13). In esophageal cancers, DiRas1 appeared to decrease ERK1/2 and MAPK-mediated signals, leading to increased cell death, decreased migration, and decreased invasion (13).
DiRas1 may function via nonproductive associations with effectors or activators of pro-oncogenic small GTPases, similar to how Rap1A (15) and Rheb (16) antagonize Ras signaling. DiRas1 was reported to bind to the effector domain of C-RAF in cells (12), but a yeast two-hybrid screen detected no association between DiRas1 and C-RAF or B-RAF (11). Interestingly, a number of Ras- and Rap-specific GEFs and GAPs did not mediate GTP exchange or hydrolysis of DiRas1, although Rap1GAP1/2 could hydrolyze GTP on DiRas1 (17). We hypothesize that rather than sequestering effectors for pro-oncogenic small GTPases, DiRas1 may act as a tumor suppressor by sequestering GEFs for these small GTPases.
Here, we identified DiRas1 as a binding partner for SmgGDS. Our in silico docking analysis predicted that DiRas1 can compete with other small GTPases, such as RhoA and K-Ras4B, for SmgGDS binding. Consistent with this prediction, DiRas1 potently inhibited interactions of SmgGDS with a broad range of pro-oncogenic small GTPases, including RhoA, K-Ras4B, and Rap1A. In addition, DiRas1 inhibited basal and RhoA-mediated NF-κB activity in HEK293T, glioblastoma, and breast cancer cell lines. Taken together, these findings identify a novel way in which the tumor suppressive GTPase DiRas1 represses signals mediated by several pro-oncogenic Ras and Rho family GTPases.
Experimental Procedures
cDNA Constructs
Constructs encoding N-terminal Myc-tagged or HA-tagged small GTPases and C-terminal HA-tagged SmgGDS constructs were created as described previously (5, 18, 19). DiRas1 cDNA constructs in the pcDNA3.1 vector were purchased from cDNA.org, and dominant-negative mutants were purchased from Top Gene Technologies. RhoA and SmgGDS cDNAs in pLIC-His were kind gifts from John Sondek (University of North Carolina) and were created as described previously (20–22). Full-length DiRas1 in pLIC-His or pETM11 was created by subcloning DiRas1 from DiRas1-pcDNA3.1 (Top Gene Technologies). All cDNA sequences were verified by DNA sequencing of the entire ORF.
Cell Lines and Transfections
HEK293T, U87, T47D, and MCF-7 cells were obtained from the American Type Culture Collection, and U251 cells were obtained from Sigma. Cells were maintained in high glucose DMEM with l-glutamine medium with 10% heat-inactivated FBS, except for MCF-7 cells, which were maintained as indicated by the American Type Culture Collection. Cell cultures were supplemented with penicillin and streptomycin (Life Technologies). All cDNAs were transfected into cells using Lipofectamine 2000 (Life Technologies) according to the manufacturer's protocol.
Docking and Modeling Studies
A model for SmgGDS-607 (UniProt P52306-1) was created using the I-TASSER 2.1 standalone modeler (23). The 607 isoform was then manually converted into the SmgGDS-558 isoform (P52306-2) followed by loop reconstructions using YASARA homology modeling (24). A model for DiRas1 (O95057, amino acids 1–195) was created using YASARA homology modeling. Global docking of DiRas1 (ligand) to SmgGDS-558 (receptor) was performed using AutoDock (25), calculating 50 docking predictions on five receptor ensembles for a total of 250 docking predictions. Following cluster analysis of the docking results in YASARA, the top 10 conformations were energy minimized using the NOVA force field (26), with water added to 0.997 g/ml, and a final energy minimization with the AMBER03 (27) force field was performed. Binding energy for the top 10 conformations was determined in kcal/mol, factoring out water. The electrostatic surface for DiRas1 was calculated with a static Poisson-Boltzmann Solver. Models for RhoA, K-Ras4B, and Rap1A were then structurally aligned against the top docking conformation of DiRas1 using the MUSTANG algorithm (28).
Co-immunoprecipitation Assays
HA-SmgGDS-558 cDNA constructs alone or in combination with cDNA constructs encoding Myc-tagged WT GTPases were transfected into HEK293T cells. Constructs encoding DiRas1 with an HA tag (rather than a Myc tag) were also used in some experiments. After 24 h, the cells were lysed and immunoprecipitated with HA-conjugated agarose beads (Sigma), and the immunoprecipitates were subjected to Western blotting.
In Vitro Transcription and Translation Assays
The indicated cDNAs were transcribed and translated using the TnT quick coupled transcription/translation system (Promega) with [35S]methionine per the manufacturer's instructions. Translated proteins were then incubated and immunoprecipitated using anti-HA antibody, separated by SDS-PAGE, and examined by autoradiography, as described previously (19).
ECL-Western Blotting
Equal numbers of transfected cells were boiled in Laemmli sample buffer and subjected to electrophoresis using precast Bis-Tris 3–20% gels (Life Technologies) or 10% SDS-PAGE gels (for transcription and translation assays). The proteins were transferred to PVDF and immunoblotted using antibodies against SmgGDS (BD Transduction Laboratories; 612511), GAPDH (Santa Cruz Biotechnology; sc-32233), Myc (Covance; PRB-150P), HA (Covance; MMS-101P), RhoA (Cytoskeleton; ARH03-A), and DiRas1 (Proteintech; 12634-AP). Bound antibodies were visualized using HRP-linked secondary antibodies (GE Healthcare), as previously described (6).
Protein Expression and Purification
SmgGDS and GTPase constructs were produced in BL21 (DE3) Escherichia coli, as previously described (4). After harvesting and lysing bacterial cells, the His6-tagged proteins were purified via Ni2+ affinity chromatography. The proteins were then concentrated, and the final protein concentration was determined using A280 or the bisinchoninic acid protein assay (Pierce) per the manufacturer's instructions prior to storage at −80 °C.
Guanine Nucleotide Exchange Assays
The ability of SmgGDS to mediate guanine nucleotide exchange was determined using the fluorescent nucleotide analog N-methylanthraniloyl (MANT)-GTP as previously described (29). Exchange assays were performed with a LS-55 fluorescence spectrometer (PerkinElmer Life Sciences) with λex = 360 nm and λem = 430 nm and slits of 5 nm. The exchange buffer contained 50 mm NaCl, 20 mm Tris-HCl (pH 8.0), 5 mm MgCl2, 5% glycerol, 1 mm DTT, and 400 nm MANT-GTP. The exchange buffer, containing either SmgGDS (final concentration, 20 μm) or EDTA (final concentration, 10 μm), was allowed to equilibrate to reach baseline before RhoA or DiRas1 was added (2 μm) at the indicated times.
Determination of SmgGDS Binding Affinity
SmgGDS-558 was biotinylated with NHS-PEG4-Biotin (Thermo Scientific) at a 1:1 molar ratio, diluted to 50 ng/μl in binding buffer (50 mm Hepes, pH 7.8, 150 mm NaCl, 2 mm TECP, 2 mm MgCl2, 20 μm GDP, and 0.25 mg/ml BSA) before binding to streptavidin probes using an Octet Red (ForteBio) 96-well biolayer interferometry analysis system. After washing in binding buffer (180 s), SmgGDS-loaded probes were cycled through wells in the following order: binding buffer (baseline, 120 s), RhoA or DiRas1 (association, 900 s), binding buffer (dissociation, 600 s), and binding buffer with 1 m NaCl (regeneration, 180 s). The cycle was repeated for each concentration of GTPase tested. After subtracting the signal from a control probe without SmgGDS, the steady-state response at equilibrium during each association phase was fit to the following equation by nonlinear regression to determine Kd: Response at equilibrium = Ymax * [GTPase]/(Kd + [GTPase]).
Normal and Tumor Tissues
Tissue microarrays of archival normal breast tissue and breast cancers were analyzed, with two cores per sample represented on the arrays. For noncancerous brain tissue, archived frozen tissue from surgery in epileptic patients was obtained from the Medical College of Wisconsin Brain and Spinal Cord Tissue Bank. All protocols were approved by the Medical College of Wisconsin Institutional Review Board.
Immunohistochemical (IHC) Staining
IHC staining was performed on tissues fixed in 10% neutral-buffered formalin. After dewaxing, the samples were treated with antigen retrieval solution (10 mm citrate buffer, pH 6.0) for 10 min at 95 °C. Endogenous peroxidase was blocked using hydrogen peroxide followed by a serum block (Vector Laboratories). DiRas1 staining (1:300; Sigma; HPA050164) or isotype control staining was performed overnight at 4 °C. A biotinylated secondary antibody, Vectastain ABC kit, and diaminobenzidine peroxidase substrate (Vector Laboratories) were used with hematoxylin counterstain (Invitrogen). For human cortex tissue, a pathologist (A. C. M.) determined the localization of positive staining. Normal human breast tissue samples were verified by a practicing breast pathologist (I. A.-B.). For human breast tissue and breast cancer samples, staining was classified as previously described (8). Briefly, the cells were assigned a score based on intensity: 0 (negative), 1 (weak), 2 (moderate), or 3 (strong). The percent positive cells in normal ductal tissue or tumor cells was scored as: 0 (0%), 1 (<10%), 2 (11–50%), 3 (51–75%), and 4 (76–100%). Each blinded sample was scored by a trained technician and a physician, and the average score was calculated. Each sample was then assigned an immunoreactive score (IRS), which is the product of the intensity and percent positive scores (8).
NF-κB Transcriptional Activation Assays
U87, U251, T47D, and MCF7 cells were transfected with the indicated cDNAs, as well as both the pNifty-Luciferase NF-κB luciferase reporter (Invivogen) and β-gal reporter plasmids in 48-well plates using Lipofectamine 2000 (Life Technologies), as previously described (6). HEK293T cells were transfected with the indicated cDNAs, as well as the pNifty-Luc NF-κB luciferase reporter plasmid in 6-well plates. After 24 h, luminescence was quantified by adding luciferin (0.15 mg/ml) to each well and measuring luminescence with a FLUOstar Omega plate reader. Transfection efficiency/cell number was normalized by cell counting (HEK293T cells) after luminescence measurements or by measuring β-gal activity by washing cells with PBS, incubating the wells with a β-gal reagent (Pierce) for 30 min, and measuring absorbance at 405 nm. NF-κB activity was calculated using the luminescence value divided by the optical density and normalized to values obtained for the vector-transfected cells.
RT-PCR
Total RNA was extracted from U87 and U251 cells transfected with vector or DiRas1 for 72 h using Quick RNA Mini Prep (Zymo Research) and was reverse transcribed with iScript cDNA synthesis kit (Bio-Rad) according to themanufacturer's instructions. PCR was carried out to detect levels of IL-8 and r18S using a GeneMate GCL-48 Thermal Cycler. Three independent experiments were performed to analyze the relative gene expression. PCR primers are as follows: IL-8, CTTGGCAGCCTTCCTGATTTCT and GTTTTCCTTGGGGTCCAGACAG and r18S, TGAGGCCATGATTAAGAGGG and AGTCGGCATCGTTTATGGTC.
Statistical Analysis
The results are the means ± S.E. Symbols above a column indicate a statistical comparison between the control and experimental group by unpaired Student's t test, Mann-Whitney U test, or one-way analysis of variance with Dunnett's multiple comparison test, as indicated in the figure legends. p values less than 0.05 were considered significant.
Results
DiRas1 Associates with the RhoGEF SmgGDS
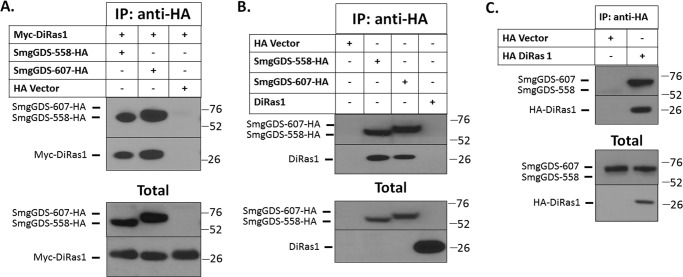
The RhoGEF SmgGDS is predominantly expressed as a long form (SmgGDS-607) or a splice variant lacking one of its armadillo domains (SmgGDS-558) (5). We aimed to identify novel proteins interacting with SmgGDS by utilizing LC/MS to detect proteins that co-immunoprecipitated with SmgGDS-558-HA and SmgGDS-607-HA in HEK293T cells. Among the proteins that co-precipitated with HA-tagged SmgGDS, we identified the small GTPase DiRas1, a poorly characterized Ras family small GTPase with tumor-suppressive functions (11–13). We confirmed that DiRas1 interacts with SmgGDS in HEK293T cells by immunoprecipitating HA-tagged SmgGDS and detecting both Myc-tagged DiRas1 (Fig. 1A) and endogenous DiRas1 (Fig. 1B) in the immunoprecipitates. The immunoprecipitation of HA-tagged DiRas1 also pulls down endogenous SmgGDS (Fig. 1C). Although the levels of endogenous DiRas1 in HEK293T cell lysates were below our limit of detection via Western blotting, we found that immunoprecipitation could enrich DiRas1 to levels that are detectable in the Western blots (Fig. 1B).
FIGURE 1.
DiRas1 binds the small GTPase-modulating protein SmgGDS. A, HEK293T cells were co-transfected with SmgGDS-HA and vector or Myc-DiRas1 cDNAs and after 24 h were lysed, immunoprecipitated with anti-HA antibodies, and subjected to Western blotting with anti-HA and anti-Myc antibodies. B, HEK293T cells were transfected with cDNAs for HA-tagged SmgGDS-558 and SmgGDS-607. After 24 h, the association of endogenous DiRas1 with SmgGDS-HA was examined as in A. Western blotting of the total cell lysates (total) shows endogenous DiRas1 and HA-tagged SmgGDS. C, HA-DiRas1 was transfected into HEK293T cells, and the association of endogenous SmgGDS was examined after 24 h via immunoprecipitation as in A. Western blotting was performed for endogenous SmgGDS and HA-tagged DiRas1. The results are representative of three independent experiments.
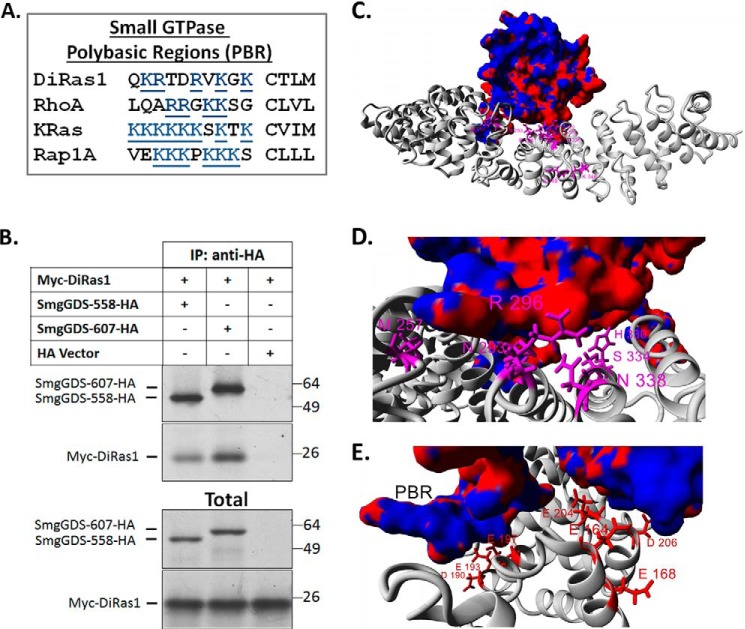
SmgGDS interacts with small GTPases containing C-terminal PBRs, which are thought to interact with an electronegative patch in SmgGDS (4, 5). DiRas1 contains basic amino acids in its C-terminal domain, indicating a PBR (Fig. 2A). Our in vitro transcription and translation assays demonstrated direct binding between SmgGDS and DiRas1 (Fig. 2B). To further examine potential binding interactions between SmgGDS-558 and DiRas1, in silico docking studies were performed using a homology model of SmgGDS-558 and DiRas1 (primarily based on the DiRas1 crystal structure; Protein Data Bank code 2gf0). In silico analysis, coupled with analysis of SmgGDS mutations that affect RhoA GTPase activity (4), revealed that DiRas1 likely binds to SmgGDS using a binding pocket similar to that used by RhoA, K-Ras4B, and Rap1A (Fig. 2, C–E). This modeling suggested that binding of DiRas1 to SmgGDS inhibits interactions with other Ras and Rho GTPases.
FIGURE 2.
DiRas1 is predicted to bind to SmgGDS in the same way that RhoA binds SmgGDS. A, the PBRs of DiRas1 and other small GTPases known to bind to SmgGDS are shown. DiRas1 has a PBR containing basic residues, similar to the other small GTPases shown. B, Myc-DiRas1 and HA-SmgGDS cDNAs were translated in the presence of [35S]methionine in in vitro transcription and translation assays. In vitro translated proteins were co-incubated and immunoprecipitated (IP) with anti-HA antibodies, and Western blotting was performed with anti-HA and anti-Myc antibodies. C–E, in silico docking studies of DiRas1 (electrostatic surface) on SmgGDS-558 (gray) suggest that DiRas1 binds to SmgGDS similarly to the way in which RhoA binds SmgGDS. The top docking conformation for DiRas1 is shown as an electrostatic surface plot (red, electronegative; blue, electropositive). SmgGDS variants known to alter RhoA guanine nucleotide exchange (4) are colored magenta (D), and electrostatic amino acids needed for RhoA binding are colored red (E). The close proximity of these amino acids to the docking interface suggests the DiRAs1 and RhoA bind to SmgGDS in a similar manner.
SmgGDS Is Not a GEF for DiRas1 and Binds DiRas1 with a Higher Affinity than RhoA
SmgGDS is a GEF for RhoA and RhoC, but not for K-Ras, Rap1A, or Rac1 in vitro (4). We sought to determine whether SmgGDS acts as a GEF for DiRas1, because there are no known GEFs for DiRas1. The GEF activity of SmgGDS for RhoA is not isoform-dependent, and SmgGDS-558 appears to promote malignant phenotypes more potently than the full-length SmgGDS protein (6). Therefore, we utilized SmgGDS-558 for the remainder of our studies.
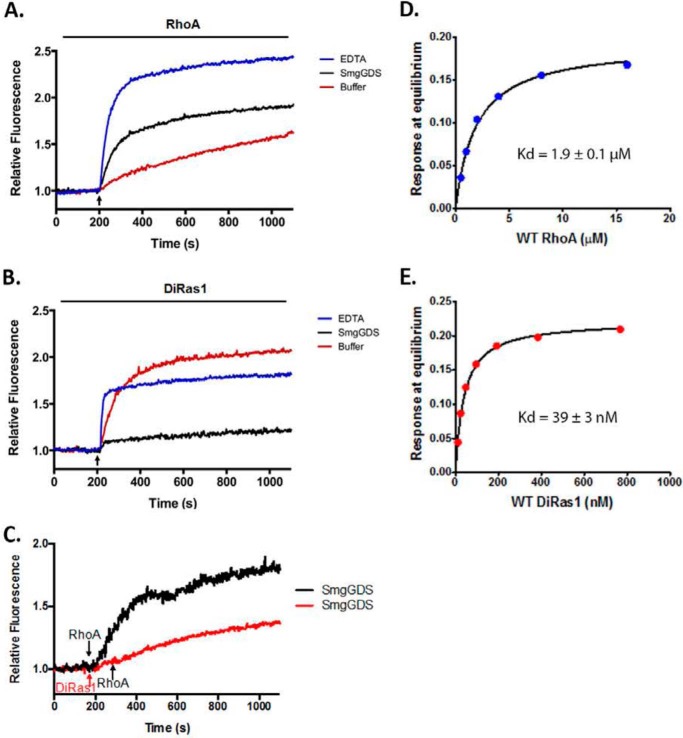
We examined whether SmgGDS promotes GDP to GTP exchange in vitro using nucleotide exchange assays. SmgGDS activates RhoA (Fig. 3A), consistent with other studies (4). In our assays, DiRas1 alone became GTP bound at a relatively high rate in the absence of EDTA or a GEF (Fig. 3B, red curve). This observation is consistent with reports that DiRas1 has a high guanine nucleotide off rate and low intrinsic GTPase activity, resulting in the high percentage of GTP-bound DiRas1 found in cells (11, 17). However, when DiRas1 was exposed to SmgGDS, GTP exchange failed to occur (Fig. 3B, black curve). This unexpected finding indicates that SmgGDS binds DiRas1 and inhibits its ability to undergo GDP/GTP exchange. We examined whether the presence of DiRas1 inhibited SmgGDS-mediated RhoA GDP/GTP exchange in vitro as well. As shown in Fig. 3C, RhoA demonstrates less GDP/GTP exchange when DiRas1 is present (red curve) than when no DiRas1 is present (black curve). Thus, DiRas1 nonproductively associates with SmgGDS, and this association can decrease RhoA GDP/GTP exchange.
FIGURE 3.
SmgGDS is not a GEF for DiRas1, and SmgGDS exhibits a higher affinity for DiRas1 than for RhoA. A and B, MANT-GTP guanine nucleotide exchange assays demonstrate that SmgGDS can mediate guanine nucleotide exchange of RhoA (A) but does not mediate guanine nucleotide exchange of DiRas1 (B). Fluorescence with exchange buffer alone or with SmgGDS (final concentration, 20 μm) or EDTA (final concentration, 10 μm) was measured. At the indicated time (arrow), RhoA or DiRas1 (2 μm) was added, and nucleotide exchange was measured. Relative fluorescent values were obtained by normalizing values to the baseline fluorescence value. C, MANT-GTP assay demonstrates that DiRas1 can inhibit guanine nucleotide exchange of RhoA in the presence of SmgGDS. Using the same methods as in A and B, fluorescence with SmgGDS (final concentration, 20 μm) was measured. For the black curve, RhoA (2 μm, black arrow) was added to SmgGDS at the indicated time. For the red curve, DiRas1 (2 μm, red arrow) was added to SmgGDS at the indicated time, and RhoA was added after an additional 60 s, as indicated (black arrow). D and E, the binding affinity of SmgGDS and RhoA (D) and SmgGDS and DiRas1 (E) was determined using a biolayer interferometry assay as described under “Experimental Procedures.” The Kd for SmgGDS and RhoA was 1.9 ± 0.1 μm, whereas the Kd for SmgGDS and DiRas1 was 39 ± 3 nm. The results are representative of two or more independent experiments.
We subsequently examined whether SmgGDS exhibited different binding affinities for DiRas1 and RhoA using a biolayer interferometry assay. These studies demonstrated a much stronger binding affinity of SmgGDS for DiRas1, with a Kd of 1.9 ± 0.1 μm for RhoA (Fig. 3D) and a Kd of 39 ± 3 nm for DiRas1 (Fig. 3E).
DiRas1 Expression Abrogates Binding of Pro-oncogenic Small GTPases to SmgGDS
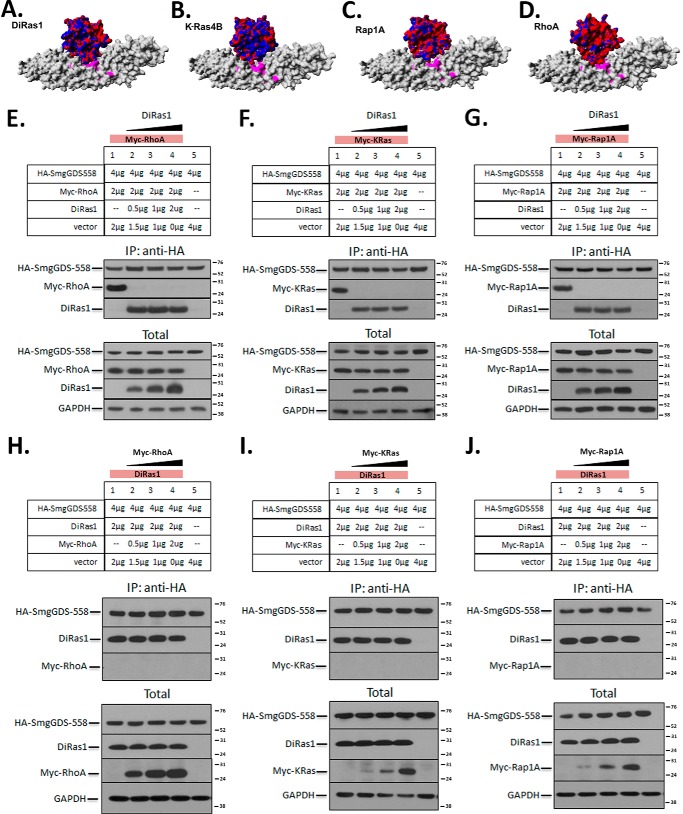
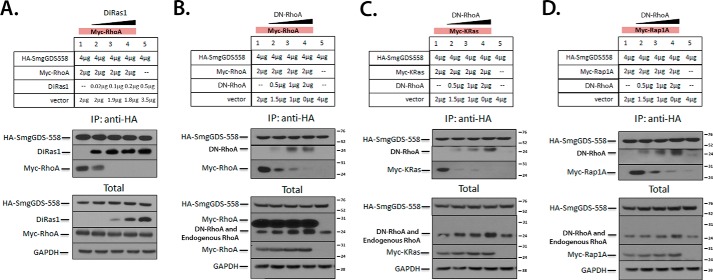
Our analyses predicted that DiRas1 binds to SmgGDS in a similar manner to a number of pro-oncogenic small GTPases (Fig. 4, A–D), which led us to examine whether DiRas1 expression decreased the binding of other small GTPases to SmgGDS. As shown in Fig. 4 (E–G), DiRas1 potently inhibited the detectable interactions of SmgGDS with RhoA, K-Ras4B, and Rap1A, even when the DiRas1 cDNA that was transfected was only 25% of the molar ratio of the cDNA encoding the pro-oncogenic small GTPases (lane 2 in Fig. 4, E–G). Increasing levels of RhoA, Kras4B, or Rap1A (Fig. 4, H–J, respectively), transfected with up to a 1:1 molar ratio of cDNA with DiRas1 (lane 4 in Fig. 4, H–J), did not cause decreased interactions of DiRas1 and SmgGDS. Progressively decreasing the amount of untagged DiRas1 transfected for co-immunoprecipitation studies reveals that transfection an ∼1% molar ratio of DiRas1:Myc-RhoA cDNA results in a detectable amount of Myc-RhoA binding to SmgGDS-558-HA (lane 2 in Fig. 5A). These results support our hypothesis that DiRas1 manifests its tumor-suppressive function in part by sequestering SmgGDS from pro-oncogenic small GTPases, much as a dominant-negative small GTPase may function (30). Indeed, increasing levels of DN-RhoA causes decreased binding of WT RhoA, WT KRas, and WT Rap1A (Fig. 5, B–D, respectively), similar to results seen with WT DiRas1 expression (Fig. 4, E–G).
FIGURE 4.
DiRas1 expression potently inhibits SmgGDS association with other small GTPases. A–D, in silico docking studies suggest that DiRas1 (A, electrostatic surface) binds to SmgGDS-558 (gray surface plot) in a similar manner to K-Ras4B (B), Rap1A (C), and RhoA (D). The color scheme is as defined in Fig. 2. E–G, SmgGDS-558-HA and Myc-tagged RhoA (E), KRas-4B (F), and Rap1A (G) cDNAs were co-transfected into HEK293T cells along with increasing amounts of DiRas1 cDNA as indicated, such that all cells were transfected with identical amounts of total cDNA. After 24 h the cells were lysed, immunoprecipitated with anti-HA antibodies, and subjected to Western blotting for HA and Myc tags, as well as DiRas1 and GAPDH (lysates). DiRas1 expression inhibited SmgGDS association with all other small GTPases examined. The results are representative of three independent experiments. H–J, SmgGDS-558-HA and increasing amounts of Myc-tagged RhoA (H), KRas-4B (I), and Rap1A (J) cDNAs were co-transfected into HEK293T cells along with a constant level of DiRas1 cDNA as indicated, such that all cells were transfected with identical amounts of total cDNA. After 24 h, the cells were lysed, immunoprecipitated with anti-HA antibodies, and subjected to Western blotting as described in E. The results are representative of two or more independent experiments.
FIGURE 5.
Diras1 potently inhibits binding of RhoA to SmgGDS and acts in a fashion similar to DN-RhoA. A, co-transfection of tagged SmgGDS and RhoA in HEK293T cells was performed as in Fig. 4E, with much lower levels of increasing untagged DiRas1 (amounts indicated), such that all cells were transfected with identical amounts of total cDNA. After 24 h the cells were lysed, immunoprecipitated with anti-HA antibodies, and subjected to Western blotting for HA and Myc tags, as well as DiRas1 and GAPDH (lysates). B–D, DN-RhoA inhibits small GTPase binding to SmgGDS in a manner similar to WT DiRas1. Co-transfections of HEK293T cells were performed as indicated in Fig. 4E, with constant levels of Myc-tagged RhoA (B), KRas (C), and Rap1A (D), and increasing levels of untagged DN-RhoA, as indicated. The cells were lysed after 24 h, and immunoprecipitation with anti-HA antibodies and Western blotting were performed as follows (top to bottom in each section): anti-HA, anti-RhoA (showing immunoprecipitated untagged DN-RhoA), and anti-Myc. In the lysate panels, from top to bottom, antibodies used were: anti-HA, untagged RhoA (which binds tagged RhoA in B, untagged DN-RhoA, and endogenous RhoA, as indicated), Myc tag, and GAPDH. The results are representative of two or more independent experiments.
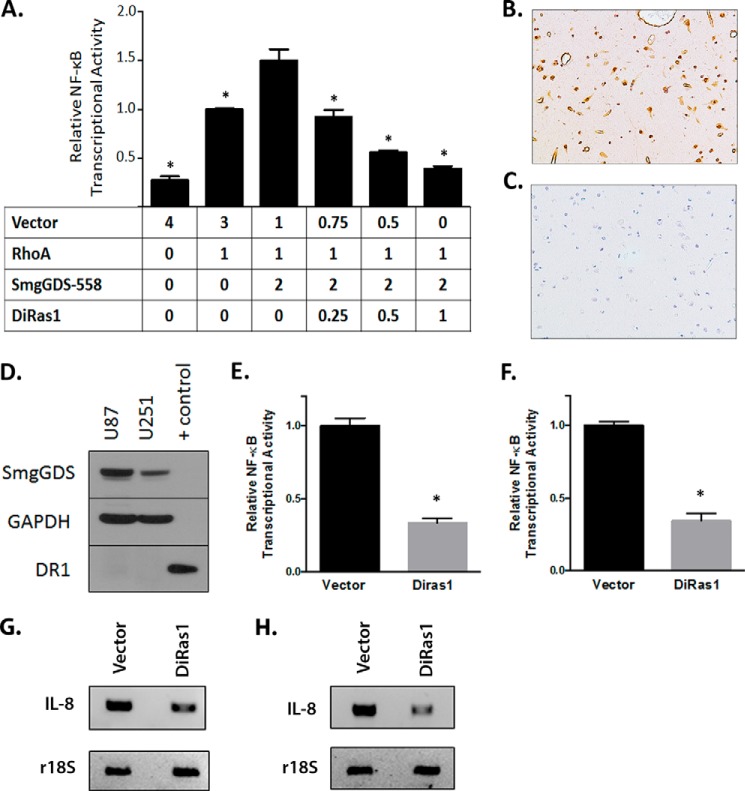
DiRas1 Expression Inhibits SmgGDS- and RhoA-mediated NF-κB Transcriptional Activity
Our observation that DiRas1 inhibits the interactions of pro-oncogenic small GTPases with SmgGDS (Figs. 4 and 5A) led us to characterize the functional consequences of this inhibition. Small GTPases, such as RhoA, can activate the NF-κB pathway (31–33). In addition, SmgGDS-558 can promote RhoA-mediated NF-κB transcriptional activity in cancer cells (6). We found that DiRas1 expression inhibits RhoA- and SmgGDS-mediated NF-κB transcriptional activity in HEK293T cells (Fig. 6A). DiRas1 expression was previously demonstrated to be absent from a large proportion of high grade gliomas (12), but expression of DiRas1 in normal gliomas cells has not been demonstrated. Using an antibody that was commercially validated and verified by our testing to be sensitive to DiRas1 expression (data not shown), we found that DiRas1 was expressed in glial cells, as well as neurons and the microvasculature of noncancerous human cerebral cortex (Fig. 6B). Using an isotype control, no staining was detectable (Fig. 6C). Consistent with a previous study showing no detectable DiRas1 expression in a large proportion of high grade human gliomas (12), the U87 and U251 glioblastoma cell lines did not express detectable amounts of DiRas1 (Fig. 6D). We next investigated whether introduced expression of DiRas1 inhibits endogenous NF-κB transcriptional activation in U87 and U251 glioblastoma tumor cells. DiRas1 inhibited basal NF-κB transcriptional activity in U87 and U251 cells (Fig. 6, E and F), suggesting that loss of this tumor-suppressive GTPase in glioblastoma tumors can enhance NF-κB transcriptional activation. We then examined whether mRNA expression of the NF-κB-regulated cytokine, IL-8, was altered with DiRas1 expression in the glioblastoma cell lines. Using RT-PCR, we found that IL-8 transcript levels were decreased when DiRas1 was expressed in U87 and U251 cells (Fig. 6, G and H). Future studies will be needed to determine the functional significance of DiRas1-mediated altered NF-κB transcriptional activity in normal tissues and tumors.
FIGURE 6.
DiRas1 inhibits RhoA- and SmgGDS-mediated NF-κB transcriptional activity. A, DiRas1 expression mitigates RhoA- and SmgGDS-induced NF-κB activation. HEK293T cells were co-transfected with NF-κB reporter plasmid, along with cDNAs encoding RhoA, SmgGDS-558, DiRas1, and empty vector in the amounts indicated in μg. After 24 h, luminescence was quantified and normalized, with RhoA-mediated activation set as 1.0. The results are from three independent experiments. *, p < 0.0001 via one-way analysis of variance with Dunnett's multiple comparison tests, when compared with the cells expressing SmgGDS and RhoA. B and C, IHC staining of nonmalignant human cortex tissue reveals DiRas1 staining (B) in glial cells and other cell types (20× magnification), with no staining using an isotype control (C). D, glioblastoma U87 and U251 cells lines (50 μg of lysate each) lack detectable DiRas1 protein expression. Control cell lysate (3 μg) is HEK293T cells expressing DiRas1 cDNA. E and F, re-expressing DiRas1 in glioblastoma cell lines reduces basal NF-κB transcriptional activity. U87 (E) and U251 (F) cells were transfected with empty vector or DiRas1 and NF-κB reporter and β-gal. After 24 h, luminescence was quantified and normalized. The results are the means ± S.E. from at least three independent experiments. *, p < 0.0001 compared with vector-transfected cells using the Mann-Whitney U test. G and H, mRNA expression of the NF-κB regulated gene IL-8 is decreased in U87 (G) and U251 (H) cells expressing DiRas1 cDNA, compared with vector-transfected cells. The cells were transfected with vector or DiRas1 cDNA for 72 h, total RNA was extracted and reverse transcribed to cDNA, and RT-PCR was performed, using r18S as a control. The results are representative of three independent experiments.
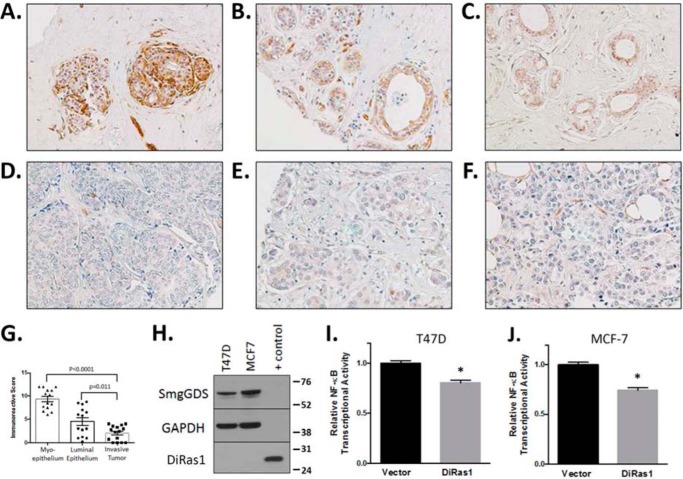
Because a separate DiRas family member, DiRas3 (ARHI), is an established tumor suppressor in breast cancer (14), and SmgGDS- and Rho-mediated NF-κB transcriptional activation has been reported in breast cancer (6), we examined DiRas1 protein expression in normal human breast epithelial tissue and in human breast cancers by performing IHC on a tissue microarray containing normal breast tissue samples (n = 15) and invasive ductal carcinoma cases (n = 15). DiRas1 expression was scored blindly by two independent observers, which demonstrated that 13 of 15 normal breast apical ductal epithelium samples stained positively, whereas the invasive ductal carcinoma samples expressed lower levels of DiRas1, with a mean IRS of 4.50 versus 2.05 for normal tissue versus tumors, respectively (p = 0.011; Table 1). Myoepithelial cells expressed even higher levels of DiRas1, with a mean IRS of 9.04 (p < 0.0001 versus invasive ductal carcinoma; Table 1). Representative images from the DiRas1 IHC demonstrate that most normal human apical ductal epithelial cells and myoepithelial cells stained positively for DiRas1 (Fig. 7, A–C), whereas breast tumor cells from human invasive ductal carcinomas do not demonstrate as intense DiRas1 staining (Fig. 7, D–F). However, examining DiRas1 expression in normal breast epithelial tissue demonstrates a range of expression scores (Fig. 7G). Future studies will characterize DiRas1 expression patterns in different molecular subtypes of invasive breast cancers and whether DiRas1 expression correlates with prognosis. To examine whether DiRas1 also affects NF-κB transcriptional activity in breast cancer cells, we re-expressed DiRas1 in the human breast cancer cell lines MCF-7 and T47D, which do not express endogenous DiRas1 protein (Fig. 7H). DiRas1 expression caused decreased NF-κB transcriptional activity in both MCF-7 and T47D cells (Fig. 7, I and J), although the decrease in transcriptional activation was not as dramatic as that exhibited in the glioblastoma cell lines (Fig. 6). These findings support our model that DiRas1 binds to and sequesters SmgGDS, diminishing signaling by pro-oncogenic small GTPases and suppressing NF-κB activity (Fig. 8).
TABLE 1.
DiRas1 is expressed in non-malignant breast epithelial tissue and to a lesser extent in breast invasive ductal carcinoma
DiRas1 protein is expressed in benign human ductal epithelial cells but largely absent in invasive breast cancers. DiRas1 IHC staining in ductal apical epithelium in benign breast samples, myoepithelial cells in benign breast samples, and invasive breast cancers was assessed by two independent observers. IRS was calculated and averaged for each sample, with aggregate results shown. The p value was obtained using an unpaired Student's t test.
| Tissue | N | Mean of DiRas1 IRS (S.E.) | p value vs. invasive ductal carcinoma |
|---|---|---|---|
| Benign apical breast epithelium | 15 | 4.50 (0.82) | 0.011 |
| Myoepithelium in benign tissue | 15 | 9.04 (0.83) | <0.0001 |
| Invasive ductal carcinoma | 15 | 2.05 (0.41) |
FIGURE 7.
DiRas1 is expressed in normal breast epithelium but absent from breast cancers, and it inhibits NF-κB transcriptional activity in breast cancer cell lines. A–F, DiRas1 is expressed in normal human ductal epithelium and myoepithelium (A–C) but is expressed to a lesser extent in most invasive breast tumors (D–F). These images are representative of the samples quantified in Table 1. G, scatter dot plot shows the IRS score for each sample, as well as the means ± the S.E. H, the human breast cancer cell lines T47D and MCF-7 (50 μg each) express no detectable DiRas1 protein, respectively. Control cell lysate (4 μg) is HEK293T cells expressing DiRas1 cDNA. I and J, DiRas1 expression diminishes basal NF-κB transcriptional activity in T47D (I) and MCF-7 (J) cells. The breast cancer cells were transfected with empty vector or DiRas1 and NF-κB reporter and β-gal. After 24 h, luminescence was quantified and normalized to vector-transfected cells. The results are the means ± S.E. from at least three independent experiments. *, p < 0.0001 compared with vector-transfected cell activity using the Mann-Whitney U test.
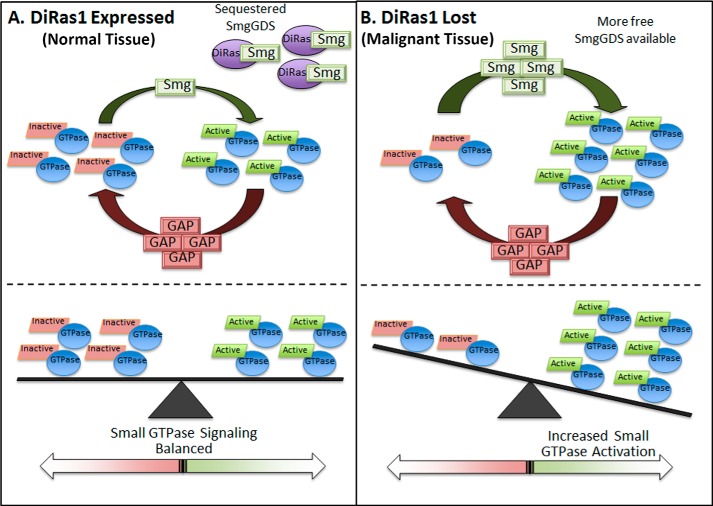
FIGURE 8.
Proposed model of DiRas1-mediated alteration in oncogenic small GTP signaling via binding to SmgGDS. A, in noncancerous tissues where DiRas1 is expressed, it can nonproductively bind SmgGDS and diminish SmgGDS interactions with other small GTPases, promoting balanced oncogenic small GTPase activity. B, in malignant tissues when DiRas1 expression is decreased or lost, more SmgGDS protein is available to interact with oncogenic small GTPases and promote their activation, resulting in increased pro-oncogenic small GTPase signaling.
Discussion
Here we identified the tumor-suppressive small GTPase DiRas1 as a novel SmgGDS-interacting protein that can limit the binding of other small GTPases to SmgGDS. This conclusion is based on our findings that SmgGDS is not a GEF for DiRas1 (Fig. 3), that SmgGDS exhibits a stronger binding affinity for DiRas1 than RhoA (Fig. 3), and that expression of low amounts of DiRas1 eliminates detectable interactions of SmgGDS with RhoA, K-Ras4B, and Rap1A (Fig. 4). This newly defined role for DiRas1 may constitute a mechanism by which normal tissues expressing DiRas1 can temper pro-oncogenic small GTPase activation. When DiRas1 expression is lost, SmgGDS can more easily bind to and activate small GTPases that promote malignancy (Fig. 8).
Our results suggest that DiRas1 binds to SmgGDS in a manner similar to the binding of both Ras family (K-Ras4B and Rap1A) and Rho family (RhoA) GTPases. DiRas1 strikingly and consistently eliminated detectable SmgGDS interactions with these Ras and Rho family pro-oncogenic small GTPases (Figs. 4 and 5A), suggesting that DiRas1 behaves like a dominant-negative small GTPase (Fig. 5, B–D) (30). In vitro, it appears that SmgGDS can directly facilitate guanine nucleotide exchange for RhoA and RhoC, but not RhoB, K-Ras4B, Rac1/2, Cdc42, or Rap1A/B (4). SmgGDS has also been found to promote the malignant phenotype in a number of cancers via its ability to increase the pro-oncogenic functions of a variety of PBR-containing small GTPases (reviewed in Ref. 9), most likely by acting as a chaperone (5) or scaffold (34).
We examined whether DiRas1 expression suppresses signaling pathways downstream of the pro-oncogenic small GTPase RhoA. In vitro, the presence of DiRas1 decreased SmgGDS-mediated GDP/GTP exchange (Fig. 3C). Recent evidence suggests that knockdown of SmgGDS-558, but not full-length SmgGDS-607, inhibits RhoA activation as well as Rho-mediated NF-κB transcriptional activity in breast cancer cells (6). Although both SmgGDS-607 and SmgGDS-558 can activate RhoA in vitro, only SmgGDS-558 promotes RhoA activation in cells (4). Consistent with the effective inhibition of RhoA and SmgGDS interactions by DiRas1, we found that DiRas1 potently inhibited RhoA- and SmgGDS-558-mediated NF-κB transcriptional activity in HEK293T cells (Fig. 6A). Activation of RhoA, as well as other small GTPases, can promote NF-κB activity in several cancers (31, 33, 35, 36). Loss of SmgGDS interactions with these GTPases because of DiRas1 expression can likely lead to decreased NF-κB activation in cancer cells (Figs. 6 and 7). DiRas1 is reported to be a tumor suppressor in gliomas, with decreased expression occurring more frequently in higher grade gliomas, such as glioblastoma (11, 12). Activated NF-κB is critical to glioblastoma cell growth and proliferation (10). We found that re-expression of DiRas1 in U87 and U251 glioblastoma cell lines inhibited NF-κB activity (Fig. 6, E and F), and mRNA levels of the NF-κB transcriptionally regulated gene IL-8 were decreased in glioblastoma cells expressing DiRas1 (Fig. 6, G and H). These results suggest that restoring DiRas expression or its downstream signals may be therapeutic strategies to diminish NF-κB activation in malignant gliomas.
It has recently been reported that SmgGDS-558 enhances RhoA activation, as well as NF-κB activation in breast cancer cell lines (6). The DiRas family member DiRas3 is expressed in normal breast tissue and absent from up to 70% of invasive breast tumors (14); here, we found that DiRas1 was expressed in normal human breast apical epithelial and myoepithelial tissue but was expressed at lower levels in invasive breast tumors (Table 1 and Fig. 7). The range of expression seen in normal breast epithelial tissue (Fig. 7G) could be due to differences in the normal mammary tissue caused by factors such as pre- or postmenopausal status, pregnancy or postpartum conditions, or even genetic changes predisposing to breast cancer, because many normal tissue breast samples are obtained from patients obtaining contralateral prophylactic mastectomies because of breast cancer in the contralateral breast. The specific clinical details of the patients from which our normal tissue samples were obtained were not available. In addition to finding that DiRas1 expression appears to be lower in breast cancers, we found that expression of DiRas1 inhibited NF-κB transcriptional activity in two breast cancer cell lines (Fig. 7, I and J). Future studies will be needed to determine the extent of the tumor-suppressive functions of DiRas1 in breast cancer and the implications of the loss of this small GTPase.
Beyond the specific inhibition of SmgGDS interactions with pro-oncogenic small GTPases, it is also possible that DiRas1 acts globally, inhibiting pro-oncogenic small GTPases through other pathways not mediated by SmgGDS. Previous data suggest that DiRas1 may antagonize Ras-mediated signaling. There are conflicting reports on whether DiRas1 binds RAF family members (11, 12), but a single study reported that DiRas1 associates (nonproductively) with the effector domain of C-RAF (12). DiRas1 was reported to decrease Elk-1 transactivation (12), and DiRas1 expression in esophageal squamous cell carcinomas diminished the phosphorylation of the Ras downstream effectors c-Raf, MEK, and ERK1/2, resulting in decreased BAD Ser-112 phosphorylation and increased apoptosis (13). BAD serine phosphorylation can also be mediated by Rho family member signaling (37, 38), although the ability of DiRas1 to antagonize Rho family GTPase signaling that directly impacts BAD serine phosphorylation has not been examined.
Several subfamilies in the Ras GTPase superfamily display tumor-suppressive effects (39), some of which may be mediated by nonproductive associations with SmgGDS that abrogate pro-oncogenic small GTPase-SmgGDS binding. However, not all tumor-suppressive GTPases have a strong PBR, suggesting that not all Ras family tumor suppressors inhibit malignancy through PBR-dependent interactions with SmgGDS. Further studies will be needed to determine whether other DiRas family members can bind to SmgGDS and inhibit the binding of pro-oncogenic small GTPases to SmgGDS. Relating to this question, it was recently reported that SmgGDS binds to DiRas2, which is a PBR-containing GTPase that is closely related to DiRas1 (40). The potential ability of DiRas2 to inhibit SmgGDS functions, including its interactions with pro-oncogenic GTPases, was not examined in this previous report (40).
Our findings further elucidate the mechanism by which DiRas1 mediates its tumor suppressive functions. These results suggest that reduced DiRas1 expression increases the availability of SmgGDS for binding to pro-oncogenic GTPases, suggesting a unique mechanism in which SmgGDS signals may be amplified in cancers. In conjunction with previous work (12), our findings demonstrate that DiRas1 can antagonize a multitude of small GTPases by nonproductive associations with activating proteins. Further characterization of DiRas1 expression in other normal and cancerous tissues will help identify additional malignancies that have enhanced SmgGDS- and oncogenic small GTPase-mediated signaling because of the loss of DiRas1.
Author Contributions
C. B. conceived and coordinated the study, evaluated the data, and wrote the paper. A. F., C. B., A. D. H., A. R., P. G., and E. L. L. performed experiments in Figs. 1–7 and Table 1. J. W. P. performed the modeling and docking studies in Figs. 2 and 4. I. A.-B. reviewed breast pathology for Table 1 and Fig. 7. A. C. M. created the TMAs and interpreted Fig. 6 (B and C). K. N. performed LC/MS and analysis. B. C. J., A. J. L., and C. A. F. performed the in vitro SmgGDS/small GTPase binding studies, and C. L. W. assisted with coordination of the study interpretation and editing of the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank John Sondek for the generous gift of the pLIC-His SmgGDS and RhoA constructs for the guanine nucleotide exchange assays, as well as for technical advice regarding these assays. In addition, we thank the Medical College of Wisconsin Brain and Spinal Cord Tissue Bank and Mona Al-Gizawiy for assistance with CNS tissue studies. We also thank Jacek Zielonka for assistance using the fluorescence spectrometer.
This work was supported, in whole or in part, by funding from the Medical College of Wisconsin Cancer Center and Research Affairs Committee, Advancing a Healthier Wisconsin, the Radiological Society of North America, and the Robert D. and Patricia E. Kern Family Foundation (to C. B.). This work was also supported by the National Center for Research Resources, the National Center for Advancing Translational Sciences, and the Office of the Director of the National Institutes of Health through Grant 8KL2TR000056 (to C. B.) and by the Nancy Laning Sobczak, Ph.D., Breast Cancer Research Award (to C. B. and C. L. W.), Institutional Research Grant 86-004-26 from the American Cancer Society (to C. B.), and National Institutes of Health Grants R01CA188871 (to C. L. W.), F32GM112317 (to B. C. J.), and R01GM040602 (to C. A. F.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- GEF
- guanine nucleotide exchange factor
- DN
- dominant negative
- GAP
- GTPase activation protein
- IRS
- immunoreactive score
- MANT
- N-methylanthraniloyl
- PBR
- polybasic region
- IHC
- immunohistochemical.
References
- 1. Alan J. K., and Lundquist E. A. (2013) Mutationally activated Rho GTPases in cancer. Small GTPases 4, 159–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bos J. L. (1989) ras oncogenes in human cancer: a review. Cancer Res. 49, 4682–4689 [PubMed] [Google Scholar]
- 3. Pylayeva-Gupta Y., Grabocka E., and Bar-Sagi D. (2011) RAS oncogenes: weaving a tumorigenic web. Nat. Rev. Cancer 11, 761–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hamel B., Monaghan-Benson E., Rojas R. J., Temple B. R., Marston D. J., Burridge K., and Sondek J. (2011) SmgGDS is a guanine nucleotide exchange factor that specifically activates RhoA and RhoC. J. Biol. Chem. 286, 12141–12148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berg T. J., Gastonguay A. J., Lorimer E. L., Kuhnmuench J. R., Li R., Fields A. P., and Williams C. L. (2010) Splice variants of SmgGDS control small GTPase prenylation and membrane localization. J. Biol. Chem. 285, 35255–35266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hauser A. D., Bergom C., Schuld N. J., Chen X., Lorimer E. L., Huang J., Mackinnon A. C., and Williams C. L. (2014) The SmgGDS splice variant SmgGDS-558 is a key promoter of tumor growth and RhoA signaling in breast cancer. Mol. Cancer Res. 12, 130–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schuld N. J., Hauser A. D., Gastonguay A. J., Wilson J. M., Lorimer E. L., and Williams C. L. (2014) SmgGDS-558 regulates the cell cycle in pancreatic, non-small cell lung, and breast cancers. Cell Cycle 13, 941–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhi H., Yang X. J., Kuhnmuench J., Berg T., Thill R., Yang H., See W. A., Becker C. G., Williams C. L., and Li R. (2009) SmgGDS is up-regulated in prostate carcinoma and promotes tumour phenotypes in prostate cancer cells. J. Pathol. 217, 389–397 [DOI] [PubMed] [Google Scholar]
- 9. Tew G. W., Lorimer E. L., Berg T. J., Zhi H., Li R., and Williams C. L. (2008) SmgGDS regulates cell proliferation, migration, and NF-κB transcriptional activity in non-small cell lung carcinoma. J. Biol. Chem. 283, 963–976 [DOI] [PubMed] [Google Scholar]
- 10. Nogueira L., Ruiz-Ontañon P., Vazquez-Barquero A., Moris F., and Fernandez-Luna J. L. (2011) The NFκB pathway: a therapeutic target in glioblastoma. Oncotarget. 2, 646–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kontani K., Tada M., Ogawa T., Okai T., Saito K., Araki Y., and Katada T. (2002) Di-Ras, a distinct subgroup of ras family GTPases with unique biochemical properties. J. Biol. Chem. 277, 41070–41078 [DOI] [PubMed] [Google Scholar]
- 12. Ellis C. A., Vos M. D., Howell H., Vallecorsa T., Fults D. W., and Clark G. J. (2002) Rig is a novel Ras-related protein and potential neural tumor suppressor. Proc. Natl. Acad. Sci. U.S.A. 99, 9876–9881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhu Y.-H., Fu L., Chen L., Qin Y.-R., Liu H., Xie F., Zeng T., Dong S.-S., Li J., Li Y., Dai Y., Xie D., and Guan X.-Y. (2013) Downregulation of the novel tumor suppressor DIRAS1 predicts poor prognosis in esophageal squamous cell carcinoma. Cancer Res. 73, 2298–2309 [DOI] [PubMed] [Google Scholar]
- 14. Yu Y., Luo R., Lu Z., Wei Feng W., Badgwell D., Issa J.-P., Rosen D. G., Liu J., and Bast R. C. Jr (2006) Biochemistry and biology of ARHI (DIRAS3), an imprinted tumor suppressor gene whose expression is lost in ovarian and breast cancers. Methods Enzymol. 407, 455–468 [DOI] [PubMed] [Google Scholar]
- 15. Sakoda T., Kaibuchi K., Kishi K., Kishida S., Doi K., Hoshino M., Hattori S., and Takai Y. (1992) smg/rap1/Krev-1 p21s inhibit the signal pathway to the c-fos promoter/enhancer from c-Ki-ras p21 but not from c-raf-1 kinase in NIH3T3 cells. Oncogene 7, 1705–1711 [PubMed] [Google Scholar]
- 16. Aspuria P.-J., and Tamanoi F. (2004) The Rheb family of GTP-binding proteins. Cell. Signal. 16, 1105–1112 [DOI] [PubMed] [Google Scholar]
- 17. Gasper R., Sot B., and Wittinghofer A. (2010) GTPase activity of Di-Ras proteins is stimulated by Rap1GAP proteins. Small GTPases 1, 133–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lanning C. C., Ruiz-Velasco R., and Williams C. L. (2003) Novel mechanism of the co-regulation of nuclear transport of SmgGDS and Rac1. J. Biol. Chem. 278, 12495–12506 [DOI] [PubMed] [Google Scholar]
- 19. Lanning C. C., Daddona J. L., Ruiz-Velasco R., Shafer S. H., and Williams C. L. (2004) The Rac1 C-terminal polybasic region regulates the nuclear localization and protein degradation of Rac1. J. Biol. Chem. 279, 44197–44210 [DOI] [PubMed] [Google Scholar]
- 20. Rossman K. L., Worthylake D. K., Snyder J. T., Siderovski D. P., Campbell S. L., and Sondek J. (2002) A crystallographic view of interactions between Dbs and Cdc42: PH domain-assisted guanine nucleotide exchange. EMBO J. 21, 1315–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Snyder J. T., Worthylake D. K., Rossman K. L., Betts L., Pruitt W. M., Siderovski D. P., Der C. J., and Sondek J. (2002) Structural basis for the selective activation of Rho GTPases by Dbl exchange factors. Nat. Struct. Biol. 9, 468–475 [DOI] [PubMed] [Google Scholar]
- 22. Stols L., Gu M., Dieckman L., Raffen R., Collart F. R., and Donnelly M. I. (2002) A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr. Purif. 25, 8–15 [DOI] [PubMed] [Google Scholar]
- 23. Roy A., Kucukural A., and Zhang Y. (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Krieger E., Joo K., Lee J., Lee J., Raman S., Thompson J., Tyka M., Baker D., and Karplus K. (2009) Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins 77, 114–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., and Olson A. J. (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Krieger E., Koraimann G., and Vriend G. (2002) Increasing the precision of comparative models with YASARA NOVA: a self-parameterizing force field. Proteins 47, 393–402 [DOI] [PubMed] [Google Scholar]
- 27. Duan Y., Wu C., Chowdhury S., Lee M. C., Xiong G., Zhang W., Yang R., Cieplak P., Luo R., Lee T., Caldwell J., Wang J., and Kollman P. (2003) A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 24, 1999–2012 [DOI] [PubMed] [Google Scholar]
- 28. Konagurthu A. S., Whisstock J. C., Stuckey P. J., and Lesk A. M. (2006) MUSTANG: a multiple structural alignment algorithm. Proteins 64, 559–574 [DOI] [PubMed] [Google Scholar]
- 29. Rojas R. J., Kimple R. J., Rossman K. L., Siderovski D. P., and Sondek J. (2003) Established and emerging fluorescence-based assays for G-protein function: Ras-superfamily GTPases. Comb. Chem. High Throughput Screen 6, 409–418 [DOI] [PubMed] [Google Scholar]
- 30. Farnsworth C. L., and Feig L. A. (1991) Dominant inhibitory mutations in the Mg2+-binding site of RasH prevent its activation by GTP. Mol. Cell. Biol. 11, 4822–4829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Montaner S., Perona R., Saniger L., and Lacal J. C. (1998) Multiple signalling pathways lead to the activation of the nuclear factor κB by the Rho family of GTPases. J. Biol. Chem. 273, 12779–12785 [DOI] [PubMed] [Google Scholar]
- 32. Montaner S., Perona R., Saniger L., and Lacal J. C. (1999) Activation of serum response factor by RhoA is mediated by the nuclear factor-κB and C/EBP transcription factors. J. Biol. Chem. 274, 8506–8515 [DOI] [PubMed] [Google Scholar]
- 33. Perona R., Montaner S., Saniger L., Sánchez-Pérez I., Bravo R., and Lacal J. C. (1997) Activation of the nuclear factor-κB by Rho, CDC42, and Rac-1 proteins. Genes Dev. 11, 463–475 [DOI] [PubMed] [Google Scholar]
- 34. Shin E.-Y., Lee C.-S., Cho T. G., Kim Y. G., Song S., Juhnn Y.-S., Park S. C., Manser E., and Kim E.-G. (2006) βPak-interacting exchange factor-mediated Rac1 activation requires smgGDS guanine nucleotide exchange factor in basic fibroblast growth factor-induced neurite outgrowth. J. Biol. Chem. 281, 35954–35964 [DOI] [PubMed] [Google Scholar]
- 35. Gastonguay A., Berg T., Hauser A. D., Schuld N., Lorimer E., and Williams C. L. (2012) The role of Rac1 in the regulation of NF-κB activity, cell proliferation, and cell migration in non-small cell lung carcinoma. Cancer Biol. Ther. 13, 647–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lin G., Tang Z., Ye Y.-B., and Chen Q. (2012) NF-κB activity is downregulated by KRAS knockdown in SW620 cells via the RAS-ERK-IκBα pathway. Oncol. Rep. 27, 1527–1534 [DOI] [PubMed] [Google Scholar]
- 37. Kang J., and Pervaiz S. (2012) Crosstalk between Bcl-2 family and Ras family small GTPases: potential cell fate regulation? Front. Oncol. 2, 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang B., Zhang Y., and Shacter E. (2004) Rac1 inhibits apoptosis in human lymphoma cells by stimulating Bad phosphorylation on Ser-75. Mol. Cell. Biol. 24, 6205–6214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Colicelli J. (2004) Human RAS superfamily proteins and related GTPases. Sci. STKE 2004, RE13. [DOI] [PMC free article] [PubMed] [Google Scholar]