Abstract
Nuclear speckles are subnuclear storage sites containing pre-mRNA splicing machinery. Proteins assembled in nuclear speckles are known to modulate transcription and pre-mRNA processing. We have previously identified nuclear speckle-related protein 70 (NSrp70) as a novel serine/arginine (SR)-related protein that co-localizes with classical SR proteins such as serine/arginine-rich splicing factor 1 (SRSF1 or ASF/SF2) and SRSF2 (SC35). NSrp70 mediates alternative splice site selection, targeting several pre-mRNAs, including CD44 exon v5. Here we demonstrated that NSrp70 interacts physically with two SR proteins, SRSF1 and SRSF2, and reverses their splicing activity in terms of CD44 exon v5 as exon exclusion. The NSrp70 RS-like region was subdivided into three areas. Deletion of the first arginine/serine-rich-like region (RS1) completely abrogated binding to the SR proteins and to target mRNA and also failed to induce splicing of CD44 exon v5, suggesting that RS1 is critical for NSrp70 functioning. Interestingly, RS1 deletion also resulted in the loss of NSrp70 and SR protein speckle positioning, implying a potential scaffolding role for NSrp70 in nuclear speckles. NSrp70 contains an N-terminal coiled-coil domain that is critical not only for self-oligomerization but also for splicing activity. Consistently, deletion of the coiled-coil domain resulted in indefinite formation of nuclear speckles. Collectively, these results demonstrate that NSrp70 acts as a new molecular counterpart for alternative splicing of target RNA, counteracting SRSF1 and SRSF2 splicing activity.
Keywords: alternative splicing, CD44, dimerization, heterogeneous nuclear ribonucleoprotein (hnRNP), RNA binding protein, NSrp70, RS-like region, SR protein, coiled-coil domain, nuclear speckle
Introduction
The mammalian cell nucleus contains non-membranous subnuclear bodies such as the nucleolus, Cajal bodies, promyelocytic leukemia bodies, and nuclear speckles. These bodies are generally classified by the presence of a distinct group of proteins and RNAs within them (1, 2). Nuclear speckles are irregular, punctuate structures that vary in size and shape at the immunofluorescence microscope level and are located in interchromatin regions of the nucleoplasm of mammalian cells (3, 4). Several reports have demonstrated that nuclear speckles serve as storage/assembly sites for pre-mRNA splicing machinery and, therefore, elaborately regulate gene expression (2). Nuclear speckles are known to be functional centers that spatially organize transcriptional and posttranscriptional mechanisms of gene regulation. RNA-binding proteins involved in transcription and pre-mRNA processing, such as small nuclear ribonucleoprotein and arginine/serine-rich family factors (SR protein, SR-related protein), are associated with nuclear speckles (5, 6). Remarkably, two classical SR proteins, SRSF1 and SRSF2, are well known components of nuclear speckles. These SR proteins modulate both constitutive and alternative splicing by assembly of the mature spliceosome (7, 8).
In a previous report, we identified a novel SR-related protein, nuclear speckle-related protein 70 (NSrp70), that modulates alternative splicing site selection of several mRNAs in vivo, including Tra2β1 v2, Fas v6, and CD44 exon v5. Interestingly, NSrp70 co-localizes and interacts physically with SRSF1 and SRSF2 in nuclear speckles through its RS-like region (9). However, its physiological functions are largely unknown. Therefore, in this study, our primary goal was to investigate whether NSrp70 synergizes the alternative splicing actions with SRSF1 and SRSF2, or whether it counteracts these functions. In addition, because the binding site in the RS-like region of NSrp70 has not yet been clearly determined, we generated deletion mutants and determined which site is critical for the function in terms of speckle localization with SRSF1 and SRSF2 and the alternative splicing of target mRNAs.
Several sources of evidence have suggested that RNA-binding proteins are related to functional aggregation to form paraspeckles in the nucleus and stress granules in the cytoplasm as RNA-protein granules (10). For example, proteins of the Drosophila behavior/human splicing (DBHS)2 family, including PSF, p54nrb, and PSPC1 (Paraspeckle component 1) are major components of paraspeckles (11). Interestingly, it has been demonstrated that the coiled-coil (CC) domain in DBHS proteins is important for self-dimerization formation and paraspeckle targeting. When DBHS proteins bind to their target DNA, the CC domain helps to form a condensational assembly, therefore enriching recruitment of other functional factors that then rapidly promote a specific response (12, 13). In silico analysis revealed that NSrp70 also contains a predicted CC domain. We therefore questioned whether this domain is involved in dimerization or multimerization of NSrp70 and whether it plays a role in condensational assembly by recruiting other factors, including SRSF1 and SRSF2, in nuclear speckles.
SRSF1 and SRSF2 contain two unique structures: one or two N-terminal RNA recognition motifs (RRMs) that determine specific sequence interactions and C-terminal arginine/serine (RS)-rich dipeptide domains that regulate protein-protein interactions and thereby mediate the recruitment of the spliceosome (14). The RS domain plays a role as a nuclear localization signal that specifically targets nuclear speckles (15). Phosphorylation at multiple serine residues influences efficient nuclear speckle assembly, pre-mRNA splicing, and mRNA nuclear export (16). However, the role of the RS-like region in NSrp70 is still unclear. Therefore, we investigated whether NSrp70 exerts its function via direct binding to target RNA or whether it binds indirectly through the RS domains of SRSF1 and SRSF2.
Here we report that NSrp70 displayed the opposite activity to SRSF1 and SRSF2 in the alternative splicing of the CD44 exon v5 minigene. Furthermore, mutagenesis studies revealed that the first RS-like region of NSrp70 is critical for its activity, interacting with the two SR proteins and directly with target mRNA. We found that the CC domain is essential for condensational assembly and that disruption of the CC domain by site-directed mutagenesis results in indefinite formation of nuclear speckles and NSrp70 dysfunction.
Experimental Procedures
Reagents and Antibodies
Anti-human NSrp70 (CCDC55) antibody, glutaraldehyde solution, and His-selected nickel affinity gel were purchased from Sigma-Aldrich (St. Louis, MO). Anti-myc and anti-β-actin antibodies were purchased from Cell Signaling Technology (Beverly, MA). Total RNA isolation reagent was purchased from Molecular Research Center, Inc. (Cincinnati, OH). Reverse transcript PCR premix and conventional PCR premix were purchased from iNtRON Biotechnology (Seongnam-si, Gyeonggi-do, Korea). The Magna RIPTM kit was purchased from EMD Millipore (Billerica, MA). Texas Red-conjugated anti-mouse IgG and Lipofectamine 2000 reagent were acquired from Life Technologies. Rabbit polyclonal anti-GFP antibody was developed in rabbit using purified recombinant full-length GFP protein. Non-muscle actin-derived from human platelets was purchased from Cytoskeleton Inc. (Denver, CO). Bis(sulfosuccinimidyl) suberate (BS3) was purchased from Thermo Scientific (Waltham, MA). All cell culture materials used in this work were from Life Technologies.
Cell Culture and Transfection
HEK293T cells (CRL-1573, ATCC) were maintained in DMEM supplemented with 10% FBS, penicillin, and streptomycin. Constructs were transfected by Lipofectamine 2000 (Life Technologies) in HEK293T cells according to the instructions of the manufacturer instructions. 48 h after transfection, cells were collected for RNA and protein purification or image analysis.
Plasmids
The mammalian expression vectors pEGFP-C1_SRSF1 (ASF/SF2) and pETv5_CD44 and the pCR3.1_MGTra minigenes were gifts from Dr. Brian J. Morris (University of Sydney, NSW, Australia). pEGFP-C1_PML3 was obtained from Dr. David J. Picketts (University of Pennsylvania, School of Medicine, Philadelphia, PA). The pEGFP-C1 or pCS4–3Myc_NSrp70, _SRSF1, and _SRSF2 constructs were generated using a method reported previously (9).
To produce His-tagged NSrp70, CCD, and RS1M recombinant proteins, the pET-28a vector was used as an expression vector. This has a T7 promoter and incorporates six His residues at the N terminus of the expressed protein. The coding sequences of NSrp70 and mutant NSrp70 were amplified by PCR, and the products were incorporated into pET-28a.
Purification of Recombinant Proteins
Expression of recombinant proteins in Escherichia coli BL21 (DE3) cells was performed as described previously (9). Expression of recombinant proteins was induced by addition of isopropyl β-d-1-thiogalactopyranoside (0.3 mm) to culture medium for 16 h at 28 °C, and then cells were collected. The cell pellets were resuspended in PBS (0.1 mm CaCl2, 1 mm MgCl2, and 4% sucrose), sonicated, and centrifuged. After centrifugation, recombinant proteins in the supernatant were purified by affinity chromatography on a His-selected nickel affinity gel (Sigma-Aldrich). The gel was equilibrated with 10 volumes of buffer (50 mm sodium phosphate (pH 8.0), and 0.3 m NaCl) and incubated with the supernatant. The gel was washed with five volumes of wash buffer (50 mm sodium phosphate (pH 8.0), 0.3 m NaCl, and 10 mm imidazole). The protein was eluted with increasing concentrations of imidazole up to 250 mm, and the collected fractions were dialyzed to remove imidazole. The proteins were assessed by SDS-PAGE and Coomassie blue staining.
In Vivo Splicing Assays
In vivo splicing assays were performed essentially as described previously (17). Briefly, a splicing reporter minigene was cotransfected with an increasing amount of wild-type or mutant GFP_NSrp70 or Myc_NSrp70 constructs together with GFP- or Myc-tagged SRSF1 and SRSF2 in HEK293T cells. Empty plasmids were added to ensure that the same amount of DNA was transfected. 48 h after transfection, total RNA was extracted from cells using Total RNA isolation reagent (Molecular Research Center, Inc.). The cDNA was transcribed from 2 μg of total RNA using reverse transcription PCR premix (iNtRON Biotechnology). PCR for the CD44 minigene was performed as described previously (9). The PCR products were analyzed with a 1–1.5% agarose gel, and the splicing pattern was quantified using ImageJ (National Institutes of Health).
Immunoprecipitation and Western Blotting
HEK293T cells (1 × 107) transfected with the indicated constructs were lysed using lysis buffer (20 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1% Triton X-100, and one tablet of EDTA-free protease inhibitor mixture (Roche)) for 30 min on ice. Cell lysates were centrifuged at 14,000 × g for 30 min at 4 °C. Approximately 0.5 mg of extract was mixed with anti-GFP antibody-conjugated Sepharose 4B (GE Healthcare Life Sciences). The immunocomplexes were incubated overnight at 4 °C with frequent mixing, washed three times each with washing buffer I (20 mm Tris-HCl (pH 7.5), 150 mm NaCl, and 1% Triton X-100) and washing buffer II (20 mm Tris-HCl (pH 7.5) and 150 mm NaCl) and resolved by 10–12% SDS-PAGE. Proteins were electroblotted on to a PVDF membrane (PerkinElmer Life Sciences) by means of a Trans-Blot SD semidry transfer cell (Bio-Rad). The membrane was blocked in 5% skim milk for 1 h, rinsed, and incubated with the indicated antibodies in TBS containing 0.1% Tween-20 (TBS-T) and 3% skim milk overnight. Excess primary antibody was removed by washing the membrane three times in TBS-T. The membrane was then incubated with 0.1 μg/ml peroxidase-labeled secondary antibody (against rabbit or mouse) for 1 h. After three washes in TBS-T, bands were visualized by ECL Western blotting detection (iNtRON Biotechnology).
RNA Immunoprecipitation
RNA immunoprecipitation (RIP) experiments were performed using a Magna RIP RNA-binding protein immunoprecipitation kit according to the instructions of the manufacturer (Millipore) using a modified version of a method described previously (18). Briefly, HEK293T cells (1 × 107) transfected with the indicated constructs were lysed on ice in complete RIP lysis buffer (Millipore). NSrp70 antibody or rabbit IgG control antibody was precoated onto magnetic bead protein A/G for 30 min at room temperature. Antibody-coated beads were washed twice with RIP wash buffer (Millipore) using a magnetic separator. Protein lysates were then incubated with rabbit anti-NSrp70- or rabbit control IgG-coated beads overnight at 4 °C. The immunoprecipitated samples were then centrifuged and washed with ice-cold RIP wash buffer six times using a magnetic separator. After the final wash, the RNA-protein complexes were dissociated from the beads by incubating in proteinase K buffer (Millipore) for 30 min at 55 °C. The supernatants were collected using a magnetic separator. RNA extraction from the supernatant was performed with phenol:chloroform:isoamyl alcohol (125:24:1) (Sigma-Aldrich) and precipitated using ethanol supplemented with salt solution I, salt solution II, and precipitation enhancer overnight. The RNA pellets were then washed and suspended with RNase-free water. The cDNA was transcribed from total RNA using reverse transcription PCR Premix (iNtRON Biotechnology).
PCR was performed with the following primers (the respective forward and reverse pairs are indicated): endogenous CD44 containing exon v5 (p1 and p2) (19), p1 5′-CTGAAGACATCTACCCCAGCAAC-3′ and p2 5′-CAGAAATGGCACCACTGCTTAT-3′; first v5, 5′-AGACAGAATCAGCACCAGTG-3′ and 5′-TTTTTTTTTTAGGCTGTGGTTCCG-3′; and second v5, 5′-CCTTTCAATAACCATGAGTATCAGG-3′ and 5′-TTTTTTTTTTTGCTTGTAGCATGTGG-3′. The amplification protocol consisted of denaturation at 95 °C for 30 s, annealing at 50–57 °C for 20 s, and extension at 72 °C for 15–17 s. The 30 cycles were preceded by denaturation at 72 °C for 7 min. The three antiparallel minigene CD44 exon v5 RNAs (L, M, and R) for the RNA competition assay and the CD44 exon v5 mRNA (first v5 and second v5) were synthesized by Bioneer (Daejeon, Korea).
Immunofluorescence Staining and Confocal Imaging Analysis
For visualization of fluorescence, transfected HEK293T cells were cultured on 18-mm non-coated round coverslips (Paul Marienfeld GmbH & Co. KG) coated with fibronectin (Sigma-Aldrich). Cells were fixed with 3.7% formaldehyde for 15 min, washed three times with PBS at room temperature, and permeabilized with 1% BSA and 0.25% Triton X-100 in PBS for 20 min. Cells were then blocked with 1% BSA in PBS for 1 h, rinsed with PBS, and incubated with primary antibodies in blocking buffer overnight at 4 °C. Secondary antibodies were added after washing and incubated for 3 h at room temperature in the dark before rinsing with PBS. The cells were then washed and mounted with fluorescent mounting medium (Dako). The samples were examined with an FV1000 confocal laser-scanning microscope (Olympus) equipped with ×40, ×60, and ×100 objectives.
Protein Cross-linking Assay
For cross-linking of cell lysates, cells were transfected with the indicated plasmids and lysed with lysis buffer as described above. Glutaraldehyde (Sigma-Aldrich) or BS3 (Thermo Scientific) was added to the lysate at the indicated concentration. After incubating the lysate on ice for 20 min (glutaraldehyde) or 50 min (BS3), the reactions were stopped by adding 4× loading buffer, and the samples were heated at 100 °C for 5 min and resolved by SDS-PAGE. Western blotting analysis was performed as described above. For cross-linking of recombinant proteins, cell lysates were replaced with His_NSrp70 protein or non-muscle actin and subsequently processed as described above.
Coiled-Coil Prediction
Coiled-coil prediction was performed using the COILS program. We inserted the amino acid sequence of wild-type human NSrp70 and mutants to analyze coiled-coil prediction in the following specific format: window width, 28; matrix, MTIDK (myosins, tropomysins, intermediate filaments, desmosomal proteins, and kinesins).
Protein Structural Modeling
Protein sequences of NSrp70 were obtained from the NCBI database (NCBI reference sequence, NP_115517.1). Structural modeling and three-dimensional prediction of NSrp70 were performed using the Iterative Threading Assembly Refinement (I-TASSER) server on the basis of their confidence score (20). The five models (supplemental Fig. 1) generated were modified and visualized using the Discovery Studio 4.0 Visualizer program.
Statistical Analysis
Mean values were calculated using data taken from at least three independent experiments conducted on different days. Where significance testing was performed, unpaired Student's t tests and one-way analysis of variance were used. Differences between groups were considered significant at p < 0.05.
Results
NSrp70 Counteracts SRSF1- and SRSF2-induced CD44 Exon v5 Exclusion
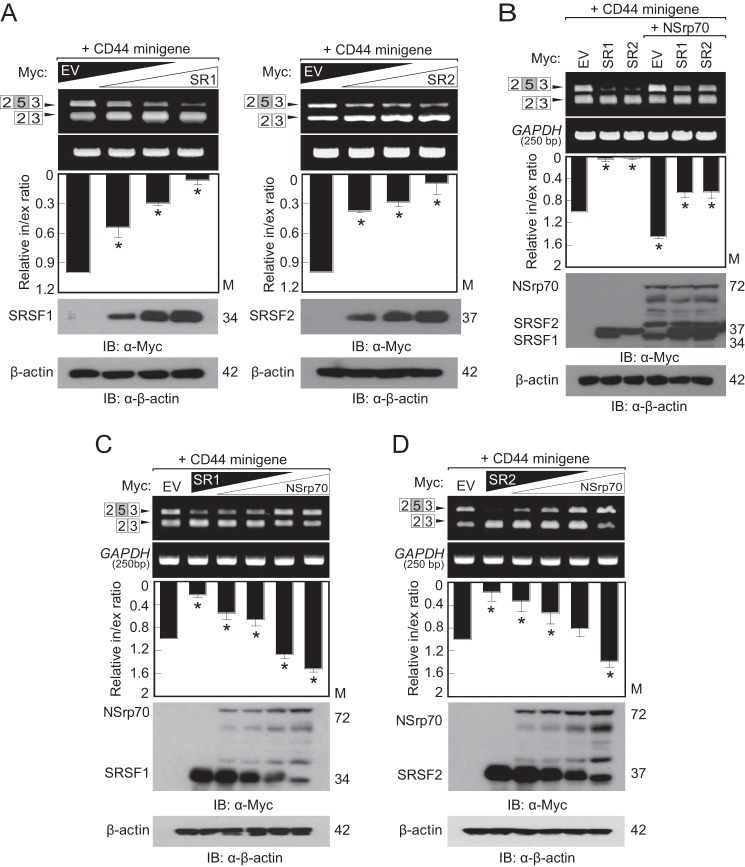
We have reported previously that NSrp70 modulates alternative splice site selection of some minigenes, including Tra2β1, Fas, and CD44. We have also found that NSrp70 interacts physically with the classical SR proteins SRSF1 and SRSF2, but its functional relationship with these two splicing factors is uncharacterized. To understand their roles in relation to the inclusion or exclusion of target mRNA exons, we tested the effects of NSrp70 and two SR proteins on the CD44 exon v5 minigene. Both Myc-tagged SRSF1 (Myc_SRSF1) and SRSF2 (Myc_SRSF2) increased CD44 exon v5 exclusion in a transfection concentration-dependent manner (Fig. 1A). On the contrary, NSrp70 increased CD44 exon v5 inclusion and blocked SRSF1- or SRSF2-mediated CD44 exon v5 exclusion (Fig. 1B). To investigate whether alternative site selection of CD44 exon v5 is dependent on the expression levels of each splicing factor, HEK293T cells were transfected with increasing or decreasing concentrations of vectors containing NSrp70 and SRSF1 or SRSF2. RT-PCR showed that NSrp70 and the two SR proteins have opposite functions and that their actions are concentration-dependent (Fig. 1, C and D). These results are interesting because NSrp70 may have an equal competitive capability against classical SR proteins for alternative splicing site selection of target pre-mRNA.
FIGURE 1.
NSrp70 counteracts SRSF1 and SRSF2 to induce alternative splicing of the CD44 exon v5 minigene. A, promotion of CD44 minigene exon v5 exclusion by SR proteins. HEK293T cells were cotransfected with increasing amounts of Myc_SRSF1 or _SRSF2 (0–2 μg) and the CD44 minigene (1 μg). A parental vector (EV, 0–2 μg) was added to ensure that similar amounts of cDNA were transfected. Exon inclusion or exclusion was determined by RT-PCR (top panel). The ratio of exclusion or inclusion of CD44 exon v5 is shown as bar graphs (center panel). Increased expression of Myc_SRSF1 or Myc_SRSF2 was confirmed by Western blotting using the indicated antibodies (bottom panel). EV, pCS4–3Myc_empty vector; SR1, SRSF1; SR2, SRSF2; Relative in/ex ratio, relative inclusion band/exclusion band ratio; IB, immunoblotting; M, molecular mass. B, promotion of CD44 minigene exon v5 inclusion by NSrp70. HEK293T cells were cotransfected with the CD44 minigene (1 μg) and Myc_SRSF1 (2 μg) or Myc_SRSF2 (2 μg) with or without Myc_NSrp70 (2 μg). The experiments were performed as described in A. C and D, NSrp70 counteracts SRSF1 and SRSF2. HEK293T cells were cotransfected with increasing amounts of Myc_NSrp70 (0–2 μg) and Myc_SRSF1 (C) or Myc_SRSF2 (0–2 μg) (D) and the CD44 minigene (1 μg). The experiments were performed as described in A. All data are representative of three independent experiments. *, p < 0.05 versus pCS4–3Myc_empty vector.
NSrp70 Interacts with SRSF1 and SRSF2 through the First RS-like Region (RS1)
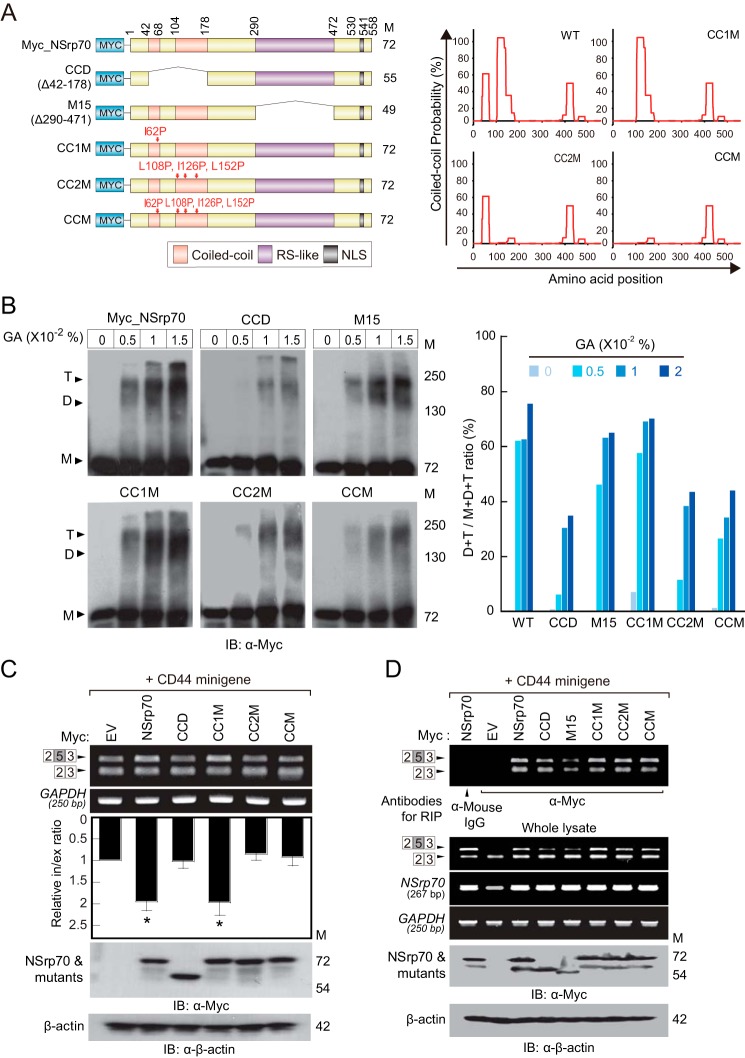
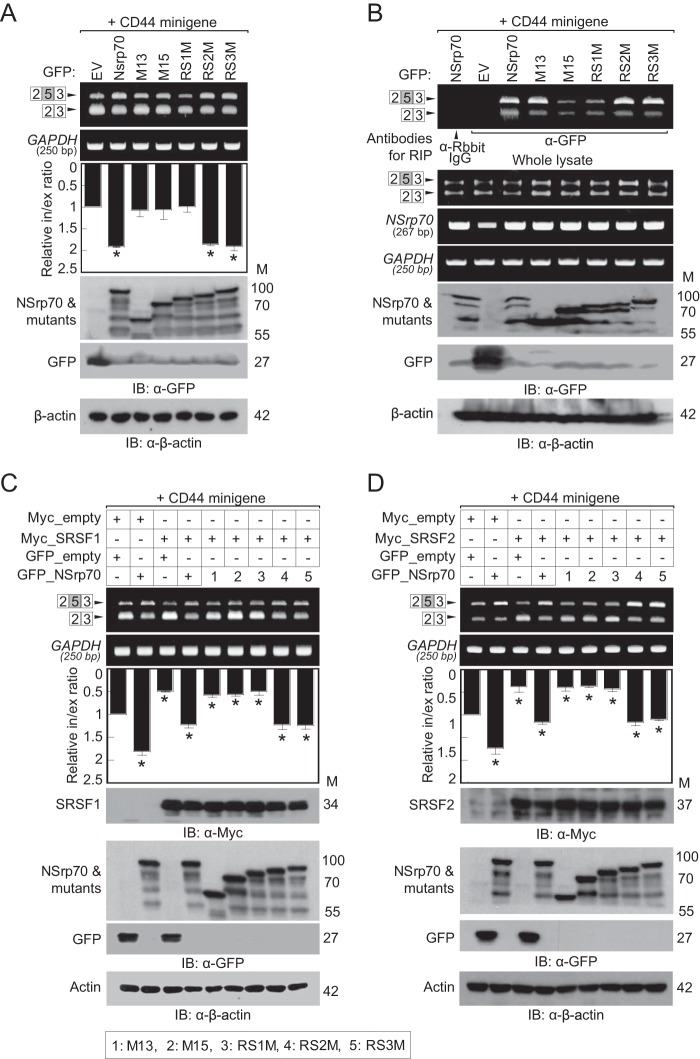
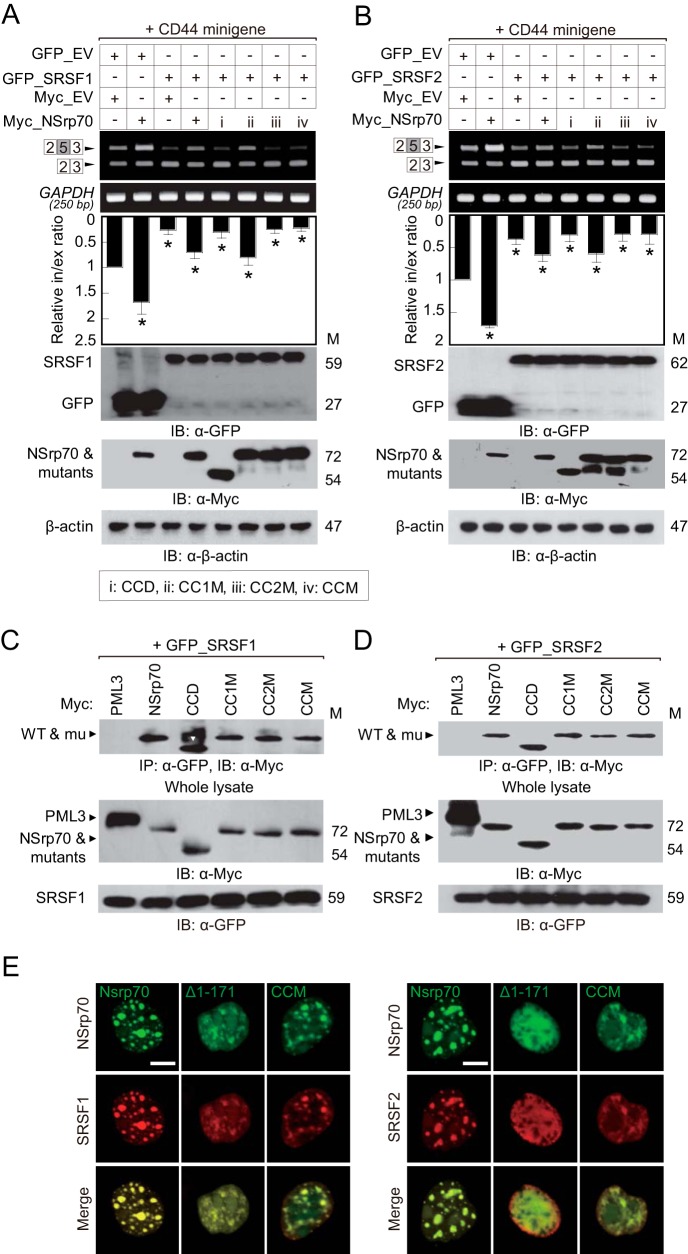
The RS domain in SR proteins has been demonstrated previously to provide a binding site for other proteins (21). Previously, we identified that NSrp70 has an RS-like region that is important for splicing activity and for interaction with SRSF1 and SRSF2 (9). To examine which specific RS-like region influences the activity of NSrp70 against SR proteins, we referred to a previous NSrp70 structure that shows a predicted coiled-coil domain in the center of the RS-like region (9). We subdivided the RS-like region into three regions on the basis of its predicted CC domain (Fig. 6A, right panel) and generated three deletion mutants, omitting one of each region: RS1M, RS2M (CC domain), and RS3M (Fig. 2A). In immunoprecipitation assays, RS1M showed no interaction with SRSF1 and SRSF2 (Fig. 2B). Furthermore, RS1M showed a marked loss of speckle localization (Fig. 2C). Interestingly, in the presence of RS1M or M15 (a mutant with the entire RS-like region deletion), SRSF1 and SRSF2 were dispersed within the nucleus, suggesting that NSrp70 may be important for the assembly of SR proteins at nuclear speckles.
FIGURE 6.
The coiled-coil domain of NSrp70 influences oligomerization and alternative splicing but not binding to mRNA. A, Schematic of wild-type and coiled-coil region point mutants of NSrp70 (left panel) and analysis of the predicted probability of oligomerization through the coiled-coil domain using the COILS program (right panel). M, molecular mass; NLS, nuclear localization signal; CCD, coiled-coil deleted NSrp70. B, HEK293T cells were transiently transfected with Myc_NSrp70 and its mutants. The cross-linking assay was performed as described in Fig. 5B. The ratio of oligomerization of NSrp70 mutants is shown as a bar graph (right panel). Data are representative of three independent experiments. M, monomer; T, trimer; D, dimer; GA, glutaraldehyde. C and D, HEK293T cells were cotransfected with Myc_NSrp70 constructs and the CD44 minigene. C, the splicing activity and protein expression were determined as described in Fig. 1A. Data are representative of three independent experiments. EV, pCS4–3Myc; Relative in/ex ratio, relative inclusion band/exclusion band ratio; IB, immunoblotting. *, p < 0.05 versus pCS4–3Myc. D, after cell lysis, the sample was immunoprecipitated with the indicated antibodies (anti-mouse IgG or anti-Myc). RNAs purified from the immunoprecipitated samples and whole lysates were determined by RT-PCR. Expression of NSrp70 and its mutants was confirmed by Western blotting with the indicated antibodies.
FIGURE 2.
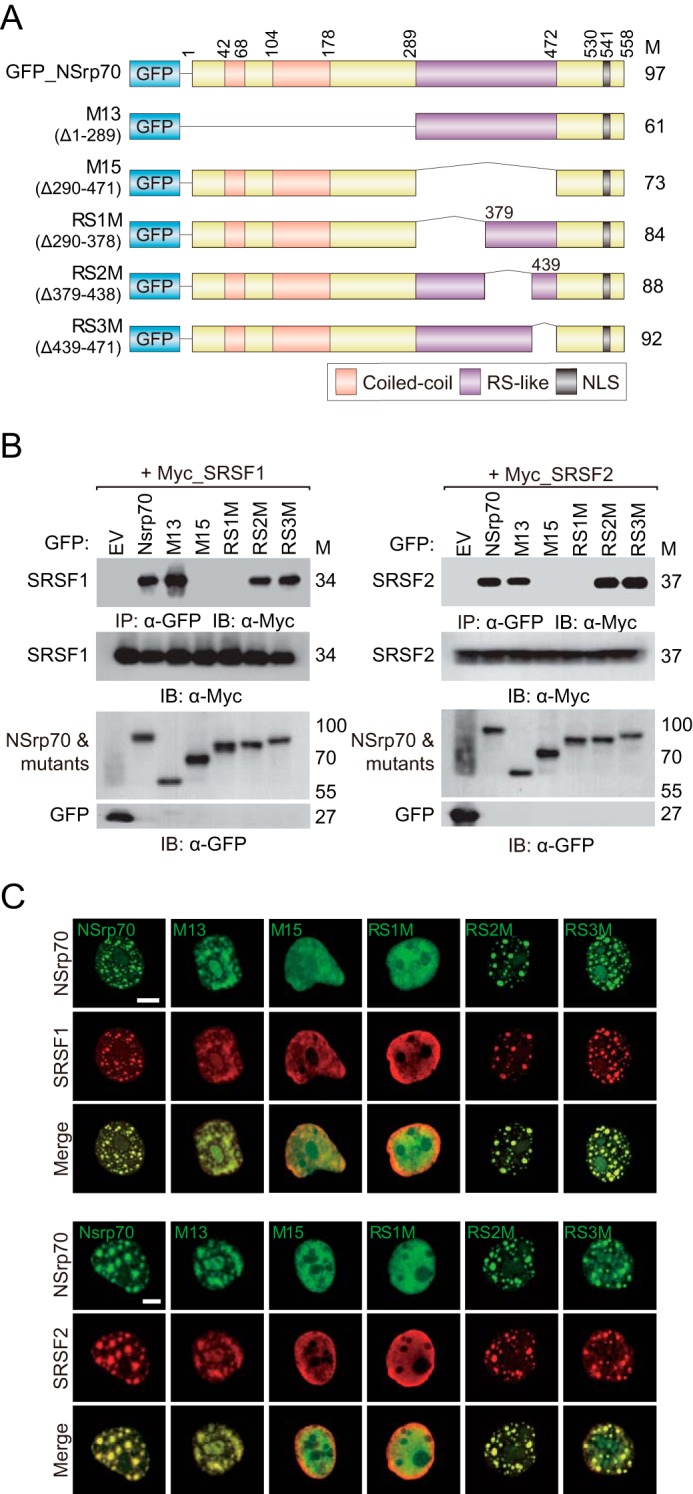
The first RS-like region of NSrp70 interacts with SRSF1 and SRSF2. A, schematic of wild-type and RS-like region deletion mutants of NSrp70. M, molecular mass; NLS, nuclear localization signal. B, HEK293T cells were cotransfected with GFP_NSrp70 constructs and Myc_SRSF1 (left panel) or _SRSF2 (right panel). After cell lysis, samples were immunoprecipitated and blotted with antibodies against the indicated proteins. EV, pEGFP-C1_empty vector; IP, immunoprecipitation; IB, immunoblotting. C, HEK293T cells were cotransfected for 48 h with GFP_NSrp70 constructs and Myc_SRSF1 or Myc_SRSF2. The cells were fixed on fibronectin-coated coverslips and stained for Myc_SRSF1 or Myc_SRSF2 (mouse polyclonal anti-Myc antibody) and visualized with a Texas Red-conjugated anti-mouse IgG under a confocal microscope equipped with a ×60 objective. Scale bars = 10 μm. Data shown are representative of three independent experiments.
NSrp70 Interacts with Target Pre-mRNA through the RS1 Region
We examined whether the RS1 region is also involved in physical interaction with target mRNA, thereby affecting splicing function. To this end, we performed an RNA immunoprecipitation assay using antibodies against GFP_NSrp70. We found that RS1M showed a complete loss of splicing activity and that, surprisingly, it was correlated with the loss of RNA binding for CD44 exon v5 (Fig. 3, A and B). These results suggest that the RS1 region provides a binding site not only for SR proteins but also for target mRNAs (Fig. 3, A and B). In agreement with this, RS1M could not counteract the alternative splicing activity of SRSF1 and SRSF2 on CD44 exon v5, whereas RS2M and RS3M retained complete activity (Fig. 3, C and D).
FIGURE 3.
The first RS-like region in NSrp70 is essential for mRNA binding and alternative splicing of CD44 exon v5. A and B, HEK293T cells were cotransfected with GFP_NSrp70 wild-type or mutants and the CD44 minigene. A, the splicing activity and protein expression were determined as described in Fig. 1A. EV, pEGFP-C1_empty vector; Relative in/ex ratio, relative inclusion band/exclusion band ratio; IB, immunoblotting; M, molecular mass. B, after cell lysis, the samples were immunoprecipitated with the indicated antibodies (anti-rabbit IgG or anti-GFP), and then the RNAs were purified from the immunoprecipitated samples for RT-PCR. Whole lysates were used as a positive control for CD44 exon v5. Expression of NSrp70, GFP, and β-actin was confirmed by Western blotting with the indicated antibodies. C and D, the CD44 minigene, GFP_NSrp70 constructs, and Myc_SRSF1 or Myc_SRSF2 were cotransfected into HEK293T cells. Splicing activity and protein expression were determined as described in Fig. 1A. All data are representative of three independent experiments. *, p < 0.05 versus pEGFP-C1_empty vector.
To extend these findings to a specific functional amino acid sequence in the RS1 region, we predicted the NSrp70 structure using the I-TASSER server. This server provided the top four predicted models on the basis of confidence scores. The RS-like regions (RS1, RS2, and RS3) of NSprp70 were cropped from the predicted structures and presented (supplemental Fig. 1). We found that all RS1 regions (amino acids 290–378) are comprised of four α-helical chains (supplemental Fig. 1). On the basis of this result, we individually deleted each α-helix in the RS1 region. However, none of deletion mutants showed a loss of interaction with SR protein and mRNA compared with the wild type (Fig. 4, B and D). Consistently, these mutants could also regulate alternative splicing of the CD44 exon v5 inclusion (Fig. 4C). These results demonstrate that the RS1 region is the minimum region required for binding to SR proteins and target mRNA and also for full alternative splicing activity.
FIGURE 4.
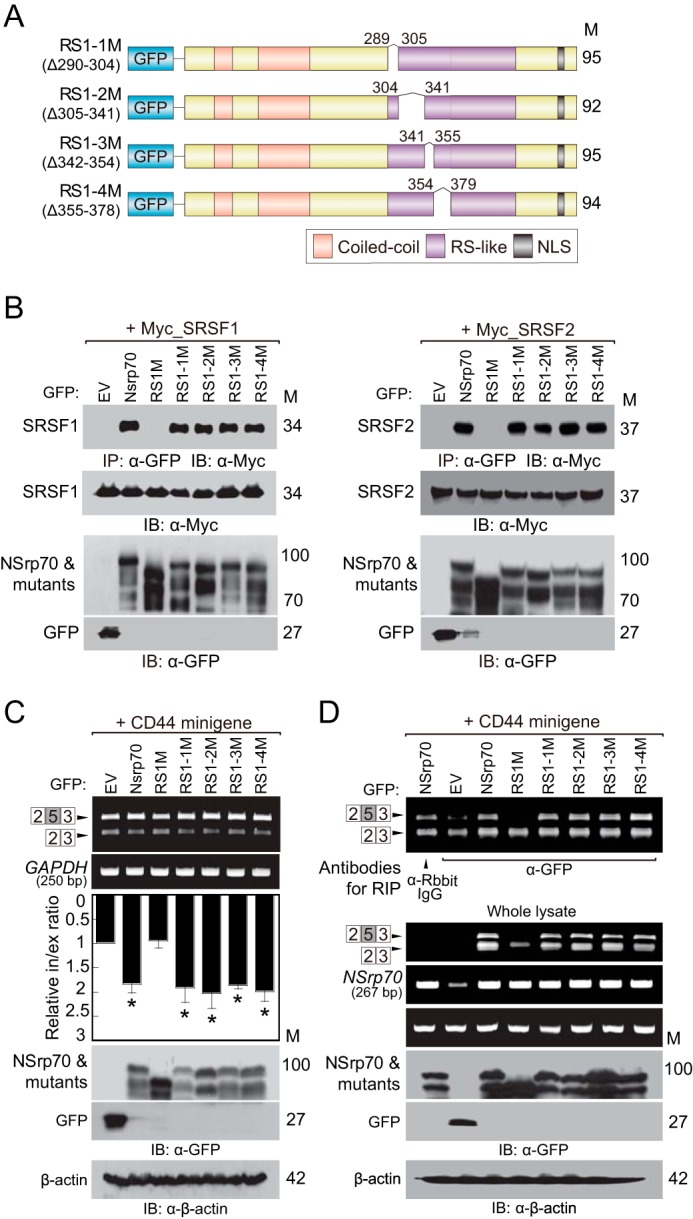
NSrp70 minimally requires the first RS-like region (amino acids 289–379) for alternative splicing of the CD44 minigene and binding to the SRSF1 and SRSF2. A, schematic of partial deletion mutants of RS1M. M, molecular mass; NLS, nuclear localization signal. B, HEK293T cells were cotransfected with partial deletion mutants of GFP_RS1M and Myc_SRSF1 (left panel) or Myc_SRSF2 (right panel). After cell lysis, samples were immunoprecipitated and blotted with antibodies against the indicated proteins. EV, pEGFP-C1_empty vector; IP, immunoprecipitation; IB, immunoblotting. C, HEK293T cells were cotransfected with partial deletion mutants of GFP_RS1M and the CD44 minigene. The splicing activity and protein expression were determined as described in Fig. 1A. Data are representative of three independent experiments. *, p < 0.05 versus pEGFP-C1_empty vector. D, HEK293T cells were cotransfected using the same conditions as in Fig. 4C. After cell lysis, the sample was immunoprecipitated with the indicated antibodies (anti-rabbit IgG or anti-GFP). RNAs purified from the immunoprecipitated samples and whole lysates were determined by RT-PCR. Expression of NSrp70 and its mutants were confirmed by Western blotting with the indicated antibodies.
The CC Domain of NSrp70 Influences Oligomerization and Splicing Activity
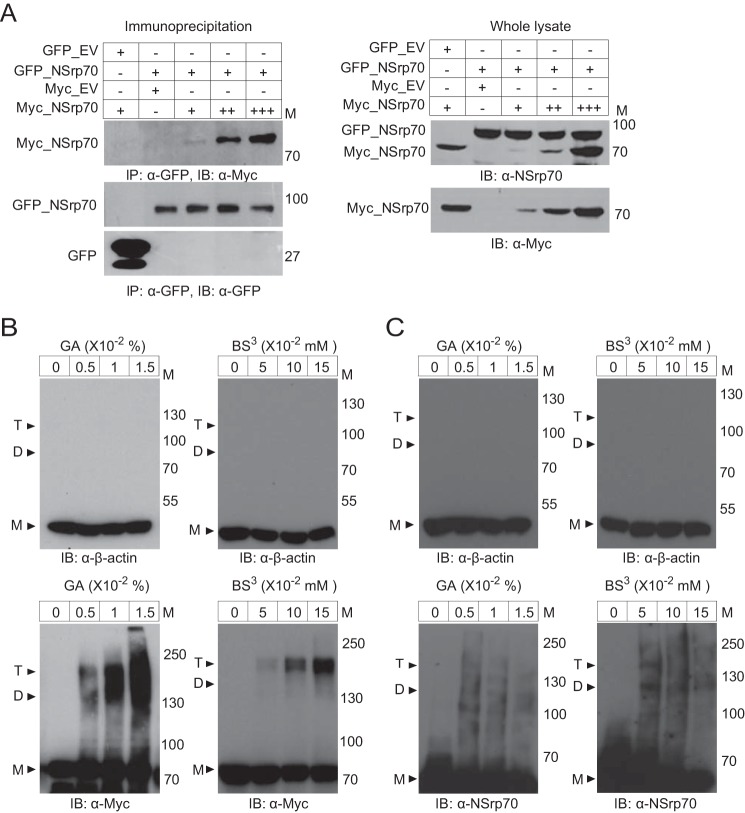
NSrp70 has an N-terminal CC domain, and we have demonstrated previously that deletion of the CC domain (amino acids 1–170) results in loss of NSrp70 alternative splicing activity. Because the CC domain is known to promote dimerization (22, 23), we questioned whether NSrp70 is a dimer, and, if so, whether dimerization is required for its alternative splicing activity. To this end, we cotransfected HEK293T cells with GFP_NSrp70 and increasing concentrations of Myc_NSrp70 and performed immunoprecipitation assays with an antibody against GFP. Western blotting analysis revealed that Myc_NSrp70 was recovered at GFP_NSrp70-transfected HEK293T cell samples but not the GFP sample, suggesting that NSrp70 is a dimer (Fig. 5A). To further confirm the dimerization or oligomerization of NSrp70, we also performed a protein cross-linking assay using glutaraldehyde and BS3. As shown in Fig. 5B, cell lysates with Myc_NSrp70, but not β-actin, were detected as dimers or trimers, suggesting that NSrp70 exists as a dimer or multimer under physiological conditions. In addition, to rule out nonspecific interactions, we performed a cross-linking assay with purified His-tagged NSrp70 recombinant protein and found that His_NSrp70 also showed dimer or trimer forms after cross-linking (Fig. 5C). To further confirm that NSrp70 dimerization is mediated by the CC domain, we predicted the structure of the CC domain-induced multimerization using the COILS program (24) (Fig. 6A, right panel) and then generated mutants of the CC domain (CC1M, CC2M, and CCM) using site-directed mutagenesis (Fig. 6A, left panel). Cross-linking assays revealed that mutation of the CC2 region (CC2M) significantly reduced dimer or multimer formation compared with the wild type or CC1M (Fig. 6B).
FIGURE 5.
NSrp70 exists as a dimer or oligomer. A, HEK293T cells were cotransfected with GFP_NSrp70 and increasing amounts of Myc_NSrp70. After cell lysis, samples were immunoprecipitated and blotted with antibodies against the indicated proteins. M, molecular mass; IP, immunoprecipitation; IB, immunoblotting. B, HEK293T cells were transiently transfected with Myc_NSrp70. The cell lysates were incubated on ice for 20 min with glutaraldehyde (0–0.015%) and for 50 min with BS3 (0–0.015 mm). The samples were resolved by SDS-PAGE and blotted with anti-β-actin or anti-Myc antibodies. Protein positions of the monomer (M), dimer (D), and trimer (T) are indicated by arrowheads. All data are representative of three independent experiments. GA, glutaraldehyde. C, recombinant His_NSrp70 and non-muscle actin were used for the cross-linking assay, and the results were determined as described above (A). All data are representative of three independent experiments.
We next questioned whether oligomerization of NSrp70 affects the alternative splicing of target mRNA. To this end, HEK293T cells were cotransfected with each construct (Fig. 6A, left panel) and with the CD44 exon v5 minigene, and exon inclusion was analyzed by RT-PCR. CC2M and CCM both showed loss of exon inclusion activity compared with the wild type (Fig. 6C). However, these mutants had a normal binding affinity for target mRNA (Fig. 6D), suggesting that NSrp70 oligomerization plays a role in splicing activity but not in binding to target mRNA. Similarly, CCM or CC2M could not block SRSF1- and SRSF2-mediated CD44 exon v5 exclusion (Fig. 7, A and B), although these mutant proteins showed no loss of binding to SRSF1 and SRSF2 (Fig. 7, C and D).
FIGURE 7.
The coiled-coil domain of NSrp70 has no interaction site for SRSF1 or SRSF2. A and B, the CD44 minigene, Myc_NSrp70 constructs, and GFP_SRSF1 or GFP_SRSF2 were cotransfected into HEK293T cells. The splicing activity and protein expression were determined as described in Fig. 1A. *, p < 0.05 versus cotransfection sample of GFP_empty vector and Myc_empty vector. Relative in/ex ratio, relative inclusion band/exclusion band ratio; M, molecular mass; IB, immunoblotting; CCD, coiled-coil deleted NSrp70. C and D, HEK293T cells were cotransfected with Myc_NSrp70 constructs and GFP_SRSF1 (left panel) or GFP_SRSF2 (right panel). After cell lysis, samples were immunoprecipitated and blotted with antibodies against the indicated proteins. mu, mutant; IP, immunoprecipitation. E, HEK293T cells were cotransfected for 48 h with GFP_NSrp70 constructs and Myc_SRSF1 or Myc_SRSF2. The cells were fixed on fibronectin-coated coverslips and stained for Myc_SRSF1 or Myc_SRSF2 (mouse polyclonal anti-Myc antibody) and visualized with a Texas Red-conjugated anti-mouse IgG under a confocal microscope equipped with a ×60× objective. All data are representative of three independent experiments. Scale bars = 10 μm.
Finally, we determined whether oligomerization of NSrp70 affects the assembly of nuclear speckles. Interestingly, although speckles were present, the presence of the CC domain deletion mutant (Δ1–171) or CCM resulted in no evident speckle boundary compared with wild-type NSrp70 (Fig. 7E). Taken together, these results suggest that the CC domain is also important for the condensational assembly of proteins in nuclear speckles.
NSrp70 Interacts Directly with Target Pre-mRNA through the RS1 Region
Our results suggested that the RS1 region of NSrp70 is important for interaction not only with SR proteins (SRSF1 and SRSF2) but also with target pre-mRNA, a result confirmed by RIP. At this point, we raised the question of whether NSrp70 binds directly to target pre-mRNA or whether this occurs via an SR protein that could interact with target pre-mRNA.
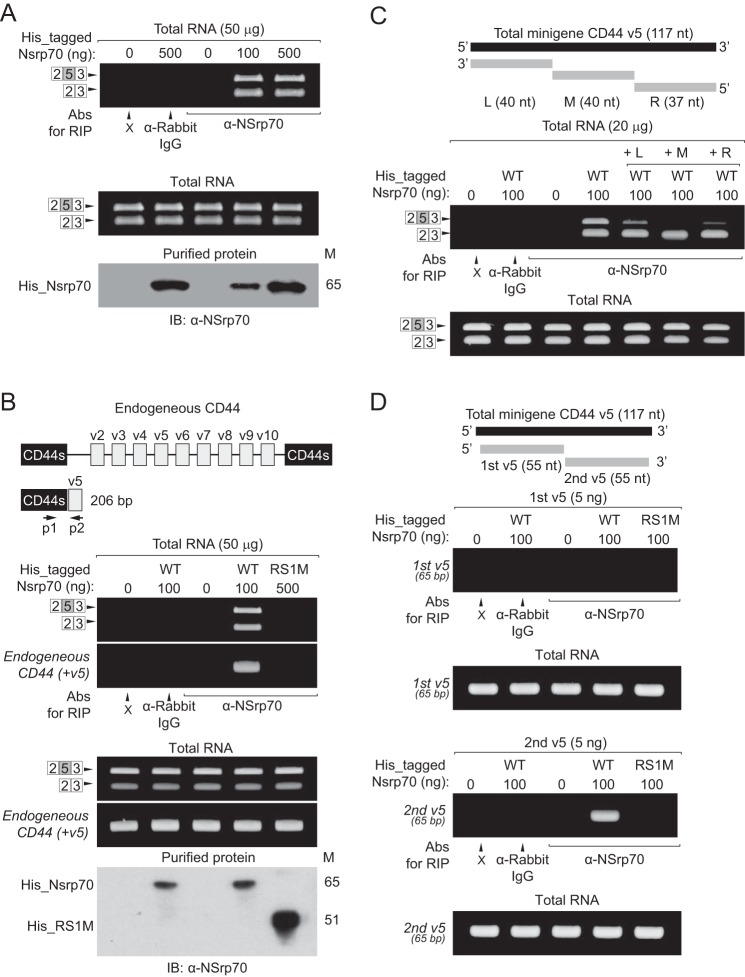
Our previous RIP assay (Figs. 3B and 4D) could not exclude endogenous SR proteins that contained a conserved RRM and also could not rule out the formation of NSrp70-SR protein complexes or SR protein-mRNA complexes. To solve this problem, we utilized recombinant RIP, a RIP method described previously (18). We incubated recombinant His-tagged NSrp70 with total RNA purified from CD44 minigene-transfected HEK293T cells (Fig. 8A). Subsequent steps were performed as for the conventional RIP method. Finally, we found that NSrp70 could interact directly with target pre-mRNA compared with the negative control (Fig. 8A). Interestingly, RS1M could not immunoprecipitate CD44 exon v5 as well as endogenous CD44 containing variable exon 5, suggesting that RS1 is a critical region for target RNA binding (Fig. 8B).
FIGURE 8.
NSrp70 binds directly to target mRNA. A, recombinant His_NSrp70 (100–500 ng) proteins were mixed with total RNA (50 μg) purified from CD44 minigene-transfected HEK293T cells. RNA-protein complexes were immunoprecipitated with the indicated antibodies (Abs, anti-rabbit IgG or anti-NSrp70). RNAs were purified from the immunoprecipitation sample, and then RT-PCR was performed for CD44. As a loading control, the existence of CD44 mRNA was confirmed from each sample by RT-PCR. Concentrations of recombinant His_NSrp70 were confirmed by Western blotting with α-NSrp70 antibody. M, molecular mass; IB, immunoblotting. B, recombinant His_NSrp70 (100 ng) and His_RS1M (500 ng) were mixed with total RNA (50 μg) purified from CD44 minigene-transfected HEK293T cells. The RIP assay was performed as described above (A). CD44s, CD44 standard isoform; endogenous CD44 (+v5), endogenous CD44 containing exon v5; p1, CD44v5 p1 primer; p2, CD44v5 p2 primer. C, for the RNA competition assay, total RNA (20 μg) purified from CD44 minigene-transfected HEK293T cells was incubated with antiparallel minigene CD44 exon v5 RNA (L, M, R). After incubation for 10 min, the RNA mixtures were mixed with His_NSrp70 (100 ng). The RIP assay was performed as described above (A). nt, nucleotide number; L, left sequence; M, middle sequence; R, right sequence. D, His_NSrp70 (100 ng) and His_RS1M (500 ng) were mixed with synthesized RNAs as shown (first and second region of CD44 exon v5). The RIP assay was performed as described above (A). All data are representative of three independent experiments. dA, adenine.
As shown in Fig. 8, A and B, NSrp70 binds the CD44 minigene not only with exon v5 (2 + 5 + 3) but also without v5 (2 + 3). These results led us to address whether the RS1 region binds directly to exon v5 of CD44 mRNA. We therefore partially mapped the specific site of CD44 exon v5 interaction with the RS1 region of NSrp70. An RNA competition assay revealed that the mid (M) region of CD44 exon v5 is most important for binding to NSrp70 but that the other regions also affect binding affinity (Fig. 8C). To further confirm the specific binding sequences in CD44 exon v5, we synthesized two separate parts (first v5 and second v5) of CD44 exon v5 RNAs tagged with 10 adenine nucleotides (Fig. 8D, top panel). Wild-type NSrp70, but not RS1M, could immunoprecipitate the second region of CD44 exon v5 (Fig. 8D). Combined with the results of the RNA competition assay (Fig. 8C), these results demonstrate that the RS1 region of NSrp70 binds directly to the mid-to-rear region of CD44 exon v5.
Discussion
Expression of the cell surface molecule CD44 is an excellent example of alternative splicing, and, therefore, CD44 exon v5 has been investigated extensively (25, 26). In this work, we found that the SR proteins SRSF1 and SRSF2 induce exclusion of CD44 exon v5 in a concentration-dependent manner. Interestingly, however, we observed that NSrp70 blocks the function of two of these SR proteins and induces inclusion of CD44 exon v5. Physical interaction with SR proteins and with target mRNA was critical for the NSrp70-induced inclusion of CD44 exon v5. The RS1 region provided a binding site for both SR proteins and CD44 mRNA. Therefore, RS1 deletion significantly diminished speckle localization of NSrp70. Moreover, RS1 deletion resulted in dispersed localization of SRSF1 and SRSF2 in the nucleus, suggesting that NSrp70 may act as a molecular scaffold and recruit other proteins for nuclear speckle organization. The N-terminal coiled-coil domain is important for the oligomerization of NSrp70 and also influences the activity of NSrp70.
A previous model of SRSF1 activity in CD44 exon v5 splicing proposed that SRSF1 has a RNA-binding specificity for the purine-rich splicing enhancer (27) and that the CD44 exon v5 region consists of a purine-rich exon splicing enhancer site (28). This model suggested that SRSF1 increases CD44 exon v5 inclusion through interference with the repressive activity of heterogeneous nuclear ribonucleoprotein, which is known to counteract the activity of SRSF1 (29, 30). However, we unexpectedly found that both SRSF1 and SRSF2 increase CD44 exon v5 exclusion in a concentration-dependent manner, suggesting that SRSF1 is not always specific for exon inclusion but may also be involved in exon exclusion. In agreement with this, a recent genome-wide analysis has revealed that SR proteins (SRSF1–4) cause similar numbers of exon inclusion and exon inclusion events (31–33). This finding suggests that the combinational contents of the splicing machinery complex are more important for alternative splicing than the specific mRNA interaction tendency of a single SR protein. Although a more detailed understanding of the mechanisms of NSrp70 function are needed, these results suggest that, after site selection, there are further, more complicated mechanisms to determine exon inclusion or exclusion of certain mRNAs. Another possibility is that the composition of the RNA splicing assembly may be another determinant for site selection.
In a previous report, we observed that NSrp70 has an RS-like region (amino acids 290–471) that is important for the alternative site selection of several minigenes. On the basis of these results, we hypothesized that binding of NSrp70 to SRSF1 through this domain is critical to suppress the negative function of SRSF1 on CD44 exon v5 inclusion. Indeed, the RS domain has been proposed to mediate protein-protein interactions for spliceosomal recruitment and nuclear speckle assembly (34, 35). Therefore, the interaction between NSrp70 and SRSF1 and SRSF2 through the RS-like region is not surprising, although this domain was not a typical RS domain containing >40% of a consecutive RS sequence (9, 36). In the results presented here, however, it is interesting to note that binding to SRSF1 and SRSF2 only involves the first RS1 region. However, further subdivision of RS1 regions had no effect, suggesting that RS1 is the minimum region required for binding to SRSF1 and SRSF2 and to target mRNA.
An important question raised at this point was how the RS1-like region binds to both SR proteins and mRNA simultaneously. Several reports have demonstrated that SR proteins bind to protein and mRNA through two separate domains: the RRM and RS domains. Of interest is that NSrp70 binds both proteins and mRNA through only one RS1-like region. Because the sequence of the RS1-like region in NSrp70 is different from the typical RRM and RS domain and because it only has a few SR dipeptides with no concomitant features of an RRM (9), we suggest here a unique function of the RS1-like region: to recruit both interacting proteins and mRNA simultaneously. To understand this, we modeled the NSrp70 structure using the I-TASSER server and found that the RS1-like region consisted of only an α helix (supplemental Fig. 1). In general, the components of most RRM- and RNA-binding domains have an α helix and β sheets together (37). Interestingly, there are two proteins that contain only an α-helix in the RNA binding domain: Pumilio and Vts1. These proteins contain a pumilio domain and a sterile alpha motif (SAM) domain, respectively (38, 39). Especially the predicted structure of the RS1-like region in NSrp70 is similar to the pumilio domain, and the sequence identity between the two domains was lower than 20% (data not shown). The property of the pumilio domain to bind the interacting proteins in addition to mRNA also suggests a similar function with NSrp70. Another study is now underway to identify whether the two proteins, NSrp70 and pumilio, have any functional similarity.
Proteins containing the CC domain are known to induce homophilic oligomerization through dimeric or trimetric interactions (40, 41). Consistent with this, we found that NSrp70 could self-interact to form multimers through the CC domain and that multimerization influenced its alternative splicing activity but had no effect on binding to SR proteins or to target mRNA. These results suggest that, although the CC domain does not directly affect the activity of NSrp70, it may affect activity indirectly by modulating nuclear speckle organization. Recently, several reports have suggested that RNA-binding proteins are involved in functional aggregation, forming paraspeckles in the nucleus and RNA-protein granules, known as stress granules, in the cytoplasm (10). For example, proteins of the DBHS family, including SFPQ (PSF), NONO (p54nrb), and PSPC1, are major components of paraspeckles and can modulate paraspeckle formation in the nucleus (11). Proteins of the DBHS family have two RRMs at the N terminus, a conserved core domain known as NONA/paraspeckle, and a CC domain at the C terminus. Crystal structures of DBHS family members have revealed that these proteins polymerize into infinite linear self-dimer structures through the CC domain, which is important for DBHS proteins targeting paraspeckles (12, 13). DBHS proteins bind to target DNA or NEAT1 as scaffolding long-noncoding RNA for paraspeckle formation, and this unique ability to polymerize in a linear pattern helps these proteins to localize to the condensational assembly binding site, providing enriched recruitment of other functional factors. This could rapidly promote a specific response, such as DNA damage repair or a transcriptional event. Indeed, these self-associated oligomers are connected with recent themes of higher-order assemblies in signaling complexes leading to signal amplification, reduction of biological noise, and temporal and spatial control of signal transduction (42).
Although we discovered that NSrp70 counteracts SRSF1 and SRSF2 in terms of CD44 exon v5 alternative splicing, we do not yet know whether NSrp70 also effectively blocks SR protein function in vivo. Interestingly, a previous report has demonstrated that CD3 ζ chain mRNA in T cells exists as two isoforms: a full sequence form and a specific 3′ UTR-deleted form that influences mRNA stability, leading to mRNA degradation (43). Interestingly, we have evidence that ectopic expression of NSrp70 reduces the expression of the CD3 ζ chain in Jurkat T cells.3 Further research is underway, but we tentatively suggest that NSrp70 may act as a negative regulator of T cell activation through control of mRNA stability against SRSF1 or SRSF2.
We also investigated potential binding proteins of NSrp70 using LC-mass methods and found that the candidate proteins show a broad spectrum related to gene regulation, including DNA helicases, small nuclear ribonucleoproteins, heterogeneous nuclear ribonucleoproteins, SR proteins, exon junction complex proteins, and ribosomal proteins.3 These results indicate that NSrp70 may be involved in many physiological processes. Interestingly, we found recently that the level of NSrp70 is reduced dramatically during T cell activation in response to anti-CD3/28 antibodies, suggesting that NSrp70 is an important regulator of T cell activation or differentiation. Further studies are now in progress to elucidate the global gene expression profiles that are affected by NSrp70 in T cells.
In conclusion, we identified that NSrp70 has two major domains. The first is the N-terminal CC domain, which is important for self-interaction, allowing NSrp70 to form multimers that influence nuclear speckle organization and NSrp70 activity. The second is the RS1 region, which provides a binding site for the SR proteins SRSF1 and SRSF2 and also for target mRNA. The opposing splicing activity of NSrp70 compared with SRSF1 and SRSF2 suggests that it acts as a molecular balancer for the expression of certain genes whose expression is controlled by SRSF1 and SRSF2. Further study, in addition to these results, of this novel splicing factor may provide us with new insights into RNA biogenesis and protein isoform regulation.
Author Contributions
C. H. K. performed and analyzed most of the experiments and wrote the manuscript. Y. D. K. and E. K. C. performed and analyzed the experiments shown in Figs. 1, 5, and 7. H. R. K. and B. R. N. carried out and analyzed the experiments involving immunofluorescence staining and confocal imaging analysis and helped with manuscript preparation. S. H. I. provided technical assistance and contributed to the preparation of the manuscript and the Figures. C. D. J. conducted and designed the research, analyzed the data, and wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
This work was supported by Basic Science Program Grant 2015R1A2A1A15052658 and Creative Research Initiative Program Grant 2015R1A3A2066253 through National Research Foundation (NRF) grants funded by the Ministry of Science, ICT, and Future Planning; by Basic Science Program Grant 2013R1A6A3A04064259 through NRF grants funded by the Ministry of Education, Korea, and the Bio Imaging Research Center at the Gwangju Institute of Science and Technology. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Figure 1.
C. H. Kim, Y. D. Kim, E. K. Choi, H. R. Kim, B. R. Na, S. H. Im, and C. D. Jun, unpublished data.
- DBHS
- Drosophila behavior/human splicing
- CC
- coiled-coil
- RRM
- RNA recognition motif
- BS3
- bis(sulfosuccinimidyl) suberate
- RIP
- RNA immunoprecipitation.
References
- 1. Lamond A. I., and Spector D. L. (2003) Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 4, 605–612 [DOI] [PubMed] [Google Scholar]
- 2. Spector D. L., and Lamond A. I. (2011) Nuclear speckles. Cold Spring Harb. Perspect. Biol. 10.1101/cshperspect.a000646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hall L. L., Smith K. P., Byron M., and Lawrence J. B. (2006) Molecular anatomy of a speckle. Anat. Rec. A. Discov. Mol. Cell Evol. Biol. 288, 664–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fakan S. (1994) Perichromatin fibrils are in situ forms of nascent transcripts. Trends Cell Biol. 4, 86–90 [DOI] [PubMed] [Google Scholar]
- 5. Tripathi V., Song D. Y., Zong X., Shevtsov S. P., Hearn S., Fu X.-D., Dundr M., and Prasanth K. V (2012) SRSF1 regulates the assembly of pre-mRNA processing factors in nuclear speckles. Mol. Biol. Cell 23, 3694–3706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mintz P. J., Patterson S. D., Neuwald A. F., Spahr C. S., and Spector D. L. (1999) Purification and biochemical characterization of interchromatin granule clusters. EMBO J. 18, 4308–4320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Long J. C., and Caceres J. F. (2009) The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 417, 15–27 [DOI] [PubMed] [Google Scholar]
- 8. Zhong X.-Y., Wang P., Han J., Rosenfeld M. G., and Fu X.-D. (2009) SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol. Cell 35, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim Y.-D., Lee J.-Y., Oh K.-M., Araki M., Araki K., Yamamura K., and Jun C.-D. (2011) NSrp70 is a novel nuclear speckle-related protein that modulates alternative pre-mRNA splicing in vivo. Nucleic Acids Res. 39, 4300–4314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li Y. R., King O. D., Shorter J., and Gitler A. D. (2013) Stress granules as crucibles of ALS pathogenesis. J. Cell Biol. 201, 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fox A. H., Lam Y. W., Leung A. K., Lyon C. E., Andersen J., Mann M., and Lamond A. I. (2002) Paraspeckles: a novel nuclear domain. Curr. Biol. 12, 13–25 [DOI] [PubMed] [Google Scholar]
- 12. Lee M., Sadowska A., Bekere I., Ho D., Gully B. S., Lu Y., Iyer K. S., Trewhella J., Fox A. H., and Bond C. S. (2015) The structure of human SFPQ reveals a coiled-coil mediated polymer essential for functional aggregation in gene regulation. Nucleic Acids Res. 10.1093/nar/gkv156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Passon D. M., Lee M., Rackham O., Stanley W. A., Sadowska A., Filipovska A., Fox A. H., and Bond C. S. (2012) Structure of the heterodimer of human NONO and paraspeckle protein component 1 and analysis of its role in subnuclear body formation. Proc. Natl. Acad. Sci. 109, 4846–4850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fu X. D. (1995) The superfamily of arginine/serine-rich splicing factors. RNA 1, 663–680 [PMC free article] [PubMed] [Google Scholar]
- 15. Cáceres J. F., Misteli T., Screaton G. R., Spector D. L., and Krainer A. R. (1997) Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol. 138, 225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Twyffels L., Gueydan C., and Kruys V. (2011) Shuttling SR proteins: more than splicing factors. FEBS J. 278, 3246–3255 [DOI] [PubMed] [Google Scholar]
- 17. Cooper T. A. (2005) Use of minigene systems to dissect alternative splicing elements. Methods 37, 331–340 [DOI] [PubMed] [Google Scholar]
- 18. Townley-Tilson W. H., Pendergrass S. A., Marzluff W. F., and Whitfield M. L. (2006) Genome-wide analysis of mRNAs bound to the histone stem-loop binding protein. RNA 12, 1853–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang C., Liang H., Zhao H., and Jiang X. (2012) CD44 variant isoforms are specifically expressed on peripheral blood lymphocytes from asthmatic patients. Exp. Ther. Med. 4, 79–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang Y. (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kohtz J. D., Jamison S. F., Will C. L., Zuo P., Lührmann R., Garcia-Blanco M. A., and Manley J. L. (1994) Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature 368, 119–124 [DOI] [PubMed] [Google Scholar]
- 22. Mason J. M., and Arndt K. M. (2004) Coiled coil domains: stability, specificity, and biological implications. ChemBioChem 5, 170–176 [DOI] [PubMed] [Google Scholar]
- 23. Mahrenholz C. C., Abfalter I. G., Bodenhofer U., Volkmer R., and Hochreiter S. (2011) Complex networks govern coiled-coil oligomerization-predicting and profiling by means of a machine learning approach. Mol. Cell Proteomics 10.1074/mcp.M110.004994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lupas A., Van Dyke M., and Stock J. (1991) Predicting coiled coils from protein sequences. Science 252, 1162–1164 [DOI] [PubMed] [Google Scholar]
- 25. Bell M. V., Cowper A. E., Lefranc M. P., Bell J. I., and Screaton G. R. (1998) Influence of intron length on alternative splicing of CD44. Mol. Cell Biol. 18, 5930–5941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Loh T. J., Moon H., Cho S., Jung D. W., Hong S. E., Kim D. H., Green M. R., Zheng X., Zhou J., and Shen H. (2014) SC35 promotes splicing of the C5-V6-C6 isoform of CD44 pre-mRNA. Oncol. Rep. 31, 273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tacke R., and Manley J. L. (1995) The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 14, 3540–3551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. König H., Ponta H., and Herrlich P. (1998) Coupling of signal transduction to alternative pre-mRNA splicing by a composite splice regulator. EMBO J. 17, 2904–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Matter N., Marx M., Weg-Remers S., Ponta H., Herrlich P., and König H. (2000) Heterogeneous ribonucleoprotein A1 is part of an exon-specific splice-silencing complex controlled by oncogenic signaling pathways. J. Biol. Chem. 275, 35353–35360 [DOI] [PubMed] [Google Scholar]
- 30. Kashima T., and Manley J. L. (2003) A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat. Genet. 34, 460–463 [DOI] [PubMed] [Google Scholar]
- 31. Änkö M.-L., Müller-McNicoll M., Brandl H., Curk T., Gorup C., Henry I., Ule J., and Neugebauer K. M. (2012) The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol. 13, R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pandit S., Zhou Y., Shiue L., Coutinho-Mansfield G., Li H., Qiu J., Huang J., Yeo G. W., Ares M. Jr., and Fu X. D. (2013) Genome-wide analysis reveals SR protein cooperation and competition in regulated splicing. Mol. Cell 50, 223–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bradley T., Cook M. E., and Blanchette M. (2014) SR proteins control a complex network of RNA-processing events. RNA 10.1261/rna.043893.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu J. Y., and Maniatis T. (1993) Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 75, 1061–1070 [DOI] [PubMed] [Google Scholar]
- 35. Shen H., Kan J. L., and Green M. R. (2004) Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol. Cell 13, 367–376 [DOI] [PubMed] [Google Scholar]
- 36. Manley J. L., and Krainer A. R. (2010) A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev. 24, 1073–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lunde B. M., Moore C., and Varani G. (2007) RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 8, 479–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang X., McLachlan J., Zamore P. D., and Hall T. M. (2002) Modular recognition of RNA by a human Pumilio-homology domain. Cell 110, 501–512 [DOI] [PubMed] [Google Scholar]
- 39. Oberstrass F. C., Lee A., Stefl R., Janis M., Chanfreau G., and Allain F. H. (2006) Shape-specific recognition in the structure of the Vts1p SAM domain with RNA. Nat. Struct. Mol. Biol. 13, 160–167 [DOI] [PubMed] [Google Scholar]
- 40. Hadley E. B., Testa O. D., Woolfson D. N., and Gellman S. H. (2008) Preferred side-chain constellations at antiparallel coiled-coil interfaces. Proc. Natl. Acad. Sci. U.S.A. 105, 530–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Parry D. A., Fraser R. D., and Squire J. M. (2008) Fifty years of coiled-coils and α-helical bundles: a close relationship between sequence and structure. J. Struct. Biol. 163, 258–269 [DOI] [PubMed] [Google Scholar]
- 42. Wu H. (2013) Higher-order assemblies in a new paradigm of signal transduction. Cell 153, 287–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moulton V. R., and Tsokos G. C. (2010) Alternative splicing factor/splicing factor 2 regulates the expression of the ζ subunit of the human T cell receptor-associated CD3 complex. J. Biol. Chem. 285, 12490–12496 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.