TET2 and TET3 redundantly regulate Foxp3 stability, and their activity can be modulated by vitamin C.
Abstract
Ten-eleven translocation (TET) enzymes oxidize 5-methylcytosine (5mC) to 5-hydroxymethylcytosine and other oxidized methylcytosines, intermediates in DNA demethylation. In this study, we examine the role of TET proteins in regulating Foxp3, a transcription factor essential for the development and function of regulatory T cells (T reg cells), a distinct lineage of CD4+ T cells that prevent autoimmunity and maintain immune homeostasis. We show that during T reg cell development in the thymus, TET proteins mediate the loss of 5mC in T reg cell–specific hypomethylated regions, including CNS1 and CNS2, intronic cis-regulatory elements in the Foxp3 locus. Similar to CNS2-deficient T reg cells, the stability of Foxp3 expression is markedly compromised in T reg cells from Tet2/Tet3 double-deficient mice. Vitamin C potentiates TET activity and acts through Tet2/Tet3 to increase the stability of Foxp3 expression in TGF-β–induced T reg cells. Our data suggest that targeting TET enzymes with small molecule activators such as vitamin C might increase induced T reg cell efficacy.
DNA methyltransferases (DNMTs) add a methyl group to cytosine to generate 5-methylcytosine (5mC); in somatic cells, this modification is typically present in the dinucleotide CpG (Ooi et al., 2009). DNA methylation is gradually lost in a replication-dependent manner during several processes of cell lineage specification, including the differentiation of naive T cells into Th2 cells (Lee et al., 2002). The three mammalian members of the ten-eleven translocation (TET) family of Fe(II) and 2-oxoglutarate–dependent dioxygenases, TET1, TET2, and TET3 (Iyer et al., 2009; Tahiliani et al., 2009), successively oxidize 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) in DNA (Tahiliani et al., 2009; He et al., 2011; Ito et al., 2011). All three oxidized methylcytosine species are intermediates in DNA demethylation, the replacement of 5mC with unmodified C (Pastor et al., 2013; Wu and Zhang, 2014).
The X chromosome–encoded transcription factor Foxp3 is essential for the development and function of regulatory T (T reg) cells, a distinct lineage of CD4+ T cells that prevent autoimmunity and maintain immune homeostasis (Sakaguchi et al., 2008; Josefowicz et al., 2012). T reg cells that gain Foxp3 expression at precursor stage in the thymus are termed thymus-derived T reg cells, whereas those that develop extrathymically in vivo are termed peripherally derived T reg cells (Sakaguchi et al., 2008; Josefowicz et al., 2012; Abbas et al., 2013); Foxp3+-induced T reg (iT reg) cells can be generated from naive T cells by stimulation through the T cell receptor in the presence of the inducer TGF-β (Chen et al., 2003; Abbas et al., 2013).
Foxp3 expression during T reg cell differentiation is regulated by three conserved noncoding sequence (CNS) elements located in the first intron of the Foxp3 gene, upstream of the first coding exon (Zheng et al., 2010; Feng et al., 2014; Li et al., 2014). Of these, CNS2 (also known as T reg cell–specific demethylated region; Floess et al., 2007) is unusual in that it controls the stability of Foxp3 expression in a manner linked to the DNA modification status of CNS2 (Floess et al., 2007; Huehn et al., 2009; Toker and Huehn, 2011; Toker et al., 2013). First, CpG sites in the Foxp3 CNS2 element are predominantly unmethylated (C/5fC/5caC) in T reg cells, but fully methylated (5mC/5hmC) in naive T cells and iT reg cells (Floess et al., 2007; Kim and Leonard, 2007; Polansky et al., 2008; Zheng et al., 2010; Toker et al., 2013). Second, cell division results in the loss of Foxp3 expression (Zheng et al., 2010; Feng et al., 2014; Li et al., 2014), a phenomenon associated with increased DNA methylation at CNS2 and other regions of the Foxp3 locus (Feng et al., 2014); this loss of Foxp3 expression is far more pronounced in iT reg cells with methylated CNS2 than in T reg cells in which CNS2 is not methylated (Floess et al., 2007). Third, inhibition of DNA methylation by the DNMT inhibitor 5-azacytidine (Kim and Leonard, 2007; Polansky et al., 2008), or genetic deletion of the gene encoding DNMT1 (Josefowicz et al., 2009), eliminated the requirement for TGF-β and promoted Foxp3 expression by naive CD4+ T cells in response to TCR stimulation alone. Fourth, T reg cells from CNS2-deleted mice lose Foxp3 expression in a proliferation-, STAT5-, and NFAT-dependent manner after adoptive transfer, and the mice develop autoimmunity relatively late in life (Zheng et al., 2010; Feng et al., 2014; Li et al., 2014).
Here, we examined the role of TET proteins in regulating the stability of Foxp3 expression (i.e., the maintenance of T reg cell identity). We show that Tet2 and Tet3 act redundantly to maintain the demethylated status of several regulatory regions in T reg cells, including CNS1 and CNS2 in the Foxp3 gene. Similar to T reg cells from CNS2-deficient animals, T reg cells from Tet2/Tet3 double-deficient mice show a marked impairment of the stability of Foxp3 expression. Conversely, we show that addition of the TET activator vitamin C during mouse and human iT reg cell differentiation maintains TET enzymatic activity and potentiates the loss of 5mC in CNS1 and CNS2, an effect correlated with a pronounced increase in the stability of Foxp3 expression. Our data indicate that TET proteins regulate the stability of Foxp3 expression by modulating the methylation status of conserved noncoding elements within the Foxp3 locus as well as potentially other regulatory regions in the T reg cell genome, suggesting that targeting TET enzymes with small molecule activators such as vitamin C might increase iT reg cell efficacy in clinical applications such as transplant rejection and autoimmune disease.
RESULTS
TET proteins mediate the loss of 5mC in T reg cell–specific regulatory regions during T reg cell development
To examine the dynamic changes of DNA modification during T reg cell differentiation, we sorted cells from male Foxp3–internal ribosome entry site (IRES)–enhanced GFP (eGFP) reporter mice at sequential stages of their differentiation toward the T reg cell lineage in the thymus and their final maturation in the periphery. In these mice, a cassette encoding IRES-eGFP had been inserted into the 3′ untranslated region of the Foxp3 gene, generating a bicistronic region encoding both Foxp3 and eGFP under the control of the Foxp3 promoter (Haribhai et al., 2007). DNA was prepared from the following purified populations: double-positive (DP) thymocytes; CD4+CD25−Foxp3− single-positive (SP; CD4 SP) T cells; CD25+Foxp3− (Lio and Hsieh, 2008) and CD25−Foxp3+ (Tai et al., 2013) cells, both thought to be T reg cell precursors; thymic CD25+Foxp3+ T reg cells; and peripheral T reg cells (Fig. S1 A). Because bisulfite (BS) sequencing cannot distinguish 5mC and 5hmC (Huang et al., 2010), we used the oxidative BS (oxBS) method, which distinguishes 5mC and 5hmC at single-base resolution by selectively oxidizing 5hmC to 5fC (Booth et al., 2012, 2013). We note that neither BS nor oxBS sequencing (BS-seq and oxBS-seq, respectively) can distinguish unmodified C from 5fC or 5caC (Booth et al., 2015), leading us to avoid the term “DNA demethylation” (which implies complete replacement of 5mC by unmodified C) in favor of “loss of 5mC” (which includes the possibility of conversion of 5mC to 5fC or 5caC). Amplicons spanning 11 CpGs in the CNS2 element (Fig. 1 A) were sequenced using the MiSeq platform, and the percentage of 5mC, 5hmC, and C/5fC/5caC at each CpG was calculated.
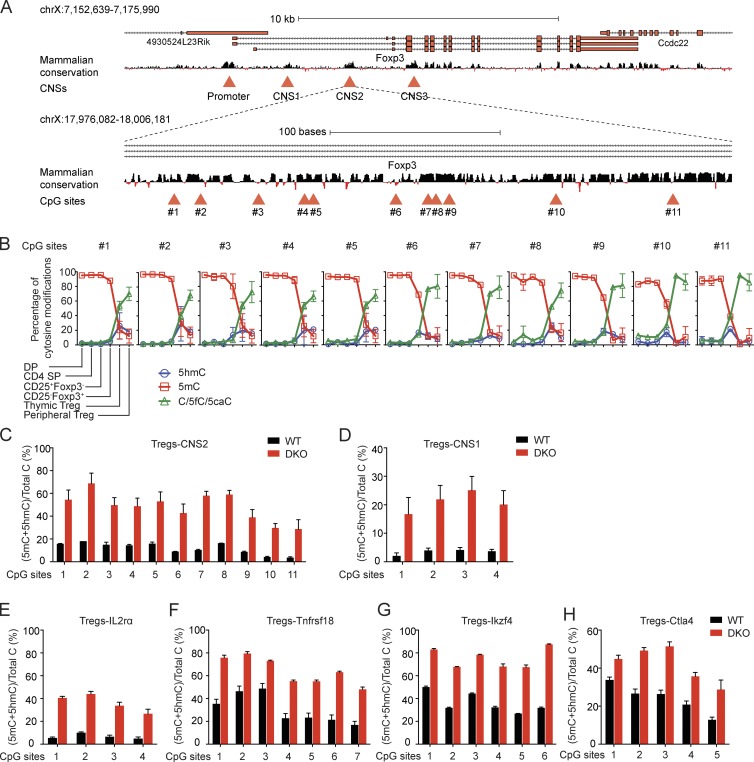
Figure 1.
Double deficiency of Tet2 and Tet3 impairs the loss of 5mC. (A) Schematic representation of the Foxp3 locus. Top: Foxp3 transcript variants and mammalian conservation track are shown, and the conserved promoter region and the three conserved noncoding sequences corresponding to the intronic enhancers CNS1, CNS2, and CNS3 are highlighted with red triangles. Bottom: 11 CpG sites in the CNS2 region that were assessed to determine their methylation-hydroxymethylation status are indicated with red triangles. (B) Dynamic changes in 5mC, 5hmC, and C/5fC/5caC in the Foxp3 CNS2 enhancer during T reg cell lineage specification. The graphs depict the cytosine modification status of the 11 CpGs in the CNS2 region as determined by BS-seq and oxBS-seq in six different T cell subsets (DP, CD4 SP, precursor T reg cells [CD25+Foxp3− and CD25−Foxp3+], thymic T reg cells, and peripheral T reg cells) that together capture the sequential steps of differentiation that lead to T reg cell specification. Error bars (which in many cases are too small to be detectable) show mean ± SD of thousands of sequencing reads from two independent experiments. (C–H) Graphs depicting the percentage of 5mC + 5hmC determined by BS-seq in peripheral T reg cells in 11 CpGs in Foxp3 CNS2 (C), 4 CpGs in Foxp3 CNS1 (D), 4 CpGs in Il2ra intron 1a (E), 7 CpGs in Tnfrsf18 exon 5 (F), 6 CpGs in Ikzf4 intron 1b (G), and 5 CpGs in Ctla4 exon 2 (H). Error bars show mean ± SD of thousands of sequencing reads from three to four independent experiments.
The results were unambiguous. As previously reported for CD4 SP cells (Toker et al., 2013), all CpGs in Foxp3 CNS2 were methylated in both DP and CD4 SP cells and progressively lost 5mC in T reg cell precursors, with thymic and peripheral T reg cells showing the lowest levels of 5mC (Fig. 1 B). CD25−Foxp3+ precursor cells displayed a clear loss of 5mC, which was most apparent at CpG motifs 6, 9, 10, and 11. In contrast, 5hmC generally reached its peak levels in either CD25−Foxp3+ precursor cells or thymic T reg cells (Fig. 1 B). These data suggest that CD25+Foxp3− precursors represent an earlier stage of T reg cell differentiation compared with CD25−Foxp3+ precursors, consistent with previous findings that CD25−Foxp3+ precursors differentiate more potently than CD25+Foxp3− precursors into CD25+Foxp3+ T reg cells both in vitro and in vivo (Tai et al., 2013).
The appearance of 5hmC at intermediate stages of T reg cell lineage specification pointed to the involvement of TET proteins, the enzymes responsible for generating 5hmC (Tahiliani et al., 2009). To test the importance of TET proteins in T reg cell function, we generated and examined Tet2−/−Tet3fl/flCD4Cre and Tet2fl/flTet3fl/flCD4Cre (here termed Tet2/3 DKO) mice. Tet2 and Tet3 were chosen because they are the major TET proteins expressed in differentiated tissues and cell types, including lymph nodes and spleen (Tsagaratou and Rao, 2013), and because of a previous study showing that 5hmC, Tet2, and Tet3 are enriched in thymic T reg cell populations (Toker et al., 2013). The strategies used for conditional disruption of the Tet2 and Tet3 genes have been described previously (Ko et al., 2011, 2015; Kang et al., 2015).
To test whether the loss of 5mC in Foxp3 CNS2 would be impaired in the absence of TET function, we examined the DNA modification status of Foxp3 regulatory regions in CD4+CD25+ peripheral T reg cells isolated from young (3–4 wk old) male Tet2/3 DKO mice. We first assessed 5mC and 5hmC levels by BS- and oxBS-seq. The 4 CpGs in CNS1 and the 11 CpGs in CNS2 were largely unmethylated (<10% 5mC) in WT peripheral T reg cells but showed increased methylation (40–60% 5mC) in peripheral T reg cells from Tet2/3 DKO mice (Fig. S1, D and E); they were fully methylated (close to 100% 5mC) in CD4 SP thymocytes from both WT and Tet2/3 DKO mice, as expected (Fig. S1, G and H). In contrast, the five CpGs within CNS3, and all CpGs within two distinct regions of an upstream CpG island located between −5 and −5.5 kb upstream of the transcription start site, were completely unmethylated (<1% 5mC), both in peripheral T reg cells and in CD4 SP thymocytes from WT mice, and did not show any increase in methylation in Tet2/3 DKO mice (Fig. S1, F, I, and J). Thus, the four regulatory regions we examined in the Foxp3 locus lose 5mC at different developmental stages in the thymus: CNS1 and CNS2 lose 5mC after the CD4 SP stage in a process that requires Tet2 and Tet3, whereas CNS3 and the upstream CpG island lose 5mC before the CD4 SP stage and remain unmethylated in thymic T reg cells, even in the absence of Tet2 and Tet3.
Because CD4+CD25+ T reg cells cannot readily be distinguished from conventional activated CD4+ T cells, we ensured T reg cell purity by crossing Tet2/3 DKO mice with Foxp3-IRES-eGFP reporter mice. By standard BS-seq, peripheral T reg cells from Tet2/3 DKO mice showed a 20–50% increase in 5mC + 5hmC levels at CNS1 and CNS2 compared with WT T reg cells (Fig. 1, C and D) and a comparable 20–40% increase at several other regulatory regions that undergo T reg cell–specific DNA hypomethylation: Il2ra intron 1a, Tnfrsf18 exon 5, Ikzf4 intron 1b, and Ctla4 exon 2 (Fig. 1, E–H; Ohkura et al., 2012).
Notably, the increased DNA methylation required profound TET loss of function. Deletion of Tet2 alone had no effect, and deletion of Tet3 alone had only a mild effect, on the DNA modification status of CNS1 and CNS2 in CD4+CD25+ peripheral T reg cells from male Tet2−/− or Tet3fl/flCD4Cre mice compared with WT mice (Fig. S1, K and L). Together, the data show that Tet2 and Tet3 function redundantly to mediate the loss of 5mC not only at the Foxp3 locus but also at other regulatory regions during T reg cell development in vivo.
The stability of Foxp3 expression is compromised in the absence of Tet2 and Tet3
Given the established connection between CNS2 methylation status and T reg cell stability (Floess et al., 2007; Huehn et al., 2009; Zheng et al., 2010; Toker and Huehn, 2011; Toker et al., 2013), we compared the stability of Foxp3 expression in T reg cells from Tet2/3 DKO versus WT mice. Young Tet2/3 DKO mice (3–5 wk old) exhibited a normal percentage of Foxp3+ cells in the thymus, but a significantly reduced frequency of Foxp3+ cells in the periphery (Fig. 2 A).
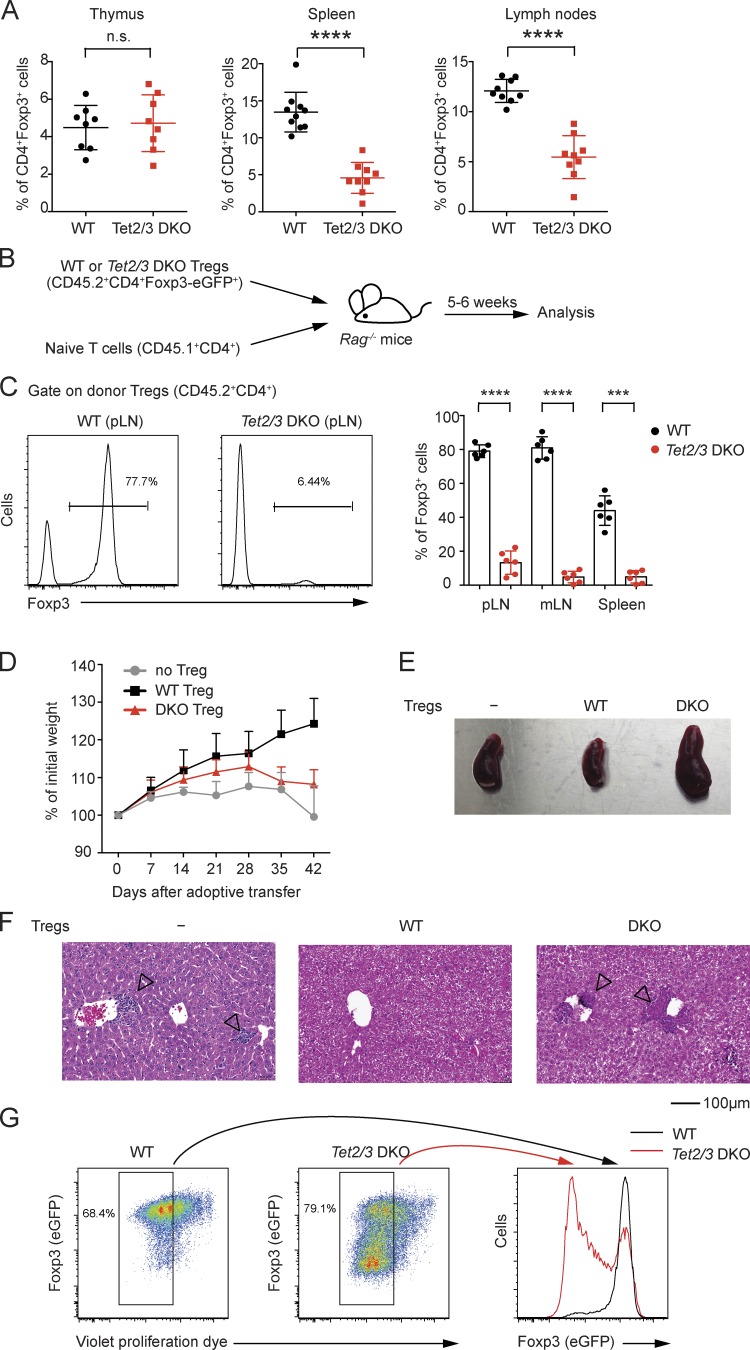
Figure 2.
The stability of T reg cells is significantly compromised with double deficiency of Tet2 and Tet3. (A) Percentage of Foxp3+ T reg cells among CD4 SP thymocytes or splenic and lymph node CD4+ T cells in Tet2/3 DKO mice and littermate controls. n = 8–9. n.s., not significant. (B) Schematic representation of the adoptive transfer experiment for assessing T reg cell stability in vivo. (C) Analysis of peripheral lymphoid organs from Rag-deficient mice 5–6 wk after adoptive transfer of peripheral T reg cells from young (3–4 wk old) WT (CD45.2+) or Tet2/3 DKO (CD45.2+) reporter mice as a CD4+Foxp3-eGFP+ population together with congenically marked naive T cells (CD45.1+CD4+). Left: Representative histograms of Foxp3+ cells within CD45.2+CD4+ cells. Right: The mean percentage of Foxp3+ cells in the CD45.2+CD4+ cell gate. Each dot represents one mouse. Error bars show mean ± SD from three independent experiments with six mice per group. pLN, peripheral lymph node. mLN, mesenteric lymph node. (D–F) The Foxp3 stability assay using T reg cells sorted from either WT or Tet2/3 DKO reporter mice as the CD4+Foxp3-eGFP+ population. The percent change in initial weight after adoptive transfer (D), the picture of spleens (E), and hematoxylin and eosin staining of livers (F) from adoptive transfer without T reg cells or with WT or Tet2/3 DKO T reg cells are shown. Arrowheads indicate mononuclear cell infiltration in the liver. Data are representative of three independent experiments. (G) WT or Tet2/3 DKO Foxp3-eGFP+ T reg cells were labeled with violet proliferation dye and cultured in vitro for 3 d and then analyzed for Foxp3-eGFP expression by flow cytometry. Left: Representative pseudocolor plots for WT or Tet2/3 DKO T reg cells. Right: Histogram overlay of Foxp3-eGFP+ cells from WT and Tet2/3 DKO T reg cells within the dividing cells. Data are representative of three independent experiments. ***, P < 0.001; ****, P < 0.0001 by Student’s t test.
We purified WT or Tet2/3 DKO T reg cells (CD45.2+CD4+Foxp3-eGFP+) by FACS and transferred them together with congenically marked WT CD45.1+CD4+ naive T cells (which serve as a source of IL-2) into Rag-deficient recipients (Fig. 2 B). When the recipient mice were examined 5–6 wk later, Tet2/3 DKO T reg cells exhibited a marked decrease of Foxp3 expression compared with WT T reg cells in peripheral lymphoid organs (Fig. 2 C). Consistent with loss of Foxp3 expression and consequent decrease of T reg cell function, recipient mice co-injected with naive T cells and Tet2/3 DKO T reg cells did not gain weight compared with mice that received WT T reg cells (Fig. 2 D) and exhibited splenomegaly (Fig. 2 E) and mononuclear cell infiltration in the liver (Fig. 2 F, arrowheads). Furthermore, when stimulated with anti-CD3/anti-CD28 in the presence of IL-2, Foxp3-eGFP+ T reg cells from Tet2/3 DKO mice progressively lost, whereas WT Foxp3-eGFP+ T reg cells maintained, Foxp3 (eGFP) expression upon cell division (Fig. 2 G). Thus, double deficiency of Tet2 and Tet3 in T cells compromises not only the loss of 5mC at T reg cell–specific regulatory regions, including CNS1 and CNS2, but also the stability of Foxp3 expression subsequent to cell division in peripheral T reg cells.
Vitamin C promotes the loss of 5mC in CNS1 and CNS2 during iT reg cell differentiation
Naive T cells can be converted into iT reg cells in vitro by TCR stimulation in the presence of TGF-β (Chen et al., 2003), and this process is potentiated by the addition of retinoic acid (RA; Benson et al., 2007). CpG sites in the Foxp3 CNS2 element are predominantly unmethylated in T reg cells but uniformly methylated in iT reg cells generated in the presence of TGF-β in vitro (Floess et al., 2007; Kim and Leonard, 2007; Polansky et al., 2008; Zheng et al., 2010; Toker et al., 2013). In cell proliferation and adoptive transfer experiments, Foxp3 expression was lost more rapidly in iT reg cells with methylated CNS2 compared with T reg cells with unmethylated CNS2 (Floess et al., 2007; Ohkura et al., 2012), reinforcing the conclusion that the DNA methylation (5mC/5hmC) status of CNS2 correlates with the stability of Foxp3 expression (Floess et al., 2007; Polansky et al., 2008; Zheng et al., 2010; Toker et al., 2013).
Naturally occurring T reg cells are difficult to isolate because they are present in very small numbers; hence, iT reg cells have been used in the clinic to treat autoimmunity and graft-versus-host disease in humans (Tang and Bluestone, 2013). In this context, the instability of Foxp3 expression in iT reg cells is a serious potential limitation. So far, our data indicated that TET proteins influenced the stability of Foxp3 expression by maintaining low levels of DNA methylation at T reg cell–specific regulatory regions, leading us to ask whether TET activators could be used pharmacologically to maintain Foxp3 expression in iT reg cells. We chose first to test vitamin C, which has been reported to increase TET activity in mouse embryonic stem cells (ESCs; Blaschke et al., 2013); in the context of this study, we have shown that vitamin C enhances the activity of recombinant TET1 in vitro. To extend this observation further, we developed a cell-based assay to examine the activity of each of the three TET proteins in transfected HEK293 cells (Fig. 3 A). The results showed that vitamin C potentiated the activity of all three TET proteins; however, the strongest apparent effect was observed in TET3-expressing cells, most likely because these cells showed the lowest level of 5hmC in control conditions in the absence of vitamin C (Fig. 3 A).
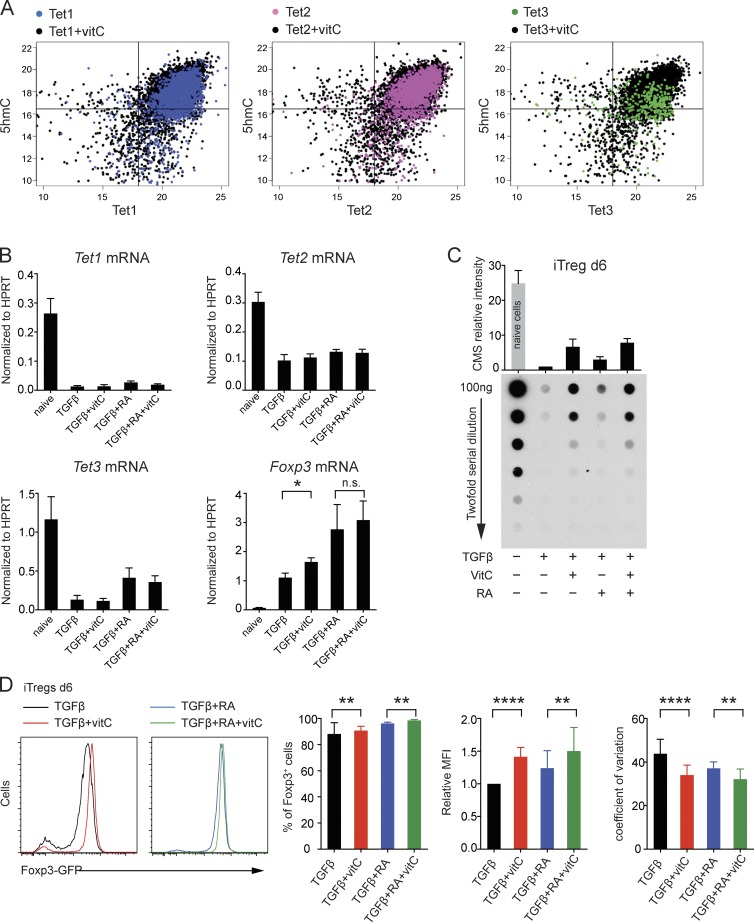
Figure 3.
The effects of vitamin C on TET activity and expression. (A) The effect of vitamin C on TET activity. HEK293T cells were transfected with HA-tagged TET1–3, cultured in the presence/absence of vitamin C, and then stained and imaged for TET expression (x axis) and 5hmC intensity (y axis). The y/x axis is shown in log2 scale. Each dot represents a TET-expressing cell, with hundreds of cells analyzed from independent wells. (B) Quantitative real-time PCR analysis of Tet1, Tet2, Tet3, and Foxp3 mRNA expression levels in naive and iT reg cells differentiated for 3 d in the presence of TGF-β, TGF-β + vitamin C (vitC), TGF-β + RA, or TGF-β + RA + vitamin C. Error bars show mean ± SD from three independent experiments. (C) Genomic DNA from naive and iT reg cells differentiated for 6 d in the presence of TGF-β, TGF-β + vitamin C, TGF-β + RA, or TGF-β + RA + vitamin C was treated with sodium BS to convert 5hmC to CMS. The relative intensity of CMS was quantified by anti-CMS dot blot assay and normalized to the amount of CMS detected in iT reg cells differentiated for 6 d with TGF-β alone. Error bars show mean ± SD from three independent experiments. (D, left) Representative histogram overlay. Right: Graphs for the percentage of Foxp3+ cells, relative MFI (normalized to the MFI in TGF-β condition), and coefficient of variation for iT reg cells differentiated for 6 d in the presence of TGF-β, TGF-β + vitamin C, TGF-β + RA, and TGF-β + RA + vitamin C. Error bars show mean ± SD from more than five independent experiments. *, P < 0.05; **, P < 0.01; ****, P < 0.0001 by Student’s t test. n.s., not significant.
Based on these data, we asked whether the addition of vitamin C during iT reg cell differentiation would promote the loss of 5mC and/or stabilize Foxp3 expression in the resulting iT reg cells. To initiate iT reg cell differentiation, naive T cells from Foxp3-IRES-eGFP reporter mice were treated with TGF-β alone (Chen et al., 2003) or with TGF-β + RA (Benson et al., 2007) in the presence or absence of vitamin C. T cell activation under any conditions—including Th1 and Th2 polarizing conditions (Tsagaratou et al., 2014)—resulted in decreased TET mRNA expression as well as a strong decrease in overall genomic 5hmC by 5–6 d (Fig. 3, B and C). RA increased Tet3 mRNA levels, as also observed in mouse ESCs (Koh et al., 2011). Vitamin C did not affect Tet mRNA levels and caused a modest increase in Foxp3 mRNA levels (Fig. 3 B) but clearly augmented TET enzymatic activity, as assessed by anti–cytosine-5-methylenesulphonate (CMS) dot blot (Ko et al., 2010) to measure overall genomic levels of 5hmC (Fig. 3 C). Thus, vitamin C counters the abrupt loss of 5hmC that occurs in activated T cells, presumably by maintaining TET activity during T cell activation and differentiation.
The presence of vitamin C during iT reg cell differentiation resulted in a small but significant increase of the percentage of Foxp3 (eGFP)-expressing cells and also increased the mean fluorescent intensity (MFI) of Foxp3 (eGFP) expression (i.e., the amount of Foxp3 per cell; Fig. 3 D). Moreover, RA and vitamin C both caused a pronounced tightening of the peak of the histogram for Foxp3-eGFP+ cells, which we quantified as a reduced coefficient of variation (Fig. 3 D, right).
When assessed by standard BS-seq, all CpGs in both CNS1 and CNS2 were essentially fully modified (5mC + 5hmC) in naive CD4+ T cells from Foxp3-IRES-eGFP reporter mice (Fig. 4, A and B, gray bars). Consistent with previous data (Floess et al., 2007; Kim and Leonard, 2007), iT reg cell differentiation for 6 d with TGF-β or TGF-β and RA led to minimal loss of 5mC + 5hmC at CNS2, with the exception of CpGs 10 and 11 at the 3′ end (Fig. 4 D, black and blue bars). In contrast, CpGs in CNS1, particularly CpG 1, lost a substantial amount of 5mC + 5hmC under these conditions (Fig. 4 C, black and blue bars). However, when vitamin C was added into the cultures, all CNS1 and CNS2 CpGs showed a progressive loss of 5mC + 5hmC that was apparent by day 3 of iT reg cell differentiation (Fig. 5, A and B) and became pronounced by day 6 (Fig. 4, C and D, red and green bars). This effect was specific for iT reg cell differentiation because the addition of vitamin C to T cell cultures activated with anti-CD3 and anti-CD28 in the absence of polarizing cytokines or neutralizing anticytokine antibodies (Th0 conditions) had little or no effect on the loss of 5mC + 5hmC at all CpGs of CNS1 and CNS2 (Fig. 4, A and B, black and red bars).
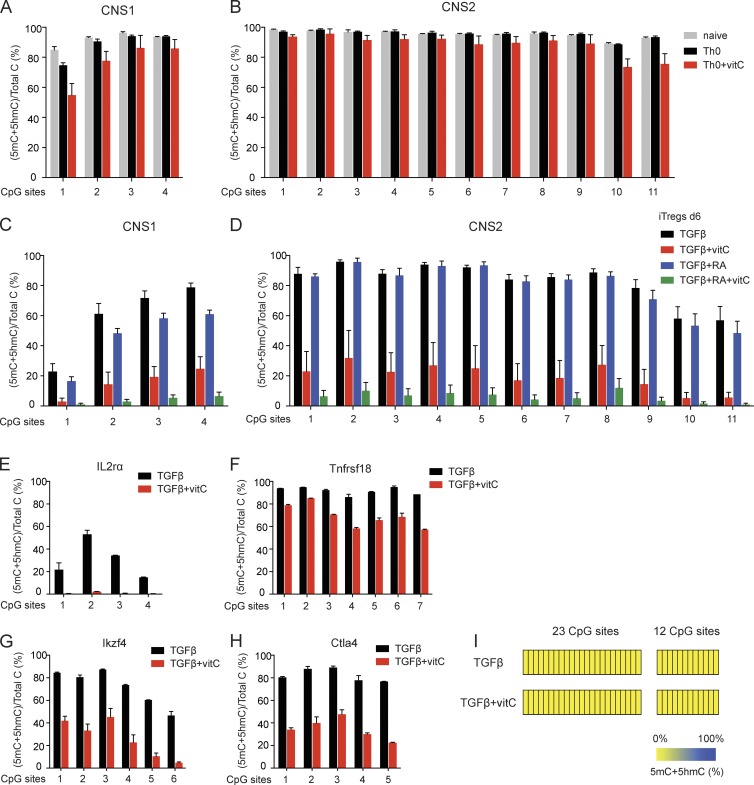
Figure 4.
Supplementation with vitamin C during iT reg cell differentiation leads to the loss of 5mC in T reg cell–specific regulatory regions. (A and B) BS-seq of the four CpG sites in Foxp3 CNS1 (A) and the 11 CpG sites in Foxp3 CNS2 (B). Graphs depict the percentage of (5mC + 5hmC)/total C in naive CD4+ T cells and naive CD4+ T cells activated under Th0 condition without or with vitamin C. Error bars show mean ± SD of thousands of sequencing reads from two to four independent experiments. (C and D) BS-seq of the four CpG sites in Foxp3 CNS1 (C) and the 11 CpG sites in Foxp3 CNS2 (D). Graphs depict the percentage of (5mC + 5hmC)/total C in iT reg cells differentiated for 6 d in the presence of TGF-β, TGF-β + vitamin C, TGF-β + RA, and TGF-β + RA + vitamin C. Error bars show mean ± SD of thousands of sequencing reads from four independent experiments. (E–H) BS-seq of four CpGs in Il2ra intron 1a (E), seven CpGs in Tnfrsf18 exon5 (F), six CpGs in Ikzf4 intron 1b (G), and five CpGs in Ctla4 exon2 (H). Graphs depict the percentage of (5mC + 5hmC)/total C in iT reg cells differentiated for 6 d in the presence of TGF-β or TGF-β + vitamin C. Error bars show mean ± SD of thousands of sequencing reads from at least two independent experiments. (I) Heat maps depicting the percentage of (5mC + 5hmC)/total C in CpGs within two distinct regions of an upstream CpG island in the Foxp3 locus in iT reg cells differentiated for 6 d in the presence of TGF-β or TGF-β + vitamin C, as determined by BS-seq. Data show mean of thousands of sequencing reads from at least two independent experiments.
Figure 5.
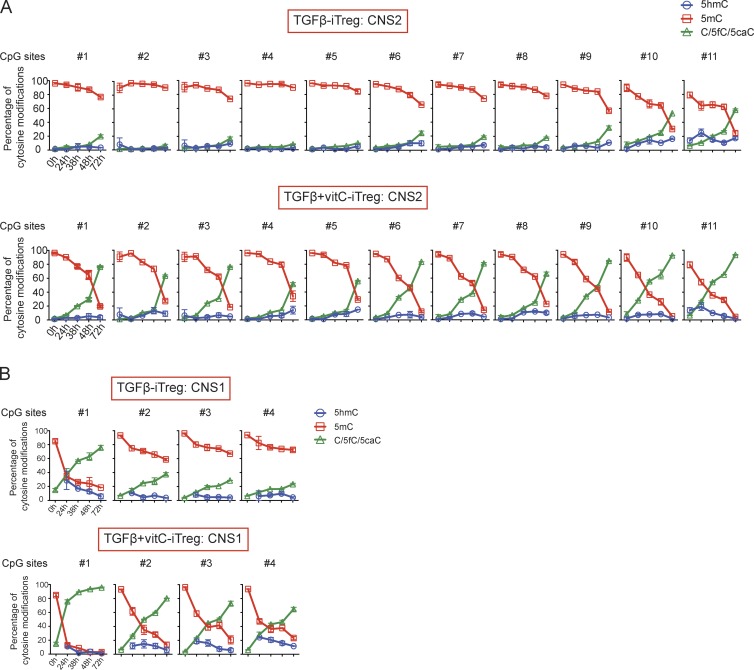
Dynamic alterations in cytosine modifications in Foxp3 CNS1 and CNS2 during iT reg cell differentiation in the presence of vitamin C. Dynamic changes in 5mC, 5hmC, and C/5fC/5caC in the Foxp3 CNS2 (A) and Foxp3 CNS1 (B) during iT reg cell in vitro differentiation. Diagrams depicting the cytosine methylation status of the 11 CpGs in CNS2 (A) and four CpGs in CNS1 (B) as determined by BS-seq and oxBS-seq at five different time points (0, 24, 38, 48, and 72 h) that together capture the sequential steps of differentiation that lead to the loss of 5mC in CNS2 (A) and CNS1 (B) in the presence of vitamin C. Data at 0 h for CNS1 locus is BS-seq. Error bars (which in many cases are too small to be detectable) show mean ± SD of thousands of sequencing reads from two independent experiments.
CpGs within other T reg cell–specific regulatory regions—Il2ra intron 1a, Tnfrsf18 exon 5, Ikzf4 intron 1b, and Ctla4 exon 2—also displayed loss of 5mC + 5hmC when vitamin C was added into the cultures during iT reg cell differentiation (Fig. 4, E–H). In contrast, CpGs within the upstream CpG island were already completely demethylated (<1% 5mC) in TGF-β–induced iT reg cells, and addition of vitamin C did not affect this demethylated state (Fig. 4 I). Together, these data indicate that vitamin C promotes the loss of 5mC + 5hmC at T reg cell–specific regulatory regions during iT reg cell differentiation. For most subsequent experiments, we focused on Foxp3 CNS1 and CNS2, which exhibited the most significant alteration of methylation in both peripheral T reg cells from Tet2/3 DKO mice and in iT reg cells differentiated in the presence of vitamin C.
To capture the dynamic alterations in cytosine modifications in CNS1 and CNS2 during iT reg cell differentiation, we performed BS-seq and oxBS-seq with samples collected at 24, 38, 48, and 72 h after activation of naive T cells with TGF-β in the presence or absence of vitamin C (we did not consider RA for these experiments because it did not affect DNA modification status; Fig. 4, C and D, black and blue bars). iT reg cells differentiating in the presence of TGF-β alone showed a progressive but relatively minor loss of 5mC at CNS2, <40–50% at 72 h except at CpGs 10 and 11 of CNS2 (Fig. 5 A, top), whereas iT reg cells induced by TGF-β + vitamin C showed a considerably more rapid loss of 5mC at CNS2 (Fig. 5 A, bottom). In cells cultured with TGF-β + vitamin C, there was a substantial (40–50%) loss of 5mC at CpGs 10 and 11 of CNS2 even at 24 h, just barely after initiation of DNA replication in S phase in a small fraction of the cells and well before the occurrence of cell division (not depicted); and at all CNS2 CpGs, there was <20% residual 5mC at 72 h (Fig. 5 A, bottom). Vitamin C also greatly potentiated the rate of loss of 5mC from the four CpG residues in CNS1, especially CpG 1 (Fig. 5 B). In all cases, 5hmC levels were low but clearly detectable, as expected for an intermediate in the conversion of 5mC to 5fC/5caC/C (Fig. 5). Altogether, the addition of the TET activator vitamin C during TGF-β–induced iT reg cell differentiation in vitro promotes the loss of 5mC in CNS1 and CNS2 in the Foxp3 locus, as well as other T reg cell–specific regulatory regions.
Vitamin C acts via TET proteins to induce the loss of 5mC
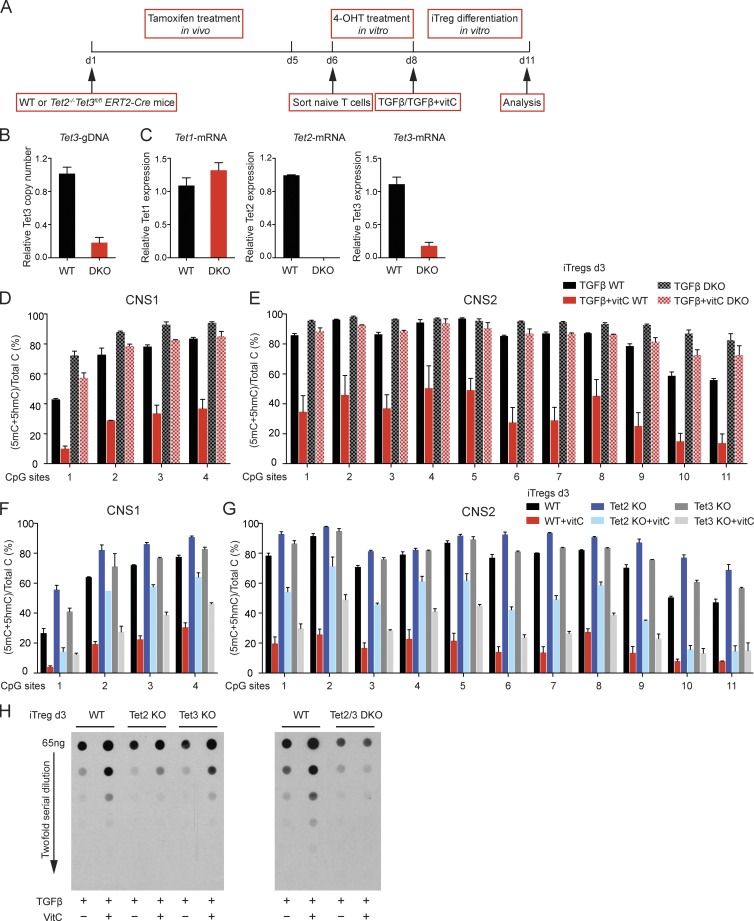
We asked whether the ability of vitamin C to potentiate the loss of 5mC in Foxp3 CNS1 and CNS2 was mediated through TET proteins. Tet2/3 DKO mice, in which Tet3 is deleted using CD4Cre, have a decreased frequency of naive T cells (see Materials and methods subsection Mice); hence, for these experiments, we chose to delete Tet3 inducibly in mice expressing tamoxifen-inducible ERT2-Cre. WT and Tet2−/−Tet3fl/flERT2-Cre mice were injected intraperitoneally with tamoxifen for five consecutive days, after which naive T cells were sorted and cultured in vitro with 4-hydroxytamoxifen (4-OHT) for two more days (Fig. 6 A). Analysis of genomic DNA showed that ∼80% of Tet3 exon 2 was deleted under these conditions (Fig. 6 B). At the mRNA level, Tet2 transcripts from the floxed exons were undetectable; consistent with the amount of undeleted Tet3 exon 2, residual amounts of Tet3 transcripts (∼20%) were observed, and the level of Tet1 transcripts increased slightly, possibly a result of cellular compensation for the loss of Tet2 and Tet3 (Fig. 6 C).
Figure 6.
Vitamin C induces the loss of 5mC in CNS1 and CNS2 during iT reg cell differentiation via TET proteins. (A) Schematic representation of the experimental strategy for Tet3 deletion using the ERT2-Cre system. (B) Quantitative RT-PCR analysis of Tet3 deletion efficiency at the genomic DNA level. The relative copy number was normalized to GAPDH. (C) Quantitative RT-PCR analysis of Tet1, Tet2, and Tet3 mRNA expression levels. The relative expression level was normalized to HPRT. Error bars show mean ± SD from three independent experiments. (D and E) BS-seq of the four CpG sites in Foxp3 CNS1 (D) and 11 CpG sites in Foxp3 CNS2 (E). Graphs depict the percentage of (5mC + 5hmC)/total C in iT reg cells differentiated for 3 d from WT or Tet2−/−Tet3fl/fl ERT2-Cre cells in the presence of TGF-β or TGF-β + vitamin C, in both cases after tamoxifen and 4-OHT treatment as shown in A. (F and G) BS-seq of the four CpG sites in Foxp3 CNS1 (F) and the 11 CpG sites in Foxp3 CNS2 (G). Graphs depict the percentage of (5mC + 5hmC)/total C in iT reg cells differentiated in the presence of TGF-β and TGF-β + vitamin C for 3 d from WT, Tet2−/−, and Tet3fl/fl CD4-Cre naive CD4+ T cells. Error bars show mean ± SD of thousands of sequencing reads from two to three independent experiments. (H) Genomic DNA from WT, Tet2 KO, Tet3 KO, and Tet2/3 DKO naive and iT reg cells differentiated for 3 d in the presence of TGF-β and TGF-β + vitamin C was treated with sodium BS to convert 5hmC to CMS. The relative intensity of CMS was quantified by anti-CMS dot blot assay. Data are representative of three independent experiments.
Naive T cells from tamoxifen-treated mice were differentiated into iT reg cells in the presence of either TGF-β or TGF-β + vitamin C, and the cytosine modification status of Foxp3 CNS1 and CNS2 was assessed by standard BS-seq. WT TGF-β–induced iT reg cells remained highly methylated (5mC + 5hmC) at CNS1 and CNS2 (Fig. 6, D and E, solid black bars), but vitamin C caused significant loss of this modification (Fig. 6, D and E, solid red bars). Tet2/3 DKO iT reg cells induced with TGF-β alone showed higher 5mC + 5hmC levels at CNS1 and CNS2 compared with WT iT reg cells, but in this case addition of vitamin C caused only a minor loss or no loss of 5mC + 5hmC (Fig. 6, D and E, compare hatched black and red bars); the small decrease may be the result of residual Tet1 or Tet3 expression (Fig. 6, B and C). Thus, Tet2 and Tet3 are major targets of vitamin C in T cells.
To ask which TET protein was the major target of vitamin C in iT reg cells, we repeated these experiments with iT reg cells lacking only Tet2 or Tet3. Vitamin C caused a substantial decrease of 5mC + 5hmC in CNS1 and CNS2 in Tet2 KO iT reg cells, which retain Tet1 and Tet3 (Fig. 6, F and G, blue bars), as well as a global increase in 5hmC assessed by anti-CMS dot blot (Fig. 6 H). Tet3 KO iT reg cells, which express Tet1 and Tet2, showed a more striking decrease of 5mC + 5hmC by vitamin C (Fig. 6, F and G, gray bars) and a more striking increase of 5hmC (Fig. 6 H) than Tet2 KO iT reg cells, suggesting a stronger effect of vitamin C on Tet2 compared with Tet3 in this experimental setting. However, the greatest decrease in 5mC + 5hmC and the greatest increase in 5hmC were observed in WT iT reg cells (Fig. 6, F–H), indicating that both Tet2 and Tet3 were targets of vitamin C.
Further attesting to the redundancy between Tet2 and Tet3, the frequency of Foxp3-expressing cells was unchanged in cells with single deletion of Tet2 or Tet3, but was substantially reduced in Tet2/3 DKO cells (Fig. S2, A and B). Moreover, vitamin C increased the MFI of Foxp3 (eGFP) expression in WT iT reg cells and in iT reg cells with single deletion of Tet2 or Tet3 (Fig. S2, A and B, right), but not in Tet2/3 DKO iT reg cells (Fig. S2 B, right). Together, these data indicate that vitamin C acts through both Tet2 and Tet3 to potentiate the loss of 5mC + 5hmC at T reg cell–specific regulatory regions, as well as to increase the MFI of Foxp3 expression in differentiating iT reg cells.
Vitamin C potentiates the stability of Foxp3 expression in mouse iT reg cells
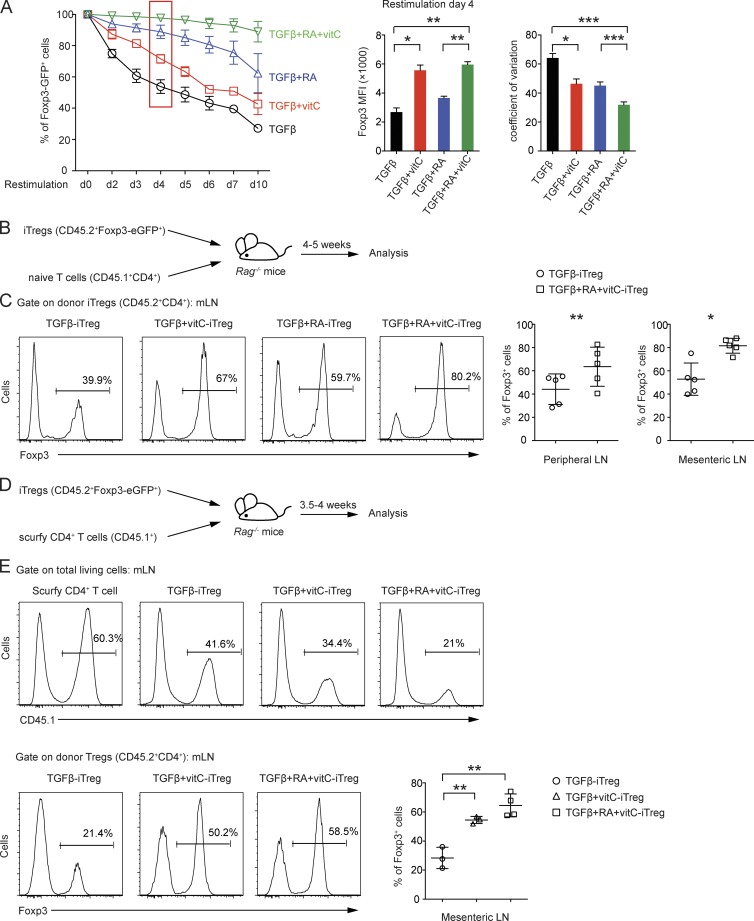
Given that vitamin C induced the loss of 5mC + 5hmC at T reg cell–specific regulatory regions including Foxp3 CNS1 and CNS2 during iT reg cell differentiation, we asked whether iT reg cells induced with TGF-β and vitamin C would display increased stability of Foxp3 expression in vitro and in vivo compared with iT reg cells induced with TGF-β alone. We differentiated purified naive T cells from Foxp3-IRES-eGFP reporter mice into iT reg cells under four conditions: (1) TGF-β alone, (2) TGF-β + vitamin C, (3) TGF-β + RA, and (4) TGF-β + RA + vitamin C. Foxp3-eGFP+ cells were sorted at day 6 and restimulated with anti-CD3 and anti-CD28 in the absence of polarizing cytokines or neutralizing antibodies, and Foxp3 (eGFP) expression was monitored daily (Fig. 7 A). Indeed, iT reg cells generated in the presence of vitamin C lost Foxp3 (eGFP) expression considerably more slowly during restimulation-induced proliferation than corresponding iT reg cells generated using TGF-β or TGF-β + RA alone (Fig. 7 A, left). In addition, iT reg cells generated using vitamin C displayed an approximately twofold increase in the amount of Foxp3 per cell, as assessed by increased MFI (Fig. 7 A, middle). Again, both RA and vitamin C reduced the coefficient of variation, with an additive effect of both agents together (Fig. 7 A, right).
Figure 7.
Supplementation of vitamin C during iT reg cell differentiation enhances the stability of iT reg cells in vitro and in vivo. (A) Naive CD4+ T cells from Foxp3-IRES-eGFP reporter mice were differentiated into iT reg cells for 6 d in the presence of TGF-β, TGF-β + vitamin C, TGF-β + RA, or TGF-β + RA + vitamin C, and then the Foxp3-eGFP+ population was sorted and restimulated with anti-CD3 and anti-CD28 antibodies. The percentage of Foxp3-eGFP+ cells was monitored daily after restimulation. Right: Geometric MFI and coefficient of variation for Foxp3-eGFP+ cells on day 4 after restimulation. Error bars show mean ± SD from three independent experiments. (B) Schematic representation of the adoptive transfer experiment for assessing iT reg cell stability in vivo. (C, left) Representative histograms of Foxp3+ cells from Rag-deficient mice 4–5 wk after adoptive transfer of sorted iT reg cells differentiated for 6 d in the presence of TGF-β, TGF-β + vitamin C, TGF-β + RA, or TGF-β + RA + vitamin C. Right: Graphs for the percentage of Foxp3+ cells from peripheral and mesenteric lymph nodes. Data are from five mice per group. (D) Schematic representation of adoptive transfer experiment of scurfy CD4+ T cells. (E) Representative histograms of CD45.1+ cells (top) and CD45.2+CD4+Foxp3+ cells (bottom) from Rag-deficient mice 3.5–4 wk after adoptive transfer of iT reg cells differentiated in the presence of TGF-β, TGF-β + vitamin C, or TGF-β + RA + vitamin C together with scurfy CD4+ T cells. Bottom right: Graph for the percentage of Foxp3+ cells from mesenteric lymph nodes. Data are from three or four mice per group. Error bars show mean ± SD. mLN, mesenteric lymph node. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by Student’s t test.
We determined the in vivo stability of Foxp3 expression on iT reg cells induced in the presence of vitamin C by transferring CD45.2+Foxp3-eGFP+ iT reg cells generated under the four conditions together with congenically marked CD45.1+CD4+ naive T cells into Rag-deficient recipients (Fig. 7 B). Analysis of the percentage of Foxp3+ cells in peripheral lymphoid organs 4–5 wk after adoptive transfer showed that iT reg cells generated in the presence of vitamin C were more resistant to the loss of Foxp3 and thus were more stable than the corresponding iT reg cells generated in the absence of vitamin C (Fig. 7 C).
Scurfy mice lack T reg cells because they carry a natural frame-shift mutation of Foxp3 that abrogates Foxp3 protein expression (Brunkow et al., 2001). Transfer of peripheral CD4+ T cells from scurfy male mice into Rag-deficient recipients leads to expansion of autoreactive scurfy CD4+ T cells and severe autoimmune inflammation because of a lack of T reg cell suppression; both features can be suppressed by cotransfer of congenically marked T reg cells (Choi et al., 2010). To examine the in vivo function of iT reg cells generated in the presence of vitamin C, we transferred scurfy CD4+ T cells (CD45.1+) into Rag-deficient recipient mice together with iT reg cells from CD45.2+ Foxp3-eGFP reporter mice generated in the presence of TGF-β alone, TGF-β + vitamin C, or TGF-β + RA + vitamin C (Fig. 7 D). iT reg cells generated in the presence of TGF-β + vitamin C or TGF-β + RA + vitamin C were superior to iT reg cells induced by TGF-β alone in suppressing scurfy CD4+ T cell expansion, as shown by analysis of the percentage of scurfy CD4+ T cells in peripheral lymphoid organs 3.5–4 wk after adoptive transfer (Fig. 7 E, top). Moreover, iT reg cells generated in the presence of vitamin C were more resistant to the loss of Foxp3 even under these inflammatory conditions (Fig. 7 E, bottom). In summary, addition of vitamin C during iT reg cell differentiation led to Tet2/3-dependent loss of 5mC in T reg cell–specific regulatory regions and increased the stability of Foxp3 expression by the resulting iT reg cells.
Vitamin C potentiates CNS2 demethylation and the stability of FOXP3 expression in human iT reg cells
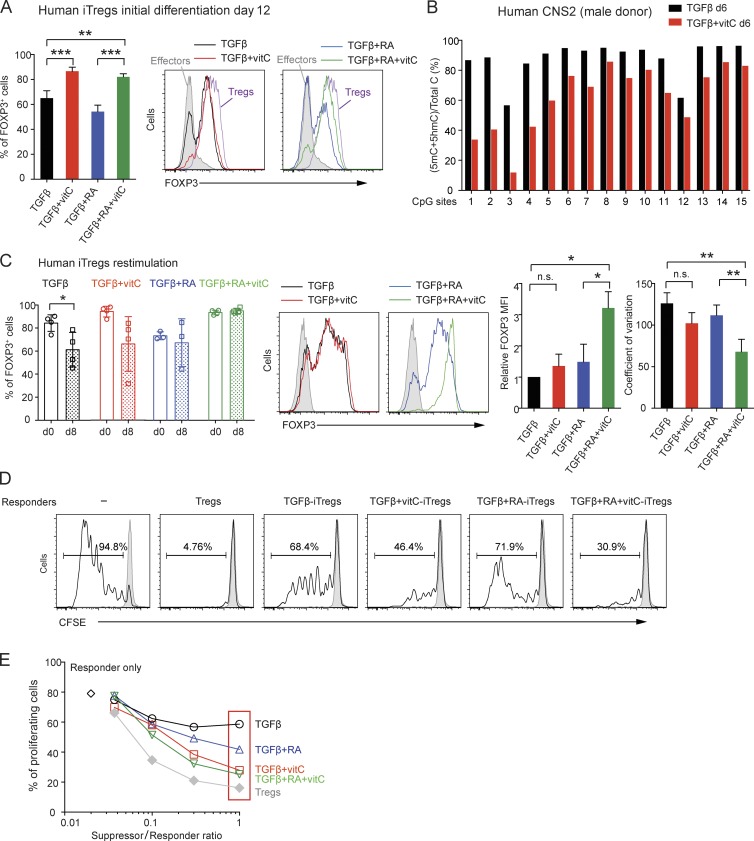
Finally, we asked whether vitamin C also worked via TET proteins to potentiate the loss of 5mC and enhance the stability of FOXP3 expression in human iT reg cells. We sorted naive CD4+ T cells from human peripheral blood, stimulated them with anti-CD3 and anti-CD28 in the presence of TGF-β, TGF-β + RA, TGF-β + vitamin C, or TGF-β + RA + vitamin C, and analyzed FOXP3 expression at different times after stimulation. Similar percentages of FOXP3+ cells were found in all TGF-β–treated cultures on day 4 (Fig. S3 A); by day 12, however, the percentage of FOXP3+ cells declined in iT reg cells stimulated in the presence of TGF-β or TGF-β + RA, but remained high in iT reg cells additionally treated with vitamin C (Figs. 8 A and S3 A). The percentage of FOXP3+ cells and the MFI of FOXP3 expression were similar in iT reg cells generated with TGF-β + RA + vitamin C compared with expanded ex vivo T reg cells (Fig. 8 A, right, compare purple and green histograms). In contrast, human CD4+ T cells activated in the presence of IL-2 only (effectors), which transiently induce low levels of FOXP3 (Allan et al., 2007; Tran et al., 2007; Wang et al., 2007), showed very low FOXP3 expression on day 12 as expected (Fig. 8 A, gray histograms).
Figure 8.
Vitamin C promotes the loss of 5mC in human FOXP3 CNS2 and enhances the stability and suppressive function of human iT reg cells. (A, left) Percentage of Foxp3+ cells in human iT reg cells differentiated from naive CD4+ T cells in the presence of TGF-β, TGF-β + vitamin C, TGF-β + RA, and TGF-β + RA + vitamin C on day 12. Error bars show mean ± SD from three to four independent donors. Right: Representative histogram overlay of Foxp3 expression for human iT reg cells differentiated for 12 d in the presence of TGF-β, TGF-β + vitamin C, TGF-β + RA, and TGF-β + RA + vitamin C. Effector CD4+ T cells (filled gray) and expanded T reg cells (purple) are shown for comparison. (B) BS-seq of 15 CpGs in human FOXP3 CNS2. Graph depicts the percentage of (5mC + 5hmC)/total C in human iT reg cells differentiated for 6 d without or with vitamin C from a male donor. (C, left) Analysis of Foxp3 expression in human iT reg cells generated as indicated in the presence of TGF-β, TGF-β + vitamin C, TGF-β + RA, and TGF-β + RA + vitamin C (d0) and restimulated with anti-CD3 and anti-CD28 antibodies for 8 d (d8). Graphs show results from three independent donors. Middle: Representative histogram overlay of Foxp3 expression for human iT reg cells on day 8 after restimulation. Right: Foxp3 MFI (normalized to the MFI in TGF-β condition) and coefficient of variation analyzed on day 8 after restimulation. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by Student’s t test. n.s., not significant. (D) Proliferation analysis by CFSE dilution in human CD8+ T cells (responders) stimulated with anti-CD3 antibody and cultured with T reg or iT reg cells (suppressors) at a responder/suppressor ratio of 1:0.3. The percentage of proliferating cells (black line) and of resting, unstimulated responders (gray shaded histogram) are shown in each plot. (E) Percentage of proliferating responder cells at different responder/suppressor ratios. Results are representative of three independent donors.
To analyze FOXP3 stability in human iT reg cells generated with and without vitamin C, we restimulated iT reg cells previously polarized under the four indicated conditions with anti-CD3 and anti-CD28 in the absence of TGF-β or RA and analyzed FOXP3 expression at different days after restimulation (Figs. 8 C and S3 B). Starting at day 4, the percentage of FOXP3+ cells declined under most conditions but was maintained in iT reg cells previously differentiated with TGF-β + RA + vitamin C (Fig. 8 C, left; and Fig. S3 B). Moreover, at the end of the restimulation period (day 8), the MFI of FOXP3 expression was significantly higher in iT reg cells generated with TGF-β + RA + vitamin C than in the other culture conditions and also displayed a tighter distribution (narrower peak and lower coefficient of variation; Fig. 8 C, middle and right, green histogram and bars). Moreover, as shown for mouse iT reg cells in Figs. 3, 4, and 5, the presence of vitamin C augmented TET enzymatic activity (Fig. S3 C) and potentiated the loss of 5mC at CNS2 (Figs. 8 B and S3 D) in human iT reg cells collected after 6 d of differentiation.
Finally, we evaluated the ability of vitamin C to potentiate the suppressor activity of human iT reg cells differentiated under the four conditions. iT reg cells differentiated with TGF-β + RA + vitamin C were superior to all the other in vitro–generated iT reg cells in their ability to suppress proliferation of CFSE-labeled heterologous CD8+ T cells (responders) over the course of a 5-d suppression assay in vitro (Fig. 8, D and E). Although this could in part reflect the increased numbers of iT reg cells with increased FOXP3 expression in the TGF-β + RA + vitamin C cultures, the results collectively show that vitamin C results in the loss of 5mC at FOXP3 CNS2 and promotes the stability of FOXP3 expression in human iT reg cells as it does in mice, and moreover potentiates the suppressor function of human iT reg cells to the levels observed in T reg cells isolated and expanded directly from human peripheral blood.
DISCUSSION
Expression of Foxp3 and establishment of a T reg cell–specific hypomethylation pattern are both indispensable for T reg cell development and function (Ohkura et al., 2012). Here, we implicate TET proteins in these processes. We focused on Tet2 and Tet3 as the major TET proteins expressed in differentiated tissues and cell types, including lymph node and spleen T cells and thymic T reg cell populations (Toker et al., 2013; Tsagaratou and Rao, 2013). By mapping 5mC and 5hmC at a single-base resolution, we show that 5mC is lost progressively during T reg cell development at several T reg cell–specific regulatory regions, including CNS1 and CNS2 in the Foxp3 gene. During this developmental progression, 5hmC levels are low but peak in T reg cell precursors or thymic T reg cells, as expected for a transient intermediate in a TET-mediated process of DNA demethylation (Pastor et al., 2013; Wu and Zhang, 2014). Furthermore, we show that the loss of 5mC from T reg cell–specific regulatory regions is significantly impaired in peripheral T reg cells from Tet2/3 DKO mice. Accompanying the loss of 5mC, peripheral T reg cells from Tet2/3 DKO mice fail to maintain Foxp3 expression after adoptive transfer in vivo or TCR stimulation in vitro.
The data point to considerable redundancy among the three TET proteins in mediating the loss of 5mC at regulatory regions and the stability of Foxp3 expression in T reg cells. In our hands, single deletion of Tet2 in peripheral T reg cells did not result in any gain of CpG methylation (5mC + 5hmC) at Foxp3 CNS1 and CNS2; single deletion of Tet3 resulted in a minimal increase, and double deletion of both Tet2 and Tet3 resulted in a more striking increase (compare Fig. 1 [C and D] with Fig. S1 [K and L]). Similarly, although the phenotypes of single Tet1- or Tet2-deficient T reg cells were not investigated, a recent paper reported increased 5mC + 5hmC/total C in Tet1/2 DKO T reg cells (Yang et al., 2015), indicating that Tet1 activity also contributes to T reg cell function. Indeed, we find that Tet1 mRNA is expressed at slightly higher levels in the absence of Tet2 and Tet3 (Fig. 6 C), consistent with the conclusion that all three TET proteins contribute to CNS2 demethylation and stabilization of Foxp3 expression. Triple depletion of all three TET proteins in T reg cells will be needed to confirm this point.
A previous study (Nair and Oh, 2014) reported a surprisingly high increase in the level of 5mC + 5hmC at CNS2 in T reg cells from mice lacking Tet2 alone (∼90% 5mC + 5hmC/total C). In contrast, we see a negligible increase (Fig. S1, K and L). The discrepancies may reflect differences in genetic background of the mice (we generated Tet2-deficient mice on a pure C57BL/6 background, whereas those of Nair and Oh [2014] were on a mixed C57BL/6-129 background) or differences in BS conversion efficiency, time of BS treatment, or BS concentration or potency, which we controlled for by spiking equivalent amounts of λ-DNA fragment into all samples (Table S2). We note that manual sequence analysis of small numbers of clones can result in sampling errors, which can be compounded by poor PCR amplification because of DNA damage resulting from prolonged BS treatment; we minimized these problems by sequencing thousands of amplicons from BS-converted DNA on a MiSeq platform and subjecting the data to rigorous statistical analysis. We also note that TET enzymes are dioxygenases that use molecular oxygen, 2-oxoglutarate (a product of the Krebs cycle), and reduced iron (Fe(II)) as substrates and cofactors (Pastor and Rao, 2013; Huang and Rao, 2014). As a result, TET enzymatic activity is likely to be sensitive to many signals, including hypoxia, metabolic state (2-oxoglutarate levels), Fe2+ availability, and the redox environment, as well as vitamin C levels as shown here. If any of these factors were suboptimal in the diet or environment of the mice analyzed by Nair and Oh (2014), it would lead to a larger apparent difference between WT and Tet2-deficient T reg cells.
Vitamin C is a known coactivator for TET proteins in several settings, including mouse ESCs (Blaschke et al., 2013), embryonic fibroblasts (Minor et al., 2013), during induced pluripotent stem cell reprogramming (Chen et al., 2013), and erythroid precursors (Ruiz et al., 2015), and as shown by our laboratory (Blaschke et al., 2013) and others (Yin et al., 2013), it acts directly on recombinant Tet1 and Tet2 in vitro. Here, we show that the presence of vitamin C (ascorbate) during iT reg cell differentiation leads to a marked loss of 5mC at CNS1 and CNS2 as judged by BS-seq and oxBS-seq (Fig. 5), as well as increased stability of Foxp3 expression after restimulation in vitro or adoptive transfer in vivo (Fig. 7). BS-seq revealed that the loss of 5mC induced by vitamin C was apparent in WT cells, in iT reg cells lacking Tet3, and to a lesser extent in iT reg cells lacking Tet2, but was completely abrogated in iT reg cells doubly deficient for both Tet2 and Tet3 (Fig. 6, D–H). These data again attest to the strong redundancy between Tet2 and Tet3. Given the likely involvement of Tet1 (Yang et al., 2015), we conclude that all three TET proteins are likely targets of vitamin C and function redundantly to regulate CpG methylation at T reg cell–specific regulatory regions, including CNS1 and CNS2 of Foxp3.
Together, vitamin C and RA potently stabilized Foxp3 expression in TGF-β–induced iT reg cells (Fig. 7). Vitamin C enhanced TET catalytic activity without altering Tet mRNA levels (Fig. 3), consistent with its well-established ability to potentiate directly the activity of many (but not all) Fe(II) and 2-oxoglutarate–dependent dioxygenases (Flashman et al., 2010), including the collagen prolyl hydroxylase, whose defective activity results in scurvy (Loenarz and Schofield, 2008). We previously showed that vitamin C acts directly on recombinant TET1 in vitro to increase its enzymatic activity (Blaschke et al., 2013); here, we have used a cell-based assay and high-throughput automated microscopy to extend this finding to all three TET family proteins (Fig. 3 A). Most likely, vitamin C potentiates TET dioxygenase function by maintaining iron in its reduced Fe(II) form (Du et al., 2012); other less likely scenarios include a postulated role of vitamin C as a direct cofactor of TET (Dickson et al., 2013) or ascorbate-induced localization of TET enzymes to the regulatory regions of T reg cell–specific genes (for which there is no evidence at present). The stabilizing effect of RA on Foxp3 protein expression has been noted before (Benson et al., 2007). RA increased Tet3 mRNA levels (Fig. 3 B) as also independently observed in mouse ESCs (Koh et al., 2011). However, RA did not affect the CpG methylation status of CNS1 and CNS2 (Fig. 4, C and D). Potentially, RA could act through its nuclear receptors to affect Foxp3 gene transcription or could regulate Foxp3 stability at the protein level as well.
In a previous study, T reg cells lacking CNS2 were shown to lose Foxp3 upon adoptive transfer into recipient mice, whereas T reg cells lacking CNS1 did not (Zheng et al., 2010). In contrast, our findings suggest that the stability of Foxp3 expression might be regulated by both these conserved elements as well as potentially other T reg cell–specific regulatory regions in vivo. Notably, CNS1 CpG 1 and CNS2 CpGs 10 and 11 displayed a particularly rapid loss of 5mC, both during T reg cell development in the thymus and iT reg cell differentiation in cell culture, suggesting that transcription factors binding in the vicinity of these CpGs might be involved in recruitment of Tet2 and/or Tet3. For instance, the presence of an NFAT binding site near CNS2 CpGs 10 and 11 (Li et al., 2014) and the rapid loss of 5mC at these CpGs during iT reg cell differentiation suggest that NFAT may recruit TET proteins to this region of CNS2. A similar activation-dependent mechanism may explain TET recruitment to CNS1, which has several consensus NFAT sites (Tone et al., 2008) and displays some loss of 5mC + 5hmC even in T cells activated under Th0 conditions. IL-2 maintains the stability of Foxp3 expression, and STAT5—the downstream transcriptional effector of IL-2 signaling—is known to bind CNS2 and so may also be involved (Burchill et al., 2007). Finally, factors specific to iT reg cell differentiation (e.g., TGF-β–activated SMAD factors; Tone et al., 2008) must also have a role because naive T cells activated under Th0 conditions failed to lose 5mC in CNS2 even in the presence of equivalent amounts of IL-2 and vitamin C.
Two recent publications using CNS2 KO mice (Feng et al., 2014; Li et al., 2014) have suggested that CNS2 deficiency stochastically affects the stability of ∼50% of mature T reg cells or selectively affects the stability of T reg cells bearing low levels of Foxp3. Regardless of which situation applies, the data suggest that the DNA modification status of CNS2 is not the sole determinant of T reg cell stability; other cis-elements and mechanisms undoubtedly have a role. The more striking effects observed at CNS1 and CNS2 compared with other regulatory regions most likely reflect the relative strength of recruitment of TET proteins to these regions by sequence-specific transcription factors.
From a therapeutic perspective, we have shown that vitamin C increases the stability and suppressive function of human iT reg cells as well, suggesting that manipulation of TET dioxygenase activity using either vitamin C or other more selective TET activators identified in small molecule screens might be a practical strategy for stabilizing iT reg cell function in the clinic.
MATERIALS AND METHODS
Mice
To evaluate the methylation status of the CNS2 locus of the Foxp3 gene, we used 4-wk-old male Foxp3-IRES-eGFP reporter mice, which were originally obtained from The Jackson Laboratory and further backcrossed to the B6/C57 background for >10 generations. Congenic mice (strain B6.SJL-PtprcaPepcb/BoyJ) were obtained from The Jackson Laboratory. Tet2−/−Tet3fl/flCD4-Cre, Tet2fl/flTet3fl/flCD4-Cre mice, and Tet2−/−Tet3fl/fl ERT2-Cre mice were generated in our laboratory using B6/Taconic background “artemis” ESCs. All WT mice used in this study are littermate controls that do not express Cre. All breeding and experiments were reviewed and approved by the Institutional Animal Care and Use Committee of the La Jolla Institute for Allergy and Immunology.
Flow cytometry and FACS
All the antibodies used for flow cytometry analyses and FACS sorting were purchased from eBioscience and BioLegend. For experiments evaluating DNA methylation and hydroxymethylation in the CNS2 locus, thymocytes were isolated from young (4 wk old) Foxp3-IRES-eGFP male mice. In these mice, a cassette encoding IRES-eGFP was inserted into the 3′ untranslated region of Foxp3 to generate a bicistronic locus encoding both Foxp3 and eGFP under the control of Foxp3 promoter (Haribhai et al., 2007). The thymocytes were stained using CD4 (GK1.5) and CD25 (PC61). Foxp3 expression was indicated by eGFP expression. CD4+CD8+ (DP), CD4+CD25−eGFP− cells (CD4 SP), CD4+CD25+eGFP− (precursor T reg cells), CD4+CD25−eGFP+ (precursor T reg cells), and CD4+CD25+GFP+ (thymic T reg cells) were isolated (Fig. S1 A).
To obtain peripheral T reg cells, spleen and lymph nodes were isolated from young (4 wk old) male Foxp3-GPP reporter mice. Subsequently, CD4+ T cells were enriched (Dynabeads untouched mouse CD4 T cells; Invitrogen) and stained with the surface markers CD4 (GK1.5), CD8 (53-6.7), CD62L (MEL-14), CD25 (PC61), and CD44 (IM7). Peripheral T reg cells were isolated as CD4+CD25+eGFP+ cells (Fig. S1 A). Post-sort purity was >99%. For Foxp3 intracellular analyses, cells were surface stained and then stained with anti-Foxp3 antibody (eBioscience) using the Foxp3 Fixation/Permeabilization kit (eBioscience) and analyzed by flow cytometry.
In vitro iT reg cell differentiation and restimulation
CD4+CD25−CD62LhiCD44lo (eGFP− for Foxp3-IRES-eGFP reporter mice) naive T cells were FACS sorted from lymph nodes of 4–6-wk-old mice and differentiated into iT reg cells with plate-bound anti-CD3 (clone 2C11) and anti-CD28 (clone 37.51) antibodies at 1 µg/ml in the presence of 2 ng/ml recombinant human TGF-β (PeproTech) and 100 U/ml recombinant human IL-2 (rhIL-2). For the conditions with RA (Sigma-Aldrich) or vitamin C (Sigma-Aldrich), 100 nM RA and 100 µg/ml vitamin C were added into the culture. For the restimulation experiments, iT reg cells differentiated for 6 d were harvested and sorted for eGFP+ (Foxp3+) population. Cells were counted and plated at 0.8 × 106 cells/ml for restimulation with plate-bound anti-CD3 at 50 ng/ml and anti-CD28 at 25 ng/ml, and the cells were then monitored for eGFP expression after restimulation.
In vivo T reg/iT reg cell stability assay
In vivo T reg cell stability assay was performed as described previously (Zheng et al., 2010). In brief, T reg cells were FACS sorted from Tet2/3 DKO mice and age-matched WT controls. 105 CD45.2+ T reg cells mixed with 106 CD4+ naive T cells FACS sorted from CD45.1+ congenic mice were injected intravenously into Rag-deficient recipients. For iT reg cell stability assay, 4 × 105 in vitro TGF-β–induced iT reg cells were mixed together with 2 × 106 CD45.1+CD4+ naive T cells and injected intravenously into Rag-deficient recipients. 4–5 wk after adoptive transfer, the CD45.2+CD4+Foxp3+ cell population was analyzed by flow cytometry.
Scurfy CD4+ T cell adoptive transfer
Scurfy CD4+ T cells were isolated from spleen and peripheral lymph nodes from male Scurfy mice and purified using Dynabeads (purity of >98%; Life Technologies). 5 × 105 CD45.1+CD4+ scurfy T cells were injected into Rag-deficient mice alone or mixed with 105 in vitro–generated iT reg cells (TGF-β alone, TGF-β + vitamin C, and TGF-β + RA + vitamin C). 3.5–4 wk after adoptive transfer, the percentage of CD45.1+CD4+ cells and CD45.2+CD4+Foxp3+ cells were analyzed by flow cytometry.
Tet3 deletion in ERT2-Cre transgenic system
To recombine the Tet3 floxed allele using the ERT2-Cre transgenic system, tamoxifen (Sigma-Aldrich) was solubilized at 10 mg/ml in corn oil (Sigma-Aldrich) and delivered into either WT control mice or Tet2−/−Tet3fl/fl ERT2-Cre mice by intraperitoneal injection at 2 mg/mouse/day for five consecutive days. Mice were sacrificed 24 h after the last injection, and naive T cells were FACS purified and treated for 2 d in vitro with 4-OHT (Sigma-Aldrich) to reinforce Tet3 deletion efficiency. The cells were then washed with cell culture medium and induced into iT reg cells with TGF-β or TGF-β + vitamin C.
Quantitative analysis of CMS levels using dot blots
Genomic DNA samples were treated with sodium BS using the Methylcode kit (Invitrogen). BS-treated DNA samples were then denatured in 0.4 M NaOH and 10-mM EDTA at 95°C for 10 min and neutralized by adding an equal volume of cold 2 M ammonium acetate, pH 7.0. Twofold serial dilutions of the denatured DNA samples were spotted on a nitrocellulose membrane in an assembled Bio-Dot apparatus (Bio-Rad Laboratories) according to the manufacturer’s instructions. The membrane was washed with 2× SSC buffer, air dried, vacuum baked at 80°C for 2 h, and then blocked with 5% nonfat milk for 1 h and incubated with anti-CMS antibody (1:3,000) overnight at 4°C. After incubating with HRP-conjugated anti–rabbit IgG secondary antibody, the membrane was visualized by enhanced chemiluminescence.
Analysis of vitamin C effect on TET proteins
HEK293T cells were transfected with hemagglutinin-tagged TET1–3 using Lipofectamine 2000 (Invitrogen) in OptiMEM (Gibco). 6 h later, DNA-Lipofectamine mixture was replaced with DMEM with or without vitamin C (Sigma-Aldrich). 48 h after transfection, cells were fixed with 4% paraformaldehyde in PBS for 15 min and permeabilized with 0.2% Triton X-100 in PBS for 15 min at room temperature. Subsequently, DNA was denatured with 2 N HCl at room temperature for 30 min and neutralized with 100 mM Tris-HCl buffer, pH 8.5, for 10 min. Cells were then washed and incubated in blocking buffer PBS, 1% BSA, and 0.05% Tween 20 at room temperature for 1 h. Next, mouse anti-5hmC antibody (diluted to 1:500; Active Motif) for 5hmC staining or goat anti-HA antibody (diluted to 1:500; Bethyl) for TET staining was added in blocking buffer at 4°C overnight. The cells were rinsed three times with 0.2% Triton X-100 in PBS and incubated at room temperature for 2 h with donkey anti–mouse Alexa Fluor 555 and donkey anti–goat Alexa Fluor 647 (diluted to 1:1,000; Invitrogen) in blocking buffer, again washed three times with 0.2% Triton X-100 in PBS, and stained with DAPI. Cells were imaged with the Image Express Micro System (Molecular Devices), and data were analyzed with R software.
In vitro human iT reg cell differentiation and restimulation
PBMCs were isolated by Lymphoprep gradient (Cosmo Biosciences) from Leuco reduction system white blood cells obtained from the San Diego Blood Bank from de-identified healthy donors. Per the Institutional Review Board (IRB) of La Jolla Institute (FWA 00000032), work with de-identified human specimens purchased from the blood bank does not meet the regulatory definition of human subject research and does not require IRB approval. CD4+ naive T cells were FACS sorted from PBMCs as a CD4+CD45RA+CCR7+ population. Cells were stimulated with Dynabeads human T cell activator CD3/CD28 (Invitrogen) for 4 d at a cell/bead ratio of 1:1 in the presence of 2 ng/ml TGF-β and 100 U/ml rhIL-2. For the conditions with RA or vitamin C, 100-nM RA and 100 µg/ml vitamin C were added into the culture. After the initial 4-d stimulation, the beads were removed and the cells were maintained in complete media (IMDM with 5% FCS, 2% human serum, and β-mercaptoethanol) supplemented with rhIL-2. For the restimulation assay, the cells were subjected to a second round of polarization under the same conditions and restimulated with Dynabeads human T cell activator CD3/CD28 for 2 d at a cell/bead ratio of 1:1 in the presence of rhIL-2 and were analyzed for Foxp3 expression by intracellular staining.
Human T reg cell suppression assay
PBMCs isolated by Lymphoprep gradient from adult peripheral blood or buffycoats were labeled with CFSE (Invitrogen). In brief, PBMCs were washed with PBS and resuspended in PBS at 107 cells/ml. CFSE was added at a final concentration of 5 µM, and cells were incubated for 8 min in the dark with gentle agitation. The reaction was quenched by the addition of an equal volume of FBS, and cells were washed twice in a complete medium. For suppression assays, suppressor T cells (iT reg cells or expanded T reg cells), at day 12 after the first round of stimulation, were rested without IL-2 or any other cytokine or factor for 6 h at 37°C. CFSE-labeled responder cells (PBMCs) were plated at a density of 4 × 104 cells/well in a 96-well U bottom plate, and suppressor cells were added at different suppressor/responder ratios. Soluble anti-CD3 agonistic antibody (OKT3 clone; eBioscience) was added at 100 ng/ml. After 5 d, cells were stained with fixable viability dye, anti-CD4, and anti-CD8 antibodies. Responder CD8+ T cells were gated as live cells (negative for the fixable viability dye), and the percentage of cells that had diluted CFSE (proliferating cells) was evaluated using unstimulated, CFSE-labeled PBMCs kept in culture for the same period of time as reference. Proliferation rate of responder cells, without suppressor cells, was used to calculate the percentage of suppression as follows: 100 − ([% of proliferating cells × 100]/% of proliferating responders alone).
Generation of heat maps
Heat maps were generated using Genesis software (Sturn et al., 2002).
Statistical analysis
Significance was determined by two-tailed Student’s t tests using Prism 6 (GraphPad Software). Asterisks indicate p-values: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; n.s., not significant. The number of mice used in each experiment is indicated in the figure legends. For the methylation analyses in Fig. 1 (C–H; WT vs. Tet2/3 DKO), Fig. 4 (C–H; TGF-β vs. TGF-β + vitamin C and TGF-β + RA vs. TGF-β + RA + vitamin C), and Fig. 6 (D–G; TGF-β WT vs. TGF-β + vitamin C WT; Tet2 KO vs. Tet2 KO + vitamin C; and Tet3 KO vs. Tet3 KO + vitamin C), all outcomes are significant with P < 0.05, and in many cases, < 0.01 or lower.
BS- and oxBS-seq
The DNA oxidation procedure was done as described previously (Booth et al., 2012, 2013). In brief, up to 1 µg of genomic DNA was ethanol precipitated before oxidation and then filtered through P6 SSC Micro bio-spin column (Bio-Rad Laboratories). DNA was denatured in 0.05 M NaOH for 30 min at 37°C and then snap cooled on ice for 5 min (total of 24 µl). 1 µl of KRuO4 solution (15 mM in 0.05-M NaOH; Sigma-Aldrich) was then added, and the denatured DNA was held on ice for 1 h, with occasional vortexing. The DNA was purified using P6 SSC Micro bio-spin column, and the oxidized and nonoxidized DNA samples were treated in parallel with sodium BS (MethylCode Bisulfite Conversion kit; Invitrogen). PCR primers (Table S1) were designed using Methyl Primer Express software (Life Technologies) or from previous studies (Lal et al., 2009; Ohkura et al., 2012). The PCR amplicons were generated using the PyroMark PCR kit (QIAGEN) and quantified using Quant-iT PicoGreen dsDNA reagent (Invitrogen). To monitor BS conversion efficiency, a 210-bp spike-in control was generated using unmethylated λ-DNA (Promega) as a template and added to the genomic DNA to a final percentage of 0.5% (Table S2). PCR amplicons were then used for library preparation using NEBNext DNA Library Modules for Illumina platform (New England Biolabs, Inc.). The final libraries were quantified using the KAPA library quantification kit for Illumina (KAPA Biosystems) and sequenced on Miseq (300 bp, paired end; Illumina). The data are based on thousands of sequence reads per amplicon, and a rigorous statistical analysis of 5mC, 5hmC, and C/5fC/5caC levels was performed as described in the following two sections.
BS-seq and oxBS-seq data analysis
The BS and oxBS reads were mapped to mouse genome mm9 and λ-phage DNA using the Bismark mapping tool. The mapping was done using the paired end Bowtie2 backend with the following parameter values: −I 0 –X 600 –N 0. For each of the samples, the bismark_methylation_extractor script in the Bismark package was used to extract the number of times each cytosine within the amplicons was converted. These counts were used to calculate the proportions for each cytosine to be nonmethylated (or formylmethylated/carboxylmethylated), methylated, or hydroxymethylated. For details, see the Bismark manual (http://www.bioinformatics.babraham.ac.uk/projects/bismark/Bismark_User_Guide.pdf).
Statistical model for BS-seq and oxBS-seq data
We used a multinomial random variable to model the methylation status of a given cytosine (index omitted for brevity). The proportions or probabilities of the possible states C, 5mC, 5hmC, 5fC, and 5caC are p(C), p(5mC), p(5hmC), p(5fC), and p(5caC) (they sum up to unity), respectively. The cytosine modifications 5fC and 5caC cannot be distinguished from C using the BS- and oxBS-seq approaches, and thus those probabilities are collapsed and denoted as p(C∪5fC∪5caC) = p(C) + p(5fC) + p(5caC). The unknown parameters are denoted as θ = p(C∪5fC∪5caC), p(5mC), and p(5hmC). BS- and oxBS-seq experiments result in convoluted observations of the aforementioned random variable. For example, the probability of sequenced thymine after BS treatment is p(C∪5fC∪5caC), whereas the probability of observing cytosine is 1 − p(C∪5fC∪5caC) = p(5mC) + p(5hmC). Similarly, the probabilities of observing thymine and cytosine after oxidation and BS treatment are p(C∪5fC∪5caC) + p(5hmC) and p(5mC), respectively. For a given cytosine, let DBS and DoxBS denote the experimental datasets from the BS- and oxBS-seq experiments, respectively. Moreover, let NBS = C and NBS = T be the number of cytosine and thymine observations in DBS. Similarly, we have NoxBS = C and NoxBS = T for DoxBS. Then the likelihood of the data are p(DBS,DoxBS|θ) = p(DBS|θ) × p(DoxBS|θ), where p(DBS|θ) = p(C∪5fC∪5caC)NBS = T × (p(5mC) + p(5hmC))NBS = C and p(DoxBS|θ) = (p(C∪5fC∪5caC) + p(5hmC))NBS = T × p(5mC)NBS = C. The model is inferred using the Bayesian framework.
Inspired by the Dirichlet-Binomial model, we set a Dirichlet prior for θ, p(θ|α). A weak prior p(θ|α) is defined such that the methylation states are equally probable a priori by setting α = (2, 2, 2)T. The posterior distribution of θ cannot be solved analytically with a Dirichlet prior (although it is a conjugate prior for the categorical likelihood) because of the convoluted observations. Therefore, we resort to the Metropolis-Hastings algorithm. A proper proposal distribution for drawing a new parameter sample θ* based on the current one θi has to fulfill the unity sum constraint Σθ* = 1. To enforce this, we choose a Dirichlet distribution as the proposal distribution. The parameter αprop of the Dirichlet proposal distribution q(θ*|θi) is the current parameter sample θi multiplied by a scalar-β. The scalar-β controls the deviation of the proposal distribution, and thus it can be used to obtain a desired acceptance ratio. Empirically, we set β = 50 to achieve an acceptance ratio varying from 30 to 50%. The Markov Chain Monte Carlo chains were run for 50,000 iterations, which was enough for the chains to converge to their equilibrium distributions. The drawn parameter samples were used to estimate the parameter posterior distributions. The parameter posterior mean was used as an estimate of the proportions of cytosine modifications p(C) ≈ p(C∪5fC∪5caC), p(5mC), and p(5hmC).
Online supplemental material
Fig. S1 shows the gating strategies and the methylation analysis of Foxp3 CNS regions in CD4 SP thymocytes and T reg cells isolated from WT, Tet2/3 DKO mice, or mice with a single deletion of Tet2 or Tet3. Fig. S2 shows the percentage and MFI of Foxp3+ cells in iT reg cells differentiated from naive T cells isolated from WT, Tet2 KO, Tet3 KO, and Tet2/3 DKO mice in the presence of TGF-β or TGF-β/vitamin C. Fig. S3 shows the percentage of Foxp3+ cells during a time course analysis of human iT reg cell differentiation and restimulation and the effects of vitamin C on global 5hmC level and methylation changes of FOXP3 CNS2 in human iT reg cells. Table S1 is a list of primer sequences used for amplicon sequencing in this study. Table S2 is a list of BS conversion efficiency assessed by spike-in control for BS or oxBS in this study. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20151438/DC1.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ryan Hastie and Susan Togher for help in maintaining mouse colonies and members of the Rao laboratory for suggestions and discussions. We thank Cheryl Kim, Kurt Gunst, and Lara Nosworthy at the La Jolla Institute flow cytometry facility for help with cell sorting experiments, Jeremy Day and Dr. Gregory Seumois of the La Jolla Institute sequencing facility for help with next-generation sequencing, and Dr. Zbigniew Mikulski, Dr. Bill Kiosses, and Margie Chadwell of the La Jolla Institute microscopy and histology facility for help with histological and microscopic analysis.
This work was supported by National Institutes of Health Research Project grants AI44432 and CA151535 (to A. Rao) and HL114093 (to A. Rao and P. Vijayanand), by a Translational Research Program Award from the Leukemia and Lymphoma Society (grant LLS TRP 6464-15 to A. Rao), and by the Academy of Finland Centre of Excellence in Molecular Systems Immunology and Physiology Research (to H. Lähdesmäki). T. Äijö was supported by a graduate student fellowship from the Finnish Doctoral Program in Computational Sciences. S. Trifari and A. Tsagaratou were supported by Irvington postdoctoral fellowships from the Cancer Research Institute. W.A. Pastor was supported by a predoctoral graduate research fellowship from the National Science Foundation. C.-W.J. Lio is supported by an Irvington Postdoctoral Fellowship from the Cancer Research Institute. X. Li was supported by a postdoctoral fellowship from California Institute for Regenerative Medicine, University of California, San Diego Interdisciplinary Stem Cell Research and Training Grant II (TG2-01154).
The authors declare no competing financial interests.
Author contributions: A. Rao conceived the project and supervised project planning and execution. X. Yue performed BS-seq and oxBS-seq for the Foxp3 locus, dot-blot assays, and experiments for mouse T reg cells. S. Trifari performed experiments with human T cells. A. Tsagaratou isolated WT and Tet2/3 DKO T reg cells and T reg cell precursors in the thymus. W.A. Pastor generated Tet3fl/fl mice. J.A. Zepeda-Martínez examined vitamin C effects on TET activity in HEK293T cells under the supervision of Y. Huang and analyzed the data. C.-W.J. Lio suggested and helped with scurfy transfer experiments. X. Li helped with amplicon sequencing. T. Äijö and H. Lähdesmäki performed bioinformatic analyses of BS-seq and oxBS-seq data. P. Vijayanand provided input for experimental design and critically evaluated the manuscript. A. Rao and X. Yue wrote the manuscript with assistance from S. Trifari.
Footnotes
Abbreviations used:
- 5caC
- 5-carboxylcytosine
- 5fC
- 5-formylcytosine
- 5hmC
- 5-hydroxymethylcytosine
- 5mC
- 5-methylcytosine
- BS
- bisulfite
- CMS
- cytosine-5-methylenesulphonate
- DNMT
- DNA methyltransferase
- DP
- double positive
- eGFP
- enhanced GFP
- ESC
- embryonic stem cell
- IRES
- internal ribosome entry site
- MFI
- mean fluorescent intensity
- oxBS
- oxidative BS
- RA
- retinoic acid
- SP
- single positive
- TET
- ten-eleven translocation
References
- Abbas A.K., Benoist C., Bluestone J.A., Campbell D.J., Ghosh S., Hori S., Jiang S., Kuchroo V.K., Mathis D., Roncarolo M.G., et al. 2013. Regulatory T cells: recommendations to simplify the nomenclature. Nat. Immunol. 14:307–308. 10.1038/ni.2554 [DOI] [PubMed] [Google Scholar]
- Allan S.E., Crome S.Q., Crellin N.K., Passerini L., Steiner T.S., Bacchetta R., Roncarolo M.G., and Levings M.K.. 2007. Activation-induced FOXP3 in human T effector cells does not suppress proliferation or cytokine production. Int. Immunol. 19:345–354. 10.1093/intimm/dxm014 [DOI] [PubMed] [Google Scholar]
- Benson M.J., Pino-Lagos K., Rosemblatt M., and Noelle R.J.. 2007. All-trans retinoic acid mediates enhanced T reg cell growth, differentiation, and gut homing in the face of high levels of co-stimulation. J. Exp. Med. 204:1765–1774. 10.1084/jem.20070719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke K., Ebata K.T., Karimi M.M., Zepeda-Martínez J.A., Goyal P., Mahapatra S., Tam A., Laird D.J., Hirst M., Rao A., et al. 2013. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 500:222–226. 10.1038/nature12362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth M.J., Branco M.R., Ficz G., Oxley D., Krueger F., Reik W., and Balasubramanian S.. 2012. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 336:934–937. 10.1126/science.1220671 [DOI] [PubMed] [Google Scholar]
- Booth M.J., Ost T.W., Beraldi D., Bell N.M., Branco M.R., Reik W., and Balasubramanian S.. 2013. Oxidative bisulfite sequencing of 5-methylcytosine and 5-hydroxymethylcytosine. Nat. Protoc. 8:1841–1851. 10.1038/nprot.2013.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth M.J., Raiber E.A., and Balasubramanian S.. 2015. Chemical methods for decoding cytosine modifications in DNA. Chem. Rev. 115:2240–2254. 10.1021/cr5002904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunkow M.E., Jeffery E.W., Hjerrild K.A., Paeper B., Clark L.B., Yasayko S.A., Wilkinson J.E., Galas D., Ziegler S.F., and Ramsdell F.. 2001. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat. Genet. 27:68–73. 10.1038/83784 [DOI] [PubMed] [Google Scholar]
- Burchill M.A., Yang J., Vogtenhuber C., Blazar B.R., and Farrar M.A.. 2007. IL-2 receptor β-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J. Immunol. 178:280–290. 10.4049/jimmunol.178.1.280 [DOI] [PubMed] [Google Scholar]
- Chen J., Guo L., Zhang L., Wu H., Yang J., Liu H., Wang X., Hu X., Gu T., Zhou Z., et al. 2013. Vitamin C modulates TET1 function during somatic cell reprogramming. Nat. Genet. 45:1504–1509. 10.1038/ng.2807 [DOI] [PubMed] [Google Scholar]
- Chen W., Jin W., Hardegen N., Lei K.J., Li L., Marinos N., McGrady G., and Wahl S.M.. 2003. Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J. Exp. Med. 198:1875–1886. 10.1084/jem.20030152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.M., Shin J.H., Sohn M.H., Harding M.J., Park J.H., Tobiasova Z., Kim D.Y., Maher S.E., Chae W.J., Park S.H., et al. 2010. Cell-permeable Foxp3 protein alleviates autoimmune disease associated with inflammatory bowel disease and allergic airway inflammation. Proc. Natl. Acad. Sci. USA. 107:18575–18580. 10.1073/pnas.1000400107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson K.M., Gustafson C.B., Young J.I., Züchner S., and Wang G.. 2013. Ascorbate-induced generation of 5-hydroxymethylcytosine is unaffected by varying levels of iron and 2-oxoglutarate. Biochem. Biophys. Res. Commun. 439:522–527. 10.1016/j.bbrc.2013.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., Cullen J.J., and Buettner G.R.. 2012. Ascorbic acid: chemistry, biology and the treatment of cancer. Biochim. Biophys. Acta. 1826:443–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Arvey A., Chinen T., van der Veeken J., Gasteiger G., and Rudensky A.Y.. 2014. Control of the inheritance of regulatory T cell identity by a cis element in the Foxp3 locus. Cell. 158:749–763. 10.1016/j.cell.2014.07.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flashman E., Davies S.L., Yeoh K.K., and Schofield C.J.. 2010. Investigating the dependence of the hypoxia-inducible factor hydroxylases (factor inhibiting HIF and prolyl hydroxylase domain 2) on ascorbate and other reducing agents. Biochem. J. 427:135–142. 10.1042/BJ20091609 [DOI] [PubMed] [Google Scholar]
- Floess S., Freyer J., Siewert C., Baron U., Olek S., Polansky J., Schlawe K., Chang H.D., Bopp T., Schmitt E., et al. 2007. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 5:e38 10.1371/journal.pbio.0050038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haribhai D., Lin W., Relland L.M., Truong N., Williams C.B., and Chatila T.A.. 2007. Regulatory T cells dynamically control the primary immune response to foreign antigen. J. Immunol. 178:2961–2972. 10.4049/jimmunol.178.5.2961 [DOI] [PubMed] [Google Scholar]
- He Y.F., Li B.Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L., et al. 2011. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 333:1303–1307. 10.1126/science.1210944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., and Rao A.. 2014. Connections between TET proteins and aberrant DNA modification in cancer. Trends Genet. 30:464–474. 10.1016/j.tig.2014.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Pastor W.A., Shen Y., Tahiliani M., Liu D.R., and Rao A.. 2010. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One. 5:e8888 10.1371/journal.pone.0008888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huehn J., Polansky J.K., and Hamann A.. 2009. Epigenetic control of FOXP3 expression: the key to a stable regulatory T-cell lineage? Nat. Rev. Immunol. 9:83–89. 10.1038/nri2474 [DOI] [PubMed] [Google Scholar]
- Ito S., Shen L., Dai Q., Wu S.C., Collins L.B., Swenberg J.A., He C., and Zhang Y.. 2011. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 333:1300–1303. 10.1126/science.1210597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer L.M., Tahiliani M., Rao A., and Aravind L.. 2009. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 8:1698–1710. 10.4161/cc.8.11.8580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefowicz S.Z., Wilson C.B., and Rudensky A.Y.. 2009. Cutting edge: TCR stimulation is sufficient for induction of Foxp3 expression in the absence of DNA methyltransferase 1. J. Immunol. 182:6648–6652. 10.4049/jimmunol.0803320 [DOI] [PubMed] [Google Scholar]
- Josefowicz S.Z., Lu L.F., and Rudensky A.Y.. 2012. Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol. 30:531–564. 10.1146/annurev.immunol.25.022106.141623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Lienhard M., Pastor W.A., Chawla A., Novotny M., Tsagaratou A., Lasken R.S., Thompson E.C., Surani M.A., Koralov S.B., et al. 2015. Simultaneous deletion of the methylcytosine oxidases Tet1 and Tet3 increases transcriptome variability in early embryogenesis. Proc. Natl. Acad. Sci. USA. 112:E4236–E4245. 10.1073/pnas.1510510112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.P., and Leonard W.J.. 2007. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. J. Exp. Med. 204:1543–1551. 10.1084/jem.20070109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M., Huang Y., Jankowska A.M., Pape U.J., Tahiliani M., Bandukwala H.S., An J., Lamperti E.D., Koh K.P., Ganetzky R., et al. 2010. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 468:839–843. 10.1038/nature09586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M., Bandukwala H.S., An J., Lamperti E.D., Thompson E.C., Hastie R., Tsangaratou A., Rajewsky K., Koralov S.B., and Rao A.. 2011. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc. Natl. Acad. Sci. USA. 108:14566–14571. 10.1073/pnas.1112317108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M., An J., Pastor W.A., Koralov S.B., Rajewsky K., and Rao A.. 2015. TET proteins and 5-methylcytosine oxidation in hematological cancers. Immunol. Rev. 263:6–21. 10.1111/imr.12239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K.P., Yabuuchi A., Rao S., Huang Y., Cunniff K., Nardone J., Laiho A., Tahiliani M., Sommer C.A., Mostoslavsky G., et al. 2011. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 8:200–213. 10.1016/j.stem.2011.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal G., Zhang N., van der Touw W., Ding Y., Ju W., Bottinger E.P., Reid S.P., Levy D.E., and Bromberg J.S.. 2009. Epigenetic regulation of Foxp3 expression in regulatory T cells by DNA methylation. J. Immunol. 182:259–273. 10.4049/jimmunol.182.1.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.U., Agarwal S., and Rao A.. 2002. Th2 lineage commitment and efficient IL-4 production involves extended demethylation of the IL-4 gene. Immunity. 16:649–660. 10.1016/S1074-7613(02)00314-X [DOI] [PubMed] [Google Scholar]
- Li X., Liang Y., LeBlanc M., Benner C., and Zheng Y.. 2014. Function of a Foxp3 cis-element in protecting regulatory T cell identity. Cell. 158:734–748. 10.1016/j.cell.2014.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lio C.W., and Hsieh C.S.. 2008. A two-step process for thymic regulatory T cell development. Immunity. 28:100–111. 10.1016/j.immuni.2007.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loenarz C., and Schofield C.J.. 2008. Expanding chemical biology of 2-oxoglutarate oxygenases. Nat. Chem. Biol. 4:152–156. 10.1038/nchembio0308-152 [DOI] [PubMed] [Google Scholar]
- Minor E.A., Court B.L., Young J.I., and Wang G.. 2013. Ascorbate induces ten-eleven translocation (Tet) methylcytosine dioxygenase-mediated generation of 5-hydroxymethylcytosine. J. Biol. Chem. 288:13669–13674. 10.1074/jbc.C113.464800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair V.S., and Oh K.I.. 2014. Down-regulation of Tet2 prevents TSDR demethylation in IL2 deficient regulatory T cells. Biochem. Biophys. Res. Commun. 450:918–924. 10.1016/j.bbrc.2014.06.110 [DOI] [PubMed] [Google Scholar]
- Ohkura N., Hamaguchi M., Morikawa H., Sugimura K., Tanaka A., Ito Y., Osaki M., Tanaka Y., Yamashita R., Nakano N., et al. 2012. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity. 37:785–799. 10.1016/j.immuni.2012.09.010 [DOI] [PubMed] [Google Scholar]
- Ooi S.K., O’Donnell A.H., and Bestor T.H.. 2009. Mammalian cytosine methylation at a glance. J. Cell Sci. 122:2787–2791. 10.1242/jcs.015123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor W.A., Aravind L., and Rao A.. 2013. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat. Rev. Mol. Cell Biol. 14:341–356. 10.1038/nrm3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polansky J.K., Kretschmer K., Freyer J., Floess S., Garbe A., Baron U., Olek S., Hamann A., von Boehmer H., and Huehn J.. 2008. DNA methylation controls Foxp3 gene expression. Eur. J. Immunol. 38:1654–1663. 10.1002/eji.200838105 [DOI] [PubMed] [Google Scholar]
- Ruiz M.A., Rivers A., Ibanez V., Vaitkus K., Mahmud N., DeSimone J., and Lavelle D.. 2015. Hydroxymethylcytosine and demethylation of the γ-globin gene promoter during erythroid differentiation. Epigenetics. 10:397–407. 10.1080/15592294.2015.1039220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi S., Yamaguchi T., Nomura T., and Ono M.. 2008. Regulatory T cells and immune tolerance. Cell. 133:775–787. 10.1016/j.cell.2008.05.009 [DOI] [PubMed] [Google Scholar]
- Sturn A., Quackenbush J., and Trajanoski Z.. 2002. Genesis: cluster analysis of microarray data. Bioinformatics. 18:207–208. 10.1093/bioinformatics/18.1.207 [DOI] [PubMed] [Google Scholar]
- Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L., and Rao A.. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 324:930–935. 10.1126/science.1170116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai X., Erman B., Alag A., Mu J., Kimura M., Katz G., Guinter T., McCaughtry T., Etzensperger R., Feigenbaum L., et al. 2013. Foxp3 transcription factor is proapoptotic and lethal to developing regulatory T cells unless counterbalanced by cytokine survival signals. Immunity. 38:1116–1128. 10.1016/j.immuni.2013.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q., and Bluestone J.A.. 2013. Regulatory T-cell therapy in transplantation: moving to the clinic. Cold Spring Harb. Perspect. Med. 3:a015552 10.1101/cshperspect.a015552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toker A., and Huehn J.. 2011. To be or not to be a Treg cell: lineage decisions controlled by epigenetic mechanisms. Sci. Signal. 4:pe4 10.1126/scisignal.2001783 [DOI] [PubMed] [Google Scholar]
- Toker A., Engelbert D., Garg G., Polansky J.K., Floess S., Miyao T., Baron U., Düber S., Geffers R., Giehr P., et al. 2013. Active demethylation of the Foxp3 locus leads to the generation of stable regulatory T cells within the thymus. J. Immunol. 190:3180–3188. 10.4049/jimmunol.1203473 [DOI] [PubMed] [Google Scholar]
- Tone Y., Furuuchi K., Kojima Y., Tykocinski M.L., Greene M.I., and Tone M.. 2008. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat. Immunol. 9:194–202. 10.1038/ni1549 [DOI] [PubMed] [Google Scholar]
- Tran D.Q., Ramsey H., and Shevach E.M.. 2007. Induction of FOXP3 expression in naive human CD4+FOXP3− T cells by T-cell receptor stimulation is transforming growth factor-β dependent but does not confer a regulatory phenotype. Blood. 110:2983–2990. 10.1182/blood-2007-06-094656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsagaratou A., and Rao A.. 2013. TET proteins and 5-methylcytosine oxidation in the immune system. Cold Spring Harb. Symp. Quant. Biol. 78:1–10. 10.1101/sqb.2013.78.020248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsagaratou A., Äijö T., Lio C.W., Yue X., Huang Y., Jacobsen S.E., Lähdesmäki H., and Rao A.. 2014. Dissecting the dynamic changes of 5-hydroxymethylcytosine in T-cell development and differentiation. Proc. Natl. Acad. Sci. USA. 111:E3306–E3315. 10.1073/pnas.1412327111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Ioan-Facsinay A., van der Voort E.I., Huizinga T.W., and Toes R.E.. 2007. Transient expression of FOXP3 in human activated nonregulatory CD4+ T cells. Eur. J. Immunol. 37:129–138. 10.1002/eji.200636435 [DOI] [PubMed] [Google Scholar]
- Wu H., and Zhang Y.. 2014. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 156:45–68. 10.1016/j.cell.2013.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R., Qu C., Zhou Y., Konkel J.E., Shi S., Liu Y., Chen C., Liu S., Liu D., Chen Y., et al. 2015. Hydrogen sulfide promotes Tet1- and Tet2-mediated Foxp3 demethylation to drive regulatory T cell differentiation and maintain immune homeostasis. Immunity. 43:251–263. 10.1016/j.immuni.2015.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin R., Mao S.Q., Zhao B., Chong Z., Yang Y., Zhao C., Zhang D., Huang H., Gao J., Li Z., et al. 2013. Ascorbic acid enhances Tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals. J. Am. Chem. Soc. 135:10396–10403. 10.1021/ja4028346 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Josefowicz S., Chaudhry A., Peng X.P., Forbush K., and Rudensky A.Y.. 2010. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 463:808–812. 10.1038/nature08750 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.