Nuvolone et al. report a new mouse model to elucidate the functional role of cellular prion protein in physiology and disease.
Abstract
Although its involvement in prion replication and neurotoxicity during transmissible spongiform encephalopathies is undisputed, the physiological role of the cellular prion protein (PrPC) remains enigmatic. A plethora of functions have been ascribed to PrPC based on phenotypes of Prnp−/− mice. However, all currently available Prnp−/− lines were generated in embryonic stem cells from the 129 strain of the laboratory mouse and mostly crossed to non-129 strains. Therefore, Prnp-linked loci polymorphic between 129 and the backcrossing strain resulted in systematic genetic confounders and led to erroneous conclusions. We used TALEN-mediated genome editing in fertilized mouse oocytes to create the Zurich-3 (ZH3) Prnp-ablated allele on a pure C57BL/6J genetic background. Genomic, transcriptional, and phenotypic characterization of PrnpZH3/ZH3 mice failed to identify phenotypes previously described in non–co-isogenic Prnp−/− mice. However, aged PrnpZH3/ZH3 mice developed a chronic demyelinating peripheral neuropathy, confirming the crucial involvement of PrPC in peripheral myelin maintenance. This new line represents a rigorous genetic resource for studying the role of PrPC in physiology and disease.
The cellular prion protein (PrPC) is a ubiquitously expressed membrane-anchored protein encoded by the Prnp gene. Misfolding of PrPC generates the scrapie prion protein (PrPSc) and leads to a class of invariably lethal, neurodegenerative conditions termed transmissible spongiform encephalopathies, or prion diseases. Despite intense investigation and the availability of at least seven independently generated lines of Prnp−/− mice, little is known about the physiological function of PrPC (Aguzzi et al., 2013).
Two key genetic features of existing Prnp−/− mouse lines constitute systematic experimental confounders that hampered the elucidation of the physiological role of PrPC (Steele et al., 2007). The first confounder stems from the design of Prnp targeting vectors. In four lines (PrnpNgsk/Ngsk, PrnpRcm0/Rcm0, PrnpRkn/Rkn, and PrnpZH2/ZH2) deletion of Prnp exon 3 spanning a splice acceptor site resulted in spurious overexpression of the Prnd-encoded Doppel protein, causing severe ataxia and Purkinje cell loss (Steele et al., 2007). The second confounder depends on the embryonic stem (ES) cells and breeding schemes used for the generation of Prnp−/− mice. All Prnp−/− lines currently available have been generated in ES cells derived from the 129 strain of the laboratory mouse and are maintained in non-129 backgrounds, with the exception of the PrnpEdbg/Edbg line. Consequently, 129-derived genomic material flanking the targeted Prnp locus on chromosome 2 represents a systematic genetic confounder when Prnp−/− and PrnpWT/WT mice are compared (Nuvolone et al., 2013; Striebel et al., 2013). In this study, we set out to overcome these limitations by generating a co-isogenic line of Prnp−/− mice in the well-characterized C57BL/6J background using transcription activator-like effector nuclease (TALEN)–based genome editing.
RESULTS AND DISCUSSION
TALEN-induced disruption of the Prnp protein-coding sequence
The complete protein-coding DNA sequence (CDS) for mouse PrPC is located within exon 3 of the Prnp gene. To eliminate PrPC expression in C57BL/6J without disrupting the Prnp gene architecture, we used a TALEN pair targeting a site within the Prnp CDS in close proximity to the start codon (Fig. 1 A). 1 of 44 F0 pups was found to carry a Prnp allele with an 8-bp deletion (termed PrnpZH3). The frame shift within PrnpZH3 introduced a premature stop codon in the sequence coding for the PrPC secretory signal peptide (Fig. 1 B). PrnpZH3 was efficiently transmitted through the germ line, and mice homozygous for PrnpZH3 were obtained in the F2 generation (C57BL/6J-PrnpZH3/ZH3, hereafter termed PrnpZH3/ZH3), as assessed by restriction fragment length polymorphism (RFLP) analysis and TaqMan-based allelic discrimination assay (Fig. 1, C and D).
Figure 1.
TALEN-based generation of C57BL/6J-PrnpZH3/ZH3 mice. (A) TALEN-binding sites within Prnp exon E3 and start codon (yellow) of the protein coding sequence. Colors indicate the code for repeat-variable diresidues. The Prnp TALEN pair incorporates second-generation heterodimeric FokI cleavage domains (FokIELD and FokIKKR). (B) Sanger sequencing reads of a PrnpWT and a PrnpZH3 allele from the founder F0 mouse. A deletion of 8 bp in the PrnpZH3 allele (highlighted by a red box on the WT sequence) introduces a T/D residue change, followed by a premature STOP codon (*) after residue 14 within the sequence encoding the PrPC signal peptide. The deletion also eliminates the Tsp45I recognition sequence (blue letters on WT sequence). As a result of sequence characteristics in this region, an alternative 8-bp deletion (ACTATGTG), shifted by 4 bp in respect to the previous deletion, is also compatible with the generation of the PrnpZH3 allele. (C) Representative image of routinely used RFLP analysis discriminating PrnpWT/WT (digested amplicons), PrnpWT/ZH3 (digested and undigested amplicons), and PrnpZH3/ZH3 mice (only undigested amplicon). Primers location, restriction site, and expected sizes for digestion products are indicated on top of the gel image. (D) Allelic discrimination genotyping using a FAM-labeled WT-specific probe and a Yakima Yellow–labeled ZH3-specific probe. NTC, no-template control. ΔRn, difference in normalized reporter fluorescence after and before amplification. Apart from NTC, each triangle denotes one mouse (n = 4 mice/genotype). The mean (triangle) and SD (blue error bars) for four technical replicates of each mouse/NTC sample are shown. (E) Immunoblot analysis of PrPC expression in different CNS regions (Cx, cortex; Sc, spinal cord; Cb, cerebellum) of PrnpWT/WT (WT) and PrnpZH3/ZH3 (ZH3) mice was performed using POM1 (against helices α1 and α3 of the PrPC globular domain). The blot was also decorated with anti-actin antibody as control. (F) Brain PrPC levels as determined by sandwich POM1-POM2 ELISA. PrnpEdbg/Edbg (Edbg) served as negative controls. Each circle denotes a mouse (n = 3 mice/genotype). Horizontal bar indicates mean. WT→KO, consecutive log2 dilutions of PrnpWT/WT into PrnpEdbg/Edbg homogenate, indicating that the threshold of detectability was 1:16. (G) Immunofluorescence staining of cerebelli. MAP2 is displayed in green, PrPC, detected with POM19 (against helices β1 and α3 of globular domain) in red, and DAPI in blue. Bar, 20 µm. (D–G) Representative data from two independent experiments.
As expected, PrnpZH3/ZH3 mice showed no detectable PrPC expression in central nervous system (CNS) tissues, as assessed by Western blotting (Fig. 1 E), by a high sensitivity sandwich ELISA (Fig. 1 F) or by immunofluorescence (Fig. 1 G). Collectively, these data indicate that a TALEN-induced deletion of 8 bp within Prnp results in a functional disruption of the Prnp CDS and abolishes the competence for PrPC expression.
Analysis of TALEN off-target cleavage and chromosomal aberrations
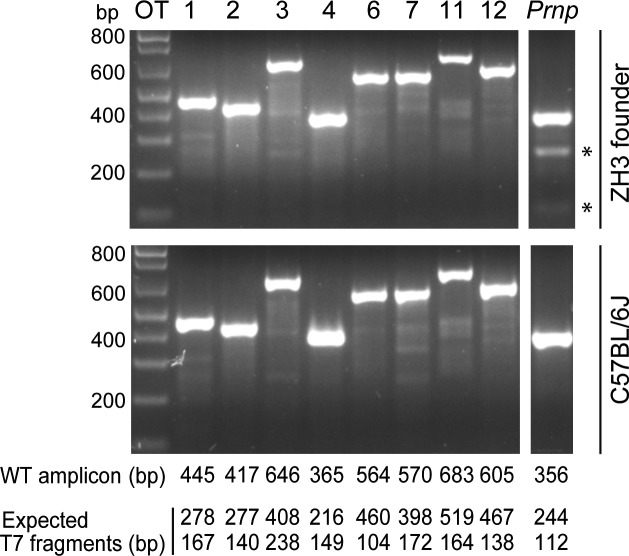
TALENs do not typically cause extensive genomic off-target modifications. However, cleavage of closely related off-target sites (OTs) can occur (Doyle et al., 2012). We PCR amplified eight potential OTs from the TALEN-targeted founder and a C57BL/6J control (Tables S1 and S2). Amplicons were subjected to an annealing protocol that enables the formation of heteroduplexes in the presence of heterozygous mutations. Treatment of these reannealed amplicons with T7 endonuclease I, which cleaves heteroduplexes, did not yield any digestion products indicative of TALEN off-target cleavage (Fig. 2).
Figure 2.
C57BL/6-PrnpZH3/ZH3 mice do not have TALEN off-target cleavage sites. T7 endonuclease I digestion of PCR products generated from predicted OTs (Table S1) and Prnp as a digestion positive control. Analyses were performed on the founder PrnpWT/ZH3 mouse and one control C57BL/6J mouse. Before enzymatic digestion, amplicons were subjected to a temperature gradient enabling the formation of heteroduplexes in the presence of heterozygous mutations in the amplified gDNA. In the presence of TALEN-induced mutations, fragments of the size indicated below the gels are expected to appear, in addition to the undigested, WT amplicon. Nonconsecutive lanes from the same gel show Prnp amplicon as a control. Only in the founder PrnpWT/ZH3 mouse T7 endonuclease I digestion of the Prnp amplicon results in the formation of the two predicted fragments (indicated by an asterisk).
We next investigated the presence of chromosomal abnormalities in the PrnpZH3/ZH3 line. Giemsa banding (G-banding) and spectral karyotyping showed a normal 40X,Y karyotype (Fig. S1 A) in 14/25 metaphases from a fibroblast cell line obtained from a PrnpZH3/ZH3 mouse. The remaining karyotyped metaphases showed some degrees of chromosomal aberrations, including six metaphases with 79 or 80 chromosomes, possibly representing cell culture artifacts (Littlefield and Mailhes, 1975). To account for this possibility, we performed G-banding analysis of primary splenocytes from another PrnpZH3/ZH3 mouse. Here, we found a normal 40X,Y karyotype in 35/35 metaphases. These analyses excluded the presence of large TALEN-induced chromosomal aberrations.
We then performed high-density array comparative genomic hybridization (aCGH). This analysis showed the presence of a relative loss of genomic DNA (gDNA) in one PrnpZH3/ZH3 mouse compared with one C57BL/6J control mouse in a 34.4-kbp region of chromosome 16 encompassing the Maats1 locus (Fig. S1 B). This could reflect either a genomic loss in the PrnpZH3/ZH3 mouse or a genomic gain in the C57BL/6J mouse. Copy number variants (CNVs) are frequently observed among different individuals of the same inbred colony of laboratory mice, including C57BL/6J, and de novo CNV occur with an incidence of 1–14% (Egan et al., 2007; Flatscher-Bader et al., 2011). Therefore the degree of genomic variation between the two analyzed PrnpZH3/ZH3 and C57BL/6J individual mice is not dissimilar to the variation seen between different individuals of the C57BL/6J strain and falls within the natural genetic variation of inbred strains of the laboratory mouse. Importantly, we did not identify any structural change linked to the targeted Prnp locus on chromosome 2, which would represent a systematic confounder in studies comparing PrnpZH3/ZH3 and PrnpWT/WT mice.
PrnpZH3/ZH3 mice lack the flanking gene problem
We next studied the genomic background of PrnpZH3/ZH3 mice. Whole-genome single-nucleotide polymorphism (SNP) analysis of PrnpZH3/ZH3 confirmed that 100% of the analyzed markers were of the C57BL/6J type (Fig. S2). To further investigate the genomic similarity of PrnpZH3/ZH3 mice with C57BL/6J mice, we performed RNA sequencing in hippocampi of 3-mo-old male mice. For additional comparison, we analyzed hippocampi from congenic aged-matched PrnpZH1/ZH1 males that had been backcrossed for >12 generations to the C57BL/6 background.
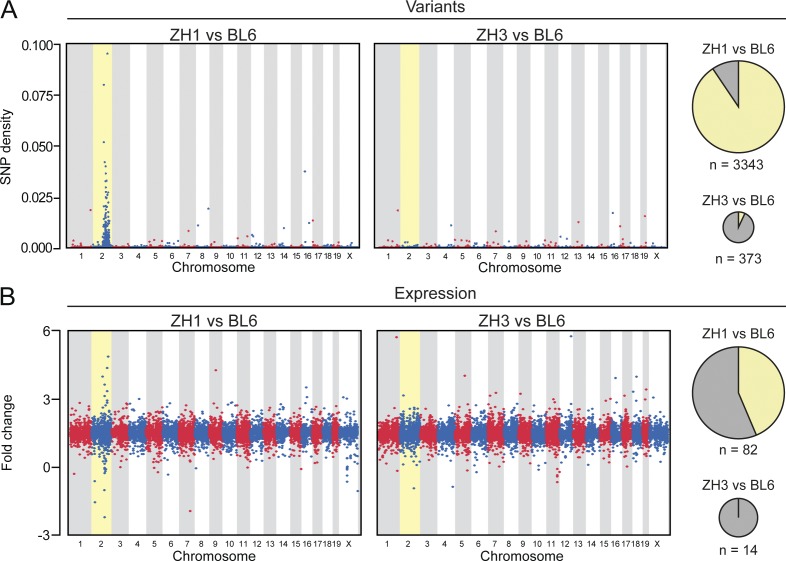
In comparison with C57BL/6J mice, PrnpZH1/ZH1 showed a significantly higher number of variants than PrnpZH3/ZH3 (mean number of variants, including SNPs and insertions/deletions, INDEL: 3,343 versus. 373; P < 0.01, Student’s t test; Fig. 3 A). Notably, the 373 variants detected in PrnpZH3/ZH3 included the 8-bp deletion within Prnp exon 3 (position chr2:131936464) characteristic of the PrnpZH3 allele, confirming the accuracy of our procedure for variant identification (Fig. S3). By plotting the density of sequence variation against its genomic location, we observed an obvious clustering of variants in chromosome 2 of PrnpZH1/ZH1 mice, with >90% of variants in this chromosome, as expected from the inevitable flanking-gene artifacts derived from the targeting and breeding strategies in these mice. Crucially, no such clustering was observed in PrnpZH3/ZH3 mice (Fig. 3 A).
Figure 3.
RNA sequencing in PrnpWT/WT, PrnpZH1/ZH1, and PrnpZH3/ZH3 hippocampi. RNA sequencing was performed on hippocampi from four mice per each group. (A) Physical distribution of sequence variant density across the genome based on variants unique to PrnpZH1/ZH1 (ZH1, left) or PrnpZH3/ZH3 (ZH3, right) compared with C57BL/6J mice (BL6). Each dot denotes the ratio of variants and total number of annotated bases in 50-kb windows throughout the genome. Pie charts depict the number (n, proportional to the area) and chromosomal distribution (yellow, chromosome 2; gray, other chromosomes) of sequence variants in the two comparisons. For variant identification, the three mice with the highest coverage within each group were considered. (B) Physical distribution of differential gene expression levels across the genome based on the comparison of PrnpZH1/ZH1 (left) or PrnpZH3/ZH3 (right) with respect to C57BL/6J mice. Each dot denotes a gene. Pie charts depict the number (n, proportional to the area) and chromosomal distribution (yellow, chromosome 2; gray, other chromosomes) of DEGs (fdr < 0.05 and absolute log2 ratio > 0.5) in the two comparisons.
We then sought to identify genes differentially expressed between PrnpZH1/ZH1 or PrnpZH3/ZH3 versus C57BL/6J hippocampi. The mean Prnp mRNA levels were significantly higher in C57BL/6J hippocampi (17,538 reads per kilobase per million mapped reads [RPKM]) than in PrnpZH1/ZH1 (5,781 RPKM, false discovery rate [fdr]: 3.98 × 10−85) and PrnpZH3/ZH3 hippocampi (13,278 RPKM, fdr < 0.01; Fig. S3). Conversely, C57BL/6J, PrnpZH1/ZH1 or PrnpZH3/ZH3 did not differ in their mean Prnd levels (4 RPKM, 5 RPKM, and 2 RPKM, respectively), nor in their mean Matts1 levels (26 RPKM, 22 RPKM, and 18 RPKM, respectively). The latter observation excludes a systematic loss of genomic material comprising the Matts1 locus in the PrnpZH3/ZH3 colony. Importantly, we did not identify any gene transcribed in C57BL/6J or PrnpZH1/ZH1 hippocampi that was not detectable in PrnpZH3/ZH3 hippocampi. This observation excludes the presence of deletions >100 bp (and as such not detectable with our variant identification strategy based on sequencing short reads) induced by TALEN in the fraction of the PrnpZH3/ZH3 genome inferred from the hippocampus transcriptome.
When plotting the differential expression levels of all detectable genes against their genomic location, PrnpZH1/ZH1 showed obvious clustering of up- and down-regulated genes versus C57BL/6J on chromosome 2 (Fig. 3 B). We then focused on differentially expressed genes (DEGs) with fdr < 0.05 and absolute log2 ratio > 0.5. Using these filters, we detected a significant enrichment of chromosome 2 genes with differential expression level in PrnpZH1/ZH1 (82 genes, 35 of which resided on chromosome 2) as opposed to PrnpZH3/ZH3 mice compared with the same C57BL/6J controls (14 genes, none of which resided on chromosome 2; P = 0.02, Fisher’s exact test, Fig. 3 B). Only two genes are shared between these two lists (Tables S3 and S4). This enrichment may reflect sequence variations in regulatory elements present in the 129-genomic region introgressed in the genome of PrnpZH1/ZH1 mice, which results in increased or reduced expression of neighboring genes (cis-expression quantitative trait loci [eQTL]; Keane et al., 2011). This phenomenon is likely to underlie the enrichment of chromosome 2 genes among the DEGs between congenic PrnpZH1/ZH1 mice and matched WT mice in previous microarray-based transcriptomic analyses (Chadi et al., 2010; Benvegnù et al., 2011). A similar phenomenon can be seen in congenic lines for other genes, where the chromosome bearing the targeted gene is the one with the highest number of DEGs between the congenic knockout and WT mice (Ejlerskov et al., 2015), even though the lack of comparison with a co-isogenic mouse does not allow concluding whether this effect is a result of the flanking gene problem or not. Also, it is plausible that this effect can become less apparent depending on the functional consequence of the gene under investigation and the experimental setup used (Lusis et al., 2007).
Next, we analyzed alternative splicing by assessing the presence of differential exon usage between PrnpZH1/ZH1 or PrnpZH3/ZH3 versus C57BL/6J hippocampi. We applied similar stringent filters used to identify DEGs (with adjusted P value < 0.05 and absolute log2 ratio > 0.5). Interestingly, we found no genes with differential exon usage in PrnpZH3/ZH3 mice compared with C57BL/6J with the applied filters. Conversely, 9 exons, belonging to 7 genes (6 of which were on chromosome 2), were differentially expressed between PrnpZH1/ZH1 and C57BL/6J mice (Table S5). This enrichment of chromosome 2 genes among genes with differential exon usage in PrnpZH1/ZH1 versus C57BL/6J hippocampi likely reflects the presence of sequence variants affecting alternative splicing of neighboring genes (Hull et al., 2007; Cool et al., 2010). Indeed, the lack of enrichment of chromosome 2 genes for loci undergoing differential expression or exon usage in PrnpZH3/ZH3 versus C57BL/6J hippocampi excludes its dependence from Prnp genetic ablation and conversely suggests that this is yet another, often neglected, genetic confounder of studies with non–co-isogenic knockout mice (Suzuki and Nakayama, 2003).
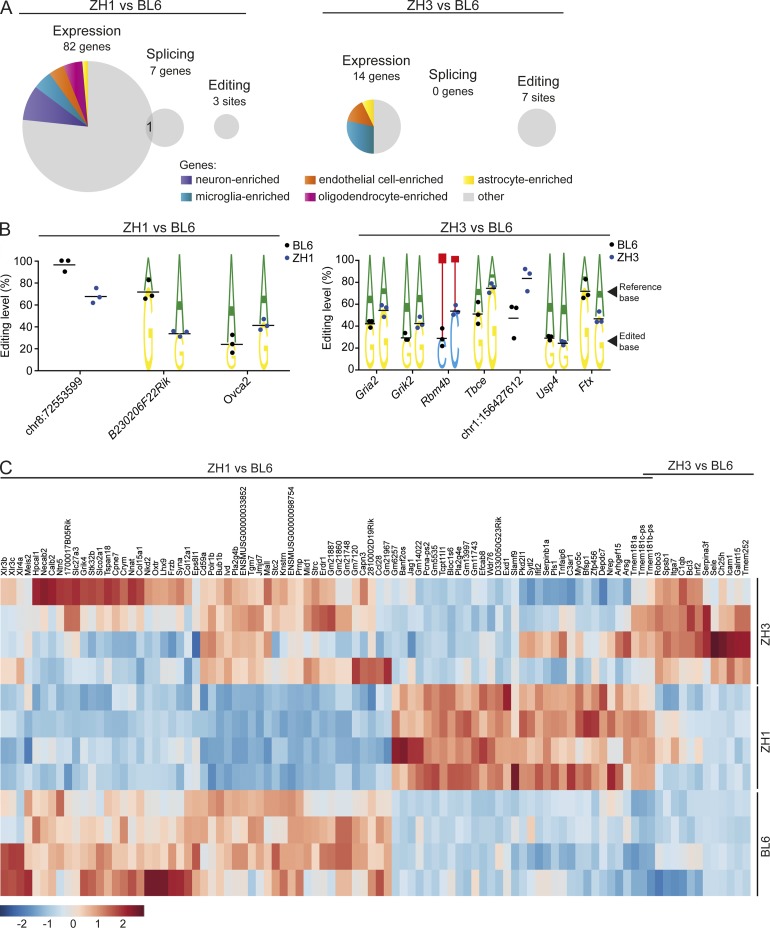
We next analyzed the impact of Prnp genetic ablation in vivo on various aspects of RNA metabolisms. Besides genes with differential expression and exon usage, we analyzed transcripts undergoing RNA editing. By comparing transcript sequences to reference genomic sequences and to published databases of RNA-editing sites in mice, we found 271 sites with evidence for RNA editing in PrnpZH1/ZH1, PrnpZH3/ZH3, and/or C57BL/6J hippocampi. Compared with C57BL/6J, three sites in PrnpZH1/ZH1 and seven sites in PrnpZH3/ZH3 hippocampi showed a significantly different level of RNA editing (P < 0.05; Fig. 4, A and B; and Tables S6 and S7). We then selected three of these sites (mapped to Gria2, Grik2, and Rbm4b) and performed Sanger sequencing of gDNA from the mice included in the RNA sequencing experiment. This analysis confirmed the presence, in homozygosity, of the expected, reference nucleotide (Fig. S4), thereby excluding that the observed mismatch in the RNA reads might be a result of DNA polymorphisms, rather than RNA editing. In all cases in which sites could be assigned to a gene, edited sites were found to be present in noncoding regions. Also, in all but one of these cases, one adenosine in the reference genome was found to be replaced by a guanosine in the sequenced reads (Fig. 4 B), which is in line with an adenosine-to-inosine RNA-editing process (Li and Church, 2013). Sites with differential RNA editing between C57BL/6J and PrnpZH3/ZH3 hippocampi included two intronic sites belonging to genes (Grik2 and Gria2) encoding for members of the kainate family of glutamate receptors (Fig. 4 B). RNA editing in these intronic positions of these genes have been previously reported (Stilling et al., 2014), whereas editing in the coding regions of Grik2 and Gria2 transcripts significantly affects protein function (Sommer et al., 1991; Silberberg et al., 2012). It will be of interest to test whether any of these changes might be related to the currently debated role of PrPC in kainate-induced excitotoxicity (Striebel et al., 2013; Carulla et al., 2015).
Figure 4.
Impact of Prnp genetic ablation on RNA metabolism in vivo. RNA sequencing was performed on hippocampi from four mice per each group. (A) Pie charts depict the number (proportional to the area) of genes showing differential expression (expression), exon usage (splicing), or RNA editing levels (editing) between PrnpZH1/ZH1 (ZH1, left) or PrnpZH3/ZH3 (ZH3, right) and C57BL/6J mice. Colors indicate genes known to be enriched in specific cell types of the central nervous system. (B) Sites with differential RNA-editing levels between PrnpZH1/ZH1 (left) or PrnpZH3/ZH3 (right) and C57BL/6J mice. Editing level indicates percentage of reads showing an edited base instead of the canonical base. Each dot denotes one mouse. This analysis is based on the three mice with the highest coverage for each group. Horizontal bar indicates mean. For sites assigned to a gene (indicated with the gene name), the lower nucleotide indicates the edited base and the upper nucleotide the reference base. The other sites are indicated with their genomic location. One site (indicated as Ovca2) is mapped to two neighboring genes (Ovca2 and Mir684-1) on the same strand. (C) Heat map of genes with differential expression levels between PrnpZH1/ZH1 (left) or PrnpZH3/ZH3 (right) and C57BL/6J mice. Each row represents one mouse. Two genes (Tmem-181b-ps and Tmem-181c-ps) are common to both comparisons.
In the case of PrnpZH1/ZH1 versus C57BL/6J hippocampi, there was one gene, Wdr76, on chromosome 2, with evidence of both differential expression and exon usage (Fig. 4 A and Tables S3 and S5). No such changes were detected between PrnpZH3/ZH3 and C57BL/6J hippocampi. Wdr76 is mapped on chromosome 2, ∼10 Mb apart from Prnp, and is highly polymorphic between C67BL/6 and 129 strains, with numerous SNPs between these strains, including a coding nonsynonymous one (rs27425186; Mouse Phenome Database). Interestingly, Wdr76 plays a role in the recovery from genotoxic stress (Gallina et al., 2015) and congenic B6.129-PrnpZH1/ZH1 mice have been recently shown to have a defective repair of induced DNA damage in the brain compared with C57BL/6N WT mice (Bravard et al., 2015). It will be of interest to investigate to what extent the observed phenotype can be influenced by differences in Wdr76 expression, splicing and coding sequence between the two examined genotypes.
A subset of genes with differential expression is represented by transcripts known to be enriched in different cell types of the CNS (Fig. 4 A). When analyzing the 82 genes found to be differentially expressed between C57BL/6J and PrnpZH1/ZH1 hippocampi, PrnpZH3/ZH3 showed an overall pattern of expression closer to the one of C57BL/6J hippocampi (Fig. 4 C). Analogously, for the 14 DEGs between C57BL/6J and PrnpZH3/ZH3 hippocampi, PrnpZH1/ZH1 showed an overall expression profile closer to the one of C57BL/6J hippocampi. Pathway analysis based on these 14 DEGs did not identify any canonical pathway associated with more than 2 DEGs. However, it is of interest that several of these 14 DEGs are associated with immunological functions (e.g., Icam1, C1qb, Itga7, Sele, and Spsb1).
Collectively, these observations highlight, at a molecular level, that congenic PrnpZH1/ZH1 and co-isogenic PrnpZH3/ZH3 mice are largely divergent and that, in PrnpZH1/ZH1 mice, a conspicuous proportion of the observed transcriptional changes is related to genes on chromosome 2, where an ES cell–derived genomic region (of 129 type) is retained. Moreover, these data confirm that PrnpZH3/ZH3 mice have a bona fide C57BL/6J genome, including the region flanking Prnp, and therefore are devoid of the flanking gene problem affecting other non–co-isogenic lines.
PrnpZH3/ZH3 macrophages do not show enhanced phagocytic activity
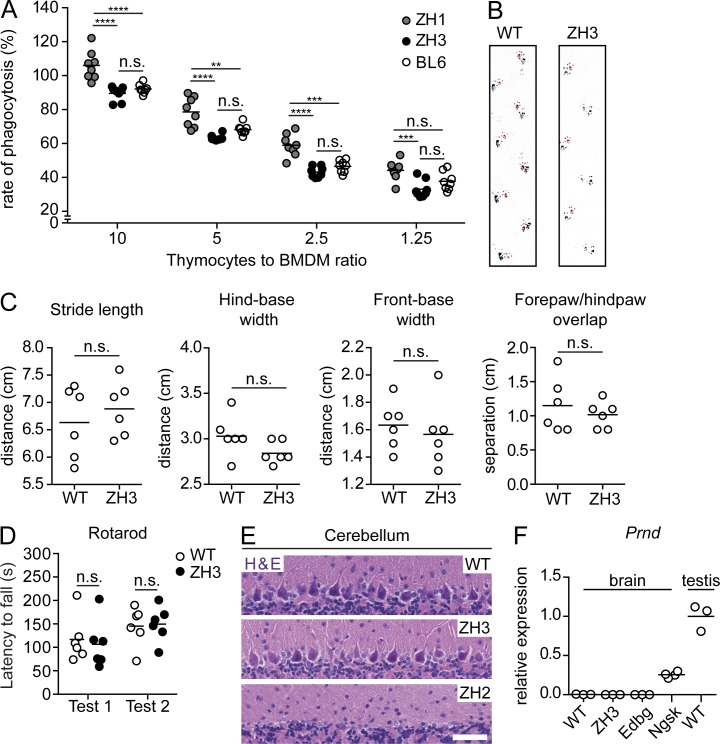
Hyperphagocytosis of apoptotic cells in primary macrophages from non–co-isogenic Prnp−/− lines was originally attributed to the absence of Prnp (de Almeida et al., 2005), but was later shown to be the result of a flanking gene problem (Nuvolone et al., 2013). We measured the phagocytic activity of primary BM-derived C57BL/6J and PrnpZH3/ZH3 macrophages exposed to apoptotic thymocytes. Congenic PrnpZH1/ZH1 macrophages were included as controls. In line with our previous observations (Nuvolone et al., 2013), we found increased phagocytic activity only in PrnpZH1/ZH1 macrophages, but not in PrnpZH3/ZH3 macrophages (Fig. 5 A and Fig. S5).
Figure 5.
C57BL/6J-PrnpZH3/ZH3 mice do not show artifactual phenotypes of other Prnp−/− lines. (A) Rate of phagocytosis of BMDM obtained from C57BL/6J (BL6) and PrnpZH3/ZH3 mice (ZH3) and exposed to different numbers of apoptotic thymocytes (indicated as thymocytes to BMDM ratios). Macrophages from PrnpZH1/ZH1 mice (ZH1) served as control. Each dot denotes a macrophage well exposed to thymocytes (n = 8 macrophage wells/genotype/condition). Values were normalized to the rate of phagocytosis in C57BL/6J mice with 10 thymocytes to BMDM ratio (mean set as 100%). Horizontal bar indicates mean. Data are from two independent experiments. n.s.: not significant; two-way ANOVA, Bonferroni’s multiple comparisons test. (B) Representative images of footprints from 10–14-mo-old PrnpWT/WT and PrnpZH3/ZH3 mice. Magenta, fore paw; blue, hind paw. (C) Quantification of different gait parameters. Each circle denotes a mouse (n = 6 mice/genotype). Horizontal bar indicates mean. n.s., not significant; Student’s t test. (B and C) are representative data from two/three trials. (D) Latency to fall at the rotarod test in PrnpWT/WT and PrnpZH3/ZH3 mice. Mice were 10–14-mo-old at test 1 and test 2 was performed 1 mo after. Each circle denotes a mouse (n = 6 mice/genotype). Horizontal bar indicates mean. n.s., no significant difference between genotypes, two-way ANOVA. (E) Representative images of hematoxylin and eosin (H & E) staining of cerebelli of 60-wk-old PrnpWT/WT and PrnpZH3/ZH3 mice (n = 3 and 8 mice, respectively). 63-wk-old PrnpZH2/ZH2 mice served as control (n = 2 mice). Bar, 50 µm. Data are from two experiments. (F) Brain Prnd mRNA levels as determined by qRT-PCR. Values were normalized to levels in testis of PrnpWT/WT mice (mean set as 1). PrnpEdbg/Edbg (Edbg) and PrnpNgsk/Ngsk (Ngsk) served as negative and positive controls, respectively. Each circle denotes a mouse (n = 4 mice for Ngsk and n = 3 mice for all other genotypes). Horizontal bar indicates mean.
PrnpZH3/ZH3 mice do not display Prnd overexpression and associated neurodegeneration
Purkinje cell degeneration in the cerebellum and associated ataxia, starting from the age of 6 mo, is a hallmark of several lines of Prnp-ablated mice and is caused by inappropriate intergenic splicing and overexpression of the neighboring Prnd gene (Moore et al., 1999). We monitored PrnpZH3/ZH3 mice for the occurrence of similar neurodegenerative changes. 10–14-mo-old PrnpWT/WT and PrnpZH3/ZH3 littermates showed similar walking patterns as assessed by the footprint test (Fig. 5, B and C), and similar performance in the rotarod test (Fig. 5 D). At 15 mo of age, no significant loss of Purkinje cells was evident (Fig. 5 E). Also, quantitative real-time PCR (qRT-PCR) showed brain levels of Prnd transcripts similar between PrnpWT/WT and PrnpZH3/ZH3 mice (Fig. 5 F), in line with the RNA sequencing data. Collectively, these data indicate that PrnpZH3/ZH3 mice did not experience any significant perturbation of Prnd expression and its associated neurodegeneration observed in four of the previously generated Prnp−/− lines.
PrnpZH3/ZH3 mice develop a chronic demyelinating peripheral neuropathy
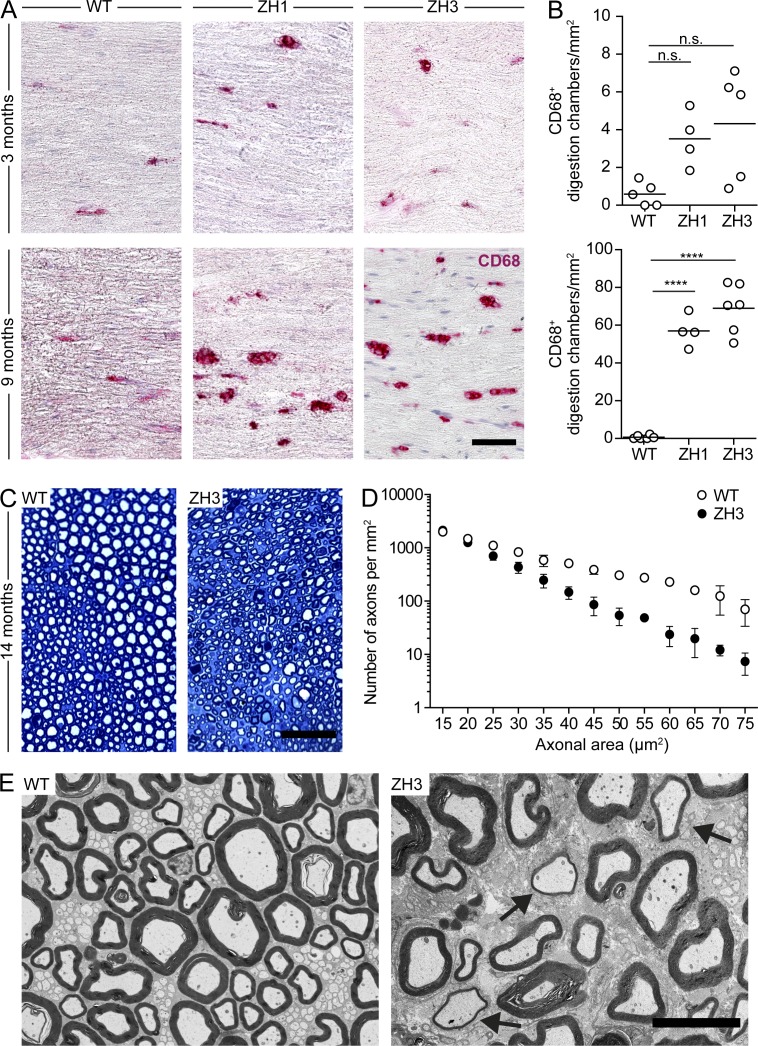
Progressive chronic demyelinating polyneuropathy (CDP) is a robust phenotype present in all examined Prnp−/− lines, including co-isogenic PrnpEdbg/Edbg mice, and results from the absence of neuronal PrPC expression (Bremer et al., 2010). We analyzed the integrity of peripheral nerves in PrnpZH3/ZH3 mice and found a trend toward increased CD68+ digestion chambers (indicative of myelin degradation and resorption by macrophages) in the sciatic nerves of 3-mo-old PrnpZH3/ZH3 mice that reached statistical significance at 9 mo (Fig. 6, A and B). At 14 mo of age, PrnpZH3/ZH3 mice showed a significant reduction of axonal density in sciatic nerves as compared with PrnpWT/WT (Fig. 6, C and D), as well as ultrastructural signs of demyelination (Fig. 6 E). All these features strongly resemble the changes described in sciatic nerves of other previously examined Prnp−/− lines and confirm the crucial involvement of PrPC in peripheral myelin maintenance (Bremer et al., 2010).
Figure 6.
C57BL/6J-PrnpZH3/ZH3 mice develop a chronic demyelinating peripheral neuropathy. (A) Representative images of CD68 staining of longitudinal sections of sciatic nerves show digestion chambers of macrophages and myelin debris in PrnpZH1/ZH1 (ZH1), PrnpZH3/ZH3 (ZH3), and PrnpWT/WT(WT) mice at 3 and 9 mo of age. Bar, 50 µm. (B) Corresponding quantification of CD68+ digestion chambers in sciatic nerves at the same time points as in A. Each dot denotes a mouse (n = 4–6 mice/genotype/time point). Horizontal bar indicates mean. n.s., not significant; ****, P < 0.0001. One-way ANOVA, Bonferroni’s multiple comparisons test. (C) Representative images of toluidine blue staining of semi-thin cross-sections of sciatic nerves from PrnpWT/WT and PrnpZH3/ZH3 mice at 14 mo of age. Bar, 50 µm. (D) Corresponding quantification of density of axons of different size in sciatic nerves of 14-mo-old mice (WT, n = 3 mice; ZH3, n = 8 mice). Symbols indicate mean, whiskers indicate minimum and maximum values. Two-way ANOVA shows a significant effect (P < 0.0001) of genotype. (E) Representative images of transmission electron microscopy of cross sections from sciatic nerves of 14-mo old mice (WT, n = 3 mice; ZH3, n = 4 mice). Arrows indicate thinly myelinated axons. Bar, 10 µm. (A–E) Data from two experiments.
PrnpZH3/ZH3 mice as a genetic resource for prion science
Overall, these data confirm that PrnpZH3/ZH3 mice lack the genetic confounders and artifactual phenotypes of non–co-isogenic Prnp−/− lines. Because of the stringent genetic controls described above, PrnpZH3/ZH3 mice constitute an unprecedented genetic tool for elucidating the physiological function of PrPC and its involvement in pathological conditions. This line will be particularly well suited to intercrossing and/or comparing with other genetically modified mice raised on the well-characterized C57BL/6 background. Our institution will be pleased to provide unrestricted access to PrnpZH3/ZH3 mice for noncommercial purposes.
The strategy exemplified in the generation and characterization of PrnpZH3/ZH3 mice carries implications beyond the prion community. The majority of studies involving knockout mice are based on non–co-isogenic, ES cell–derived knockout lines. However, unmatched genetic background between such knockout mice and WT counterparts (even when using littermates from heterozygous breedings) is unavoidable in these cases and this situation often gives rise to serious and systematic genetic confounders (Vanden Berghe et al., 2015). Common efforts to target each gene of the mouse genome in C57BL/6-derived ES cells (Collins et al., 2007) represent a valid strategy to prevent these problems. Moreover, the recent availability of genome-editing techniques, including the TALEN technique presented here, is now enabling the rapid and simple manipulation of the mouse genome. Our study demonstrate that this approach may also be applicable to genes for which knockout mice are already available (and extensively characterized), thereby generating improved genetic models and disproving potentially misattributed physiological functions.
MATERIALS AND METHODS
Ethical statement
Animal care and experimental protocols were performed in compliance with the Swiss Ethical Principles and Guidelines for Experiments on Animals, the Swiss Animal Protection Law, and with the internal guidelines of the University of Zurich, under the approval of the Veterinary Office of the Canton Zurich (animal permits Versuchstierhaltung 123, 90/2013). All efforts were made to minimize animal discomfort and suffering.
Mice
The following mice were studied: PrnpZH1/ZH1 mice (Büeler et al., 1992) backcrossed to C57BL/6 for >12 generations; PrnpZH2/ZH2 mice (Rossi et al., 2001) on a mixed B6129 background; PrnpEdbg/Edbg mice on a pure 129/Ola background (Manson et al., 1994) and co-isogenic WT 129/Ola mice, both provided by J. Manson, H. Baybutt, and N.A. Mabbott (University of Edinburgh, Edinburgh, Scotland, UK); PrnpNgsk/Ngsk mice (Sakaguchi et al., 1995) extensively backcrossed to C57BL/6; PrnpGFP/GFP mice (Heikenwalder et al., 2008) backcrossed to C57BL/6 for 10 generations provided by W. Jackson (Deutsches Zentrum für Neurodegenerative Erkrankungen, Bonn, Germany) and S. Lindquist (Massachusetts Institute of Technology, Cambridge, MA); WT C57BL/6J and Crl:CD1(ICR) mice purchased from The Jackson Laboratory, Charles River, or bred in-house. Genetically vasectomized mice (Haueter et al., 2010) were maintained in house. Genotypes were verified by PCR on DNA obtained from ear punches as indicated in the original description of each line. All animals were maintained in temperature- and light-controlled rooms (12 light/12 dark) with food and water ad libitum in a high-hygienic grade facility. Mice were euthanized by cervical dislocation, followed by decapitation, or anesthetized with ketamine-xylazine and transcardially perfused with a solution of PBS containing heparin, unless otherwise specified. Archival material, including paraffin-embedded tissues and gDNA samples originally described in previous studies (Genoud et al., 2004; Nuvolone et al., 2013), was also analyzed. Archival gDNA samples included material provided by the Institute of Physical and Chemical Research (Saitama, Japan).
Genome editing in mouse embryos
Design of TALEN-targeting strategy was based on the employment of second-generation heterodimeric FokI cleavage domains fused to a truncated TALE C terminus for improved specificity and cleavage efficiency and microinjections were performed as previously described (Hermann et al., 2014).
The TALEN target sites 5′-TGGCTGCTGGCCCTCT-3′ and 5′-TGCAGAGGCCGACATCA-3′ within the Prnp protein coding sequence were identified using the TALEN-NT algorithm (Doyle et al., 2012). TALE-NT was also used to predict potential OTs for the Prnp TALEN pair and the 8 PCR-accessible out of the 12 top-scoring sites were selected for further analysis. TALENs were assembled using the Golden Gate TALEN and TAL Effector kit (plasmid kit 1000000024; Addgene; Cermak et al., 2011) and the pCAG-T7 heterodimeric TALEN destination vectors (plasmids 40131 and 40132; Addgene; Hermann et al., 2014). In vitro mRNA transcription, capping, and polyadenylation were performed using the mMESSAGE mMACHINE T7 Ultra kit. Before injection, the mRNAs were purified using NucAway Spin Columns (Ambion). mRNA quality was verified by denaturing gel electrophoresis; concentration was determined by spectrophotometry. Microinjection of C57BL/6J embryos with Prnp TALEN mRNAs at a concentration of 100 ng/µl resulted in gene modification in >40% of founder animals as previously described (Hermann et al., 2014). In the present study, a concentration of 10 ng/µl was employed. C57BL/6J female mice underwent ovulation induction by i.p. injection of 5 IU equine chorionic gonadotrophin (PMSG; Folligon–InterVet), followed by i.p. injection of 5 IU human chorionic gonadotropin (Pregnyl–Essex Chemie) 48 h later. For the recovery of zygotes, C57BL/6J females were mated with males of the same strain immediately after the administration of human chorionic gonadotropin. All zygotes were collected from oviducts 24 h after the human chorionic gonadotropin injection, and were then freed from any remaining cumulus cells by a 1–2 min treatment of 0.1% hyaluronidase (Sigma-Aldrich) dissolved in M2 medium (Sigma-Aldrich). Mouse embryos were cultured in M16 (Sigma-Aldrich) medium at 37°C and 5% CO2. For micromanipulation, embryos were transferred into M2 medium.
All microinjections were performed using a microinjection system comprised of an inverted microscope equipped with Nomarski optics (Nikon), a set of micromanipulators (Narashige), and a FemtoJet microinjection unit (Eppendorf). TALEN mRNAs were injected at a concentration of 10 ng/µl into the cytoplasm of fertilized mouse oocytes. Embryos that survived the microinjection were transferred on the same day into the oviducts of 8–16-wk-old pseudopregnant Crl:CD1(ICR) females (0.5 d used after coitus) that had been mated with sterile genetically vasectomized males (Haueter et al., 2010) the day before embryo transfer. Pregnant females were allowed to deliver and raise their pups until weaning age.
Identification of nonhomologous end-joining–modified alleles
gDNA was extracted from tissue biopsies using a buffer containing 10 mM Tris-HCl, pH 9.0, 50 mM KCl, 0.45% Nonident p40, 0.45% Tween-20, and Proteinase K. For detecting nonhomologous end-joining–mediated insertions/deletions, amplicons of the Prnp locus and selected potential TALEN off-target cleavage sites were generated using appropriate primers (listed in Table S2) and Herculase II Fusion DNA Polymerase (Agilent Technologies) using the following conditions: 95°C for 2 min, 35 cycles of 95°C for 30 s, 63°C for 30 s (with a ramp of −0.75°C/cycle for the first 20 cycles and a constant temperature of 48°C for the remaining 15 cycles), 72°C for 20 s, and 72°C for 3 min. PCR products were purified using the QIAquick PCR Purification kit (QIAGEN) and subjected to heteroduplex formation using the following conditions in a thermocycler: 95°C 2 min, 95°C to 85°C (−2°C/s), and 85°C to 25°C (−0.1°C/s), and then digested with T7 endonuclease (NEB) for 20 min at 37°C. Digestion products were resolved on a 2% agarose gel. For Sanger sequencing, PCR products were cloned into pGEM-T easy (Promega).
Allelic discrimination assay
To distinguish WT from ZH3 alleles of Prnp, an amplicon encompassing the 8 bp deletion was generated in the presence of two TaqMan probes, each one specific for the WT or the ZH3 allele and carrying a different fluorophore at the 5′ end (FAM and Yakima Yellow, respectively) and the BHQ1 quencher at the 3′ end (primers and probes sequences in Table S2) using TaqMan Universal PCR Master Mix, No AmpErase UNG on a ViiA 7 Real-Time PCR System following a standard genotyping protocol (Life Technology).
RFLP analysis
To distinguish WT versus ZH3 alleles of Prnp, gDNA was PCR amplified using primers listed in Table S2. Amplicons were digested with Tsp45I, and digestion products were separated by electrophoresis on an agarose gel.
G-banding and spectral karyotyping
Mouse ear biopsies were cut into small pieces, and then incubated first with collagenase (Sigma-Aldrich) at 37°C for 1 h, and then with 0.05% Trypsin/0.53 mM EDTA (Wisent) at 37°C for 30 min. Cells were then cultured in α-MEM, 15% FBS, and antibiotics (GE Healthcare) for 1 wk, subcultured into 60-mm dishes until 80% confluence, incubated in 0.05 µg/ml colcemid (Invitrogen) for 20 min, trypsinized, pelleted, incubated in 75 mM KCl hypotonic solution at 37°C for 1 h, and fixed in 3:1 methanol/glacial acetic acid fixative. Mouse spleen was flushed with RPMI 1640, 250 mM Hepes, 10% FBS, 2 mM l-glutamine, and antibiotics (GE Healthcare), splenocytes were filtered, and cultured in the presence of 50 µg/ml LPS (Sigma-Aldrich) at 37°C for 72 h. Cells were then incubated in 0.05 µg/ml colcemid (Invitrogen) for 15 min, pelleted, incubated in 75 mM KCl hypotonic solution at 37°C for 1 h and fixed in 3:1 methanol/glacial acetic acid fixative. Cell suspensions were dropped onto slides in a Thermotron, aged overnight, G-banded and imaged, destained, denatured, and then hybridized overnight with denatured mouse spectral karyotyping reagent (Applied Spectral Imaging), washed, and counterstained with DAPI. For splenocytes, the steps for spectral karyotyping were omitted.
Image acquisition was performed with an Olympus BX61 microscope (Olympus) equipped with a SpectraCube SD300 (Applied Spectral Imaging) consisting of an optical head with a special Fourier-transform spectrometer, and a cooled CCD camera COOL-1300QS (VDS Vosskühler GmbH). Samples were illuminated with a xenon lamp Lambda LS (Sutter Instrument Company) through a Lambda 10-B optical filter changer with SmartShutter (Sutter Instrument Company) and imaged with a 60×/N.A. 1.42 oil immersion objective (Olympus). Images, typically consisting of a built from 128 frames of 600 ms, were acquired using Spectral Imaging acquisition software Version 4.5 (Applied Spectral Imaging). DAPI images were acquired separately. G-banded samples were imaged with a CCD camera (VDS Vosskühler GmbH) on the Olympus BX61 microscope set in bright-field mode with a 100×/N.A. 1.40 oil immersion objective (Olympus). To optimize contrast, a green filter was inserted in the illumination pathway. Spectral karyotyping images were analyzed using SkyView Version 2.1.1 (Applied Spectral Imaging).
Array comparative genome hybridization
gDNA was extracted from mouse tissue using QIAamp DNA mini kit (QIAGEN) and 1 µg of purified gDNA was labeled using the CytoSure Genomic DNA Labeling kit (Oxford Gene Technology), according to the manufacturers’ instructions. Cy3-labeled C57BL/6J reference and cy5-labeled PrnpZH3/ZH3 experimental sample were hybridized with SurePrint G3 Mouse CGH Microarray kit, 1 × 1 M (Agilent Technologies) according to manufacturer’s instructions. The 1 × 1 M array consists of 963,261 distinct 60-mer oligonucleotide probes, plus 1,000 replicates and an additional 6,745 quality control features, resulting in an 1.8-kb overall median spacing (1.5 kb in Ref-Seq genes; Agilent Technologies). Data were analyzed using Agilent Genomic Workbench 7.0.4.0 software (Agilent Technologies). Probes were annotated against the UCSC mm9 (NCBI Build 37) genome build. After a quality-control step, variant calling was performed using the aberration algorithm ADM-2 with the following filters: threshold 10.0, windows size 2 kb, diploid peak centralization ON, fuzzy zero ON, GC correction ON, combine replicates (intra-assay) ON, minimum number of probes 10, minimum average absolute log2 ratio ≥0.3.
Whole-genome SNP analysis
gDNA was purified from tail biopsies or ear punches using the Gentra Puregene Mouse Tail kit (QIAGEN) according to manufacturer’s instructions. Whole-genome SNP analysis (Taconic Laboratories) was performed using the Illumina Mouse MD Linkage Panel array consisting of 1,449 strain-informative SNP markers spanning the whole genome (at least three SNPs every 5 Mb, with at least one SNP informative for C57BL/6J versus other strains; Illumina). Results were compared with data from reference strains (129S6/SvEvTac, C57BL/6JBomTac, and C57BL/6NTac).
qRT-PCR
RNA was isolated from snap frozen tissues using the RNeasy mini kit (QIAGEN). RNA concentration and quality were determined with NanoDrop (Thermo Fisher Scientific). Reverse transcription was performed using the QuantiTect Reverse Transcription kit (QIAGEN), according to the manufacturer’s instructions. qRT-PCR was performed on a ViiA 7 Real-Time PCR System (Life Technology). Amplification was performed with Fast Start SYBR Green Master (ROX; Roche) with 0.5 µM of each forward and reverse primers for the target of interest or appropriate normalization genes (Table S2) and cDNA as template, using the following conditions: 50°C for 2 min, 95°C for 10 min, 40 cycles of 95°C for 15 s, and 60°C for 1 min. After each run, a melting curve analysis was performed, using the following conditions: 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s. For all steps, a ramp rate of 1.6°C/s was used. Raw Ct values were used to calculate relative expression levels of target genes, after normalization to three internal control genes (Eif2a, Utpc6, and Gapdh) according to geNorm (Vandesompele et al., 2002). Eif2a and Utpc6 primer sequences were originally reported in (Kosir et al., 2010); Gapdh primer sequences were obtained from the Genomics Platform–University of Geneva.
RNA sequencing
Mice under deep anesthesia were transcardially perfused with ice-cold PBS heparin treated with diethylpyrocarbonate. Hippocampi were immediately dissected with the help of an adult mouse brain slicer matrix (Zivic Instruments), snap frozen, and kept –80°C until homogenization. Total RNA was isolated using the RNeasy Plus universal mini kit (QIAGEN), snap frozen, and kept at –80°C until further analysis. Library preparation for RNA sequencing was performed, as previously described (Nuvolone et al., 2013) with modifications. RNA quality was assessed using Bioanalyzer 2100 (Agilent Technologies) and Qubit (1.0) Fluorometer (Life Technologies). Only samples with a 28S/18S ratio between 1.5 and 2 and a 260 nm/280 nm ratio between 1.8 and 2.1 were further processed. The TruSeq RNA Sample Prep kit v2 (Illumina) was used in the subsequent steps. In brief, 1 µg of total RNA per sample was poly A–enriched, reverse transcribed into double-stranded cDNA and ligated with TruSeq adapters. PCR was performed to selectively enrich for fragments containing TruSeq adapters on both ends. Quality and quantity of enriched libraries were analyzed using Qubit (1.0) Fluorometer and Caliper GX LabChip GX (Caliper Life Sciences). The resulting product is a smear with a mean fragment size of ∼260 bp. Libraries were normalized to 10 nM in Tris-Cl 10 mM, pH 8.5, with 0.1% (vol/vol) Tween 20.
TruSeq PE Cluster kit v4-cBot-HS (Illumina) was used for cluster generation using 2 pM of pooled normalized libraries on the cBOT. Sequencing was performed on Illumina HiSeq 2500 at 1 × 100 bp using the TruSeq SBS kit v4-HS (Illumina).
RNA sequencing data analysis was performed as previously described (Nuvolone et al., 2013), with minor modifications. Reads were quality-checked using FastQC. Low-quality ends were clipped (5′, 3 bases; 3′, 10 bases). Trimmed reads were aligned to the reference genome and transcriptome (FASTA and GTF files, respectively, downloaded from the Ensembl GRCm38) with STAR version 2.3.0e_r291 (Dobin et al., 2013) with default settings. Loci with mismatches with respect to the reference genome were detected following the best practice workflow to identify variants from RNA sequencing data using GATK version 3.4.0 (DePristo et al., 2011). This analysis was performed on the three mice with the highest sequencing coverage for each group and only mismatches in common to all three mice in each group were considered. Sequence mismatches were dichotomized as either RNA-editing sites if present in a previously published catalog of 17`831 RNA editing sites in mice (Stilling et al., 2014) or otherwise as variants. Variants were annotated using snpEFF version 3.4 (Cingolani et al., 2012), and distribution of the reads across genomic isoform expression was quantified using the R package GenomicRanges (Lawrence et al., 2013) from Bioconductor Version 3.0. DEGs were identified based on fdr values using the R package edgeR (Robinson et al., 2010) from Bioconductor Version 3.0. Differential exon usage was detected based on adjusted P values with the DEXSeq R-package (Anders et al., 2012) using default parameters. Visualization of RNA sequencing coverage was performed using Integrative Genome Viewer (Robinson et al., 2011). Pathway analysis was performed using Ingenuity Pathway Analysis (Ingenuity Systems). Data from RNA sequencing analyses have been deposited to GEO under the dataset code GSE75510.
A list of genes enriched in neurons, astrocytes, oligodendrocytes, microglia and CNS endothelial cells (500 genes per cell type) was retrieved using the cell type enrichment query from a transcriptomic based database, (Zhang et al., 2014), searching for genes enriched in one cell type respect to all others. In the case of oligodendrocytes, oligodendrocytes precursor cells, newly formed oligodendrocytes and myelinating oligodendrocytes were considered together. The resulting, nonoverlapping lists of CNS cell type enriched genes were used for comparisons with the lists of genes with differential gene expression, exon usage, and RNA editing in the present study.
Sequencing of gDNA
GDNA was extracted as from tissue biopsies using a buffer containing 10 mM Tris-HCl, pH 9.0, 50 mM KCl, 0.45% Nonident p40, 0.45% Tween-20 and Proteinase K. Target regions were amplified by PCR using appropriate primers (listed in Table S2) under the following conditions: 2 min at 94°C; 35 cycles of 30 s at 94°C, 30 s at 58°C, and 45 s at 72°C; 5 min at 72°C. Amplicons were subjected to agarose gel electrophoresis and DNA was purified from excised bands using the Nucleospin Gel and PCR Cleanup kit (Macherey-Nagel), according to manufacturer’s instructions. Sanger sequencing was performed at Microsynth AG, using the same primer pair used for PCR amplification.
Western blotting
Snap-frozen tissues were used to prepare 10% (w/vol) homogenates in RIPA buffer (25 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1% Nonident p40, 1% sodium deoxycholate, and 0.1% SDS) + cOmplete Mini Protease Inhibitor Cocktail (Roche) using 5-mm stainless steel beads (QIAGEN) and Tissue Lyser LT (QIAGEN). Total protein concentration was measured using the BCA Protein Assay (Thermo Fisher Scientific) according to the manufacturer’s instructions. 20 µg of total protein homogenates in NuPAGE LDS sample buffer (Invitrogen) with β-mercaptoethanol as reducing agent were separated on a NuPAGE 12% Bis-Tris (Invitrogen) and transferred onto a Protran Nitrocellulose Transfer Membrane (Whatman) using the NuPAGE Gel Electrophoresis System (Invitrogen) according to the manufacturer’s instructions. Precision Plus Protein Dual Color Standards (Bio-Rad Laboratories) and MagicMark XP Standard to Antibodies (Invitrogen) were used as molecular weight ladder. Membranes were subsequently blocked with 5% w/vol Top-block (Fluka) in TBS supplemented with 0.1% vol/vol Tween-20 (TBST). Antibodies were incubated in 1% wt/vol Top-block (Fluka) in TBST. Primary antibodies used were: anti-PrPC mouse monoclonal antibodies POM1 (280 ng/ml; Polymenidou et al., 2008) and anti-actin mouse monoclonal antibody, Clone C4 (1:8,000; EMD Millipore). HRP-conjugated goat anti–mouse IgG (H+L; 1:17,000 dilution; Invitrogen) was used as secondary antibody. Blots were developed using Luminata Western HRP Substrates (EMD Millipore) and visualized using the Stella detector (Raytest).
PrPC ELISA
PrPC was quantified in tissue homogenates by sandwich ELISA using POM1 and POM2 antibodies as previously described (Polymenidou et al., 2008).
Footprint test
The footprint test was performed as previously described (Carter et al., 1999), with minor modifications. To obtain footprints, the fore and hind feet of the mice were painted with magenta and blue nontoxic inks, respectively. Mice were then allowed to walk along a 90-cm long, 7-cm wide, 20-cm high corridor on a fresh white paper. Mice had up to three trials. Footprints were analyzed for the following parameters, as previously described (Carter et al., 1999): stride length (mean distance of footprints from the same paw in consecutive strides); hind-base width (mean distance between right and left hind footprints); front-base width (mean distance between right and left front footprints); and front/hind footprint overlap (mean distance between hind footprint and the preceding front footprint). Footprints at the beginning and at the end of the corridor were excluded. For each step parameter, 4–12 values were measured and the mean was calculated and used for analysis. The operator was blind to mouse genotype.
Rotarod test
The rotarod test was performed as previously described (Sorce et al., 2014), with minor modifications. The rotarod apparatus consisted of a rotating cylinder (ø 3 cm) subdivided into five 57-mm-wide lanes by dividers (ø 25 cm; Ugo Basile). Each test comprised a habituation phase and an experimental phase. The habituation phase consisted of three sessions of 1 min each, at a constant speed of 5 rotations per minute (rpm), with inter-session intervals of at least 15 min. The test phase, which started at least 15 min after the last habituation trial, consisted of three sessions of maximum 5 min each, at a constant acceleration from 5 rpm to maximum of 40 rpm, with inter-session intervals of at least 15 min. Falling from the drum or clinging to the rod and passively rotating with it were equally considered to assess latency to fall. Rotarod tests were performed at the same time of the day (between 2 p.m. and 4 p.m.), with a 1-mo interval. In each cage, mice were randomly tested and the operator was blind to their genotype.
Histology, immunohistochemistry, and immunofluorescence
Hematoxylin and eosin stainings were performed on sections from formalin-fixed, paraffin-embedded tissue using standard protocols. CD68 staining was performed on 7–10 µm cryo-sections from snap-frozen sciatic nerves embedded in OCT medium as previously described (Bremer et al., 2010). After fixation with formalin and acetone solutions, sections were incubated with rat anti–mouse CD68 monoclonal antibody (1:100 dilution, clone FA-11; Serotec), followed by incubation with goat anti–rat IgG (1:150 dilution; Antibodies Online), and donkey anti–goat IgG conjugated with alkaline phosphatase (1:80 dilution; Jackson ImmunoResearch Laboratories). Immunoreactivity was visualized using the mix Fast Red staining kit (Sigma-Aldrich). Toluidine blue staining was performed using standard procedures on semi-thin sections (500 nm) from epon-embedded sciatic nerves that had been fixed in situ with 3.9% glutaraldehyde in 0.1 M PBS, pH 7.4, after transcardial perfusion with PBS. In all cases, slides were scanned with NanoZoomer and images were visualized using the NanoZoomer Digital Pathology System (NDPview; Hamamatsu Photonics). For morphometric analysis of CD68+ digestion chambers in sciatic nerves, at least 10 regions of interest per section were selected from a total of two to four sections per mouse. For morphometric analysis of axonal density in sciatic nerves, at least three regions of interest per section were selected from three to nine sections per mouse. Images were analyzed using semiautomatized softwares developed in-house. Operators were blind to the genotype of the analyzed cases.
Immunofluorescence stainings were performed on sections from formalin-fixed, paraffin-embedded tissue. After deparaffinization through graded alcohols and heat-induced antigen retrieval in citrate buffer (0.01 M, pH 6.0), sections were incubated with anti-POM19 (20 µg/ml; Polymenidou et al., 2008) and MAP2 (1:500; Abcam) antibodies, followed by incubation with fluorescently labeled secondary antibodies (1:1,000; Alexa Fluor 488 or 555; Invitrogen) and with DAPI (Life technologies) for nuclear staining. Images were acquired with the fluorescence microscope (BX-61; Olympus) equipped with a cooled black/white charge-coupled device camera, using identical acquisition settings.
In all cases, images were prepared using Photoshop and Illustrator software (Adobe).
Transmission electron microscopy
Mice under deep anesthesia were transcardially perfused with PBS heparin and sciatic nerves were then fixed in situ with 2.5% glutaraldehyde + 2% paraformaldehyde in 0.1 M PBS, pH 7.4. Tissues were then embedded in epon, and semi-thin sections (500 nm) were stained with toluidine blue using standard procedures. Ultrathin sections were mounted on copper grids coated with Formvar membrane and contrasted with uranyl acetate/lead citrate. Specimens were examined using a Hitachi H-7650 electron microscope (Hitachi High-Tech) operating at 80 kV.
Phagocytosis assay with BM-derived macrophages (BMDMs)
On day 1, femurs of age-matched adult males were flushed with culture medium (RPMI 1640, 10% FBS, 1% Glutamax and antibiotics; all from GE Healthcare). BM cells were and plated into 6-well plates (TPP) at a density of 2 × 106/well in 3-ml culture medium enriched with 10 ng/ml of murine macrophage colony-stimulation factor (Invitrogen) and cultured overnight at 37°C and 5% CO2. On day 2, nonadhering cells were transferred to a Nunc UpCell Surface cell culture dish (Sigma-Aldrich). On day 6, the resulting BMDMs were harvested, adjusted to 5 × 105 cells in 500 µl, and plated into 24-well plates. On the same day, thymocytes were harvested from 6–12-wk-old C57BL/6 mice and incubated overnight at 37°C and 5% CO2 in culture medium in the presence of 0.1 µM dexamethasone to induce apoptosis. On day 7, thymocyte apoptosis was assessed using the FITC Annexin V Apoptosis Detection kit II (BD). Apoptotic thymocytes were washed, suspended at 106/ml in PBS, and labeled with 20 ng/ml of the pH-sensitive dye pHrodo Red, SE (Invitrogen) for 30 min at room temperature. Labeled thymocytes were washed and resuspended in culture medium to achieve different cellular densities, and then 500 µl of cell suspension were added to each well of the BMDM culture for 1 h at 37°C. After washing, BMDMs were harvested with Stem Pro Accutase (Invitrogen) and gentle scraping, and subsequently stained with FITC-labeled anti-CD11b or isotype control antibody (BD). Flow cytometry was performed using a FACSCalibur (BD) with CellQuestPro software. At least 10,000 events were acquired in the living gate. Rate of phagocytosis was determined with FlowJo software (Tree Star) as the percentage of pHrodo positivity among CD11b+ cells.
Statistical analysis
Statistical significance was assessed using GraphPad Prism software with two-tailed unpaired Student’s t test or one- or two-way ANOVA and Bonferroni’s multiple comparison post-test, as appropriate. α level was set at 0.05. For each statistical analysis, the statistical test, the group size and the resulting p-value are indicated in the corresponding figure legends.
Online supplemental material
Fig. S1 shows that C57BL/6J-PrnpZH3/ZH3 mice do not have chromosomal aberrations. Fig. S2 shows whole-genome SNP analysis. Fig. S3 shows Prnp mRNA coverage. Fig. S4 shows Sanger sequencing of selected genomic sites undergoing RNA editing. Fig. S5 shows flow cytometry analysis of phagocytic activity. Table S1 lists Predicted TALEN OTs analyzed. Table S2 lists primers and probes used. Table S3 lists DEGs between PrnpZH1/ZH1 and C57BL/6J hippocampi. Table S4 lists DEGs between PrnpZH3/ZH3 and C57BL/6J hippocampi. Table S5 lists differentially expressed exons between PrnpZH1/ZH1 and C57BL/6J hippocampi. Table S6 lists loci with differential RNA editing level between PrnpZH1/ZH1 and C57BL/6J hippocampi. Table S7 lists loci with differential RNA editing level between PrnpZH3/ZH3 and C57BL/6J hippocampi. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20151610/DC1.
Supplementary Material
ACKNOWLEDGMENTS
We thank W. S. Jackson, S. Lindquist, H. Baybutt, N. A. Mabbott, J. Manson, and the Institute of Physical and Chemical Research, Japan, for providing mice and DNA samples; N. Schmid for help with behavioral tests; C. Aquino Fournier for RNA sequencing; Taconic for whole-genome SNP analysis; the Center for Applied Genomics at the Sick Children Hospital, Toronto, and in particular R. Wong and J.A. Herbrick for karyotyping; and C. Lu for aCGH, J. Tchinda, and H. Rehrauer for help with aCGH data interpretation; A. Reyes for help with DEXSeq; M. Bieri and N. Wey for software development; Y. Fuhrer, M. Delic, R. Moos, A. Varol, K. Arroyo, B. Piccapietra, M. König, M. Tarnowska, E. Skoczylas, and C. Albrecht for excellent technical help; and members of the Aguzzi laboratory for critical discussions.
This work was supported by grants from Collegio Ghislieri, Pavia, Italy (to M. Nuvolone). A. Aguzzi was supported by two Advanced Grants from the European Research Council (250356 and 670958); the European Union (NEURINOX, 278611); the Swiss National Foundation (31003A_141193, Sinergia grant, CRSII3_147660 and R'Equip, 316030_157745); the E-Rare JTC, 31ER30_160672; the Novartis Research Foundation; the Clinical Research Priority Programs “Small RNAs” and “Mechanisms and Models of Primary Human Hemato-Lymphatic Diseases” of the University of Zurich; and the Swiss Initiative in System Biology SystemsX.ch (2014/260, “Systems biology of prion diseases”).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- aCGH
- array comparative genomic hybridization
- BMDM
- BM-derived macrophage
- CDS
- protein-coding DNA sequence
- CNV
- copy number variant
- DEG
- differentially expressed gene
- eQTL
- expression quantitative trait loci
- fdr
- false discovery rate
- gDNA
- genomic DNA
- OT
- off target
- PrPC
- cellular prion protein
- PrPSc
- scrapie prion protein
- qRT-PCR
- quantitative real-time PCR
- RFLP
- restriction fragment length polymorphism
- RPKM
- reads per kilobase per million mapped reads
- SNP
- single-nucleotide polymorphism
- TALEN
- transcription activator-like effector nuclease
References
- Aguzzi A., Nuvolone M., and Zhu C.. 2013. The immunobiology of prion diseases. Nat. Rev. Immunol. 13:888–902. 10.1038/nri3553 [DOI] [PubMed] [Google Scholar]
- Anders S., Reyes A., and Huber W.. 2012. Detecting differential usage of exons from RNA-seq data. Genome Res. 22:2008–2017. 10.1101/gr.133744.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benvegnù S., Roncaglia P., Agostini F., Casalone C., Corona C., Gustincich S., and Legname G.. 2011. Developmental influence of the cellular prion protein on the gene expression profile in mouse hippocampus. Physiol. Genomics. 43:711–725. 10.1152/physiolgenomics.00205.2010 [DOI] [PubMed] [Google Scholar]
- Bravard A., Auvré F., Fantini D., Bernardino-Sgherri J., Sissoëff L., Daynac M., Xu Z., Etienne O., Dehen C., Comoy E., et al. 2015. The prion protein is critical for DNA repair and cell survival after genotoxic stress. Nucleic Acids Res. 43:904–916. 10.1093/nar/gku1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer J., Baumann F., Tiberi C., Wessig C., Fischer H., Schwarz P., Steele A.D., Toyka K.V., Nave K.A., Weis J., and Aguzzi A.. 2010. Axonal prion protein is required for peripheral myelin maintenance. Nat. Neurosci. 13:310–318. 10.1038/nn.2483 [DOI] [PubMed] [Google Scholar]
- Büeler H., Fischer M., Lang Y., Bluethmann H., Lipp H.P., DeArmond S.J., Prusiner S.B., Aguet M., and Weissmann C.. 1992. Normal development and behaviour of mice lacking the neuronal cell-surface PrP protein. Nature. 356:577–582. 10.1038/356577a0 [DOI] [PubMed] [Google Scholar]
- Carter R.J., Lione L.A., Humby T., Mangiarini L., Mahal A., Bates G.P., Dunnett S.B., and Morton A.J.. 1999. Characterization of progressive motor deficits in mice transgenic for the human Huntington’s disease mutation. J. Neurosci. 19:3248–3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carulla P., Llorens F., Matamoros-Angles A., Aguilar-Calvo P., Espinosa J.C., Gavín R., Ferrer I., Legname G., Torres J.M., and del Río J.A.. 2015. Involvement of PrP(C) in kainate-induced excitotoxicity in several mouse strains. Sci. Rep. 5:11971 10.1038/srep11971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T., Doyle E.L., Christian M., Wang L., Zhang Y., Schmidt C., Baller J.A., Somia N.V., Bogdanove A.J., and Voytas D.F.. 2011. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39:e82 10.1093/nar/gkr218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadi S., Young R., Le Guillou S., Tilly G., Bitton F., Martin-Magniette M.L., Soubigou-Taconnat L., Balzergue S., Vilotte M., Peyre C., et al. 2010. Brain transcriptional stability upon prion protein-encoding gene invalidation in zygotic or adult mouse. BMC Genomics. 11:448 10.1186/1471-2164-11-448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., and Ruden D.M.. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 6:80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins F.S., Rossant J., and Wurst W.; International Mouse Knockout Consortium . 2007. A mouse for all reasons. Cell. 128:9–13. 10.1016/j.cell.2006.12.018 [DOI] [PubMed] [Google Scholar]
- Cool B.H., Chan G.C., Lee L., Oshima J., Martin G.M., and Hu Q.. 2010. A flanking gene problem leads to the discovery of a Gprc5b splice variant predominantly expressed in C57Bl/6J mouse brain and in maturing neurons. PLoS One. 5:e10351 10.1371/journal.pone.0010351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida C.J., Chiarini L.B., da Silva J.P.E., E Silva P.M., Martins M.A., and Linden R.. 2005. The cellular prion protein modulates phagocytosis and inflammatory response. J. Leukoc. Biol. 77:238–246. 10.1189/jlb.1103531 [DOI] [PubMed] [Google Scholar]
- DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M., et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43:491–498. 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle E.L., Booher N.J., Standage D.S., Voytas D.F., Brendel V.P., Vandyk J.K., and Bogdanove A.J.. 2012. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 40(W1):W117-22 10.1093/nar/gks608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan C.M., Sridhar S., Wigler M., and Hall I.M.. 2007. Recurrent DNA copy number variation in the laboratory mouse. Nat. Genet. 39:1384–1389. 10.1038/ng.2007.19 [DOI] [PubMed] [Google Scholar]
- Ejlerskov P., Hultberg J.G., Wang J., Carlsson R., Ambjørn M., Kuss M., Liu Y., Porcu G., Kolkova K., Friis Rundsten C., et al. 2015. Lack of Neuronal IFN-β-IFNAR Causes Lewy Body- and Parkinson’s Disease-like Dementia. Cell. 163:324–339. 10.1016/j.cell.2015.08.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatscher-Bader T., Foldi C.J., Chong S., Whitelaw E., Moser R.J., Burne T.H., Eyles D.W., and McGrath J.J.. 2011. Increased de novo copy number variants in the offspring of older males. Transl. Psychiatry. 1:e34 10.1038/tp.2011.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallina I., Colding C., Henriksen P., Beli P., Nakamura K., Offman J., Mathiasen D.P., Silva S., Hoffmann E., Groth A., et al. 2015. Cmr1/WDR76 defines a nuclear genotoxic stress body linking genome integrity and protein quality control. Nat. Commun. 6:6533 10.1038/ncomms7533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genoud N., Behrens A., Miele G., Robay D., Heppner F.L., Freigang S., and Aguzzi A.. 2004. Disruption of Doppel prevents neurodegeneration in mice with extensive Prnp deletions. Proc. Natl. Acad. Sci. USA. 101:4198–4203. 10.1073/pnas.0400131101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haueter S., Kawasumi M., Asner I., Brykczynska U., Cinelli P., Moisyadi S., Bürki K., Peters A.H., and Pelczar P.. 2010. Genetic vasectomy-overexpression of Prm1-EGFP fusion protein in elongating spermatids causes dominant male sterility in mice. Genesis. 48:151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikenwalder M., Kurrer M.O., Margalith I., Kranich J., Zeller N., Haybaeck J., Polymenidou M., Matter M., Bremer J., Jackson W.S., et al. 2008. Lymphotoxin-dependent prion replication in inflammatory stromal cells of granulomas. Immunity. 29:998–1008. 10.1016/j.immuni.2008.10.014 [DOI] [PubMed] [Google Scholar]
- Hermann M., Cermak T., Voytas D.F., and Pelczar P.. 2014. Mouse genome engeineering using designer nucleases. J. Vis. Exp. 86:e5093 10.3791/50930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull J., Campino S., Rowlands K., Chan M.S., Copley R.R., Taylor M.S., Rockett K., Elvidge G., Keating B., Knight J., and Kwiatkowski D.. 2007. Identification of common genetic variation that modulates alternative splicing. PLoS Genet. 3:e99 10.1371/journal.pgen.0030099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane T.M., Goodstadt L., Danecek P., White M.A., Wong K., Yalcin B., Heger A., Agam A., Slater G., Goodson M., et al. 2011. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 477:289–294. 10.1038/nature10413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosir R., Acimovic J., Golicnik M., Perse M., Majdic G., Fink M., and Rozman D.. 2010. Determination of reference genes for circadian studies in different tissues and mouse strains. BMC Mol. Biol. 11:60 10.1186/1471-2199-11-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M., Huber W., Pagès H., Aboyoun P., Carlson M., Gentleman R., Morgan M.T., and Carey V.J.. 2013. Software for computing and annotating genomic ranges. PLOS Comput. Biol. 9:e1003118 10.1371/journal.pcbi.1003118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.B., and Church G.M.. 2013. Deciphering the functions and regulation of brain-enriched A-to-I RNA editing. Nat. Neurosci. 16:1518–1522. 10.1038/nn.3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlefield L.G., and Mailhes J.B.. 1975. Observations of de novo clones of cytogentically aberrant cells in primary fibroblast cell strains from phenotypically normal women. Am. J. Hum. Genet. 27:190–197. [PMC free article] [PubMed] [Google Scholar]
- Lusis A.J., Yu J., and Wang S.S.. 2007. The problem of passenger genes in transgenic mice. Arterioscler. Thromb. Vasc. Biol. 27:2100–2103. 10.1161/ATVBAHA.107.147918 [DOI] [PubMed] [Google Scholar]
- Manson J.C., Clarke A.R., Hooper M.L., Aitchison L., McConnell I., and Hope J.. 1994. 129/Ola mice carrying a null mutation in PrP that abolishes mRNA production are developmentally normal. Mol. Neurobiol. 8:121–127. 10.1007/BF02780662 [DOI] [PubMed] [Google Scholar]
- Moore R.C., Lee I.Y., Silverman G.L., Harrison P.M., Strome R., Heinrich C., Karunaratne A., Pasternak S.H., Chishti M.A., Liang Y., et al. 1999. Ataxia in prion protein (PrP)-deficient mice is associated with upregulation of the novel PrP-like protein doppel. J. Mol. Biol. 292:797–817. 10.1006/jmbi.1999.3108 [DOI] [PubMed] [Google Scholar]
- Nuvolone M., Kana V., Hutter G., Sakata D., Mortin-Toth S.M., Russo G., Danska J.S., and Aguzzi A.. 2013. SIRPα polymorphisms, but not the prion protein, control phagocytosis of apoptotic cells. J. Exp. Med. 210:2539–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M., Moos R., Scott M., Sigurdson C., Shi Y.Z., Yajima B., Hafner-Bratkovic I., Jerala R., Hornemann S., Wuthrich K., et al. 2008. The POM monoclonals: a comprehensive set of antibodies to non-overlapping prion protein epitopes. PLoS One. 3:e3872 10.1371/journal.pone.0003872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., and Mesirov J.P.. 2011. Integrative genomics viewer. Nat. Biotechnol. 29:24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi D., Cozzio A., Flechsig E., Klein M.A., Rülicke T., Aguzzi A., and Weissmann C.. 2001. Onset of ataxia and Purkinje cell loss in PrP null mice inversely correlated with Dpl level in brain. EMBO J. 20:694–702. 10.1093/emboj/20.4.694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi S., Katamine S., Shigematsu K., Nakatani A., Moriuchi R., Nishida N., Kurokawa K., Nakaoke R., Sato H., Jishage K., et al. 1995. Accumulation of proteinase K-resistant prion protein (PrP) is restricted by the expression level of normal PrP in mice inoculated with a mouse-adapted strain of the Creutzfeldt-Jakob disease agent. J. Virol. 69:7586–7592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberberg G., Lundin D., Navon R., and Öhman M.. 2012. Deregulation of the A-to-I RNA editing mechanism in psychiatric disorders. Hum. Mol. Genet. 21:311–321. 10.1093/hmg/ddr461 [DOI] [PubMed] [Google Scholar]
- Sommer B., Köhler M., Sprengel R., and Seeburg P.H.. 1991. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 67:11–19. 10.1016/0092-8674(91)90568-J [DOI] [PubMed] [Google Scholar]
- Sorce S., Nuvolone M., Keller A., Falsig J., Varol A., Schwarz P., Bieri M., Budka H., and Aguzzi A.. 2014. The role of the NADPH oxidase NOX2 in prion pathogenesis. PLoS Pathog. 10:e1004531 10.1371/journal.ppat.1004531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele A.D., Lindquist S., and Aguzzi A.. 2007. The prion protein knockout mouse: a phenotype under challenge. Prion. 1:83–93. 10.4161/pri.1.2.4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stilling R.M., Benito E., Gertig M., Barth J., Capece V., Burkhardt S., Bonn S., and Fischer A.. 2014. De-regulation of gene expression and alternative splicing affects distinct cellular pathways in the aging hippocampus. Front. Cell. Neurosci. 8:373 10.3389/fncel.2014.00373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striebel J.F., Race B., Pathmajeyan M., Rangel A., and Chesebro B.. 2013. Lack of influence of prion protein gene expression on kainate-induced seizures in mice: studies using congenic, coisogenic and transgenic strains. Neuroscience. 238:11–18. 10.1016/j.neuroscience.2013.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., and Nakayama M.. 2003. Differential profiles of genes expressed in neonatal brain of 129X1/SvJ and C57BL/6J mice: A database to aid in analyzing DNA microarrays using nonisogenic gene-targeted mice. DNA Res. 10:263–275. 10.1093/dnares/10.6.263 [DOI] [PubMed] [Google Scholar]
- Vanden Berghe T., Hulpiau P., Martens L., Vandenbroucke R.E., Van Wonterghem E., Perry S.W., Bruggeman I., Divert T., Choi S.M., Vuylsteke M., et al. 2015. Passenger Mutations Confound Interpretation of All Genetically Modified Congenic Mice. Immunity. 43:200–209. 10.1016/j.immuni.2015.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., and Speleman F.. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:H0034 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen K., Sloan S.A., Bennett M.L., Scholze A.R., O’Keeffe S., Phatnani H.P., Guarnieri P., Caneda C., Ruderisch N., et al. 2014. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34:11929–11947. 10.1523/JNEUROSCI.1860-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.