Abstract
Cholangiocellular carcinoma (CCC) and pancreatic ductal adenocarcinoma (PDAC) are two highly aggressive cancer types that arise from epithelial cells of the pancreatobiliary system. Owing to their histological and morphological similarity, differential diagnosis between CCC and metastasis of PDAC located in the liver frequently proves an unsolvable issue for pathologists. The detection of biomarkers with high specificity and sensitivity for the differentiation of these tumor types would therefore be a valuable tool. Here, we address this problem by comparing microdissected CCC and PDAC tumor cells from nine and eleven cancer patients, respectively, in a label-free proteomics approach. The novel biomarker candidates were subsequently verified by immunohistochemical staining of 73 CCC, 78 primary, and 18 metastatic PDAC tissue sections. In the proteome analysis, we found 180 proteins with a significantly differential expression between CCC and PDAC cells (p value < 0.05, absolute fold change > 2). Nine candidate proteins were chosen for an immunohistochemical verification out of which three showed very promising results. These were the annexins ANXA1, ANXA10, and ANXA13. For the correct classification of PDAC, ANXA1 showed a sensitivity of 84% and a specificity of 85% and ANXA10 a sensitivity of 90% at a specificity of 66%. ANXA13 was higher abundant in CCC. It presented a sensitivity of 84% at a specificity of 55%. In metastatic PDAC tissue ANXA1 and ANXA10 showed similar staining behavior as in the primary PDAC tumors (13/18 and 17/18 positive, respectively). ANXA13, however, presented positive staining in eight out of eighteen secondary PDAC tumors and was therefore not suitable for the differentiation of these from CCC. We conclude that ANXA1 and ANXA10 are promising biomarker candidates with high diagnostic values for the differential diagnosis of intrahepatic CCC and metastatic liver tumors deriving from PDAC.
The majority of malignant neoplasms located in the liver are metastases originating from primary tumor sites in other organs, most commonly the colon or the pancreas (1). In many cases, a histological or immunohistological examination by an experienced pathologist can specify the type and origin of the underlying cancer. Hepatocellular carcinoma or hepatic metastasis from primaries such as pulmonary adenocarcinoma, colorectal adenocarcinoma, and breast carcinoma are usually easily distinguishable by morphology and means of known immunohistochemical markers. For primary cholangiocellular carcinoma (CCC)1 and metastases of pancreatic ductal adenocarcinoma (PDAC), however, the distinction in a liver biopsy is basically an unsolvable task because of their high similarity. This is an important and frequently asked clinical question though, because treatment options differ significantly for the two cancer types. In the case of CCC, a surgical approach can be beneficial if the diagnosis is made at an early stage of tumor progression. In contrast, palliation is often the only option if the cancer's origin is the pancreas (2).
In most cases, pathologists rely on supporting information gained from radiologic or sonographic examinations to enable a differential diagnosis. High-quality imaging may detect the majority of pancreatic masses. However, these techniques, especially MRI, which would be the most suitable, are not available in every clinic. Furthermore, radiologic examinations are higher in cost than e.g. an immunohistochemical analysis and if a metastatic PDAC in the liver is suspected, a biopsy is mandatory before palliative chemotherapy in any case (3). Hence, immunohistochemical biomarkers supporting pathologists in the differential diagnosis of CCC and PDAC would be of vital importance.
Although there are genomic differences in CCC and PDAC, the value of these in daily practice is uncertain. For example, mutations in isocitrate dehydrogenase IDH1 and IDH2 were found only in cholangiocarcinomas of intrahepatic origin (about one third). Extrahepatic cholangiocarcinomas do not show these mutations (4). Furthermore, 20–54% of intrahepatic CCC tumors harbor k-ras-mutations in contrast to 90% of PDAC cases (5). Nevertheless, neither IDH nor KRAS mutations appear to be suitable for distinction between the two tumor entities.
Although several immunohistochemical markers have been tested in regard to this challenge, so far, none have presented results sufficient for a clinical implementation. In 2007, Ney et al. described the use of podocalyxin-like protein 1 (PODXL-1) for differentiating PDAC from adenocarcinomas of the biliary and gastrointestinal tracts. The immunohistochemical study revealed the expression of PODXL-1 in 44% of the PDAC cases (71/160), whereas none of the intrahepatic (0/18) and only one of the extrahepatic CCC (1/13) were stained (6). The protein agrin, on the other hand, was proposed to aid in differential diagnosis of primary and metastatic cancers to the liver because of different expression patterns. Although in PDAC it showed a faint staining over broad areas, CCC tissue was stained stronger and more extensively (2). A combination of two markers, N-cadherin and the antibody human pancreatic cancer fusion protein #2 (HPC2), was suggested by Hooper et al. who showed that N-cadherin is expressed predominantly in CCC tissue whereas HPC2 stained a higher number of PDAC cases. Combining both markers significantly increased specificity, but at the expense of sensitivity (7). Likewise, a biomarker panel consisting of four proteins (S100P, pVHL, MUC5AC, and CK17) was recently described to be helpful in distinguishing intrahepatic CCC from metastatic PDAC. There, the staining pattern S100P-/pVHL+/MUC5AC-/CK17- was observed in 59% of the 41 CCC and none of the 60 PDAC cases. However, PDAC metastases were not tested (8). It becomes clear, that despite the effort, there is to date no biomarker or marker panel that can distinguish reliably between these two entities.
While in all of the above mentioned studies the selection of the candidates was hypothesis-driven, we here conducted a biomarker discovery study comparing CCC and PDAC cells by highly sensitive and accurate proteomics techniques to identify novel biomarker candidates. To overcome possible bias because of the high heterogeneity of CCC and PDAC tumor tissue, the malignant cells were isolated specifically by laser capture microdissection. Promising candidate proteins resulting from the quantitative proteomics analysis were subsequently verified by immunohistochemistry (IHC) in a cohort of 73 CCC and 78 PDAC sections as well as thirteen metastatic liver tumors from PDAC.
EXPERIMENTAL PROCEDURES
Clinical Data
In total, tumor tissue from 189 cancer patients was collected (supplemental Table S1). Intrahepatic CCC tissue and corresponding nonmalignant liver tissue from 84 patients was excised during partial hepatectomy at the University Hospital of Essen, Department of General, Visceral, and Transplantation Surgery, Germany. All tumors were diagnosed according to current WHO-criteria (9). All cases were intrahepatic cholangiocarcinoma, grossly the mass-forming type (MF-type) with adenocarcinoma morphology. Rare histologic variants and combined hepatocellular-cholangiocarcinoma were not included. Tissue from 87 primary PDAC tumors and eighteen metastatic liver tumors from PDAC was collected during surgery at University Hospital of Essen and University Hospital of Tübingen, Germany. Informed consent was obtained from each patient and the study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki. The local ethics committee approved the study (11–4839-BO).
Experimental Design
For this study, three sample cohorts were assembled. The set for the proteome analysis comprised 11 CCC and nine PDAC samples (cohort 1). Experience from similar studies conducted previously has proven sample sizes in this range sufficient to detect reproducible proteome patterns and therefore reveal potential biomarkers that were successfully verified in subsequent experiments (10–12). From each sample, three technical replicates were analyzed by LC-MS/MS to improve overall protein identification and quantification accuracy.
A set of 10 CCC and 10 PDAC tumors was composed for a first immunohistochemical screening of candidates in a time and material saving way (cohort 2). This set included four of the CCC samples used for the proteome analysis and the other 16 from cohort 3, assembled for the verification (supplemental Table S1). They were chosen according to availability of material. Cohort 3, which was independent from cohort 1, was used for the verification of candidate proteins by immunohistochemistry. It comprised 73 CCC, 78 PDAC, and 18 PDAC metastases from the liver (mPDAC). The aim of the immunohistochemical study was the first-time estimation and assessment of the diagnostic accuracy of the selected markers in a large cohort. Therefore, the highest number of samples available to us was included in order to improve the estimates and decrease the size of the confidence intervals. The number of PDAC metastases available was heavily limited because only cases with a definite diagnosis were included.
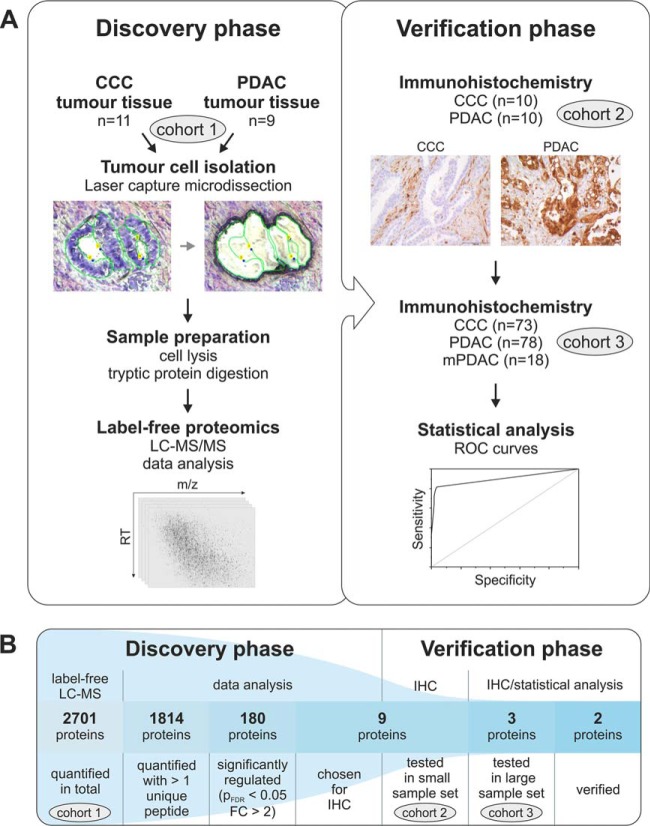
More detailed information concerning the sample cohort composition is summarized in Table I. Fig. 1 visualizes the study design.
Table I. Composition of the three sample cohorts assembled for this study. n = total number of patients within a subgroup; f = number of female patients; m = number of male patients; min = age of the youngest patient at the time of operation in years; max = age of the oldest patient.
| Cohort | Analysis method | CCC |
PDAC |
mPDAC |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Sex |
Age |
n | Sex |
Age |
n | Sex |
Age |
|||||||||||
| f | m | min | max | mean | f | m | min | max | mean | f | m | min | max | mean | |||||
| 1 | LC-MS | 11 | 9 | 2 | 31 | 77 | 59 | 9 | 5 | 4 | 54 | 81 | 66 | - | - | - | - | - | - |
| 2 | IHC | 10 | 9 | 1 | 47 | 74 | 61 | 10 | 3 | 7 | 48 | 81 | 65 | - | - | - | - | - | - |
| 3 | IHC | 73 | 47 | 26 | 28 | 81 | 63 | 78 | 46 | 32 | 48 | 94 | 69 | 18 | 8 | 10 | 49 | 80 | 67 |
Fig. 1.
The discovery and verification of biomarker candidates for the differential diagnosis of CCC and PDAC. The experimental workflow used for this study can be divided into two phases (A). During the discovery phase, the protein list resulting from the LC-MS experiment was filtered in several steps (B). This led to the selection of proteins for the verification by immunohistochemistry (IHC). Abbreviations: m/z, mass to charge ratio; RT, retention time; ROC, receiver operating characteristic; pFDR, t test p value after adjustment (Benjamini, Hochberg); FC, fold change.
Proteome Analysis
Tissue Preparation
Directly after surgery tissue samples were frozen in liquid nitrogen and stored at −80 °C. For laser capture microdissection the tissue was cryosectioned at −20 °C. The 10 μm sections were placed on 1.0 polyethylene naphthalate (PEN) membrane-covered slides (Carl Zeiss Microscopy, Thornwood, NY) and dehydrated by dipping into 70% ethanol for 5 s. Cell nuclei were stained for 30 s with 1% (w/v) cresyl violet in 50% ethanol. To remove excess cresyl violet and dehydrate the tissue sections, slides were dipped five times in 70% ethanol, once in 100% ethanol and then air dried.
Laser Capture Microdissection
Selective isolation of malignant cells from the cryo-sectioned tumor tissue was achieved by laser capture microdissection (LCM) on a Palm MicroBeam (Carl Zeiss Microscopy). A total area of 1 mm2 was dissected from each sample and collected in 15 μl 50 mm ammonium bicarbonate with 0.1% RapiGest SF Surfactant (Waters, Eschborn, Germany). Until further processing, samples were stored at −80 °C.
In-solution Digestion
Cell lysis was performed by sonicating twice for 1 min 20 s on ice in an ultrasonic bath and subsequent incubation at 95 °C for 5 min. Proteins' disulfide bonds were reduced with 5 mm DTT at 60 °C for 30 min and alkylated with 15 mm iodoacetamide for 30 min at ambient temperature in the dark. For enzymatic protein digestion, 50 ng trypsin (SERVA Electrophoresis, Heidelberg, Germany) was added and samples were incubated at 37 °C for 16 h. Acidification with 0.5% TFA for 30 min at 37 °C ensured quenching of enzyme activity and precipitation of hydrolyzed RapiGest. The latter was removed by centrifugation. The supernatant was dried in a centrifugal evaporator and resolved in 0.1% TFA. From each sample containing 1 mm2 tumor cell area 1/10 (corresponding to 100,000 μm2) was used for a sample pool (used as a reference sample later). The remaining material of every sample was split into three equal amounts (corresponding to about 300,000 μm2 each) for triplicate measurement of each biological replicate.
LC-MS/MS Analysis
For the LC-MS/MS study the 60 samples were randomized within three blocks corresponding to the three technical replicates, with CCC and PDAC samples alternating. In addition, six pools of all samples were analyzed, one after every tenth sample. For each measurement, tryptic peptides originating from ∼300,000 μm2 of tumor cells dissolved in 15 μl 0.1% TFA were injected into an Ultimate 3000 RSLCnano system (Dionex, Idstein, Germany) online coupled to an LTQ Orbitrap Elite mass spectrometer (Thermo Scientific, Bremen, Germany). The peptides were preconcentrated for 7 min on a trap column (Acclaim® PepMap 100, 75 μm × 2 cm, C18, 5 μm, 100 Å) using 30 μl/min 0.1% TFA and subsequently separated on an analytical column (Acclaim® PepMap RSLC, 75 μm × 50 cm, C18, 5 μm, 100 Å) by applying a gradient from 5–40% solvent B over 98 min (solvent A, 0,1% formic acid; solvent B, 0,1% formic acid, 84% acetonitrile; 400 nl/min; column oven temperature 60 °C). Peptide spectra were acquired in the Orbitrap analyzer at a resolution of 60,000. Using a data-dependent acquisition mode, the 20 most abundant peptides were then fragmented by collision-induced dissociation and measured in the linear ion trap. Further details concerning MS operating parameters have been described previously (13).
Data Analysis
Protein identification was performed using Proteome Discoverer (ver. 1.4) (Thermo Scientific, Bremen, Germany) searching the UniProt database (Release 2014_05; 545,388 entries; 20,339 human entries) via Mascot (ver. 2.3.0.2) (Matrix Sciences Ltd., London, UK). Search parameters were as follows: enzyme, trypsin; maximum missed cleavage sites, 1; taxonomy, Homo sapiens; mass tolerance, 5 ppm for precursor and 0.4 Da for fragment ions; dynamic modification, oxidation (M); static modification, carbamidomethyl (C). On peptide level, search results were filtered for a false discovery rate (FDR) < 1% using the PSM Validator tool implemented in Proteome Discoverer. Protein grouping was not applied.
Ion intensity-based label-free quantification was performed using Progenesis QI for proteomics (ver. 2.0.5387.52102, Nonlinear Dynamics Ltd., Newcastle upon Tyne, UK). LC-MS runs were aligned to account for retention time shifts. As a reference, one of the sample pool's measurements was used. Detected ions were filtered for those eluting between 18 and 112 min of the LC run, charged positively two, three, or fourfold and exhibiting more than two isotopes. For ion intensity quantification, an aggregate peak map containing the m/z and retention time information of all sample files was created and applied to each sample. This ensures no missing values. Peptide and protein identifications from Proteome Discoverer were imported into Progenesis QI for proteomics and matched to peptide spectra. Protein abundances were calculated by averaging normalized ion intensities of all unique peptides of one protein – including those measured at different charge states. Proteins quantified with only one unique peptide were removed from the study. Further details about peptide quantification have been described previously (10).
The mass spectrometric data has been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (14) with the dataset identifier PXD001833 and 10.6019/PXD001833. In addition, the peptide and protein identification and quantification data is listed in supplemental Tables S2, S3, and S4.
Statistical Rationale for Proteome Analysis
Normalized protein abundances were obtained from the software Progenesis and transformed by arcsinh to reduce the skewness of the data and meet the assumptions of the t test applied later on. Resulting values were averaged for each triple of technical replicates, and used to calculate the p value of Student's t test (independent samples, two-sided, assuming equal variances). Furthermore, the fold changes between groups were determined on the scale of normalized abundances. Coefficients of variation (CV) of technical replicates were calculated, using the transformed normalized protein abundances, to show reproducibility of the LC-MS system. A significantly differential expression was defined as an absolute fold change > 2 and an FDR-corrected p value (pFDR) < 0.05. The FDR was controlled by adjusting the raw p values of the t test using the method of Benjamini and Hochberg (15). For all applications of statistical inference, the significance level equals 0.05. The described analysis was conducted with the software R (R Core Team (2014). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria).
Immunohistochemistry
Preparation and Staining of Tissue Samples
Tissue microarrays were constructed using three cores per case with a diameter of 1 mm. In addition, tissue sections from metastatic PDAC tumors excised from the liver were prepared. Tissue was formalin fixed, paraffin embedded and sectioned at 4 μm thickness before dewaxing and pretreatment in EDTA buffer (pH 9) for 20 min at 95 °C. All staining reactions were carried out with a Dako Autostainer (Dako, Glostrup, Denmark). Details regarding the applied primary antibodies and the respective reaction conditions are listed in supplemental Table S5. Negative controls were prepared by incubation with nonimmune immunoglobulin instead of the primary antibody at the same concentration and included in every run. Tumor cases that presented with a specific staining during antibody establishment were included as positive controls in every subsequent run. Exemplary control stainings are depicted in supplemental Fig. S1.
Evaluation of Immunohistochemical Stains
The immunohistochemical staining of tissue sections was evaluated using an immuno-reactive score (IRS) as described previously (11). Briefly, the proportion of positively stained cells was graded as 0 (0%), 1 (1–5%), 2 (6–10%), 3 (11–50%), or 4 (51–100%) and the staining intensity was expressed as 0 (none), 1 (weak), 2 (intermediate), or 3 (strong). Both values were multiplied to obtain an IRS between 0 and 12. The evaluation was carried out independently by two observers. Cases with slight discrepancies concerning staining intensity or proportion of stained cells were additionally evaluated by a third independent observer. The values given by two out of three observers were used for further calculations. There were no cases with both values differing by more than one level. The inter-rater variability was determined by calculating Cohen's kappa with squared weights.
Statistical Rationale for Immunohistochemical Analysis
Previous to the analysis of the distributions of the IR scores, the interobserver variability was assessed by means of the simple percentage of exact agreement as well as by weighted Kappa, where disagreements are weighted according to their squared distance from perfect agreement. For these computations, the R package irr was used (16).
In contrast to the (transformed) abundancies in the proteome study, the IRS data set is analyzed using a nonparametric approach owing to the underlying distribution. The significance of the difference in immunohistochemical staining between all three groups (CCC, PDAC, and mPDAC) was assessed for each candidate, using Kruskal-Wallis' test based on the IRS. After adjustment of resulting p values for multiple testing according to Benjamini-Hochberg, the maximum p value was below 0.001. Subsequently, the post-hoc pairwise comparisons using Mann-Whitney's U test (again adjusted using Benjamini-Hochberg) were performed to determine which groups differ significantly in their staining patterns.
Diagnostic characteristics of the univariate marker candidates were assessed by means of ROC (receiver operating characteristics) analyses performed with the R package pROC (17). First, the overall discriminative power of the candidates was compared based on the area under the ROC curve (AUC). Along with the AUC values, corresponding 95% confidence intervals were calculated as defined by DeLong (18). For each antibody, the optimal IRS cut-off value was determined in order to best discriminate the two groups according to Youden's criterion. This is equivalent to the maximization of the sum of sensitivity and specificity. It is noted that the obtained diagnostic values are overoptimistic because of the optimization process. However, they are considered to be suitable for the comparison of different candidates within this study. A remark regarding the notation: In the direct comparison of CCC and PDAC, we considered PDAC as “cases” and CCC as “controls,” thus, in the following, sensitivity is the (true) probability of classifying a PDAC sample correctly whereas specificity is the (true) probability of a correct result for CCC.
When comparing diagnostic accuracy of two markers based on the same sample cohort, the McNemar test is used to account for the dependence of observations. Given p values correspond to the two-sided case.
RESULTS
In order to identify potential biomarkers for the differentiation of CCC and PDAC, the proteins extracted from the respective tumor cells were analyzed by label-free LC-MS/MS. The malignant cells were previously isolated from tissue sections by laser capture microdissection to account for the heterogeneity of these desmoplastic neoplasms. After analysis of the proteomics data set, suitable candidate proteins were evaluated by IHC—first in a small test set, then in a large cohort of primary CCC and PDAC as well as metastatic PDAC tumors from the liver. An overview of the described study design is depicted in Fig. 1.
Discovery Phase
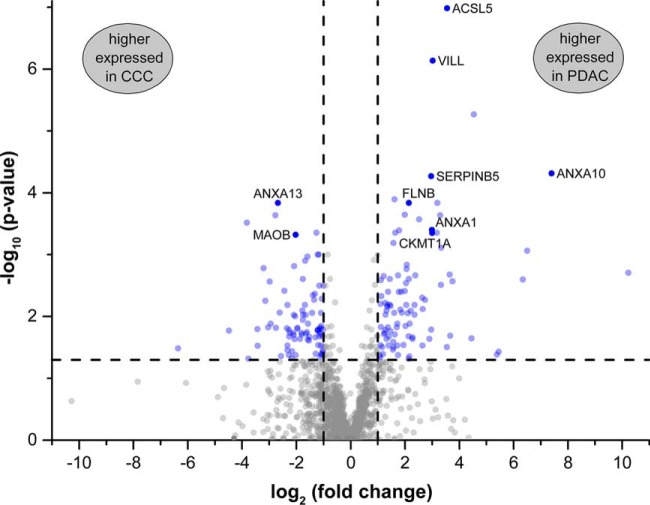
In the label-free LC-MS/MS data, a total of 61,775 peptide ions were quantified in an ion intensity-based manner. This led to the identification and quantification of 2701 proteins of which 1814 were quantified with more than one unique peptide. This analysis was performed using three technical replicates of each sample. In comparison, using only one replicate each, on average 2395 proteins could be quantified in total, and 1519 with more than one unique peptide. Technical replicates presented a very high repeatability, which was assessed by calculating the coefficient of variation (CV) of each triplicate measurement of every protein. For 92% of all protein abundances a CV of less than 10% was observed (39% < 1% CV; 85% < 5% CV), and only for 5% it was above 20%. A significantly differential expression between the two experimental groups (absolute fold change > 2, FDR-corrected p value of t test < 0.05) was observed for 180 proteins in which 85 were more abundant in CCC and 95 in PDAC cells (supplemental Table S6). Analyzing only one technical of each biological replicate, an average of 148 significantly differential proteins were identified. This means that the triplicate measurements increased the number of potentially relevant proteins by almost 22%. In order to select the most suitable candidates from the 180 differentially regulated proteins for the subsequent verification, the Euclidian distance was calculated for each protein. This considers both fold change and p value and is visualized in Fig. 2.
Fig. 2.
Volcano plot visualizing p values and fold changes of the 1814 proteins quantified with more than one unique peptide. Cut-off values are indicated by dashed lines. Significantly differential proteins (absolute fold change > 2, FDR-corrected p value < 0.05) are highlighted in blue. The nine proteins ACSL5 (Long-chain-fatty-acid-CoA ligase 5), ANXA1 (Annexin A1), ANXA10, ANXA13, CKMT1A (Creatine kinase U-type, mitochondrial), FLNB (Filamin-B), MAOB (Amine oxidase B), SERPINB5 (Serpin B5), and VILL (Villin-like protein) were chosen for immunohistochemical verification.
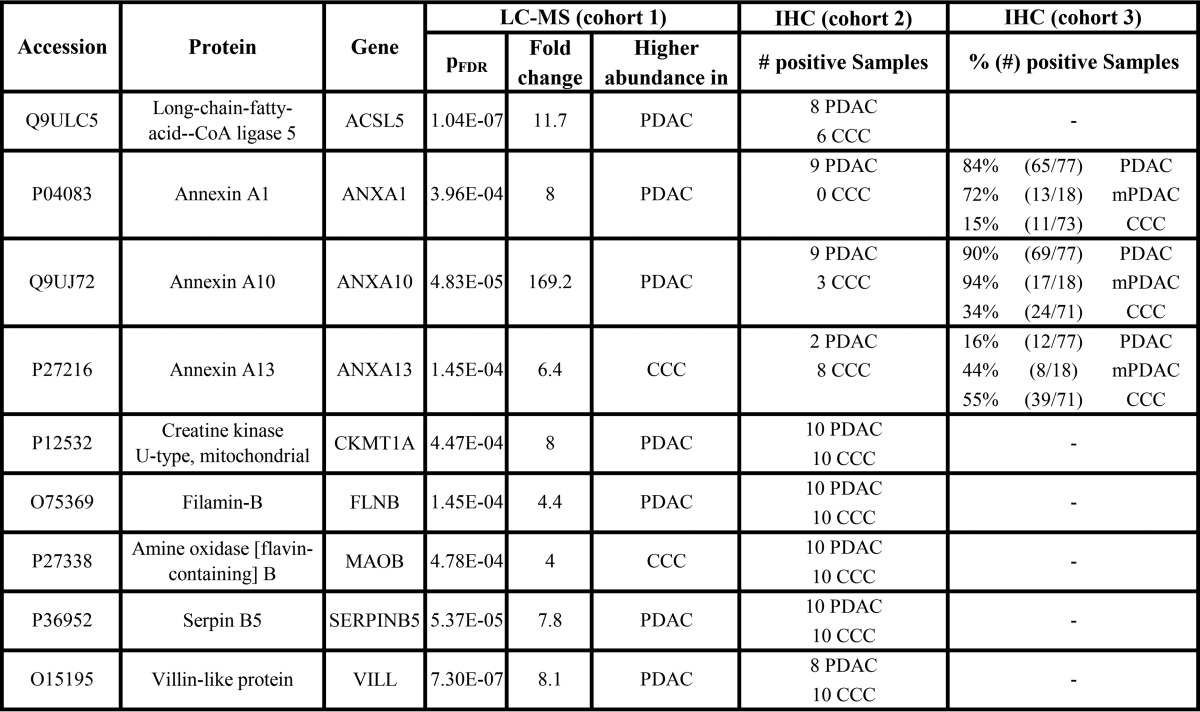
In addition, the expression profiles—the relative peptide and protein abundances in all samples—were observed. These should show little variation between samples from one group and of different peptides of one protein as well as a clear separation of the two groups. Especially promising results were achieved for nine proteins that were, therefore, selected for immunohistochemical verification (Table II).
Table II. Expression of candidate proteins in CCC and PDAC samples determined by label-free proteomics and IHC. For sample set 2, immunohistochemical staining was considered to be positive if at least 10% of the tumour cells were stained, regardless of the staining intensity. For sample cohort 3, individual cut-off values for each antibody were established by ROC analysis (Fig. 4).
Verification Phase
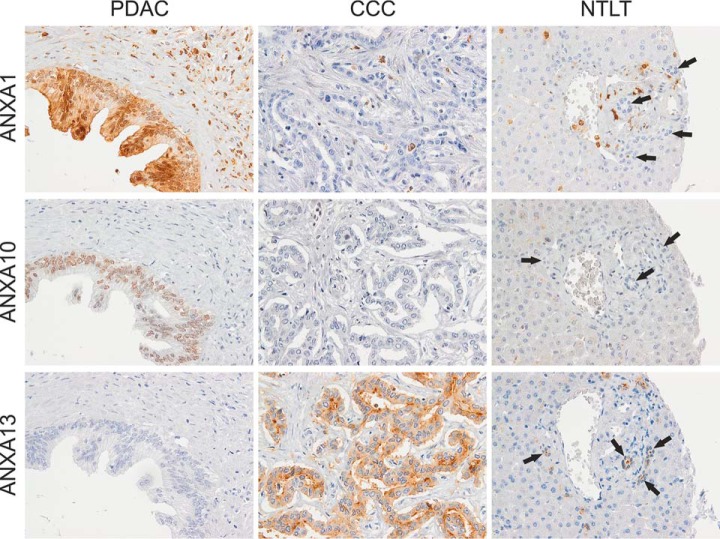
The nine candidates were first tested in a tissue microarray including ten CCC and ten PDAC tumor samples to assess cell type specificity of the proteins and the performance of the antibodies in a time and material saving fashion. Here, the annexins ANXA1, ANXA10, and ANXA13 showed the most promising results (Table II) and were therefore transferred to the next verification step in the form of a tissue microarray consisting of tumor sections from 73 CCC and 78 PDAC patients. Corresponding nontumorous liver tissue (NTLT) from the CCC patients was also analyzed to determine the expression of the candidate proteins in normal bile duct epithelium. Representative staining patterns are shown in Fig. 3 (and in supplemental Fig. S2 for the remaining six proteins).
Fig. 3.
Representative immunohistochemical staining of PDAC, CCC, and nontumorous liver tissue (NTLT) with antibodies against the annexins A1, A10, and A13. ANXA1 presented with a strong nuclear and diffuse cytoplasmic staining of PDAC cells whereas most tumorous and all nontumorous cholangiocytes (indicated by arrows) were unstained. In addition, stromal and inflammatory cells often expressed high amounts of ANXA1. In the case of ANXA10, a nuclear staining of PDAC cells varying in intensity was observed. CCC cells showed none or weak staining and nonmalignant cholangiocytes were completely unstained. ANXA13 was more often expressed in CCC than PDAC cells as well as in nontumorous bile duct cells presenting low to moderate expression, especially in the luminal membranes but also often in the cytoplasm. Hepatocytes did generally not express ANXA1, ANXA10, or ANXA13. Original magnification 400x.
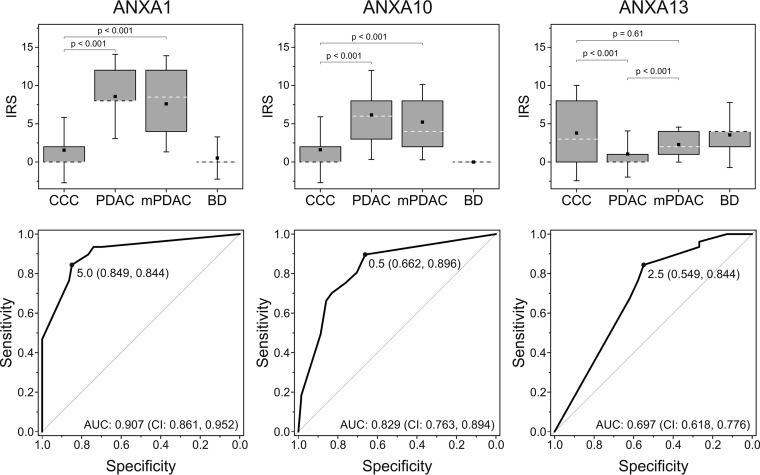
For all of the following statistical analyses the immuno-reactive scores (IRS) that take the percentage of stained cells and the staining intensity into account were considered. The scoring was performed by two independent observers with a good to very good agreement as reflected by high values of weighted kappa: 0.836 for ANXA1, 0.782 for ANXA10, and 0.852 for ANXA13. Concordance of IRS was achieved in 77% of all cases for ANXA1 staining, 64% for ANXA10, and 71% for ANXA13. The remaining samples, which showed only slight discrepancies, were scored by a third observer resulting in final IRS for all samples. These confirmed the significantly differential expression of ANXA1, ANXA10, and ANXA13 in primary PDAC cells as compared with CCC cells (p < 0.001, Mann-Whitney, adjusted) observed in the proteomics experiment (Fig. 4, upper panel).
Fig. 4.
Statistical evaluation of the immunohistochemical analysis of annexin A1, A10, and A13 in sample cohort 3. Box plots (upper panel) visualize immuno-reactive scores (IRS) for CCC (n = 73), PDAC (n = 78), and mPDAC (n = 13) tissue as well as nontumorous bile duct cells (BD; n = 67). Boxes represent the 25th and 75th percentiles. White dashed lines indicate the median, squares within the boxes the mean value and whiskers the 1.5-fold standard deviation. p values reflecting the significance of differences between two groups were calculated using the Mann-Whitney test after adjustment. Receiver operating characteristic (ROC) curves (lower panel) were calculated to determine the optimal IRS cut-off value for each potential biomarker. These are indicated on the curve including the sensitivity and specificity values at this point in brackets. Abbreviations: AUC, area under the curve; CI: 95% confidence interval.
To evaluate optimal cut-off values for the IRS for each antibody, ROC analysis was performed comparing primary PDAC and CCC samples. Here, sensitivity was defined as the percentage of correctly classified PDAC samples that, in this case, means a positive staining for ANXA1 and ANXA10 and negative immuno-reactivity for ANXA13. Specificity is therefore to be understood as the proportion of correctly classified CCC samples. ROC curves including AUC values, optimized IRS cut-offs as well as the sensitivity and specificity at this threshold are depicted in Fig. 4 (lower panel). The highest AUC was thereby obtained for ANXA1 (0.907). It exhibits a sensitivity of 84% at a specificity of 85%. The latter is lower for ANXA10 with 66% specificity, however, sensitivity reaches almost 90%. Analysis of ANXA13 resulted in an AUC of 0.697 with a sensitivity of 84% and a specificity of 55%. In addition, NTLT was evaluated regarding staining of nonmalignant cholangiocytes. Here, 7% of all samples showed positive staining for ANXA1, whereas none were positive for ANXA10. On the other hand, in 70% of the samples positive ANXA13 staining in cholangiocytes was visible (Fig. 4, upper panel). However, for none of these marker candidates, significant correlation between the expression in nonmalignant cholangiocytes and CCC cells was verifiable (for more detailed data see supplemental Table S7).
Analysis of Metastatic PDAC Tumor Tissue
Because all three potential markers performed well in the comparison of primary PDAC and CCC tumors, they were also tested in eighteen cases of metastatic PDAC. Here, ANXA1 and ANXA10 showed similar staining as in the primary PDAC samples. Applying the previously established IRS cut-off values, 13 out of 18 and 17 out of 18 tissue sections were positive for ANXA1 and ANXA10, respectively (Table II). These markers, therefore, show a significantly differential expression between mPDAC and CCC tissue (p < 0.001) (Fig. 4, upper panel) with sensitivities and specificities of 72 and 85% for ANXA1 and 94 and 66% for ANXA10 (specificity refers to the cohort of CCC samples previously described). ANXA13, on the other hand, was expressed in 8 out of 18 (56% Sensitivity) mPDAC cases (Table II). This means that, in terms of ANXA13 expression, mPDAC cells behave differently than PDAC cells that hardly express this protein (p < 0.001). Thus, ANXA13 is in this case not suitable to differentiate between CCC and mPDAC (p = 0.61; Fig. 4, upper panel).
Combination of ANXA1 and ANXA10 in a Biomarker Panel
In many cases the combination of single biomarkers in form of a biomarker panel can be beneficial. Because ANXA1 and ANXA10 showed the most promising results so far, a multivariate analysis including these two proteins was performed. Table III summarizes the congruency and discrepancies of CCC and PDAC sample classification by ANXA1 and ANXA10 scoring. Although there is no significant difference in the classification accuracy regarding the PDAC samples, ANXA1 outperforms ANXA10 in the CCC cohort. Overall, the combination of ANXA1 and ANXA10 did not improve the diagnostic values of the single marker ANXA1. Considering the panel to be positive for PDAC if at least one marker classifies the samples as PDAC (applying above mentioned cut-off values) resulted in sensitivity of 96% for PDAC and 100% for mPDAC but specificity of only 61%. If the panel is considered to be positive for PDAC only if both antibodies show positive staining, the specificity is improved to 90%, whereas the sensitivity for PDAC drops to 78% and for mPDAC even to 69%.
Table III. Correlation of positive or negative status of tumour samples classified according to ANXA1 or ANXA10 staining. The p values of the McNemar test are 0.424 for the PDAC samples and 0.007 for the CCC samples.
| PDAC-Samples | ANXA10 |
CCC-Samples | ANXA10 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Classified as CCC | Classified as PDAC | Classified as CCC | Classified as PDAC | ||||||||
| ANXA1 | Classified as CCC | 3 | 4% | 9 | 12% | ANXA1 | Classified as CCC | 43 | 61% | 17 | 24% |
| Classified as PDAC | 5 | 7% | 59 | 78% | Classified as PDAC | 4 | 6% | 7 | 10% | ||
DISCUSSION
The issue of classifying a liver tumor correctly regarding a possible pancreatic or cholangiocellular origin is equally challenging because of morphologic similarities as well as crucial for therapeutic decisions. It is a question pathologists are regularly confronted with. Since, up to now, no reliable diagnostic method such as the application of tissue markers is available, this study aimed at identifying novel biomarkers for differentiating CCC from PDAC. We therefore chose a global unbiased approach in form of LC-MS-based shotgun proteomics for the discovery phase. This has previously proven to be a suitable method for biomarker discovery studies (10, 11) and incidentally holds the advantage of enabling subsequent re- or meta-analysis of the data for, e.g. functional studies.
Here, each sample was analyzed in technical triplicates. This increased the total number of quantifiable proteins by 13% and those quantified by at least two unique peptides by 19% highlighting the benefit of technical replicate measurements. Tabb et al. showed a repeatability of protein identifications by LC-MS/MS on LTQ Orbitrap instruments of 70–80%. Supposedly, especially peptides with an abundance at the threshold of triggering an MS/MS can be lost if measuring only once (19). This also leads to a lower number of peptides per protein resulting in the exclusion of some proteins and a less exact quantification of others. Repeated measurement of an ion intensity also improves the confidence of the quantification for each peptide although our results revealed a very high repeatability between multiple LC-MS/MS measurements of the same sample. The latter has also been described in a previous study published by our group (20). The number of significantly differential proteins was increased by 22%. These additional proteins of interest can be of high importance for future studies such as pathway analyses. For example, they include several proteins known to be associated with cancer and metastasis formation such as protein kinase C delta (21), CD44 (22), cathepsin E (23), or Ras-related protein Rab-34 (24). Also serpin B5, which was chosen as a candidate protein in this study, would not have been identified as differentially expressed using only one technical replicate.
For this project, the malignant cells were isolated from the heterogeneous tumor tissue by LCM. We were able to achieve solid and reproducible LC-MS/MS data while applying as little as 300,000 μm2 of tissue that corresponds to only few thousand tumor cells. Our results underline the necessity of this step because other cell types within the tumor show different protein expression patterns than the actual tumor cells. ANXA1 e.g. was often strongly detectable in stromal or inflammatory cells, also in CCC tissue, but not in tumorous cholangiocytes. Analysis of the whole tumor tissue would have therefore falsified quantitative results.
The verification of selected biomarker candidates was performed via IHC because this is a well-established method used in pathological routine diagnostic. Out of the nine proteins tested in the first verification step, three could be transferred to the second stage involving a larger sample cohort. The reasons for discrepancies between proteomics and IHC results as seen for the remaining six candidates can be diverse. On the one hand, different patient cohorts were used for the identification and the subsequent verification. Therefore, differing protein expressions can be due to biological variances. On the other hand, the reason might be related to the difference in methodologies. While LC-MS allows continuous protein quantification, IHC – although characterized by low limits of detection – is merely a semiquantitative method. Especially for proteins of rather high abundance it is often not possible to assess subtler quantitative variations. This might be the case for CKMT1A, FLNB, MAOB, and Serpin B5 for which positive reactivity was observed in all twenty tissue samples from cohort 2. Also, some antibodies used for IHC show unspecific binding, or bind only to certain protein isoforms that often cannot be distinguished in shotgun LC-MS experiments, in most cases. Nevertheless, these proteins might still be of relevance in a different context, e.g. in relation with tumor biology or as noninvasive biomarkers detectable in body fluids. This was, however, not within the scope of this study.
Here, we identified and verified the three annexin proteins ANXA1, ANXA10, and ANXA13 as being differentially expressed between primary CCC and PDAC cells. They belong to a highly conserved protein family comprising twelve members in vertebrates. They are characterized by the ability of binding negatively charged phospholipids in a calcium-dependent manner. This can lead to a rapid translocation from the cytosol to plasma or intracellular membranes and is important for their interaction with binding partners (25). They are involved in several cellular processes such as calcium signaling, vesicle trafficking, cell division, growth regulation, and apoptosis, which is why their differential expression in tumor cells that has been reported for many of the annexins in several cancer types is plausible (26).
ANXA1 is the best studied annexin, also in association with cancer. For example, it has been shown to be decreased in breast carcinoma, prostatic adenocarcinoma, and esophageal squamous carcinoma, but increased in esophageal adenocarcinoma and hairy cell leukemia (26). In PDAC, a study comparing tumorous to matched normal pancreas tissue showed a higher ANXA1 expression in the malignant tissue that was correlated with poor tumor differentiation (27). Our study confirmed the abundance of ANXA1 in PDAC cells while it furthermore provided the best diagnostic values for distinguishing PDAC from CCC (84% sensitivity, 85% specificity). In contrast to our findings, Hongsrichan et al. discovered an overexpression of ANXA1 in 64 out of 68 CCC cases. This was in comparison to hepatocellular carcinoma and healthy liver tissue (28). However, all of these CCC tumors were associated to an Opisthorchis viverrini infection that, consequently, seems to have an effect on the protein expression profile of the cancer cells. ANXA1 has also been linked to tumorigenesis and metastasis formation on the molecular level as it is a substrate for several serine/threonine and tyrosine kinases (25). Upon phosphorylation, for instance, it migrates to the cell membrane where it interacts with epidermal growth factor receptor (EGFR) (29). It has been shown that upon nontransient EGFR activation with consequential tumor growth, both ANXA1 expression and phosphorylation were increased (30). Furthermore, silencing ANXA1 in human CCC cell lines by siRNA induced down-regulation of transforming growth factor (TGF)-β, matrix metalloproteinases (MMP) 2 and MMP 9 as well as up-regulation of NF-κB (28), all of which are proteins involved in cancer development and metastasis.
Next to ANXA1, in the present study, ANXA10 received promising diagnostic values with a sensitivity of 90% and a specificity of 66% for the identification of PDAC versus CCC. Recently, an immunohistochemical study examining ANXA10 expression in several types of tumorous and nontumorous tissue was published. Here, normal expression was observed in the gastric mucosa, the duodenum, the kidney, and the urinary bladder with a mainly nuclear and weak cytoplasmic staining, and none in normal bile ducts and pancreatic ducts. Within their PDAC cohort, 78% (57/73) of the primary and 83% (19/23) of the metastatic tumors showed positive ANXA10 staining, while it was only 17% (10/59) of the intrahepatic CCC cases. Taking the general biological variance into consideration, these results are very similar to ours. Concerning the pancreatobiliary system, this study also included adenocarcinomas of the ampulla of Vater, the extrahepatic bile ducts and the gallbladder. Interestingly, this indicated that the closer the primary tumor site is located to the pancreas, the more likely a positive ANXA10 staining is and, conversely, the closer it is to the liver, the less primary tumors express ANXA10 (31). Thus, although ANXA10 expression is not visible in healthy cells of the pancreatobiliary system, their localization has an effect on the ANXA10 abundance in tumor cells deriving from them. This knowledge is also important for the potential establishment of ANXA10 as a biomarker.
For ANXA13 our observations were different. The abundance of this protein did not seem to correlate with the primary tumor site but with the actual localization of the malignancy because it was detected in 55% of the CCC and only 16% of the PDAC sections, but it was also expressed by 44% of the mPDAC cases. ANXA13 is thought to be the probable common ancestor of all vertebrate annexins (32). It is supposed to have a high tissue specificity in healthy tissue, restricted only to intestinal and kidney epithelial cells (33), although it was also detected in nontumorous cholangiocytes in the present study. In malignant cells, the decreased ANXA13 expression has been described to cause protection against Rapamycin, an anticancer drug (34). Because of our findings, it is not suitable for distinguishing mPDAC in the liver from CCC. However, it might be of relevance in the context of the molecular mechanisms of PDAC metastasis formation because of the significantly differential expression between the primary and the secondary tumors.
The combination of biomarkers in the form of a panel can often improve diagnostic values. For ANXA1 and ANXA10 this was not the case. Nevertheless, in comparison to marker panels described in previous publications, ANXA1 delivers better diagnostic values even on its own. The panel S100P-/pVHL+/MUC5AC-/CK17-, for example, has a specificity of 100% for CCC when compared with PDAC but only a sensitivity of 59% (8) and combining MUC1 and CK17 led to positive predictive values of 76% for PDAC and 58% for CCC (35). It could therefore be advantageous to implement ANXA1, or also ANXA10, into these panels or construct a new panel with the most suitable proteins. In conclusion, ANXA1 has a high potential of becoming a valuable marker for the differential diagnosis of CCC and metastatic PDAC in the liver.
Supplementary Material
Acknowledgments
We thank Laura Malkus, Birgit Korte, Stephanie Tautges, Kristin Rosowski, Dorothe Möllmann, and Don Marvin Voss for their excellent technical assistance.
Footnotes
Author contributions: J.P., D.A.M., T.B., H.E.M., B. Sipos, H.A.B., and B. Sitek designed research; J.P., J.K., S.B., and H.A.B. performed research; J.P., M.A., and M.E. analyzed data; J.P., M.A., J.K., D.A.M., T.B., H.E.M., H.A.B., and B. Sitek wrote the paper; P.K. and B. Sipos provided clinical samples.
* The PROFILE project is cofunded by the European Union (European Regional Development Fund - Investing in your future) and the German federal state North Rhine-Westphalia (NRW), project number z0911bt004d/e. A part of this study was funded by P.U.R.E. (Protein Unit for Research in Europe), a project of North Rhine-Westphalia, a federal state of Germany and by the Mercator Research Center Ruhr (MERCUR), project number GZ An-2013-0056.
 This article contains supplemental Figs. S1 and S2 and Tables S1 to S7.
This article contains supplemental Figs. S1 and S2 and Tables S1 to S7.
1 The abbreviations used are:
- CCC
- cholangiocellular carcinoma
- ANXA
- annexin
- AUC
- area under the curve
- BH
- Benjamini-Hochberg
- CKMT1A
- creatine kinase U-type mitochondrial
- CV
- coefficient of variation
- EGFR
- epidermal growth factor receptor
- FDR
- false discovery rate
- FLNB
- filamin-B
- HPC2
- human pancreatic cancer fusion protein #2
- IHC
- immunohistochemistry
- IRS
- immuno-reactive score
- LCM
- laser capture microdissection
- MAOB
- Amine oxidase [flavin-containing] B
- MMP
- matrix metalloproteinases
- (m)PDAC
- (metastases of) pancreatic ductal adenocarcinoma
- NTLT
- nontumorous liver tissue
- PODXL-1
- podocalyxin-like protein 1
- ROC
- receiver operating characteristics
- TGF
- transforming growth factor.
REFERENCES
- 1. Kasper H. U., Drebber U., Dries V., and Dienes H. P. (2005) [Liver metastases: incidence and histogenesis]. Z. Gastroenterol. 43, 1149–1157 [DOI] [PubMed] [Google Scholar]
- 2. Somoracz A., Tatrai P., Horvath G., Kiss A., Kupcsulik P., Kovalszky I., and Schaff Z. (2010) Agrin immunohistochemistry facilitates the determination of primary versus metastatic origin of liver carcinomas. Hum. Pathol. 41, 1310–1319 [DOI] [PubMed] [Google Scholar]
- 3. German S3-Guideline “S3-Leitlinie zum exokrinen Pankreaskarzinom” (2013) Version 1.0, AWMF registration number: 032/010OL.
- 4. Borger D. R., Tanabe K. K., Fan K. C., Lopez H. U., Fantin V. R., Straley K. S., Schenkein D. P., Hezel A. F., Ancukiewicz M., Liebman H. M., Kwak E. L., Clark J. W., Ryan D. P., Deshpande V., Dias-Santagata D., Ellisen L. W., Zhu A. X., and Iafrate A. J. (2012) Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist 17, 72–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blechacz B., and Gores G. J. (2008) Cholangiocarcinoma: advances in pathogenesis, diagnosis, and treatment. Hepatology 48, 308–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ney J. T., Zhou H., Sipos B., Buttner R., Chen X., Kloppel G., and Gutgemann I. (2007) Podocalyxin-like protein 1 expression is useful to differentiate pancreatic ductal adenocarcinomas from adenocarcinomas of the biliary and gastrointestinal tracts. Hum. Pathol. 38, 359–364 [DOI] [PubMed] [Google Scholar]
- 7. Hooper J. E., Morgan T. K., Grompe M., Sheppard B. C., Troxell M. L., Corless C. L., and Streeter P. R. (2012) The novel monoclonal antibody HPC2 and N-cadherin distinguish pancreatic ductal adenocarcinoma from cholangiocarcinoma. Hum. Pathol. 43, 1583–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lok T., Chen L., Lin F., and Wang H. L. (2014) Immunohistochemical distinction between intrahepatic cholangiocarcinoma and pancreatic ductal adenocarcinoma. Hum. Pathol. 45, 394–400 [DOI] [PubMed] [Google Scholar]
- 9. Bosman F. T., World Health Organization, and International Agency for Research on Cancer. (2010) WHO classification of tumours of the digestive system, 4th Ed., International Agency for Research on Cancer, Lyon. [Google Scholar]
- 10. Megger D. A., Bracht T., Kohl M., Ahrens M., Naboulsi W., Weber F., Hoffmann A. C., Stephan C., Kuhlmann K., Eisenacher M., Schlaak J. F., Baba H. A., Meyer H. E., and Sitek B. (2013) Proteomic differences between hepatocellular carcinoma and nontumorous liver tissue investigated by a combined gel-based and label-free quantitative proteomics study. Mol. Cell. Proteomics 12, 2006–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Padden J., Megger D. A., Bracht T., Reis H., Ahrens M., Kohl M., Eisenacher M., Schlaak J. F., Canbay A. E., Weber F., Hoffmann A. C., Kuhlmann K., Meyer H. E., Baba H. A., and Sitek B. (2014) Identification of novel biomarker candidates for the immunohistochemical diagnosis of cholangiocellular carcinoma. Mol. Cell. Proteomics 13, 2661–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Molleken C., Sitek B., Henkel C., Poschmann G., Sipos B., Wiese S., Warscheid B., Broelsch C., Reiser M., Friedman S. L., Tornoe I., Schlosser A., Kloppel G., Schmiegel W., Meyer H. E., Holmskov U., and Stuhler K. (2009) Detection of novel biomarkers of liver cirrhosis by proteomic analysis. Hepatology 49, 1257–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bracht T., Schweinsberg V., Trippler M., Kohl M., Ahrens M., Padden J., Naboulsi W., Barkovits K., Megger D. A., Eisenacher M., Borchers C. H., Schlaak J. F., Hoffmann A. C., Weber F., Baba H. A., Meyer H. E., and Sitek B. (2015) Analysis of disease-associated protein expression using quantitative proteomics-fibulin-5 is expressed in association with hepatic fibrosis. J. Proteome Res. 14, 2278–2286 [DOI] [PubMed] [Google Scholar]
- 14. Vizcaino J. A., Cote R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Perez-Riverol Y., Reisinger F., Rios D., Wang R., and Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Benjamini Y., and Hochberg Y. (1995) Controlling the false discovery rate – a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B Met. 57, 289–300 [Google Scholar]
- 16. Gamer M., Lemon J., Fellows I., and P S. (2010) irr: Various coefficients of interrater reliability and agreement (Version 0.83) [Software]. Available from http://CRAN.R-project.org/package=irr.
- 17. Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J. C., and Muller M. (2011) pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. DeLong E. R., DeLong D. M., and Clarke-Pearson D. L. (1988) Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44, 837–845 [PubMed] [Google Scholar]
- 19. Tabb D. L., Vega-Montoto L., Rudnick P. A., Variyath A. M., Ham A. J., Bunk D. M., Kilpatrick L. E., Billheimer D. D., Blackman R. K., Cardasis H. L., Carr S. A., Clauser K. R., Jaffe J. D., Kowalski K. A., Neubert T. A., Regnier F. E., Schilling B., Tegeler T. J., Wang M., Wang P., Whiteaker J. R., Zimmerman L. J., Fisher S. J., Gibson B. W., Kinsinger C. R., Mesri M., Rodriguez H., Stein S. E., Tempst P., Paulovich A. G., Liebler D. C., and Spiegelman C. (2010) Repeatability and reproducibility in proteomic identifications by liquid chromatography-tandem mass spectrometry. J. Proteome Res. 9, 761–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Megger D. A., Pott L. L., Ahrens M., Padden J., Bracht T., Kuhlmann K., Eisenacher M., Meyer H. E., and Sitek B. (2014) Comparison of label-free and label-based strategies for proteome analysis of hepatoma cell lines. Biochim. Biophys. Acta 1844, 967–976 [DOI] [PubMed] [Google Scholar]
- 21. Garg R., Benedetti L. G., Abera M. B., Wang H., Abba M., and Kazanietz M. G. (2014) Protein kinase C and cancer: what we know and what we do not. Oncogene 33, 5225–5237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li X. P., Zhang X. W., Zheng L. Z., and Guo W. J. (2015) Expression of CD44 in pancreatic cancer and its significance. Int. J. Clin. Exp. Pathol. 8, 6724–6731 [PMC free article] [PubMed] [Google Scholar]
- 23. Zaidi N., Hermann C., Herrmann T., and Kalbacher H. (2008) Emerging functional roles of cathepsin E. Biochem. Biophys. Res. Commun. 377, 327–330 [DOI] [PubMed] [Google Scholar]
- 24. Wang H. J., Gao Y., Chen L., Li Y. L., and Jiang C. L. (2015) RAB34 was a progression- and prognosis-associated biomarker in gliomas. Tumour Biol. 36, 1573–1578 [DOI] [PubMed] [Google Scholar]
- 25. Hoque M., Rentero C., Cairns R., Tebar F., Enrich C., and Grewal T. (2014) Annexins – scaffolds modulating PKC localization and signaling. Cell. Signal. 26, 1213–1225 [DOI] [PubMed] [Google Scholar]
- 26. Mussunoor S., and Murray G. I. (2008) The role of annexins in tumor development and progression. J. Pathol. 216, 131–140 [DOI] [PubMed] [Google Scholar]
- 27. Bai X. F., Ni X. G., Zhao P., Liu S. M., Wang H. X., Guo B., Zhou L. P., Liu F., Zhang J. S., Wang K., Xie Y. Q., Shao Y. F., and Zhao X. H. (2004) Overexpression of annexin 1 in pancreatic cancer and its clinical significance. World J. Gastroenterol. 10, 1466–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hongsrichan N., Rucksaken R., Chamgramol Y., Pinlaor P., Techasen A., Yongvanit P., Khuntikeo N., Pairojkul C., and Pinlaor S. (2013) Annexin A1: a new immunohistological marker of cholangiocarcinoma. World J. Gastroenterol. 19, 2456–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Radke S., Austermann J., Russo-Marie F., Gerke V., and Rescher U. (2004) Specific association of annexin 1 with plasma membrane-resident and internalized EGF receptors mediated through the protein core domain. FEBS Lett. 578, 95–98 [DOI] [PubMed] [Google Scholar]
- 30. de Coupade C., Gillet R., Bennoun M., Briand P., Russo-Marie F., and Solito E. (2000) Annexin 1 expression and phosphorylation are upregulated during liver regeneration and transformation in antithrombin III SV40 T large antigen transgenic mice. Hepatology 31, 371–380 [DOI] [PubMed] [Google Scholar]
- 31. Lu S. H., Yuan R. H., Chen Y. L., Hsu H. C., and Jeng Y. M. (2013) Annexin A10 is an immunohistochemical marker for adenocarcinoma of the upper gastrointestinal tract and pancreatobiliary system. Histopathology 63, 640–648 [DOI] [PubMed] [Google Scholar]
- 32. Iglesias J. M., Morgan R. O., Jenkins N. A., Copeland N. G., Gilbert D. J., and Fernandez M. P. (2002) Comparative genetics and evolution of annexin A13 as the founder gene of vertebrate annexins. Mol. Biol. Evol. 19, 608–618 [DOI] [PubMed] [Google Scholar]
- 33. Turnay J., Lecona E., Fernandez-Lizarbe S., Guzman-Aranguez A., Fernandez M. P., Olmo N., and Lizarbe M. A. (2005) Structure-function relationship in annexin A13, the founder member of the vertebrate family of annexins. Biochem. J. 389, 899–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reiske H., Sui B., Ung-Medoff H., Donahue R., Li W. B., Goldblatt M., Li L., and Kinch M. S. (2010) Identification of annexin A13 as a regulator of chemotherapy resistance using random homozygous gene perturbation. Anal. Quant. Cytol. Histol. 32, 61–69 [PubMed] [Google Scholar]
- 35. Chu P. G., Schwarz R. E., Lau S. K., Yen Y., and Weiss L. M. (2005) Immunohistochemical staining in the diagnosis of pancreatobiliary and ampulla of Vater adenocarcinoma: application of CDX2, CK17, MUC1, and MUC2. Am. J. Surg. Pathol. 29, 359–367 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.