Abstract
The aryl hydrocarbon receptor (AHR) is a ligand-activated transcription factor that is regulated by environmental toxicants that function as AHR agonists such as 2,3,7,8 tetrachlorodibenzo-p-dioxin (TCDD). L-Type Amino Acid Transporter 1 (LAT1) is a leucine transporter that is overexpressed in cancer. The regulation of LAT1 by AHR in MCF-7 and MDA-MB-231 breast cancer cells (BCCs) was investigated in this report. Ingenuity pathway analysis (IPA) revealed a significant association between TCDD-regulated genes (TRGs) and molecular transport. Overlapping the TCDD-RNA-Seq dataset obtained in this study with a published TCDD-ChIP-seq dataset identified LAT1 as a primary target of AHR-dependent TCDD induction. Short interfering RNA (siRNA)-directed knockdown of AHR confirmed that TCDD-stimulated increases in LAT1 mRNA and protein required AHR expression. TCDD-stimulated increases in LAT1 mRNA was also inhibited by the AHR antagonist CH-223191. Upregulation of LAT1 by TCDD coincided with increases in leucine uptake by MCF-7 cells in response to TCDD. Chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) assays revealed increases in AHR, AHR nuclear translocator (ARNT) and p300 binding and histone H3 acetylation at an AHR binding site in the LAT1 gene in response to TCDD. In MCF-7 and MDA-MB-231 cells, endogenous levels of LAT1 mRNA and protein were reduced in response to knockdown of AHR expression. Knockdown experiments demonstrated that proliferation of MCF-7 and MDA-MB-231 cells is dependent on both LAT1 and AHR. Collectively, these findings confirm the dependence of cancer cells on leucine uptake and establish a mechanism for extrinsic and intrinsic regulation of LAT1 by AHR.
Keywords: Aryl Hydrocarbon Receptor (AHR), L-Type Amino Acid Transporter 1 (LAT-1), TCDD, gene expression, breast cancer
Graphical abstract
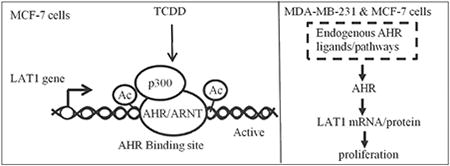
1. Introduction
Halogenated aromatic hydrocarbons (HAHs) are environmental toxicants that are formed as byproducts of industry and municipal waste incineration [1, 2]. 2,3,7,8 tetrachlorodibenzo-p-dioxin (TCDD) exhibits the highest affinity for AHR compared with other HAHs [3]. In the absence of ligand, AHR is associated with chaperone proteins including heat shock protein 90 (HSP90) [4, 5], Aryl Hydrocarbon Receptor Interacting Protein (also known as XAP2) [6-8] and the co-chaperone protein p23 [9] in the cytoplasm. Upon binding to TCDD, AHR translocates from the cytoplasm into the nucleus and binds AHR nuclear translocator (ARNT) [10-12]. TCDD-induced AHR/ARNT dimers confer transcriptional activity specifically to AHR response elements (AHR-REs) that cluster near the promoter regions of TCDD target genes [12]. CYP1A1 and CYP1B1 are phase I xenobiotic metabolizing enzymes that are transcriptionally induced by TCDD via AHR [12]. The induction of CYP1A1 and CYP1B1 transcription by TCDD also requires several transcriptional coactivators including steroid receptor coactivator 2 (SRC1), steroid receptor coactivator 2 (SRC2), p300 and BRG-1 [13-15].
In addition to xenobiotic metabolism, immune responses are modulated by AHR and the outcome is dependent on the AHR ligand. For instance, T regulatory cells (Tregs) suppress excessive immune responses and their differentiation is promoted by TCDD or kynurenine (Kyn), and both of these AHR ligands are immunosuppressive [16, 17]. Th17 cells are proinflammatory T cells and their expansion and differentiation is enhanced by the endogenous AHR ligand 6-formylindolo [3, 2-b] carbazole (FICZ), but suppressed by TCDD [17, 18]. Developmental and functional immunity is dependent on AHR and the dietary AHR ligands indolo [3,2-b] carbazole (ICZ) and 3,3-diidolylmethane (DIM) [19, 20]. Finally, cytokine and chemokine gene expression in dendritic cells (DC) and macrophages is increased by AHR ligands, and the transcription of AHR is increased by nuclear factor kappa B in response to lipopolysaccharide (LPS) in innate immune cells [21-23].
TCDD-RNA-Seq analysis described herein identified 137 TCDD-regulated genes (TRGs) in MCF-7 breast cancer cells (BCCs) among which is L-Type Amino Acid Transporter 1 (LAT1). The uptake of large neutral acids including: leucine, arginine, phenylalanine, tyrosine, and tryptophan is mediated by LAT1 [24-26]. Breast, colorectal, head and neck, leukemia, lymphoma, melanoma, prostate and parathyroid cancers express higher levels of LAT1 compared with corresponding normal tissue [27]. LAT1 promotes proliferation of cancer cells by stimulating the uptake of amino acids that are important for protein synthesis [27]. In addition, its ability to promote cellular uptake of leucine would also increase the activity of mTORC1 which has been reported to be important for the growth and survival of some cancers [28]. TCDD has been reported to increase LAT1 mRNA in HEPG2 cells, which are a model of hepatocellular carcinoma [29]. These observations strongly suggest that LAT1 is critical to cancer cell growth and survival. However, the mechanism by which TCDD or AHR regulates LAT1 expression has not been determined.
The objective of this report was to investigate extrinsic regulation of LAT1 by TCDD/AHR and intrinsic (endogenous) regulation of LAT1 by AHR. Extrinsic regulation of LAT1 by TCDD/AHR was investigated in MCF-7 cells. Intrinsic regulation of LAT1 by AHR was investigated in MCF-7 and MDA-MB-231 cells because these BCC lines have been reported to exhibit endogenous AHR activity [30-33]. Based on our findings, we report a new role for AHR as an extrinsic and intrinsic regulator of LAT1 expression in BCCs and show that AHR binds to LAT1 AHR-REs as part of a transcriptional activator complex.
2. Methods
2.1. Materials
Dulbecco's Modified Eagle Medium/High glucose (DMEM) with L-glutamine and sodium pyruvate, phenol red-free DMEM, phosphate buffered saline (PBS), fetal bovine serum (FBS), penicillin, streptomycin, and dimethyl sulfoxide (DMSO) were purchased from Thermo Fisher Scientific (Pittsburgh, PA). Sodium dodecyl sulfate (SDS), 30% acrylamide/bis solution, ammonium persulfate, Tween-20, 2-mercaptoethanol and polyvinylidene difluoride (PVDF) membranes were obtained from BIO-RAD (Hercules, CA). Non-targeting short interfering RNA (siRNA) (cat # D-001810-01-20), ON-TARGET plus human siRNA's against AHR (cat # J-004990-08-0010, and cat # J-004990-06-0010) and LAT1 (cat # J-004953-09-0010) were purchased from GE Dharmacon (Lafayette, CO). 2,3,7,8 tetrachlorodibenzo-p-dioxin (TCDD) was obtained from Cambridge Isotopes Laboratory (Andover, MA). The AHR antagonist CH-223191 was purchased from Sigma-Aldrich (St. Louis, MO). MCF-7 and MDA-MB-231 BCCs were purchased from ATCC (Manassas, VA) and maintained in DMEM, 10% FBS, with penicillin (100 IU/mL) and streptomycin (100 IU/mL).
2.2. TCDD RNA-Seq
250,000 MCF-7 cells were seeded in 35 mm plates in DMEM supplemented with 10 % FBS for 24 hr, followed by overnight serum-starvation in phenol red-free DMEM, and then treated with vehicle (DMSO) or 10 nM TCDD for 6 hrs. RNA-Seq analysis was based on 4 biological replicates in each experimental group. Total RNA purification kits (Qiagen, Valencia, CA) were used to extract total RNA. RNA sample quality was assessed using Bioanalyzer RNA Nano chips (Agilent); all RNA samples had an RNA Integrity Number greater than or equal to 8. RNA-Seq libraries were prepared from 1 μg of total RNA using a TruSeq RNA Prep Kit (Illumina Inc., San Diego, CA). RNA-Seq was performed using an Illumina HiSeq1000 in a 2 × 100 base paired end design yielding a minimum of 50 million reads per sample. Differentially expressed genes were identified at a False Discovery Rate (FDR) of 5% as detailed in our prior report [33]. Raw reads and processed data (unnormalized and normalized read counts by gene) were deposited in the Gene Expression Omnibus (GEO) at the National Center for Biotechnology Information and are accessible via accession number GSE76608.
2.3. Ingenuity pathway analysis (IPA)
TCDD-regulated genes (TRGs) were expressed as a ratio of TCDD/DMSO and loaded into IPA software (Ingenuity Systems, Redwood City, CA). Of the 137 TRGs identified by RNA-Seq, 116 were mapped to known functions and pathways by IPA. The Core Analysis tool and the Fisher Exact Test in IPA were used to identify statistically significant associations between TRGs and cellular and molecular pathways. We configured the core analysis to report Benjamini-Hochberg corrected p-values.
2.4. Reverse transcription and real-time polymerase chain reaction (RT-qPCR)
RT-qPCR assays were carried out as described in our prior reports [33, 34]. In brief, total RNA was extracted using RNA purification columns (Qiagen) and 100-300 ng of extracted RNA was reverse transcribed to cDNA using High Capacity Reverse Transcription kits (Thermo Fisher Scientific). Realtime qPCR reactions were performed in triplicate using SYBR Green Master Mix according to the manufacturer's instructions (Thermo Fisher Scientific). Relative changes in gene expression were calculated using the 2−ΔΔCT formula as described by Livak and Schmittgen [35]. Glyceraldehyde-3-phosphate (GAPDH) mRNA levels served as the internal control. The sequences of the qPCR primers used to amplify GAPDH and AHR mRNA have been published [33]. LAT1 mRNA qPCR primers were: forward, 5′-ccgaggagaaggaagaggc-3′; reverse, 5′-gaagatgcccgagccgataa-3′. The Student-Newman–Keuls (SNK) post-hoc test was used to determine statistically significant differences among groups following one-way analysis of variance (ANOVA).
2.5. Short interfering RNA (siRNA) assays and Western blotting
The siRNA knockdowns were performed as detailed in our prior reports [33, 34]. Briefly, 200,000 cells (MCF-7 or MDA-MB-231) in 1 mL of DMEM supplemented with 10% FBS were mixed directly with 100 nM of siRNA that was non-targeting, AHR-targeting or LAT1-targeting and 3 μL of Lipofectamine RNAiMAX transfection reagent (Thermo Fisher Scientific) and immediately plated in 35 mm tissue culture plates for 48 hrs. MCF-7 cells were then treated with vehicle (DMSO) or 10 nM TCDD for 16 hrs. Treatments were removed and cells were rinsed once with PBS. For Western blotting, total protein was extracted by scraping cells in 2× Laemmli Sample Buffer containing β-mercaptoethanol (BME). Laemmli sample buffer and BME were purchased from BIO-RAD. Standard Western blotting techniques were used to analyze ∼ 10 μg of protein per sample (please refer to our prior reports for technical details [33, 34]). Western blot analysis of GAPDH was used to confirm equal protein loading. Blots were probed with anti-GAPDH antibody (diluted 1:10,000), anti-AHR antibody (diluted 1:5,000) or anti-LAT1 antibody (diluted 1:2,000) overnight at 4oC, followed by incubation with anti-HRP secondary antibody (1:5000) for 1 hr at room temperature. The blots were then rinsed with PBS + 0.1% tween 20, and then developed with enhanced chemiluminescent substrate Millipore Corp., (Billerica, MA). The anti-GAPDH antibody was purchased from Millipore (Cat #MAB374). The anti-AHR antibody was obtained from Santa Cruz Biotechnology (Santa Cruz, CA, Cat #H-211) and the anti- LAT1 antibody was purchased from Cell Signaling Technology (Danvers, MA, Cat #5347). Densitometry was calculated with ImageJ PC-based software (National Institute of Health). The Student-Newman–Keuls (SNK) post-hoc test was used to determine statistically significant differences among groups following one-way analysis of variance (ANOVA).
2.6. Chromatin immunoprecipitation followed by qPCR (ChIP-qPCR)
The ChIP-qPCR assays were carried out as described in our previous report [34]. In brief, nonspecific IgG, and antibodies that were specific for AHR, ARNT or p300 were obtained from Santa Cruz Biotechnology. The antibodies against acetylated lysine 9 or lysine 14 in histone H3 were purchased from Cell Signaling Technology. The magnetic protein A beads and proteinase K were purchased from Life Technologies (Carlsbad, CA). A recent TCDD-ChlP-Seq report identified an AHR binding site within a 900 bp region in LAT1 corresponding to coordinates 87,840,300 to 87,841,199 (human genome version 19 (Hg19)) [36]. This AHR binding site was investigated in this report by ChIP-qPCR with primers that span coordinates 87,840,403 to 87,840,544, which were: [forward 5′-GCACGTACCTGTAGGGGTTG -3′ and reverse 5-ATGCTCTCTCCCCGGTGATT-3′]. The ChIP-qPCR primers used to amplify the AHR binding sites in the CYP1B1 gene have been published [37]. ChIP-qPCR data was expressed as % input in which signals obtained from the ChIP are divided by signals obtained from an input sample. Statistical differences among groups were determined by the SNK post-hoc test following one-way analysis of variance (ANOVA)
2.7. Leucine uptake experiments
Leucine uptake experiments were performed in MCF-7 cells grown to confluence on 24 well plates. The cells were first washed twice with Na-free buffer (130 mM TMACl, 4.7 mM KCl, 1 mM MgSO4, 1.25 mM CaCl2, 20 mM HEPES; pH 7.4) and incubated with the same for 10 min at room temperature. The uptake was then initiated by incubating the cells for 30 secs with Na-HEPES buffer (130 mM NaCl, 4.7 mM KCl, 1 mM MgSO4, 1.25 mM CaCl2, 20 mM HEPES; pH 7.4) with 10 μCi of 3H-L-Leucine (PerkinElmer; Waltham, MA) and 10 μM L-Leucine (Sigma-Aldrich). The reaction was stopped with ice cold Na-HEPES buffer after which the cells were washed twice with the same ice-cold buffer. The cells were then lysed in 500 μl of 1 N NaOH followed by incubation for 20 min at 70° C. The lysed contents of each well was collected in a 7ml scintillation tube and mixed with 5 ml Ecoscint A (National Diagnostics; Atlanta, GA). Leucine uptake experiments were conducted using chemicals obtained from Sigma-Aldrich. The vials were kept in the dark for 48h and the radioactivity was determined in a Beckman 6500 scintillation counter.
2.8. MDA-MB-231 proliferation experiments
20,000 MDA-MB-231 or 10,000 MCF-7 cells in 1mL of DMEM + 10% FBS were mixed directly with siRNA's (100 nM) that were non-targeting, AHR-targeting or LAT1-targeting and 3 μL of RNAiMax Transfection Reagent. The cells were then plated into 96 well plates at a density of 2000 MDA-MB-231 or 1000 MCF-7 cells per well. After 3 days, cell proliferation was assayed with the Aqueous One Solution Cell Proliferation Assay (Promega, Madison, WI) in accordance with the manufacturer's protocol.
3. Results
3.1 TCDD RNA-Seq
RNA-Seq analysis revealed that TCDD-regulated the expression of 137 genes in MCF-7 BCCs at an FDR < 5%, with all fold changes greater than or equal to 1.3 compared with vehicle. Of these 137 TCDD-regulated genes (TRGs), 116 were mapped to known functions by IPA. Comparison of TRGs with a published TCDD/AHR-ChIP-Seq data [36] revealed that 47 genes that were shared between the two gene sets (Fig. 1). These 47 genes included known TCDD target genes such as CYP1A1 [38], CYP1B1 [38] and ALDH3A1 [39] (Fig. 1). Bioinformatic analysis revealed that the 47 TCDD target genes were significantly associated with metabolic pathways including: lipid metabolism, carbohydrate metabolism, nucleic acid metabolism, vitamin and mineral metabolism and energy production (Table 1). The 47 TCDD target genes were also associated with cancer processes including: cell death and survival, cell cycle, cellular growth and proliferation, and molecular transport (Table 1).
Fig. 1. Genes common between TCDD RNA-Seq data and TCDD-ChIP-Seq data.
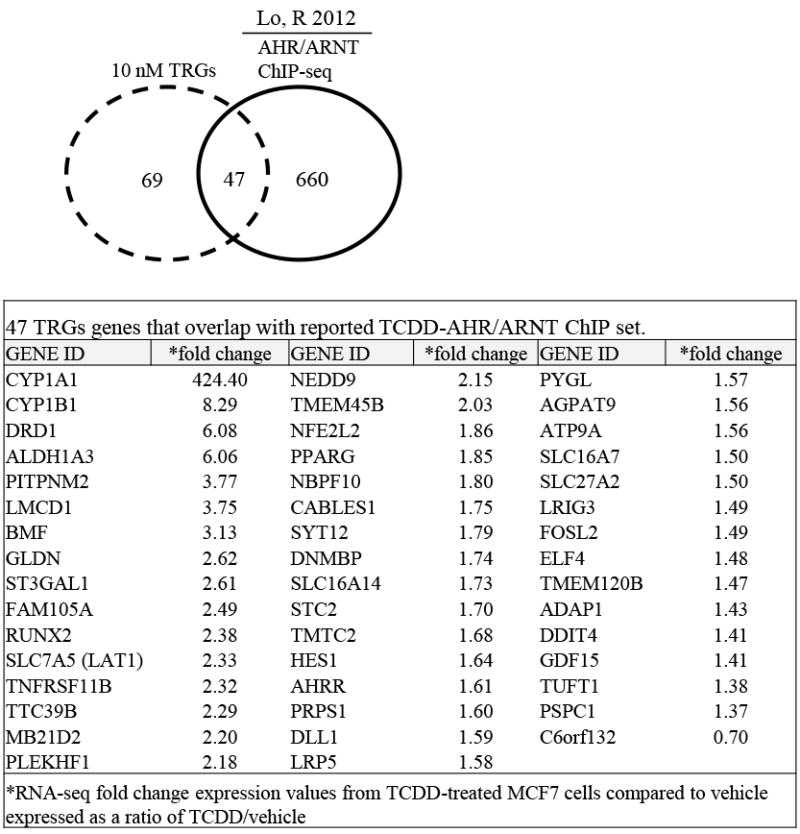
Analysis of reported MCF-7 TCDD-ChIP-Seq data revealed that 47 of the 116 TCDD-RNA-Seq genes were TCDD-AHR/ARNT bound genes. The specific 47 TCDD-regulated genes and their associated fold changes are shown in the table.
Table 1.
IPA cellular and molecular functions associated with the 47 TRGs that overlap with reported TCDD-AHR/ARNT ChlP-seq.
| Category | *B-H p-value | Target molecules in dataset |
|---|---|---|
| Lipid Metabolism | 8.55E-03-3.52E-02 | 14 |
| Small Molecule Biochemistry | 8.55E-03-4.11E-02 | 26 |
| Cell Death and Survival | 8.55E-03-4.11E-02 | 30 |
| Gene Expression | 8.55E-03-3.2E-02 | 12 |
| Cell Cycle | 8.55E-03-3.91E-02 | 16 |
| Cellular Development | 8.55E-03-3.54E-02 | 37 |
| Cellular Growth and Proliferation | 8.55E-03-3.93E-02 | 42 |
| Carbohydrate Metabolism | 8.55E-03-3.2E-02 | 9 |
| Drug Metabolism | 8.55E-03-3.71E-02 | 10 |
| Energy Production | 8.55E-03-3.27E-02 | 8 |
| Molecular Transport | 8.55E-03-3.71E-02 | 15 |
| Vitamine and Mineral Metabolism | 9.07E-03-3.2E-02 | 8 |
| Nucleic Acid Metabolism | 9.68E-03-3.2E-02 | 4 |
p-values are calculated by Fishers exact test and corrected for multiple testing by the Benjamini- Hochberger p-values (B-H) method (B-H p-value). Column 2 shows the range of B-H corrected p-values for the biofunctions in a given category. Target molecules in dataset are the number of RNA-Seq TRGs in a given biofunction.
3.1 TCDD/AHR regulation of LAT1 and leucine uptake
The TCDD-RNA-Seq indicated that LAT1 (also known as SLC7A5) was induced 2.33-fold by TCDD (Table 1), while TCDD-ChIP-Seq evidence suggested that AHR could directly bind to intron 2 of the LAT1 gene [36]. We decided to further investigate extrinsic regulation of LAT1 by TCDD, considering that prior reports indicate that upregulation of LAT1 could be important for breast cancer progression [40–42]. To investigate whether TCDD increases LAT1 expression through AHR, MCF-7 cells were transfected with short interfering RNA against AHR (AHRi). Control MCF-7 cells were transfected with non-targeting siRNA (cRNAi). siRNA-treated cells were then exposed to either vehicle or TCDD (10 nM) for 6 hr. Reductions in AHR mRNA were confirmed in MCF-7 cells transfected with AHRi compared with those transfected with cRNAi (Fig. 2A). As expected, TCDD-stimulated increases (3-fold) in LAT1 mRNA compared with vehicle in MCF-7 cells transfected with cRNAi (Fig. 2B). In contrast, AHRi significantly reduced the stimulatory effect of TCDD on LAT1 expression (P < 0.01; Fig. 2B). These results indicate that LAT1 regulation by TCDD is mediated by AHR.
Fig. 2. AHR mediates TCDD-stimulated increases in LAT1 mRNA.
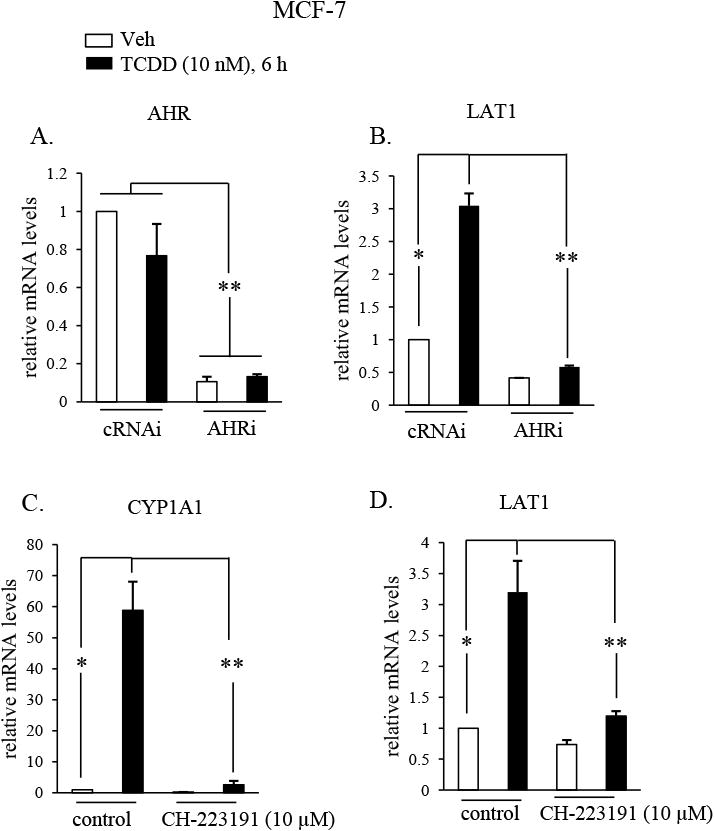
A & B, MCF-7 cells were transfected with siRNAs that were either non-targeting (cRNAi) or AHR targeting (AHRi) and then treated with DMSO vehicle (Veh) or TCDD (10 nM) for 6 hrs. C & D, MCF-7 cells were treated vehicle or TCDD (10 nM) in the absence (controls) or presence of CH-223191 (10 μM) for 6 hrs. A-D, AHR, LAT1, CYP1A1 or GAPDH mRNA were quantified by qRT-PCR from total RNA. GAPDH mRNA levels were used to normalize samples. *, P < 0.05; **, P > 0.01. Data shown are the means ± S.E. of three independent experiments.
To investigate whether TCDD increases LAT1 expression by binding to AHR, MCF-7 cells were treated with CH-221391, which is an AHR antagonist that specifically inhibits the binding of TCDD to AHR [43]. TCDD induction of CYP1A1 transcription is a commonly used readout of TCDD-induced AHR activity that requires TCDD to bind AHR. To verify that CH-221391 is an AHR antagonist in MCF-7 cells, its ability to suppress induction of CYP1A1 by TCDD was measured by RT-qPCR. The findings revealed that TCDD-induced increases in CYP1A1 were indeed reduced by CH-221391, indicating that it is an effective AHR antagonist in MCF-7 cells (Fig. 2C). As expected, TCDD-stimulated increases (∼3-fold) in LAT1 mRNA in control MCF-7 cells not treated with CH-221391 (Fig. 2D). In contrast, CH-221391 significantly (P < 0.01) suppressed the stimulatory effect of TCDD on LAT1 expression (Fig. 2D). These results indicate that regulation of LAT1 by TCDD requires TCDD to bind AHR.
Western blot experiments were conducted to confirm that TCDD induction of LAT1 mRNA leads to increases in LAT1 protein. Exposure to TCDD stimulated robust increases (∼10-fold) in LAT1 protein in MCF-7 cells transfected with cRNAi (Fig. 3A & B). Induction of LAT1 protein by TCDD was completely suppressed in MCF-7 cells transfected with AHRi (Fig. 3A & B). As expected, AHR protein levels in MCF-7 cells transfected with AHRi was lower than those transfected with cRNAi (Fig. 3A & C). Leucine uptake experiments were performed to investigate if the induction of LAT1 protein by TCDD coincided with increases in leucine uptake by MCF-7 cells. Significant increases in leucine uptake by cells was observed in response to TCDD exposure compared with vehicle-treated cells (Fig. 4). Taken together, these results indicate that the induction of LAT1 protein by TCDD leads to a functional increase in leucine uptake by MCF-7 cells.
Fig. 3. AHR mediates TCDD-stimulated increases in LAT1 protein.
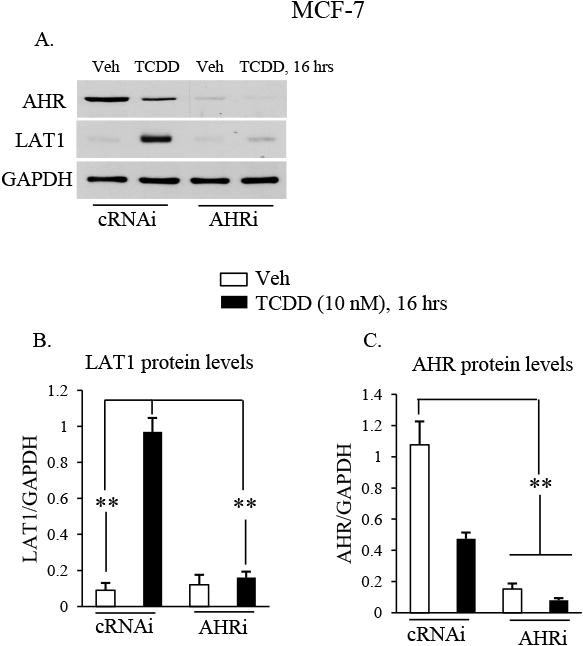
A-C, MCF-7 cells were transfected with siRNAs that were either non-targeting (cRNAi) or AHR targeting (AHRi) and then treated with vehicle (Veh) or TCDD (10 nM) for 16 hrs. Total cellular protein was then isolated and subjected to Western blot analysis. The blot was then probed with the indicated antibodies. Relative levels of AHR or LAT1 protein was expressed as a ratio to GAPDH loading control. *, P < 0.05; **, P > 0.01. Data shown are the means ± S.E. of three independent experiments.
Fig. 4. TCDD increases leucine uptake.
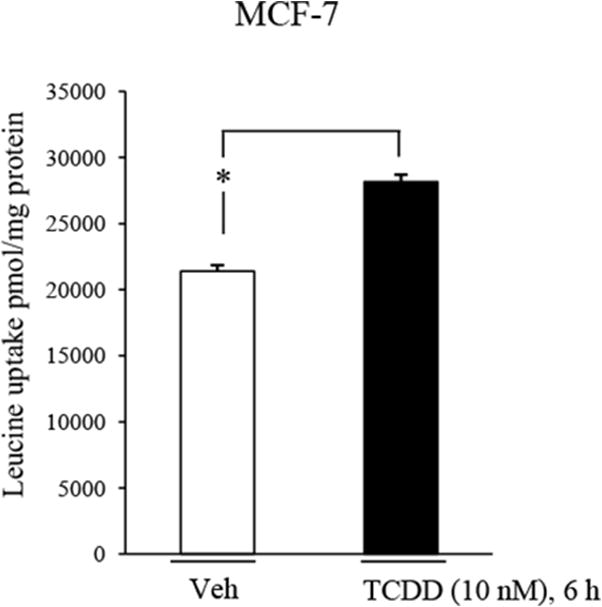
MCF-7 cells were treated with vehicle or 10 nM TCDD for 6 hrs, followed by analysis of leucine uptake as detailed in the material and methods. *, P < 0.05. Data shown are the means ± S.E. of three independent experiments.
3.2 TCDD-induced AHR/ARNT/p300 recruitment to an AHR binding site in the LAT1 gene
A prior report by Lo and Matthews (2012) identified an AHR binding site in intron 2 of the LAT1 gene by TCDD-ChIP-seq analysis in MCF-7 cells [36]. In order to characterize the mechanism of LAT1 induction, we measured AHR recruitment to the AHR response elements (AHR-REs) in LAT1 intron 2 by ChIP-qPCR analysis. The results showed a significant (P < 0.05) 4-fold increase in AHR binding to the intron 2 site in response to TCDD compared with vehicle (Fig. 5A). In accordance with its known mechanism of action, TCDD-increased (by 37-fold) AHR binding to AHR-REs that are located upstream from the CYP1B1 transcription start site (Fig. 5D).
Fig. 5. TCDD-stimulated AHR/ARNT/p300 recruitment and histone H3 acetylation at an AHR binding site in the LAT1 gene.
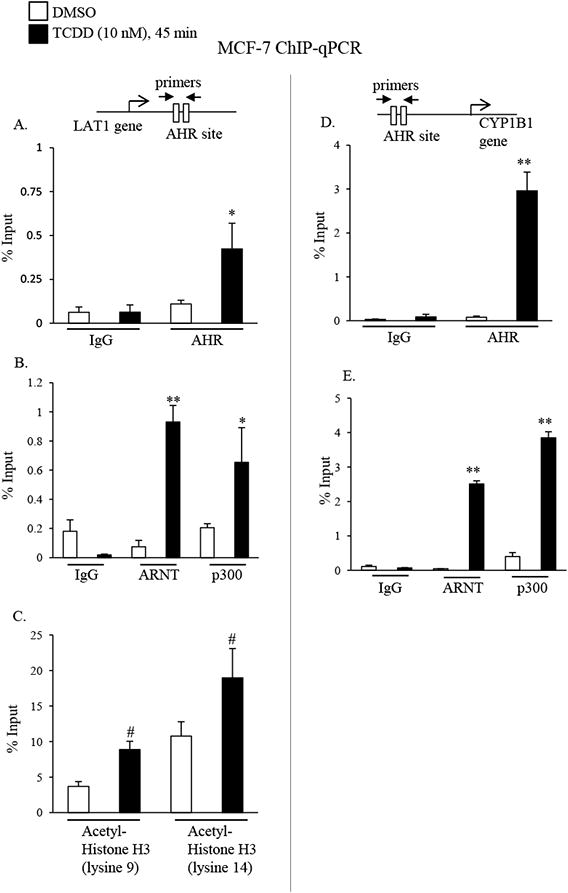
A-E, MCF-7 cells were treated with DMSO or 10 nM TCDD for 45 minutes. Cells were then subjected to ChIP with non-specific IgG (IgG), or AHR, ARNT, p300, acetylated histone H3 (lysine 9 or 14) targeting antibodies, followed by qPCR amplification of the AHR binding site in the LAT1 (A-C) or CYP1B1 (D & E) genes. Significant increases in AHR, ARNT, or p300 binding to AHR binding sites in the LAT1 (A-C) or CYP1B1 (D & E) gene by TCDD are indicated by *, P < 0.05, or **, P < 0.01. C, Significant increases in the acetylation of lysine 9 or 14 in histone H3 at the AHR binding site in the LAT1 gene by TCDD are indicated by #, P < 0.05. Data shown are the means ± S.E. of three independent experiments.
By binding to AHR, the ARNT transcription factor promotes AHR recruitment to AHR-REs in TCDD target genes [10, 12]. We assessed ARNT binding to the LAT1 intron 2 and CYP1B1 using ChIP-qPCR analysis and the findings revealed its recruitment to AHR-REs in the LAT1 and CYP1B1 genes was increased by 12- and 55-fold, respectively in response to TCDD (Fig. 5B & E). Prior reports indicate that AHR/ARNT heterodimers recruit the p300 transcription complex to TCDD target genes [14]. The AHR-REs in LAT1 and CYP1B1 genes exhibited 3-fold and 10-fold increases, respectively, in p300 binding in response to TCDD compared with vehicle (Fig. 5B & E).
The p300 complex has inherent histone acetylase activity and is known to increase in histone H3 acetylation at lysine 9 and lysine 14, both of which are markers of active transcription [44]. Consistent with its physical recruitment to the LAT1 AHR-RE, we observed increases in the acetylation of histone H3 at lysine 9 and lysine 14 (by 2.4 and 1.8-fold, respectively) at the AHR-RE in response to TCDD (Fig. 5C). Collectively, these data indicate that extrinsic regulation of LAT1 by TCDD is mediated via the AHR binding and recruitment of p300 to the AHR-RE in the LAT1 gene.
3.3 Endogenous regulation of LAT1 by AHR promotes MCF-7 and MDA-MB-231 proliferation
Intrinsic regulation of LAT1 by AHR was investigated in MCF-7 and MDA-MB-231 cells because these BCC lines have been reported to exhibit endogenous AHR activity [30-33]. Western blot analysis revealed basal AHR and LAT1 protein expression in MCF-7 (Fig. 6A) and MDA-MB-231 (Fig. 6C) cells transfected with cRNAi. Transfection with AHRi reduced AHR protein in MCF-7 (Fig. 6A) and MDA-MB-231 cells (Fig. 6C) to levels that were not detected by standard Western blot analysis. Reducing AHR protein expression with AHRi also suppressed the levels of LAT1 protein in MCF-7 (by ∼70%) (Fig. 6A) and MDA-MB-231 (by ∼60%) (Fig. 6C) and the levels of LAT1 mRNA in MCF-7 (by ∼50%) (Fig. 6B) or MDA-MB-231 (by ∼40%) (Fig. 6D) compared with controls. These data indicate that endogenous AHR activity regulates LAT1 expression in MCF-7 and MDA-MB-231 cells.
Fig. 6. Endogenous regulation of LAT1 by AHR in MCF-7 and MDA-MB-231 cells.
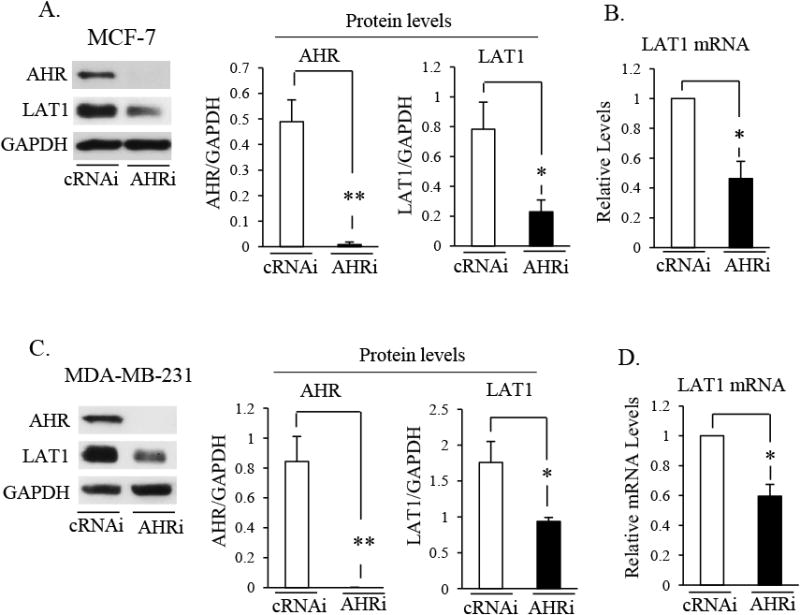
A-D, MCF-7 (A & B) or MDA-MB-231 (C & D) cells were transfected with control (cRNAi) or AHR (AHRi) siRNA for 72 or 48 hrs, respectively. A & C, Total cellular protein was then isolated and subjected to Western blot analysis. Blots were then probed with the indicated antibodies. Relative levels of AHR or LAT1 protein was expressed as a ratio of AHR/GAPDH or LAT1/GAPDH, respectively. *, P < 0.05; **, P >0.01. B & D, RT-qPCR analyses of LAT1 mRNA levels in MCF-7 (B) or MDA-MB-231 (D) cells transfected with cRNAi or AHRi. GAPDH mRNA levels were used to normalize samples. *, P < 0.05. A-D, Data shown are the means ± S.E. of three independent experiments.
Since LAT1 is known to promote cancer cell proliferation by stimulating the amino acid uptake [27], we sought to determine if AHR expression and its regulation of LAT1 in MCF-7 and MDA-MB-231 are important for proliferation. To this end, MCF-7 and MDA-MD-231 cells were transfected with cRNAi, AHRi or LAT1-targeting siRNA (LAT1i). After 3 days, significant reductions in proliferation was observed in MCF-7 (Fig. 7A) and MDA-MB-231 (Fig. 7B) cells expressing LAT1i or AHRi compared with those transfected with cRNAi. These findings suggest that AHR regulation of LAT1 and LAT1 expression are important for the proliferation of MCF-7 and MDA-MB-231 cells.
Fig. 7. AHR and LAT1 promote MCF-7 and MDA-MB-231 proliferation.
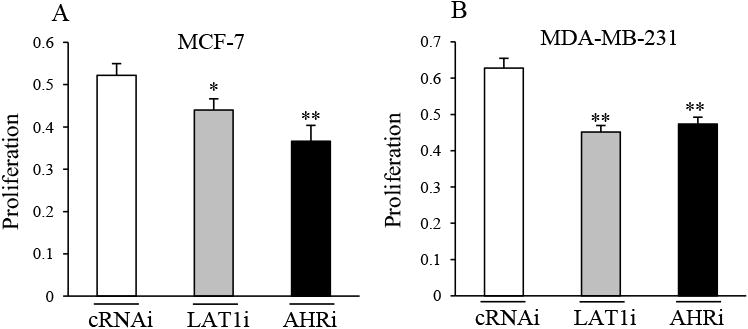
A & B, MCF-7 (A) or MDA-MB-231 (B) cells were transfected with cRNAi, LAT1i or AHRi for 3 days, followed by analysis of proliferation (please see methods for details). Significant decreases in proliferation by LAT1i or AHRi are indicated by *P < 0.05, or **P < 0.01. Data shown are the means ± S.E. of nine replicates.
4. Discussion
The findings of this report provide new insight into extrinsic and intrinsic regulation of LAT1 by AHR. Reducing AHR with AHRi suppressed extrinsic regulation of LAT1 by TCDD in MCF-7 cells (Fig. 2 & 3) and intrinsic regulation of LAT1 in MCF-7 and MDA-MB-231 cells (Fig. 6). These findings indicate that AHR regulates LAT1 expression. ChIP-qPCR results indicate that extrinsic regulation of LAT1 by TCDD is mediated via the AHR binding site in the LAT1 gene (Fig. 5). Indeed, the binding of AHR/ARNT/p300 and the acetylation of histone H3 at the AHR site in the LAT1 gene was increased by TCDD (Fig. 5). Consistent with reports showing that AHR promotes MCF-7 and MDA-MB-231 cancer processes [30, 34], our proliferation assays indicate that reducing AHR suppressed their proliferation (Fig. 7).
Prior reports have provided important insights into amino acid uptake by other transporters in MCF-7 and MDA-MB-231 cells. Karunakaran et al (2011) demonstrated that SLC6A14 (also known as Atbo,+) is a Na+ dependent, estrogen-induced transporter that mediates the uptake of all essential amino acids, including leucine, in MCF-7 cells [45]. The SLC6A14 inhibitor α-methyl-DL-tryptophan stimulated apoptosis of MCF-7, but not MDA-MB-231 cells, which was attributed to selective expression of SLC6A14 in MCF-7, but not MDA-MB-231 cells [45]. Shennan et al (2003) established that MCF-7, but not MDA-MB-231 cells express LAT2, which is an isoform of LAT1 that also mediates leucine uptake [46]. Our observation that MCF-7 cells exhibit high basal leucine uptake activity in the absence of TCDD can be explained by the transporter activity of SLC6A14 and LAT2 as well as basal LAT1 activity (Fig. 4). Since TCDD induction of SLC6A14 and LAT2 mRNA was not observed in our RNA-Seq data, we conclude that the increase in leucine uptake in the presence of TCDD in MCF-7 cells is mediated via increased expression of LAT1.
CYP1A1 and CYP1B1 harbor upstream AHR-REs within 1 kb of their transcription start sites [37, 47, 48]. Reported TCDD-ChIP-seq data indicated that the LAT1 gene lacks promoter AHR-REs [36], but its expression is regulated by an AHR binding site located in intron 2 (Fig. 5), which is 29 kb from the LAT1 promoter. Although long distance regulation of gene promoters by AHR-REs is relatively novel for TCDD, it is not uncommon for gene promoters to be regulated by distal enhancers [49].
AHR stimulation of transcription may rely on endogenous ligands. D'Amato et al (2015) demonstrated that MDA-MB-231 cells synthesize kynurenine, which is a tryptophan metabolite and a known endogenous AHR ligand [30]. Production of kynurenine by MDA-MB-231 cells is mediated by tryptophan 2,3-dioxygenase (TDO2), which is the first and rate-limiting enzyme in the kynurenine synthesis pathway [30]. Thus, the observed AHR activity that is required for LAT1 expression in MDA-MB-231 cells could be attributed to kynurenine interacting with AHR. However, endogenous AHR activity could also reflect its interaction with the other tryptophan metabolites that also function as AHR agonists such as kynurenic acid or xanthurenic acid [21]. Regulation of LAT1 by unliganded AHR is also a possibility, considering that AHR can be regulated by cyclic AMP [50, 51].
Previous reports have provided important evidence that breast cancer progression may require LAT1. For instance, LAT1 expression is upregulated in human breast tumors compared with normal breast tissue [40]. Shennan et al (2008) demonstrated that 2-aminobicyclo-(2,2,1)-heptane-2-carboxylic acid (BCH), which inhibits L type transporters, suppressed the proliferation of MCF-7, MDA-MB-231 and ZR-75-1 BCCs [41]. Our findings now establish that specifically knocking down the expression of LAT1 with siRNA inhibits proliferation of MCF-7 and MDA-MB-231 cells (Fig. 7).
In addition to breast cancer cells, normal cells that require high levels of amino acid uptake for their proliferation and differentiation have been reported to express LAT1 and AHR. In this regard, leucine uptake by activated T cells is mediated through LAT1 and LAT1 null T cells exhibit defects in proliferation and effector activity [52-54]. As noted in the introduction, AHR promotes the differentiation of Tregs and Th17 cells in response to TCDD or FICZ, respectively [16-18]. Given these prior reports, it is possible that in addition to carcinogenesis, the induction of LAT1 by AHR may promote the proliferation and differentiation of Tregs or Th17 cells depending on which AHR ligand is present.
Acknowledgments
This work was supported by a grant from the Appalachian Clinical and Translational Science Institute, Marshall University Joan C Edwards School of Medicine, the WV-INBRE grant (P20GM103434) and support from the Marshall University Genomics and Bioinformatics Core. This work was also supported by the NASA-WV Space Grant Consortium Graduate Research Fellowship Program (J.K.T).
Footnotes
Competing Interests: The authors declare that they have no competing interests
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Justin K. Tomblin, Email: tomblin36@marshall.edu.
Subha Arthur, Email: arthursu@marshall.edu.
Donald A. Primerano, Email: primeran@marshall.edu.
Ateeq R. Chaudhry, Email: chaudhry@marshall.edu.
Jun Fan, Email: fanj@marshall.edu.
James Denvir, Email: denvir@marshall.edu.
References
- 1.Sorg O. AhR signalling and dioxin toxicity. Toxicology letters. 2014;230:225–33. doi: 10.1016/j.toxlet.2013.10.039. [DOI] [PubMed] [Google Scholar]
- 2.Poland A, Knutson JC. 2,3,7,8-tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: examination of the mechanism of toxicity. Annual review of pharmacology and toxicology. 1982;22:517–54. doi: 10.1146/annurev.pa.22.040182.002505. [DOI] [PubMed] [Google Scholar]
- 3.Van den Berg M, Birnbaum LS, Denison M, De Vito M, Farland W, Feeley M, et al. The 2005 World Health Organization reevaluation of human and Mammalian toxic equivalency factors for dioxins and dioxin-like compounds. Toxicological sciences : an official journal of the Society of Toxicology. 2006;93:223–41. doi: 10.1093/toxsci/kfl055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perdew GH. Association of the Ah receptor with the 90-kDa heat shock protein. The Journal of biological chemistry. 1988;263:13802–5. [PubMed] [Google Scholar]
- 5.Kazlauskas A, Sundstrom S, Poellinger L, Pongratz I. The hsp90 chaperone complex regulates intracellular localization of the dioxin receptor. Molecular and cellular biology. 2001;21:2594–607. doi: 10.1128/MCB.21.7.2594-2607.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carver LA, Bradfield CA. Ligand-dependent interaction of the aryl hydrocarbon receptor with a novel immunophilin homolog in vivo. The Journal of biological chemistry. 1997;272:11452–6. doi: 10.1074/jbc.272.17.11452. [DOI] [PubMed] [Google Scholar]
- 7.Meyer BK, Pray-Grant MG, Vanden Heuvel JP, Perdew GH. Hepatitis B virus X-associated protein 2 is a subunit of the unliganded aryl hydrocarbon receptor core complex and exhibits transcriptional enhancer activity. Molecular and cellular biology. 1998;18:978–88. doi: 10.1128/mcb.18.2.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer BK, Perdew GH. Characterization of the AhR-hsp90-XAP2 core complex and the role of the immunophilin-related protein XAP2 in AhR stabilization. Biochemistry. 1999;38:8907–17. doi: 10.1021/bi982223w. [DOI] [PubMed] [Google Scholar]
- 9.Kazlauskas A, Poellinger L, Pongratz I. Evidence that the co-chaperone p23 regulates ligand responsiveness of the dioxin (Aryl hydrocarbon) receptor. The Journal of biological chemistry. 1999;274:13519–24. doi: 10.1074/jbc.274.19.13519. [DOI] [PubMed] [Google Scholar]
- 10.Reyes H, Reisz-Porszasz S, Hankinson O. Identification of the Ah receptor nuclear translocator protein (Arnt) as a component of the DNA binding form of the Ah receptor. Science (New York, NY) 1992;256:1193–5. doi: 10.1126/science.256.5060.1193. [DOI] [PubMed] [Google Scholar]
- 11.Denison MS, Soshilov AA, He G, DeGroot DE, Zhao B. Exactly the same but different: promiscuity and diversity in the molecular mechanisms of action of the aryl hydrocarbon (dioxin) receptor. Toxicological sciences : an official journal of the Society of Toxicology. 2011;124:1–22. doi: 10.1093/toxsci/kfr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beischlag TV, Luis Morales J, Hollingshead BD, Perdew GH. The aryl hydrocarbon receptor complex and the control of gene expression. Critical reviews in eukaryotic gene expression. 2008;18:207–50. doi: 10.1615/critreveukargeneexpr.v18.i3.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beischlag TV, Wang S, Rose DW, Torchia J, Reisz-Porszasz S, Muhammad K, et al. Recruitment of the NCoA/SRC-1/p160 family of transcriptional coactivators by the aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator complex. Molecular and cellular biology. 2002;22:4319–33. doi: 10.1128/MCB.22.12.4319-4333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor RT, Wang F, Hsu EL, Hankinson O. Roles of coactivator proteins in dioxin induction of CYP1A1 and CYP1B1 in human breast cancer cells. Toxicological sciences : an official journal of the Society of Toxicology. 2009;107:1–8. doi: 10.1093/toxsci/kfn217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang S, Hankinson O. Functional involvement of the Brahma/SWI2-related gene 1 protein in cytochrome P4501A1 transcription mediated by the aryl hydrocarbon receptor complex. The Journal of biological chemistry. 2002;277:11821–7. doi: 10.1074/jbc.M110122200. [DOI] [PubMed] [Google Scholar]
- 16.Mezrich JD, Fechner JH, Zhang X, Johnson BP, Burlingham WJ, Bradfield CA. An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. Journal of immunology (Baltimore, Md : 1950) 2010;185:3190–8. doi: 10.4049/jimmunol.0903670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quintana FJ, Basso AS, Iglesias AH, Korn T, Farez MF, Bettelli E, et al. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008;453:65–71. doi: 10.1038/nature06880. [DOI] [PubMed] [Google Scholar]
- 18.Veldhoen M, Hirota K, Westendorf AM, Buer J, Dumoutier L, Renauld JC, et al. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–9. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- 19.Kiss EA, Vonarbourg C, Kopfmann S, Hobeika E, Finke D, Esser C, et al. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science (New York, NY) 2011;334:1561–5. doi: 10.1126/science.1214914. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Innocentin S, Withers DR, Roberts NA, Gallagher AR, Grigorieva EF, et al. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell. 2011;147:629–40. doi: 10.1016/j.cell.2011.09.025. [DOI] [PubMed] [Google Scholar]
- 21.DiNatale BC, Murray IA, Schroeder JC, Flaveny CA, Lahoti TS, Laurenzana EM, et al. Kynurenic acid is a potent endogenous aryl hydrocarbon receptor ligand that synergistically induces interleukin-6 in the presence of inflammatory signaling. Toxicological sciences : an official journal of the Society of Toxicology. 2010;115:89–97. doi: 10.1093/toxsci/kfq024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lahoti TS, Boyer JA, Kusnadi A, Muku GE, Murray IA, Perdew GH. Aryl Hydrocarbon Receptor Activation Synergistically Induces Lipopolysaccharide-Mediated Expression of Proinflammatory Chemokine (c-c motif) Ligand 20. Toxicological sciences : an official journal of the Society of Toxicology. 2015;148:229–40. doi: 10.1093/toxsci/kfv178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vogel CF, Khan EM, Leung PS, Gershwin ME, Chang WL, Wu D, et al. Cross-talk between aryl hydrocarbon receptor and the inflammatory response: a role for nuclear factor-kappaB. The Journal of biological chemistry. 2014;289:1866–75. doi: 10.1074/jbc.M113.505578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanai Y, Segawa H, Miyamoto K, Uchino H, Takeda E, Endou H. Expression cloning and characterization of a transporter for large neutral amino acids activated by the heavy chain of 4F2 antigen (CD98) The Journal of biological chemistry. 1998;273:23629–32. doi: 10.1074/jbc.273.37.23629. [DOI] [PubMed] [Google Scholar]
- 25.Segawa H, Fukasawa Y, Miyamoto K, Takeda E, Endou H, Kanai Y. Identification and functional characterization of a Na+-independent neutral amino acid transporter with broad substrate selectivity. The Journal of biological chemistry. 1999;274:19745–51. doi: 10.1074/jbc.274.28.19745. [DOI] [PubMed] [Google Scholar]
- 26.Yanagida O, Kanai Y, Chairoungdua A, Kim DK, Segawa H, Nii T, et al. Human L-type amino acid transporter 1 (LAT1): characterization of function and expression in tumor cell lines. Biochimica et biophysica acta. 2001;1514:291–302. doi: 10.1016/s0005-2736(01)00384-4. [DOI] [PubMed] [Google Scholar]
- 27.Wang Q, Holst J. L-type amino acid transport and cancer: targeting the mTORC1 pathway to inhibit neoplasia. American journal of cancer research. 2015;5:1281–94. [PMC free article] [PubMed] [Google Scholar]
- 28.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–93. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarkar D, Kambe F, Hirata A, Iseki A, Ohmori S, Seo H. Expression of E16/CD98LC/hLAT1 is responsive to 2,3,7,8-tetrachlorodibenzo-p-dioxin. FEBS letters. 1999;462:430–4. doi: 10.1016/s0014-5793(99)01574-4. [DOI] [PubMed] [Google Scholar]
- 30.D'Amato NC, Rogers TJ, Gordon MA, Greene LI, Cochrane DR, Spoelstra NS, et al. A TDO2-AhR Signaling Axis Facilitates Anoikis Resistance and Metastasis in Triple-Negative Breast Cancer. Cancer research. 2015;75:4651–64. doi: 10.1158/0008-5472.CAN-15-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goode G, Ballard BR, Manning HC, Freeman ML, Kang Y, Eltom SE. Knockdown of aberrantly upregulated aryl hydrocarbon receptor reduces tumor growth and metastasis of MDA-MB-231 human breast cancer cell line. International journal of cancer Journal international du cancer. 2013 doi: 10.1002/ijc.28297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goode G, Pratap S, Eltom SE. Depletion of the aryl hydrocarbon receptor in MDA-MB-231 human breast cancer cells altered the expression of genes in key regulatory pathways of cancer. PloS one. 2014;9:e100103. doi: 10.1371/journal.pone.0100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salisbury TB, Tomblin JK, Primerano DA, Boskovic G, Fan J, Mehmi I, et al. Endogenous aryl hydrocarbon receptor promotes basal and inducible expression of tumor necrosis factor target genes in MCF-7 cancer cells. Biochemical pharmacology. 2014 doi: 10.1016/j.bcp.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomblin JK, Salisbury TB. Insulin like growth factor 2 regulation of aryl hydrocarbon receptor in MCF-7 breast cancer cells. Biochemical and biophysical research communications. 2014;443:1092–6. doi: 10.1016/j.bbrc.2013.12.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif) 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Lo R, Matthews J. High-resolution genome-wide mapping of AHR and ARNT binding sites by ChIP-Seq. Toxicological sciences : an official journal of the Society of Toxicology. 2012;130:349–61. doi: 10.1093/toxsci/kfs253. [DOI] [PubMed] [Google Scholar]
- 37.Matthews J, Wihlen B, Thomsen J, Gustafsson JA. Aryl hydrocarbon receptor-mediated transcription: ligand-dependent recruitment of estrogen receptor alpha to 2,3,7,8-tetrachlorodibenzo-p-dioxin-responsive promoters. Molecular and cellular biology. 2005;25:5317–28. doi: 10.1128/MCB.25.13.5317-5328.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spink DC, Spink BC, Cao JQ, DePasquale JA, Pentecost BT, Fasco MJ, et al. Differential expression of CYP1A1 and CYP1B1 in human breast epithelial cells and breast tumor cells. Carcinogenesis. 1998;19:291–8. doi: 10.1093/carcin/19.2.291. [DOI] [PubMed] [Google Scholar]
- 39.Vasiliou V, Reuter SF, Williams S, Puga A, Nebert DW. Mouse cytosolic class 3 aldehyde dehydrogenase (Aldh3a1): gene structure and regulation of constitutive and dioxin-inducible expression. Pharmacogenetics. 1999;9:569–80. [PubMed] [Google Scholar]
- 40.Liang Z, Cho HT, Williams L, Zhu A, Liang K, Huang K, et al. Potential Biomarker of L-type Amino Acid Transporter 1 in Breast Cancer Progression. Nuclear medicine and molecular imaging. 2011;45:93–102. doi: 10.1007/s13139-010-0068-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shennan DB, Thomson J. Inhibition of system L (LAT1/CD98hc) reduces the growth of cultured human breast cancer cells. Oncology reports. 2008;20:885–9. [PubMed] [Google Scholar]
- 42.Shennan DB, Thomson J, Gow IF, Travers MT, Barber MC. L-leucine transport in human breast cancer cells (MCF-7 and MDA-MB-231): kinetics, regulation by estrogen and molecular identity of the transporter. Biochimica et biophysica acta. 2004;1664:206–16. doi: 10.1016/j.bbamem.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 43.Kim SH, Henry EC, Kim DK, Kim YH, Shin KJ, Han MS, et al. Novel compound 2-methyl-2H-pyrazole-3-carboxylic acid (2-methyl-4-o-tolylazo-phenyl)-amide (CH-223191) prevents 2,3,7,8-TCDD- induced toxicity by antagonizing the aryl hydrocarbon receptor. Molecular pharmacology. 2006;69:1871–8. doi: 10.1124/mol.105.021832. [DOI] [PubMed] [Google Scholar]
- 44.Grant PA. A tale of histone modifications. Genome biology. 2001;2 doi: 10.1186/gb-2001-2-4-reviews0003. Reviews0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karunakaran S, Ramachandran S, Coothankandaswamy V, Elangovan S, Babu E, Periyasamy-Thandavan S, et al. SLC6A14 (ATB0,+) protein, a highly concentrative and broad specific amino acid transporter, is a novel and effective drug target for treatment of estrogen receptor-positive breast cancer. The Journal of biological chemistry. 2011;286:31830–8. doi: 10.1074/jbc.M111.229518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shennan DB, Thomson J, Barber MC, Travers MT. Functional and molecular characteristics of system L in human breast cancer cells. Biochimica et biophysica acta. 2003;1611:81–90. doi: 10.1016/s0005-2736(03)00028-2. [DOI] [PubMed] [Google Scholar]
- 47.Kress S, Reichert J, Schwarz M. Functional analysis of the human cytochrome P4501A1 (CYP1A1) gene enhancer. European journal of biochemistry / FEBS. 1998;258:803–12. doi: 10.1046/j.1432-1327.1998.2580803.x. [DOI] [PubMed] [Google Scholar]
- 48.Tsuchiya Y, Nakajima M, Yokoi T. Critical enhancer region to which AhR/ARNT and Sp1 bind in the human CYP1B1 gene. Journal of biochemistry. 2003;133:583–92. doi: 10.1093/jb/mvg075. [DOI] [PubMed] [Google Scholar]
- 49.Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–4. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oesch-Bartlomowicz B, Oesch F. Role of cAMP in mediating AHR signaling. Biochemical pharmacology. 2009;77:627–41. doi: 10.1016/j.bcp.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 51.Oesch-Bartlomowicz B, Huelster A, Wiss O, Antoniou-Lipfert P, Dietrich C, Arand M, et al. Aryl hydrocarbon receptor activation by cAMP vs. dioxin: divergent signaling pathways. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:9218–23. doi: 10.1073/pnas.0503488102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hayashi K, Jutabha P, Endou H, Sagara H, Anzai N. LAT1 is a critical transporter of essential amino acids for immune reactions in activated human T cells. Journal of immunology (Baltimore, Md : 1950) 2013;191:4080–5. doi: 10.4049/jimmunol.1300923. [DOI] [PubMed] [Google Scholar]
- 53.Nii T, Segawa H, Taketani Y, Tani Y, Ohkido M, Kishida S, et al. Molecular events involved in up-regulating human Na+-independent neutral amino acid transporter LAT1 during T-cell activation. The Biochemical journal. 2001;358:693–704. doi: 10.1042/0264-6021:3580693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sinclair LV, Rolf J, Emslie E, Shi YB, Taylor PM, Cantrell DA. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nature immunology. 2013;14:500–8. doi: 10.1038/ni.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]


