Abstract
Restoring chromatin structure with high fidelity after mitosis is critical for cell survival. Transcriptional reactivation of genes is the first step toward establishing identity of the daughter cell. During mitosis, chromatin bookmarking factors associated with specific chromatin regions ensure the restoration of the original gene expression pattern in daughter cells. Recent findings have provided new insights into the mechanisms, regulation, and biological significance of gene bookmarking in eukaryotes. In this review, we discuss how epigenetic factors, such as Poly(ADP-ribose) Polymerase-1, establish epigenetic memory in mitotic chromatin.
Keywords: Cellular memory, Mitotic bookmarking, PARP1, Epigenetic factor, Transcription
Introduction
In order to function, cells regulate gene expression in a temporospatial manner. This process depends on cell memory. Since each cell division removes the imprint of transcription factors from DNA, certain mechanisms must be in place to initiate and direct the post-mitotic transcription reactivation in the daughter cells at mitotic exit. This process is called mitotic bookmarking.
During the mitotic stage of cell cycle, chromosomes are separated into two identical sets in two cells. Both daughter cells are genetically identical to the parent cell. During interphase, transcription occurs at multiple loci. However, when a cell enters mitosis, increased phosphorylation of histones forces the majority of transcription factors to dissociate from chromatin [1]. Chromatin condenses, thus preventing nuclear transcription machinery from accessing DNA. Transcription is shut off completely. All cellular functions progressing during mitosis depend completely on the pre-existing mRNAs [2]. To record genomic sites required for transcriptional activation after mitosis, specific histone modifications and some transcription factors are retained inside condensed chromatin, a phenomenon known as “mitotic bookmarking” [3,4] (Figure 1). Genome-wide studies have shown that these protein factors regulate the reactivation of gene transcription throughout the genome [5, 6]. Mitotic-specific knockdown and downregulation of these bookmarking genes have demonstrated their essential role in prompting transcription reactivation after mitosis is completed. In this review, we summarize recent progress in research on mitotic bookmarking mechanisms and highlight our finding that Poly(ADP-ribose) polymerase 1 (PARP1) is an important factor for chromatin bookmarking [5].
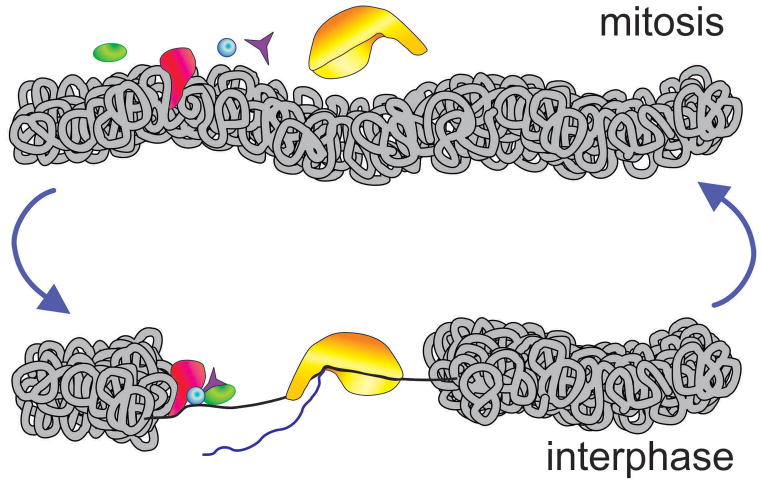
Figure 1. Epigenetic memory factors that maintain transcription-competent state following mitotic exit.
During mitosis, the extreme condensation of chromatin excludes most regulatory proteins from the contact with DNA, shutting off gene transcription. Epigenetic memory factors (green) are responsible for marking mitotic chromatin so that proper transcriptional patterns are restored precisely, without even the slightest deviation.
Mechanisms of mitotic bookmarking
Increasing phosphorylation of chromatin proteins at the beginning of mitosis pushes transcription factors away from DNA and leads to chromatin condensation [7, 8, 9, 10]. The major force driving chromatin condensation is phosphorylation of histone H3 at Ser-10, Ser-28, and other sites [4]. Histone modification causes most proteins, including RNA PolII, chromatin proteins (HP1), and chromatin remodeling proteins (BMI1), to dissociate from chromatin during for the duration of mitosis [11]. However, despite its condensation, chromatin retains more than 20% of factors as functional memory machinery for transcription bookmarking [11]. The two major mechanisms for mitotic bookmarking involve marking chromatin by exchange of histone variants and histone modifications [3] and retaining transcription factors as mitotic bookmarking proteins on mitotic chromatin [4].
Chromatin bookmarking by histone modifications and exchange
Histone modifications serve as landmarks to provide cellular memory after cell division [4]. These histone marks facilitate the recruitment of chromatin reader proteins, transcription factors and other regulatory proteins during interphase [4]. Some histone modifications, such as H3-K4 methylation, H3-K79 methylation, and H3 and/H4 acetylation, are stably maintained on the parental nucleosomes throughout mitosis [12], marking the proper positions of active chromatin during and just after mitosis [4]. Selective repressive histone modifications such as H3 lysine 9 trimethylation (H3K9me3) and H3K27 trimethylation (H3K27me3) [13] can also be retained throughout mitosis where they play an epigenetic role in keeping target genes silent in the subsequent interphase.
The incorporation of histone variants and the control of nucleosome positioning also provides mechanisms for mitotic bookmarking. For example, a nucleosome containing lysine 4-methylated H3.3 histone variant appears to be required for transcription activation at the MyoD promoter after mitosis [14]. Nucleosome positioning preference during mitosis also helps determine whether certain genes keep post-mitotic silencing (GRP78 and WFS1 genes) or post-mitotic expression (PLK1 gene) [15]. To silence gene expression in mitosis, nucleosome at +1 position shifts downstream to mask the transcription start site and returns to its initial spot after mitosis [15]. It has been further shown that nucleosome containing H2A.Z at the +1 position of active gene promoter shifts upstream to mark the transcription initiation site during mitosis and rapidly reactivates GRP78 gene when mitosis is completed [15]. H3K4 trimethylation is also maintained at these promoters during mitosis [15]. Little is known about how histone exchange regulates post-mitotic chromatin dynamics and chromatin accessibility to transcription initiation machinery.
Chromatin bookmarking by retaining transcription factors
Multiple transcription factors have been identified as mitotic bookmarks for epigenetic memory [3]. CTCF [16], PARP1 [5], FoxA1 [6], GATA1 [17], p300 [18], Brd4 [19], TFIID [20], RUNX2 [21], Hsp70 [22], and TBP-PP2A [23] have all been found to mark specific regions of mitotic chromatin. All these mitotic bookmarking proteins are capable of transmitting transcription information to the daughter cells, hence are often dubbed as epigenetic factors. Acting either alone or with other proteins, these factors are physically attached to the tightly condensed mitotic chromatin for bookmarking. While the majority of regulatory proteins are forced out of the condensed chromatin, the physical presence of epigenetic factors during mitosis marks the promoters of different genes for rapid reactivation of transcription after mitosis.
For many years, there has been a consensus that these transcription factors have conserved functions. The RUNX2 protein remains associated with chromosomes during mitosis through selective sequence-specific DNA binding [21]. RUNX2 was initially identified as a bone-specific nuclear matrix protein that interacts with the osteocalcin promoter [24,25]. The family of RUNX proteins (RUNX1, RUNX2 and RUNX3) is composed of lineage-specific transcription factors essential for hematopoietic, neuronal, gastrointestinal, and osteogenic cell fates. Increased evidence showing that RUNX2 is retained at mitotic chromosomes reflects its epigenetic function in bookmarking of target genes in different cancer cells [26]. The FoxA1 protein was originally identified as a pioneer transcription factor by initial binding to DNA elements for transcription induction [27]. However, it was also found that FoxA1 binds to mitotic chromatin, both specifically and nonspecifically [6]. In mitosis, FoxA1 is specifically bound to the promoters of important live genes which have very high expression in the interphase. In addition, the nonspecific binding of FoxA1 to the vicinity of other target genes may contribute to increase chromtin mobility for reinitiation of gene expression in the interphase. Both types of binding regulate genes responsible for liver differentiation in interphase [6]. It is well documented that the hematopoietic transcription factor GATA1 is required to establish lineage-specific expression during hematopoiesis [28]. A recent study also showed that hematopoietic gene expression programs are faithfully transferred through mitosis to the daughter cells by GATA1 binding to mitotic chromatin [17]. In contrast, two other protein transcription coregulators, FOG1 and TAL1, dissociate from chromatin upon mitosis entry [17]. Upon entering interphase, mitotic GATA1 target genes were reactivated more rapidly than those genes dissociated with GATA1 [17], suggesting that bookmarking facilitates rapid gene expression. Together, these studies imply that different transcription factors bookmark different sets of genes for tissue- or developmental-specific transcription regulation during cell cycle.
Apart from transcription factors directly involved in the bookmarking of specific genes, as described above, several proteins related to transcription have been shown to bookmark chromatin domains locally. For example, CTCF, an insulator protein, contributes to three-dimensional organization of Drosophila genome [29]. Recently, it was reported that CTCF also partially maintains binding to different chromatin domains during mitosis, suggesting that it may mark local chromatin domains in order to reinitiate gene transcription after mitosis [16]. Similarly, chromatin remodeling complex protein p300 forms a preinitiation complex with the transcription factors TBP, RNA polII, Brd4 and Mediator, and it is retained on Fos promoter and enhancer in mitotic chromatin [18]. After mitosis, p300 facilitates rapid reconstitution of long-range chromatin interactions and enhances loading of Brd4 [18]. Brd4, a double bromodomain protein, plays a significant role in regulating transcriptional elongation by binding to and recruiting transcriptional elongation factors to active genes [30]. It has been shown that Brd4 marks transcriptionally activated genes just before a cell enters mitosis and ensures their early expression after mitosis [19]. Wong et al. found that this holds true for transcription activation of Fos promoter because P300-deficient cells showed significant loss of Brd4 [18]. Future studies will elucidate how individual transcription factors coordinate with chromatin-regulating proteins, such as CTCF, to bookmark chromatin for transcription activation.
PARP1: a major protein to bookmark mitotic chromatin
For 50 years, research on PARP1, which generates poly(ADP-ribose) (PAR) polymer, has focused on DNA damage and repair [31,32]. However, recent findings have expanded the functions of PARP1 to include the regulation of chromatin structure [33], gene expression [34], and RNA processing [35,36], in a wide range of biological contexts, such as reproduction [37,38], development [39], aging [40], stem cells [39,41], inflammation [42], metabolism [43] and cancer [44]. PARP1 also acts as an epigenetic factor for mitotic bookmarking [5]. In mitotic synchronized cell division, PARP1 is associated with chromatin and binds to specific genes in different pathways including Cell adhesion, intercellular interaction, receptor-mediated signaling and protein secretion [5]. For example, through ChIP-seq it has been established that PARP1 binds to the promoter of genes in mitosis and colocalizes with nucleosomes containing H2A.Z and H2A.X histone variants [5]. ChIP-seq for histone the variants H2Ax and/or H2A.Z will help establishing global interaction of PARP1 with H2A.X or H2A.Z.
Information also flows from chromatin to PARP1. PARP1 can function as the transcription repressor or activator, depending on its enzymatic activity [34]. On the one hand, enzymatically silent PARP1 binds to the nucleosomes to increase chromatin condensation for transcription inhibition [34]. On the other hand, nucleosomes also contain histone variants which can stimulate PARP1 enzymatic activity in chromatin, leading to further modification of histones and chromatin decondensation for transcription activation [33]. In Drosophila, Jil1-phosphorylated H2Av activates PARP1 enzymatically which poly(ADP-ribosyl)ates histone proteins. This, in turn decondenses chromatin for transcription activation of PARP1-dependent genes [45]. Comparison of RNA-seq data of interphase cells [46] with PARP1 ChIP-seq data [5] revealed that interphase-specific PARP1 binding correlates with gene silencing. This observation fits a model in which PARP1 protein binding to promoters memorizes the site for gene expression, while transcription activation depends on PARP1 enzymatic activity in the interphase (Figure 2).
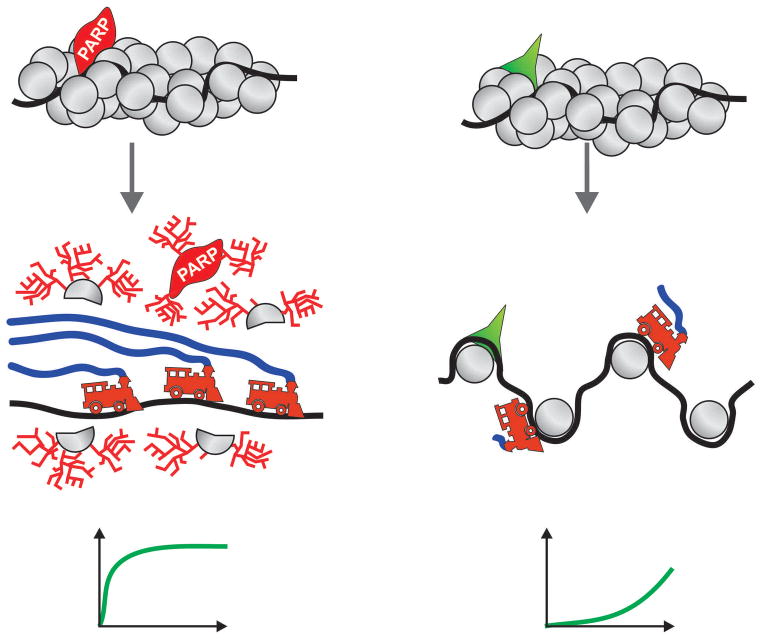
Figure 2. Model showing how PARP1 serves as an epigenetic memory factor to facilitate post-mitotic transcription.
PARP1 remains bound to chromatin during mitosis, but facilitates transcription after mitosis by poly(ADP-ribosyl)ating chromatin, effectively opening it. Upon depletion or inhibition of PARP1, chromatin remains condensed after mitosis, thereby blocking or delaying transcriptional restart. The charts at the bottom illustrate RNA transcripts accumulation rate typical for PARP1 dependent and PARP1 independent transcription respectively.
In addition, it has been found that there exists biasedness of PARP1 binding during mitosis [5]. In malignant cells, PARP1 binds to the majority of tumor suppressor genes (RB1, APC, MLH1, MBD2, NF1, SMAD2, NFKB1, FOXP1, AR and EZH2) during interphase, while in mitosis, it binds to proto-oncogenes (PDGFRB, EGFR/HER1, ERBB2/HER2, c-Src/CSK, SYK, Bruton’s tyrosine kinase, Abl2, MAP3K13, CDK18 and c-Myc) [5]. These results suggest that expression of proto-oncogenes is PARP1-dependent after mitosis and that interphase-specific PARP1 binding silences tumor suppressor genes in interphase cells. This new finding further expands the new function of PARP1 to cancer research and raises the possibility of a paradigm shift relative to the effect of PARP1 on cancer cells, as very different from its role in DNA repair.
Another interesting finding was that PARP1 specifically bookmarks genes determined by NFATC2 protein during mitosis [5]. NFATC2, a transcriptional factor, can regulate gene expression during oncogenic transformation [47]. Notably, approximately 65% of all sites of PARP1 localization in mitosis and interphase are proximal to NFATC2 [5]. Similar to PARP1, NFATC2 remains associated with mitotic chromatin [5]. However, whether post-mitotic transcription initiation depends on NFATC2 is unclear. Therefore, genome-wide transcriptome studies in NFATC2 depleted cells are needed to establish its role in post-mitotic transcription initiation.
In summary, the action mechanism of PARP1 as a bookmarking factor is very different from that of other reported factors. First, it marks the genomic sites that need to be rapidly reactivated immediately after mitosis. Secondly, in the interphase, activated PARP1 PARylates itself and other chromatin factors, causing them dissociated with chromatin. This action opens chromatin to allow transcription machinery to access DNA [Figure 2].
Perspectives
Recent research on mitotic bookmarking has demonstrated the importance of precise activation of genes for cell identity and survival after mitosis. Despite the growing list of bookmarking factors and their functions, our current understanding remains minimal. It remains unclear how mitotic bookmarking factors recognize the appropriate chromatin regions in whole genome. Second, all reported studies suggest that bookmarking factors target genes for their rapid post-mitotic activation, yet it is not fully understood how the nonspecific binding of these factors to chromatin contributes to gene reactivation. Third, we should investigate the three-dimensional interaction between bookmarking proteins and chromatin or genes during mitosis to determine the implications of these interactions in post-mitotic gene reactivation. Fourth, if such proteins control post-mitotic transcription reactivation of long-distance genes, the underlying mechanism should be investigated. Finally, further studies are needed to determine whether manipulation of transcription reactivation of bookmarked pathology-related genes has clinical relevance.
In our study, we observed that PARP1 bookmarks cell identity genes involved in cell niche interactions, metabolic and developmental pathways, cell adhesion, communication, proliferation and apoptosis and tumorigenic genes, including proto-oncogenes and tumor suppressor genes. We propose the manipulation of post-mitotic transcription activation of proto-oncogenes and tumor suppressor genes to control the growth of cancerous cells by compromising PARP1 bookmarking using PARP1 inhibitors. In addition, it will be interesting to examine how the cell coordinates PARP1 action with other bookmarking factors such as histone modifications for mitotic bookmarking. It has been shown that PARP1 can inhibit activity of the histone demethylase KDM5B through poly(ADP-ribosyl)ation to maintain H3K4me3 status of many PARP1-dependent genes in MCF-7 cells [48]. Thus, it is possible that PARP1 may also facilitate maintaining specific histone modifications of the target gene prompters for bookmarking through interaction with the histone modifying enzymes during mitosis. Recent studies have suggested that non-coding RNAs [ncRNAs] are also involved in epigenetic regulation of gene expression [49]. Indeed, PARP1 can modulate ncRNA-mediated pathways such as microRNA-dependent translation repression [50] and production of heat shock-induced long non-coding RNA(hnrù-n) [35]. Therefore, further results should be pressured to explore if the interplay bewtween PARP1 and ncRNAs plays any significant role in mitotic bookmarking.
In addition to bookmarking, we might want to examine whether other mechanisms control post-mitotic transcriptional reactivation such as the recent finding that cohesin marks the condensed chromatin [51. In addition, it has been established that the surrounding environment makes heritable epigenetic changes in response to the external or internal environment [52]. For example, cancer cell density, cell contact and stress level increased after each mitotic division, eventually leading to changes in the internal or external environment. Thus, it would be interesting to know whether environmental changes affect mitotic bookmarking.
Acknowledgments
We thank David Martin and Kate Pechenkina for critical reading of the manuscript and valuable comments. The expenses were defrayed by a grant from the National Institutes of Health (R01DK082623) (to A.V.T.).
Footnotes
Conflict of Interest
Niraj Lodhi, Yingbiao Ji and Alexei Tulin declare that they have no conflict of interest.
Compliance with Ethics Guidelines
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
Papers of particular interest, published recently, have been highlighted as:
* Of importance
** Of major importance
- 1.McCloy RA, Parker BL, Rogers S, Chaudhuri R, Gayevskiy V, Hoffman NJ, Ali N, Watkins DN, Daly RJ, James DE, Lorca T, Castro A, Burgess A. Global Phosphoproteomic Mapping of Early Mitotic Exit in Human Cells Identifies Novel Substrate Dephosphorylation Motifs. Mol Cell Proteomics. 2015;14:2194–212. doi: 10.1074/mcp.M114.046938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanenbaum ME, Stern-Ginossar N, Weissman JS, Vale RD. Regulation of mRNA translation during mitosis. Elife. 2015:4. doi: 10.7554/eLife.07957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kadauke S, Blobel GA. Mitotic bookmarking by transcription factors. Epigenetics Chromatin. 2013;6(6) doi: 10.1186/1756-8935-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang F, Higgins JM. Histone modifications and mitosis: countermarks, landmarks, and bookmarks. Trends in cell biology. 2013;23:175–184. doi: 10.1016/j.tcb.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 5**.Lodhi N, Kossenkov AV, Tulin AV. Bookmarking promoters in mitotic chromatin: poly(ADP-ribose)polymerase-1 as an epigenetic mark. Nucleic Acids Res. 2014;42:7028–38. doi: 10.1093/nar/gku415. This paper showed that PARP1 serves as a genome-wide bookmark in mitotic chromatin. It was demonstrated that PARP-1 is colocalized with H2A.Z-containing nucleosome in the mitosis and its enzymatic activity is required for transcription reactivation in the interphase. This paper revealed how dual functions of PARP1 as either repressor or activator establishes cellular memory. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6**.Caravaca JM, Donahue G, Becker JS, He X, Vinson C, Zaret KS. Bookmarking by specific and nonspecific binding of FoxA1 pioneer factor to mitotic chromosomes. Genes Dev. 2013;27:251–60. doi: 10.1101/gad.206458.112. This paper demonstrated that the pioneer factor FoxA1 specifically binds to the target genes which are crucial for liver differentiation in both mitosis and interphase. In addition, FoxA1 has the nonspecific binding ability to mitotic chromaion, which is also important for gene reactivation after mitosis. This paper showed a role of the nonspecific binding in bookmarking. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martínez-Balbás MA, Dey A, Rabindran SK, Ozato K, Wu C. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell. 1995;83:29–38. doi: 10.1016/0092-8674(95)90231-7. [DOI] [PubMed] [Google Scholar]
- 8.Gottesfeld JM, Forbes DJ. Mitotic repression of the transcriptional machinery. Trends Biochem Sci. 1997;22:197–202. doi: 10.1016/s0968-0004(97)01045-1. [DOI] [PubMed] [Google Scholar]
- 9.Dovat S, Ronni T, Russell D, Ferrini R, Cobb BS, Smale AST. Common mechanism for mitotic inactivation of C2H2 zinc finger DNA-binding domains. Genes Dev. 2002;16:2985–2990. doi: 10.1101/gad.1040502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delcuve GP, He S, Davie JR. Mitotic partitioning of transcription factors. J Cell Biochem. 2008;105:1–8. doi: 10.1002/jcb.21806. [DOI] [PubMed] [Google Scholar]
- 11.Egli D, Birkhoff G, Eggan K. Mediators of reprogramming: transcription factors and transitions through mitosis. Nat Rev Mol Cell Biol. 2008;9:505–516. doi: 10.1038/nrm2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kouskouti A, Talianidis I. Histone modifications defining active genes persist after transcriptional and mitotic inactivation. EMBO J. 2005;24:347–357. doi: 10.1038/sj.emboj.7600516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13*.Follmer NE, Wani AH, Francis NJ. A polycomb group protein is retained at specific sites on chromatin in mitosis. PLoS Genet. 2012;8:e1003135. doi: 10.1371/journal.pgen.1003135. This paper found that PcG proteins remain bound to some specific sites on mitotic chromatin in Drosophila S2 cells. The specific binding by a PcG protein (Posterior Sex Combs) may facilitate reestablishing gene silencing in the interphase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ng RK, Gurdon JB. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat Cell Biol. 2008;10:102–9. doi: 10.1038/ncb1674. [DOI] [PubMed] [Google Scholar]
- 15.Kelly TK, Jones PA. Role of nucleosomes in mitotic bookmarking. Cell Cycle. 2011;10:370–1. doi: 10.4161/cc.10.3.14734. [DOI] [PubMed] [Google Scholar]
- 16.Shen W, Wang D, Ye B, Shi M, Zhang Y, Zhao Z. A possible role of Drosophila CTCF in mitotic bookmarking and maintaining chromatin domains during the cell cycle. Biol Res. 2015;27:48–27. doi: 10.1186/s40659-015-0019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17**.Kadauke S, Udugama MI, Pawlicki JM, Achtman JC, Jain DP, Cheng Y, Hardison RC, Blobel GA. Tissue-specific mitotic bookmarking by hematopoietic transcription factor GATA1. Cell. 2012;150:725–37. doi: 10.1016/j.cell.2012.06.038. This paper showed that a transcription factor GATA1 remains associated with the main hematopoietic regulatory genes in mitosis of erythroid cells, which is critical for gene reactivation in the interphase. This study suggested that bookmarking by a specific factor is important for maintaining the cell type identity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18*.Wong MM, Byun JS, Sacta M, Jin Q, Baek S, Gardner K. Promoter-bound p300 complexes facilitate post-mitotic transmission of transcriptional memory. PLoS One. 2014;9:e99989. doi: 10.1371/journal.pone.0099989. This paper demonstrated that p300 bookmarks target genes with the pre-initiation complexes (PICs) for rapid transcription activation in the interphase. This paper established a connection of histone modifications with transcriptional memory. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dey A, Nishiyama A, Karpova T, McNally J, Ozato K. Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription. Mol Biol Cell. 2009;20:4899–4909. doi: 10.1091/mbc.E09-05-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christova R, Oelgeschläger T. Association of human TFIID-promoter complexes with silenced mitotic chromatin in vivo. Nat Cell Biol. 2002;4:79–82. doi: 10.1038/ncb733. [DOI] [PubMed] [Google Scholar]
- 21.Young DW, Hassan MQ, Yang XQ, Galindo M, Javed A, Zaidi SK, Furcinitti P, Lapointe D, Montecino M, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic retention of gene expression patterns by the cell fate-determining transcription factor Runx2. Proc Natl Acad Sci U S A. 2007;104:3189–94. doi: 10.1073/pnas.0611419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xing H, Wilkerson DC, Mayhew CN, Lubert EJ, Skaggs HS, Goodson ML, Hong Y, Park-Sarge OK, Sarge KD. Mechanism of hsp70i gene bookmarking. Science. 2005;307:421–3. doi: 10.1126/science.1106478. [DOI] [PubMed] [Google Scholar]
- 23.Xing H, Vanderford NL, Sarge KD. The TBP-PP2A mitotic complex bookmarks genes by preventing condensin action. Nat Cell Biol. 2008;10:1318–23. doi: 10.1038/ncb1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bidwell JP, Van Wijnen AJ, Fey EG, Dworetzky S, Penman S, Stein JL, Lian JB, Stein GS. Osteocalcin gene promoter-binding factors are tissue-specific nuclear matrix components. Proc Natl Acad Sci U S A. 1993;90:3162–6. doi: 10.1073/pnas.90.8.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merriman HL, van Wijnen AJ, Hiebert S, Bidwell JP, Fey E, Lian J, Stein J, Stein GS. The tissue-specific nuclear matrix protein, NMP-2, is a member of the AML/CBF/PEBP2/runt domain transcription factor family: interactions with the osteocalcin gene promoter. Biochemistry. 1995;34:13125–32. doi: 10.1021/bi00040a025. [DOI] [PubMed] [Google Scholar]
- 26.Pockwinse SM, Kota KP, Quaresma AJC, Imbalzano AN, Lian JB, van Wijnen AJ, Stein JL, Stein GS, Nickerson JA. Live Cell Imaging of the Cancer-related Transcription Factor RUNX2 during Mitotic Progression. J Cell Physiol. 2011;226:1383–1389. doi: 10.1002/jcp.22465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes & Development. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferreira R, Ohneda K, Yamamoto M, Philipsen S. GATA1 function, a paradigm for transcription factors in hematopoiesis. Molecular and Cellular Biology. 2005;25:1215–1227. doi: 10.1128/MCB.25.4.1215-1227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishihara K, Oshimura M, Nakao M. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol Cell. 2006;23:733–42. doi: 10.1016/j.molcel.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 30.Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. Journal of Biological Chemistry. 2007;282:13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- 31.Thomas C, Tulin AV. Poly-ADP-Ribose Polymerase: Machinery for Nuclear Processes. Mol Aspects Med. 2013;34:1124–1137. doi: 10.1016/j.mam.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kraus WL, Hottiger MO. PARP1 and gene regulation: progress and puzzles. Molecular Aspects of Medicine. 2013;34:1109–1123. doi: 10.1016/j.mam.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 33.Kotova E, Lodhi N, Jarnik M, Pinnola AD, Ji Y, Tulin AV. Drosophila histone H2A variant (H2Av) controls poly (ADP-ribose) polymerase 1 (PARP1) activation in chromatin. Proceedings of the National Academy of Sciences. 2011;108:6205–6210. doi: 10.1073/pnas.1019644108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ji Y, Tulin AV. The roles of PARP1 in gene control and cell differentiation. Current Opinion in Genetics & Development. 2010;20:512–518. doi: 10.1016/j.gde.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ji Y, Tulin AV. Poly(ADP-ribosyl)ation of heterogeneous nuclear ribonucleoproteins modulates splicing. Nucleic Acids Res. 2009;37:3501–13. doi: 10.1093/nar/gkp218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ji Y, Tulin AV. Post-transcriptional regulation by poly (ADP-ribosyl) ation of the RNA-binding proteins. International Journal of Molecular Sciences. 2013;14:16168–16183. doi: 10.3390/ijms140816168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dantzer F, Mark M, Quenet D, Scherthan H, Huber A, Liebe B, Ménissier-de Murcia J. Poly (ADP-ribose) polymerase-2 contributes to the fidelity of male meiosis I and spermiogenesis. Proceedings of the National Academy of Sciences. 2006;103:14854–14859. doi: 10.1073/pnas.0604252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyer-Ficca ML, Lonchar J, Credidio C, Ihara M, Li Y, Wang ZQ, Meyer RG. Disruption of poly (ADP-ribose) homeostasis affects spermiogenesis and sperm chromatin integrity in mice. Biology of reproduction. 2009;81:46–55. doi: 10.1095/biolreprod.108.075390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ji Y, Tulin AV. Poly(ADP-ribose) controls DE-cadherin-dependent stem cell maintenance and oocyte localization. Nat Communications. 2012;3:760. doi: 10.1038/ncomms1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bürkle A. DNA repair and PARP in aging. Free radical research. 2006;40:1295–1302. doi: 10.1080/10715760600915288. [DOI] [PubMed] [Google Scholar]
- 41.Lai YS, Chang CW, Pawlik KM, Zhou D, Renfrow MB, Townes TM. SRY (sex determining region Y)-box2 (Sox2)/poly ADP-ribose polymerase 1 (Parp1) complexes regulate pluripotency. Proceedings of the National Academy of Sciences. 2012;109:3772–3777. doi: 10.1073/pnas.1108595109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ba X, Garg NJ. Signaling mechanism of poly (ADP-ribose) polymerase-1 (PARP1) in inflammatory diseases. The American journal of pathology. 2011;178:946–955. doi: 10.1016/j.ajpath.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bai P, Cantó C. The role of PARP1 and PARP-2 enzymes in metabolic regulation and disease. Cell metabolism. 2012;16:290–295. doi: 10.1016/j.cmet.2012.06.016. [DOI] [PubMed] [Google Scholar]
- 44.Schiewer MJ, Goodwin JF, Han S, Brenner JC, Augello MA, Dean JL, Knudsen KE. Dual roles of PARP1 promote cancer growth and progression. Cancer Discovery. 2012;2:1134–1149. doi: 10.1158/2159-8290.CD-12-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas CJ, Kotova E, Andrake M, Adolf-Bryfogle J, Glaser R, Regnard C, Tulin AV. Kinase-mediated changes in nucleosome conformation trigger chromatin decondensation via poly(ADP-ribosyl)ation. Mol Cell. 2014;53:831–42. doi: 10.1016/j.molcel.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 47.Tie X, Han S, Meng L, Wang Y, Wu A. NFAT1 Is Highly Expressed in, and Regulates the Invasion of, Glioblastoma Multiforme Cells. PLoS ONE. 2013;8:e66008. doi: 10.1371/journal.pone.0066008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krishnakumar R, Kraus WL. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Molecular Cell. 2010;39:736–749. doi: 10.1016/j.molcel.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peschansky VJ, Wahlestedt C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics. 2014;9:3–12. doi: 10.4161/epi.27473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leung AK, Vyas S, Rood JE, Bhutkar A, Sharp PA, Chang P. Poly (ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Molecular Cell. 2011;42:489–499. doi: 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yan J, Enge M, Whitington T, Dave K, Liu J, Sur I, Taipale J. Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell. 2013;154:801–813. doi: 10.1016/j.cell.2013.07.034. [DOI] [PubMed] [Google Scholar]
- 52.Szyf M. Nongenetic inheritance and transgenerational epigenetics. Trends Mol Med. 2015;21:134–44. doi: 10.1016/j.molmed.2014.12.004. [DOI] [PubMed] [Google Scholar]