Abstract
Liposome-based drug delivery systems hold great potential for cancer therapy. The aim of this study was to design a nanodevice for targeted anchoring of liposomes (with and without cholesterol) with encapsulated anticancer drugs and antisense N-myc gene oligonucleotide attached to its surface. To meet this main aim, liposomes with encapsulated doxorubicin, ellipticine and etoposide were prepared. They were further characterized by measuring their fluorescence intensity, whereas the encapsulation efficiency was estimated to be 16%. The hybridization process of individual oligonucleotides forming the nanoconstruct was investigated spectrophotometrically and electrochemically. The concentrations of ellipticine, doxorubicin and etoposide attached to the nanoconstruct in gold nanoparticle-modified liposomes were found to be 14, 5 and 2 µg·mL−1, respectively. The study succeeded in demonstrating that liposomes are suitable for the transport of anticancer drugs and the antisense oligonucleotide, which can block the expression of the N-myc gene.
Keywords: liposome, gold nanoparticles, N-myc, doxorubicin, ellipticine, etoposide
1. Introduction
Liposomes are artificial spherical vesicles, which sizes range from tens of nanometres to several micrometres. They consist of one or more phospholipid bilayers surrounding an inner cavity. Synthetic or natural phospholipid molecules consist of polar head groups and hydrophobic hydrocarbon tails connected by a linker, such as glycerol [1]. Due to the fact that liposomes are primarily synthesized from biologically degradable materials, they are non-toxic and non-immunogenic. Due to their biocompatibility, size and hydrophobic and hydrophilic character, liposomes represent superb “micro-sized containers” suitable not only for delivery of drugs, but also for other functional biomolecules, such as nucleic acids and proteins [2,3,4,5]. Hydrophobic molecules can be loaded into the bilayer and hydrophilic ones are loaded into the cavity, where they are protected from degradation processes [6,7]. In addition, changes in the lipid composition, surface charge, and the method of preparation significantly affect their properties [1]. Due to this fact, liposomes have found application in medicine and pharmacy as delivery vehicles for drugs, antisense oligonucleotides, cloned genes, and recombinant proteins [8,9,10]. Moreover, it has been well documented that numerous anti-cancer agents were less-toxic in the liposomal form in comparison with conventional free drugs [11,12,13,14]. Besides delivery purposes, liposomes can be also used as a cell membrane model to study the function of membrane proteins in biophysics and in biochemistry [15,16].
Numerous scientists have been solving the issue how to deliver drugs selectively to the site of action. This issue is closely connected with lowering of doses, reducing drug waste, and eliminating the unwanted side effects of therapy. Nanogold-modified liposomes have found an application in many areas, especially in chemistry for free DNA sensing [17]. Moreover, gold has a great affinity to thiol groups and the self-assembly of an organic monolayer on gold is one of the best methods for surface modification of various materials. Some thiol-containing compounds can be easily removed from the Au surface, thus, they have found applications where temporary nanoscale coatings were required [18]. Utilization the gold nanoparticles (AuNPs) modified liposome as a nanotransporter device with attraction to thiol modified oligonucleotides has been published, too [19].
The aim of this study was to suggest and develop a modular nanodevice consisting of AuNPs-modified liposomes with enclosed anticancer drugs and the antisense oligonucleotide, which can block the expression of the selected gene. Doxorubicin, ellipticine and/or etoposide were used as examples (due to their fluorescent properties), however, any other drug may be used. N-myc gene was used as a model antisense oligonucleotide to demonstrate the concept.
2. Experimental Section
2.1. Chemicals and Material
Anticancer drugs (doxorubicin, ellipticine and etoposide), cholesterol, 1,2-dioleoyl-sn-glycero-3-phospho-rac-(1-glycerol) sodium salt, chloroform, sodium citrate, HAuCl4·3H2O and other reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA) in ACS purity (i.e., purity meeting the standards of the American Chemical Society), unless noted otherwise. Hydrogenated phosphatidylcholine from soybean was a gift from Lipoid GmbH (Ludwigshafen, Germany). Magnetic particles (MPs) oligo(DT)25 were purchased from Invitrogen (Oslo, Norway). The deionised water was prepared using Aqual 25 reverse osmosis equipment (Aqual, Brno, Czech Republic). The deionised water was further purified by using a MilliQ Direct QUV apparatus equipped with an UV lamp. The resistance was 18 MΩ. The pH was measured using a WTW inoLab pH meter (WTW GmbH, Weilheim, Germany).
2.2. Preparation of Gold Nanoparticles (AuNPs)
Gold nanoparticles were prepared by citrate method at room temperature according to Kimmling et al. and Polte et al. [20,21]. Briefly, an aqueous solution of sodium citrate (0.5 mL, 40 mM) was added to a solution of HAuCl4·3H2O (10 mL, 1 mM). The colour of the solution slowly changed from yellow to violet. The mixture was stirred overnight.
2.3. Preparation of Liposome Film and Liposome Encapsulating Doxorubicin, Ellipticine and/or Etoposide
Liposomes were prepared according to a published method [22] with some modifications. Briefly, cholesterol (100 mg), 1,2-dioleoyl-sn-glycero-3-phospho-rac-(1-glycerol) sodium salt (100 mg) and phosphatidylcholine (100 mg) were dissolved in chloroform (4.5 mL). Cholesterol-free liposomes were prepared in the same manner and dissolved in 3.5 mL of chloroform. A lipid film was obtained by rotary evaporation of chloroform. Residual chloroform was removed by a stream of nitrogen. Solutions containing 500 µg of the anticancer drugs and water (500 µL) were added to the lipid film (10 mg). Samples were homogenized with an Ultra-Turrax T8 (IKA Werke GmbH, Staufen, Germany) for 10 min. The homogenized mixtures were then heated and shaken for 15 min at 60 °C at a Thermomixer Comfort (Eppendorf, Hamburg, Germany). The samples were then washed several times with MiliQ water on an Amicon 3k (Merck Millipore, Merck KgaA, Darmstadt, Germany). The final volume of samples was 1 mL.
2.4. Modification of Liposome Surface by AuNPs
Modification of liposomes was done according to Bhuvana et al. [17]. Liposomes containing anticancer drugs prepared as described in the previous section were used. After cooling, a 1 mM solution (500 µL) of gold nanoparticles was added. The mixture was shaken for 3 h on a Biosan Orbital Shaker OS-10 (Biosan Ltd., Riga, Latvia). Subsequently, the volume was filtered through Amicon 3K (Merck Millipore, Merck KgaA, Darmstadt, Germany) under the following conditions: 3500 rpm, 20 °C, and 15 min. The mixture was washed several times with MilliQ water and finally diluted to 1 mL.
2.5. Amplification of Exon 2 of Human N-myc Gene
Neuroblastoma cells were obtained from the Faculty of Science, Masaryk University (Brno, Czech Republic). Isolation of genomic DNA of neuroblastoma cells were performed using a MagNA Pure Compact, Nucleic Acid Isolation Kit I (Roche, Mannheim, Germany), according to the manufacturer’s instructions. The sequence was obtained from the GenBank database; with the following accession number X03294.1. One part of the exon 2 of human N-myc gene was obtained by polymerase chain reaction (PCR) amplification of genomic DNA, using a set of primers (5-ATGCCGGGCATGATCTGC (h-N-myc-exon2-fw) and 5-TGCAGCTTCTCGCTCA (h-N-myc-exon2-rev)). The PCR products were purified using the MinElute PCR Purification Kit (Quiagen, Hilden, Germany).
2.6. Isolation of AuNPs-Modified Liposomes Using Magnetic Microparticles
For isolation of nanogold-modified liposomes, magnetic particles modified by polythymine chain (25 thymines) (Invitrogen, Oslo, Norway) were used. The solution used for washing MPs contained 0.1 M NaCl and 0.05 M phosphate buffer. The hybridization solution was as follows: 0.1 M Na2HPO4; 0.1 M NaH2PO4; 0.6 M guanidinium thiocyanate (Amresco, Solon, OH, USA); 0.15 M Tris-HCl (pH 7.5); and 0.2 M NaCl. For the first hybridization 10 µL of MPswas pipetted to an Eppendorf tube placed in a magnetic holder and washed three times with 100 µL of the phosphate buffer. Afterwards, 10 µL of oligodeoxynucleotide (100 µM) 5′ AAAA AAAA AAGA CTGG GTAG TTAA CCTT ACGT CT-SH 3′ and 10 µL of hybridization buffer was added and incubated (30 min, 25 °C) on a Multi RS-60 (Biosan Ltd.). Sample was washed three times with 100 µL of phosphate buffer using the magnetic holder. Subsequently, the elongated antisense N-myc oligonucleotide (as-N-myc) (5′-GTTC-TTGC-AGAT-CATG-CCCG-CTGA-CCCA-TCAA-3′) was attached to magnetic particle oligonucleotide probe (MPs-ODN probe). To 10 µL of MPs-ODN probe in the phosphate buffer 10 µL of as-N-myc ODN (100 µM) was pipetted and washed three times with 100 µL of phosphate buffer. Subsequently, the N-myc gene from exon 2 was attached in the same way. The 10 µL of N-myc gene sequence (100 µM) was incubated (30 min, and 25 °C) on Multi RS-60 (Biosan Ltd.) and afterwards washed three times with 100 µL of the phosphate buffer. The last step was labelling of 10 µL of nanoconstruct by 10 µL of anticancer drugs encapsulated into AuNPs modified liposome. The mixture was incubated (30 min, ad 25 °C) and washed using the magnetic holder three times with the phosphate buffer (3 × 100 µL). Afterwards, 10 µL of water in ACS purity was added and incubated (5 min, 95 °C, and 14,000 rpm) on a Thermomixer Comfort (Eppendorf). Finally, samples were rapidly cooled on ice. Magnetic particles were separated by a magnet and the solution was used for following measurements. Other experimental details can be found in [23,24,25,26,27].
2.7. UV/VIS Spectroscopy
Fluorescence spectra were acquired by a Tecan Infinite 200 PRO multifunctional microplate reader (TECAN, Mannedorf, Switzerland). Excitation wavelength for ellipticine, doxorubicin and etoposide was 420, 480 and 250 nm, respectively. The fluorescence scan of ellipticine, doxorubicin and etoposide was measured within the range from 450 to 850 nm, 510–850 nm and 280–850 nm, respectively, per 2-nm steps. The detector gain was set to 100. Absorbance of ssDNA was measured at λ = 260 and 280 nm. Each absorbance value is an average of three measurements. The samples for both measurements (2 µL) were placed in a 16 well Tecan NanoQuant plate. All measurements were performed at 30 °C controlled by the Tecan Infinite 200 PRO.
2.8. Determination of ODN-CA Peak
Determinations of ODN by square wave voltammetry were performed by a 663 VA Stand instrument (Metrohm, Herisau, Switzerland) connected with an AUTOLAB Analyzer (Metrohm) using the adsorptive transfer technique and a standard cell with three electrodes. A hanging mercury drop electrode (HMDE) with a drop area of 0.4 mm2 was the working electrode. An Ag/AgCl/3M KCl electrode was the reference and platinum electrode was auxiliary. For data processing GPES 4.9 software by Metrohm was employed. The analysed samples were deoxygenated prior to measurements by purging with argon (99.999%). For CA peak analysis the Tris buffer (10 mM Trizma base adjusted to pH = 5 with acetic acid) was used as a supporting electrolyte. The parameters of the measurement were as it follows: initial potential 0 V, end potential −1.7 V, deoxygenating with argon 30 s, accumulation time 120 s, step potential 5 mV, modulation amplitude 25 mV, volume of injected sample: 5 µL, and volume of measurement cell: 2 mL (5 μL of sample and 1995 µL supporting electrolyte).
2.9. Descriptive Statistics
Data were processed using EXCEL® (Microsoft, Redmond, WA, USA). Results are expressed as mean ± standard deviation unless noted otherwise (EXCEL®).
3. Results and Discussion
In this study, we focused on the preparation of a modular nanodevice combining the transport of an anticancer drug and silencing oligonucleotide able to block the selected gene. To prove this concept, three fluorescent anticancer drugs (doxorubicin, etoposide and ellipticine) as well as N-myc gene were used, however due to the modularity of the construct any other drug/gene may be used. To effectively couple the components and remove the unreacted molecules, the benefits of attachment to magnetic particles were exploited. The manipulation of the nanoconstruct by the external magnetic field simplifies the preparation and purification.
3.1. Characterization of Fluorescence Behaviour of Enclosed Drugs in Liposomes
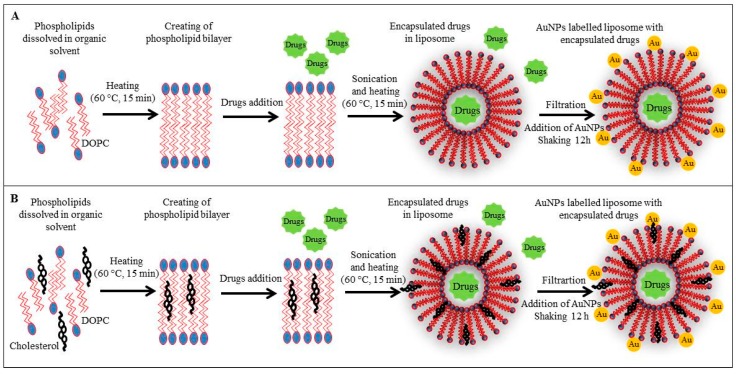
The first step consisted of enclosing of the anticancer drugs in liposomes with subsequent modification of their surface with AuNPs. First, the liposome without cholesterol (LIP-10, Figure 1A) and cholesterol-containing liposome (LIP-8, Figure 1B) were used for encapsulation of the anticancer drugs doxorubicin, ellipticine or etoposide exhibiting fluorescence properties. Both liposome variants were modified by AuNPs for specific bonding to thiolated oligonucleotide used as a carrier of antisense N-myc gene.
Figure 1.
The experimental scheme of encapsulation of drugs in AuNPs-labelled liposomes. (A) Phosphatidylcholine liposome; (B) Phosphatidylcholine-cholesterol liposome. Other experimental conditions is in Experimental Section.
We used fluorescence spectroscopy for analysis of the intensity of doxorubicin, etoposide and ellipticine fluorescence. Firstly, we determined the dependence of fluorescence intensity on drug concentration.
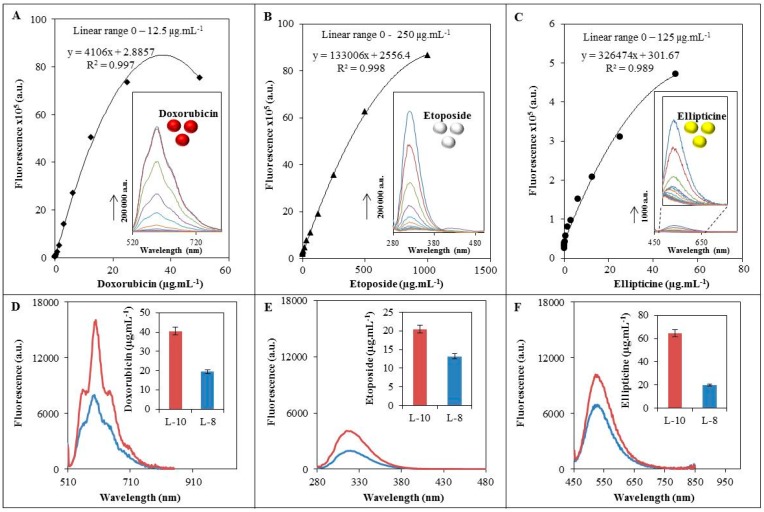
The excitation wavelengths of doxorubicin, etoposide and ellipticine were λ = 480, 250 and 420 nm, respectively, and emission was recorded within the ranges of λ = 510–850, 280–850 and 450–850 nm, respectively. In the case of doxorubicin, the calibration curve within the range of 0.05–50 µg·mL−1 showed a polynomial trend fitting the following equation in the linear part (0.05–12.5 µg·mL−1): y = 4106x + 2.8857 and R2 = 0.997 (Figure 2A). For etoposide, a polynomial calibration curve within the range of 1–1000 µg·mL−1 was obtained. The linear part within the range from 1 to 250 µg·mL−1 fitted the following equation y = 133006x + 2556.4 and R2 = 0.998 was determined (Figure 2B). Finally, the dependence on the ellipticine concentration within the 0.05–50 µg·mL−1 range showed a polynomial trend and the regression equation of the linear part in the 0.05–30 µg·mL−1 range y = 326474x + 301.67 and R2 = 0.9893 was obtained, see Figure 2C. The observed polynomial trend of the calibration curve in all fluorescence label cases is caused by quenching of the fluorescence due to the high concentration [28,29]. To ensure that the fluorescence quenching did not occur in the linear part of the calibration curve, we diluted the anticancer drugs accordingly for further experiments.
Figure 2.
Dependence of fluorescence signal on drug concentration. The corresponding fluorescence spectra are given in the insets. (A) Doxorubicin within the 0.05–50 µg·mL−1 concentration range; (B) etoposide within the 1–1000 µg·mL−1 concentration range and (C) ellipticine within the 0.05–50 µg·mL−1 concentration range. Fluorescence spectra of encapsulated drugs in cholesterol-free liposomes (red line) and liposomes with cholesterol (blue line): (D) doxorubicin; (E) etoposide; and (F) ellipticine. The calculated encapsulated concentration of drug in liposomes is given in the inserted graphs. Excitation wavelengths for doxorubicin, etoposide and ellipticine were 480, 250 and 420 nm, respectively.
Drug loading and encapsulation efficiency is very important in a drug delivery system [30]. We evaluated the encapsulation efficiency of the studied anticancer drugs in the liposomes prepared with cholesterol (LIP-8) and cholesterol free liposomes (LIP-10) by comparison of the anticancer drugs fluorescence encapsulated and non-encapsulated in the liposome before and after addition of 3% sodium dodecyl sulphate (SDS), which caused the drug release from the liposomes. Firstly, samples were diluted to the concentration in the linear range of each anticancer drug to avoid any fluorescence quenching. The concentration of each drug encapsulated into the liposome was estimated to be 160 µg·mL−1 which is 16% of the applied concentration of 1000 µg·mL−1 as described in the Experimental Section (data not shown). Cholesterol is often included in liposome formulations to give further rigidity to the bilayer that may improve the in vivo and in vitro stability of the liposomes [31]. We studied the fluorescence behaviour of liposomes with cholesterol and cholesterol-free liposomes. The fluorescence spectra in Figure 2D–F show different fluorescence intensities of anticancer drugs for both varieties of liposomes. The intensity of emission of the doxorubicin encapsulated in LIP-8 or LIP-10 at λ = 510 nm was 16,000 arbitrary units (a.u.) and 8000 a.u., respectively. The differences in emissions between LIP-8 and LIP-10 corresponded to 50% of LIP-10 (Figure 2D). From Figure 2E it is evident that the intensity of emission of the etoposide encapsulated in LIP-8 or LIP-10 at λ = 250 nm was 4000 a.u. or 1900 a.u., respectively. The difference in emissions corresponded to 47% of LIP-10. Finally, the intensity of emission of the ellipticine encapsulated in LIP-8 or LIP-10 at λ = 510 nm was 10,000 a.u. or 7000 a.u., respectively. The difference in emissions corresponded to 68% of LIP-10. In the case of the unmodified liposomes and liposomes modified with AuNPs, no change in emission (fluorescence) was observed (data not shown). The differences in fluorescence between both types of liposomes are probably caused by cholesterol addition, which is in good agreement with previously reported data [32]. Quenching of fluorescence by cholesterol was also described e.g., for human serum albumin [33].
3.2. Hybridization of ODN
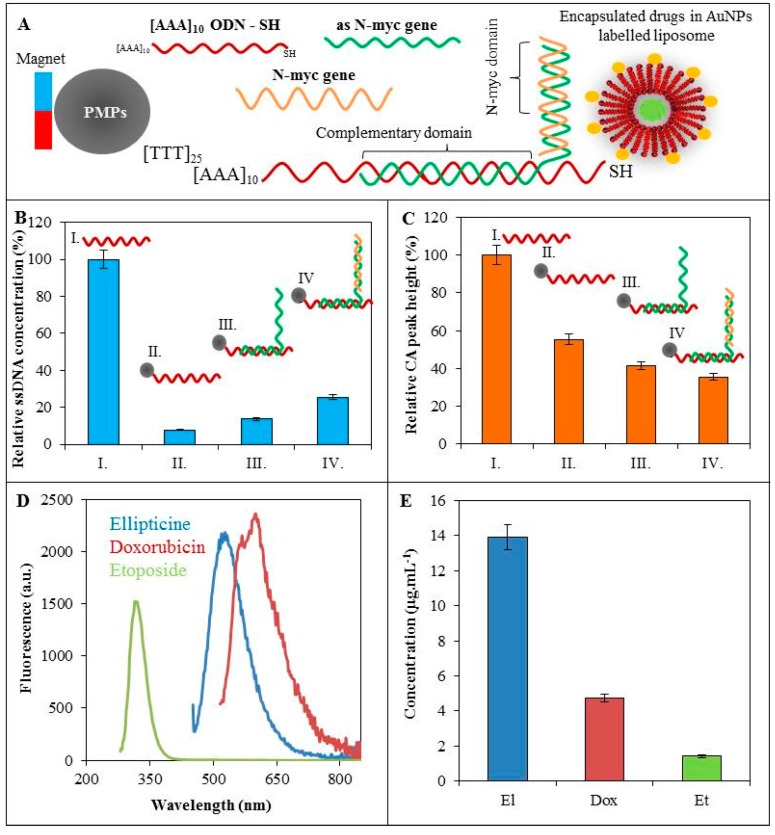
MPs are a suitable tool for DNA isolation and manipulation allowing for the purification of DNA from various matrices [34,35,36,37]. Various modifications of the magnetic microparticles could provide the specific binding to analytes [38,39,40]. In our case, we used thymine-labelled magnetic particles for coupling of the (AAA)10ODN-SH. To this sequence, the oligonucleotide (as-N-myc gene) was hybridized. ODN sequence is specially designed to bind partially to (AAA)10ODN-SH and partially to N-myc gene. The scheme of the nanoconstruct is shown in Figure 3A. To be effective, a high oligonucleotide hybridization yield is required. Hybridization yield depends on numerous factors, including nucleotide sequence, temperature, salt concentration and the time of hybridization. Based on our previously published results, we estimated the optimal conditions for hybridization process [23,24,25,26,27], which was confirmed spectrophotometrically and electrochemically. Primarily, we investigated the relative ssDNA concentration using spectrophotometric analysis (Figure 3B). The 100% is expressed as 50 µg/mL (AAA)10ODN-SH concentration before isolation by magnetic particles (I, Figure 3B). Afterwards, the 90% decrease of relative ssDNA concentration occurred after the isolation of the oligonucleotide (II, Figure 3B). The isolated ODN concentration is limited by binding capacity of magnetic particles. Binding the as-N-myc sequence (III, Figure 3B) caused a relative concentration increase to 13% and finally, the nanoconstruct was tested to be capable of binding of N-myc gene (50 µg/mL), which caused a 26% relative concentration increase.
Figure 3.
(A) Scheme of the nanoconstruct of the thymine modified paramagnetic particles (PMPs), which were bound with (AAA)10ODN-SH based on their complementarity. On the thiolated ends of oligonucleotides the AuNPs were tied to modified liposome with encapsulated fluorescence label. On the complementary domain of (AAA)10ODN-SH the as-N-myc sequence was hybridized for binding to N-myc gene; (B) Spectrophotometric analysis of relative ssDNA concentration and (C) electrochemical analysis (square wave voltammetry) of relative CA peak height during hybridization of: (I.) 50 µg·mL−1 (AAA)10ODN-SH isolated by (II.) thymine labelled MPs and (III.) attached 50 µg·mL−1 elongated as-N-myc sequence for (IV) binding of 50 µg·mL−1 N-myc gene. During the individual hybridization steps, samples were washed with phosphate buffer; (D) Fluorescence spectra of isolated drugs (ellipticine, doxorubicin, or etoposide, all in concentrations of 160 µg·mL−1). Excitation wavelengths for ellipticine, doxorubicin and etoposide were 420, 480 and 250 nm, respectively; (E) Concentration yield of encapsulated ellipticine, doxorubicin, or etoposide (160 µg·mL−1) in the liposome after nanoconstruct isolation.
Subsequently, electrochemical analysis of hybridization process was performed (Figure 3C). The nucleic acids have been reported to give redox common signals of adenine and cytosine (CA peak) at −1.4 V potential. It is commonly known that the ssDNA provides a higher electrochemical signal in comparison with dsDNA on HMDE. The highest signal of CA peak was found for (AAA)10ODN-SH (I). After the isolation of the ODN by paramagnetic particles (II), the relative CA peak height decreased to 55%. The attachment of as-N-myc sequence (III) caused the relative peak height decrease to 41%. Relative CA peak height of nanoconstruct with bonded N-myc gene was estimated to 35%.
3.3. Labelling of ODN
Due to the high affinity of gold for thiol groups (-SH), (AAA)10ODN-SH was attached to AuNPs-modified liposome with various encapsulated anticancer drugs, which were characterized in Section 3.1. Fluorescent drugs such as doxorubicin are suitable tools for in vivo imaging of cell systems [41]. Since the fluorescent labels encapsulated into LIP-8 showed a lower fluorescence we decided to use the LIP-10 variant for further experiments. Using the protocol mentioned above we built a nanoconstruct with the attached fluorescence label, which was purified using paramagnetic particles. Due to the limited binding capacity of MPs, only a fraction of the total liposomal drug (160 µg·mL−1) is bound to the MPs and is able to go through the whole magnetic field isolation process. It clearly follows from the results obtained and shown in Figure 3C that the emission intensity of the doxorubicin encapsulated in LIP-10 at λ = 600 nm corresponded to an isolated doxorubicin concentration of 5 µg·mL−1. The ellipticine emission intensity corresponded to an isolated concentration ellipticine of 14 µg·mL−1. Finally, the emission intensity of etoposide at λ = 320 nm corresponded to an isolated etoposide concentration of 2 µg·mL−1.
4. Conclusions
In this paper, we presented a nanoconstruct which carries the as-N-myc gene and a fluorescent label formed by anticancer drugs with fluorescence properties (doxorubicin, ellipticine, or etoposide) encapsulated into the liposome modified by AuNPs. Due to its specific structure, this construct has the ability to turn off the N-myc gene, which promotes the progression of neuroblastoma and the presence of fluorescent drugs encapsulated into liposomes enables in vivo imaging of nanoconstruct transport into the target cell. Connectivity of nanoconstruct is mediated by repeating adenine sequence, which is complementary to thymine labelled magnetic particle and enables the purification of resulting nanoconstruct. Our results confirmed the decrease of the fluorescence of doxorubicin, ellipticine and etoposide by cholesterol, which naturally occurs in biological lipid membranes of eukaryotic cells. The concentrations of doxorubicin, ellipticine, and etoposide were found to be in the range of 5, 2 and 14 µg·mL−1 respectively, which is sufficient for in vivo imaging of treated cancer cells. Because of its properties, our construct should be a useful tool for gene therapy as well as for cell labelling.
Acknowledgments
The financial support from the project GA CR NANOCHEMO 14-18344S and from the Ministry of Health of the Czech Republic for conceptual development of research organization 00064203 (University Hospital Motol, Prague, Czech Republic) is highly acknowledged. Authors thank to Dagmar Uhlirova for technical support.
Author Contributions
S.S. performed conjugation experiments and drafted the manuscript; L.N. and J.K. performed fluorescence measurements and data analyses; B.R.-N. corrected manuscript; A.M.J.J. performed gene experiments; P.K. prepared nanoparticles and liposomes; M.K. was involved in voltammetry measurements; M.M., M.S. and T.E were involved in the design of the study and manuscript writing; V.A. and R.K. coordinated experiments and participated in design of the study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Akbarzadeh A., Rezaei-Sadabady R., Davaran S., Joo S.W., Zarghami N., Hanifehpour Y., Samiei M., Kouhi M., Nejati-Koshki K. Liposome: Classification, preparation, and applications. Nanoscale Res. Lett. 2013;8:1–9. doi: 10.1186/1556-276X-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schafer J., Hobel S., Bakowsky U., Aigner A. Liposome-polyethylenimine complexes for enhanced DNA and sirna delivery. Biomaterials. 2010;31:6892–6900. doi: 10.1016/j.biomaterials.2010.05.043. [DOI] [PubMed] [Google Scholar]
- 3.Chen X.A., Wang X.H., Wang Y.S., Yang L., Hu J., Xiao W.J., Fu A., Cai L.L., Li X., Ye X., et al. Improved tumor-targeting drug delivery and therapeutic efficacy by cationic liposome modified with truncated bfgf peptide. J. Control. Release. 2010;145:17–25. doi: 10.1016/j.jconrel.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Villares G.J., Zigler M., Wang H., Melnikova V.O., Wu H., Friedman R., Leslie M.C., Vivas-Mejia P.E., Lopez-Berestein G., Sood A.K., et al. Targeting melanoma growth and metastasis with systemic delivery of liposome-incorporated protease-activated receptor-1 small interfering rna. Cancer Res. 2008;68:9078–9086. doi: 10.1158/0008-5472.CAN-08-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martins S., Sarmento B., Ferreira D.C., Souto E.B. Lipid-based colloidal carriers for peptide and protein delivery-liposomes versus lipid nanoparticles. Int. J. Nanomed. 2007;2:595–607. [PMC free article] [PubMed] [Google Scholar]
- 6.Milla P., Dosio F., Cattel L. Pegylation of proteins and liposomes: A powerful and flexible strategy to improve the drug delivery. Curr. Drug Metab. 2012;13:105–119. doi: 10.2174/138920012798356934. [DOI] [PubMed] [Google Scholar]
- 7.Bochot A., Fattal E. Liposomes for intravitreal drug delivery: A state of the art. J. Control. Release. 2012;161:628–634. doi: 10.1016/j.jconrel.2012.01.019. [DOI] [PubMed] [Google Scholar]
- 8.Gregoriadis G. The carrier potential of liposomes in biology and medicine. N. Engl. J. Med. 1976;295:765–770. doi: 10.1056/NEJM197609302951406. [DOI] [PubMed] [Google Scholar]
- 9.Allen T.M. Long-circulating (sterically stabilized) liposomes for targeted drug-delivery. Trends Pharmacol. Sci. 1994;15:215–220. doi: 10.1016/0165-6147(94)90314-X. [DOI] [PubMed] [Google Scholar]
- 10.Bangham A.D. Review of lasic, liposomes: From physics to applications. Biophys. J. 1994;67:1358–1359. doi: 10.1016/S0006-3495(94)80607-1. [DOI] [Google Scholar]
- 11.Ding Z.Y., Zhou L., Liu Y.M., Lu Y. Safety and efficacy of paclitaxel liposome for elderly patients with advanced non-small cell lung cancer: A multi-center prospective study. Thorac. Cancer. 2013;4:14–19. doi: 10.1111/j.1759-7714.2012.00134.x. [DOI] [PubMed] [Google Scholar]
- 12.Xu X., Wang L., Xu H.Q., Huang X.E., Qian Y.D., Xiang J. Clinical comparison between paclitaxel liposome (lipusu®) and paclitaxel for treatment of patients with metastatic gastric cancer. Asian Pac. J. Cancer Prev. 2013;14:2591–2594. doi: 10.7314/APJCP.2013.14.4.2591. [DOI] [PubMed] [Google Scholar]
- 13.Zhao C., Feng Q., Dou Z.P., Yuan W., Sui C.G., Zhang X.H., Xia G.M., Sun H.F., Ma J. Local targeted therapy of liver metastasis from colon cancer by galactosylated liposome encapsulated with doxorubicin. PLoS ONE. 2013;8:290. doi: 10.1371/journal.pone.0073860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gabizon A., Shmeeda H., Barenholz Y. Pharmacokinetics of pegylated liposomal doxorubicin—Review of animal and human studies. Clin. Pharmacokinet. 2003;42:419–436. doi: 10.2165/00003088-200342050-00002. [DOI] [PubMed] [Google Scholar]
- 15.Dick R.A., Goh S.L., Feigenson G.W., Vogt V.M. Hiv-1 gag protein can sense the cholesterol and acyl chain environment in model membranes. Proc. Natl. Acad. Sci. USA. 2012;109:18761–18766. doi: 10.1073/pnas.1209408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jonsson F., Beke-Somfai T., Andreasson J., Norden B. Interactions of a photochromic spiropyran with liposome model membranes. Langmuir. 2013;29:2099–2103. doi: 10.1021/la304867d. [DOI] [PubMed] [Google Scholar]
- 17.Bhuvana M., Narayanan J.S., Dharuman V., Teng W., Hahn J.H., Jayakumar K. Gold surface supported spherical liposome-gold nano-particle nano-composite for label free DNA sensing. Biosens. Bioelectron. 2013;41:802–808. doi: 10.1016/j.bios.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 18.Dilimon V.S., Rajalingam S., Delhalle J., Mekhalif Z. Self-assembly mechanism of thiol, dithiol, dithiocarboxylic acid, disulfide and diselenide on gold: An electrochemical impedance study. Phys. Chem. Chem. Phys. 2013;15:16648–16656. doi: 10.1039/c3cp51804c. [DOI] [PubMed] [Google Scholar]
- 19.Nejdl L., Rodrigo M.A.R., Kudr J., Ruttkay-Nedecky B., Konecna M., Kopel P., Zitka O., Hubalek J., Kizek R., Adam V. Liposomal nanotransporter for targeted binding based on nucleic acid anchor system. Electrophoresis. 2014;35:393–404. doi: 10.1002/elps.201300197. [DOI] [PubMed] [Google Scholar]
- 20.Kimling J., Maier M., Okenve B., Kotaidis V., Ballot H., Plech A. Turkevich method for gold nanoparticle synthesis revisited. J. Phys. Chem. B. 2006;110:15700–15707. doi: 10.1021/jp061667w. [DOI] [PubMed] [Google Scholar]
- 21.Polte J., Ahner T.T., Delissen F., Sokolov S., Emmerling F., Thunemann A.F., Kraehnert R. Mechanism of gold nanoparticle formation in the classical citrate synthesis method derived from coupled in situ xanes and saxs evaluation. J. Am. Chem. Soc. 2010;132:1296–1301. doi: 10.1021/ja906506j. [DOI] [PubMed] [Google Scholar]
- 22.Kunjachan S., Blauz A., Mockel D., Theek B., Kiessling F., Etrych T., Ulbrich K., van Bloois L., Storm G., Bartosz G., et al. Overcoming cellular multidrug resistance using classical nanomedicine formulations. Eur. J. Pharm. Sci. 2012;45:421–428. doi: 10.1016/j.ejps.2011.08.028. [DOI] [PubMed] [Google Scholar]
- 23.Huska D., Hubalek J., Adam V., Vajtr D., Horna A., Trnkova L., Havel L., Kizek R. Automated nucleic acids isolation using paramagnetic microparticles coupled with electrochemical detection. Talanta. 2009;79:402–411. doi: 10.1016/j.talanta.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 24.Krejcova L., Dospivova D., Ryvolova M., Kopel P., Hynek D., Krizkova S., Hubalek J., Adam V., Kizek R. Paramagnetic particles coupled with an automated flow injection analysis as a tool for influenza viral protein detection. Electrophoresis. 2012;33:3195–3204. doi: 10.1002/elps.201200304. [DOI] [PubMed] [Google Scholar]
- 25.Krizkova S., Jilkova E., Krejcova L., Cernei N., Hynek D., Ruttkay-Nedecky B., Sochor J., Kynicky J., Adam V., Kizek R. Rapid superparamagnetic-beads-based automated immunoseparation of zn-proteins from staphylococcus aureus with nanogram yield. Electrophoresis. 2013;34:224–234. doi: 10.1002/elps.201200234. [DOI] [PubMed] [Google Scholar]
- 26.Krizkova S., Ryvolova M., Hynek D., Eckschlager T., Hodek P., Masarik M., Adam V., Kizek R. Immunoextraction of zinc proteins from human plasma using chicken yolk antibodies immobilized onto paramagnetic particles and their electrophoretic analysis. Electrophoresis. 2012;33:1824–1832. doi: 10.1002/elps.201100638. [DOI] [PubMed] [Google Scholar]
- 27.Zitka O., Krizkova S., Krejcova L., Hynek D., Gumulec J., Masarik M., Sochor J., Adam V., Hubalek J., Trnkova L., et al. Microfluidic tool based on the antibody-modified paramagnetic particles for detection of 8-hydroxy-2′-deoxyguanosine in urine of prostate cancer patients. Electrophoresis. 2011;32:3207–3220. doi: 10.1002/elps.201100430. [DOI] [PubMed] [Google Scholar]
- 28.Blazkova I., Nguyen V.H., Dostalova S., Kopel P., Stanisavljevic M., Vaculovicova M., Stiborova M., Eckschlager T., Kizek R., Adam V. Apoferritin modified magnetic particles as doxorubicin carriers for anticancer drug delivery. Int. J. Mol. Sci. 2013;14:13391–13402. doi: 10.3390/ijms140713391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sikorski M., Krystkowiak E., Steer R.P. The kinetics of fast fluorescence quenching processes. J. Photochem. Photobiol. A Chem. 1998;117:1–16. doi: 10.1016/S1010-6030(98)00318-9. [DOI] [Google Scholar]
- 30.Narang A.S., Delmarre D., Gao D. Stable drug encapsulation in micelles and microemulsions. Int. J. Pharm. 2007;345:9–25. doi: 10.1016/j.ijpharm.2007.08.057. [DOI] [PubMed] [Google Scholar]
- 31.Cagdas F.M., Ertugral N., Bucak S., Atay N.Z. Effect of preparation method and cholesterol on drug encapsulation studies by phospholipid liposomes. Pharm. Dev. Technol. 2011;16:408–414. doi: 10.3109/10837451003774401. [DOI] [PubMed] [Google Scholar]
- 32.Kominkova M., Guran R., Rodrigo M.A.R., Kopel P., Blazkova I., Chudobova D., Nejdl L., Heger Z., Ruttkay-Nedecky B., Zitka O., et al. Study of functional qualities of different types of tailored liposomes with encapsulated doxorubicin using electrochemical and optical methods. Int. J. Electrochem. Sci. 2014;9:2993–3007. [Google Scholar]
- 33.Peng L., Minbo H., Fang C., Xi L., Chaocan Z. The interaction between cholesterol and human serum albumin. Protein Pept. Lett. 2008;15:360–364. doi: 10.2174/092986608784246542. [DOI] [PubMed] [Google Scholar]
- 34.Smerkova K., Dostalova S., Skutkova H., Ryvolova M., Adam V., Provaznik I., Kizek R. Isolation of xis gen fragment of lambda phage from agarose gel using magnetic particles for subsequent enzymatic DNA sequencing. Chromatographia. 2013;76:329–334. doi: 10.1007/s10337-012-2326-1. [DOI] [Google Scholar]
- 35.Kremplova M., Fialova D., Nejdl L., Hynek D., Trnkova L., Hubalek J., Kizek R., Adam V. Influence of magnetic microparticles isolation on adenine homonucleotides structure. Materials. 2014;7:1455–1472. doi: 10.3390/ma7031455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zitka O., Skalickova S., Merlos M.A.R., Krejcova L., Kopel P., Adam V., Kizek R. Sequences of pandemic-causing viruses isolated and detected by paramagnetic particles coupled with microfluidic system and electrochemical detector. Int. J. Electrochem. Sci. 2013;8:12628–12642. [Google Scholar]
- 37.Saiyed Z.M., Bochiwal C., Gorasia H., Telang S.D., Ramchand C.N. Application of magnetic particles (Fe3O4) for isolation of genomic DNA from mammalian cells. Anal. Biochem. 2006;356:306–308. doi: 10.1016/j.ab.2006.06.027. [DOI] [PubMed] [Google Scholar]
- 38.Gai L.G., Han X.Y., Hou Y.H., Chen J., Jiang H.H., Chen X.C. Surfactant-free synthesis of Fe3O4@pani and Fe3O4@ppy microspheres as adsorbents for isolation of pcr-ready DNA. Dalton Trans. 2013;42:1820–1826. doi: 10.1039/C2DT32164E. [DOI] [PubMed] [Google Scholar]
- 39.Chen X.W., Mao Q.X., Liu J.W., Wang J.H. Isolation/separation of plasmid DNA using hemoglobin modified magnetic nanocomposites as solid-phase adsorbent. Talanta. 2012;100:107–112. doi: 10.1016/j.talanta.2012.07.095. [DOI] [PubMed] [Google Scholar]
- 40.Prodelalova J., Rittich B., Spanova A., Petrova K., Benes M.J. Isolation of genomic DNA using magnetic cobalt ferrite and silica particles. J. Chromatogr. A. 2004;1056:43–48. doi: 10.1016/S0021-9673(04)01448-7. [DOI] [PubMed] [Google Scholar]
- 41.Dai X.W., Yue Z.L., Eccleston M.E., Swartling J., Slater N.K.H., Kaminski C.F. Fluorescence intensity and lifetime imaging of free and micellar-encapsulated doxorubicin in living cells. Nanomed. Nanotechnol. Biol. Med. 2008;4:49–56. doi: 10.1016/j.nano.2007.12.002. [DOI] [PubMed] [Google Scholar]