Abstract
Aim:
Malic enzymes are oxidative decarboxylases with NAD+ or NAD(P)+ as cofactor that catalyze the conversion of L-malate to pyruvate and CO2. The aim of this study was to discover and characterize a potent inhibitor of human NAD(P)+-dependent malic enzyme 2 (ME2).
Methods:
Recombinant human ME2-His-Tag fusion protein was overexpressed in E coli and purified with Ni-NTA resin. A high-throughput screening (HTS) assay was developed to find ME2 inhibitors. Detergent Brij-35 was used to exclude false positives. The characteristics of the inhibitor were analyzed with enzyme kinetics analysis. A thermal shift assay for ME2 was carried out to verify the binding of the inhibitor with the enzyme.
Results:
An HTS system for discovering ME2 inhibitors was established with a Z′ factor value of 0.775 and a signal-to-noise ratio (S/N) of 9.80. A library containing 12 683 natural products was screened. From 47 hits, NPD387 was identified as an inhibitor of ME2. The primary structure-activity relationship study on NPD387 derivatives showed that one derivative NPD389 was more potent than the parent compound NPD387 (the IC50 of NPD389 was 4.63±0.36 μmol/L or 5.59±0.38 μmol/L, respectively, in the absence or presence of 0.01% Brij-35 in the assay system). The enzyme kinetics analysis showed that NPD389 was a fast-binding uncompetitive inhibitor with respect to the substrate NAD+ and a mixed-type inhibitor with respect to the substrate L-malate.
Conclusion:
NPD389 is a potent ME2 inhibitor that binds to the enzyme in a fast-binding mode, acting as an uncompetitive inhibitor with respect to the substrate NAD+ and a mixed-type inhibitor with respect to the substrate L-malate.
Keywords: malic enzyme, ME2, inhibitor, natural product, NPD389, high-throughput screening, structure-activity relationship, enzyme kinetics, thermal shift assay
Introduction
Malic enzymes (ME) are oxidative decarboxylases that catalyze the conversion of L-malate to pyruvate and CO2 and use NAD+ or NAD(P)+ as a cofactor1,2,3. In mammals, there are three malic enzyme isoforms with different cofactor specificities: the cytosolic NADP+-dependent ME1, the mitochondrial NAD(P)+-dependent ME2, and the mitochondrial NADP+-dependent ME3. Although ME2 can use either NAD+ or NADP+as a cofactor, it has been demonstrated that this enzyme favors NAD+as a cofactor under physiological conditions4.
Pyruvate is an intracellular hub in the conversion of three major nutrient substrates: carbohydrates, fatty acids and amino acids. NADPH, which is necessary for the maintenance of intracellular glutathione, acts as a coenzyme in many biochemical reactions in vivo. In tumor cells, glutaminolysis is upregulated to meet the needs of NADPH and pyruvate5. ME2 catalyzes the conversion from malate to pyruvate and NAD(P)H. Therefore, ME2 plays an important role in glutaminolysis6,7.
Previous studies indicate that ME2 activity increases as cells progress towards neoplasia in a rat tracheal epithelial line8, and similar findings have been reported for Morris hepatomas9. The knockdown of endogenous ME2 levels impairs the proliferation of K562 cells, leads to apoptosis in K562 cells and suppresses tumor growth in vivo4. Silencing ME2 severely impairs the growth of the HCT-116 and U2OS cancer cell lines. Furthermore, overexpression of ME2 enhances HCT-116 and U2OS cell growth. It is known that ME2 is involved in the regulation of p53 during tumor metabolism, senescence and growth6. Thus, ME2 is believed to be a potential anticancer target. In light of the scarcity of ME2 inhibitors10, establishing a high-throughput screening system for the discovery of ME2 inhibitors may lay the foundation for novel anti-tumor drug discovery.
In this study, we developed a molecular-level high-throughput screening assay for ME2 inhibitors. We screened 12 683 natural products and identified a novel ME2 inhibitor, NPD387. From a structure-activity relationship study, we obtained a more potent derivative, NPD389, and we then investigated the characteristics of NPD389.
Materials and methods
Materials and instruments
The plasmid pRH281-ME2 was a kind gift from Prof Liang TONG of Columbia University (New York, NY, USA). The Escherichia coli strains BL21-CodonPlus (DE3) and JM109 were purchased from Stratagene (La Jolla, CA, USA) and Promega (Madison, WI, USA), respectively. 3-Indoleacrylic acid (IAA), β-nicotinamide adenine dinucleotide hydrate (NAD+) and SYPRO orange protein gel stain were purchased from Sigma Aldrich (St Louis, MO, USA). Ni-NTA His-Bind Resin was obtained from Merck Millipore (Billerica, MA, USA). L-malate was obtained from MP Biomedicals LLC (Santa Ana, CA, USA). The other reagents and solvents used in the experiments were of analytical grade.
The Spectra Max 340 PC 384 microplate reader was from Molecular Devices (Sunnyvale, CA, USA). The Fisher Scientific Sonic Dismembrator Model 500 was from Bio Logics, Inc (Manassas, VA, USA). The transparent, 384-well, medium protein-binding plates were from PerkinElmer (Seattle, WA, USA). The SAGIAN core integrated robotic system was from Beckman Coulter (Fullerton, CA, USA). The Light Cycler® 480 System was from Roche (Basel, BS, Switzerland).
Expression and purification of ME2
The plasmid pRH281-ME2 was transformed into E coli BL21-CodonPlus (DE3) cells for expression. BL21-CodonPlus (DE3) cells containing the recombinant plasmid were grown in 1 L of Luria-Bertani (LB) medium in the presence of ampicillin (100 mg/L) at 37 °C with agitation at 250 rounds per minute. Protein expression was induced at 18 °C and 180 rounds per minute by adding 400 μmol/L of 3-Indoleacrylic acid (IAA) when the cultures reached an optical density of 0.4–0.6 at 600 nm (OD600). After 15 h of induction, the cells were harvested by centrifugation for 5 min at 4 °C and 6000 rounds per minute, washed with phosphate-buffered saline (PBS) buffer, and resuspended in lysis buffer (20 mmol/L Tris-HCl pH=7.5, 200 mmol/L NaCl, 1 mmol/L β-mercaptoethanol, 1% Triton X-100, 1 mmol/L PMSF, and Cocktail). The cells were lysed by sonication for 6 min on ice. After centrifugation at 12 000 rounds per minute for 20 min, the supernatant was incubated with Ni-NTA Resin for 1.5 h at 4 °C. After washing with lysis buffer containing 50 mmol/L imidazole, the proteins were eluted with lysis buffer containing 250 mmol/L imidazole, dialyzed with buffer (20 mmol/L Tris-HCl pH=7.5, 200 mmol/L NaCl, 1 mmol/L β-mercaptoethanol, 1 mmol/L EDTA, and 10% glycerol) at 4 °C to remove the imidazole and then stored at −70 °C until use.
pH optimization
An activity assay for ME2 was established using NAD+ and L-malate as substrates; the NADH catalyzed by ME2 was monitored by measuring absorbance at a wavelength of 340 nm.
We prepared nine assay buffers including 50 mmol/L Bis-Tris, 50 mmol/L HEPES, 50 mmol/L Citric acid, 10 mmol/L MgCl2, 12 mmol/L L-malate, and 1 mmol/L NAD+, with different pH values ranging from 4.0 to 8.0. The reactions were started by adding 37 nmol/L of ME2, and the absorbance of NADH at 340 nm was monitored for 4 min using a Spectra Max 340 PC 384 microplate reader.
The Km and Kcat of ME2
To calculate the Km of NAD+, the reactions were started by adding 15 nmol/L ME2 to enzyme reaction mixtures that contained 50 mmol/L MES pH=6.5, 10 mmol/L MgCl2, 24 mmol/L L-malate, and different concentrations of NAD+. To determine the Km of L-malate, the enzyme reaction mixtures contained 50 mmol/L MES pH=6.5, 10 mmol/L MgCl2, 1 mmol/L NAD+, 15 nmol/L ME2, and different concentrations of L-malate. For all reactions, absorbance at 340 nm was monitored for 4 min using a Spectra Max 340 PC 384 microplate reader. Kcat was calculated as Vmax/[E concentration]; its units are 1/s. The formula to calculate Km has been described previously11.
The IC50 and Ki of ATP
The enzyme reaction mixtures contained 50 mmol/L MES pH=6.5, 10 mmol/L MgCl2, 3 mmol/L L-malate, 0.2 mmol/L NAD+, 133 nmol/L ME2, and different concentrations of ATP. The IC50 was calculated using Prism 5 software (Graph Pad, San Diego, CA, USA) from the non-linear curve fitting of the percent inhibition (% inhibition) versus the inhibitor concentration [I] using the following equation: % Inhibition=100/(1+[IC50/[I]]k), where k represents the Hill coefficient and IC50=Ki (1+[S]/Km), where [S] represents the concentration of substrate. When [S] is equal to Km, then Ki=IC50/2.
Z′ factor and S/N of HTS assay of ME2
Both DMSO and ATP were utilized on each plate to calculate the Z′ factor. The enzyme reaction mixtures contained 50 mmol/L MES pH=6.5, 10 mmol/L MgCl2, 3 mmol/L L-malate, 0.2 mmol/L NAD+, 133 nmol/L ME2, and 1 mmol/L ATP or 4% DMSO. The quality control parameter (Z′ factor) was calculated in real time. The Z′ factor was defined in terms of four parameters: the means and standard deviations of both the positive (p) and negative (n) controls (μp, δp, and μn, δn, respectively); Z′=1-3(δp+δn)/(μn−μp). In our assay, ATP and DMSO were the positive and negative controls, respectively.
The signal-to-noise ratio (S/N) is the mean of the positive signal from reactions containing DMSO (ie, 100% enzyme activity) over the mean of the background in the reactions treated with known inhibitors. The background is determined by measuring the replicon levels in wells treated with ATP at 1 mmol/L12.
HTS screening for ME2 inhibitors and exclusion of false positive hits using Brij-35
A total of 12 683 natural products collected from different resources (deposited in Chinese National Compound Library) with wide structural diversity were randomly screened. One microliter of a stock solution of each compound in DMSO was transferred to a well of transparent 384-well medium protein-binding plates; this yielded a final compound concentration of 40 μg/mL and 4% DMSO in a final reaction volume of 25 μL. The assay buffer contained 50 mmol/L MES pH=6.5, 10 mmol/L MgCl2, 3 mmol/L L-malate, and 0.2 mmol/L NAD+. The reactions were initiated by adding 133 nmol/L of ME2, and the absorbance of 340 nm was monitored for 4 min using a Spectra Max 340 PC 384 microplate reader. We used ATP as a positive control and DMSO as a negative control to evaluate our HTS system. Both the negative and positive controls were added to each plate for the calculations. The inhibition effect of each compound was represented by the percentage of the slope of the linear portion of its kinetic curve relative to that of the negative control. The quality control parameters [coefficient of variation (CV) and Z′ factor] were calculated in real time13. All screening operations were performed using a SAGIAN core integrated robotic system (Beckman Coulter, Fullerton, CA, USA).
To calculate the 50% inhibition concentration (IC50), we performed inhibition assays with 133 nmol/L ME2, 50 mmol/L MES pH=6.5, 10 mmol/L MgCl2, 3 mmol/L L-malate, and 0.2 mmol/L NAD+; the inhibitors were diluted to around the estimated IC50 values. To exclude false-positive results from HTS screening, all the hits were tested again using the same assay in an assay buffer containing 0.01% (w/v) Brij-35.
NPD389 is a more potent inhibitor than NPD387
To elucidate the SARs between ME2 and the derivatives, we tested the inhibitory strength of the derivatives of NPD387. The reaction buffer contained 133 nmol/L ME2, 50 mmol/L MES pH=6.5, 10 mmol/L MgCl2, 3 mmol/L L-malate, and 0.2 mmol/L NAD+; the inhibitors were diluted to around the estimated IC50 values. We also included 0.01% (w/v) Brij-35 in the reactions to exclude false positives.
Determination of the characteristics of inhibition of ME2 by NPD389
To investigate whether the inhibition of ME2 by NPD389 was reversible, incubation solution containing 1.3 μmol/L ME2 with 70 μmol/L NPD389 was dialyzed to dissociate the inhibitor from the enzyme for different periods of time, and then the enzyme was dialyzed into the assay buffer (which included the substrates) to initiate the reaction. In the time-independent inhibition experiment, ME2 at a concentration of 1.3 μmol/L was pre-incubated with 70 μmol/L NPD389 on ice for the indicated times, and then added to the assay system. For the inhibitory characterization study, the assay was conducted in the absence or presence of the inhibitor (NPD389 or ATP) at various concentrations; when L-malate was the variable, the concentration of NAD+ was fixed at 1 mmol/L, and when NAD+ was the variable, the concentration of L-malate was fixed at 12 mmol/L. The kinetic data were analyzed using Graph Pad Prism 5.00 and presented as the mean±SD from at least three independent experiments unless otherwise specified.
Thermal shift assay of ME2 in combination with inhibitors
To investigate the binding between ATP and ME2, we used an assay buffer containing 50 mmol/L MES pH=6.5, 10× dilution Sypro Orange, 2.5 μmol/L ME2, and varying concentrations of ATP. The thermal shift assay was conducted using a Light Cycler 480 System. Assay buffer was added to the wells of the 384-well LightCycler 480 Multi-well Plate. The plate was heated from 30 to 80 °C with a heating rate of 0.11 °C/s. The fluorescence intensity was measured with Ex/Em: 465/580 nm.
To evaluate the potential binding between NPD389 and ME2, we used an assay buffer containing 50 mmol/L MES pH=6.5, 10× Sypro Orange, 2.5 μmol/L ME2, and varying concentrations of NPD389. To study the influence of NAD+ on the binding between NPD389 and ME2, we used buffer containing 20 mmol/L NAD+ in the same assay. To determine the effect of L-malate on the binding of the inhibitor to the enzyme, 30 mmol/L L-malate was added to the thermal shift assay buffer.
Statistical analysis
All data shown are representative of three independent experiments. The data are presented as the mean±SD.
Results
Establishment of a high-throughput assay to identify ME2 inhibitors
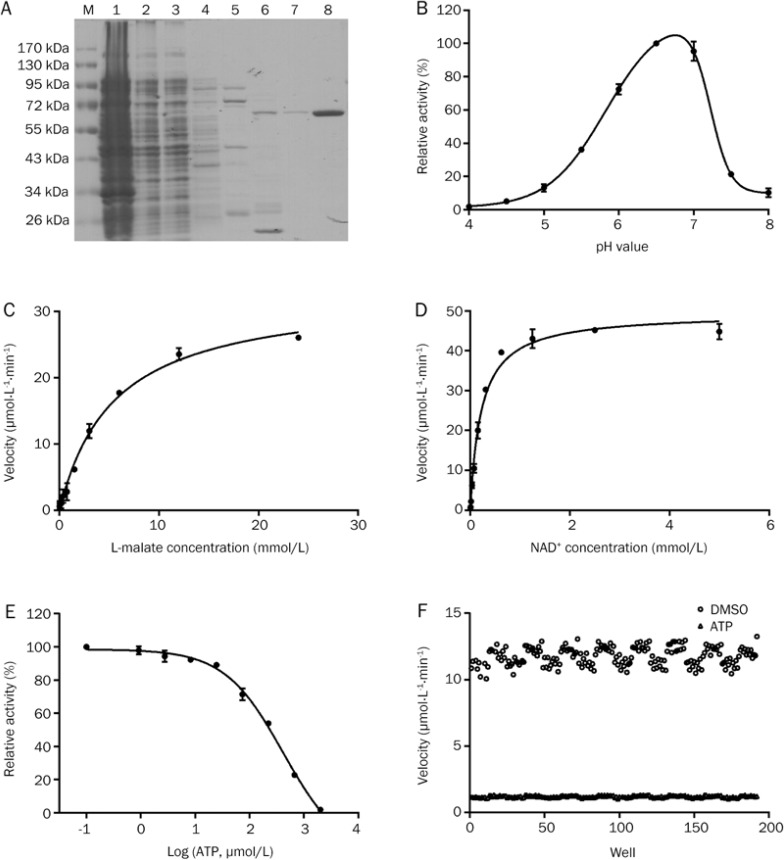
We used E coli strain BL21-CodonPlus to overexpress ME2. Because the recombinant human ME2 protein contains a His-tag, Ni-NTA His-binding resin was applied to purify the recombinant protein. After washing with 10, 50, and 100 mmol/L imidazole solutions, the target proteins were obtained by elution with 250 mmol/L imidazole solution (Figure 1A) and dialyzed at 4 °C to remove the imidazole. SDS-PAGE indicated that the mass of the protein was approximately 60 kDa, which is consistent with previously published results7. The enzyme was purified 142-fold with a yield of 16% from whole lysate, and had a specific activity of 1652.25±11.69 Units·min−1·mg−1 of protein (Table 1).
Figure 1.
Establishment of a high-throughput screening system to identify inhibitors of ME2. (A) SDS-PAGE analysis of purified ME2 separated using a 10% polyacrylamide gel and stained with Coomassie Brilliant Blue. M, protein marker. Lanes 1–8 are the precipitate, supernatant, flow-through, and elution fractions with 10, 50, 100, 250, and 250 mmol/L imidazole, respectively. (B) Optimization of the pH of the screening system. (C, D) Determination of Km and Kcat of L-malate (C) and NAD+ (D). (E) Dose-response curve of inhibition of ME2 by ATP. (F) Determination of the Z′ factor and S/N of the HTS system for ME2 inhibitors. Error bars represent SD. n=3.
Table 1. Summary of ME2 purification process from BL21-CodonPlus.
| Fractions | Total protein (mg) | Specific activity (Units/mg prot) | Total activity (Units) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|
| Supernatant | 1331.09±31.45 | 11.62±0.82 | 15468.63±1091.47 | 1.00 | 100 |
| Flow-through | 1049.77±71.85 | 0.28±0.01 | 294.64±8.35 | 0.02 | 2 |
| Elution (0) | 9.46±0.41 | 1.66±0.09 | 15.70±0.83 | 0.14 | 0 |
| Elution (50) | 50.52±3.09 | 3.19±0.13a | 161.27±6.38a | 0.27a | 1a |
| Elution (100) | 0.79±0.02 | 521.73±7.16a | 414.62±5.69a | 44.90a | 3a |
| Elution (250) | 1.52±0.04 | 1158.04±16.40a | 1764.86±25.00a | 99.65a | 11a |
| Elution (−) | 1.52±0.04 | 1652.25±11.69 | 2511.43±17.76 | 142.19 | 16 |
Note: 1 Unit presents 1 nmol NADH product per minute; Data marked with symbol a represents the catalytic activity determined including imidazole in the elution buffer; Elution (0–250) means the elution fraction using buffer containing 0–250 mmol/L imidazole; The fraction of Elution (−) was the dialyzed fraction of Elution (250) from the imidazole.
The pH value of a solution has a great effect on enzyme activity. To obtain the maximal activity of ME2, we optimized the pH value. We measured enzyme activity in buffers with pH values ranging from 4.0 to 8.0. ME2 had the highest activity at a pH of 6.5 (Figure 1B). We then determined the Km of NAD+ and L-malate in this enzymatic system. The Km and Kcat of L-malate were 5.47±0.40 mmol/L and 35.6 s−1, respectively, whereas the Km and Kcat of NAD+ were 0.22±0.02 mmol/L and 55.12 s−1, respectively (Figure 1C and 1D); these values are consistent with the previously reported Kms of L-malate and NAD+ (8.39±1.09 mmol/L and 0.35±0.02 mmol/L, respectively14).
It is important to optimize the concentrations of the substrates before eslishing a high-throughput screening system. When a substrate concentration less than the Km value is used, the screening system can easily identify competitive inhibitors. When a substrate concentration greater than the Km value is used, the screening system can much more easily identify uncompetitive inhibitors. Non-competitive inhibitors of the enzyme will not be greatly affected by the substrate concentration15,16. To acquire as many novel inhibitors of ME2 as possible, we used 0.2 mmol/L NAD+ and 3 mmol/L L-malate in the HTS assay; both of these concentrations are close to the respective Km values.
Determination of IC50 and Ki of the physiological inhibitor ATP
ATP, the physiological inhibitor of ME2, was utilized to evaluate the stability and quality of our HTS assay. The IC50 of ATP in our assay system was determined to be 414±23.92 μmol/L (Figure 1E). Because ATP is a competitive inhibitor with regard to NAD+, the concentration of substrate applied in our assay system should be equal to Km, so the Ki of ATP was calculated to be 0.2 mmol/L using the formula IC50=Ki(1+[S]/Km); this value was close to the Ki value of ATP of 0.2±0.02 mmol/L published previously7.
Validation of the HTS assay
Because a large-scale screen would require large amounts of materials and compounds, we began by carrying out a pilot screen to assess the quality of the HTS assay. The Z′ factor has been proposed for reference in high-throughput screening to represent the response of an inhibitor or activator in a particular assay. If the value of the Z′ factor exceeds 0.5, it indicates that the assay is viable17,18,19,20,21. We used ATP as the positive control and DMSO as the negative control to evaluate our HTS system. The Z′ factor of our HTS system was calculated to be 0.775 (Figure 1F), which indicates that our assay is appropriate for high-throughput screening.
The second criterion that can be used to measure the robustness of an assay is the signal-to-noise ratio (S/N). S/N indicates the mean of the positive signal over the mean of the background22. In our system, S/N was calculated to be 9.80, suggesting that our ME2 assay has robust and reproducible window for the inhibitor discovery.
Discovery of a novel inhibitor of ME2 by HTS
A total of 12 683 natural products from the Chinese National Compound Library were screened. We obtained 47 natural products as hits from the primary screening at a concentration of 40 μg/mL; the hit rate was 0.37%. We then determined the dose-response curves of these 47 inhibitors.
HTS is a primary technique for the identification of novel leads in drug discovery and chemical biology. Unfortunately, it is susceptible to false-positive hits. One common cause of false-positives is the accumulation of organic molecules into colloidal aggregates, which inhibit enzymes nonspecifically23. Detergents such as Brij-35 are often used in drug discovery research to rule out the presence of promiscuous small molecules and non-specific inhibitors that act by aggregating in solution or through undesirable precipitation in aqueous assay buffers24,25. As expected, we excluded 32 promiscuous compounds using 0.01% Brij-35 in the subsequent dose-response curve determination, leaving 15 ME2 inhibitors with different structures. The final hit rate of the HTS assay was therefore 0.12%.
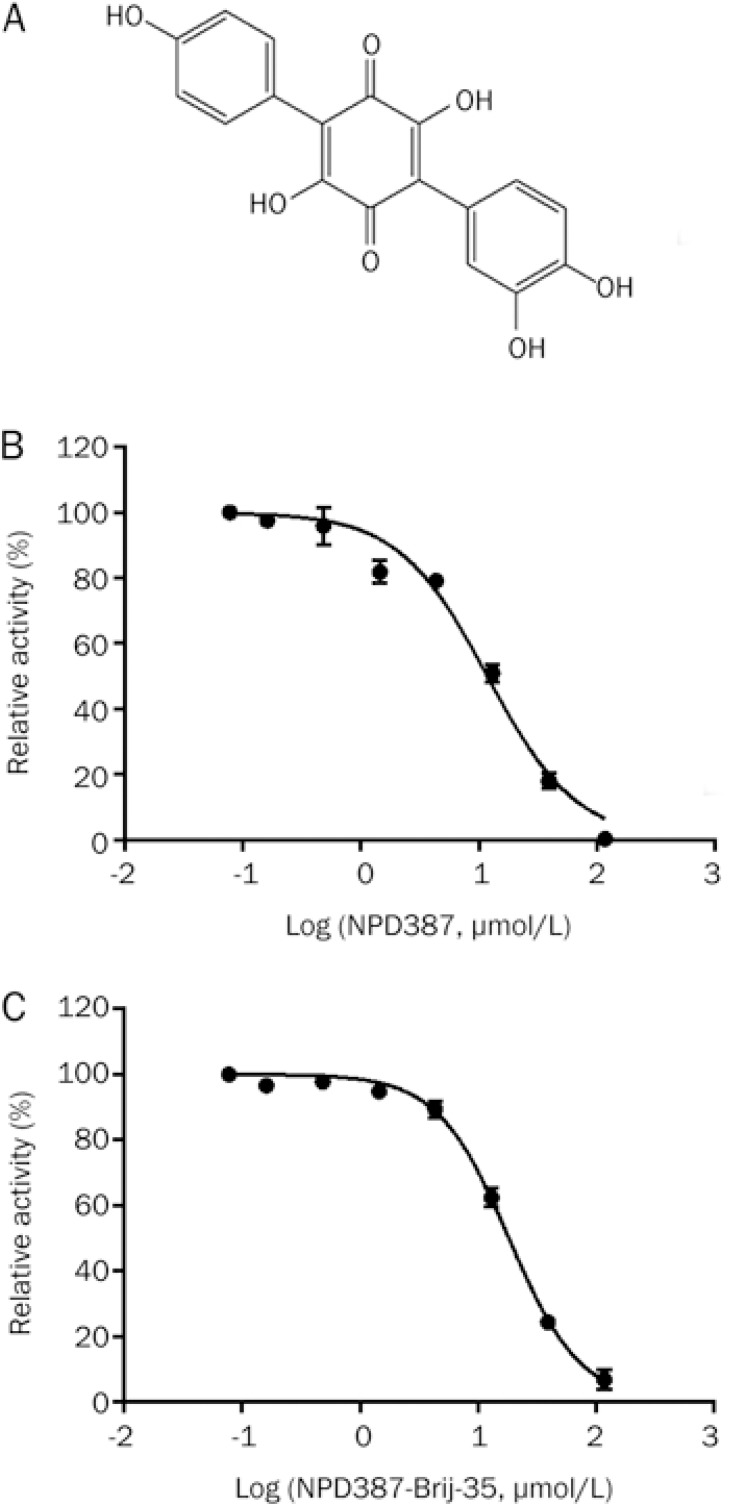
After the hits were validated and dose-response curves were generated, a compound named NPD387 with a novel structure (Figure 2A) (chemical name: 3,3′,4,4″″,6′-pentahydroxy-[1,1′:4′,1″″-terphenyl]-2′,5′-dione) was identified as a moderate inhibitor of ME2, with IC50 values of 11.84±0.57 μmol/L and 18.27±1.46 μmol/L in the absence and presence of 0.01% Brij-35, respectively (Figure 2B, 2C, Table 2).
Figure 2.
NPD387 is a novel inhibitor of ME2. (A) The structure of NPD387. (B) Dose-response curve of inhibition of ME2 by NPD387. (C) Dose-dependent inhibition of ME2 by NPD387 with 0.01% Brij-35. Error bars represent SD. n=3.
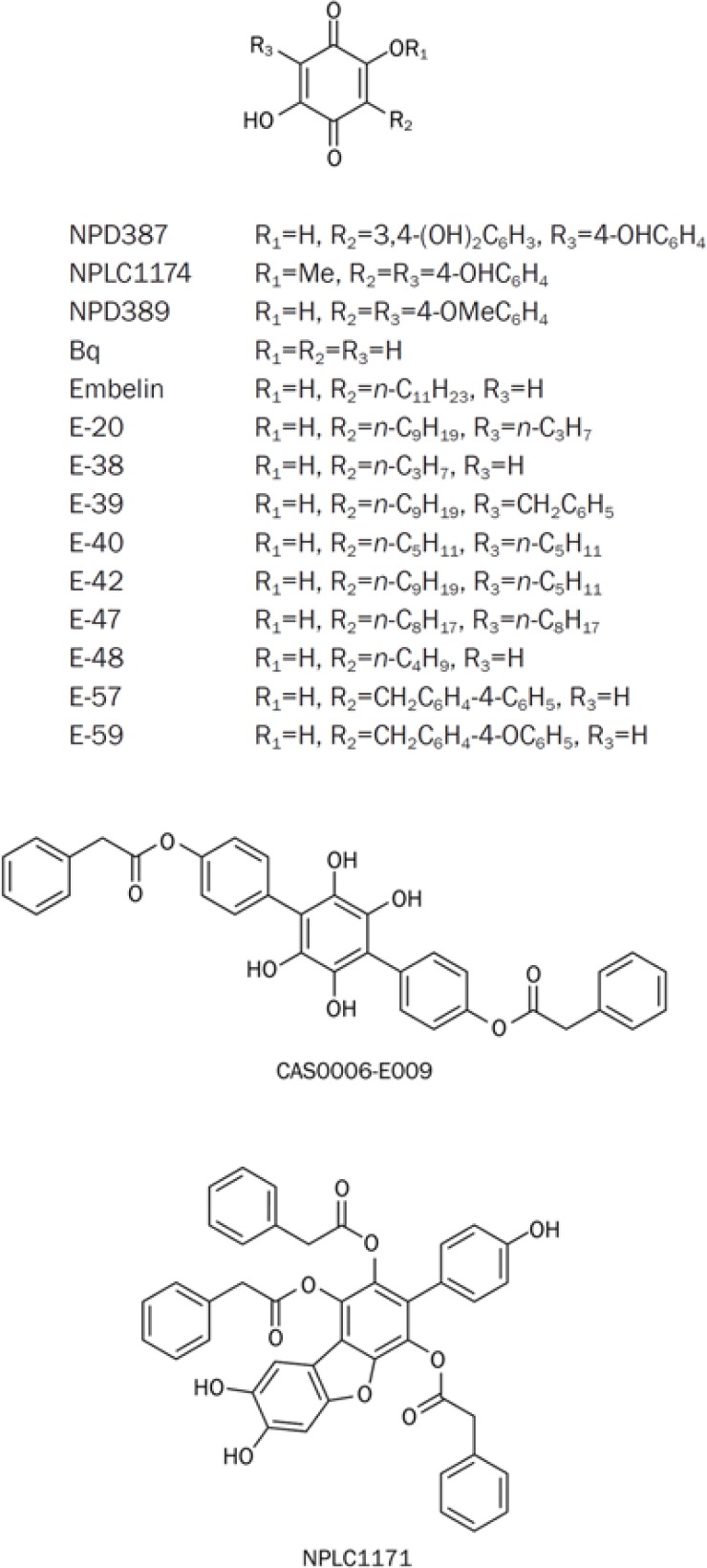
Table 2. Screening structural analogs of NPD387.
| Compound | R1 | R2 | R3 | IC50 (μmol/L) |
|
|---|---|---|---|---|---|
| No Brij-35 | 0.01% Brij-35 | ||||
| NPD387 | H | 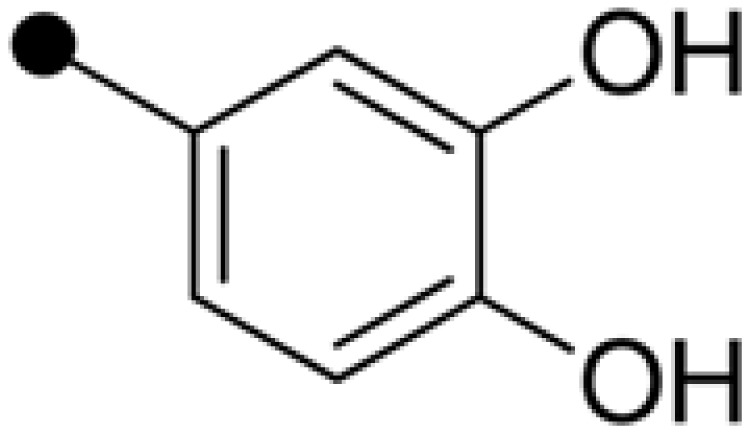 |
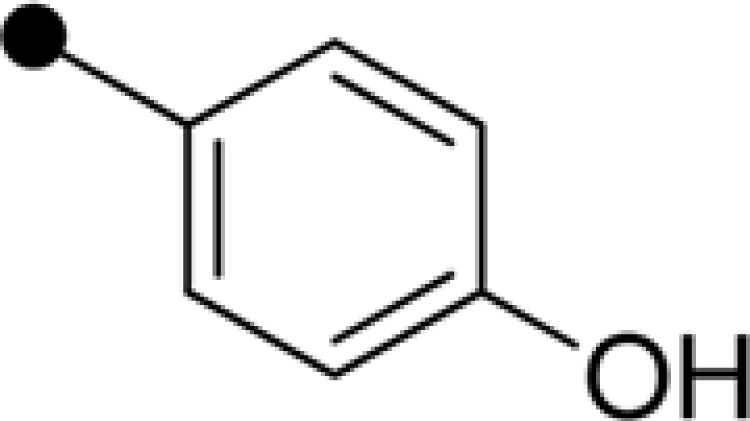 |
11.84±0.57 | 18.27±1.46 |
| CAS0006-E009 | * | * | * | 2.13±0.20 | 31.02±1.54 |
| NPLC1171 | # | # | # | 48.63±2.36 | – |
| NPLC1174 | Me |  |
 |
– | – |
| NPD389 | H | 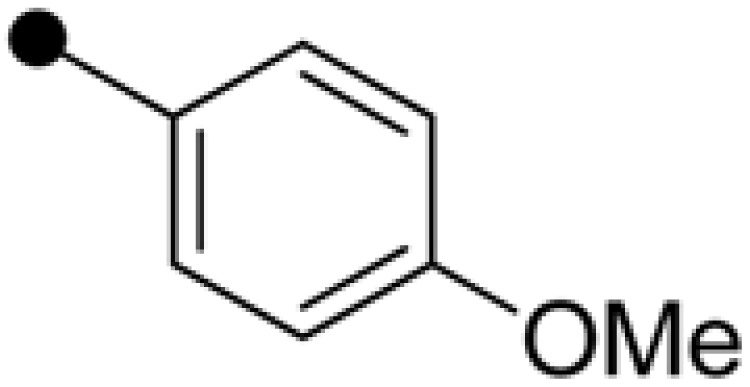 |
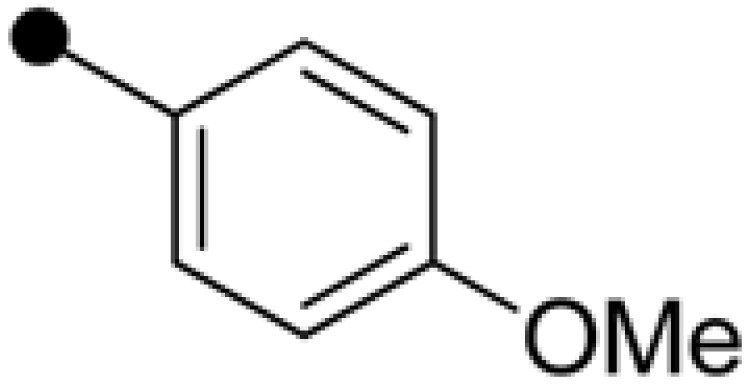 |
4.63±0.36 | 5.59±0.38 |
| Bq | H | H | H | 8.34±0.71 | 7.32±2.31 |
| Embelin | H |  |
H | 52.40±3.88 | 110.68±4.77 |
| E-20 | H |  |
 |
– | – |
| E-38 | H |  |
H | 184.61±6.97 | 103.78±5.60 |
| E-39 | H |  |
 |
69.23±3.09 | – |
| E-40 | H |  |
 |
– | – |
| E-42 | H |  |
 |
– | – |
| E-47 | H |  |
 |
– | – |
| E-48 | H |  |
H | – | 158.74±8.24 |
| E-57 | H |  |
H | 17.43±1.17 | 23.17±1.50 |
| E-59 | H |  |
H | 18.59±1.25 | 34.09±3.28 |
SARs between ME2 and derivatives
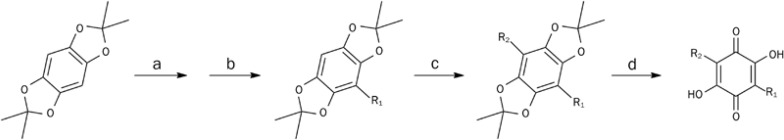
After the identification of NPD387 as a lead compound in the search for inhibitors of ME2, we synthesized a series of NPD387 derivatives and measured their inhibition of ME2 to determine their SARs (Figure 3). Bq and Embelin were purchased from Alfa Aesar (Shanghai, China) and used without any further purification, and the other derivatives were prepared as described previously26 and as detailed in Scheme 1. The preliminary SARs are shown below.
Figure 3.
The structures and formulae of derivatives of NPD387.
Scheme 1.
The synthesis of derivatives of NPD387. Reagent: (a) n-BuLi, THF, −10 °C; (b) R1, Br, −10 °C to RT; (c) n-BuLi, THF, −10 °C; R2, Br, −10 °C to RT; (d) 1,4-dioxane, 6 N HCl, air, reflux.
It is clear that the original compound, NPD387, and one derivative, NPD389, showed strong ME2 inhibitory activities, but NPLC1174 exhibited no potency, which indicates that the 2,5-dihydroxyl groups are indispensable for the activity of the compound. In addition, when the carbonyl groups of the 2,5-dihydroxyl benzoquinone skeleton were reduced to phenolic hydroxyl, the compound displayed decreased activity (CAS0006-E009). Therefore, the 2,5-dihydroxyl benzoquinone skeleton is essential for the ME2 inhibitory activity of these compounds.
In addition, we tested a series of 3-alkyl-substituted 2,5-dioxo-benzoquinones and 3,6-dialkyl substituted 2,5-dioxo-benzoquinones. We found that the activities of these compounds were decreased to different extents. Specifically, the C3 alkyl chain-substituted compounds showed remarkable decreases in activity, and the C3,C6 dialkyl-substituted compounds exhibited a complete loss of activity. Among these compounds, only compounds E-57 and E-59 retained some inhibitory activity. This phenomenon indicates that the presence of flexible alkyl groups at C3 or C6 is incompatible with inhibition of ME2.
Among the compounds we screened, NPD387, NPD389 and Bq displayed the strongest inhibition of ME2. The common feature of these three compounds is their 2,5-dihydroxy benzoquinone skeleton. By comparing the IC50s of NPD387 and NPD389 with that of Bq, we found that substituted phenyl groups at C3 and C6 had some impact on inhibitory activity towards ME2. Specially, 4-OMe-substituted phenyl groups at C3 and C6 could improve the activity (NPD389), but 4-OH substituted phenyl groups at C3 and C6 could weaken the interaction between the compound and ME2 (NPD387), decreasing the inhibitory activity of the compound.
Characterization of NPD389 as a more potent ME2 inhibitor
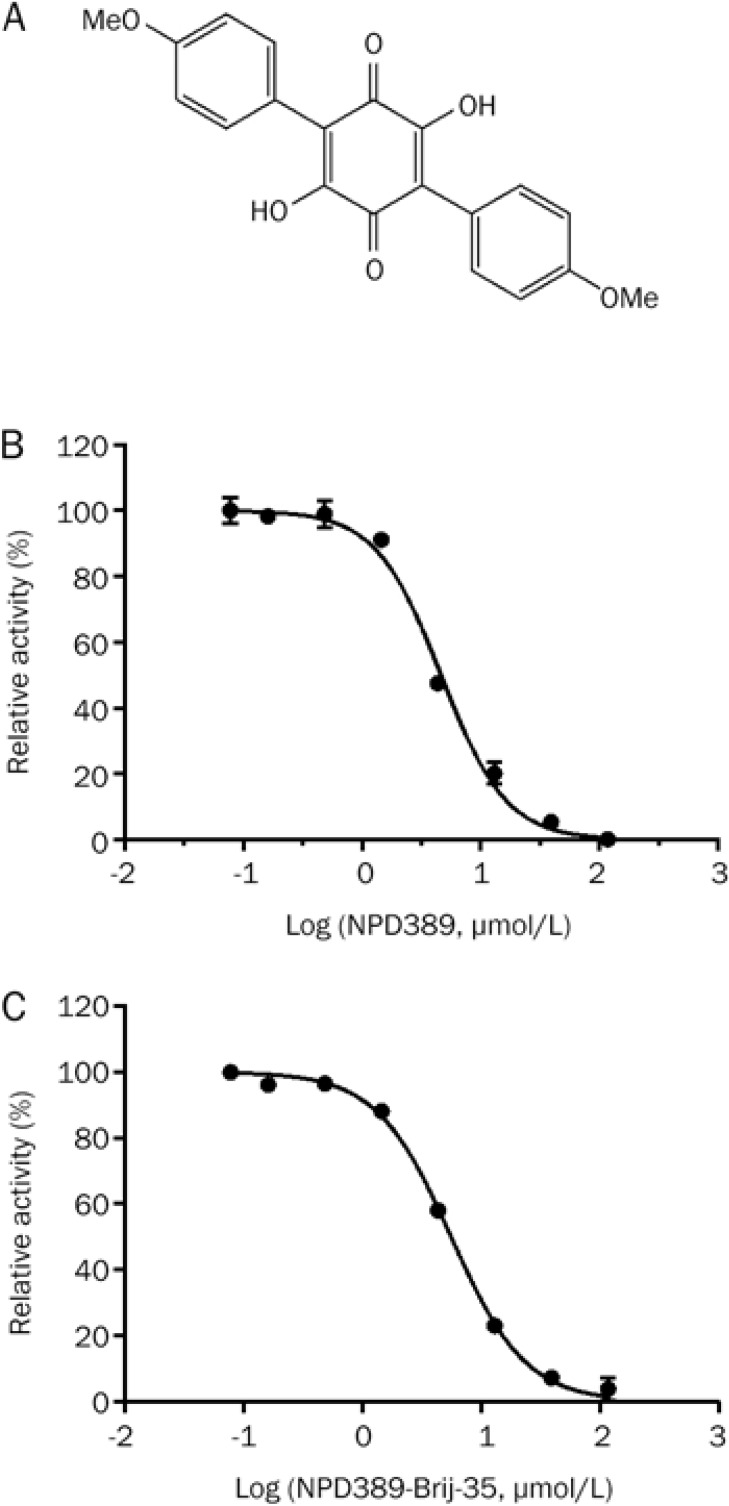
Through structural modification, we generated NPD389, which is a more potent inhibitor of ME2 than NPD387 (Figure 4A). The IC50 of NPD389 was 4.63±0.36 μmol/L or 5.59±0.38 μmol/L when 0.01% Brij-35 was absent from or present in the assay system, respectively (Figure 4B and 4C, Table 2).
Figure 4.
NPD389 was identified as a more potent inhibitor of ME2 than NPD387. (A) The structure of NPD389. (B) Dose-response curve of inhibition of ME2 by NPD389. (C) Dose-dependent inhibition of ME2 by NPD389 with 0.01% Brij-35. Error error bars represent SD. n=3.
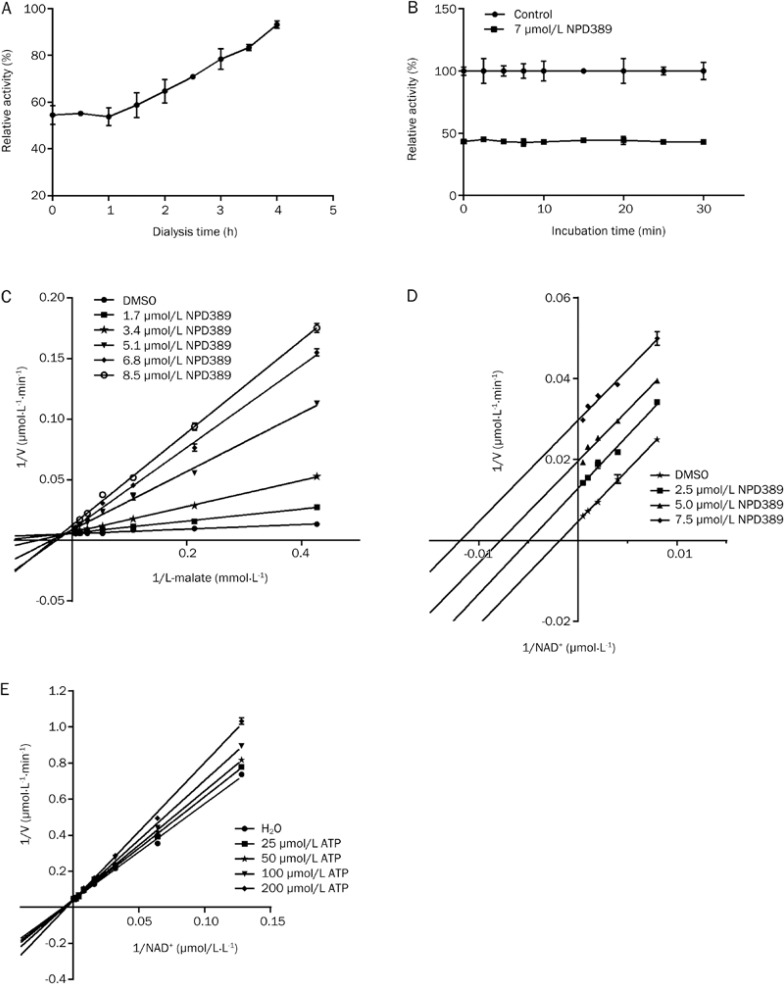
We further investigated the mode by which NPD389 inhibits ME2. NPD389 is a reversible inhibitor of ME2, as shown by the restored activity of ME2 when the dialysis time was extended (Figure 5A). The unchanged inhibitory rate over incubation time indicated that NPD389 binds with ME2 in a fast-binding mode (Figure 5B). NPD389 is a mixed-type competitive inhibitor for the substrate L-malate, as shown by the increased Km values and decreased Vmax values that are seen when the concentration of inhibitor is increased; the Lineweaver-Burk plot of different concentrations of NPD389 intersected in the second quadrant of the reciprocal plot (Figure 5C). Moreover, NPD389 is an uncompetitive inhibitor for the other substrate NAD+ because both the Km and Vmax values decreased when the concentration of inhibitor was increased, and the Lineweaver-Burk plots of four different inhibitor concentrations showed parallel lines (Figure 5D). In this enzyme kinetic characterization study, our data showed that ATP (the positive control) is a competitive inhibitor with respect to NAD+ (Figure 5E), consistent with previously reports7.
Figure 5.
The mode of inhibition of ME2 by NPD389. (A) The effect of NPD389 on enzyme activity after incubation with the enzyme and subsequent dialysis. (B) Time-independent inhibition of ME2 by NPD389. (C) Double-reciprocal plot with changes in the concentrations of the inhibitor NPD389 and the substrate L-malate. (D) Double-reciprocal plot with changes in the concentrations of the inhibitor NPD389 and the substrate NAD+. (E) Double-reciprocal plot with changes in the concentrations of the inhibitor ATP and the substrate NAD+. Error bars represent SD. n=3.
Validation of NPD389 binding to ME2 through thermal shift assay
The fluorescence-based thermal shift assay is a general method for the identification of inhibitors of target proteins from compound libraries. This method is based on the energetic coupling between ligand binding and protein unfolding. Using an environmentally sensitive fluorescent dye (Sypro Orange) to monitor protein thermal unfolding, the ligand-binding affinity can be assessed from the shift in the unfolding temperature (ΔTm) obtained in the presence of ligands relative to that obtained in the absence of ligands27,28. A shift in the melting temperature (Tm) of an enzyme during incubation with a given test compound indicates that the test compound specifically binds to the enzyme.
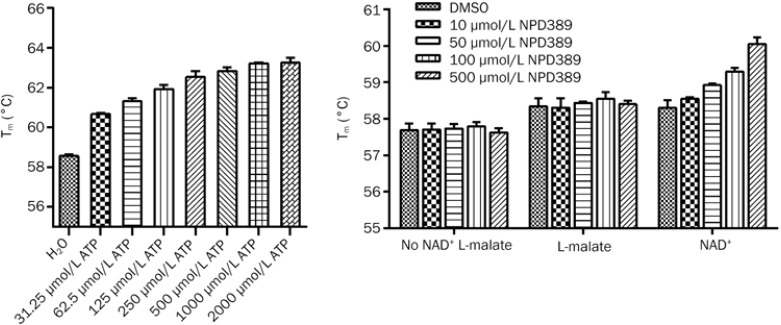
After optimization with ATP as positive control inhibitor, we used a 10× dilution of Sypro Orange and 2.5 μmol/L ME2 protein in the thermal shift assay; under these conditions, a perfect melting curve could be obtained. As shown in Figure 6A, there was a dose-dependent shift in Tm with the increasing concentrations of ATP, and a ΔTm of 4.26±0.18 °C was calculated by comparing Tm in the absence of ATP to that with 500 μmol/L of ATP, which was close to its IC50 value (Figure 6A). This thermal shift result confirmed that ATP is a competitive inhibitor of ME2 with respect to NAD+.
Figure 6.
The effect of ATP and NPD389 on the Tm of ME2 probed with Sypro Orange. (A) The effect of varying concentrations of ATP on Tm. (B) The effect of NPD389 on Tm. From left to right, the groups are: without NAD+ and L-malate; in the presence of L-malate; and in the presence of NAD+, respectively. Error bars represent SD. n=3.
We then determined whether there was a shift in the Tm of ME2 with NPD389. The result indicated that there was a dose-dependent shift Tm in the presence of NAD+. However, there was no significant difference when the assay included only L-malate or neither substrate (Figure 6B). This result demonstrated that NPD389 is an uncompetitive inhibitor of ME2 with respect to NAD+ because the shift in the Tm of ME2 induced by NPD389 occurred when the substrate NAD+ was present.
Discussion
Evidence from previous studies has supported the concept that ME2 may be a promising target for cancer therapy. No inhibitor of human ME2 has been reported previously except for the physiological inhibitor ATP; this lack of an inhibitor makes it difficult to validate the role of ME2 in tumorigenesis and to develop cancer therapies based on ME2. The purpose of this study was to discover a novel inhibitor of human ME2.
Since being developed approximately 20 years ago, HTS has become a key technique in drug discovery. HTS provides a fast, effective, and convenient way to test hundreds of thousands of structurally diverse compounds for their ability to modulate disease-relevant targets and to identify novel compounds with interesting biological activities; this approach yields molecules that can be optimized into drugs. In this study, an HTS assay was established to identify small-molecule inhibitors of ME2. The average Z′ factor of this assay was 0.775, which demonstrates satisfactory HTS quality control.
Through large-scale screening and structural modification, we identified NPD389 as the first potent ME2 inhibitor. Inhibition of ME2 by NPD389 is independent of incubation time, which indicates that NPD389 is a fast-binding inhibitor of ME2. We also demonstrated that NPD389 is a mixed type inhibitor with respect to L-malate and an uncompetitive inhibitor with respect to NAD+ using a double reciprocal plot.
A key problem in HTS is the prevalence of non-specific inhibitors. These molecules, which have peculiar properties, act on unrelated targets and dominate the results of screening experiments. Part of the explanation for these nonspecific effects is that some organic molecules form large colloid-like aggregates that sequester and thereby inhibit enzymes23. Detergents such as Brij-35 and Tween are often used in drug discovery to weed out promiscuous small molecules and non-specific inhibitors that cause aggregation in solutions and undesirable precipitation in aqueous assay buffers24,25. Compounds CAS0006-E009 and NPLC1171 might aggregate in a solution, interfering with binding between the enzyme and the substrate and thereby resulting in remarkable differences in IC50 values measured with or without Brij-35.
To further investigate the interaction between the inhibitor and the enzyme, we used a thermal shift assay to test the combination of inhibitors and ME2. The fluorescence-based thermal shift assay is a general method for identifying inhibitors of target proteins from compound libraries. It has the advantages of simple operation, short experimental cycle, and ease of further evaluation of compounds. The thermal shift assay result showed that there was a dose-dependent shift in Tm as the concentration of ATP increased. After testing the combination of NPD389 and ME2, we found that there was a dose-dependent increase in Tm in the presence of NAD+. However, the change in the Tm of ME2 with NPD389 disappeared when there was no NAD+ in the system. These results showed that NPD389 binds to the complex of ME2 and NAD+ and that NAD+ is necessary for the binding of NPD389 to ME2. These results also confirmed that NPD389 was an uncompetitive inhibitor with respect to NAD+. Our study illustrated that the substrate of an enzyme should not be omitted in a thermal shift assay when the inhibitor is uncompetitive.
In summary, we identified a novel 2,5-dihydroxyl benzoquinone skeleton as an inhibitor of ME2 and identified a potent derivative of this inhibitor, NPD389, from a preliminary SAR study. A kinetic characterization study demonstrated that NPD389 is a fasting-binding inhibitor of ME2 and acts as an uncompetitive inhibitor with respect to NAD+ and a mixed-type competitive inhibitor with respect to L-malate.
Author contribution
Jia LI, Li-hong HU, Jing GAO, and Jing-ya LI designed the study; Yi WEN and Lei XU performed the experiments and prepared the manuscript; and Fang-lei CHEN synthesized the derivatives of NPD387.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81125023 and 81021062), the National Science and Technology Major Projects for Major New Drugs Innovation and Development (2013ZX09507002), and the Shanghai Commission of Science and Technology (11DZ2292200 and 13DZ2290300). We also thank Yan-yun FU and Bei-ying QIU for their help in editing the manuscript.
References
- Hsu RY, Lardy HA. Pigeon liver malic enzyme. II. Isolation, crystallization, and some properties. J Biol Chem 1967; 242: 520–6. [PubMed] [Google Scholar]
- Loeber G, Infante AA, Maurer-Fogy I, Krystek E, Dworkin MB. Human NAD(+)-dependent mitochondrial malic enzyme. cDNA cloning, primary structure, and expression in Escherichia coli. J Biol Chem 1991; 266: 3016–21. [PubMed] [Google Scholar]
- Xu Y, Bhargava G, Wu H, Loeber G, Tong L. Crystal structure of human mitochondrial NAD(P)+-dependent malic enzyme: a new class of oxidative decarboxylases. Structure 1999; 7: R877–89. [PubMed] [Google Scholar]
- Ren JG, Seth P, Everett P, Clish CB, Sukhatme VP. Induction of erythroid differentiation in human erythroleukemia cells by depletion of malic enzyme 2. PLoS One 2010; 5: e12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Garcia S, Lopez-Gonzalez JS, Baez-Viveros JL, Aguilar-Cazares D, Prado-Garcia H. Tumor cell metabolism: an integral view. Cancer Biol Ther 2011; 12: 939–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Du W, Mancuso A, Wellen KE, Yang X. Reciprocal regulation of p53 and malic enzymes modulates metabolism and senescence. Nature 2013; 493: 689–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Lanks CW, Tong L. Molecular mechanism for the regulation of human mitochondrial NAD(P)+-dependent malic enzyme by ATP and fumarate. Structure 2002; 10: 951–60. [DOI] [PubMed] [Google Scholar]
- Wasilenko WJ, Marchok AC. Malic enzyme and malate dehydrogenase activities in rat tracheal epithelial cells during the progression of neoplasia. Cancer Lett 1985; 28: 35–42. [DOI] [PubMed] [Google Scholar]
- Sauer LA, Dauchy RT, Nagel WO, Morris HP. Mitochondrial malic enzymes. Mitochondrial NAD(P)+-dependent malic enzyme activity and malate-dependent pyruvate formation are progression-linked in Morris hepatomas. J Biol Chem 1980; 255: 3844–8. [PubMed] [Google Scholar]
- Hou S, Liu W, Ji D, Zhao ZK. Efficient synthesis of triazole moiety-containing nucleotide analogs and their inhibitory effects on a malic enzyme. Bioorg Med Chem Lett 2011; 21: 1667–9. [DOI] [PubMed] [Google Scholar]
- Cleland WW. Statistical analysis of enzyme kinetic data. Methods Enzymol 1979; 63: 103–38. [DOI] [PubMed] [Google Scholar]
- Mondal R, Koev G, Pilot-Matias T, He Y, Ng T, Kati W, et al. Development of a cell-based assay for high-throughput screening of inhibitors against HCV genotypes 1a and 1b in a single well. Antiviral Res 2009; 82: 82–8. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 1999; 4: 67–73. [DOI] [PubMed] [Google Scholar]
- Hung HC, Chien YC, Hsieh JY, Chang GG, Liu GY. Functional roles of ATP-binding residues in the catalytic site of human mitochondrial NAD(P)+-dependent malic enzyme. Biochemistry 2005; 44: 12737–45. [DOI] [PubMed] [Google Scholar]
- Cornish-Bowden A. Why is uncompetitive inhibition so rare? A possible explanation, with implications for the design of drugs and pesticides. FEBS Lett 1986; 203: 3–6. [DOI] [PubMed] [Google Scholar]
- Dixon M. The determination of enzyme inhibitor constants. Biochem J 1953; 55: 170–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 1999; 4: 67–73. [DOI] [PubMed] [Google Scholar]
- Sui YX, Wu ZJ. Alternative statistical parameter for high-throughput screening assay quality assessment. J Biomol Screen 2007; 12: 229–34. [DOI] [PubMed] [Google Scholar]
- Zhang XD, Espeseth AS, Johnson EN, Chin J, Gates A, Mitnaul LJ, et al. Integrating experimental and analytic approaches to improve data quality in genome-wide RNAi screens. J Biomol Screen 2008; 13: 378–89. [DOI] [PubMed] [Google Scholar]
- Zhang XD. A pair of new statistical parameters for quality control in RNA interference high-throughput screening assays. Genomics 2007; 89: 552–61. [DOI] [PubMed] [Google Scholar]
- Zhang XD. Novel analytic criteria and effective plate designs for quality control in genome-scale RNAi screens. J Biomol Screen 2008; 13: 363–77. [DOI] [PubMed] [Google Scholar]
- Mondal R, Koev G, Pilot-Matias T, He YP, Ng T, Kati W, et al. Development of a cell-based assay for high-throughput screening of inhibitors against HCV genotypes 1a and 1b in a single well. Antiviral Res 2009; 82: 82–8. [DOI] [PubMed] [Google Scholar]
- McGovern SL, Helfand BT, Feng B, Shoichet BK. A specific mechanism of nonspecific inhibition. J Med Chem 2003; 46: 4265–72. [DOI] [PubMed] [Google Scholar]
- Feng BY, Shelat A, Doman TN, Guy RK, Shoichet BK. High-throughput assays for promiscuous inhibitors. Nat Chem Biol 2005; 1: 146–8. [DOI] [PubMed] [Google Scholar]
- Walters WP, Namchuk M. Designing screens: how to make your hits a hit. Nat Rev Drug Discov 2003; 2: 259–66. [DOI] [PubMed] [Google Scholar]
- Gan X, Jiang W, Wang W, Hu L. An approach to 3,6-disubstituted 2,5-dioxybenzoquinones via two sequential Suzuki couplings. three-step synthesis of leucomelone. Org Lett 2009; 11: 589–92. [DOI] [PubMed] [Google Scholar]
- Pantoliano MW, Petrella EC, Kwasnoski JD, Lobanov VS, Myslik J, Graf E, et al.High-density miniaturized thermal shift assays as a general strategy for drug discovery. J Biomol Screen 2001; 6: 429–40. [DOI] [PubMed] [Google Scholar]
- Lo MC, Aulabaugh A, Jin G, Cowling R, Bard J, Malamas M, et al. Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Anal Biochem 2004; 332: 153–9. [DOI] [PubMed] [Google Scholar]