Abstract
Few studies have focused on the early colonization of New Caledonia by insects, after the re-emergence of the main island, 37 Myr ago. Here we investigate the mode and tempo of evolution of a new endemic cricket genus, Pixibinthus, recently discovered in southern New Caledonia. First we formally describe this new monotypic genus found exclusively in the open shrubby vegetation on metalliferous soils, named ‘maquis minier’, unique to New Caledonia. We then reconstruct a dated molecular phylogeny based on five mitochondrial and four nuclear loci in order to establish relationships of Pixibinthus within Eneopterinae crickets. Pixibinthus is recovered as thesister clade of the endemic genus Agnotecous, mostly rainforest-dwellers. Dating results show that the island colonization by their common ancestor occurred around 34.7 Myr, shortly after New Caledonia re-emergence. Pixibinthus and Agnotecous are then one of the oldest insect lineages documented so far for New Caledonia. This discovery highlights for the first time two clear-cut ecological specializations between sister clades, as Agnotecous is mainly found in rainforests with 19 species, whereas Pixibinthus is found in open habitats with a single documented species. The preference of Pixibinthus for open habitats and of Agnotecous for forest habitats nicely fits an acoustic specialization, either explained by differences in body size or in acoustic properties of their respective habitats. We hypothesize that landscape dynamics, linked to major past climatic events and recent change in fire regimes are possible causes for both present-day low diversity and rarity in genus Pixibinthus. The unique evolutionary history of this old New Caledonian lineage stresses the importance to increase our knowledge on the faunal biodiversity of ‘maquis minier’, in order to better understand the origin and past dynamics of New Caledonian biota.
Introduction
Understanding the origin of island biota requires that the evolutionary history of clades is reconstructed in details and linked to the evolution of local environments [1,2]. This task is both especially interesting and demanding in hotspots of biodiversity, such as New Caledonia in the Southwest Pacific, where richness and endemism reach outstanding levels [3,4,5]. New Caledonia has a long and complex environmental history since the main island re-emerged 37 ± 2 Myr ago [6,7], after having been largely covered with an ophiolithic layer during its obduction under the Pacific plate [7,8]. This layer has been progressively reduced to only one third of the island surface after several intense weathering events under climate control. These geological and climatic events led to the establishment of a mosaic of soils, including metalliferous soils (rich in heavy metals such as magnesium, manganese and nickel), habitats and landscapes (e.g., [9]). The present-day vegetation on metalliferous soils is constituted by two main native types, rainforest and open shrubby vegetation named ‘maquis minier’, both harboring many endemic species [10,11]. Many studies refer to the interaction between metalliferous soils and the corresponding vegetation as a peculiar and original system dating back to the first ages of the island colonization. Pre-adaptation or adaptation to these specific soils on ultramafic rocks would have been an early engine of diversification of endemism for at least a major part of New Caledonian biota with the subsequent prediction that island old groups have narrow relationships to metalliferous soils [10,12,13]. This theory has both been corroborated and refuted depending on group of organism studied (e.g., [14,15,16,17]) and a dated phylogenetic framework is now clearly required to clarify this subject. The present-day vegetation dynamics can also obscure the reconstruction of past processes. The balance between ‘maquis minier’ and forest is controlled by climatic variations, fire regimes and human influences leading to a present increase and over-representation of this open vegetation type [18,19,20]. Therefore, ‘maquis minier’ is often seen as a secondary and disturbed vegetation type when it is actually a rich original endemic formation potentially indicative of early New Caledonian ecosystems [21,22].
Our study is aimed at unraveling the evolution of an insect lineage endemic to ‘maquis minier’, to date its origin, to emphasize on its adaptation to the habitat and to suggest future research directions for better understanding its relationships to landscape dynamics and human disturbance. We focused on the insect group of crickets (Orthoptera, Grylloidea) that gained increasing attention from the scientific community as they both play a significant role in ecosystem functioning [23]and are relatively sensitive to environmental disturbances [24]. In New Caledonia, the cricket fauna is rich and has been recently studied, providing some insights on the dynamics of the micro-endemism and of the adaptation to the environment [25,26,27,28]. This fauna was still insufficiently known in ‘maquis minier’ where we concentrated our efforts, permitting the discovery and description of a new genus, PixibinthusRobillard and Anso,gen. nov., that was used to ask the present questions.
Material and Methods
Study sites and cricket sampling
Intensive cricket samplings were performed from October 2013 to March 2014 to study the composition of cricket species along a gradient from ‘maquis minier’ to rainforest (S3 Table). We selected our study sites on the uniform metalliferous soil located in the southern part of Grande Terre. We subsequently recorded for each sampling site the absence / presence of each cricket species. For each cricket sampled in the field, time of day and habitats were noted. Crickets were identified as morphospecies at the Institut de Recherche pour le Développement (IRD) in Noumea, and identifications were checked by comparisons with the collections of the Muséum national d’Histoire naturelle, Paris (MNHN). Specimens were deposited in the collections of the MNHN, and in collections of the IRD, Noumea (ONNC), and in collection of the Institut Agronomique néo-Calédonien, Pocquereux (CXMNC). Field Permit: the Province Sud Environment Office (DENV- Direction de l’Environnement de la Province Sud).
Taxon description
Morphological descriptions follow the pattern and terminology of recent publications (e.g., [29,30]). Direct observations and dissections have been made using a binocular microscope Leica MZ16 at magnifications up to 115×. Male tegminal veins and cells follow terminology of Ragge [31] and Robillard & Desutter-Grandcolas [32]. Male and female genitalia have been dissected in softened or fresh specimens. Male genitalia were dissected by cutting the membranes between the paraprocts and the subgenital plate; in females, copulatory papilla were dissected by cutting the membranes between the ovipositor and the subgenital plate. Genitalia have been cleared with cold KOH and preserved in glycerine in vials pinned under specimens. Male genitalia terminology follows Desutter [33], modified in Desutter-Grandcolas [34] and Robillard & Desutter-Grandcolas [32]. Photographs of male genitalia were obtained using an AmScope MU1000 digital camera (www.Amscope.com). Genitalia were stained with a drop of Punktol (JLB, Germany).
Nomenclatural Acts
The electronic edition of this article conforms to the requirements of the amended International Code of Zoological Nomenclature, and hence the new names contained herein are available under that Code from the electronic edition of this article. This published work and the nomenclatural acts it contains have been registered in ZooBank, the online registration system for the ICZN. The ZooBank LSIDs (Life Science Identifiers) can be resolved and the associated information viewed through any standard web browser by appending the LSID to the prefix “http://zoobank.org/”. The LSID for this publication is: urn:lsid:zoobank.org:pub: AFAE2A4F-7161-4100-A1DB-D420FA5C0435. The electronic edition of this work was published in a journal with an ISSN, and has been archived and is available from the following digital repositories: PubMed Central, LOCKSS.
Bioacoustic study
Collected crickets from the field were recorded with a modified condenser microphone (CM16 Avisoft Bioacoustics, Berlin, frequency range: 3–150 kHz ± 6 dB, R. Specht pers. comm.) in a sound attenuated room controlled for temperature in the MNHN. The insects were placed over night alone in a cage made of mosquito net to avoid echoes. The microphone was placed 30 cm from the cage. Automatic recordings were made using the program Avisoft Triggering Harddisk Recorder version 2.97 and a 8-Pre MOTU sound card at a sampling frequency of 96 kilo-samples per second (16 bit). The temporal acoustic activity of crickets was obtained in the field by ambient acoustic recordings with a SongMeter SM2 Bat sensor (Wildlife Acoustics, concord, NY, U.S.A.) at Pic du Grand Kaori during 72 hours.
Cricket song terminology follows Ragge & Reynolds [35]: one song unit is called a syllable and corresponds to one opening-closing cycle of the male forewings; a group of syllables forming a cricket call is named an echeme. We used the computer software Avisoft-SASLab pro V. 5.2.06 (Avisoft bioacoustics; Specht 2012) to automatically analyze cricket calls. We used automatic analyses to calculate syllable duration and period, number of syllable per echeme, duty-cycle, and dominant frequency. Eight calling songs from 4 different males were used for the analysis (two per male). Measurements acquired from automatic analyses were verified manually in order to avoid errors or aberrant values. Sound tracks were deposited in the Sound Library of the MNHN, Paris (MNHN-SO-2015-9, 10, 11, 12, 13, 14).
Abbreviations
General morphology: FW: forewing; Tarsomere III-1: basal segment of hind leg tarsomere; T: tibiae.
Male genitalia: ect ap: ectophallic apodeme; ect arc: ectophallic arc; ect f: ectophallic fold; ect lat exp: ectophallic lateral expansion; end ap: endophallic apodeme; end s: endophallic sclerite; pse p: pseudepiphallic paramere.
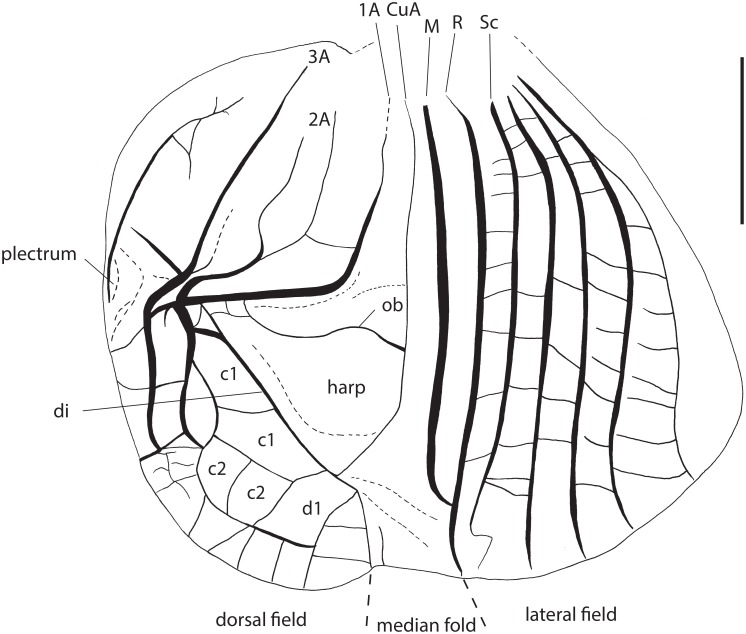
Tegminal venation: 1A-4A: first to fourth anal veins; CuA: anterior cubitus; CuP: posterior cubitus; M: median vein; Sc: subcostal vein; R: radial vein; di: diagonal vein; ob: oblique veins, c1-3: first to third cells of C alignment; d1 cell (mirror): first cell(s) of D alignment; d2: second cell of D alignment; e1: first cell of E alignment.
Morphological measurements: FIIIL: length of hind femora; FIIIW: width of hind femora; FWL: forewing length; FWW: forewing width (at the level of maximal width); Ias: inner spines on TIII dorsal side, above the spurs; Ibs: inner spines on TIII dorsal side, between the spurs; Oas: outer spines on TIII dorsal side, above the spurs; Obs: outer spines on TIII dorsal side, between the spurs; OL: ovipositor length; PronL: pronotum length; PronW: pronotum width; TIIIL: length of hind tibiae; TaIIIs: spines on outer edge of third hind tarsomere, not including the apical spine.
Acoustics: fd: dominant frequency; TSR: tooth-strike rate.
Molecular phylogeny
Taxon sampling
Two molecular datasets were built aiming both to investigate the phylogenetic placement of Pixibinthus within crickets (= dataset 1), and to generate a molecular dating to estimate divergence times of Pixibinthus and its close relatives (= dataset 2).
The molecular sampling of dataset 1 comprised six individuals of Pixibinthus (from three different locations). As Pixibinthus shares the typical morphology of the Lebinthini tribe within the subfamily Eneopterinae (see below), a special emphasis has been put on the taxonomical sampling of this tribe, by incorporating representatives of the genera Agnotecous Saussure, 1878 (nine species; 14 samples), Lebinthus Stål, 1877 (six species; seven samples), Cardiodactylus Saussure, 1878 (four species; five samples), and Swezwilderia Chopard, 1929 (one species). The sampling was then completed with other species belonging to the other four current tribes of the Eneopterinae subfamily (sensu [36]): Eurepini (one species), Eneopterini (one species), Nisitrini (two species; three samples), and Xenogryllini (one species). A total of 26 species were then used in the ingroup sampling of dataset 1 (corresponding to 39 samples; Tables 1 and 2). Two species from a different cricket subfamily (Gryllinae: Acheta domesticus (Linnaeus, 1758) and Gryllus bimaculatus De Geer, 1773) were finally used as most external outgroups.
Table 1. List of taxa investigated in the molecular study, with voucher information, and country and locality origin of samples, taxonomical classification. molecular dataset.
The phylogeny-based taxonomies of Chintauan-Marquier et al. [37] for Gryllidea and Robillard & Desutter-Grandcolas [36] for Eneopterinae were used; superfamilies and families are indicated for all taxa, and subfamilies and tribes for ingroup taxa only. Abbreviations used for subfamilies: ENEO = Eneopterinae, GRYL = Gryllinae; Tribes: Ene = Eneopterini, Eur = Eurepini, Gry = Gryllini, Leb = Lebinthini, Nis = Nisitrini, Xen = Xenogryllini. N/A corresponds to non-available data.
| Species | Superfamily / Family | Ingroup: subfamily / tribe | Country | Locality | Vouchers |
|---|---|---|---|---|---|
| Acheta domesticus (Linnaeus, 1758) | GRYLLOIDEA / Gryllidae | GRYL / Gry | France | Laboratory strain | MNHN-EO-ENSIF3523 |
| Afrophaloria amani Desutter-Grandcolas, 2015 | GRYLLOIDEA / Phalangopsidae | Tanzania | Amani | MNHN-EO-ENSIF3341 | |
| Agnotecous albifrons Desutter-Grandcolas, 1997 CT | GRYLLOIDEA / Gryllidae | ENEO/ Leb | New Caledonia | Col Toma | MNHN-EO-ENSIF2767 |
| Agnotecous albifrons Desutter-Grandcolas, 1997 Ge | GRYLLOIDEA / Gryllidae | ENEO/ Leb | New Caledonia | Gelima | MNHN-EO-ENSIF1771 |
| Agnotecous azurensis Desutter-Grancolas, 2006 GK | GRYLLOIDEA / Gryllidae | ENEO/ Leb | New Caledonia | Grand Kaori | MNHN-EO-ENSIF2777 |
| Agnotecous azurensis Desutter-Grancolas, 2006 RB | GRYLLOIDEA / Gryllidae | ENEO/ Leb | New Caledonia | Rivière Bleue (Pourina) | MNHN-EO-ENSIF2780 |
| Agnotecous clarus Desutter-Grandcolas, 2006 | GRYLLOIDEA / Gryllidae | ENEO/ Leb | New Caledonia | Pic du Pin | MNHN-EO-ENSIF2788 |
| Agnotecous meridionalis Desutter-Grandcolas, 2006 IDP | GRYLLOIDEA / Gryllidae | ENEO/ Leb | New Caledonia | Ile des Pins | MNHN-EO-ENSIF2772 |
| Agnotecous meridionalis Desutter-Grandcolas, 2006 PB | GRYLLOIDEA / Gryllidae | ENEO/ Leb | New Caledonia | Port Boisé | MNHN-EO-ENSIF2771 |
| Agnotecous obscurus (Chopard, 1970) Ao | GRYLLOIDEA / Gryllidae | ENEO/ Leb | New Caledonia | Aoupinié | MNHN-EO-ENSIF2786 |
| Agnotecous obscurus (Chopard, 1970) Ma | GRYLLOIDEA / Gryllidae | ENEO/ Leb | New Caledonia | Mandjélia | MNHN-EO-ENSIF2785 |
| Agnotecous occidentalis Desutter-Grandcolas, 2006 | GRYLLOIDEA / Gryllidae | ENEO/ Leb | New Caledonia | Col des Roussettes | MNHN-EO-ENSIF2765 |
| Agnotecous robustus (Chopard, 1915) | GRYLLOIDEA / Gryllidae | ENEO/ Leb | New Caledonia | Aoupinié | MNHN-EO-ENSIF2752 |
| Agnotecous sarramea Desutter-Grancolas, 1997 | GRYLLOIDEA / Gryllidae | ENEO/ Leb | New Caledonia | Mé Aréto | MNHN-EO-ENSIF2764 |
| Agnotecous yahoue Otte, 1987 Ko | GRYLLOIDEA / Gryllidae | ENEO/ Leb | New Caledonia | Monts Koghis | MNHN-EO-ENSIF2766 |
| Agnotecous yahoue Otte, 1987 Mo | GRYLLOIDEA / Gryllidae | ENEO/ Leb | New Caledonia | Mont Mou | MNHN-EO-ENSIF2773 |
| Anaxipha sp. affinis nitida (Chopard, 1925) | GRYLLOIDEA / Trigonidiidae | French Guiana | Arataye | MNHN-EO-ENSIF3260 | |
| Bullita sp. | GRYLLOIDEA / Trigonidiidae | New Caledonia | Yahoue | MNHN-EO-ENSIF3393 | |
| Cardiodactylus enkraussi Otte, 2007 | GRYLLOIDEA / Gryllidae | ENEO/ Leb | Vanuatu | Espiritu Santo, Vathé | MNHN-EO-ENSIF2366 |
| Cardiodactylus guttulus (Matsumura, 1913) | GRYLLOIDEA / Gryllidae | ENEO/ Leb | Japan | MNHN-EO-ENSIF1193 | |
| Cardiodactylus novaeguineae (Haan, 1842) | GRYLLOIDEA / Gryllidae | ENEO/ Leb | Vanuatu | Espiritu Santo, Peavot | MNHN-EO-ENSIF2030 |
| Cardiodactylus novaeguineae (Haan, 1842) NC | GRYLLOIDEA / Gryllidae | ENEO/ Leb | New Caledonia | Lifou | MNHN-EO-ENSIF1921 |
| Cardiodactylus tankara Robillard, 2009 | GRYLLOIDEA / Gryllidae | ENEO/ Leb | Vanuatu | Espiritu Santo, Butmas | MNHN-EO-ENSIF2410 |
| Cearacesa sp. | GRYLLOIDEA / Gryllidae | Brazil | Pernambuco | MNHN-EO-ENSIF3270 | |
| Diatrypa sp. | GRYLLOIDEA / Gryllidae | French Guiana | Arataye | MNHN-EO-ENSIF3261 | |
| Ectecous sp. | GRYLLOIDEA / Phalangopsidae | French Guiana | Arataye | MNHN-EO-ENSIF3384 | |
| Eneoptera guyanensis Chopard, 1931 | GRYLLOIDEA / Gryllidae | ENEO / Ene | French Guiana | Montagne de Kaw | MNHN-EO-ENSIF27411 / MNHN-EO-ENSIF36872 |
| Eurepini sp. | GRYLLOIDEA / Gryllidae | ENEO / Eur | Australia | Northern Territory, Litchfield National Park | MNHN-EO-ENSIF3155 |
| Fryerius sp. | GRYLLOIDEA / Gryllidae | Comoros | Moheli | MNHN-EO-ENSIF3378 | |
| Gryllotalpa sp. | GRYLLOTALPOIDEA / Gryllotalpidae | Mozambique | Cabo Delgado | MNHN-EO-ENSIF3315 | |
| Gryllus bimaculatus De Geer, 1773 | GRYLLOIDEA / Gryllidae | GRYL / Gry | France | Laboratory strain | MNHN-EO-ENSIF3524/3404 |
| Homeogryllus orientalis Desutter, 1985 | GRYLLOIDEA / Phalangopsidae | Mozambique | Cabo Delgado | MNHN-EO-ENSIF3603 | |
| Lebinthus lifouensis Desutter-Grandcolas, 1997 | GRYLLOIDEA / Gryllidae | ENEO/ Leb | New Caledonia | Lifou | MNHN-EO-ENSIF1346 |
| Lebinthus luae Robillard & Tan, 2013 | GRYLLOIDEA / Gryllidae | ENEO/ Leb | Singapore | Labrador park | MNHN-EO-ENSIF2740 |
| Lebinthus nattawa Robillard, 2009 | GRYLLOIDEA / Gryllidae | ENEO/ Leb | Vanuatu | Nattawa, Santo | MNHN-EO-ENSIF2564 |
| Lebinthus santoensis Robillard, 2009 P | GRYLLOIDEA / Gryllidae | ENEO/ Leb | Vanuatu | Espiritu Santo, Peavot | MNHN-EO-ENSIF2484 |
| Lebinthus santoensis Robillard, 2009 V | GRYLLOIDEA / Gryllidae | ENEO/ Leb | Vanuatu | Espiritu Santo, Vathé | MNHN-EO-ENSIF2437 |
| Lebinthus sp. PNG | GRYLLOIDEA / Gryllidae | ENEO/ Leb | Papua New Guineae | New Ireland | MNHN-EO-ENSIF1171 / MNHN-EO-ENSIF1572 |
| Lebinthus villemantae Robillard, 2010 | GRYLLOIDEA / Gryllidae | ENEO/ Leb | Indonesia | Sulawesi, Bulu Saraun | MNHN-EO-ENSIF2739 |
| Luzaridella obscura Desutter-Grandcolas, 1992 | GRYLLOIDEA / Phalangopsidae | French Guiana | Arataye | MNHN-EO-ENSIF3253 | |
| Microlandreva sp. Chopard, 1958 | GRYLLOIDEA / Gryllidae | Mayotte | MNHN-EO-ENSIF3307 | ||
| Nisitrus vittatus (Haan, 1842) Se | GRYLLOIDEA / Gryllidae | ENEO / Nis | Malaysia | Selangor, Mount Kira | MNHN-EO-ENSIF3134 |
| Nisitrus vittatus (Haan, 1842) Si | GRYLLOIDEA / Gryllidae | ENEO / Nis | Singapore | Bukit Timah Natural Reserve | MNHN-EO-ENSIF2742 |
| Ornebius xanthopterus Guérin-Méneville, 1844 | GRYLLOIDEA / Mogoplistidae | Mauritius | Le morne | Collection SH-2011-016 | |
| Paranisitra longipes Chopard, 1925 | GRYLLOIDEA / Gryllidae | ENEO / Nis | Philippines | Luzon, Mount Makiling | MNHN-EO-ENSIF3157 |
| Pixibinthus sonicus Anso & Robillard, sp. nov. FN1 | GRYLLOIDEA / Gryllidae | ENEO/ Leb | New Caledonia | Forêt Nord | MNHN-EO-ENSIF99 |
| Pixibinthus sonicus Anso & Robillard, sp. nov. FN2 | GRYLLOIDEA / Gryllidae | ENEO/ Leb | New Caledonia | Forêt Nord | MNHN-EO-ENSIF83 |
| Pixibinthus sonicus Anso & Robillard, sp. nov. GK1 | GRYLLOIDEA / Gryllidae | ENEO/ Leb | New Caledonia | Grand Kaori | MNHN-EO-ENSIF150 |
| Pixibinthus sonicus Anso & Robillard, sp. nov. GK2 | GRYLLOIDEA / Gryllidae | ENEO/ Leb | New Caledonia | Grand Kaori | MNHN-EO-ENSIF125 |
| Pixibinthus sonicus Anso & Robillard, sp. nov. RB1 | GRYLLOIDEA / Gryllidae | ENEO/ Leb | New Caledonia | Rivière Bleue (Rivière Blanche) | MNHN-EO-ENSIF99 |
| Pixibinthus sonicus Anso & Robillard, sp. nov. RB2 | GRYLLOIDEA / Gryllidae | ENEO/ Leb | New Caledonia | Rivière Bleue (Rivière Blanche) | MNHN-EO-ENSIF133 |
| Pteroplistes masinagudi Jaiswara, 2014 | GRYLLOIDEA incertae sedis | India | Tamil Nadu | Collection BNHS | |
| Swezwilderia sp. | GRYLLOIDEA / Gryllidae | ENEO /? | Fiji | Viti Levu | MNHN-EO-ENSIF2737 |
| Xenogryllus marmoratus Bolívar, 1890 | GRYLLOIDEA / Gryllidae | ENEO / Xen | Japan | Honshu, Nara City | MNHN-EO-ENSIF3161 |
Table 2. Genbank accession numbers of studied taxa.
Newly generated sequences are indicated with an asterisk (and would be very soon referred to a genbank accession number). The superscript numbers refer to the publication where the sequences were first published: 1[27], 2[38], 3[39], 4[40], 5[41], 6[42], 7[43], 8[37], 9[44].
| Species | dataset 1 | dataset 2 | 16S | 12S | Cytb | CO1 | CO2 | 28SA | EF1a | H3 | 18S |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Acheta domesticus (Linnaeus, 1758) | x | x | AF2486982 | Z976119 | AF2486822 | JX8974031 | JX8974391 | JX8974651 | GQ8866923 | KR903150* | AD18SITS1* |
| Afrophaloria amani Desutter-Grandcolas, 2015 | x | KR9036968 | KR9038588 | KR9033538 | N/A | N/A | KR9035248 | N/A | KR9031738 | KR9040498 | |
| Agnotecous albifrons Desutter-Grandcolas, 1997 CT | x | x | JX8973531 | JX8973941 | JX8973141 | JX8974181 | JX8974461 | JX8974901 | JX8975271 | JX8975721 | JX8975831 |
| Agnotecous albifrons Desutter-Grandcolas, 1997 Ge | x | JX8973541 | JX8973961 | JX8973161 | JX8974161 | JX8974451 | N/A | JX8975291 | JX8975741 | JX8975851 | |
| Agnotecous azurensis Desutter-Grancolas, 2006 GK | x | JX8973601 | JX8973891 | JX8973311 | JX8974261 | N/A | JX8974721 | JX8975051 | JX8975651 | JX8975931 | |
| Agnotecous azurensis Desutter-Grancolas, 2006 RB | x | x | JX8973581 | JX8973761 | JX8973291 | JX8974231 | JX8974531 | JX8974751 | JX8975021 | JX8975661 | JX8975951 |
| Agnotecous clarus Desutter-Grandcolas, 2006 | x | x | JX8973471 | JX8974001 | JX8973241 | JX8974071 | JX8974511 | JX8974921 | JX8975231 | JX8975541 | JX8975901 |
| Agnotecous meridionalis Desutter-Grandcolas, 2006 IDP | x | JX8973491 | JX8974011 | JX8973111 | JX8974201 | N/A | JX8974881 | JX8975191 | JX8975531 | JX8975791 | |
| Agnotecous meridionalis Desutter-Grandcolas, 2006 PB | x | x | JX8973501 | JX8974021 | JX8973131 | JX8974101 | JX8974421 | JX8974891 | JX8975201 | JX8975501 | JX8975971 |
| Agnotecous obscurus (Chopard, 1970) Ao | x | JX8973561 | JX8973981 | JX8973191 | JX8974151 | JX8974491 | N/A | JX8975101 | N/A | JX8975911 | |
| Agnotecous obscurus (Chopard, 1970) Ma | x | x | JX8973571 | JX8973931 | JX8973201 | JX8974121 | JX8974501 | JX8974871 | JX8975251 | JX8975761 | JX8975871 |
| Agnotecous occidentalis Desutter-Grandcolas, 2006 | x | x | JX8973621 | JX8973861 | JX8973221 | JX8974341 | JX8974611 | N/A | JX8975121 | JX8975701 | JX8975891 |
| Agnotecous robustus (Chopard, 1915) | x | x | JX8973591 | JX8973751 | JX8973331 | JX8974061 | JX8974431 | N/A | JX8974981 | JX8975551 | JX8975881 |
| Agnotecous sarramea Desutter-Grancolas, 1997 | x | x | JX8973721 | JX8973801 | JX8973421 | JX8974301 | JX8974561 | JX8974711 | JX8975111 | JX8975611 | JX8975981 |
| Agnotecous yahoue Otte, 1987 Ko | x | x | JX8973661 | JX8973871 | JX8973341 | JX8974381 | JX8974631 | JX8974781 | JX8975001 | JX8975491 | JX8976021 |
| Agnotecous yahoue Otte, 1987 Mo | x | JX8973671 | JX8973881 | JX8973371 | JX8974371 | JX8974621 | JX8974791 | JX8975011 | N/A | N/A | |
| Anaxipha sp. affinis nitida (Chopard, 1925) | x | KR9037358 | KR9039068 | KR9033968 | N/A | N/A | KR9035708 | N/A | KR9032208 | KR9040948 | |
| Bullita sp. | x | KR9038108 | KR9040018 | KR9034738 | N/A | N/A | KR9036498 | N/A | KR9033078 | KR9041838 | |
| Cardiodactylus enkraussi Otte, 2007 | x | x | JF972518 / JF9724864 | JF9725024 | N/A | KU705561* | KU705550* | N/A | KU705611* | KU705595* | JF9725334 |
| Cardiodactylus guttulus (Matsumura, 1913) | x | x | JF9725194 | JF9725034 | JF9724874 | KU705562* | N/A | KU705580* | KU705612* | KU705596* | JF9725344 |
| Cardiodactylus novaeguineae (Haan, 1842) | x | x | JF9725214 | JF9725064 | JF9724904 | KU705563* | KU705551* | KR903500* | KU705613* | KR903151* | JF9725374 |
| Cardiodactylus novaeguineae (Haan, 1842) NC | x | x | AY9052995 | AY9052705 | AY9053535 | KU705564* | KU705552* | KU705588* | KU705614* | KU705597* | AY9053295 |
| Cardiodactylus tankara Robillard, 2009 | x | x | JF9725224 | JF9725084 | JF9724914 | KU715286* | KU726590* | KU705581* | N/A | KU705598* | JF9725384 |
| Cearacesa sp. | x | KR9037988 | KR9039858 | KR9034598 | N/A | N/A | KR9036388 | N/A | KR9032948 | KR9041708 | |
| Diatrypa sp. | x | KR9037288 | KR9038998 | KR9033898 | N/A | N/A | KR9035648 | N/A | KR9032148 | KR9040878 | |
| Ectecous sp. | x | KR9037268 | KR9038978 | KR9033878 | N/A | N/A | KR9035628 | N/A | KR9032128 | KR9040858 | |
| Eneoptera guyanensis Chopard, 1931 | x | x | AY9053015 | AY9052725 | AY9053555 | JX8974041 | KU705553* | KU705582* | JX8974951 | JX8975471 | AY9053315 |
| Eurepini sp. | x | x | KR903674* | KR903834* | KR903331* | KU705565* | KU705554* | KR903503* | N/A | KR903153* | KR904028* |
| Fryerius sp. | x | KR9037308 | KR9039018 | KR9033918 | N/A | N/A | KR9035668 | N/A | KR9032168 | KR9040898 | |
| Gryllotalpa sp. | x | KR9037838 | KR9039618 | KR9034438 | N/A | N/A | KR9036218 | N/A | KR9032698 | KR9041478 | |
| Gryllus bimaculatus De Geer, 1773 | x | x | AF2486852 | AY9052925 | AF2486592 | N/A | KU705555* | KR903504* | N/A | KR903154* | AF5145096 |
| Homeogryllus orientalis Desutter, 1985 | x | KR9037528 | KR9039258 | KR9034138 | N/A | N/A | KR9035868 | N/A | KR9032358 | KR9041128 | |
| Lebinthus lifouensis Desutter-Grandcolas, 1997 | x | x | AY9053095 | AY9052795 | AY9053645 | KU705566* | KU705556* | KU705583* | N/A | KU705599* | AY9053365 |
| Lebinthus luae Robillard & Tan, 2013 | x | x | JF9725244 | KR904017* | JF9724934 | KU705567* | KU705557* | KR903665* | KU705615* | KR903321* | KR904199* |
| Lebinthus nattawa Robillard, 2009 | x | x | JF9725254 | JF9725104 | JF9724944 | KU705568* | KU705558* | KU705584* | N/A | KU705600* | JF9725414 |
| Lebinthus santoensis Robillard, 2009 P | x | KU705528* | KU708011* | KU5535* | KU705569* | N/A* | KU705585* | N/A | KU705601* | KU705543* | |
| Lebinthus santoensis Robillard, 2009 V | x | x | JF9725274 | JF9725114 | JF9724954 | JX8974051 | JX8974411 | JX8974671 | N/A | JX8975481 | JF9725424 |
| Lebinthus sp. PNG | x | x | JF9725284 | JF9725134 | KU705536* | KU715289* | KU715288* | KU715290* | KU715292* | KU715291* | JF9725444 |
| Lebinthus villemantae Robillard, 2010 | x | x | JF9725264 | JF9725124 | JF9724964 | KU705570* | KU705559* | KU705586* | N/A | KU705602* | JF9725434 |
| Luzaridella obscura Desutter-Grandcolas, 1992 | x | KR9037088 | KR9038718 | KR9033658 | N/A | N/A | KR9035368 | N/A | KR9031868 | KR9040618 | |
| Microlandreva sp. Chopard, 1958 | x | KR9037828 | KR9039608 | KR9034428 | N/A | N/A | KR9036208 | N/A | KR9032688 | KR9041468 | |
| Nisitrus vittatus (Haan, 1842) Se | x | AY9053145 | AY905284 | AY9053695 | KU705571* | N/A | KU705587* | N/A | KU705603* | AY9053405 | |
| Nisitrus vittatus (Haan, 1842) Si | x | x | AY9053145 | AY905284 | AY9053695 | KU705572* | N/A | KR903667* | JN8878837 | JX8975461 | KR9042015 |
| Ornebius xanthopterus Guérin-Méneville, 1844 | x | KR9037698 | KR9039448 | KR9034308 | N/A | N/A | KR9036058 | N/A | KR9032548 | KR9041318 | |
| Paranisitra longipes Chopard, 1925 | x | x | KR903827* | KR904020* | N/A | KU715287* | N/A | KR903668* | N/A | KR903325* | KR904202* |
| Pixibinthus sonicus Anso & Robillard, sp. nov. FN1 | x | KU705531* | KU708014* | KU705539* | KU705573* | N/A | KU705589* | N/A | KU705604* | N/A* | |
| Pixibinthus sonicus Anso & Robillard, sp. nov. FN2 | x | KU705532* | KU708015* | KU705540* | KU705574* | N/A | KU705590* | N/A | KU705605* | KU705547* | |
| Pixibinthus sonicus Anso & Robillard, sp. nov. GK1 | x | x | KU705529* | KU708017* | KU705537* | KU705575* | N/A | KU705591* | N/A | KU705606* | KU705545* |
| Pixibinthus sonicus Anso & Robillard, sp. nov. GK2 | x | KU705534* | KU708016* | KU705542* | KU705576* | N/A | KU705593* | N/A | KU705607* | KU705549* | |
| Pixibinthus sonicus Anso & Robillard, sp. nov. RB1 | x | KU705530* | KU708012* | KU705538* | KU705577* | N/A | KU705592* | N/A | KU705608* | KU705546* | |
| Pixibinthus sonicus Anso & Robillard, sp. nov. RB2 | x | KU705533* | KU708013* | KU705541* | KU705578* | N/A | KU705594* | N/A | KU705609* | KU705548* | |
| Pteroplistes masinagudi Jaiswara, 2014 | x | KR9036938 | KR9038548 | KR9033498 | N/A | N/A | KR9035218 | N/A | KR9031708 | KR9040458 | |
| Swezwilderia sp. | x | x | JF9725294 | JF9725144 | JF9724984 | KU705579* | N/A | N/A | N/A | KR903327* | JF9725454 |
| Xenogryllus marmoratus Bolívar, 1890 | x | x | KR903830* | KR904024* | KR903491* | N/A* | KU705560* | N/A | KU705610* | KR903329* | KR904206* |
The molecular sampling of dataset 2 was composed of a sub-sampling of dataset 1 to reduce each ingroup species to a single individual, except Cardiodactylus novaeguineae that occurs in New Caledonia and other Pacific islands (Tables 1 and 2). We also increased the species sampling of the Grylloidea superfamily in order both to better estimate divergence times and to use an appropriate calibration point (see below). We consequently added representatives of the major Gryllidea lineages delimited in Chintauan-Marquier et al. [37]:Trigonidiidae (two species), Pteroplistinae (one species), Phalangopsidae (four species), and Gryllidae (six species). We finally used as most external outgroups, one species belonging to Mogoplistidae (Grylloidea) and one species of Gryllotalpidae (Gryllotalpoidea; Tables 1 and 2).
Selection of DNA regions
We selected nine DNA loci, five mitochondrial and four nuclear, proved to have been useful in previous phylogenetic studies on Grylloidea [16,27,37,40,41]. These are fragments of the small (12S rRNA, ~ 400 bp) and large (16S rRNA, ~ 500 bp) mitochondrial ribosomal subunits, the mitochondrial gene coding for cytochrome b protein (cytb, ~ 400 bp), fragments of the mitochondrial cytochrome oxidase subunits 1 (CO1, ~ 750 bp) and 2 (CO2, ~ 400 bp) genes, a fragment of the small nuclear ribosomal subunit (18S rRNA, ~ 650 bp), a fragment of the large nuclear ribosomal subunit (28S rRNA, ~ 400 bp), and fragments of the genes coding for H3 protein (H3, ~ 330 bp) and elongation factor 1-α (EF1α, ~ 950 bp). We used these nine DNA loci for analyses of dataset 1, to refine and improve the internal phylogenetic resolution. However we observeda high level of missing sequences for the CO1 and CO2 regions, especially for outgroup species of dataset 2. We therefore used only the seven following loci for phylogenetic analyses of this dataset: 12S, 16S, 18S, 28SA, cytb, EF1a, and H3.
Laboratory procedures and Preparation of datasets
Molecular work was performed at the Service de Systématique Moléculaire of the MNHN. All sequences were generated by using the DNA extraction, amplification and sequencing protocols of the nine DNA loci, described in Nattier at al. [27]. Some DNA extractions were made from median legs of crickets using the Epmotion 5075 robot (Eppendorf). In total we generated 106 new DNA sequences (33% of the total samplings) for this study (Tables 1 and 2), while other sequences were generated in previous studies and downloaded from Genbank [27,37,38,39,40,41,42,43,44]. Primers and annealing temperatures for each DNA locus are given in S1A Table. Newly generated sequences were edited in Sequencher v.4.9 (Gene Codes Co.) and BioEdit v.7.0.5.3 [45], blasted with NCBI blast tools, and submitted to GenBank (Tables 1 and 2).
Multiple alignments were generated for each DNA locus using the software Muscle v.3.7 [46] with default parameter settings, by using the online portal Phylogeny.fr [47]. Subsequently we adjusted them manually. We concatenated the different DNA locus partitions with SequenceMatrix v.1.7.7 [48]. The datasets 1 and 2 resulted in a combined alignment of ~4750 bp and 3789 bp, respectively (S1B and S1C Table). Several ambiguous and large indel regions in the alignments of loci 12S, 16S, 18S, and EF1α were found within the dataset 2, and attributed to the inclusion of several distantly related Grylloidea taxa. This induced problematic homology hypotheses, and we consequently removed them from the DNA alignments (S1 Table).
Phylogenetic analyses
Preliminary single-locus and combined phylogenetic inferences were carried out using maximum likelihood (ML) and Bayesian Markov Chain Monte Carlo (MCMC) analyses for both datasets 1 and 2. Best-fit model for each DNA locus was identified using jModelTest v.2.1.3 [49] based on the Akaike Information criterion (S1B and S1C Table). The combined datasets were partitioned to allow each locus to have its specific model parameters [50,51].
The ML analyses were performed in RAxML v.7.4.2. [52,53] by running 500 likelihood searches. To evaluate node support, a supplementary bootstrap analysis (BS; [54]) was performed using 500 replicates. A clade with a BS value > 95% was considered as well supported.
The Bayesian MCMC analyses were performed using MrBayes v.3.2.6 [51]. The single-locus and combined analyses were set as follows: four Metropolis-coupled Markov chains with an incremental heating temperature of 0.2 were run for 50 and 100 million generations with a tree sampled every 10000th generation for dataset 1 and dataset 2, respectively. The analysis was repeated four times for both datasets, all starting with random trees. Bayesian MCMC analyses were carried out using the CIPRES Science Gateway v.3.3 [55]. The MCMC sampling was considered sufficientwhen the effective sampling size (ESS) was higher than 200, as verified in Tracer v.1.6 [56], or when the potential scale reduction factor was reasonably close to 1.0 for each parameter (PSRF; [57]). After a burn-in period of 5× 106 and 10 × 106 for dataset 1 and dataset 2, respectively, the remaining trees were used to construct a majority rule consensus tree and its associated Bayesian posterior probabilities (PP). A clade with a PP value higher than 0.95 was considered as well supported. Trees were finally visualized with FigTree v.1.4.0. (http://tree.bio.ed.ac.uk/software/figtree/).
Bayesian divergence times estimates
The temporal evolution of Pixibinthus and its close relatives was estimated using the Bayesian MCMC approach implemented in BEAST v.1.8.0 for dataset 2 [58]. DNA loci were combined and partitions were set as in the preliminary Bayesian MCMC analyses (see above). We unlinked all DNA site models to analyze each locus under separate substitution models. An uncorrelated relaxed molecular clock model was selected to allow estimates of independent rate variation across branches following a lognormal distribution. The Birth-Death speciation process was implemented for the tree prior, assuming constant speciation and extinction rates per lineage. The mean of the branch rates (UCLD.mean) was set to follow a lognormal distribution with a log (mean) set to -4 and a log (standard deviation) set to 1. We started our analysis with a random tree. The MCMC was run for 100 million generations and sampled every 10000th generations. The analysis for each dataset was conducted three times, and was carried out using the CIPRES Science Gateway v.3.3 [55]. Convergence of runs and adequate MCMC sampling were checked using Tracer v.1.6 [56]. The first 10 × 106 generations of each run were manually discarded as burn-in. The remaining trees were summarized using a Maximum Clade Credibility target tree in TreeAnnotator v.1.8.0 [58], as well as Bayesian posterior probability (PP), median height (= age estimate) and the 95% highest posterior density heights interval (95% HPD) of each node.
Calibration point
Divergence dates were previously estimated to test biogeographical hypotheses in Eneopterinae crickets by Nattier et al. [40]. Age estimates were mainly obtained with several geological calibration points, considering that few fossils or dated trees were available at that time. Since that study, Song et al. [59] produced a large dated tree for Orthoptera which provided a secondary calibration for the present study in order to avoid the generally less accurate geological calibrations. We choose to use the secondary calibration point which corresponds to the divergence time between the Grylloidea and Gryllotalpoidea superfamilies. This calibration was assigned to the root node of our majority rule consensus tree generated in the preliminary Bayesian MCMC analyses of dataset 2. It corresponds to the divergence time between our most external outgroup species, Gryllotalpa sp. and all the other Grylloidea species of our dataset 2 sampling. This calibration was set to follow a uniform distribution, and by using a lower bound of 220 Myr and an upper bound of 250 Myr, which correspond to the 95% HPD of this divergence time, with a median value of 233.99 Myr [59].
Species richness and relative diversification rates
We tested the difference in species richness between Pixibinthus and its sister-clade (see below) using Equation (3) of Slowinski & Guyer [60]. To allow calculating this estimate and to ensure that the first basal split of this sister-clade in dating tree was assessed, we preliminary estimated its species richness, according to Desutter-Grandcolas & Robillard [26], Robillard et al. [61], and our own knowledge.
Ensemble species distribution modelling
Ensemble Species Distribution Model (ESDM) analyses were carried out using combined R packages, and based on nine algorithms (GLM, GAM, MARS, GBM, CTA, RF, MAXENT, ANN and SVM) in order to test if the contemporary geographical range of Pixibinthus could have been more expanded, and would have been constrained by environmental changes. Dataset was composed of the coordinates of field sampling sites where Pixibinthussonicus has been recorded. Nine GIS ecological layers were used for the modelling analyses. Uncorrelated variables were selected among the 19 bioclimatic variables available in New Caledonia from worldclim database [62]leading to the selection of seven bioclimatic variables (BIO1, 2, 3, 4, 7, 12 and 15) at 30arc-second [62]. We also included altitude from Digital Elevation Model at 10m resolution, and main vegetation units distribution map available at 300m resolution. All layers were rescaled with a 300 m resolution. We used 30% randomly selected occurrence data as a testing dataset. The consensus model was built by summing algorithms suitability maps weighted with their area under the receiving operating characteristic (ROC) curve (AUC), only for model with an AUC > 0.75 [63]. To evaluate the relative contribution of each variable to the model Pearson's correlation coefficient between the full model and one without each variable is measured [64].
Results
Systematics and taxon description
The new genus and the new species are described here. Habitat, life history traits and calling song are also precisely defined.
Order: Orthoptera
Family: Gryllidae
Subfamily: Eneopterinae
Tribe: Lebinthini
Genus:Pixibinthus Robillard and Anso, gen. nov.
urn:lsid:zoobank.org:act:94405C62-C382-407F-BAC0-4B631EC5A0D3
Type species: Pixibinthussonicus Anso & Robillard, sp. nov.
Etymology: Genus named after ‘Pixie’, the diminutive mythical creatures of folklore, hidden by nature, close but unseen, as these crickets which were never encountered by previous authors despite past extensive studies on crickets in New Caledonia.
Diagnosis: Among Eneopterinae and Lebinthini genera, Pixibinthus is characterized by a diminutive size, smaller than all previously described species of Lebinthus from the Loyalties and Vanuatu (L. lifouensis Desutter-Grandcolas, 1997 and L. santoensis Robillard, 2009 respectively). For numerous aspects, the new genus appears as an intermediate form between Lebinthus and Agnotecous. It resembles these genera by microptery in both sexes (FW short and hind wings absent), but differs by the following characters: head shape more rounded with smaller eyes and wider fastigium; in dorsal view, combined width represents 35% of head width, against 50% in L. santoensis and 46% in L. bitaeniatus; head rounded in facial view, contrary to the triangular shape in Lebinthus; ocelli very small compared to Lebinthus; male FWs with dorsal field and lateral field of similar length, unlike in Agnotecous, but as in Lebinthus; female FWs as long as in males, unlike in most Lebinthus species, slightly overlapping.
Description: Genus characterized by its small size. Head shape uncommon, fastigium wider than long, three times as wide as scape. Eyes small, little prominent; Ocelli very small. Pronotum dorsal disk almost rectangular, wider than long, its posterior margin straight. Legs rather short. Fore tibiae with two tympana; inner tympanum covered by a sclerotized expansion (Fig 1A), its membrane visible along a small longitudinal slit only; outer tympanum ellipsoidal, transversally plicate. Fore tibiae with two inner and one outer apical spurs. Tibiae II with two inner and two outer spurs. Hind femora very wide and muscular. Hind tibiae serrulated on their whole length, not furrowed longitudinally, with four pairs of subapical spurs and three pairs of apical spurs; inner spurs long and curved, outer spurs short and straight. Tarsomeres III-1 with two dorso-apical spines and a row of spines on outer dorsal edge; one lateral outer spine.FWs short, not reaching abdomen mid-length; hindwings absent.
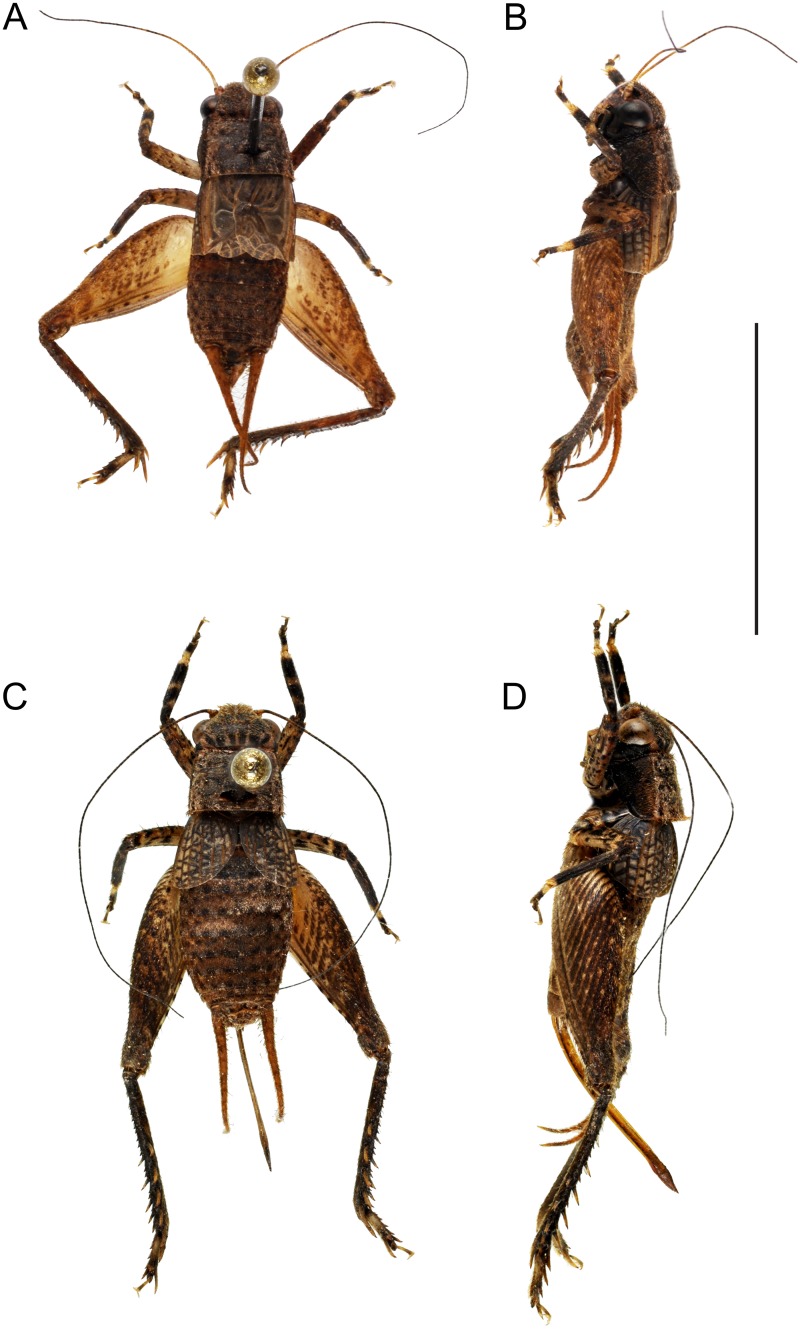
Fig 1. Dorsal and lateral views of Pixibinthus sonicus.
(A) & (B) Male. (C) & (D) Female. Scale bar = 5 mm.
Male: Metanotal glands absent. Lateral and dorsal fields of FWs of similar lengths (Figs 2 and 3). 1A vein (file) with a marked angle (only curved in Lebinthus). Harp with one bisinuated oblique vein. Mirror and d2 cell not differentiated from other cells of D alignment. Stridulatory file with teeth both on transverse and longitudinal parts of 1A. Diagonal vein well visible. CuP absent but claval fold visible. CuA faint, slightly curved inward. Median fold small, located on dorsum as in Lebinthus and Agnotecous. Chord cells differentiated. Apical field including only few cells posterior to mirror (alignment E). Subgenital plate clog-shaped, twice as long as sternites; inner side of subgenital plate with lateral swellings.
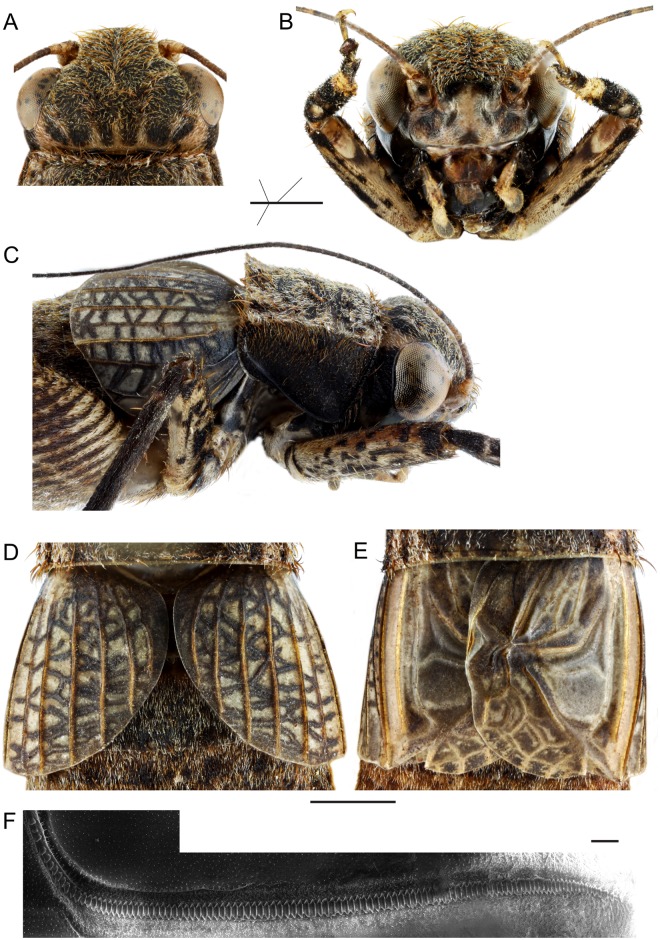
Fig 2. Detailed morphology of Pixibinthus sonicus.
(A)Dorsal view of the female face. (B) Facial of the female face. (C) Lateral view of the anterior body part of female. (D) Dorsal view of detailed FWs for female. (E) Dorsal view of detailed FWs for male (E). (F) Stridulatory file with SEM. Scale bars = 1 mm for (A), (B), (C), (D), (E); 50 μm for (F).
Fig 3. Male forewing venation of Pixibinthus sonicus.
For abbreviations and symbols, see Material and methods. Scale bar = 1 mm.
Male genitalia: Pseudepiphallus triangular, basal margin straight, posterior apex elongate, without paired lophi and forming a wide gutter (Fig 4). Rami straight, parallel and short. Ventral pseudepiphallic plate wide. Pseudepiphallic parameres sclerotized, convergent, their basis strong, with two posterior lobes, one oriented dorsally and one forming a rounded ventral plate, and a basal membranous lobe. Ectophallic apodemes parallel and long, their apex lamellate. Ectophallic arc well sclerotized, wide and slightly curved posteriorly, with a small medio-posterior expansion. Ectophallic fold long, with elongate lateral sclerites forming a “)(“pattern; its apex triangular and membranous. Endophallic sclerite large, comprising posteriorly a short median expansion and lateral arms; sclerite very long anteriorly, exceeding pseudepiphallic sclerite. Endophallic apodeme with well-developed lateral lamellas and a narrow dorsal crest.
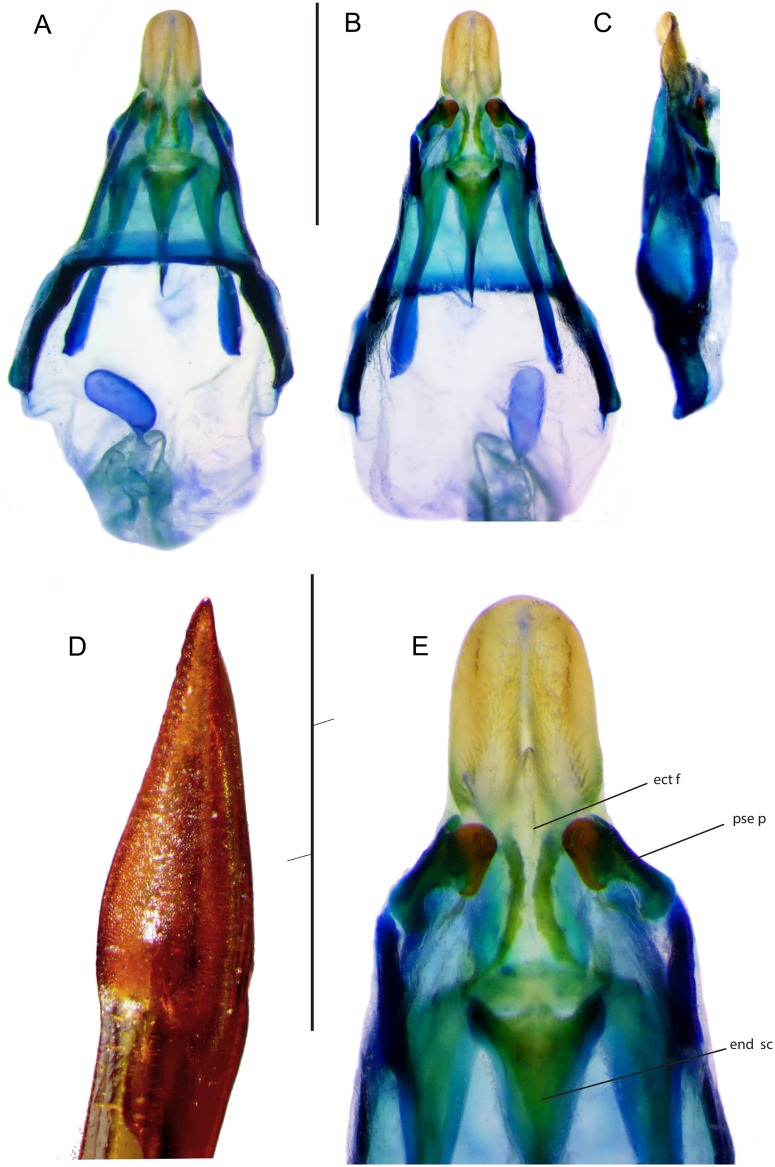
Fig 4. Genitalia of Pixibinthus sonicus.
(A) Dorsal, (B) ventral, and (C) lateral views of male genitalia. (D) Lateral view of apex of female ovipositor. (E) Ventral view of male genitalia. For abbreviations and symbols, see Material and methods. Scale bars = 1 mm.
Female: FWs as long as in males (Fig 2), slightly overlapping, dark brown with whitish veins; dorsal field with 7 strong longitudinal veins; lateral field with 4 longitudinal veins; Sc without bifurcations. Ovipositor rather short, as long as hind femur, laterally flattened, apex lanceolate and slightly denticulate dorsally (Fig 4D).
Female genitalia: Copulatory papilla little sclerotized, with a basal sclerite forming a wide ring; apex little differentiated, slightly indented andfolded ventrally.
Pixibinthus sonicus Anso and Robillard, sp. nov.
urn:lsid:zoobank.org:act:CA5D7BDC-7504-435C-8B67-5BDCF11987BB
Type material: Holotype male: New Caledonia, Grande Terre, Forêt Nord, milieu paraforestier, 22.191554 S 166.5608 E, 100 m, 1.XII.2013-12.II.2014, PAFN1-24, jour, litière, J. Anso (MNHN-EO-ENSIF244). Allotype female: Same locality, date and collector as HT, PAFN2-4, jour, litière (MNHN-EO-ENSIF71). Paratypes: New Caledonia, Grande Terre, Pic du Grand Kaori, 22.16471 S 166.53399 E, 260 m, 11.II.2014, 1♂, AGN1, jour, litière, J. Anso (MNHN-EO-ENSIF98); XI.2013, 1♀, Lfem1, J. Anso (MNHN-EO-ENSIF124). Forêt Nord, milieu paraforestier, 22.191554 S 166.5608 E, 100 m, 1.XII.2013-12.II.2014, J. Anso: 1♂, PAFN1-46, jour, litière (MNHN-EO-ENSIF70); 1♀, PAFN2-23, (MNHN-EO-ENSIF90); 1♀, PAFN2-2, jour, litière (IAC); 1♀, PAFN2-3, jour, litière (MNHN-EO-ENSIF144); 1♂, PAFN2-12, nuit, litière (IAC); 1♂, PAFN2-24, jour, litière (Nouméa); 1♀, PAFN2-33, nuit, litière (MNHN-EO-ENSIF361); 1♀, PAFN1-37, nuit, litière (MNHN-EO-ENSIF142); 1♂, PAFN1-38, nuit, litière (MNHN-EO-ENSIF81); 1♀, PAFN2-52, nuit, litière (MNHN-EO-ENSIF123); 1♀, PAFN1-1, jour, litière (MNHN-EO-ENSIF77); 1♂, PAFN1-13, jour, litière, molecular sample L66LeNCFN2 (MNHN-EO-ENSIF83); 1♀, PAFN1-15, jour, litière (Nouméa); 1♂, PAFN1-23, jour, litière, molecular sample L60LeNCFN1 (MNHN-EO-ENSIF99); 1♀, PAFN1-22, jour, litière (MNHN-EO-ENSIF127); 1♀, PAFN1-40, jour, litière(MNHN-EO-ENSIF362); 1♀, PAFN1-42, jour, litière (MNHN-EO-ENSIF118); 1♂, PAFN1-51, nuit, litière (MNHN-EO-ENSIF128). Nouvelle-Calédonie, Grande Terre, Pic du Grand Kaori, 22.16471 S 166.53399 E, 260 m, maquis, VI.2013, adultes en élevage, J. Anso: 3 males (MNHN-EO-ENSIF149, 150, 153); 4 males, enregistrement appel F0-males1-4 (MNHN-EO-ENSIF158, 154, 152, 125), 4 females, (MNHN-EO-ENSIF129, 132, 139, 140).
Additional material examined: Parc de la Rivière Bleue, maquis, 22.08375 S 166.424776 E, 200 m, 11-19.III.2014: 1 male, MARIV1-1, nuit, litière, molecular sample L59LeNCRBa1 (MNHN-EO-ENSIF105); 1 juvenile, MARIV1-15, jour, litière, molecular sample L67LeNCRBa2 (MNHN-EO-ENSIF133); 7 juveniles, MARIV1-3, 6, 7, 12, 13, 14, 19, litière (MNHN); 5 juveniles, MARIV2-2, 3, 7, 8, 9, litière (MNHN). Forêt Nord, milieu paraforestier, 22.191554 S 166.5608 E, 100 m, 1.XII.2013-26.II.2014: 12 juveniles, PAFN2-9,14,25,48,51,53,56, PAFN1-47,52,58,59,60, nuit, litière; 3 juveniles, PAFN2-49, PAFN1-16,45, jour, litière; 1♀, PAFN1-14, jour, litière; 1♀, PAFN1-7, nuit, litière (MNHN). Chute de la Madeleine, maquis, 22.23568 S 166.85268 E, 255m, 10-26.II.2014, J. Anso: 1 juvenile, MAMAD1-24, molecular sample L116PixMad1 (MNHN-EO-ENSIF65); 4 juveniles, MAMAD1-11, 24, 26, litière (MNHN); 3 juveniles, MAMAD2-7, 9, 22, litière (MNHN).
Type locality: New Caledonia, Grande Terre, Forêt Nord.
Etymology: Named after the very high dominant frequency of the species’ calling song.
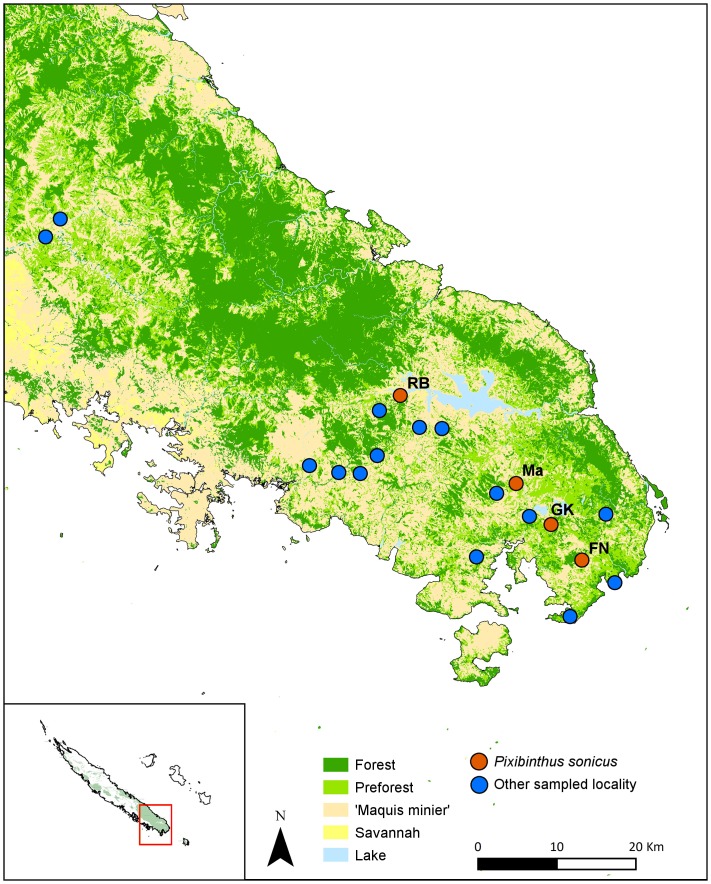
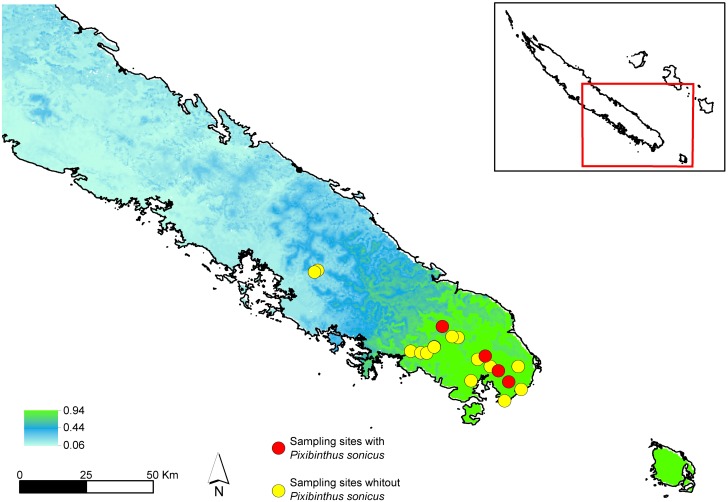
Distribution: New Caledonia, Grande Terre, Province Sud: Rivière Blanche, Chute de la Madeleine, Pic du Grand Kaori, Forêt Nord (Fig 5).
Fig 5. Map of sampled localities in the southern part of New Caledonian Grande Terre.
Blue dots indicate localities where populations of Pixibinthus sonicus were not found. Orange dots indicate localities where P. sonicus was found.
Diagnosis
Pixibinthus sonicus is recognized by its small size, face light brown with seven black spots, male genitalia, and high dominant frequency of calling song.
Description: In addition to the characters of the genus: Size very small, stocky shape. Coloration: No contrasted sexual dimorphism (Fig 1A & 1C). Body mostly brown to dark brown, slightly contrasted with lighter parts on legs. Head dorsum with six wide dark brown bands visible basally and fused between the eyes; fastigium dark brown; area posterior to eyes yellow; cheeks entirely shiny black;face light brown, with seven characteristic black spots: four forming a square on the front head, one above epistomal suture, and two bellow antennal pits; spots variable in size and connectivity, sometimes more or less fused together in a complex pattern. Maxillary palpi: article 1–3 black, articles 4–5 lighter with apical darker ring on article 5. Pronotum brown to dark brown, with lighter lateral margins with small black dots; lateral lobes of pronotum homogeneously black. Pronotum dorsal disk and lateral lobes separated by a short concave yellow line. Legs: Tibiae I black with three small lighter flecks; tibiae II entirely black with a whitish apical ring; tibiae III dark brown; femora III yellowish with dark brown diagonal stripes strongly contrasted on external face; femora I and II yellowish with small brown flecks; first and third tarsomeres I-III dark yellowish basally, their apex dark brown. Abdomen brown, darker in apical part, with one yellowish small dot for each segment. Cerci yellowish, with small scattered darker flecks.
Male
FW mostly gray to dark brown with a pale area including bases of CuA, M and R veins. Area between M and R whitish, veins orange brown. FW venation: 1A with 110 stridulatory teeth, 12 on the basal angle and 98 on the transverse part of the file (n = 1; Fig 2F). Cell c1 with a faint median transverse vein; c2 rectangular. M yellow brown, curved posteriorly, fused to R near FW apex, very thin near fusion; R straight, strong along its whole length. FW lateral field including Sc and 3–4 strong longitudinal veins. Hind tibia inner serrulation (n = 3): no spine between the apical and the first subapical spur; one spine between subapical spurs 1 and 2; two spines (1–2) between spurs 2 and 3; two spines between subapical spurs 3 and 4; seven spines (4–10) above subapical spur 4; outer serrulation: no spine between apical and subapical spurs; one spine between subapical spurs 1 and 2; two spines (1–3) between subapical spurs 2 and 3; two spines (2–3) between subapical spurs 3 and 4; seven spines (6–8) above subapical spur 4. Tarsomeres III-1: four outer (3–4) and no inner spines, in addition to apical spines.
Female
Hind tibia inner serrulation (n = 3): no spine between the apical and the first subapical spur; one spine between subapical spurs 1 and 2; two spines (1–2) between spurs 2 and 3; one spine (1–2) between subapical spurs 3 and 4; four (4–5) spines (4–10) above subapical spur 4; outer serrulation: no spine between apical and subapical spurs; one spine between subapical spurs 1 and 2; two spines between subapical spurs 2 and 3; two spines between subapical spurs 3 and 4; eight spines above subapical spur 4. Tarsomeres III-1: three outer and no inner spines, in addition to apical spines.
Measurements
See Table 3.
Table 3. Measurements of Pixibinthus sonicus sp. nov.(in mm).
| PronL | PronW | FWL | FWW | FIIIL | FIIIW | TIIIL | TIII spines | TaIIIs | OL | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ias | Ibs | Oas | Obs | ||||||||||
| Male holotype | 2 | 2.3 | 3.3 | 2.6 | 6.8 | 2.4 | 5.3 | 5 | 6 | 8 | 5 | ||
| Males paratype (n = 5) | 1.8–2.1 | 2.4–2.7 | 2.8–3.5 | 1.8–2.0 | 6.1–7.1 | 2.1–2.5 | 4.5–5.5 | 4–6 | 3–6 | 8–11 | 5–6 | 3–6 | |
| (Male mean) | (2.0) | (2.5) | (3.1) | (2.0) | (6.7) | (2.3) | (5.2) | (5) | (4) | (9) | (5) | (3) | |
| Allotype female | 1.7 | 2.5 | 2.1 | - | 6.5 | 2.4 | 5.3 | 3 | 4 | 8 | 5 | 5.5 | |
| Females paratype (n = 5) | 1.7–2.1 | 2.1–2.8 | 2.0–2.4 | - | 6.5–8.1 | 2.2–2.7 | 5.1–5.6 | 4–6 | 3–5 | 7–9 | 4–6 | 3–4 | 5.5–7.0 |
| (Female mean) | (1.9) | (2.5) | (2.2) | - | (7.0) | (2.4) | (5.4) | (5) | (4) | (8) | (5) | (4) | (6.4) |
Juvenile
Body color light brown mottled with dark brown to black spots. Head, including face, pronotum and legs with similar color patterns as in adults.
Habitat and life history traits: Pixibinthus sonicus was clearly identified as a new monotypic genus and new species after intensive sampling on various habitats, in the southern part of New Caledonia, representing an area of about 2150 km2. Among 19 sampled localities, including shrubby vegetation, preforest and rainforest, P. sonicus was only found in low to tall sclerophyllous shrubland and was clearly absent from forested areas (Fig 6). Four populations of P. sonicus were identified in our study sites. Three populations were found in low sclerophyllous ‘maquis minier’, with dominant strata lower than 5–6 meters. One population was found in tall sclerophyllous ‘maquis miniers’ at Forêt Nord (22.32259 S 166.93134 E), 6 km away from the population of Pic du Grand Kaori, separated by high ridges. Adults and juveniles were both collected during day and night, in the leaf litter. In these three localities, abundances were relatively small with a dozen of specimens collected in each site.
Fig 6. Habitat of Pixibinthus sonicus.
(A)Low sclerophyllous ‘maquis minier’ around les Chutes de la Madeleine. (B) Detailed view of habitat in leaf litter. (C) Juvenile of P. sonicus (size: approximately 1cm).
Behavior: Males of Pixibinthus sonicus emit their calling song starting from 16:30 PM until next morning at 03:00 AM, as attested by 72 hours ambient recording with SM2 Bat; this activity would characterize P. sonicus as a nocturnal species.
Acoustic description
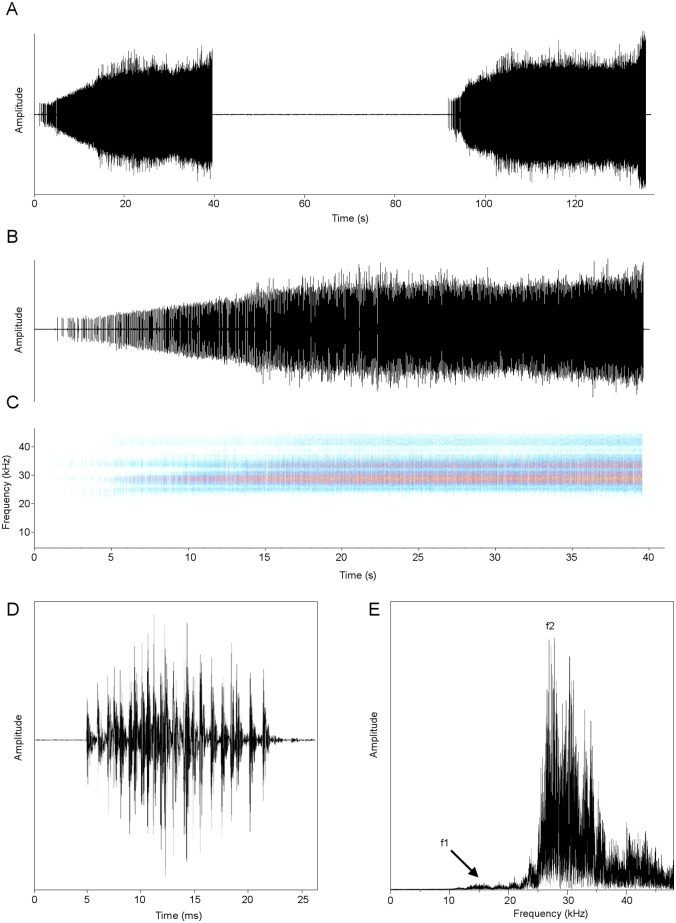
Four males were recorded under laboratory controlled condition. Each call of Pixibinthus sonicus is a trill lasting 38.2 ± 7.4 s (Fig 7A) and made of 918 ± 130 syllables, with a period of 152.8 ± 72.2 s (as recorded at 25°C). Syllables last18.4 ± 2.5 ms, with a period of 41.7 ± 16 ms, and with a duty cycle of 44.7%. The sound amplitude gradually increases during the 300 to 400 first syllables (Fig 7B), and remains twice higher and constant until the end of the trill. The dominant frequency is 27.9 ± 2.8 kHz, and corresponds to the second harmonic of the spectrum (Fig 7C and 7E), the fundamental frequency f1 being almost silent.
Fig 7. Calling song of Pixibinthus sonicus.
(A) Oscillogram showing two successive trills; (B) detailed oscillogram and (C) sonogram of one trill; (D) oscillogram of one syllable; (E) and linear spectrogram of one echeme. Symbols: f1, fundamental frequency; f2, second harmonic (dominant frequency).
Phylogenetic relationships of Pixibinthus
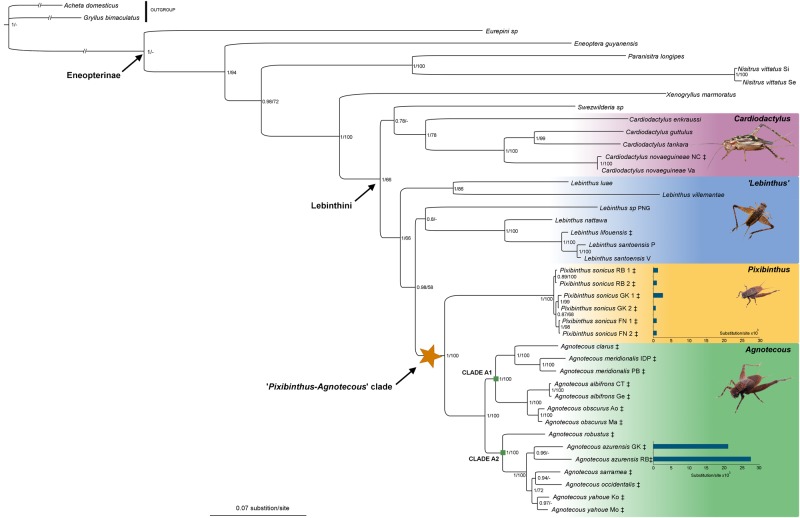
The ML and Bayesian MCMC single-locus phylogenetic topologies yield no supported incongruence for both datasets 1 and 2 (with a PP > 0.95 or BS > 85%; data not shown) and are less resolved than the combined phylogenetic analyses. The ML (data not shown), Bayesian MCMC and BEAST combined analyses provide no supported incongruence between topologies (Fig 8; S1 and S2 Figs). Since the Bayesian MCMC combined analysis allows us together to take into account uncertainty only the results of this analysis are discussed hereafter (Fig 8).
Fig 8. Bayesian majority rule consensus tree of Eneopterinae crickets based on dataset 1, including 28 species and nine DNA markers.
Bayesian posterior probability (PP) andML bootstrap (BS) support values are indicated for each node on the right. A clade with a BS < 50% or not recovered in the ML analyses is indicated with a dash. Major clades are shaded with a color scale. Horizontal bars represent the scales of substitution rates (substitution per site). Species occurring in New Caledonia are represented with the ‡ symbol.
These analyses confirm that the genus Pixibinthus belongs to the Lebinthini tribe (PP = 1; Fig 8). The species P. sonicus is monophyletic, since all of the individuals sampled in this study group together with very short branch lengths (PP = 1, see below). Moreover P. sonicus is placed as sister to the monophyletic genus Agnotecous (PP = 1; Fig 8), and both form the newly-defined ‘Pixibinthus-Agnotecous’ clade, with a high support (PP = 1). This latter lineage is nested with high support within the genus Lebinthus (PP = 0.98), a paraphyletic group under revision (Robillard et al. in prep). Within Agnotecous two main clades are recovered as sister groups. The Agnotecous clade A1 includes the A. clarus, A. meridionalis, A. albifrons and A. obscurus samples (PP = 0.99). The Agnotecous clade A2 comprises the A. robustus, A. azurensis, A. sarramea, A. occidentalis, and A. yahoue individuals (PP = 1).
The phylogenetic branch lengths (BL) estimated in this study are short in all Pixibinthus individuals (between 0.69 and 2.8 × 103 substitutions/site; Fig 8), and larger in Agnotecous samples (mostly between 3.23 and 27.51 × 103 substitutions/site, except A. albifrons with BL comprised between 1.38 and 1.69 × 103 substitutions/site). Moreover the BL of Pixibinthus samples from the localities Grand Kaori and Rivière Bleue are 12 and 23 times shorter, respectively, than the BL of A.azurensis from the same places (compare P. sonicus RB and P. sonicus GK with A. azurensis RB and A. azurensis GK; Fig 8).
Divergence time estimates
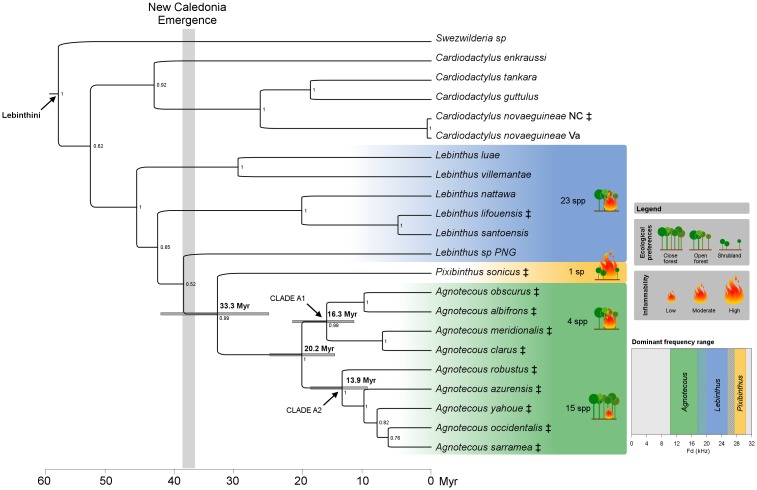
The BEAST analyses showed that the most recent common ancestor of the ‘Pixibinthus-Agnotecous’ clade appeared around the Eocene-Oligocene boundary (with a median divergence time estimate of 33.3 Myr, with a 95% HPD of 24.4–43.1 Myr; Fig 9; Table 4). The crown ages of the genus Agnotecous, clade A1, and clade A2 were estimated around 20.2 Myr, 16.3 Myr, and 13.9 Myr, respectively (with 95% HPD of 14.9–25.82, 11.6–21.6, and 9.9–18.5 Myr, respectively; S2 Fig).
Fig 9. BEAST maximum credibility clade chronogram of the main lineages of Lebinthini crickets, based on dataset 2, including 41 species and seven DNA markers (only the 21 species of Lebinthini tribe are shown).
Divergence times, ecological preferences and species richness (inferred from [26,61] and from our personal observations in the field), and relative habitat inflammability (inferred from [65])are indicated for the three main lineages: ‘Lebinthus’,Pixibinthus, and Agnotecous. Bayesian posterior probabilities (PP) are indicated for each node on the right. Gray node bars correspond to the 95% highest posterior density of median age estimates and are only provided for clades that are discussed. Dominant frequency range for the three mains lineages are indicated, with stripped area for overlapping frequency ranges. Species occurring in New Caledonia are represented with the ‡ symbol.
Table 4. Ecological, biological, morphological and dating characteristics for the three Lebinthini groups.
Acoustic recording impaired by inadequate recording material are indicate with an asterisk. Body sizes of species were inferred from the FIIIL femur length measurements. Abbreviation: N/A corresponds to non-available data.
| Pixibinthus | Agnotecous clade A1 | Agnotecous clade A2 | Agnotecous | 'Lebinthus' | |
|---|---|---|---|---|---|
| Characteristics | |||||
| Number of species | 1 | 4 | 15 | 19 | 23 |
| Habitat | Shrubland, low to tall = 'maquis minier' | Secondary and open forest, savannah | Forest | Forest, secondary forest and savannah | Coastal and secondary forest, open forest |
| Habitat openness | Open | Open | Closed | Mainly closed to open | Open |
| Habitat inflammability | High | Moderate | Low | Mainly low to moderate | Moderate |
| Diets | Detritivorous | Detritivorous | Detritivorous | Detritivorous | Detritivorous |
| Dominant frequency range (kHz) | 25.5–30.5 | 15–18 | 11–19.2 | 11–19.2 | (12) 16.7–27.5 |
| Body size (mm) | 6.1–7.1 | 10.9–15.2 | 9.5–16.4 | 9.5–16.4 | 7–15.7 |
| Substrate | Ultramafic | Ultramafic and Non-ultramafic | Ultramafic and Non-ultramafic | Ultramafic and Non-ultramafic | Non-ultramafic |
| Age estimates | |||||
| Stem age (Myr) | 33.3 | 20.2 | 20.2 | 33.3 | N/A |
| 95% HPD stem age (Myr) | [24.4–43.1] | [14.9–25.8] | [14.9–25.8] | [24.4–43.1] | N/A |
| Crown age (Myr) | N/A | 16.3 | 13.9 | 20.2 | N/A |
| 95% HPD crown age (Myr) | N/A | [11.6–21.6] | [9.9–18.5] | [14.9–25.8] | N/A |
Species richness estimates and relative diversification rates
The application of Equation (3) of Slowinsky and Guyer (1993) showed that Pixibinthus had significantly less species than its Agnotecous sister-clade (p = 0.0526*; one species vs. 19; Fig 8).
Ensemble species distribution modelling
ESDM analyses revealed that the suitable habitat of Pixibinthussonicus was not restricted to the four occurrence sites (AUC = 0.986), but also covered a large area of the southern of Grande Terre, exclusively on metalliferous soils (Fig 10).
Fig 10. Predicted ecological distribution of Pixibinthus sonicus using ESDM.
Warmer colors indicate areas with higher habitat suitability for P. sonicus. Reddots show sampling sites where Pixibinthus sonicus was recorded; yellowdots show sampling sites where P. sonicus was not observed.
Discussion
1. The discovery of a new original cricket genus in ‘maquis minier’
In this study we described a new monotypic cricket genus, Pixibinthus, endemic to New Caledonia. Its single species P. sonicus, has only been observed in the southern part of New Caledonian main island, living in specific open shrubby vegetation on ultramafic rocks (i.e. ‘maquis minier’; Table 4). Unexpectedly, P. sonicus exhibits the highest calling frequency ever recorded for any Grylloidea species, a record previously heldby Lebinthus santoensis at 26.5 kHz in Vanuatu [66]. Our phylogenetic study placed Pixibinthus as sister to the genus Agnotecous, also endemic to Grande Terre (Fig 8). This study confirms that the genus Lebinthus is paraphyletic as suggested by recent taxonomic [30] and phylogenetic studies [27,40].
2. Origin of the ‘Pixibinthus-Agnotecous’ clade in New Caledonia
The origin of the ‘Pixibinthus-Agnotecous’ clade in New Caledonia was estimated in our study to ca. 33.3 Myr (with a 95% HPD of 24.4–43.1 Myr; Fig 9; Table 4). Before the discovery of the close relationship between these two genera, the early colonization of New Caledonia by Agnotecous was previously estimated between 10.6 and 15.3 Myr (with a 95% HPD of 5.3–21.9 Myr; [40]). The inclusion of Pixibinthus in our DNA sampling also helped refining the age estimate of Agnotecous in New Caledonia. Our dating results pushed back the crown age of the ‘Pixibinthus-Agnotecous clade’ up to ca. 23 Myr. [40] also tentatively incorporated an extreme fossil calibration using a very old but dubious fossil of Grylloidea in order to test some specific biogeographical hypotheses. Their second analysis dated Agnotecous at approximately 24 Myr, which already permitted to reject the assumption of a clade older than the island. However this result was not retained as a robust estimate within that study, given the uncertainty on the fossil used for calibration. Strikingly our results however gave very similar estimates, despite the differences in dating methods, calibrations, and taxonomic and gene samplings.
According to our results, the ‘Pixibinthus-Agnotecous clade’is an old New Caledonian lineage that arrived not long after the postulated re-emergence of New Caledonia, estimated by geologists around 37 ± 2 Myr [6,7], similarly to some other groups of terrestrial arthropods such as fig wasps [67] or caddisflies [68]. Although Pixibinthus presently appears as a relict, actually a unique taxon on a long phylogenetic branch [69], its origin does not seem to predate the emergence of the island as for some harvestmen [70] tentatively assumed to have jumped over now-drowned islands earlier than 37 My ago [40]. This provisional relict status depends on further sampling that could reveal other extant species belonging to the same genus, though several and extensive previous studies focusing on forest habitats did not find any species [25,26,27].
According to classical accounts on New Caledonia, old clades should primarily occupy habitats on metalliferous soils which were dominant at the time of emergence of the island [10,12,71,72]. The relationship between Pixibinthus and ‘maquis minier’ is consistent with this assumption, though its sister-group Agnotecous is not restricted to metalliferous soils [27].
The common ancestor of this cricket clade probably settled in early New Caledonian open habitats, a preference which can be inferred as ancestral for this lineage, also considering the preference for open forests of close relatives such as Indo-Pacific Lebinthus species [28,66,73] (Fig 9). A similar pattern has already been observed in Cardiodactylus novaeguineae, whose ongoing colonization is documented in open backshore habitats of New Guinean and Melanesian archipelagoes, including New Caledonia [74]. New Caledonia would then have been successively colonized three times by crickets of the subfamily Eneopterinae: the ‘Pixibinthus-Agnotecous clade’, C. novaeguineae, which has a wide distribution in the Western Pacific, and L. lifouensis, restricted to the Loyalty Islands ([26,40]; Fig 8).
3. Two different ecological specializations within the ‘Pixibinthus-Agnotecous’ clade
Interestingly, the Pixibinthus lineage would have persisted in the seemingly ancestral choice of open habitats since its origin in New Caledonia. This species appears to have specialized upon the emblematic and peculiar sclerophyllous shrubby‘maquis minier’ growing on ultramafic rocks ([11,75]; Table 4). The exact timing of this specialization could not be inferred in this study, but only estimated between around 33 Myr to present, as this species is the only representative of the genus.
The sister-group Agnotecous differs, as 16 out of its 19 species are rainforest-dwellers (Agnotecous clade A2 plus A. meridionalis), while only three other species (clade A1) are known from more open forests ([26,27,61]; Table 4; S2 Table). According to the topology, occurring in closed habitatsis however a secondary specialization within Agnotecous(clade A2: 13.9 Myr; Fig 9). We can confidently hypothesize that the postulated radiation of Agnotecous in forest habitats started around 20 Myr (crown age of the genus Agnotecous; Table 4).
4. Ecological specializations suggest mutual evolution of body size and airborne signal
To the best of our knowledge, the clear-cut ecological specializations in two vegetation types of two sister-clades (Agnotecous mostly in closed forest and Pixibinthus in open ‘maquis minier’; Table 4) is undocumented within the New Caledonian insect fauna. In this unusual case, the study of acoustic behavior could be a relevant feature to understand such different ecological preferences.
Before the discovery of Pixibinthus, [76]described the few Lebinthini as ‘living paradoxes’, since they were producing high-frequency calls in forested habitats (S2 Table), which are characterized by greater attenuation and absorption on those frequencies [77,78]. But now, the significantly lower frequency calls observed in Agnotecous species (compared to Pixibinthus) might be explained by new acoustic and ecological constraints inherent to forest habitats (Fig 9, dominant frequency range diagram). The colonization of forested areas would have allowed more favorable living conditions for the ancestor of Agnotecous, with a release of ecological pressures. Resources, including food and shelter from predators, were presumably greater than in open habitats [79,80], which is usually associated with an increase of body size([81,82]; Table 4). Consequently, the Agnotecous lineage would have undergone a relative reduction of their call frequencies as a consequence to a larger body size [83], whereas Pixibinthus would have conserved a smaller body size and a high-frequency song (S2 Table). A formal test of this hypothesis will necessitate further studies taking into account precise acoustic analyses at the scale of whole clade analyzed with phylogenetic comparative methods, especially to identify the role of body size and habitat characteristics on the evolution of call frequencies (e.g. [84]).
In crickets, calling songs are crucial to attract females, and are strongly influenced by environmental conditions [85]. Pixibinthus sonicus exhibits a high-frequency call compared to Agnotecous, and this species lives in open habitats, where sound transmission is strikingly different than in forest areas: amplitude fluctuations are higher and reverberation is lower, which tends to facilitate high-frequency communication [78,86,87]. In addition, the open ‘maquis minier’ where P. sonicus lives is usually composed of patchy ‘leaf litter islands’, where conspecific potential receivers are likely to be concentrated in the close vicinity of male signalers (as according to our field observations). These circumstances may have concurred to maintain, or favor, high-frequency communication in Pixibinthus.
5. Pixibinthus: failing to diversify?
Despite a large temporal window, we observed imbalanced species richness between Pixibinthus and Agnotecous (as shown by a significant p-value of the Slowinsky and Gruyer test). We noticed also important dissimilarities in species richness with a single species for Pixibinthus vs. 19 for Agnotecous (Table 4). Both genera are detritivorous, as most crickets, meaning that diet might not explain this difference. Furthermore, although the knowledge on their predation pressure and biotic interactions is scarce, similar categories of potential predators seem present in both environments (mainly spiders and lizards). Based on that statement, we propose a working hypothesis to explain these uneven diversifications considering past and recent abiotic events, relative to climatic oscillations and fire regimes.
5.1. Impact of past climatic fluctuations?
Climatic fluctuations are greatly suspected to have been an important factor responsible for the natural expansion and contraction of New Caledonian rainforests over time [15,88,89]. As an example, allopatric speciation has been documented as one of the main diversification driver for the rainforest-dwelling Agnotecous [27]. Past expansion-contractions of this vegetation have been shown in southern main land through palynological records [65,90,91]. If such weather shifts had been critical in open shrubby habitats, leading to the isolation of ‘maquis minier’ patches, we should observe as many allopatric speciations in Pixibinthus as in Agnotecous a situation that is not supported by our data. On the contrary, the southern plain ‘maquier miniers’ could have been permanently connected [90,92]. Pixibinthus may then have failed to diversify because of relatively high habitat connectivity, regardless to its dispersal ability. The gene flux between distant populations could have been maintained over time, limiting allopatric speciation, as suggested by short internal phylogenetic branches within P. sonicus. The contrast with some Agnotecous species, whose isolated forested populations begin to be genetically isolated (such as illustrated by long branches of A. azurensis; Fig 8), could then be particularly significant.
5.2. Impact of fire regimes?
Fire regime may have equally played a key role in the apparent lack of diversification of Pixibinthus. Before human arrival (ca. 3000 year ago), New Caledonian landscapes were shaped by low natural fire regime [21,90]. As the exclusive ecological preference of Pixibinthus matches the most flammable habitat of New Caledonia [65], which are now exposed to repeated fires [93], could have led to occasional and localized extinctions, enhancing isolation of its populations in a first phase. However, these newly burnt areas would in time have constituted a favorable matrix for recolonization, ultimately leading to population reconnections. By contrast, fire damages are expected to be buffered by dense forested areas [21,94]. The specialization of Agnotecous in forest habitats would have preserved its populations, allowing the emergence of many species according to the hypothesis proposed by Nattier et al. [27]. In this likely scenario, we might assume that Pixibinthus would be more sensitive to fire damages than Agnotecous.
6. Pixibinthus: on the way to extinction?
More recently, fire regime was dramatically increased by human’s arrival in New Caledonia, as documented in the southern region by palynological and charcoal records from Quaternary sediments [65,90,91]. According to McCoy et al. [65], fire frequency has increased from one fire to five every century since human settlement. This recent (less than 3000 years) and abnormally high fire regime would have increased the effect of natural fire on native vegetation, and especially in shrubby ‘maquis minier’. Though, repeated occurrence of dramatic fire events would have led to the loss of 10.000 ha of vegetation per year in New Caledonia, of which 10% in the southern part (civilian security of New Caledonia, pers. comm.). These recent human-made fires are mostly sudden and destroy very large areas in short time. For example, 4300 and 800 ha were burnt in 2005 and 2013, respectively (civilian security of New Caledonia, pers. comm.). This increasing change in fire regime created a large vegetation mosaic of maquis-forest, dominated by flammable maquis, especially in the southern part of New Caledonia. This mosaic influence fire regime with more maquis prone to burn rather than forest remnants [93,95]. Before man arrival, the low regime of fire was related to a maquis-forest mosaic dominated by forest, which we hypothesize offer more opportunity of stable open shrubby maquis areas available for Pixibinthus population with a low turn over (a mosaic of maquis minier of differents ages in a matrix of forest). According to the actual fire regime, Curt et al. [93] estimate that more than 10 000 ha of maquis have burnt over the period 1999–2010. These authors also estimate that with a such fire regime it would take 34 years to burn an area equivalent to the whole area of maquis in New Caledonia. The known distribution of Pixibinthus being ca. 1.200 ha (Fig 5), means that its geographical range could have been virtually entirely burnt within one year. Furthermore, since the potential sustainable habitat of Pixibinthus is larger than its actual distribution, as inferred by both niche modelling analyses (Fig 10), the geographical expansion of this cricket lineage could have been held back by environmental perturbations, like drastic fire regimes, rather than by limited living conditions. As a consequence, the probability of extinction of Pixibinthus populations may have been greatly enhanced by human arrival, preventing recolonization processes and reconnection between isolated populations, even if the favorable habitat of this Pixibinthus is considerably enlarged.
7. Conclusion
Our study highlights an original discovery for the cricket fauna in emblematic south-eastern ‘maquis miniers’ from New Caledonia. The new endemic monotypic genus Pixibinthus appears to have originated from an old lineage contemporary to the 37 Myr-re-emergence of the main island. Its single species would have conserved ancestral traits for open habitats with high-frequency calls (up to 30 kHz). This lineage could be the result of a tumultuous history involving two mutually non-exclusive evolutionary patterns: (1) an apparent non-speciation linked to the past dynamics of its particular habitat, and (2) extinctions caused by a recent high human-made fire regime. Nevertheless, the long-branch phylogenetic signature of the new genus is difficult to interpret, which could be elucidated by a relevant genetic population study on the entire distribution range of this species. So far, the scarceness and the observed distribution of the species, restricted to the south raises issues about its presence in the other parts of New Caledonia. Additional sampling efforts should be done in other localities, well distant from the current known distribution, as for example in ‘maquis minier’ located at high-elevations or those located in north-western isolated mountains, on which allopatric speciation events have occurred in plants [96,97], geckos [98,99], skinks [100,101] and grasshoppers [16]. In New Caledonia clear-cut specializations between two sister lineages have been frequently highlighted for different geological substrates in insects (e.g. in caddisflies [14], or in grasshopper [16,102]), in squamata [99,100,101] or in plants (e.g. in Codia sp. [103], or Diospyros sp. [13]). However ecological specialization in contrasted vegetation types, such as shown in our study, was undocumented for New Caledonia, even if it is strongly suspected in some insect groups (e.g. in some Chrysomelid beetles or weevil beetles, in some Mogoplistidae lineages, or in the Bullita-Koghiella clade) or plant groups (e.g. in the Morierina–Thiolliera clade or within the large Psychotria). Our findings stressed the importance to study animal or plant diversifications by considering all stages of ecological succession, such as disturbed habitat (e.g. shrubby vegetation), which could provide valuable information to understand the origins of New Caledonian biodiversity, massively threatened by anthropogenic pressures.
Supporting Information
Bayesian posterior probability (PP) / ML bootstrap (BS) support values are indicated for each node on the right. A clade with a BS < 50% or not recovered in the ML analyses is indicated with a dash.
(TIF)
Bayesian posterior probability (PP) / bootstrap (BS) support values are indicated for each node on the right. A clade with a BS < 50% or not recovered in the ML analyses is indicated with a dash. Gray node bars correspond to the 95% highest posterior density of median age estimates.
(TIF)
A) Characteristics of primers (names, sequences, annealing temperatures, references where they were first designed). Characteristics of the DNA partitions, used in the phylogenetic analyses: B) ‘dataset 1’: for phylogenetic relationships; C) ‘dataset 2’: for dating estimates.
(XLS)
Acoustic recording obtained with inadequate recording material are indicated with an asterisk. Unpublished data are indicated with two asterisks. Species only known from the type specimen are indicated with three asterisks. Abbreviation: N/A corresponds to unavailable data.
(XLSX)
(XLSX)
Acknowledgments
We would like to thank Frédéric Rigault & Edouard Bourget (IMBE/IRD Noumea), Léa Colmas, and Alexia Omont who provided help for field work, habitat characterization and for mapping our sampled localities. We also thank Céline Gomez (IMBE/IRD Noumea) and Sylvain Schmitt (AMAP/IAC) who performed Ensemble Species Distribution Modelling. We are also grateful to Eric Vidal (IMBE/IRD Noumea) for interesting discussion and logistic support. We thank the New Caledonian Direction de l’Environnement de la Province Sud, for collecting permits. We thank the Grand Observatoire du Pacifique Sud (GOPS) for its contribution to funding through the project entitled ‘Bioacoustique des grillons de Nouvelle-Calédonie' in the context of their call for proposal 2013. We also thank the Labex BCDIV (Diversités biologiques et culturelles) for funding a part of this study, under the project entitled "A diachronic study of biodiversity and a test of biogeographic scenarios in New Caledonia as based on field inventories of fossil and present-day insects".
Data Availability
All relevant data are within the paper and its Supporting Information files. All audio files are available from the Sonothèque du Muséum national d'Histoire naturelle database (http://sonotheque.mnhn.fr/) (accession number(s): MNHN‑SO‑2015-9, 10, 11, 12, 13, 14).
Funding Statement
This study was partly funded by a PhD grant, ‘Bourse d’encouragement à la recherche universitaire, attributed to JA by the Government of New Caledonia. The bioacoustic study was supported by a grant from the Grand Observatoire du Pacifique Sud: AAP GOPS 2013 / ‘Bioacoustique des grillons de Nouvelle-Calédonie’. The laboratory procedures were realized through the two research programs: Action Transversale du Muséum (ATM) ‘Biodiversité actuelle et fossile. Crises, stress, restaurations et panchronisme: le message systématique’, and ATM ‘Barcode’. The Labex BCDIV (Diversités biologiques et culturelles) funded a part of this study, under the project entitled "A diachronic study of biodiversity and a test of biogeographic scenarios in New Caledonia as based on field inventories of fossil and present-day insects". The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Carlquist SJ (1965) Island life; a natural history of the islands of the world. [Google Scholar]
- 2.Whittaker RJ, Fernández-Palacios JM (2007) Island biogeography: ecology, evolution, and conservation: Oxford University Press. [Google Scholar]
- 3.Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 403: 853–858. [DOI] [PubMed] [Google Scholar]
- 4.Kier G, Kreft H, Lee TM, Jetz W, Ibisch PL, Nowicki C, et al. (2009) A global assessment of endemism and species richness across island and mainland regions. Proceedings of the National Academy of Sciences 106: 9322–9327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grandcolas P, Murienne J, Robillard T, Desutter-Grandcolas L, Jourdan H, Guilbert E, et al. (2008) New Caledonia: a very old Darwinian island? Philosophical Transactions of the Royal Society B: Biological Sciences 363: 3309–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schellart W, Lister G, Toy V (2006) A Late Cretaceous and Cenozoic reconstruction of the Southwest Pacific region: tectonics controlled by subduction and slab rollback processes. Earth-Science Reviews 76: 191–233. [Google Scholar]
- 7.Pelletier B (2006) Geology of the New Caledonia region and its implications for the study of the New Caledonian biodiversity. Compendium of marine species from New Caledonia: 17–30. [Google Scholar]
- 8.Cluzel D, Maurizot P, Collot J, Sevin B (2012) An outline of the geology of New Caledonia; from Permian-Mesozoic Southeast Gondwanaland active margin to Cenozoic obduction and supergene evolution. Episodes 35: 72–86. [Google Scholar]
- 9.Chevillotte V, Chardon D, Beauvais A, Maurizot P, Colin F (2006) Long-term tropical morphogenesis of New Caledonia (Southwest Pacific): Importance of positive epeirogeny and climate change. Geomorphology 81: 361–375. [Google Scholar]
- 10.Morat P, Jaffré T, Veillon J-M, MacKee H (1986) Affinités floristiques et considérations sur l’origine des maquis miniers de la Nouvelle-Calédonie. Adansonia 2: 133–182. [Google Scholar]
- 11.Jaffré T (1980) Étude écologique du peuplement végétal des sols dérivés de roches ultrabasiques en Nouvelle Calédonie. [Google Scholar]
- 12.Pillon Y, Munzinger J, Amir H, Lebrun M (2010) Ultramafic soils and species sorting in the flora of New Caledonia. Journal of Ecology 98: 1108–1116. [Google Scholar]
- 13.Paun O, Turner B, Trucchi E, Munzinger J, Chase MW, Samuel R (2015) Processes Driving the Adaptive Radiation of a Tropical Tree (Diospyros, Ebenaceae) in New Caledonia, a Biodiversity Hotspot. Systematic biology: syv076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Espeland M, Johanson KA, Hovmöller R (2008) Early Xanthochorema (Trichoptera, Insecta) radiations in New Caledonia originated on ultrabasic rocks. Molecular Phylogenetics and Evolution 48: 904–917. 10.1016/j.ympev.2008.06.006 [DOI] [PubMed] [Google Scholar]
- 15.Pouteau R, Trueba S, Feild TS, Isnard S (2015) New Caledonia: a Pleistocene refugium for rain forest lineages of relict angiosperms. Journal of Biogeography. [Google Scholar]
- 16.Nattier R, Grandcolas P, Pellens R, Jourdan H, Couloux A, Poulain S, et al. (2013) Climate and Soil Type Together Explain the Distribution of Microendemic Species in a Biodiversity Hotspot. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murienne J, Pellens R, Budinoff R, Wheeler W, Grandcolas P (2008) Phylogenetic analysis of the endemic New Caledonian cockroach Lauraesilpha. Testing competing hypotheses of diversification. Cladistics 24: 802–812. [Google Scholar]
- 18.Delcourt PA, Delcourt HR (1983) Late-Quaternary vegetational dynamics and community stability reconsidered. Quaternary Research 19: 265–271. [Google Scholar]
- 19.Prentice IC (1986) Vegetation responses to past climatic variation. Vegetatio 67: 131–141. [Google Scholar]
- 20.Byrne M, Steane DA, Joseph L, Yeates DK, Jordan GJ, Crayn D, et al. (2011) Decline of a biome: evolution, contraction, fragmentation, extinction and invasion of the Australian mesic zone biota. Journal of Biogeography 38: 1635–1656. [Google Scholar]
- 21.Jaffré T, Rigault F, Dagostini G (1998) Impact des feux de brousse sur les maquis ligno-herbacés des roches ultramafiques de Nouvelle-Calédonie. Adansonia 20: 173–189. [Google Scholar]
- 22.Morat P, Jaffré T, Tronchet F, Munzinger J, Pillon Y, Veillon J-M, et al. (2012) Le référentiel taxonomique Florical et les caractéristiques de la flore vasculaire indigène de la Nouvelle-Calédonie. Adansonia 34: 179–221. [Google Scholar]
- 23.Wellstein C, Schröder B, Reineking B, Zimmermann NE (2011) Understanding species and community response to environmental change—A functional trait perspective. Agriculture, ecosystems & environment 145: 1–4. [Google Scholar]
- 24.Gerlach J, Samways M, Pryke J (2013) Terrestrial invertebrates as bioindicators: an overview of available taxonomic groups. Journal of Insect Conservation 17: 831–850. [Google Scholar]
- 25.Desutter-Grandcolas L (1997) Le peuplement de grillons (Orthoptères, Grylloidae): espèces et données nouvelles. Mémoire du Muséum national d'Histoire naturelle 171: 165–177. [Google Scholar]
- 26.Desutter-Grandcolas L, Robillard T (2006) Phylogenetic systematics and evolution of Agnotecous in New Caledonia (Orthoptera: Grylloidea, Eneopteridae). Systematic Entomology 31: 65–92. [Google Scholar]
- 27.Nattier R, Grandcolas P, Elias M, Desutter-Grandcolas L, Jourdan H, Couloux A, et al. (2012) Secondary sympatry caused by range expansion informs on the dynamics of microendemism in a biodiversity hotspot. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robillard T, Yap S, Yngente MV (2013) Systematics of cryptic species of Lebinthus crickets in Mount Makiling (Grylloidea, Eneopterinae). Zootaxa 3693: 049–063. [DOI] [PubMed] [Google Scholar]
- 29.Robillard T, Gorochov AVe, Poulain S, Suhardjono YR (2014) Revision of the cricket genus Cardiodactylus (Orthoptera, Eneopterinae, Lebinthini): the species from both sides of the Wallace line, with description of 25 new species. Zootaxa 3854: 1–104. 10.11646/zootaxa.3854.1.1 [DOI] [PubMed] [Google Scholar]
- 30.Vicente NM, Olivero P, Lafond A, Dong J, Robillard T (2015) Gnominthus gen. nov., a new genus of crickets endemic to Papua New Guinea with novel acoustic and behavioral diversity (Insecta, Orthoptera, Gryllidae, Eneopterinae). Zoologischer Anzeiger-A Journal of Comparative Zoology 258: 82–91. [Google Scholar]
- 31.Ragge DR (1955) The wing venation of the Orthoptera Saltatoria. British Museum (Natural History), London: 159 [Google Scholar]
- 32.Robillard T, Desutter-Grandcolas L (2004) Phylogeny and the modalities of acoustic diversification in extant Eneopterinae (Insecta, Orthoptera, Grylloidea, Eneopteridae). Cladistics 20: 271–293. [DOI] [PubMed] [Google Scholar]
- 33.Desutter L (1987) Structure et évolution du complexe phallique des Gryllidea (orthoptères) et classification des genres néotropicaux de Grylloidea. I. Bulletin de la Société entomologique de France 23: 213–239. [Google Scholar]
- 34.Desutter-Grandcolas L (2003) Phylogeny and the evolution of acoustic communication in extant Ensifera (Insecta, Orthoptera). Zoologica Scripta 32: 525–561. [Google Scholar]
- 35.Ragge DR, Reynolds WJ, editors (1998) The songs of the grasshoppers and crickets from Western Europe. 591 p. [Google Scholar]
- 36.Robillard T, Desutter-Grandcolas L (2008) Clarification of the taxonomy of extant crickets of the subfamily Eneopterinae (Orthoptera: Grylloidea; Gryllidae). Zootaxa 1789: 66–68. [Google Scholar]
- 37.Chintauan-Marquier IC, Legendre F, Hugel S, Robillard T, Grandcolas P, Nel A, et al. (2015) Laying the foundations of evolutionary and systematic studies in crickets (Insecta, Orthoptera): a multilocus phylogenetic analysis. Cladistics. [DOI] [PubMed] [Google Scholar]
- 38.Huang Y, Ortí G, Sutherlin M, Duhachek A, Zera A (2000) Phylogenetic relationships of North American field crickets inferred from mitochondrial DNA data. Molecular Phylogenetics and Evolution 17: 48–57. [DOI] [PubMed] [Google Scholar]
- 39.Regier JC, Shultz JW, Zwick A, Hussey A, Ball B, Wetzer R, et al. (2010) Arthropod relationships revealed by phylogenomic analysis of nuclear protein-coding sequences. Nature 463: 1079–1083. 10.1038/nature08742 [DOI] [PubMed] [Google Scholar]
- 40.Nattier R, Robillard T, Desutter-Grandcolas L, Couloux A, Grandcolas P (2011) Older than New Caledonia emergence? A molecular phylogenetic study of the eneopterine crickets (Orthoptera: Grylloidea). Journal of Biogeography 38: 2195–2209. [Google Scholar]
- 41.Robillard T, Desutter-Grandcolas L (2006) Phylogeny of the cricket subfamily Eneopterinae (Orthoptera, Grylloidea, Eneopteridae) based on four molecular loci and morphology. Molecular Phylogenetics and Evolution 40: 643–661. [DOI] [PubMed] [Google Scholar]
- 42.Jost M, Shaw K (2006) Phylogeny of Ensifera (Hexapoda: Orthoptera) using three ribosomal loci, with implications for the evolution of acoustic communication. Molecular Phylogenetics and Evolution 38: 510–530. [DOI] [PubMed] [Google Scholar]
- 43.Jaiswara R, Balakrishnan R, Robillard T, Rao K, Cruaud C, Desutter-Grandcolas L (2012) Testing concordance in species boundaries using acoustic, morphological, and molecular data in the field cricket genus Itaropsis (Orthoptera: Grylloidea, Gryllidae: Gryllinae). Zoological Journal of the Linnean Society 164: 285–303. [Google Scholar]
- 44.Flook P, Klee S, Rowell C (1999) Combined molecular phylogenetic analysis of the Orthoptera (Arthropoda, Insecta) and implications for their higher systematics. Systematic biology 48: 233–253. [DOI] [PubMed] [Google Scholar]
- 45.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT; 1999. pp. 95–98. [Google Scholar]
- 46.Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, et al. (2008) Phylogeny. fr: robust phylogenetic analysis for the non-specialist. Nucleic acids research 36: W465–W469. 10.1093/nar/gkn180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vaidya G, Lohman DJ, Meier R (2011) SequenceMatrix: concatenation software for the fast assembly of multi‐gene datasets with character set and codon information. Cladistics 27: 171–180. [DOI] [PubMed] [Google Scholar]
- 49.Posada D (2008) jModelTest: phylogenetic model averaging. Molecular biology and evolution 25: 1253–1256. 10.1093/molbev/msn083 [DOI] [PubMed] [Google Scholar]
- 50.Nylander JA, Ronquist F, Huelsenbeck JP, Nieves-Aldrey J (2004) Bayesian phylogenetic analysis of combined data. Systematic biology 53: 47–67. [DOI] [PubMed] [Google Scholar]
- 51.Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 52.Silvestro D, Michalak I (2012) raxmlGUI: a graphical front-end for RAxML. Organisms Diversity & Evolution 12: 335–337. [Google Scholar]
- 53.Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- 54.Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution: 783–791. [DOI] [PubMed] [Google Scholar]
- 55.Miller M, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees In: Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA: 1–8. [Google Scholar]
- 56.Rambaut A, Drummond AJ (2009) Tracer v.1.5. In: http://beast.bio.ed.ac.uk/tracer/.
- 57.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic biology 61: 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC evolutionary biology 7: 214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Song H, Amédégnato C, Cigliano MM, Desutter-Grandcolas L, Heads SW, Huang Y, et al. (2015) 300 million years of diversification: elucidating the patterns of orthopteran evolution based on comprehensive taxon and gene sampling. Cladistics. [DOI] [PubMed] [Google Scholar]
- 60.Slowinski JB, Guyer C (1993) Testing whether certain traits have caused amplified diversification: an improved method based on a model of random speciation and extinction. American Naturalist: 1019–1024. 10.1086/285586 [DOI] [PubMed] [Google Scholar]
- 61.Robillard T, Nattier R, Desutter-Grandcolas L (2010) New species of the New Caledonian endemic genus Agnotecous (Orthoptera, Grylloidea, Eneopterinae, Lebinthini). Zootaxa 2559: 17–35. [Google Scholar]
- 62.Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A (2005) Very high resolution interpolated climate surfaces for global land areas. International journal of climatology 25: 1965–1978. [Google Scholar]
- 63.Marmion M, Parviainen M, Luoto M, Heikkinen RK, Thuiller W (2009) Evaluation of consensus methods in predictive species distribution modelling. Diversity and Distributions 15: 59–69. [Google Scholar]
- 64.Thuiller W, Lafourcade B, Engler R, Araújo MB (2009) BIOMOD—a platform for ensemble forecasting of species distributions. Ecography 32: 369–373. [Google Scholar]
- 65.McCoy S, Jaffré T, Rigault F, Ash J (1999) Fire and succession in the ultramafic maquis of New Caledonia. Journal of Biogeography 26: 579–594. [Google Scholar]
- 66.Robillard T (2009) Eneopterinae crickets (Insecta, Orthoptera, Grylloidea) from Vanuatu. Zoosystema 31: 577–618. [Google Scholar]
- 67.Cruaud A, Jabbour-Zahab R, Genson G, Ungricht S, Rasplus J-Y (2012) Testing the Emergence of New Caledonia: Fig Wasp Mutualism as a Case Study and a Review of Evidence. PLoS ONE 7: e30941 10.1371/journal.pone.0030941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Espeland M, Johanson KA (2010) The diversity and radiation of the largest monophyletic animal group on New Caledonia (Trichoptera: Ecnomidae: Agmina). Journal of evolutionary biology 23: 2112–2122. 10.1111/j.1420-9101.2010.02072.x [DOI] [PubMed] [Google Scholar]
- 69.Grandcolas P, Nattier R, Trewick S (2014) Relict species: a relict concept? Trends in Ecology & Evolution 29: 655–663. [DOI] [PubMed] [Google Scholar]
- 70.Sharma PP, Giribet G (2012) Out of the Neotropics: Late Cretaceous colonization of Australasia by American arthropods. Proceedings of the Royal Society of London B: Biological Sciences: rspb20120675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Espeland M, Johanson KA (2010) The effect of environmental diversification on species diversification in New Caledonian caddisflies (Insecta: Trichoptera: Hydropsychidae). Journal of Biogeography 37: 879–890. [Google Scholar]
- 72.Pillon Y (2012) Time and tempo of diversification in the flora of New Caledonia. Botanical Journal of the Linnean Society 170: 288–298. [Google Scholar]
- 73.Robillard T (2010) New species of the genus Lebinthus (Orthoptera, Grylloidea, Eneopterinae, Lebinthini) from Indonesia and the Solomon islands. Zootaxa 2386: 25–48. [Google Scholar]
- 74.Robillard T, Ichikawa A (2009) Redescription of two Cardiodactylus species (Orthoptera, Grylloidea, Eneopterinae): the supposedly well-known C. novaeguineae (Haan, 1842), and the semi-forgotten C. guttulus (Matsumura, 1913) from Japan. Zoological science 26: 878–891. 10.2108/zsj.26.878 [DOI] [PubMed] [Google Scholar]
- 75.Proctor J (2003) Vegetation and soil and plant chemistry on ultramafic rocks in the tropical Far East. Perspectives in plant ecology, evolution and systematics 6: 105–124. [Google Scholar]
- 76.Robillard T, Desutter-Grandcolas L (2004) High-frequency calling in Eneopterinae crickets (Orthoptera, Grylloidea, Eneopteridae): adaptive radiation revealed by phylogenetic analysis. Biological Journal of the Linnean Society 83: 577–584. [Google Scholar]
- 77.Richards DG, Wiley RH (1980) Reverberations and Amplitude Fluctuations in the Propagation of Sound in a Forest: Implications for Animal Communication. The American Naturalist 115: 381–399. [Google Scholar]
- 78.Wiley RH, Richards DG (1978) Physical constraints on acoustic communication in the atmosphere: Implications for the evolution of animal vocalizations. Behavioral Ecology and Sociobiology 3: 69–94. [Google Scholar]
- 79.Fartmann T, Krämer B, Stelzner F, Poniatowski D (2012) Orthoptera as ecological indicators for succession in steppe grassland. Ecological Indicators 20: 337–344. [Google Scholar]
- 80.Langellotto GA, Denno RF (2004) Responses of invertebrate natural enemies to complex-structured habitats: a meta-analytical synthesis. Oecologia 139: 1–10. [DOI] [PubMed] [Google Scholar]
- 81.Atkinson D (1994) Temperature and organism size: a biological law for ectotherms? Advances in ecological research 25: 1–1. [Google Scholar]
- 82.Gardner JL, Peters A, Kearney MR, Joseph L, Heinsohn R (2011) Declining body size: a third universal response to warming? Trends in Ecology & Evolution 26: 285–291. [DOI] [PubMed] [Google Scholar]
- 83.Bennet-Clark HC (1998) Size and scale effects as constraints in insect sound communication. Philosophical Transactions of the Royal Society of London Series B: Biological Sciences 353: 407–419. [Google Scholar]
- 84.Garamszegi LZ (2014) Modern phylogenetic comparative methods and their application in evolutionary biology: Springer. [Google Scholar]
- 85.Forrest T (1994) From sender to receiver: propagation and environmental effects on acoustic signals. American Zoologist 34: 644–654. [Google Scholar]
- 86.McCracken KG, Sheldon FH (1997) Avian vocalizations and phylogenetic signal. Proceedings of the National Academy of Sciences 94: 3833–3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ryan MJ, Brenowitz EA (1985) The Role of Body Size, Phylogeny, and Ambient Noise in the Evolution of Bird Song. The American Naturalist 126: 87–100. [Google Scholar]
- 88.Poncet V, Munoz F, Munzinger J, Pillon Y, Gomez C, Couderc M, et al. (2013) Phylogeography and niche modelling of the relict plant Amborella trichopoda (Amborellaceae) reveal multiple Pleistocene refugia in New Caledonia. Molecular Ecology 22: 6163–6178. 10.1111/mec.12554 [DOI] [PubMed] [Google Scholar]
- 89.Pintaud J-C, Jaffré T, Puig H (2001) Chorology of New Caledonian palms and possible evidence of Pleistocene rain forest refugia. Comptes Rendus de l'Académie des Sciences-Series III-Sciences de la Vie 324: 453–463. [DOI] [PubMed] [Google Scholar]
- 90.Hope G, Pask J (1998) Tropical vegetational change in the late Pleistocene of New Caledonia. Palaeogeography, Palaeoclimatology, Palaeoecology 142: 1–21. [Google Scholar]
- 91.Stevenson J (2004) A late-Holocene record of human impact from the southwest coast of New Caledonia. The Holocene 14: 888–898. [Google Scholar]
- 92.Stevenson J, Hope G (2005) A comparison of late Quaternary forest changes in New Caledonia and northeastern Australia. Quaternary Research 64: 372–383. [Google Scholar]
- 93.Curt T, Borgniet L, Ibanez T, Moron V, Hély C (2015) Understanding fire patterns and fire drivers for setting a sustainable management policy of the New-Caledonian biodiversity hotspot. Forest Ecology and Management 337: 48–60. [Google Scholar]
- 94.Jaffré T (1995) Distribution and ecology of the conifers of New Caledonia. [Google Scholar]
- 95.Gomez C, Mangeas M, Curt T, Ibanez T, Munzinger J, Dumas P, et al. (2015) Wildfire risk for main vegetation units in a biodiversity hotspot: modeling approach in New Caledonia, South Pacific. Ecology and Evolution 5: 377–390. 10.1002/ece3.1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pillon Y (2008) Biodiversité, origine et évolution des Cunoniaceae: implications pour la conservation de la flore de Nouvelle-Calédonie: Citeseer. [Google Scholar]
- 97.Schmid M (1982) Endémisme et spéciation en Nouvelle-Calédonie. Compte Rendu des Séances de la Société de Biogéographie 48: 52–60. [Google Scholar]
- 98.Bauer AM, Jackman T, Sadlier RA, Whitaker AH (2006) A revision of the Bavayia validiclavis group (Squamata: Gekkota: Diplodactylidae), a clade of New Caledonian geckos exhibiting microendemism. Proceedings—California Academy of Sciences 57: 503. [Google Scholar]
- 99.Skipwith P, Jackman T, Whitaker AH, Bauer AM, Sadlier RA (2014) New data on Dierogekko (Squamata: Gekkota: Diplodactylidae), with the description of a new species from Île Baaba, Province Nord, New Caledonia. Mémoires du Muséum national d'histoire naturelle 206: 13–30. [Google Scholar]
- 100.Sadlier RA, Smith SA, Bauer AM, Whitaker AH (2009) Three new species of skink in the genus Marmorosphax Sadlier (Squamata: Scincidae) from New Caledonia. Mémoires du Muséum national d'histoire naturelle 198: 373–390. [Google Scholar]
- 101.Sadlier RA, Bauer AM, Wood PL, Smith SA, Whitaker AH, Jackman TR (2014) Cryptic speciation in the New Caledonian lizard genus Nannoscincus (Reptilia: Scincidae) including the description of a new species and recognition of Nannoscincus fuscus Günther. Mémoires du Muséum national d'histoire naturelle 206: 45–68. [Google Scholar]
- 102.Murienne Jm, Grandcolas P, Piulachs MD, Bellés X, D'Haese C, Legendre F, et al. (2005) Evolution on a shaky piece of Gondwana: is local endemism recent in New Caledonia? Cladistics 21: 2–7. [DOI] [PubMed] [Google Scholar]
- 103.Pillon Y, Munzinger J, Amir H, Hopkins HC, Chase MW (2009) Reticulate evolution on a mosaic of soils: diversification of the New Caledonian endemic genus Codia (Cunoniaceae). Molecular Ecology 18: 2263–2275. 10.1111/j.1365-294X.2009.04178.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bayesian posterior probability (PP) / ML bootstrap (BS) support values are indicated for each node on the right. A clade with a BS < 50% or not recovered in the ML analyses is indicated with a dash.
(TIF)
Bayesian posterior probability (PP) / bootstrap (BS) support values are indicated for each node on the right. A clade with a BS < 50% or not recovered in the ML analyses is indicated with a dash. Gray node bars correspond to the 95% highest posterior density of median age estimates.
(TIF)
A) Characteristics of primers (names, sequences, annealing temperatures, references where they were first designed). Characteristics of the DNA partitions, used in the phylogenetic analyses: B) ‘dataset 1’: for phylogenetic relationships; C) ‘dataset 2’: for dating estimates.
(XLS)
Acoustic recording obtained with inadequate recording material are indicated with an asterisk. Unpublished data are indicated with two asterisks. Species only known from the type specimen are indicated with three asterisks. Abbreviation: N/A corresponds to unavailable data.
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. All audio files are available from the Sonothèque du Muséum national d'Histoire naturelle database (http://sonotheque.mnhn.fr/) (accession number(s): MNHN‑SO‑2015-9, 10, 11, 12, 13, 14).