Abstract
Bacteria use trans-translation and the alternative rescue factors ArfA (P36675) and ArfB (Q9A8Y3) to hydrolyze peptidyl-tRNA on ribosomes that stall near the 3' end of an mRNA during protein synthesis. The eukaryotic protein ICT1 (Q14197) is homologous to ArfB. In vitro ribosome rescue assays of human ICT1 and Caulobacter crescentus ArfB showed that these proteins have the same activity and substrate specificity. Both ArfB and ICT1 hydrolyze peptidyl-tRNA on nonstop ribosomes or ribosomes stalled with ≤6 nucleotides extending past the A site, but are unable to hydrolyze peptidyl-tRNA when the mRNA extends ≥14 nucleotides past the A site. ICT1 provided sufficient ribosome rescue activity to support viability in C. crescentus cells that lacked both trans-translation and ArfB. Likewise, expression of ArfB protected human cells from death when ICT1 was silenced with siRNA. These data indicate that ArfB and ICT1 are functionally interchangeable, and demonstrate that ICT1 is a ribosome rescue factor. Because ICT1 is essential in human cells, these results suggest that ribosome rescue activity in mitochondria is required in humans.
Author Summary
Ribosomes can stall during protein synthesis on truncated or damaged mRNAs that lack a stop codon. In bacteria, these “non-stop” ribosomes are rescued by trans-translation or by an alternative rescue factor, ArfA or ArfB. Most eukaryotes do not have trans-translation, but mammals have a homolog of ArfB named ICT1. ICT1 is targeted to mitochondria, and is essential in human cells. Here, we show that human ICT1 and ArfB from the bacterium Caulobacter crescentus have the same biochemical activity and specificity. We also demonstrate that ICT1 and ArfB are functionally interchangeable in both bacteria and human cells. Collectively, this work demonstrates a new essential function in human cells—rescue of mitochondrial non-stop translation complexes.
Introduction
The presence of a stop codon at the end of an open reading frame signals that the nascent protein is complete. Decoding of the stop codon by a release factor results in peptidyl-tRNA hydrolysis, releasing the completed protein and allowing the ribosome to be recycled [1]. Specific contacts between the release factors and bases in the stop codon are required for efficient catalysis of peptidyl-tRNA hydrolysis [2]. This stop codon recognition is necessary to prevent release factors from acting at sense codons and prematurely terminating translation. However, ribosomes can sometimes translate to the end of an mRNA without terminating at an in-frame stop codon. Translation cannot terminate normally at these “non-stop” complexes, because there is no stop codon in the decoding center to promote release factor activity. Ribosomes must be rescued from non-stop complexes so they can be recycled for productive protein synthesis [3,4].
In bacteria, non-stop complexes are rescued primarily by trans-translation. During trans-translation, a small protein, SmpB (P0A832), and a specialized RNA, tmRNA (EG30100), recognize a non-stop complex and release the ribosome at a stop codon within tmRNA. trans-Translation also targets the nascent polypeptide and mRNA from the non-stop complex for degradation [3,5]. Genes encoding tmRNA or SmpB have been identified in >99.9% of sequenced bacterial genomes [4]. trans-Translation is essential in some bacteria [6–8], but other species can survive without ssrA (encoding tmRNA) and smpB [9–11]. Some species, such as C. crescentus, have a severe growth defect when ssrA is deleted [12]. In other species, such as Escherichia coli, there is a relatively mild phenotype [13]. Synthetic-lethal screens have identified two alternative rescue factors, ArfA and ArfB, that can rescue non-stop complexes in the absence of trans-translation [14–16]. ArfA, found in E. coli and closely related bacteria, allows the release factor RF-2 to hydrolyze peptidyl-tRNA on non-stop ribosomes [17–20]. ArfB, found in C. crescentus and species from many phyla, contains a catalytic domain similar to release factors but does not include domains required for stop codon recognition. ArfB catalyzes hydrolysis of peptidyl-tRNA on non-stop ribosomes [15,16,21]. The C-terminal tail of ArfB is important for its activity [22], and structural studies suggest that it binds in the empty mRNA channel of non-stop ribosomes, similar to the C-terminal tail of SmpB [23]. arfA is essential in E. coli ΔssrA cells [14], and arfB is essential in C. crescentus ΔssrA cells [21], indicating that these species require at least one ribosome rescue mechanism. These observations have led to the suggestion that ribosome rescue activity may be essential for most or all bacteria [5].
Eukaryotes use Dom34/Pelota (P33309/Q9BRX2) and Hbs1 (P32769) to rescue ribosomes from non-stop mRNAs during translation in the cytoplasm [24,25], but factors required for this system are not present in mitochondria. Mammals have an ArfB homolog, ICT1, which is encoded in the nucleus and transported to mitochondria [26]. Knockdown experiments have demonstrated that ICT1 is essential in human cells [26,27], but conflicting models have been proposed to explain why ICT1 is essential [28–30]. Like ArfB, ICT1 can hydrolyze peptidyl-tRNA on E. coli non-stop ribosomes in vitro [22]. Because ICT1 can also hydrolyze peptidyl-tRNA on E. coli ribosomes assembled on short mRNAs with a stop or sense codon in the A site, it has been proposed to act as a general release factor that can terminate translation at any codon [26]. ICT1 has also been proposed to act on ribosomes stalled in the middle of an mRNA based on its ability to promote protein synthesis in reactions stalled by omission of a cognate release factor or tRNA [31]. Two mRNAs encoded in human mitochondria terminate with an AGA or AGG codon, so if ICT1 can act as non-specific release factor it might terminate translation of these messages. However, the sequence similarity between ICT1 and ArfB suggests that these factors are likely to have the same activity. ICT1 and ArfB share the conserved GGQ motif found in release factor catalytic domains, as well as residues in the C-terminal tail that are required for ArfB activity on non-stop ribosomes (Fig 1) [22,31]
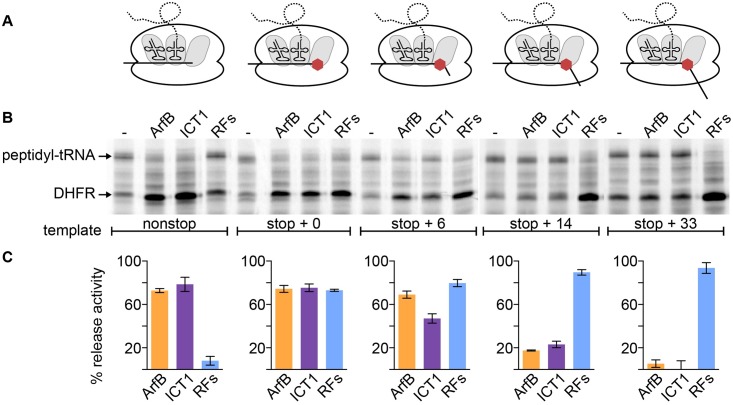
Fig 1. ICT1 and ArfB share conserved residues that are required for release activity.
Clustal Omega alignment of human ICT1 and ArfB proteins from E. COLI and C. CRESCENTUS. Blue stars indicate residues required for ≥ 60% ICT1 peptidyl-tRNA hydrolysis activity on non-stop ribosomes [22]. Red stars indicate residues required for ≥ 60% E. COLI ArfB hydrolysis activity on non-stop ribosomes [22]. The N-terminal extension of ICT1 contains the mitochondrial localization signal. The remaining 143 C-terminal residues, thought to constitute the active portion of ICT1, share 26% sequence identity with C. CRESCENTUS ArfB.
Using a direct assay for peptidyl-tRNA hydrolysis in vitro, we find that the substrate specificity of ICT1 is almost identical to that of ArfB. Both factors catalyze peptidyl-tRNA hydrolysis on ribosomes stalled with no mRNA in the A site, or with mRNA extending a short distance past the A site, but have little activity on ribosomes stalled in the middle of an intact mRNA. In addition, we find that ArfB and ICT1 are interchangeable in vivo, both in C. crescentus and in human cells. These data indicate that ICT1 is a ribosome rescue factor and cannot terminate translation in the middle of mRNA, and suggest that mitochondrial ribosome rescue activity is essential in humans.
Results
Human ICT1 hydrolyzes peptidyl-tRNA on non-stop translation complexes
To evaluate the substrate specificity of ICT1, we used a gel-based assay to measure peptidyl-tRNA hydrolysis on E. coli ribosomes translating protein from mRNA. In these experiments, protein is produced using in vitro transcription-translation reactions and the components are separated on Bis-Tris gels that preserve the ester bond in peptidyl-tRNA. The fraction of protein that remains in the peptidyl-tRNA band indicates the extent of peptidyl-tRNA hydrolysis during the reaction [21].
To confirm that ICT1 can hydrolyze peptidyl-tRNA on non-stop ribosomes in a manner similar to ArfB, a coupled transcription-translation system lacking RF1, RF2, and RF3 was used to express a folA gene (encoding DHFR) that lacked a stop codon. Consistent with previous observations of peptidyl-tRNA hydrolysis on non-stop ribosomes [15,16,21], addition of a release factor mixture containing RF-1, RF-2, and RF-3 to the reaction had little effect on the percentage of DHFR found in peptidyl-tRNA, but when ArfB was included 74 ± 1% of the DHFR was released (Fig 2). When ICT1 was included in the reaction, 78 ± 8% DHFR was released, indicating that ICT1 has a similar activity to ArfB on non-stop ribosomes.
Fig 2. ICT1 and ArfB hydrolyze peptidyl-tRNA on ribosomes near the 3’ end of an mRNA.
In vitro transcription/translation reactions were performed with template lacking a stop codon, or with template with 0, 6, 14, or 33 bases past the stop codon. (A) Cartoons depicting the expected result of translation in the absence of added rescue or release factors. (B) Representative autoradiograms of reactions resolved on Bis-Tris gels. (C) Column graphs show average release activity from ≥3 replicates with error bars indicating the standard deviation.
To determine if ICT1 and ArfB can also release ribosomes stalled with mRNA extending into or past the A site, the peptidyl-tRNA hydrolysis assay was repeated with longer folA templates. A stop codon was added to the end of the non-stop template, and 0, 6, 14, or 33 nucleotides were added after the stop codon. Each template was designed to produce an mRNA with a stem-loop at the 3’ end to limit exonuclease activity during the reaction. Translation of these mRNAs will result in a ribosome stalled at the stop codon with peptidyl-tRNA in the P site. As expected, addition of release factors to reactions with any of these templates resulted in release of most of the peptidyl-tRNA (Fig 2). ICT1 and ArfB hydrolyzed peptidyl-tRNA as efficiently as release factors when the stop + 0 template was used. Substantial peptidyl-tRNA hydrolysis activity by ICT and ArfB was also observed with the stop + 6 template, but significantly less activity was observed when the template had a longer sequence past the stop codon (p < 0.001). Almost no hydrolysis was observed with ICT1 or ArfB on the stop + 33 template. These results indicate that ICT1 and ArfB can hydrolyze peptidyl-tRNA on ribosomes stalled near the 3’ end of an mRNA, and that a codon in the A site does not interfere with ribosome rescue. However, mRNAs that extend ≥ 14 bases past the A site substantially decrease activity of both ICT1 and ArfB.
Human ICT1 has ArfB activity in C. crescentus
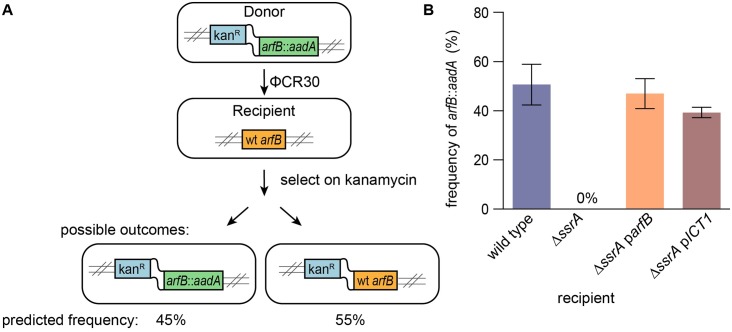
The activity of ICT1 and ArfB in vitro was similar enough to suggest that ICT1 might substitute for ArfB in C. crescentus. In a previous study, we used genetic linkage analysis to demonstrate that deletions of ssrA and arfB are synthetically lethal in C. crescentus [21]. A phage ΦCR30 lysate was prepared from strain KCK 428. In this strain, arfB is replaced with a copy of the aadA gene (conferring resistance to spectinomycin) and a copy of the nptII gene (conferring resistance to kanamycin) is inserted in the chromosome 22 kb from the arfB locus. The co-transduction frequency of the two antibiotic resistance markers could then be measured by infecting recipient cells with phage, selecting for transductants on kanamycin, and screening the kanamycin-resistant cells for spectinomycin resistance. Based on their relative locations on the chromosome, the predicted frequency with which the aadA and nptII genes should co-transduce when arfB is not essential is 45% [32]. Consistent with this prediction, when lysate is used to infect wild-type cells, arfB::aadA is recovered at a frequency of 51 ± 8% When this lysate was used to infect ΔssrA cells, transductants with the arfB deletion were not recovered, indicating that arfB is essential in the absence of ssrA. To determine if ICT1 could provide ribosome rescue activity to support viability of C. crescentus, the co-transduction experiment was repeated using ΔssrA cells that contained ICT1 (Fig 3). When the ΔssrA strain expressed ICT1 from a plasmid, arfB could be deleted, as indicated by the co-transduction of arfB::aadA with the nptII gene. The co-transduction frequency of aadA and nptII into ΔssrA cells expressing ArfB or ICT1 was not significantly different (p = 0.12), indicating that ICT1 can suppress the synthetic-lethal phenotype of deleting arfB and ssrA. In contrast, when ΔssrA cells harboring empty plasmid were used as the recipient, none of >500 kanamycin-resistant transductions were resistant to spectinomycin (Fig 3).
Fig 3. ICT1 ribosome release activity supports viability in c. crescentus.
A co-transduction experiment was used to test whether ICT1 complements the synthetic lethal phenotype of deleting ARFB and SSRA. (A) Cartoon depicting the co-transduction experiment and predicted frequency of the outcomes if ARFB were not essential. (B) Column graph indicating the average co-transduction frequency from 3 independent experiments, with error bars indicating the standard deviation.
ICT1 suppresses the growth defect in C. crescentus ΔssrA cells
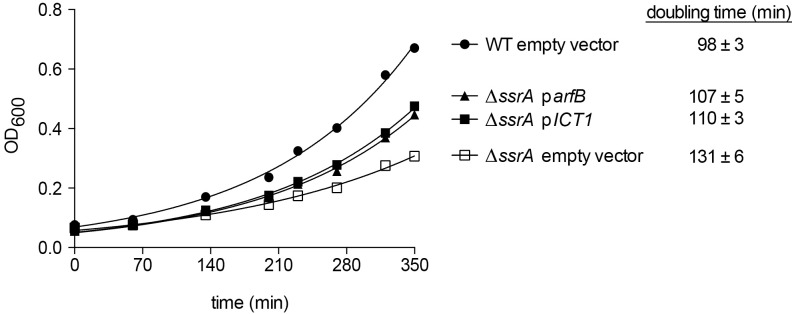
Previous characterization of trans-translation and ArfB activity in C. crescentus showed that cells lacking trans-translation grow slowly [12], and over-expression of ArfB partially suppresses this growth defect [21]. To determine if ICT1 can perform the same function as ArfB in this assay, the growth rate of the ΔssrA strain expressing ICT1 was measured and compared to control strains. ΔssrA cells expressing ICT1 cells grew at a rate similar to ΔssrA cells expressing ArfB, and significantly faster than ΔssrA cells (p <0.01) (Fig 4). These results indicate that ICT1 activity supports the same rate of growth as ArfB activity, and suggest that human ICT1 is functionally interchangeable with ArfB in C. crescentus.
Fig 4. expression of ICT1 partially complements the growth defect of δssra cells.
Growth of wild-type cells, ΔSSRA cells, or ΔSSRA cells expressing ArfB or ICT1 from a plasmid was monitored during exponential phase. The average doubling times (± standard deviation) from ≥3 experiments are shown.
ArfB rescues human cells when ICT1 is silenced
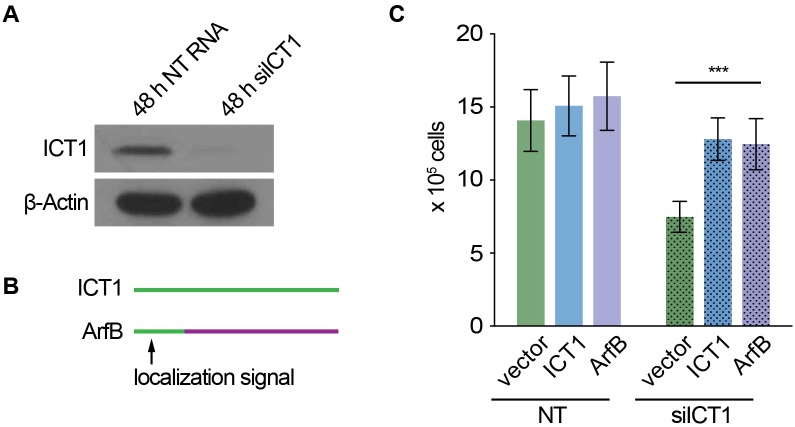
Mutant versions of ICT1 that lack the GGQ motif required for peptidyl-tRNA hydrolysis do not support viability in human cells when the endogenous ICT1 is silenced [26], indicating that ICT1-mediated peptidyl-tRNA hydrolysis is essential. If the essential activity of ICT1 is rescue of non-stop ribosomes, C. crescentus ArfB might be able to support viability in the absence of ICT1. To test this possibility, a gene encoding the N-terminal sequence of ICT1 containing the mitochondrial localization signal [33] fused to ArfB was constructed. HEK293 cells were transfected with a vector to express ArfB or ICT1, or with an empty vector. Eight hours later, these cells were transfected with siRNA that silenced ICT1 or with control siRNA, and after 6 days the number of living cells was determined. Consistent with previous results, ICT1 silencing decreased the number of viable cells, but cells expressing a version of ICT1 that was not silenced by the siRNA were not affected. Expression of ArfB prior to ICT1 silencing also prevented loss of viability. Cells expressing ArfB prior to ICT1 silencing grew to significantly higher numbers than cells transfected with vector alone (p < 0.0001) (Fig 5). There was no significant difference in viable cell number when cells were rescued with ArfB versus ICT1 (p = 0.75). These results indicate that ArfB can perform the essential function of ICT1 in human cells.
Fig 5. c. crescentus ArfB rescues human cells from ICT1 silencing.
Viability of HEK293 cells expressing ICT1 or ArfB was determined after silencing endogenous ICT1. (A) Western blot showing depletion of ICT1 after silencing with siICT1. Non-targeting siRNA (siNT) was used as a negative control. (B) Schematic diagram showing ArfB with ICT1 localization signal that was used for rescue. (C) Column graphs showing average viable cell numbers from 5 independent experiments. Error bars indicate standard deviation. *** indicates p < 0.0001.
Discussion
Rescue of ribosomes from non-stop translation complexes is a critical function for most bacteria, and the results presented here indicate that rescue of mitochondrial ribosomes is also essential in some eukaryotes. C. crescentus cells can survive without tmRNA and SmpB because they have ArfB to rescue ribosomes in the absence of trans-translation [21]. We have previously shown that C. crescentus ArfB can hydrolyze peptidyl-tRNA from non-stop ribosomes in vitro [21]. The data described here show that ArfB is also active on ribosomes stalled with a full codon in the A site or with 6 nucleotides past the A site, but longer mRNA extensions strongly inhibit ArfB activity. This substrate specificity is similar to that observed for tmRNA-SmpB [34–36], and indicates that ArfB is unlikely to interfere with translation elongation or with ribosomes paused during translation of full-length mRNAs. Instead, ArfB activity is consistent with a role in rescuing ribosomes that have translated to the 3’ end of an mRNA without terminating, ribosomes that have stalled after cleavage of the mRNA in the A site by a nuclease such as RelE [37], and stalled ribosomes that have had the 3’ portion of the mRNA removed by exonuclease activity [36,38].
Several lines of evidence demonstrate that human ICT1 is a ribosome rescue factor like ArfB, and not a non-specific release factor that can act on ribosomes stalled in the middle of an mRNA. First, the specificity of ICT1 in vitro for non-stop ribosomes or ribosomes with short mRNA extensions past the A site is similar to that of ArfB. Second, ICT1 can functionally replace ArfB in C. crescentus. Expression of ICT1 suppresses the synthetic lethality of deleting ssrA and arfB, and over-expression of ICT1 increases the growth rate in ΔssrA cells to the same extent as over-expression of ArfB. Third, expression of ArfB in human cells suppresses the lethal effects of silencing ICT1, indicating that ArfB can functionally replace ICT1 in human cells. Finally, ICT1 would have little opportunity to act as a non-specific release factor during translation of intact transcripts in the mitochondria because mammalian mitochondrial transcripts are polyadenylated with ~50 nucleotides [39–44]. Based on the substrate specificity of ICT1 in vitro, this poly(A) tail would block ICT1 activity unless the mRNA was truncated, so ICT1 substrates for in vivo are likely to be non-stop complexes. Because ICT1 is essential in human cells [26,27], these results suggest that ribosome rescue in mitochondria is essential for human cell viability. The activity of ICT1 as a rescue factor and not a non-specific release factor would also explain why ICT1 does not interfere with translation elongation in mitochondria.
ICT1 substrate specificity has important implications for translation termination in mitochondria. Two human mitochondrial genes end in an AGG (ND6) or AGA (COI) codon. There are no cognate mitochondrial tRNAs for these codons and the mitochondrial release factor mtRF1a is unable terminate translation at AGA or AGG [45], so it is unclear how translation is terminated for these genes. ICT1 has been proposed to function as the termination factor at these codons based on analysis of activity in vitro on ribosomes stalled with an mRNA extending up to 14 nucleotides past the A site [30,31]. Our data show that ICT1 activity is greatly reduced on ribosomes stalled with an mRNA extending 14 nucleotides past the A site, and ICT1 activity is completely absent when the mRNA extends 33 nucleotides past the A site. Because the COI AGA codon is 72 nucleotides from the end of the transcript and the ND6 AGG codon is 500–550 nucleotides from the end of the transcript [39,42], ICT1 should have no activity at these codons in either their unmodified or polyadenylated form. In addition, ICT1 can support viability in C. crescentus, so it cannot have an intrinsic ability to terminate translation at AGA or AGG because these codons encode arginine in bacteria. Likewise, ArfB does not recognize AGA or AGG in C. crescentus, so the ability of ArfB to replace ICT1 in human mitochondria suggests that termination at AGA or AGG in the middle of a transcript is not an essential function for ICT1. One possible mechanism for both ICT1 and ArfB to terminate translation at these codons is that stalling of the ribosomes leads to truncation of the mRNA 3’ of the ribosome, thereby producing a substrate for the rescue factors. A second possible mechanism for termination at AGA or AGG by ICT1 and ArfB would be that mitochondrial ribosomes might respond differently when they stall in the middle of an mRNA than do bacterial ribosomes. Mitochondrial ribosomes are descended from bacterial ribosomes, but are highly specialized for translating a small number of mRNAs encoded in the mitochondrial genome. The decoding center and peptidyl transfer center of mitochondrial ribosomes are very similar to bacterial ribosomes, but other architectural features are highly diverged [46]. For example, mammalian mitochondria have dramatically reduced rRNAs and lack 5S rRNA, but contain 36 proteins not found in bacteria and incorporate a tRNA as a structural component of the large subunit [47]. It is possible that this different architecture causes mitochondrial ribosomes to adopt a conformation that promotes rescue by ICT1 and ArfB when they are stalled in the middle of an mRNA. If ICT1 does not terminate translation at these codons, one of the other members of the mitochondrial RF family might perform this function.
Mitochondria descend from a progenitor of α-proteobacteria [48,49], the bacterial class that includes C. crescentus. Most α-proteobacterial species contain both trans-translation and ArfB, and some protist mitochondria encode ssrA and smpB [4], suggesting that the primordial mitochondrion had both ribosome rescue systems. Why did mitochondria keep ArfB and discard trans-translation, whereas almost all bacteria have kept trans-translation whether they have an alternative rescue factor or not? Mitochondria encode only 13 proteins, all of which are integral membrane proteins. Perhaps this limited proteome decreases the selective advantages of trans-translation, for example by enabling proteases to recognize incomplete versions of proteins without the tmRNA-encoded tag. Alternatively, the constraints of importing factors encoded in the nucleus might have favored ICT1 over tmRNA-SmpB, because ICT1 acts as a single protein. Interestingly, some plants encode an ArfB/ICT1 homolog with a chloroplast-targeting signal, (Q84JF2, e.g.) suggesting that ribosome rescue activity may be found in other organelles, and that the ArfB-type ribosome rescue may be generally favored in eukaryotic organelles.
Materials and Methods
Bacterial growth
Strains are described in Table 1. C. crescentus strains were grown at 30°C [50] in peptone-yeast extract (PYE) medium supplemented with tetracycline (2 μg/ml), streptomycin (50 μg/mL), spectinomycin (100 μg/mL), kanamycin (20 μg/mL), or xylose (0.3%) where appropriate. Growth was monitored by measuring optical density at 600 nm.
Table 1. Strains, plasmids, primers and RNAs used in this study.
| Name | Description | Reference |
|---|---|---|
| Strains | ||
| CB15N | Wild-type C. crescentus | [50] |
| CB15NΔssrA | In-frame deletion of ssrA | [12] |
| KCK426 | E. coli BL21(DE3) carrying pET28arfB | [21] |
| KCK428 | CB15N arfB::Ω with Kan marker 22.4 kb downstream | [21] |
| Plasmids | ||
| pET28arfB | Used to express C. crescentus ArfB from T7 promoter | [21] |
| pET28ICT1 | Used to express H. sapiens ICT1 from T7 promoter | This study |
| pMSCVarfB | pMSCVneo expressing ArfB with localization signal | This study |
| pMSCVICT1 | pMSCVneo expressing ICT1 | This study |
| parfB | encodes ArfB under the control of a xylose promoter | [21] |
| pICT1 | encodes ICT1 under the control of a xylose promoter | This study |
| Primers | ||
| HAF_T7 | CGAAATTAATACGACTCACTATAGGG | [21] |
| DHFR_NS_R | AAACCCCTCCGTTTAGAGAGGGGTTTTGCTAGTATCCGCCGCTCCAGAATCTCAAAGCAA | [21] |
| DHFR_stop_0 | TTAACCCCTCCGTTTTAGAGAGGGGTTAATTGCTAGCCGCCGCTCCAGAATCTCAAAGCA | This study |
| DHFR_stop_6 | AAACCCTTACTCCGTAGAGAGTAAGGGTTTTGCTAGCCGCCGCTCCAGAATCTCAAAGCA | This study |
| DHFR_stop_14 | AAAAACCCCTCCGTTTAAGAGAGGGGTTTTGCTAGTATCCGCCGCTCCAGAATCTCAAAG | This study |
| DHFR_stop_33 | AAAACCCCTCCGTTTAGAGAGGGGTTTTGCTAGTTACCGCCGCTCCAGAATCTCAAAGCA | This study |
| RNAs | ||
| siNT | AGGUAGUGUAAUCGCCUUG dtdt | Eurofins Genomics |
| siICT1 | GCCGCUAUCAGUUCCGGAA dtdt | [26] |
Plasmid construction
To construct pET28ICT1 for expression and purification of mature ICT1, the coding sequence (codon-optimized for E. coli) ICT1 lacking the mitochondrial localization signal was purchased as a gBlock Gene Fragment (IDT) and inserted into pET28b by Gibson assembly [51]. Construction of parfB (formerly pCC1214) has been described previously [26]. pICT1 was constructed by digesting pET28ICT1 with NdeI and BamHI and ligating the resulting fragment into parfB digested with the same enzymes. pMCSVICT1 was constructed by Gibson assembly of the human ICT1 sequence into the EcoRI and NotI sites of pMSCVneo. Alternative codons were selected for the region of ICT1 targeted by the siRNA to ensure that only endogenous ICT1 would be silenced. pMCSVarfB was similarly constructed by Gibson assembly of a gBlock Gene Fragment into pMSCVneo at the EcoRI and NotI sites. The arfB construct encodes the 62 residue N-terminal extension of human ICT1 to target it to mitochondria followed by the 142 residues of C. crescentus ArfB sequence codon-optimized for expression in human cells. The ArfB coding sequence was codon-optimized for expression in human cells.
Purification of ICT1 and ArfB
Purification of C. crescentus ArfB has been described previously [21]. ICT1 was purified using a similar protocol. Strain KCK477 was grown to OD600 ~ 0.8 and expression was induced by addition of isopropyl-ß-D-thiogalactopyranoside (IPTG) to 1 mM. Cells were grown for 3 h at 37°C, harvested by centrifugation at 4°C, and resuspended in 30 ml lysis buffer (6 M guanidine hydrochloride, 20 mM sodium phosphate, 400 mM NaCl) [pH 7.8]. Cells were lysed by sonication and the lysates were cleared by centrifugation at 11,000 g for 30 min and applied to a column packed with 500 μl Ni-nitrilotriacetic acid (NTA) agarose (Qiagen) slurry equilibrated in DB buffer (8 M urea, 20 mM sodium phosphate 500 mM NaCl) [pH 7.8]. The column was washed 3X by rocking with 10 bed volumes DB buffer, washed 3X by rocking with 20 bed volumes DW buffer (8 M urea, 20 mM sodium phosphate, 500 mM NaCl) [pH 6.0], and washed 3X with 20 bed volumes of DW buffer [pH 5.3]. Protein was eluted in 1 ml fractions in elution buffer (8 M urea, 20 mM sodium phosphate, 500 mM NaCl) [pH 4.0]. Fractions containing ICT1 were dialyzed against ICT1 dialysis buffer (10 mM HEPES [pH 7.6], 150 mM NaCl). The 6 histidine tag was cleaved with Thrombin CleanCleave Kit (Sigma) at 4°C for 3 h according to the manufacturer’s instructions. Residual 6x-His tagged protein was removed by incubation with Ni-NTA agarose.
Peptidyl-tRNA hydrolysis assays
Peptidyl-tRNA hydrolysis by ArfB, ICT1, and release factors was assayed using the PURExpressΔRF1,2,3 kit (New England Biolabs). Template used for in vitro transcription and translation was generated by PCR using the primers listed in Table 1. Each template was designed to produce an mRNA with a stem-loop at the 3’ end to limit exonuclease activity during the reaction. PURExpressΔRF1,2,3 kit components were mixed according to the manufacturer’s instructions and incubated with 200 nM ArfB, 200 nM ICT1, or 100 nM RF1, RF2 and RF3 for 1 h at 37°C. Anti-ssrA oligonucleotide was added to 5 μM to inhibit any trans-translation activity from tmRNA in the kit components. Samples were precipitated in 20 μl cold acetone, resuspended in loading buffer pH 6.5 (5 mM sodium bisulfite, 50 mM MOPS [morphonlinepropanesulfonic acid], 50 mM Tris, 1 μM EDTA, 0.1% SDS, 5% glycerol, 0.01% xylene cyanol, and 0.01% bromophenol blue), heated to 65°C for 5 minutes, and resolved on Bis-Tris gels with MOPS running buffer (250 mM MOPS, 250 mM Tris, 5 mM EDTA and 0.5% SDS).
Phage transduction
ΦCR30 lysate was prepared from strain KCK428 as described previously [32]. The resulting lysate was used to transduce wild-type or ΔssrA cells harboring pICT1, parfB, or empty vector. Overnights of each strain were grown in PYE supplemented with tetracycline and xylose to mid log phase. 25 μl ΦCR30 prepared from KCK428 was added and cultures were incubated at 30°C for 2.5 h with shaking. Cells were then plated on PYE with kanamycin and xylose to select for transductants. The resulting colonies were tested for spectinomycin and streptomycin resistance to determine the frequency with which arfB:aadA co-transduced.
ICT1 silencing
Pre-annealed non-targeting control siNT, or targeting siICT1[26] RNAs were purchased from Eurofins MWG Operon. To demonstrate efficient knockdown, 7.5 × 105 HEK293 cells were seeded in a 6-well plate and transfected once every 24 h with siNT and siICT1 as follows: a mixture containing 90 μl serum-free DMEM (Corning), 3.4 μl siRNA (20 μM stock), and 4 μl TransIT-siQUEST (Mirus Bio) was incubated at room temperature for 30 min and added to the cells. After 48 h, cells were lysed by addition of buffer containing 150 mM sodium chloride, 50 mM Tris [pH 8.0], 5 mM EDTA, 2 mM phenylmethylsulfonyl fluoride, 2 mM sodium orthovanadate, 10 mM sodium fluoride, and 1% Igepal TM CA-360 (USB). The efficacy of ICT1 silencing was then determined by western blot using polyclonal mouse anti-human ICT1 (Sigma).
ICT1 rescue in HEK293 cells
HEK293 cells (ATCC) were seeded in 6-well plates at 5 × 104 cells per well and allowed to adhere for 18 h. Cells were transfected by combining 1 μg pMSCV vector, pMSCVICT1, or pMSCVarfB, 90 μl serum-free DMEM, and 3.8 μl TransIT-293 (Mirus Bio) and allowing the mixture to incubate for 30 min at room temperature. 8 h after transfecting with plasmid, cells were transfected with siNT or siICT1 according to the siRNA transfection protocol described in the previous section. Viable cell numbers were determined by 0.4% trypan blue staining of trypsinized cells 6 days after silencing.
Acknowledgments
We thank members of the Keiler lab for critical review of the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grant GM068720 from the National Institute of General Medical Sciences to KCK. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461: 1234–1242. 10.1038/nature08403 [DOI] [PubMed] [Google Scholar]
- 2.Ito K, Uno M, Nakamura Y. A tripeptide “anticodon” deciphers stop codons in messenger RNA. Nature. 2000;403: 680–684. 10.1038/35001115 [DOI] [PubMed] [Google Scholar]
- 3.Keiler KC. Mechanisms of ribosome rescue in bacteria. Nat Rev Microbiol. 2015;13: 285–297. 10.1038/nrmicro3438 [DOI] [PubMed] [Google Scholar]
- 4.Hudson CM, Lau BY, Williams KP. Ends of the line for tmRNA-SmpB. Front Microbiol. 2014;5: 421 10.3389/fmicb.2014.00421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keiler KC, Feaga HA. Resolving nonstop translation complexes is a matter of life or death. J Bacteriol. 2014;196: 2123–2130. 10.1128/JB.01490-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang C, Wolfgang MC, Withey J, Koomey M, Friedman DI. Charged tmRNA but not tmRNA-mediated proteolysis is essential for Neisseria gonorrhoeae viability. EMBO J. 2000;19: 1098–1107. 10.1093/emboj/19.5.1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramadoss NS, Zhou X, Keiler KC. tmRNA is essential in Shigella flexneri. PLoS ONE. 2013;8: e57537 10.1371/journal.pone.0057537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thibonnier M, Thiberge J-M, De Reuse H. Trans-translation in Helicobacter pylori: essentiality of ribosome rescue and requirement of protein tagging for stress resistance and competence. PLoS ONE. 2008;3: e3810 10.1371/journal.pone.0003810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Svetlanov A, Puri N, Mena P, Koller A, Karzai AW. Francisella tularensis tmRNA system mutants are vulnerable to stress, avirulent in mice, and provide effective immune protection. Mol Microbiol. 2012;85: 122–141. 10.1111/j.1365-2958.2012.08093.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Julio SM, Heithoff DM, Mahan MJ. ssrA (tmRNA) plays a role in Salmonella enterica serovar Typhimurium pathogenesis. J Bacteriol. 2000;182: 1558–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shin J-H, Price CW. The SsrA-SmpB ribosome rescue system is important for growth of Bacillus subtilis at low and high temperatures. J Bacteriol. 2007;189: 3729–3737. 10.1128/JB.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keiler KC, Shapiro L. TmRNA is required for correct timing of DNA replication in Caulobacter crescentus. J Bacteriol. 2003;185: 573–580. 10.1128/JB.185.2.573-580.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oh BK, Apirion D. 10Sa RNA, a small stable RNA of Escherichia coli, is functional. Mol Gen Genet. 1991;229: 52–56. [DOI] [PubMed] [Google Scholar]
- 14.Chadani Y, Ono K, Ozawa S-I, Takahashi Y, Takai K, Nanamiya H, et al. Ribosome rescue by Escherichia coli ArfA (YhdL) in the absence of trans-translation system. Mol Microbiol. 2010;78: 796–808. 10.1111/j.1365-2958.2010.07375.x [DOI] [PubMed] [Google Scholar]
- 15.Handa Y, Inaho N, Nameki N. YaeJ is a novel ribosome-associated protein in Escherichia coli that can hydrolyze peptidyl-tRNA on stalled ribosomes. Nucleic Acids Res. 2011;39: 1739–1748. 10.1093/nar/gkq1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chadani Y, Ono K, Kutsukake K, Abo T. Escherichia coli YaeJ protein mediates a novel ribosome-rescue pathway distinct from SsrA- and ArfA-mediated pathways. Mol Microbiol. 2011;80: 772–785. 10.1111/j.1365-2958.2011.07607.x [DOI] [PubMed] [Google Scholar]
- 17.Chadani Y, Ito K, Kutsukake K, Abo T. ArfA recruits release factor 2 to rescue stalled ribosomes by peptidyl-tRNA hydrolysis in Escherichia coli. Mol Microbiol. 2012;86: 37–50. 10.1111/j.1365-2958.2012.08190.x [DOI] [PubMed] [Google Scholar]
- 18.Shimizu Y. ArfA recruits RF2 into stalled ribosomes. J Mol Biol. 2012;423: 624–631. 10.1016/j.jmb.2012.08.007 [DOI] [PubMed] [Google Scholar]
- 19.Kurita D, Chadani Y, Muto A, Abo T, Himeno H. ArfA recognizes the lack of mRNA in the mRNA channel after RF2 binding for ribosome rescue. Nucleic Acids Res. 2014;42: 13339–13352. 10.1093/nar/gku1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng F, Jin H. Peptide release promoted by methylated RF2 and ArfA in nonstop translation is achieved by an induced-fit mechanism. RNA. 2016;22: 49–60. 10.1261/rna.053082.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feaga HA, Viollier PH, Keiler KC. Release of nonstop ribosomes is essential. MBio. 2014;5: e01916 10.1128/mBio.01916-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kogure H, Handa Y, Nagata M, Kanai N, Guntert P, Kubota K, et al. Identification of residues required for stalled-ribosome rescue in the codon-independent release factor YaeJ. Nucleic Acids Res. 2014;42: 3152–3163. 10.1093/nar/gkt1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gagnon MG, Seetharaman SV, Bulkley D, Steitz TA. Structural basis for the rescue of stalled ribosomes: structure of YaeJ bound to the ribosome. Science. 2012;335: 1370–1372. 10.1126/science.1217443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Passos DO, Doma MK, Shoemaker CJ, Muhlrad D, Green R, Weissman J, et al. Analysis of Dom34 and its function in no-go decay. Mol Biol Cell. 2009;20: 3025–3032. 10.1091/mbc.E09-01-0028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen L, Muhlrad D, Hauryliuk V, Cheng Z, Lim MK, Shyp V, et al. Structure of the Dom34-Hbs1 complex and implications for no-go decay. Nat Struct Mol Biol. 2010;17: 1233–1240. 10.1038/nsmb.1922 [DOI] [PubMed] [Google Scholar]
- 26.Richter R, Rorbach J, Pajak A, Smith PM, Wessels HJ, Huynen MA, et al. A functional peptidyl-tRNA hydrolase, ICT1, has been recruited into the human mitochondrial ribosome. EMBO J. 2010;29: 1116–1125. 10.1038/emboj.2010.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kogure H, Hikawa Y, Hagihara M, Tochio N, Koshiba S, Inoue Y, et al. Solution structure and siRNA-mediated knockdown analysis of the mitochondrial disease-related protein C12orf65. Proteins. 2012;80: 2629–2642. 10.1002/prot.24152 [DOI] [PubMed] [Google Scholar]
- 28.Chrzanowska-Lightowlers ZM, Lightowlers RN. Response to “Ribosome Rescue and Translation Termination at Non-standard Stop Codons by ICT1 in Mammalian Mitochondria.” PLoS Genet. 2015;11: e1005227–4. 10.1371/journal.pgen.1005227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeuchi N, Nierhaus KH. Response to the Formal Letter of Z. Chrzanowska-Lightowlers and R. N. Lightowlers Regarding Our Article "Ribosome Rescue and Translation Termination at Non-Standard Stop Codons by ICT1 in Mammalian Mitochondria". PLoS Genet. 2015;11: e1005218 10.1371/journal.pgen.1005218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lind C, Sund J, Aqvist J. Codon-reading specificities of mitochondrial release factors and translation termination at non-standard stop codons. Nat Commun. 2013;4: 2940 10.1038/ncomms3940 [DOI] [PubMed] [Google Scholar]
- 31.Akabane S, Ueda T, Nierhaus KH, Takeuchi N. Ribosome Rescue and Translation Termination at Non-Standard Stop Codons by ICT1 in Mammalian Mitochondria. PLoS Genet. 2014;10: e1004616–14. 10.1371/journal.pgen.1004616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.West L, Yang D, Stephens C. Use of the Caulobacter crescentus genome sequence to develop a method for systematic genetic mapping. J Bacteriol. 2002;184: 2155–2166. 10.1128/JB.184.8.2155-2166.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Handa Y, Hikawa Y, Tochio N, Kogure H, Inoue M, Koshiba S, et al. Solution structure of the catalytic domain of the mitochondrial protein ICT1 that is essential for cell vitality. J Mol Biol. 2010;404: 260–273. 10.1016/j.jmb.2010.09.033 [DOI] [PubMed] [Google Scholar]
- 34.Ivanova N, Pavlov MY, Felden B, Ehrenberg M. Ribosome rescue by tmRNA requires truncated mRNAs. J Mol Biol. 2004;338: 33–41. 10.1016/j.jmb.2004.02.043 [DOI] [PubMed] [Google Scholar]
- 35.Moore SD, Sauer RT. Ribosome rescue: tmRNA tagging activity and capacity in Escherichia coli. Mol Microbiol. 2005;58: 456–466. 10.1111/j.1365-2958.2005.04832.x [DOI] [PubMed] [Google Scholar]
- 36.Hayes CS, Sauer RT. Cleavage of the A site mRNA codon during ribosome pausing provides a mechanism for translational quality control. Mol Cell. 2003;12: 903–911. [DOI] [PubMed] [Google Scholar]
- 37.Pedersen K, Zavialov AV, Pavlov MY, Elf J, Gerdes K, Ehrenberg M. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell. 2003;112: 131–140. [DOI] [PubMed] [Google Scholar]
- 38.Garza-Sánchez F, Shoji S, Fredrick K, Hayes CS. RNase II is important for A-site mRNA cleavage during ribosome pausing. Mol Microbiol. 2009;73: 882–897. 10.1111/j.1365-2958.2009.06813.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bobrowicz AJ, Lightowlers RN, Chrzanowska-Lightowlers Z. Polyadenylation and degradation of mRNA in mammalian mitochondria: a missing link? Biochm Soc Trans. 2008;36: 517–519. 10.1042/BST0360517 [DOI] [PubMed] [Google Scholar]
- 40.Chang JH, Tong L. Mitochondrial poly(A) polymerase and polyadenylation. Biochim Biophys Acta. 2012;1819: 992–997. 10.1016/j.bbagrm.2011.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomecki R, Dmochowska A, Gewartowski K, Dziembowski A, Stepien PP. Identification of a novel human nuclear-encoded mitochondrial poly(A) polymerase. Nucleic Acids Res. 2004;32: 6001–6014. 10.1093/nar/gkh923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slomovic S, Laufer D, Geiger D, Schuster G. Polyadenylation and degradation of human mitochondrial RNA: the prokaryotic past leaves its mark. Mol Cell Biol. 2005;25: 6427–6435. 10.1128/MCB.25.15.6427-6435.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagaike T, Suzuki T, Katoh T, Ueda T. Human mitochondrial mRNAs are stabilized with polyadenylation regulated by mitochondria-specific poly(A) polymerase and polynucleotide phosphorylase. J Biol Chem. 2005;280: 19721–19727. 10.1074/jbc.M500804200 [DOI] [PubMed] [Google Scholar]
- 44.Ojala D, Attardi G. Expression of the mitochondrial genome in HeLa cells. XIX. Occurrence in mitochondria of polyadenylic acid sequences, “free” and covalently linked to mitochondrial DNA-coded RNA. J Mol Biol. 1974;82: 151–174. [DOI] [PubMed] [Google Scholar]
- 45.Nozaki Y, Matsunaga N, Ishizawa T, Ueda T, Takeuchi N. HMRF1L is a human mitochondrial translation release factor involved in the decoding of the termination codons UAA and UAG. Genes Cells. 2008;13: 429–438. 10.1111/j.1365-2443.2008.01181.x [DOI] [PubMed] [Google Scholar]
- 46.Ott M, Amunts A, Brown A. Organization and Regulation of Mitochondrial Protein Synthesis. Annu Rev Biochem. 2016;85: annurev–biochem–060815–014334. 10.1146/annurev-biochem-060815-014334 [DOI] [PubMed] [Google Scholar]
- 47.Amunts A, Brown A, Toots J, Scheres SHW, Ramakrishnan V. Ribosome. The structure of the human mitochondrial ribosome. Science. 2015;348: 95–98. 10.1126/science.aaa1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fitzpatrick DA, Creevey CJ, McInerney JO. Genome phylogenies indicate a meaningful alpha-proteobacterial phylogeny and support a grouping of the mitochondria with the Rickettsiales. Mol Biol Evol. 2006;23: 74–85. 10.1093/molbev/msj009 [DOI] [PubMed] [Google Scholar]
- 49.Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Pontén T, Alsmark UC, Podowski RM, et al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396: 133–140. 10.1038/24094 [DOI] [PubMed] [Google Scholar]
- 50.Evinger M, Agabian N. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol. 1977;132: 294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6: 343–345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.