Abstract
Introduction
Schizophrenia is a genetically heterogeneous disorder that is associated with several common and rare genetic variants. As technology involved, cost advantages of chip based genotyping was combined with information about rare variants, resulting in the Infinium HumanExome Beadchip. Using this chip, a sample of 493 patients with schizophrenia or schizoaffective disorder and 484 healthy controls was genotyped.
Results
From the initial 242901 SNVs, 88306 had at least one minor allele and passed quality control. No variant reached genomewide-significant results (p<10-8). The SNP with the lowest p-value was rs1230345 in WISP3 (p = 3.05*10−6), followed by rs9311525 in CACNA2D3 (p = 1.03*10−5) and rs1558557 (p = 3.85*10−05) on chromosome 7. At the gene level, 3 genes were of interest: WISP3, on chromosome 6q21, a signally protein from the extracellular matrix. A second candidate gene is CACNA2D3, a regulator of the intracerebral calcium pathway. A third gene is TNFSF10, associated with p53 mediated apoptosis.
Introduction
Schizophrenia is a psychiatric disorder characterized by the presence of psychotic and negative symptoms and has a heterogeneous presentation and prognosis. Combined with schizoaffective disorder, it has an estimated lifetime prevalence of approximately 1%.[1,2] Schizophrenia has a high heritability (estimated between 65 and 81%),[3,4] and evidence suggests a polygenic inheritance, with an established role of both rare variants with large effects, as well as common Single Nucleotide Polymorphisms (SNPs) with small effects.[5,6] Given this complexity, early genetic studies failed to replicate previous associations, leading to a pessimistic outlook on schizophrenia genetics.[7]
Technological advances such as chip-based genotyping made large-scale studies using genome-wide information affordable and technically possible. Genome-wide association studies (GWAS) use tagging SNPs to identify common risks alleles, based upon the principle of linkage disequilibrium. Thus, by using only between 250 000 and 1 million SNPs, the whole genome is scanned for risk loci.
Although initial GWAS studies had limited (less than 500 cases) sample sizes and power,[8] subsequent studies with increasing sample sizes led to several common SNPs associated with schizophrenia.[9–22] Due to the nature of GWAS studies, incorporating many common variants, a stringent correction for multiple testing has to be applied (typically, Bonferroni correction with genome-wide significance defined as a p-value below 10−8), as well as independent replications.[23] The most recent study by the Psychiatric Genomics Consortium found 108 loci that obtained sufficiently low p-values to be associated with schizophrenia.[22]
A second technology that contributed to the knowledge of the genetic architecture of schizophrenia was next generation sequencing, which enables the identification of rare variants with minor allele frequencies below 5%. Sequencing allows for SNP genotyping, as well as for the detection of copy number variants,[24] but is expensive, slower than chip genotyping and requires additional techniques for data analysis. An increased burden of rare mutations has been found in schizophrenia.[25] Due to the high cost associated with sequencing studies, several studies were limited to either specific target regions,[26,27] or whole exome sequencing.[25,28–32]
The Human Exome consortium, incorporating researchers from different research domains such as schizophrenia and autism genetics,[33] jointly developed a SNP chip incorporating >240 000 putatively functional variants within the human exome. This chip was then marketed by Illumina, as the HumanExome Beadchip.[34] This chip was designed to be efficient towards genotyping cost and analysis burden, yet incorporating a large number of rare SNPs without adding the need for sequencing.
The current study used the HumanExome beadchip to detect rare variants in a sample of 484 patients with schizophrenia or schizoaffective disorder and 493 healthy volunteers recruited from the general population.
Methods
Sample
The current investigation uses samples from three sources: two different patient sets were used, and one set of controls. The patient sample consisted of 650 patients with psychotic spectrum disorder. Part of this sample was previously used for pharmacogenetic research [35–40]. Initial inclusion in this sample was based upon clinician diagnosis, and diagnosis was confirmed using the OPCRIT v4 questionnaire before inclusion in the current study.[41] These patients come from five different hospitals in Belgium (UPC St. Jozef, Kortenberg; Psychosociaal centrum St. Alexius, Elsene; UPC St. Kamillus, Bierbeek; Broeders Alexianen, Tienen and St. Amedeus, Mortsel).
A healthy control sample of both mentally and physically healthy plasma donors of Caucasian descent. They have never had any mental illness, have not been treated for mental illness and have never taken medication for mental disorders. This sample was obtained in collaboration with the Belgian Red Cross Flanders.
All patients and healthy controls gave written informed consent for genetic testing. After obtaining approval by the “Commissie medische ethiek” of the UZ Leuven, Leuven, Belgium, the study was approved by the local ethics committees of the coordinating and sampling hospitals and the Red Cross Belgium. The study was conducted in accordance with the current revision of the Helsinki declaration [42].
DNA analysis
DNA was extracted from peripheral blood lymphocytes using a Chemagen MSMI (Perkin Elmer–Chemagen). Samples were genotyped using the Illumina HumanExome v1.1 chip. Extraction, storage and analysis was conducted in the Center for Human Genetics in Leuven, Belgium.
DNA quality control was done following the manufacturers' guidelines using GenomeStudio software (v2010.3).[43] Genotypes were called using the supplied cluster file, with automatic re-clustering of all genotypes with a call rate below 100%. After this re-clustering, all remaining genotypes with call rates below 100% were manually verified. All samples with exactly 12 rare allele homozygote or 12 heterozygote cases (12 equals the number of samples per chip) were manually checked to exclude chip effects. All SNPs on the X and Y chromosome were manually verified. Genotypes were automatically clustered using the OPTICALL software, and Single Nucleotide Variants (SNV, both SNP and indels or deletions) with major differences in call rates between both methods were manually verified and excluded when after manual verification, no consensus calling was obtained.[44]
Further quality control was done using PLINK v1.07.[45] Ethnicity and relatedness was verified using the MDS algorithm in the KING software v1.4, as with the build-in functions of PLINK.[46] Based upon the eigenvalues, the 3 first principal components were retained. The 08/2010 release of the 1000-genomes project was used to check population membership, and samples of non-Caucasian descent were excluded.
Analysis of autosomal SNPs was done using logistic regression with the first 3 principal components (PCA) generated by KING as covariates. A combined analysis of rare (MAF<0.03) and common variants was done using the CommonRare function implemented in SKAT version 0.91.[47] A multilevel logistic regression, using sex and the first three principal components generated by KING as covariates was used to assess associations on the X-chromosome.
Results
Sample descriptive
A sample of 1023 volunteers consisting of 525 cases with DSM-IV schizophrenia or schizoaffective disorder and 496 healthy controls was genotyped.
After exclusion of samples with call rate below 98% (n = 2), duplicate samples (n = 6), samples related up to the second degree (n = 11), sex errors (n = 6), samples with excess heterozygosity (n = 2) and samples of non-Caucasian descent or other problems (n = 29), a total of 977 samples consisting of 493 cases and 484 controls remained. S1 Fig plots the ethnicity of the current sample compared with the 1000 genomes database. An overview of the first and second principal component of the MDS algorithm is given in S2 Fig.
There was no significant difference in mean age between patients and controls (resp.44.8 vs. 44, t = -1.1341, p = .26), but there were significantly more males amongst the patients than controls (resp. 70.2 vs. 57.9% male, χ2 = 15.6, p = 7.82*10−05).
DNA quality control
From the initial 242901 markers, a total of 242,401 SNVs (99.8%) passed all quality control measures. 129,453 (53.3%) SNVs were re-clustered using the built-in tool of GenomeStudio, resulting in 159,523 (65.7%) SNVs with 0 missing SNVs. The remaining 83,378 SNVs were manually verified. After this stage, QC according to Illumina’s guidelines was applied. In a final stage all SNVs on the X-chromosome were manually verified. In this QC phase, 282 SNPs were excluded from further analysis.
Using PLINK, we excluded 401 SNVs with > 2% missing alleles, 84 SNPs due to Hardy-Weinberg deviation (p < .0005), with 242,416 (99.8%) SNVs remaining for further analysis. Of these, 88,306 (36.4%) had at least one minor allele in one or more participants and were used in the subsequent analysis. S1 Data contains the cleaned PLINK data files after quality control.
Single SNP analysis
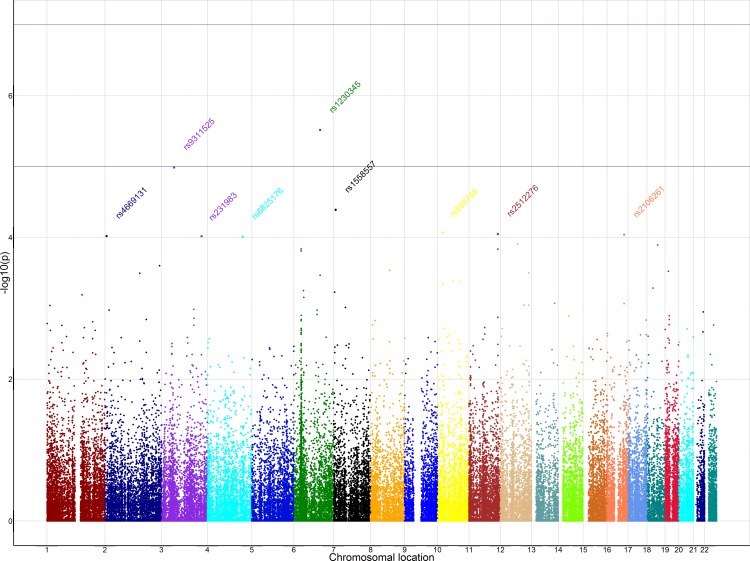
No SNP reached genomewide significance (all p> 10−8). Three SNPs had p values lower than 5*10−5 in the corrected logistic regression. Table 1 lists all SNPs that obtained a p-value below 10−4. Fig 1 shows the Manhattan plot of the logistic regression. The QQ plot of the PCA corrected logistic regression is shown in S3 Fig.
Table 1. Autosomal SNPs that had a p-value below 10−4 after logistic regression using the first 3 principal components.
| SNP | CHR | BP | n | OR | β | P | major/minor | FreqCases | FreqControls | Gene |
|---|---|---|---|---|---|---|---|---|---|---|
| rs1230345 | 6 | 112382313 | 977 | 1.639 | 4.668 | 3.05E-06 | C/A | 0.330 | 0.237 | WISP3 |
| rs9311525 | 3 | 54183550 | 976 | 0.663 | -4.411 | 1.03E-05 | G/A | 0.372 | 0.470 | CACNA2D3 |
| rs1558557 | 7 | 8308993 | 977 | 0.688 | -4.103 | 4.08E-05 | G/A | 0.394 | 0.489 | |
| rs838759 | 10 | 22498468 | 977 | 0.654 | -3.929 | 8.54E-05 | G/A | 0.217 | 0.293 | |
| rs2512276 | 11 | 124115370 | 975 | 0.692 | -3.918 | 8.92E-05 | C/G | 0.406 | 0.494 | |
| rs2106261 | 16 | 73051620 | 977 | 0.616 | -3.912 | 9.15E-05 | G/A | 0.140 | 0.206 | ZFHX3 |
| rs4669131 | 2 | 7232478 | 977 | 0.697 | -3.902 | 9.56E-05 | A/G | 0.374 | 0.464 | |
| rs231983 | 3 | 172236440 | 977 | 0.689 | -3.900 | 9.63E-05 | A/C | 0.334 | 0.421 | |
| rs6825176 | 4 | 150990695 | 977 | 0.696 | -3.896 | 9.77E-05 | A/G | 0.443 | 0.531 |
Fig 1. Manhattan plot of the autosomal chromosomes after logistic regression corrected for differences in ethnicity.
Names of the top 9 SNPs were included in the plot.
A primary SNP, the missense variant rs1230345 in the WNT1-inducible-signaling pathway protein 3 (WISP3) gene at 6q21 had the smallest p-value (n = 977, OR = 1.64, β = 4.67, p = 3.05*10−6). A second SNP was an intronic variant in the Calcium Channel, Voltage-Dependent, Alpha 2/Delta Subunit 3 (CACNA2D3) gene, rs9311525. (n = 976, OR = 0.66, β = -4.41, p = 1.03*10−5). The last SNP found was in non-coding RNA at chromosome 7, rs1558557 (n = 977, OR = 0.67, β = -4.116, p = 3.85*10−05).
On the X-chromosome, a single SNP (rs41503949), an intergenic variant near Patatin-Like Phospholipase Domain Containing 4 (PNPLA4) reached a p-value below 10−4 (β = 0.68, SE = 0.17, p>5.56*10−6). When only males were concerned, the p value kept below nominal significance (β = 0.89, SE = 0.23, p>0.0001), but not when only females were considered (β = 0.28, SE = 0.299, p = 0.34).
Additionally, SNPs investigated in previous GWAS and present on the current chip are reported in Table 2. Only five of these SNPs reached nominal significance: an intronic variant in the vaccinia related kinase 2 (VRK2) gene, rs2312147 on chromosome 2.[48] A second SNP was also intronic in Neurogenic locus notch homolog 4 (NOTCH4) on chromosome 6 (rs2071286). A single intronic SNP, rs7914558 on chromosome 10 in cyclin M2 (CNNM2) also reached nominal significance. On chromosome 10, 2 SNPs reached nominal significance. The first one is the intergenic rs1602565, and finally rs12807809 near neurogranin (NRGN).
Table 2. Replication of previously reported autosomal SNPs associated with schizophrenia in GWAS studies.[9–22].
p-values were obtained using logistic regression with the first 3 principal components as covariates.
| SNP | CHR | BP | n | OR | β | p | major/minor | FreqCases | FreqControls | Gene |
|---|---|---|---|---|---|---|---|---|---|---|
| rs4846033 | 1 | 11788564 | 977 | 1.058 | 0.121 | 0.904 | G/A | 0.010 | 0.009 | |
| rs1625579 | 1 | 98502934 | 976 | 0.870 | -1.162 | 0.245 | A/C | 0.164 | 0.184 | MIR137 |
| rs10911902 | 1 | 186632317 | 977 | 0.952 | -0.424 | 0.672 | G/A | 0.183 | 0.191 | |
| rs2312147 | 2 | 58222928 | 977 | 0.828 | -2.037 | 0.042 | G/A | 0.361 | 0.407 | VRK2 |
| rs1344706 | 2 | 185778428 | 976 | 0.924 | -0.859 | 0.391 | A/C | 0.410 | 0.429 | ZNF804A |
| rs17662626 | 2 | 193984621 | 974 | 0.910 | -0.571 | 0.568 | A/G | 0.086 | 0.094 | |
| rs10520163 | 4 | 170626552 | 977 | 1.023 | 0.253 | 0.801 | A/G | 0.501 | 0.496 | CLCN3 |
| rs13194053 | 6 | 27143883 | 977 | 0.988 | -0.092 | 0.927 | A/G | 0.165 | 0.174 | |
| rs6932590 | 6 | 27248931 | 976 | 1.116 | 1.019 | 0.308 | A/G | 0.269 | 0.257 | |
| rs928824 | 6 | 30224889 | 977 | 1.060 | 0.329 | 0.742 | G/A | 0.074 | 0.068 | HCG17 |
| rs2071286 | 6 | 32179896 | 977 | 0.780 | -2.100 | 0.036 | G/A | 0.237 | 0.267 | NOTCH4 |
| rs10503253 | 8 | 4180844 | 977 | 0.843 | -1.514 | 0.130 | C/A | 0.192 | 0.219 | CSMD1 |
| rs1155204 | 8 | 13334842 | 977 | 0.994 | -0.037 | 0.970 | A/G | 0.089 | 0.091 | DLC1 |
| rs7004633 | 8 | 89760311 | 976 | 1.143 | 1.137 | 0.255 | A/G | 0.190 | 0.171 | |
| rs7914558 | 10 | 104775908 | 976 | 0.803 | -2.410 | 0.016 | G/A | 0.387 | 0.440 | CNNM2 |
| rs11191580 | 10 | 104906211 | 977 | 0.833 | -1.069 | 0.285 | A/G | 0.075 | 0.087 | NT5C2 |
| rs1602565 | 11 | 29162136 | 977 | 1.355 | 2.155 | 0.031 | A/G | 0.137 | 0.105 | |
| rs12807809 | 11 | 124606285 | 977 | 0.771 | -2.144 | 0.032 | A/G | 0.156 | 0.193 | |
| rs548181 | 11 | 125461709 | 977 | 0.848 | -1.145 | 0.252 | G/A | 0.096 | 0.114 | STT3A |
| rs1006737 | 12 | 2345295 | 977 | 1.123 | 1.194 | 0.232 | G/A | 0.333 | 0.307 | CACNA1C |
| rs11064768 | 12 | 119818509 | 977 | 0.770 | -1.726 | 0.084 | A/G | 0.088 | 0.111 | CCDC60 |
| rs7336332 | 13 | 28058404 | 977 | 0.975 | -0.206 | 0.837 | A/G | 0.150 | 0.156 | |
| rs915071 | 14 | 32433858 | 977 | 0.960 | -0.453 | 0.651 | A/G | 0.486 | 0.496 | |
| rs8042374 | 15 | 78908032 | 977 | 0.914 | -0.821 | 0.412 | A/G | 0.222 | 0.238 | CHRNA3 |
| rs7192086 | 16 | 13061611 | 975 | 1.106 | 0.927 | 0.354 | T/A | 0.256 | 0.237 | SHISA9 |
| rs12966547 | 18 | 52752017 | 977 | 1.038 | 0.397 | 0.692 | G/A | 0.403 | 0.395 | |
| rs17512836 | 18 | 53194961 | 977 | 1.041 | 0.158 | 0.875 | A/G | 0.034 | 0.033 | TCF4 |
Combination of rare and common variation
When analysing the combined effect of common and rare variation using the CommonRare function implemented in SKAT, one gene resulted in a p value below 10−5. Using one common SNP and 2 rare SNPs in WISP3 on chromosome 6, a p-value of 4.34*10−6 was obtained. The second best p-value is obtained by the tumor necrosis factor (ligand) superfamily, member 10 gene (TNFSF10, p = 3.49*10-5), followed by CACNA2D3 (p = 1.29*10−4). The top 5 genes with at least 3 SNPs contributing to the results are displayed in Table 3.
Table 3. Top 5 genes with at least 3 SNPs per gene from the SKAT CommonRare analysis, using the first 3 principal components as covariates.
| Gene | CHR | BP | p | ntotal | ntest | nrare | ncommon |
|---|---|---|---|---|---|---|---|
| WISP3 | 6 | 112375275–112392171 | 4.719E-06 | 3 | 3 | 2 | 1 |
| TNFSF10 | 3 | 172223298–172241297 | 3.492E-05 | 3 | 3 | 2 | 1 |
| CACNA2D3 | 5 | 54908632–54935282 | 1.285E-04 | 10 | 10 | 8 | 2 |
| EBLN1 | 10 | 22497743–22498950 | 2.253E-04 | 3 | 3 | 1 | 2 |
| CD97 | 19 | 14491313–14519537 | 2.283E-04 | 5 | 5 | 4 | 1 |
Discussion
The current study evaluated exonic variation in a group of patients with schizophrenia and schizoaffective disorder. No SNP reached genome-wide significance levels (p< 10−8). At the level of genes, no gene reached genome-wide significance. These results are comparable to those of the Swedish Schizophrenia Cohort, who were also unable to find genome-wide significant results using the HumanExome Beadchip in 13000 individuals.[49] Although none of the currently investigated SNPs reached genome-wide significance, several SNPs obtained low p-values, which, combined with data from previous research, warrants further investigation.
WISP3
The rs1230345 in the WISP3 gene had the smallest p-value of all SNPs tested. As a gene, WISP3 also had the smallest p-value from a combination of one common and 2 rare common SNPs. The WISP3 gene lies within the 6q21 region, within a larger region on chromosome 6 previously associated with schizophrenia or bipolar disorder.[50–54] Although neither WISP3 nor the neighbouring TUBE1 or LAMA4 genes, have been associated with schizophrenia, the more distant FYN gene was.[55]
The WISP3 gene belongs to the CCN family of extracellular matrix associated signalling proteins. It is mainly known for its contribution to progressive pseudo-rheumatoid dysplasia and poly-articular juvenile idiopathic arthritis. It has been linked to the intracellular accumulation of reactive oxygen species in connective tissues.[56]
Animal models have not highlighted further evidence for WISP3 as a schizophrenia candidate gene: Although WISP3 is expressed in the developing midbrain of zebrafish,[57] altering the expression of WISP3 does not affect the phenotype of mice.[58] No association of WISP3 with schizophrenia has so far been described in the current literature. Further research is needed to confirm the possible role of this gene or variant in schizophrenia.
CACNA2D3
The CACNA2D3 gene forms a subunit of the L-type gated calcium channel, where it influences the trafficking and kinetic or voltage-dependent properties.[59] CACNA2D3 lies within the 3p14.3 region. The 3p14 region has been associated with schizophrenia in a single study,[60] and another study found an association the 3p14 region and the antisaccade endophenotype in schizophrenia.[61]
As one of the regulators in the calcium pathway, CACNA2D3 is an interesting candidate gene for schizophrenia as the calcium pathway is thought to be a major contributor to the genetic risk of schizophrenia or bipolar disorder,[62–64] with several studies linking genes of this pathway to both disorders.[19,21,22,65–67]
Several studies of the CACNA2D3 gene reported associations with symptoms of schizophrenia. The CACNA2D3 gene has been shown to alter pain sensitivity in both animals and humans.[68] Patients with schizophrenia display a diminished pain sensitivity, as was shown in a meta-analysis of experimental studies, independent of treatment status.[69] Knockout mice for CACNA2D3 have a decreased startle reflex. [70] The startle reflex modulation, as measured by prepulse inhibition, is a putative endophenotypes of schizophrenia.[71] In an exome sequencing study of autism, a single subject suffering from autism also had a mutation in CACNA2D3.[72] Given the evidence for members of the calcium pathway in schizophrenia, this variant could be of interest for further research.
TNFSF10
Although no single SNP emerged from the TNFSF10 gene, a joint analysis of rare and common variants resulted in a gene with the second lowest gene-wide p-value. The TNFSF10 gene plays a role in the p53-mediated programmed cell death, which is activated after cells get exposed to DNA damage.[73] Previous research has implicated modulations in cell apoptosis in schizophrenia,[74,75] but no direct link between the current gene, apoptosis and schizophrenia was found.
Only a single reference of this gene was found in connection with schizophrenia. In a study based on the dataset of the Stanley Neuropathology Consortium, comparing gene expression in bipolar disorder and schizophrenia versus controls, TNFSF10 had a significant contribution to the support vector machine algorithm for classification of schizophrenia or bipolar versus controls.[76].
Replication of previous findings
Table 2 contains the results of SNPs previously associated with schizophrenia in large GWAS studies.
Limitations
The current study has a moderate sample size. This disadvantage is partially offset by using samples with clear diagnosis of schizophrenia and schizoaffective disorder, and having an ethnically homogeneous sample. DNA was of high quality, and quality control resulted in 99.8% of designed SNVs available for analysis.
Conclusion
The current investigation of 493 patients with schizophrenia or schizoaffective disorders versus 484 healthy controls did not reveal any variant with genome-wide significant p-values. Amongst the lowest p-values were 2 genes that might be of theoretical interest: CACNA2D3, directly involved in regulating the intracerebral calcium homeostasis, and TNFSF10, a gene that is involved in apoptosis in schizophrenia. However, given the limited sample size and thus limited power, these results are preliminary at best and should be treated with caution.
Supporting Information
(DOCX)
Comparison of ethnicity of the current sample with the 1000 genomes August 2010 release. Study subjects are colored black (code OUR), and lie within the Caucasian cluster together with the Utah residents (CEU), British subjects (GBR) and Italians (TSI). The upper right cluster is formed by Americans of African descent (ASW), Puerto Rica (PUR) and Nigerians (YRI). In the lower cluster, Han Chinese (CHS and CHB) and Japanse (JPT) subjects cluster together. Lastly, Finns (FIN) are between the European and Asian clusters. Based upon this figure, 3 additional samples to the right were removed. (final n = 977).
(TIF)
First and second principal component generated by the MDS algorithm of KING in the current dataset.
(PNG)
(TIF)
Acknowledgments
Stephan Claes is a Senior Clinical Investigator of the Fund for Scientific Research Flanders (FWO Vlaanderen; 1800411N)". We would like to thank all patients and volunteers that participated in the current study, as well as the clinicians involved in their treatment.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors have no support or funding to report.
References
- 1.Perala J, Suvisaari J, Saarni SI, Kuoppasalmi K, Isometsa E, Pirkola S et al. Lifetime prevalence of psychotic and bipolar I disorders in a general population. Arch Gen Psychiatry. 2007; 64: 19–28. 64/1/19 [pii]; 10.1001/archpsyc.64.1.19 [DOI] [PubMed] [Google Scholar]
- 2.Tandon R, Keshavan MS, Nasrallah HA. Schizophrenia, "just the facts" what we know in 2008. 2. Epidemiology and etiology. Schizophr Res. 2008; 102: 1–18. S0920-9964(08)00168-0 [pii]; 10.1016/j.schres.2008.04.011 [DOI] [PubMed] [Google Scholar]
- 3.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003; 60: 1187–1192. 10.1001/archpsyc.60.12.1187 60/12/1187 [pii]. [DOI] [PubMed] [Google Scholar]
- 4.Lichtenstein P, Yip BH, Bjork C, Pawitan Y, Cannon TD, Sullivan PF et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet. 2009; 373: 234–239. S0140-6736(09)60072-6 [pii]; 10.1016/S0140-6736(09)60072-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mowry BJ, Gratten J. The emerging spectrum of allelic variation in schizophrenia: current evidence and strategies for the identification and functional characterization of common and rare variants. Mol Psychiatry. 2013; 18: 38–52. mp201234 [pii]; 10.1038/mp.2012.34 [DOI] [PubMed] [Google Scholar]
- 6.Gratten J, Wray NR, Keller MC, Visscher PM. Large-scale genomics unveils the genetic architecture of psychiatric disorders. Nat Neurosci. 2014; 17: 782–790. nn.3708 [pii]; 10.1038/nn.3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Owen MJ. Will schizophrenia become a graveyard for molecular geneticists? Psychol Med. 1992; 22: 289–293. [DOI] [PubMed] [Google Scholar]
- 8.Bergen SE, Petryshen TL. Genome-wide association studies of schizophrenia: does bigger lead to better results? Curr Opin Psychiatry. 2012; 25: 76–82. 10.1097/YCO.0b013e32835035dd [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008; 40: 1053–1055. ng.201 [pii]; 10.1038/ng.201 [DOI] [PubMed] [Google Scholar]
- 10.Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009; 460: 748–752. nature08185 [pii]; 10.1038/nature08185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, Pe'er I et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009; 460: 753–757. nature08192 [pii]; 10.1038/nature08192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D et al. Common variants conferring risk of schizophrenia. Nature. 2009; 460: 744–747. nature08186 [pii]; 10.1038/nature08186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikeda M, Aleksic B, Kinoshita Y, Okochi T, Kawashima K, Kushima I et al. Genome-wide association study of schizophrenia in a Japanese population. Biol Psychiatry. 2011; 69: 472–478. S0006-3223(10)00717-1 [pii]; 10.1016/j.biopsych.2010.07.010 [DOI] [PubMed] [Google Scholar]
- 14.Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011; 43: 969–976. ng.940 [pii]; 10.1038/ng.940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steinberg S, de JS, Andreassen OA, Werge T, Borglum AD, Mors O et al. Common variants at VRK2 and TCF4 conferring risk of schizophrenia. Hum Mol Genet. 2011; 20: 4076–4081. ddr325 [pii]; 10.1093/hmg/ddr325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yue WH, Wang HF, Sun LD, Tang FL, Liu ZH, Zhang HX et al. Genome-wide association study identifies a susceptibility locus for schizophrenia in Han Chinese at 11p11.2. Nat Genet. 2011; 43: 1228–1231. ng.979 [pii]; 10.1038/ng.979 [DOI] [PubMed] [Google Scholar]
- 17.Genome-wide association study implicates HLA-C*01:02 as a risk factor at the major histocompatibility complex locus in schizophrenia. Biol Psychiatry. 2012; 72: 620–628. S0006-3223(12)00554-9 [pii]; 10.1016/j.biopsych.2012.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rietschel M, Mattheisen M, Degenhardt F, Muhleisen TW, Kirsch P, Esslinger C et al. Association between genetic variation in a region on chromosome 11 and schizophrenia in large samples from Europe. Mol Psychiatry. 2012; 17: 906–917. mp201180 [pii]; 10.1038/mp.2011.80 [DOI] [PubMed] [Google Scholar]
- 19.Hamshere ML, Walters JT, Smith R, Richards AL, Green E, Grozeva D et al. Genome-wide significant associations in schizophrenia to ITIH3/4, CACNA1C and SDCCAG8, and extensive replication of associations reported by the Schizophrenia PGC. Mol Psychiatry. 2013; 18: 708–712. mp201267 [pii]; 10.1038/mp.2012.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lencz T, Guha S, Liu C, Rosenfeld J, Mukherjee S, DeRosse P et al. Genome-wide association study implicates NDST3 in schizophrenia and bipolar disorder. Nat Commun. 2013; 4: 2739 ncomms3739 [pii]; 10.1038/ncomms3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ripke S, O'Dushlaine C, Chambert K, Moran JL, Kahler AK, Akterin S et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013. ng.2742 [pii]; 10.1038/ng.2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014; 511: 421–427. nature13595 [pii]; 10.1038/nature13595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sullivan PF. Spurious genetic associations. Biol Psychiatry. 2007; 61: 1121–1126. S0006-3223(06)01470-3 [pii]; 10.1016/j.biopsych.2006.11.010 [DOI] [PubMed] [Google Scholar]
- 24.Abel HJ, Duncavage EJ. Detection of structural DNA variation from next generation sequencing data: a review of informatic approaches. Cancer Genet. 2013; 206: 432–440. S2210-7762(13)00157-9 [pii]; 10.1016/j.cancergen.2013.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014; 506: 185–190. nature12975 [pii]; 10.1038/nature12975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomson PA, Parla JS, McRae AF, Kramer M, Ramakrishnan K, Yao J et al. 708 Common and 2010 rare DISC1 locus variants identified in 1542 subjects: analysis for association with psychiatric disorder and cognitive traits. Mol Psychiatry. 2014; 19: 668–675. mp201368 [pii]; 10.1038/mp.2013.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jouan L, Girard SL, Dobrzeniecka S, Ambalavanan A, Krebs MO, Joober R et al. Investigation of rare variants in LRP1, KPNA1, ALS2CL and ZNF480 genes in schizophrenia patients reflects genetic heterogeneity of the disease. Behav Brain Funct. 2013; 9: 9 1744-9081-9-9 [pii]; 10.1186/1744-9081-9-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu B, Roos JL, Dexheimer P, Boone B, Plummer B, Levy S et al. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat Genet. 2011; 43: 864–868. ng.902 [pii]; 10.1038/ng.902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCarthy SE, Gillis J, Kramer M, Lihm J, Yoon S, Berstein Y et al. De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol Psychiatry. 2014; 19: 652–658. mp201429 [pii]; 10.1038/mp.2014.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takata A, Xu B, Ionita-Laza I, Roos JL, Gogos JA, Karayiorgou M. Loss-of-function variants in schizophrenia risk and SETD1A as a candidate susceptibility gene. Neuron. 2014; 82: 773–780. S0896-6273(14)00358-4 [pii]; 10.1016/j.neuron.2014.04.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Timms AE, Dorschner MO, Wechsler J. Support for the n -methyl-d-aspartate receptor hypofunction hypothesis of schizophrenia from exome sequencing in multiplex families. JAMA Psychiatry. 2013; 70: 582–590. 10.1001/jamapsychiatry.2013.1195 [DOI] [PubMed] [Google Scholar]
- 32.Girard SL, Gauthier J, Noreau A, Xiong L, Zhou S, Jouan L et al. Increased exonic de novo mutation rate in individuals with schizophrenia. Nat Genet. 2011; 43: 860–863. ng.886 [pii]; 10.1038/ng.886 [DOI] [PubMed] [Google Scholar]
- 33.Exome Chip Design Team Exome Chip Design. 2014.
- 34.Illumina Inc. Human Exome BeadChips. 2014.
- 35.De Hert M, van Winkel R, Van ED, Hanssens L, Wampers M, Scheen A et al. Prevalence of diabetes, metabolic syndrome and metabolic abnormalities in schizophrenia over the course of the illness: a cross-sectional study. Clin Pract Epidemiol Ment Health. 2006; 2: 14 1745-0179-2-14 [pii]; 10.1186/1745-0179-2-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Winkel R, van OJ, Celic I, Van ED, Wampers M, Scheen A et al. Psychiatric diagnosis as an independent risk factor for metabolic disturbances: results from a comprehensive, naturalistic screening program. J Clin Psychiatry. 2008; 69: 1319–1327. ej08m04030 [pii]. [DOI] [PubMed] [Google Scholar]
- 37.van Winkel R, Moons T, Peerbooms O, Rutten B, Peuskens J, Claes S et al. MTHFR genotype and differential evolution of metabolic parameters after initiation of a second generation antipsychotic: an observational study. Int Clin Psychopharmacol. 2010; 25: 270–276. 10.1097/YIC.0b013e32833bc60d [DOI] [PubMed] [Google Scholar]
- 38.van Winkel R, Rutten BP, Peerbooms O, Peuskens J, van Os J, De Hert M. MTHFR and risk of metabolic syndrome in patients with schizophrenia. Schizophr Res. 2010; 121: 193–198. S0920-9964(10)01351-4 [pii]; 10.1016/j.schres.2010.05.030 [DOI] [PubMed] [Google Scholar]
- 39.Moons T, Claes S, Martens GJ, Peuskens J, Van Loo KM, Van Schijndel JE et al. Clock genes and body composition in patients with schizophrenia under treatment with antipsychotic drugs. Schizophr Res. 2011; 125: 187–193. S0920-9964(10)01583-5 [pii]; 10.1016/j.schres.2010.10.008 [DOI] [PubMed] [Google Scholar]
- 40.Yu W, De Hert M, Moons T, Claes S, Correll CU, van Winkel R CNR1 and risk of the metabolic syndrome in patients with schizophrenia. 2012. [DOI] [PubMed]
- 41.Craddock M, Asherson P, Owen MJ, Williams J, McGuffin P, Farmer AE. Concurrent validity of the OPCRIT diagnostic system. Comparison of OPCRIT diagnoses with consensus best-estimate lifetime diagnoses. The British Journal of Psychiatry. 1996; 169: 58–63. [DOI] [PubMed] [Google Scholar]
- 42.[Anonymous] WMA Declaration of Helsinki—Ethical Principles for Medical Research Involving Human Subjects. 2008. [PubMed]
- 43.Illumina Inc. Infinium® Genotyping Data Analysis. 2014.
- 44.Shah TS, Liu JZ, Floyd JA, Morris JA, Wirth N, Barrett JC et al. optiCall: a robust genotype-calling algorithm for rare, low-frequency and common variants. Bioinformatics. 2012; 28: 1598–1603. bts180 [pii]; 10.1093/bioinformatics/bts180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007; 81: 559–575. S0002-9297(07)61352-4 [pii]; 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010; 26: 2867–2873. btq559 [pii]; 10.1093/bioinformatics/btq559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ionita-Laza I, Lee S, Makarov V, Buxbaum JD, Lin X. Sequence Kernel Association Tests for the Combined Effect of Rare and Common Variants. Am J Hum Genet. 2013. S0002-9297(13)00176-6 [pii]; 10.1016/j.ajhg.2013.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steinberg S, de JS, Andreassen OA, Werge T, Borglum AD, Mors O et al. Common variants at VRK2 and TCF4 conferring risk of schizophrenia. Hum Mol Genet. 2011; 20: 4076–4081. ddr325 [pii]; 10.1093/hmg/ddr325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neale B Analysis of low-frequency, Protein Altering variation in 13,000 Individuals from a Swedish Schizophrenia Cohort on the Exome Array. XXth World congress of psychiatric genetics .2012.
- 50.Lindholm E, Ekholm B, Shaw S, Jalonen P, Johansson G, Pettersson U et al. A schizophrenia-susceptibility locus at 6q25, in one of the world's largest reported pedigrees. Am J Hum Genet. 2001; 69: 96–105. S0002-9297(07)61449-9 [pii]; 10.1086/321288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cao Q, Martinez M, Zhang J, Sanders AR, Badner JA, Cravchik A et al. Suggestive evidence for a schizophrenia susceptibility locus on chromosome 6q and a confirmation in an independent series of pedigrees. Genomics. 1997; 43: 1–8. S0888-7543(97)94815-1 [pii]; 10.1006/geno.1997.4815 [DOI] [PubMed] [Google Scholar]
- 52.Martinez M, Goldin LR, Cao Q, Zhang J, Sanders AR, Nancarrow DJ et al. Follow-up study on a susceptibility locus for schizophrenia on chromosome 6q. Am J Med Genet. 1999; 88: 337–343. [pii]. [DOI] [PubMed] [Google Scholar]
- 53.Owen MJ, Williams NM, O'Donovan MC. The molecular genetics of schizophrenia: new findings promise new insights. Mol Psychiatry. 2004; 9: 14–27. 10.1038/sj.mp.4001444 4001444 [pii]. [DOI] [PubMed] [Google Scholar]
- 54.Park N, Juo SH, Cheng R, Liu J, Loth JE, Lilliston B et al. Linkage analysis of psychosis in bipolar pedigrees suggests novel putative loci for bipolar disorder and shared susceptibility with schizophrenia. Mol Psychiatry. 2004; 9: 1091–1099. 10.1038/sj.mp.4001541 4001541 [pii]. [DOI] [PubMed] [Google Scholar]
- 55.Ishiguro H, Saito T, Shibuya H, Toru M, Arinami T. Mutation and association analysis of the Fyn kinase gene with alcoholism and schizophrenia. Am J Med Genet. 2000; 96: 716–720. [pii]. [DOI] [PubMed] [Google Scholar]
- 56.Miller DS, Sen M. Potential role of WISP3 (CCN6) in regulating the accumulation of reactive oxygen species. Biochem Biophys Res Commun. 2007; 355: 156–161. S0006-291X(07)00179-9 [pii]; 10.1016/j.bbrc.2007.01.114 [DOI] [PubMed] [Google Scholar]
- 57.Fernando CA, Conrad PA, Bartels CF, Marques T, To M, Balow SA et al. Temporal and spatial expression of CCN genes in zebrafish. Dev Dyn. 2010; 239: 1755–1767. 10.1002/dvdy.22279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakamura Y, Cui Y, Fernando C, Kutz WE, Warman ML. Normal growth and development in mice over-expressing the CCN family member WISP3. J Cell Commun Signal. 2009; 3: 105–113. 10.1007/s12079-009-0040-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davies A, Kadurin I, Alvarez-Laviada A, Douglas L, Nieto-Rostro M, Bauer CS et al. The alpha2delta subunits of voltage-gated calcium channels form GPI-anchored proteins, a posttranslational modification essential for function. Proc Natl Acad Sci U S A. 2010; 107: 1654–1659. 0908735107 [pii]; 10.1073/pnas.0908735107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paunio T, Arajarvi R, Terwilliger JD, Hiekkalinna T, Haimi P, Partonen T et al. Linkage analysis of schizophrenia controlling for population substructure. Am J Med Genet B Neuropsychiatr Genet. 2009; 150B: 827–835. 10.1002/ajmg.b.30905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Greenwood TA, Swerdlow NR, Gur RE, Cadenhead KS, Calkins ME, Dobie DJ et al. Genome-wide linkage analyses of 12 endophenotypes for schizophrenia from the consortium on the genetics of schizophrenia. Am J Psychiatry. 2013; 170: 521–532. 1669964 [pii]; 10.1176/appi.ajp.2012.12020186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Berridge MJ. Calcium signalling and psychiatric disease: bipolar disorder and schizophrenia. Cell Tissue Res. 2014; 357: 477–492. 10.1007/s00441-014-1806-z [DOI] [PubMed] [Google Scholar]
- 63.Nurnberger JI Jr., Koller DL, Jung J, Edenberg HJ, Foroud T, Guella I et al. Identification of pathways for bipolar disorder: a meta-analysis. JAMA Psychiatry. 2014; 71: 657–664. 1859133 [pii]; 10.1001/jamapsychiatry.2014.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hall J, Trent S, Thomas KL, O'Donovan MC, Owen MJ. Genetic Risk for Schizophrenia: Convergence on Synaptic Pathways Involved in Plasticity. Biol Psychiatry. 2014. S0006-3223(14)00519-8 [pii]; 10.1016/j.biopsych.2014.07.011 [DOI] [PubMed] [Google Scholar]
- 65.Kwon E, Wang W, Tsai LH. Validation of schizophrenia-associated genes CSMD1, C10orf26, CACNA1C and TCF4 as miR-137 targets. Mol Psychiatry. 2013; 18: 11–12. mp2011170 [pii]; 10.1038/mp.2011.170 [DOI] [PubMed] [Google Scholar]
- 66.Ferreira MA, O'Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008; 40: 1056–1058. ng.209 [pii]; 10.1038/ng.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Green EK, Hamshere M, Forty L, Gordon-Smith K, Fraser C, Russell E et al. Replication of bipolar disorder susceptibility alleles and identification of two novel genome-wide significant associations in a new bipolar disorder case-control sample. Mol Psychiatry. 2013; 18: 1302–1307. mp2012142 [pii]; 10.1038/mp.2012.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Neely GG, Hess A, Costigan M, Keene AC, Goulas S, Langeslag M et al. A genome-wide Drosophila screen for heat nociception identifies alpha2delta3 as an evolutionarily conserved pain gene. Cell. 2010; 143: 628–638. S0092-8674(10)01133-5 [pii]; 10.1016/j.cell.2010.09.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Potvin S, Marchand S. Hypoalgesia in schizophrenia is independent of antipsychotic drugs: a systematic quantitative review of experimental studies. Pain. 2008; 138: 70–78. S0304-3959(07)00676-8 [pii]; 10.1016/j.pain.2007.11.007 [DOI] [PubMed] [Google Scholar]
- 70.Abo-Dalo B, Kim HG, Roes M, Stefanova M, Higgins A, Shen Y et al. Extensive molecular genetic analysis of the 3p14.3 region in patients with Zimmermann-Laband syndrome. Am J Med Genet A. 2007; 143A: 2668–2674. 10.1002/ajmg.a.32034 [DOI] [PubMed] [Google Scholar]
- 71.Thaker GK. Neurophysiological endophenotypes across bipolar and schizophrenia psychosis. Schizophr Bull. 2008; 34: 760–773. sbn049 [pii]; 10.1093/schbul/sbn049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012; 74: 285–299. S0896-6273(12)00340-6 [pii]; 10.1016/j.neuron.2012.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kuribayashi K, Krigsfeld G, Wang W, Xu J, Mayes PA, Dicker DT et al. TNFSF10 (TRAIL), a p53 target gene that mediates p53-dependent cell death. Cancer Biol Ther. 2008; 7: 2034–2038. 7460 [pii]. [DOI] [PubMed] [Google Scholar]
- 74.Glantz LA, Gilmore JH, Lieberman JA, Jarskog LF. Apoptotic mechanisms and the synaptic pathology of schizophrenia. Schizophr Res. 2006; 81: 47–63. S0920-9964(05)00368-3 [pii]; 10.1016/j.schres.2005.08.014 [DOI] [PubMed] [Google Scholar]
- 75.Jarskog LF, Glantz LA, Gilmore JH, Lieberman JA. Apoptotic mechanisms in the pathophysiology of schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2005; 29: 846–858. S0278-5846(05)00095-3 [pii]; 10.1016/j.pnpbp.2005.03.010 [DOI] [PubMed] [Google Scholar]
- 76.Struyf J, Dobrin S, Page D. Combining gene expression, demographic and clinical data in modeling disease: a case study of bipolar disorder and schizophrenia. BMC Genomics. 2008; 9: 531 1471-2164-9-531 [pii]; 10.1186/1471-2164-9-531 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Comparison of ethnicity of the current sample with the 1000 genomes August 2010 release. Study subjects are colored black (code OUR), and lie within the Caucasian cluster together with the Utah residents (CEU), British subjects (GBR) and Italians (TSI). The upper right cluster is formed by Americans of African descent (ASW), Puerto Rica (PUR) and Nigerians (YRI). In the lower cluster, Han Chinese (CHS and CHB) and Japanse (JPT) subjects cluster together. Lastly, Finns (FIN) are between the European and Asian clusters. Based upon this figure, 3 additional samples to the right were removed. (final n = 977).
(TIF)
First and second principal component generated by the MDS algorithm of KING in the current dataset.
(PNG)
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.