Abstract
The thrombotic microangiopathies (TMAs) and C3 glomerulopathies (C3Gs) include a spectrum of rare diseases such as atypical hemolytic uremic syndrome, thrombotic thrombocytopenic purpura, C3GN, and dense deposit disease, which share phenotypic similarities and underlying genetic commonalities. Variants in several genes contribute to the pathogenesis of these diseases, and identification of these variants may inform the diagnosis and treatment of affected patients. We have developed and validated a comprehensive genetic panel that screens all exons of all genes implicated in TMA and C3G. The closely integrated pipeline implemented includes targeted genomic enrichment, massively parallel sequencing, bioinformatic analysis, and a multidisciplinary conference to analyze identified variants in the context of each patient’s specific phenotype. Herein, we present our 1-year experience with this panel, during which time we studied 193 patients. We identified 17 novel and 74 rare variants, which we classified as pathogenic (11), likely pathogenic (12), and of uncertain significance (68). Compared with controls, patients with C3G had a higher frequency of rare and novel variants in C3 convertase (C3 and CFB) and complement regulator (CFH, CFI, CFHR5, and CD46) genes (P<0.05). In contrast, patients with TMA had an increase in rare and novel variants only in complement regulator genes (P<0.01), a distinction consistent with differing sites of complement dysregulation in these two diseases. In summary, we were able to provide a positive genetic diagnosis in 43% and 41% of patients carrying the clinical diagnosis of C3G and TMA, respectively.
Keywords: Genetic testing, Thrombotic Microangiopathy, C3 Glomerulopathy
Advances in next generation sequencing (NGS), coupled with targeted genomic enrichment (TGE), have had a profound impact on the clinical application of genetic testing in the diagnosis and treatment of a large number of human diseases.1–3 As a replacement for Sanger sequencing-based testing protocols, NGS-based panels are able to generate competitive results at a lower cost and in a shorter time; however, NGS panel validation is a requisite to ensure a diagnostic level of accuracy and sensitivity that is appropriate for clinical decision making. In addition, because NGS generates a huge number of variants, an additional requirement arises, namely the appropriate annotation and interpretation of the variants that are discovered. As a research tool, TGE&NGS is also widely used to identify novels genes and genetic modifiers of Mendelian and complex genetic diseases.2,4,5
Atypical hemolytic uremic syndrome (aHUS), thrombotic thrombocytopenic purpura (TTP), C3 glomerulonephritis (C3GN), and dense deposit disease (DDD, previously known as membranoproliferative glomerulonephritis type II) are rare diseases that share phenotypic similarities and underlying genetic commonalities, making them suitable for NGS-based genetic testing. aHUS and TTP are prototypical thrombotic microangiopathies (TMAs)—diseases characterized by thrombosis in capillaries and arterioles often due to an antecedent endothelial injury and typically seen in association with thrombocytopenia, anemia, purpura, and renal failure.6
aHUS follows event-triggered over-activation of the alternative pathway (AP) of complement at the level of the endothelial cell surface and has been causally related to permissive mutations in several complement genes like CFH, CD46, C3, CFB, and CFI, and thrombosis-related genes like THBD, DGKE, and PLG.7,8 Additional genetic risk factors include homozygosity for the deletion of CFHR3-CFHR1, a common copy number variation (CNV) that is associated with the development of an acquired risk factor, autoantibodies to the protein Factor H, which compromise its function and thereby lead to disease.9 Risk haplotypes of CFH and CD46 have been identified that modify disease penetrance and severity.7,8 TTP, in comparison, is caused by homozygous or compound heterozygous mutations in the ADAMTS13 gene10 or autoantibodies against the ADAMTS13 protein that reduce its functional activity to less than 10%.11
C3GN and DDD are subtypes of C3 glomerulopathy (C3G), a disease classification defined by predominance in the renal glomerulus of C3 deposits as resolved by immunofluorescence (C3 must be at least two orders of magnitude greater than any other immunoreactant, including immunoglobulin). Underlying fluid-phase dysregulation of the AP driven by inherited or acquired defects is heralded by C3 consumption, often with massive breakdown of C3, the fragments of which accumulate in the glomerular basement membrane.12,13 Proteinuria, hematuria, and renal failure ensue. C3GN and DDD are differentiated by differences in deposition patterns of complement debris as resolved by electron microscopy, although complement biomarker distinctions also exist consistent with underlying differences in the relative degree of AP and terminal pathway dysregulation.14 Mutations in C3, CFB, CFH, and CFHR5, and chromosomal rearrangements of the CFHR genes have been implicated in C3GN and DDD.15
In an earlier study, we applied TGE&NGS technology to identify genetic contributors to aHUS.16 Based on this experience, we have developed a clinically useful gene panel to facilitate genetic testing in aHUS, TTP, C3GN, and DDD. In this paper, we review our one-year experience with this panel, and define the steps that lead to high-quality, patient-orientated, clinically useful genetic data.
Results
Subjects
During the 11-month period beginning on January 1, 2014, 193 patients were screened using the Genetic Complement-Mediated Renal Disease Panel (GRP), a TGE&NGS panel that includes CFH, CFI, CFB, C3, CFHR5, CD46, DGKE, ADAMTS13, THBD, PLG, and CNV analysis of CFHR3-CFHR1. The GRP process and variants interpretation are illustrated in Figure 1 and Supplementary Figure 1. We included all patients on whom testing was requested with clinical information and classified patients based on the requesting physician’s clinical impression. Physician-based clinical diagnoses included: (1) TMA: 118 aHUS, six TTP and 11 other TMA patients; (2) C3G: 30 C3GN and five DDD patients; and (3) Other: nine patients with untargeted diseases, such as systemic lupus erythematosus (SLE) and IgA nephropathy (IgAN). Fourteen patients with an ambiguous diagnosis (12 aHUS/TTP patients and two C3GN/DDD patients) were also included (Table 1). The Institutional Review Board of Carver College of Medicine at the University of Iowa approved this study.
Figure 1.
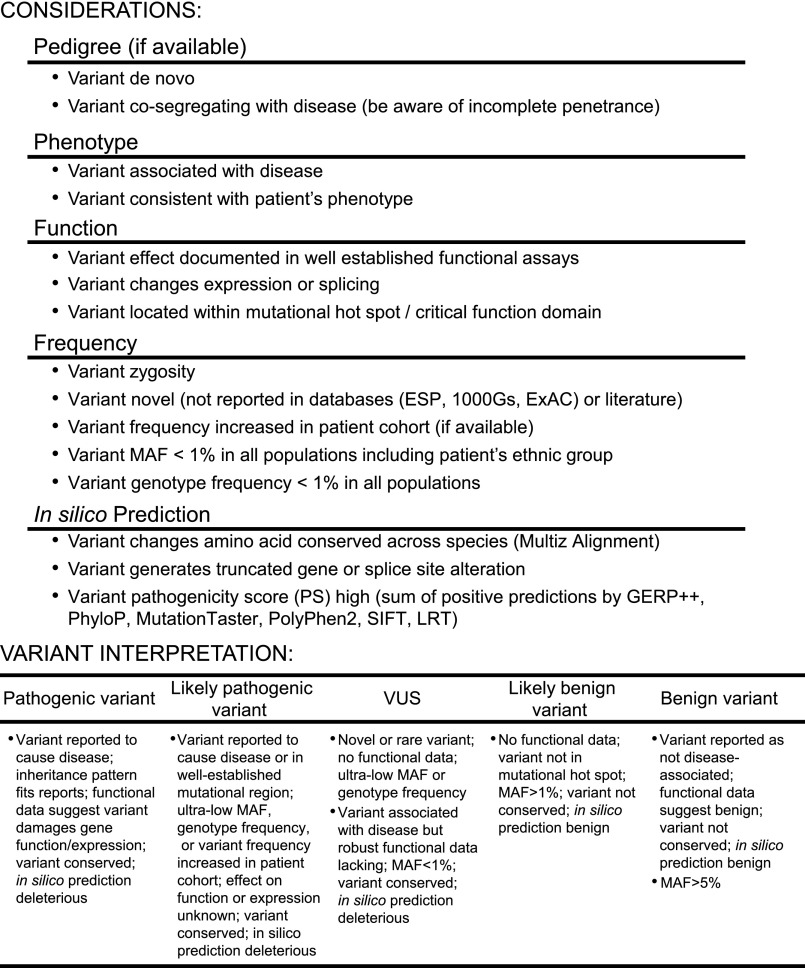
Variant interpretation is often challenging and generally requires multidisciplinary knowledge and the integration of information from multiple sources. Genetic data from all patients studied in our TGE&NGS pipeline are reviewed in a multidisciplinary care conference (Renal Group Meeting) during which time genetic data are discussed in light of all phenotypic (clinical) data available to generate a consensus Final Report (see Supplemental Figure 1). Variants are labeled as: pathogenic, likely pathogenic, variant of uncertain significance (VUS), likely benign, or benign based on predetermined metrics.
Table 1.
Patient demographic information
| TMA | C3G | Othera | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aHUS | TTP | aHUS/TTPb | Other TMA | C3GN | DDD | C3GN/DDDb | SLE | ARF | IgAN | Acute GN | ||
| Age | Years | |||||||||||
| <5 | 22 (18.6%) | 0 | 1 (8.3%) | 3 (27.3%) | 0 | 1 (20.0%) | 0 | 0 | 0 | 0 | 0 | |
| 5–18 | 27 (22.9%) | 0 | 3 (25.0%) | 2 (18.2%) | 11 (36.7%) | 2 (40.0%) | 1 (50.0%) | 0 | 0 | 0 | 1 (100.0%) | |
| >18 | 69 (58.5%) | 6 (100.0%) | 8 (66.7%) | 6 (54.5%) | 19 (63.3%) | 2 (40.0%) | 1 (20.0%) | 4 (100.0%) | 2 (100.0%) | 2 (100.0%) | 0 | |
| Sex | ||||||||||||
| Male | 50 (42.4%) | 2 (33.3%) | 1 (8.3%) | 5 (45.5%) | 19 (63.3%) | 1 (20.0%) | 1 (50.0%) | 1 (25.0%) | 2 (100.0%) | 0 | 1 (100.0%) | |
| Female | 68 (57.6%) | 4 (66.7%) | 11 (91.7%) | 6 (54.5%) | 11 (36.7%) | 4 (80.0%) | 1 (50.0%) | 3 (75.0%) | 0 | 2 (100.0%) | 0 | |
| Total | 118 | 6 | 12 | 11 | 30 | 5 | 2 | 4 | 2 | 2 | 1 | |
IgAN, IgA nephropathy.
Ambiguous diagnostic information was provided for some patients.
Validating the GRP
Prior to clinical implementation, the GRP was validated using 92 archived and de-identified TMA or C3G patients who underwent Sanger sequencing for one or more of the following genes: CFH, CFI, CFB, C3, CFHR5, CD46, and THBD. In aggregate, in the validation process, we compared genotype data from 55 exonic and 14 intronic common/rare variants. For NGS, samples were divided into two batches, each of which was sequenced on one lane of an Illumina HiSeq 2000 (Illumina Inc., San Diego, CA). To test reproducibility, three samples were sequenced twice on the HiSeq instrument by including them in both batches, and 13 samples were sequenced on both the HiSeq and MiSeq (Illumina Inc.) instruments. In all cases, results of duplicate runs were 100% concordant. Average Qvar (Phred-like quality score), Depth, and QD (Phred-like quality score divided by depth) of variant callings were 20596, 1172, and 18.30, respectively.
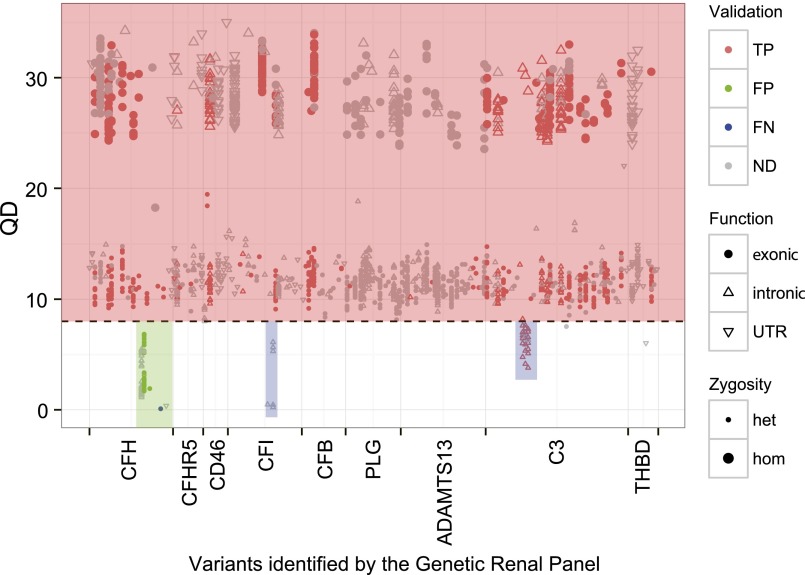
As shown in Figure 2, in total, 622 high-quality (QD>8) and 26 fair-quality (5<QD≤8) variants were Sanger-validated with 100% and 65% concordance. Four of 19 (21%) poor-quality variants (QD<5) were Sanger-confirmed. The positive predictive value based on this validation process was 96%. We observed a low-quality gap in exons 20 and 21 of CFH. Specifically, c.3138C>T (rs61822181) and c. 3150T>C (rs113347629) in exon 20 were false-positives and had QDs between 1.7 and 6.8; and c.3572C>T, p.Ser1191Leu (rs460897) in exon 22 was a false-negative and was omitted in the filtering process due to a very low QD score of 0.09 (Figure 2). Other low-quality gaps were in introns of CFI and C3, remote from targeted coding sequence and splicing sites, and were considered insignificant. On the basis of these results, we retain all variants with QD>5 and Depth>10 and Sanger confirm positive findings. In addition, we always Sanger sequence exons 20–22 of CFH.
Figure 2.
Ninety-two subjects were analyzed using both TGE&NGS and Sanger sequencing as a validation step for the GRP. Nearly all exonic variants have very high quality (red box), reflecting an effective enrichment strategy. The single exception was a small portion of CFH (green box), where we observed both false-positive and false-negative calls. For this reason, exons 20–22 of CFH are always Sanger-sequenced (see Supplemental Figure 1). Some low QD variants in introns of CFI and C3 (blue box) were ignored because they did not impact exons and splice sites. TP, true positive; FP, false positive; FN, false negative; ND, no Sanger sequencing data; UTR, untranslated region.
NGS Summary
NGS was completed on 193 samples in 50 runs of varying sample number on the MiSeq. The average number of total reads per sample was 4.01 million, with a range from 2.00 to 11.89 million (Supplementary Figure 2). Average coverage was 723× with 99.37% of the total target region covered by more than 30 reads. The lowest coverage (297×) was associated with low total reads (2.00 million). Homozygosity for del(CFHR3-CFHR1) was associated with a reduction in 30× coverage to less than 94% in all 17 homozygous deletion carriers.
Variants Calling and Filtering
A mean of 217 variants were identified per patient, which then were filtered by quality (QD>5 and Depth>10), minor allele frequency (MAF), and nucleotide effect. In total, we identified 17 novel variants not reported in the literature or any database (see CONCISE METHODS) and 74 rare variants (MAF<1% in all populations) (Supplementary Table 1, Table 2). All novel and rare variants were Sanger-validated (100% confirmation).
Table 2.
Rare and novel functional variants identified
| Disease (n) | CFH | CD46 | CFI | CFB | C3 | CFHR5 | ADAMTS13 | THBD | DGKE | PLG | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|
| TMA (147) | 14 | 9 | 10 | 1 | 8 | 4 | 10 | 2 | 3 | 4 | 65 |
| aHUS (118) | 12 (4)a | 9 (3) | 8 (2) | 1 | 7 (1) | 2 | 6 | 3 (2) | 3 | 51 | |
| TTP (6) | 1 (1) | 2 | 1 | 4 | |||||||
| aHUS/TTPb (12) | 1 | 1 | 2 | 1 | 1 | 6 | |||||
| Other (11) | 1 | 1 (1) | 2 (1) | 4 | |||||||
| C3G (37) | 6 | 0 | 2 | 3 | 5 | 1 | 1 | 1 | 3 | 1 | 23 |
| C3GN (30) | 5 | 1 | 3 | 3 (2) | 1 | 1 | 1 | 2 | 1 | 18 | |
| DDD (5) | 1 | 1 | 2 | 4 | |||||||
| C3GN/DDDb (2) | 1 | 1 | |||||||||
| Other (9) | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | 3 |
| SLE (4) | 0 | ||||||||||
| ARF (2) | 0 | ||||||||||
| IgAN (2) | 1 | 2 | 3 | ||||||||
| Acute GN (1) | 0 |
Variant count: total (novel); novel variants are not reported in any database or literature.
Ambiguous diagnostic information was provided for some patients.
Detecting the CFHR3-CFHR1 CNV
The CFHR3-CFHR1 CNV was detected by multiplex ligation-dependent probe amplification (MLPA). Homozygosity for this deletion was identified in approximately 12% of aHUS patients (Table 3), a prevalence significantly higher than that seen in 314 healthy controls (3%; Fisher’s exact test, P<0.01). All del(CFHR3-CFHR1) homozygotes were also identifiable by NGS as total target region coverage was reduced to <94% at 30× (Supplementary Figure 2). We were able to identify del(CFHR3-CFHR1) heterozygous in validation batches sequenced on the HiSeq; however, their unambiguous identification on the MiSeq was not possible due to smaller pool size and lower total coverage (data not shown).
Table 3.
Frequency of homozygous deletion of CFHR3-CFHR1
| Disease | n | Copy number of CFHR3-R1 | Percentage (homozygous deletion) | ||
|---|---|---|---|---|---|
| 0 | 1 | 2 | |||
| TMA | |||||
| aHUS | 111 | 13 | 34 | 64 | 11.71 |
| TTP | 6 | 0 | 4 | 2 | – |
| aHUS/TTPa | 12 | 0 | 3 | 9 | 0.00 |
| Other | 11 | 1 | 5 | 5 | 9.09 |
| C3G | |||||
| C3GN | 28 | 2 | 9 | 17 | 7.14 |
| DDD | 5 | 0 | 0 | 5 | – |
| C3GN/DDDa | 2 | 0 | 1 | 1 | – |
| Other | |||||
| SLE | 4 | 1 | 3 | 0 | – |
| ARF | 2 | 0 | 1 | 1 | – |
| IgAN | 2 | 0 | 2 | 0 | – |
| Acute GN | 1 | 0 | 1 | 0 | – |
Ambiguous diagnostic information was provided for some patients.
Variant Interpretation
To provide a clinically relevant report, a multidisciplinary board (Renal Group Meeting) reviewed all genetic data in the context of the available clinical data (Figure 1, Supplementary Figure 1). Variants with MAF>1% known to be unrelated to disease were classified as benign. Ultra-rare variants reported as pathogenic in the literature with supporting functional evidence were classified as pathogenic. Novel or rare variants that change protein sequence but have an unknown impact on protein function were classified as either likely pathogenic or variant of uncertain significance (VUS), a distinction that reflected two additional calculations. Likely pathogenic variants were also: (1) missense variants with pathogenicity scores ≥5 (based on GERP++,17 PhyloP,18 MutationTaster,19 PolyPhen2,20 SIFT,21 and LRT22); ultra-rare or disease-associated (based on a comparison of reported MAF to disease-associated prevalence in the Renal Variant Database, an in-house variants database of 1085 de-identified patients with TMAs or C3Gs); and found in disease-related functional domains/loci; or (2) novel and caused loss of function. Note that known pathogenic and likely pathogenic variants have similar pathogenicity scores and that likely pathogenic variants have higher pathogenicity scores than VUSs (Supplementary Figure 3). Based on genotypic findings and the clinical phenotype, additional testing was occasionally recommended. The Renal Group Meeting also served as the forum for discussion of all email inquiries regarding patient management of these ultra-rare diseases.
Variant Burden and Distribution
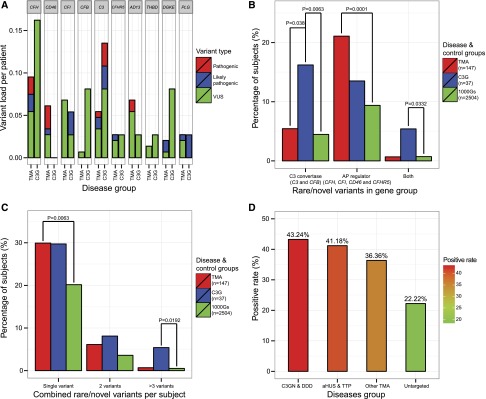
Variant burden per disease group was calculated as the sum of VUS/likely pathogenic/pathogenic variants divided by the number of patients. For example, in the group of 147 patients with TMA, eight VUSs, three likely pathogenic, and three pathogenic variants were identified in CFH, for a rare variant load of 0.095. As shown in Figure 3A, the variant load in CFH, CFI, and C3 was high in both TMA and C3G patients, while for CD46 and CFB, the variant load was high in only TMA or C3G, respectively. ADAMTS13 variants were found mainly in patients with aHUS or TTP (variant load, 0.073), suggesting a common mechanism driving these two diseases. The variant load in DGKE was high in C3G patients but all DGKE variants were present in heterozygosity; the function impact of this finding is not known. A single patient, a newborn with aHUS, was homozygous for a DGKE variant (c.465–2A>G) predicted to disrupt exon splicing.
Figure 3.
VUS/likely pathogenic/pathogenic variants distributed unevenly across genes and gene groups in TMA and C3G patients. (A) Variant load in CFH, CFI, and C3 was high for both patients with TMA and C3G. (B) Variants accumulated in C3 convertase genes (C3 and CFB) in patients with C3G and in AP regulator genes (CFH, CFI, CD46, and CFHR5) in patients with TMA. (C) Patients with C3G and TMA were more likely to carry single rare/novel variants than control samples retrieved from 1000 Genomes Project (1000Gs). (D) More patients with C3G and TMA carried VUS/likely pathogenic/pathogenic variants than did patients with untargeted diseases (see Table 2). (Fisher’s exact test was used to compare intergroup differences).
The distribution of VUS/likely pathogenic/pathogenic variants was significantly different between C3G and TMA patients at the level of functionally grouped genes. As shown in Figure 3B, 16.2% (n=6), 13.5% (n=5), and 5.4% (n=2) of patients with C3G, respectively, carried variants only in C3 convertases genes (C3 and CFB), only in AP regulators (CFH, CFI, CD46 and CFHR5), or in both gene groups, as compared with 5.4% (n=8), 21.1% (n=31), and 0.7% (n=1) of patients with TMA. We also assessed single/combined variants per patient and found that 43.2% C3G and 36.7% of patients with TMA carry at least one VUS/likely pathogenic/pathogenic variant, both prevalences significantly higher (P<0.05) than controls. As show in Figure 3C, the prevalence of multiple VUS/likely pathogenic/pathogenic variants per patient was also higher in the C3G group as compared with the TMA group and controls (13.5% [n=5] versus 6.8% [n=10] versus 4.1% [n=103], respectively; P<0.05). Control data were retrieved and filtered from the 1000 Genomes Project (1000Gs), Phase 3 (n=2504), an unphenotyped control group unlikely (P<0.01) to include patients with TMA or C3G.
Positive Rate
Identification of VUS/likely pathogenic/pathogenic variants was a significant finding. Additionally, we also considered the deletion of CFHR3-CFHR1 as a positive result in patients with aHUS and recommended screening for anti-CFH autoantibodies if that test had not been considered.23 Across all patient categories (aHUS, TTP, C3GN and DDD), we had a diagnostic rate of approximately 40%; however, if we consider only CFH, CD46, CFI, CFB, and C3 in the aHUS group of patients (118 patients), the positive rate was 27.1%, which is lower than the commonly reported rate of approximately 45%.24 The diagnostic rate was also low in patients with other diagnoses (Figure 3D). More than half of patients had a negative genetic screen, suggesting the presence of unrecognized genetic and/or acquired drivers of disease.
Molecular Modeling
To better understand the functional impact of variants in C3 convertase genes, refined molecular modeling was completed for VUS/likely pathogenic/pathogenic variants identified in C3 and factor B. Using the AMOEBA polarizable force field as a part of the Force Field X software package, four of the five C3 variants and both factor B variants identified in C3G patients were predicted to have molecular interactions that might impact convertase function and/or regulation. Two of the four C3 variants in TMA patients were also predicted to destabilize folding (Supplementary Figure 4, Supplementary Table 2).
Discussion
Advances in DNA sequencing technologies have changed medical practice by making it feasible and cost-effective to include comprehensive, disease-specific genetic testing in the initial phases of a patient’s clinical evaluation. This technology, typically some variation of TGE&NGS, requires a well-developed and well-supported infrastructure to implement. Requisite equipment includes dedicated robotics and NGS machines to facilitate high throughput and to minimize opportunities for human error.
After sequencing, a patient-specific genetic variant list must be created from the millions of reads generated for each patient. This step requires a well-established bioinformatics pipeline that is easy to use, runs batch files, detects CNVs, and is readily updated as improved tools become available. However, arguably the most important aspect in the overall process comes next—the interpretation of the generated variant list in light of the patient’s phenotype. This step requires a multidisciplinary meeting at which time each patient’s genotypic data can be considered in an open forum in light of their clinical presentation. Integrating genotype and phenotype requires clinicians with expertise both in the disease in question and human genetics, scientists with expertise in the wet-lab techniques, bioinformaticians with expertise in the analysis of NGS data, genetic counselors, and clinical geneticists. The end product should be a clinical report that is easy for the clinician to understand and interpret, and that is helpful in enhancing better patient care.
The GRP analytic pipeline has been built with these goals in mind (Figure 1, Supplementary Figure 1). Validation studies show that the technical performance of the GRP is outstanding. It provides comprehensive coverage of the targeted genes and detects an important CNV in the CFH-related region. As part of the validation testing, we did identify three exons in CFH with low-quality coverage metrics (exons 20–22), reflecting the high sequence homology of this region of CFH to exons 4–6 in CFHR1. This finding is important because the three carboxy-terminal short consensus repeats of factor H harbor the majority of the aHUS-causing CFH mutations. In addition to TGE&NGS, exons 20–22 of CFH are therefore Sanger-sequenced in all patients. However, as this paper and our earlier work confirm16,25 (Figure 2), Sanger sequence is not required to validate NGS-identified variants of high quality. With appropriate metrics, NGS data universally agree with Sanger sequencing data (QD>8), although confirmatory Sanger sequencing is mandatory for variants with QD<8. As compared with Sanger sequencing panels, the GRP panel significantly reduces patient cost (from approximately $6000 to approximately $3000) and turnaround time (from several months to 4 weeks, although 1-week turnaround is possible in emergent cases).
In the 193 patients we have tested to date, we identified 11 pathogenic variants, 12 likely pathogenic variants, and 68 VUSs. Included in this number are 17 novel variants, for which further functional confirmation is required. It is notable that VUS/likely pathogenic/pathogenic variants cluster in AP regulator genes in patients with TMA, consistent with an abnormality in AP regulation as a driving factor in this disease (Figure 3B). In patients with C3G, in contrast, these variants were abundant in both C3 convertase and AP regulator genes. Molecular remodeling suggests these variants likely affect the function and/or regulation of the convertase (Supplemental Figure 4). Additionally, the prevalence of multiple variants was higher in patients with C3G, suggesting a multifactorial genetic contribution to C3GN and DDD (Figure 3, B and C). An unexpected finding was the increased DGKE variant load in patients with C3G and IgAN and its possible association with hematuria,26–28 which may reflect a role for DGKE in endothelial cell activation and damage.29 CNV analysis over the CFHR3-CFHR1 region confirmed the increased frequency of homozygosity for del(CFHR3-CFHR1) in aHUS patients (Table 3). We also noted an important limitation that the relatively small sample size reduces the statistical power of this study. Therefore, those findings should be validated in a large patient cohort in a future study.
In aggregate, we provided a positive genetic diagnosis in 43% and 41% of patients with reported clinical diagnoses of C3G and TMA, respectively. Our data suggest that rare and novel genetic variants are more frequent in both C3 convertase and complement regulator genes in patients with C3G, while in patients with TMA, they are more frequent primarily in complement regulator genes. This distinction further refines our understanding of these diseases. Small sample size, however, reduces statistical power and, as such, these findings should be validated in a large patient cohort.
Concise Methods
Sample Preparation, Target Genomic Enrichment, and Next Generation Sequencing
For each sample, genomic DNA was extracted from peripheral blood using the Gentra Puregene Kit (Qiagen Inc., Valencia, CA) and integrity was evaluated by 1% agarose gel electrophoresis. The absorbance at 230:260:280 was measured using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE) to ensure DNA samples met our minimal quality metrics of 1.8 for 260/280 and 260/230 ratios. DNA concentration was determined using the Qubit dsDNA HS Assay Kit (Life Technologies, Carlsbad, CA).
For each GRP gene, the coding sequence and flanking splice sites were captured using the Agilent SureSelect Target Enrichment System (Agilent Technologies, Santa Clara, CA). Library preparation was performed with SureSelect TGE baits and SureSelectXT Reagent Kits (Agilent Technologies) following the manufacturer’s protocol. Preparation was automated using a Zephyr Workstation (PerkinElmer, Waltham, MI). Library quality and concentration were evaluated using a Bioanalyzer 2100 (Agilent Technologies). Libraries passing this quality control step were pooled and sequenced with a 100-bp paired-end module in one lane on a HiSeq 2000 (Illumina Inc.) or a 150-bp paired-end module on a MiSeq Sequencer (Illumina Inc.).
Next-Generation Sequencing Data Analysis
NGS data storage and analysis were performed on dedicated computing resources maintained by the Iowa Institute of Human Genetics at the University of Iowa. Sequencing data were archived as fastq files on a secured storage server and then analyzed using locally implemented open-source Galaxy software on a high-performance computing cluster. The workflow for variant calling integrated publicly available tools: reads were mapped using Burrows–Wheeler Alignment against human reference genome GRCh37/hg19; duplicates were removed by Picard; realignment, calibration, and variant calling were performed with GATK; variant annotation was performed with a CLCG Annotation and Reporting Tool developed by our bioinformatics team.
Variant Prioritization and Sanger Validation
Total number of reads per sample varied as a function of the number of samples per run and DNA input per sample. Low-quality variants (Depth<10 or QD<5) were filtered out by quality control. Common variants with MAF>1% in any population were excluded (based on the NHLBI GO Exome Sequencing Project [ESP, evs.gs.washington.edu], the 1000 Genomes Project [1000Gs, www.1000genomes.org], and most recently, the Exome Aggregation Consortium [ExAC, exac.broadinstitute.org]). Variants also were filtered based on predicted effect, retaining nonsynonymous single nucleotide variants, canonical splicing changes, and indels. These variants were prioritized based on MAF, nucleotide conservation, reported functional/expressional impact, and phenotype correlation. Other reference databases routinely queried included the aHUS Mutation Database (www.fh-hus.org), Human Gene Mutation Database, and our in-house Renal Variant Database. GERP++,17 PhyloP,18 MutationTaster,19 PolyPhen2,20 SIFT,21 and LRT22 were used to calculate variant-specific pathogenicity scores, which we based on the sum of tools predicting a given variant to be deleterious. All reported variants were Sanger-validated.
Copy Number Variation
CNVs across the CFHR3-CFHR1 region were identified using an MLPA set of 13 probes and six control probes, all designed following the MRC-Holland synthetic probe design protocol. Patients with deletion in CFHR3-CFHR1 were further tested for CNV of CFH, CFHR4, CFHR2, and CFHR5. MLPA was performed using the SALSA MLPA Reagent Kit (MRC-Holland, Amsterdam, The Netherlands), resolved on a 3130xl Genetic Analyzer (Life Technologies), and analyzed with GeneMapper software (Life Technologies). In each MLPA run, we included eight control samples (five normal controls [three females, two males], two controls with a heterozygous deletion of CFHR3-CFHR1 [one female, one male], and one control with a homozygous deletion for CFHR3-CFHR1).
Molecular Modeling
Homology models were acquired and refined using the AMOEBA polarizable force field as a part of the Force Field X software package.30,31 The model refinement consisted of local minimization followed by rotamer optimization around the mutation and then a second minimization step. The first minimization step eliminated obvious steric clashes in the protein; rotamer optimization allowed side chain atoms of residues near the mutation to be altered into a specific set of discrete conformations (rotamers) with low energy,32 and the final minimization step allowed rigid conformations in side chains to relax. This protocol was used to refine the wild-type model to remove model bias before modeling the identified variants; wild-type and mutant models were superimposed using the PyMOL molecular visualization program.
Statistical Analyses
Data were analyzed using R (v2.15.1). Fisher exact test was used to analyze categorical data. All tests were two-tailed, and P values less than 0.05 were considered significant.
Disclosures
None.
Supplementary Material
Acknowledgments
This study was supported in part by the Foundation for Children with Atypical HUS. We are grateful to the many clinicians who have entrusted us with the genetic analysis of their patients.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2015040385/-/DCSupplemental.
References
- 1.Biesecker LG, Green RC: Diagnostic clinical genome and exome sequencing. N Engl J Med 370: 2418–2425, 2014 [DOI] [PubMed] [Google Scholar]
- 2.Renkema KY, Stokman MF, Giles RH, Knoers NV: Next-generation sequencing for research and diagnostics in kidney disease. Nat Rev Nephrol 10: 433–444, 2014 [DOI] [PubMed] [Google Scholar]
- 3.Boyd SD: Diagnostic applications of high-throughput DNA sequencing. Annu Rev Pathol 8: 381–410, 2013 [DOI] [PubMed] [Google Scholar]
- 4.Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, Shendure J: Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet 12: 745–755, 2011 [DOI] [PubMed] [Google Scholar]
- 5.Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, Braxton A, Beuten J, Xia F, Niu Z, Hardison M, Person R, Bekheirnia MR, Leduc MS, Kirby A, Pham P, Scull J, Wang M, Ding Y, Plon SE, Lupski JR, Beaudet AL, Gibbs RA, Eng CM: Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med 369: 1502–1511, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.George JN, Nester CM: Syndromes of thrombotic microangiopathy. N Engl J Med 371: 654–666, 2014 [DOI] [PubMed] [Google Scholar]
- 7.Rodríguez de Córdoba S, Hidalgo MS, Pinto S, Tortajada A: Genetics of atypical hemolytic uremic syndrome (aHUS). Semin Thromb Hemost 40: 422–430, 2014 [DOI] [PubMed] [Google Scholar]
- 8.Kavanagh D, Goodship TH, Richards A: Atypical hemolytic uremic syndrome. Semin Nephrol 33: 508–530, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhattacharjee A, Reuter S, Trojnár E, Kolodziejczyk R, Seeberger H, Hyvärinen S, Uzonyi B, Szilágyi Á, Prohászka Z, Goldman A, Józsi M, Jokiranta TS: The major autoantibody epitope on factor H in atypical hemolytic uremic syndrome is structurally different from its homologous site in factor H-related protein 1, supporting a novel model for induction of autoimmunity in this disease. J Biol Chem 290: 9500–9510, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levy GG, Nichols WC, Lian EC, Foroud T, McClintick JN, McGee BM, Yang AY, Siemieniak DR, Stark KR, Gruppo R, Sarode R, Shurin SB, Chandrasekaran V, Stabler SP, Sabio H, Bouhassira EE, Upshaw JD, Jr, Ginsburg D, Tsai HM: Mutations in a member of the ADAMTS gene family cause thrombotic thrombocytopenic purpura. Nature 413: 488–494, 2001 [DOI] [PubMed] [Google Scholar]
- 11.Tsai HM, Rice L, Sarode R, Chow TW, Moake JL: Antibody inhibitors to von Willebrand factor metalloproteinase and increased binding of von Willebrand factor to platelets in ticlopidine-associated thrombotic thrombocytopenic purpura. Ann Intern Med 132: 794–799, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barbour, TD, Ruseva, MM, Pickering, MC: Update on C3 glomerulopathy. Nephrology, dialysis, transplantation: official publication of the European Dialysis and Transplant Association - European Renal Association, 2014. [DOI] [PMC free article] [PubMed]
- 13.Barbour TD, Pickering MC, Terence Cook H: Dense deposit disease and C3 glomerulopathy. Semin Nephrol 33: 493–507, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Nester CM, Martin B, Skjoedt MO, Meyer NC, Shao D, Borsa N, Palarasah Y, Smith RJ: Defining the complement biomarker profile of C3 glomerulopathy. Clin J Am Soc Nephrol 9: 1876–1882, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao X, Pickering MC, Smith RJ: C3 glomerulopathy: the genetic and clinical findings in dense deposit disease and C3 glomerulonephritis. Semin Thromb Hemost 40: 465–471, 2014 [DOI] [PubMed] [Google Scholar]
- 16.Bu F, Maga T, Meyer NC, Wang K, Thomas CP, Nester CM, Smith RJ: Comprehensive genetic analysis of complement and coagulation genes in atypical hemolytic uremic syndrome. J Am Soc Nephrol 25: 55–64, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S: Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLOS Comput Biol 6: e1001025, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A, NISC Comparative Sequencing Program : Distribution and intensity of constraint in mammalian genomic sequence. Genome Res 15: 901–913, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwarz JM, Cooper DN, Schuelke M, Seelow D: MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 11: 361–362, 2014 [DOI] [PubMed] [Google Scholar]
- 20.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR: A method and server for predicting damaging missense mutations. Nat Methods 7: 248–249, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar P, Henikoff S, Ng PC: Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4: 1073–1081, 2009 [DOI] [PubMed] [Google Scholar]
- 22.Chun S, Fay JC: Identification of deleterious mutations within three human genomes. Genome Res 19: 1553–1561, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore I, Strain L, Pappworth I, Kavanagh D, Barlow PN, Herbert AP, Schmidt CQ, Staniforth SJ, Holmes LV, Ward R, Morgan L, Goodship TH, Marchbank KJ: Association of factor H autoantibodies with deletions of CFHR1, CFHR3, CFHR4, and with mutations in CFH, CFI, CD46, and C3 in patients with atypical hemolytic uremic syndrome. Blood 115: 379–387, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bresin E, Rurali E, Caprioli J, Sanchez-Corral P, Fremeaux-Bacchi V, Rodriguez de Cordoba S, Pinto S, Goodship TH, Alberti M, Ribes D, Valoti E, Remuzzi G, Noris M, European Working Party on Complement Genetics in Renal Diseases : Combined complement gene mutations in atypical hemolytic uremic syndrome influence clinical phenotype. J Am Soc Nephrol 24: 475–486, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shearer AE, Black-Ziegelbein EA, Hildebrand MS, Eppsteiner RW, Ravi H, Joshi S, Guiffre AC, Sloan CM, Happe S, Howard SD, Novak B, Deluca AP, Taylor KR, Scheetz TE, Braun TA, Casavant TL, Kimberling WJ, Leproust EM, Smith RJ: Advancing genetic testing for deafness with genomic technology. J Med Genet 50: 627–634, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zand L, Lorenz EC, Cosio FG, Fervenza FC, Nasr SH, Gandhi MJ, Smith RJ, Sethi S: Clinical findings, pathology, and outcomes of C3GN after kidney transplantation. J Am Soc Nephrol 25: 1110–1117, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reich HN, Troyanov S, Scholey JW, Cattran DC, Toronto Glomerulonephritis Registry : Remission of proteinuria improves prognosis in IgA nephropathy. J Am Soc Nephrol 18: 3177–3183, 2007 [DOI] [PubMed] [Google Scholar]
- 28.Sellier-Leclerc AL, Fremeaux-Bacchi V, Dragon-Durey MA, Macher MA, Niaudet P, Guest G, Boudailliez B, Bouissou F, Deschenes G, Gie S, Tsimaratos M, Fischbach M, Morin D, Nivet H, Alberti C, Loirat C, French Society of Pediatric Nephrology : Differential impact of complement mutations on clinical characteristics in atypical hemolytic uremic syndrome. J Am Soc Nephrol 18: 2392–2400, 2007 [DOI] [PubMed] [Google Scholar]
- 29.Bruneau S, Néel M, Roumenina LT, Frimat M, Laurent L, Frémeaux-Bacchi V, Fakhouri F: Loss of DGKε induces endothelial cell activation and death independently of complement activation. Blood 125: 1038–1046, 2015 [DOI] [PubMed] [Google Scholar]
- 30.Shi Y, Xia Z, Zhang J, Best R, Wu C, Ponder JW, Ren P: The Polarizable Atomic Multipole-based AMOEBA Force Field for Proteins. J Chem Theory Comput 9: 4046–4063, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ren P, Wu C, Ponder JW: Polarizable Atomic Multipole-based Molecular Mechanics for Organic Molecules. J Chem Theory Comput 7: 3143–3161, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shapovalov MV, Dunbrack RL, Jr: A smoothed backbone-dependent rotamer library for proteins derived from adaptive kernel density estimates and regressions. Structure 19: 844–858, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.