Abstract
Evaluating the mRNA profile of podocytes in the diabetic kidney may indicate genes involved in the pathogenesis of diabetic nephropathy. To determine if the podocyte-specific gene information contained in mRNA profiles of the whole glomerulus of the diabetic kidney accurately reflects gene expression in the isolated podocytes, we crossed Nos3−/− IRG mice with podocin-rtTA and TetON-Cre mice for enhanced green fluorescent protein labeling of podocytes before diabetic injury. Diabetes was induced by streptozotocin, and mRNA profiles of isolated glomeruli and sorted podocytes from diabetic and control mice were examined 10 weeks later. Expression of podocyte-specific markers in glomeruli was downregulated in diabetic mice compared with controls. However, expression of these markers was not altered in sorted podocytes from diabetic mice. When mRNA levels of glomeruli were corrected for podocyte number per glomerulus, the differences in podocyte marker expression disappeared. Analysis of the differentially expressed genes in diabetic mice also revealed distinct upregulated pathways in the glomeruli (mitochondrial function, oxidative stress) and in podocytes (actin organization). In conclusion, our data suggest reduced expression of podocyte markers in glomeruli is a secondary effect of reduced podocyte number, thus podocyte-specific gene expression detected in the whole glomerulus may not represent that in podocytes in the diabetic kidney.
Keywords: diabetic nephropathy, gene transcription, glomerulus, podocyte, signaling
A large body of evidence suggests that podocyte injury is a key event in diabetic nephropathy (DN). The reduction in podocyte density is the strongest predictor of progressive DN1 and its extent correlates directly with the magnitude of proteinuria.2 Apoptosis, detachment of podocytes from the glomerular basement membrane, and epithelial-mesenchymal transition (EMT) are potential mechanisms for podocyte injury and loss in DN3–5; however, its exact mechanism of loss remains unclear. Gene expression profiling of glomeruli or cortex of animal or human diabetic kidneys have been performed to ascertain differential regulation of genes involved in DN pathogenesis.6,7 However, due to the heterogeneity in cell types, these data provide limited information specifically on podocyte injury. Recently, mRNA profiles obtained directly from podocytes have been reported,8,9 but such information from the diabetic kidney has not been determined.
In this study we compared the mRNA profiles of glomeruli and podocytes between diabetic and control mice. We employed streptozotocin (STZ)-induced diabetes in eNOS−/− mice, resulting in a more pronounced DN phenotype than STZ induction alone.10 In order to specifically label and isolate podoctyes, we crossed the eNOS−/− mice with the IRG mice11 that ubiquitously express a red fluorescent protein prior to Cre-mediated recombination and an enhanced green fluorescent protein (EGFP) following recombination. These mice were further bred with podocin-rtTA and TetON-Cre (LC1) transgenic mice for inducible podocyte-specific EGFP expression. Mice were fed with doxycycline to induce EGFP expression permanently in podocytes prior to diabetes induction by at 8 weeks of age (STZ-eNOS−/−). Body weight, blood glucose, and urine excretion of albumin were monitored every 2 weeks (Supplemental Figure 1, A–D). Urine albumin-to-creatinine ratio steadily increased in the diabetic mice starting from 2 weeks post-STZ injection, and by 8 weeks a 10-fold increase was observed in comparison to the control mice (Supplemental Figure 1D). By 10 weeks STZ-eNOS−/− mice developed typical histologic findings of DN, including mesangial expansion and foot process effacement (Supplemental Figure 2, A and B).
We obtained whole glomeruli or sorted podocytes from diabetic and control mice at 10 weeks post-STZ injection for mRNA sequencing (RNA-seq). Sorted podocytes were viable and had enriched expression of podocyte differentiation markers (Supplemental Figure 3, A and B). Each glomerular RNA sample was from an individual mouse (n=4), while RNA samples from sorted podocytes were pooled from five mice per sample (n=3 diabetic, n=2 controls; one sample from a control mouse was discarded from analysis due to a technical issue). The top 50 differentially expressed genes (DEGs) in diabetic glomeruli or podocytes are listed in Supplemental Table 1. The principle component analysis and the heatmap of DEGs are shown in Figure 1, A–C. Consistent with the previous studies, the expression of podocyte-specific markers WT-1, nephrin, and synaptopodin was significantly downregulated in the diabetic glomeruli (Table 1). To our surprise, this downregulation was not observed in sorted podocytes of diabetic mice (Table 1). The discrepancy in podocyte marker expression level between isolated glomeruli and sorted podocytes was further validated by real-time quantitative PCR (qPCR) (Figure 2A). We confirmed by immunostaining that EGFP labeling co-localizes with WT-1 (Supplemental Figure 4A), and consistent with the previous findings3 podocyte number per glomerulus was significantly lower in the diabetic kidneys (Supplemental Figure 4B). When the RNA-seq data were corrected for podocyte number per glomerulus, the apparent downregulation of podocyte marker expression in diabetic glomeruli also disappeared (Table 1), consistent with the observation in the sorted podocytes.
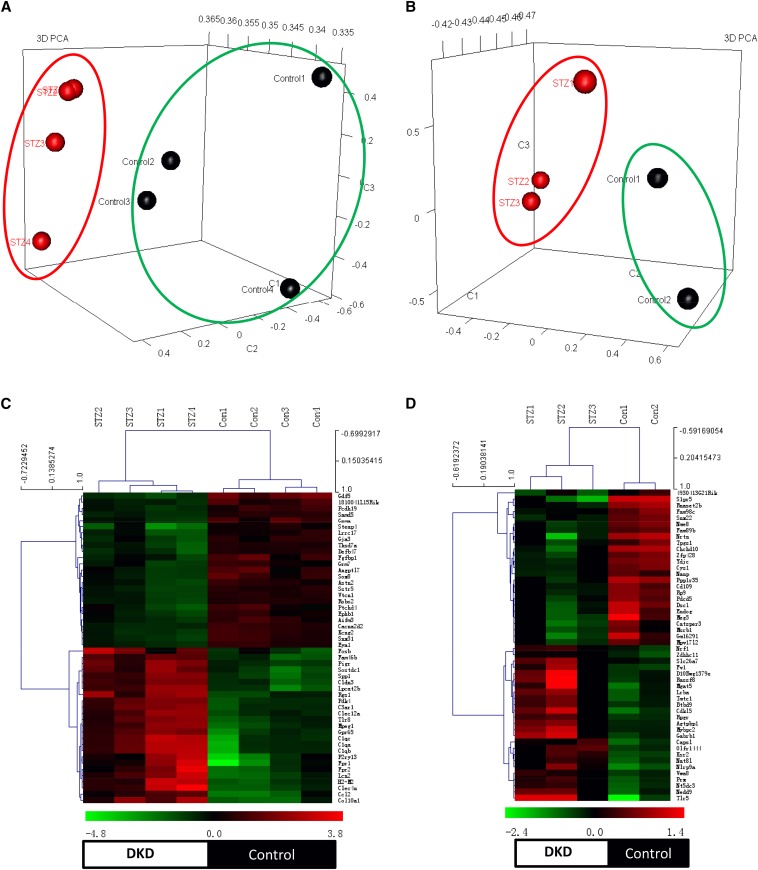
Figure 1.
Heat map and PCA analysis between glomeruli and sorted podocytes from control and diabetic mice. The PCA was performed for transcriptomic data from glomeruli (A) and podocytes (B) between diabetic and non-diabetic mice. Genes with the highest loadings in the first three principal components were plotted in 3D visualization. For both isolated glomeruli and podocytes, PCA revealed that the diabetic and non-diabetic samples form distinct clusters, indicating that samples within each biologic group have more similarity. Each symbol represents each biologic sample (green circles, non-diabetic mice; red circles, diabetic mice). Heatmaps of the top 50 up- or downregulated genes from isolated glomeruli (C) and sorted podocytes (D) between diabetic and control mice (green marks, downregulation; red marks, upregulation). There were four glomerular RNA samples in each group, two podocyte RNA samples from control mice and three samples from diabetic mice. Unsupervised hierarchical clustering was performed for gene expression between samples. Four clusters in each heatmap represented upregulated genes in diabetic mice (top, left cluster), upregulated genes in control mice (top, right cluster), downregulated genes in diabetic mice (bottom, left cluster), and downregulated genes in control mice (bottom right cluster).
Table 1.
Relative expression levels of podocyte markers in sorted podocytes and isolated glomeruli between control and diabetic mice
| Control 1 | Control 2 | Control 3 | Control 4 | STZ 1 | STZ 2 | STZ 3 | STZ 4 | Fold Change | P Value | |
|---|---|---|---|---|---|---|---|---|---|---|
| Relative gene expression in sorted podocytes (normalized to Control 1) | ||||||||||
| Nphsl | 1.0000 | 1.4010 | 1.4496 | 1.4010 | 1.0136 | 1.0729 | 0.73 | |||
| Nphs2 | 1.0000 | 1.0510 | 1.1061 | 1.2399 | 1.3092 | 1.1881 | 0.09 | |||
| Synpo | 1.0000 | 1.2015 | 1.2964 | 0.9701 | 1.1626 | 1.0384 | 0.794 | |||
| WT1 | 1.0000 | 1.2140 | 1.1817 | 0.9231 | 0.8920 | 0.9023 | 0.50 | |||
| Relative gene expression in isolated glomeruli (normalized to Control 1) | ||||||||||
| Nphsl | 1.0000 | 0.9028 | 0.8721 | 1.0153 | 0.4458 | 0.6045 | 0.7450 | 0.5020 | 0.6061 | 0.0021 |
| Nphs2 | 1.0000 | 0.8765 | 1.2160 | 1.2981 | 0.7768 | 0.6394 | 1.2565 | 0.5793 | 0.7407 | 0.17 |
| Synpo | 1.0000 | 0.8482 | 0.9441 | 1.2078 | 0.5581 | 0.6555 | 0.9655 | 0.4299 | 0.6522 | 0.04 |
| WT1 | 1.0000 | 0.9631 | 0.9858 | 1.0115 | 0.4076 | 0.5153 | 0.7412 | 0.4816 | 0.5418 | 0.001 |
| Relative gene expression in isolated glomeruli corrected for podocyte number (normalized to Control 1) | ||||||||||
| Nphsl | 1.0000 | 0.9028 | 0.8721 | 1.0153 | 0.7402 | 1.0036 | 1.2370 | 0.8015 | 0.9979 | 0.99 |
| Nphs2 | 1.0000 | 0.8765 | 1.2160 | 1.2981 | 1.2898 | 1.0617 | 1.0863 | 0.8298 | 0.9720 | 0.83 |
| Synpo | 1.0000 | 0.8482 | 0.9441 | 1.2078 | 0.9266 | 1.0883 | 1.3030 | 0.7138 | 1.0079 | 0.96 |
| WT1 | 1.0000 | 0.9631 | 0.9858 | 1.0115 | 0.6767 | 0.8556 | 1.2306 | 0.6676 | 0.8662 | 0.35 |
The expression of selected podocyte-specific markers obtained from the RNA-seq data were normalized to a control sample (Control 1) and compared between control and diabetic mice. The top panel shows the selected gene expression in sorted podocytes; the middle panel shows their expression in isolated glomeruli; and the bottom panel shows their expression in isolated glomeruli when first corrected by the number of podocytes. The relative gene expression was calculated by dividing normalized reads value (reads per kilobase per million reads) for each sample by the average value of Control 1. The fold change in gene expression in diabetic mice was calculated by dividing average normalized STZ value by control value. P values were calculated using Student’s unpaired t test.
Figure 2.
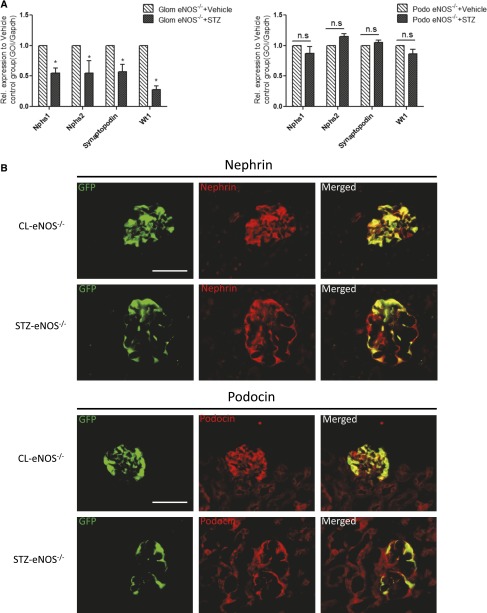
qPCR analysis and immunostaining of podocyte marker between diabetic and control mice. (A) qPCR was performed for podocyte-specific genes, Neph1, Neph2, synaptopodin, and WT-1, in isolated glomeruli and sorted podocytes from both diabetic and non-diabetic mice. (*P<0.05 compared with citrate-treated control glomeruli, n=3; GOI, gene of interest). (B) Immunostaining of podocyte markers was performed in the kidney of these mice. EGFP was visualized in the podocytes of both STZ-eNOS−/− and CL-eNOS−/−mice. Immunofluorescence staining was performed for nephrin (top panel) and podocin (bottom panel) in the kidney of these mice. The representative images taken from in each group are shown (n=3, original magnification ×400, bar=50 μm).
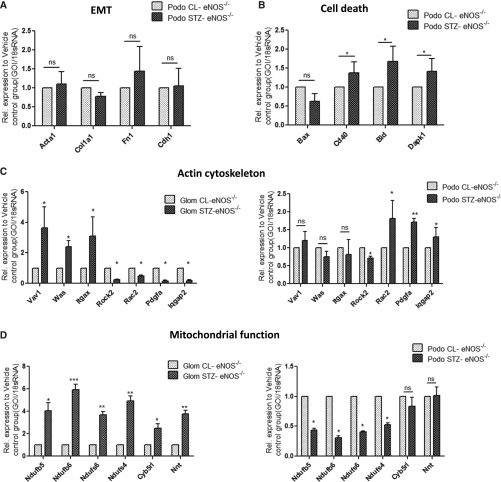
A significant loss of nephrin immunostaining has been previously reported in the glomeruli of patients with advanced DN.12 This may be due to the cleavage of the extracellular domain of nephrin molecule which becomes undetectable (as all available antibodies recognize only the extracellular domain of nephrin) and/or to the loss of podocytes. Immunostaining of diabetic kidneys for nephrin and podocin in fact showed a reduction in staining area, rather than of staining intensity (Figure 2B). We also did not detect any changes in EMT marker expression in podocytes by qPCR analysis (Figure 3A) and by immunostaining (Supplemental Figure 5). RNA-seq data further revealed that the expression of cell death-related genes Bid, Dapk1, and Cd40 were significantly increased in diabetic podocytes, which was confirmed by qPCR analysis (Figure 3B). In addition, a modest increase in cleaved Caspase-3 expression was observed by immunostaining in EGFP-positive cells, suggesting an increased podocyte apoptosis in the diabetic kidney (Supplemental Figure 6).
Figure 3.
qPCR validation of gene expression related to the major pathways in isolated glomeruli and podocytes between diabetic and control mice. (A) RT-PCR analysis was performed to assess the levels of mRNA for EMT-related genes expression in podocytes sorted from STZ-eNOS−/− and CL-eNOS−/− mice (*P<0.05 compared with those in CL-eNOS−/−, n=3). (B) RT-PCR analysis was performed to assess the levels of mRNA for cell-death–related gene expression in podocytes sorted from STZ-eNOS−/− and CL-eNOS−/− mice (*P<0.05 compared with those in CL-eNOS−/−, n=3). (C) RT-PCR analysis was performed to assess the levels of mRNA for actin cytoskeleton-related gene expression in glomeruli and podocytes isolated from STZ-eNOS−/− and CL-eNOS−/− mice (*P<0.05 compared with those in CL-eNOS−/−, n=3). (D) RT-PCR analysis of mitochondria and oxidative stress-related genes in isolated glomeruli and sorted podocytes from both STZ-eNOS−/− and CL-eNOS−/− mice (n=3, *P<0.05 compared with CL-eNOS−/−). GOI, gene of interest.
The above findings, from an unbiased approach, suggest that the decreased podocyte marker expression in diabetic glomeruli is likely a secondary effect of podocyte loss, rather than a direct result of podocyte dedifferentiation or EMT. This is an important step in better understanding DN pathogenesis, and it also indirectly supports the notion that terminally differentiated podocytes do not further dedifferentiate, similar to neurons. Nevertheless, we cannot rule out the possibility that podocytes may undergo dedifferentiation or EMT in other diabetic animal models with more advanced DN.
Analysis of the DEGs in isolated glomeruli revealed that the upregulated genes in diabetic glomeruli were mostly involved in the regulation of mitochondrial function and the oxidative stress pathway, whereas the downregulated genes were in involved in cell-cell signaling/communication, growth factor receptor-mediated pathways, and angiogenesis (Supplemental Figures 7 and 8). Consistent with the previous observations,13 these data suggest that mitochondrial dysfunction and oxidative stress are key events in the diabetic kidney leading to glomerular injury. Alteration of growth factor-mediated pathways may be related to cell survival. Changes in genes related to angiogenesis may be reflective of endothelial cell injury in DN. However, whether these processes occur mainly in podocytes, mesangial cells, or glomerular endothelial cells cannot be concluded.
Analysis of the DEGs in sorted podocytes showed that many upregulated genes were involved in the actin organization (Supplemental Figure 9), suggesting that significant alteration of the actin cytoskeleton occurs in the early stage of diabetes-induced podocyte injury, which is consistent with the foot process effacement (Supplemental Figure 2B). In persistent or aggravated injury, the early cytoskeletal changes may eventually lead to podocyte detachment or death, ensuing in their loss in DN. It also suggests that intervention to prevent podocyte detachment or death in the early stages of injury may be an effective therapy against diabetes-induced podocyte loss. Most of the downregulated genes in diabetic podocytes were involved in the RNA processing and endoplasmic reticulum function (Supplemental Figure 10), suggesting a possible alteration in the mTOR pathway and autophagy/ER stress response in podocytes, which are known to be involved in podocyte injury in DN.14,15
The above pathway analyses of DEGs in diabetic mice indicate significant differences in the altered pathways between isolated glomeruli versus podocytes. Interestingly, the regulation of actin cytoskeleton-related genes differs between glomeruli and podocytes (i.e., some gene expression changes in opposite directions), suggesting that changes of actin cytoskeleton-related genes in other glomerular cells may mask their changes in podocytes. Our observation that actin cytoskeleton and mitochondrial function/oxidative stress are major pathways differentially regulated in podocytes and glomeruli, respectively, were further validated by qPCR analysis of select genes in each pathway (Figure 3, C and D). 8-Oxoguanine (8-oxoG) immunostaining further confirmed that the oxidative stress in the diabetic kidney is increased mostly in glomerular endothelial cells rather than in podocytes (Supplemental Figure 11, A–C), which is consistent with a recently published study.16 These findings raise an important question as to whether mitochondrial dysfunction and increased oxidative stress occur in diabetic podocytes. However, many previous studies, mostly from cultured cells, suggest that oxidative stress is a key event in podocyte injury in diabetic kidney. In addition, recent studies from Dr. Kumar Sharma’s laboratory suggest that mitochondrial-generated superoxide is reduced in the diabetic kidney.17 Therefore, it would be important to further determine whether mitochondrial-generated reactive oxygen species and NADPH oxidase (NOX)-generated reactive oxygen species are differentially regulated in specific glomerular cell types and at various stages during DN.
This study has a few limitations. First, it is limited to a model of type 1 diabetes induced by STZ at a single time point. Although we used multiple low doses of STZ to induce diabetes, a direct toxic effect of STZ in kidney cells cannot be excluded.18 Future studies are required to determine whether similar observations would be made in other animal models of DN (i.e., type 2 diabetes, non-toxin–induced diabetes, and in a more susceptible genetic background) and to compare the podocyte gene expression profiles between mice with early and late DN. We anticipate that pathways related to actin cytoskeleton disorganization in podocytes might occur in early DN. Second, although we used sorted podocytes from control mice to normalize for possible altered gene expression in sorting and digestion processes, the degree of such change may not be identical between diabetic and control mice. A recent approach of translating ribosome affinity purification which avoids the digestion and sorting processes may provide better information on mRNA profiles of podocytes.9 Future studies are required to compare the two approaches and to determine whether the translating ribosome affinity purification method may yield more information on the mechanism of podocyte injury in DN. Third, although each podocyte sample contained pooled podocytes sorted from five mice, the number of sample size per group used for sequencing was still small. Therefore, it is difficult to control for experimental variation that might contribute to the altered gene expressions between groups.
In conclusion, podocyte mRNA profiles provide more precise information on the mechanism of podocyte injury in comparison to the glomerular mRNA profiles. Because it is not possible to sort podocytes from human diabetic kidney, we believe that our data from whole glomeruli and sorted podocytes may be valuable in interpreting the gene expression profiles obtained from human diabetic glomeruli.
Concise Methods
Generation of Transgenic Mice
Animal studies were performed in accordance with the guidelines of and approved by the Institutional Animal Care and Use Committee at the Icahn School of Medicine at Mount Sinai (New York, NY). Mice were housed in a specific pathogen-free facility with free access to chow and water and a 12-hour day/night cycle. Breeding and genotyping were performed according to the standard procedures. NPHS2.rtTA19 and IRG mice11 (B6;C3-Tg(CAG-DsRed,-EGFP)5Gae/J) were purchased from The Jackson Laboratory (Bar Harbor, ME). The LC1 transgenic mouse was a generous gift from M.J. Moeller (University of Aachen, Germany).20 To generate diabetic mice with inducible EGFP labeling in podocytes, we first crossed the IRG mice with eNOS–/– in C57BL/6 background (Jackson Laboratory), and then with NPHS2.rtTA and LC1 mice in order to generate the experimental NPHS2.rtTA;LC1;IRG;eNOS–/– mice. For the induction Cre-mediated EGFP expression, animals received doxycycline hydrochloride (Sigma-Aldrich, St Louis, MO) via drinking water (2 mg/ml with 5% sucrose) during pregnancy and nursing up to 4 weeks of age. To increase the induction efficiency, the mice received doxycycline (2 mg/ml) orally from P6 to P18 three times per week.21 For induction of diabetes, male NPHS2.rtTA;LC1;IRG;eNOS–/– mice at 8 weeks of age were injected over 5 consecutive days with low-dose STZ (50 μg/g per day intraperitoneally; Sigma-Aldrich). Body weight and hind-limb blood glucose levels were monitored bi-weekly by glucometer readings. Diabetes was confirmed by fasting blood glucose level >300 mg/dl. The age- and sex-matched littermates injected with vehicle (CL-eNOS−/−) served as non-diabetic controls. Urine samples were collected bi-weekly. The mice were sacrificed at 10 weeks post-STZ injection.
Measurement of Urinary Albumin-to-Creatinine Ratio
Urine creatinine was quantified using commercial kits from BioAssay Systems (Hayward, CA). Urine albumin was determined using a commercial assay from Bethyl Laboratory Inc. (Houston, TX). Urine albumin excretion was expressed as the ratio of urine albumin to creatinine.
Kidney Histology
Harvested kidney samples for histology were fixed in 10% formalin, embedded in paraffin, and cut into 4 µm sections. Periodic acid-Schiff stained sections were used for assessment of kidney histology. Assessment of the mesangial expansion was performed by pixel counts on a minimum of 15 glomeruli per section in a blinded manner, under 400× magnifications (Zeiss AX10 microscope; Carl Zeiss Canada, Toronto, ON, Canada). For transmission electron microscopy, kidney cortex samples fixed in 2.5% glutaraldehyde were sectioned, mounted on a copper grid, and then images were photographed using a Hitachi H7650 microscope (Tokyo, Japan) as described previously.22
Immunofluorescence Staining
Frozen sections were used for immunofluorescence staining for WT-1, podocin, and nephrin as described previously23 and images were taken using the Carl Zeiss Axioplan 2 IE microscope. 8-OxoG staining on formaldehyde-perfused frozen sections was done as previously described16 using anti–8-oxoG monoclonal antibody (N45.1; Japan Institute for the Control of Aging). Sections were also stained with rabbit anti-podocin antibody (a gift from Dr. Peter Mundel) and antibodies for CD31 (MEC 7.46; Abcam, Inc.), E-cadherin (4065; Cell Signaling Technology), Alpha SMA (5694; Abcam, Inc.), and Cleaved caspase3 (9664; Cell Signaling Technology).
Isolation of Glomeruli and Sorting of Podocytes
Glomeruli were isolated by Dynabead perfusion and EGFP-labeled podocytes were sorted as described recently.8 Briefly, animals were perfused with prewarmed 8 ml bead solution and 2 ml bead solution with enzymatic digestion buffer (Collagenase type II 300U/ml, Proteinase E 1 mg/ml and Dnase I 50 U/ml). Kidneys then were removed, decapsulated, minced into 1 mm3 pieces, and digested in 3 ml digestion buffer at 37°C for 15 min on a rotator (100 rpm). Digested tissues were passed through a 100-μm cell strainer and collected by centrifugation. The pellet was resuspended in 2 ml of Hanks’ buffered salt solution and glomeruli were washed three times and collected using a magnet. The separated glomeruli were resuspended in 2 ml digestion buffer and incubated at 37°C for 40 min at 1400 rpm/min on a thermomixer. During the second digestion period, the solution was vortexed every 10 min and sheared with a 27G needle every 15 min. Then, the solution was put on a magnetic particle concentrator and the supernatant was pooled. The suspension was then sieved through a 40μm cell strainer and centrifuged at 1500 rpm for 5 min at 4°C. After a two-step approach for primary cell purification, single cells were resuspended in 0.5 ml of Hanks’ buffered salt solution supplemented with 2% fetal bovine serum, 25 mM HEPES and 4′6-diamidino-2-phenylindole (1 mg/ml). The single-cell suspension was then sorted into EGFP-positive and EGFP-negative populations with a BD Aria II cell sorter with a laser excitation at 488 nm and a sheath pressure of 30 PSI. On average, 450,000 podocytes were sorted per mouse (see Supplementary Figure 1).
mRNA Isolation for RNA Sequencing
Total RNA was isolated from either isolated glomeruli or sorted podocytes by using the RNeasy mini kit (Qiagen 74104) according to the manufacturer’s protocol. RNA concentrations were quantified using a Nano-drop Spectrophotometer at a wavelength of 260 nm. RNA samples were analyzed by Bioanalyzer at a concentration of 100–200 ng/μl to verify the concentration and the purity of samples. Only the samples with RNA integrity values of >7.0 were used for mRNA sequencing at the Genomic Core Facility at Icahn School of Medicine at Mount Sinai School.
Bioinformatics Analysis of mRNA Sequencing (RNA-seq) Data
The RNA-seq data were analyzed by following the procedure described below. Briefly, after sequence quality filtering at a cutoff of a minimum quality score Q20 in at least 90% bases, the good-quality reads aligned to Reads were processed and aligned to the University of California Santa Cruz (UCSC) Mus musculus reference genome and transcriptome (build mm10) using the Burrows-Wheeler Aligner (bwa).24 The reads that are uniquely aligned to the exon and splicing junction sites for each transcript were combined to calculate an expression level for a corresponding transcript and further normalized based on reads per kilobase per million reads25 in order to compare transcription levels among samples. The transcripts with a low raw read count <100 in all the samples were excluded for downstream analysis. Gene expression value was transformed to the log 2 base scale. Principle Component Analysis (PCA) was first performed to assess the sample correlations using the expression data of all the genes. The differentially expressed genes in STZ mice compared with control mice were identified by the R package DEGseq for sorted podocytes26 and we selected the genes based on DEGseq adjusted P<0.05 and 1.5-fold change in this 2:3 comparison on which a standard statistic test was not applicable. The limma test27 was applied for analysis of data in isolated glomeruli. A specific gene was considered differentially expressed if the P value given by these methods was ≤0.05. The Gene Ontology and pathway analysis for the differentially expressed genes were then performed with fold change cutoff of ≥1.5 using INGENUITY© IPA (www.ingenuity.com/products/ipa) and the online tool Enrichr.28 The read coverage of gene functional elements was also visualized by the Integrative Genome Viewer tool (www.broadinstitute.org/igv/) from the genome alignment file. Heatmap analysis was performed for the top 50 differentially expressed genes using the DEGseq test after the median center was transformed using Multi-Experiment Viewer software.29
Real-Time PCR
Primers for RT-PCR were designed by using Primer-Blast (NCBI) (Supplemental Table 2). Gene expression was normalized to Gapdh, and fold change in expression relative to the control group was calculated using the 2–∆∆Ct method. Two technical replicates per gene were used.
Statistical Analysis
Data are expressed as mean±SEM. For comparison of means between three or more groups, two-way ANOVA with Bonferroni post-test was applied. For comparisons of means between two groups, two-tailed, unpaired t tests were performed. Prism 5 (GraphPad, La Jolla, CA) was used for statistical analyses.
Disclosures
None.
Supplementary Material
Acknowledgments
J.F., Z.H.L., and J.C.H. are supported by Chinese 973 fund 2012CB517601; J.C.H. is supported by National Institutes of Health (NIH) 1R01-DK078897, NIH 1R01-DK088541, VA Merit Award, and NIH P01-DK56492; P.Y.C. is supported by NIH 1R01-DK098126-01A1.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2015040421/-/DCSupplemental.
References
- 1.Steffes MW, Schmidt D, McCrery R, Basgen JM, International Diabetic Nephropathy Study Group : Glomerular cell number in normal subjects and in type 1 diabetic patients. Kidney Int 59: 2104–2113, 2001 [DOI] [PubMed] [Google Scholar]
- 2.Pagtalunan ME, Miller PL, Jumping-Eagle S, Nelson RG, Myers BD, Rennke HG, Coplon NS, Sun L, Meyer TW: Podocyte loss and progressive glomerular injury in type II diabetes. J Clin Invest 99: 342–348, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Susztak K, Raff AC, Schiffer M, Böttinger EP: Glucose-induced reactive oxygen species cause apoptosis of podocytes and podocyte depletion at the onset of diabetic nephropathy. Diabetes 55: 225–233, 2006 [PubMed] [Google Scholar]
- 4.Petermann AT, Pippin J, Krofft R, Blonski M, Griffin S, Durvasula R, Shankland SJ: Viable podocytes detach in experimental diabetic nephropathy: potential mechanism underlying glomerulosclerosis. Nephron, Exp Nephrol 98: e114–e123, 2004 [DOI] [PubMed] [Google Scholar]
- 5.Kang YS, Li Y, Dai C, Kiss LP, Wu C, Liu Y: Inhibition of integrin-linked kinase blocks podocyte epithelial-mesenchymal transition and ameliorates proteinuria. Kidney Int 78: 363–373, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hodgin JB, Nair V, Zhang H, Randolph A, Harris RC, Nelson RG, Weil EJ, Cavalcoli JD, Patel JM, Brosius FC, 3rd, Kretzler M: Identification of cross-species shared transcriptional networks of diabetic nephropathy in human and mouse glomeruli. Diabetes 62: 299–308, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woroniecka KI, Park AS, Mohtat D, Thomas DB, Pullman JM, Susztak K: Transcriptome analysis of human diabetic kidney disease. Diabetes 60: 2354–2369, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boerries M, Grahammer F, Eiselein S, Buck M, Meyer C, Goedel M, Bechtel W, Zschiedrich S, Pfeifer D, Laloë D, Arrondel C, Gonçalves S, Krüger M, Harvey SJ, Busch H, Dengjel J, Huber TB: Molecular fingerprinting of the podocyte reveals novel gene and protein regulatory networks. Kidney Int 83: 1052–1064, 2013 [DOI] [PubMed] [Google Scholar]
- 9.Grgic I, Hofmeister AF, Genovese G, Berhardy AJ, Sun H, Maarouf OH, Bijol V, Pollak MR, Humphreys BD: Discovery of new glomerular disease-relevant genes by translational profiling of podocytes in vivo. Kidney Int 86: 1116–1129, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuen DA, Stead BE, Zhang Y, White KE, Kabir MG, Thai K, Advani SL, Connelly KA, Takano T, Zhu L, Cox AJ, Kelly DJ, Gibson IW, Takahashi T, Harris RC, Advani A: eNOS deficiency predisposes podocytes to injury in diabetes. J Am Soc Nephrol 23: 1810–1823, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Gasperi R, Rocher AB, Sosa MA, Wearne SL, Perez GM, Friedrich VL, Jr, Hof PR, Elder GA: The IRG mouse: a two-color fluorescent reporter for assessing Cre-mediated recombination and imaging complex cellular relationships in situ. Genesis 46: 308–317, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jim B, Ghanta M, Qipo A, Fan Y, Chuang PY, Cohen HW, Abadi M, Thomas DB, He JC: Dysregulated nephrin in diabetic nephropathy of type 2 diabetes: a cross sectional study. PLoS One 7: e36041, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reidy K, Kang HM, Hostetter T, Susztak K: Molecular mechanisms of diabetic kidney disease. J Clin Invest 124: 2333–2340, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gödel M, Hartleben B, Herbach N, Liu S, Zschiedrich S, Lu S, Debreczeni-Mór A, Lindenmeyer MT, Rastaldi MP, Hartleben G, Wiech T, Fornoni A, Nelson RG, Kretzler M, Wanke R, Pavenstädt H, Kerjaschki D, Cohen CD, Hall MN, Rüegg MA, Inoki K, Walz G, Huber TB: Role of mTOR in podocyte function and diabetic nephropathy in humans and mice. J Clin Invest 121: 2197–2209, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cunard R, Sharma K: The endoplasmic reticulum stress response and diabetic kidney disease. Am J Physiol Renal Physiol 300: F1054–F1061, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daehn I, Casalena G, Zhang T, Shi S, Fenninger F, Barasch N, Yu L, D’Agati V, Schlondorff D, Kriz W, Haraldsson B, Bottinger EP: Endothelial mitochondrial oxidative stress determines podocyte depletion in segmental glomerulosclerosis. J Clin Invest 124: 1608–1621, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dugan LL, You YH, Ali SS, Diamond-Stanic M, Miyamoto S, DeCleves AE, Andreyev A, Quach T, Ly S, Shekhtman G, Nguyen W, Chepetan A, Le TP, Wang L, Xu M, Paik KP, Fogo A, Viollet B, Murphy A, Brosius F, Naviaux RK, Sharma K: AMPK dysregulation promotes diabetes-related reduction of superoxide and mitochondrial function. J Clin Invest 123: 4888–4899, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Breyer MD, Böttinger E, Brosius FC, 3rd, Coffman TM, Harris RC, Heilig CW, Sharma K, AMDCC : Mouse models of diabetic nephropathy. J Am Soc Nephrol 16: 27–45, 2005 [DOI] [PubMed] [Google Scholar]
- 19.Shigehara T, Zaragoza C, Kitiyakara C, Takahashi H, Lu H, Moeller M, Holzman LB, Kopp JB: Inducible podocyte-specific gene expression in transgenic mice. J Am Soc Nephrol 14: 1998–2003, 2003 [DOI] [PubMed] [Google Scholar]
- 20.Smeets B, Uhlig S, Fuss A, Mooren F, Wetzels JF, Floege J, Moeller MJ: Tracing the origin of glomerular extracapillary lesions from parietal epithelial cells. J Am Soc Nephrol 20: 2604–2615, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wanner N, Hartleben B, Herbach N, Goedel M, Stickel N, Zeiser R, Walz G, Moeller MJ, Grahammer F, Huber TB: Unraveling the role of podocyte turnover in glomerular aging and injury. J Am Soc Nephrol 25: 707–716, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mallipattu SK, Liu R, Zhong Y, Chen EY, D’Agati V, Kaufman L, Ma’ayan A, Klotman PE, Chuang PY, He JC: Expression of HIV transgene aggravates kidney injury in diabetic mice. Kidney Int 83: 626–634, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, Chuang PY, D’Agati VD, Dai Y, Yacoub R, Fu J, Xu J, Taku O, Premsrirut PK, Holzman LB, He JC: Nephrin Preserves Podocyte Viability and Glomerular Structure and Function in Adult Kidneys [published online ahead of print February 2, 2015]. J Am Soc Nephrol 10.1681/ASN.2014040405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Durbin R: Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B: Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628, 2008 [DOI] [PubMed] [Google Scholar]
- 26.Wang L, Feng Z, Wang X, Wang X, Zhang X: DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26: 136–138, 2010 [DOI] [PubMed] [Google Scholar]
- 27.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK: limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A: Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14: 128, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J: TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378, 2003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.