Cryo–electron tomography and structural labeling show that the intermediate and light chains of the outer dynein arm (ODA) form a distinct complex, designated ODA-Beak, which can transmit mechanosignals from the nexin–dynein regulatory complex to the heavy chains of ODA.
Abstract
The outer dynein arm (ODA) is a molecular complex that drives the beating motion of cilia/flagella. Chlamydomonas ODA is composed of three heavy chains (HCs), two ICs, and 11 light chains (LCs). Although the three-dimensional (3D) structure of the whole ODA complex has been investigated, the 3D configurations of the ICs and LCs are largely unknown. Here we identified the 3D positions of the two ICs and three LCs using cryo–electron tomography and structural labeling. We found that these ICs and LCs were all localized at the root of the outer-inner dynein (OID) linker, designated the ODA-Beak complex. Of interest, the coiled-coil domain of IC2 extended from the ODA-Beak to the outer surface of ODA. Furthermore, we investigated the molecular mechanisms of how the OID linker transmits signals to the ODA-Beak, by manipulating the interaction within the OID linker using a chemically induced dimerization system. We showed that the cross-linking of the OID linker strongly suppresses flagellar motility in vivo. These results suggest that the ICs and LCs of the ODA form the ODA-Beak, which may be involved in mechanosignaling from the OID linker to the HCs.
INTRODUCTION
Cilia and flagella are conserved motile organelles that play important roles in cellular motility and development of vertebrates (Gibbons, 1981; Hirokawa et al., 2006). The beating motions of cilia and flagella are driven by the outer and inner dynein arms (ODAs and IDAs, respectively). The Chlamydomonas ODA is an ∼2-MDa protein complex composed of three heavy chains (HCs), two ICs, and 11 light chains (LCs; Sakato and King, 2004). The three-dimensional (3D) structure of the ODA complex and the nucleotide-dependent conformational changes in the HCs have been intensively studied by cryo–electron microscopy and tomography (Nicastro et al., 2006; Ishikawa et al., 2007; Oda et al., 2007; Movassagh et al., 2010; Ueno et al., 2012; Lin et al., 2014), and the molecular interactions among ICs and LCs have been investigated using genetics and chemical cross-linking (King et al., 1991, 1995; Mitchell and Kang, 1993; Dibella et al., 2004, 2005). However, the 3D architecture of ICs and LCs remains to be fully elucidated. Although we roughly determined the 3D positions of IC1 and IC2 in the ODA-microtubule cross-bridging complex in a previous study (Oda et al., 2013), it is necessary to locate the ODA subunits in situ in order to determine the precise molecular architecture of ODA in cilia and flagella.
In this study, we found that the ICs and LCs constitute the root of the outer–inner dynein (OID) linker, using cryo–electron tomography and structural labeling. We also investigated the molecular mechanisms responsible for OID linker–mediated regulation of flagellar motility using the rapamycin-based cross-linking of FK506-binding protein 1A (FKBP) and FKBP-rapamycin binding domain (FRB). Our results suggest a possible mechanosignaling pathway from the OID linker to the HCs through a complex of ICs and LCs.
RESULTS AND DISCUSSION
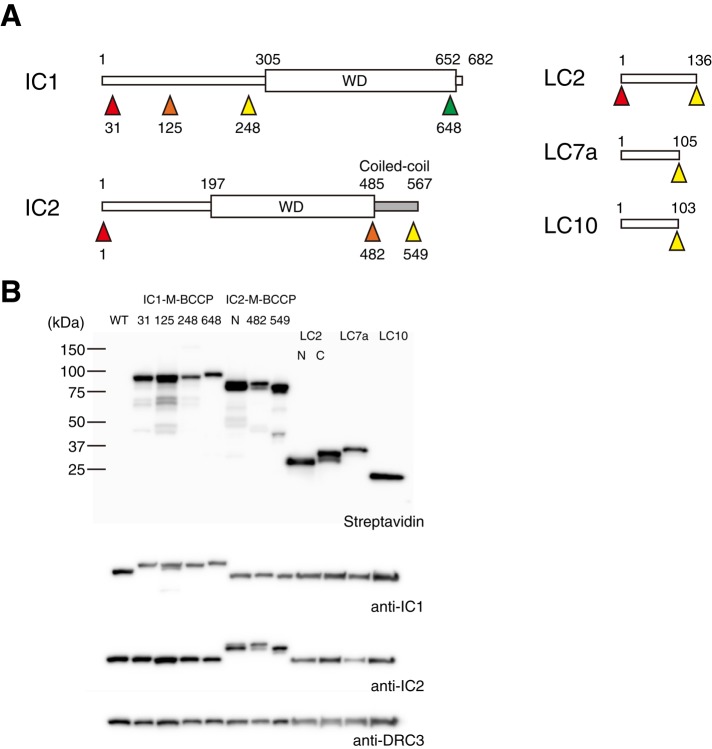
Biotinylation tagging of ICs and LCs
To investigate the 3D configuration of ODA ICs and LCs, we labeled four positions in IC1, three positions in IC2, two positions in LC2, and one position in LC7a and LC10 with acetyl-CoA carboxylase biotin carboxyl carrier protein (BCCP) tags (Figure 1A and Table 1; Furuta et al., 2009; Oda et al., 2013; Oda and Kikkawa, 2013). Positions of the tags on ICs were determined based on the domain organization; IC1 is divided into the amino-terminal (N-terminal) domain and carboxyl-terminal (C-terminal) WD-repeat domain (King et al., 1995; Wilkerson et al., 1995), and IC2 is divided into the N-terminal domain, the middle WD-repeat domain, and the C-terminal coiled-coil domain (Mitchell and Kang, 1991; Ogawa et al., 1995; DiBella et al., 2005). Biotinylation of and streptavidin-binding to BCCP tags were confirmed using immunoblotting and immunofluorescence (Figure 1B and Supplemental Figure S1A). The varied signal intensities of the immunoblots and immunofluorescence among the rescued strains suggest substoichiometric expression and/or labeling of tagged ICs and LCs (Figure 1B and Supplemental Figure S1A). However, wild-type motility of the rescued strains (Table 1) suggests that the expression of tagged ICs and LCs functionally restored defects of IC- and LC-missing mutants.
FIGURE 1:
BCCP-tagging of ICs and LCs. (A) Domain organization of ICs and LCs. Arrowheads indicate positions of BCCP tags. Numbers indicate amino acid residues. Both IC1 and IC2 have WD repeat domains (WD), and the C-terminal domain of IC2 is predicted to form a coiled-coil (Lupas et al., 1991). Colors of arrowheads correspond to colors of label densities in Figure 2B. (B) Immunoblots of axonemal proteins separated by SDS–PAGE and probed with various antibodies. All BCCP-tagged proteins were properly expressed and biotinylated.
TABLE 1:
Strains used in this study.
| Strain | Abbreviation | Mutated gene | Missing structure | Swimming speed (μm/s) | Beat frequency (Hz) | ODA occupancy (%) | Reference |
|---|---|---|---|---|---|---|---|
| Wild-type CC-125 | 174.8 ± 12.4 | 60 ± 7 | 98.9 | ||||
| oda4-s7 | βHC (truncated) | 65.3 ± 8.1 | 35 ± 4 | ND | |||
| oda6 | IC2 | ODA | 60.9 ± 5.6 | 29 ± 3 | 0 | Kamiya (1988), Mitchell and Kang (1991) | |
| oda9 | IC1 | ODA | 62.1 ± 6.4 | 30 ± 4 | ND | Kamiya (1988), Wilkerson et al. (1995) | |
| oda12-1 | LC2, LC10 | ODA (reduced) | 64.9 ± 4.2 | 31 ± 4 | ND | Pazour et al. (1999), Furuta et al. (2009) | |
| oda12-2 | LC2 | ODA (reduced) | 81.5 ± 6.1 | 40 ± 5 | ND | Pazour et al. (1999) | |
| oda15 | LC7a | ODA (reduced), IDA f (reduced) | 73.8 ± 5.8 | 36 ± 4 | 19a | DiBella et al. (2004) | |
| ida6 | DRC2 | N-DRC (partial) | 77.6 ± 4.7 | 71 ± 8 | ND | Kato et al. (1993) | |
| oda9-IC1-M31BCCP | IC1-M31 | 170.2 ± 15.1 | 60 ± 7 | 99.3 | |||
| oda9-IC1-M125BCCP | IC1-M125 | 176.4 ± 13.3 | 60 ± 6 | 99.0 | |||
| oda9-IC1-M248BCCP | IC1-M248 | 175.1 ± 15.9 | 61 ± 6 | 99.5 | |||
| oda9-IC1-M648BCCP | IC1-M648 | 171.7 ± 19.2 | 60 ± 7 | 99.1 | |||
| oda6-IC2-NBCCP | IC2-N | 98.1 ± 10.6 | 74 ± 7 | 99.4 | Oda et al. (2013) | ||
| oda6-IC2-M482BCCP | IC2-M482 | 169.5 ± 14.2 | 60 ± 5 | 99.0 | |||
| oda6-IC2-M549BCCP | IC2-M549 | 171.3 ± 15.5 | 60 ± 5 | 99.3 | |||
| oda12-2-LC2-NBCCP | LC2-N | 167.8 ± 11.4 | 59 ± 6 | 98.8 | |||
| oda12-2-LC2-CBCCP | LC2-C | 170.9 ± 18.7 | 61 ± 5 | 99.3 | |||
| oda15-LC7a-CBCCP | LC7a-C | 178.0 ± 12.4 | 60 ± 5 | 99.1 | |||
| oda12-1-LC2-LC10-CBCCP | LC10-C | 170.6 ± 16.4 | 60 ± 5 | 99.4 | |||
| oda6-IC2-NFRB | IC2-NFRB | 120.7 ± 11.0 | 65 ± 6 | ND | |||
| ida6-DRC2-MFKBP | DRC2-MFKBP | 181.0 ± 19.7 | 60 ± 5 | ND | |||
| oda6-IC2-NFRB ida6-DRC2-MFKBP | IC2-NFRB/DRC2-MFKBP | 116.9 ± 13.9 | 65 ± 6 | ND | |||
| oda6-IC2-NFKBP ida6-DRC2-MFKBP | IC2-NFKBP/DRC2-MFKBP | 115.6 ± 10.2 | 65 ± 6 | ND |
Swimming speed: means ± SEM were calculated from 20 cells. Beat frequency: means ± SEM were calculated from >500 cells. ODA occupancy: presence of ODAs along DMTs was examined in tomograms, and ODA occupancy was calculated as (number of ODA)/(number of ODA + number of ODA-missing gaps). The total of >5-μm-long axonemes was examined for each strain. ND, not determined.
aODA occupancy of oda15 was estimated from the biochemically determined value in DiBella et al. (2004).
ICs and LCs form the ODA-Beak complex
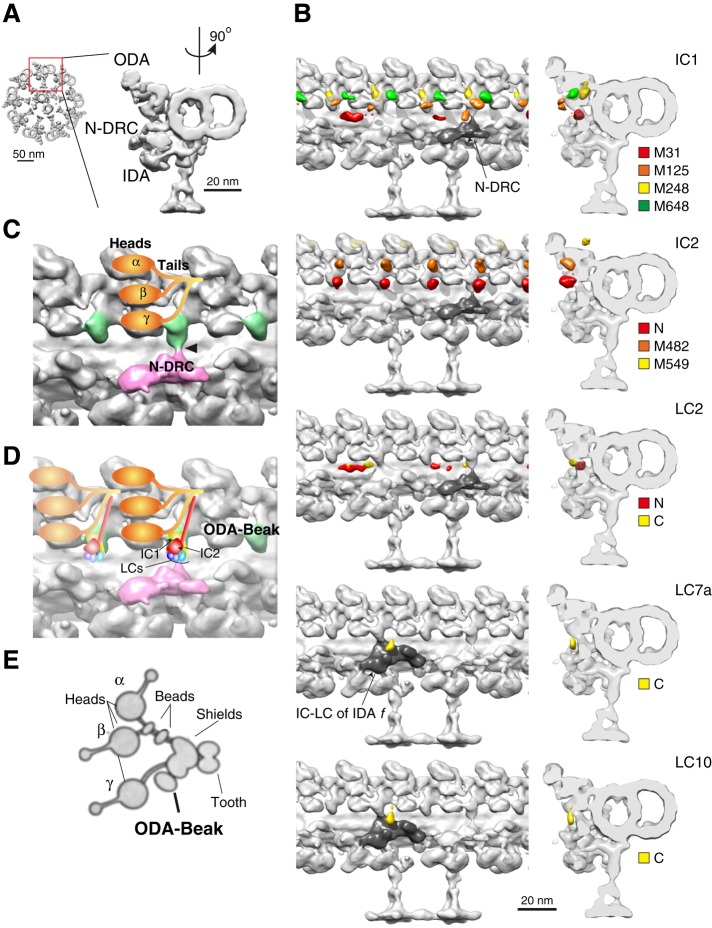
Next we identified the 3D positions of the BCCP tags on ICs and LCs, using cryo–electron tomography and structural labeling (Figure 2, A and B; Oda et al., 2013; Oda and Kikkawa, 2013). Surprisingly, all of the ICs and LCs were located around the root of the OID linker (Figure 2C, green). We designated this region the ODA-Beak complex, based on the bouquet structure of the isolated ODA (Figure 2, D and E, modified from Figure 2a of Goodenough and Heuser, 1984).
FIGURE 2:
Structural labeling of ICs and LCs. (A) The 3D structures of the axoneme. Left, tip-to-base view of the 9+2 structure. Right, enlarged view of one of the DMTs. The 90°-rotated views of the DMT are shown on the left of B. (B) The 3D localizations of the labels on ICs and LCs. Arrowheads indicate positions of slices on the right. Colored densities indicate positions of streptavidin labels. Colors of the label densities correspond to colors of arrowheads in Figure 1A. Position of IC2-M549 differed from that in our previous result (Oda et al., 2013). We believe that our previous localization of the C-terminus of IC2 was an artifact due to flexibility of the ODA–microtubule complex. N-DRC and IC-LC complex of IDA f are indicated (gray). (C, D) Structural configuration of ODA and N-DRC. (C) Approximate positions of α, β, and γ HCs (orange) based on previous reports (Nicastro et al., 2006; Ishikawa et al., 2007; Oda et al., 2007; Movassagh et al., 2010; Ueno et al., 2012; Lin et al., 2014). Ovals and lines indicate head and tail domains of HCs, respectively. Arrowhead indicates one of the OID linkers (OID linker 3a) bridging between ODA and N-DRC (pink). (D) Possible 3D configuration of ICs and LCs. Red structure represents IC2, composed of WD repeat (yellow frustum) and coiled-coil (yellow rod). Yellow structure behind IC2 represents IC1, composed of WD repeat (red frustum) and the N-terminal domain (red rod). Three small ovals below IC2 represent LCs. (C, D) ICs and LCs form the ODA-Beak complexes (green). (E) Diagram modified from Figure 2a of Goodenough and Heuser (1984), showing the bouquet structure of the isolated ODA. Annotations of subunits were taken from the same figure, except for assignments of the α, β, and γ HCs.
The position of IC1 is of interest because IC1 reportedly binds to the outer doublet microtubules (DMTs; King et al., 1991, 1995). Although we were unable to detect densities bridging between the IC1 and DMT (Supplemental Figure S2A and Supplemental Movie S1), the label densities of IC1-M248 were located on the DMT-facing side of the ODA-Beak (Figure 2B, IC1, yellow). Because the junction between the N-terminal domain and WD-repeat domain of IC1 has been proposed to be a possible DMT-binding region (King et al., 1995), the ODA-Beak can be connected to DMT via the middle segment of IC1, which is not visible on our electron density map probably because the DMT-binding domain of IC1 is either flexible or thin.
The N-terminal domain of IC2 is reported to be essential for the assembly of LC2, LC6, and LC9, as the oda6-r88 mutant, which has a sequence alteration in residues 31–54 of IC2 (Mitchell and Kang, 1993), forms an ODA that lacks the three LCs (DiBella et al., 2005). In agreement with these previous biochemical and genetic analyses, labels on LC2 were located in close proximity to the N-terminus of IC2 (Figure 2B, Supplemental Movie S2, IC2, red; Oda et al., 2013; Oda and Kikkawa, 2013). Considering the LC deficiency in oda6-r88 (DiBella et al., 2005), our results suggest that the ODA-Beak is composed of IC1, IC2, LC2, LC6, LC7a, LC9, and LC10.
In contrast, the densities of IC2-M549 were located away from the ODA-Beak and were observed on the outer surface of the ODA (Figure 2B, IC2, yellow). Because the C-terminal domain of IC2 is predicted to form a coiled-coil (Lupas et al., 1991; DiBella et al., 2005), the C-terminal domain of IC2 is likely to take an extended conformation from the ODA-Beak to the tail domains of α and β HCs (Figure 2, C and D; Movassagh et al., 2010; Lin et al., 2014). This model is intriguing because the OID linker was previously shown to modulate ODA activity (Oda et al., 2013). Our results suggest that the ODA-Beak transmits signals from the OID linker to HCs via the C-terminal coiled-coil domain of IC2.
We attempted to investigate the role of the C-terminal domain of IC2 by generating a partial deletion mutant of IC2, but we were unable to rescue the motility and ODA-assembly defects of the oda6 (IC2-deficient mutant) strain by expressing the coiled-coil-deleted IC2 (unpublished data), suggesting that the C-terminal coiled-coil of IC2 is essential for ODA assembly.
Because one ODA contains one copy of IC1, IC2, LC2, and LC7a and two copies of LC10 (King and Witman, 1989; King and Kamiya, 2009; King, 2011; Bowman et al., 1999; DiBella et al., 2004), there should be at least four label densities within one 96-nm repeat of DMT (e.g., labels on IC2 in Figure 2B). However, some labels on IC1 and LCs were invisible in a subset of ODAs, which seems to depend on the relative position to OID linkers (Figure 2B). For example, the label densities of IC1-M31 (Figure 2B, IC1, red) and LC2-N (Figure 2B, LC2, red) appeared on the distal side of the ODA-Beaks that were not connected to OID linkers. On the other hand, the label densities of LC2-C (Figure 2B, LC2, yellow) appeared on the proximal side of the ODA-Beaks that were connected to the OID linkers. The label densities of LC7a-C and LC10-C (Figure 2B, LC7a, LC10, yellow) appeared only on the distal side of the ODA-Beak that were connected to the ODA-IDA f linker (the OID linker 1; Bui et al., 2012).
To examine whether these variations in labeling result from substoichiometric expression of tagged IC and LC and, we quantified the occupancy of ODAs along DMTs of wild-type and rescued strains (Table 1). Because absence of IC and LC results in ODA assembly defects (Kamiya, 1988; Mitchell and Kang, 1999; Wilkerson et al., 1995; Pazour et al., 1999; DiBella et al., 2004), insufficient expression of tagged ICs and LCs must be detected as gaps in the arrays of ODAs in tomograms. However, we observed few gaps in the rescued strains, as well as in wild-type cells. These results suggest that tagged ICs and LCs were sufficiently expressed to rescue ODA assembly defects.
We suppose that these variations in labeling can be attributed to flexibility of the labeled domains, substoichiometric labeling caused by steric hindrance, and/or limited signal-to-noise ratio of tomograms. Precise positions of the unlabeled domains of ICs and LCs remain to be determined with other methods.
Cross-linking of ODA-Beak and nexin–dynein regulatory complex using the FKBP-rapamycin-FRB system
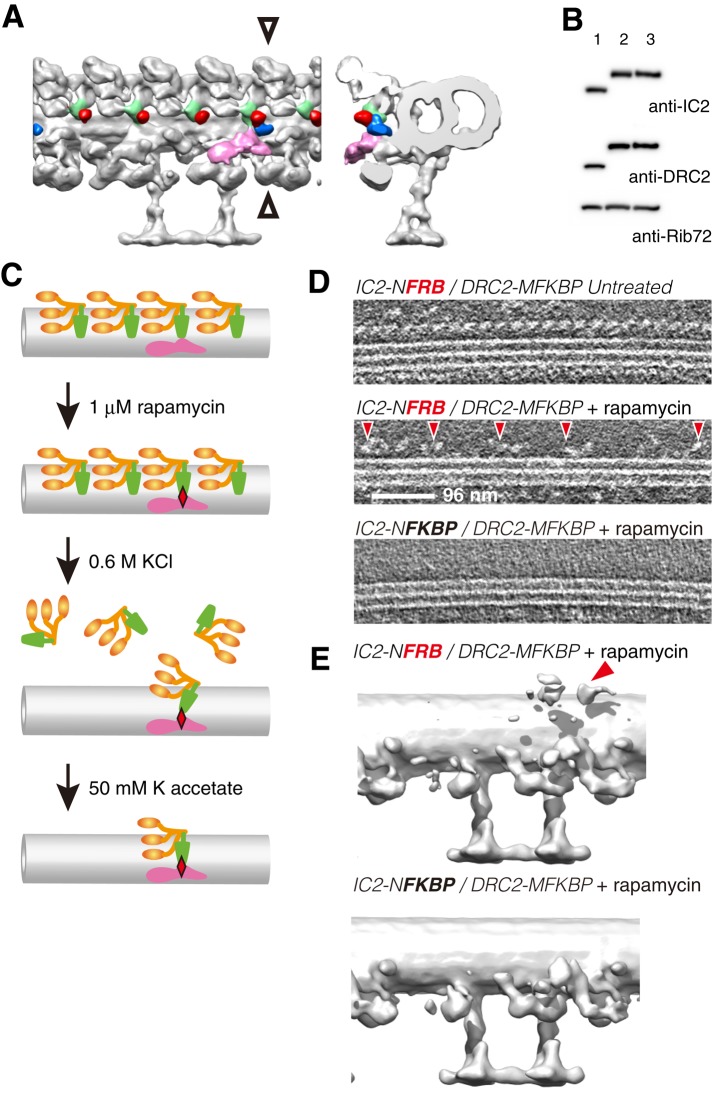
To investigate the role of the ODA-Beak in the regulation of flagellar motility, we focused on the OID linker between ODA-Beak and nexin–dynein regulatory complex (N-DRC; Figure 2C, arrowhead, OID linker 3a; Bui et al., 2012). We examined our previous electron microscopy data and found that the N-terminus of IC2 is located in close proximity to the middle segment of DRC2 (Figure 3A; Oda et al., 2013, 2015; Oda and Kikkawa, 2013). We hypothesized that we could manipulate the signal transmission from N-DRC to ODA-Beak in vivo if we cross-linked IC2 and DRC2 using rapamycin-based cross-linking of FKBP and FRB (Rivera et al., 1996).
FIGURE 3:
Cross-bridging of ODA-Beak and N-DRC using FKBP-rapamycin-FRB system. (A) Positions of N-terminus of IC2 (red) and middle segment of DRC2 (blue) reported previously (Oda et al., 2013, 2015). ODA-Beak and N-DRC are colored green and pink, respectively. (B) Immunoblots of axonemal proteins probed with various antibodies. Lane 1, wild-type; lane 2, IC2-NFRB/DRC2-MFKBP; and lane 3, IC2-NFKBP/DRC2-MFKBP. Both IC2 and DRC2 were properly tagged with either FRB or FKBP. (C) Schematic of verification of cross-linking between ODA and N-DRC. Axonemes were treated with 1 μM rapamycin to cross-link ODA-Beak (green) and N-DRC (pink). Red diamond represents cross-linking with rapamycin between IC2-NFRB and DRC2-MFKBP molecules. Next most ODAs were extracted with 0.6 M KCl, but one-fourth of ODAs were expected to remain anchored to DMTs via NFRB-rapamycin-FKBP ternary complexes. Decreasing ionic strength to 50 mM potassium acetate allowed reattachment of anchored ODAs to DMTs (Takada et al., 1992). (D) Tomogram slices showing ODAs on DMTs (top) In untreated axonemes, ODAs were aligned along DMTs with 24-nm periodicity. Center, in IC2-NFRB/DRC2-MFKBP axonemes, rapamycin treatment followed by 0.6 M KCl extraction removed most ODAs, but a subset of ODAs remained attached to DMTs with ∼96-nm periodicity (red arrowheads). Bottom, in IC2-NFKBP/DRC2-MFKBP axonemes, rapamycin treatment followed by 0.6 M KCl extraction removed all the ODAs, as rapamycin does not induce homodimerization of FKBP. (E) Averaged subtomograms of axonemes treated with rapamycin and KCl. Top, DMT structure of IC2-NFRB/DRC2-MFKBP axonemes showed densities of ODA anchored to N-DRC (red arrowhead). In IC2-NFKBP/DRC2-MFKBP axonemes, all ODAs were dissociated from DMTs.
We inserted human FKBP (Harding et al., 1989) after His-245 of DRC2 and added the FRB of human rapamycin target 1 (RAPT1; Chiu et al., 1994) to the N-terminus of IC2. We generated the IC2-NFRB/DRC2-MFKBP strain, in which rapamycin cross-links IC2 and DRC2. As a negative control, we also generated the IC2-NFKBP/DRC2-MFKBP strain, which has FKBP tags on both IC2 and DRC2, so that rapamycin treatment does not cross-link IC2 and DRC2 (Figure 3B and Table 1). To verify that ODA and N-DRC were cross-linked by the FKBP-rapamycin-FRB ternary complex (Choi et al., 1996), we extracted ODAs after cross-linking IC2-NFRB and DRC2-MFKBP with rapamycin (Figure 3, C and E, and Supplemental Figure S3A). We found that one ODA every 96 nm remained attached to the DMT (Figure 3D, arrowheads), and cryo–electron tomography confirmed that the remaining ODA is located adjacent to N-DRC (Figure 3D, arrowhead). These results indicate that we successfully cross-linked ODA-Beak and N-DRC by IC2-NFRB-rapamycin-DRC2-MFKBP heterodimerization.
In vivo cross-linking of ODA-Beak and N-DRC suppressed flagellar motility
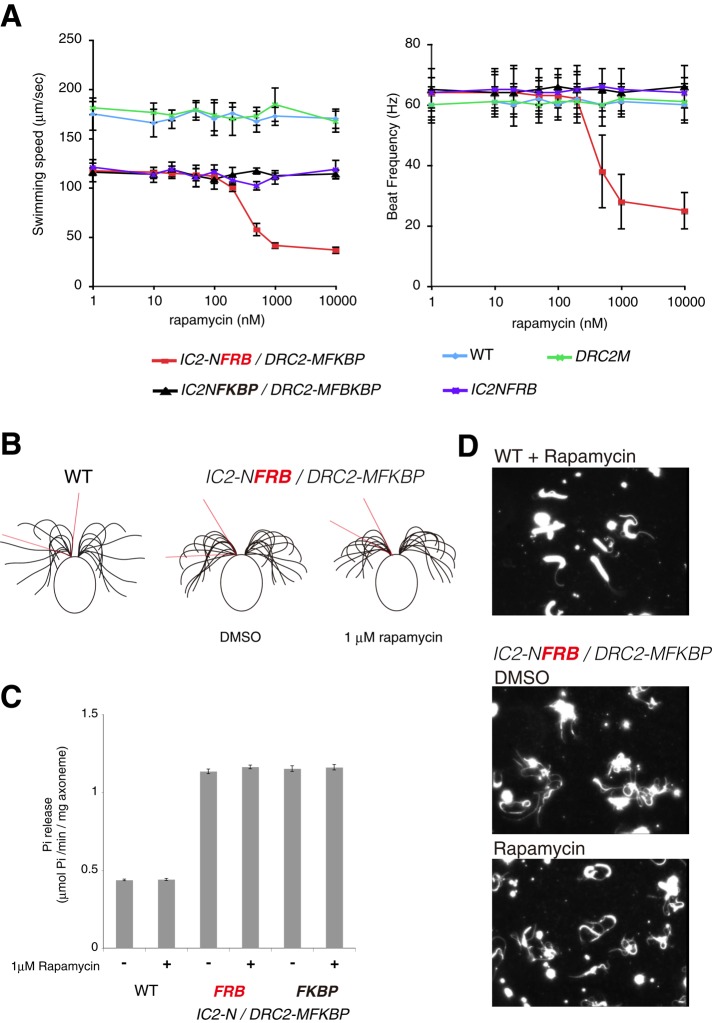
We examined the effects of ODA-Beak and N-DRC cross-linking on cell motility by treating live cells of IC2-NFRB/DRC2-MFKBP with rapamycin (Figure 4A). Although rapamycin is known to suppress the growth of Chlamydomonas (Crespo et al., 2005), it did not affect the motility of wild-type and IC2-NFKBP/DRC2M-FKBP cells within 5–30 min of observation (Figure 4A). Of interest, rapamycin treatment suppressed the motility of IC2-NFRB/DRC2-MFKBP cells in a dose-dependent manner. At 1 μM rapamycin, swimming speed and beat frequency decreased by 65 and 55%, respectively. The half-maximal inhibitory concentrations of rapamycin on the swimming speed and beat frequency were ∼360 and ∼460 nM, respectively. Rapamycin-dependent decreases in beat frequency suggest that cross-linking between ODA-Beak and N-DRC affected ODA activity (Kamiya and Okamoto, 1985; Brokaw and Kamiya, 1987). Next we analyzed the flagellar waveforms of IC2-NFRB/DRC2-MFKBP cells and found that rapamycin-treatment decreased the amplitude of beating (Figure 4B), suggesting that the cross-linking also affects IDA activity (Brokaw and Kamiya, 1987). These results agreed with our previous results that the OID linker works as a hub controller for ODA and IDA activities (Oda et al., 2013). We suppose that the interaction between ODA-Beak and N-DRC needs to be dynamic in order to allow constitutive cross-linking of the two structures to disrupt the regulation of ODA and IDA activities.
FIGURE 4:
Effects of cross-linking between ODA-Beak and N-DRC. (A) Motility analyses of live cells. Only IC2-NFRB/DRC2-MFKBP cells showed rapamycin-dependent decreases in motility. Means ± SEM were calculated from 20 cells. (B) Waveforms of wild-type and IC2-NFRB/DRC2-MFKBP cells after dimethyl sulfoxide (DMSO) and rapamycin treatments. Rapamycin treatment reduced wave amplitude (red lines) compared with DMSO-treated control. (C) ATPase activities of axonemes. In accordance with previous results (Oda et al., 2013), addition of tags to the N-terminus of IC2 alone caused hyperactivation of axonemal ATPase activities. Rapamycin treatment did not affect elevated ATPase activities. Means ± SEM were calculated from 10 measurements. (D) Sliding disintegration assays. Axonemes were disintegrated by incubation with 1 mM ATP and 0.3 μg/ml nagarse for 1 min. In accordance with previous results (Oda et al., 2013), addition of tags to the N-terminus of IC2 enhanced sliding disintegration. Rapamycin treatment (1 μM) did not affect DMT sliding activity.
Note that the N-terminal tagging of IC2 alone reduced the swimming speed by 33% and slightly increased beat frequency by 8% (Figure 4A, IC2-NFRB), probably due to changes in the interaction between ODA and N-DRC (Oda et al., 2013). We analyzed the effects of rapamycin on ATPase and microtubule-sliding activities of IC2-NFRB/DRC2-MFKBP axonemes, but both activities were up-regulated, regardless of rapamycin treatment (Figure 4, C and D, and Supplemental Figure S3B; Oda et al., 2013). These results suggest that the effects of cross-linking between the ODA-Beak and N-DRC are detectable in in vivo live-cell experiments, but, in contrast, the effects are masked by hyperactivation of ODA activity caused by the N-terminal tagging of IC2 on in vitro biochemical assays.
Remaining questions about the DMT-binding region of ODA
Localization of IC1, IC2, LC2, LC7a, and LC10 in ODA-Beak indicates that the rest of the ODA subdomains are composed of the remaining LCs, three HCs, and docking complex (DC). Although DC is reportedly to form a 24-nm-long, oval-shaped structure (Owa et al., 2014), our recent results suggest that DC takes on a flexible conformation and does not create a clear density on the averaged subtomogram of DMT (Oda et al., 2016). In addition, Ichikawa et al. (2015) recently reported that LC1 locates at the stalk head domain of γ HC. Considering the low molecular weights of other LCs, it is likely that most of the remaining ODA subdomains are composed of the three HCs. The approximate positions of the head domains of HCs have been defined based on their characteristic ring-shaped structure (Burgess et al., 2003; Nicastro et al., 2006; Movassagh et al., 2010; Lin et al., 2014). However, the positions of HC tail domains are not well defined, particularly their N-terminal segments. We observed the oda4-s7 axoneme, which lacks the head domain of β HC (Sakakibara, 1993), and found a distinct DMT-binding region (Supplemental Figure S2, blue), which may correspond to the “density B” found in the mouse respiratory cilia structure (Ueno et al., 2014). We believe that this DMT-binding region is mainly composed of the HC tail domain, based on the assumption stated earlier. It is of great interest which HCs/LCs comprise the DMT-binding site (Supplemental Figure S2, B and C, arrowheads).
MATERIALS AND METHODS
Strains and reagents
Chlamydomonas reinhardtii wild-type strain CC-125 cells were grown in Tris-acetate-phosphate (TAP) medium. To screen transformants, cells were grown on TAP agar supplemented with paromomycin (10 μg/ml; Sigma-Aldrich, St. Louis, MO) or hygromycin B (20 μg/ml: Nacalai Tesque, Kyoto, Japan). The C. reinhardtii strains used in this study are listed in Table 1. Anti-IC2 antibody 1869A was purchased from Sigma-Aldrich, anti-DRC3 antibody was generated in a previous study (Oda et al., 2015), and anti-Rib72 was a kind gift from R. Kamiya (Gakushuin University, Tokyo, Japan; Ikeda et al., 2003). cDNA sequence encoding the amino acids 1–261 of IC1 was inserted into the pGEX-6p-2 plasmid, and polypeptides were expressed in Escherichia coli cells. Anti-IC1 rabbit polyclonal antibodies were then raised against the purified proteins.
Preparation of axonemes
Chlamydomonas cells were deflagellated with dibucaine-HCl (Wako Pure Chemical Industries, Tokyo, Japan), and axonemes were collected by centrifugation (Piperno et al., 1977). Flagella were demembranated with 1% Nonidet P-40 in HMDENa buffer or HMDEK buffer composed of 30 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid–NaOH, pH 7.2, 5 mM MgCl2, 1 mM dithiothreitol, 1 mM ethylene glycol tetraacetic acid, 50 mM NaCl or 50 mM CH3COOK, and 1× Protease Inhibitor Cocktail (Nacalai Tesque).
Electrophoresis and immunoblotting
Axonemal proteins were resolved by SDS–PAGE on 5–15% polyacrylamide gradient gels (Nacalai Tesque) and blotted onto polyvinylidene difluoride membranes. Blots were probed with streptavidin conjugated with horseradish peroxidase (Thermo Scientific, Rockford, IL) or the indicated primary antibodies.
Construction of the expression vectors
Expression plasmids for IC2, DRC2, and IC2-NBCCP were as described previously (Oda et al., 2013, 2015). Fragments spanning from the start codon to immediately before the stop codon for genes encoding IC1, LC2, LC7a, and LC10 were amplified with genomic PCR using genomic DNA from the wild-type strain CC-125 and then inserted into pIC2 plasmids (Oda et al., 2015). We inserted the tag sequence corresponding to amino acids 141–228 of Chlamydomonas BCCP in the middle of the sequences of IC1 (between Ile-31 and Pro-32, Asp-125 and Met-126, Val-248 and Pro-249, and Pro-648 and Glu-649) and IC2 (between Thr-482 and Gly-483, and Thr-549 and Thr-550) and added tags to the N-terminus of LC2 and the C-termini of LC2, LC7a, and LC10. For expression plasmids of IC2-NFRB and IC2-NFKBP, we added the codon-optimized cDNA sequence of human FKBP and FRB domain of human RAPT1 (residues 2021–2113), respectively, to the N-terminus of IC2. For expression plasmid of DRC2-MFKBP, we inserted the codon-optimized cDNA sequence of FKBP between His-245 and Arg-246 of DRC2. At the junctions between the DRC2 and FKBP-tag sequence, we inserted eight–amino acid linker sequences (Lys-Gly-Ser-Gly-Ser-Gly-Ser-Gly and Lys-Ser-Ala-Lys-Ala-Ser-Ala-Ser).
Fluorescence microscopy detection of axonemes
For fluorescence staining using streptavidin, demembranated axonemes were attached to glass slides and blocked with 1 mg/ml bovine serum albumin (BSA) in HMDEK buffer. Axonemes were incubated with 1 μg/ml Alexa Fluor 546–conjugated streptavidin (Invitrogen, Carlsbad, CA) for 1 min. Labeled axonemes were washed three times with HMDEK buffer and observed using a fluorescence microscope (IX60; Olympus, Tokyo, Japan). Images were recorded using a charge-coupled device (CCD) camera (ORCA-R2; Hamamatsu Photonics, Hamamatsu, Japan).
Sample preparation for cryo–electron tomography
Streptavidin–cytochrome c labeling of BCCP-tagged axonemes was carried out as described previously (Oda et al., 2014). Demembranated axonemes were incubated with 0.05 mg/ml streptavidin for 15 min at 4°C in HMDENa buffer. Axonemes were then washed with HMDENa buffer and incubated with 0.05 mg/ml biotinylated cytochrome c for 15 min at 4°C in the presence of 1 mg/ml BSA. Next axonemes were washed and again incubated with streptavidin. Labeled or unlabeled axonemes were resuspended in HMDEK buffer at a concentration of 0.02 mg/ml and mixed with an equal amount of 15-nm colloidal gold suspension conjugated with BSA (Aurion, Wageningen, Netherlands).
For rapamycin cross-linking experiments, axonemes were incubated with 1 μM rapamycin (Wako Pure Chemicals) in HMDEK for 10 min. Treated axonemes were centrifuged and resuspended in HMDE plus 0.6 M KCl buffer and incubated for 30 min. Axonemes were then centrifuged and resuspended in HMDEK plus 0.1 μM rapamycin.
Home-made holey carbon grids were glow-discharged for 20 s. Suspended axonemes plus colloidal gold (5 μl) were loaded onto the grids and plunge-frozen in liquid ethane at −180°C with a Leica EM GP automated plunge-freezing device (Leica Microsystems, Wetzlar, Germany).
Image acquisition
Grids were transferred to a JEM-3100FEF transmission electron microscope (JEOL, Tokyo, Japan) with a Gatan 914 high-tilt liquid nitrogen cryo-transfer holder (Gatan, Pleasanton, CA). Tilt series images were recorded at −180°C using a TemCam-F416 CMOS camera (TVIPS, Gauting, Germany), and automated acquisition was performed using the EM-TOOLs program (TVIPS). The angular range of the tilt series was from −60 to 60° with 2.0° increments. The total electron dose was limited to ∼100 e−/Å2. Images were recorded at 300 keV, with 6- to 9-μm defocus, at a magnification of 25,700× and a pixel size of 6 Å. An in-column omega energy filter was used to enhance image contrast in the zero-loss mode with a slit width of 20 eV.
Image processing
Image processing for subtomogram averaging of DMT structures was carried out as described previously (Oda and Kikkawa, 2013; Oda et al., 2014). Tilt series images were aligned and back-projected to reconstruct 3D tomograms using the IMOD software package (Kremer et al., 1996). Tomograms of intact axonemes with a high signal-to-noise ratio were selected and used for subtomogram averaging of the 96-nm repeats of DMTs. Alignment and averaging of subtomograms were conducted using custom Ruby-Helix scripts (Metlagel et al., 2007) and the PEET software suite (Nicastro et al., 2006). The numbers of DMT subtomograms averaged were as follows: 755 for wild type; 848 for IC1-M31; 1104 for IC1-M125; 984 for IC1-M482; 504 for IC1-M648; 940 for IC2-M482; 1168 for IC2-M549; 1160 for LC2-C; 720 for LC7a-C; and 948 for LC10-C. The effective resolutions determined by Fourier shell correlation with a cutoff value of 0.5 were within 4.5–5.0 nm (Supplemental Figure S1B).
Surface renderings were generated using UCSF Chimera (Pettersen et al., 2004). The electron microscopy maps of averaged DMT are available at the EM Data Bank (www.emdatabank.org) under the accession numbers EMD-6515-6524.
Statistical analysis
To identify statistically significant differences, we applied Student’s t test to compare wild-type and streptavidin-labeled axonemes as described previously (Oda and Kikkawa, 2013; Oda et al., 2014). First, wild-type and streptavidin-labeled subtomograms were randomly divided into three data sets. Subtomograms for each data set were aligned and averaged, and a total of six averaged subtomograms were created. We calculated the t value for each voxel and presented it as a single t-value map. The isosurface threshold values were t > 7.17, with a one-tailed probability of <0.1%.
Measurements of swimming velocity and beat frequency
The swimming velocity of Chlamydomonas cells was recorded using an inverted CKX41 microscope (Olympus) at a total magnification of 100×. A red filter with a cutoff wavelength of 630 nm was inserted before the condenser lens to suppress the cellular response to light. The beat frequency of flagella was measured as described previously (Kamiya, 2000). Briefly, fluctuations in the intensity of microscopic images of swimming cells were analyzed using a photodetector, and spectra were generated using fast Fourier transform (FFT). The position of the peak was considered to be the average value, and the SD was obtained from the shape of the peak, fitted with a Gaussian curve. In a typical experiment at a total magnification of 100×, ∼500–1000 cells contributed to one FFT spectrum.
Waveform analysis
Chlamydomonas cells were observed using a dark-field microscope (BX53; Olympus), and images were captured using a high-speed digital camera (EXILIM EX-F1; Casio, Tokyo, Japan) at 600 frames/s. Cells whose flagella were clearly in focus were selected, and the shapes of flagella were manually traced using Illustrator (Adobe).
ATPase assay
The rate of phosphate release by axonemes was measured using Biomol Green reagent (Enzo Life Sciences, Farmingdale, NY). Axonemes (0.1 mg/ml) were incubated for 5 min in HMDEK buffer in the presence of 1 mM ATP. Released phosphate concentrations were calculated based on changes in absorbance at 620 nm.
Sliding disintegration of the axoneme
Axonemes were absorbed onto a glass slide, and sliding disintegration was initiated with HMDEK buffer containing 1 mM ATP and 0.3 μg/ml nagarse. Sliding of doublet microtubules was observed using a dark-field microscope (BX53; Olympus) equipped with a 40× oil-immersion objective lens and a 100-W mercury lamp. Image sequences were recorded using an electron multiplying CCD (ADT-33S; FLOVEL, Tokyo, Japan).
Supplementary Material
Acknowledgments
This work was supported by CREST, the Japan Science and Technology Agency (to M.K.), the Kazato Research Foundation (to T.O.), the Takeda Science Foundation (to M.K. and T.O.), Japan Society for the Promotion of Science KAKENHI Grant 15642352 (to T.O.), and the Institute for Fermentation, Osaka (to T.O.).
Abbreviations used:
- HC
heavy chain
- LC
light chain
- N-DRC
nexin-dynein regulatory complex
- ODA
outer dynein arm
- OID linker
outer-inner dynein linker.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-10-0723) on February 10, 2016.
REFERENCES
- Bowman AB, Patel-King RS, Benashski SE, McCaffery JM, Goldstein LS, King SM. Drosophila roadblock and Chlamydomonas LC7: a conserved family of dynein-associated proteins involved in axonal transport, flagellar motility, and mitosis. J Cell Biol. 1999;146:165–180. [PMC free article] [PubMed] [Google Scholar]
- Brokaw CJ, Kamiya R. Bending patterns of Chlamydomonas flagella: IV. Mutants with defects in inner and outer dynein arms indicate differences in dynein arm function. Cell Motil Cytoskeleton. 1987;8:68–75. doi: 10.1002/cm.970080110. [DOI] [PubMed] [Google Scholar]
- Bui KH, Yagi T, Yamamoto R, Kamiya R, Ishikawa T. Polarity and asymmetry in the arrangement of dynein and related structures in the Chlamydomonas axoneme. J Cell Biol. 2012;198:913–925. doi: 10.1083/jcb.201201120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess SA, Walker ML, Sakakibara H, Knight PJ, Oiwa K. Dynein structure and power stroke. Nature. 2003;421:715–718. doi: 10.1038/nature01377. [DOI] [PubMed] [Google Scholar]
- Chiu MI, Katz H, Berlin V. RAPT1, a mammalian homolog of yeast Tor, interacts with the FKBP12/rapamycin complex. Proc Natl Acad Sci USA. 1994;91:12574–12578. doi: 10.1073/pnas.91.26.12574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Chen J, Schreiber SL, Clardy J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- Crespo JL, Diaz-Troya S, Florencio FJ. Inhibition of target of rapamycin signaling by rapamycin in the unicellular green alga Chlamydomonas reinhardtii. Plant Physiol. 2005;139:1736–1749. doi: 10.1104/pp.105.070847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiBella LM, Gorbatyuk O, Sakato M, Wakabayashi K, Patel-King RS, Pazour GJ, Witman GB, King SM. Differential light chain assembly influences outer arm dynein motor function. Mol Biol Cell. 2005;16:5661–5674. doi: 10.1091/mbc.E05-08-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiBella LM, Sakato M, Patel-King RS, Pazour GJ, King SM. The LC7 light chains of Chlamydomonas flagellar dyneins interact with components required for both motor assembly and regulation. Mol Biol Cell. 2004;15:4633–4646. doi: 10.1091/mbc.E04-06-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta A, Yagi T, Yanagisawa HA, Higuchi H, Kamiya R. Systematic comparison of in vitro motile properties between Chlamydomonas wild-type and mutant outer arm dyneins each lacking one of the three heavy chains. J Biol Chem. 2009;284:5927–5935. doi: 10.1074/jbc.M807830200. [DOI] [PubMed] [Google Scholar]
- Gibbons IR. Cilia and flagella of eukaryotes. J Cell Biol. 1981;91:107s–124s. doi: 10.1083/jcb.91.3.107s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U, Heuser J. Structural comparison of purified dynein proteins with in situ dynein arms. J Mol Biol. 1984;180:1083–1118. doi: 10.1016/0022-2836(84)90272-9. [DOI] [PubMed] [Google Scholar]
- Harding MW, Galat A, Uehling DE, Schreiber SL. A receptor for the immunosuppressant FK506 is a cis-trans peptidyl-prolyl isomerase. Nature. 1989;341:758–760. doi: 10.1038/341758a0. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Tanaka Y, Okada Y, Takeda S. Nodal flow and the generation of left-right asymmetry. Cell. 2006;125:33–45. doi: 10.1016/j.cell.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Ichikawa M, Saito K, Yanagisawa HA, Yagi T, Kamiya R, Yamaguchi S, Yajima J, Kushida Y, Nakano K, Numata O, et al. Axonemal dynein light chain-1 locates at the microtubule binding domain of the gamma heavy chain. Mol Biol Cell. 2015;26:4236–4247. doi: 10.1091/mbc.E15-05-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Brown JA, Yagi T, Norrander JM, Hirono M, Eccleston E, Kamiya R, Linck RW. Rib72, a conserved protein associated with the ribbon compartment of flagellar A-microtubules and potentially involved in the linkage between outer doublet microtubules. J Biol Chem. 2003;278:7725–7734. doi: 10.1074/jbc.M210751200. [DOI] [PubMed] [Google Scholar]
- Ishikawa T, Sakakibara H, Oiwa K. The architecture of outer dynein arms in situ. J Mol Biol. 2007;368:1249–1258. doi: 10.1016/j.jmb.2007.02.072. [DOI] [PubMed] [Google Scholar]
- Kamiya R. Mutations at twelve independent loci result in absence of outer dynein arms in Chlamydomonas reinhardtii. J Cell Biol. 1988;107:2253–2258. doi: 10.1083/jcb.107.6.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya R. Analysis of cell vibration for assessing axonemal motility in Chlamydomonas. Methods. 2000;22:383–387. doi: 10.1006/meth.2000.1090. [DOI] [PubMed] [Google Scholar]
- Kamiya R, Okamoto M. A mutant of Chlamydomonas reinhardtii that lacks the flagellar outer dynein arm but can swim. J Cell Sci. 1985;74:181–191. doi: 10.1242/jcs.74.1.181. [DOI] [PubMed] [Google Scholar]
- Kato T, Kagami O, Yagi T, Kamiya R. Isolation of two species of Chlamydomonas reinhardtii flagellar mutants, ida5 and ida6, that lack a newly identified heavy chain of the inner dynein arm. Cell Struct Funct. 1993;18:371–377. doi: 10.1247/csf.18.371. [DOI] [PubMed] [Google Scholar]
- King SM. Composition and assembly of axonemal dyneins. In: King SM, editor. Dyneins: Structure, Biology and Disease. Boston: Academic Press; 2011. pp. 208–243. [Google Scholar]
- King SM, Kamiya R. Axonemal dyneins: assembly, structure, and force generation. In: Witman GB, editor. The Chlamydomonas Sourcebook. 2nd ed. Vol. 3. Oxford, UK: Academic Press; 2009. pp. 144–145. [Google Scholar]
- King SM, Patel-King RS, Wilkerson CG, Witman GB. The 78,000-M(r) intermediate chain of Chlamydomonas outer arm dynein is a microtubule-binding protein. J Cell Biol. 1995;131:399–409. doi: 10.1083/jcb.131.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SM, Wilkerson CG, Witman GB. The Mr 78,000 intermediate chain of Chlamydomonas outer arm dynein interacts with alpha-tubulin in situ. J Biol Chem. 1991;266:8401–8407. [PubMed] [Google Scholar]
- King SM, Wilkerson CG, Witman GB. The Mr 78,000 intermediate chain of Chlamydomonas outer arm dynein interacts with alpha-tubulin in situ. J Biol Chem. 1991;266:8401–8407. [PubMed] [Google Scholar]
- King SM, Witman GB. Molecular structure of Chlamydomonas outer arm dynein. In: Warner FD, Satir P, Gibbons IR, editors. Cell Movement. The Dynein ATPases. Vol. 1. New York: Alan R. Liss; 1989. pp. 61–75. [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Lin J, Okada K, Raytchev M, Smith MC, Nicastro D. Structural mechanism of the dynein power stroke. Nat Cell Biol. 2014;16:479–485. doi: 10.1038/ncb2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Metlagel Z, Kikkawa YS, Kikkawa M. Ruby-Helix: an implementation of helical image processing based on object-oriented scripting language. J Struct Biol. 2007;157:95–105. doi: 10.1016/j.jsb.2006.07.015. [DOI] [PubMed] [Google Scholar]
- Mitchell DR, Kang Y. Identification of oda6 as a Chlamydomonas dynein mutant by rescue with the wild-type gene. J Cell Biol. 1991;113:835–842. doi: 10.1083/jcb.113.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DR, Kang Y. Reversion analysis of dynein intermediate chain function. J Cell Sci. 1993;105:1069–1078. doi: 10.1242/jcs.105.4.1069. [DOI] [PubMed] [Google Scholar]
- Movassagh T, Bui KH, Sakakibara H, Oiwa K, Ishikawa T. Nucleotide-induced global conformational changes of flagellar dynein arms revealed by in situ analysis. Nat Struct Mol Biol. 2010;17:761–767. doi: 10.1038/nsmb.1832. [DOI] [PubMed] [Google Scholar]
- Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313:944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- Oda T, Abe T, Yanagisawa H, Kikkawa M. Docking complex-independent alignment of outer dynein arms with 24-nm periodicity in vitro. J Cell Sci. 2016 doi: 10.1242/jcs.184598. (in press) [DOI] [PubMed] [Google Scholar]
- Oda T, Hirokawa N, Kikkawa M. Three-dimensional structures of the flagellar dynein-microtubule complex by cryoelectron microscopy. J Cell Biol. 2007;177:243–252. doi: 10.1083/jcb.200609038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T, Kikkawa M. Novel structural labeling method using cryo-electron tomography and biotin-streptavidin system. J Struct Biol. 2013;183:305–311. doi: 10.1016/j.jsb.2013.07.003. [DOI] [PubMed] [Google Scholar]
- Oda T, Yagi T, Yanagisawa H, Kikkawa M. Identification of the outer-inner dynein linker as a hub controller for axonemal dynein activities. Curr Biol. 2013;23:656–664. doi: 10.1016/j.cub.2013.03.028. [DOI] [PubMed] [Google Scholar]
- Oda T, Yanagisawa H, Kikkawa M. Detailed structural and biochemical characterization of the nexin-dynein regulatory complex. Mol Biol Cell. 2015;26:294–304. doi: 10.1091/mbc.E14-09-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T, Yanagisawa H, Yagi T, Kikkawa M. Mechanosignaling between central apparatus and radial spokes controls axonemal dynein activity. J Cell Biol. 2014;204:807–819. doi: 10.1083/jcb.201312014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa K, Kamiya R, Wilkerson CG, Witman GB. Interspecies conservation of outer arm dynein intermediate chain sequences defines two intermediate chain subclasses. Mol Biol Cell. 1995;6:685–696. doi: 10.1091/mbc.6.6.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owa M, Furuta A, Usukura J, Arisaka F, King SM, Witman GB, Kamiya R, Wakabayashi K. Cooperative binding of the outer arm-docking complex underlies the regular arrangement of outer arm dynein in the axoneme. Proc Natl Acad Sci USA. 2014;111:9461–9466. doi: 10.1073/pnas.1403101111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Koutoulis A, Benashski SE, Dickert BL, Sheng H, Patel-King RS, King SM, Witman GB. LC2, the chlamydomonas homologue of the t complex-encoded protein Tctex2, is essential for outer dynein arm assembly. Mol Biol Cell. 1999;10:3507–3520. doi: 10.1091/mbc.10.10.3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Piperno G, Huang B, Luck DJ. Two-dimensional analysis of flagellar proteins from wild-type and paralyzed mutants of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1977;74:1600–1604. doi: 10.1073/pnas.74.4.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera VM, Clackson T, Natesan S, Pollock R, Amara JF, Keenan T, Magari SR, Phillips T, Courage NL, Cerasoli F, Jr, et al. A humanized system for pharmacologic control of gene expression. Nat Med. 1996;2:1028–1032. doi: 10.1038/nm0996-1028. [DOI] [PubMed] [Google Scholar]
- Sakakibara H. A Chlamydomonas outer arm dynein mutant with a truncated beta heavy chain. J Cell Biol. 1993;122:653–661. doi: 10.1083/jcb.122.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakato M, King SM. Design and regulation of the AAA+ microtubule motor dynein. J Struct Biol. 2004;146:58–71. doi: 10.1016/j.jsb.2003.09.026. [DOI] [PubMed] [Google Scholar]
- Takada S, Sakakibara H, Kamiya R. Three-headed outer arm dynein from Chlamydomonas that can functionally combine with outer-arm-missing axonemes. J Biochem. 1992;111:758–758. doi: 10.1093/oxfordjournals.jbchem.a123832. [DOI] [PubMed] [Google Scholar]
- Ueno H, Bui KH, Ishikawa T, Imai Y, Yamaguchi T, Ishikawa T. Structure of dimeric axonemal dynein in cilia suggests an alternative mechanism of force generation. Cytoskeleton (Hoboken) 2014;71:412–422. doi: 10.1002/cm.21180. [DOI] [PubMed] [Google Scholar]
- Ueno H, Ishikawa T, Bui KH, Gonda K, Ishikawa T, Yamaguchi T. Mouse respiratory cilia with the asymmetric axonemal structure on sparsely distributed ciliary cells can generate overall directional flow. Nanomedicine. 2012;8:1081–1087. doi: 10.1016/j.nano.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Wilkerson CG, King SM, Koutoulis a, Pazour GJ, Witman GB. The 78,000 M(r) intermediate chain of Chlamydomonas outer arm dynein is a WD-repeat protein required for arm assembly. J Cell Biol. 1995;129:169–178. doi: 10.1083/jcb.129.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.