Clathrin adaptors link cargo to the clathrin coat. The clathrin adaptor Ent5 is also required for the maturation of clathrin coats at the trans-Golgi network or endosome, suggesting that it plays a key mechanistic role in coat formation. This function requires only the Ent5 clathrin-binding sites and not its interaction with other endosomal adaptors.
Abstract
Clathrin is a ubiquitous protein that mediates membrane traffic at many locations. To function, clathrin requires clathrin adaptors that link it to transmembrane protein cargo. In addition to this cargo selection function, many adaptors also play mechanistic roles in the formation of the transport carrier. However, the full spectrum of these mechanistic roles is poorly understood. Here we report that Ent5, an endosomal clathrin adaptor in Saccharomyces cerevisiae, regulates the behavior of clathrin coats after the recruitment of clathrin. We show that loss of Ent5 disrupts clathrin-dependent traffic and prolongs the lifespan of endosomal structures that contain clathrin and other adaptors, suggesting a defect in coat maturation at a late stage. We find that the direct binding of Ent5 with clathrin is required for its role in coat behavior and cargo traffic. Surprisingly, the interaction of Ent5 with other adaptors is dispensable for coat behavior but not cargo traffic. These findings support a model in which Ent5 clathrin binding performs a mechanistic role in coat maturation, whereas Ent5 adaptor binding promotes cargo incorporation.
INTRODUCTION
Clathrin-dependent traffic is a central facet of all eukaryotic cell biology. It mediates traffic at multiple locations, including endocytic traffic that originates at the plasma membrane and endosomal traffic that originates at trans-Golgi network (TGN) or at the endosomes (reviewed in Brodsky et al., 2001). It regulates nearly every aspect of cellular behavior through effects on the localization of transmembrane, extracellular, and organellar proteins (reviewed in McMahon and Boucrot, 2011). To perform these many different functions, the clathrin coat must bind to many different protein cargoes and package these into transport carriers. This cargo selection is performed by clathrin adaptors.
There are more than a dozen different clathrin adaptors encoded in the genomes of most eukaryotes (reviewed in Owen et al., 2004). Each clathrin adaptor acts as a complex interaction hub. In addition to binding to transmembrane cargo, most bind phospholipids, small GTPases, or other membrane-associated proteins that confer specificity to adaptor recruitment. Adaptors also directly interact with clathrin, the major structural component of the clathrin coat. This interaction links transmembrane cargo to the forming transport carrier. This function is the minimum definition of a clathrin adaptor. Many adaptors also perform additional mechanistic roles in the coat, such as bending membranes or stimulating clathrin polymerization (Ahle and Ungewickell, 1986; Morgan et al., 2000; Ford et al., 2002; Kalthoff et al., 2002; Kelly et al., 2014; Miller et al., 2015; Skruzny et al., 2015). However, it is unclear whether all adaptors perform such central mechanistic roles or whether some act solely as linkers between cargo and clathrin. Determining which adaptors perform mechanistic roles and what those roles are is important to understanding the regulation of clathrin function in vivo.
Elucidating the roles of clathrin adaptors at the TGN and early and late endosomes has been particularly challenging due, in part, to difficulties in imaging individual transport events and because cells can adapt to disruption in clathrin-dependent traffic by up-regulating other pathways (Seeger and Payne, 1992). In the yeast Saccharomyces cerevisiae, five clathrin adaptors are known to function at the TGN and endosomes: the heteromeric AP-1 complex; the Golgi localized γ-adaptins, which are encoded by the paralogues GGA1 and GGA2; Ent3, a protein belonging to the ENTH-A subfamily of epsins; and Ent5, a protein belonging to the ENTH-D subfamily of epsins (Rad et al., 1995; Boman et al., 2000; Dell’Angelica et al., 2000; Hirst et al., 2000; Duncan et al., 2003; Costaguta et al., 2006; De Craene et al., 2012). Genetic studies suggest that AP-1 and Gga proteins act in distinct pathways (Boman et al., 2000). Ent3 appears to act exclusively with Ggas (Costaguta et al., 2006; Daboussi et al., 2012). In contrast, the role of Ent5 has been unclear. Deletion of Ent5 causes only minor defects in traffic, suggesting that its role may be minor or cargo specific (Costaguta et al., 2006). However, it localizes to every Gga2 or AP-1 structure in vivo and is required for maximal Gga2 interaction with clathrin (Daboussi et al., 2012; Hung et al., 2012). These data suggest that it has a more central role.
To clarify the function of Ent5, we reexamined the role of Ent5 in endosomal/TGN traffic using new approaches. Using a quantitative assay of endosomal traffic, we find that loss of Ent5 impairs endosomal traffic. In addition, removal of Ent5 prolongs the lifespan of clathrin coats after Gga2 and clathrin are recruited, indicating a defect after coat assembly initiates. We find that the direct interaction of Ent5 with clathrin is required for its role in coat behavior and cargo traffic, whereas direct interaction of Ent5 with Gga2 or AP-1 is important for Ent5′s function but not the turnover of Gga2-containing structures. Taken together, these results suggest that clathrin binding by Ent5 plays a key mechanistic role in the maturation of Gga2-containing transport carriers, whereas adaptor binding by Ent5 does not.
RESULTS
Ent5 provides a central function in endosomal clathrin-dependent traffic
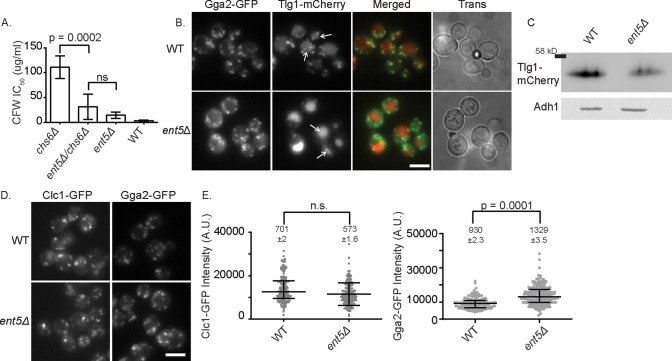
Previous research implicated Ent5 functions in endosomal/TGN traffic; however, it was unclear whether Ent5 was a specialized cargo-specific adaptor or played a central mechanistic role in endosomal/TGN clathrin-dependent traffic (ECT; Costaguta et al., 2006; Copic et al., 2007). To better understand its role in ECT, we tested whether the deletion of ENT5 impaired ECT, using a quantitative calcofluor white (CFW) sensitivity assay. This assay measures the fidelity of ECT by making intracellular retention of the chitin synthase Chs3 dependent on ECT. When ECT is defective, some Chs3 is found at the cell surface in cells lacking CHS6, whereas, in otherwise wild-type cells, all Chs3 is retained intracellularly when CHS6 is deleted (Valdivia et al., 2002). Cell surface Chs3 makes the cells sensitive to CFW. We found that deletion of ENT5 increased the CFW sensitivity of cells lacking CHS6 (Figure 1A). As a further test of the role of Ent5 in ECT, we examined the localization of the soluble N-ethylmaleimide–sensitive factor attachment protein receptor Tlg1 in cells lacking Ent5. In wild-type cells, Tlg1-mCherry was found in the vacuole and in punctate structures that colocalized with Gga2-GFP, consistent with the known localization of Tlg1 at the TGN. In contrast, in cells lacking Ent5, Tlg1-mCherry puncta were rarer and dimmer than in wild-type cells (Figure 1B). Furthermore, steady-state levels of Tlg1 are lower in cells lacking Ent5, suggesting that Tlg1 is missorted to the vacuole and degraded in these cells (Figure 1C). These results are consistent with the loss of Ent5 causing a defect in ECT but do not distinguish between cargo-specific or central mechanistic roles for Ent5.
FIGURE 1:
Loss of Ent5 disrupts traffic at the TGN and/or endosomes. (A) Loss of Ent5 disrupts traffic of Chs3. Loss of Ent5 increases the sensitivity of cells lacking Chs6 to the cell wall–binding toxin CFW. The IC50 for indicated cells. Error bars indicate SD; p values were determined by Student’s t test. (B) Loss of Ent5 disrupts localization of Tlg1-mCherry. Micrographs show Z-stack projections of Gga2-GFP, expressed from its endogenous locus, and Tlg1-mCherry, expressed from its own promoter on a 2-μ plasmid in wild-type and ent5Δ cells. Arrows indicate vacuoles visible in phase-contrast image. (C) Loss of Ent5 reduces steady-state expression levels of Tlg1-mCherry. Lysates prepared from cells expressing Tlg1-mCherry from its own promoter on a 2-μ plasmid in wild-type cells and ent5Δ cells were subjected to immunoblot analysis. (D) Loss of Ent5 does not alter the fluorescence intensity of Chc1-GFP structures and increases the fluorescence intensity of Gga2-GFP. Z-stack projections of indicated cells expressing Chc1-GFP and Gga2-GFP from their endogenous loci. (E) Fluorescence intensity measurements of structures in C. Scatterplots display mean value and SEM; p values were calculated using a two-tailed Mann–Whitney U test for the null hypothesis that the medians were equal. Horizontal bars indicate median and interquartile ranges. Scale bars, 5 μm.
To distinguish between a cargo-specific or central mechanistic role, we investigated whether Ent5 alters the maximal recruitment of Gga2 or clathrin heavy chain (Chc1). To do this, we monitored the intensity of Clc1–green fluorescent protein (GFP) and Gga2-GFP expressed from their endogenous loci in wild type and cells lacking Ent5. Surprisingly, the intensity of Clc1-GFP was unaffected by deletion of ENT5, suggesting that Ent5 is not required for the maximal recruitment of clathrin (Figure 1D). Similarly, Gga2-GFP was recruited to punctate structures in the absence of Ent5. However, the intensity of Gga2 structures was increased 1.4-fold in cells lacking Ent5. These results suggest that Ent5 is not required for the association of clathrin and Gga2 with membranes.
The increased intensity of Gga2 caused by loss of Ent5 is similar to the increased intensity of endocytic proteins caused by loss of endocytic epsins (Maldonado-Baez et al., 2008). In endocytosis, this increased intensity is caused by the stalling of endocytic events after the initiation of coat formation. To test whether the increased intensity of Gga2 could be explained by a stalling of endosomal coat formation, we monitored the kinetics of coat assembly. To do this, we performed time-lapse microscopy on fluorescently labeled Chc1 and Gga2. In wild-type cells, these structures show a relatively stereotypical behavior: fluorescence intensity of Gga2 and Chc1 increased steadily to a maximum point and then decreased rapidly (Figure 2, B and C). This behavior is believed to reflect the assembly of one or a coordinated assembly of multiple clathrin coats on an endosomal organelle followed by the rapid disassembly of the coat(s) or the rapid movement of the vesicle(s) after the complete formation of the vesicle(s) (Daboussi et al., 2012; Hung et al., 2012). The mean lifespan of such structures, defined as the time point when the structure was first visible over background to when it was no longer visible, was 24 s for Gga2-GFP and 23 s for Chc1–red fluorescent protein (RFP; Figure 2D). In contrast, in cells lacking Ent5, this stereotypical behavior was perturbed. The rate of fluorescence intensity increase was less uniform, and the mean lifespan of the structures was 11 s longer for Gga2 and 14 s longer for Chc1. This increase in lifespan was largely due to an increase in the fraction of events with very long lifespans (>30 s; Figure 2E). The increased lifespan is unlikely to be caused by reduced movement of the structures into or out of the plane of focus, because the mean speed of movement of structures determined from mean square displacement measurements was indistinguishable from that for wild type (unpublished data). These results suggest that loss of Ent5 causes a defect in coat formation after clathrin and Gga2 recruitment but before coat disassembly.
FIGURE 2:
Loss of ENT5 prolongs coat lifespan. (A) Representative images of structures analyzed by live-cell microscopy. Boxed regions indicate representative structures selected for analysis. (B) Kymographs of representative structures. Each time frame is 3 s. (C) Intensity vs. time plots of selected structures. Gray lines are the traces of each individual structure. Red lines correspond to Spot1 and blue lines to Spot2 in B. Black lines are the average of 10 independent structures. (D) Quantification of the lifespan of Gga2-containing structures (top) and classification of structures based on their lifespan (bottom). Scatter charts display mean value and SEM; horizontal bars indicate median and interquartile ranges. Data are representative of two independent experiments. p values reflect a two-tailed Mann–Whitney test. Scale bar, 5 μm.
Multiple domains of Ent5 contribute to its function and localization
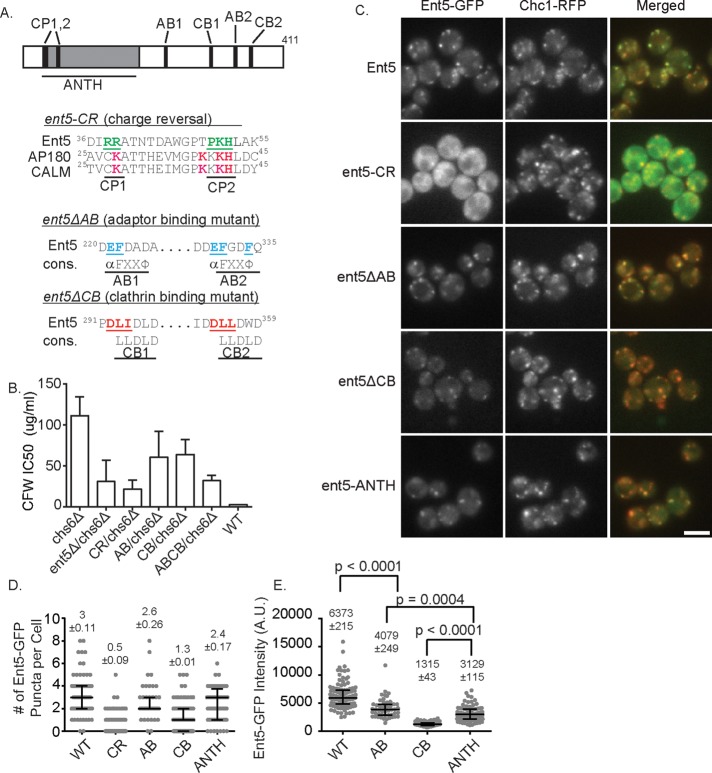
The extension in lifespan of Gga2 and Chc1 structures in cells lacking Ent5 suggests that Ent5 acts as more than a cargo linker. To better understand the role of Ent5 in ECT, we investigated the importance of different Ent5 domains and motifs in Ent5 function. To do this, we mutated each of the known domains and/or motifs in Ent5 (Figure 3A). ENT5 encodes an N-terminal ANTH domain. This domain is believed to bind cargo and/or lipids. To disrupt the function of the ANTH domain, we mutated several positively charged residues that are predicted to lie on the surface of the ANTH domain and are similar to residues that interact with lipids in other ANTH domains to generate a Ent5 ANTH-domain charge-reversal mutant (Ent5-CR; Ford et al., 2002; Sun et al., 2005). Ent5 also binds the γ-ear of clathrin adaptors Gga2 and AP-1. We disrupted this activity by mutating key acidic and hydrophobic residues of the highly conserved γ-ear interaction motif to generate an Ent5 adaptor-binding mutant (Ent5ΔAB; Nogi et al., 2002; Duncan et al., 2003; Mills et al., 2003). Finally, Ent5 contains a pair of clathrin box motifs that mediate interaction with clathrin in many proteins (Dell’Angelica, 2001). We disrupted clathrin binding by mutating key residues of each clathrin box to generate an Ent5 clathrin-binding mutant (Ent5ΔCB; Figure 4A). When expressed from the endogenous ENT5 locus, each of the mutant proteins was expressed at the same level as wild-type Ent5 (Supplemental Figure S1, A–C). However, each of the mutant alleles reduced the functional activity of Ent5 as assessed using the quantitative CFW assay (Figure 3B), suggesting that each activity contributes to Ent5 function in ECT.
FIGURE 3:
Schematic of Ent5 and mutations generated. (A) Top, ANTH domain is indicated in gray, charged patches predicted to be important for ANTH function are indicated as CP1 and CP2, adaptor-binding sites are indicated as AB1 and AB2, and clathrin boxes are indicated as CB1 and CB2. Bottom, residues mutated are underlined. For ent5-CR, alignment with rat AP180 and CALM is shown. Residues important for phosphoinositide-binding residues in AP180 and CALM are red; underlined residues were mutated to glutamic acid. For ent5ΔAB and ent5ΔCB alignment, the consensus sequence is shown (cons); underlined residues were mutated to alanine. (B) CFW sensitivity of indicated mutants. p = 0.0034 for chs6Δ vs. ent5Δ, p = 0.0004 for chs6Δ vs. LB, p = 0.0374 for chs6Δ vs. AB, p = 0.0174 for chs6Δ vs. CB, p = 0.0005 for chs6Δ vs. ABCB, and p < 0.0001 for chs6Δ vs. wild type. Error bars indicate SD; p values were determined using Student’s t test. (C) Z-stack projection of Ent5-GFP and Chc1-RFP in indicated mutants. (D) Quantification of the number of Ent5-GFP puncta per cell in a central plane. (E) Fluorescence intensity measurements of individual puncta in indicated cells. Scatterplots display mean value and SEM; horizontal bars indicate median and interquartile ranges. Scale bar, 5 μm. p values reflect a two-tailed Mann–Whitney test.
FIGURE 4:
Clathrin is required for maximal interaction of Ent5 with Gga2 and localization of Ent5 to membranes. (A) A representative immunoprecipitation reaction. GFP-tagged Ent5 was immunoprecipitated from cell lysates, and the immunoprecipitates were probed with antibodies to Ent5, Gga2, and clathrin to monitor the effect of Ent5 mutations on the Ent5–Gga2 and Ent5–clathrin interactions. Apparent size shift in mutant proteins may be due to differences in surfactant binding, as previously observed for other mutations that alter charge (Shi et al., 2008; Rath et al., 2009; De Zutter et al., 2013). Quantification of the effects of Ent5 mutations on (B) Ent5–Gga2 interaction or (C) Ent5–Chc1 interaction (n = 3). Error bars indicate SD; p values determined with Student’s t test. (D) Chc1-box mutation blocks interaction of Ent5 with clathrin. Clc1 was immunoprecipitated from lysates of cells lacking CHC1 transformed with plasmids that contain wild-type CHC1 or chc1Δbox. (E) Live-cell imaging of CHC1-deleted cells transformed with plasmids that contain wild-type CHC1 or chc1Δbox mutant. Scale bar, 5 μm.
We next investigated the effects of each mutation on the localization of Ent5. To do this, we expressed GFP-fusion proteins from the endogenous loci. We first confirmed that the addition of the GFP tag did not interfere with protein function, using the quantitative CFW assay (Supplemental Figure S1D). We then assessed Ent5 localization by counting the GFP puncta per cell in a central plane (Figure 3, C and D). Each mutation altered the number of Ent5 puncta per cell in the central plane; however, the magnitude of the effect differed substantially. The ent5-CR mutation had the strongest effect. It increased the percentage of cells with no central-plane Ent5 puncta from 1.4% in wild-type to 69% in mutant cells (Figure 3C). Furthermore, the mean number of puncta per cell in a central plane was reduced to less than one in the mutant cells from three in wild-type cells. To determine whether the loss of Ent5 puncta reflected a loss of endosomes, we monitored endosomes using clathrin as an endosomal marker. Clathrin structures were abundant in these cells, suggesting the mutation prevents endosomal localization of Ent5 rather than disrupts endosomal structures.
Similar to the effects of ent5-CR, the ent5ΔCB mutation increased the percentage of cells with no puncta to 32% and reduced the mean number of puncta per cell to one without altering clathrin localization. The ent5ΔAB mutation, which mutates the two motifs that interact with Gga2 and AP-1, had the weakest defect. It did not significantly alter the percentage of cells with no puncta and reduced the mean number of puncta per cell only slightly, from 3 to 2.6. Taken together, these results demonstrate that the ANTH domain and clathrin-binding motifs play important roles in Ent5 localization to clathrin-rich structures.
We next analyzed the effect of the mutations on the amount of Ent5 recruited to each puncta (Figure 3E). We were unable to perform this analysis on the Ent5-CR cells due the low fluorescence intensity of this mutant allele. We found that the mean intensity of Ent5ΔCB structures was reduced 4.8-fold compared with wild type. In contrast, the mean intensities of Ent5ΔAB structures were only modestly reduced 1.6-fold. Taken together, these results demonstrate that the ent5-CR and ent5ΔCB mutations perturb either the recruitment or persistence of Ent5 in clathrin-rich structures, whereas the ent5ΔAB mutation does not substantially perturb its localization.
The strong effect of the ent5ΔCB mutation on Ent5 localization suggests that clathrin binding plays a key role in Ent5 localization. The finding that the effect of ent5ΔAB mutation was less severe than the effect of the ent5ΔCB mutation is surprising because the affinity of Ent5 for Gga2 is predicted to be at least five times higher than the affinity of Ent5 for clathrin (Miele et al., 2004; Fang et al., 2010; Zhuo et al., 2015). Therefore, based on just the predicted protein interaction affinity, the ent5ΔAB allele should have a stronger effect than the ent5ΔCB allele. However, we previously reported that clathrin binding by Gga2 enhances Gga2 binding to Ent5, suggesting that clathrin binding can promote or stabilize the interaction between adaptors (Hung et al., 2012). To test whether clathrin stabilizes the interaction between Ent5 and Gga2, we performed coimmunoprecipitation analysis. We found that Ent5ΔCB coimmunoprecipitated less Gga2 than wild-type Ent5. This suggests that the ent5ΔCB allele reduces the interaction of Ent5 with Gga2, in addition to reducing the interaction with clathrin (Figure 4, A–C). In contrast, the ent5ΔAB allele only reduced the interaction with Gga2 and did not interfere with the interaction with clathrin. The stronger effect of the ent5ΔCB allele on localization is thus consistent with a stronger effect of this allele on physical interactions with the coat.
Surprisingly, we found that Ent5ΔCB coimmunoprecipitated a small amount of clathrin, whereas Ent5ΔAB coimmunoprecipitated a small amount of Gga2. Because these alleles were designed to ablate each activity entirely, we suspected that this interaction might be indirect. For example, Ent5ΔAB could interact with Gga2 indirectly via clathrin. To test this possibility, we investigated the interactions of Ent5 that lacked both adaptor- and clathrin-binding motifs (ABCB) with clathrin and Gga2. This allele reduced clathrin and Gga2 binding to undetectable levels, suggesting that, for the Ent5ΔAB and Ent5ΔCB proteins, the unexpected interactions are indirect.
The strong effect of the clathrin binding on Ent5 localization is not consistent with a previous report that the N-terminal ANTH domain of Ent5 was sufficient for localization (Costaguta et al., 2006). We investigated this apparent contradiction in two ways. First, we reanalyzed the localization of the ANTH domain. We found that the ANTH domain was found in fewer puncta per cell in a central plane and that these puncta are dimmer than wild-type Ent5 (Figure 3, C–E). This suggests that the ANTH domain is not sufficient for maximal Ent5 localization. Second, as an independent confirmation of the importance of clathrin binding in Ent5 localization, we monitored Ent5 localization in cells expressing a version of clathrin that lacks the interaction site for proteins like Ent5 (chc1Δbox) as the only version of clathrin (Collette et al., 2009). We first confirmed that the chc1Δbox protein does not interact with Ent5 by coimmunoprecipitation (Figure 4D). We then investigated the localization of Ent5 in this strain. We found that Ent5 was localized to fewer puncta in the chc1Δbox cells and that the puncta were dimmer (Figure 4E). This reduction of Ent5 localization was not due to a gross defect in endosomal structures, since Gga2 structures were abundant in these cells. Taken together, these results strongly suggest that ANTH-domain functions and clathrin binding play a pivotal role in Ent5 function and localization, whereas adaptor binding is important for Ent5 function but is less important for localization.
Ent5 clathrin binding promotes turnover of Gga2 structures in vivo
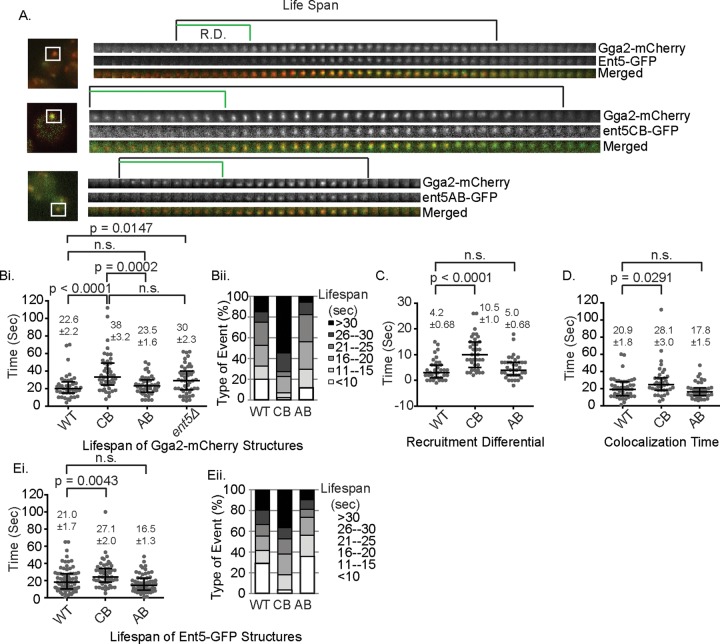
To better understand how the Ent5 mutations perturb ECT, we monitored the effect of ENT5 mutations on the behavior of endosomal clathrin coats using Gga2-mCherry. The ent5ΔCB allele extended the lifespan of Gga2 structures by a mean of 5 s, which is indistinguishable from lifespan extension observed in cells lacking Ent5 entirely (Figure 5, A and B). In contrast, the ent5ΔAB allele did not alter the lifespan of Gga2 structures. These results indicate that the interaction between Ent5 and clathrin regulates the behavior of endosomal clathrin coats, whereas the interaction between Ent5 and Gga2 is dispensable for the normal lifespan of clathrin coats.
FIGURE 5:
Mutation of clathrin-binding or adaptor-binding domains affects the lifespan of Ent5-Gga2 structures and the timing of the recruitment of Ent5. (A) Representative images of the events analyzed. R.D., recruitment differential. The region highlighted by the box indicates the area in the associated kymograph. Each frame is 1 s. (B, i) Quantification of the lifespan of Gga2 in indicated Ent5 mutants. (ii) Each event was classified based on its lifespan. (C) Quantification of the recruitment differential and (D) colocalization time in indicated Ent5 mutants. (E, i) Quantification of the lifespan of Gga2 in indicated mutants. (ii) Each event was classified based on its lifespan. Scatter charts display mean value and SEM; horizontal bars indicate median and interquartile ranges. p values reflect a two-tailed Mann–Whitney test.
We next investigated the recruitment of the Ent5 mutant proteins in relation to Gga2. In wild-type cells, Ent5 is recruited after Gga2 with a mean “recruitment differential” of 4 s (Figure 5C). Because the Ent5-CR mutant protein is barely detectable in these structures, we did not perform this analysis for this allele. The ent5ΔCB mutation extended the mean of recruitment differential by 6 s, whereas the ent5ΔAB mutation did not alter the recruitment differential. Because the ent5ΔCB mutation increased both the lifespan of Gga2 structures and the recruitment differential between Ent5 and Gga2, we asked whether the lifespan extension of Gga2 structures could be explained exclusively by the delay in Ent5 recruitment. However, even after Ent5 was recruited, the lifespan was extended, as reflected in an increase in the average time that Ent5 and Gga2 colocalize from 20 s in wild-type cells to 28 s in mutant cells and an increase in the lifespan of Ent5 structures (Figure 5, D and E). Taken together, these results demonstrate that the lifespan extension of the Gga2 structures is due to both a delay in the recruitment of Ent5 and a delay in the maturation of the structure after Ent5 recruitment. This suggests that even after Ent5 is recruited, the inability of Ent5 to bind clathrin delays endosomal coat formation. This suggests that in addition to regulating recruitment of Ent5 to the endosomal clathrin coat, clathrin binding of Ent5 performs an important function that promotes ECT.
DISCUSSION
Epsins are an ancient family of proteins that are found in every known eukaryote. They are important for both endosomal and endocytic clathrin coats. This conservation suggests that epsins perform a critical function that has been retained during evolution. However, the nature of this function is unknown. Initial studies of epsins either failed to reveal a strong defect in membrane traffic upon depletion or loss of endocytic or endosomal epsins or revealed a cargo-selective function (Duncan et al., 2003; Aguilar et al., 2006; Maldonado-Baez et al., 2008; Chen et al., 2009; Pasula et al., 2012). This led to the suggestion that epsins act as cargo-specific adaptors and do not play a key role in the formation of the transport carrier. Recent work has clearly established that endocytic epsins play fundamental mechanistic roles in the formation of the transport carrier (Maldonado-Baez et al., 2008; Messa et al., 2014; Miller et al., 2015; Skruzny et al., 2015). We now revealed that Ent5 plays a fundamental mechanistic role at the endosome. Based on the lifespan extension observed in cells lacking Ent5, we propose that Ent5 plays a pivotal role in the formation of endosomal clathrin coats rather than a cargo-specific function. These data further suggest that Ent5 plays a role in the late stages of coat assembly, after the recruitment of Gga2 and clathrin. A late-acting function is also supported by the observation that wild-type Ent5 is recruited after clathrin (Daboussi et al., 2012).
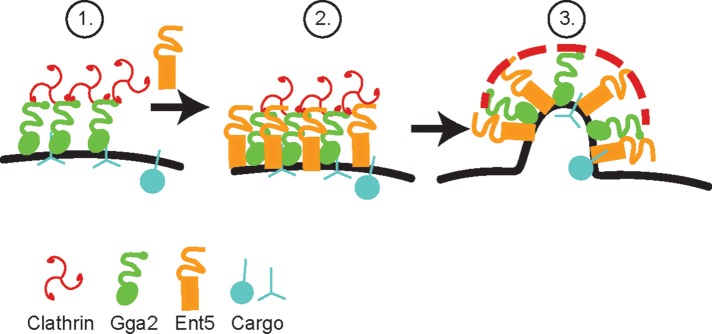
We propose that the key function performed by Ent5 is the stimulation of clathrin assembly (Figure 6). This is consistent with the extended lifespan of coats containing Ent5, Gga2, and clathrin in cells expressing Ent5-CB. In contrast, despite reduced Ent5 recruitment, Ent5-AB–containing coats do not stall. We speculate that in wild-type and in Ent5-AB–expressing cells, Ent5 binding to clathrin promotes a conformational organization of clathrin that enables clathrin polymerization. This suggests that Gga2 on its own is not capable of promoting this conformational organization. This may be because Gga2 contains only one copy of the clathrin- binding motif known as a clathrin box (Drake and Traub, 2001). A single clathrin box can bind clathrin; however, two clathrin boxes appear to be required for clathrin assembly in vitro (Holkar et al., 2015). Although Gga2 contains many clathrin-binding sites in addition to the clathrin box, these additional sites are not clathrin boxes. They are low-affinity DLL-type binding sites, which may not promote rapid clathrin polymerization (Dannhauser et al., 2015; Zhuo et al., 2015). Thus assembly may rely on Ent5 alone, which contains two clathrin boxes, or on the cooperation of the three clathrin boxes available when both Ent5 and Gga2 are present. Our findings are similar to a recent study showing that acute inhibition of EpsinR, the closest orthologue of Ent5 in mammalian cells, inhibits clathrin coat assembly at a stage after clathrin recruitment but before disassembly (Hirst et al., 2015). However, these results and ours are also consistent with other models, such as a role for Ent5 and EpsinR in disassembling clathrin coats. Therefore resolving the exact molecular mechanism by which Ent5 and EpsinR interfere with traffic will require an in vitro system that monitors assembly and organization directly using these proteins.
FIGURE 6:
Model of coat assembly. 1) Gga2 accumulates on membranes and recruits clathrin. 2) Ent5, initially recruited via its ANTH domain, is stabilized by interaction with clathrin and Gga2. This allows the recruitment of Ent5-specific cargo. 3) Ent5 stimulates the formation of the transport carrier, possibly by inducing clathrin polymerization or forming a subclathrin coat. Gga2 helps to increase the amount of Ent5 in the carrier either through initial recruitment of more Ent5 or, as illustrated, by Gga2 indirectly linking Ent5 to the clathrin coat.
A pivotal late-acting role may be a universal characteristic for epsin-related proteins. Loss of all three mammalian endocytic epsins (Eps1–3) stalls coat maturation at a stage after clathrin recruitment (Messa et al., 2014). Similarly, loss of yeast endocytic epsins stalls endocytosis at a stage long after clathrin is normally recruited (Maldonado-Baez et al., 2008). This suggests that epsins may have a universal role in the clathrin coat after clathrin recruitment. However, it is unclear whether Ent5 and endocytic epsins perform the same molecular activity to promote coat maturation. Mammalian endocytic epsins appear to promote endocytosis by linking actin to the clathrin coat (Messa et al., 2014). However, Ent5 lacks the sequences important for actin interaction, suggesting that Ent5 is unlikely to act via this mechanism. On the other hand, endocytic epsins could promote a late stage by both actin binding and stimulation of clathrin assembly. Indeed, recent in vitro work demonstrates that isolated endocytic epsins are potent stimulators of clathrin assembly, suggesting that clathrin assembly is a primary function of epsins (Dannhauser and Ungewickell, 2012; Dannhauser et al., 2015; Holkar et al., 2015).
Although the data presented here clearly indicate a late-acting and pivotal role for the clathrin-binding motifs of Ent5, the functional importance of adaptor binding is less clear. The loss of adaptor binding by Ent5 impairs Chs3 traffic as much as loss of clathrin binding, yet loss of adaptor binding does not alter the lifespan of Gga2 structures. This means that although Ent5–Gga2 interaction is important, it does not play a pivotal role in the maturation of the coat at the level detectable by fluorescence microscopy. One possible function of the Ent5–Gga2 interaction is to dictate how much Ent5 is recruited. Because Gga2 can bind both clathrin and Ent5 at the same time, it may act as a bridge to recruit more Ent5 than clathrin could recruit unaided. In support of such a model, substantially less Ent5ΔAB is recruited to Gga2 structures than wild-type Ent5. This reduced recruitment of Ent5 could impair traffic of Chs3 by reducing the amount of Ent5–Chs3 complexes in the coat. Alternatively, Ent5 binding to Gga2 may be required to productively couple clathrin polymerization to membranes. The requirement for productive coupling in coat assembly was demonstrated with in vitro assays of endocytic adaptors (Dannhauser and Ungewickell, 2012). In these assays, the endocytic adaptor AP180 could promote clathrin cage assembly, but these cages lacked membrane. Intriguingly, Ent5 and AP180 share an N-terminal ANTH domain, which differs from the ENTH domain in epsin in its ability to bind lipids with an extended interface (Duncan and Payne, 2003). Thus Ent5 may require Gga2 to productively couple clathrin assembly to the membrane. Resolving the molecular requirement for Ent5-Gga2 interaction will require examination of the formation of Ent5-Gga2 coats on liposomes.
These results also confirm a crucial role for the ANTH domain in Ent5 localization, consistent with previous reports (Costaguta et al., 2006). Mutation of the ANTH domain disrupts Ent5 localization, suggesting that the ANTH domain is required for localization. However, how the ent5-CR mutation disrupts Ent5 localization is unclear. Although many ANTH domains bind phosphoinositides, they can also bind proteins (Koo et al., 2011; Miller et al., 2011). Thus the ent5-CR allele could interfere with either lipid or protein interaction. We have been unable to show specific phosphoinositide-binding defects caused by the ent5-CR mutation. However, in our hands, Ent5 shows weak nonspecific binding to many phosphoinositides, consistent with previous surface plasmon resonance and vesicle centrifugation analysis of Ent5 (Narayan and Lemmon, 2006). This may be because in vivo Ent5 binds lipids only when associated with a cofactor similar to some endocytic ANTH domains, which require heterodimerization for efficient lipid binding (Skruzny et al., 2015). Alternatively, the ent5-CR mutation may abolish an unknown protein interaction. However, the ability of the ANTH domain alone to localize further confirms that the ANTH domain plays a major role in Ent5 localization.
In summary, these results demonstrate that Ent5 plays a pivotal role late in clathrin coat formation. All known domains and motifs of Ent5 contribute to the function of Ent5 in Chs3 traffic. Furthermore, all three activities are required for maximal Ent5 recruitment to membranes, although the ANTH domain is the most important for localization. Clathrin binding appears to be uniquely important for the maturation of clathrin coats, whereas adaptor binding appears dispensable for coat maturation but may be important for linking cargo to coats. Together with recent reports of key mechanistic roles for endocytic epsins, these results suggest that epsin family members are key mechanistic drivers of clathrin coat function.
MATERIALS AND METHODS
Yeast strains and plasmids
Yeast strains and plasmids are listed in Table 1. Fluorescent tags and gene deletions were introduced by a standard PCR-based method (Longtine et al., 1998). Strains containing multiple genomic modifications were generated by standard yeast genetics. For point mutations, plasmids containing the genomic region of ENT5 were mutated using QuikChange mutagenesis (Stratagene). The resulting plasmids were linearized and transformed into SEY 6210, SEY 6211, and ent5Δ cells to replace the ENT5 locus. pSL6 and pSL6-box encoding wild-type or chc1-box mutant were described previously (Collette et al., 2009).
TABLE 1:
Yeast strains and plasmids.
| Description | Reference/source | |
|---|---|---|
| Yeast strains | ||
| SEY 6210 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 suc2-Δ9 lys2-801; GAL | Robinson et al. (1988) |
| SEY6211 | MATA leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 suc2-Δ9 lys2-801; GAL | Robinson et al. (1988) |
| BY4742 | MATα his3Δ0 leu2Δ0 ura3Δ0 lys2Δ0 | Invitrogen |
| MDY421 | SEY6211 ENT5::URA3 | This study |
| MDY507 | SEY6210 ent5CR(R17E, R18E, K51E, H52E)::URA3 | This study |
| MDY 476 | SEY6211 ent5CB(D292A, L293A, I294A, D354A, L355A, I356A)::URA3 | This study |
| MDY477 | SEY6210 ent5CB(D292A, L293A, I294A, D354A, L355A, I356A)::URA3 | This study |
| MDY523 | SEY6210 ent5AB(E221A, F222A, E330A, F331A, F334A)::URA3 | This study |
| MDY524 | SEY6211 ent5AB(E221A, F222A, E330A, F331A, F334A)::URA3 | This study |
| MDY481 | SEY6210 ent5ABCB(E221A, F222A, D292A, L293A, E330A, F331A, F334A, D354A, L355A, I356A)::URA3 | This study |
| MDY 249 | SEY6211 ent5D::TRP1 | This study |
| MDY250 | SEY6210 ent5D::TRP1 | This study |
| DLY1489 | SEY6211chs6Δ:TRP1 | This study |
| DLY 497 | SEY6210 ent5D::TRP1, chs6Δ:TRP1 | This study |
| DLY 498 | SEY6210 ent5CR(R17E, R18E, P50E, K51E, H52E)::URA3 chs6Δ:TRP1 | This study |
| DLY 1409 | SEY6210 ent5CR(R17E, R18E, P50E, K51E, H52E)::URA3chs6Δ:TRP1 | This study |
| DLY 499 | SEY6211 ent5AB(E221A, F222A, E330A, F331A, F334A)::URA3 chs6Δ:TRP1 | This study |
| DLY 1406 | SEY6211 ent5AB(E221A, F222A, E330A, F331A, F334A)::URA3 chs6Δ:TRP1 | This study |
| DLY 500 | SEY6210 ent5ABCB(E221A, F222A, D292A, L293A, E330A, F331A, F334A, D354A, L355A, I356A)::URA3 MDY481 chs6Δ:TRP1 | This study |
| DLY 501 | SEY6210 ent5ABCB(E221A, F222A, D292A, L293A, E330A, F331A, F334A, D354A, L355A, I356A)::URA3 MDY481 chs6Δ:TRP1 | This study |
| DLY 502 | SEY6210 ent5CB(D292A, L293A, I294A, D354A, L355A, I356A)::URA3 chs6Δ:TRP1 | This study |
| DLY 1408 | SEY6211 ent5CB(D292A, L293A, I294A, D354A, L355A, I356A)::URA3 chs6Δ:TRP1 | This study |
| MDY551 | SEY6211 ENT5-GFP-HIS3Mx:URA3 | This study |
| MDY 552 | SEY6210 ENT5-GFP-HIS3Mx:URA3 | This study |
| MDY 556 | SEY6210 ent5CR(R17E, R18E, P50E, K51E, H52E)-GFP-HIS3MX:URA3 | This study |
| MDY 562 | SEY6210 ent5AB(E221A, F222A, E330A, F331A, F334A)::URA3-GFP-HIS3MX:URA3 | This study |
| MDY 566 | SEY6210 ent5CB(D292A, L293A, I294A, D354A, L355A, I356A)-GFP-HIS3MX:URA3::URA3 | This study |
| MDY 577 | SEY6210 ent5ABCB(E221A, F222A, D292A, L293A, E330A, F331A, F334A, D354A, L355A, I356A)-GFP-HIS3MX:URA3::URA3 | This study |
| MDY553 | SEY6211 ENT5-GFP-HIS3Mx:URA3 CHC1-RFP::KanMx | This study |
| MDY554 | SEY6210 ENT5-GFP-HIS3Mx:URA3 CHC1-RFP::KanMx | This study |
| MDY557 | SEY6210 ent5CR(R17E, R18E, P50E, K51E,H52E)-GFP-HIS3MX:URA3 CHC1-RFP::KanMx | This study |
| MDY564 | SEY6210 ent5AB(E221A, F222A, E330A, F331A, F334A)::URA3-GFP-HIS3MX:URA3 CHC1-RFP::KanMx | This study |
| MDY565 | SEY6210 ent5AB(E221A, F222A, E330A, F331A, F334A)::URA3-GFP-HIS3MX:URA3 CHC1-RFP::KanMx | This study |
| MDY567 | SEY6210 ent5ABCB(E221A, F222A, D292A, L293A, E330A, F331A, F334A, D354A, L355A, I356A)-GFP-HIS3MX:URA3::URA3 CHC1:RFP::KanMx | This study |
| MDY568 | SEY6210 ent5CR(R17E, R18E, P50E, K51E, H52E)-GFP-HIS3MX:URA3 CHC1-RFP::KanMx | This study |
| MDY367 | SEY6211 ent5ANTH(1-184)-GFP::TRP1 CHC1::RFP::KanMX | This study |
| MDY 370 | SEY6210 ent5ANTH(1-184)-GFP::TRP1 CHC1::RFP::KanMX | This study |
| DLY1491 | SEY6210 ent5AB(E221A, F222A, E330A, F331A, F334A)::URA3-GFP-HIS3MX:URA3Gga2-:mCherry:KanMx | This study |
| DLY1492 | SEY6210 ent5AB(E221A, F222A, E330A, F331A, F334A)::URA3-GFP-HIS3MX:URA3 GGA2-:mCherry:KanMx | This study |
| DLY1493 | SEY6210 ent5CR(R17E, R18E, P50E, K51E, H52E)-GFP-HIS3MX:URA3 GGA2-:mCherry:KanMx | This study |
| DLY1494 | SEY6210 ent5CR(R17E, R18E, P50E, K51E, H52E)-GFP-HIS3MX:URA3 GGA2-:mCherry:KanMx | This study |
| DLY 886 | MATA his3Δ0 leu2Δ0 ura3Δ0 lys2Δ0 Chc1-RFP::KanMX Gga2::GFP-KanMX | This study |
| DLY 887 | MATα his3Δ0 leu2Δ0 ura3Δ0 lys2Δ0 Chc1-RFP::KanMX Gga2::GFP-KanMX | This study |
| DLY 888 | MATα his3Δ0 leu2Δ0 ura3Δ0 met15Δ0 lys2Δ0 Chc1-RFP::KanMX Gga2::GFP-KanMX ent5D:KanMX | This study |
| DLY 787 | MATA his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 lys2Δ0 chc1Δ:His3 pSL6-Box (F26W, I79S Q89M) | This study |
| DLY 788 | MATA his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 lys2Δ0 chc1Δ:His3 pSL6 (URA3, CEN, CHC1) | This study |
| DLY1609 | SEY6210 ent5D::TRP1 Clc1-GFP- HIS3Mx | This study |
| DLY1610 | SEY6210 ENT5::URA3 Clc1-GFP- HIS3Mx | This study |
| DLY1622 | SEY6210 Ent5 Δ::TRP1 Gga2-GFP-His3Mx | This study |
| DLY1624 | SEY6211 L Gga2-GFP-His3Mx | This study |
| Plasmids | ||
| pSL6 | URA3, CEN, CHC1 | Collette et al. (2009) |
| pSL6-Box | URA3, CEN, chc1(F26W, I79S Q89M) | Collette et al. (2009) |
| pRS416-mCherry-Tlg1 | Tlg1-tagged mCherry | David Katzmann (Biochemistry and Molecular Biology Department, Mayo Clinic College of Medicine, Rochester, MN) |
Quantitative calcofluor white sensitivity assay
To determine the effect of CFW on cells growth, log-phase cells were diluted 100-fold into yeast extract/peptone/dextrose medium supplemented with different concentrations of CFW and then incubated at 30°C for 10 h before measurement of the optical intensity. Growth inhibition was calculated by normalizing to the absorbance reading of untreated cells. The IC50 values—halfway between the maximal and the minimal inhibition—were derived by a sigmoidal dose-response curve (variable slope, four parameters) using GraphPad Prism.
Media, antibodies, and reagents
Yeast cells were grown in yeast/peptone (YP) medium supplemented with 2% glucose (D) or synthetic medium (SM) supplemented with 2% glucose and an amino acid mix (Lang et al., 2014). Antibodies against Ent5 and Gga2 were described previously (Aoh et al., 2011). Antibodies against GFP and monomeric RFP were from Santa Cruz Biotechnology (Dallas, TX). Alexa Fluor secondary antibodies were from Invitrogen (Carlsbad, CA). Peroxidase conjugate antibodies were from Sigma (St. Louis, MO). ECL plus reagent was from Advansta (Menlo Park, CA). Calcofluor white fluorescent brightener and lyticase were obtained from Sigma-Aldrich.
Whole-cell yeast extracts
To generate lysates, log-phase cells were pelleted, resuspended in Laemmli sample buffer (2% SDS, 1% 2-mercaptoethanol), boiled, and lysed by glass-bead mechanical disruption. The lysates were collected after centrifugation. After SDS–PAGE, samples were transferred to nitrocellulose, blocked with 4% milk in TBS-T (137 mM NaCl, 15.2 mM Tris-HCl, 4.54 mM Tris, 0.896 mM Tween 20), and then probed with primary and fluorescent secondary antibodies. Fluorescence signals were detected on a Typhoon imaging system (Amersham Biosciences, Piscataway, NJ) chemiluminescence signals were detected on a Chemi-Doc-It system (UVP, Upland, CA). Protein intensities were quantitated by ImageJ (National Institutes of Health, Bethesda, MD). To determine the correction factor for the antibody against Ent5, lysates of cells expressing GFP-Ent5 were collected. After the described SDS–PAGE and immunoblotting procedures, parallel samples were detected with antibody against Ent5 and GFP. The correction factor was calculated by determining the ratio between GFP and Ent5 signals.
Immunoprecipitation
For immunoprecipitation, spheroplasts were first generated by resuspending cells in 100 mM Tris-SO4, pH 9.5, 2% glucose, and 5 mM dithiothreitol for 10 min. Cells were then resuspended in YP medium supplemented with 0.5% glucose, 10 mM Tris-HCl, 1.2 M sorbitol, and 120 U of lyticase. Cells were gently agitated for 30 min at 30°C and then washed in 1.2 M sorbitol and resuspended in buffer A (100 mM 2-(N-morpholino)ethanesulfonic acid, pH 6.5, 0.5 mM MgCl2, 1 mM ethylene glycol tetraacetic acid) with protease inhibitors cocktail (Sigma-Aldrich). Cells were lysed by glass-bead lysis, followed by the addition of 1% Triton X-100. The lysates were clarified by centrifugation at 13,000 rpm for 10 min at 4°C. The lysates were incubated overnight at 4°C with 100 μl of 20% protein A–agarose slurry and 3 μl of antibody. The lysates were washed three times with ice-cold buffer A, and the bound proteins were eluted and mixed with SDS sample buffer.
Microscopy
Before imaging, cells were grown to log phase in SM supplemented with 2% glucose and amino acids. Cells were briefly centrifuged and mounted on untreated coverslides for imaging. Images were collected as previously described with a Nikon Ti-E inverted microscope with a 1.4 numerical aperture/100× oil immersion objective. The Lumencor LED light engine (472/20 nm for GFP and 543/20 or 575/20 nm for mCherry) was used for fluorophore excitation. Filters used for imaging were, for fluorescence resonance energy transfer and high-resolution colocalization, dual-band excitation filter ET/GFP-mCherry (59002x), excitation dichroic (89019bs), and emission-side dichroic (T560lpxr); and for emission filters, ET525/50 m and ET595/50 m (Chroma). The puncta per cell were quantitated by counting the foci in a single central plane as described previously (Aoh et al., 2013).
Measurement of fluorescence intensities of endosomal puncta was conducted using a previously described method (Aravamudhan et al., 2014). Briefly a 6 × 6–pixel box was centered on the brightest four pixels of a manually selected spot in the micrograph. The pixels within this box were treated as the signal region. To estimate the background fluorescence, a larger 8 × 8–pixel box was placed concentrically with the signal region. The median value of pixel intensities in the outermost ring of this box was used as the background intensity and subtracted from each pixel in the signal region. The fluorescence signal was then calculated as the sum of all 36 pixels in the 6 × 6 box. These operations were accomplished using a custom graphical user interface written in MATLAB. Intensity analysis was performed on structures from a minimum of 50 cells. Data shown come from analysis performed on a single day; intensity variance between days was minimal.
For time-lapse microscopy, single-plane images were taken every second for a total duration of 2 min. Nonoverlapping structures from multiple cells were identified. The first frame in which structures were visible was considered t = 0. The lifespan was monitored until the median intensity dropped to background levels. Analysis was performed on structures from a minimum of 10 cells/genotype. Data shown come from analysis performed on a single day and are representative of lifespans determined on more than three different days.
Supplementary Material
Acknowledgments
We thank A. P. Joglekar, K. S. Bloom, and E. D. Salmon for access to and help with microscopes used in this study and S. K. Lemmon for reagents. We acknowledge members of the Duncan laboratory for helpful comments and technical support. This work was supported by National Institutes of Health Research Grant GM-092741 (M.C.D.).
Abbreviations used:
- CFW
calcofluor white
- ECT
endosomal clathrin-dependent traffic
- TGN
trans-Golgi network.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-08-0588) on February 3, 2016.
REFERENCES
- Aguilar RC, Longhi SA, Shaw JD, Yeh LY, Kim S, Schon A, Freire E, Hsu A, McCormick WK, Watson HA, Wendland B. Epsin N-terminal homology domains perform an essential function regulating Cdc42 through binding Cdc42 GTPase-activating proteins. Proc Natl Acad Sci USA. 2006;103:4116–4121. doi: 10.1073/pnas.0510513103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahle S, Ungewickell E. Purification and properties of a new clathrin assembly protein. EMBO J. 1986;5:3143–3149. doi: 10.1002/j.1460-2075.1986.tb04621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoh QL, Graves LM, Duncan MC. Glucose regulates clathrin adaptors at the trans-Golgi network and endosomes. Mol Biol Cell. 2011;22:3671–3683. doi: 10.1091/mbc.E11-04-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoh QL, Hung CW, Duncan MC. Energy metabolism regulates clathrin adaptors at the trans-Golgi network and endosomes. Mol Biol Cell. 2013;24:832–847. doi: 10.1091/mbc.E12-10-0750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravamudhan P, Felzer-Kim I, Gurunathan K, Joglekar AP. Assembling the protein architecture of the budding yeast kinetochore-microtubule attachment using FRET. Curr Biol. 2014;24:1437–1446. doi: 10.1016/j.cub.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman AL, Zhang C, Zhu X, Kahn RA. A family of ADP-ribosylation factor effectors that can alter membrane transport through the trans-Golgi. Mol Biol Cell. 2000;11:1241–1255. doi: 10.1091/mbc.11.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky FM, Chen CY, Knuehl C, Towler MC, Wakeham DE. Biological basket weaving: formation and function of clathrin-coated vesicles. Annu Rev Cell Dev Biol. 2001;17:517–568. doi: 10.1146/annurev.cellbio.17.1.517. [DOI] [PubMed] [Google Scholar]
- Chen H, Ko G, Zatti A, Di Giacomo G, Liu L, Raiteri E, Perucco E, Collesi C, Min W, Zeiss C, et al. Embryonic arrest at midgestation and disruption of Notch signaling produced by the absence of both epsin 1 and epsin 2 in mice. Proc Natl Acad Sci USA. 2009;106:13838–13843. doi: 10.1073/pnas.0907008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collette JR, Chi RJ, Boettner DR, Fernandez-Golbano IM, Plemel R, Merz AJ, Geli MI, Traub LM, Lemmon SK. Clathrin functions in the absence of the terminal domain binding site for adaptor-associated clathrin-box motifs. Mol Biol Cell. 2009;20:3401–3413. doi: 10.1091/mbc.E08-10-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copic A, Starr TL, Schekman R. Ent3p and Ent5p exhibit cargo-specific functions in trafficking proteins between the trans-Golgi network and the endosomes in yeast. Mol Biol Cell. 2007;18:1803–1815. doi: 10.1091/mbc.E06-11-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costaguta G, Duncan MC, Fernandez GE, Huang GH, Payne GS. Distinct roles for TGN/endosome epsin-like adaptors Ent3p and Ent5p. Mol Biol Cell. 2006;17:3907–3920. doi: 10.1091/mbc.E06-05-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daboussi L, Costaguta G, Payne GS. Phosphoinositide-mediated clathrin adaptor progression at the trans-Golgi network. Nat Cell Biol. 2012;14:239–248. doi: 10.1038/ncb2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannhauser PN, Platen M, Boning H, Ungewickell H, Schaap IA, Ungewickell EJ. Effect of clathrin light chains on the stiffness of clathrin lattices and membrane budding. Traffic. 2015;16:519–533. doi: 10.1111/tra.12263. [DOI] [PubMed] [Google Scholar]
- Dannhauser PN, Ungewickell EJ. Reconstitution of clathrin-coated bud and vesicle formation with minimal components. Nat Cell Biol. 2012;14:634–639. doi: 10.1038/ncb2478. [DOI] [PubMed] [Google Scholar]
- De Craene JO, Ripp R, Lecompte O, Thompson JD, Poch O, Friant S. Evolutionary analysis of the ENTH/ANTH/VHS protein superfamily reveals a coevolution between membrane trafficking and metabolism. BMC Genomics. 2012;13:297. doi: 10.1186/1471-2164-13-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Angelica EC. Clathrin-binding proteins: got a motif? Join the network. Trends Cell Biol. 2001;11:315–318. doi: 10.1016/s0962-8924(01)02043-8. [DOI] [PubMed] [Google Scholar]
- Dell’Angelica EC, Puertollano R, Mullins C, Aguilar RC, Vargas JD, Hartnell LM, Bonifacino JS. GGAs: a family of ADP ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex. J Cell Biol. 2000;149:81–94. doi: 10.1083/jcb.149.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Zutter JK, Levine KB, Deng D, Carruthers A. Sequence determinants of GLUT1 oligomerization: analysis by homology-scanning mutagenesis. J Biol Chem. 2013;288:20734–20744. doi: 10.1074/jbc.M113.469023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake MT, Traub LM. Interaction of two structurally distinct sequence types with the clathrin terminal domain beta-propeller. J Biol Chem. 2001;276:28700–28709. doi: 10.1074/jbc.M104226200. [DOI] [PubMed] [Google Scholar]
- Duncan MC, Costaguta G, Payne GS. Yeast epsin-related proteins required for Golgi-endosome traffic define a gamma-adaptin ear-binding motif. Nat Cell Biol. 2003;5:77–81. doi: 10.1038/ncb901. [DOI] [PubMed] [Google Scholar]
- Duncan MC, Payne GS. ENTH/ANTH domains expand to the Golgi. Trends Cell Biol. 2003;13:211–215. doi: 10.1016/s0962-8924(03)00076-x. [DOI] [PubMed] [Google Scholar]
- Fang P, Li X, Wang J, Niu L, Teng M. Structural basis for the specificity of the GAE domain of yGGA2 for its accessory proteins Ent3 and Ent5. Biochemistry. 2010;14:7949–7955. doi: 10.1021/bi1010255. [DOI] [PubMed] [Google Scholar]
- Ford MG, Mills IG, Peter BJ, Vallis Y, Praefcke GJ, Evans PR, McMahon HT. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- Hirst J, Edgar JR, Borner GH, Li S, Sahlender DA, Antrobus R, Robinson MS. Contributions of epsinR and gadkin to clathrin-mediated intracellular trafficking. Mol Biol Cell. 2015;26:3085–3103. doi: 10.1091/mbc.E15-04-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J, Lui WW, Bright NA, Totty N, Seaman MN, Robinson MS. A family of proteins with gamma-adaptin and VHS domains that facilitate trafficking between the trans-Golgi network and the vacuole/lysosome. J Cell Biol. 2000;149:67–80. doi: 10.1083/jcb.149.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holkar SS, Kamerkar SC, Pucadyil TJ. Spatial control of epsin-induced clathrin assembly by membrane curvature. J Biol Chem. 2015;290:14267–14276. doi: 10.1074/jbc.M115.653394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung CW, Aoh QL, Joglekar AP, Payne GS, Duncan MC. Adaptor autoregulation promotes coordinated binding within clathrin coats. J Biol Chem. 2012;287:17398–17407. doi: 10.1074/jbc.M112.349035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalthoff C, Alves J, Urbanke C, Knorr R, Ungewickell EJ. Unusual structural organization of the endocytic proteins AP180 and epsin 1. J Biol Chem. 2002;277:8209–8216. doi: 10.1074/jbc.M111587200. [DOI] [PubMed] [Google Scholar]
- Kelly BT, Graham SC, Liska N, Dannhauser PN, Honing S, Ungewickell EJ, Owen DJ. Clathrin adaptors. AP2 controls clathrin polymerization with a membrane-activated switch. Science. 2014;345:459–463. doi: 10.1126/science.1254836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo SJ, Markovic S, Puchkov D, Mahrenholz CC, Beceren-Braun F, Maritzen T, Dernedde J, Volkmer R, Oschkinat H, Haucke V. SNARE motif-mediated sorting of synaptobrevin by the endocytic adaptors clathrin assembly lymphoid myeloid leukemia (CALM) and AP180 at synapses. Proc Natl Acad Sci USA. 2011;108:13540–13545. doi: 10.1073/pnas.1107067108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang MJ, Martinez-Marquez JY, Prosser DC, Ganser LR, Buelto D, Wendland B, Duncan MC. Glucose starvation inhibits autophagy via vacuolar hydrolysis and induces plasma membrane internalization by down-regulating recycling. J Biol Chem. 2014;289:16736–16747. doi: 10.1074/jbc.M113.525782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Maldonado-Baez L, Dores MR, Perkins EM, Drivas TG, Hicke L, Wendland B. Interaction between Epsin/Yap180 adaptors and the scaffolds Ede1/Pan1 is required for endocytosis. Mol Biol Cell. 2008;19:2936–2948. doi: 10.1091/mbc.E07-10-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon HT, Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2011;12:517–533. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- Messa M, Fernandez-Busnadiego R, Sun EW, Chen H, Czapla H, Wrasman K, Wu Y, Ko G, Ross T, Wendland B, De Camilli P. Epsin deficiency impairs endocytosis by stalling the actin-dependent invagination of endocytic clathrin-coated pits. Elife. 2014;3:e03311. doi: 10.7554/eLife.03311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele AE, Watson PJ, Evans PR, Traub LM, Owen DJ. Two distinct interaction motifs in amphiphysin bind two independent sites on the clathrin terminal domain beta-propeller. Nat Struct Mol Biol. 2004;11:242–248. doi: 10.1038/nsmb736. [DOI] [PubMed] [Google Scholar]
- Miller SE, Mathiasen S, Bright NA, Pierre F, Kelly BT, Kladt N, Schauss A, Merrifield CJ, Stamou D, Honing S, Owen DJ. CALM regulates clathrin-coated vesicle size and maturation by directly sensing and driving membrane curvature. Dev Cell. 2015;33:163–175. doi: 10.1016/j.devcel.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SE, Sahlender DA, Graham SC, Honing S, Robinson MS, Peden AA, Owen DJ. The molecular basis for the endocytosis of small R-SNAREs by the clathrin adaptor CALM. Cell. 2011;147:1118–1131. doi: 10.1016/j.cell.2011.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills IG, Praefcke GJ, Vallis Y, Peter BJ, Olesen LE, Gallop JL, Butler PJ, Evans PR, McMahon HT. EpsinR: an AP1/clathrin interacting protein involved in vesicle trafficking. J Cell Biol. 2003;160:213–222. doi: 10.1083/jcb.200208023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JR, Prasad K, Hao W, Augustine GJ, Lafer EM. A conserved clathrin assembly motif essential for synaptic vesicle endocytosis. J Neurosci. 2000;20:8667–8676. doi: 10.1523/JNEUROSCI.20-23-08667.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan K, Lemmon MA. Determining selectivity of phosphoinositide-binding domains. Methods. 2006;39:122–133. doi: 10.1016/j.ymeth.2006.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi T, Shiba Y, Kawasaki M, Shiba T, Matsugaki N, Igarashi N, Suzuki M, Kato R, Takatsu H, Nakayama K, Wakatsuki S. Structural basis for the accessory protein recruitment by the gamma-adaptin ear domain. Nat Struct Biol. 2002;9:527–531. doi: 10.1038/nsb808. [DOI] [PubMed] [Google Scholar]
- Owen DJ, Collins BM, Evans PR. Adaptors for clathrin coats: structure and function. Annu Rev Cell Dev Biol. 2004;20:153–191. doi: 10.1146/annurev.cellbio.20.010403.104543. [DOI] [PubMed] [Google Scholar]
- Pasula S, Cai X, Dong Y, Messa M, McManus J, Chang B, Liu X, Zhu H, Mansat RS, Yoon SJ, et al. Endothelial epsin deficiency decreases tumor growth by enhancing VEGF signaling. J Clin Invest. 2012;122:4424–4438. doi: 10.1172/JCI64537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rad MR, Phan HL, Kirchrath L, Tan PK, Kirchhausen T, Hollenberg CP, Payne GS. Saccharomyces cerevisiae Apl2p, a homologue of the mammalian clathrin AP beta subunit, plays a role in clathrin-dependent Golgi functions. J Cell Sci. 1995;108:1605–1615. doi: 10.1242/jcs.108.4.1605. [DOI] [PubMed] [Google Scholar]
- Rath A, Glibowicka M, Nadeau VG, Chen G, Deber CM. Detergent binding explains anomalous SDS–PAGE migration of membrane proteins. Proc Natl Acad Sci USA. 2009;106:1760–1765. doi: 10.1073/pnas.0813167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M, Payne GS. A role for clathrin in the sorting of vacuolar proteins in the Golgi complex of yeast. EMBO J. 1992;11:2811–2818. doi: 10.1002/j.1460-2075.1992.tb05348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Huang G, Kandror KV. Self-assembly of Glut4 storage vesicles during differentiation of 3T3-L1 adipocytes. J Biol Chem. 2008;283:30311–30321. doi: 10.1074/jbc.M805182200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skruzny M, Desfosses A, Prinz S, Dodonova SO, Gieras A, Uetrecht C, Jakobi AJ, Abella M, Hagen WJ, Schulz J, et al. An organized co-assembly of clathrin adaptors is essential for endocytosis. Dev Cell. 2015;33:150–162. doi: 10.1016/j.devcel.2015.02.023. [DOI] [PubMed] [Google Scholar]
- Sun Y, Kaksonen M, Madden DT, Schekman R, Drubin DG. Interaction of Sla2p’s ANTH domain with PtdIns(4,5)P2 is important for actin-dependent endocytic internalization. Mol Biol Cell. 2005;16:717–730. doi: 10.1091/mbc.E04-08-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia RH, Baggott D, Chuang JS, Schekman RW. The yeast clathrin adaptor protein complex 1 is required for the efficient retention of a subset of late Golgi membrane proteins. Dev Cell. 2002;2:283–294. doi: 10.1016/s1534-5807(02)00127-2. [DOI] [PubMed] [Google Scholar]
- Zhuo Y, Cano KE, Wang L, Ilangovan U, Hinck AP, Sousa R, Lafer EM. Nuclear magnetic resonance structural mapping reveals promiscuous interactions between clathrin-box motif sequences and the N-terminal domain of the clathrin heavy chain. Biochemistry. 2015;54:2571–2580. doi: 10.1021/acs.biochem.5b00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.