Abstract
Phosphine (PH3) fumigation is the primary method worldwide for controlling insect pests of stored commodities. Over-reliance on phosphine, however, has led to the emergence of strong resistance. Detailed genetic studies previously identified two loci, rph1 and rph2, that interact synergistically to create a strong resistance phenotype. We compared the genetics of phosphine resistance in strains of Rhyzopertha dominica and Tribolium castaneum from India and Australia, countries having similar pest species but widely differing in pest management practices. Sequencing analysis of the rph2 locus, dihydrolipoamide dehydrogenase (dld), identified two structurally equivalent variants, Proline49>Serine (P49S) in one R. dominica strain and P45S in three strains of T. castaneum from India. These variants of the DLD protein likely affect FAD cofactor interaction with the enzyme. A survey of insects from storage facilities across southern India revealed that the P45/49S variant is distributed throughout the region at very high frequencies, in up to 94% of R. dominica and 97% of T. castaneum in the state of Tamil Nadu. The abundance of the P45/49S variant in insect populations contrasted sharply with the evolutionary record in which the variant was absent from eukaryotic DLD sequences. This suggests that the variant is unlikely to provide a strong selective advantage in the absence of phosphine fumigation.
Introduction
Phosphine (PH3) fumigation has been used across the world for more than four decades as the primary method to disinfest stored grain from insect pests. Widespread and frequent use of PH3 from the time of the United Nations Food and Agriculture Organisation (FAO) global survey in the 1970s (FAO, 1975), and which subsequently increased because of methyl bromide phase out led to the development of high levels of PH3 resistance in several pest species including Rhyzopertha dominica (F.), Tribolium castaneum (Herbst) and Cryptolestes ferrugineus (Stephens) (Tyler et al., 1983; Rajendran, 1992; Ren et al., 1994; Acda et al., 2000; Collins et al., 2002; Benhalima et al., 2004; Lorini et al., 2007; Opit et al., 2012; Nayak et al., 2013). In Australia, strong resistance in R. dominica (600 × ) was first detected in 1997 (Collins et al., 2002) and in T. castaneum strong resistance (431 × ) was found in 2000 (Jagadeesan et al., 2012).
Detailed genetic analysis of these strains identified two loci conferring strong resistance in both R. dominica and T. castaneum (Jagadeesan et al., 2012; Mau et al., 2012a, b). The first, rph1 (tc-rph1 for T. castaneum), is responsible for weak resistance, whereas the second, rph2, acts synergistically with rph1 to confer strong resistance (Schlipalius et al., 2002). Complementation analysis of R. dominica strains collected from widely geographically separated locations in Australia (Queensland, New South Wales and South Australia) confirmed that the genes responsible for high-level resistance are highly conserved, in that rph1 and rph2 contribute to the resistance phenotype in all the outbreaks studied (Mau et al., 2012a, b). This raises the question of whether this is the case for samples from different countries.
Following the above studies, Schlipalius et al. (2012) recently identified rph2 as dihydrolipoamide dehydrogenase (DLD), a flavin-dependent oxidoreductase essential for energy metabolism (Patel and Roche, 1990) that contains a reactive disulfide and a FAD cofactor, which are directly involved in electron transfer (Williams, 1992). Resolution of the structure of DLD from human and yeast showed that the three-dimensional protein folding was highly conserved across taxa (Brautigam et al., 2005). The protein structure also revealed that protein variants causing E3 deficiency diseases in humans occur at three general locations in the human enzyme: the dimer interface, the reactive disulfide and the FAD/NAD+-binding sites. Schlipalius et al. (2012) also showed that the variants that cause resistance in R. dominica, T. castaneum and the nematode Caenorhabditis elegans (Maupas) also occurred at these general sites. This information now enables direct detection of resistance variants at the rph2 locus in field populations (Kaur et al., 2013).
Although high levels of PH3 resistance have been reported in the key grain insect pests in India (Rajendran et al., 2004), there has been no previous investigation into the genetics of resistance in that country. The observation that the same two genes mediate resistance in independently arising Australian strains (Mau et al., 2012a, b) indicated us that there may be a similar situation prevailing in Indian populations.
The present study investigates the extent to which rph2 mediates PH3 resistance in two the key stored product pests, R. dominica and T. castaneum, from two different geographic regions of the world with radically different grain storage and pest management practices. The widespread distribution and abundance of a single resistance allele in India led us to search the evolutionary record for evidence of it being neutral or under positive or negative selection in species that had not been exposed to PH3.
Materials and methods
Insect strains
Two strongly PH3-resistant R. dominica strains (an Australian strain, QRD569 and an Indian strain IRDMdu) were used in this study. QRD569 was collected near Millmerran, Queensland, in 1997 (Collins et al., 2002) and IRDMdu generated from samples collected in 2009 from central and state grain storage reserves in Madurai, Tamil Nadu and southern India. Four strains of T. castaneum (an Australian strain QTC931 and three Indian strains, ITCMdu, ITCCbe and ITCND) were used in this study. QTC931 was collected from a central storage at Natcha, Queensland, in 2000 (Jagadeesan et al., 2012), whereas ITCMdu, ITCCbe and ITCND were established from samples collected in 2009 from central and state storage reserves in Madurai (Tamil Nadu, southern India), Coimbatore (Tamil Nadu, southern India) and New Delhi (Delhi, northern India), respectively. The Australian strains had been selected previously for homozygosity, whereas the Indian strains had undergone three successive selections, soon after their initial collection to promote homozygosity of the resistance genes within the populations. Following preliminary resistance bioassays (FAO, 1975), R. dominica strain IRDMdu had been diagnosed as strongly resistant; therefore, this strain was purified using a discriminating dose for strong resistance of 0.25 mg l−1 of PH3 for 48 h. Between selections, the survivors of PH3 selection were allowed to mate freely and their progenies were collected for further selection. Similarly, discriminating dose testing for the T. castaneum strains showed that ITCMdu was diagnosed as strongly resistant, whereas ITCCbe and ITCND were diagnosed as weakly resistant. Therefore, ITCMdu was purified using a discriminating dose for strong resistance of 0.25 mg l−1 of PH3 for 20 h, and a discriminating dose for weak resistance of 0.03 mg l−1 for 20 h was used for ITCCbe and ITCND. R. dominica strains were maintained on organically grown wheat and T. castaneum on whole-wheat flour and yeast 20:1 at controlled regimes of 30 °C and 55% relative humidity (RH).
Insect survey collection from India
R. dominica and T. castaneum insect samples were collected in 2011–2012 at 30 and 31 different locations, respectively, from the southern Indian states of Tamil Nadu, Karnataka and Andhra Pradesh (Supplementary Figure S1,Supplementary Table S1). Grain samples were sieved for insects, which were identified, counted and subsequently cultured on whole-wheat flour at 30 °C and 55% RH.
Fumigation
All fumigations were undertaken according to the published standard method (FAO, 1975) for 20 h (T. castaneum) and 48 h (R. dominica) at 25 °C and 70% RH to enable more accurate probit analysis (Daglish et al., 2002). PH3 was generated in a collection tube containing aluminium phosphide introduced into a solution of sulphuric acid (5%). PH3 concentration was determined with gas chromatography using a thermal conductivity detector with nitrogen (N2) as the standard (Winks and Waterford, 1986).
Mortality responses to PH3 of the resistant strains were measured against a range of PH3 concentrations, that is, 0.025–5.0 mg l−1 for R. dominica and 0.008–15 mg l−1 for T. castaneum. Fumigation was undertaken by placing 50 adult beetles (1- to 2-week-old) in a 30-ml ventilated plastic cup containing 10 g whole grain as one replicate inside the gas-tight desiccators and injecting PH3 through a rubber septum in the lid using a gas-tight syringe. Insects were exposed to PH3 for 20 (T. castaneum) or 48 h (R. dominica) at 25 °C and 70% RH, and then removed from the desiccators and held at 25 °C and 55% RH until end point mortality was assessed after 7 days. All experiments were replicated three times.
Data analysis
All mortality data were corrected using Abbott's correction for control mortality (⩽10% Abbott, 1925) before the probit analysis (Finney, 1971). The analysis was performed using GenStat11 statistical package (Payne et al., 2008). The resistance ratio for the resistant strains was calculated by dividing the LC50 of resistant strain by the LC50 value of a reference Queensland strain, a susceptible R. dominica, QRD14 (Collins et al., 2002) and T. castaneum strain, QTC4 (Bengston et al., 1999). The 95% confidence interval (CI) for the resistance ratio at LC50 was calculated following the method established by Robertson and Preisler. (1992). The resulting CI was used to test the equality of the two LC50 values (that is, if the value 1 is contained in the CI for the resistance ratio, and then the LC50 are not significantly different).
Sequencing of the rph2 resistance gene (DLD)
cDNA synthesis and sample preparation for sequencing
Total RNA was extracted from 20 mixed-sex adult beetles of the selected resistant IRDMdu, ITCMdu, ITCCbe and ITCND strains using the QIAGEN (Melbourne, VIC, Australia) RNA extraction kit as per the manufacturer's protocol. First-strand cDNA synthesis was generated from 1 μg of total RNA (Bioline) according to the manufacturer's protocol. The DLD fragment was amplified in a reaction volume of 20 μl containing 4 μl 5 × PCR buffer, 1.5 μl 50 mM MgCl2, 1.0 μl 10 mM dNTPs, 0.2 μl 1.25 U μl−1 Taq DNA polymerase, 2.5 μl 10 μM of forward (5′-TCCCGGTCCAACATTTTAGTA-3′) and reverse (5′-CGGCCTAACCTTAAAAATAC-3′) primers and 2 μl of 1:10 diluted template cDNA stock. The PCR conditions were as follows: denaturation for 3 min at 95 °C, and then 35 cycles of 95 °C for 30 s, 55 °C for 30 s, 72 °C for 1 min and 30 s, with a final extension at 72 °C for 2 min. The PCR products were visualised using gel electrophoresis (1.5% agarose in 1 × TAE, 100 V for 45 min). The resulting PCR product was purified and sequenced.
Genomic DNA extraction
Genomic DNA of R. dominica and T. castaneum samples collected from different storage sites in India was extracted using a CTAB DNA extraction method as described by Doyle and Doyle (1987). Briefly, the DNA extraction buffer contained 100 mM Tris-HCl (pH 8), 10 mM EDTA, 1.4 M NaCl, 2% CTAB and 5% β-mercaptoethanol. Individual insect samples were homogenised with 200 μl of DNA extraction buffer and incubated at 65 °C for 1 h. Chloroform: isoamyl alcohol (24:1) was added and mixed by inversion for 10 min, and subsequently centrifuged at 15 000 g at 4 °C for 10 min. The clear aqueous phase was then transferred into a new sterile microfuge tube and 200 μl of ice-cold isopropanol was added, mixed gently by inversion and kept at −20 °C for overnight incubation. The precipitate was then pelleted using centrifugation at 15 000 g at 4 °C for 10 min, after which the pellet was washed with 70% of ethanol and resuspended in 20 μl TE buffer. The DNA was left to dissolve overnight at 4 °C before use.
Resistance marker visualisation
A cleaved amplified polymorphic sequence marker assay was designed on the basis of the single-nucleotide polymorphism (SNP)that confers a substitution in the DLD gene found in the both species collected in India. In R. dominica, the DLD gene region was amplified using PCR in a reaction volume of 25 μl containing 2.5 μl of 10 × PCR buffer, 1 μl each of 10 μM forward (5′-CTTAATTTCAGCGACAGACT-3′) and reverse (5′-GCCATGTGATAATAGTGTGAG-3′) primer, 0.5 μl of Taq polymerase (1.5 U) and 2 μl (25–50 ng) of template DNA. The PCR conditions were as follows: initial denaturation for 5 min at 95 °C, followed by 35 cycles of 95 °C for 1 min, 57 °C for 30 s and 72 °C for 1 min and a final extension at 72 °C for 7 min. The resistance polymorphism causes a C to T substitution at position 111 of the PCR fragments. The amplified 315-bp product was digested with MboI at 37 °C for 4 h and, the presence of the resistant variant results in two fragments of 108 and 207 bp, whereas the sensitive genotype results in no cleavage.
The DLD gene fragment of T. castaneum was amplified using PCR in a reaction volume of 25 μl containing 2.5 μl of 10 × PCR buffer, 1 μl of 10 μM forward (5′-GCCCTGACTGTCTTCCACCA-3′) and reverse (5′-AGCCTTGACAGCATTTTCCT-3′) primer, 0.5 μl (1.5 U) of Taq polymerase and 2 μl (25–50 ng) of template DNA. The PCR conditions consisted of 5 min at 95°C, followed by 35 cycles of 95 °C for 1 min, 55 °C for 30 s and 72 °C for 1 min and a final extension at 72 °C for 7 min. The amplified 368-bp product was digested with 1 U of MboI at 37 °C for 4 h in a reaction volume of 15 μl containing 5 μl of PCR product, 1 μl of 10 × buffer and 9 μl of nuclease-free water. The digested product was visualised as above. The polymorphism causes a G to A substitution at position 296 of PCR fragment, which causes the formation of an MboI restriction enzyme recognition site, similar to that found in R. dominica. Thus, presence of the resistant variant results in cleavage of the PCR product into 296- and 72-bp fragments, whereas the wild-type sequence does not digest.
Evolutionary analysis of the DLD Proline>Serine variant by BLAST
Natural variants corresponding to T. castaneum DLD proline 45 (P45) were identified through BLAST search against the nonredundant database at NCBI (http://www.ncbi.nlm.nih.gov) using CLC Genomics Workbench 6.5 (http://www.clcbio.com). An E-value of 1 × 10−58 was observed to be the threshold between DLD orthologues and the closest paralogue of DLD, glutathione reductase. Sequences near the threshold were tested by BLAST homology search to confirm their identity. The resulting 669 sequences were aligned and scanned visually to identify variants of P45/49.
Results
Comparative analysis of Australian and Indian R. dominica and T. castaneum strains
Two genes have previously been identified, rph1 and rph2, that explain the PH3 resistance phenotype of a strongly resistant strain of R. dominica (QRD569) and T. castaneum (QTC931) from Queensland, Australia. The response to PH3 of each strain of R. dominica and T. castaneum from Australia and India were analysed using the probit mortality model.
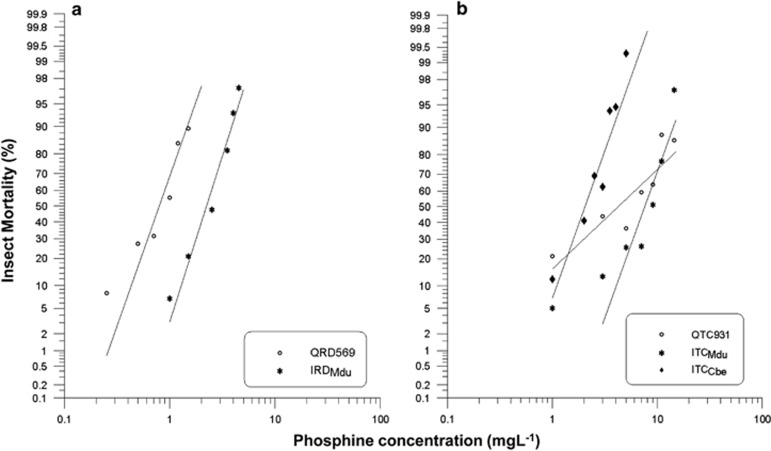
The observed response curve of R. dominica strains shows that IRDMdu was more resistant to PH3 than QRD569 (Figure 1). The resistance ratio of QRD569 is 435 × that of QRD14 parent, whereas, for IRDMdu the resistance ratio is 1283 × greater than that of QRD14 (LC50= 0.00174 mg l−1), which is ~3x greater than that of QRD569 (Supplementary Table S2). We hypothesised that the additional resistance in IRDMdu could be due to stronger resistance effect from different alleles of rph1 and/or rph2 genes.
Figure 1.
Response to phosphine strongly resistant insect strains from India and Australia. Results are presented as log-dose mortality of the resistant strains; (a) QRD569 and IRDMdu, (b) QTC931, ITCMdu and ITCCbe.
Likewise, the observed response curve of T. castaneum strains shows that the resistant phenotype of ITCMdu was double, whereas the ITCCbe strain was half that of the strong resistant reference strain from Queensland, QTC931 (Figure 1 and Supplementary Table S2). The resistance ratio of ITCMdu is 1081 × that of the QTC4 parent, whereas for ITCCbe the resistance ratio is 280 × greater than that of QTC4 (LC50=0.007 mg l−1).
Sequencing, protein prediction and alignment
DNA sequence of the dihydrolipoamide dehydrogenase gene in the IRDMdu (GenBank accession KP843188) strain revealed a P49S polymorphism, which corresponds to a previously reported allele present in a strongly resistant Australian strain, NNRD2864 (Table 1; Schlipalius et al., 2012). This polymorphism is near the active site of the protein and is equivalent to amino acid 15 of the yeast DLD protein (1JEH) and participates in FAD cofactor-binding (Figure 2; Brautigam et al., 2005). The previously described K142E polymorphism (Schlipalius et al., 2012) of QRD569 is present in a different region of the protein that affects the accessibility of the active site to the lipoamide cofactor and is not involved in FAD cofactor binding. There were several other polymorphisms detected in the strain; however, none of these were characterised for association with resistance (see Table 1).
Table 1. Table of amino-acid variants sequenced from resistant and susceptible strains of R. dominica and T. castaneum.
| Species | Strain | Phenotype | rph2 |
AA positiona |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 (3) | 49 (45) | 81 (77) | 135 (131) | 142 (138) | 334 (330) | 361 (357) | 376 (372) | 411 (407) | 416 (412) | 476 (472) | ||||
| R. dominica | QRD14 | Susceptible | − | Y | P | V | G | K | R | A | K | S | V | E |
| R. dominica | QRD369 | Weak | − | Y | P | V | G | K | C | A | K | G | V | E |
| R. dominica | QRD569 | Strong | + | Y | P | V | G | E | C | A | K | G | V | E |
| R. dominica | NNRD2864 | Strong | + | C | S | V | G | K | R | A | K | G | V | E |
| R. dominica | IRDMdu | Strong | + | Y | S(het) | D(het) | G | K | R | G | E | G | V | G |
| T. castaneum | QTC4 | Susceptible | − | S | P | V | G | K | R | A | T | G | I | E |
| T. castaneum | QTC931 | Strong | + | S | P | V | S | K | R | A | T | G | T | E |
| T. castaneum | ITCCbe | Strong | + | S | S | V | G | K | R | A | T | G | I | E |
| T. castaneum | ITCMdu | Strong | + | S | S | V | G | K | R | A | T | G | I | E |
| T. castaneum | ITCND | Strong | + | S | S | V | G | K | R | A | T | G | I | E |
Candidate resistance mutations are highlighted in grey.
Position numbers are given in native sequence position with the R. dominica position and the aligned T. castaneum position in brackets.
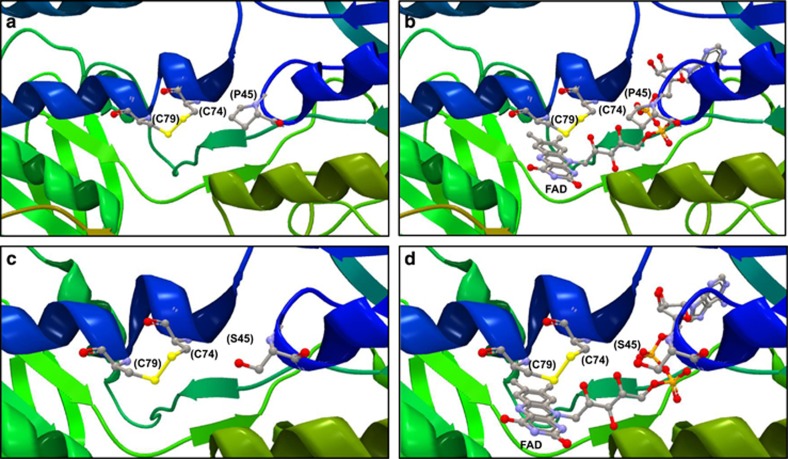
Figure 2.
Structure of the DLD and glutathione reductase protein-active sites. The protein structures have not been determined for either T. castaneum or R. dominica; therefore, the highly conserved orthologues of the DLD and glutathione reductase proteins from Saccharomyces cerevisiae (1JEH and 2HQM; Brautigam et al., 2005; Yu and Zhou, 2007) are shown to illustrate the location of the P>S variant amino acids relative to the active sites of the respective proteins. The extreme degree of sequence conservation between the paralogous enzymes is reflected in easily recognisable structural similarity between the DLD protein (a, b) and glutathione reductase (c, d). The placement of the FAD cofactor shown in b, d indicates that the P>S change is unlikely to affect the electron transfer reaction that occurs between the cysteine residues and the three-ring structure of the FAD. The cysteine (as the disulfide) and proline residues are highlighted and numbered according to the equivalent T. castaneum coordinates.
Sequencing of the two southern Indian T. castaneum strains (ITCMdu and ITCCbe, GenBank accession KP843190 and KP843189) revealed a shared polymorphism that causes a missense amino-acid substitution P45S (Table 1), which corresponds to the P49S substitution reported for the Indian R. dominica strain and NNRD2864 from Australia (Schlipalius et al., 2012). To determine whether the observed allele was present in northern India, we selected the ITCND strain from New Delhi for strong resistance with a discriminating dose of 0.25 mg l−1 (20 h), and sequenced a pool of the survivors. We found the same P45S polymorphism homozygous in the DLD sequence (KP843191). No other polymorphisms that may be related to resistance were detected in the Indian T. castaneum strains. To test the possibility whether this mutation arose once and spread throughout India, we compared the synonymous SNP variants that reside within the coding sequence of the gene (Table 2). This revealed unique polymorphisms for each strain within the gene, suggesting that the resistant strains are likely to be independently arising.
Table 2. Table of SNP variants in the coding region of DLD sequenced from Indian T. castaneum compared with Australian reference strain QTC4.
| Strain |
Positiona |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 133 | 219 | 222 | 339 | 390 | 741 | 810 | 966 | 987 | 1278 | 1344 | |
| QTC4 | C | C | C | C | G | C | A | G | A | C | C |
| ITCCbe | T | T | T | T | A | C | G | A | G | T | T |
| ITCMdu | T | C | T | T | G | C | G | A | G | C | T |
| ITCND | T | T | T | T | A | T | G | A | G | C | T |
Abbreviations: DLD, dihydrolipoamide dehydrogenase; SNP, single-nucleotide polymorphism.
Denotes position referenced to the start of the coding region for QTC4 DLD. Nucleotide variations that are different to the QTC4 reference are highlighted in grey.
Resistant allelic frequency for R. dominica and T. castaneum populations in India
The results of the rph2-resistant marker allele frequency survey are given in Table 3. In R. dominica, the highest allele frequency was found in the state of Tamil Nadu, in south India, where more than 94% of individuals carried a resistance allele at rph2, followed by those recorded for the other two south Indian states, Karnataka (78% of individuals) and Andhra Pradesh (76% of individuals). In T. castaneum, the resistance allele frequency was quite high in all three states investigated, with >95% of individuals observed carrying the resistance allele.
Table 3. Frequency and distribution of rph2 P45/49S resistance variant in Indian populations of R. dominica and T. castaneum.
| Species | Location by state | No. of insect tested |
Total of each genotype |
R alleles (%) | S alleles (%) | Individuals with R allele (%) | ||
|---|---|---|---|---|---|---|---|---|
| R | S | H | ||||||
| R. dominica | Tamil Nadu | 35 | 8 | 2 | 25 | 58.6 | 41.4 | 94.3 |
| Andhra Pradesh | 21 | 3 | 5 | 13 | 45.2 | 54.8 | 76.2 | |
| Karnataka | 32 | 7 | 7 | 18 | 50.0 | 50.0 | 78.1 | |
| T. castaneum | Tamil Nadu | 39 | 25 | 1 | 13 | 80.8 | 19.2 | 97.4 |
| Andhra Pradesh | 45 | 13 | 2 | 30 | 62.2 | 37.8 | 95.6 | |
| Karnataka | 40 | 19 | 1 | 20 | 72.5 | 27.5 | 97.5 | |
Abbreviations: H, heterozygous; R, homozygous resistant; S, homozygous susceptible.
Evolutionary analysis of the DLD Proline>Serine variant
The abundance and widespread distribution of the P45/49S variant suggested to us that it may have been a common pre-existing variant in India before the use of PH3 that gained a selective advantage once PH3 fumigation became common practice. We reasoned that if it was indeed a common variant, we would find it in DLD sequences of other insects in the NCBI database. When we carried out BLASTp analysis, however, we found that not only was the P>S variant not found in any of 64 arthropod DLD sequences, other than reported PH3-resistant pest insects of stored grain, but neither was the variant found in any of 605 non-insect eukaryotic sequences (Supplementary Table S3). The only variants seen in eukaryotes of the proline equivalent to P45 of T. castaneum was a valine substitution in some plants and fungi and a single example of a tyrosine substitution in an amoeboid with a cyanobacterium endosymbiont. We do not show the numbers, but a fraction of eubacterial and archaeal DLD proteins varied at the equivalent proline, a small proportion of which carried the P>S substitution. The scarcity of the P45/49S variant across all eukaryotic taxa provides no evidence to support the hypothesis that it was either neutral or under positive selection in insects or other species that had not been exposed to PH3 fumigation. The complete absence of the variant in eukaryotes other than PH3-resistant grain pests suggests that it was more likely to have been under strong negative selection under natural conditions. Despite the paucity of this variant in the DLD protein, it is present in two close paralogues, glutathione reductase and thioredoxin reductase. The three-dimensional structure of the three related sequences are extremely similar, two of which, DLD and glutathione reductase, are shown in Figure 2 (Rice et al., 1984; Brautigam et al., 2005; Yu and Zhou, 2007). As can be seen, the active site disulfide, the FAD cofactor and the proline or serine residues are similarly aligned between the two proteins. This similarity suggests that the P>S substitution is unlikely to eliminate enzyme activity, but may well change the activity of the variant DLD.
Discussion
In each of eight previously tested strains of R. dominica and T. castaneum from Australia, strong resistance to PH3 was because of alleles of rph2/dld together with resistance alleles at a second locus alternatively named rph1 or tc-rph1 (Jagadeesan et al., 2012; Schlipalius et al., 2012; Mau et al., 2012a, b). This indicated that only a limited number of genes could be modified to generate resistance, but the analysis was limited to one country with well-defined national guidelines for the management of post-harvest grain pests. The current study extends the analysis of PH3 resistance factors to India, a country with quite different post-harvest grain storage system and pest management practices, different climate conditions and a different emphasis on the types of grain produced. In addition, factors such as type of storages (for example, bag stacks), typical length of storage and domestic mobility of grains within India are significantly different to Australia. These differences alter the spectrum of insect pests of primary and secondary importance between the two countries, although R. dominica and T. castaneum are significant pests in both countries. Interestingly, the currently studied Indian strains that we tested had a resistance phenotype nearly twice of that observed in Australia, with R. dominica 1283 × and T. castaneum 1081 × more resistant than the fully susceptible reference strains from Australia, QRD14 and QTC4, respectively. Despite these differences and the potentially large number of targets of PH3 action, our analysis revealed that the previously identified resistance locus rph2 is also responsible for the strong resistance phenotype in India.
The identity of rph1 is unknown, but rph2 encodes DLD (2012), a key enzyme of energy metabolism (Patel and Roche, 1990) that contains a reactive disulfide and a FAD cofactor that are directly involved in electron transfer (Williams, 1992). Sequencing of the dld gene showed a P45S variant in all three of the Indian T. castaneum strains. This serine substitution for the original proline occurs in the FAD cofactor-binding site in the enzyme. Interestingly, a P49S variant exists at the equivalent position in the protein of an Australian R. dominica strain that is the most strongly resistant strain of this species in Australia (Mau et al., 2012a).
DNA sequencing of the dld gene from eight strongly PH3-resistant strains of R. dominica and T. castaneum from Australia resulted in identification of five different resistance-causing variants (Mau et al., 2012a; Schlipalius et al., 2012), whereas in this study all four Indian strains (one R. dominica and three T. castaneum) carried the same resistance-causing amino-acid variant. The T. castaneum strains, in particular ITCMdu and ITCND, came from widely geographically separated regions (2500 km apart), making it likely that the resistance variants were independently derived, a hypothesis that we confirmed by DNA sequencing of the dld gene from each strain. The dld gene sequences of each strain contained unique nucleotide variants in addition to the common variant responsible for resistance. This supports the notion that the resistance mutations occurred independently in pre-existing unique gene sequence contexts. The actual mutation in the proline codon was the same in each of the T. castaneum strains as well as the R. dominica strain from India and two previously published R. dominica strains from Australia (CCT to TCT); however, that is the only single-nucleotide change that can result in a proline to serine change.
The observation that the P45/49S variant was responsible for resistance in all four strains from India motivated us to survey the distribution and abundance of this allele in southern India more broadly. We found that at grain storage sites across three states of southern India (Table 3, Supplementary Table S1), the P45/49S allele is present in ⩾95% of T. castaneum and 76–94% of R. dominica that were tested. The most obvious conclusion is that the high frequency of this resistance variant is the result of pest management practice in India that differs from practices in Australia where strong resistance in general and this variant in particular are rare.
While PH3 exposure clearly selects for DLD variants that result in strong resistance, it was not possible to determine a priori whether DLD variants that confer PH3 resistance existed in populations of pest insects before PH3 exposure because of being neutral or under positive selection or whether they were unlikely to persist in the population because of negative selection. It is quite possible that variants in such a key enzyme of energy metabolism as DLD could significantly affect the fitness of the animal carrying the variant. If metabolism was altered in a way that provided a selective advantage under natural environmental conditions, it could favour the persistence of the variant in wild populations. If this were the case, the low frequency of the P45/49S variant in Australia as opposed to its abundance across southern India could reflect climatic differences between the two countries. This is an important consideration as it could not only influence the development of resistance as a pest management problem, but may also provide information for the effective management of resistance. This is a difficult problem to address, as any pre-existing variants still could be quite rare before PH3 use and therefore difficult to detect.
To resolve this issue, we pursued a bioinformatic analysis of DLD sequences present in GenBank to scan for variants across the span of evolutionary time. This analysis revealed that the P45/49S variant was not found in any insect, other than reported PH3-resistant pest insects of stored grain. Neither was the variant found in any other eukaryotic gene sequence (Supplementary Table S3). The complete absence of the variant across the span of eukaryotic evolution, throughout diverse environments and diverse species contexts, makes it highly unlikely that the variant is either neutral or under positive selection in natural insect populations. The absence of the variant in eukaryotes that have not been exposed to PH3 makes it much more likely that, under natural conditions, the variant is under strong negative selection. The only other variants at P45/49 observed in multicellular eukaryotes were a change to valine (V) in fungi and plants (Supplementary Table S3) and to tyrosine (Y) in an amoeboid with a cyanobacterium endosymbiont. Thus, it appears that there are extreme constraints on changes to the proline at position 45/49, probably due to a disruption to the normal function of DLD. The appearance and seemingly rapid proliferation of the P45/49S variant in insects exposed to PH3 suggest that fumigation is providing extreme positive selective pressure for change of the DLD protein. It is interesting to note that two paralogues of DLD, glutathione reductase and thioredoxin reductase, not only have structures that are extremely similar to that of DLD, but they each have a serine amino acid at the position corresponding to P45/49 from DLD. This may explain why DLD, but not glutathione reductase or thioredoxin reductase, is uniquely sensitive to PH3 as indicated by the absence of resistance variants in the other two proteins. A more detailed understanding of the modified enzyme function that gives rise to PH3 resistance may help to identify an as yet undiscovered vulnerability of PH3-resistant insects.
Data archiving
DLD sequences have been deposited in GenBank, accession numbers KP843188–KP843191.
Acknowledgments
We would like to acknowledge the support of the Australian Government's Cooperative Research Centres Program and the Australia–India Strategic Research Grand Challenge Fund (GCF010006), jointly sponsored by the Indian Department of Science and Technology (DST), and the Australian Department of Innovation, Industry, Science and Research (DIISR). RK and RJ received an International Postgraduate Research Scholarship from the University of Queensland. We specially thank Hervoika Pavic, Linda Bond and Andrew Tuck for technical assistance and managers of various state and central grain reserves of India for their assistance in sample collection.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Abbott WS. (1925). A method of computing the effectiveness of an insecticide. J Econ Entomol 18: 265–267. [Google Scholar]
- Acda MA, Bengston M, Daglish GJ. (2000). Response to phosphine of susceptible and resistant strains of Rhyzopertha dominica (Fabricius)(Coleoptera: Bostrichidae) from the Philippines. Asia Life Sci 9: 103–113. [Google Scholar]
- Bengston M, Collins PJ, Daglish GJ, Hallman VL, Kopittke R, Pavic H. (1999). Inheritance of phosphine resistance in Tribolium castaneum (Coleoptera: Tenebrionidae). J Econ Entomol 92: 17–20. [Google Scholar]
- Benhalima H, Chaudhry MQ, Mills KA, Price NR. (2004). Phosphine resistance in stored-product insects collected from various grain storage facilities in Morocco. J Stored Prod Res 40: 241–249. [Google Scholar]
- Brautigam CA, Chuang JL, Tomchick DR, Machius M, Chuang DT. (2005). Crystal structure of human dihydrolipoamide dehydrogenase: NAD(+)/NADH binding and the structural basis of disease-causing mutations. J Mol Biol 350: 543–552. [DOI] [PubMed] [Google Scholar]
- Collins PJ, Daglish GJ, Bengston M, Lambkin TM, Pavic H. (2002). Genetics of resistance to phosphine in Rhyzopertha dominica (Coleoptera: Bostrichidae). J Econ Entomol 95: 862–869. [DOI] [PubMed] [Google Scholar]
- Daglish GJ, Collins PJ, Pavic H, Kopittke RA. (2002). Effects of time and concentration on mortality of phosphine-resistant Sitophilus oryzae (L) fumigated with phosphine. Pest Manage Sci 58: 1015–1021. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19: 11–15. [Google Scholar]
- FAO. (1975). Recommended methods for the detection and measurement of resistance of agricultural pests to pesticides. Tentative method for adults of some major pest species of stored cereals, with methyl bromide and phosphine - FAO Method no. 16. Plant Protect Bull 23: 12–25. [Google Scholar]
- Finney DJ. (1971) Probit Analysis, 3rd edn. Cambridge University Press: Cambridge. [Google Scholar]
- Jagadeesan R, Collins PJ, Daglish GJ, Ebert PR, Schlipalius DI. (2012). Phosphine resistance in the Rust red flour beetle, Tribolium castaneum (Coleoptera: Tenebrionidae): Inheritance, gene interactions and fitness costs. PLoS ONE 7: e31582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur R, Daniels EV, Nayak MK, Ebert PR, Schlipalius DI. (2013). Determining changes in the distribution and abundance of a Rhyzopertha dominica phosphine resistance allele in farm grain storages using a DNA marker. Pest Manage Sci 69: 685–688. [DOI] [PubMed] [Google Scholar]
- Lorini I, Collins PJ, Daglish GJ, Nayak MK, Pavic H. (2007). Detection and characterisation of strong resistance to phosphine in Brazilian Rhyzopertha dominica (F.) (Coleoptera: Bostrychidae). Pest Manage Sci 63: 358–364. [DOI] [PubMed] [Google Scholar]
- Mau YS, Collins PJ, Daglish GJ, Nayak MK, Ebert PR. (2012. a). The rph2 gene is responsible for high level resistance to phosphine in independant field strains of Rhyzopertha dominica. PLoS ONE 7: e34027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mau YS, Collins PJ, Daglish GJ, Nayak MK, Pavic H, Ebert PR. (2012. b). The rph1 gene is a common contributor to the evolution of phosphine resistance in independent field isolates of Rhyzopertha dominica. PLoS ONE 7: e31541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak MK, Holloway JC, Emery RN, Pavic H, Bartlet J, Collins PJ. (2013). Strong resistance to phosphine in the rusty grain beetle, Cryptolestes ferrugineus (Stephens) (Coleoptera: Laemophloeidae): its characterisation, a rapid assay for diagnosis and its distribution in Australia. Pest Manage Sci 69: 48–53. [DOI] [PubMed] [Google Scholar]
- Opit GP, Phillips TW, Aikins MJ, Hasan MM. (2012). Phosphine resistance in Tribolium castaneum and Rhyzopertha dominica from stored wheat in Oklahoma. J Econ Entomol 105: 1107–1114. [DOI] [PubMed] [Google Scholar]
- Patel MS, Roche TE. (1990). Molecular biology and biochemistry of pyruvate dehydrogenase complexes. FASEB J 4: 3224–3233. [DOI] [PubMed] [Google Scholar]
- Payne RW, Murray DA, Harding SA, Baird DB, Soutar DM. (2008) GenStat for Windows (11th Edition) Introduction. VSN International: Hemel Hempstead. [Google Scholar]
- Rajendran S. (1992). Selection for resistance to phosphine or methyl bromide in Tribolium castaneum (Coleoptera, Tenebrionidae). Bull Entomol Res 82: 119–124. [Google Scholar]
- Rajendran S, Parveen H, Begum K, Chethana R. (2004). Influence of phosphine on hatching of Cryptolestes ferrugineus (Coleoptera: Cucujidae), Lasioderma serricorne (Coleoptera: Anobiidae) and Oryzaephilus surinamensis (Coleoptera: Silvanidae). Pest Manage Sci 60: 1114–1118. [DOI] [PubMed] [Google Scholar]
- Ren YL, O'Brien IG, Whittle GP. (1994). Stored Products Protection. Proceeding of the 6th International Conference on Stored Product Protection Highley EWE, Banks HJ, Champ BR (ed.) CAB: Wallingford, UK: Canberra. pp 173–177. [Google Scholar]
- Rice DW, Schulz GE, Guest JR. (1984). Structural relationship between glutathione reductase and lipoamide dehydrogenase. J Mol Biol 174: 483–496. [DOI] [PubMed] [Google Scholar]
- Robertson JL, Preisler HK. (1992) Pesticide Bioassays with Arthropods. CRC Press: Boca Raton, FL. [Google Scholar]
- Schlipalius DI, Cheng Q, Reilly PEB, Collins PJ, Ebert PR. (2002). Genetic linkage analysis of the lesser grain borer Rhyzopertha dominica identifies two loci that confer high level resistance to the fumigant phosphine. Genetics 161: 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlipalius DI, Valmas N, Tuck AG, Jagadeesan R, Ma L, Kaur R et al. (2012). A core metabolic enzyme mediates resistance to phosphine gas. Science 338: 807–810. [DOI] [PubMed] [Google Scholar]
- Tyler PS, Taylor RW, Rees DP. (1983). Insect resistance to phosphine fumigation in food warehouses in Bangladesh. International Pest Control 25: 10–13. [Google Scholar]
- Williams CHJ. (1992). Lipoamide dehydrogenase, glutathione reductase, thioredoxin reductase, and mercuric ion reductase-a family of flavoenzyme transhydrogenases. In: Muller F (ed). Chemistry and Biochemistry of Flavoenzymes. CRC Press: Boca Raton. [Google Scholar]
- Winks RG, Waterford CJ. (1986). The relationship between concentration and time in the toxicity of phosphine to adults of a resistant sdtrain of Tribolium castaneum (Herbst). J Stored Prod Res 22: 85–92. [Google Scholar]
- Yu J, Zhou C-Z. (2007). Crystal structure of glutathione reductase Glr1 from the yeast Saccharamyces cerevisiae. Protein Struct Funct Bioinform 68: 972–979. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.