Abstract
Small mammal dispersal is strongly affected by geographical barriers. However, commensal small mammals may be passively transported over large distances and strong barriers by humans (often with agricultural products). This pattern should be especially apparent in topographically complex landscapes, such as mountain ranges, where valleys and/or peaks can limit dispersal of less vagile species. We predict that commensal species would have lower genetic differentiation and higher migration rates than related non-commensals in such landscapes. We contrasted population genetic differentiation in two sympatric Rattus species (R. satarae and R. rattus) in the Western Ghats mountains in southern India. We sampled rats from villages and adjacent forests in seven locations (20–640 km apart). Capture-based statistics confirmed that R. rattus is abundant in human settlements in this region, whereas R. satarae is non-commensal and found mostly in forests. Population structure analyses using ~970-bp mitochondrial control region and 17 microsatellite loci revealed higher differentiation for the non-commensal species (R. satarae F-statistics=0.420, 0.065, R. rattus F-statistics=0.195, 0.034; mitochondrial DNA, microsatellites, respectively). Genetic clustering analyses confirm that clusters in R. satarae are more distinct and less admixed than those in R. rattus. R. satarae shows higher slope for isolation-by-distance compared with R. rattus. Although mode of migration estimates do not strongly suggest higher rates in R. rattus than in R. satarae, they indicate that migration over long distances could still be higher in R. rattus. We suggest that association with humans could drive the observed pattern of differentiation in the commensal R. rattus, consequently impacting not only their dispersal abilities, but also their evolutionary trajectories.
Introduction
Humans present a strong evolutionary force, driving the trajectories of several animal and plant species closely associated with them, called commensals or synanthropes (Matisoo-Smith, 2009; Zeder, 2012). These species are believed to be intermediate between being wild and being domesticated (Vigne, 2011; Zeder, 2012) and include some of the world's most cosmopolitan species such as the house mouse (Mus musculus), brown rat (Rattus norvegicus), house sparrow (Passer domesticus) and house crow (Corvus splendens).
Commensals benefit from human interactions through exploitation of anthropogenic ecosystems for food and shelter. In addition, human-mediated transport of founding individuals over long distances results in large geographical ranges for these species and colonisation of new areas, which would otherwise be inaccessible (Cucchi and Vigne, 2006; Jones et al., 2012). Consequently, human activities could promote their diffusion by eliminating ecological, as well as landscape barriers, thereby aiding in ‘jump dispersal' across longer distances and potent barriers (Schrey et al., 2014).
Of the prominent landscape features that act as barriers to dispersal, mountain ranges have been the most extensively studied. Dispersal across such topographically complex areas is believed to be more restricted than across flat areas of similar size because of spatial and ecological constraints associated with elevational gradients (Trenel et al., 2008). We would expect small mammal populations to be more isolated in mountain ranges because of their inherently lower dispersal ability and also the relatively disconnected habitats in the mountains (Ferro, 2013).
Although several studies have assessed the genetic structure of small mammal populations either in the context of anthropogenically influenced dispersal (Kajdacsi et al., 2013) or in complex mountainous landscapes (Schulte-Hostedde et al., 2001) separately, very few studies have addressed the dispersal abilities of commensal and non-commensal species that coexist in a naturally fragmented, discontinuous landscape (see Brouat et al., 2007). We hypothesised that in the context of montane regions, a commensal small mammal species should show less genetic differentiation and higher migration rates between different commensal populations than a related non-commensal species in the same region. The commensal should also show no effect of geographic distance on genetic isolation. In this study, we compared two Rattus species in the Western Ghats mountain range in India. First, we investigated whether there is habitat partitioning between the two species in the context of commensalism. Further, using mitochondrial and nuclear microsatellite markers, we tested (a) if genetic differentiation is higher and migration rates are lower in the non-commensal species as compared with the commensal species and (b) if the non-commensal species shows a stronger correlation between genetic isolation and geographic distance than the commensal species.
Materials and methods
Study area
The study was performed in the Western Ghats, a 1600 km long mountain range in the southern Indian subcontinent. Average elevation ranges from 500 m in the north to over 2500 m in the south. This biogeographically important region is characterised by rolling hills with pristine forests, interspersed with human settlements. The habitat ranges from dry and moist deciduous forests (between an elevational range of 500–1000 m), montane evergreen forests (above 1000 m) and shola forest-grassland complexes (above 2000 m).
There are three major biogeographical breaks in the Western Ghats: the Goa, Palghat and Shencottah gaps. The oldest and widest among these is Palghat gap (Robin et al., 2010), which is 40 km across at its widest point. The Palghat gap has been identified as a barrier for several taxa including plants (Subramanyam and Nayar, 1974), birds (Robin et al., 2010) and even large mammals like elephants (Vidya et al., 2004).
Study species
The species Rattus satarae commonly known as the white-bellied wood rat or Sahyadri forest rat, is endemic to the Western Ghats of India and is distributed throughout the mountain range from the northern lower elevation dry deciduous habitats to the southern high elevation tropical montane forests (Molur and Singh, 2009; Pagès et al., 2011). The closest relatives of this species are R. norvegicus and R. nitidus both of which occur in the Indochinese region (Pagès et al., 2011). It is the most commonly trapped species in this region and is thought to be non-commensal with humans (Molur and Singh, 2009). However, very little is known about the ecology of this species.
The black rat, Rattus rattus (lineage I as described in Aplin et al., 2011) is found in all the continents with the possible exception of Antarctica. The ancestral distribution of this species was probably India from where it has spread globally through passive transportation on ships (Aplin et al., 2011). Phylogenetically, this species is representative of the ‘rattus–exulans' sister group as described in Pagès et al., 2010. Its ubiquitous presence in even the smallest of human settlements has earned it the reputation of a pest, necessitating frequent eradication attempts. It is a native species in the Western Ghats and described as the dominant species in the small mammal community (Shanker and Sukumar, 1998; Ramachandran, 2013).
Sampling
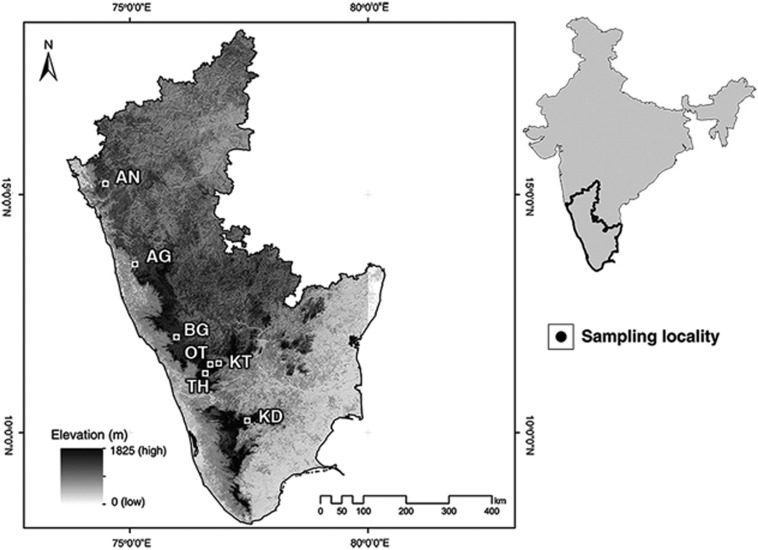
We sampled over a period of 2 years from May 2011 to April 2013 in small to medium-sized villages and towns (1000–90 000 inhabitants) and in forests <2 km away from these settlements. A ‘sampling locality' consists of three ‘habitats'—the village, the adjoining forest and the forest edge, that is, a trapping site in the ecotone between the village and the forest, which we selected in order to determine where R. rattus and R. satarae coexist. We sampled seven such ‘sampling localities' in the Western Ghats (distance 17–640 km; Figure 1). All the forest and edge habitats sampled in this study were either a part of or connected to some larger undisturbed forest, which did not have other human settlements for the most part.
Figure 1.
Sampling locations. Physical map of the eight sampling localities in the Western Ghats. The mountain range is represented by the high elevation regions along the western coast. Sampling localities include seven villages sampled in this study (name of the forest mentioned in brackets): AN—Joida (Anshi National Park), AG—Agumbe (Agumbe Rainforest), BG—Kutta (Brahmagiri Wildlife Sanctuary), OT—Ooty (Ooty Reserve Forest), KT—Kotagiri (Longwood shola), TH—Thiashola tea plantation (Thiashola), KD—Kodaikanal (Vattakanal shola).
We used Sherman traps baited with peanut butter or dried fish to live-trap rats. We captured rats inside houses, shops and storage buildings, and in the forests in each sampling locality. We deployed 50–100 traps in each sampling locality for 3–5 days along lines with 10-m intervals. We placed traps on the ground in both houses and forests. We identified species in field based on the body measurements and morphological descriptions in Pagès et al. (2011) and later confirmed the identities using genetics. In a second sampling effort from July to September 2014, we concentrated on increasing the sample size in the ‘village' habitat. We utilised 25 snap traps and 35 Sherman traps in the previously described sampling framework (ethical permission from Institutional Animal Ethics Committee). We obtained tissue biopsies from the tail of the captured individuals, which were subsequently released, and stored the biopsies in 100% ethanol before transporting to the laboratory where we transferred them to −20 °C cold storage. We obtained liver biopsies from the snap-trapped individuals.
Habitat partitioning between species
Previous studies have suggested that R. rattus and R. satarae are sympatric species (Molur and Singh, 2009; Pagès et al., 2011). In such a scenario, we would expect both the species to have equivalent capture probabilities in either villages or forests. To statistically assess whether the two species occupied same or different habitats, we performed a binomial test to determine if both species have equal probability of capture in either habitat. In order to graphically visualise our results, we simulated a distribution of the number of captures of any one species expected if 1000 trials were performed for the total number of captures in that habitat and assuming an equal capture probability for either species (P=0.5). We then plotted our observed number of successes (that is, actual number of captures) for either species in each habitat. For this analysis, we used our capture data from the sampling effort made in May 2011 to April 2013 in forest and village habitats only. All analyses were performed with the software R v3.0.2 (R Core Team, 2013).
DNA extraction and amplification
We extracted total genomic DNA from the tissue samples using the DNeasy Blood and Tissue kit (QIAGEN, New Delhi, India) following the manufacturer's protocol. We confirmed the genetic species identity of 150 out of 324 samples using the mitochondrial cytochrome b gene using the Rattus-specific primers RGlu2L-RCb9H, which amplify ~762 bp of cyt b (Robins et al., 2007). This was performed in order to test if field identification is reliable. For all the other samples, we used the mitochondrial control region to confirm species identity. Results presented are based on species identification using a combination of either mitochondrial loci.
We amplified 970 bp of the mitochondrial control region consisting of the hypervariable regions I and II and the flanking tRNA region in two parts with primers pairs EGL4L-RJ3R and RJ2F-EGL3H as described in (Robins et al., 2008) for all samples. We purified all PCR products by the single-step enzymatic process with exonuclease 1 and shrimp alkaline phosphatase (from New England Biolabs, Bangalore, India). All sequencing reactions were performed on ABI3100xl Automated system (Bangalore, India). We performed sequence assembly using the program Geneious v7 (Geneious created by Biomatters: available from http://www.geneious.com/.Geneiousv.7). All haplotypes have been deposited in GenBank database with the accession numbers KJ603317–KJ603409 and KP159524–KP159565.
We selected 22 microsatellite loci from different genetic studies on the Rattus genus (Abdelkrim et al., 2005; Loiseau et al., 2008; Russell et al., 2009). Out of these, five did not amplify in R. satarae. We used the following 17 microsatellite loci: Rr14, Rr17, Rr21, Rr22, Rr54, Rr67, Rr68, D10Rat20, D11Mgh5, D7Rat13, D15Rat77, D5Rat83, D2Rat234, D18Rat96, D20Rat46, D11Rat56 and D10Mit5. The forward primer for each locus was tagged with one of four fluorescent dyes: 6-FAM, VIC, NED, PET and we amplified each locus separately with the MasterMix (QIAGEN) kit and then pooled all loci in five separate sets for genotyping. We visualised 2 μl of each PCR product on a 2% agarose gel and used the band intensity to determine the volume of product to be genotyped. A typical genotyping mixture consisted of 3–4 μl PCR product, 9.5 μl formamide and 0.5 μl size standard (LIZ 500, Applied Biosystems, Bangalore, India). We scored allele sizes manually using the GeneMapper software version 4 (Applied Biosystems). For binning the scored allele sizes, we used the program TANDEM (Matschiner and Salzburger, 2009). Individuals with similar genotypes were identified using GIMLET (Valière, 2002). We trimmed pairs of individuals that had identical genotypes at >15 loci. In addition, average within-group relatedness, genotyping errors including false alleles and allelic dropouts, test for selective neutrality of loci and test for presence of null alleles were also performed (Supplementary Methods).
Genetic diversity and population differentiation
For mitochondrial control region, we calculated genetic diversity including the number of haplotypes and polymorphic sites, as well as nucleotide and haplotype diversity with DnaSP v5 (Librado and Rozas, 2009). We quantified genetic differentiation among populations by calculating the ‘FST' statistics computed with ARLEQUIN v3.5 (Excoffier and Lischer, 2010) using the pairwise distance matrix with a gamma correction of α=0.39 (Hingston et al., 2005) on populations with high sample sizes (AG, KT, TH and KD). We constructed median joining networks for the haplotypes by estimating the pairwise differences between the haplotypes with the program PopArt (developed by Jessica Leigh: available from http://popart.otago.ac.nz/index.shtml).
For microsatellite data, we calculated allelic richness (corrected for sample size) and the number of alleles using FSTAT v2.9.3.2 (Goudet, 2003) and tested whether allelic richness and genetic diversity differed significantly between the two species (1000 permutations). We also calculated observed (HO) and expected heterozygosities (HE) and deviations from Hardy–Weinberg equilibrium with ARLEQUIN v3.5, correcting for multiple comparisons using the false discovery rate approach (Benjamini and Hochberg, 1995) implemented in R v3.0.2.
We estimated pairwise genetic differentiation among populations using RST-based methods, which are more suitable for the high mutation rates of microsatellite data, as they assume a stepwise mutation model, which takes into account the difference in allele sizes of the locus. We used populations with high sample sizes (AG, KT, TH and KD) for this purpose and used the program ARLEQUIN v3.5 to calculate RST. We also performed analysis of molecular variance for each species separately to understand how the variance was partitioned across the locations.
In order to determine the number of genetic clusters present in the geographically partitioned data sets, we used a clustering algorithm used in the program STRUCTURE v2.3.4 (Falush et al., 2003). First, we used our relatedness estimates to trim both the data sets using a threshold of 0.2 (indicating half-sibship) to retain a single representative genotype. Then, we performed 10 iterations each for K=1–10 for R. rattus and R. satarae data sets. We edited each parameter set to run for 2 million steps in the Markov-Chain-Monte-Carlo simulation chain with 500 000 burn, assuming correlated allele frequencies under an admixture model. For each individual, we obtained the membership coefficient (q value) for each cluster. We collated the output from STRUCTURE and compared the likelihoods for each run to select the most likely value of K using the Evanno method (Evanno et al., 2005) as implemented in the web version of STRUCTURE HARVESTER (Earl and vonHoldt, 2011). After summarising the runs with CLUMPP (Jakobsson and Rosenberg, 2007), we graphically visualised membership coefficients (q) for all individuals using DISTRUCT (Rosenberg, 2003).
Isolation-by-distance
In order to determine the effect of geographic distance on genetic isolation, we first linearised the pairwise RST values as RST=RST/1 – RST (Slatkin, 1995). We next obtained geographic distances between the sampling locations using latitude-longitude coordinates from our sampling sites. We then performed Mantel tests by regressing pairwise estimates of linearised RST with the ln-transformed geographic distances (10 000 permutations) with the web version of GenePop (Rousset, 2008). We then visualised the transformed values from GenePop graphically using R 3.0.2. As having low sample sizes in the data set is likely to increase variability of the correlation among pairs being tested (Landguth et al., 2012), we performed this analysis after removing three populations (AN, BG and OT) from both data sets.
Estimation of migration rates
We also estimated effective population size (Θ=4x effective population size, Ne x mutation rate, μ) and migration rates (M=migration rate, m/mutation rate, μ) through Bayesian inference in the program MIGRATE-N v3.2.1 (Beerli, 2006) for both species separately. We used the microsatellite data set for this analysis. We used only populations, which had >10 individuals of both species for this analysis in order to avoid pooling populations, with low sample sizes and also to prevent over-parameterisation of the analysis. Thus we had four populations: AG, KT, TH and KD. Initial Θ and M values were generated from calculation of FST and the next parameter values were proposed by Metropolis sampling. To generate priors, we first calculated the Theta (Hom) and Theta (k) for each data set with the program ARLEQUIN. We then used these estimates as the conservative median of a uniform prior distribution for Θ in the program MIGRATE. We used the default prior distribution for M. We ran the data sets initially with these prior distributions for a short Markov-Chain-Monte-Carlo simulation with a single chain (5000 recorded steps with 100 increments and 10% burn-in). Finally, with the results of the shorter chain as prior distribution, longer chains were then run with three heated Markov-Chain-Monte-Carlo simulation chains (temperature 1.5, 3, 100 000) and one cold chain with chain swapping at every 10th interval (50 000 recorded steps with 200 increments and 5000 burn-in).
In order to assess jump dispersal, we regressed the mode of our estimates of (asymmetric) migration rates with distance in kilometres between the source and sink populations. In a scenario of jump dispersal, migration rates of R. rattus would be much higher than R. satarae for more distant populations. To assess overlap between the parameter estimates in a one-tailed framework, we obtained the 95th percentile of the R. satarae posterior distribution and tabulated the proportion above this cutoff in R. rattus distribution for the corresponding parameter. Larger proportion of values above the cutoff would suggest higher migration in R. rattus.
Results
A total of 319 rats (143 R. satarae and 176 R. rattus) were captured in seven sampling localities from all the three habitats (village, forest and forest edge) in 4209 trap nights (Figure 1). In the forest, we captured 76 rats that were all identified genetically as R. satarae. In villages, we captured 174 rats, 168 of which were R. rattus and 6 were R. satarae. From the forest edge, we captured 69 rats (61 R. satarae and 8 R. rattus).
Habitat partitioning between the two rattus species
We used the capture information from the sampling effort in May 2011 to April 2013. Total trapping success was slightly higher in forests (7.5%) than in villages (6.3% in villages). Binomial tests for observed number of captures in both habitats confirmed that both species have unequal capture probability (P=0.0001). Capture data revealed that in forests, R. satarae has much higher capture probability as compared with R. rattus, whereas in villages, R. rattus has higher probability of capture (Supplementary Figures F1a and b). This suggests that R. rattus is more abundant in the commensal, village habitat, whereas R. satarae is more abundant in the non-commensal, forested habitat.
Genetic diversity
The genetic data set included 172 R. rattus and 132 R. satarae (Table 1). Relatedness analysis on microsatellite markers revealed average within-group relatedness values between 0.04 and 0.17 (Supplementary Table S2). Individuals with relatedness values higher than 0.5 were removed from further analyses (13 in R. satarae data set and none in R. rattus).
Table 1. Sample table with summary statistics and genetic diversity indices.
| Locality | AN | AG | BG | OT | KT | TH | KD | Total |
|---|---|---|---|---|---|---|---|---|
| R. satarae | ||||||||
| N | 6 | 24 | 4 | 4 | 28 | 32 | 34 | 132 |
| h | 4 | 13 | 4 | 2 | 13 | 14 | 20 | 70 |
| Na | 5.71 | 11.29 | 5.29 | 4.06 | 12.47 | 12.77 | 14.06 | |
| HO | 0.75 | 0.75 | 0.81 | 0.61 | 0.70 | 0.71 | 0.73 | |
| HE | 0.84 | 0.86 | 0.87 | 0.78 | 0.88 | 0.87 | 0.89 | |
| R. rattus | ||||||||
| N | 32 | 21 | 25 | 27 | 25 | 23 | 19 | 172 |
| h | 16 | 7 | 11 | 7 | 10 | 10 | 10 | 81 |
| Na | 15.12 | 12.77 | 13.88 | 11.29 | 12.24 | 12.29 | 11.88 | |
| HO | 0.84 | 0.77 | 0.87 | 0.75 | 0.76 | 0.76 | 0.77 | |
| HE | 0.90 | 0.88 | 0.90 | 0.83 | 0.87 | 0.88 | 0.89 | |
Abbreviations: h, number of mitochondrial D-loop haplotypes; N, number of samples; Na, number of alleles; HO, observed heterozygosity; HE, expected heterozygosity.
Total mitochondrial haplotypes were lower in R. satarae (70 and 81 haplotypes in R. satarae and R. rattus, respectively, Table 1, accession numbers KJ603317–KJ603409 and KP159524–KP159565) as expected from unequal sample sizes; however, haplotypic diversity was comparable (0.981 and 0.969 for R. satarae and R. rattus, respectively). Nucleotide diversity was higher for R. satarae (π=0.017) compared with R. rattus (π=0.009).
Number of microsatellite alleles, observed and expected heterozygosities were comparable for either species (Table 1,Supplementary Table S1). One microsatellite locus (Rr21) did not amplify in two populations of R. satarae, AN and AG. It was treated as missing data (missing data: R. satarae=2.13%, R. rattus=0.5%). All loci were tested for deviation from Hardy–Weinberg equilibrium. Allelic richness estimates (corrected for sample size) were much higher for R. rattus than for R. satarae. However, despite the unequal sample sizes, there was no difference between the group means for allelic richness and gene diversity (allelic richness, gene diversity: R. satarae=1.86, 0.86, R. rattus=1.88, 0.88; P>0.05). Test for neutrality of loci indicated that microsatellite loci were selectively neutral in both species (Supplementary Table S3). Presence of null alleles was indicated for several loci; the overall frequency of null alleles was higher in R. satarae than in R. rattus (null allele frequencies in Supplementary Table S4 for R. satarae and R. rattus data sets). However, the mean null allele frequencies per population were low for both the data sets.
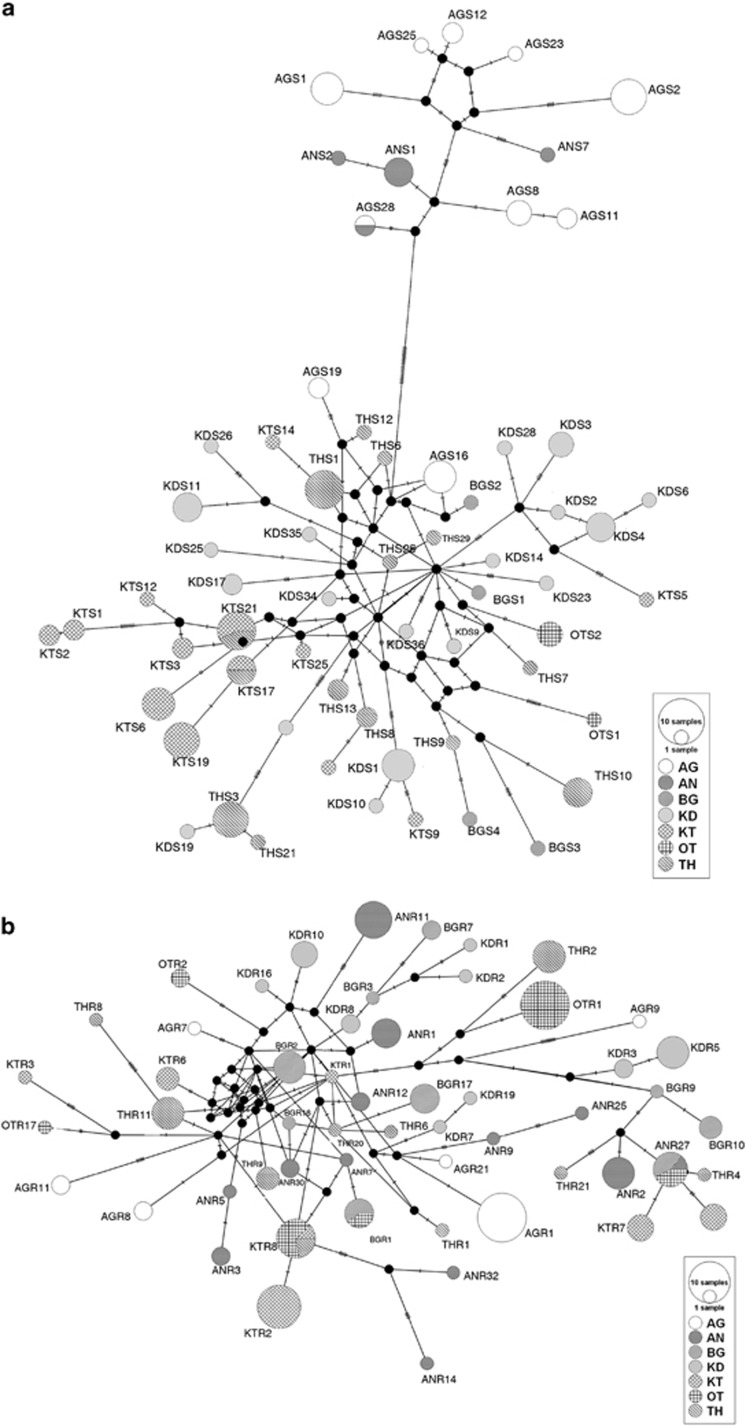
The median joining network for the 81 haplotypes of R. satarae (Figure 2a) showed a clear distinction between the northern (AN and AG) populations and the central-southern populations. The central-southern group haplotypes were mostly unstructured for the populations BG, OT and TH, but the populations KT and KD emerge as distinct haplogroups. Haplotype sharing was apparent only between the geographically closer populations AN–AG and KT–TH. On the other hand, R. rattus network for the 70 haplotypes (Figure 2b) had no clear structure in haplotype clustering and showed haplotype sharing between distant populations such as AN and OT.
Figure 2.
Haplotype network constructed using the median joining method. (a) Median joining network for 70 haplotypes of R. satarae reveal clear differentiation of the populations into geographical north–south groups and further structuring within the south cluster. Haplotype sharing is rare and present only between close populations. (b) Median joining network for 81 haplotypes of R. rattus indicates haplotype sharing between the central populations. There is no well-defined geographical structure.
Population structure
FST calculated using the mitochondrial locus was much higher for R. satarae (0.420; P=0) than for R. rattus (0.195; P=0) for four populations (AG, KT, TH and KD). Statistically significant pairwise FST values (P<0.05) ranged from 0.111 to 0.529 for R. satarae and 0.098 to 0.262 for R. rattus (Table 2a and b). Analysis of molecular variance based on mitochondrial DNA indicated that nearly 9.0% of the total variation was attributed to diversity among populations in R. satarae (df=1, sum of squares=37.94, variance components=1.01) and 8.7% in R. rattus (df=1, sum of squares=16.60, variance components=0.5).
Table 2. Estimates of population differentiation in four population pairs: F-statistics.
| AG | KT | TH | KD | |
|---|---|---|---|---|
| (a) R. satarae | ||||
| AG | 0 | 0.529* | 0.474* | 0.482* |
| KT | 0.065* | 0 | 0.170* | 0.206* |
| TH | 0.073* | 0.072* | 0 | 0.111* |
| KD | 0.126* | 0.054* | 0.030* | 0 |
| (b) R. rattus | ||||
| AG | 0 | 0.262* | 0.206* | 0.223* |
| KT | 0.026* | 0 | 0.098* | 0.168* |
| TH | 0.033 | 0.031* | 0 | 0.136* |
| KD | 0.025 | 0.031* | 0.054* | 0 |
Values above diagonal are pairwise FST values calculated using mitochondrial control region (970 bp in R. satarae, 973 bp in R. rattus). Values below diagonal are pairwise RST values calculated using 17 microsatellite loci.
*P<0.05.
Global RST values for microsatellite loci (four population pairs for each species) also suggested higher levels of genetic differentiation in R. satarae (RST=0.065, P=0) and R. rattus (RST=0.034, P=0). As the mean null allele frequencies were low and as such would have only a marginal effect on estimates of population differentiation (Chapuis and Estoup, 2007), we did not correct for null alleles in our data sets. Analysis of molecular variance for microsatellite loci suggested that very low level of total variation was attributable to diversity among populations (8.31% in R. satarae: df=1, sum of squares=1741.4, variance components=24.5; 3.3% in R. rattus: df=1, sum of squares=1008.1, variance components=13.0). Significant pairwise RST values in R. satarae ranged from 0.030 to 0.126 and in R. rattus from 0.026 to 0.054 (Table 2a and b) suggesting low to high level of genetic differentiation among the different populations.
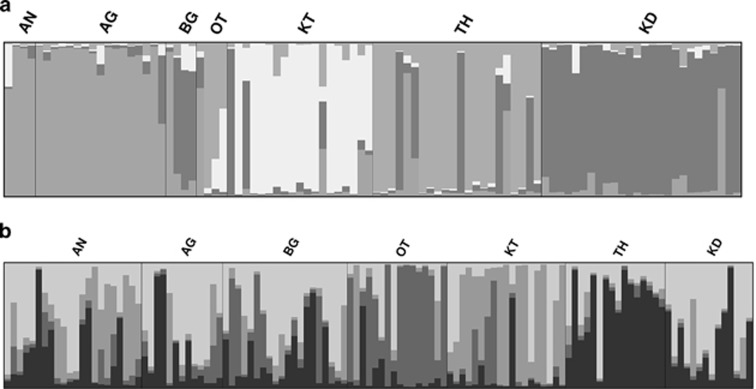
For STRUCTURE analysis, we included only unrelated individuals (96 R. satarae and 120 R. rattus). Structure identified K=4 as the most likely number of Hardy–Weinberg clusters (ΔK=107.4 ) in the R. satarae data set. The histogram of the individual q values for four clusters (Figure 3a) suggested that the northern populations of AN and AG, the central populations of KT, TH and the southern population KD form four distinct clusters. The central population of BG clustered with mostly KD with one individual showing similarity with the northern cluster. OT was overall admixed with northern, KT and TH. TH and KT had some individuals with the southern KD genotypes indicating some level of gene flow. Four clusters were also suggested in R. rattus (ΔK=51.107), but the membership coefficients indicated high level of admixture between the clusters (Figure 3b).
Figure 3.
Genetic cluster analyses of the data sets with microsatellite loci for both species. Each vertical bar represents a single individual and colour of the individual represents membership coefficient of the individual for a cluster. (a) Histogram of individual membership coefficients in R. satarae for K=4 (ΔK=107.4 ). (b) Histogram of individual membership coefficients in R. rattus for K=4 (ΔK=51.107). The full colour version of this figure is available at the Heredity online.
Isolation-by-distance
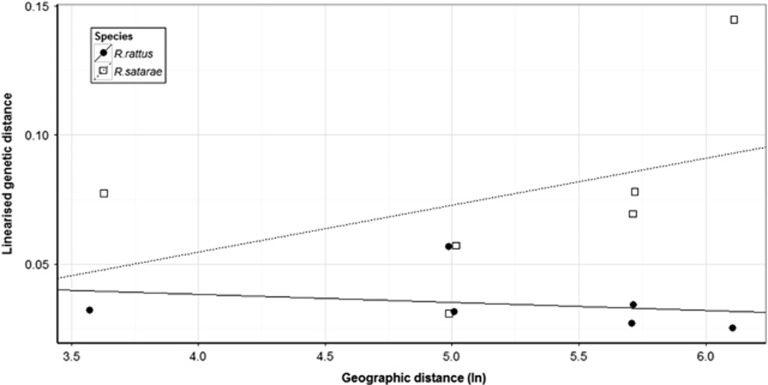
Mantel tests based on four populations (Figure 4) revealed nonsignificant isolation-by-distance in R. satarae (slope=0.018, P>0.1), whereas there was no correlation in R. rattus (slope=−0.003, P>0.1) for the similar population pairs.
Figure 4.
Relationship between logarithm of geographic distance and linearised genetic distance. RST=RST/1–RST as estimated using the microsatellite data set (closed circles, solid line: R. rattus, open squares, dotted line: R. satarae). No correlation is significant at 95% confidence level.
Migration rate and effective population size estimation
The Bayesian estimates for Θ (4Neμ) indicated that the effective population size was high but similar for both species (Table 3). The populations TH and KD had much higher estimate for Ne in R. satarae.
Table 3. Estimates of mutation scaled effective population size.
| Population | R. satarae | R. rattus |
|---|---|---|
| AG | 0.13 (0.10, 1.90) | 0.13 (0.10, 1.97) |
| KT | 0.13 (0.10, 1.84) | 0.13 (0.10, 1.90) |
| TH | 0.93 (0.10, 2.70) | 0.13 (0.10, 2.03) |
| KD | 0.53 (0.10, 2.63) | 0.13 (0.10, 2.36) |
Mode of the posterior distribution of Θ (Θ=4Neμ, where Ne is effective population size and μ is mutation rate) for the non-commensal R. satarae and commensal R. rattus. Values in brackets represent the 95% confidence limits of the mode.
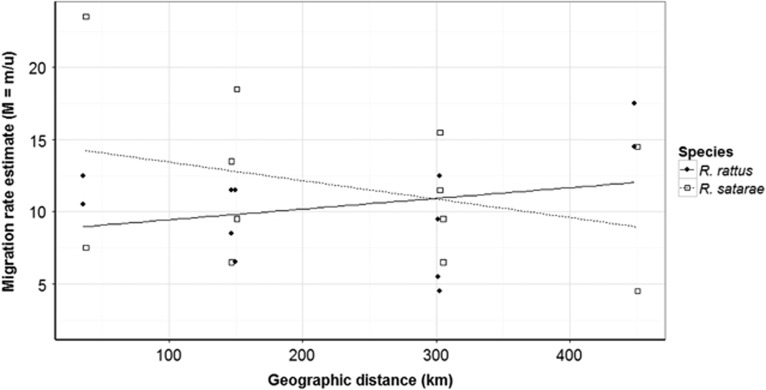
We estimated 12 asymmetric migration rates, M (m/μ) between the four populations AG, KT, TH and KD. Although we observed higher modes for the posterior distribution of migration rates in R. satarae than in R. rattus (Table 4), larger proportion of the posterior distribution in R. rattus was higher than that of R. satarae (Supplementary Table S5). This indicates that the distribution of the estimated migration rate values in R. rattus are higher than in R. satarae. In addition, migration rates had a negative correlation with distance between the source populations in R. satarae (Figure 5). There was no such effect of distance on migration rate estimates in R. rattus, and in fact, the farthest populations (AG–KD, 400 km apart) had very high migration rates.
Table 4. Estimates of the mutation scaled migration rate.
| Source-sink | Distance | R. rattus | R. satarae |
|---|---|---|---|
| KT–TH | 38 | 12.5 (0, 36) | 7.5 (0, 32) |
| TH–KT | 10.5 (0, 35) | 23.5 (0, 47) | |
| TH–KD | 147 | 11.5 (0, 36) | 13.5 (0, 38) |
| KD–TH | 8.5 (0, 34) | 6.5 (0, 32) | |
| KT–KD | 151 | 6.5 (0, 31) | 9.5 (0, 34) |
| KD–KT | 11.5 (0, 36) | 18.5 (0, 42) | |
| AG–KT | 303 | 9.5 (0, 35) | 11.5 (0, 36) |
| KT–AG | 5.5 (0, 30) | 15.5 (0, 39) | |
| AG–TH | 305 | 12.5 (0, 38) | 6.5 (0, 31) |
| TH–AG | 4.5 (0, 30) | 9.5 (0, 34) | |
| AG–KD | 450 | 17.5 (0, 42) | 4.5 (0, 29) |
| KD–AG | 14.5 (0, 38) | 14.5 (0, 39) |
Mode of the posterior distribution of M (M=m/μ, where m is the immigration rate per generation and μ is mutation rate) for the non-commensal R. satarae and commensal R. rattus. Values in brackets represent the 95% confidence limits of the mode.
Figure 5.
Estimation of asymmetric migration rates for four populations. Comparing migration rates with distance (in km) between source populations for R. rattus and R. satarae (closed circles, solid line: R. rattus, open squares, dotted line: R. satarae). Modes of posterior distribution of migration rates were used to construct this plot.
Discussion
In this study, we compared population genetic structure of two species R. rattus and R. satarae in a topographically complex region of the Western Ghats, a mountain range in southern India.
Commensal R. rattus, non-commensal R. satarae?
We found that the two species occupied different types of habitat in the same region: R. rattus seemed mostly associated with human settlements and R. satarae appeared abundant in forested habitats. Pagès et al. (2011) have suggested that R. rattus and R. satarae are sympatric and syntopic in the montane tropical forests of southern Western Ghats. We found possible co-occurrence of the two species in forest edge and to a much lesser extent, in villages. Therefore, we propose that although the two species are sympatric, there is allopatry at the microhabitat scale.
As a genus, Rattus is the most speciose among rodents (Musser and Carleton, 2005). Several Rattus species are described as being obligate or opportunistic commensals with large geographic ranges (Singleton et al., 2003). Our ecological sampling data reveal that some species are not commensal. R. satarae belongs to the 'norvegicus-nitidus' clade of the Rattus phylogeny (Pagès et al., 2011). The other members of this clade, R. norvegicus and R. nitidus are both described to be commensal (Taylor et al., 1982). R. norvegicus although worldwide in distribution, is believed to have originated in China (Ness et al., 2012) and R. nitidus is also distributed in southern China (Aplin et al., 2008). Thus, it is interesting to investigate the disjointed distribution and distinctive habitat preference of R. satarae in this context. Such cases could be highlighted as adaptive perspectives into understanding evolution in Rattus as this genus has reportedly undergone rapid speciation (Rowe et al., 2011).
Population structure and genetic differentiation may be higher in non-commensal R. satarae than the commensal R. rattus
Measures of population differentiation depend on both effective population size and the magnitude of gene flow. As a result of their small body size, most rodents are expected (Carbone and Gittleman, 2002) and observed to have large effective population sizes (Ness et al., 2012). Estimates for our study species R. rattus and R. satarae also indicated high effective population sizes. It is therefore possible that the high effective size in both species leads to the overall low levels of observed genetic differentiation.
Although the observed genetic differentiation was low, all population genetic analyses (haplotype network, analysis of molecular variance, FST) based on mitochondrial, as well as nuclear markers indicated higher genetic differentiation for non-commensal R. satarae than the commensal R. rattus. Also, population clusters in the non-commensal species showed better partitioning, whereas those in the commensal showed admixture. In addition, isolation-by-distance was higher in the non-commensal than in the commensal. FST for mitochondrial markers is higher than that of nuclear regions, possibly because of male biased dispersal and female philopatry (reviewed in Zhang and Hewitt, 2003).
It would be interesting to test the effect of known biogeographic divides on genetic differentiation within the Western Ghats. The Palghat gap, which is the oldest and widest divide in this mountain range, has been identified as a barrier for several taxa including plants (Subramanyam and Nayar, 1974), amphibians (Nair et al., 2012), birds (Robin et al., 2010) and even large mammals like elephants (Vidya et al., 2004). Sampling designed to investigate the special effects of the barriers may help elucidate the effect of this biogeographic barrier on the dispersal of Rattus species. It would also be interesting to investigate patterns of genetic differentiation (as driven by biogeographical barriers) in other rodent species.
Higher rates for long-distance migration in commensal R. rattus
Our results suggest high gene flow over long distance (MIGRATE and isolation-by-distance analyses) for the commensal R. rattus. These observations could support models invoking passive transportation by humans of commensal species over long distances and the resultant jump dispersal. Long distance migration has been reported in other commensal species such as Rattus norvegicus (Gardner-Santana et al., 2009; Kajdacsi et al., 2013) and has been attributed to human-mediated dispersal (Baker, 1994) and disturbance effects such as neighbourhood restoration (Taylor and Quy, 2009).
Can we conclude that commensals generally have lower population differentiation and higher gene flow than non-commensals? Brouat et al. (2007), using a similar pairwise sampling method as ours, found that the commensal species has higher genetic structure and stronger correlation between genetic and geographic distances than non-commensals. The landscape they studied was topographically homogeneous with large savannas interspersed. Although commensal habitats were relatively isolated in their study, our study area has high human population density and large urban/rural settlements. We suggest that whether commensals have higher dispersal and migration over large distance may depend specifically on the landscape, and additional study may be necessary to elucidate generalisable patterns.
It is also possible that rats in the commensal habitat are exposed to intermittent control and eradication measures along with constant anthropogenic disturbance. Such processes may lead to repeated events of local extinction through instability in food and nesting resources, and subsequent recolonisation through either active or passive dispersal. This could result in complex and very location-specific patterns of population genetic variation and structure for the commensal species.
Possible evolutionary implications of commensalism
Jump dispersal can have several possible evolutionary consequences. A continuously high level of gene flow would potentially lead to a complete breakdown of isolation between populations and hence, of local adaptation (Bridle and Vines, 2007). In such a scenario, it would be interesting to study how commensals adapt to changes in the environment that are not human associated.
On the other hand, it has been suggested that the ‘commensal' environment itself may provide buffering against the environmental and climatic changes, through availability of protected nesting, roosting and breeding sites, and through reduced predation pressures (Wyncoll and Tangri, 1991). Therefore, the commensal species may be expected to show effects of release from natural selection in the commensal environment, which would greatly affect the evolutionary trajectories of these species. Alternatively, the commensal habitat itself may present a different host of selective forces such as increased pathogen loads because of crowding, pest control measures through rodenticides and recurrent anthropogenic disturbance, which may elicit a different pattern of adaptation in the commensal species. In addition, as these commensals are being increasingly recognised as sources of zoonotic diseases (Firth et al., 2014), their increased dispersal ability would have tremendous impact on the spread of such diseases.
Summary
In summary, we present a population genetic comparison of two co-occurring rats in the Southern Western Ghats. The commensal R. rattus reveals lower population structure than the non-commensal R. satarae. In addition, differences in gene flow between the two species are accentuated for longer distances, potentially indicating jump dispersal. Our results are significant because (1) they identify a non-commensal species of Rattus, a speciose group where most species appear commensal and (2) they support long-distance dispersals of commensals, a possible mechanistic basis for their geographical success. Our results suggest that on evolutionary time scales, local adaptation may be difficult to maintain in commensal populations of species, such as those in the genus Rattus.
Data archiving
All haplotypes have been deposited in GenBank database with the accession numbers KJ603317–KJ603409 and KP159524–KP159565. Raw microsatellite genotype data available from the Dryad Digital Repository. 10.5061/dryad.8265 (http://dx.doi.org/10.5061/dryad.8265).
Acknowledgments
This study constitutes part of the doctoral thesis of AV. Field sampling was performed with research permissions from the state forest departments of Karnataka and Tamil Nadu. We acknowledge Ganesh Mane, Deepak Kamble for helping in field work and Abhishek Shukla in generating genetic data. We thank Dr Fiona Savory, Krishnapriya Tamma and other lab members at the Ecology and Evolution group at National Centre for Biological Sciences for inputs in manuscript preparation, and Aditya Joshi for help in preparing maps. This work was funded by Ramanujan grant to URK and NCBS internal support to URK. All molecular work was performed at the National Centre for Biological Sciences, Bangalore.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Abdelkrim J, Pascal M, Samadi S. (2005). Island colonization and founder effects: the invasion of the Guadeloupe islands by ship rats (Rattus rattus. Mol Ecol 14: 2923–2931. [DOI] [PubMed] [Google Scholar]
- Aplin K, Lunde D, Molur S. (2008). Rattus nitidus. The IUCN Red List of Threatened Species. Version 2014.3. Available at www.iucnredlist.org (downloaded on 16 April 2015.
- Aplin KP, Suzuki H, Chinen AA, Chesser RT, Have J, ten, Donnellan SC et al. (2011). Multiple geographic origins of commensalism and complex dispersal history of black rats. PLoS One 6: e26357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker AEM. (1994). Stowaway transport rates of house mice (Mus domesticus) and deermice (Peromyscus maniculatus)*. In: Proceedings of the Sixteenth Vertebrate Pest Conference (1994). University of California: Davis. [Google Scholar]
- Beerli P. (2006). Comparison of Bayesian and maximum likelihood inference of population genetic parameters. Bioinformatics 22: 341–345. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57: 289–300. [Google Scholar]
- Bridle JR, Vines TH. (2007). Limits to evolution at range margins: when and why does adaptation fail? Trends Ecol Evol 22: 140–147. [DOI] [PubMed] [Google Scholar]
- Brouat C, Loiseau A, Kane M, Bâ K, Duplantier J-M. (2007). Population genetic structure of two ecologically distinct multimammate rats: the commensal Mastomys natalensis and the wild Mastomys erythroleucus in southeastern Senegal. Mol Ecol 16: 2985–2997. [DOI] [PubMed] [Google Scholar]
- Carbone C, Gittleman JL. (2002). A common rule for the scaling of carnivore density. Science 295: 2273–2276. [DOI] [PubMed] [Google Scholar]
- Chapuis MP, Estoup A. (2007). Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol 24: 621–631. [DOI] [PubMed] [Google Scholar]
- Cucchi T, Vigne J-D. (2006). Origin and diffusion of the house mouse in the Mediterranean. Hum Evol 21: 95–106. [Google Scholar]
- Earl DA, vonHoldt BM. (2011). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4: 359–361. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10: 564–567. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro I. (2013). Rodent endemism, turnover and biogeographical transitions on elevation gradients in the northwestern Argentinian Andes. Mamm Biol—Zeitschrift für Säugetierkd 78: 322–331. [Google Scholar]
- Firth C, Bhat M, Firth MA, Williams SH, Frye MJ, Simmonds P et al. (2014). Detection of zoonotic pathogens and characterization of novel viruses carried by commensal Rattus norvegicus in New York City. mBio 5: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner-Santana LC, Norris DE, Fornadel CM, Hinson ER, Klein SL, Glass GE. (2009). Commensal ecology, urban landscapes, and their influence on the genetic characteristics of city-dwelling Norway rats (Rattus norvegicus). Mol Ecol 18: 2766–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet J. (2003). FSTAT Version 2.9.4.: A Program to Estimate and Test Population Genetics Parameters. Updat. from Goudet (1995). Available from http://www.unil.ch/izea/softwares/fstat.html.
- Hingston M, Goodman SM, Ganzhorn JU, Sommer S. (2005). Reconstruction of the colonization of southern Madagascar by introduced Rattus rattus. J Biogeogr 32: 1549–1559. [Google Scholar]
- Jakobsson M, Rosenberg NA. (2007). CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23: 1801–1806. [DOI] [PubMed] [Google Scholar]
- Jones EP, Skirnisson K, McGovern TH, Gilbert MTP, Willerslev E, Searle JB. (2012). Fellow travellers: a concordance of colonization patterns between mice and men in the North Atlantic region. BMC Evol Biol 12: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajdacsi B, Costa F, Hyseni C, Porter F, Brown J, Rodrigues G et al. (2013). Urban population genetics of slum-dwelling rats (Rattus norvegicus) in Salvador, Brazil. Mol Ecol 22: 5056–5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landguth EL, Fedy BC, OYLER-McCANCE SJ, Garey AL, Emel SL, Mumma M et al. (2012). Effects of sample size, number of markers, and allelic richness on the detection of spatial genetic pattern. Mol Ecol Resour 12: 276–284. [Google Scholar]
- Librado P, Rozas J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- Loiseau A, Rahelinirina S, Rahalison L, Konečný A, Duplantier JM, Brouat C. (2008). Isolation and characterization of microsatellites in Rattus rattus. Mol Ecol Resour 8: 916–918. [DOI] [PubMed] [Google Scholar]
- Matisoo-Smith E. (2009). The commensal model for human settlement of the Pacific 10 years on—what can we say and where to now? J Isl Coast Archaeol 4: 151–163. [Google Scholar]
- Matschiner M, Salzburger W. (2009). TANDEM: integrating automated allele binning into genetics and genomics workflows. Bioinformatics 25: 1982–1983. [DOI] [PubMed] [Google Scholar]
- Molur S, Singh M. (2009). Non-volant small mammals of the Western Ghats of Coorg District, southern India. Area 1: 589–608. [Google Scholar]
- Musser GG, Carleton MD. (2005). Superfamily Muroidea. In: Wilson DE, Reeder DM (eds) Mammal Species of the World A Taxonomic and Geographic Reference. The John Hopkins University Press, Baltimore, MD, USA, pp 894–1531. [Google Scholar]
- Nair A., Gopalan S V., George S, Kumar KS, Teacher a. GF, Merilä J. (2012). High cryptic diversity of endemic Indirana frogs in the Western Ghats biodiversity hotspot (T Garner and J Austin, Eds.). Anim Conserv 15: 489–498. [Google Scholar]
- Ness RW, Zhang Y-H, Cong L, Wang Y, Zhang J-X, Keightley PD. (2012). Nuclear gene variation in wild brown rats. G3 (Bethesda) 2: 1661–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagès M, Chaval Y, Herbreteau V, Waengsothorn S, Cosson J-F, Hugot J-P et al. (2010). Revisiting the taxonomy of the Rattini tribe: a phylogeny-based delimitation of species boundaries. BMC Evol Biol 10: 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagès M, Corbet G, Orth A, Volobouev V, Michaux J, Catzeflis F. (2011). Morphological, chromosomal, and genic differences between sympatric Rattus rattus and Rattus satarae in South India. J Mammal 92: 659–670. [Google Scholar]
- R Core Team. (2013) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria. Available from http://www.R-project.org/. [Google Scholar]
- Ramachandran V. (2013). Effect of habitat alteration on canopy bird and small mammal communities in the wet evergreen forests of the Western Ghats, PhD thesis. Manipal University: Manipal, Karnataka, India.
- Robin V V, Sinha A, Ramakrishnan U. (2010). Ancient geographical gaps and paleo-climate shape the phylogeography of an endemic bird in the sky islands of southern India. In: S Gadagkar (ed). PLoS One 5: e13321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins JH, Hingston M, Matisoo- Smith E, Ross HA. (2007). Identifying Rattus species using mitochondrial DNA. Mol Ecol Notes 7: 717–729. [Google Scholar]
- Robins JH, McLenachan PA, Phillips MJ, Craig L, Ross HA, Matisoo- Smith E. (2008). Dating of divergences within the Rattus genus phylogeny using whole mitochondrial genomes. Mol Phylogenet Evol 49: 460–466. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA. (2003). DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4: 137–138. [Google Scholar]
- Rousset F. (2008). GENEPOP'007: a complete reimplementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour 8: 103–106. [DOI] [PubMed] [Google Scholar]
- Rowe KC, Aplin KP, Baverstock PR, Moritz C. (2011). Recent and rapid speciation with limited morphological disparity in the genus Rattus. Syst Biol 60: 188–203. [DOI] [PubMed] [Google Scholar]
- Russell JC, Abdelkrim J, Fewster RM. (2009). Early Colonisation population structure of a Norway rat island invasion. Biol Invasions 11: 1557–1567. [Google Scholar]
- Schrey AW, Liebl AL, Richards CL, Martin LB. (2014). Range expansion of house sparrows (Passer domesticus) in Kenya: evidence of genetic admixture and human-mediated dispersal. J Hered 105: 60–69. [DOI] [PubMed] [Google Scholar]
- Schulte-Hostedde AI, Gibbs HL, Millar JS. (2001). Microgeographic genetic structure in the yellow-pine chipmunk (Tamias amoenus). Mol Ecol 10: 1625–1631. [DOI] [PubMed] [Google Scholar]
- Shanker K, Sukumar R. (1998). Community structure and demography of small-mammal populations in insular montane forests in southern India. Oecologia 116: 243–251. [DOI] [PubMed] [Google Scholar]
- Singleton GR, Hinds LA, Krebs CJ, Spratt DM (eds). (2003) Rats, Mice and People: Rodent Biology and Management. Australian Centre for International Agricultural Research: Canberra. [Google Scholar]
- Slatkin M. (1995). A Measure of population subdivision based on microsatellite allele frequencies. Genetics 139: 457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanyam K, Nayar M. (1974). Vegetation and phytogeography of the Western Ghats. In: Mani MS (ed) Ecology and Biogeography in India, Monographiae Biologicae. Springer Netherlands: DordrechtVol 23. [Google Scholar]
- Taylor JM, Calaby JH, Deusen HMV a N. (1982). A revision of the Genus Rattus (Rodentia, Muridae ) in the New Guinean Region. Bull Am Museum Nat Hist 173: 177–136. [Google Scholar]
- Taylor K, Quy R. (2009). Long distance movements of a common rat (Rattus norvegicus) revealed by radio-tracking. Mammalia 42: 63–72. [Google Scholar]
- Trenel P, Hansen MM, Normand S, Borchsenious F. (2008). Landscape genetics, historical isolation and cross-Andean gene flow in the wax palm, Ceroxylon echinulatum (Arecaceae). Mol Ecol 17: 3528–3540. [DOI] [PubMed] [Google Scholar]
- Valière N. (2002). GIMLET: a computer program for analysing genetic individual identification data. Mol Ecol Notes 2: 377–379. [Google Scholar]
- Vidya TNC, Fernando P, Melnick DJ, Sukumar R. (2004). Population differentiation within and among Asian elephant (Elephas maximus) populations in southern India. Heredity (Edinb) 94: 71–80. [DOI] [PubMed] [Google Scholar]
- Vigne J-D. (2011). The origins of animal domestication and husbandry: a major change in the history of humanity and the biosphere. C R Biol 334: 171–181. [DOI] [PubMed] [Google Scholar]
- Wyncoll G, Tangri D. (1991). The origins of commensalism and human sedentism. Paléorient 17: 157–159. [Google Scholar]
- Zeder MA. (2012). Pathways to animal domestication. In: Gepts P, Famula T, Bettinger R (eds) Biodiversity in Agriculature: Domestication, Evolution, and Sustainability. Cambridge University Press: New York. [Google Scholar]
- Zhang D-X, Hewitt GM. (2003). Nuclear DNA analyses in genetic studies of populations: practice, problems and prospects. Mol Ecol 12: 563–584. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.