Abstract
Epithelial–mesenchymal transition (EMT) has been closely related with invasive and metastatic properties of cancer. Recently, the convergence of DNA damage response and EMT in cancer development has received a great amount of scientific attention. Here, we showed that EMT is induced by the downregulation of RAP80, a well‐known regulator for DNA damage response. The knockdown of RAP80 leads to EMT‐like morphological changes and the increase of tumor sphere formation in non‐adhesive culture. Mechanistically, RAP80 controls a reciprocal regulatory axis of ZEB1 (for EMT activation) and miR200c (for EMT inhibition). The downregulation of RAP80 increases ZEB1 protein and decreases miR200c expression to activate EMT signaling in the form of drastic inhibitions of E‐cadherin, p16 and p21 expression. Using in vivo metastasis analysis, RAP80 knockdown cells are shown to dramatically metastasize into the lung and generate more malignant phenotype compared to controls. Interestingly, the expression level of RAP80 was positively correlated with the survival rate in lung adenocarcinoma and breast cancer patients. These findings indicate that RAP80 is a critical gatekeeper in impeding EMT‐induced metastasis and malignant phenotypes of cancer as well as preserving DNA integrity.
Keywords: Cancer, epithelial–mesenchymal transition, metastasis, RAP80, ZEB1
DNA damage response (DDR) is essential to genomic integrity by preventing the accumulation and transmission of mutations.1 DDR is frequently found in precancerous lesions and believed to be a barrier to tumorigenesis by induction of senescence or death in cells with excessive DNA damage.2, 3 Although the tumor‐suppressive roles of DDR in early premalignant lesions are well established,4, 5 the function of DDR is unclear in tumor progression during later stages, such as metastasis and tumor malignancy.
Receptor‐associated protein 80 (RAP80 or UIMC1) is an ubiquitin interaction motif‐containing (UIMC) nuclear protein that facilitates the recruitment of the BRCA1‐BARD1‐Abraxas/CCDC98‐MERIT‐BRCC36 complex to DNA damage sites, which initiates DDR.6, 7, 8, 9, 10, 11, 12, 13 Although previous studies indicate that defective RAP80 function/expression could lead to genomic instability and subsequent increased early stage cancer risk,14, 15, 16 the possible role of RAP80 on tumor progression and malignancy in later stages, including EMT‐derived metastasis, has not been investigated.
Recently, the convergence of DDR and epithelial–mesenchymal transition (EMT) in cancer development was reported.17, 18 EMT trans‐differentiation can generate cells with stem‐like properties.18 Cancer stem cells have been known to be resistant against DNA damage through activation of DDR.19, 20 Notably, the EMT regulator ZEB1 also promotes DDR and tumor resistance to DNA damage, suggesting that ZEB1 could be a convergent regulator for both EMT and DDR.17
In this study, we investigate the effect of the downregulation of RAP80 expression on EMT and cancer metastasis. We show the decrease of RAP80 expression to activate EMT through increasing the protein level of ZEB1, a convergent regulator for DDR and EMT, which drastically promotes pulmonary metastasis of cancer cells. Our results indicate that RAP80 is a critical gatekeeper that can impede EMT‐derived metastasis of cancer as well as secure DNA integrity.
Materials and Methods
Cell cultures
The HeLa cells were purchased from American Type Culture Collection and cultured in DMEM (HyClone, SH30243.01) with high glucose and l‐glutamine, supplemented with 10% FBS (HyClone, GE Healthcare Life Science, South Logan, USA, SH30919.03), 10 000 U/mL penicillin and 10 000 μg/mL streptomycin (GIBCO Invitrogen, Grand Island, NY, USA, 15140‐122), and maintained in humidified incubators at 37°C with 5% CO2.
Antibodies and reagents
The following antibodies were used for western blot: RAP80,6 vimentin (#5741; Cell Signaling Technology, Inc. Danvers, MA, USA), E‐cadherin (#3195; Cell Signaling Technology) and TCF8/ZEB1 (#3396; Cell Signaling Technology, Inc. Danvers, MA, USA), c‐myc (sc‐40; Santa Cruz Biotechnology, Texas, USA) and β‐actin (sc‐477778; Santa Cruz Biotechnology, Texas, USA). Indocyanine Green (ICG) was purchased from Dongindang Pharm (Siheung, Korea).
RAP80 knockdown HeLa cell line generation
To produce lentivirus expressing shRNA for RAP80, pLKO‐shRAP80 or pLKO‐empty plasmids were co‐transfected with the lentivirus packaging plasmids (psPAX2, pMD2G and VSV‐G) into 293T cells. The virus containing cell culture supernatant was harvested, filtered through a 0.22‐μm pore‐size filter and used to infect HeLa cells. To generate stable RAP80 knockdown cells, infected cells were selected by puromycin (2 μg/mL) for 1 week.
Tumor sphere formation assay
Tissue culture dishes were coated with polyhydroxyethylmethacrylate polymer (polyHEMA; Sigma‐Aldrich, St. Louis, MO, USA) to facilitate sphere formation.21, 22 Briefly, polyHEMA was dissolved in 95% ethanol at 12% (w/v). A working solution was made by a further dilution of 1:10 in 95% ethanol and was added to 12‐well tissue culture plates at 0.3 mL per well. A hydrophobic surface was formed after the polyHEMA solution dried out at room temperature in a tissue culture hood. Approximately, 1 × 105 cells were plated into the polyHEMA‐coated 12‐well plates and incubated for 1 day or 2 days for sphere formation. Tumor sphere were counted at 1 and 2 days.
MTT assays
Cells seeded on 24‐well micro‐plates at 1 × 104 cells/well were incubated with suspended cells for the indicated time periods. Following incubation with the suspended culture, the medium was removed, and the cells were then incubated with 10‐μL MTT (M5655; Sigma Aldrich, St. Louis, MO, USA) solution (5 mg/mL MTT in PBS) for 1 h. The samples were then solubilized in DMSO. The purple formazan dye was quantified by absorbance at 540 nm.
Immunoblotting
Total proteins were extracted using a NETN lysis buffer (0.5% NP40, 150 mM NaCl, 0.5 mM EDTA, 20 mM Tris) with protease inhibitor cocktail, phosphatase inhibitor cocktail and 1 mM DTT and subjected to western blot analysis with specific antibodies. Fusion Solo ChemiDoc system (Fisher Biotech, Wembley, Australia) and Bio1D software (Fisher Biotech, Wembley, Australia) were used for detecting chemiluminescence.
Quantitative RT‐PCR
Total RNA were isolated and 1 μg of total RNA used for cDNA synthesis. The SYBR green qPCR assay kit (Toyobo, Osaka, Japan) was used with diluted cDNA for each sample. The samples were amplified with the CFX96 (Bio‐Rad, Hercules, California, USA). Primers used for qRT‐PCR are listed in Table S1.
Animal care and xenograft experiments
Balb/C nude mice (5 weeks, male) were used in all studies. All mice were housed at the Chungnam National University mouse facilities and all procedures were approved by the Institutional Animal Care and Use Committee (CNU‐00540). RAP80 knockdown and control HeLa cells were i.v. injected at 1 × 105 cells/mouse. On days 1, 3, 7, 14 and 25 after i.v. injection, lungs were harvested to check metastasis under light microscope. The same lung samples were fixed with 10% formalin and embedded in paraffin. H&E staining was performed using the Leica Jung Autostainer XL (Leica Microsystems, Buffalo Grove, IL, USA).
Labeling and in vivo tracking of cells using near‐infrared fluorescence imaging
For the labeling of cells with ICG, control and shRAP80‐2 HeLa cells suspended in cell culture medium were incubated with ICG (1 mg/mL) at 37°C for 3 h. Unlabeled ICG was removed by washing the cell mixture with PBS (GIBCO Invitrogen, Grand Island, NY, USA). After washing, the ICG labeled cells were re‐suspended into PBS and a fixed number of cells (1 × 106 cells/100 μL) were injected via tail vein. For near‐infrared (NIR) fluorescence imaging, lungs were dissected at 1, 2 and 7 days after injection. The lungs were placed in a chamber sealed against light and connected to the cold charge‐coupled device (CCD) camera (Orca ERG; Hamamatsu Photonics, Hamamatsu City, Japan). NIR images of lungs were acquired using a 760‐nm LED light as the excitation light source and an 845/55 emission filter.
Statistical analysis
Student's t‐tests were applied for comparisons. All data are expressed as mean ± SD. The significance threshold was at a level of 5% (*P < 0.05; **P < 0.01, ***P < 0.001). All the experiments were repeated three times.
Results
Downregulation of RAP80 induces epithelial–mesenchymal transition phenotypes
To investigate the role of RAP80 on EMT, RAP80 knockdown cells were generated by the stable expression of RAP80 shRNA in HeLa cells using shRAP80 lentivirus to test if EMT signal is induced. In RAP80 knockdowned cells, vimentin and c‐myc (mesenchymal specific proteins) are increased; in contrast, E‐cadherin (a epithelial specific protein) is markedly decreased (Fig. 1a). We also show that vimentin mRNA transcript is increased by RAP80 knockdown (Fig. 1b). However, N‐cadherin is barely detected and not changed by Rap80 knockdown (Fig. 1a). Among four independent Rap80 knockdown cells, we selected two independent Rap80 knockdown cells (shRap80‐1 and shRap80‐2) for further analysis. First, RAP80 knockdown cell lines were tested for morphological changes induced by EMT. As shown in Figure 2(a), RAP80 knockdown cells tend to shrink and detach from surfaces to become globular, indicating that loss of RAP80 induces EMT‐like morphological changes. To find further evidence for the induction of EMT in RAP80 knockdown cells, we tested anchorage independent growth and tumor sphere formation, another characteristic change of EMT.23 Anchorage independent growth and tumor sphere formation were tested with cells grown in poly‐HEMA (poly‐hydroxyethyl methacrylate)‐treated surfaces to prevent cell adhesion. Under non‐adhesive culture conditions, the number and morphology of tumor spheres with control and RAP80 knockdown cells were analyzed. As shown in Figure 2(b,c), the number of tumor spheres is greatly increased in the shRAP80‐1 and the shRAP80‐2 cell lines, indicating EMT induction in RAP80 knockdown cells. In addition, the morphology of spheres is different in control compared to RAP80 knockdown cells. The control vector‐infected cells formed small spheres that were round with tightly connected cells (Fig. 2b, left panel). In contrast, RAP80 knockdown cells (shRAP80‐1 and shRAP80‐2) formed large spheres that were perfectly round and, instead, were loosely connected and easily dissociated (Fig. 2b middle and right panel). To confirm cell viability under non‐adhesive culture conditions, MTT assay was performed. As shown in Figure 2(d), RAP80 knockdown cells showed significant increase of cell viability under non‐adhesive culture conditions, indicating that loss of RAP80 induces anchorage‐independent cell growth. Together, the loss of RAP80 expression induces EMT‐related cellular phenotypes, such as tumor sphere formation and anchorage‐independent growth.
Figure 1.
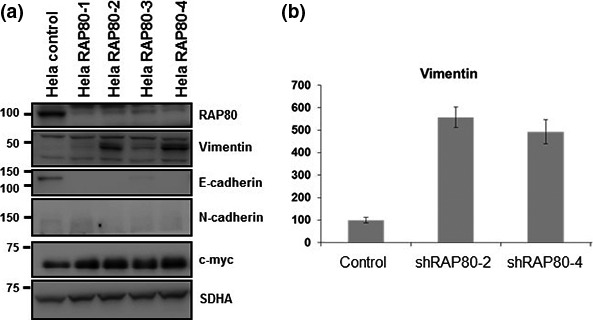
Epithelial–mesenchymal transition is induced by RAP80 knockdown. (a) Control and RAP80 knockdown HeLa cell lysates subjected to immunoblotting analysis using anti‐RAP80, vimentin, E‐cadherin, N‐cadherin, c‐myc and succinate dehydrogenase (SDHA) as loading control.31 (b) Total RNA extracted from control and RAP80 knockdown Hela cells subjected to RT‐PCR with vimentin specific RT‐PCR primers. Averages of three independent experiments were presented with error bars.
Figure 2.
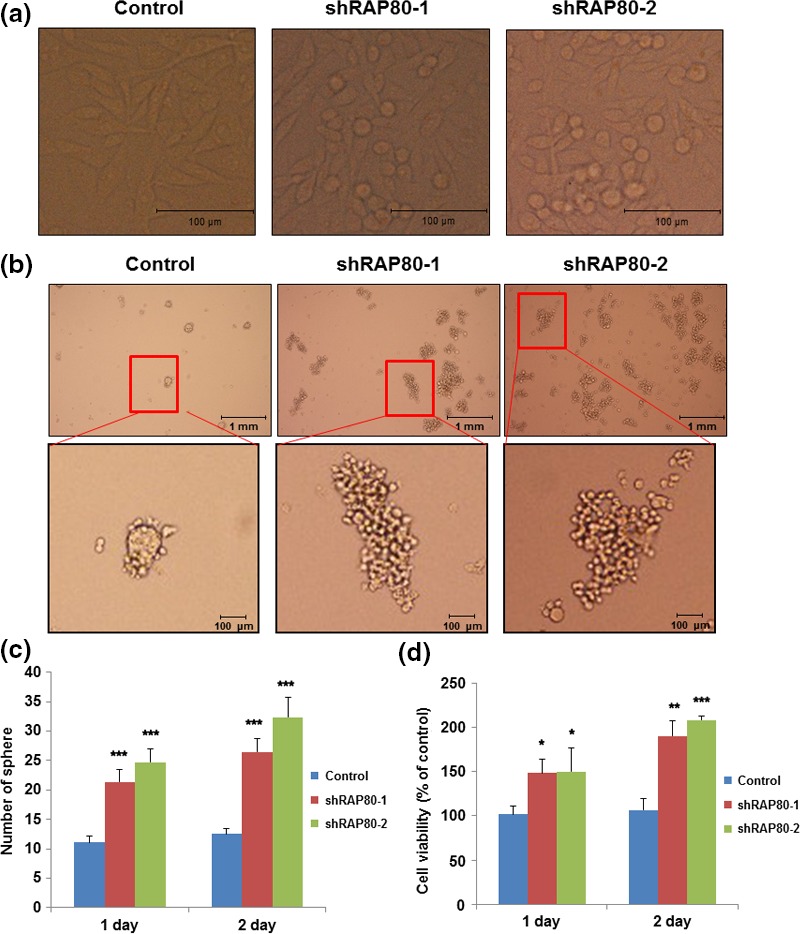
Epithelial–mesenchymal transition‐like morphology and tumor sphere formation are increased in RAP80 knockdown cells. (a) Control and two independent RAP80 knockdown HeLa cells (shRAP80‐1 and shRAP80‐2) imaged by light microscopy. (b) Control and RAP80 knockdown HeLa cells incubated in poly‐HEMA coated plates for 1 day to generate tumor spheres and imaged by light microscopy. (c) The number of tumor spheres in the control and RAP80 knockdown HeLa cells counted after 1 and 2 days of non‐adhesive culture and presented as a mean of three independent experiments with error bars. (d) Cell viability under non‐adhesive culture condition determined by MTT assay and presented as the percentage of relative absorbance to control from three independent experiments with error bars.
ZEB1 is increased in RAP80 knockdown cells inducing epithelial–mesenchymal transition signaling
Epithelial–mesenchymal transition is an initial process of metastasis for malignant cancer progression, governed by a variety of regulators. Among EMT regulators, ZEB1 has been recently identified as an important regulator of EMT but also a regulator of radiosensitivity and DNA damage response.17 Because RAP80 is a well‐known regulator of DNA damage response, ZEB1 may be the intersection between EMT and DNA damage response. As shown in Figure 3(a), the protein level of ZEB1 increases in two independent RAP80 knockdown cell lines (shRAP80‐1 and shRAP80‐2). ZEB1 is a critical transcriptional regulator for EMT signaling proteins such as E‐cadherin, p16 and p21.24, 25 As shown in Figure 3(a), RAP80 knockdown causes the downregulation of E‐cadherin, p16 and p21 protein levels. Together, RAP80 regulates the EMT process through ZEB1 and its downstream effectors, such as E‐cadherin, p16 and p21. The mRNA level of E‐cadherin, p16 and p21 were determined using a quantitative real‐time PCR, since ZEB1 directly regulates the transcription of these effectors. As shown in Figure 3(b), two independent RAP80 knockdown cell lines showed significant decrease of E‐cadherin, p16 and p21 mRNA, correlated with relatively high expression of ZEB1 in RAP80 knockdown cell lines. Importantly, ZEB1 has been recently reported as a reciprocal repressor of miR200c, promoting EMT and cancer cell invasion.26 As shown in the bottom panel of Figure 3(b), miR200c is also significantly repressed in RAP80 knockdown cell lines. To consolidate negative correlation of RAP80 and ZEB1, we analyze the expression of RAP80 and ZEB1 in 42 lung cancer cell lines using the Body Atlas database from Nextbio Research. As shown in Figure 4(a) and Table S2, RAP80 expression is negatively correlated with ZEB1 expression, suggesting a RAP80‐ZEB1 regulatory axis for EMT. To support this axis, we analyze the protein level of RAP80, ZEB1 and vimentin in lung cancer cell lines (H1650, H1975 and H2030). As shown in Figure 4(b), H1975 show the highest expression of RAP80, with lowest expression of ZEB1 and vimentin, indicating that RAP80 would suppress ZEB1 and vimentin expression. However, E‐cadherin and N‐cadherin expression is not correlated with RAP80 and ZEB1, probably due to cell type variability. To further support the RAP80‐ZEP1 regulatory axis for EMT, we test if overexpressed RAP80 could suppress ZEB1 and vimentin expression in H1650 that has the lowest RAP80 protein level. As shown in Figure 4(c), ZEB1 and vimentin are decreased and E‐cadherin is increased by ectopic expressed RAP80, indicating that RAP80 plays an inhibitory role on EMT signaling. However, N‐cadherin is not affected by ectopic expressed RAP80. We also show that E‐cadherin (epithelial marker) is decreased by RAP80 knockdown in MCF7, a breast cancer cell line (Fig. S1). Taken together, the RAP80‐miR200c‐ZEB1 regulatory axis plays an important role in EMT signaling.
Figure 3.
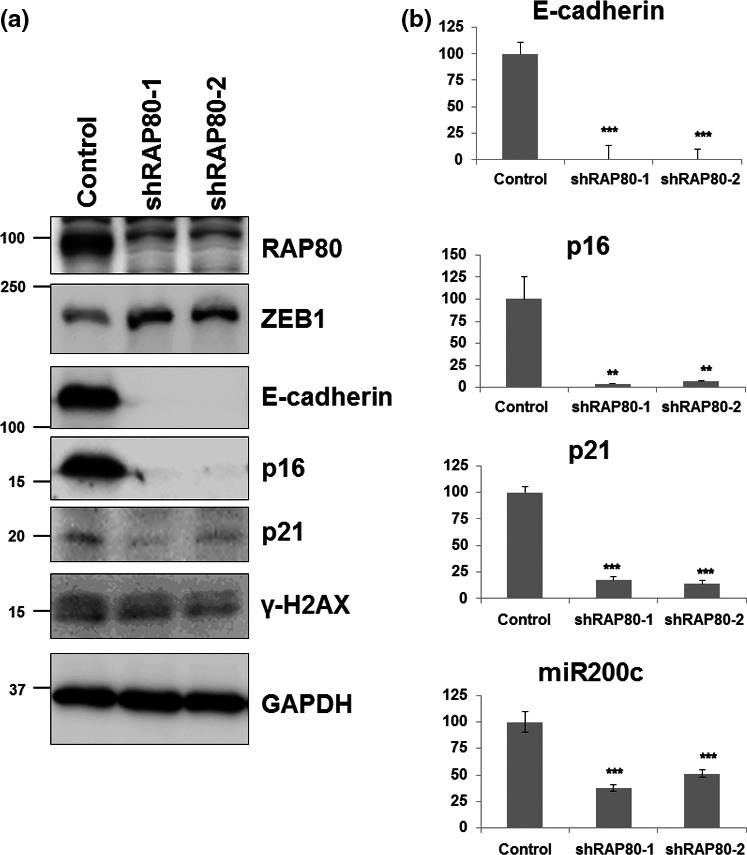
Epithelial–mesenchymal transition signals are activated in RAP80 knockdown cells. (a) Control and RAP80 knockdown HeLa cell lysates subjected to immunoblotting analysis using anti‐RAP80, ZEB1, E‐cadherin, N‐cadherin, p16, p21, γ‐H2AX and GAPDH. (b) Total RNA extracted from control and RAP80 knockdown Hela cells subjected to RT‐PCR with E‐cadherin, p16, p21 and miR200c‐specific RT‐PCR primers. Averages of three independent experiments are presented with error bars.
Figure 4.
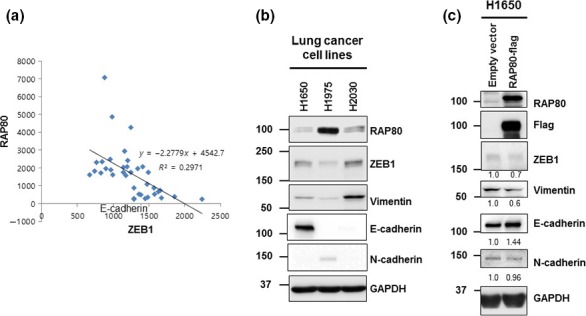
RAP80 suppress ZEB1 expression and epithelial–mesenchymal transition signal. (a) Negative correlation of RAP80 with ZEB1 in 42 lung cancer cell lines. Transcript levels of RAP80 and ZEB1 from Body Atlas expression database of Nextbio are plotted and the correlation coefficient has been calculated. (b) H1650, H1975 and H2030 lung cancer cell lysate was subjected to western blotting with anti‐RAP80, ZEB1, vimentin, E‐cadherin, N‐cadherin and GAPDH antibodies. (c) H1650 cells were transfected with RAP80‐flag expression plasmid and cell lysates were subjected to western blotting with anti‐RAP80, Flag, ZEB1, vimentin, E‐cadherin, N‐cadherin and GAPDH antibodies. Signal intensity is quantified and presented at the bottom of the figure.
Downregulation of RAP80 induces metastasis and makes cancer cell malignant
Metastasis and malignancy of cancer has been closely related with EMT activation. Because we show that EMT is activated by RAP80 knockdown, it is important to determine whether metastasis is also regulated by RAP80. To investigate the role of RAP80 on metastasis, control and RAP80 knockdown cells were labeled with ICG for NIR imaging and injected i.v. into Balb/C nude mice to analyze the amount of metastasis into the lung. As shown in Figure 5, RAP80 knockdown cells largely accumulated in the lung at 1, 2 and 7 days after injection, compared with control vector‐infected cell injected mice. These data also indicate that the downregulation of RAP80 activates metastasis to the lung.
Figure 5.
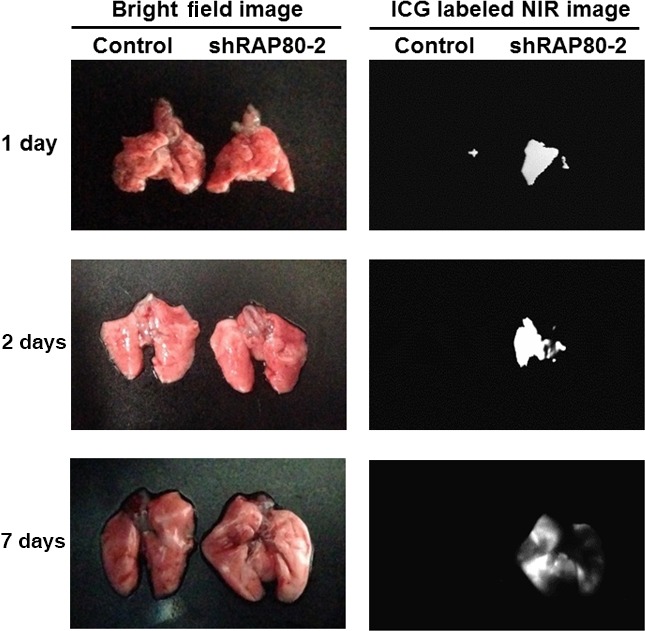
RAP80 knockdown cells massively metastasize into lung after i.v. injection. Bright field (left) and near‐infrared (NIR) fluorescence (right) images of mice lungs after injection of Indocyanine Green (ICG)‐labeled control and shRAP80‐2 HeLa cells (1 × 105 cells/injection). Lungs were dissected and imaged at 1, 2 and 7 days after injection.
To test malignancy of RAP80 knockdown HeLa cells, we i.v. injected control and RAP80 knockdown HeLa cells into Balb/C nude mice, killing mice at 1, 2, 7, 15 and 25 days after injection to analyze cancer malignancy in the lung. As shown in Figure 6(a), the lung of RAP80 knockdown cell‐injected mice showed increase in metastatic nodule number at 7, 14 and 25 days after injection and completely collapsed at 25 days after injection. Histological analysis (H&E staining) showed a large accumulation of non‐alveolar cells in alveoli structure at 1, 2, 7 and 14 days after injection of RAP80 knockdown cells, indicating that shRAP80‐2 cells accumulated in the lung (Fig. 6b). Importantly, the size and number of metastasized cancer nodules are greatly increased in the lung of shRAP80‐2‐injected mice (Fig. 6b). To consolidate the conclusion, we performed the same Xenograft experiment and periodically checked body weight for 15 days after injection. As shown in Figure 6(c), shRAP80‐2 injected mice showed significant decrease of body weight, indicating health problems. At 15 days after injection, all mice are killed for further analysis. As shown in Figure S2(a), shRAP80‐2‐injected mice showed a hunchback phenotype, indicating severe muscle weakness, probably due to cancer cachexia. For a statistical analysis of metastatic nodules, we dissected the lungs (Fig. S2b) and counted the number of metastatic nodules. As shown in Figure 6(c), the shRAP80‐2‐injected group showed a significant increase of metastatic nodules in the lung. Together, the downregulation of RAP80 induces metastasis and makes cancer cells more malignant.
Figure 6.
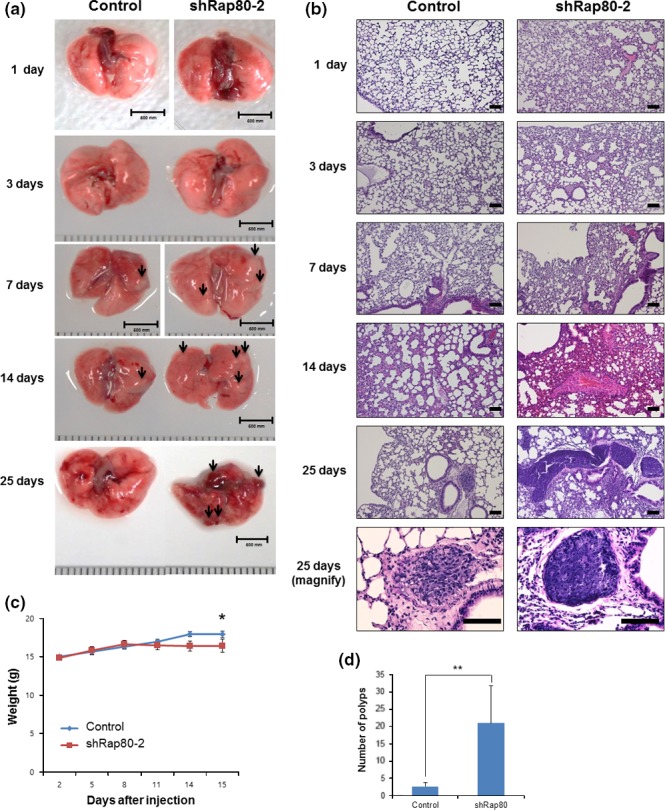
RAP80 knockdown cells generate malignant tumor in lung after i.v. injection. (a) Bright field images of mice lungs after injection of control and shRAP80‐2 HeLa cells (1 × 105 cells/injection). Lungs were dissected and imaged at 1, 3, 7, 14 and 25 days after injection. Arrows indicate the metastatic nodules. (b) The same lung samples subjected to H&E staining for histological analysis. Scale bar: 100 μm. (c) Body weight was measured and presented as an average and error bar (standard error). *Student's t‐test P < 0.05. (d) The quantitative analysis of metastatic nodules in lung at 15 days after injection. Data presented as an average with error bar (SD). **Student's t‐test P < 0.01.
RAP80 expression level is positively correlated with survival rate in breast and lung cancer patients
To investigate the prognostic significance of RAP80 in human cancer, a survival analysis was performed with large clinical datasets from The Cancer Genome Atlas. For survival analysis, the samples were divided into two groups (low and high) according to the expression of RAP80. By Kaplan–Meier analysis, the group expressing low levels of RAP80 showed significantly worse outcomes for survival than the high‐expressing groups in breast cancer (Fig. 7a) and lung adenocarcinoma patients (Fig. 7b). Together, the expression level of RAP80 is positively associated with overall survival in breast cancer and lung adenocarcinoma patients.
Figure 7.
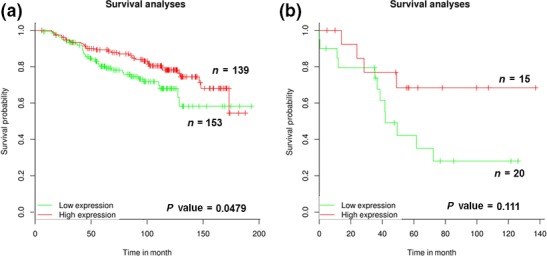
Kaplan–Meier (KM) survival curves of breast cancer and lung adenocarcinoma patients are stratified by expression levels of RAP80. KM overall survival curve of RAP80 from The Cancer Genome Atlas database analysis. Green or red box indicates RAP80 low expression or RAP80 high expression patient groups in (a) breast cancer and (b) lung adenocarcinoma, respectively. n, number of patients.
Discussion
For several decades, radiation therapy and DNA‐damaging drugs have been used as an effective treatment for many different cancers, which cause single‐strand and double‐strand DNA breaks leading to cancer cell death.3 Although DNA damage‐based cancer treatments are effective, after treatment, resistant tumor cells arise and show metastatic properties with more malignant phenotypes. Despite the clear association of DNA damage response and EMT‐induced metastasis, the conversing molecular mechanism of DDR and EMT is largely unknown.
In the present study, we show that EMT is induced by the downregulation of the well‐known DNA damage response regulator, RAP80. Although RAP80 is a critical regulator for DNA damage response, RAP80 knockdown itself did not cause a detectable amount of DNA damage response in previous studies27 and our result (Fig. 3a, γ‐H2AX) suggests another regulatory mechanism. Of note, RAP80 has been reported as the nuclear transcriptional regulator28 and we confirmed the nuclear localization of RAP80 (Fig. S3). ATM was found to stabilize ZEB1, in response to ionizing radiation‐induced DNA damage, promoting DNA damage response and radioresistance.17 Of note, ZEB1 has been extensively studied as a critical transcriptional regulator for EMT signaling.29, 30 Here, we first show that ZEB1 is increased and its reciprocal inhibitor miR200c is decreased in RAP80 knockdown cells, indicating that RAP80 regulates the miR200c/ZEB1 regulatory axis to activate the EMT signaling pathway (Fig. 3).
Epithelial–mesenchymal transition has often been associated with invasive metastasis and cancer malignancy. Along this line, we show that RAP80 knockdown cells invasively metastasize to the lung and generate more malignant tumors compared with control vector‐infected cells (Figs 5 and 6). Importantly, the patient group with low level RAP80 expression showed poor prognosis in clinical datasets of lung adenocarcinoma and breast cancer, suggesting that the downregulation of RAP80 is correlated with progression and malignancy of cancer (Fig. 7).
In summary, we demonstrate the downregulation of RAP80 activating EMT through reciprocal regulators, miR200c and ZEB1, promoting pulmonary metastasis of cancer cells. Of note, the level of RAP80 was found to negatively correlate with the survival rate in lung adenocarcinoma and breast cancer patients. These findings indicate that RAP80 is a critical gatekeeper to impede metastasis of cancer as well as to secure DNA integrity.
Disclosure Statement
The other authors have no conflict of interest to declare.
Supporting information
Fig. S1. The effects of RAP80 in MCF7, a breast cancer cell line. MCF7 cells were transfected with shRAP80‐1 and Sh‐RAP80‐2 and subjected to western analysis using anti‐RAP80, E‐cadherin and β‐actin.
Fig. S2. RAP80 knockdown cells generate malignant tumor in lung at 15 days after i.v. injection. (a) The image of mice after 15 days injection of control and shRAP80‐2 Hela cells (1 × 105 cells/injection). (b) The bright field image of mouse lungs at 15 days after injection of control and shRAP80‐2 Hela cells (1 × 105 cells/injection).
Fig. S3. RAP80 localized in nuclear. HeLa cells were immunostained with anti‐RAP80 antibody (green) and DAPI (blue).
Table S1. Primer sequences for RT‐qPCR.
Table S2. RAP80 and ZEB1 expression in various lung cancer cell lines from Body Atlas expression database of Nextbio.
Acknowledgments
This work is supported by the National Research Foundation of Korea (NRF‐2012M3A9C6050087, 2013R1A6A9067028 and 2013R1A1A2008576 to J.Y. Lee) and by a grant from the National R&D Program for Cancer Control, Ministry for Health, Welfare and Family Affairs, Republic of Korea (HI11C1596) and National Research Foundation of Korea (NRF) grant funded by the Korea government (MEST) (NRF‐2015R1A2A2A01003975) to H. Kim. The funders had no role in the study design, data collection and analysis, the decision to publish or the preparation of the manuscript.
Cancer Sci 107 (2016) 267–273
Funding Information
This work is supported by the National Research Foundation of Korea (NRF‐2012M3A9C6050087, 2013R1A6A9067028 and 2013R1A1A2008576 to J.Y. Lee) and by a grant from the National R&D Program for Cancer Control, Ministry for Health, Welfare and Family Affairs, Republic of Korea (HI11C1596) and National Research Foundation of Korea (NRF) grant funded by the Korea government (MEST) (NRF‐2015R1A2A2A01003975) to H. Kim. The funders had no role in the study design, data collection and analysis, the decision to publish or the preparation of the manuscript.
References
- 1. Lukas J, Lukas C, Bartek J. More than just a focus: the chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol 2011; 13: 1161–9. [DOI] [PubMed] [Google Scholar]
- 2. Wang H, Zhang X, Teng L, Legerski RJ. DNA damage checkpoint recovery and cancer development. Exp Cell Res 2015; 334: 350–8. [DOI] [PubMed] [Google Scholar]
- 3. Khanna A. DNA damage in cancer therapeutics: a boon or a curse? Cancer Res 2015; 75: 2133–8. [DOI] [PubMed] [Google Scholar]
- 4. Gorgoulis VG, Vassiliou LV, Karakaidos P et al Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 2005; 434: 907–13. [DOI] [PubMed] [Google Scholar]
- 5. Bartkova J, Horejsi Z, Koed K et al DNA damage response as a candidate anti‐cancer barrier in early human tumorigenesis. Nature 2005; 434: 864–70. [DOI] [PubMed] [Google Scholar]
- 6. Kim H, Chen J, Yu X. Ubiquitin‐binding protein RAP80 mediates BRCA1‐dependent DNA damage response. Science 2007; 316: 1202–5. [DOI] [PubMed] [Google Scholar]
- 7. Kim H, Huang J, Chen J. CCDC98 is a BRCA1‐BRCT domain‐binding protein involved in the DNA damage response. Nat Struct Mol Biol 2007; 14: 710–5. [DOI] [PubMed] [Google Scholar]
- 8. Liu Z, Wu J, Yu X. CCDC98 targets BRCA1 to DNA damage sites. Nat Struct Mol Biol 2007; 14: 716–20. [DOI] [PubMed] [Google Scholar]
- 9. Shao G, Patterson‐Fortin J, Messick TE et al MERIT40 controls BRCA1‐Rap80 complex integrity and recruitment to DNA double‐strand breaks. Genes Dev 2009; 23: 740–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sobhian B, Shao G, Lilli DR et al RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 2007; 316: 1198–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang B, Hurov K, Hofmann K, Elledge SJ. NBA1, a new player in the Brca1 A complex, is required for DNA damage resistance and checkpoint control. Genes Dev 2009; 23: 729–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang B, Matsuoka S, Ballif BA et al Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science 2007; 316: 1194–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yan J, Kim YS, Yang XP et al The ubiquitin‐interacting motif containing protein RAP80 interacts with BRCA1 and functions in DNA damage repair response. Cancer Res 2007; 67: 6647–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yin Z, Menendez D, Resnick MA, French JE, Janardhan KS, Jetten AM. RAP80 is critical in maintaining genomic stability and suppressing tumor development. Cancer Res 2012; 72: 5080–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nikkila J, Coleman KA, Morrissey D et al Familial breast cancer screening reveals an alteration in the RAP80 UIM domain that impairs DNA damage response function. Oncogene 2009; 28: 1843–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rebbeck TR, Mitra N, Domchek SM et al Modification of BRCA1‐associated breast and ovarian cancer risk by BRCA1‐interacting genes. Cancer Res 2011; 71: 5792–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang P, Wei Y, Wang L et al ATM‐mediated stabilization of ZEB1 promotes DNA damage response and radioresistance through CHK1. Nat Cell Biol 2014; 16: 864–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mani SA, Guo W, Liao MJ et al The epithelial–mesenchymal transition generates cells with properties of stem cells. Cell 2008; 133: 704–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bao S, Wu Q, McLendon RE et al Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006; 444: 756–60. [DOI] [PubMed] [Google Scholar]
- 20. Baumann M, Krause M, Hill R. Exploring the role of cancer stem cells in radioresistance. Nat Rev Cancer 2008; 8: 545–54. [DOI] [PubMed] [Google Scholar]
- 21. Folkman J, Moscona A. Role of cell shape in growth control. Nature 1978; 273: 345–9. [DOI] [PubMed] [Google Scholar]
- 22. Kim Y, Lin Q, Zelterman D, Yun Z. Hypoxia‐regulated delta‐like 1 homologue enhances cancer cell stemness and tumorigenicity. Cancer Res 2009; 69: 9271–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guha M, Srinivasan S, Ruthel G et al Mitochondrial retrograde signaling induces epithelial–mesenchymal transition and generates breast cancer stem cells. Oncogene 2014; 33: 5241–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu Y, El‐Naggar S, Darling DS, Higashi Y, Dean DC. Zeb1 links epithelial–mesenchymal transition and cellular senescence. Development 2008; 135: 579–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yu JM, Sun W, Hua F et al BCL6 induces EMT by promoting the ZEB1‐mediated transcription repression of E‐cadherin in breast cancer cells. Cancer Lett 2015; 365: 190–200. [DOI] [PubMed] [Google Scholar]
- 26. Park SM, Gaur AB, Lengyel E, Peter ME. The miR‐200 family determines the epithelial phenotype of cancer cells by targeting the E‐cadherin repressors ZEB1 and ZEB2. Genes Dev 2008; 22: 894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yin Z, Menendez D, Resnick MA, French JE, Janardhan KS, Jetten AM. RAP80 is critical in maintaining genomic stability and suppressing tumor development. Cancer Res 2012; 72: 5081–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yan Z, Kim Y‐S, Jetten AM. RAP80, a novel nuclear protein that interacts with the retinoid‐related testis‐associated receptor. J Biol Chem 2002; 277: 32384–5. [DOI] [PubMed] [Google Scholar]
- 29. Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial–mesenchymal transition. Nat Rev Mol Cell Biol 2014; 15: 178–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Takeyama Y, Sato M, Horio M et al Knockdown of ZEB1, a master epithelial‐to‐mesenchymal transition (EMT) gene, suppresses anchorage‐independent cell growth of lung cancer cells. Cancer Lett 2010; 296: 216–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Janer A, Antonicka H, Lalonde E et al An RMND1 mutation causes encephalopathy associated with multiple oxidative phosphorylation complex deficiencies and a mitochondrial translation defect. Am J Hum Genet 2012; 91: 741. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. The effects of RAP80 in MCF7, a breast cancer cell line. MCF7 cells were transfected with shRAP80‐1 and Sh‐RAP80‐2 and subjected to western analysis using anti‐RAP80, E‐cadherin and β‐actin.
Fig. S2. RAP80 knockdown cells generate malignant tumor in lung at 15 days after i.v. injection. (a) The image of mice after 15 days injection of control and shRAP80‐2 Hela cells (1 × 105 cells/injection). (b) The bright field image of mouse lungs at 15 days after injection of control and shRAP80‐2 Hela cells (1 × 105 cells/injection).
Fig. S3. RAP80 localized in nuclear. HeLa cells were immunostained with anti‐RAP80 antibody (green) and DAPI (blue).
Table S1. Primer sequences for RT‐qPCR.
Table S2. RAP80 and ZEB1 expression in various lung cancer cell lines from Body Atlas expression database of Nextbio.


