Abstract
Cis-regulatory elements (CREs) are crucial links in developmental gene regulatory networks, but in many cases, it can be difficult to discern whether similar CREs are functionally equivalent. We found that despite similar conservation and binding capability to upstream activators, different GATA cis-regulatory motifs within the promoter of the C. elegans endoderm regulator elt-2 play distinctive roles in activating and modulating gene expression throughout development. We fused wild-type and mutant versions of the elt-2 promoter to a gfp reporter and inserted these constructs as single copies into the C. elegans genome. We then counted early embryonic gfp transcripts using single-molecule RNA FISH (smFISH) and quantified gut GFP fluorescence. We determined that a single primary dominant GATA motif located -527 bp upstream of the elt-2 start codon was necessary for both embryonic activation and later maintenance of transcription, while nearby secondary GATA motifs played largely subtle roles in modulating postembryonic levels of elt-2. Mutation of the primary activating site increased low-level spatiotemporally ectopic stochastic transcription, indicating that this site acts repressively in non-endoderm cells. Our results reveal that CREs with similar GATA factor binding affinities in close proximity can play very divergent context-dependent roles in regulating the expression of a developmentally critical gene in vivo.
Keywords: endoderm, cis-regulation, promoter, smFISH, transcription, GATA
Introduction
For the nematode C. elegans and its close relatives, early embryonic development is characterized by a tight link between cell lineage and cell fate that is largely determined by transcriptional gene regulatory networks (GRNs). Determining how transcription factors activate their respective targets within a GRN at the cis-regulatory level is key to understanding how multicellular organisms develop robustly.
However, understanding cis-regulation has been complicated by the fact that the vast majority of eukaryotic transcription factors have very short DNA binding domains, often leading to vastly more potential cognate binding sites than real functional targets (Mirny & Wunderlich, 2008). Additionally, individual transcription factors can co-occur with one or more paralogous factors from the same family leading to multiple transcription factors sharing individual cis-regulatory sites. Although recent years have seen vast advances in the mapping of transcription factors to their binding sites though techniques such as ChIP-seq (Gerstein et al., 2010), such techniques do not necessarily reveal whether bound sites are functionally equivalent. Many transcription factors are known to act as both activators and repressors depending on context.
Low target specificity, gene duplications, and contextual role switching have the potential to play a role in advancing developmental robustness. During early embryonic development, transcriptional networks must be robust to extrinsic insults as well as intrinsic variability at the molecular level. Cell divisions need to be spatially and temporally coordinated in the face of environmental variability and stochastic fluctuations of key molecules.
The transcriptional regulation of the C. elegans endoderm specifying gene elt-2 is a good model for studying how cis-regulatory mechanisms impact developmental robustness. The gene elt-2 is an essential switch for the endoderm cell fate decision and a fundamental developmental bottleneck: failure to activate elt-2 results in a lethal absence of endoderm. The major trans-activators of elt-2 are well characterized and have been demonstrated to contribute to developmental robustness at the trans-level. END-3, END-1, and ELT-7 are closely related GATA transcription factors that redundantly activate elt-2 during early embryonic development(Lowry et al., 2009; Raj, Rifkin, Andersen, & van Oudenaarden, 2010; Sommermann, Strohmaier, Maduro, & Rothman, 2010; Zhu et al., 1997; Zhu, Fukushige, McGhee, & Rothman, 1998), and ELT-2 maintains its own transcription through larval and adult stages by autoregulation (T Fukushige, Hendzel, Bazett-Jones, & McGhee, 1999) (Fig. 1A). Single null mutants of END-1, END-3, or ELT-7 are largely viable - with only transient developmental anomalies occurring in end-1 and end-3 single null mutants and a low (5-9%) rate of developmental failure in end-3 mutants (Boeck et al., 2011; Maduro et al., 2005; Sommermann et al., 2010). A paralogous pair of redundant and nearly identical GATA factors - med-1,2 - also helps to activate end-3 and end-1 (Maduro, Broitman-Maduro, Mengarelli, & Rothman, 2007; Maduro, Meneghini, Bowerman, Broitman-Maduro, & Rothman, 2001). Raj et al. (2010) demonstrated that particular mutations in the upstream maternal activating factor skn-1 result in failure to activate med-1,2 and end-3 and highly variable expression of end-1. Noisy end-1 expression, in turn, leads to bimodal elt-2 expression states (Raj et al., 2010). The presence of redundant trans-activating factors effectively buffers the activation of elt-2 from variability in levels of any single activator.
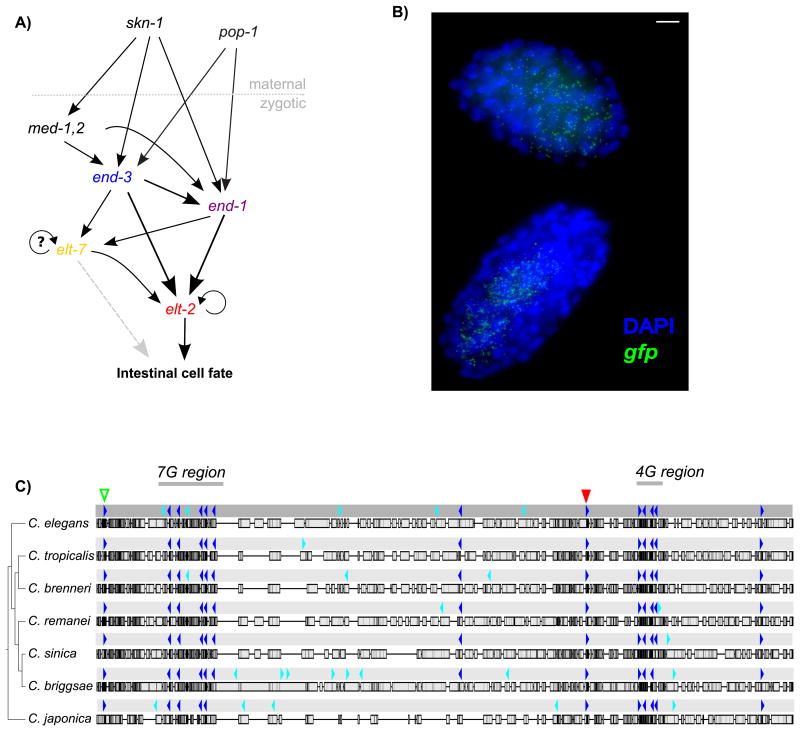
Figure 1.
The elt-2 activation network and upstream promoter region. A) Network diagram of elt-2 activators. B) Maximum Z-stack projection of smFISH image from strain 1879:WT. C) 1879 bp promoter region upstream of elt-2 aligned to orthologous regions in Elegans supergroup members. Dark blue indicates a heavily conserved HGATAR motif and cyan indicates a weakly conserved HGATAR motif. The red triangle indicates position of -527 bp ACTGATAAGA “A-site” motif. The green triangle indicates position of a -1857 bp ACTGATAAGG motif.
Despite our good understanding of elt-2's trans-activators, little is known about how these trans-activators operate at the cis-regulatory level. The exact sequences and relative positions of the cis-regulatory motifs necessary for driving elt-2 expression have not been determined, nor are there apparent TATA box (GTATAWWAG) or Sp1 core promoter motifs in the immediate region upstream of the elt-2 transcriptional start site (TSS) (WormBase release WS220) (Harris et al., 2010; Saito et al., 2013)). Furthermore, we could not identify any sequence similarities to the known basal promoter fragment of the pes-10 gene. Based on the fact that all the known embryonic activators of elt-2 are GATA transcription factors, we can narrow down the candidate cis-regulatory sequences considerably. The ∼2 kb region upstream of the elt-2 start codon in C. elegans contains eighteen of the consensus GATA factor binding motif HGATAR, with thirteen conserved in sequence and relative spacing throughout the Elegans supergroup (sequence data from C. elegans, C. tropicalis, C. brenneri, C. remanei, C. sinica, C. briggsae, and C. japonica) (Félix, Braendle, & Cutter, 2014; Huang, Ren, Qiu, & Zhao, 2014) (Fig. 1C).
This conglomeration of potential GATA factor binding sites in the elt-2 upstream region suggests several possible ways that these motifs might interact with the trans-factors END-3, END-1, ELT-7, and ELT-2 itself to control elt-2 transcriptional activation. Perhaps many independently dispersed transcription start sites, driven or aided by GATA factor binding, contribute additively and redundantly to overall gene expression levels and noise (Juven-Gershon & Kadonaga, 2010). Under this dispersed promoter scenario, mutation of single HGATAR motifs might be expected to reduce transcription activation proportionate to the number of motifs mutated(Davidson, 2001; Flores et al., 2000). Alternatively, but not exclusively, elt-2 cis-activation could also be driven by a combinatorial code involving binding of different GATA factors with different specificities. Under a combinatorial control scenario, mutation of any single HGATAR motif in a larger combinatorial code should result in an equivalent impact on gene expression as mutating any single motif in the same code. Finally, the contributions of different sites may instead be unequal, with one or a few key sites responsible for the majority of expression and the rest playing minor supporting roles.
In addition, few studies have deeply examined the relationship between between cis-regulatory elements (CREs) and gene expression variability. For a gene like elt-2, variability in early embryonic activation might be expected to have severe consequences for viability.
In this study, we dissect the elt-2 promoter in a reporter transgene context to determine how conserved transcription factor binding sites drive activation and maintenance of a key developmental regulator. Using a technique for labeling individual mRNAs, we also precisely capture the relationship between conserved features of the elt-2 promoter with gene expression variability in early embryonic development.
Materials and Methods
Strain Generation
To control for copy number variations that could affect gene expression levels, we generated single copy insertions of promoter gfp reporter lines using MosSCI (Frøkjaer-Jensen et al., 2008; Frøkjær-Jensen et al., 2012). To generate the reporter, we used PCR fusion to join the elt-2 3′ UTR to a gfp fragment amplified from pPD95.81 (gift from Andrew Fire; Addgene kit # 1000000001). We generated mutant promoter variants by PCR fusion of synthetic oligonucleotides containing mutated HGATAR motifs to PCR amplified elt-2 promoter sequence. GIBSON assembly was used to fuse wild-type and mutant promoter constructs to the gfp∷elt-2 3′UTR DNA fragment and a pCFJ350 MosSCI backbone vector (gift from Erik Jorgenson; Addgene plasmid #34866). All Pelt-2∷gfp∷elt-2 3′UTR reporters were integrated at site ttTi5605 (strain EG6699) with stable integration confirmed by PCR screening of insert junctions into the C. elegans genome.
In vitro binding assays
For EMSAs, DNA binding domain sequences for GATA transcription factors were cloned into pET His6 TEV LIC (1B) for N-terminal His6 tagging (gift from Scott Gradia; Addgene plasmid # 29653). Constructs were transformed into Rosetta 2(DE3) pLysS competent cells, and fusion protein expression was induced by addition of 0.1 mM IPTG at 18°C for 12 hours. His6 tagged proteins were purified by use of TALON metal affinity beads (Clontech). Binding assays were done at room temperature in EMSA buffer (10mM HEPES, 200mM NH4 OAc, 30 mM NaCl, 1.5mM MgCl2, 0.5 uM Zn(OAc)2, 0.2 mg/ml BSA, 1mM DTT, 8% glycerol, pH 7.0). EMSAs were run on a 8% 29:1 Acrylamide/Bis-acrylamide 8% glycerol 0.5X TBE gel and stained with Sybr Green Nucleic acid gel stain (Molecular Probes S-7563). (See supplemental methods for more detail).
smFISH data collection
For imaging of embryonic development, we collected and fixed embryos collected from adults 52-53 hours post-synchronization in 4% formaldehyde. Collected embryos were hybridized to Cy3∷gfp, ATTO 647N∷elt-2, and Alexa 594∷end-1 smFISH probes (Raj et al., 2010, 2008) (Stellaris). Nuclei were labeled with DAPI. For each smFISH data set, we generated Z-stack images of at least 170 embryos with a maximum spacing of 0.4 uM at 100× magnification on an epifluorescence microscope. For quantifying spot counts, we used an inhouse developed machine learning spot classification tool, AroSpotFinding Suite, to automatically collect spot data counts (Rifkin, 2011; Wu & Rifkin, 2015). Nuclei were counted by hand and used as a measure of developmental stage.
Gut fluorescence quantification
To obtain consistent gut reporter gene fluorescence data, we determined that it was important to control for food density. We found that different food concentrations could affect both mean gut fluorescence intensity and overall size. To control for food levels, we grew all strains in liquid culture at controlled food concentrations. We aliquoted and pelleted 2 L of overnight OP50 grown in LB culture and aspirated off all excess liquid. We then weighed the mass of each pellet, and froze each pellet at -80 °C. For each data collection day, an OP50 food pellet would be thawed and resuspended in S-medium at a concentration of 40 mg/ml. Worms synchronized overnight S-medium would then be placed in a solution of OP50 and S-medium for 22 hours at a concentration of approximately 500 worms per ml. For imaging, the synchronized worms were mounted onto 3% agarose pad slides, and anaesthetized with 10 mM levamisole. Images were taken on an AxioImager R1 at 10× magnification. Quantification was performed using a custom MATLAB script (available upon request).
Statistical analysis of smFISH data
To determine whether the trajectories of gene expression for different strains were significantly different across time, we calculated the difference between smoothing splines fit to the data for each strain. We constructed a null distribution for this difference trajectory by randomly assigning the strain labels for each datapoint (permuting within a developmental stage: 0-1E, 2E, 4E, 8E), fitting splines to the shuffled dataset, and calculating the difference between these shuffled splines. We repeated this 10,000 times for each pair of trajectories to form null distributions. Because we were looking for significant differences in expression at any point along the trajectories, we adjusted the significance level cutoff to account for multiple testing. We estimated the number of parameters used in the splines and used the Dunn-Šidák method to determine a conservative single-test significance level that would yield an experiment-wise significance level of 0.05 (Ury, 1976). These adjusted cutoffs varied slightly between comparisons but were around 0.006.
Results
Since elt-2 is an essential gene, we used a reporter construct to investigate the relationship between conserved and putatively functional features of the elt-2 promoter and elt-2 promoter-driven gene expression levels and noise. We integrated wild-type and mutant elt-2 promoter reporter constructs (Pelt-2∷gfp∷elt-2 3′ UTR; see methods) into the C. elegans genome at a defined location using Mos1-mediated single copy insertion (MosSCI) (Frøkjaer-Jensen et al, 2008; Frøkjær-Jensen, Davis, Ailion, & Jorgensen, 2012)). We then measured gfp transcript levels across early embryonic development using single-molecule RNA Fluorescence In Situ Hybridization (smFISH) (Raj, van den Bogaard, Rifkin, van Oudenaarden, & Tyagi, 2008) (Fig. 1B). We also confirmed that transgene insertion did not impact endogenous elt-2 levels significantly by labeling elt-2 as a control (Supplemental Fig S10).
Previous research had shown that a 5.1 kb region upstream of the elt-2 start codon is sufficient to drive gut reporter expression when introduced to wild-type worms in the form of an extrachromosomal array (T Fukushige, Hawkins, & McGhee, 1998; Tetsunari Fukushige, Hendzel, Bazett-Jones, & McGhee, 1999). We found that this large region contained a putative coding region (C39B10.7) and non-coding RNA (C33D3.6). In order avoid duplicating these potential trans-regulatory elements, we initially looked at the shorter 1879 bp region between the putative non-coding RNA C33D3.6 and the elt-2 start codon. This 1879 bp region contains 18 motifs matching to the GATA transcription factor binding consensus sequence HGATAR with 10 of the 18 motifs matching to TGATAA - a more specific motif often found in the promoters of gut expressed genes (McGhee et al., 2009).
Fluorescence microscopy of the 1879:WT reporter strain showed that the 1879 bp upstream fragment was sufficient to drive production of gfp transcripts during embryonic development (smFISH of gfp transcripts) as well as the remainder of the worm's life span (GFP fluorescence) (Table 1, Supplemental Fig S1). Although this reporter produced lower levels of expression than endogenous elt-2 in worms with between approximately 70 and 120 nuclei, the overall expression trajectory was similar to endogenous elt-2 (Supplemental Fig. S1)(Nair, Walton, Murray, & Raj, 2013; Raj et al., 2010), suggesting that many if not most of the critical CREs for the majority of elt-2 expression fall within this region and that targeted mutation of the reporter's promoter would give valuable insights into the logic and functional organization of elt-2 cis-regulation.
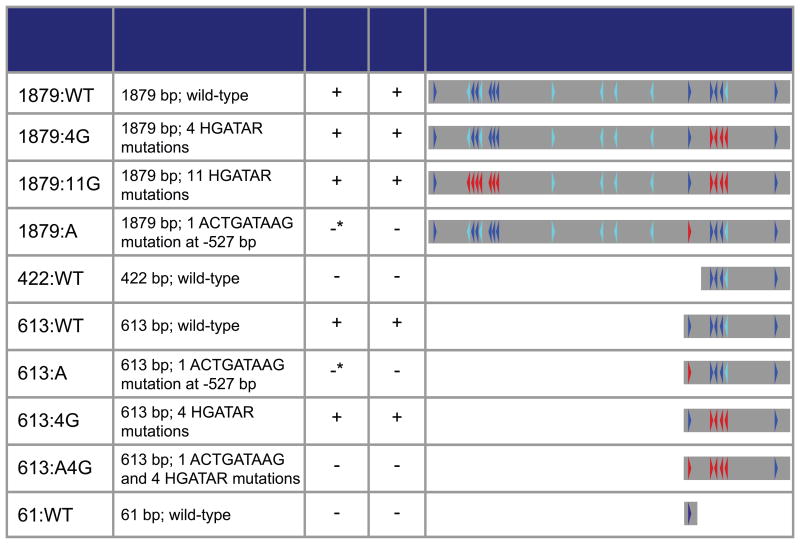
Table 1.
Table of wild-type and HGATAR mutant strains mutated for this study. Dark blue indicates a heavily conserved HGATAR motif and cyan indicates a weakly conserved HGATAR motif. Red indicates an artificially mutated HGATAR motif. *Asterisk indicates low-level transcription observed.
The promoter region from -613 to -422 bp upstream of the elt-2 start codon is necessary for gene expression
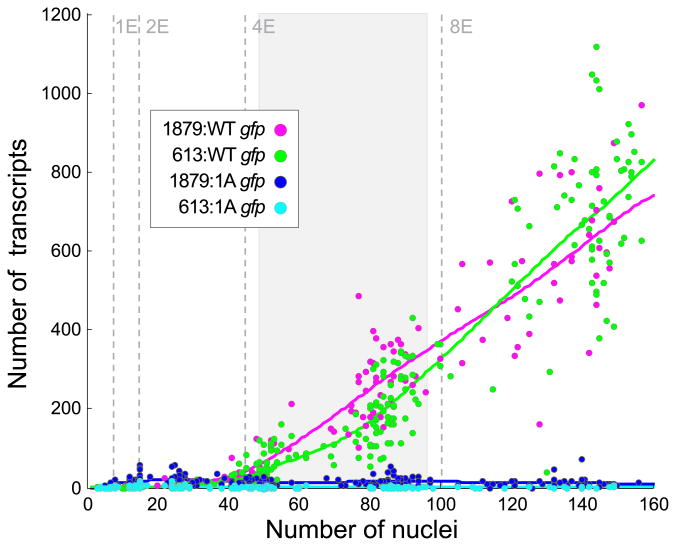
To cut down on the number of candidate HGATAR motifs that could be responsible for elt-2 promoter-driven gene activation, we tested whether a more minimal promoter could drive gut gfp expression. We first generated a reporter construct encompassing 613 bp upstream of the elt-2 start codon (Table 1 613:WT). This shortened promoter produced gfp expression levels only about 50 transcripts lower than the 1879 bp promoter during the 4E stage, suggesting that this fragment contains the primary CREs needed for driving endoderm gene expression (Figs. 2, S2).
Figure 2.
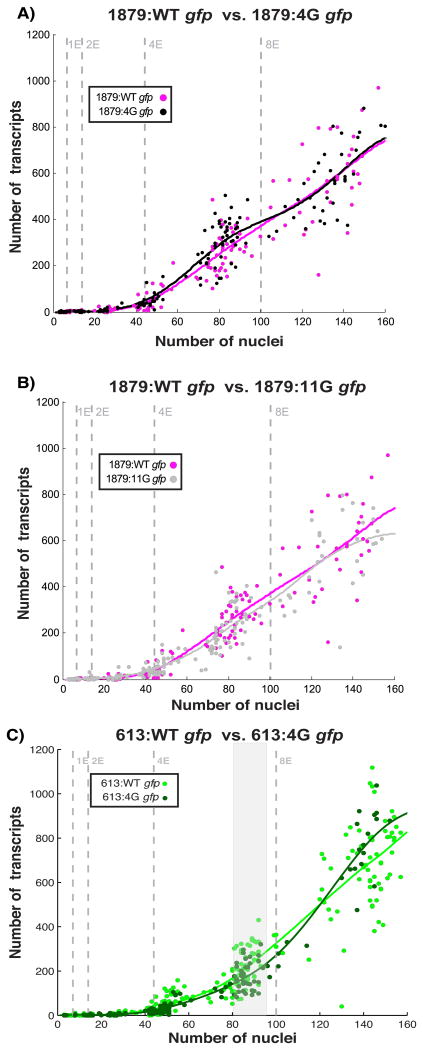
The majority of elt-2 promoter-driven expression is dependent upon a single ACTGATAAG motif at -527 bp. A) gfp expression levels of 1879:WT (n=187), 613:WT (n=459), 1879:A (n=253), and 613:A (n=217). Lines represent smoothing splines of data. The shaded grey area represents the part of the developmental trajectories that are significantly different between the longer and shorter WT promoters (Supplemental Fig. S2).
To determine whether presence of the 4G motif cluster was sufficient for driving reporter expression, we generated a 422 bp un-mutated elt-2 promoter reporter strain (Table 1 422:WT). The 422:WT strain failed to produce any reporter expression during embryogenesis (assessed by smFISH) or in adults (assessed by GFP fluorescence). These results demonstrate that one or more CREs located between -613 and -422 upstream of the elt-2 start codon are necessary for promoter-driven gene expression. Within this region, we identified a single highly conserved ACTGATAAGA motif at -527 bp (Fig. 1C; red arrow).
An ACTGATAAG sequence -527 bp from elt-2 start codon is necessary for elt-2 promoter-driven gene expression
The -527 bp ACTGATAAGA sequence matched perfectly to the highest scoring sequence found to be associated with intestinally expressed genes in McGhee et al. (2009). Furthermore, the ACTGATAAG portion of this sequence is almost perfectly conserved in the Elegans supergroup (in C. remanei the sequence is AGTGATAAG) (Fig. 1C; red arrow). Close examination of TSS data from Saito et al. 2013 revealed both dispersed low-level transcription along the 1879 bp elt-2 upstream region and a distinct peak of transcriptional activity at -482 bp (Fig S3).
To test whether the -527 bp ACTGATAAG motif (hereafter A-site) is necessary for reporter activity, we generated mutant 613 bp and 1879 bp reporters with the ACTGATAAG motif mutated to ACTCTGTAG (Table 1 613:A, 1879:A). Embryonic transcript expression revealed near total loss of embryonic reporter expression in both mutant strains (Figs. 2, 3, S4). Additionally, we did not observe any embryonic, larval, or adult GFP expression distinguishable from regular gut autofluorescence in either mutant strain (Table 1).
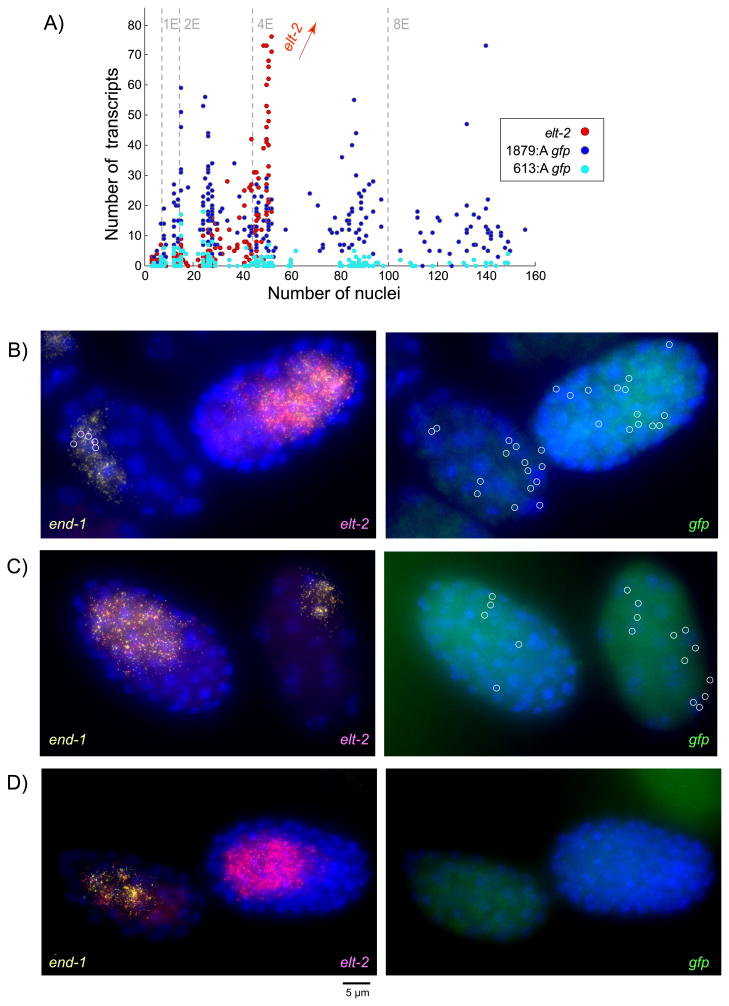
Figure 3.
Stochastic low-level ectopic transcription occurs in the absence of the -527 bp ACTGATAAG element with an increase in transcription during the 1E and 2E stages. A) gfp transcript counts for A-site mutants 1879:A (n=253) and 613:A (n=217) compared to elt-2 in the same embryos (elt-2 data pooled from 1879:A and 613:A (n=470); elt-2 transcript counts greater than 80 not shown). (B-D) Left column: end-1 in yellow elt-2 in red. Right column: gfp in green; all gfp transcripts circled for ease of visualization. B) Ectopic expression of gfp outside of end-1 and elt-2 expressing E-cells in strain 1879:A (five early elt-2 transcripts circled in left column). C) Ectopic early gfp expression in 613:A. D) Mutation of 4G HGATAR sites eliminates low-level stochastic gfp reporter expression.
These extreme drops in gene expression demonstrate that a single, non-redundant ACTGATAAG at -527 bp is necessary for the vast majority of gene expression driven by the elt-2 promoter during and after embryonic development.
Mutation of -527 bp ACTGATAAG element results in precocious and spatially ectopic stochastic low-level transcription
Although reporter strains with mutations in the A-site motif exhibited greatly reduced gfp expression, in many embryos reporter mRNA expression was not zero. Instead, strains 613:A and 1879:A exhibited low-level stochastic transcription during early embryonic development - an observation made possible by our highly sensitive smFISH assay. The 613:A mutant produced variable numbers of transcripts ranging from 0 to 18 during early embryonic development. Similarly, the 1879:A mutant exhibited stochastic, low-level transcription ranging from 0 to 73 transcripts over the same time span (Fig. 3A, S4). Total absence of gfp expression in strains 422:WT and 613:A4G confirmed that low-level expression in strains 613:A and 1879:A was not the result of leaky expression due to insertion site effects (Table 1 and Fig. 3D).
Surprisingly, the peak expression in strains 613:A and 1879:A exceeded both endogenous elt-2 levels and gfp levels in wild-type reporter constructs 613:WT and 1879:WT during the 1E and 2E stages, (Figs. 3A, S4A-C). Peak expression in 1879:A also exceeded peak expression in 613:A across embryonic development, indicating that regions of the promoter upstream of -613 bp play a role in driving low-level transcription independent of the A-site (Fig. S4D).
Close examination of embryo images at the 1E and 2E stages revealed that in many cases, precocious stochastic transcription could occur in both E cells and other lineages. By using elt-2, end-1, and gfp smFISH probes in the same embryos, we were able to determine that gfp expression occurs in cells not expressing elt-2 or end-1 during very early development (Fig. 3B, C). This ectopic expression suggests that the A-site not only serves as a primary activator, but also as a transcriptional repressor against stochastic low-level transcription in non-E-cells during very early embryonic development.
We roughly estimated the level of ectopic expression at the 2E stage in both wild-type and mutant reporter strains by quantifying the amount of gfp not directly overlapping end-1 and elt-2 expression (Supplementary Fig. S5). Under our estimates, 1879:A was found to have significantly greater levels of ectopic expression (p-value < 0.0001) than its wild-type counterpart.
ELT-2, END-1, END-3, and ELT-7 have an in vitro binding affinity for the A-site as well as secondary GATA site regions
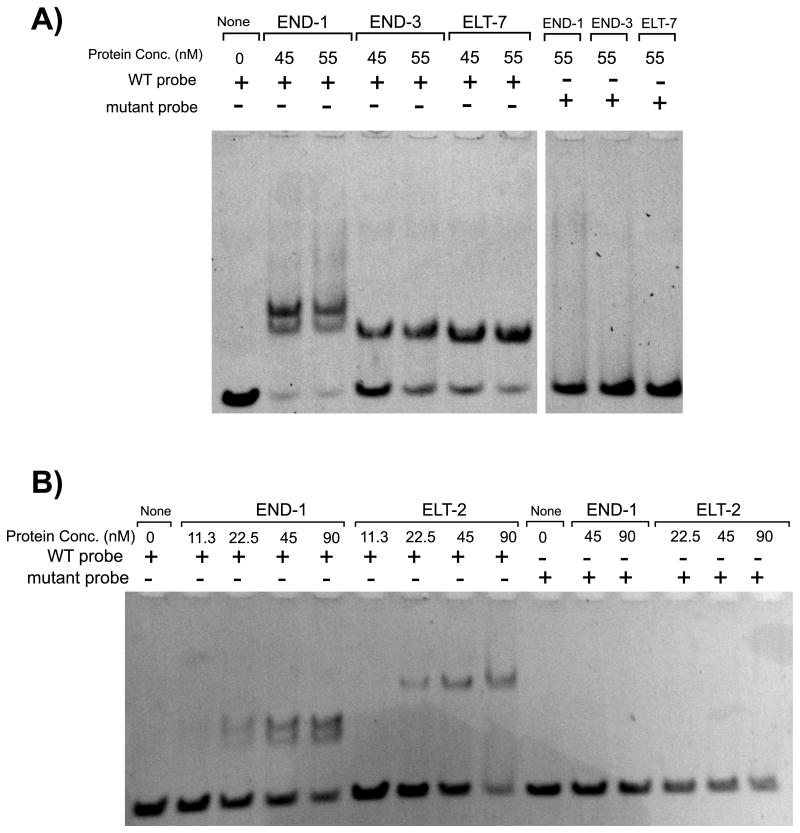
To assess in vitro binding of the END-1, END-3, ELT-7, and ELT-2 DNA binding domains to the A-site we performed EMSAs using DNA probes corresponding to the 50 bp region containing the A-site (corresponding to sequence -549 to -499 bp upstream of the endogenous elt-2 start codon). The END-1, END-3, ELT-7, and ELT-2 DNA binding domains all showed in vitro binding affinity for the A-site containing region which could be eliminated by mutating the core motif from ACTGATAAG to ACTCTGTAG (Fig. 4). This mutation was identical to the mutations we generated to create mutant strains 1879:A and 613:A. The capability of essentially all known embryonic upstream activators of elt-2 to bind to the A-site is consistent with the largely redundant roles these upstream activators play in driving elt-2 expression. In a separate paper, we report that END-1, END-3, and ELT-7 also all bind with greatest affinity to a TGATAA sequence – essentially the core component of the A-site (Tracy and Rifkin; not yet published).
Figure 4.
END-1, END-3, ELT-2, and ELT-7 DNA binding domains all bind in vitro to an ACTGATAAG motif. (A-B) All EMSAs were performed using 50 bp synthetic oligonucleotide probe corresponding to the sequence -549 to -499 bp upstream of the elt-2 start codon containing an ACTGATAAG motif (WT probe) or a ACTCTGTAG (mut probe). Probe concentration in all lanes is 70 nM. A) END-1, END-3, and ELT-7 binding. Non-adjacent lanes from the same gel. B) END-1 and ELT-2 binding.
Interestingly, we found END-1/DNA complexes running as a doublet at all protein concentrations tested when using an A-site wild-type probe. This double banding requires the ACTGATAAG motif and can be due to END-1 fragment multimer binding on the A-site, conformational changes to DNA structure induced by END-1 binding to the A-site, or END-1 binding at the A-site potentiating binding to non-A-site sequences on the DNA probe.
Conserved secondary GATA motif clusters have weak impact on promoter-driven gene expression during early embryogenesis
The abundance of conserved HGATAR motifs within the elt-2 upstream region initially suggested that elt-2 activation might dependent on multiple GATA motifs in either a combinatorial or additive manner. In a combinatorial activation scenario for a focused promoter, transcription would be dependent upon one or more clusters of cis-regulatory motifs, and mutation of cluster motifs should result in failure to activate gene expression. In an additive scenario, each binding site or cluster of sites would contribute towards the overall rate of transcription, and mutation of some cluster motifs would lower, but not necessarily eliminate, elt-2 expression. Partial redundancy within this additive scenario would mean that more sites would need to be mutated before an effect would be apparent.
Within the 1879 bp elt-2 upstream element we identified two clusters of HGATAR motifs hereafter “secondary GATA sites”) that show conservation in both sequence and spacing in the Elegans supergroup. Between -400 bp and -338 bp there are four HGATAR motifs (the 4G region) that are heavily conserved between C. elegans and five other members of the Elegans supergroup, including C. japonica (Fig. 1C). Between -1679 bp and -1525 bp upstream of the elt-2 start codon there are seven HGATAR motifs (the 7G region), with four conserved between C. elegans and six other members of the Elegans supergroup (Fig. 1C).
To determine whether these HGATAR motif cluster play a role in elt-2 promoter-driven gene activation, we performed site directed mutagenesis on HGATAR motifs within the 4G and 7G regions while keeping the A-site intact (Fig. 5). Surprisingly, mutating as many as eleven HGATAR motifs across two clusters had insignificant impacts on gene expression levels and noise during embryonic development, despite the heavy conservation of motifs within these clusters (Supplemental Figs. S6, S7, Fig. 1C). The only strain exhibiting statistically significant differences from its wild-type counterpart was 613:4G, and this was only observed as a seemingly minor temporary drop in mean expression during the 4E stage (Fig. 5C, Supplemental Fig. S7).
Figure 5.
Mutation of secondary 4G and 7G HGATAR motifs have minimal impact on embryonic gene expression levels or variability. (A-B) Mutation of 4G and 7G sites in a 1879 bp promoter fragment does not significantly alter embryonic expression (Fig. S7). gfp transcript counts shown for 1879:WT (n=187), 1879:4G (n=217), 1879:11G(n=214). C) Mutation of 4G sites in a minimal 613 bp promoter does not significantly alter embryonic expression (Supplemental Fig. S8). gfp transcript counts shown for 613:WT (n=459) and 613:4G (n=217). Shaded grey area represents timepoints where mean expression trajectory of 613:WT is significantly different from that of 613:4G (Supplemental Fig. S7).
We confirmed that known upstream activators of elt-2 could bind in vitro to the 4G region by performing an EMSA using a DNA probe corresponding to an 80 bp region encompassing the 4G region GATA sites (located -410 to -330 bp upstream of the elt-2 start codon) (Supplemental Fig. S8). We observed distinct band shifts for ELT-7, END-3, and END-1 binding domains, confirming that elt-2 activators can bind to regions containing secondary GATA motifs. Our results also indicate that the END-1, END-3, and ELT-7 DNA binding domains have distinct binding characteristics to the 80 bp region probed. At high protein concentrations, we observed three band shifts for ELT-7, four for END-3, and more than five for END-1. The band shift pattern for ELT-7 suggests that the ELT-7 binding domain binds to three out of the four GATA motifs within the 4G region at the protein concentrations tested. Interestingly, although five primary band shifts were visible for END-1, we clearly observed double banding on the three fastest migrating primary bands similar to the double banding observed in Fig. 4. These doublets are consistent with multimerization of the END-1 protein fragment we used on individual DNA motifs, and suggest that in vitro END-1 behavior seen in Fig. 4 is seen with GATA motifs in general rather than specific to the A-site motif.
These results indicate that although secondary GATA motif clusters have a minimal impact on early embryonic gene activation in vivo, they exhibit similar binding behavior to early embryonic elt-2 activators in vitro.
Reporter GFP fluorescence in larval and adult worms reveal secondary GATA motifs modulate post-embryonic expression
To investigate whether secondary GATA site mutations had impacts on elt-2 expression later in life, we imaged GFP fluorescence in larval and adult worms. The expression of elt-2 persists though larval stages and adult life via autoregulation (Tetsunari Fukushige et al, 1999) and positive feedback from elt-2 targets (Zhang, Judy, Lee, & Kenyon, 2013). We found that only strains with an intact A-site, except for the super minimal promoter strain 61:WT, expressed gut GFP during larval and adult stages (Table 1). The A-site was indispensable for both early embryonic activation as well as a larval and adult maintenance of gene expression.
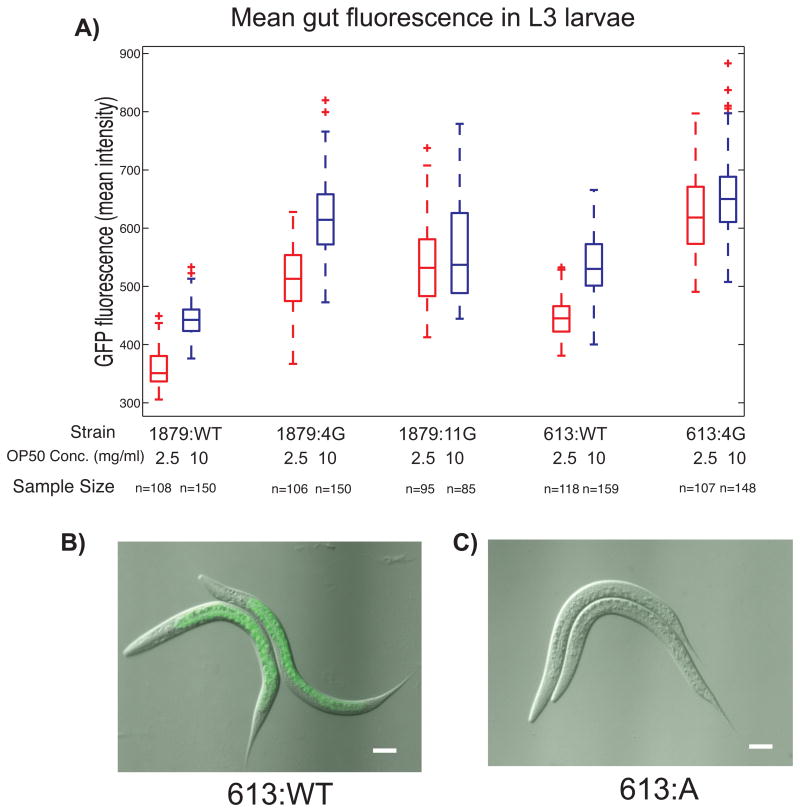
To quantify this expression we looked at L3 larvae 22 hours post-synchronization (later stages introduced gut autofluorescence complications). We found significant differences in mean gut fluorescence between wild-type promoter reporter strains and strains with mutations in the 4G and 7G regions (Fig. 6, Supplemental Fig. S9). Mean fluorescence and worm size were strongly coupled with food concentrations in some strains, indicating that diet has a positive effect on elt-2 promoter driven gene expression. In 1879:WT, 613:WT, 1879:4G, and 613:4G increasing the concentration of OP50 food from 2.5 mg/ml to 10 mg/ml significantly increased mean gut fluorescence (Supplemental Fig S9). This response is consistent with ELT-2's role as the activator of genes involved in digestion (McGhee et al, 2009).
Figure 6.
Secondary HGATAR motifs play a role in modulating post-embryonicgene expression. A) Mean gut GFP fluorescence levels for wild-type and mutant elt-2 promoter-reporter strains 22 hours post-synchronization. Red boxes represent worms grown at 2.5 mg/ml OP50 liquid media and blue boxes represent worms grown at 10 mg/ml OP50 liquid media. Red crosses represent outliers. B) Gut GFP fluorescence in 613:WT. C) Absence of gut fluorescence in 613:A mutants.
Mutating the 4G GATA motifs immediately downstream of the A-site in both long and short promoter contexts (1879:4G, 613:4G), resulted in significant increases in mean gut fluorescence and variability across different dietary contexts compared to wild-type counterparts (Supplemental Fig. S9). These increases demonstrate that some or all of the GATA motifs within the 4G region play a transcriptionally repressive role during post-embryonic development. The location of these sites immediately downstream of the A-site and TSS (Supplemental Fig. S3) suggest a steric mechanism.
We found that the apparent function of 7G region GATA motifs is dependent on context, and may be related to dietary response. Strains 1879:11G and 613:4G both lack many GATA motifs and both exhibit greater variability and less distinction between dietary regimes in post-embryonic reporter expression. In contrast to all other strains, 1879:11G did not show a statistically significant difference in mean gut fluorescence between groups raised at 10 mg/ml OP50 and groups raised at 2.5 mg/ml OP50 (p=0.0451; αSID = 0.00465). 1879:11G also experiences a slight but significant drop in mean gut intensity at 10 mg/ml compared to 1879:4G (p=1.78E-10; αSID = 0.00465), suggesting 7G region GATA sites may be activating under certain contexts. 613:WT, however, maintains a significant dichotomy in dietary response (p=2.85E-36; αSID = 0.00465) despite loss of many upstream GATA motifs. These results indicate that 7G and 4G clusters (upstream and downstream of the A-site respectively) may not necessarily function independently of each other.
Overall, many of the 7G and 4G GATA sites modulate post-embryonic elt-2 levels in both repressive and activating ways, dependent upon dietary context. Although many of these GATA motifs have similar sequence and conservation to the A-site, our functional dissection reveals that the roles of secondary motifs can be highly divergent and not necessarily related to activation. In contrast to the A-site, the secondary GATA motifs we examined play dispensable yet important roles in setting elt-2 expression levels in different environmental contexts.
An ACTGATAAG motif is not sufficient for driving gene expression to near-wild-type levels
Because mutation of the A-site produced a disproportionate impact on gene expression levels, we hypothesized that the presence of an ACTGATAAG motif might be sufficient for driving gene expression. In the 1879:A mutant strain, we kept a second naturally occurring ACTGATAAGG motif at -1857 bp intact (Fig. 1C green arrow). Despite the presence of an intact ACTGATAAG sequence at -1857 bp, this construct could not rescue reporter gene expression to wild-type promoter levels in A-site mutants (Figs. 2, 3B).
To determine whether a more minimal promoter element could be generated from the A-site, we generated a promoter reporter construct consisting of a 61 bp region containing the A-site (Table 1 61:WT). This construct failed to produce either embryonic gfp transcripts as assessed by smFISH or adult gut GFP fluorescence. This failure to drive even low-level reporter expression demonstrates that an ACTGATAAG motif alone is not sufficient for driving gene expression, and that a secondary CRE or some kind of positional information is still necessary to trigger transcription. Our results indicate that such a secondary CRE is not one of the GATA sites mutated in this study, and is unlikely to be a GATA motif.
4G region HGATAR motifs play a role in driving low-level ectopic expression
Having identified the 613 bp elt-2 upstream region as a minimal promoter fragment, we sought to determine if mutation of 4G region sites would have a more obvious impact on gene expression in a minimal promoter context. Mutation of 4G region HGATAR sites in strain 613:4G resulted in a slightly altered, but near-wild-type embryonic expression profile (Fig. 5C, Supplemental Fig. S7).
Mutation of the A-site along with 4G HGATAR motifs (Table 1 613:A4G) resulted in total loss of gene expression, effectively bringing the low-level stochastic expression observed in strain 613:A to zero for all embryos sampled (Fig. 3C). This indicates that low-level stochastic transcription resulting from mutation of the A-site was dependent upon one or more of the 4G HGATAR motifs.
Discussion
In this study, we performed the first in vivo single-molecule resolution investigation of the relation between CREs and transcriptional output during C. elegans development. We found that CREs in close proximity with similar sequences, similar conservation, and capable of binding to the same transcription factors can have wildly different functional roles in development.
Rather than multiple HGATAR sites in the elt-2 promoter serving as redundant transcriptional activators, a single critically positioned key motif is necessary for both early END-1/END-3 driven embryonic activation as well as later ELT-2/ELT-7 driven larval and adult maintenance of gene expression. This key GATA site is used for earliest activation, later maintenance, and, surprisingly, repression of spatiotemporally ectopic early embryonic expression. Our results suggest that the critically positioned A-site acts as a GATA responsive “core” or “proximal” promoter element distinct from nearby conserved motifs with similar HGATAR sequences.
In contrast, the majority of GATA motifs mutated in this study appear to be dispensable for embryonic activation, but important for tuning postembryonic expression levels. We found that in wild-type promoter contexts, gene expression levels were dependent upon dietary food concentrations, with less food leading to less elt-2 promoter activation. Furthermore, despite binding capability to upstream activators (Supplemental Fig. S8, Tracy and Rifkin, unpublished), mutation of the HGATAR motifs within the 4G region paradoxically resulted in elevated gene expression levels and variability in both long and short promoter contexts (1879:4G, 613:4G) compared to wild-type reporters. This implies that some or all of these GATA sites play repressive roles in postembryonic development. One possible explanation for this behavior is the formation of repressive or simply non-activating multimers on 4G region sites by otherwise activating GATA factors. Mammalian GATA factors have been reported to form homo and hetero-oligmers (Chen et al. 2012), and the conservation of spacing and positioning of three of the 4G region sites within the Elegans supergroup suggest that orientation is important for this region of the promoter. Another possibility is that these sites may bind post-embryonically expressed non-GATA transcription factors (see below).
Although the extended DNA motif sequence of each individual GATA CRE may contribute to establishing the distinctive roles of the dominant primary A-site versus secondary auxiliary GATA CREs, we found that the extended sequence motif does not completely explain the relative importance of the A-site. The A-site's extended sequence is ACTGATAAGA, but mutation of the key A-site motif at -527 bp could not be rescued to near-wild-type levels by any of 9 remaining TGATAA or 17 remaining HGATAR motifs present in strain 1879:A (Figs. 2, 3, S2, S4). Furthermore, one of the alternate TGATAA motifs is a very similar ACTGATAAGG located at -1857 bp that remains intact in strain 1879:A (Fig 1C; green arrow). However, this similar sequence is insufficient to activate promoter-reporter gene expression. The presence of an A-site is also not sufficient for driving even low-level expression in a 61 bp minimal promoter (Table 1 61:WT) context suggesting that more cis-information may be needed for gene activation. This additional information could come in the form of a second motif, the local chromatin context, or even the relative abundance of T-rich sequence (Grishkevich, Hashimshony, & Yanai, 2011).
Despite this insufficiency, evidence from a wide array of studies indicate that the ACTGATAAG sequence can act as a binding site for multiple gut-associated transcription factors, some of which are not even GATA factors. Several studies previously demonstrated that similar or identical sequences are present and functionally necessary in the promoters of elt-2 regulated gut genes (Egan et al., 1995; Tetsunari Fukushige, Goszczynski, Yan, & McGhee, 2005; MacMorris et al., 1992; MacMorris, Spieth, Madej, Lea, & Blumenthal, 1994; McGhee et al., 2009). Furthermore, previous research into the FOXO transcription factor DAF-16 revealed that the promoters of DAF-16 targets are enriched for the sequence TGATAAG (also known as “Daf-16 associated element” or DAE) and that DAF-16 can bind to the TGATAAG motif in vitro (Murphy et al., 2003; Zhang, Judy, Lee, & Kenyon, 2013). A recent study, Mueller et al. 2014, also showed that TGATAAG sequences, particularly the specific sequence ACTGATAAGA, are heavily over-represented in the promoters of genes that are upregulated in response to ultraviolet light exposure, and downregulated in response to starvation. This study implicated the GATA factor EGL-27 as the primary effector of UV stress-responsive gene activation.
These studies combined with our results suggest that in the worm's adult life, regulatory signals from different pathways may converge on and compete for access of a single dominant primary cis-regulatory motif in the elt-2 promoter, effectively forming a regulatory information bottleneck. Such a bottleneck can be useful in setting an upper limit on the level of transcriptional activation that can occur while simultaneously allowing responsiveness to signals related to processes such as aging(Zhang et al., 2013), stress (Schieber & Chandel, 2014), and disease resistance (Head & Aballay, 2014). Once the primary activation site is saturated with binding factors, further activation may be impeded.
The wide affinity of GATA-family motifs combined with the large number of GATA related transcription factors may partially explain our observations of low-level stochastic ectopic transcription in A-site mutants in both E cells and non-E-cells. The absence of transcriptional activators such as med-1/2, end-3, or end-1 in non-E-cells implies that there may be other GATA-binding transcriptional activators present outside of the nascent endoderm during early embryogenesis (Budovskaya et al., 2008; Gilleard, Shafi, Barry, & McGhee, 1999; Mueller et al., 2014; Murphy et al., 2003). We found that in a minimal 613 bp promoter context, secondary auxiliary HGATAR motif(s) in the 4G region are necessary for producing low-level stochastic transcription (Table 1 613:A4G; Fig 3D). Including more proximal regions of the elt-2 promoter (-1879 bp to -613 bp; contains 13 HGATAR motifs) in an A-site mutant background increases low-level transcription levels even further. Our analysis of deep sequencing data from Saito et al. 2013 revealed small numbers of TSS reads overlying proximal promoter regions including the 7G and 4G regions, suggesting many ectopic transcripts may originate from secondary GATA sites (Supplemental Fig. S3) (Saito et al., 2013).
These non-specific mini-activation events reveal a layer of transcriptional regulation not previously observed. For multicellular organisms, off target activation events can result in ectopic expression that can have negative consequences for cell fate specification, particularly if the gene being ectopically expressed is an autoregulatory transcription factor. Here we observe that an intact A-site plays a role in suppressing this ectopic expression in non-E-cells. The same site used for activation in the endoderm is used for repression in non-endoderm fated cells.
Conclusions
Our study shows that C. elegans can serve as a powerful model for single-molecule cell-type-specific cis-regulatory studies. We determined that a single key cis-regulatory site - in the midst of a host of similarly conserved sites - is used for the earliest activation and post-embryonic maintenance of the expression of an essential regulatory gene, and that this same CRE helps repress early stochastic expression in non-E cells. We identified diverse roles for secondary sites in tuning postembryonic gene expression. The wildly different roles served by CREs examined in this study illustrate the diverse functions that similar CREs can take on and reveal a distinction between early and post-embryonic elt-2 activation and function.
Supplementary Material
Highlights.
Promoter activation and maintenance of the C. elegans endoderm-specifying gene elt-2 is dependent upon a single dominant ACTGATAAG motif located -527 upstream of the elt-2 start codon.
Mutation of the dominant ACTGATAAG motif leads to early ectopic stochastic transcription.
Many secondary GATA motifs play quantitatively negligible roles in embryonic activation of elt-2, but play roles in tuning post-embryonic expression.
The DNA binding domains of the known elt-2 activating transcription factors ELT-7, END-1, END-3, and ELT-2 itself are able to bind to the both the crucial -527 bp motif as well as other regions of the elt-2 promoter in vitro.
Acknowledgments
Special thanks to George Kassavetis and Jim Kadonaga for help with in vitro assays, Jim McGhee for elt-2 plasmids and discussion of the elt-2 promoter, Emily Troemel for microscope time, and Allison Wu for help in generating smFISH probes and initial processing of the data. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). This work was supported in part by Graduate Assistance in Areas of National Need (GAANN), NIH predoctoral training grant T32 GM008666, and NIH grant R01GM103782 to SAR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Boeck ME, Boyle T, Bao Z, Murray J, Mericle B, Waterston R. Specific roles for the GATA transcription factors end-1 and end-3 during C. elegans E-lineage development. Developmental Biology. 2011;358(2):345–355. doi: 10.1016/j.ydbio.2011.08.002. http://doi.org/10.1016/j.ydbio.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budovskaya YV, Wu K, Southworth LK, Jiang M, Tedesco P, Johnson TE, Kim SK. An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell. 2008;134(2):291–303. doi: 10.1016/j.cell.2008.05.044. http://doi.org/10.1016/j.cell.2008.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH. Genomic Regulatory Systems: Development and Evolution. Academic Press; 2001. [Google Scholar]
- Egan CR, Chung MA, Allen FL, Heschl MFP, Van Buskirk CL, McGhee JD. A Gut-to-Pharynx/Tail Switch in Embryonic Expression of the Caenorhabditis elegans ges-1 Gene Centers on Two GATA Sequences. Developmental Biology. 1995;170(2):397–419. doi: 10.1006/dbio.1995.1225. http://doi.org/10.1006/dbio.1995.1225. [DOI] [PubMed] [Google Scholar]
- Félix MA, Braendle C, Cutter AD. A Streamlined System for Species Diagnosis in Caenorhabditis (Nematoda: Rhabditidae) with Name Designations for 15 Distinct Biological Species. PLoS ONE. 2014;9(4):e94723. doi: 10.1371/journal.pone.0094723. http://doi.org/10.1371/journal.pone.0094723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores GV, Duan H, Yan H, Nagaraj R, Fu W, Zou Y, Banerjee U. Combinatorial Signaling in the Specification of Unique Cell Fates. Cell. 2000;103(1):75–85. doi: 10.1016/s0092-8674(00)00106-9. http://doi.org/10.1016/S0092-8674(00)00106-9. [DOI] [PubMed] [Google Scholar]
- Frøkjær-Jensen C, Davis MW, Ailion M, Jorgensen EM. Improved Mos1- mediated transgenesis in C. elegans. Nature Methods. 2012;9(2):117–118. doi: 10.1038/nmeth.1865. http://doi.org/10.1038/nmeth.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Jorgensen EM. Single-copy insertion of transgenes in Caenorhabditis elegans. Nature genetics. 2008;40(11):1375–1383. doi: 10.1038/ng.248. http://doi.org/10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushige T, Hawkins MG, McGhee JD. The GATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Developmental Biology. 1998;198(2):286–302. [PubMed] [Google Scholar]
- Fukushige T, Hendzel MJ, Bazett-Jones DP, McGhee JD. Direct visualization of the elt-2 gut-specific GATA factor binding to a target promoter inside the living Caenorhabditis elegans embryo. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(21):11883–11888. doi: 10.1073/pnas.96.21.11883. http://doi.org/10.1073/pnas.96.21.11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushige Tetsunari, Goszczynski B, Yan J, McGhee JD. Transcriptional control and patterning of the pho-1 gene, an essential acid phosphatase expressed in the C. elegans intestine. Developmental Biology. 2005;279(2):446–461. doi: 10.1016/j.ydbio.2004.12.012. http://doi.org/10.1016/j.ydbio.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Ikegami K. Integrative Analysis of the Caenorhabditis elegans Genome by the modENCODE Project. Science (New York, NY) 2010;330(6012):1775–1787. doi: 10.1126/science.1196914. http://doi.org/10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilleard JS, Shafi Y, Barry JD, McGhee JD. ELT-3: A Caenorhabditis elegans GATA Factor Expressed in the Embryonic Epidermis during Morphogenesis. Developmental Biology. 1999;208(2):265–280. doi: 10.1006/dbio.1999.9202. http://doi.org/10.1006/dbio.1999.9202. [DOI] [PubMed] [Google Scholar]
- Grishkevich V, Hashimshony T, Yanai I. Core promoter T-blocks correlate with gene expression levels in C. elegans. Genome Research. 2011;21(5):707–717. doi: 10.1101/gr.113381.110. http://doi.org/10.1101/gr.113381.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TW, Antoshechkin I, Bieri T, Blasiar D, Chan J, Chen WJ, Sternberg PW. WormBase: a comprehensive resource for nematode research. Nucleic Acids Research. 2010;38(Database issue):D463–467. doi: 10.1093/nar/gkp952. http://doi.org/10.1093/nar/gkp952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head B, Aballay A. Recovery from an Acute Infection in C. elegans Requires the GATA Transcription Factor ELT-2. PLoS Genet. 2014;10(10):e1004609. doi: 10.1371/journal.pgen.1004609. http://doi.org/10.1371/journal.pgen.1004609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RE, Ren X, Qiu Y, Zhao Z. Description of Caenorhabditis sinica sp. n. (Nematoda: Rhabditidae), a nematode species used in comparative biology for C. elegans. PloS One. 2014;9(11):e110957. doi: 10.1371/journal.pone.0110957. http://doi.org/10.1371/journal.pone.0110957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T, Kadonaga JT. Regulation of Gene Expression via the Core Promoter and the Basal Transcriptional Machinery. Developmental Biology. 2010;339(2):225–229. doi: 10.1016/j.ydbio.2009.08.009. http://doi.org/10.1016/j.ydbio.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry JA, Gamsjaeger R, Thong SY, Hung W, Kwan AH, Broitman-Maduro G, Mackay JP. Structural analysis of MED-1 reveals unexpected diversity in the mechanism of DNA recognition by GATA-type zinc finger domains. The Journal of Biological Chemistry. 2009;284(9):5827–5835. doi: 10.1074/jbc.M808712200. http://doi.org/10.1074/jbc.M808712200. [DOI] [PubMed] [Google Scholar]
- MacMorris M, Broverman S, Greenspoon S, Lea K, Madej C, Blumenthal T, Spieth J. Regulation of vitellogenin gene expression in transgenic Caenorhabditis elegans: short sequences required for activation of the vit-2 promoter. Molecular and Cellular Biology. 1992;12(4):1652–1662. doi: 10.1128/mcb.12.4.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMorris M, Spieth J, Madej C, Lea K, Blumenthal T. Analysis of the VPE sequences in the Caenorhabditis elegans vit-2 promoter with extrachromosomal tandem array-containing transgenic strains. Molecular and Cellular Biology. 1994;14(1):484–491. doi: 10.1128/mcb.14.1.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro MF, Broitman-Maduro G, Mengarelli I, Rothman JH. Maternal deployment of the embryonic SKN-1-->MED-1,2 cell specification pathway in C. elegans. Developmental biology. 2007;301(2):590–601. doi: 10.1016/j.ydbio.2006.08.029. http://doi.org/10.1016/j.ydbio.2006.08.029. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Hill RJ, Heid PJ, Newman-Smith ED, Zhu J, Priess JR, Rothman JH. Genetic redundancy in endoderm specification within the genus Caenorhabditis. Developmental Biology. 2005;284(2):509–522. doi: 10.1016/j.ydbio.2005.05.016. http://doi.org/10.1016/j.ydbio.2005.05.016. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Meneghini MD, Bowerman B, Broitman-Maduro G, Rothman JH. Restriction of Mesendoderm to a Single Blastomere by the Combined Action of SKN-1 and a GSK-3β Homolog Is Mediated by MED-1 and -2 in C. elegans. Molecular Cell. 2001;7(3):475–485. doi: 10.1016/s1097-2765(01)00195-2. http://doi.org/10.1016/S1097-2765(01)00195-2. [DOI] [PubMed] [Google Scholar]
- McGhee JD, Fukushige T, Krause MW, Minnema SE, Goszczynski B, Gaudet J, Kalb JM. ELT-2 Is the Predominant Transcription Factor Controlling Differentiation and Function of the C. elegans Intestine, from Embryo to Adult. Developmental Biology. 2009;327(2):551–565. doi: 10.1016/j.ydbio.2008.11.034. http://doi.org/10.1016/j.ydbio.2008.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirny LA, Wunderlich Z. Fundamentally different strategies for transcriptional regulation are revealed by information-theoretical analysis of binding motifs. Trends in Genetics. 2008;25(10):4. doi: 10.1016/j.tig.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller MM, Castells-Roca L, Babu V, Ermolaeva MA, Müller RU, Frommolt P, Schumacher B. DAF-16/FOXO and EGL-27/GATA promote developmental growth in response to persistent somatic DNA damage. Nature Cell Biology. 2014;16(12):1168–1179. doi: 10.1038/ncb3071. http://doi.org/10.1038/ncb3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, … Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424(6946):277–283. doi: 10.1038/nature01789. http://doi.org/10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Nair G, Walton T, Murray JI, Raj A. Gene transcription is coordinated with, but not dependent on, cell divisions during C. elegans embryonic fate specification. Development (Cambridge, England) 2013;140(16):3385–3394. doi: 10.1242/dev.098012. http://doi.org/10.1242/dev.098012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Rifkin SA, Andersen E, van Oudenaarden A. Variability in gene expression underlies incomplete penetrance. Nature. 2010;463(7283):913–918. doi: 10.1038/nature08781. http://doi.org/10.1038/nature08781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nature Methods. 2008;5(10):877–879. doi: 10.1038/nmeth.1253. http://doi.org/10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifkin SA. Identifying fluorescently labeled single molecules in image stacks using machine learning. Methods in molecular biology (Clifton, NJ) 2011;772:329–348. doi: 10.1007/978-1-61779-228-1_20. http://doi.org/10.1007/978-1-61779-228-1_20. [DOI] [PubMed] [Google Scholar]
- Saito TL, Hashimoto S, Gu SG, Morton JJ, Stadler M, Blumenthal T, Morishita S. The transcription start site landscape of C. elegans. Genome Research. 2013;23(8):1348–1361. doi: 10.1101/gr.151571.112. http://doi.org/10.1101/gr.151571.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieber M, Chandel NS. TOR signaling couples oxygen sensing to lifespan in C. elegans. Cell Reports. 2014;9(1):9–15. doi: 10.1016/j.celrep.2014.08.075. http://doi.org/10.1016/j.celrep.2014.08.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommermann EM, Strohmaier KR, Maduro MF, Rothman JH. Endoderm development in Caenorhabditis elegans: the synergistic action of ELT-2 and -7 mediates the specification→differentiation transition. Developmental Biology. 2010;347(1):154–166. doi: 10.1016/j.ydbio.2010.08.020. http://doi.org/10.1016/j.ydbio.2010.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ury HK. A Comparison of Four Procedures for Multiple Comparisons among Means (Pairwise Contrasts) for Arbitrary Sample Sizes. Technometrics. 1976;18(1):89–97. http://doi.org/10.2307/1267921. [Google Scholar]
- Wu AC, Rifkin SA. Aro: a machine learning approach to identifying single molecules and estimating classification error in fluorescence microscopy images. BMC Bioinformatics. 2015;16(1):102. doi: 10.1186/s12859-015-0534-z. http://doi.org/10.1186/s12859-015-0534-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Judy M, Lee SJ, Kenyon C. Direct and Indirect Gene Regulation by a Life-Extending FOXO Protein in C. elegans: Roles for GATA Factors and Lipid Gene Regulators. Cell Metabolism. 2013;17(1):85–100. doi: 10.1016/j.cmet.2012.12.013. http://doi.org/10.1016/j.cmet.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Fukushige T, McGhee JD, Rothman JH. Reprogramming of early embryonic blastomeres into endodermal progenitors by a Caenorhabditis elegans GATA factor. Genes & Development. 1998;12(24):3809–3814. doi: 10.1101/gad.12.24.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Hill RJ, Heid PJ, Fukuyama M, Sugimoto A, Priess JR, Rothman JH. end-1 encodes an apparent GATA factor that specifies the endoderm precursor in Caenorhabditis elegans embryos. Genes & Development. 1997;11(21):2883–2896. doi: 10.1101/gad.11.21.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.