Abstract
Background
Lithium remains a first-line treatment in bipolar disorder, but individual response is variable. Previous studies have suggested that lithium response is a heritable trait. However, no genetic markers have been reproducibly identified.
Methods
Here we report the results of a genome-wide association study of lithium response in 2,563 patients collected by 22 participating sites from the International Consortium on Lithium Genetics (ConLiGen); the largest attempted so far. Data from over 6 million common single nucleotide polymorphisms (SNPs) were tested for association with categorical and continuous ratings of lithium response of known reliability.
Findings
A single locus of four linked SNPs on chromosome 21 met genome-wide significance criteria for association with lithium response (rs79663003: p=1·37×10−8; rs78015114: p=1·31×10−8; rs74795342: p=3·31×10−9; rs75222709: p=3·50×10−9). In an independent, prospective study of 73 patients treated with lithium monotherapy for a period of up to two years, carriers of the response-associated alleles had a significantly lower rate of relapse than carriers of the alternate alleles (p=0·03, hazard ratio = 3·8).
Interpretation
The response-associated region contains two genes coding for long non-coding RNAs (lncRNAs), AL157359.3 and AL157359.4. LncRNAs are increasingly appreciated as important regulators of gene expression, particularly in the CNS. Further studies are needed to establish the biological context of these findings and their potential clinical utility. Confirmed biomarkers of lithium response would constitute an important step forward in the clinical management of bipolar disorder.
INTRODUCTION
Bipolar disorder (BD) is an often devastating psychiatric illness characterized by disruptive mood swings, with intervals of partial or full recovery. BD type I and II affect at least 2% of the world’s population; subthreshold forms afflict another 2%1. BD consumes a substantial portion of mental health resources. Worldwide, the direct and indirect costs are large, with an estimated US$151 billion spent in the US alone in 20092. Moreover, up to 15% of sufferers die by suicide3.
Mood stabilizers are a primary mode of medication treatment for BD4. Among these drugs, lithium stands out as a preventive agent for manic episodes5 and suicide6. Consequently, lithium is still recommended as a first-line treatment for BD, even though individual response is variable. Many patients show a robust improvement with lithium and a subset is highly responsive7–9, with near total resolution of symptoms. On the other hand, at least 30% of patients are only partially responsive, and more than 30% have no clinical response to lithium.
Evidence suggests that some of the variability in lithium response has a genetic basis, but sample sizes in such studies have been limited. Good responders are more likely to have a family history of BD than poor responders10. Patients who stabilized on lithium tend to aggregate within families11, 12. A twin study reported better lithium prophylaxis in twins whose co-twin also had BD13.
Genetic markers of lithium response could provide insight into the biological mechanism of lithium action and might be valuable for treatment planning. However, few pharmacogenetic studies of lithium have been published, and those have generally employed small samples and variable definitions of response. Candidate gene studies have focused on genes purported to be involved in the therapeutic action of lithium, but replicable results have not emerged14, 15. Three genome-wide association studies (GWAS) of lithium response have been published. The first was from the Systematic Treatment Enhancement Program for Bipolar Disorder (STEP-BD) cohort16, in which 458 BD-I/II patients were treated with lithium and response was evaluated as time to recurrence during lithium treatment. No genome-wide significant results were identified. A second GWAS17 was carried out in 204 Sardinian BD patients (only 52 were genotyped with SNP arrays). No SNPs reached genome-wide significance. Most recently, Chen and colleagues18 performed a GWAS on 294 highly treatment adherent individuals of Asian ancestry selected from a larger set of about 2000 treated for BD-I with lithium monotherapy. The authors reported genome-wide significant association with a cluster of SNPs at 3p24.1. However, to date, all reported studies have failed to replicate these findings in either Asian or European ancestry samples19–21.
In order to overcome the problems inherent in smaller sample sizes, we established the international Consortium on Lithium Genetics (www.ConLiGen.org) in 200822. Here we report the results of an initial GWAS of lithium response in 2563 BD patients – by far the largest sample to date – using phenotype and genotype data from 22 ConLiGen sites from four continents (Europe, America, Asia and Australia; for details, see Table S1; Supplementary Materials). The Taiwanese sample included is independent from the sample studied by Chen and colleagues18. Four linked SNPs met genome-wide significance criteria for association with a quantitative measure of lithium response. The associated locus has been annotated with two long, non-coding RNA (lncRNA) genes. If replicated, these findings would constitute a novel genetic marker and could implicate lncRNAs in the mechanism of lithium response.
MATERIALS AND METHODS
Participants
Written informed consent was obtained from all participants. Study sites were largely non-overlapping (Table S1; Supplementary Materials). Over the timeframe of this study (phenotyping between 2008 and 2013), available samples were collected and genotyped in two distinct phases. We thus analyzed the data as two distinct GWAS, referred to herein as ‘GWAS 1’ and ‘GWAS 2’ (for a detailed rationale, see Supplementary Materials). The analysis pipeline is shown in Figure S1 (Supplementary Materials). Briefly, a total of 3193 participants were genotyped; 2935 remained after quality control (QC).
A DSM-III or DSM-IV diagnosis of a bipolar spectrum disorder (see Supplementary Materials) was required, along with data on gender and total score on the Alda scale (see below). We included all patients in whom response could be reliably evaluated: patients were required to have taken lithium for a minimum of 6 months with no additional mood stabilizer added. Comorbid conditions were not among the exclusion criteria. After this step, 1162 individuals were included in GWAS 1 and 1401 were included in GWAS 2.
Phenotypes
We used the “Retrospective Criteria of Long-Term Treatment Response in Research Subjects with Bipolar Disorder” (Alda scale12) for the evaluation of long-term treatment response to lithium. This scale measures the change in illness episodes in the course of treatment with lithium. Briefly, the Alda scale quantifies symptom improvement in the course of treatment (A score, range 0–10), which is then weighted against five criteria (B score) that assess confounding factors, each scored 0, 1 or 2. The total score is then derived by subtracting the total B score from the A score. Negative scores are set to 0 by default so that the total score ranges from 0 to 10.
ConLiGen previously conducted a multi-stage inter-rater reliability study23 aimed at finding the optimal way in which Alda subscale values can be combined for response evaluation. We evaluated two main phenotypes for lithium response: a dichotomous phenotype (good/poor response to lithium), that has been successfully used in previous studies9, 13, and a continuous phenotype (range 0 to 10). We found the most reliable dichotomous phenotype to be that which designated all subjects with a total score ≥ 7 as “responders”. The most reliable continuous phenotype was found to be one that used the A score but excluded all subjects with a total B score > 4. Descriptive statistics of the phenotypes of the total sample analyzed in the present study can be found in Table 1; excluded participants are detailed in Table S2 (Supplementary Materials).
Table 1.
Phenotypic characteristics of individuals used for the analyses
| GWAS 1 | GWAS 2 | |
|---|---|---|
| All subjects | ||
| N | 1162 | 1401 |
| Percent female | 59·30 | 56·17 |
| Age at interview | 47·80 (13·99) | 46·84 (13·83) |
| ALDA scale A score | 6·03 (3·14) | 6·35 (2·90) |
| ALDA scale total B score | 2·11 (1·63) | 2·86 (1·68) |
| ALDA scale total score | 4·29 (3·32) | 3·90 (3·02) |
| Dichotomous phenotype: Good response (ALDA scale total score 7 or greater) | ||
| N | 361 | 342 |
| Age at interview | 51·72 (14·27) | 48·92 (14·80) |
| Percent female | 56·23 | 51·75 |
| ALDA scale A score | 9·21 (0·82) | 9·36 (0·77) |
| ALDA scale total B score | 0·88 (0·84) | 1·38 (0·96) |
| ALDA scale total score | 8·33 (1·10) | 7·99 (1·01) |
| Dichotomous phenotype: Poor response (ALDA scale total score 6 or less) | ||
| N | 801 | 1059 |
| Age at interview | 45·86 (13·44) | 46·17 (13·44) |
| Percent female | 60·67 | 57·60 |
| ALDA scale A score | 4·60 (2·71) | 5·38 (2·66) |
| ALDA scale total B score | 2·66 (1·59) | 3·34 (1·58) |
| ALDA scale total score | 2·47 (2·19) | 2·58 (2·14) |
| Continuous phenotype (ALDA scale A score, all subjects with a total B score greater 4 excluded) | ||
| N | 1065 | 1168 |
| Age at interview | 48·12 (14·00) | 46·97 (13·84) |
| Percent female | 59·91 | 56·34 |
| ALDA scale A score | 6·13 (3·13) | 6·52 (2·87) |
| ALDA scale total B score | 1·78 (1·26) | 2·35 (1·16) |
| ALDA scale total score | 4·59 (3·28) | 4·40 (2·94) |
Genotyping, quality control and imputation
DNA was extracted from peripheral blood samples. Samples were genotyped at the NIMH, Life & Brain Center at the University of Bonn, or Broad Institute using either Affymetrix or Illumina SNP arrays (Table S1; Supplementary Materials), according to the manufacturers’ protocols.
Quality control and imputation were carried out in batches corresponding to distinct SNP arrays and ethnicities. Six batches of data were used in GWAS 1, including five of European ancestry (Affymetrix 6·0, Human610/660W, HumanOmniExpress, HumanOmni1-Quad, HumanOmni2.5), and one of Japanese ancestry (HumanOmni2.5). Five batches of data were used in GWAS2, including four European-ancestry data sets (Affymetrix 6·0, Human660W, HumanOmni1-Quad, HumanOmniExpress), and one Taiwanese data set (HumanOmniExpress) not overlapping with the sample studied by Chen and colleagues18. Quality control parameters for retaining SNPs and subjects, including relatedness checking and population stratification analysis are detailed in the Supplementary Materials.
Genotype imputation was performed using the prephasing/imputation strategy24 implemented by SHAPEIT225 and minimac26. The full 1000 Genomes Project data set was used as the reference panel. Imputation was performed separately for each SNP array and ancestry group. Gene dosages for all markers with imputation r2 ≥ 0.5 in all batches were used for the final association tests.
Statistical analysis
Association testing was carried out separately in European-ancestry and Asian-ancestry samples. We analyzed both the categorical and quantitative response phenotypes. Using PLINK v1·0727, the association between allele dosages and the dichotomous phenotype was evaluated by logistic regression and the association between allele dosages and the quantitative phenotype was evaluated by linear regression. Genotyping platform was used as a covariate and, in the European-ancestry samples, the first four principal components of population structure were also included in the model to control for population stratification (Supplementary Materials). Site of collection was not included as a covariate because it was highly co-linear with genotyping platform. Results across GWAS 1 and GWAS 2 were combined by meta-analysis using METAL28, under a fixed-effects model with heterogeneity testing.
Overall results in GWAS 1 were compared to those in GWAS 2 by use of the Sign Test (Supplementary Materials). If there were no association between SNPs and traits, the expectation is that 50% of the β coefficients would have the same sign. The significance of the observed proportion was evaluated under the binomial distribution. To investigate the contribution of the BD risk profile scores (RPS) to lithium response, we used the LD clumped complete result file of 108 835 SNPs from the PGC bipolar GWAS29 to calculate –log (OR) weighted RPS in each of the two European ancestry samples. Regression was then used to test whether the calculated RPS scores had any effect on the association between SNP dosages and lithium response by adding the RPS scores as an additional covariate in the regression model.
ROLE OF THE FUNDING SOURCES
The funding bodies had no role in study design, data collection, data analysis, data interpretation, or writing of the report. LH, UH, FJM, and TGS had full access to all the data. The corresponding authors FJM and TGS had final responsibility for the decision to submit for publication.
RESULTS
Our principal goal was to identify common genetic variants associated with differential response to lithium. Neither GWAS 1 nor GWAS 2 alone detected a genome-wide significant (p<5×10−8) result. However, there was greater than chance consistency between GWAS 1 and GWAS 2 in the overall direction of association. For the continuous phenotype, of 606 independent SNPs in GWAS 1 with p<0·001, 326 (54%) had the same sign in GWAS 2. This represents a significantly greater agreement than chance alone (p=0·028). For the dichotomous phenotype, of 555 independent SNPs in GWAS 1 with p<0·001, 317 (57%) had the same sign in GWAS 2, significantly (p=0·0003) greater than chance (for the complete list of SNPs used in this test, see Table S4, Supplementary Materials).
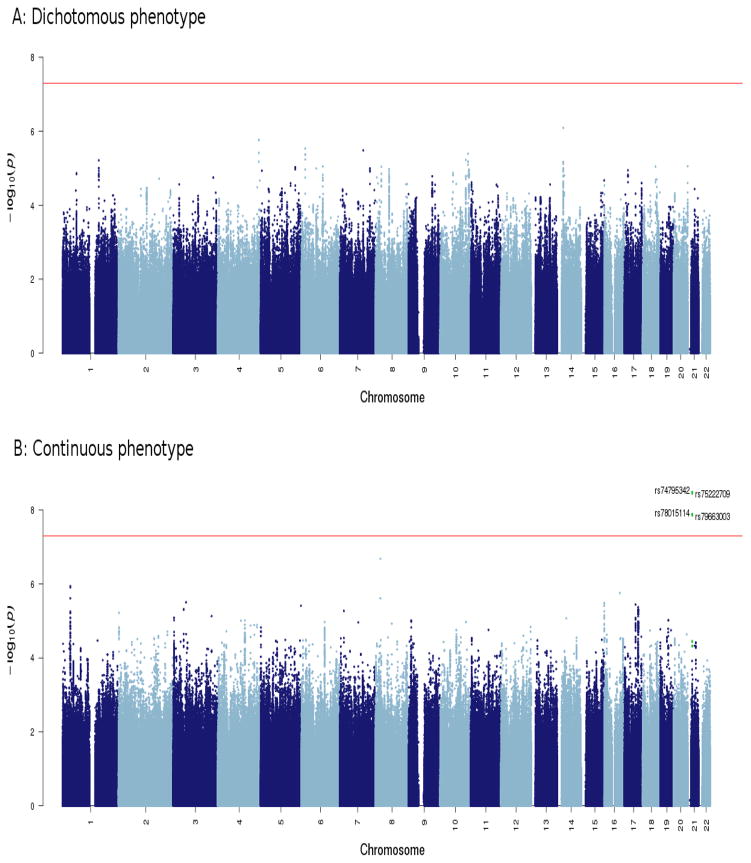
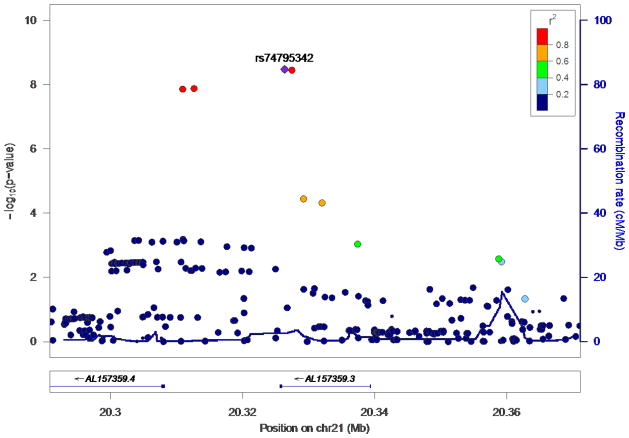
When both studies were combined by meta-analysis, genome-wide significance was attained for the continuous phenotype (Figures 1–3). Table 2 summarizes the top results (p<1×10−6) with the continuous phenotype (see Table S5 for the top results with the dichotomous phenotype). A region on chromosome 21 contained four SNPs that showed genome-wide significant association with lithium response (Figure 2) (minimum p=3·31×10−9). These four SNPs are in very strong LD with each other and have similar minor allele frequencies. The same 4 SNPs were associated with the dichotomous definition of lithium response at a p~0·01. These four SNPs also reached significance when only the European-ancestry population was considered (Table 2). The imputation quality for these 4 SNPs was excellent and was supported by direct genotyping in a subset of the total sample (see assay validation on page 2 of Supplementary Materials and Table S3).
Figure 1.
Meta-analysis results of dichotomous and continuous lithium response phenotypes in all participants (CEU plus Asian samples). Genome-wide significant association can be detected with the continuous phenotype. SNPs in green are in linkage disequilibrium (r2>0·6) with the index SNPs (rs74795342).
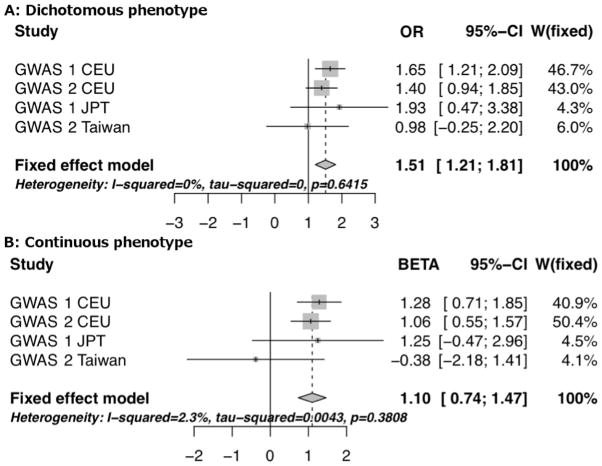
Figure 3.
Forest plots for the most significant SNP, rs74795342. [Panel A, dichotomous phenotype; Panel B, continuous phenotype].
Table 2.
Regionsa of the genome showing the strongest association signals with the continuous trait
| Chr | Positionb | SNP | A1c | A2d | FRQe | Gene | P | Directionsf | BETA (CI)g | Heterogeneity P-valh |
|---|---|---|---|---|---|---|---|---|---|---|
| Both Populations | ||||||||||
| 21 | 20310893 | rs79663003 | T | C | 0·94 | AL157359.4 | 1·37E-8 | +++- | 1·04 (0·68 – 1·40) | 0·30 |
| 21 | 20312612 | rs78015114 | T | C | 0·94 | AL157359.4 | 1·31E-8 | +++- | 1·04 (0·68 – 1·40) | 0·31 |
| 21 | 20326336 | rs74795342 | G | A | 0·94 | AL157359.3 | 3·31E-9 | +++- | 1·10 (0·74 – 1·47) | 0·46 |
| 21 | 20327427 | rs75222709 | T | G | 0·94 | AL157359.3 | 3·50E-9 | +++- | 1·10 (0·73 – 1·46) | 0·46 |
|
| ||||||||||
| European ancestry only | ||||||||||
| 1 | 34604567 | rs9662615 | T | C | 0·44 | CSMD2 | 5·26E-7 | ++ | 0·45 (0·27 – 0·62) | 0·83 |
| 1 | 34608545 | rs771148 | C | T | 0·40 | CSMD2 | 7·01E-7 | ++ | 0·45 (0·27 – 0·63) | 0·64 |
| 7 | 18444419 | rs61549860 | T | A | 0·22 | HDAC9 | 5·44E-7 | ++ | 0·59 (0·36 – 0·83) | 0·92 |
| 21 | 20310893 | rs79663003 | T | C | 0·94 | AL157359.4 | 1·30E-8 | ++ | 1·10 (0·72 – 1·48) | 0·41 |
| 21 | 20312612 | rs78015114 | T | C | 0·94 | AL157359.4 | 1·25E-8 | ++ | 1·10 (0·72 – 1·48) | 0·41 |
| 21 | 20326336 | rs74795342 | G | A | 0·94 | AL157359.3 | 3·00E-9 | ++ | 1·16 (0·78 – 1·54) | 0·63 |
| 21 | 20327427 | rs75222709 | T | G | 0·94 | AL157359.3 | 3·14E-9 | ++ | 1·16 (0·78 – 1·54) | 0·64 |
|
| ||||||||||
| Asian ancestry only | ||||||||||
| 8 | 21662848 | rs7833426 | A | G | 0·92 | GFRA2 | 2·10E-7 | ++ | 3·66 (2·37 – 4·94) | 0·23 |
Regions with at least one SNP with a P value of less than 1x10−6 for European, Asian, or both populations.
hg19
A1: Effect allele
A2: Reference allele
Frequency of the effect allele
Summary of effect direction for each study (‘+’ means subjects who carry the A1 allele have better lithium response)
BETA: beta coefficient for continuous trait (mean difference in ALDA scale A score for each effect allele); CI: 95% confidence interval
P-value for the meta-analysis heterogeneity test
Figure 2.
Regional association plot of the region on chromosome 21 in which the genome-wide significant SNPs are located. Imputation quality for the top 4 SNPs was excellent (Table S3, Supplementary Materials).
The associated chromosomal region contains no known protein coding genes. Two lncRNAs have been identified in the region, ENSG00000232193 (AL157359.4) and ENSG00000226204 (AL157359.3). Two of the SNPs (rs74795342 and rs75222709) are located in the intronic region of the gene, AL157359.3. The other two SNPs (rs79663003 and rs78015114) lie between these two lncRNA genes.
In the smaller Asian-ancestry samples, only rs7833426 on chromosome 8 had a p-value <10−6. This SNP lies within an intron of GFRA2, which codes for a glial cell line-derived neurotrophic factor (GDNF) receptor. This SNP did not pass QC in the European samples due to a minor allele frequency < 5%.
When the GWAS 1 and GWAS 2 were meta-analyzed under the dichotomous phenotype definition, there were no genome-wide significant results (Figure 1). The SNP with the lowest p-value (p=8·10×10−7) lies near an annotated lncRNA (ENSG00000258081) on chromosome 14 (Table S5, Supplementary Materials).
We calculated the power of this study to detect the observed association findings under an additive genetic model using Quanto (v1.2.4, http://biostats.usc.edu/Quanto.html). The sample had ~65% power to detect the reported association signal at an alpha of 5·0×10−8. For the dichotomous trait, however, the sample size still lacked power to identify genome-wide significant association, even for common SNPs (MAF=0·2) with relatively large effect sizes (OR=2).
It is possible that lithium response is related to the overall genetic risk burden for BD rather than to lithium per se. To assess this, we reevaluated the association between the most significant SNPs in a model that corrected for differences in overall BD risk burden (Risk Profile Score, RPS) in the European-ancestry samples. Similar results were obtained (Table S6, Supplementary Materials). The four SNPs on chromosome 21 continued to show genome-wide significant association with lithium response. There was also no detectable relationship between RPS and Alda Score in this sample (data not shown). These results demonstrate that the findings are specific to lithium response, and do not reflect genetic risk for BD.
We assessed genetic association of lithium response in the subset of patients diagnosed with bipolar I disorder. This narrower phenotype comprised about 79% of all participants. Results showed robust association of the same four SNPs on chromosome 21 with the continuous lithium response trait, suggesting that these SNPs play a role in lithium response in subjects with more narrowly defined BD.
Retrospective assessment of lithium response, while reliable in previous studies and when assessed within ConLiGen23, is limited by recall bias, incomplete information, and other sources of unmeasured variance. To evaluate the potential impact of these sources of error and test the identified SNPs in an independent sample, we genotyped all four SNPs in samples of BD patients treated with lithium monotherapy and assessed prospectively. The sample was recruited entirely from the San Diego Veterans Affairs Medical Center. A total of 89 bipolar disorder patients participated in the prospective study. After screening for eligibility and initial assessment, patients were started on lithium and entered the stabilization phase. The goal in this phase was to stabilize patients within three months on lithium monotherapy. Following this, patients were observed for one month to assure stabilization after discontinuation of other medications. Patients then entered the maintenance phase and were followed at two to four month intervals for two years. Basic characteristics of this sample can be found in Table S7 (for details on genotyping and statistical analysis, see Supplementary Materials). After excluding 16 subjects due to screening failure, diagnosis change, voluntary withdrawal, and non-compliance, 73 bipolar patients (BPI:65, BPII:8) were used for the final data analyses.
After correction for several factors known to influence relapse (see Supplementary Materials.) heterozygote carriers of the alleles associated with poorer lithium response showed a significantly higher rate of relapse compared to carriers of the alternate alleles (p=0·03, hazard ratio = 3·8; Figure S5 & table S8).
DISCUSSION
This is the largest GWAS of lithium response in BD published to date. In a sample of more than 2500 individuals, we detected genome-wide significant evidence of association with SNPs at a locus on chromosome 21. Further support for this finding was detected in a small, independent, prospectively ascertained sample of patients on lithium monotherapy. The associated locus is thought to encode two long, non-coding RNA genes. This finding could have important implications for our understanding of lithium’s mechanism of action in BD. Replication in independent samples is needed. Personal treatment planning based on genetic data depends on identification of additional markers and their total contribution to differences between individuals in response to treatment. Detection of genome-wide significant markers for a phenotype is the first step in demonstrating if such a goal is achievable.
This study has several limitations. ConLiGen relies on retrospective ratings of treatment response, which lack precision and are subject to recall bias. However, response was rated using a well-validated instrument12, previously shown to be reliable by members of the ConLiGen Consortium23, and the results were supported in a prospectively assessed, independent sample. The ConLiGen sample encompassed a variety of patients from a range of ancestries and clinical settings. This is more representative of real-world clinical situations, where patients present at various stages of BD and with a range of illness severity and underlines the robustness of our results. As for any GWAS of a complex trait, sample size is critical. The ConLiGen sample size appears small when compared to GWAS of categorical disease phenotypes, where samples on the order of 10,000 are often required. However, common alleles have been found to exert larger effects upon pharmacogenomic traits30, 31, where samples of 2500 cases are relatively large. On the other hand, the statistically significant excess agreement in the direction of association between GWAS 1 and GWAS 2 that we observed suggests that additional genome-wide significant associations might emerge from larger sample sizes.
Our results do not support previous reports of individual genes strongly associated with lithium response16, 18. Some of those reports were based on smaller samples that may not be comparable to those we studied. They may also represent false positives. Much larger sample sizes would be needed to exclude any particular genes in a GWAS, however.
The main findings seem to implicate lncRNA genes. This implication is causally uncertain, since we have not yet linked allelic variation at the associated SNPs to expression or function of either transcript. There has been an increasing appreciation of the role of lncRNAs in gene regulation, especially in the CNS. An ongoing study of gene expression in peripheral blood during and after acute episodes of BD found a trend toward decreased expression of one of the lncRNAs identified within the association region (AL157359.3; p=0·08, fold change=1·17) after an acute manic episode (Po-Hsiu Kuo, personal communication, 12/2014).
Even if confirmed, the clinical importance of these findings may be limited. The relatively low frequency of the response-associated alleles means that genetic testing would be uninformative in most patients. These and additional genetic markers from future studies could ultimately lead to a clinically informative test, but additional information from established predictors such as family history may be needed, as has been observed for other phenotypes32. In line with similar approaches in the field, polygenic score analyses to predict lithium response may prove to be especially informative, provided that larger, adequately phenotyped samples become available. Clinical utility is a high bar, but the current dearth of good biomarkers of lithium response means that any robust genetic markers could constitute a real step forward.
Any GWAS is subject to experimental error. Type I error can occur, although stringent levels of genome-wide significance keep this to a minimum. Association findings might reflect unobserved variables. The alleles found to be associated with poor lithium response in this study could actually reflect something else, such as treatment adherence. Supportive results in a longitudinal, prospectively-rated sample are encouraging, but due to distinct methods of rating lithium response this cannot be viewed as a replication of the ConLiGen results. On the other hand, relapse over the course of two years on lithium monotherapy is in some ways a better phenotype than that assessed by the Alda Scale, which relies on retrospective ratings. The fact that the same alleles were associated with both retrospective response and prospective relapse may actually increase the importance of the findings and their potential clinical relevance.
GWAS are best viewed as an important starting point for additional investigations. Before embarking on functional studies, future work will need to replicate and extend these findings using comparable ratings of lithium response in large samples. As we may have missed some additional true positive markers due to power constraints, such studies should also target the longer list of SNPs that were associated with lithium response at less significant p-values than were formally reported here. Summary results for SNPs with p values <5x10−5 are posted at www.conligen.org; the corresponding authors can be contacted for more complete summary results. Additional experimental work is needed to establish the functional SNP(s) and their biological impact, if any, in cellular or animal models. Such models could facilitate screening for other drugs that mimic lithium, thus generating novel therapeutic candidates suitable for further study.
Supplementary Material
RESEARCH IN CONTEXT.
Evidence before this study
Lithium is a mainstay in the treatment of bipolar disorder (BD), also known as manic-depressive illness, and may exert neuroprotective effects in neurodegenerative disorders. However, little is known about lithium’s mechanism of action. Individual response in BD varies from excellent to very poor, with about 30% of patients considered good responders7–9. Many genetic association studies of lithium response have been performed, but samples were small, and replicable findings have not emerged14, 15. Three genome-wide association studies (GWAS) of lithium response have been published to date, each implicating different loci.16–18
Added value of this study
The international Consortium on Lithium Genetics (www.ConLiGen.org) has assembled the largest GWAS on lithium response in BD to date, totaling over 2,500 individuals. We now present genome-wide significant evidence of association between lithium response and common genetic variants on chromosome 21. The genetic region associated with response contains two long non-coding RNA genes, which are increasingly appreciated as important regulators of gene expression, particularly in the CNS. These findings suggest a novel potential mechanism of action for lithium. In an independent, prospectively followed clinical sample, the identified genetic markers also helped predict relapse during lithium treatment.
Implications of all the available evidence
This study suggests that a better understanding of drug mechanisms and response can be achieved through international cooperative efforts that leverage clinical expertise with large-scale genomics. The genetic markers identified here show predictive value in a prospective clinical sample, but further studies are needed to establish the potential clinical utility of these findings and their biological context. Confirmed biomarkers of lithium response would constitute an important step forward in the clinical management of BD.
Acknowledgments
We are greatly indebted to all the study participants without whom this research would not have been possible.
We are grateful to the members of our Scientific Advisory Board (www.conligen.org/sab.html) for critical input over the course of the project.
This work was in part funded by the Deutsche Forschungsgemeinschaft (DFG; grant no. RI 908/7-1) and the Intramural Research Program of the National Institute of Mental Health (ZIA-MH00284311; NCT00001174).
The genotyping was in part funded by the German Federal Ministry of Education and Research (BMBF) through the Integrated Network IntegraMent (Integrated Understanding of Causes and Mechanisms in Mental Disorders), under the auspices of the e:Med Programme (grants awarded to Thomas G. Schulze, Marcella Rietschel, and Markus M. Nöthen).
Markus M. Nöthen received support from the Alfried Krupp von Bohlen und Halbach-Stiftung. Franziska Degenhardt received support from the BONFOR Programme of the University of Bonn, Germany.
Some data and biomaterials were collected as part of eleven projects (Study 40) that participated in the National Institute of Mental Health (NIMH) Bipolar Disorder Genetics Initiative. From 2003–2007, the Principal Investigators and Co-Investigators were: Indiana University, Indianapolis, IN, R01 MH59545, John Nurnberger, M.D., Ph.D., Marvin J. Miller, M.D., Elizabeth S. Bowman, M.D., N. Leela Rau, M.D., P. Ryan Moe, M.D., Nalini Samavedy, M.D., Rif El-Mallakh, M.D. (at University of Louisville), Husseini Manji, M.D. (at Johnson and Johnson), Debra A. Glitz, M.D. (at Wayne State University), Eric T. Meyer, Ph.D., M.S. (at Oxford University, UK), Carrie Smiley, R.N., Tatiana Foroud, Ph.D., Leah Flury, M.S., Danielle M. Dick, Ph.D (at Virginia Commonwealth University), Howard Edenberg, Ph.D.; Washington University, St. Louis, MO, R01 MH059534, John Rice, Ph.D, Theodore Reich, M.D., Allison Goate, Ph.D., Laura Bierut, M.D. K02 DA21237; Johns Hopkins University, Baltimore, M.D., R01 MH59533, Melvin McInnis, M.D., J. Raymond DePaulo, Jr., M.D., Dean F. MacKinnon, M.D., Francis M. Mondimore, M.D., James B. Potash, M.D., Peter P. Zandi, Ph.D, Dimitrios Avramopoulos, and Jennifer Payne; University of Pennsylvania, PA, R01 MH59553, Wade Berrettini, M.D., Ph.D.; University of California at San Francisco, CA, R01 MH60068, William Byerley, M.D., and Sophia Vinogradov, M.D.; University of Iowa, IA, R01 MH059548, William Coryell, M.D., and Raymond Crowe, M.D.; University of Chicago, IL, R01 MH59535, Elliot Gershon, M.D., Judith Badner, Ph.D., Francis McMahon, M.D., Chunyu Liu, Ph.D., Alan Sanders, M.D., Maria Caserta, Steven Dinwiddie, M.D., Tu Nguyen, Donna Harakal; University of California at San Diego, CA, R01 MH59567, John Kelsoe, M.D., Rebecca McKinney, B.A.; Rush University, IL, R01 MH059556, William Scheftner, M.D., Howard M. Kravitz, D.O., M.P.H., Diana Marta, B.S., Annette Vaughn-Brown, M.S.N., R.N., and Laurie Bederow, M.A.; NIMH Intramural Research Program, Bethesda, MD, 1Z01MH002810-01, Francis J. McMahon, M.D., Layla Kassem, Psy.D., Sevilla Detera-Wadleigh, Ph.D, Lisa Austin, Ph.D, Dennis L. Murphy, M.D.; Howard University, William B. Lawson, M.D., Ph.D., Evarista Nwulia, M.D., and Maria Hipolito, M.D. This work was supported by the NIH grants P50CA89392 from the National Cancer Institute and 5K02DA021237 from the National Institute of Drug Abuse.
The Canadian part of the study was supported by a grant #64410 from the Canadian Institutes of Health Research to MA. We wish to thank Ms. Joanne Petite and Ms. Giselle Kraus for assistance with data collection.
Collection and phenotyping of the Australian UNSW sample, by Philip B. Mitchell, Peter R. Schofield, Janice M. Fullerton and Adam Wright, was funded by an Australian NHMRC Program Grant (No. 1037196)
The collection of the Barcelona sample was supported by the Centro de Investigación en Red de Salud Mental (CIBERSAM) and IDIBAPS (grant numbers PI080247, PI1200906, PI12/00018, 2014SGR1636, and 2014SGR398). Jean-Michel Aubry and Alexandre Dayer were supported by the Swiss National Science Foundation (grant number 32003B_125469. David Cousins was supported by a Medical Research Council Clinician Scientist Fellowship Award (MR/L006642/1). Louise Frisén was supported by the Swedish Research Council (grant no. 523-2011-3807). Maria Grigoroiu-Serbanescu was supported by UEFISCDI, Romania, grant no. 89/2012. Po-Hsiu Kuo was funded by the Taiwan Ministry of Science and Technology (grant no. MST 99-2314-B-002-140-MY3 and 102-2314-B-002-117-MY3). Carlos A. López-Jaramillo was funded by by the “Estrategia de Sostenibilidad 2014-2015” program of the University of Antioquia. Tomas Novak was supported by the Ministry of Health of the Czech Republic (grant no. IGA NT1389). James B. Potash (Funding): James Wah Fund and Project MATCH. Thomas G. Schulze and Urs Heilbronner received support from the Dr.-Lisa-Oehler-Foundation (Kassel, Germany). Alessio Squassina has a post-doctoral fellowship funded by the Sardinia Regional Government POR Sardegna FSE Operational Program of the Autonomous Region of Sardinia, European Social Fund 2007–2013—Axis IV Human Resources, Objective l.3, Line of Activity l.3.1 Naomi R Wray was funded by Australian NHMRC Fellowships 613602 and 1078901.
This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD. (http://biowulf.nih.gov).
Genotyping for part of the Swedish sample was funded by the Stanley Center for Psychiatric Research at the Broad Institute.
Primary sources of funding: Deutsche Forschungsgemeinschaft (DFG), National Institute of Mental Health (NIMH) Intramural Research Program
Footnotes
Declaration of interests
Adli M has received a grant from Servier, speaker’s fees from Servier, Lundbecck, Aristo, Parexel, Gilead, ViiV, and Deutsche Bank plus a non-financial support from Lundbeck. Akiyama K has received speaker’s fees from Taisho Toyama Pharmaceutical. Alda M is funded by a grant of the Canadian Institutes of Health Research. Bauer M has received speaker’s fees from Alkermes, Astra Zeneca, BristolMyersSquibb, and Ferrer Internacional. Etain B received non-financial support from Labex Biopsy and Fondation Fondamental. Hashimoto R received grants and speaker honoraria from Dainippon Sumitomo Pharma and Novartis plus speaker honoraria from Eli Lilly Japan, GlaxoSmithKline, Hisamitsu Pharmaceutical, Janssen Pharmaceutical, Nippon Zoki Pharmaceutical, Otsuka Pharmaceutical, Astellas Pharma, Pfizer, and the Yoshitomiyakuhin Corporation. Kato T received a grant from Takeda Pharmaceutical and fees from Kyowa Hakko Kirin, Eli Lilly Japan, Otsuka Pharmaceutical, GlaxoSmithKline, Taisho Toyama Pharmaceutical, Dainippon Sumitomo Pharma, Meiji Seika Pharma, Pfizer Japan, Mochida Pharmaceutical, Shionogi & Co, Janssen Pharmaceutical, Yoshitomiyakuhin Corporation, Agilent Technologies, Astellas Pharma, and Wako Pure Chemical Industries. Kusumi I received grants and fees from Dainippon Sumitomo Pharma, Eisai, Eli Lilly, GlaxoSmithKline, Kyowa Hakko Kirin, Meiji Seika Pharma, MSD, Novartis, Otsuka, Ono Pharmaceutical, Pfizer, Tanabe Mitsubishi Pharma, Takeda Pharmaceutical, Shionogi, and Yoshitomi Pharmaceutical; he received grants from AbbVie GK, Asahi Kasei Pharma, Boehringer Ingelheim, Chugai Pharmaceutical, and Daiichi Sankyo and fees from Astellas Pharma and Janssen Pharmaceutical. McCarthy MJ served as unpaid consultant for Pathway Genomic (San Diego, USA). McElroy received a grant and fees from Naurex and Shire, further grants from Alkermes, Cephalon, Forest, Marriott Foundation, Orexigen Therapeutics, and Takeda Pharmaceutical, he further has served on the advisory boards for Bracket, Hoffmann-La Roche, MedAvante, Sunovion and received fees from Novo Nordisk. Perlis RH received personal fees from RID Ventures, Genomind LLC, Healthrageous, Pfizer, Perfect Health, Proteus and Psybrain. Schofield PR received a grant from NHMRC. Schulze TG received a grant and fees from Roche Pharmaceuticals. Stamm TJ received personal fees from Servier, Lundbeck and BristolMyerSquibb. All above listed interests are outside of the submitted work. All other authors declare no conflict of interests.
Contributors
Hou L, Heilbronner U, Degenhardt F, Alda M, Rietschel M, McMahon FJ, and Schulze TG designed the study, contributed to analysis and interpretation of data and wrote the first draft of the manuscript. Akula N, Chen HC, Cichon S, Forstner AJ, Frye MA, Herms S, Hoffman P, Jamain S, Mattheisen M, Nöthen M, Shekhtman T, Wray NR, and Zandi PP provided further data analyses. Hou L did the statistical analyses and prepared the tables and figures. Adli M, Akula N, Alda M, Bauer M, Cichon S, Czersiki P, Degenhardt F, Del Zompo M, Hauser J, Heilbronner U, Landén M, McMahon FJ, Hou L, Perlis RH, Reininghaus E, Rietschel M, Rybakowski JK, Schalling M, Schofield PR, Schulze TG, Shilling PD, Smoller JW, Squassina A were responsible for study design. Responsible for patient recruitment were Adli M, Akiyama K, Alda M, Ardau R, Arias B, Aubry JM, Backlund L, Banzato EM, Bauer M, Baune BT, Bellivier F, Benabarre A, Colom F, Jiménez E, Vieta E, Bengesser SA, Bhattacharjee AK, Birner A, Cervantes P, Chilotti C, Clark SR, Cruceanu C, Czerski P, Dayer A, Del Zompo M, DePaulo JR, Etain B, Falkai P, Frisén L, Frye MA, Fullerton JM. Gard S, Garnham J. Goes FS, Grigoroiu-Serbanescu M, Grof P, Gruber O, Hashimoto R, Hauser J, Jamain S, Jiménez E, Kahn JP, Kassem L, Kato T, Kelsoe J, Reif A, Kittel-Schneider S, Volkert J, Kliwicki S, König B, Kuo PH, Kusumi I, Lackner N, Laje G, Landén M, Leboyer M, Leckband SG, MacQueen G, Maj M, Manchia M, Martinsson L, McElroy SL, Mitchell PB, Mitjans M, Mondimore FM, Monteleone P, Novak T, Ösby U, Ozaki N, Pfennig A, Potash J, Reich-Erkelenz D, Reininghaus E, Rietschel M, Rouleau GA, Rybakowski JK, Schofield PR, Schubert KO, Schulze TG, Schweizer B, Severino G, Shilling PD, Shimoda K, Simhandl C, Slaney C, Stamm TJ, Stopkova P, Tighe SK, Tortorella A, Turecki G, Witt S, Wright A, Young TL, and Zandi PP. Patient in-depth phenotyping was carried out by Adli M, Akiyama K, Alda M, Ardau R, Arias B, Backlund L, Bauer M, Baune BT, Bellivier F, Benabarre A, Colom F, Jiménez E, Vieta E, Bengesser SA, Birner A, Brichant-Petitjean C, Bui E, Chilotti C, Cruceanu C, Dantas CR, Del Zompo M, DePaulo JR, Frye MA, Goes FS, Grigoroiu-Serbanescu M, Grof P, Hashimoto R, Jiménez E, Kassem L, Kliwicki S, Kuo PH, Lackner N, Landén M, and Lavebratt C. All authors contributed to drafting the work or revising it critically for important intellectual content and made substantial contributions to the concept and design of the study and acquisition, analysis and interpretation of data.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Merikangas KR, Akiskal HS, Angst J, et al. Lifetime and 12-month prevalence of bipolar spectrum disorder in the National Comorbidity Survey replication. Archives of general psychiatry. 2007;64(5):543–52. doi: 10.1001/archpsyc.64.5.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dilsaver SC. An estimate of the minimum economic burden of bipolar I and II disorders in the United States: 2009. Journal of affective disorders. 2011;129(1–3):79–83. doi: 10.1016/j.jad.2010.08.030. [DOI] [PubMed] [Google Scholar]
- 3.Goodwin FK, Jaison KR. Manic-Depressive Illness: Bipolar Disorders and Recurrent Depression. 2. 2007. [Google Scholar]
- 4.Geddes JR, Miklowitz DJ. Treatment of bipolar disorder. Lancet. 2013;381(9878):1672–82. doi: 10.1016/S0140-6736(13)60857-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauer MS, Mitchner L. What is a "mood stabilizer"? An evidence-based response The American journal of psychiatry. 2004;161(1):3–18. doi: 10.1176/appi.ajp.161.1.3. [DOI] [PubMed] [Google Scholar]
- 6.Tondo L, Hennen J, Baldessarini RJ. Lower suicide risk with long-term lithium treatment in major affective illness: a meta-analysis. Acta psychiatrica Scandinavica. 2001;104(3):163–72. doi: 10.1034/j.1600-0447.2001.00464.x. [DOI] [PubMed] [Google Scholar]
- 7.Baldessarini RJ, Tondo L. Does lithium treatment still work? Evidence of stable responses over three decades Archives of general psychiatry. 2000;57(2):187–90. doi: 10.1001/archpsyc.57.2.187. [DOI] [PubMed] [Google Scholar]
- 8.Rybakowski JK, Chlopocka-Wozniak M, Suwalska A. The prophylactic effect of long-term lithium administration in bipolar patients entering treatment in the 1970s and 1980s. Bipolar disorders. 2001;3(2):63–7. doi: 10.1034/j.1399-5618.2001.030203.x. [DOI] [PubMed] [Google Scholar]
- 9.Garnham J, Munro A, Slaney C, et al. Prophylactic treatment response in bipolar disorder: results of a naturalistic observation study. Journal of affective disorders. 2007;104(1–3):185–90. doi: 10.1016/j.jad.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 10.Kleindienst N, Engel R, Greil W. Which clinical factors predict response to prophylactic lithium? A systematic review for bipolar disorders. Bipolar disorders. 2005;7(5):404–17. doi: 10.1111/j.1399-5618.2005.00244.x. [DOI] [PubMed] [Google Scholar]
- 11.Duffy A, Alda M, Milin R, Grof P. A consecutive series of treated affected offspring of parents with bipolar disorder: is response associated with the clinical profile? Canadian journal of psychiatry Revue canadienne de psychiatrie. 2007;52(6):369–76. doi: 10.1177/070674370705200606. [DOI] [PubMed] [Google Scholar]
- 12.Grof P, Duffy A, Cavazzoni P, et al. Is response to prophylactic lithium a familial trait? The Journal of clinical psychiatry. 2002;63(10):942–7. doi: 10.4088/jcp.v63n1013. [DOI] [PubMed] [Google Scholar]
- 13.Mendlewicz J, Verbanck P, Linkowski P, Wilmotte J. Lithium accumulation in erythrocytes of manic-depressive patients: an in vivo twin study. The British journal of psychiatry : the journal of mental science. 1978;133:436–44. doi: 10.1192/bjp.133.5.436. [DOI] [PubMed] [Google Scholar]
- 14.McCarthy MJ, Leckband SG, Kelsoe JR. Pharmacogenetics of lithium response in bipolar disorder. Pharmacogenomics. 2010;11(10):1439–65. doi: 10.2217/pgs.10.127. [DOI] [PubMed] [Google Scholar]
- 15.Can A, Schulze TG, Gould TD. Molecular actions and clinical pharmacogenetics of lithium therapy. Pharmacology, biochemistry, and behavior. 2014 doi: 10.1016/j.pbb.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perlis RH, Smoller JW, Ferreira MA, et al. A genomewide association study of response to lithium for prevention of recurrence in bipolar disorder. The American journal of psychiatry. 2009;166(6):718–25. doi: 10.1176/appi.ajp.2009.08111633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Squassina A, Manchia M, Borg J, et al. Evidence for association of an ACCN1 gene variant with response to lithium treatment in Sardinian patients with bipolar disorder. Pharmacogenomics. 2011;12(11):1559–69. doi: 10.2217/pgs.11.102. [DOI] [PubMed] [Google Scholar]
- 18.Chen CH, Lee CS, Lee MT, et al. Variant GADL1 and response to lithium therapy in bipolar I disorder. The New England journal of medicine. 2014;370(2):119–28. doi: 10.1056/NEJMoa1212444. [DOI] [PubMed] [Google Scholar]
- 19.Consortium on Lithium Genetics. Hou L, Heilbronner U, et al. Variant GADL1 and response to lithium in bipolar I disorder. The New England journal of medicine. 2014;370(19):1857–9. doi: 10.1056/NEJMc1401817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikeda M, Kondo K, Iwata N. Variant GADL1 and response to lithium in bipolar I disorder. The New England journal of medicine. 2014;370(19):1856–7. doi: 10.1056/NEJMc1401817. [DOI] [PubMed] [Google Scholar]
- 21.Cruceanu C, Alda M, Dion PA, Turecki G, Rouleau GA. No evidence for GADL1 variation as a bipolar disorder susceptibility factor in a Caucasian lithium-responsive cohort. The American journal of psychiatry. 2015;172(1):94–5. doi: 10.1176/appi.ajp.2014.14070855. [DOI] [PubMed] [Google Scholar]
- 22.Schulze TG, Alda M, Adli M, et al. The International Consortium on Lithium Genetics (ConLiGen): an initiative by the NIMH and IGSLI to study the genetic basis of response to lithium treatment. Neuropsychobiology. 2010;62(1):72–8. doi: 10.1159/000314708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manchia M, Adli M, Akula N, et al. Assessment of Response to Lithium Maintenance Treatment in Bipolar Disorder: A Consortium on Lithium Genetics (ConLiGen) Report. PloS one. 2013;8(6):e65636. doi: 10.1371/journal.pone.0065636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nature genetics. 2012;44(8):955–9. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nature methods. 2013;10(1):5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 26.Fuchsberger C, Abecasis GR, Hinds DA. minimac2: faster genotype imputation. Bioinformatics. 2015;31(5):782–4. doi: 10.1093/bioinformatics/btu704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Psychiatric GWAS Consortium Bipolar Disorder Working Group. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nature genetics. 2011;43(10):977–83. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aberg K, Adkins DE, Bukszar J, et al. Genomewide association study of movement-related adverse antipsychotic effects. Biological psychiatry. 2010;67(3):279–82. doi: 10.1016/j.biopsych.2009.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malhotra AK, Correll CU, Chowdhury NI, et al. Association between common variants near the melanocortin 4 receptor gene and severe antipsychotic drug-induced weight gain. Archives of general psychiatry. 2012;69(9):904–12. doi: 10.1001/archgenpsychiatry.2012.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Do CB, Hinds DA, Francke U, Eriksson N. Comparison of family history and SNPs for predicting risk of complex disease. PLoS genetics. 2012;8(10):e1002973. doi: 10.1371/journal.pgen.1002973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.