Abstract
Aberrant Notch signaling has been linked to many cancers including choroid plexus (CP) tumours, a group of rare and predominantly pediatric brain neoplasms. We developed animal models of CP tumours by inducing sustained expression of Notch1 that recapitulate properties of human CP tumours with aberrant NOTCH signaling. Whole transcriptome and functional analyses showed that tumour cell proliferation is associated with Sonic Hedgehog (Shh) in the tumour microenvironment. Unlike CP epithelial cells, which have multiple primary cilia, tumour cells possess a solitary primary cilium as a result of Notch-mediated suppression of multiciliate diffferentiation. A Shh-driven signaling cascade in the primary cilium occurs in tumour cells but not in epithelial cells. Lineage studies show that CP tumours arise from mono-ciliated progenitors in the roof plate characterized by elevated Notch signaling. Abnormal SHH signaling and distinct ciliogenesis are detected in human CP tumours, suggesting SHH pathway and cilia differentiation as potential therapeutic avenues.
Choroid plexus (CP) neoplasms represent rare primary brain tumours found predominantly in children. While CP papillomas (CPP) are benign, CP carcinomas (CPC) are malignant1, 2. These tumours are believed to originate from CP epithelium, which differentiate from the roof plate to form the CP, a specialized tissue that produces cerebrospinal fluid (CSF) in each ventricle of the brain3. Surgical resection remains the primary treatment for CPPs and is associated with excellent prognosis. However, clinical outcomes for patients with incompletely resected tumors, recurrent tumors, metastatic spread, or CPCs, can be devastating4, 5. Notch signaling, tumour protein p53 (TP53) mutations, genetic and epigenetic changes have been described6–15.
Sonic hedgehog (Shh) signaling, a crucial pathway in development and cancers, is mediated by Patched (Ptch1) and Smoothened (Smo) receptors in the primary cilium where they orchestrate a signaling cascade that activates the expression of downstream targets, including Gli1, Mycn, and cyclin D1 (Ccnd1)16, 17. By inducing sustained Notch 1 expression, we developed mouse models of CP tumours that closely resemble human CP tumours with abnormal NOTCH signaling. We show that Notch-induced CP tumour relies on Shh from the tumour microenvironment through their primary cilium. Aberrant SHH signaling and unique cilia patterns found in human CP tumours may serve as potential therapeutic targets.
RESULTS
Notch pathway activation leads to CP tumours
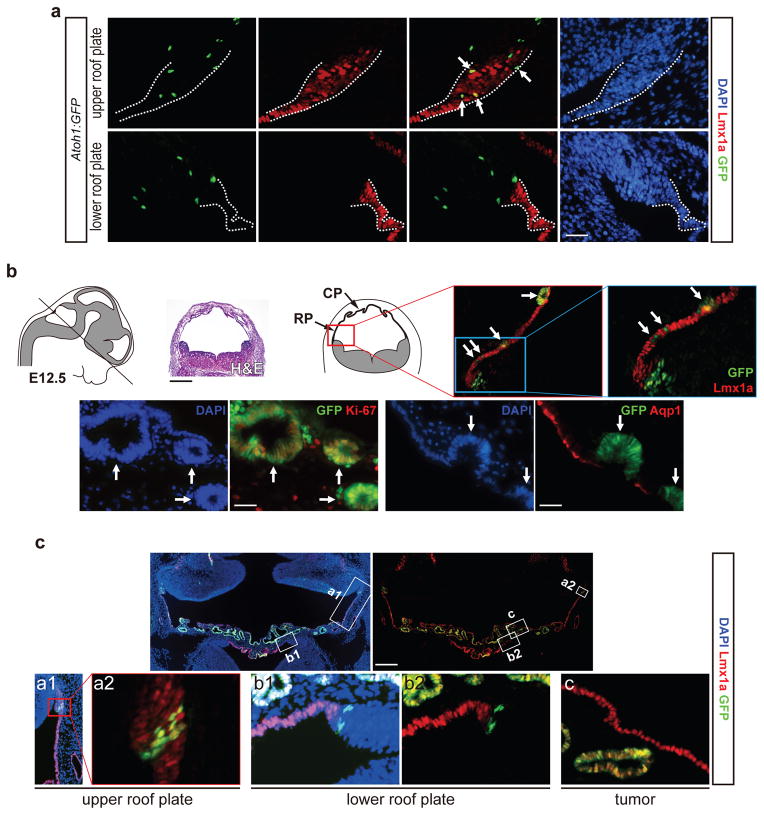
A molecularly-defined boundary exists between the rhombic lip consisting of neural progenitors expressing the transcription factor Atonal Homolog 1 (Atoh1, also known as Math1), and the roof plate, characterized by the expression of Wnt1, Gdf7, and the transcription factor Lmx1a18–21. Some Lmx1a+ cells are present in the rhombic lip and contribute to the cerebellum20, 21. To determine whether rhombic lip progenitors contribute to the roof plate/CP lineage, we utilized Math1-Cre to drive Cre expression in Atoh1+ progenitors22 (Fig. 1a). When crossed with Rosa26-EYFP Cre reporter strain23, the resulting Math1-Cre;Rosa26-EYFP mice have cells expressing enhanced yellow fluorescent protein (EYFP) in the CP in addition to cerebellum (Fig. 1b). Though these EYFP+ cells comprise < 0.5% of hindbrain CP epithelium, they express CP markers Lmx1a, orthodenticle homeobox 2 (Otx2), cytokeratins, and Aquaporin 1 (Aqp1) (Fig. 1c, 1d, Supplementary Fig. 1a), indicating some Atoh1+ progenitors contribute to hindbrain roof plate/CP lineage.
Figure 1.
Constitutive Notch 1 signaling leads to CP tumour. (a) Schematic illustration of the strategy for Notch 1 signaling activation in vivo. (b) Brain hemispheres and CPs from postnatal day 7 (P7) mice are shown. Notice that the enlarged CP from Math1-Cre;Rosa26-NICD1;Rosa26-EYFP (Mcre;NICD1;EYFP) animal contains many EYFP+ cells (arrows), while EYFP+ cells (arrowheads) are sparsely found in the CP from Math1-Cre;Rosa26-EYFP (Mcre;EYFP) animals. Scale bar: 1 mm. (c) Analysis of gene expression in CP of Math1-Cre;Rosa26-EYFP mice. EYFP expression (green) labels cells derived from Atoh1+ progenitors. The expression of Lmx1a (red), Otx2 (red), cytokeratins (red), and Aqp1 (red) labels CP epithelial cells. Scale bar: 25μm. (d) Quantitation of the percentage of EYFP+ cells or NICD1+/GFP+ cells in Otx2+ hindbrain CP epithelium of Mcre;EYFP mice or Math1-Cre;Rosa26-NICD1 (Mcre;NICD1) mice, respectively, at different time points (n=3 tumours from 3 Mcre;NICD1 animals/time point; CPs from Mcre;EYFP mice: n=4 (P0, P14), n=6 (P7, P21), n=5 (P90), data from a single experiment are shown, raw data are available in Supplementary Table 9; mean ± SEM, two-way ANOVA, **, P<0.01; ***, P<0.001; ns, non-significant). (e) H&E staining of CPs from Mcre;NICD1 and wild type (WT) mice at P0 and P14. Notice that CP epithelium exhibits the “hobnail” configuration (arrowhead), while the enlarged CP in Mcre;NICD1 mice appear “flattened” on ventricular surfaces (arrow). Vesicular sac with accumulated CSF is shown (asterisk). Scale bars: white, 500μm; black, 25μm. (f) H&E staining of human CP papilloma (CPP) and normal CP. Scale bar: 25μm. (g) Quantitative RT-PCR (Q-RT-PCR) analysis of Hes1 and Hes5 expression in CP tumours (black circles) and wild type CPs (white circles) at P0, P7, P14, and P21 (data from technical replicates of each specimen set in a single experiment are shown; experiment was not repeated. Raw data can be found in Supplementary Table 9). (h) The expression of Ki-67 is shown in CP tumours from Mcre;NICD1 mice, Lmx1a-Cre;Rosa26-NICD1 (Lcre;NICD1) mice, and normal CPs from wild type mice. Dotted lines mark the boundary of lateral ventricles. The expression of Ki-67 in human CPP and normal CP is shown. Scale bar: 50μm.
To determine the effects of Notch signaling on CP, Math1-Cre;Rosa26-EYFP mice were crossed with a Rosa26-NICD1 strain that conditionally expresses the intracellular domain of Notch 1 (NICD1) and green fluorescent protein24 (GFP, Fig. 1a). In Math1-Cre;Rosa26-NICD1;Rosa26-EYFP animals, hindbrain CP is significantly enlarged with many EYFP+ cells (Fig. 1b). Sustained NICD1 expression in Atoh1+ lineage leads to > 50-fold increase (~50%) in its contribution to hindbrain CP epithelium at birth that peaks at postnatal day 14 (P14) (~80%) (Fig. 1d, Supplementary Fig. 1a). While wild type CP epithelium exhibits an orderly cobblestone-like appearance with a rough, “hobnail” configuration on apical surfaces, CPs from Math1-Cre;Rosa26-NICD1 (Mcre;NICD1) mice are characterized by papillary projections lined by flattened epithelium with cell crowding, elongation, and stratification, reminiscent of human CP papilloma (Fig. 1e, 1f, Supplementary Fig. 1b). When the Rosa26-NICD1 strain was crossed with Lmx1a-Cre transgenic mice that express Cre in the roof plate/CP lineage20, abnormal CP growth with identical characteristics developed in the lateral ventricles and hindbrain of Lmx1a-Cre;Rosa26-NICD1 (Lcre;NICD1) animals (Supplementary Fig. 2). Though these abnormal CP growths don’t invade surrounding regions, CSF accumulation and ventricular dilation are present in these animals (Fig. 1e, Supplementary Fig. 1b, Supplementary Fig. 2), and 40–50% of Lcre;NICD1 animals die from hydrocephalus. CP tumours of Mcre;NICD1 mice exhibit increased expression of Hes1 and Hes5, indicating Notch pathway activation (Fig. 1g, Supplementary Fig. 1c, 1d). While no Ki-67 expression is detected in wild type CP epithelium at P7, abundant Ki-67+ cells are present in CPs from age-matched Mcre;NICD1 or Lcre;NICD1 mice, resembling cell proliferation in human CP tumours (Fig. 1h), indicating that Notch pathway activation causes aberrant growth of CP into tumours.
Enhanced proliferation in Notch-induced CP tumour
While CP epithelial cells in wild type and Math1-Cre;Rosa26-EYFP mice (EYFP+ or EYFP−) remain post-mitotic after birth, ~40% of NICD1+/GFP+ cells in Mcre;NICD1 mice are Ki-67+. This percentage gradually decreases to ~1% after 3 weeks of age (Fig. 2a, 2b, Supplementary Fig. 3a). 5-ethynyl-2′-deoxyuridine (EdU) incorporation assays also revealed enhanced tumour cell proliferation in Mcre;NICD1 mice (Supplementary Fig. 4a). As tumour cells exit the cell cycle, Ccnd1 expression is downregulated, while Cdkn1b (p27 Kip1) expression is upregulated (Fig. 2c, Supplementary Fig. 4b, 4c). Cleaved caspase-3 expression is not detected (Supplementary Fig. 4d). Together, these results indicate that Notch-induced CP tumor undergoes enhanced proliferation transiently after birth.
Figure 2.
Notch-induced CP tumours undergo enhanced proliferation and exhibit defects in differentiation. (a) The expression of Ki-67 is shown in CP tumours from Mcre;NICD1 mice at P0, P7, and P14; CPs from P0 wild type (WT) and Mcre;EYFP mice, respectively, serve as control. Ki-67 expression (red) labels proliferating cells, GFP expression (green) marks NICD1+ tumour cells, expression of Lmx1a (green) labels CP epithelium in wild type mice, and EYFP expression (green, arrowheads) detected with a GFP antibody demarcates CP epithelial cells derived from Atoh1+ progenitors in Mcre;EYFP mice. DAPI staining (blue) marks nuclei. Scale bar: 25μm. (b) Quantitation of the percentage of Ki-67+ cells in NICD1+/GFP+ tumour cells from Mcre;NICD1 mice and Lmx1a+ wild type CP epithelial cells at different time points (n=3 CPs from 3 animals/genotype/time point, data from a single experiment are shown, raw data are available in Supplementary Table 9; mean ± SEM, two-way ANOVA, ****, P<0.0001). (c) The expression of Ki-67 and Cdkn1b (p27 Kip1) is shown in tumours from Mcre;NICD1 mice and wild type (WT) CPs at P7. Notice that staining for Ki-67 and p27 is mutually exclusive. And p27 or Ki-67 expression is absent in wild type CP epithelium despite positive staining in the cerebellum (Cb). Scale bar: 25μm. (d) The expression of Lmx1a (red), Otx2 (red), Ttr (red), Aqp1 (red), and cytokeratins (red) is shown in CP tumours. GFP expression (green) labels NICD1+ tumour cells. Dotted lines mark the boundary between GFP+ tumour cells and epithelium. DAPI staining (blue) marks nuclei. Scale bar: 25μm. (e) Q-RT-PCR analysis of Ttr and Aqp1 expression in tumours from Mcre;NICD1 mice (black circles) and wild type (WT) CPs (white circles) at different time points (data from technical replicates in a single experiment are shown and available in Supplementary Table 9. Experiment was not repeated for Ttr expression; Aqp1 expression analysis was repeated in one independent experiment). (f) Western blot analysis of Aqp1 and Otx2 expression in CP tumours and wild type (WT) CPs at P14. β-actin serves as loading control. Molecular size marker and representative image of unprocessed blots can be found in Supplementary Figure 9.
Tumour cells express the CP marker Lmx1a, but not mesenchymal marker MafB25, and the expression of Aqp1, Transthyretin (Ttr), cytokeratins, and Otx2 is consistently reduced compared to CP epithelial cells26 (Fig. 2d–f, Supplementary Fig. 3b, Supplementary Fig. 4e, 4f), indicating that sustained Notch signaling interferes with differentiation of tumour cells even after they become post-mitotic.
Abnormal Shh signaling in CP tumour cells
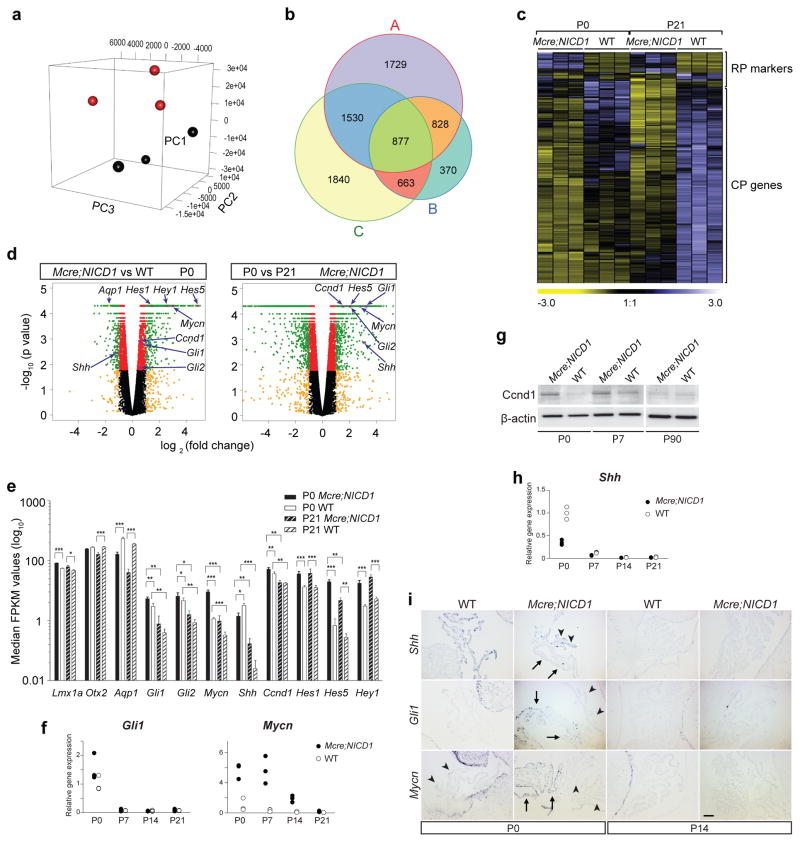
To identify signals that drive tumour cell proliferation, we compared transcriptional profiles of tumours and wild type CPs at P0 (tumour cells are proliferative) and P21 (tumour cells are post-mitotic) using RNA-seq. Tumours and CPs clustered separately in principal component analysis, indicating distinct molecular profiles (Fig. 3a). Tumour cells exhibit gene expression profiles defined by differential expression of 2,738 (P0) and 4,964 (P21) transcripts (Supplementary Table 1, Fig. 3b). Study of these differentially expressed transcripts identified 1,705 common targets, including Hes1 and Hes5, Aqp1, cytokeratins, and Otx2, all of which show significant differential expression by Q-RT-PCR and immunostaining analyses, validating RNA-seq results (Fig. 1g, Fig. 2d–f, Fig. 3d, 3e, Supplementary Fig. 1c, 1d, Supplementary Fig. 3b, Supplementary Fig. 4e, Supplementary Table 1). Though tumour cells express higher mRNA levels for roof plate markers Lmx1a, Gdf7, Zic3, Zic4, and Msx2, the expression of many genes found in CP epithelium is significantly lower27–32 (Fig. 3c, 3e, Supplementary Table 1).
Figure 3.
Notch-induced CP tumours display aberrant Shh signaling. (a) Principal component analysis of CP tumours (black dots) and wild type CPs (WT, red dots) at P21 (n=3 specimens/genotype). (b) Venn diagram of differential genes (FDR < 0.05) between tumours and CPs at P21 (A, n=3 specimens/genotype), at P0 (B, n=3 specimens/genotype), and of tumour cells between P0 and P21 (C, n=3 specimens/time point). (c) Hierarchical clustering of tumours and CPs (n=3 specimens/genotype/time point) based on genes expressed in roof plate (RP) and CP (one-way ANOVA, FDR < 0.05, fold change is shown). (d) Volcano-plot analysis of gene expression of tumours and CPs at P0 (n=3 specimens/genotype), and of tumours at P0 and P21 (n=3 specimens/time point). Differential transcripts with statistical significance (FDR < 0.05, -log10 of p-value, y-axis) are shown in red (< 2 fold change) or green (> 2 fold change) dots. Non-significant genes (FDR > 0.05) are shown in black or yellow dots (2-fold cutoff). Arrows label select genes with significant differential expression. (e) Median FKPM values of differential genes between tumours and CPs (n=3 specimens/genotype/time point, mean ± SEM, two-way ANOVA, *, P< 0.05; **, P<0.01; ***, P<0.001). Q-RT-PCR analysis of the expression of Gli1 and Mycn (f), and Shh (h) in tumours (black circles) and CPs (white circles) at different time points (data from technical replicates of each specimen set in a single experiment are shown; experiment was repeated independently once with similar results. Raw data can be found in Supplementary Table 9). (g) Western blot analysis of Ccnd1 expression in tumours and CPs. β-actin serves as loading control (n=1 specimen/genotype/time point, representative blot image from one of three independent repeated experiments is shown). Molecular size marker and representative image of unprocessed blots can be found in Supplementary Figure 9. (i) In-situ hybridization analysis of Shh, Gli1, and Mycn expression in tumours and wild type CPs. Shh expression is detected in epithelium (arrowheads), but absent in tumour cells (arrows) at P0, a pattern complementary to those of Gli1 and Mycn. Scale bar: 25μm.
Comparison of tumour expression profiles at P0 and P21 uncovered 4,910 differentially expressed transcripts (Fig. 3b, Supplementary Table 2). We reasoned that differential genes unique to P0 tumour cells may include those involved in proliferation. To identify these genes, we excluded the 1,705 common differential targets between tumours and CPs at each time point to obtain 1,033 differential transcripts in tumours at P0 (Fig. 3b). We overlapped these 1,033 unique targets with the 4,910 differential genes between tumours of P0 and P21, further narrowing it down to 663 genes (Fig. 3b, Supplementary Table 3). Interrogating this shortened list of genes using ingenuity pathway analysis led us to a promising candidate: Shh signaling, which is also identified in analysis of larger dataset (Supplementary Tables 1, 3). Tumour cells exhibit increased expression of Gli1, Gli2, Mycn and Ccnd1 compared to wild type CP epithelium at P0 and P7. After P7, when tumour cells start to exit the cell cycle, the expression of these genes decreases to levels of those in control CPs, suggesting that decreased proliferation correlates with attenuated Shh signaling (Fig. 3d–g, 3i, Supplementary Fig. 4b). In addition, while the expression of p27 and Cdkn2b is upregulated in non-proliferating tumour cells, Cdkn1c (p57 Kip2) is expressed at higher levels in mature CP (Fig. 2c, Supplementary Fig. 4c, Supplementary Table 1). Among hedgehog ligands, only Shh is abundantly expressed in hindbrain CP epithelial cells at birth and declines to undetectable levels after P14, resulting in lower expression levels in tumours than in wild type CPs at P0 (Fig. 3d, 3e, 3h, 3i).
Shh drives CP tumour cell proliferation
To determine the role of Shh signaling in Notch-induced CP tumour, we treated tumour cells with recombinant N-terminal fragment of Shh (ShhN). Tumour cells formed spheres under serum-free conditions and continued to express Lmx1a, indicating intact lineage characteristics under these conditions (Fig. 4a, 4c). After 96 hours, more Ki-67+ cells were detected in tumour spheres treated with ShhN than in untreated tumour spheres (Fig. 4b). While epithelial cells formed aggregates that remained unresponsive, the size of tumour spheres and number of tumour cells were increased by ShhN, including post-mitotic tumours at P21 or later, whereas Smo inhibitor cyclopamine abolished such effects, indicating ShhN stimulates tumour cell proliferation (Fig. 4c, 4d). To determine whether tumour growth requires Shh, we treated Lcre;NICD1 and Mcre;NICD1 animals with Smo inhibitor vismodegib (100 mg/kg) or vehicle daily from embryonic day 17.5 (E17.5) through P7, or from day E15.5 for 4 days, respectively33,34. Vismodegib treatment shrank hindbrain CP tumours, significantly reduced the number of tumour cells, and improved the survival of Lcre;NICD1 mice (Fig. 4e, 4f). Vismodegib also decreased tumour cell proliferation and suppressed Mycn expression in Mcre;NICD1 animals without affecting CP development in wild type littermates (Fig. 4g–i, Supplementary Fig. 5). Together, these results indicate that Shh drives the growth of Notch-induced CP tumours.
Figure 4.
Shh drives the proliferation of Notch-induced CP tumours. Lmx1a (a) (red) and Ki-67 (b) (red) expression is shown in cultured tumour cells from Mcre;NICD1 mice. GFP (green) marks tumour cells, DAPI staining (blue) labels nuclei. Scale bar: 10μm. (c) Images of tumour cells and wild type (WT) epithelial cells. Scale bar: 50μm. (d) Quantitation of tumour cells after 96-hour treatments (n=3 specimens/treatment/time point, raw data are available in Supplementary Table 9; mean ± SEM, two-way ANOVA, ****, P<0.0001; ns, non-significant). (e) Kaplan-Meier curve depicting the survival of Lcre;NICD1 mice treated with vismodegib (n=22 animals) or vehicle (n=19 animals). (f) Hindbrain CPs from day P7 Lcre;NICD1 mice treated with vismodegib (lower) or vehicle (upper). Scale bar: 1 mm. Quantitation of tumour cells in treated animals is shown (n=7 animals/treatment, raw data are available in Supplementary Table 9; mean ± SEM, two-tailed unpaired t test, **, P<0.01). (g) H&E staining of tumours from treated Mcre;NICD1 mice. Ki-67 expression (red) labels proliferating cells, while GFP (green) marks tumour cells. DAPI staining (blue) labels nuclei. Scale bar: 25μm. (h) Analysis of tumour cell proliferation in animals shown in (g) (n=10 animals/treatment; mean ± SEM, two-tailed unpaired t test, **, P<0.01). (i) In-situ hybridization analysis of Mycn expression in tumour (arrows) or epithelial cells (arrowheads) in animals shown in (g). Scale bar: 25μm. (j) Principal component analysis of human CPPs (red dots, n=7 tumours from 7 individuals) and normal CPs (blue dots, n=8 CPs from 8 individuals). (k) MetaCore analysis of differential genes in human CPPs. (l) Venn diagram shows the overlap of differential genes (FDR < 0.05) between human CPPs and normal CPs (A), with those between murine tumours and wild type CPs at P21 (B, n=3 specimens/genotype) and P0 (C, n=3 specimens/genotype). (m) MetaCore analysis of overlapping genes between mouse and human CPPs shown in (l). (n) Q-RT-PCR analysis of gene expression in human CP tumours (CPP: n=7 tumours from 7 individuals; CPC: n=2 tumours from 2 individuals). Values represent fold changes relative to human CP epithelium. Data are derived from single experiment shown in Supplementary Table 9 and not repeated.
SHH and NOTCH signaling in human CP tumours
We examined NOTCH and SHH signaling in human CP tumours. First, we utilized published datasets and analyzed tumour transcriptomes, epigenomes, and genomes. Principal component analysis of human CPPs and normal CPs revealed distinct molecular profiles for CPPs13 (Fig. 4j). MetaCore enrichment analysis of differential genes in human CPPs placed NOTCH and SHH signaling among significantly enriched pathways (Fig. 4k, Supplementary Table 4). Second, we repeated gene expression profiling of CP tumours in Mcre:NICD1 animals and wild type CPs at P0 and P21 using a similar microarray approach. The differentially expressed genes in murine CPPs overlapped with those identified in human CPPs (Fig. 4l). MetaCore analysis of these common differential transcripts between murine and human CPPs revealed significant enrichment for genes in both pathways, indicating murine CPPs display striking resemblance to human CPPs (Fig. 4m, Supplementary Table 5). Third, we examined datasets from recently published studies of human CP tumours and found higher expression levels of roof plate markers in CPPs compared to CPCs14, 15 (Supplementary Fig. 6a). MetaCore gene expression analysis showed significant enrichment for SHH and NOTCH pathways in human CPCs, an observation also supported by analysis of methylation and copy number variation data (Supplementary Fig. 6b–d, Supplementary Table 6). Q-RT-PCR analysis of human CP tumours revealed increased expression for genes of both pathways in CP tumours compared to normal human CP epithelial cells (Fig. 4n). In situ hybridization demonstrated SHH mRNA in both CPPs and CPCs (Percentage of SHH-positive cells in CPP: 24.82 ± 3.32, n=11 specimens examined from 10 individuals; CPC: 36.33 ± 5.40, n=10; Disease-free control: 100%; n=5; Of note, signal strength varied among samples, likely due to brain regions sampled, specimen age and storage) (Supplementary Fig. 6e). Together, these results indicate that human CP tumours display aberrant NOTCH and SHH signaling, suggesting that both pathways may play a similarly important role in human CP tumourigenesis.
Notch-induced CP tumour cells are mono-ciliated
In vertebrates, Shh-driven signaling occurs via the primary cilium16, 17. To understand the mechanisms by which tumour cells with sustained Notch signaling respond to Shh, we examined primary cilia in CP tumour and epithelial cells. Transmission electron microscopy and staining for cilia markers ADP-ribosylation factor-like 13b (Arl13b)35, γ-tubulin, and acetylated α-tubulin revealed multiple short primary cilia in wild type and EYFP+ epithelial cells in Math1-Cre;Rosa26-EYFP mice, whereas a single, longer primary cilium is present in tumour cells (epithelial cells: 1.70 ± 0.11 μm, n=10; tumour cells: 3.78 ± 0.13 μm, n=12; two-tailed unpaired t test, p<0.0001; data from a single experiment are shown, raw data are available in Supplementary Table 9) (Fig. 5a–c, Supplementary Fig. 7a, 7b). Gene expression profiling showed that many genes involved in ciliogenesis are downregulated in tumour cells36–38 (Fig. 5d). The expression of forkhead box J1 (Foxj1) and multiciliate differentiation and DNA synthesis associated cell cycle protein (Mcidas), two crucial regulators of the differentiation of cells with numerous motile cilia39–41, is consistently reduced in tumour cells (Fig. 5e, Supplementary Fig. 7c), indicating that Notch signaling suppresses Mcidas and Foxj1 expression, and blocks multiciliate differentiation of tumour cells. Similar to tumour cells, progenitors in hindbrain roof plate display increased expression of Hes1 and Hes5 at day E14.5, and possess single primary cilium (Fig. 5f, 5g, Supplementary Fig. 7d, 7e), suggesting that active Notch signaling preserves the single primary cilium of the progenitors during development. We characterized cilia pattern in human CP tumours (17 CPPs and 13 CPCs). Compared to normal human CP epithelial cells with multiple primary cilia, CPPs comprise either mono-ciliated tumour cells only or mixed populations of mono-ciliated and multi-ciliated cells, whereas all CPCs consist exclusively of cells that exhibit solitary primary cilium (Fig. 5h, 5i, Supplementary Fig. 7f).
Figure 5.
Notch-induced CP tumours possess solitary primary cilium. (a) Transmission electron micrographs of primary cilia in tumour cells (arrow) from Mcre;NICD1 animals and wild type (WT) CP epithelium (arrowheads). Scale bar: 0.5μm. (b) The expression of cilia markers Arl13b (red) and γ-tubulin (red) in tumour cells is shown. Dotted lines mark the boundary between GFP+ (green) tumour cells and Aqp1+ (yellow) epithelium. DAPI staining (blue) labels nuclei. Primary cilia in epithelial (arrowheads, upper) or tumour cells (arrows, lower) are magnified in inset pictures. Scale bar: 10μm. (c) The expression of Arl13b (red), γ-tubulin (red), and acetylated α-tubulin (act-α-tubulin, red) is shown in cultured CP cells. GFP (green) labels tumour cells, while Aqp1 (yellow) labels epithelial cells. DAPI staining (blue) labels nuclei. Scale bar: 5μm. (d) Hierarchical clustering of tumours and normal CPs at P0 based on 147 genes involved in cilia differentiation (n=3 specimens/genotype; one-way ANOVA, FDR < 0.05, fold change is shown). (e) Q-RT-PCR analysis of the expression of Mcidas and Foxj1 in tumours (black circles) and CPs (white circles) at different time points (data from technical replicates of each specimen in a single experiment are shown and available in Supplementary Table 9. Experiment was not repeated for Mcidas expression; Foxj1 expression analysis was repeated in one independent experiment). (f) In-situ hybridization analysis of the expression of Hes1 and Hes5 is shown in hindbrain roof plate (red dotted lines) and tumour cells (arrows) in Mcre;NICD1 or wild type animals at day E14.5. Scale bar: 100μm. (g) Primary cilia are shown for cells in hindbrain roof plate, CP epithelium, and tumours shown in (f). Arl13b (red) and γ-tubulin (red) expression marks primary cilia and basal body, respectively. DAPI staining (blue) labels nuclei. RP: roof plate. Scale bar: 10μm. (h) Representative images show ARL13B expression (red) in human CP tumour cells (arrows) or normal CP epithelium (arrowhead). Primary cilia are magnified in inset pictures. DAPI staining (blue) labels nuclei. Scale bar: 10μm. (i) Summary of cilia pattern in human CPPs (n=17 tumours from 16 individuals), CPCs (n=13 tumours from 13 individuals), and normal CPs (n=6 CPs from 6 disease-free individuals).
Mono-ciliated CP tumour cells are uniquely capable of transducing Shh signals
To determine whether the distinct cilia pattern of tumour cells affects Shh signaling, we characterized Shh-driven Smo ciliary translocation. Both CP epithelial and tumour cells express Lmx1a when grown with serum; however, only tumour cells can proliferate (Fig. 6a, 6b, Supplementary Fig. 7g). After serum removal, cells were treated with ShhN or SAG, a Shh pathway agonist42 (Fig. 6a). While such treatment failed to promote ciliary accumulation of Smo in epithelial cells, it led to translocation of Smo into the solitary primary cilium of tumour cells (Shh: n=3, 71.78±6.93%, p<0.0001; SAG: n=3, 74.99±3.03%, p<0.0001; two-way ANOVA) (Fig. 6c, 6d), even though both cell types express similar levels of Ptch1 and Smo (Fig. 6e), indicating that Shh signaling in the primary cilium is preserved in tumour cells, but lost in epithelial cells despite their multiple primary cilia. Indeed, ShhN or SAG restored the percentage of Ki-67+ tumour cells after serum removal, an effect that can be reversed by cyclopamine, indicating that tumour cells are uniquely capable of responding to Shh through proliferation (Fig. 6f–h).
Figure 6.
Mono-ciliated CP tumour cells are uniquely capable of transducing Shh signals. (a) Dissociated CP tumour cells from Mcre;NICD1 animals were initially cultured in the presence of 10% fetal bovine serum (FBS). Cultured CP tumour cells were then switched to serum-free conditions for 2 days followed by treatment with ShhN or SAG for 1.5 or 3 days for signaling and proliferation analysis, respectively. (b) The expression of Lmx1a (red) and Ki-67 (red) is shown in CP tumour cells cultured with FBS. GFP (green) labels NICD1+ CP tumour cells, while Aqp1 labels CP epithelial cells. DAPI staining (blue) labels nuclei. Scale bar: 25μm. After treatment with ShhN, SAG, or vehicle, Smo (red) localization is shown in CP tumour (c) or epithelial cells (d). Arl13b expression (green) labels primary cilia, while DAPI staining (blue) labels nuclei. Scale bar: 10μm. (e) Q-RT-PCR analysis of Ptch1 and Smo expression in CP tumours from Mcre;NICD1 animals (black circles) and wild type CPs (WT, while circles) at P7 (data from technical replicates of each set of specimen in a single experiment are shown; experiment was not repeated. Raw data can be found in Supplementary Table 9). (f) The expression of Ki-67 (green) in CP tumour cells from Mcre;NICD1 animals treated with ShhN for 48 hours treatment is shown. Arl13b expression (red) labels primary cilia, DAPI staining (blue) labels nuclei. Notice that Ki-67 expression is present in mono-ciliated CP tumour cell, while multi-ciliated CP epithelial cell lacks Ki-67 expression. Scale bar: 10μm. (g) The expression of Ki-67 (red) in CP tumour cells from Mcre;NICD1 animals treated as indicated is shown. GFP (green) labels NICD1+ tumour cells. DAPI staining (blue) labels nuclei. Scale bar: 25μm. (h) Quantitation of the percentage of Ki-67+ cells in NICD1+/GFP+ tumour cells after 72-hour treatment as indicated (n=4 specimens/treatment, data from a single experiment are shown, raw data are available in Supplementary Table 9; mean ± SEM, two-way ANOVA, ***, P< 0.001; ****, P<0.0001).
Notch-induced CP tumour arises from roof plate progenitors
The similarity in gene expression between tumour and roof plate cells, together with elevated Notch signaling and solitary primary cilium in the latter, suggests that Notch-induced CP tumour is related to the roof plate. To delineate the developmental origin of CP tumour, we first analyzed the distribution of Atoh1+ progenitors in hindbrain roof plate using Math1M1GFP mice with enhanced green fluorescent protein (EGFP) fused to the C-terminal of Atoh143. In addition to rhombic lip, Atoh1:EGFP+ cells are present in the Lmx1+/Otx2+ upper roof plate but largely absent from lower roof plate (Fig. 7a, Supplementary Fig. 8a). Second, we analyzed tumour formation in Mcre;NICD1 animals during development. At day E12.5, though many NICD1+/GFP+ cells are located in rhombic lip bordering Lmx1a+ roof plate, some NICD1+/GFP+ cells are present within CP forming into papillary structures (Fig. 7b). These prospective Lmx1a+ tumour cells undergo proliferation (Ki-67+), and remain undifferentiated (Aqp1−) (Fig. 7b, Supplementary Fig. 8b). At day E14.5, most NICD1+/GFP+ tumour cells are found in Lmx1a+ upper roof plate and rostral half of the CP (Fig. 7c). Third, proliferative progenitors (Lmx1a+/Ki-67+/Aqp1−) in hindbrain roof plate differentiate into post-mitotic epithelial cells that form the CP epithelium (Lmx1a+/Ki-67−/Aqp1+) during embryogenesis44, 45. The shape and size roof plate are similar between wild type and Mcre;NICD1 mice; however, tumour cells remain proliferative and undifferentiated (Ki-67+/Aqp1−) even after their incorporation into CP epithelium (Fig. 8a, Supplementary Fig. 8c). Together, these results indicate that CP tumours in these animals arise from roof plate progenitors and migrate into the CP where they continue to undergo Shh-driven proliferation throughout development (Fig. 8b, 8c).
Figure 7.
Notch-induced CP tumours arise from progenitors in the roof plate. (a) Atoh1:EGFP expression in Math1M1GFP mice at day E14.5 is detected with a GFP antibody and shown in green signals. Lmx1a expression (red) marks the roof plate (marked by dotted lines), while DAPI staining (blue) labels nuclei. Notice that Atoh1:EGFP+ progenitors (arrows) are present only in Lmx1a+ upper roof plate. Scale bar: 50μm. (b) Schematic illustration of hindbrain roof plate (RP) and CP at day E12.5. Solid line marks the transverse plane across the roof plate shown in the H&E staining and diagram. Boxed region in the diagram representing the interface of the rhombic lip and roof plate is shown in Mcre:NICD1 mice at day E12.5. GFP (green) marks Atoh1+ rhombic lip progenitors and prospective tumour cells (arrows) in the roof plate and CP. The expression of Lmx1a (red) marks the roof plate/CP lineage. While Ki-67 expression (red) labels proliferating cells, Aqp1 expression (red) marks differentiated epithelial cells. DAPI staining (blue) labels nuclei. Scale bars: black, 1 mm; white, 30μm. (c) Gene expression in the roof plate and CP in Mcre:NICD1 mice at day E14.5. Boxed regions of progenitors within upper (a1, a2) and lower (b1, b2) roof plate as well as tumour cells (c) in CP are shown in higher magnification. GFP expression (green) marks NICD1+ tumour cells in the roof plate and CP. Lmx1a expression (red) labels the roof plate and CP lineage, while DAPI staining (blue) marks nuclei. Scale bar: 300μm.
Figure 8.
Notch-induced CP tumour cells retain properties of roof plate progenitor. (a) Analysis of gene expression and cell proliferation in tumour cells and progenitors within hindbrain roof plate in Mcre;NICD1 and wild type (WT) animals, respectively, at day E12.5. The expression of Lmx1a (red) marks the roof plate and CP lineage. Ki-67 expression (green) labels proliferating cells, and Aqp1 expression (green) marks differentiated epithelial cells. White dotted lines demarcate domain of progenitors in hindbrain roof plate. Boxed regions of Lmx1a+/Ki-67+/Aqp1− progenitors (a, c, e, f), tumour cells (b, d) (arrows), and Lmx1a+/Ki-67−/Aqp1+ differentiated epithelial cells (b, d) (red bracket lines) are shown in higher magnification. Scale bar: 100μm. (b) Schematic diagram of interaction between Notch and Shh pathways during the roof plate/CP morphogenesis and tumour formation. Epithelial progenitors in the roof plate (RP, yellow) next to the rhombic lip (RL) exhibits active Notch signaling, possess a solitary primary cilium, and proliferate in response to Shh (green dots) secreted from multiciliated epithelial cells (grey). The self-renewal will cease as Notch pathway activity in progenitor cells is attenuated to allow for multiciliate differentiation, thereby abolishing the response of the progenitors to Shh that drives their expansion during development. (c) Epithelial progenitors with constitutive Notch pathway activity (red) remain mono-ciliated and undergo aberrant proliferation to become tumour cells that retain the ability to respond to Shh signals in the local environment and undergo Shh-driven proliferation.
DISCUSSION
In this study, we examined Notch signaling in CP tumours, a group of rare brain neoplasms most commonly found in children. Consistent with a role for NOTCH pathway in these tumours8–11, analysis of human CP tumour datasets revealed significant enrichment for NOTCH signaling. Using animal models, we showed that sustained Notch 1 signaling leads to CP tumours that, similar to human CP tumours, express CP markers and undergo increased proliferation. Cross-species molecular analysis demonstrates that these CP tumours closely resemble their human counterpart, validating our animal models as accurate representation of human disease.
CP tumours are thought to originate from CP epithelium14, 15. Analysis of Math1-Cre;Rosa26-EYFP mice indicates that EYFP+ epithelial cells derived from Atoh1+ progenitors in roof plate exhibit properties similar to the rest of CP epithelium. Indeed, the identical CP tumours in Lcre;NICD1 and Mcre:NICD1 mice indicate that both Atoh1+ and Atoh1− lineages in CP are sensitive to Notch signaling activation. However, despite the expression of CP markers, expression of many genes found in mature epithelium fails to be upregulated in tumour cells, suggesting a related but distinct developmental origin, or a block in differentiation, or both. The presence of Atoh1+ progenitors and nascent tumour cells from Mcre:NICD1 mice in roof plate suggest CP tumours arise from progenitors within this region44, 45. In agreement, tumour cells and these progenitors display similar characteristics: increased expression of roof plate markers, elevated Notch signaling, and single primary cilium. The development of CP tumour induced by sustained Notch signaling may reflect its inherent role in roof plate/CP morphogenesis46–49: Notch pathway suppresses multiciliate and epithelial differentiation of progenitors, thereby preserving the primary cilium-based Shh signaling (Fig. 8b). Compared to CPCs, human CPPs express higher levels of roof plate markers (Supplementary Fig. 5a), consistent with their developmental origin.
We provide evidence that CP epithelial cells secrete Shh that drives the proliferation of a distinct population of Shh-responsive progenitors in upper roof plate. Consistent with previous reports, we showed Shh expression in hindbrain CP epithelium at birth that gradually disappear within two weeks50, 51. Interestingly, low-level cell proliferation and Mycn expression is detected in tumour cells from Mcre;NICD1 animals at P14, presumably driven by residual or trans-vesicular Shh from other brain regions, as shown for CP-derived signals regulating distant cell populations in the brain52–54. Tissue-specific deletion may delineate the role of Shh from these sources in tumour growth25, 50.
Analysis of published datasets demonstrated enrichment for SHH signaling, suggesting a role for deregulated SHH expression in human CP tumours, including CPCs13–15. Consistently, tumours from Mcre;NICD1 or Lcre;NICD1 mice exhibit active proliferation simiar to that observed in CPCs at P0 when Shh is robustly expressed. Our results suggest that Shh pathway inhibition may represent a viable strategy for targeted therapy for CP tumours. SHH pathway inhibitors, clinically approved for treating other more common cancer types, may be “repurposed” for treating patients with CP tumours exhibiting abnormal SHH signaling. Indeed, vismodegib treatment interevenes with tumour progression in our models, though further validation with xenograft models would be necessary. Tumour becomes quiescent as Shh expression decreases, suggesting that additional changes are present to support aggressive growth of CP tumours as seen in human CPCs with aberrant NOTCH signaling. Tumour cells proliferate in the presence of serum (Fig. 7b), indicating that serum may contain factors capable of driving tumour growth. Amplifications of TAF12, NFYC, and RAD54L play an important role in CPCs15. Their expression is not significantly changed in our CP tumours; however, deregulation of these genes may alter the behavior of Notch-induced CP tumours. TP53 alterations play a cruical role in human CPCs6, 14. CP tumours in our models retain the wild type Tp53 and exhibit signs of Tp53 signaling and DNA damage response (Supplementary Tables 1, 2, 3, and 5, Fig. 4m), suggesting that Tp53 loss may facilitate malignant transformation of Notch-induced CP tumours.
Aberrant NOTCH signaling has been linked to different cancers in humans and therapeutically targeted55–57. The single primary cilium on tumour cells with constitutive Notch signaling proves to be essential for Shh signaling, while multiple primary cilia may interfere with cilium-dependent signaling activities58–61. A Notch-regulated signaling cascade involving Mcidas, Myb and Foxj1 plays an important role in the differentiation of cells with multiple motile cilia39–41, 62–64. Our results reveal the interaction between Notch signaling and primary cilia crucial for CP tumourigenesis (Fig. 8c): sustained Notch signaling preserves single primary cilium by suppressing Mcidas and Foxj1 expression to block multiciliate differentiation, transforming progenitors into tumour cells that undergo Shh-driven proliferation. Targeting Notch-mediated multiciliate differentiation may represent a rational strategy for CP tumour treatment. Human CP epithelial cells exhibit a “hobnail” contour on the apical side, whereas the surface of CP tumour cells is more flattened9, 65. Our results suggest that cilia pattern of tumour cells may mediate this distinct morphology. Indeed, most human CP tumours (including all CPCs) display solitary primary cilium. Understanding the interaction between Notch signaling, ciliogenesis, and epithelial differentiation in CP tumours is essential to validate the therapeutic potential of Notch inhibitors.
METHODS
Mice
Gt(ROSA)26Sortm1(Notch1)Dam/J (Rosa26-NICD1) mice, B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J (Rosa26-EYFP) mice, B6.129S-Atoh1tm4.1Hzo/J (Math1M1GFP) mice, B6.Cg-Tg(Atoh1-cre)1Bfri/J (Math1-Cre) transgenic mice, and C57BL/6 mice (all from Jackson Laboratory, Bar Harbor, ME), and Tg(Lmx1a-cre)1Kjmi (Lmx1a-Cre) transgenic mice were maintained by breeding with C57BL/6 mice. Experimental procedures on animals housed at Sanford Research were approved by Sanford Research Institutional Animal Care and Use Committee and performed in compliance with national regulatory standards. No statistical method was used to predetermine sample size in animal experiments. The animal experiments were not randomized. The investigators were not blinded to group allocation during experiments and outcome assessment. Experimental animals were administered 100 mg/kg vismodegib (LC laboratories, Woburn, MA, V-4050) or vehicle following two regimens: daily treatment from day E15.5 through E18.5 (Mcre;NICD1 mice: 10 animals for each treatment); or from day E17.5 through day P7 (Lcre;NICD1 mice: 29 animals for vismodegib, 26 animals for vehicle), by gastric gavage of pregnant or nursing females.
Human Samples
All human CP specimens were procured with informed consent from human subjects following the requirements by institutional review boards at Sanford Research, Sanford Burnham Prebys Medical Discovery Institute, and Ludwig-Maximilans-University. All CP specimens from Boston Children’s Hospital were obtained with informed consent from human subjects under an approved institutional review board protocol (Supplementary Table 7). Normal human CP epithelial cells (ScienCell, San Diego, CA) were used as control in gene expression analysis. All tissues were handled in accordance with guidelines and regulations for the research use of human brain tissue set forth by the NIH (http://bioethics.od.nih.gov/humantissue.html). Diagnoses of human CP specimens from Boston Children’s Hospital were reviewed by two neuropathologists (H.G.W.L., S.S.) using standard WHO criteria66.
Isolation and culture of primary CP cells
Multiple sets of CP specimens from Mcre;NICD1 and/or wild type mice were collected. To obtain sufficient number of cells, each set of specimens included tissues pooled from multiple animals of the same genotype. Gender information is not available for animals at P0 and P7. Both male and female animals were used at other time points (see Supplementary Table 9 for information on animals used for each experiment). Dissected CP specimens were dissociated with forceps under stereoscope followed by enzymatic digestion at 37°C for 20 minutes in 0.7 mg/ml of hyaluronic acid (H3506, Sigma-Aldrich, St. Louis, MO), 0.2 mg/ml of kynurenic acid (Sigma-Aldrich K3375), and 1 mg/ml of trypsin in Hank’s balanced salt solution (HBSS, 14170-112; Life Technologies, Grand Island, NY) supplemented with 2 mM glucose. Trypsin inhibitor ovomucoid (LS003085, Worthington Biochemical Corporation, Lakewood, NJ) was added to stop enzymatic digestion and dissociated CP cells were centrifuged at 200 g for 5 minutes at 4°C. Cell pellets were resuspended in Dulbecco’s modified Eagle’s medium/Nutrient Mixture F-12 Ham’s-Liquid Media (DMEM/F12, SH30271; Thermo Fisher Scientific, Waltham, MA), and cultured in DMEM/F12 supplemented with 30 ng/ml of EGF (Sigma-Aldrich E4127), 30 ng/ml of FGF2 (Sigma-Aldrich F0291), B27 supplement, 2 mM glutamine, and 100U/mL penicillin/streptomycin (all from Life Technologies, Grand Island, NY). After treatment under serum-free conditions for 96 hours, cells were dissociated mechanically by pipetting and quantified. For culture with serum, medium was supplemented with 10% fetal bovine serum. Cytosine β-D-arabinofuranoside (Ara-C, 20 μM; Sigma-Aldrich C1768) was added the following day to eliminate contaminating fibroblasts. Cultured cells were treated with ShhN67 (200 ng/ml), or SAG (200 nM) (11914, Cayman Chemical Company, Ann Arbor, Michigan), with or without cyclopamine (10 μM) (LC laboratories C-8700). Primary CP tumour or epithelial cells were not listed in the database of commonly misidentified cell lines maintained by ICLAC and NCBI Biosample. Analyses of gene expression, proliferation, and signal transduction were performed in cultured primary CP cells (Fig. 4a–4d, Fig. 6b–6d, 6f–6h, Supplementary Fig. 7g). Results from these studies confirmed their identity. Given the short time (< 8 days) during which CP cells were maintained as primary cultures supplemented with antibiotics, we did not perform test for mycoplasma contamination.
Histology, Immunohistochemistry, Immunofluorescence, and Immunocytochemistry
Whole brains were fixed overnight in 4% paraformaldehyde (PFA) and processed for paraffin embedding and sectioning. Tissue sections were deparaffinized with CitriSolv (Decon Labs, King of Prussia, PA), then rehydrated with graded ethanol series. For frozen tissues, samples were further equilibrated in 20% sucrose at 4°C for 24–48 hours, embedded in TissuTek-Optimal Cutting Temperature (O.C.T.) compound (Sakura Finetek, Torrance, CA), and sectioned (15–20 μm thickness) on a cryostat. Cultured cells were fixed in 4% PFA at room temperature for 10 minutes and stored in phosphate buffered saline (PBS). For primary cilia staining, cells were fixed with cold methanol for 10 minutes and stored in PBS at 4°C.
No randomization was used to determine how samples were allocated to experimental groups and processed. Immunostaining was carried out as described previously67. Heat-induced eptiope retrieval was performed for paraffin-embedded tissue sections in Rodent Decloaker (Biocare Medical, Concord, CA). For immunohistochemistry, endogenous peroxidase activities were inactivated in 3% H2O2 for 10 minutes at room temperature. Tissue sections were blocked with 10% normal sera in PBS-0.1% Triton X-100 for 1 hour at room temperature and incubated with primary antibodies for 1 hour. After washes with PBS, biotinylated secondary antibodies were applied for 1 hour, followed by treatment with Avidin/Biotinylated enzyme Complex and substrate/chromogen incubation (Vector laboratories, Burlingame, CA). Slides were counterstained with hematoxylin. For immunofluorescence, tissue sections or cells were sequentially probed with primary antibodies and fluorescently labeled secondary antibodies (Jackson Immunoresearch, West Grove, PA). Primary antibodies used and dilution ratios are: mouse monoclonal anti-Acetylated α-Tubulin (1:500, ab24610, clone 6-11B-1, abcam, Cambridge, MA), mouse monoclonal anti-Acetylated α-Tubulin (1:500, Sigma-Aldrich T7451, clone 6-11B-1), mouse monoclonal anti-Arl13b (1:500, clone N295B/66, NeuroMab, Davis, CA), rabbit anti-Arl13b (1:500, 17711-1-AP, Proteintech, Chicago, IL), mouse monoclonal anti-Aquaporin 1 (1:1000, clone 1/22, abcam ab9566), rabbit anti-Aquaporin 1 (1:1000, AB2219, EMD Millipore, Billerica, MA), rabbit monoclonal anti-Cleaved Caspase-3 (Asp175) (clone 5A1E) (1:800, 9664, Cell Signaling Technology, Danvers, MA), mouse monoclonal anti-Ccnd1 (1:100, sc-450, clone 72-13G, Santa Cruz Biotechnology, Dallas, TX), mouse monoclonal anti-Cdkn1b (p27 Kip1) (1:100, 610242, clone 57/Kip1/p27, BD Biosciences, San Jose, CA), rabbit anti-Cytokeratins (1:100, Z0622, Dako, Carpinteria, CA), mouse monoclonal anti-Foxj1 (1:50, 14-9965, Clone 2A5, eBioscience, San Diego, CA), chicken anti-GFP (1:1000, GFP-1010, Aves Lab, Tigard, OR), mouse monoclonal anti-γ-Tubulin (Sigma-Aldrich T6557, clone GTU-88, 1:10000), guinea pig anti-Hes1 (1:150, from Dr. Ryoichiro Kageyama), rabbit monoclonal anti-Ki-67 (abcam ab16667, clone SP6, 1:100), goat anti-Lmx1a (Santa Cruz sc-54273, 1:100), rabbit anti-MafB (1:200, IHC-00351, Bethyl Laboratories, Montgomery, TX), rabbit anti-Otx2 (EMD Millipore AB9566, 1:500), rabbit anti-Smo68 (1:100), sheep anti-Transthyretin (abcam ab9015, 1:500).
The investigator was blinded to group allocation in the following two experiments. First, EYFP+ or GFP+ cells in 300 Otx2+ cells were assessed by analyzing 3 distinct tissue regions of each sample. The percentage of EYFP+ or GFP+ cells was obtained by averaging the numbers EYFP+ or GFP+ cells per 100 Otx2+ cells of all samples for each genotype at each time point. Second, in analysis of cell proliferation, the number of Ki-67+ cells in 300 Lmx1a+ or GFP+ cells were assessed from 3 distinct tissue regions for each animal. The percentage of Ki-67+ cells in total Lmx1a+ or GFP+ cell population was calculated by averaging the numbers of Ki-67+ cells per 100 Lmx1a+ or 100 GFP+ cells of all samples for each genotype at each time point or each treatment. For proliferation analysis of cultured cells, Ki-67 expression was assessed by analyzing 3 distinct fields: the percentage of Ki-67+ cells was calculated by averaging the numbers of Ki-67+ cells per 100 cells of all samples for each treatment.
For primary cilia staining of CP specimens from mice, cilia pattern was assessed by analyzing 3 distinct tissue regions of each animal. To measure the length of primary cilia, 12 primary cilia from tumour cells and 10 primary cilia from epithelial cells were examined. Smo ciliary translocation was assessed by analyzing 3 distinct fields: the percentage of cells with ciliary accumulation of Smo was calculated by averaging the number of cells with Smo translocation per 100 cells of all samples for each treatment. Information on animals used for each experiment can be found in Supplementary Table 9.
For staining of primary cilia in human CP tumours and normal human CPs, the investigator was blinded to group allocation. Tissues used include: normal CP: 6 disease-free individuals; CPP: 17 tumour specimens from 16 individuals; CPC: 13 tumours from 10 individuals (Supplementary Table 9). Cilia staining was assessed by analyzing 5 distinct tissue regions.
5-ethynyl-2′-deoxyuridine (EdU) incorporation assay
One hour after intraperitoneal injection of 100mg/kg EdU (7180, Setareh Biotech, Eugene, OR), whole brains were harvested from animals. Tissue sections were rinsed with Tris-buffered saline (TBS) and then incubated for 20 minutes with solution consisting of 100 mM pH 8.5 Tris, 1 mM CuSO4, 10 mM Cy5-azide (B3030, Lumiprobe, Hallandale Beach, FL) and 100 mM ascorbic acid. Information on animals used is provided in Supplementary Table 9.
Image Acquisition
Whole-mount images were obtained using a Nikon SMZ1000 Stereomicroscope. Light and fluorescent microscopic images were obtained by a Nikon Eclipse 90i microscope system or a Nikon confocal microscope system A1+.
Transmission Electron Microscopy
CP specimens were collected at different time points. To ensure collection of sufficient amount of normal CP specimens, each normal CP specimen includes tissues pooled from 2 wild type animals for P7, P14 and P22. Information on animals used is available in Supplementary Table 9. The investigator was blinded to group allocation. Tissues were fixed in 4% PFA, 1% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4) at 4°C overnight. After washing with cacodylate buffer supplemented with 10% sucrose, tissues were post-fixed with 1% osmium tetroxide (OsO4), followed by incubation with 1% uranyl acetate in 30% ethanol. Tissue samples were dehydrated, then transferred to propylene oxide, and embedded in Eponate-12 epoxy resin (Ted Pella, Redding, CA). Tissue samples were sectioned (85 nm thickness) with a Leica UC-6 ultramicrotome (Wein, Austria). Sections were counterstained with uranyl acetate and lead citrate, and observed under the JEOL 1400 transmission electron microscope (JEOL Ltd, Tokyo, Japan). Images were taken with a Gatan UltraScan 1000 CCD digital camera (Gatan, Inc., Pleasanton, CA).
Immunoblotting
Immunoblotting was carried out as described previously67. Multiple sets of CP specimens were collected at each time point. To obtain sufficient quantity of protein, each set of specimens may contain tissues pooled from several animals of the same genotype. Gender information is not available for animals at P0 and P7 (see Supplementary Table 9 for information on animals used). Tissues were homogenized in protein lysis buffer (Tris/HCl pH 7.4, 1 mM EDTA, 150 mM NaCl, 1% NP40, 0.25% sodium deoxycholate, 1X protease inhibitor cocktails (Research Product International, Mount Prospect, IL). Protein concentrations were measured by a standard BCA assay (Life Technologies). Equal amounts of protein were resuspended in a reduced sample buffer, separated by SDS-PAGE, and then transferred to a polyvinylidene difluoride membrane. After incubation with 5% nonfat milk in TBST (TBS, 0.5% Tween 20) for 1 hour, the membrane was incubated with primary antibodies for 1 hour. The membrane was washed and incubated with horse radish peroxidase (HRP)-conjugated secondary antibodies (GE Healthcare, Piscataway, NJ) for 1 hour. After washes, chemiluminescence detection was performed using Western Lightning Plus-ECL (PerkinElmer, Waltham, MA). β-actin was used as a protein loading control. Primary antibodies used include: mouse monoclonal anti-β-actin (Sigma-Aldrich A5441, clone AC-15, 1:2000), rabbit anti-Aquaporin 1 (EMD Millipore AB2219, 1:1000), mouse monoclonal anti-Ccnd1 (Santa Cruz sc-450, clone 72-13G, 1:200), rabbit anti-Hes1 (Dr. Ryoichiro Kageyama, 1:1000), rabbit anti-Otx2 (EMD Millipore AB9566, 1:500).
RNA preparation, Quantitative RT-PCR, In-situ hybridization, microarray, and RNA-seq
For murine tissues, three independent sets of tumour specimens from Mcre;NICD1 mice and three sets CP specimens from wild type mice were collected at each time point. To ensure that sufficient amount of RNA can be extracted, each set of specimens included tissues pooled from several animals of the same genotype. Gender information was not available for animals at P0 and P7 (see Supplementary Table 9 for information on animals used for each set of specimens). For human tissues, seven CPPs from seven individuals, two CPCs from two individuals, and normal human CP epithelial cells (ScienCell) were used (Supplementary Table 9). Total RNA was extracted using Trizol and PureLink RNA Mini Kit (Life Technologies) according to the manufacturer’s instructions. For quantitative RT-PCR (Q-RT-PCR), total RNA samples were converted to cDNA using GoScript Reverse Transcription System (Promega, Madison, WI). All reactions were set up in triplicate with ABsolute Blue QPCR Mix (Thermo Fisher Scientific) and run on an ABI 7000 Sequence Detection System (Life Technologies). Gene-specific primers and probes for mouse Aqp1, Ttr, Gli1, Mycn, Shh, Ptch1, Smo, Hes1, Hes5, Foxj1, Mcidas, and control Actinb were used (Supplementary Table 8). The following Taqman assays for human genes were used (Life Technologies): GLI1 (Hs01110766_m1), PTCH1 (Hs00181117_m1), MYCN (Hs00232074_m1), HES5 (Hs01387463_g1), HES1 (Hs00172878_m1), HEY1 (Hs01114113_m1), ACTB (Hs99999903_m1). Data were analyzed using ABI Fast System SDS software. Transcript levels were determined as the number of transcripts for each gene relative to those of Actb (mouse) or ACTB (human). The results for each set of specimens were obtained by averaging transcript levels of technical triplicates and used for subsequent analyses. Data from Q-RT-PCR experiments can be found in Supplementary Table 9. Exclusion was applied only on rare occasions when one of the three wells of the triplicate was a significant outlier; however, for each sample in question, Q-RT-PCRs were repeated in independent experiments, which validated the exclusion.
In-situ hybridization was performed by RNA In Situ Hybridization Core facility at Baylor College of Medicine as described previously69. For analysis of Shh, Mycn, and Gli1 expression, CP specimens from multiple animals were collected at P0 and P14. For analysis of Mycn expression in Mcre;NICD1 animals treated with vismodegib or vehicle, three tumours from three vismodegib-treated mice, and five tumours from five vehicle-treated mice were examined at P0. For Hes1 and Hes5 expression analysis, two Mcre;NICD1 mice and one wild type mouse were collected at day E14.5. Gender information was not available for embryos, P0 animals, and the P8 animal for sense probe controls (see Supplementary Table 9 for information animals used for each experiment). For human tissue samples, SHH (probe #600951), PPIB (positive control, probe #313901), dapB (negative control, probe #310043) RNA were visualized using RNAscope 2.0 HD detection kit-BROWN (Advanced Cell Diagnostics, Hayward, CA) according to manufacturer’s protocol. Human tissues used include: normal CP: 5 disease-free individuals; CPP: 11 tumour specimens from 10 individuals; CPC: 10 tumours from 10 individuals (Supplementary Tables 7, 9). SHH expression was assessed in 5 distinct tissue regions: percentage of SHH-expressing cells was calculated by averaging the numbers of SHH+ cells per 100 cells in 5 distinct tissue regions of each specimen.
Microarray and RNA-seq studies were performed at the DNA Analysis Core at Sanford Burnham Prebys Medical Discovery Institute (La Jolla, CA). For microarray experiments, labeled cRNA was prepared from 500 ng RNA using the Illumina® RNA Amplification Kit from Ambion (Life Technologies). The labeled cRNA (1500 ng) was hybridized overnight at 58°C to the Sentrix ® MouseWG-6 Expression BeadChip (>46,000 gene transcripts; Illumina, San Diego, CA) according to the manufacturer’s instructions. BeadChips were subsequently washed and developed with fluorolink streptavidin-Cy3 (GE Healthcare). BeadChips were then scanned with an Illumina BeadArray Reader. tumourPathway analysis using the GeneGo MetaCore Analytical Suite (http://genego.com; GeneGo, St. Joseph, MI) was used to score and rank pathways enriched in datasets by the proportion of pathway associated genes with significant expression values.
For RNA-seq experiments, total RNA samples were ribo-depleted using the Ribominus Eukaryote System (Life Technologies), and used to generate sequencing libraries of barcoded fragment using the Ion Total RNA-Seq Kit V2 (Life Technologies). Libraries were sequenced on the Ion Proton sequencer, three libraries per Ion Proton PI Chip, using 200 bp sequencing reagents. Reads were aligned to the mouse genome (mm10) using a combination of Tophat2, and Bowtie2 (http://tophat.cbcb.umd.edu/). Differentially expressed transcripts were detected using the Cufflinks Cuffdiff package (http://cufflinks.cbcb.umd.edu/) and transcripts with a q-value <0.05 (Cuffdiff) were analyzed by ingenuity pathway analysis (Ingenuity Systems, Redwood City, CA). Principal component analysis was performed using R package rgl (http://cran.r-project.org/web/packages/rgl/index.html), a 3D visualization system based on OpenGL. Hierarchical clustering was performed using Genesis (http://genome.tugraz.at/genesisclient/genesisclient_description.shtml).
Statistical analysis & Reproducibility
Statistical analyses were performed with GraphPadPrism 6.0 (GraphPad Software Inc., La Jolla, CA). All pooled data were expressed as the mean ± SEM. Differences between two groups were compared using an unpaired two-tailed t-test. Differences between multiple groups were analyzed with one-way or two-way ANOVA followed by Tukey’s multiple comparisons test.
Results were considered significant at p < 0.05 (*); p<0.01 (**); p<0.001 (***); p<0.0001 (****). For western blot analysis, the expression of Aqp1 and Otx2 at P14, and Hes1 expression at P0 and P7, were repeated in one independent experiment. Analyses of the expression of Hes1 at P90, and Ccnd1 expression were repeated in two independent experiments with similar results. RNA-seq and microarray studies were performed in single experiments. For in-situ hybridization, analyses of Mycn expression in visomodegib-treated animals, and Mycn and Gli1 expression at P0 and P14, were repeated in one independent experiment with similar results. Analysis of Shh, Hes1, and Hes5 expression was performed in single experiments (Supplementary Table 9). SHH expression analysis in human tissues was conducted in one experiment with samples divided into two groups. For Q-RT-PCR analysis, the expression of Hes1, Hes5, Ttr, Ptch1, Smo, and Mcidas was examined in single experiments, while Aqp1, Gli1, Mycn, Shh, and Foxj1 expression analysis was repeated in one independent experiment with similar results. Q-RT-PCR analysis of human samples was performed in single experiment. Analyses of cell proliferation (Ki-67 or EdU staining), percentage of EYFP+ or GFP+ cells in CP epithelium, tumour cell numbers, and Smo translocation were performed in single experiments. The measurement of cilia length was repeated in one independent experiment with similar results. Representative images were selected from two independent experiments.
Supplementary Material
Acknowledgments
We thank all members of the laboratory for helpful discussions. We are grateful to Drs. Kathleen Millen, James Kim, and Ryoichiro Kageyama for providing Lmx1a-Cre transgenic mouse strain, Smo antibody, and Hes1 antibodies, respectively, and Drs. Charles Eberhart, Michael Taylor, and Sandro Santagata for providing human CP tumour samples. We wish to acknowledge the Labatt Brain Tumour Research Centre Tumour and Tissue Repository, which is supported by b.r.a.i.n child and Meagan’s Walk. We are indebted to Claire Evans and Emily Grandprey for excellent technical assistance. We thank Drs. W. Keith Miskimins and Kameswaran Surendran for helpful suggestions, Drs. Jianning Tao and Diane Maher for critical reading of the manuscript and helpful discussions. This project is supported by: Boston Children’s Hospital IDDRC P30 HD18655; Sanford Research, and Institutional Development Awards (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health (NIH) under grant number 5P20GM103548 (Cancer), which also supports Cores at Sanford Research together with NIH grant 1P20GM103620-01A1 (Pediatrics). The RNA In Situ Hybridization Core facility at Baylor College of Medicine is supported by a Shared Instrumentation grant from the NIH (1S10OD016167). Additional support was provided by the National Brain Tumor Society (R.W.R.).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interest
Accession numbers
Published human CP tumour datasets (GSE14098, GSE60886) were downloaded from GEO database for analysis. Datasets from our studies were deposited at NCBI. Sequencing: BioProject ID, PRJNA282889; Microarray: GSE77123.
AUTHOR CONTRIBUTIONS
L.L. and H.Z. conceived and planned the project, and wrote the manuscript. H.G.W.L. and M.L.C. reviewed diagnoses of human tissue samples. J.W., J.O., R.W.R. and U.S. analyzed morphological characters and gene expression pattern of human tumour samples. M.P.L and M.K.L. performed cilia and gene expression analyses in human tissue samples. K.B.G. provided assistance with gene expression analysis. E.Z. conduced RNA-seq data processing and analysis. J.S. provided technical advice, support and data analysis for electron microscopy studies.
References
- 1.Gopal P, Parker JR, Debski R, Parker JC., Jr Choroid plexus carcinoma. Archives of pathology & laboratory medicine. 2008;132:1350–1354. doi: 10.5858/2008-132-1350-CPC. [DOI] [PubMed] [Google Scholar]
- 2.Ogiwara H, Dipatri AJ, Jr, Alden TD, Bowman RM, Tomita T. Choroid plexus tumors in pediatric patients. British journal of neurosurgery. 2012;26:32–37. doi: 10.3109/02688697.2011.601820. [DOI] [PubMed] [Google Scholar]
- 3.Lun MP, Monuki ES, Lehtinen MK. Development and functions of the choroid plexus-cerebrospinal fluid system. Nature reviews Neuroscience. 2015;16:445–457. doi: 10.1038/nrn3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun MZ, et al. Current management of choroid plexus carcinomas. Neurosurgical review. 2014;37:179–192. doi: 10.1007/s10143-013-0499-1. discussion 192. [DOI] [PubMed] [Google Scholar]
- 5.Safaee M, et al. Surgical outcomes in choroid plexus papillomas: an institutional experience. Journal of neuro-oncology. 2013;113:117–125. doi: 10.1007/s11060-013-1097-3. [DOI] [PubMed] [Google Scholar]
- 6.Tabori U, et al. TP53 alterations determine clinical subgroups and survival of patients with choroid plexus tumors. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28:1995–2001. doi: 10.1200/JCO.2009.26.8169. [DOI] [PubMed] [Google Scholar]
- 7.Nupponen NN, et al. Platelet-derived growth factor receptor expression and amplification in choroid plexus carcinomas. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2008;21:265–270. doi: 10.1038/modpathol.3800989. [DOI] [PubMed] [Google Scholar]
- 8.Beschorner R, Waidelich J, Trautmann K, Psaras T, Schittenhelm J. Notch receptors in human choroid plexus tumors. Histology and histopathology. 2013;28:1055–1063. doi: 10.14670/HH-28.1055. [DOI] [PubMed] [Google Scholar]
- 9.Dang L, et al. Notch3 signaling initiates choroid plexus tumor formation. Oncogene. 2006;25:487–491. doi: 10.1038/sj.onc.1209074. [DOI] [PubMed] [Google Scholar]
- 10.Fouladi M, et al. Phase I trial of MK-0752 in children with refractory CNS malignancies: a pediatric brain tumor consortium study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:3529–3534. doi: 10.1200/JCO.2011.35.7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Safaee M, et al. Choroid plexus papillomas: advances in molecular biology and understanding of tumorigenesis. Neuro-oncology. 2013;15:255–267. doi: 10.1093/neuonc/nos289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruland V, et al. Choroid plexus carcinomas are characterized by complex chromosomal alterations related to patient age and prognosis. Genes, chromosomes & cancer. 2014;53:373–380. doi: 10.1002/gcc.22148. [DOI] [PubMed] [Google Scholar]
- 13.Hasselblatt M, et al. TWIST-1 is overexpressed in neoplastic choroid plexus epithelial cells and promotes proliferation and invasion. Cancer research. 2009;69:2219–2223. doi: 10.1158/0008-5472.CAN-08-3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merino DM, et al. Molecular characterization of choroid plexus tumors reveals novel clinically relevant subgroups. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21:184–192. doi: 10.1158/1078-0432.CCR-14-1324. [DOI] [PubMed] [Google Scholar]
- 15.Tong Y, et al. Cross-Species Genomics Identifies TAF12, NFYC, and RAD54L as Choroid Plexus Carcinoma Oncogenes. Cancer cell. 2015;27:712–727. doi: 10.1016/j.ccell.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barakat MT, Humke EW, Scott MP. Learning from Jekyll to control Hyde: Hedgehog signaling in development and cancer. Trends in molecular medicine. 2010;16:337–348. doi: 10.1016/j.molmed.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang J, Hui CC. Hedgehog signaling in development and cancer. Developmental cell. 2008;15:801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Machold R, Fishell G. Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron. 2005;48:17–24. doi: 10.1016/j.neuron.2005.08.028. [DOI] [PubMed] [Google Scholar]
- 19.Wang VY, Rose MF, Zoghbi HY. Math1 expression redefines the rhombic lip derivatives and reveals novel lineages within the brainstem and cerebellum. Neuron. 2005;48:31–43. doi: 10.1016/j.neuron.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 20.Chizhikov VV, et al. The roof plate regulates cerebellar cell-type specification and proliferation. Development. 2006;133:2793–2804. doi: 10.1242/dev.02441. [DOI] [PubMed] [Google Scholar]
- 21.Chizhikov VV, et al. Lmx1a regulates fates and location of cells originating from the cerebellar rhombic lip and telencephalic cortical hem. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10725–10730. doi: 10.1073/pnas.0910786107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matei V, et al. Smaller inner ear sensory epithelia in Neurog 1 null mice are related to earlier hair cell cycle exit. Developmental dynamics : an official publication of the American Association of Anatomists. 2005;234:633–650. doi: 10.1002/dvdy.20551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC developmental biology. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nielsen CM, Dymecki SM. Sonic hedgehog is required for vascular outgrowth in the hindbrain choroid plexus. Developmental biology. 2010;340:430–437. doi: 10.1016/j.ydbio.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johansson PA, et al. The transcription factor Otx2 regulates choroid plexus development and function. Development. 2013;140:1055–1066. doi: 10.1242/dev.090860. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y, Helms AW, Johnson JE. Distinct activities of Msx1 and Msx3 in dorsal neural tube development. Development. 2004;131:1017–1028. doi: 10.1242/dev.00994. [DOI] [PubMed] [Google Scholar]
- 28.Elsen GE, Choi LY, Millen KJ, Grinblat Y, Prince VE. Zic1 and Zic4 regulate zebrafish roof plate specification and hindbrain ventricle morphogenesis. Developmental biology. 2008;314:376–392. doi: 10.1016/j.ydbio.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McMahon AR, Merzdorf CS. Expression of the zic1, zic2, zic3, and zic4 genes in early chick embryos. BMC research notes. 2010;3:167. doi: 10.1186/1756-0500-3-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marques F, et al. Transcriptome signature of the adult mouse choroid plexus. Fluids and barriers of the CNS. 2011;8:10. doi: 10.1186/2045-8118-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowyer JF, et al. Comparison of the global gene expression of choroid plexus and meninges and associated vasculature under control conditions and after pronounced hyperthermia or amphetamine toxicity. BMC genomics. 2013;14:147. doi: 10.1186/1471-2164-14-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janssen SF, Gorgels TG, Ten Brink JB, Jansonius NM, Bergen AA. Gene expression-based comparison of the human secretory neuroepithelia of the brain choroid plexus and the ocular ciliary body: potential implications for glaucoma. Fluids and barriers of the CNS. 2014;11:2. doi: 10.1186/2045-8118-11-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie J, Bartels CM, Barton SW, Gu D. Targeting hedgehog signaling in cancer: research and clinical developments. OncoTargets and therapy. 2013;6:1425–1435. doi: 10.2147/OTT.S34678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Smaele E, Ferretti E, Gulino A. Vismodegib, a small-molecule inhibitor of the hedgehog pathway for the treatment of advanced cancers. Curr Opin Investig Drugs. 2010;11:707–718. [PubMed] [Google Scholar]
- 35.Caspary T, Larkins CE, Anderson KV. The graded response to Sonic Hedgehog depends on cilia architecture. Developmental cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 36.Choksi SP, Babu D, Lau D, Yu X, Roy S. Systematic discovery of novel ciliary genes through functional genomics in the zebrafish. Development. 2014;141:3410–3419. doi: 10.1242/dev.108209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas J, et al. Transcriptional control of genes involved in ciliogenesis: a first step in making cilia. Biology of the cell/under the auspices of the European Cell Biology Organization. 2010;102:499–513. doi: 10.1042/BC20100035. [DOI] [PubMed] [Google Scholar]
- 38.Hoh RA, Stowe TR, Turk E, Stearns T. Transcriptional program of ciliated epithelial cells reveals new cilium and centrosome components and links to human disease. PloS one. 2012;7:e52166. doi: 10.1371/journal.pone.0052166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stubbs JL, Vladar EK, Axelrod JD, Kintner C. Multicilin promotes centriole assembly and ciliogenesis during multiciliate cell differentiation. Nature cell biology. 2012;14:140–147. doi: 10.1038/ncb2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu X, Ng CP, Habacher H, Roy S. Foxj1 transcription factors are master regulators of the motile ciliogenic program. Nature genetics. 2008;40:1445–1453. doi: 10.1038/ng.263. [DOI] [PubMed] [Google Scholar]
- 41.Gomperts BN, Gong-Cooper X, Hackett BP. Foxj1 regulates basal body anchoring to the cytoskeleton of ciliated pulmonary epithelial cells. Journal of cell science. 2004;117:1329–1337. doi: 10.1242/jcs.00978. [DOI] [PubMed] [Google Scholar]
- 42.Chen JK, Taipale J, Young KE, Maiti T, Beachy PA. Small molecule modulation of Smoothened activity. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:14071–14076. doi: 10.1073/pnas.182542899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rose MF, et al. Math1 is essential for the development of hindbrain neurons critical for perinatal breathing. Neuron. 2009;64:341–354. doi: 10.1016/j.neuron.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hunter NL, Dymecki SM. Molecularly and temporally separable lineages form the hindbrain roof plate and contribute differentially to the choroid plexus. Development. 2007;134:3449–3460. doi: 10.1242/dev.003095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Awatramani R, Soriano P, Rodriguez C, Mai JJ, Dymecki SM. Cryptic boundaries in roof plate and choroid plexus identified by intersectional gene activation. Nature genetics. 2003;35:70–75. doi: 10.1038/ng1228. [DOI] [PubMed] [Google Scholar]
- 46.Broom ER, Gilthorpe JD, Butts T, Campo-Paysaa F, Wingate RJ. The roof plate boundary is a bidirectional organiser of dorsal neural tube and choroid plexus development. Development. 2012;139:4261–4270. doi: 10.1242/dev.082255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Imayoshi I, Shimogori T, Ohtsuka T, Kageyama R. Hes genes and neurogenin regulate non-neural versus neural fate specification in the dorsal telencephalic midline. Development. 2008;135:2531–2541. doi: 10.1242/dev.021535. [DOI] [PubMed] [Google Scholar]
- 48.Bill BR, et al. Development and Notch signaling requirements of the zebrafish choroid plexus. PloS one. 2008;3:e3114. doi: 10.1371/journal.pone.0003114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garcia-Lecea M, Kondrychyn I, Fong SH, Ye ZR, Korzh V. In vivo analysis of choroid plexus morphogenesis in zebrafish. PloS one. 2008;3:e3090. doi: 10.1371/journal.pone.0003090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang X, et al. Sonic hedgehog signaling regulates a novel epithelial progenitor domain of the hindbrain choroid plexus. Development. 2009;136:2535–2543. doi: 10.1242/dev.033795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lun MP, et al. Spatially heterogeneous choroid plexus transcriptomes encode positional identity and contribute to regional CSF production. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2015;35:4903–4916. doi: 10.1523/JNEUROSCI.3081-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang X, et al. Transventricular delivery of Sonic hedgehog is essential to cerebellar ventricular zone development. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:8422–8427. doi: 10.1073/pnas.0911838107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lehtinen MK, et al. The choroid plexus and cerebrospinal fluid: emerging roles in development, disease, and therapy. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:17553–17559. doi: 10.1523/JNEUROSCI.3258-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spatazza J, et al. Choroid-plexus-derived Otx2 homeoprotein constrains adult cortical plasticity. Cell reports. 2013;3:1815–1823. doi: 10.1016/j.celrep.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.South AP, Cho RJ, Aster JC. The double-edged sword of Notch signaling in cancer. Seminars in cell & developmental biology. 2012;23:458–464. doi: 10.1016/j.semcdb.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koch U, Radtke F. Notch signaling in solid tumors. Current topics in developmental biology. 2010;92:411–455. doi: 10.1016/S0070-2153(10)92013-9. [DOI] [PubMed] [Google Scholar]
- 57.Groth C, Fortini ME. Therapeutic approaches to modulating Notch signaling: current challenges and future prospects. Seminars in cell & developmental biology. 2012;23:465–472. doi: 10.1016/j.semcdb.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mahjoub MR, Stearns T. Supernumerary centrosomes nucleate extra cilia and compromise primary cilium signaling. Current biology : CB. 2012;22:1628–1634. doi: 10.1016/j.cub.2012.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mahjoub MR. The importance of a single primary cilium. Organogenesis. 2013;9:61–69. doi: 10.4161/org.25144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stasiulewicz M, et al. A conserved role for Notch signaling in priming the cellular response to Shh through ciliary localisation of the key Shh transducer Smo. Development. 2015;142:2291–2303. doi: 10.1242/dev.125237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kong JH, et al. Notch activity modulates the responsiveness of neural progenitors to sonic hedgehog signaling. Developmental cell. 2015;33:373–387. doi: 10.1016/j.devcel.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marcet B, et al. Control of vertebrate multiciliogenesis by miR-449 through direct repression of the Delta/Notch pathway. Nature cell biology. 2011;13:693–699. doi: 10.1038/ncb2241. [DOI] [PubMed] [Google Scholar]
- 63.Morimoto M, et al. Canonical Notch signaling in the developing lung is required for determination of arterial smooth muscle cells and selection of Clara versus ciliated cell fate. Journal of cell science. 2010;123:213–224. doi: 10.1242/jcs.058669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tan FE, et al. Myb promotes centriole amplification and later steps of the multiciliogenesis program. Development. 2013;140:4277–4286. doi: 10.1242/dev.094102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Imai M, Tominaga J, Matsumae M. Choroid plexus papilloma originating from the cerebrum parenchyma. Surgical neurology international. 2011;2:151. doi: 10.4103/2152-7806.86228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Louis DNOH, Wiestler OD, Cavenee WK, editors. WHO Classification of tumours of the central nervous system. IARC Press; Lyon: 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao H, Ayrault O, Zindy F, Kim JH, Roussel MF. Post-transcriptional down-regulation of Atoh1/Math1 by bone morphogenic proteins suppresses medulloblastoma development. Genes & development. 2008;22:722–727. doi: 10.1101/gad.1636408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim J, Lee JJ, Kim J, Gardner D, Beachy PA. Arsenic antagonizes the Hedgehog pathway by preventing ciliary accumulation and reducing stability of the Gli2 transcriptional effector. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:13432–13437. doi: 10.1073/pnas.1006822107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yaylaoglu MB, et al. Comprehensive expression atlas of fibroblast growth factors and their receptors generated by a novel robotic in situ hybridization platform. Developmental dynamics : an official publication of the American Association of Anatomists. 2005;234:371–386. doi: 10.1002/dvdy.20441. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.