Summary
Perturbation in the transcriptional control of genes driving differentiation is an established paradigm whereby oncogenic fusion proteins promote leukemia. From a retinoic acid (RA) sensitive Acute Promyelocytic Leukemia (APL) cell line, we derived an RA-resistant clone characterized by a block in transcription initiation, despite maintaining wild-type PML/RARA expression. We uncovered an aberrant interaction between PML/RARA, Nucleophosmin (NPM) and Topoisomerase II Beta (TOP2B). Surprisingly, RA stimulation in these cells results in enhanced chromatin association of the nucleosome remodeler BRG1. Inhibition of NPM or TOP2B abrogated BRG1 recruitment. Furthermore, NPM inhibition and targeting BRG1 restored differentiation when combined with RA. Here, we demonstrate a role for NPM and BRG1 in obstructing RA-differentiation and implicate chromatin remodeling in mediating therapeutic resistance in malignancies. NPM mutations are the most common genetic change in patients with acute leukemia (AML) therefore, our model may be applicable to other more common leukemias driven by NPM.
Introduction
Nuclear receptors are ligand-activated transcription factors that transduce messages carried by signaling molecules into transcriptional responses. The retinoid receptor alpha (RARA) finely tunes granulocytic differentiation by acting as a transcriptional regulator of genes involved in this program (Kastner et al., 2001).
In the absence of ligand, RARA is bound to DNA along with its partner receptor, the retinoid X receptor (RXR), and co-repressors (Heinzel et al., 1997; Horlein et al., 1995; Kurokawa et al., 1995). Upon binding ligand, RARA undergoes a conformational change, releasing co-repressors, and recruiting an arsenal of co-activator proteins that facilitate the recruitment of RNA polymerase II (RNAPII) and the general transcription factors (GTFs) to the promoter (Dilworth and Chambon, 2001; Shao et al., 2000). Several chromatin-remodeling complexes make direct physical interactions with RARA and carry out structural modifications of chromatin to regulate transcription. BRG1, the ATPase subunit of the SWI/SNF complex, plays a critical role in differentiation through regulation of gene expression and is required for transactivation by many nuclear receptors, including RARA (Dilworth et al., 2000).
The importance of RARA in granulopoiesis is evident in Acute Promyelocytic Leukemia (APL). APL is a form of acute myeloid leukemia (AML) characterized clinically by an accumulation of immature promyelocytes in the bone marrow and peripheral blood, stemming from a blockage in myeloid differentiation (Collins et al., 1990; Melnick and Licht, 1999). The majority of APL patients respond to the differentiating action of pharmacological concentrations of all-trans retinoic acid (RA), a vitamin A derivative. In fact, this treatment was the first example of a successful therapeutic approach inducing differentiation rather than cytotoxicity, and it has since become the prototype for differentiation therapy in cancer. Although treatment with RA alone results in a complete remission, a significant proportion of patients relapse and subsequently develop RA resistance, a phenomenon that can be modeled in vitro (Gallagher, 2002; Rosenauer et al., 1996).
At the molecular level, APL blasts harbor a chromosomal translocation involving the RARA gene located on chromosome 17 (Melnick and Licht, 1999; Rowley et al., 1977). Numerous fusion partners of RARA have been identified, but the PML gene of chromosome 15 is the most common translocation site. Approximately 95% of affected individuals have the (15;17) translocation, producing the PML/RARA chimera (Jurcic et al., 2007). PML/RARA acts as a dominant negative inhibitor of retinoid receptor function. The fusion protein still binds DNA, heterodimerizes with RXR, and binds RA (Benedetti et al., 1997; Dong et al., 1996; Jansen et al., 1995; Perez et al., 1993). However, PML/RARA is a much more potent repressor than RARA, as it is unresponsive to physiological concentrations of ligand, such that co-repressors are not released and RA target genes remain unexpressed (Grignani et al., 1998; He et al., 1998; Lin et al., 1998).
APL cell lines are a useful model to study the conversion of transcription factors into oncogenic facilitators in other hematological malignancies. Additionally, in vitro-derived RA-resistant cells provide clues into the mechanisms of RA resistance in APL. We have isolated 3 RA-resistant clones from the parental RA-sensitive cell line NB4, denoted MR2, MR4 and MR6 (Rosenauer et al., 1996).
We previously reported that RA-resistance in the MR2 cell line, which retains wild-type PML/RARA, is associated with an altered pattern of high-molecular weight complexes binding to PML/RARA (Rosenauer et al., 1996). We furthered this observation by identifying 8 members of these complexes. These include Nucleophosmin (NPM), a nucleolar protein (Prestayko et al., 1974) intimately linked with the development of AML (Falini et al., 2005), and another was Topoisomerase II Beta (TOP2B), which we characterized as playing a central role in RA-resistance (McNamara et al., 2008). NPM plays important roles in the regulation of cell proliferation and apoptosis and is more highly expressed in malignant and proliferating cells than in normal cells (Chan et al., 1989; Feuerstein and Mond, 1987). Conversely, NPM expression is down-regulated in cells undergoing differentiation (Hsu and Yung, 1998; Hsu and Yung, 2003). Strikingly, NPM1 is commonly mutated in leukemic blasts in a high proportion of patients with AML with the most common mutations leading to aberrant cytoplasmic translocation of this nucleolar phosphoprotein (termed NPMc+). However, it remains unclear how cells harboring elevated NPM achieve malignant properties. Here we characterize the role of NPM as a transcriptional co-repressor of the PML/RARA oncoprotein and a key mediator of the differentiation block observed in RA-resistant APL cells.
Results
Levels of PML/RARA-interacting proteins are elevated in the MR2 resistant line
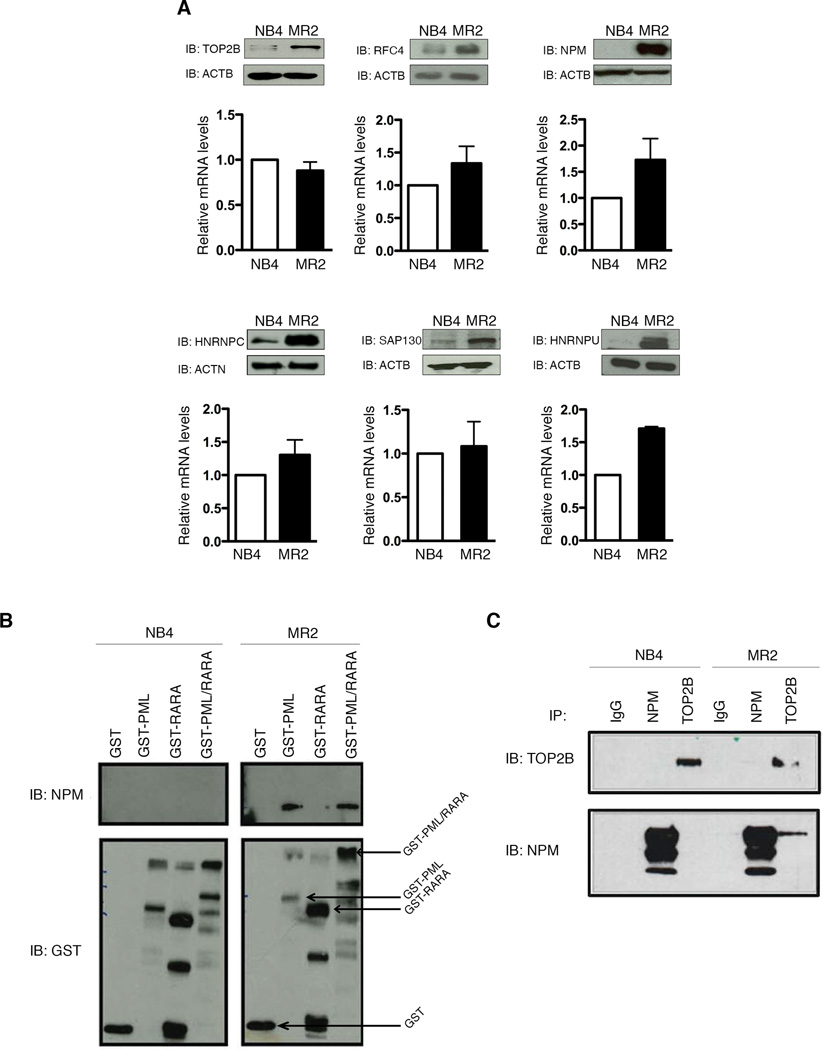
Previous work using high-performance liquid chromatography (HPLC) analysis of PML/RARA in the RA-resistant MR2 line revealed the formation of higher molecular-weight PML/RARA complexes, not evident in the parental NB4 line (Rosenauer et al., 1996). Using mass spectrometry, we identified 8 members of these complexes with increased interaction with PML/RARA in MR2, compared to NB4 (Table S1). Examination of the expression profile of 6 of these reveals a strong up-regulation at the protein level in the MR2 versus the NB4, without a corresponding robust increase in mRNA (Figure 1A). This up-regulation is selective, as several other known nuclear receptor co-regulators have the same, or even decreased, expression, in MR2 (Figure S1A).
Figure 1. NPM interacts with PML/RARA and TOP2B in the MR2 resistant cells only.
(A) Differential protein (top panels) and mRNA (bottom panels) levels of six proteins identified as being differentially associated with a GST-PML/RARA fusion in MR2 cells versus NB4 cells. (B) GST pull-down assay defines PML/RARA domains that mediate interaction with NPM. Purified GST, GST-PML, GST-RARA and GST-PML/RARA protein were incubated with nuclear extracts from NB4 and MR2 cells (C) Endogenous co-immunoprecipitation with TOP2B and NPM antibodies followed by immunoblotting indicates an interaction between NPM and TOP2B solely in MR2 cells.
We examined in more detail the upregulation of NPM levels, given this protein’s central role in a large proportion of adult leukemia (Falini et al., 2005). Despite higher levels of NPM protein in the MR2 line, expression of Nucleolin (NCL), another major nucleolar protein overexpressed in rapidly dividing cells, remained the same between NB4 and MR2 (Figure S1B). The differences in NPM levels between the two cell lines are not due to differences in the solubility of NPM (Figure S1C). Furthermore, our resistant MR2 cells do not have the NPMc+ mutation, as direct sequencing of NPM by traditional Sanger sequencing revealed no differences in the NPM1 sequence (data not shown) and NPM is located exclusively within the nucleus, with no evidence of cytoplasmic NPM (Figures S1D–E). Finally, an RNA-seq based transcriptome analysis of both cell lines confirmed the minimal difference in NPM transcript levels, and showed no differences in promoter, terminator or exon usage that would account for the dramatic increase in NPM protein levels (Figure S1F). However, we also called variants from our RNA-Seq data to identify genomic differences between our two cell lines (Table S2). There are 3 variants in the NPM1 gene (one in the 5’ UTR and two intronic) in the MR2 cell line, which might be involved in the enhanced translation of NPM.
NPM interacts with PML/RARA only in the resistant cell line
We confirmed our mass spectrometry results by performing a GST pull-down with untreated NB4 and MR2 nuclear extracts (Figure 1B). An association between NPM and PML/RARA is detectable in MR2 only, and this interaction is mediated through the PML moiety of the fusion.
We previously validated TOP2B as another aberrantly associated PML/RARA protein in the MR2 line (McNamara et al., 2008). We therefore used endogenous co-immunoprecipitation (co-IP, Figure 1C) to show that NPM and TOP2B also associate. Consistent with the previous results, pull-down with a TOP2B antibody results in pull down of NPM from MR2 cellular extracts only.
NPM localizes to the CEBPB gene locus in MR2 cells
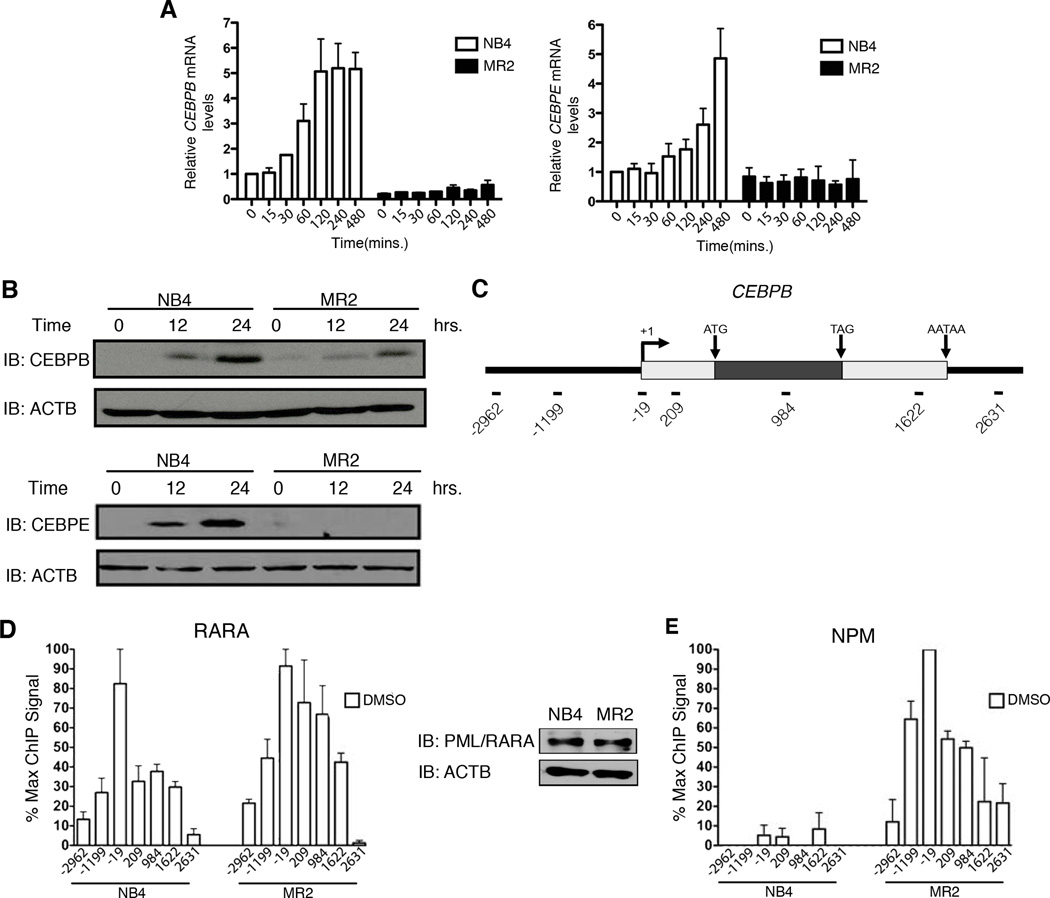
The PML/RARA fusion protein blocks myeloid differentiation by repressing RA-mediated gene expression and neutrophil maturation. Quantitative real-time PCR (Q-RT-PCR) analyses over an 8 hour RA treatment reveal induction of 4 RARA target genes in NB4 cells, while the same accumulation is not observed in the RA-resistant cell line, MR2 (Figure 2A and Figure S2A).
Figure 2. Defective transcriptional activation of RARA target genes upon RA treatment in MR2 correlates with increased NPM at the promoter.
(A) Q-RT-PCR analysis of CEBPB and CEBPE mRNA induction following 8h RA treatment expressed as fold induction over untreated cells after normalization to 18S rRNA levels. Error bars represent the standard error of the mean. (B) Immunoblot analysis demonstrating differential CEBPB and CEBPE protein expression in response to RA treatment (C) Schematic of the CEBPB locus indicating the overall gene structure. Amplicons used in real-time PCR quantification of ChIP-enriched DNA are named according to their relative distance (bps) to the transcription start site. (D) High-density ChIP tiling of RARA at the CEBPB locus under basal conditions in NB4 and MR2 cells and immunoblot analysis of PML/RARA expression levels in the two cell lines. (E) High-density ChIP tiling of NPM at the CEBPB locus under basal conditions in NB4 and MR2 cells.
Two well established RA-target genes, CEBPbeta (CEBPB) (Duprez et al., 2003) and CEBPepsilon (CEBPE) (Chih et al., 1997) both play a critical role in the differentiation of the myeloid lineage. The mRNA levels of these genes correlate with their respective protein levels, as assessed by Western blotting (Figure 2B).
Normal transcriptional regulation in response to RA requires the coordinated action of RARA and a variety of cofactor complexes (Bastien and Rochette-Egly, 2004). In order to study the exact steps of transcriptional activation in the presence of the PML/RARA fusion and at which step a transcriptional blockade occurs in the resistant cells, we performed high-resolution quantitative ChIP analysis. This assay enables us to generate detailed maps of protein occupancy on the CEBPB locus after stimulating the transactivation process. Figure 2C shows the most relevant features of this gene including the location of 7 amplicons used for Q-RT-PCR quantification of the ChIP-enriched DNA. Both NB4 and MR2 express similar levels of the fusion protein, and initial ChIP profiling of the CEBPB locus under basal conditions shows similar recruitment levels and patterns of localization for RARA in both cell lines (Figure 2D). However, a dramatically enhanced association of NPM is observed exclusively in the resistant cells (Figure 2E).
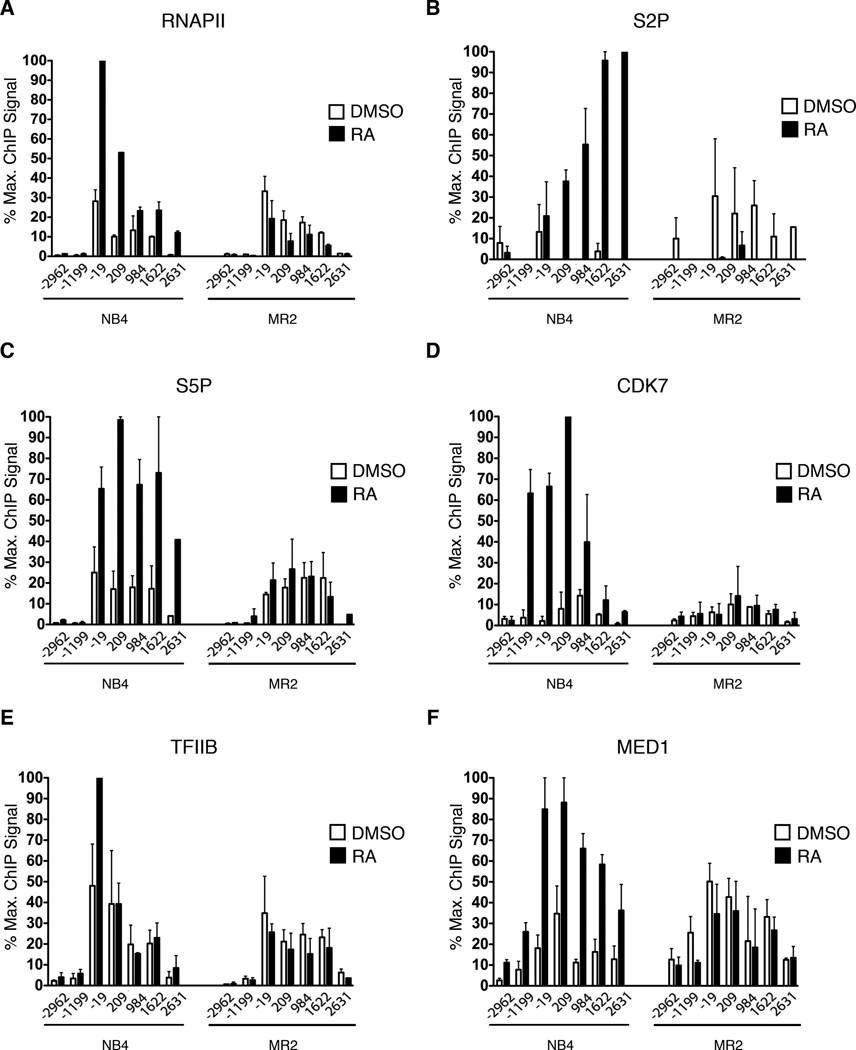
Differential RNAPII patterning and pre-initiation complex recruitment between sensitive and resistant cells at CEBPB in the RA-activated state
RNAPII activity can be stimulated at various stages of transcription by the action of myriad regulators of recruitment and post-recruitment steps. CEBPB shows little preloaded RNAPII in both the sensitive and resistant cells (Figure 3A). Upon transcriptional activation with RA, the total amount of RNAPII associated with the proximal promoter increases in NB4 cells, but not in MR2 cells (Figure 3A). This suggests that RA activates this gene, at least in part, by promoting RNAPII recruitment. RNAPII phosphorylation of the Serine 5 and 2 residues (S5P and S2P) of the carboxy-terminal domain (CTD) repeats occurs at post-recruitment steps and is catalyzed by protein kinases in the initiation and elongation complexes, respectively (Sims et al., 2004). In the sensitive cells, S5P and S2P patterning on both CEBPB and CEBPE increases strongly upon activation. S5P, a mark of active transcription initiation is increased upon treatment at the 5’ end of the gene. S2P, a mark of actively elongating RNAPII, increases towards the 3’ end of the gene (Figure 3B–C and Figures S2B–D). In contrast, neither of these signals increases in response to RA treatment in the resistant cells.
Figure 3. Differential recruitment of transcription initiating factors to CEBPB in response to RA treatment.
(A–F) High-density ChIP tiling of RNAPII, RNAPII phosphorylation at Serine 2 (S2P) and Serine 5 (S5P), CDK7, TFIIB and MED1 at CEBPB before and after RA treatment (1h) of NB4 and MR2 cells.
The failure of MR2 cells to recruit and activate RNAPII in response to RA stimulation led us to examine the recruitment of subunits of the Pre-Initiation Complex (PIC). MR2 cells fail to recruit CDK7 (Figure 3D), the kinase subunit of the general transcription TFIIH, a key component of the PIC, and which is also the principal S5 kinase (Hengartner et al., 1998). Recruitment of additional components of the PIC, such as TFIIB and TFIIF, is also stimulated by RA in NB4, but not in MR2 (Figures 3E and Figure S2E). Upon activation by ligand, RARA interacts with and recruits the Mediator co-activator complex, which stimulates RNAPII activity by diverse mechanisms, including positive effects on PIC formation, enhancer-promoter chromatin looping and transcription elongation. The subunit of the Mediator complex that is responsible for this interaction is MED1 (Yuan et al., 1998). We have shown that MED1 interacts with the PML/RARA found in NB4 in a ligand-dependent manner (Shao et al., 2000). Furthermore, the MR2 cell line maintained normal MED1 complexes that interacted with retinoid receptors in a ligand-dependent manner, when assessed by GST pull-down assay. However, by conducting ChIP tiling analyses, we now show that RA fails to recruit MED1 to the endogenous CEBPB promoter in MR2 (Figure 3F). Together, these data demonstrate that the molecular mechanism of RA resistance involves a defect in RNAPII recruitment and activation at the promoter of key differentiation genes.
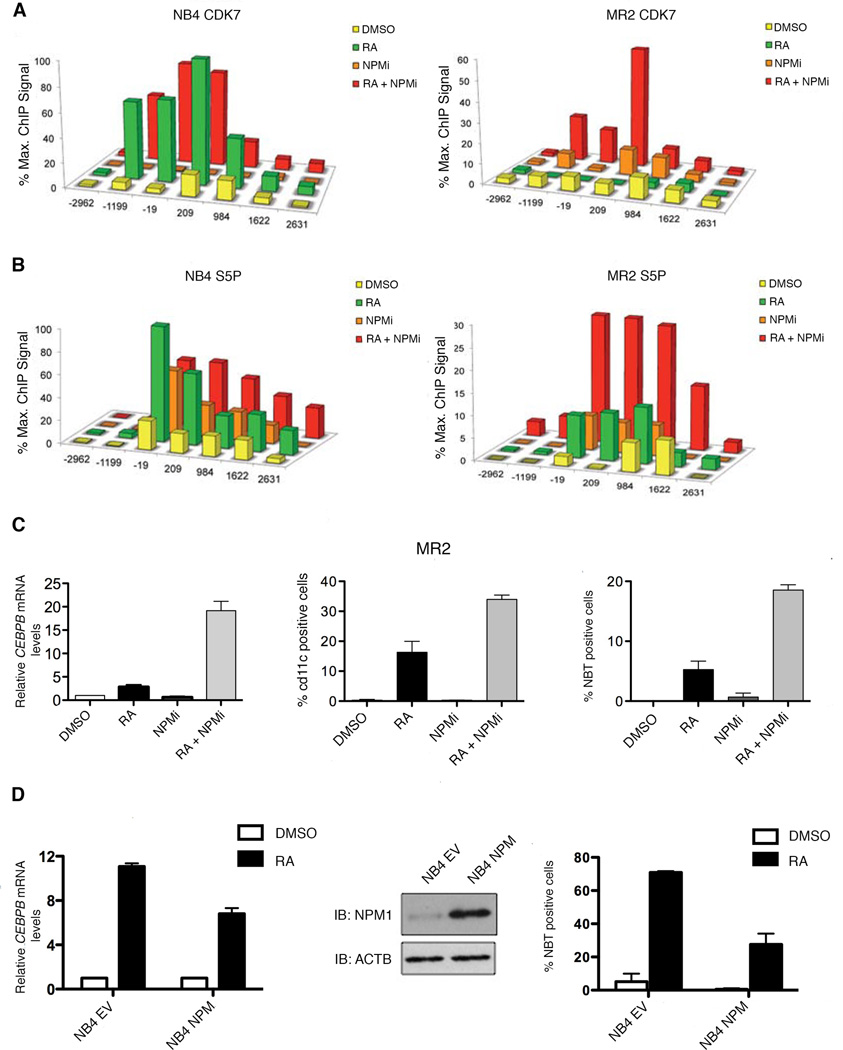
Modulation of NPM levels mediates sensitivity to RA-induced gene transcription and differentiation
To examine whether NPM overexpression and aberrant interaction with PML/RARA is responsible for the repression of RA-induced gene expression in MR2, we treated both NB4 and MR2 with the NPM inhibitor NSC348884, which has been shown to disrupt NPM’s higher order structures (Qi et al., 2008). Strikingly, in the resistant cells, NPM inhibition, in combination with RA, leads to increases in CDK7 recruitment (Figure 4A), S5 phosphorylation (Figure 4B) and restored mRNA expression of the RA-target genes (Figure 4C, left and Figures S3A–B).
Figure 4. Inhibition of NPM restores myeloid lineage differentiation in resistant cells and overexpression of NPM impairs differentiation in sensitive cells.
(A) ChIP analysis was carried out using protein extracts of DMSO, RA (1h), NPMi (16h) or NPMi and RA-treated (16h + 1h) MR2 and NB4 cells using an antibody recognizing CDK7. (B) ChIP analysis was carried out using protein extracts of DMSO, RA (1h), NPMi (16h) or NPMi and RA-treated (16h + 1h) MR2 and NB4 cells using antibodies against RNAPII S5P. (C) Q-RT-PCR analysis of CEBPB mRNA induction following DMSO, RA, NPMi or RA and NPMi treatment of MR2 cells. Data is expressed as fold induction over DMSO treated cells and normalized to 18S ribosomal RNA levels. Error bars represent the standard error (left panel). Percentages of MR2 cells expressing the differentiation marker cd11c in response to 3-day exposure to RA, NPMi, or a combination of both (middle panel). Results ofNBT reduction assay performed on MR2 cells treated with RA, NPMi, or the combination for 5 days (right panel). (D) NB4 cells were stably transfected with empty vector (NB4 EV) or the NPM expression plasmid (NB4 NPM). Overexpression of NPM was verified by separating total nuclear proteins via SDS-PAGE and followed by Western blotting using NPM antibody (middle panel). Quantitative real-time PCR analysis of CEBPB mRNA induction following 8h RA treatment expressed as fold induction over DMSO treated cells after normalization to 18S rRNA levels. Error bars represent the standard error of the mean (left panel). NBT reduction results in response to 5-day exposure to RA. Results are representative of three experiments performed in triplicate. Error bars represent standard error of the mean (right panel).
The above data led to the hypothesis that this inhibition might also abolish the differentiation block in the MR2 line. Treatment of sensitive cells with RA induces expression of the myeloid-specific cell surface marker, cd11c. We first assessed cd11c expression by FACS analysis in NB4 and MR2 cells treated for three days either with RA, NPM inhibitor (NPMi) or the combination (Figure 4C, middle and Figure S3C, top). We observed that RA alone and the combination of RA and NPMi are both sufficient to cause increased cd11c cell surface expression in NB4. However, in MR2, only the combination treatment led to the re-establishment of this differentiation marker.
To confirm that NPMi overcomes the differentiation block in APL cells, we performed a nitro-blue-tetrazolium (NBT) reduction analysis, which assesses terminal granulocytic differentiation (Klein et al., 1998). While NPMi alone had little discernible effect on differentiation in either cell line, RA and NPMi co-treatment leads to significant NBT reduction within 5 days (Figure 4C, right and Figure S3C, bottom). This is consistent with the ChIP analysis, mRNA expression and cd11c data, where only a modest response to treatment was observed in the MR2 cell line in response to 10−6 M RA.
In agreement with these data, the converse experiment, whereby overexpression of NPM is induced in the RA-sensitive line, led to RA-resistance. Figure 4D confirms overexpression of NPM. We first examined whether NPM overexpression would affect mRNA expression of RA target genes. Compared to the empty vector control, the NPM-overexpressing clone has reduced CEBPB, CEBPE, ICAM1 and CDKN1A mRNA induction (Figure 4D, left and Figure S3D). This translates to a robust decrease in NBT reduction compared to that of the NB4 control (Figure 4D, right). Cumulatively, these results support evidence that increased expression of NPM is necessary and sufficient for the inhibition of RA-induced gene expression and differentiation in an APL cell line.
RA induces recruitment of the chromatin remodeler BRG1 to CEBPB in a TOP2B/NPM-dependent fashion
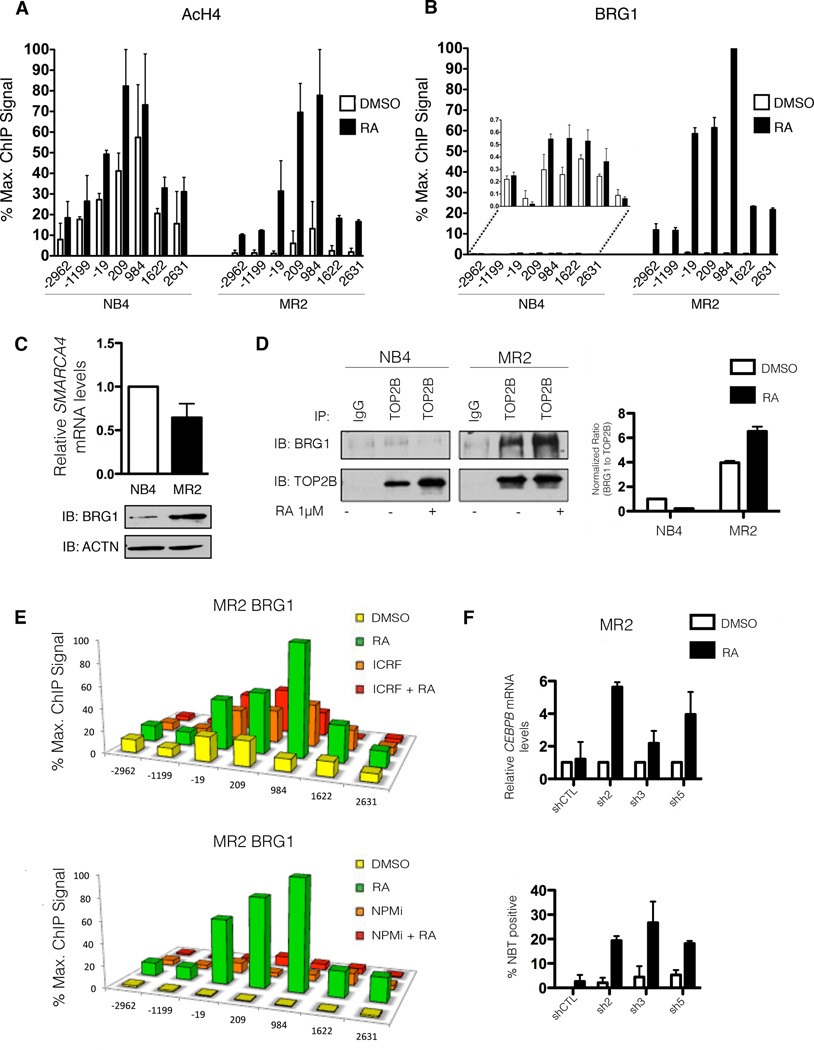
In order to activate gene expression, RARs must override repressive chromatin structures. To this end, ligand-induced conformational changes in the receptors will cause the dissociation of co-repressors and the concomitant recruitment of co-activators necessary for RNAPII recruitment (Chen et al., 1999). In the resistant and sensitive cells, there is an increase in histone H4 acetylation (Figure 5A) throughout the gene body, as well as in the promoter in response to RA. This indicates that activator-induced histone acetylation is not sufficient for RNAPII activation, an observation also made in other systems (Galbraith et al., 2013).
Figure 5. Targeting BRG1 restores sensitivity to RA-induced gene transcription and differentiation.
(A) High-density ChIP tiling of acetylated H4 (AcH4) at the CEBP locus before and after RA treatment (1h) of NB4 and MR2 cells. (B) High-density ChIP tiling of BRG1 at the CEBPB locus before and after RA treatment (1h) of NB4 and MR2 cells. (C) Immunoblot analysis demonstrating differential basal BRG1 protein expression in NB4 and MR2 cells (bottom) with no corresponding increase in BRG1 mRNA levels (top). SMARCA4 is the gene that encodes for the protein known as BRG1. (D) Endogenous co-immunoprecipitation with TOP2B antibody followed by immunoblotting for BRG1 and TOP2B indicates an interaction between BRG1 and TOP2B solely in MR2 cells (left panel). The densitometry values of immunoprecipitated BRG1 were normalized to the amount of immunoprecipitated TOP2B and reported as fold induction over DMSO treated NB4 cells. Densitometry analyses were performed with ImageJ software (right panel). (E) High-density ChIP tiling of BRG1 at CEBPB after treatment with DMSO, RA (1h), the TOP2B inhibitor ICRF (overnight), or a combined treatment of ICRF (overnight pretreatment) and RA (1h) in MR2 cells (top). High-density ChIP tiling of BRG1 at the CEBPB locus after treatment with DMSO, RA (1h), NPM inhibitor (NPMi, overnight), or a combined treatment of NPMi (overnight pretreatment) and RA (1h) in MR2 cells (bottom). (F) Q-RT-PCR analysis of CEBPB mRNA induction in BRG1 MR2-knockdown clones following 8h RA treatment expressed as fold induction over untreated cells after normalization to 18S rRNA levels. Error bars represent the standard error (top) Results of NBT reduction assay performed on BRG1 knockdown MR2 cells treated with RA for 5 days, with retreatment at day 3 (bottom).
Surprisingly, ChIP tiling upstream of transcription initiation revealed a more robust recruitment of the chromatin-remodeler ATPase subunit BRG1 to the CEBPB locus in response to RA-treatment in the resistant cells compared to the sensitive cells (Figure 5B). Examination of the BRG1 expression profile discloses a strong up-regulation of BRG1 protein in MR2 cells compared to NB4 cells (Figure 5C, bottom), without a corresponding biologically significant increase in mRNA (Figure 5C, top).
Having shown that TOP2B interacts with both PML/RARA and NPM in the resistant cells, we next used endogenous co-IP (Figure 5D, left) to investigate whether BRG1 and TOP2B associate. Pull down with TOP2B followed by immunoblotting for BRG1 reveals a basal interaction between the two proteins that is enhanced with RA treatment (Figure 5D, right and Figure S4A). In agreement with the ChIP analysis, this interaction is limited to the resistant line. To examine whether this interaction is necessary for the aberrant recruitment of BRG1 to the CEBPB locus in MR2, we treated both lines with the TOP2B inhibitor, ICRF, which induces TOP2B degradation (Xiao et al., 2003). ChIP analyses performed on cells treated with RA, ICRF or the combination (Figure 5E, top) demonstrated a direct correlation between TOP2B inhibition and reduced BRG1 recruitment in the MR2 cell line. Interestingly, treatment with ICRF in NB4 cells enhanced BRG1 recruitment, suggesting a differential functional outcome of BRG1 recruitment between the cell lines (Figure S4B, top). Similar results were obtained when both lines were treated with the NPM inhibitor alone or in combination with RA (Figure 5E, bottom and Figure S4B, bottom). Treatment with ICRF for 24 hours was sufficient to degrade TOP2B but had no discernible effect on NPM levels, nor were levels of HNRNPU, another member identified in our mass spectrometry screen, affected (Figures S4C and D). Together these results indicate that the resistant cell line expressed elevated levels of BRG1 and that recruitment of BRG1 to the promoter of RA-target genes was dependent on the repressive factors TOP2B and NPM in MR2 cells only. These data suggest NPM, TOP2B and BRG1 cooperate as a repressive complex, effectively suppressing PML/RARA-mediated transcription.
BRG1 knockdown restores sensitivity to RA
We next investigated whether BRG1 functioned as a repressor of RA-induced transcriptional activation in the MR2 cell line. Five stable BRG1 knockdown clones, along with a stable clone expressing a non-targeting shRNA were created in both the NB4 and MR2 lines. Based on the efficacy of BRG1 knockdown in MR2 (Figure S5A), we selected 3 clones (sh2, sh3 and sh5) for further analysis.
Q-RT-PCR analysis of RA-target gene expression revealed BRG1 knockdown restored sensitivity to RA treatment in the MR2 cell line, in all 3 clones tested (Figure 5F, top and Figure S5B–C). We assessed cd11c expression by FACS analysis in NB4 and MR2 clones treated for 3 days with RA (Figure S5D). RA alone and the combination of RA and BRG1 knockdown were both sufficient to cause increased cd11c cell surface expression in the RA-sensitive NB4 cells. However, in the MR2 clones, only the combination of RA with knockdown of BRG1 led to the re-establishment of this differentiation marker.
To confirm that BRG1 knockdown overcomes the differentiation block in APL cells, we again performed an NBT reduction analysis. While BRG1 knockdown alone had little effect on differentiation of any of the cell lines, RA treatment after BRG1 knockdown lead to significant NBT reduction within 5 days (Figure 5F, bottom and Figure S5E). This is consistent with the mRNA expression and cd11c data, where only a modest response to treatment was observed in the MR2 cell line in response to 10−6 M RA. Cumulatively, these results indicate that knockdown of BRG1 in resistant cells restored sensitivity to RA-mediated differentiation, from early gene expression to terminal functional capacity. Moreover, these results substantiate the hypothesis that BRG1 acts as a transcriptional repressor in the MR2 cell line.
Discussion
We observed that PML/RARA associated with a higher molecular weight complex in MR2 cells than NB4 cells. Subsequent mass spectrometry identified 8 proteins that had previously undescribed interactions with PML/RARA. We found that 2 of these, TOP2B and NPM, mediate repression in MR2, as inhibition of these proteins resulted in a loss of resistance to RA. We thus hypothesize the existence of a co-repressor complex in MR2 that interacts with PML/RARA and represses genes critical for cellular differentiation. We support this by demonstrating that both TOP2B and NPM are necessary for the recruitment of BRG1 and that functional inhibition of these proteins abates BRG1’s presence at CEBPB in MR2 cells. Similarly, many of the other proteins identified in the MR2 PML/RARA complex (Table S1) have been shown to interact with BRG1. For example, SAP130 associates with the co-repressor complex mSin3A, which incorporates BRG1 under certain conditions (Fleischer et al., 2003). HNRNPU has been shown to form a complex with BRG1 that is necessary for RNAPII mediated transcriptional activity and interactions between HNRNPC1/C2 and the SWI/SNF complex, which includes BRG1, have also been reported (Mahajan et al., 2005; Vizlin-Hodzic et al., 2011). Most interestingly, PML itself has been shown to mediate recruitment of BRG1 to the Oct4 promoter (Chuang et al., 2011).
Our data define a function for NPM as a negative regulator of RA-induced gene regulation and differentiation toward granulocytes. In RA-resistant cells, we show an interaction between PML/RARA, the fusion most commonly underlying APL, and NPM that is mediated through the PML portion of the chimera. The presence of NPM is inversely correlated with PIC formation at the CEBPB locus, and pharmacological targeting of NPM in combination with RA relieved the inhibition of transcription exerted by this protein.
NPM is irrefutably linked to human tumorigenesis. Disruption of the NPM gene by translocation is found in human hematopoietic malignancies, and NPM appears to contribute to oncogenesis by activating the oncogenic potential of the fused protein partner (Bischof et al., 1997; Fujimoto et al., 1996). Moreover, NPM acts as a transcriptional co-repressor during RA-induced differentiation of HL60 cells (Liu et al., 2007). NPM enhances the proliferative potential of hematopoietic stem cells, and NPM overexpression increases HSC survival upon DNA damage and oxidative stress (Li et al., 2006). Variant calling from our RNA-Seq data identified 3 variants in the NPM1 gene in the MR2 cell line. These data are especially intriguing given the recent report of two AML patients with NPM mutations in the 3’UTR (Abraham et al., 2014). Coupled with our data, these interesting findings highlight the importance of untranslated regions in NPM expression in leukemia.
The RNA-Seq variant calling also revealed a putative mutation in the Ligand Binding Domain in PML/RARA in the MR2 cell line. However, we have several lines of evidence that lead us to believe that this variant is not a significant contributor to RA-resistance. First, the genomic sequence of PML/RARA in MR2 is not altered. Our group has previously published a characterization of 3 RA-resistant subclones, including this MR2 clone, derived from NB4 (Shao et al., 1997). As we reported, these 3 resistant clones were subjected at that time to DNA sequencing. Sequencing revealed a point mutation in the PML/RARA sequence of only 1 of the 3 sub-clones, MR4. To confirm this finding, we recently re-sequenced the region surrounding the identified variant with Sanger sequencing and verified that the sequence in MR2 is indeed wild-type (Figure S1G). Second, the 3H-t[RA] HPLC profiles (Rosenauer et al., 1996) showed no peak at a high a higher molecular weight in MR4, the clone with the genomic alteration in PML/RARA sequence, indicating that RA binding to PML/RARA does not occur in this clone. In contrast, the MR2 clone not only shows a high molecular weight peak, this peak is higher than what corresponds to PML/RARA in NB4. This interesting piece of data, showing that RA binds to an aberrant PML/RARA complex in MR2, is the starting point for our studies to identify the members of that complex and how those impact RA resistance. Finally, if the mutation in the LBD of the PML/RARA in MR2 cells affected the functionality of the PML/RARA, we would hypothesize that even after removing all the other barriers to the RA response (such as TOP2B, NPM and BRG1), we would see no transactivation in response to RA. As is shown in this manuscript and our previous publication (McNamara and Miller, 2008), this is clearly not the case. Inhibition of TOP2B, NPM or BRG1 enables the RA transactivation function of the PML/RARA in the MR2 cell line, as clearly demonstrated by ChIP assays, RA-target gene mRNA levels and ultimately, cellular differentiation.
Transcriptional profiling by ChIP analysis surprisingly revealed that RA treatment of our resistant cells was associated with enhanced recruitment of BRG1. BRG1 is an ATPase helicase subunit of the SWI/SNF family of proteins that serves to regulate gene expression by altering the chromatin landscape surrounding genes (Wilson and Roberts, 2011). Thus, BRG1 can function both to activate and repress gene expression (Murphy et al., 1999). We hypothesized that transcriptional repression by BRG1 mediates resistance to RA-induced differentiation in MR2. Abrogation of BRG1 activity restored sensitivity to RA as observed by early gene expression, cd11c surface expression, and functional capacity of differentiated cells.
BRG1 as a transcriptional repressor has been demonstrated in other contexts. For example, Ooi et al. showed that BRG1 enhanced REST-mediated repression by recruiting and stabilizing binding of this repressor complex to chromatin (Ooi et al., 2006). BRG1 also interacts with and recruits DNA methyltransferases, thus promoting gene silencing by DNA methylation (Datta et al., 2005). Additionally, BRG1 interacts with co-repressive complexes such as mSin3A/HDAC2/PRMT5, which promote histone tail deacetylation and methylation and subsequent chromatin condensation (Pal et al., 2003). Our results are congruent with the above findings and provide a setting in which BRG1 serves as a mediator of transcriptional repression. However, whether or not this is dependent upon its nucleosomal repositioning capacity remains a topic of current investigation. Based on the data presented herein, we hypothesize that BRG1 might act to further contribute to a more heterochromatic architecture at the promoters of RA target genes in the MR2 cell line.
Cellular mechanisms underlying resistance to chemotherapeutic agents have been carefully studied, as experimental models can be easily generated via in vitro selection. Resistance to the differentiating effects of RA in APL cells in vitro and in vivo is traditionally associated with a mutation in the ligand-binding domain of the PML/RARA oncogenic fusion protein (Shao et al., 1997). In contrast, we report that stable resistance to RA in an acute leukemia cell line, in the context of a wild-type PML/RARA, is driven by an aberrant association with a putative co-repressor complex containing NPM and TOP2B, leading to recruitment of BRG1 to RA-target genes.
In summary, while the challenge of overcoming the maturation block in APL resistant cells has been met in vitro, it will be important to determine the relevance of our findings to other forms of leukemia. Additionally, while RA is validated as an effective frontline strategy in APL, our understanding of the molecular underpinnings of APL pathology and the response to RA remains tenuous. Elucidation of these may shed light onto common etiologies for other AML subtypes, particularly those with NPM mutations and fusions, and lead to the development of therapeutic strategies for the less curable forms of this disease.
Experimental Procedures
Materials
RPMI 1640 and fetal bovine serum were purchased from Wisent. All-trans RA and the Nucleophosmin inhibitor, NSC 348884, were purchased from Sigma-Aldrich and Axon Medchem BV respectively. The TOP2B inhibitor ICRF-193 was obtained from Biomol.
Cell Culture
The RA-resistant cell line MR2 was derived from the parental APL cell line NB4 (Rosenauer et al., 1996). All cell lines were maintained in RPMI-1640 supplemented with 10% fetal bovine serum and penicillin/streptomycin (Wisent). RNA Extraction and Analysis. mRNA was isolated using the Absolutely RNA® Miniprep kit (Agilent Technologies) and cDNA was generated from 1µg total RNA using the iScript cDNA Synthesis Kit (Bio-Rad Laboratories). We determined relative mRNA levels, normalized to endogenous 18S rRNA (TaqMan® hs99999901_s1 probe, Applied Biosystems), using SYBR Green I chemistry in an ABI 7500 Fast Real-time PCR machine (Applied Biosystems). Table S3 shows primer sequences. Western Blotting. See Supplemental Experimental Procedures.
Chromatin Immunoprecipitation (ChIP)
ChIP analyses were performed as detailed in Gomes et al. (Gomes et al., 2006); for details see Supplemental Experimental Procedures.
Co-Immunoprecipitation
Approximately 7×106 cells were harvested and lysed in IP lysis buffer (20 mM Tris-HCL pH 7.5, 420 mM NaCl, 2 mM MgCl2, 10% Glycerol, 0.5% Glycerol, 0.5% NP-40 and 0.5% Triton-X) completed with protease inhibitors. 2 mg of protein per condition, diluted in 0.5% Tritron-X IP buffer, were pre-cleared for two hours with protein G sepharose, after which the beads were removed. TOP2B or IgG antibody were added overnight at 4°C. Immune complexes were recovered with protein G sepharose, washed 3 times in 0.5% Triton-X IP buffer, once in 0.1% Triton-X IP buffer, then boiled in 2× SDS sample buffer, and subsequently analyzed by Western blot.
Stable NPM Transfectants
Transfection of lentiviral constructs encoding NPM1-HA and the Puro-HA-empty vector control (Applied Biological Materials Inc.) were performed using Lipofectamine 2000 (Carlsbad) with Opti-MEM media (Invitrogen). Plasmids were co-transfected with three lentivirus packaging constructs, PLP1, VSVG, PLP2, into 293FT cells. Forty-eight hours post-transfection, the virus supernatant was collected, centrifuged, and filtered through a 0.45 µm filter. Two million cells were transduced by centrifugation at 800g for 30 min. and maintained for 48h at 37 °C with 5% CO2 before beginning selection with puromycin.
shRNA-Mediated Knockdown
Cell lines stably transduced with shRNAs targeting BRG1 were established using a 24h polybrene (5 mg/ml) transduction of NB4 and MR2 cells with Mission™ TRC lentiviral particles (Sigma-Aldrich). Approximately 1×106 cells were infected with 200,000 lentiviral units in 2 ml of complete RPMI 1640 media. At Day 5 post-transduction, the cells were given fresh media supplemented with 1 µg/ml puromycin (Invitrogen) for selection purposes.
Differentiation Assays
Cell surface expression of cd11c (BD Biosciences, Franklin Lakes, NJ, USA) was determined by flow-assisted cell sorting according to the antibody manufacturer's specifications using the FACSCalibur flow cytometer. Background staining was controlled using an isotype control PE-conjugated mouse IgG1 kappa (BD Biosciences). For each sample, viable cells were gated, and expression of cd11c surface markers of 1×104 cells was evaluated. Nitro-blue-tetrazolium (NBT) reduction assays were performed as described (Momparler et al., 1990). The fraction of NBT-positive (blue) cells was determined by counting using a hemocytometer.
Supplementary Material
Acknowledgments
We are grateful to Michael Witcher, Sonia del Rincon, Audrey Emond and the Eukaryotic Gene Expression course at CSHL for critical reading of the paper and insightful scientific discussions. We also thank Ryan Henry and Jennie Sims for technical assistance. This work was supported by the Canadian Institutes of Health Research (MOP-12863), and NIH grants 2R01CA117907-08 and 5P30CA046934-27. W.H. Miller, Jr., was a Chercheur National of Fonds de la Recherche en Santé du Québec. J.N. Nichol was supported by a student fellowship from the Cole Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions:
J.N,N, M.D.G., J.M.E. and W.H.M. designed the study. J. N.N. and M.D.G. performed the experiments. C.L.K. performed the analyses of the RNA-Seq. J.N.N. and W.H.M. wrote the manuscript.
Statistical Analyses
All results are an average of three independent experiments. Error bars represent the standard error of the mean determined using GraphPad Prism software. The results of the ChIP experiments in Figures 4, S4 and 5 are the average of at least two independent PCRs from 2 separate immunoprecipitations from two independent cell cultures.
Accession Numbers
The accession number for the RNA-Seq data reported in this paper is SRA: SRP070180.
References
- Abraham A, Karathedath S, Kumaraswamy V, Jayavelu AK, Mani S, Srivastava VM, Zhang W, Zhou T, George B, Srivastava A, et al. Novel NPM1 mutation in the 3'-untranslated region identified in two patients with acute myeloid leukemia. Leukemia & lymphoma. 2014;55:1421–1424. doi: 10.3109/10428194.2013.838235. [DOI] [PubMed] [Google Scholar]
- Bastien J, Rochette-Egly C. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene. 2004;328:1–16. doi: 10.1016/j.gene.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Benedetti L, Levin AA, Scicchitano BM, Grignani F, Allenby G, Diverio D, Lo Coco F, Avvisati G, Ruthardt M, Adamo S, et al. Characterization of the retinoid binding properties of the major fusion products present in acute promyelocytic leukemia cells. Blood. 1997;90:1175–1185. [PubMed] [Google Scholar]
- Bischof D, Pulford K, Mason DY, Morris SW. Role of the nucleophosmin (NPM) portion of the non-Hodgkin's lymphoma-associated NPM-anaplastic lymphoma kinase fusion protein in oncogenesis. Molecular and cellular biology. 1997;17:2312–2325. doi: 10.1128/mcb.17.4.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan WY, Liu QR, Borjigin J, Busch H, Rennert OM, Tease LA, Chan PK. Characterization of the cDNA encoding human nucleophosmin and studies of its role in normal and abnormal growth. Biochemistry. 1989;28:1033–1039. doi: 10.1021/bi00429a017. [DOI] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Xie W, Wilpitz D, Evans RM. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- Chih DY, Chumakov AM, Park DJ, Silla AG, Koeffler HP. Modulation of mRNA expression of a novel human myeloid-selective CCAAT/enhancer binding protein gene (C/EBP epsilon) Blood. 1997;90:2987–2994. [PubMed] [Google Scholar]
- Chuang YS, Huang WH, Park SW, Persaud SD, Hung CH, Ho PC, Wei LN. Promyelocytic leukemia protein in retinoic acid-induced chromatin remodeling of Oct4 gene promoter. Stem cells. 2011;29:660–669. doi: 10.1002/stem.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SJ, Robertson KA, Mueller L. Retinoic acid-induced granulocytic differentiation of HL-60 myeloid leukemia cells is mediated directly through the retinoic acid receptor (RAR-alpha) Mol Cell Biol. 1990;10:2154–2163. doi: 10.1128/mcb.10.5.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta J, Majumder S, Bai S, Ghoshal K, Kutay H, Smith DS, Crabb JW, Jacob ST. Physical and functional interaction of DNA methyltransferase 3A with Mbd3 and Brg1 in mouse lymphosarcoma cells. Cancer research. 2005;65:10891–10900. doi: 10.1158/0008-5472.CAN-05-1455. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dilworth FJ, Chambon P. Nuclear receptors coordinate the activities of chromatin remodeling complexes and coactivators to facilitate initiation of transcription. Oncogene. 2001;20:3047–3054. doi: 10.1038/sj.onc.1204329. [DOI] [PubMed] [Google Scholar]
- Dilworth FJ, Fromental-Ramain C, Yamamoto K, Chambon P. ATP-driven chromatin remodeling activity and histone acetyltransferases act sequentially during transactivation by RAR/RXR In vitro. Molecular cell. 2000;6:1049–1058. doi: 10.1016/s1097-2765(00)00103-9. [DOI] [PubMed] [Google Scholar]
- Dong S, Zhu J, Reid A, Strutt P, Guidez F, Zhong HJ, Wang ZY, Licht J, Waxman S, Chomienne C, et al. Amino-terminal protein-protein interaction motif (POZ-domain) is responsible for activities of the promyelocytic leukemia zinc finger-retinoic acid receptor-alpha fusion protein. Proc Natl Acad Sci U S A. 1996;93:3624–3629. doi: 10.1073/pnas.93.8.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duprez E, Wagner K, Koch H, Tenen DG. C/EBPbeta: a major PML-RARA-responsive gene in retinoic acid-induced differentiation of APL cells. The EMBO journal. 2003;22:5806–5816. doi: 10.1093/emboj/cdg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, La Starza R, Diverio D, Colombo E, Santucci A, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352:254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- Feuerstein N, Mond JJ. "Numatrin," a nuclear matrix protein associated with induction of proliferation in B lymphocytes. The Journal of biological chemistry. 1987;262:11389–11397. [PubMed] [Google Scholar]
- Fleischer TC, Yun UJ, Ayer DE. Identification and characterization of three new components of the mSin3A corepressor complex. Molecular and cellular biology. 2003;23:3456–3467. doi: 10.1128/MCB.23.10.3456-3467.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto J, Shiota M, Iwahara T, Seki N, Satoh H, Mori S, Yamamoto T. Characterization of the transforming activity of p80, a hyperphosphorylated protein in a Ki-1 lymphoma cell line with chromosomal translocation t(2;5) Proc Natl Acad Sci U S A. 1996;93:4181–4186. doi: 10.1073/pnas.93.9.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith MD, Allen MA, Bensard CL, Wang X, Schwinn MK, Qin B, Long HW, Daniels DL, Hahn WC, Dowell RD, Espinosa JM. HIF1A employs CDK8-mediator to stimulate RNAPII elongation in response to hypoxia. Cell. 2013;153:1327–1339. doi: 10.1016/j.cell.2013.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher RE. Retinoic acid resistance in acute promyelocytic leukemia. Leukemia. 2002;16:1940–1958. doi: 10.1038/sj.leu.2402719. [DOI] [PubMed] [Google Scholar]
- Gomes NP, Bjerke G, Llorente B, Szostek SA, Emerson BM, Espinosa JM. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes & development. 2006;20:601–612. doi: 10.1101/gad.1398206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grignani F, De Matteis S, Nervi C, Tomassoni L, Gelmetti V, Cioce M, Fanelli M, Ruthardt M, Ferrara FF, Zamir I, et al. Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia. Nature. 1998;391:815–818. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]
- He LZ, Guidez F, Tribioli C, Peruzzi D, Ruthardt M, Zelent A, Pandolfi PP. Distinct interactions of PML-RARalpha and PLZF-RARalpha with co-repressors determine differential responses to RA in APL. Nature genetics. 1998;18:126–135. doi: 10.1038/ng0298-126. [DOI] [PubMed] [Google Scholar]
- Heinzel T, Lavinsky RM, Mullen TM, Soderstrom M, Laherty CD, Torchia J, Yang WM, Brard G, Ngo SD, Davie JR, et al. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- Hengartner CJ, Myer VE, Liao SM, Wilson CJ, Koh SS, Young RA. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Molecular cell. 1998;2:43–53. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- Horlein AJ, Naar AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass CK, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- Hsu CY, Yung BY. Down-regulation of nucleophosmin/B23 during retinoic acid-induced differentiation of human promyelocytic leukemia HL-60 cells. Oncogene. 1998;16:915–923. doi: 10.1038/sj.onc.1201615. [DOI] [PubMed] [Google Scholar]
- Hsu CY, Yung BY. Involvement of nucleophosmin/B23 in TPA-induced megakaryocytic differentiation of K562 cells. British journal of cancer. 2003;89:1320–1326. doi: 10.1038/sj.bjc.6601100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen JH, Mahfoudi A, Rambaud S, Lavau C, Wahli W, Dejean A. Multimeric complexes of the PML-retinoic acid receptor alpha fusion protein in acute promyelocytic leukemia cells and interference with retinoid and peroxisome-proliferator signaling pathways. Proc Natl Acad Sci U S A. 1995;92:7401–7405. doi: 10.1073/pnas.92.16.7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurcic JG, Soignet SL, Maslak AP. Diagnosis and treatment of acute promyelocytic leukemia. Curr Oncol Rep. 2007;9:337–344. doi: 10.1007/s11912-007-0045-9. [DOI] [PubMed] [Google Scholar]
- Kastner P, Lawrence HJ, Waltzinger C, Ghyselinck NB, Chambon P, Chan S. Positive and negative regulation of granulopoiesis by endogenous RARalpha. Blood. 2001;97:1314–1320. doi: 10.1182/blood.v97.5.1314. [DOI] [PubMed] [Google Scholar]
- Klein MB, Hayes SF, Goodman JL. Monocytic differentiation inhibits infection and granulocytic differentiation potentiates infection by the agent of human granulocytic ehrlichiosis. Infection and immunity. 1998;66:3410–3415. doi: 10.1128/iai.66.7.3410-3415.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa R, Soderstrom M, Horlein A, Halachmi S, Brown M, Rosenfeld MG, Glass CK. Polarity-specific activities of retinoic acid receptors determined by a co-repressor. Nature. 1995;377:451–454. doi: 10.1038/377451a0. [DOI] [PubMed] [Google Scholar]
- Li J, Sejas DP, Rani R, Koretsky T, Bagby GC, Pang Q. Nucleophosmin regulates cell cycle progression and stress response in hematopoietic stem/progenitor cells. The Journal of biological chemistry. 2006;281:16536–16545. doi: 10.1074/jbc.M601386200. [DOI] [PubMed] [Google Scholar]
- Lin RJ, Nagy L, Inoue S, Shao W, Miller WH, Jr, Evans RM. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature. 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- Liu H, Tan BC, Tseng KH, Chuang CP, Yeh CW, Chen KD, Lee SC, Yung BY. Nucleophosmin acts as a novel AP2alpha-binding transcriptional corepressor during cell differentiation. EMBO reports. 2007;8:394–400. doi: 10.1038/sj.embor.7400909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan MC, Narlikar GJ, Boyapaty G, Kingston RE, Weissman SM. Heterogeneous nuclear ribonucleoprotein C1/C2, MeCP1, and SWI/SNF form a chromatin remodeling complex at the β-globin locus control region. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15012–15017. doi: 10.1073/pnas.0507596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara S, Miller WH., Jr Expanding the use of retinoids in acute myeloid leukemia: spotlight on bexarotene. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:5311–5313. doi: 10.1158/1078-0432.CCR-08-1081. [DOI] [PubMed] [Google Scholar]
- McNamara S, Wang H, Hanna N, Miller WH., Jr Topoisomerase IIbeta negatively modulates retinoic acid receptor alpha function: a novel mechanism of retinoic acid resistance. Molecular and cellular biology. 2008;28:2066–2077. doi: 10.1128/MCB.01576-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnick A, Licht JD. Deconstructing a disease: RARalpha, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood. 1999;93:3167–3215. [PubMed] [Google Scholar]
- Momparler RL, Dore BT, Momparler LF. Effect of 5-aza-2'-deoxycytidine and retinoic acid on differentiation and c-myc expression in HL-60 myeloid leukemic cells. Cancer letters. 1990;54:21–28. doi: 10.1016/0304-3835(90)90086-d. [DOI] [PubMed] [Google Scholar]
- Murphy DJ, Hardy S, Engel DA. Human SWI-SNF component BRG1 represses transcription of the c-fos gene. Molecular and cellular biology. 1999;19:2724–2733. doi: 10.1128/mcb.19.4.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi L, Belyaev ND, Miyake K, Wood IC, Buckley NJ. BRG1 chromatin remodeling activity is required for efficient chromatin binding by repressor element 1-silencing transcription factor (REST) and facilitates REST-mediated repression. The Journal of biological chemistry. 2006;281:38974–38980. doi: 10.1074/jbc.M605370200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Yun R, Datta A, Lacomis L, Erdjument-Bromage H, Kumar J, Tempst P, Sif S. mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Molecular and cellular biology. 2003;23:7475–7487. doi: 10.1128/MCB.23.21.7475-7487.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez A, Kastner P, Sethi S, Lutz Y, Reibel C, Chambon P. PMLRAR homodimers: distinct DNA binding properties and heteromeric interactions with RXR. The EMBO journal. 1993;12:3171–3182. doi: 10.1002/j.1460-2075.1993.tb05986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestayko AW, Klomp GR, Schmoll DJ, Busch H. Comparison of proteins of ribosomal subunits and nucleolar preribosomal particles from Novikoff hepatoma ascites cells by two-dimensional polyacrylamide gel electrophoresis. Biochemistry. 1974;13:1945–1951. doi: 10.1021/bi00706a026. [DOI] [PubMed] [Google Scholar]
- Qi W, Shakalya K, Stejskal A, Goldman A, Beeck S, Cooke L, Mahadevan D. NSC348884, a nucleophosmin inhibitor disrupts oligomer formation and induces apoptosis in human cancer cells. Oncogene. 2008;27:4210–4220. doi: 10.1038/onc.2008.54. [DOI] [PubMed] [Google Scholar]
- Rosenauer A, Raelson JV, Nervi C, Eydoux P, DeBlasio A, Miller WH., Jr Alterations in expression, binding to ligand and DNA, and transcriptional activity of rearranged and wild-type retinoid receptors in retinoid-resistant acute promyelocytic leukemia cell lines. Blood. 1996;88:2671–2682. [PubMed] [Google Scholar]
- Rowley JD, Golomb HM, Dougherty C. 15/17 translocation, a consistent chromosomal change in acute promyelocytic leukaemia. Lancet. 1977;1:549–550. doi: 10.1016/s0140-6736(77)91415-5. [DOI] [PubMed] [Google Scholar]
- Shao W, Benedetti L, Lamph WW, Nervi C, Miller WH., Jr A retinoid-resistant acute promyelocytic leukemia subclone expresses a dominant negative PML-RAR alpha mutation. Blood. 1997;89:4282–4289. [PubMed] [Google Scholar]
- Shao W, Rosenauer A, Mann K, Chang CP, Rachez C, Freedman LP, Miller WH., Jr Ligand-inducible interaction of the DRIP/TRAP coactivator complex with retinoid receptors in retinoic acid-sensitive and -resistant acute promyelocytic leukemia cells. Blood. 2000;96:2233–2239. [PubMed] [Google Scholar]
- Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes & development. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- Vizlin-Hodzic D, Runnberg R, Ryme J, Simonsson S, Simonsson T. SAF-A forms a complex with BRG1 and both components are required for RNA polymerase II mediated transcription. PloS one. 2011;6:6. doi: 10.1371/journal.pone.0028049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nature reviews Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- Xiao H, Mao Y, Desai SD, Zhou N, Ting C-Y, Hwang J, Liu LF. The topoisomerase IIβ circular clamp arrests transcription and signals a 26S proteasome pathway. Proceedings of the National Academy of Sciences. 2003;100:3239–3244. doi: 10.1073/pnas.0736401100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan CX, Ito M, Fondell JD, Fu ZY, Roeder RG. The TRAP220 component of a thyroid hormone receptor- associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc Natl Acad Sci U S A. 1998;95:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.