Abstract
Long noncoding RNAs (lncRNAs) have emerged as critical regulators in various cellular processes. However, the potential involvement of lncRNAs in kinase signaling remains largely unknown. AMP-activated protein kinase (AMPK) acts as a critical sensor of cellular energy status. Here we show that lncRNA NBR2 (neighbor of BRCA1 gene 2) is induced by the LKB1-AMPK pathway under energy stress. Upon energy stress, NBR2 in turn interacts with AMPK and promotes AMPK kinase activity, thus forming a feed-forward loop to potentiate AMPK activation during energy stress. Depletion of NBR2 attenuates energy stress-induced AMPK activation, resulting in unchecked cell cycling, altered apoptosis/autophagy response, and increased tumor development in vivo. NBR2 is down-regulated and its low expression correlates with poor clinical outcomes in some human cancers. Together, our study uncovers a mechanism coupling lncRNAs with metabolic stress response, and provides a broad framework to further understand the regulation of kinase signaling by lncRNAs.
Introduction
Mammalian genomes encode more than 10,000 long noncoding RNAs (lncRNAs), the RNA molecules which are longer than 200 nucleotides and do not appear to encode proteins1, 2. Although lncRNAs were traditionally viewed as the products that are generated from the background noise of transcription and thus exert little fitness advantage to the cells, it has become increasingly clear that these lncRNAs play important biological functions, and their dysregulation has been connected to various human diseases, including cancer3–6.
Most current studies focus on lncRNA function in the nucleus, partly because most of the best-understood lncRNAs, such as XIST7, HOTAIR8, HOTTIP9, are all chromatin-associated lncRNAs, which are mainly localized in the nucleus. These studies have illustrated a diverse range of functions of lncRNAs in the regulation of chromatin status, transcription, and RNA processing, among others1, 10. Many lncRNAs have also been identified in the cytosol11. In fact, it has been suggested that the majority of lncRNAs probably spend most of their life time in the cytoplasm1. However, the exact functions of cytoplasmic localized lncRNAs, particularly their potential functions in the regulation of kinase signaling in the cytoplasm, remain poorly understood. In addition, although lncRNAs have been shown to regulate diverse biological processes, the role of lncRNAs in mediating metabolic checkpoint remains largely unexplored.
The AMP-activated protein kinase (AMPK) serves as a critical sensor of cellular energy status and is activated under energy stress conditions with an increased cellular AMP/ATP ratio12. AMP binding to AMPK and subsequent AMPK phosphorylation at Thr172 by the upstream kinase LKB1 leads to AMPK activation13–15. Activated AMPK then phosphorylates a number of downstream targets to inactivate ATP-consuming anabolic processes and to activate ATP-generating catabolic processes16. Thus, AMPK mainly functions as a metabolic checkpoint to restore energy balance in response to energy stress. One major anabolic process inhibited by AMPK in response to energy stress is mammalian target of rapamycin complex 1 (mTORC1)-mediated protein synthesis and cell growth17. In response to energy stress, AMPK inactivates mTORC1 and represses protein synthesis via AMPK phosphorylation of Raptor, a component of mTORC1, and the TSC1-TSC2 complex, a negative regulator of mTORC118, 19. AMPK also functions to promote autophagy and cell survival under energy stress via its phosphorylation of autophagy regulators, such as ULK120, 21. As anabolic processes, such as protein and lipid synthesis, often exert pro-growth effects in tumor development, it is well documented that AMPK activation serves to inhibit tumor development in many cancers22. Consistent with this, both the upstream kinase Lkb1 and downstream effectors of AMPK, such as TSC1 and TSC2, are bona fide tumor suppressors and are mutated in hamartoma tumor syndromes and various sporadic cancers23–25. Although the biological functions of AMPK and its downstream effectors involved in cancer development have been extensively studied22, 26, the regulatory mechanisms of AMPK activation by energy stress remain incompletely understood. In particular, it remains completely unknown whether any lncRNA is involved in AMPK-mediated metabolic checkpoint.
In this study, we identify neighbor of BRCA1 gene 2 (NBR2) as an energy stress-induced lncRNA and show that NBR2 interacts with AMPK and potentiates AMPK activation under energy stress. Consistent with the tumor suppression function of AMPK, NBR2 deficiency promotes unchecked cell cycling under energy stress and enhances tumor development in vivo, and NBR2 is down-regulated in human cancers. Our study thus reveals a previously unappreciated regulatory mechanism by lncRNAs to regulate kinase function and to mediate cellular energy responses.
Results
Energy stress induces NBR2 expression through the LKB1-AMPK pathway
To identify energy stress-induced lncRNAs, we conducted an RNA sequencing experiment in 786-O cells that had been cultured in glucose-containing or glucose-free medium. Subsequent computational analysis identified NBR2 as one of the long intergenic noncoding RNAs (lincRNAs) induced by glucose starvation. NBR2 gene encodes different splicing isoforms ranging from 1 to 2 kb (Supplemental Fig. 1). It has been shown that NBR2 is expressed in most of the tissues examined27. However, NBR2 gene does not appear to encode a protein, and its potential function remains unknown.
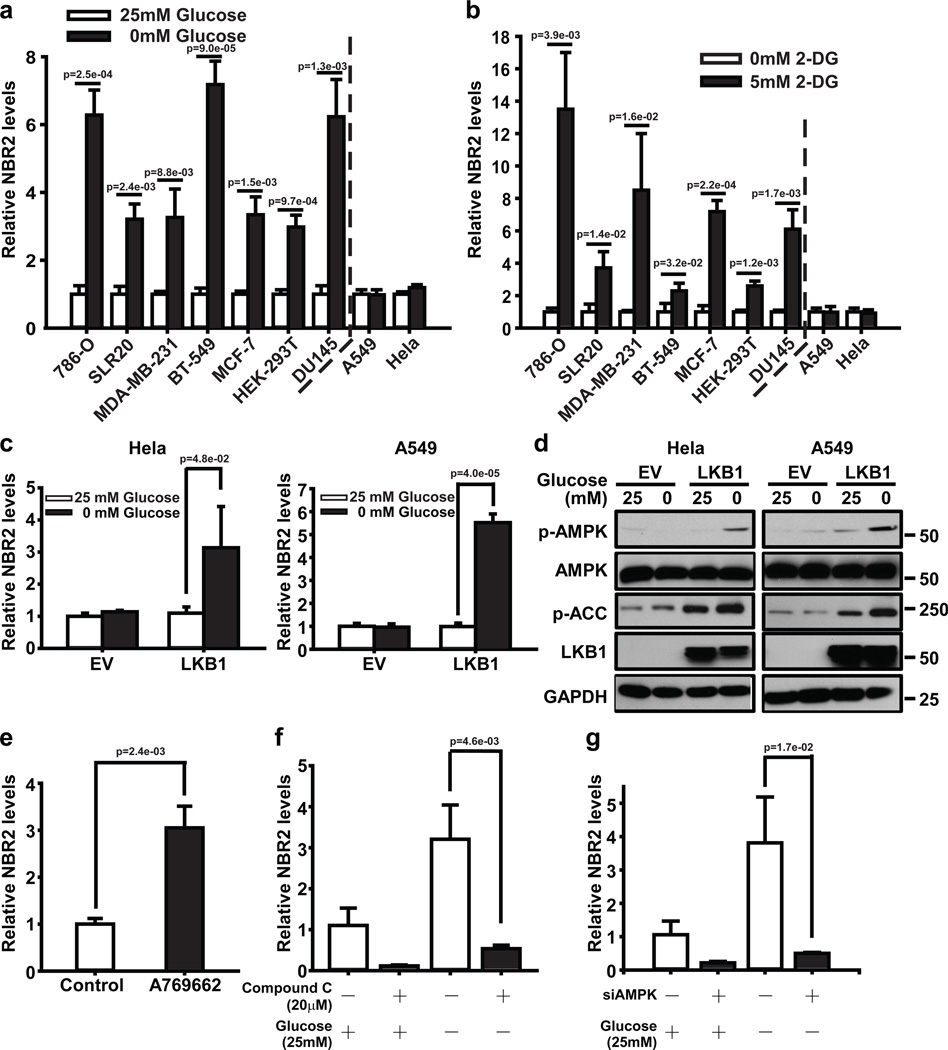
Real-time PCR revealed that glucose starvation induced NBR2 expression in different cancer cell lines, except Hela and A549 cells, which are Lkb1 deficient (Fig. 1a). Treatment with the glucose analog 2-deoxy-glucose (2DG), another energy stress inducer that inhibits hexokinase and blocks glycolysis, yielded similar results (Fig. 1b). Importantly, re-expression of Lkb1 in these Lkb1-deficient cells restored energy stress-induced NBR2 expression (Fig. 1c, d). In addition, treatment of A769662 (an AMPK activator) induced NBR2 expression (Fig. 1e), while AMPK inactivation by compound C (an AMPK inhibitor) treatment or siRNA-mediated AMPKα knockdown significantly attenuated glucose starvation-induced NBR2 expression (Fig. 1f, g, and Supplemental Fig. 2). Together, our results revealed that energy stress induces NBR2 expression at least partly through the LKB1-AMPK pathway.
Figure 1. Energy stress induces NBR2 expression through the LKB1-AMPK pathway.
(a, b) Various cell lines were cultured in 0 or 25 mM glucose-containing medium (a), or 0 or 5 mM 2DG-containing medium (b) for 12–24 hours, and then subjected to real-time PCR analysis to measure NBR2 expression (Mean ± s.d., n=3 biologically independent extracts, two-tailed paired Student’s t-test). (c, d) Hela or A549 cells stably expressing EV (empty vector) or Lkb1 expression vectors were cultured in 25 or 0 mM glucose-containing medium, and then subjected to real-time PCR (c) (Mean ± s.d., n=3 biologically independent extracts, two-tailed paired Student’s t-test) and Western blotting analyses (d). (e) MDA-MB-231 cells treated with 100 µM A769662 were subjected to real-time PCR analysis to measure NBR2 (Mean ± s.d., n=3 biologically independent extracts, two-tailed paired Student’s t-test). (f) MDA-MB-231 cells were treated with 20 µM Compound C in 25 or 0 mM glucose-containing medium for 24 hours, and then subjected to real-time PCR analysis to measure NBR2 expression (Mean ± s.d., n=3 biologically independent extracts, two-tailed paired Student’s t-test). (g) MDA-MB-231 cells transfected with AMPKα or control (Ctrl) siRNA were cultured in 25 or 0 mM glucose-containing medium for 24 hours, and then subjected to real-time PCR analysis to measure NBR2 (Mean ± s.d., n=3 biologically independent extracts, two-tailed paired Student’s t-test). Source data for a, b, c, e, f, g can be found in Supplementary Table 1. Unprocessed original scans of blots are shown in Supplemental Fig. 8.
NBR2 regulates AMPK-mTORC1 signaling under energy stress
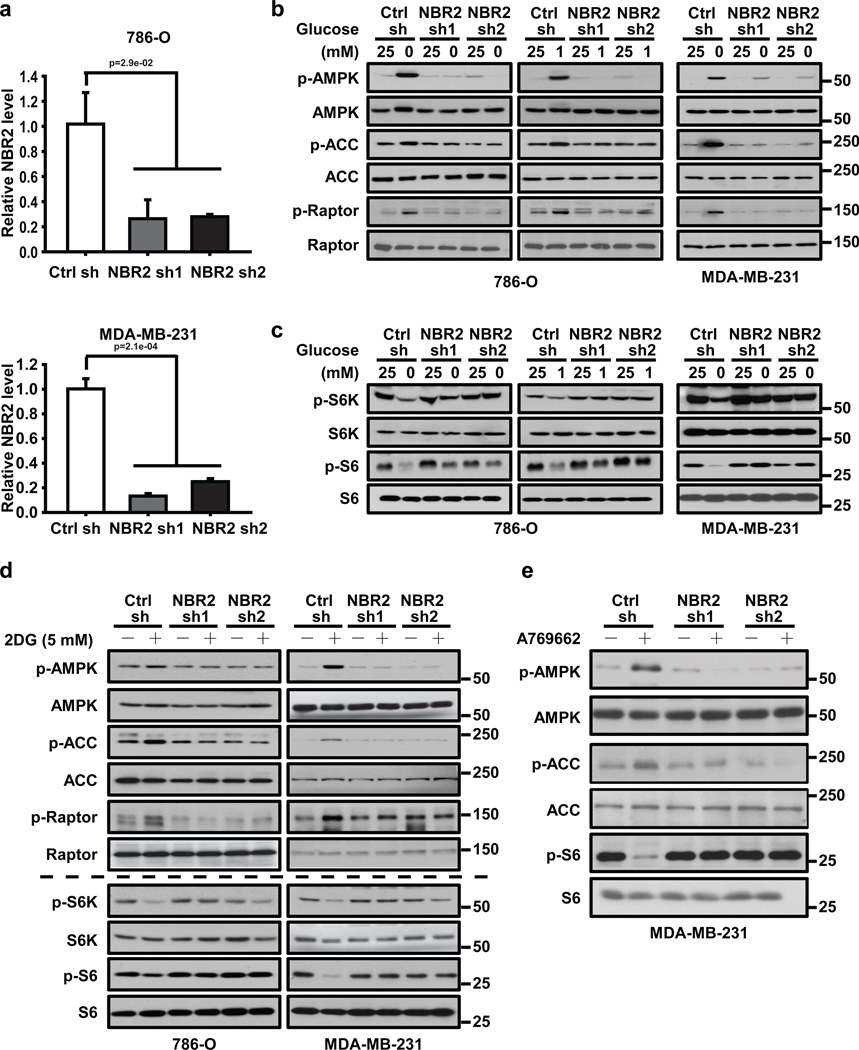
To study the potential function of NBR2 in mediating energy stress response, we generated 786-O cells (a kidney cancer cell line) and MDA-MB231 cells (a breast cancer cell line) with stable knockdown of NBR2 (Fig. 2a). We then analyzed whether knockdown of NBR2 affected any biochemical signaling surrogate induced by energy stress, including AMPK activation. As shown in Fig. 2b, glucose starvation potently induced phosphorylation of AMPK, or AMPK substrates acetyl CoA carboxylase (ACC) and Raptor18, 28. Notably, NBR2 knockdown significantly attenuated glucose starvation-induced phosphorylation of AMPK, ACC and Raptor. Accordingly, S6 and S6K de-phosphorylation induced by glucose deprivation was significantly compromised in NBR2 knockdown cells compared with control shRNA-infected cells (Fig. 2c). Finally, NBR2 knockdown also attenuated 2DG or A769662 treatment-induced AMPK activation and mTORC1 inactivation (Fig. 2d, e). Our results thus revealed that NBR2 depletion attenuates energy stress-induced AMPK activation and mTORC1 inactivation, and suggested a feed-forward mechanism on NBR2-AMPK regulation, in which AMPK initially promotes NBR2 expression in response to energy stress and NBR2 in turn regulates AMPK activation under energy stress (see Discussion).
Figure 2. NBR2 regulates AMPK-mTORC1 signaling under energy stress.
(a) Bar graph showing NBR2 shRNA-mediated knockdown efficiency by real-time PCR analysis in 786-O and MDA-MB-231 cells (Mean ± s.d., n=3 biologically independent extracts, two-tailed paired Student’s t-test). (b, c) 786-O or MDA-MB-231 cells infected with either control shRNA or NBR2 shRNA were cultured in medium with different concentrations of glucose for 24 hours. Cell lysates were then analyzed by Western blotting. (d) 786-O or MDA-MB-231 cells infected with either control shRNA or NBR2 shRNA were cultured in 0 or 5 mM 2DG-containing medium for 12 (for MDA-MB-231 cells) or 16 (for 786-O cells) hours. Cell lysates were then analyzed by Western blotting. (e) MDA-MB-231 cells infected with either control shRNA or NBR2 shRNA were cultured in 0 or 100 µM A769662-containing medium for 12 hours. Cell lysates were then analyzed by Western blotting. Source data for a can be found in Supplementary Table 1. Unprocessed original scans of blots are shown in Supplemental Fig. 8.
NBR2 regulates cell proliferation, apoptosis, and autophagy in response to energy stress
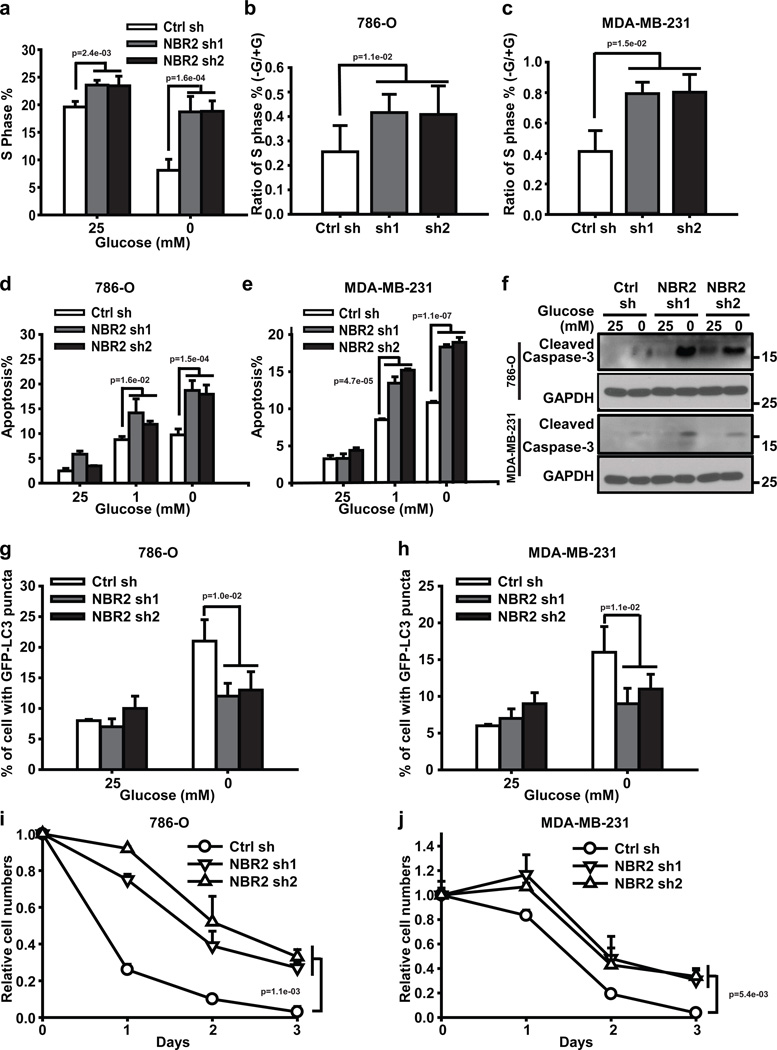
AMPK functions as a critical metabolic checkpoint; defective AMPK signaling leads to increased cell proliferation yet decreased autophagy under conditions of energy stress, leading to apoptosis12, 20. The aforementioned data prompted us to examine the impact of NBR2 deficiency on cell proliferation, apoptosis, and autophagy in response to energy stress. Glucose starvation dramatically decreased S phase entry as measured by BrdU incorporation, and knockdown of NBR2 significantly attenuated the reduction of S phase entry upon glucose starvation (Fig. 3a–c). Thus, similar to cells with defective AMPK signaling18, NBR2 deficient cells continue cycling under energy stress.
Figure 3. NBR2 regulates cell proliferation, apoptosis, and autophagy in response to energy stress.
(a) Bar graph showing the percentages of S phase (Brdu positive) cells in control shRNA or NBR2 shRNA-infected MDA-MB-231 cells which were cultured in 25 or 0 mM glucose-containing medium for 24 hours (Mean ± s.d., n=3 biologically independent extracts, two-tailed paired Student’s t-test). (b, c) Bar graph showing the –Glucose/+Glucose ratio of S phase percentages in control shRNA or NBR2 shRNA-infected 786-O cells (b) or MDA-MB-231 cells (c) (Mean ± s.d., n=3 biologically independent extracts, two-tailed paired Student’s t-test). (d–f) Control shRNA or NBR2 shRNA-infected 786O cells or MDA-MB-231 cells were cultured in medium with different concentrations of glucose for 24 hours, then subjected to Annexin V/PI staining followed by FACS analysis to measure the percentages of Annexin V positive/PI negative cells (d for 786O cells, e for MDA-MB-231 cells, Mean ± s.d., n=3 biologically independent extracts, two-tailed paired Student’s t-test), or to Western blotting analysis to measure Caspase-3 cleavage (f). (g–h) Bar graph showing the percentages of cells with LC3-GFP punctate localization in control shRNA or NBR2 shRNA-infected 786-O cells (g) or MDA-MB-231 cells (h), which were transfected with GFP-LC3 and then cultured in 25 or 0 mM glucose-containing medium for 12 (for MDA-MB-231 cells) or 18 (for 786-O cells) hours (Mean ± s.d., n=5 fields per group, each field was assessed from an independent experiment, two-tailed paired Student’s t-test). (i, j) 786-O (i) or MDA-MB-231 (j) cells infected with either control shRNA or NBR2 shRNA were cultured in glucose free medium for different days as indicated, and then subjected to cell proliferation analysis (Mean ± s.d., n=3 biologically independent extracts, two-tailed paired Student’s t-test). Source data for a, b, c, d, e, i, j can be found in Supplementary Table 1. Unprocessed original scans of blots are shown in Supplemental Fig. 8.
Although NBR2 depletion did not affect apoptosis under normal culture condition, NBR2 deficiency induced more apoptosis under glucose starvation, as evidenced by both Annexin V staining (Fig. 3d, e) and cleaved caspase-3 Western blotting (Fig 3f). In response to energy stress, AMPK activates autophagy, a cellular adaptive response to promote cell survival under stress conditions20, 21. Accordingly, glucose starvation-induced GFP-LC3 puncta formation, p62 degradation, and ULK1 phosphorylation were significantly compromised in NBR2 deficient cells (Fig. 3g, h, and Supplemental Fig. 3a, b), suggesting that energy stress-induced autophagy was defective in NBR2 deficient cells. Despite enhanced apoptosis, the number in NBR2 deficient cells increased under glucose deprived conditions because of the increase in cycling in NBR2 deficient cells (Fig. 3i, j, and Supplemental Fig. 3c, d). Collectively, our results showed that NBR2 deficiency leads to enhanced cell cycling yet decreased autophagy and increased apoptosis under energy stress, which is in line with the phenotypes from cells with defective AMPK signaling, including AMPK, Lkb1, TSC1, TSC2 deficient cells or cells reconstituted with a Raptor mutant which is non-phosphorylatable by AMPK15, 18, 19, 29, 30.
NBR2 inhibits tumor development and is down-regulated in human cancers
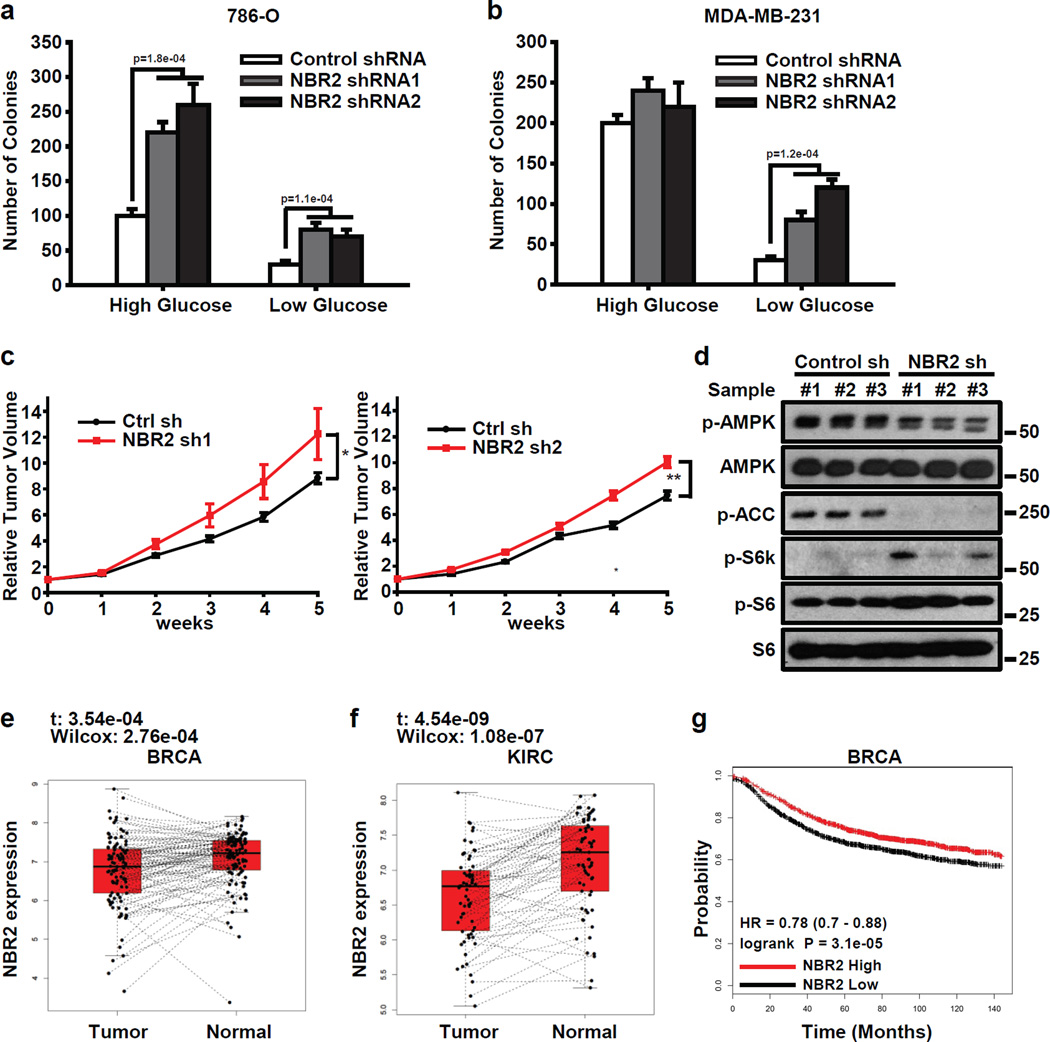
Given the important functions of AMPK in the regulation of human cancers22, we next examined the potential roles of NBR2 in tumor development. NBR2 deficiency led to increased anchorage-independent growth, one of the hallmarks of cell transformation, with a more prominent effect under glucose starvation conditions (Fig. 4a, b). In vivo experiments using the xenograft model showed that NBR2 deficiency increased tumor development (Fig. 4c). Further analyses of the tumor samples by Western blotting confirmed down-regulation of AMPK and up-regulation of mTORC1 signaling in NBR2 deficient tumors (Fig. 4d).
Figure 4. NBR2 inhibits tumor development.
(a, b) 786-O (a) or MDA-MB-231 cells (b) infected with either control shRNA or NBR2 shRNA were seeded in soft agar containing high or low concentrations of glucose as indicated. Bar graph showing the mean colony numbers from the soft agar assay (Mean ± s.d., n=5 fields per group, each field was assessed from an independent experiment, two-tailed paired Student’s t-test). (c) Relative tumor volumes of MDA-MB-231 xenograft tumors infected with either control shRNA or NBR2 shRNA at different weeks (Mean ± s.e.m., n = 5 xenograft tumors, *: P < 0.05; **: P < 0.01 two-tailed paired Student’s t-test). (d) Protein lysates obtained from xenograft tumors infected with either control shRNA or NBR2 shRNA at the end point were subjected to Western blotting analysis as indicated. (e, f) The box plot showing the expression pattern of NBR2 for each pair of tumor and normal samples in BRCA (e, n=104 matched pairs, Sutdent's t-test and Wilcoxon test) and KIRC (f, n=65 matched pairs, Sutdent's t-test and Wilcoxon test). The boxes show the median ±1 quartile, with whiskers extending to the most extreme data point within 1.5 interquartile range from the box boundaries. (g) Kaplan Meier plots of breast cancer patients stratified by the expression levels of NBR2 (nhigh = 1767, nlow = 1787, Log-Rank Test). Unprocessed original scans of blots are shown in Supplemental Fig. 8.
Consistent with the experimental results from breast and renal cancer cell lines, a survey of the RNA-seq data across different cancer types from the TCGA (The Cancer Genome Atlas) datasets revealed down-regulation of NBR2 expression in breast (BRCA) and renal (KIRC) cancer samples compared with paired normal tissue samples (Fig. 4e, f). Kaplan Meier analysis showed that breast cancer patients with NBR2-low tumors had significantly worse overall survival than those with NBR2-high tumors (Fig. 4g). Together, our data showed that NBR2 deficiency promotes tumor development, and NBR2 is down-regulated in human breast and renal cancers, suggesting that NBR2 may function as a tumor suppressor in these cancers.
Energy stress induces NBR2 interaction with AMPK
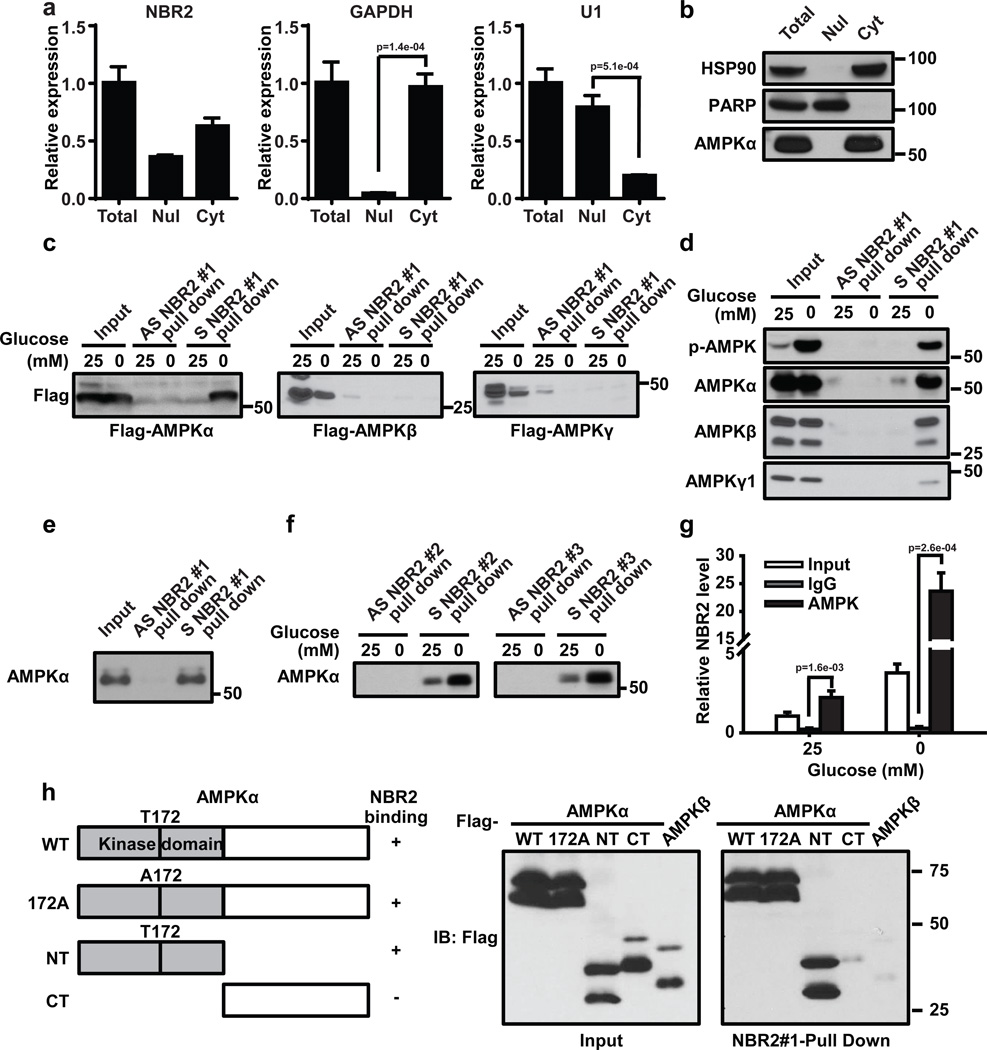
The aforementioned biological data prompted us to further study how NBR2 regulates AMPK function. Real-time PCR analyses of fractionated nuclear and cytoplasmic RNA revealed that NBR2 localized in both nucleus and cytoplasm (Fig. 5a). As expected, AMPK α showed predominant localization in the cytoplasm (Fig. 5b). AMPK exists as a heterotrimeric complex which consists of a catalytic α subunit and two regulatory β and γ subunits31. We thus examined whether NBR2 can interact with any of the subunits of AMPK by RNA-pulldown assay using in vitro-synthesized biotinylated NBR2. Such analysis revealed that NBR2 interacted with overexpressed AMPK α under glucose starvation condition, with minimal binding with overexpressed β or γ subunit (Fig. 5c). The RNA pulldown assay also revealed that glucose starvation significantly increased the interaction of NBR2 with endogenous AMPKα (Fig. 5d). Since AMPK α, β and γ subunits form a very stable complex at the endogenous level, we also observed a glucose starvation-induced binding between NBR2 and endogenous AMPK β and γ subunits (Fig. 5d), likely mediated by NBR2 interaction with endogenous AMPK α subunit. In vitro binding assay using purified AMPK α and in vitro-synthesized biotinylated NBR2 confirmed the direct binding between NBR2 and AMPK α (Fig. 5e). There exist at least three splicing isoforms of NBR2 gene (named as NBR2 #1, #2 and #3, see Supplemental Fig. 1). In the RNA pulldown experiments described above, we utilized NBR2 #1 splicing isoform. The RNA pulldown experiments showed that NBR2 #2 and #3 splicing isoforms also interacted with AMPK α upon glucose starvation (Fig. 5f). Finally, RNA immunoprecipitation (RIP) assay (using the primers which can detect all three NBR2 splicing isoforms) revealed an enrichment of NBR2 in the precipitates of AMPK α compared with IgG control, and glucose starvation substantially increased the enrichment of NBR2 in AMPK α precipitates (Note that glucose starvation resulted in much more fold increase of NBR2 level in AMPK α precipitates than of NBR2 input level) (Fig. 5g).
Figure 5. Energy stress induces NBR2 interaction with AMPK.
(a, b) Nuclear and cytoplasmic fractions of 786O cells were subjected to either real-time PCR (a, Mean ± s.d., n=3 biologically independent extracts, two-tailed paired Student’s t-test) or Western blotting analysis (b). (c) In vitro-synthesized biotinylated sense (S) or antisense (AS) NBR2 #1 were incubated with protein lysates from HEK293T cells transfected with various vectors as indicated. Precipitation reactions were conducted using streptavidin beads and then subjected to Western blotting. (d, f) In vitro-synthesized biotinylated sense (S) NBR2 or antisense (AS) NBR2 with different splicing isoforms were incubated with protein lysates from 786-O cells which had been cultured in 25 or 0 mM glucose-containing medium for 24 hours. Precipitation reactions were conducted using streptavidin beads and then subjected to Western blotting. (e) In vitro-synthesized biotinylated sense (S) or antisense (AS) NBR2 #1 were incubated with purified human AMPK α protein. Precipitation reactions were conducted using streptavidin beads and then subjected to Western blotting. (g) 786-O cells were cultured in 0 or 25 mM glucose-containing medium for 24 hours. Protein lysates were prepared and immunoprecipitated with AMPK α antibody or IgG. The RNA levels of NBR2 in immunoprecipitates or cell lysates (input) were measured by real-time PCR (Mean ± s.d., n=3 biologically independent extracts, two-tailed paired Student’s t-test). (h) In vitro-synthesized biotinylated NBR2 #1 were incubated with protein lysates from HEK293T cells transfected with various vectors and subjected to glucose starvation. Precipitation reactions were conducted using streptavidin beads and then subjected to Western blotting. Source data for a, g can be found in Supplementary Table 1. Unprocessed original scans of blots are shown in Supplemental Fig. 8.
In the experiment to map the region(s) of AMPK α which mediates AMPK interaction with NBR2, we showed that kinase domain-containing N terminal region, but not the C terminal region of AMPK α, interacted with NBR2 (Fig. 5h). Mutation of threonine 172 to alanine in AMPK α did not affect AMPK α interaction with NBR2 (Fig. 5h), indicating that AMPK phosphorylation at threonine 172 is not required for AMPK-NBR2 interaction. Together, our data revealed that glucose starvation not only induces NBR2 expression, but also enhances NBR2 interaction with AMPK, which is likely mediated by NBR2 interaction with the kinase domain of AMPK α.
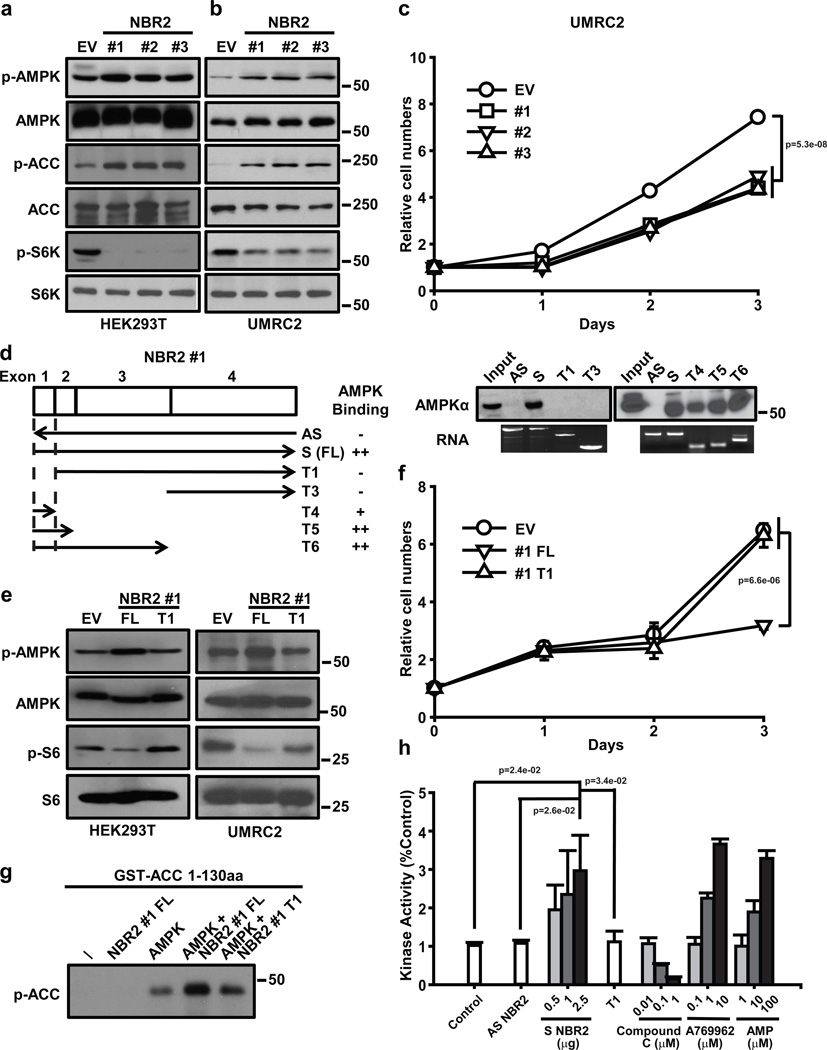
NBR2 promotes AMPK kinase activity
Next we studied the underlying mechanisms by which NBR2 regulates AMPK function. To this end, we first examined whether overexpression of NBR2 exerts any biological effect in cells. Our experiments revealed that overexpression of any of the three NBR2 splicing isoforms resulted in AMPK activation, mTORC1 inactivation (Fig. 6a, b), and decreased cell proliferation without affecting apoptosis under normal culture condition (Fig. 6c). All three splicing isoforms of NBR2 share the same first two exons located at the 5’ end of NBR2 with distinctive exons located toward the 3’ end (Supplemental Fig. 1). Our data thus indicate that the common exons in all NBR2 splicing isoforms might be important in mediating NBR2 interaction with AMPK. Consistent with this, our binding mapping experiments revealed that the first exon shared by all three NBR2 splicing isoforms is both required and sufficient to mediate NBR2 interaction with AMPK α (Fig. 6d). Furthermore, overexpression of T1 fragment of NBR2 #1, which lacks the first exon (with 159 bp out of 1045 bp full length NBR2 #1) and thus is incapable of interacting with AMPK, did not affect AMPK and mTORC1 activation status or cell proliferation, while in the parallel experiments, overexpression of full length (FL) NBR2 #1 exerted the expected effects on AMPK signaling (Fig. 6e, f). It seems that overexpression of the first exon alone (T4 fragment of NBR2 #1) was not sufficient to activate AMPK (Supplemental Fig. 4a), suggesting that other regions in NBR2 may be also important for NBR2 function in the regulation of AMPK. Together, our results showed that deletion of the first exon of NBR2 abolishes its interaction with AMPK and regulation of AMPK activation, suggesting that NBR2 regulation of AMPK activation and downstream cellular processes is likely mediated through NBR2 interaction with AMPK.
Figure 6. NBR2 promotes AMPK kinase activity.
(a, b) Protein lysates were prepared from HEK293T (a) or UMRC2 cells (b) with overexpression of EV or NBR2 expression vectors, and analyzed by Western blotting. (c) UMRC2 cells stably expressing EV or NBR2 expression vectors were cultured in 25 mM glucose-containing medium for different days as indicated, and then subjected to cell proliferation analysis (Mean ± s.d., n=3 biologically independent extracts, two-tailed paired Student’s t-test). (d) Left panel: Schematic diagram showing different truncation mutants of NBR2 #1 and the summary of their binding capabilities to AMPK α. Right panel: In vitro-synthesized biotinylated sense (S), antisense (AS), or different truncation (T) mutants of NBR2 #1 were incubated with protein lysates from 786-O cells which had been cultured in glucose free medium for 24 hours. Precipitation reactions were conducted using streptavidin beads and then subjected to Western blotting. (e) Protein lysates were prepared from HEK293T or UMRC2 cells with overexpression of EV, NBR2 #1 full length (FL), or T1 mutant expression vectors, and analyzed by Western blotting. (f) UMRC2 cells stably expressing EV, NBR2 #1 FL, or T1 mutant expression vectors were cultured in 25 mM glucose-containing medium for different days as indicated, and then subjected to cell proliferation analysis (Mean ± s.d., n=3 biologically independent extracts, two-tailed paired Student’s t-test). (g) AMPK complex precipitated from HEK293T cells was subjected to the kinase assay in the presence of ATP, in vitro synthesized RNAs and GST-ACC 1–130 aa fusion proteins as indicated. The kinase activity of AMPK was measured by phosphorylation of ACC at S79 site. (h) In vitro purified active human AMPK complex was subjected to in vitro kinase assays in the presence of ATP, SAMS peptide and in vitro synthesized biotinylated sense (S)/antisense (AS)/T1 mutant (T1) NBR2 #1 or several chemical compounds (Compound C, A769662, AMP) as indicated (see Materials & Methods for details). The Kinase activity was measured by the luminescence with a plate-reading illuminometer (Mean ± s.d., n=3 biologically independent extracts, two-tailed paired Student’s t-test). Source data for c, f, h can be found in Supplementary Table 1. Unprocessed original scans of blots are shown in Supplemental Fig. 8.
Since LKB1 functions as the major upstream kinase of AMPK in response to energy stress13–15, we examined whether NBR2 regulates LKB1 interaction with AMPK. Our results showed that NBR2 overexpression or knockdown did not affect AMPK-LKB1 interaction under either basal or glucose starvation condition (Supplemental Fig. 4b, c). In addition, we found that overexpression of NBR2 in Lkb1-deficient Hela cells could still promote AMPK activation, and co-expression of NBR2 and LKB1 in Hela cells led to synergistic increase of AMPK activation (Supplemental Fig. 4d). Together, our data suggest that NBR2 does not regulate AMPK-LKB1 interaction and it is likely that NBR2 operates in parallel to LKB1 to regulate AMPK activation.
Our data that NBR2 interacts with the kinase domain of AMPKα (Fig. 5h) prompted the hypothesis that NBR2 may directly regulate the kinase activity of the AMPK complex. Our data showed that bacterial purified GST-ACC (aa 1–130) could be readily phosphorylated by the AMPK complex precipitated from cell lysates of HEK293T cells co-transfected with AMPKα/β/γ constructs (Fig. 6g). While in vitro synthesized NBR2 alone did not lead to ACC phosphorylation, the addition of NBR2 (but not the T1 fragment of NBR2, the AMPK non-binding mutant) to the AMPK complex significantly increased ACC phosphorylation by AMPK (Fig. 6g). The in vitro kinase assay using purified AMPKα/β/γ complex and SAMS peptide as the AMPK substrate further confirmed that NBR2 promoted AMPK in vitro kinase activity (Fig. 6h). Together, our data suggest that NBR2 functions to promote AMPK kinase activity likely through its interaction with AMPK kinase domain.
The functional effects of NBR2 are partially mediated by AMPK
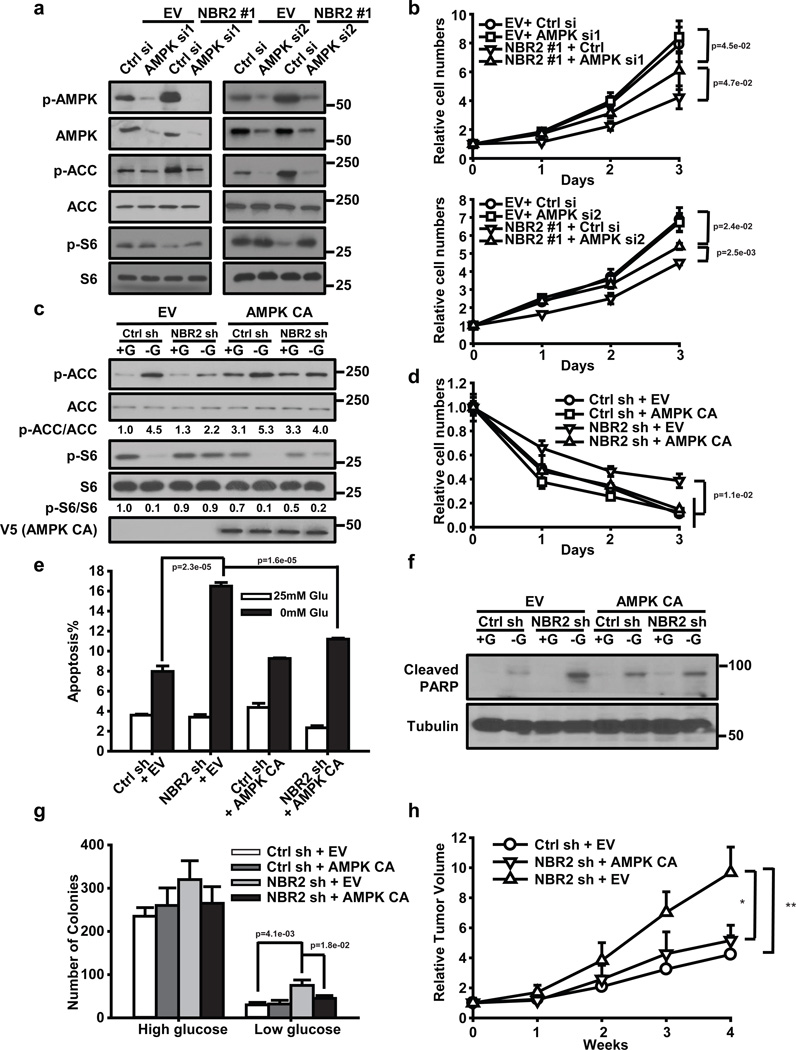
We next sought to determine the extent to which the functional effects of NBR2 are mediated by NBR2 regulation of AMPK activation. We first examined whether overexpression of NBR2 still exerted its functional effects in AMPKα knockdown cells. Such analyses revealed that, while overexpression of NBR2 increased ACC phosphorylation, decreased S6 phosphorylation, and suppressed cell proliferation in control siRNA (Ctrl si) transfected cells, such effects were attenuated in AMPKα knockdown (AMPK si) cells (Fig. 7a, b). As a complementary approach, we also examined whether restoration of constitutively active (CA) AMPK (1–312 a.a. of AMPK α1) would rescue any of the defects observed in NBR2 deficient cells. Our data revealed that overexpression of AMPK CA in NBR2 knockdown cells restored ACC or S6 phosphorylation under glucose starvation condition as expected (Fig. 7c), and correspondingly, significantly rescued cell proliferation, apoptosis, and anchorage independence growth under glucose starvation conditions in NBR2 deficient cells (Fig. 7d–g). Importantly, restoration of AMPK CA in NBR2 deficient background significantly attenuated the enhanced xenograft tumor development caused by NBR2 deficiency (Fig. 7h). Taken together, our data strongly suggested that the functional effects of NBR2 are at least partially dependent on AMPK.
Figure 7. The functional effects of NBR2 are partially mediated by AMPK.
(A and B) UMRC2 cells stably expressing EV or NBR2 expression vectors were transfected with AMPK siRNA (AMPK si1 or si2) or control siRNA (Ctrl si). Protein lysates were prepared and analyzed by Western blotting (a), or cells were cultured in 25 mM glucose-containing medium for different days as indicated, and then subjected to cell proliferation analysis (b) (Mean ± s.d., n=3 biologically independent extracts, two-tailed paired Student’s t-test). (c–g) MDA-MB-231 cells with stable expression of control shRNA (Ctrl sh) or NBR2 shRNA (NBR2 sh) were infected with empty vector (EV) or constitutively active AMPK (AMPK CA). These cells were cultured in 25 or 0 mM glucose-containing medium for 24 hours, and protein lysates were prepared and analyzed by Western blotting (c); The cells were cultured in 0 mM glucose-containing medium for different days as indicated, and then subjected to crystal violet staining to measure cell number (d) (Mean ± s.d., n=3 biologically independent extracts, two-tailed paired Student’s t-test); The cells were cultured in 25 or 0 mM glucose-containing medium for 24 hours, then subjected to Annexin V/PI staining followed by FACS analysis to measure the percentages of Annexin V positive/PI negative cells cells (e) (Mean ± s.d., n=5 fields per group, each field was assessed from an independent experiment, two-tailed paired Student’s t-test), or to Western blotting analysis to measure PARP cleavage (f); The cells were seeded in soft agar containing high or low concentrations of glucose as indicated. Bar graph showing the mean colony numbers from the soft agar assay (g) (Mean ± s.d., n=5 fields per group, each field was assessed from an independent experiment, two-tailed paired Student’s t-test). (h) Relative tumor volumes of MDA-MB-231 xenograft tumors of different genotypes at different weeks (Mean ± s.d., n = 5 xenograft tumors, *: P < 0.05, **: P < 0.01, two-tailed paired Student’s t-test). Source data for b, d, e can be found in Supplementary Table 1. Unprocessed original scans of blots are shown in Supplemental Fig. 8.
Discussion
AMPK exists as a heterotrimeric complex comprising of a catalytic α subunit and two regulatory β and γ subunits, in which γ subunit directly binds to AMP in response to energy stress31. It has been proposed that AMP activates AMPK via at least three mechanisms: (i) AMP binding to AMPK causes allosteric activation of AMPK; AMP binding leads to conformational change of AMPK complex which (ii) promotes Thr172 phosphorylation in AMPK α subunit by LKB1 and (iii) inhibits Thr172 de-phosphorylation by protein phosphatases31. Our study reveals that lincRNA NBR2 regulation of AMPK represents another important regulatory mechanism to control AMPK activation in response to energy stress. Here we propose a feed-forward model on NBR2-AMPK regulation. Specifically, energy stress-induced initial AMPK activation does not require NBR2. Activated AMPK then up-regulates NBR2 expression in response to energy stress. NBR2 in turn interacts with AMPK and promotes AMPK kinase activity under energy stress, forming a feed-forward loop to potentiate AMPK activation during chronic energy stress conditions (Supplemental Fig. 5a). NBR2 deficiency leads to AMPK inactivation during long periods of energy stress, which promotes mTORC1 activation, cell proliferation and tumor development (Supplemental Fig. 5b). Since transcription regulation in general takes longer time than allosteric regulation and phosphorylation events, we reason that cells may have evolved this lincRNA-involved regulatory mechanism to maintain AMPK activation during long periods of energy stress and to help cells adapt better to chronic stress conditions. In support of this model, our time course experiments revealed that NBR2 deficiency compromised AMPK activation at later, but not earlier, time points upon glucose starvation (Supplemental Fig. 6a) (Note that all the energy stress experiments shown in our studies used 12 hour or longer treatment time points). This mirrors well with the kinetics of NBR2 expression induction upon glucose starvation (Supplemental Fig. 6b). Since glucose starvation also significantly promotes NBR2 binding to AMPK (Fig. 5), this presumably further amplifies the effect of NBR2 to promote AMPK activation.
NBR2 gene was originally identified as a gene which is located near to the breast cancer associated gene BRCA1. Both genes lie head to head with each other on human chromosome 17, and the physical distance between the transcription start sites of the two genes is only 218 bp (Supplemental Fig. 1)27. Given the frequent mutation/deletion rates of BRCA1 in human breast and ovarian cancers and the close proximity of NBR2 gene to BRCA1 gene, it was initially postulated that NBR2 should be co-deleted/mutated with BRCA1 in certain cancers (for example, see32), and NBR2 may also play a role in tumor suppression. However, later it became clear that NBR2 does not appear to encode a protein, and it was proposed that NBR2 simply is a “junk gene”33. Since then, its potential function in tumor biology has remained unknown. In this study, we identified NBR2 as a lincRNA induced by energy stress, and showed that NBR2 indeed functions to inhibit tumor development, at least in part through its regulation of AMPK activation. It is of note that NBR2 overexpression in AMPK deficient cells can still exert moderate cell proliferation suppressive effect (Fig. 7b), suggesting that NBR2 may have other AMPK-independent function(s) to regulate cell proliferation. Identification and characterization of other NBR2 binding proteins or RNAs will further clarify its function.
The most popular model proposed for lncRNA function probably is the one that lncRNAs regulate gene expression, either in cis or in trans, via recruiting other chromatin-modification complexes or transcription factors to specific loci34, 35. This raises the possibility that NBR2 may regulate the transcription of BRCA1 gene, which resides right next to NBR2 gene. However, our data showed that BRCA1 expression was not affected by either glucose starvation or NBR2 knockdown (Supplemental Fig. 7). We should mention that, although initially it was proposed that lincRNAs mainly function to regulate neighboring gene transcription, other studies have shown that many lincRNAs do not exert such function1. Whether NBR2 regulates any other gene transcription awaits further investigation.
Methods
Cell culture studies
Human kidney cancer cell lines, human breast cancer cell lines, human prostate cancer cell lines, human embryonic Kidney 293 cells used in this study were mostly obtained from American Type Culture Collection (ATCC). All of the cell lines were free of mycoplasma contamination (tested by the vendors using the MycoAlert kit from Lonza). No cell lines used in this study are found in the database of commonly misidentified cell lines (ICLAC and NCBI Biosample) based on short tandem repeats (STR) profiling performed by vendors. Hela or A549 cells with expression of empty vector or Lkb1 expression vectors were described in36. siRNA and plasmid transfections were performed using Lipofectamine 3000 (Life Technologies). Lentiviruses or retroviruses were produced in HEK293T cells with packing mix (ViraPower Lentiviral Expression System, Invitrogen) and used to infect target cells as per manufacturer's instruction. For glucose starvation experiments, cells were cultured in DMEM with different concentrations of glucose (0, 1, or 25 mM) + 10% dialyzed FBS. To measure apoptosis, the cells were stained by Annexin V kit per manufacturer instruction (BD Bioscience)37. Briefly, treated cells were washed with PBS twice and then 1×106 cells were resuspended in 100µL of 1× binding buffer. FITC-labeled Annexin V and propidium iodide were added to samples and incubated in dark for 15 minutes at room temperature. Subsequently cells were subjected to FACS analysis. Cell cycle analysis was carried out as previously reported using FITC BRDU Flow Kit (BD Bioscience)38,39. Cell growth and soft agar assays were conducted as described in our previous publications40,41. Briefly, for cell growth assay, cells were plated in 24 well plates and were determined by crystal violet staining. Cells were stained with 0.1% crystal violet (Sigma) solution for 15 min at room temperature. Stained crystal violet was then extracted with 10% acetic acid and the intensity of color was measured by photospectormetry at OD595. To assess anchorage-independent growth, 10,000 cells per well in 0.4% agarose on top of a bottom layer of 0.7% agarose were seed triplicate wells of 6-well plates. Upon the formation of colonies, soft agar plates were stained with iodonitrotetrazolium chloride (Sigma) and the colonies were counted manually.
Constructs and reagents
shRNAs targeting human NBR2 (NM_005821.2-615s1c1, NM_005821.2-514s1c1) were purchased from Sigma (Note that these two shRNAs target splicing isoforms #1 and #3 of NBR2, while can still achieve good knockdown efficiency when measured by real time PCR primer set designed to detect all three splicing isoforms of NBR2). siRNA targeting AMPKα were purchased from Origene (SR303721, SR303722). All three splicing isoforms of NBR2 were obtained from Thermo Fisher Scientific (MGC human NBR2 sequence-verified cDNAs, clone ID: 6452095, 4339497, 4826858) and then were subcloned into Lentiviral vector pLVX (Clontech). AMPK α, AMPK β and AMPK γ entry plasmids were obtained from Human ORFeome V5.1 library. The entry clones were subsequently recombined into gateway-compatible destination expression vectors with Flag tag through LR Gateway Technology (Invitrogen). cDNA corresponding to 1–312 a.a. of AMPK α1 was cloned into entry vector, and was subsequently recombined into gateway-compatible destination expression vectors with V5 tag through LR Gateway Technology (Invitrogen). Active human AMPK α2 protein and active human AMPK α1+AMPK β1+AMPK γ1 protein were purchase form Abcam (ab79803, ab126916). 2-Deoxy-D-glucose and compound C were purchased from Sigma (D6134, P5499). A-769662 was purchased from LC laboratories (A-1803).
Quantitative real-time PCR and RIP assay
Total RNA was extracted from cells using RNeasy (Qiagen) and 1st strand cDNA was prepared with High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, ABI). Real-time PCR was performed using QuantiTect SYBR Green PCR kit (Qiagen) or TaqMan Universal PCR Master Mix (ABI), and was run on Stratagene MX3000P. For quantification of gene expression, the 2−ΔΔCt method was used. GAPDH expression was used for normalization. RIP assay was performed with Magna RIP RNA-Binding Protein Immuno-precipitation Kit (Millipore). Briefly, cells were lysed in RIP lysis buffer. Then the lysates were immuno-precipitated with antibody or IgG along with protein magnetic beads. After proteinase K digestion, the RNAs pulled down with proteins were purified by phenol chloroform extraction and precipitated in ethanol. The RNAs were then re-suspended in RNAse-free water and cDNA was synthesized and subjected to real-time-PCR to detect NBR2 or GAPDH (internal control) transcripts. The RNA level was normalized with input (10%).
RNA pull-down assays
Biotin labeled RNAs were synthesized by Scientific TranscriptAid T7 High Yield Transcription Kit (Thermo). PCR primers with T7 promoters were used to amplify DNA templates for RNA synthesis, RNA transcribed in vitro with biotin RNA labelling mix and T7 RNA polymerase, treated with RNase-free DNase I (Roche), and purified with the RNeasy Mini Kit (Qiagen). Cells lysates or purified proteins were incubated with biotin-labeled RNAs overnight. The proteins associated with biotin-labeled RNAs were then pulled down with Streptavidin Magnetic Beads (Thermo) after 1-hour incubation. The proteins was then washed and used for Western blot analysis.
Western blot analysis
Tissues were lysed with RIPA buffer (20 mM Tris pH 7.5, 150 mM NaCl, 1% Nonidet P-40, 0.5% Sodium Deoxycholate, 1 mM EDTA, 0.1% SDS) containing complete mini protease inhibitors (Roche) and phosphatase inhibitor cocktail (Calbiochem). Cultured cells were lysed with NP40 buffer (150 mM sodium chloride, 1.0% NP-40, 50 mM Tris, pH 8.0) containing complete mini protease inhibitors (Roche) and phosphatase inhibitor cocktail (Calbiochem). Western blots were obtained utilizing 20 to 40 µg of lysate protein. The following antibodies were used in this study: anti-FLAG tag (M2) mouse monoclonal antibody (Sigma-Aldrich, F3165, 1:5000 dilution), Monoclonal Anti-Vinculin antibody (Sigma-Aldrich, V4505, 1:5000 dilution), Phospho-Acetyl-CoA Carboxylase (Ser79) Antibody (Cell Signaling Technology, 3661S, 1:1,000 dilution), Acetyl-CoA Carboxylase Antibody (Cell Signaling Technology, 3662S, 1:1,000 dilution), Phospho-p70 S6 Kinase (Thr389) Antibody (Cell Signaling Technology, 9205S, 1:1,000 dilution), Phospho-AMPKα (Thr172) (40H9) Rabbit mAb (Cell Signaling Technology, 2535S, 1:1,000 dilution), AMPKα (D63G4) Rabbit mAb (Cell Signaling Technology, 5832S, 1:1,000 dilution), AMPKα (F6) Mouse mAb (Cell Signaling Technology, 2793S, 1:1,000 dilution), AMPKβ1/2 (57C12) Rabbit mAb (Cell Signaling Technology, 4150S, 1:1,000 dilution) AMPKγ1 Antibody (Cell Signaling Technology, 4187S, 1:1,000 dilution), Phospho-Raptor (Ser792) Antibody (Cell Signaling Technology, 2083S, 1:1,000 dilution), Raptor (24C12) Rabbit mAb (Cell Signaling Technology, 2280S, 1:1,000 dilution), Phospho-S6 Ribosomal Protein (Ser240/244) (D68F8) XP® Rabbit mAb (Cell Signaling Technology, 5364S, 1:5,000 dilution), S6 Ribosomal Protein (5G10) Rabbit mAb (Cell Signaling Technology, 2217S, 1:5,000 dilution), ULK1 (D8H5) Rabbit mAb (Cell Signaling Technology, 8054 S, 1:1,000 dilution), Phospho-ULK1 (Ser555) (D1H4) Rabbit mAb (Cell Signaling Technology, 5869S, 1:1,000 dilution), Phospho-ULK1 (Ser757) Antibody (Cell Signaling Technology, 6888S, 1:1,000 dilution), Cleaved PARP (Asp214) Antibody (Human Specific) (Cell Signaling Technology, 9541S, 1:2,000 dilution), PARP Antibody (Cell Signaling Technology, 9542S, 1:2,000 dilution), Cleaved Caspase-3 (Asp175) (5A1E) Rabbit mAb (Cell Signaling Technology, 9664S, 1:500 dilution), FLCN (D14G9) Rabbit mAb (Cell Signaling Technology, 3697S, 1: 2,000 dilution), HSP90 (C45G5) Rabbit mAb #4877 (Cell Signaling Technology, 4877S, 1: 1,000 dilution), GAPDH (D16H11) XP® Rabbit mAb (Cell Signaling Technology, 5174S, 1: 5,000 dilution), p70 S6 kinase α Antibody (C-18) (Santa Cruz Biotechnology, sc-230, 1:1,000 dilution), SQSTM1 Antibody (H-290) (Santa Cruz Biotechnology, sc-52275, 1:2,000 dilution), LKB1 Antibody (E-9) (Santa Cruz Biotechnology, sc-374334, 1:2,000 dilution).
Subcellular fractionation
Cells were harvested by trypsin and washed twice with PBS. Cell pellets were lysed in buffer I containing 20 mM HEPES, 10 mM KCL, 2 mM MgCl2 and 0.5% NP40. After centrifugation, supernatants were collected as cytoplasmic lysis. Pellets were further lysed in buffer II containing 0.5 M NaCl, 20 mM HEPES, 10 mM KCL, 2 mM MgCl2 and 0.5% NP40. Supernatants were collected as nuclear lysis by centrifugation. Cytoplasmic and nuclear fractions were split for RNA extraction and real-time PCR or protein extraction and Western blotting. HSP90 and PARP were used as markers of cytoplasm and nucleus in Western blotting. GAPDH and U1 were used as markers of cytoplasm and nucleus in real-time PCR.
Immunofluorescence
Cells were cultured on glass coverslips in six-well plates, washed once with PBS, and fixed in 4% paraformaldehyde for 30 min. Coverslips were mounted using Immu-mount (Thermo Shandon) and images were captured using Olympus confocal microscope. For quantification of autophagic cells, cells with > 10 GFP-LC3 punctuate dots were considered positive. Positive cells was counted and expressed as a percentage of total autophagic cells.
AMPK kinase assay
in vitro AMPK kinase activity was assessed using Promega AMPK (A1/B1/G1) Kinase Enzyme System (V1921) according to the manufacturer’s instructions. In the kinase assay using ACC fragment, bacterial purified GST- ACC 1–130 aa protein was dialyzed against 20 mM Tris-HCl, pH8.0 and 10% glycerol at 4°C overnight. For AMPK kinase assay, SFB-AMPKα1, Flag-AMPKβ2 and Flag-AMPKγ1 plasmids were co-transfected into HEK293T cells. Cells were lysed 48 hours after transfection. AMPK complex was pulled down by S protein beads and subjected to the kinase assay in the presence of 500 µM cold ATP, 10µg in vitro synthesized RNAs and 1 µg GST fusion proteins mentioned above. The reaction mixture was incubated at 30°C for 30 min, terminated with SDS-loading buffer and subjected to SDS-PAGE for Western blot analysis. Phosphorylation of ACC at S79 site was determined by ACC S79 phospho-specific antibody.
Xenograft model
All Female athymic Nude-Foxn1nu mice animal (6-week-old) experiments were performed in accordance with a protocol approved by the Institutional Animal Care and Use Committee of MD Anderson Cancer Center which is in full compliance with policies of the Institutional Animal Core and Use Committee (IACUC). Animals arriving in our facility were randomly put into cages with five mice each. They were implanted with respective tumor cells in the unit of cages, which were randomly selected. MDA-MB-231 cells were counted and suspended at 1.0×107/ml in PBS, approximately one million cells were injected subcutaneously into the flank of each mouse with different genotypes, 5 mice per group. Tumor volume measurement was initiated at two weeks after injection (defined as starting time point: week 0). Tumor progression was then monitored by bi-dimensional tumor measurements every five days using a caliper until the endpoint. Mice were sacrificed at the endpoint and the tumors were excised for further experiments. The tumor volume was calculated according to the equation v = length*width2*1/2. The tumor volume at week n is expressed as Relative Tumor Volume (RTV) and calculated according to the following formula: RTV = TVn/TV0, where TVn is the tumor volume at week n and TV0 is the tumor volume at week 0. The investigators were not blinded to allocation during experiments and outcome assessment.
RNA-seq and computational analysis
RNA-seq was performed at Sequencing and Non-Coding RNA Program at the MD Anderson Cancer Center using Applied Biosystems SOLiD™ Next Generation Sequencing platform. The LifeScope v2.5.1 was used to align the reads to the genome, generate raw counts corresponding to each known gene (total 23080 genes, including 4325 non-coding genes), and calculate the RPKM (reads per kilobase per million) values. We considered only non-coding genes that expressed at relative high levels (RPKM > 2) and showed >2 or < 0.5 fold changes between control and treatment cells. This identified a list of 17 up-regulated and 39 down-regulated non-coding RNAs.
Kaplan Meier survival analysis of cancer patients
We utilized datasets of 4,142 breast tumors which had previously been profiled by Affymetrix microarray analysis (www.kmplot.com)42. NBR2 expression (probe set ID: 207631_at) was divided by the median into high vs. low expression. Survival analysis by Kaplan-Meier and Cox Proportional Hazard analysis were performed.
TCGA data analysis
We downloaded the level-3 gene expression data for NBR2 from TCGA pan-cancer project Synapse (Synapse ID: syn300013) for breast (BRCA) and kidney cancer (KIRC). We used paired student t-test to detect the statistical difference between matched tumor and normal samples. We used long-rank tests to detect the overall survival difference between patient groups.
Accession numbers
RNA seq datasets (786O cells with or without glucose treatment) have been deposited in the Gene Expression Omnibus website with accession code GSE77415. The datasets used in Figure.4g were generated from ref.43
Oligonucleotide sequences, probes and primers (forward and reverse)
qPCR primers for gene expression and RIP:
NBR2-Forward: 5’-GGAGGTCTCCAGTTTCGGTA-3’
-
NBR2-Reverse: 5’-TTGATGTGTGCTTCCTGGG-3’
(Note that this real time PCR primer set for NBR2 is designed to detect all three splicing isoforms of NBR2)
GAPDH-Forward: 5’- CCATGGGGAAGGTGAAGGTC-3’
GAPDH -Reverse: 5’-GAAGGGGTCATTGATGGCAAC-3’
U1-Forward: 5’-TCCCAGGGCGAGGCTTATCCATT-3’
U1-Reverse: 5’-GAACGCAGTCCCCCACTACCACAAAT-3’
BRCA1-Forward: 5’-TGTGCTTTTCAGCTTGACACAGG-3’
BRCA1-Reverse: 5’-CGTCTTTTGAGGTTGTATCCGCTG-3’
Primers for RNA pull down assay:
NBR2#1-Forward: 5’-TAATACGACTCACTATAGGG AGGGGTCCAGTTGCGGCTTAT -3’
NBR2#1-Reverse: 5’-AGTTT ACTTA CTATT GCTGA -3’
NBR2#1Anti-sence-Forward: 5’- TAATACGACTCACTATAGGG AGTTTACTTACTATT GCTGA -3’
NBR2#1Anti-sence -Reverse: 5’- GGGTCCAGTTGCGGCTTAT -3’
NBR2#2-Forward: 5’- TAATACGACTCACTATAGGG GTTGCGGCTTATTGCATCACA-3’
NBR2#2-Reverse: 5’- ACTATTGCTGATTTATTACAAAGGA -3’
NBR2#2Anti-sence-Forward: 5’-TAATACGACTCACTATAGGG ACTATTGCTGATTTA TTACAAAGGA -3’
NBR2#2Anti-sence -Reverse: 5’- GTTGCGGCTTATTGCATCACA-3’
NBR2#3-Forward: 5’- TAATACGACTCACTATAGGG AGCGGGGTTGCGGCTTATT-3’
NBR2#3-Reverse: 5’- TGGGATTGAGGAGGATCTTT -3’
NBR2#3Anti-sence-Forward: 5’- TAATACGACTCACTATAGGG TGGGATTGAGGAGGA TCTTT -3’
NBR2#3Anti-sence -Reverse: 5’- GGGTTGCGGCTTATTGCATC-3’
NBR2#T1-Forward: 5’- TAATACGACTCACTATAGGG GTAAAAGTTTTCATTTGATCTG AA-3’
NBR2#T1-Reverse: 5’- AGTTT ACTTA CTATT GCTGA-3’
NBR2#T3-Forward: 5’- TAATACGACTCACTATAGGG TTTGCTGAGGATAATGGCCT -3’
NBR2#T3-Reverse: 5’- AGTTT ACTTA CTATT GCTGA-3’
NBR2#T4-Forward: 5’- TAATACGACTCACTATAGGG AGGGGTCCAGTTGCGGCTTAT -3’
NBR2#T4-Reverse: 5’- TTCAGATCAAATGAAAACTTTTAC -3’
NBR2#T5-Forward: 5’- TAATACGACTCACTATAGGG AGGGGTCCAGTTGCGGCTTAT -3’
NBR2#T5-Reverse: 5’- CTTCCTGGGCTTCCAGCAC -3’
NBR2#T6-Forward: 5’- TAATACGACTCACTATAGGG AGGGGTCCAGTTGCGGCTTAT -3’
NBR2#T6-Reverse: 5’- AGGCCATTATCCTCAGCAAA -3’
Statistics and reproducibility
Most experiments were repeated 3–5 times to be eligible for the indicated statistical analyses, and the data exhibited normal distribution. There was no estimation of group variation before experiments. For gene expression and RIP assays, relative quantities of gene expression level were normalized. The relative quantities of RIP samples were normalized by individual inputs, respectively. All results are presented as mean ± the standard deviation (SD) of at least three independent experiments, unless otherwise noted. Each exact n values are indicated in the corresponding figure legend. Comparisons were performed using two-tailed paired Student's t-test (*P <0.05, **P <0.01, and ***P <0.001), as indicated in the individual figures. For animal studies, five mice per group is the standard sample size for tumor xenograft experiments, and no statistical method was used to predetermine sample size. None of the samples/animals was excluded from the experiment, and the animals were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment. For western blotting, representative images are shown. Each of these experiments was independently repeated 3–5 times. For survival analysis, the expression of NBR2 was treated as a binary variant and divided into `high' and `low' level. Kaplan-Meier survival curves were compared using the Gehan-Breslow test with GraphPad Prism (GraphPad Software). The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment.
Supplementary Material
Acknowledgments
We thank all members of the Gan laboratory for their advice and technical assistance. This research has been supported by grants from MD Anderson Cancer Center, US Department of Defense (TS093049), Cancer Prevention & Research Institute of Texas (RP130020), National Institutes of Health (CA181196 and CA190370), Ellison Medical Foundation (AG-NS-0973-13), and Gabrielle’s Angel Foundation for Cancer Research (to B. G.). B. G. is a Kimmel Scholar and an Ellison Medical Foundation New Scholar. H. L. is supported by the National Institutes of Health (CA143883, CA175486); the R. Lee Clark Fellow Award from The Jeanne F. Shelby Scholarship Fund; a grant from the Cancer Prevention and Research Institute of Texas (RP140462); and the Mary K. Chapman Foundation and the Lorraine Dell Program in Bioinformatics for Personalization of Cancer Medicine. L. H. is supported by Cancer Prevention & Research Institute of Texas (RR150085). H. L. is supported by the National Institutes of Health (CA182424 and CA193813). B. G., J. C., J. W. and H. L. are members of the M.D. Anderson Cancer Center, and are supported by the National Institutes of Health Core Grant CA016672.
Footnotes
Author Contributions
Z. X. initiated the project and identified NBR2 as an energy stress-induced lncRNA. X. L. performed most of the experiments with the assistance from Z. X., H. L., and L. Z.. J. Z. and J. W. analyzed RNA-seq dataset. L. H. and H. L. conducted computational analysis on NBR2 expression and status in human cancers. W. W, J. C., S. L., H. L. provided reagents. B. G. supervised the study. X. L., Z. X., and B. G. designed the experiments and wrote the manuscript. All authors commented on the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huarte M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prensner JR, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nature biotechnology. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engreitz JM, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Heesch S, et al. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome biology. 2014;15:R6. doi: 10.1186/gb-2014-15-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13:251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hawley SA, et al. Complexes between the LKB1 tumor suppressor, STRAD alpha/beta and MO25 alpha/beta are upstream kinases in the AMP-activated protein kinase cascade. Journal of biology. 2003;2:28. doi: 10.1186/1475-4924-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woods A, et al. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr Biol. 2003;13:2004–2008. doi: 10.1016/j.cub.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 15.Shaw RJ, et al. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc Natl Acad Sci U S A. 2004;101:3329–3335. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardie DG, Schaffer BE, Brunet A. AMPK: An Energy-Sensing Pathway with Multiple Inputs and Outputs. Trends in cell biology. 2015 doi: 10.1016/j.tcb.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laplante M, Sabatini DM. mTOR Signaling in Growth Control and Disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gwinn DM, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 20.Egan DF, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang J, Manning BD. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem J. 2008;412:179–190. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hezel AF, Bardeesy N. LKB1; linking cell structure and tumor suppression. Oncogene. 2008;27:6908–6919. doi: 10.1038/onc.2008.342. [DOI] [PubMed] [Google Scholar]
- 25.Alessi DR, Sakamoto K, Bayascas JR. LKB1-dependent signaling pathways. Annual review of biochemistry. 2006;75:137–163. doi: 10.1146/annurev.biochem.75.103004.142702. [DOI] [PubMed] [Google Scholar]
- 26.Faubert B, et al. AMPK is a negative regulator of the Warburg effect and suppresses tumor growth in vivo. Cell Metab. 2013;17:113–124. doi: 10.1016/j.cmet.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu CF, et al. Isolation and characterisation of the NBR2 gene which lies head to head with the human BRCA1 gene. Hum Mol Genet. 1997;6:1057–1062. doi: 10.1093/hmg/6.7.1057. [DOI] [PubMed] [Google Scholar]
- 28.Sim AT, Hardie DG. The low activity of acetyl-CoA carboxylase in basal and glucagon-stimulated hepatocytes is due to phosphorylation by the AMP-activated protein kinase and not cyclic AMP-dependent protein kinase. FEBS letters. 1988;233:294–298. doi: 10.1016/0014-5793(88)80445-9. [DOI] [PubMed] [Google Scholar]
- 29.Bungard D, et al. Signaling kinase AMPK activates stress-promoted transcription via histone H2B phosphorylation. Science. 2010;329:1201–1205. doi: 10.1126/science.1191241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corradetti MN, Inoki K, Bardeesy N, DePinho RA, Guan KL. Regulation of the TSC pathway by LKB1: evidence of a molecular link between tuberous sclerosis complex and Peutz-Jeghers syndrome. Genes Dev. 2004;18:1533–1538. doi: 10.1101/gad.1199104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hardie DG. AMPK-Sensing Energy while Talking to Other Signaling Pathways. Cell Metab. 2014;20:939–952. doi: 10.1016/j.cmet.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gad S, et al. Characterisation of a 161 kb deletion extending from the NBR1 to the BRCA1 genes in a French breast-ovarian cancer family. Human mutation. 2003;21:654. doi: 10.1002/humu.9148. [DOI] [PubMed] [Google Scholar]
- 33.Jin H, et al. Structural evolution of the BRCA1 genomic region in primates. Genomics. 2004;84:1071–1082. doi: 10.1016/j.ygeno.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 34.Pandey RR, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 35.Feng J, et al. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee SW, et al. Skp2-dependent ubiquitination and activation of LKB1 is essential for cancer cell survival under energy stress. Mol Cell. 2015;57:1022–1033. doi: 10.1016/j.molcel.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin A, et al. The FoxO-BNIP3 axis exerts a unique regulation of mTORC1 and cell survival under energy stress. Oncogene. 2014;33:3183–3194. doi: 10.1038/onc.2013.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gan B, et al. Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature. 2010;468:701–704. doi: 10.1038/nature09595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gan B, et al. mTORC1-dependent and -independent regulation of stem cell renewal, differentiation, and mobilization. Proc Natl Acad Sci U S A. 2008;105:19384–19389. doi: 10.1073/pnas.0810584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gan B, et al. FoxOs enforce a progression checkpoint to constrain mTORC1-activated renal tumorigenesis. Cancer Cell. 2010;18:472–484. doi: 10.1016/j.ccr.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin A, et al. FoxO transcription factors promote AKT Ser473 phosphorylation and renal tumor growth in response to pharmacological inhibition of the PI3K-AKT pathway. Cancer Res. 2014 doi: 10.1158/0008-5472.CAN-13-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PloS one. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gyorffy B, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast cancer research and treatment. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.