Letter to Editor
Large granular lymphocyte (LGL) leukemia is a rare, clonal disease characterized by a persistent increase in the number of CD8+ cytotoxic T cells or CD16/56+ natural killer (NK) cells (1). Patients are prone to recurrent infections and often suffer from severe cytopenias and autoimmune diseases that are thought to be mediated by cytotoxic LGL lymphocytes. LGL leukemia is believed to begin as an antigen-driven immune response followed by the constitutive activation of cytotoxic T lymphocytes or NK cells. Overall, studies have highlighted the dysregulation of different apoptotic pathways (e.g. sphingolipid and FAS/FAS ligand) and the activation of survival signaling pathways (e.g. PI3K/AKT and RAS)(1). Previously, activating mutations in the Src-like homology 2 (SH2) domain of the signal transducer and activator of transcription 3 (STAT3) gene were discovered in 40 % of LGL leukemia patients (2). These findings have later been validated in other cohorts, reporting STAT3 mutation frequency to be even up to 73% in selected patient groups (3). Interestingly, aberrant STAT3 signaling is seen in almost all LGL leukemia patients (4), indicating that the JAK/STAT pathway is activated through other mechanisms as well. In addition to LGL leukemia, activating somatic STAT3 mutations have recently been found in other diseases including T-, NK- and B-cell lymphomas (5–7), hepatocellular adenomas (8), and chronic lymphoproliferative disorders of NK cells (CLPD-NKs)(9). In a clinical phase II study, it was shown that the strongest activating STAT3 mutation, Y640F, predicted therapeutic response to immunosuppressive methotrexate (MTX) in LGL leukemia, as all patients with this mutation responded to therapy after at least 4 cycles of MTX (10).
The diagnosis of T-LGL leukemia is often challenging as clinical symptoms and immunophenotypical findings bear a resemblance to other reactive conditions, making the distinction between clear malignant lymphoproliferation and reactive processes difficult. To discover novel genetic markers, we collected samples from LGL leukemia patients (n=106) who were confirmed to have no STAT3 and STAT5 hotspot mutations in the SH2-domain by exome or amplicon sequencing (11). 88 patients had T-LGL leukemia and 18 cases NK-LGL leukemia. All patients met the criteria of LGL leukemia as defined by the World Health Organization (WHO) in 2008. The study was undertaken in compliance with the principles of the Helsinki declaration and was approved by the ethics committees in the Helsinki University Central Hospital (Helsinki, Finland), Cleveland Clinic (Cleveland, Ohio), University of Virginia Cancer Center (Charlottesville, Virginia) and the Penn State Hershey Cancer Institute (Hershey, Pennsylvania). All patients and healthy controls gave written informed consents.
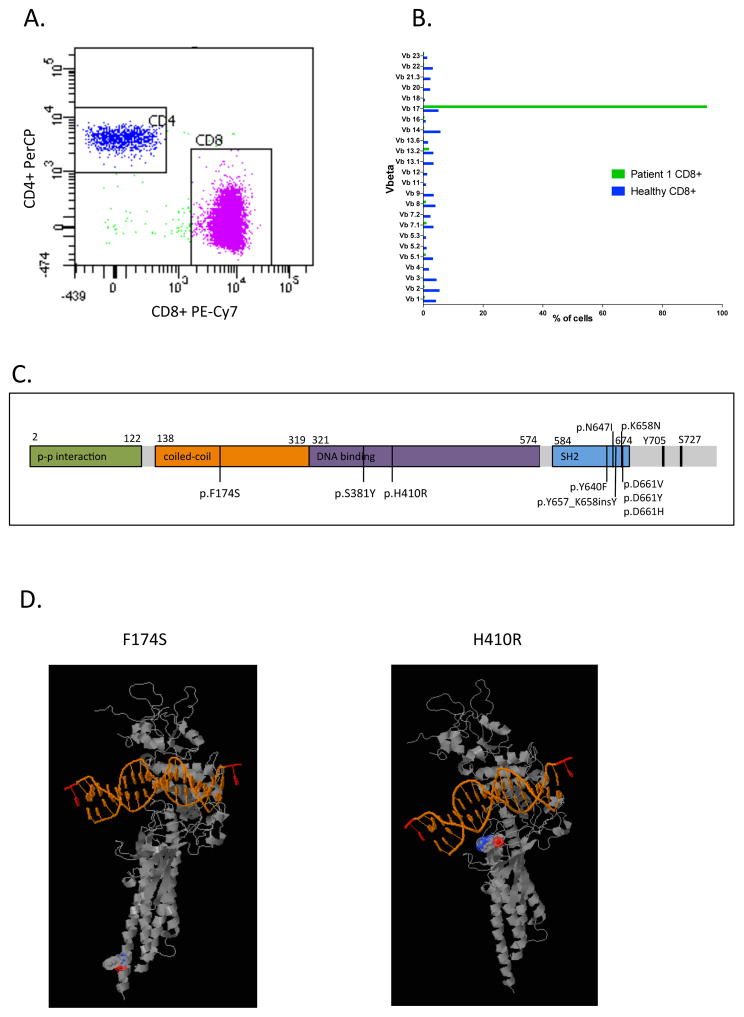
Exome sequencing of seven LGL leukemia patients was performed on CD8+ T-cells (tumor) and matched CD4+ T-cells as control. The exome was captured with the Nimblegen SeqCap EZ Exome Library v2.0 and the sequencing was performed with the Illumina HiSeq2000 sequencing platform. Candidate somatic mutations were identified with a bioinformatics pipeline as described earlier (2). In one of the 7 patients analyzed, a novel STAT3 mutation was found outside the SH2-hotspot area (patient #1, Supplemental table S1). This patient harbored a heterozygous missense mutation H410R in exon 13 of the DNA-binding domain (Supplemental table S2). The variant allele frequency (VAF) was 49% in the CD8+ fraction and the H410R variant was not observed in the CD4+ fraction. The CD8+ population consisted of one major clone (vb.17: 95%) according to the TCR Vβ results (Figure 1A–B). The H410 position is highly conserved and results in conversion of histidine to arginine (Figure 1C), which would predict for a slight increase in hydrophilicity. Patient 1 also harbored a missense mutation P1324R in FANCA and a mutation giving rise to a truncated form of MLL2 (Q1893*) (Supplemental table S2). In other 6 exome sequenced patient samples no additional novel STAT3 mutations were found (Supplemental table S2). As the mutation spectrum outside the SH2-domain hotspot in STAT3 has not been explored systematically before, we developed a targeted deep amplicon sequencing platform that allows for the sensitive analysis of all 23 coding exons of the STAT3 gene. A variant was called when variant base frequency was 0.5 % of all reads covering a given a position. Using this system, we sequenced mononuclear cell samples from 99 LGL-leukemia patients previously confirmed to be STAT3 hotspot mutation negative. From these, 3 additional patients were discovered to have STAT3 missense mutations outside of the SH2-domain (Figure 1C). Patient 2 exhibited the same H410R mutation seen in the DNA-binding domain (MNC VAF: 8.8%) and patient 3 had a S381Y mutation (MNC VAF 7%). Patient 4 had a novel F174S mutation (VAF 54% in sorted CD8+ cells, not detected in CD4+ cells) in exon 6 of the coiled-coil domain of STAT3 (Figure 1D). The clinical phenotype of patients carrying STAT3 DNA-binding and coiled coil domain mutations did not differ from other typical LGL leukemia cases (Supplemental Table S1).
Figure 1. Sorting, vbeta and sequencing results from patient 1.
A. The index patient is a 60-year-old male diagnosed with T-LGL leukemia in 2002 suffering from anemia, neutropenia, B-cell dyscrasia and hypergamma-globulinemia (Supplemental table S1). Mononuclear cells (MNCs) were separated from whole blood with Ficoll-Paque™ PLUS (GE Healthcare). The MNCs were then sorted by flow cytometry using markers for CD45+ (PerCP), CD3+ (FITC), CD4+ (APC) and CD8+ (PE) (Becton Dickinson). Based on flow cytometry the CD8+ tumor cells (90% of CD3+ population) and the CD4+ control cells (9% of the CD3+ population) were sorted.
B. Peripheral blood from the patient was used to determine the TCR Vβ repertoire of human T lymphocytes with the IO Test® Beta Mark TCR Vβ Repertoire Kit (Beckman-Coulter Immunotech). Flow cytometry measurement of the TCR repertoire of patient 1 revealed a 95% Vb.17 clone in the CD8+ cells.
C. Location of novel mutations in the coiled-coil alpha domain and DNA-binding domain in addition to the previously described SH2 domain mutations in STAT3.
D. 3D visualization of the location of the variants F174S and H410R in STAT3 (Polyphen-2).
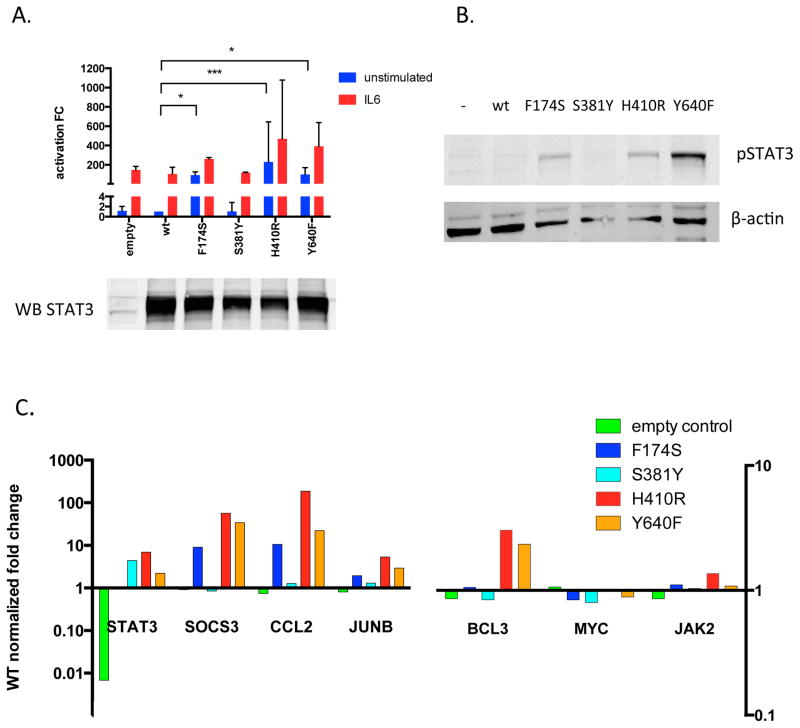
To explore the functional implications of the coiled-coil and DNA-binding domain mutations, we generated expression constructs for wild-type, F174S, S381Y, H410R, and Y640F variants of the STAT3 protein. Luciferase measurements of SIE-reporter HEK-293 cells transfected with these constructs revealed a noticeable increase in both basal and IL6-stimulated transcriptional activation induced by the coiled-coil domain F174S and DNA-binding domain H410R variants compared to wild type STAT3 (Figure 2A). The S381Y variant on the other hand did not show a similar increase. The phosphorylation status of the variants was investigated with Western blotting and revealed F174S and H410R to induce a phosphorylation pattern comparable to the known activating Y640F SH2-domain mutation (Figure 2B). However, the S381Y variant did not seem to be highly phosphorylated as the level of pSTAT3 was similar as seen with the wild-type STAT3. Furthermore, expression levels of six known STAT3 target genes were measured by qPCR, and SOCS3, CCL2, JUNB and BCL3 were revealed to be upregulated by the F174S, H410R and Y640F mutations but not by the S381Y variant when compared to the wild-type (Figure 2C).
Figure 2. Functional characterization of the variants outside of the SH2-domain in STAT3.
A. HEK cells harboring an sis-inducible (SIE) response element luciferase reporter were transfected with vector alone (empty), wild type STAT3 (wt), the three identified mutants (F174S, S381Y and H410R) or the most prevalently seen SH2-domain mutant (Y640F). The cells were either left unstimulated or treated for 3h with IL-6 to induce STAT3 phoshorylation and activation. Luciferase activity was determined with ONE-Glo™ Luciferase Assay System and results are reported as fold change (FC) compared to unstimulated wild type STAT3. Each condition was tested in triplicate and the statistical significance was calculated with Kruskal-Wallis and Dunn’s multiple comparison tests (*=p<0.05, ***=p<0.0001, error bars representing SD).
B. To investigate the phosphorylation status of the variants, HEK-293 cells transfected with the above mentioned variants were analyzed by western blot with a phosphoSTAT3 specific antibody. Protein lysates of the different variants were separated on an SDS-PAGE gel and transferred to a nitrocellulose membrane. STAT3 protein levels of the different variants were used to normalize for the transfection efficacy. β-actin was used as a loading control.
C. Total RNA of the transfected HEK293 cells was extracted and converted to cDNA using random primers. Expression levels of six known STAT3 target genes (SOCS3, JAK2, MYC, JUNB, BCL3, CCL2) were measured and the results were normalized against four housekeeping genes (NONO, PGK-1, GAPDH, LDHA), which showed uniform expression across all samples. All reactions were run in triplicate wells and gene expression was quantified using the delta-delta Cq method.
The H410 position is highly conserved and results in conversion of histidine to arginine. Arginine residues within the DNA binding domain of STAT3 have been found to promote intracellular shuttling and phosphorylation of STAT3 (12). An arginine–glutamine-exchange at the STAT3 moieties R414 and R417 reduces cytokine-dependent tyrosine phosphorylation of STAT3. The H410R variant seen in two LGL-leukemia patients is located near this area and results in one extra arginine residue. It is plausible that this increase in hydrophilicity within the DNA binding domain mediates the activation of STAT3. The other found mutation in the DNA binding domain (S381Y) was not confirmed to be activating as no increased transcriptional activity, phosphorylation or increased expression of target genes was observed. Previously, patients with severe autoimmune diseases including type 1 diabetes and immunodysregulation polyendocrinopathy enteropathy X-linked–like syndrome (IPEX) were found to harbor activating heterozygous germline mutations in the DNA-binding domain (13).
The F174S coiled-coiled domain mutation causes a conversion from hydrophobic, nonpolar phenylalanine to uncharged, polar serine. The coiled-coil domain has previously been shown to be essential in receptor binding mediated by the SH2-domain as well as subsequent activation. In concordance with our results, the α1 region encompassing amino acids 130–198 have been shown to be essential for STAT3 tyrosine phosphorylation as the deletion of α1 abolished tyrosine phosphorylation of STAT3 in EGF-induced COS-1 cells (14). One possible mechanism for the involvement of the coiled-coil domain in STAT3 phosphorylation is through augmentation of the protein-protein interaction between STAT3 and the IL6 receptor during recruitment of STAT3 to the activated receptor following ligand binding. Mutations in the coiled coil domain have previously been seen in patients with inflammatory hepatocellular adenomas (IHCA) where the E166Q mutant was shown to be constitutively phosphorylated on Tyr705, hypersensitive to IL6-stimulation and able to translocate to the nucleus(8). Similarly in diffuse large B cell lymphoma (DLBCL)(15) the M206K STAT3 mutation was able to activate STAT3 signaling.
In conclusion, T-LGL leukemia patients without STAT3 SH2-domain mutations harbor novel activating mutations in the DNA-binding and coiled-coil domain of STAT3. The frequency of mutations was 3.8% (4 of 106 patients) in patients with no previously detected hotspot mutations in STAT3 or STAT5. These findings strengthen the evidence for a central role of STAT3 in the pathogenesis of LGL-leukemia. Subsequently, the importance of screening the entire STAT3 gene in the diagnostic workup of LGL-leukemia, as well as in other lymphoid malignancies where STAT3 mutations have previously been discovered, is further emphasized.
Supplementary Material
Acknowledgments
This work was supported by the Academy of Finland, the Finnish Cancer Institute, the Finnish Cancer Societies, Sigrid Juselius Foundation, Instrumentarium Science Foundation, State funding for university-level health research in Finland, European Regional Development Fund, Swedish Cultural Foundation, Blood Disease Foundation, Signe and Ane Gyllenberg foundation and the Finnish Cultural Foundation. Work of J.P.M. and M.J.C. was supported in part by the National Institutes for Health (grants 2K24HL077522, R01 CA127264A and R01AI085578). The work of T.P.L and T.O. was supported by the National Cancer Institute (RO1 CA 098472) and the National Institutes for Health (T32CA009109).
Personnel at the Hematology Research Unit Helsinki and FIMM are acknowledged for their expert clinical and technical assistance.
Footnotes
Supplementary information is available at Leukemia’s website.
Disclosure of conflicts of interest
H.R. has received honoraria from Novartis. S.M. has received honoraria and research funding from Novartis, Pfizer and Bristol-Myers Squibb.
References
- 1.Sokol L, Loughran TP., Jr Large granular lymphocyte leukemia. Current hematologic malignancy reports. 2007;2(4):278–82. doi: 10.1007/s11899-007-0038-7. [DOI] [PubMed] [Google Scholar]
- 2.Koskela HL, Eldfors S, Ellonen P, van Adrichem AJ, Kuusanmaki H, Andersson EI, et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. The New England journal of medicine. 2012;366(20):1905–13. doi: 10.1056/NEJMoa1114885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fasan A, Kern W, Grossmann V, Haferlach C, Haferlach T, Schnittger S. STAT3 mutations are highly specific for large granular lymphocytic leukemia. Leukemia. 2013;27(7):1598–600. doi: 10.1038/leu.2012.350. [DOI] [PubMed] [Google Scholar]
- 4.Epling-Burnette PK, Liu JH, Catlett-Falcone R, Turkson J, Oshiro M, Kothapalli R, et al. Inhibition of STAT3 signaling leads to apoptosis of leukemic large granular lymphocytes and decreased Mcl-1 expression. The Journal of clinical investigation. 2001;107(3):351–62. doi: 10.1172/JCI9940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kucuk C, Jiang B, Hu X, Zhang W, Chan JK, Xiao W, et al. Activating mutations of STAT5B and STAT3 in lymphomas derived from gammadelta-T or NK cells. Nature communications. 2015;6:6025. doi: 10.1038/ncomms7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohgami RS, Ma L, Monabati A, Zehnder JL, Arber DA. STAT3 mutations are present in aggressive B-cell lymphomas including a subset of diffuse large B-cell lymphomas with CD30 expression. Haematologica. 2014;99(7):e105–7. doi: 10.3324/haematol.2013.101543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohgami RS, Ma L, Merker JD, Martinez B, Zehnder JL, Arber DA. STAT3 mutations are frequent in CD30+ T-cell lymphomas and T-cell large granular lymphocytic leukemia. Leukemia. 2013;27(11):2244–7. doi: 10.1038/leu.2013.104. [DOI] [PubMed] [Google Scholar]
- 8.Pilati C, Amessou M, Bihl MP, Balabaud C, Nhieu JT, Paradis V, et al. Somatic mutations activating STAT3 in human inflammatory hepatocellular adenomas. The Journal of experimental medicine. 2011;208(7):1359–66. doi: 10.1084/jem.20110283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jerez A, Clemente MJ, Makishima H, Koskela H, Leblanc F, Peng Ng K, et al. STAT3 mutations unify the pathogenesis of chronic lymphoproliferative disorders of NK cells and T-cell large granular lymphocyte leukemia. Blood. 2012;120(15):3048–57. doi: 10.1182/blood-2012-06-435297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loughran TP, Jr, Zickl L, Olson TL, Wang V, Zhang D, Rajala HL, et al. Immunosuppressive therapy of LGL leukemia: prospective multicenter phase II study by the Eastern Cooperative Oncology Group (E5998) Leukemia. 2015;29(4):886–94. doi: 10.1038/leu.2014.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajala HL, Eldfors S, Kuusanmaki H, van Adrichem AJ, Olson T, Lagstrom S, et al. Discovery of somatic STAT5b mutations in large granular lymphocytic leukemia. Blood. 2013;121(22):4541–50. doi: 10.1182/blood-2012-12-474577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ginter T, Fahrer J, Krohnert U, Fetz V, Garrone A, Stauber RH, et al. Arginine residues within the DNA binding domain of STAT3 promote intracellular shuttling and phosphorylation of STAT3. Cellular signalling. 2014;26(8):1698–706. doi: 10.1016/j.cellsig.2014.03.033. [DOI] [PubMed] [Google Scholar]
- 13.Flanagan SE, Haapaniemi E, Russell MA, Caswell R, Lango Allen H, De Franco E, et al. Activating germline mutations in STAT3 cause early-onset multi-organ autoimmune disease. Nature genetics. 2014;46(8):812–4. doi: 10.1038/ng.3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang T, Kee WH, Seow KT, Fung W, Cao X. The coiled-coil domain of Stat3 is essential for its SH2 domain-mediated receptor binding and subsequent activation induced by epidermal growth factor and interleukin-6. Molecular and cellular biology. 2000;20(19):7132–9. doi: 10.1128/mcb.20.19.7132-7139.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu G, Witzig TE, Gupta M. A novel missense (M206K) STAT3 mutation in diffuse large B cell lymphoma deregulates STAT3 signaling. PloS one. 2013;8(7):e67851. doi: 10.1371/journal.pone.0067851. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.