Abstract
Metallo-beta-Lactamases (MBLs) are class B β-lactamases that hydrolyze almost all clinically-available β-lactam antibiotics. MBLs feature the distinctive αβ/βα sandwich fold of the metallo-hydrolase / oxidoreductase superfamily and possess a shallow active-site groove containing one or two divalent zinc ions, flanked by flexible loops. According to sequence identity and zinc ion dependence, MBLs are classified into three subclasses (B1, B2 and B3), of which the B1 subclass enzymes have emerged as the most clinically significant. Differences among the active site architectures, the nature of zinc ligands, and the catalytic mechanisms have limited the development of a common inhibitor. In this review, we will describe the molecular epidemiology and structural studies of the most prominent representatives of class B1 MBLs (NDM-1, IMP-1 and VIM-2) and describe the implications for inhibitor design to counter this growing clinical threat.
Keywords: β-lactams, Metallo-beta-Lactamases, β-lactamase inhibitors, antibiotic resistance
Introduction
Eighty-five years after the discovery of the “mould broth filtrate” better known as penicillin [1, 2], β-lactams still remain the most common antibiotics used. Their antibacterial effect is based on disruption of the cell wall [3, 4], yet their molecular mechanisms have remained somewhat enigmatic. The cell wall consists of a peptidoglycan polymer comprised of alternating N-acetylmuramic acid (NAM) and N-acetylglucosamine (NAG) units. These units are linked by transglycosidases, and further crosslinked through modified peptides attached to each NAM unit, forming a truly “giant macromolecule” [5] that completely surrounds the cell, providing shape and rigidity that protects the cell from lysis due to osmotic pressure.
β-Lactams are defined by their four membered cyclic amide ring, and have been proposed to structurally mimic the terminal D-Ala-D-Ala bond normally cleaved by transpeptidases during peptidoglycan crosslinking [6]. After binding to the active site of transpeptidases, the β-lactam bond is attacked by the same active-site serine nucleophile used by transpeptidases during normal catalysis, but the resulting covalent ester intermediate is long lived and not easily displaced by water, or by the usual substrate, an incoming amine nucleophile from a neighboring NAM-linked peptide [7]. The resulting inhibition decreases crosslinking, mechanically weakening the cell wall.
In addition to inhibition of transpeptidases, β-lactams also covalently modify a number of different proteins, collectively termed penicillin-binding proteins (PBPs) [8]. The inhibition of multiple PBPs and loss of cell wall integrity leads to a futile cycle whereby the peptidoglycan is synthesized and degraded, depleting pools of peptidoglycan precursors and leading to enzyme-mediated lysis as well as other lysis-independent effects [9, 10]. The polypharmacology (multiple targets) of β-lactams as well as their “dominant negative” effect inhibiting non-targeted enzymes due to depleting peptidoglycan precursors, may both contribute to the efficacy and the endurance of this class of drugs [10].
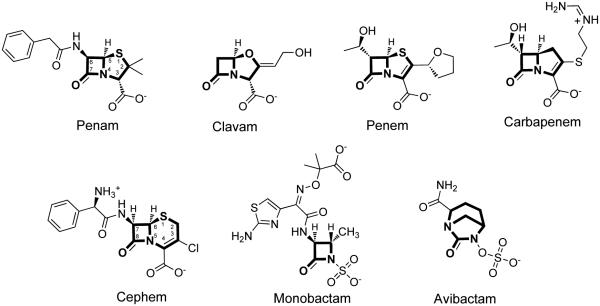
Driven by the success of penicillin and the rise of resistance mechanisms, the β-lactam scaffold of penicillin was extensively modified, and is now incorporated into a diversity of structures encompassed by at least six different clinically-used subtypes including penams, clavams, penems, carbapenems, cephems, and monobactams (Figure 1) [11]. The penams (e.g. benzylpenicillin) are bicyclic β-lactams in which the fused ring contains a sulfur at position 1, and a carboxylate moiety at the C-3 position. The clavams (e.g. clavulanic acid) have a similar bicyclic core and carboxylate substituent, but the ring sulfur is replaced by oxygen. The penems (e.g. faropenem) have the same bicyclic ring system, heteroatom substitution, and carboxylate substitutent as penams, but the bond between C-2 and C-3 is oxidized to an alkene. Carbapenems (e.g. imipenem) have the same core structure as penems, but the ring sulfur is replaced by a methylene. Derivatives of the β-lactam scaffold also include deviation from the original bicyclic ring structure of penicillin. Fused to the β-lactam ring, the cephems (e.g. cefaclor) instead have an expanded six-member ring with an alkene placed between positions C-3 and C-4, yet these derivatives still maintain both the sulfur at position 1 and an anionic carboxylate substituent alpha to the lactam nitrogen (now numbered position C-4). In contrast, the monobactams (e.g. aztreonam) lack the second fused ring altogether, yet aztreonam also incorporates an alternative anionic group (a sulfamate) directly to the lactam nitrogen.
Figure 1.
Chemical structures of selected examples from six β-lactam structural categories. Examples include a penam (benzylpenicillin), clavam (clavulanic acid), penem (faropenem), carbapenem (imipenem), cephem (cefaclor), monobactam (aztreonam), and the γ-lactam avibactam. The core scaffold for each category is shown in bold.
Although these categories represent major classes of β-lactams, many other structural modifications are present in both experimental and clinical compounds. For example, the recent development of avibactam (Figure 1) illustrates how the β-lactam ring itself can be expanded to produce a bridged γ-lactam derivative that nevertheless maintains a similar covalent mechanism of action [12]. However, in general, the β-lactam bond and the neighboring anionic group are conserved throughout these drugs, presumably because they are required to mimic the scissile amide bond and the terminal carboxylate, respectively, of transpeptidase substrates.
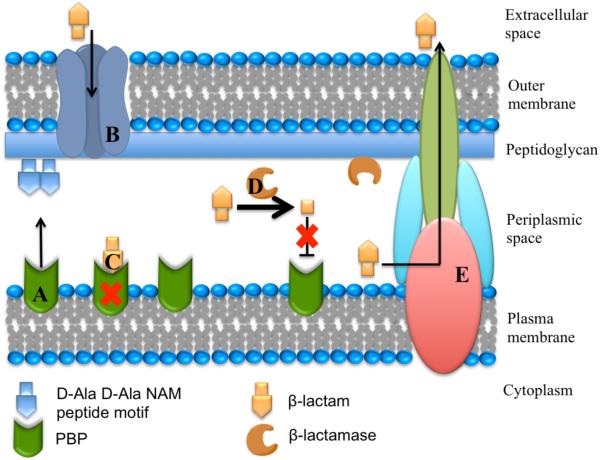
In response to selective pressures imposed by either clinically-used or naturally-occurring β-lactams, bacteria have developed multiple resistance mechanisms. Some of these mechanisms include expression of efflux pumps, reduced membrane permeability, mutated transpeptidases, and most importantly, expression of β-lactamases (Figure 2) [13, 14]. β-Lactamases catalyze the hydrolysis of the β-lactam amide bond to generate ring-opened products that are no longer capable of transpeptidase inhibition. Based on amino acid sequence identity, these enzymes are classified into four classes A, B, C and D [15]. Classes A, C and D contain enzymes that use an active-site serine nucleophile and a transient covalent reaction intermediate to catalyze the hydrolysis of β-lactam drugs. In contrast, class B contains the Metallo-Beta-Lactamases (MBLs), which catalyze hydrolysis of β-lactam drugs through a non-covalent mechanism in which one or two equivalents of bound zinc ions promote formation of a nucleophilic hydroxide that substitutes for the Ser residue used by class A, C, and D β-lactamases [16]. Details of the proposed hydrolysis mechanism for MBLs will be discussed below.
Figure 2.
Schematic representation of the Gram-negative bacterial membrane structures, including the interaction of β-lactam antibiotics and β-lactamases. A. Penicillin-binding proteins (PBPs) with transpeptidase activity are involved in cell wall biosynthesis by catalyzing the cross-linking of adjacent peptides at the terminal D-Ala-D-Ala portion of the peptidoglycan stem peptide. B. β-Lactams typically enter the periplasmic space through porins. C. β-Lactams covalently inhibit transpeptidases by mimicking transpeptidase substrates, but leading instead to a long-lived inhibitory covalent ester adduct. D. β-Lactamase enzymes are exported to the periplasm (two examples are known to attach to the inner leaflet of the outer membrane by an N-terminal lipid modification – see text for references) and catalytically hydrolyze β-lactam antibiotics to products that no longer inhibit transpeptidases. E. Bacteria can also avoid the action of antibiotics by actively transporting them to the external environment through efflux pumps.
MBLs
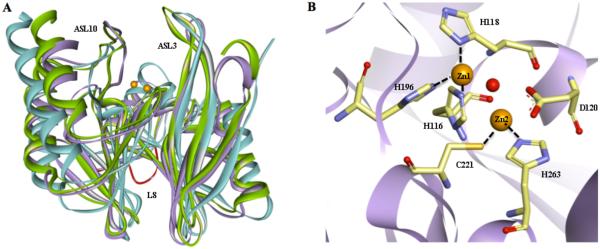
Class B β-lactamases (MBLs) are further divided into three subclasses defined primarily by differences in the primary zinc coordination shell [17]. Subclass B1 binds one zinc ion (Zn1) with three His residues (H116, H118, H196) and a second zinc ion (Zn2) with three different residues, notably including a Cys (D120, C221, H263). The B1 β-lactamases contain the largest number of clinically-relevant members, including VIMs (Verona integrin-encoded MBL), IMPs (imipenemase) and NDMs (New Delhi MBL), among others. Subclass B2 β-lactamases have a Zn1 binding site with one altered residue (N116, H118, H196), but retain a similar Zn2 site (D120, C221, H263). This subclass has the fewest members and includes enzymes produced by different species of Aeromonas, such as A. hydrophila CphA (Aeromonas carbapenem-hydrolyzing β-lactamase) [18], A. veronii ImiS (imipenem hydrolyzing metallo-β-lactamase from A. veronni bv. Sobria) [19], and Serratia fonticola Sfh-I (MBL hydrolase from Serratia fonticola) [20], among others. Finally, subclass B3 β-lactamases have a varied Zn1 binding site (H/Q116, H118, H196) and a distinctive Zn2 binding site that lacks a Cys residue (D120, H121, H263). This subclass includes Stenotrophomonas maltophilia L1 (β-lactamase 1) and Elizabethkingia meningosepticum (Chryseobacterium meningosepticum) GOB-1 (class B β-lactamase of E. meningosepticum) MBLs [21, 22], among others. Despite the low sequence conservation of MBLs (typically < 25% amino acid identities), they all exhibit a similar protein fold, and share mechanistic similarities, as described below.
In contrast to most class A, C, and D enzymes, B1 MBLs have unusually broad substrate profiles and can inactivate virtually all clinically-used bicyclic β-lactams. Notably, the MBLs’ use of a non-covalent catalytic mechanism renders commercially available serine β-lactamase-inhibiting drugs, such as sulbactam, tazobactam, clavulanic acid and avibactam, ineffective since these inhibitors work by trapping a covalent adduct with the active-site Ser residue of serine β-lactamases, which is absent in the non-homologous MBL enzymes. The β-lactam substrate spectrum of MBLs, their resistance to all approved drugs, the rapid pace at which new variants are isolated, the transferability of their genes and their isolation from environmental (not just nosocomial) sources are their most worrisome features [23-25]. Herein, we describe three prominent subgroups of class B1 MBLs, the NDMs, IMPs and VIMs, by detailing their molecular epidemiology and structural features. From these studies, we also draw implications for the design of more effective MBL inhibitors. The clinical challenge is to develop effective broad-spectrum MBL inhibitors that are still selective enough to avoid toxicity due to inhibition of off-target enzymes, and that will not easily be overcome by resistance-conferring point mutations.
Molecular epidemiology
Twenty-five years after the detection of the first serine-β-lactamase [26], Sabbath and Abraham reported a new class of β-lactamase that was chromosomally encoded in Bacillus cereus. This β-lactamase was unusually inhibited upon addition of the metal ion chelator ethylenediaminetetraacetic acid (EDTA), marking the first discovery of a metal-dependent β-lactamase [27]. Little work developing more selective MBL inhibitors was done during the next two decades, possibly because MBLs were not considered to be a serious clinical problem. This situation drastically changed in the late 1980’s, however, with the emergence of MBLs in clinically important species such as the β-lactamase CcrA (carbapenem and cephamycin resistance A) from Bacteroides fragilis [28], CphA from Aeromonas hydrophila [18] and the first transferable MBL, IMP-1 from Pseudomonas aeruginosa [29]. The discovery of the corresponding blaIMP-1 gene in Serratia marcescens [30], Achromobacter xylosoxidans, Pseudomonas putida, and Klebsiella pneumoniae clinical isolates in Japan [31], and the subsequent isolation of IMP-2 in Italy [32] and IMP-5 in Portugal [33], marked the beginning of the forthcoming expansion of the IMP MBLs.
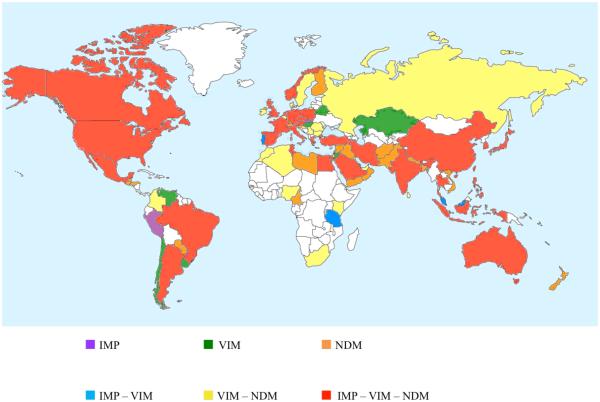
Presently, 47 variants of IMP-1 are reported from more than 30 countries, spanning all continents, except Antarctica (Figure 3). IMP-type β-lactamases are the predominant MBLs in Japan [34] and in south-east Asian regions, where at least 22 different IMP variants are identified (http://www.lahey.org/studies) [23, 35]. Sequences encoding IMP-type MBLs are mostly found in P. aeruginosa (especially belonging to the clonal complex, CC111 and CC235), Acinetobacter baumannii, and Enterobacteriaceae isolates, although they have also been occasionally identified in other organisms (e.g., P. putida) [36]. Analysis of the genetic environment of blaIMP genes has shown that most of them occur as cassettes in class 1 integrons also harboring other resistance genes, such as aac (resistance to aminoglycosides), aad (resistance to streptomycin and spectinomycin) or blaOXA-1,-2,-10, (class D serine β-lactamase) [35], and are often carried by broad host-range plasmids of incompatibility IncL/M and IncA/C groups [37, 38].
Figure 3.
Global distribution of IMP, VIM and NDM MBLs. Publications were retrieved from PubMed (http://www.ncbi.nlm.nih.gov/) using search queries such as “metallo β lactamase”, “NDM”, “VIM” or “IMP.” Only articles reporting occurrences of IMP, VIM or NDM were included. Note that as of July 2014, the only country reporting just IMP and NDM was Lebanon (not indicated).
A second major subgroup in the B1 subclass are the VIM-type MBLs, which were first described in Italy (Verona) from a P. aeruginosa strain isolated in 1997 [39], an observation that was followed by the report of an allelic variant, VIM-2, in France from a P. aeruginosa strain isolated in 1996 [40]. Presently, 41 different allotypes are identified [41], belonging to three sublineages: VIM-1 like, VIM-2 like, and VIM-7 like [24] (see below). VIM-type enzymes are the predominant MBLs in Europe, especially in Mediterranean countries such as Italy and Greece where VIM-producing P. aeruginosa strains (CC235 and CC111) have been involved in nosocomial outbreaks [23, 42]. VIM-2-like enzymes have been associated mostly with P. aeruginosa [43], whereas VIM-1-like enzymes, in particular VIM-4, have been frequently reported in strains of the Enterobacteriaceae [44-46]. Similar to blaIMP, blaVIM genes have been strongly associated with class I integrons, and integrated within either chromosomes or plasmids. Interestingly, many of the VIM-1/4 genes found in K. pneumoniae (sequence type, ST, 147 and 11) are carried on plasmids with compatibility group N, whereas reports of these genes in E. coli are often associated to IncFI/II plasmids [47]. A strong link between VIM MBLs, extended spectrum β-lactamases (ESBLs, especially CTX-M-15) and plasmid-mediated AmpC β-lactamases was also reported [48].
A third major type of B1 β-lactamases are the NDMs, which have been recognized for a shorter time than either the IMPs or VIMs. The first documented case of infection caused by NDM-producing bacteria occurred in 2008, when the blaNDM gene was detected in K. pneumoniae and E. coli from a patient returning to Sweden from India [49]. Retrospective analyses of stored cultures identified blaNDM in Enterobacteriaceae isolates dating back to 2006 [50]. An epidemiological link to the Indian subcontinent, the Balkans and the Middle East has been demonstrated in most of the reports of the global spread of NDM producers, identifying these geographic regions as endemic for NDM [51-54] (Figure 3). At the time of writing, eleven minor variants of NDM-1 (NDM-2 to -12) are identified (http://www.lahey.org/studies/), with NDM-1 being the most prevalent [54].
The blaNDM gene was also reported in numerous Enterobacteriaceae species (importantly in the epidemic clones of K. pneumoniae ST 147, 11 and 14, and E. coli 131 and 405) as well as in other Gram-negative bacteria, such as Vibrio cholerae, Pseudomonas spp. and A. baumannii [52, 55-57]. Analyses of the genetic environment revealed, in most cases, the presence of ISAba125 and a bleomycin resistance gene, up- and down-stream of blaNDM, respectively. Given that this insertion sequence is principally found in A. baumannii, this occurrence suggests that the dissemination of blaNDM-1 may have originated from this species [58]. The recently reported association of blaNDM-1 with a class 1 integron ISCR1, one of the most commonly used mechanisms for the spread of antibiotic resistance across species, may also explain the rapid expansion of this gene [59]. Lastly, despite reports of chromosomal location in certain isolates, blaNDM has been primarily associated with multiple separate acquisition events mediated by plasmids of different sizes and from incompatibility groups with both broad and narrow host ranges, such as IncF, IncA/C, IncL/M, IncH, IncN and IncX3 [51-53].
Common structural features
MBLs belong to a larger superfamily of metalloproteins with diverse biological functions beyond β-lactam hydrolysis, designated as the metallo-hydrolase/oxidoreductase superfamily in the Structural Classifications of Proteins (SCOP) database [60-62]. A common structural feature conserved in this superfamily is the overall αβ/βα protein fold, in which an internal mixed β-sheet sandwich is surrounded by solvent-exposed α-helices. Structural and sequence comparisons are consistent with the proposal that serine β-lactamases evolved from ancestral transpeptidases [63], but the evolutionary history of MBLs, which are not homologous to transpeptidases, is more ambiguous. Pseudo two-fold symmetry of the N- and C-terminal halves of MBLs suggests that the two halves may have arisen from an ancient gene duplication event [63, 64].
In the MBLs, the zinc-containing active site is positioned within a shallow groove formed at the interface of the central β sheets (Figure 4). Crystal structures of B1 β-lactamases reveal a mono or dinuclear metal ion cluster in which Zn1 is usually coordinated with tetrahedral geometry, and Zn2 with trigonal bipyramidal geometry when substrate or product ligands are absent (e.g. [64, 65]). In the structures of monozinc B1 MBLs, the Zn1 site is occupied by the sole zinc ion. In structures of dizinc B1 MBLs with differing affinities for each site, Zn1 is typically refined with higher average occupancy than Zn2, consistent with biochemical studies assigning Zn2 as the more weakly binding site [66-69]. In other B1 MBLs, the dizinc site has been shown to bind both zinc ions with positive cooperativity, with binding of the first equivalent enhancing binding of the second (reviewed in [70]). Some crystal structures of the B1 β-lactamase NDM-1 also reveal a third zinc ion (Zn3) bound at the edge of the active site, positioned approximately 8 Å from the dinculear zinc cluster [71]. However, the functional relevance of Zn3 has not been established and it may be a crystallization artifact. “Second shell zinc ligands,” residues that directly interact with the primary zinc ligands, are more varied than the primary zinc ligands and may contribute to the differing properties found among the zinc sites of B1 MBLs [16, 72-74].
Figure 4.
Common structural features among B1 MBLs. A. Superimposition of crystal structures of IMP-1 (blue, PDB code 1DD6), VIM-2 (purple, PDB code 2YZ3), and NDM-1 (green, PDB code 3Q6X) are shown as ribbon diagrams. Prominent loops surrounding the active are labeled. The L8 loop, exclusive of NDM-1, is highlighted in red. Zinc atoms are shown as orange spheres. Active site ligands and water molecules have been removed for sake of clarity. B. The active site of VIM-2 is shown as an example of B1 MBLs. Zinc liganding residues are shown as sticks, with carbons in tan, oxygens in red, sulfurs in yellow and nitrogens in blue. The bridging hydroxide is shown as a red sphere.
In addition to the primary zinc ligands, several notable active-site features appear to be structurally conserved among the representative B1 MBLs IMP-1, VIM-1 and NDM-1, despite the relatively low overall amino acid sequence identities (VIM-1:IMP-1: 31%; NDM-1:IMP-1: 30%; VIM-1:NDM-1: 31%). Firstly, a positively-charged side chain (Lys224 in IMP-1 and NDM-1 or an Arg228 supplied from a different sequence position in VIM-2) is located approximately 6 Å from from Zn2 and likely provides a counter ion for the conserved carboxylate found in all β-lactam drugs. Secondly, an Asn residue is structurally conserved in each of these three B1 MBLs, and this position (Asn233), found in active-site loop 10 (ASL10), is shown to be within H-bonding distance of the newly formed carboxylate in ring-opened β-lactam products. All of the residue numbering in this review is standardized according to the standard BBL numbering [75] and an NDM-1 alignment published elsewhere [76] (Figure 5). Thirdly, in all three β-lactamases, a conserved extended β-hairpin loop (active-site loop 3, ASL3) towers over one side of the active site, providing a broad hydrophobic surface for binding the core ring structures of the β-lactam substrates. In general, this loop is more disordered than the remainder of the protein [77-79] and was demonstrated to become rigid and close down upon binding active-site ligands [80-83]. Finally, in all three examples, a shallow groove in the protein surface extends away from either side of the dizinc cluster, allowing sufficient space to accommodate the varied substitutions found in the broad array of β-lactam substrates that contain structurally diverse substituents appended to either the four-membered lactam ring in one direction (away from Zn1), or extending from position 2 (in penams, clavams, penems and carbapenems) or position 3 (in cephems) in the other direction (away from Zn2). Recent efforts toward understanding how changes to these and other structural features of IMP, VIM and NDM MBLs impact catalysis, stability, and subsequent antibiotic resistance are addressed below.
Figure 5.
Structural alignment of IMP-1, VIM-2 and NDM-1. Alignment was constructed based on a previously published one by Cadag et al. and the standard BBL numbering scheme [75, 76]. Conserved residues are highlighted in bold, with zinc binding residues label with a “z” and key residues for substrate binding with an *. Residues that are mutated in the different variants, as well as those unique for each enzyme are bolded and colored in red, blue and green in the IMP-1, VIM-2 and NDM-1 sequence, respectively. Active site loops are denoted by pink boxes.
IMP-type MBLs
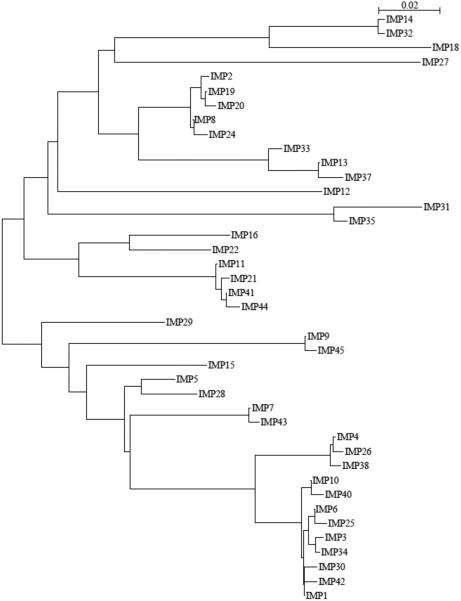
Full-length IMP-type enzymes usually consist of 246 amino acid residues, except IMP-9, -11, -21, -31, -41, -44, and -45, which contain 245 residues, and IMP-37, which is 248 amino acids in length. The mature IMP-1 protein contains 228 residues and has a molecular mass of ca. 25 kDa [30]. To date, 47 variants of IMP-1 are assigned (http://www.lahey.org/studies), although only 41 protein sequences are publicly available. The phylogenetic tree, based on amino acid identity, shows the diversity among the IMP-type MBLs (Figure 6). In accordance with previous analysis, IMP-1 and IMP-2 cluster separately, suggesting different phylogenetic origins [24, 35, 84]. Table 1 and Figures 7A, B and C provide a summary of the information of some of the variants and key residues that are discussed below.
Figure 6.
Phylogenetic tree of currently known IMP enzymes. Available amino acid sequences including the leader sequence were retrieved from GenBank at http://www.ncbi.nlm.nih.gov/. Alignments and phylogenetic trees were generated using SeaView V.3.2 [169], using Clustal X function and BioNJ algorithm [170] (Kimura, 100 bootstraps), respectively. The bar at the upper right corner gives a measure for amino acid sequence diversity.
Table 1.
Summary of the structure-function information available for the IMP-1 variants.
| Variant | Closest variant for reference |
Mutation(s) of interest |
Localization | Comments a |
|---|---|---|---|---|
| IMP-6 | IMP-1 | S262G | Second-shell ligand | Increased activity towards meropenem b and doripenem (3.2 vs. 1.1 μM−1 s−1), compared to IMP-1 |
| IMP-25 | IMP-1 | S262G G235S |
Second-shell ligand ASL10 |
Increased activity towards meropenem, compared to IMP-1 and IMP-6 b. |
| IMP-10 | IMP-1 | V67F | ASL3 | Decreased activity toward penicillins (e.g. ampicillin: 0.06 vs. 1.1 μM−1 s−1), compared to IMP-1 [93]. |
| IMP-43 | IMP-7 | V67F | ASL3 | Increased activity towards meropenem (0.34 vs. 0.044 μM−1 s−1), compared to IMP-7 [94]. |
| IMP-44 | IMP-11 | V67F F87S |
ASL3 Hydrophobic patch near the active site |
Increased activity toward imipenem (1.4 vs. 0.078 μM−1 s−1), compared to IMP-7; increased activity towards doripenem (2.3 vs. 0.11 μM−1 s−1), compared to IMP-11 [94]. |
| IMP-30 | IMP-1 | E59K | β-strand of ASL3 | Increased activity towards ceftazidime (1.6 vs. 0.25 μM−1 s−1), compared to IMP-1 [96]. |
| IMP-28 | IMP-1 | Q306H | ASL10 | Overall decreased activity compared to IMP-1 (e.g. ampicillin: 1.8 x 10−3 vs. 4.8 μM−1 s−1; ceftazidime: 1 × 10−3 vs. 0.18 μM−1 s−1; meropenem: 0.21 vs. 5.0 μM−1 s−1) [98]. |
| IMP-2 | IMP-1 | 25 mutations (85% identity) |
Distributed throughout the protein |
Decreased activity towards ampicillin (0.21 vs. 4.8 μM−1 s−1) and cephaloridine (0.27 vs. 2.4 μM−1 s−1); higher hydrolysis of carbenicillin (0.36 vs. 0.02 μM−1 s−1) and meropenem (3.3 vs. 0.5 μM−1 s−1), compared to IMP-1 [84]. |
| IMP-19 | IMP-2 | R38A |
N-terminus, on top of ASL3 |
Decreased activity toward meropenem (0.14 vs. 3.3μM−1 s−1) and imipenem (0.26 vs. 0.92 μM−1 s−1), compared to IMP-2 [99]. |
| IMP-13 | IMP-1 | G177E V266K |
β-strand 7 β-strand 11 |
Increased activity towards cefuroxime (23 vs 0.22 μM−1 s−1) and imipenem (2.5 vs.0.92 μM−1 s−1), compared to IMP-1 and IMP2, respectively [100]. |
| IMP-12 | IMP-1 | N62S | ASL3 | Decreased activity toward penicillins (e.g. ampicillin: 0.012 vs. 4.8 μM−1 s−1), compared to IMP-1 [97]. |
| IMP-18 | IMP-1 | W242G | α-helix 4 | Overall decreased activity, (e.g. ampicillin: 0.63 vs. 4.75 μM−1 s−1; cefotaxime: 0.23 vs. 0.32 μM−1 s−1; meropenem: 0.006 vs. 0.5 μM−1 s−1), compared to IMP-1. |
Based on kcat/KM values, unless noted.
Resistance towards meropenem (MIC values of 16 mg/L for IMP-1, 64 mg/L for IMP-6, and 128 mg/L for IMP-25) do not correlate with the kcat/KM values, but with the kcat (22 s−1, 60 s−1, and 100 s−1, respectively) [88].
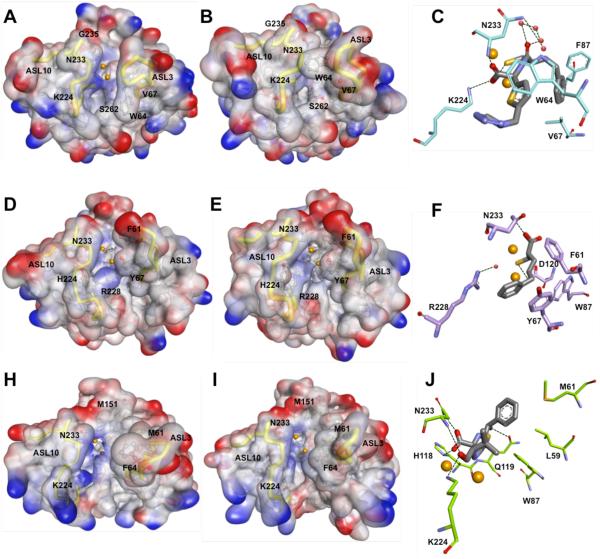
Figure 7.
Structural comparison between IMP-1 (top; panels A – C), VIM-2 (middle; panels D-F) and NDM-1 (bottom; panels G-I). Panels A, B, D, E, G and H are surface representations of the electrostatic potential of IMP-1 D120E (A; PDB 1WUP), IMP-1 in complex with a mercaptocarboxylate inhibitor (2-mercaptomethyl-4-phenylbutyrylimino)-(5-tetrazol-1-ylmethylthiophen-2-yl)-acetic acid) (B; PDB 1DD6), VIM-2 (D; PDB 1KO3), VIM-2 in complex with a mercaptocarboxylate inhibitor ((S)-2-(mercaptomethyl)-5-phenylpentanoic acid) (E; PDB 2YZ3), NDM-1 (G; PDB 3SPU), and NDM-1 in complex with hydrolyzed ampicillin (H; PDB 3Q6X). Important residues are shown as balls and sticks with carbons in gray, oxygens in red, sulfurs in yellow and nitrogens in blue. Zinc ions are represented as orange spheres. Active site loops are highlighted as yellow tubes. Panels C, F and I show major interactions in the structures B, E, H, respectively. Key residues are represented as sticks with carbons colored in cyan (IMP-1), light purple (VIM-2), and light green (NDM-1), oxygens in red, sulfurs in yellow and nitrogens in blue. Interacting active site waters are shown as red spheres; hydrogen bonds are represented as black dash lines; and zinc atoms as orange spheres.
Four natural variants from the IMP-1 cluster (IMP-6, -25, -10, and -30) are characterized extensively. IMP-6 differs from IMP-1 by an S262G mutation in the active site, and IMP-25 differs from IMP-6 by an additional G235S mutation on the ASL10. Interestingly, IMP-1 confers higher resistance levels to imipenem and several other antibiotics [85, 86], whereas IMP-6 confers higher resistance levels to meropenem [87] and doripenem [88]. IMP-25, which might be a derivative of IMP-6, confers the greatest resistance towards meropenem. Given the increasing activity relationship among these enzymes, IMP-6 is hypothesized to be the ancestor enzyme, from which IMP-1 and IMP-25 evolved. Thus, the accumulative mutations G262S to G262S / G235S represents a possible evolutionary pathway in response to the increasing clinical use of meropenem [88].
Ser262, a second shell residue, is located proximal to the primary zinc ligand His263. The change in substrate specificity conferred by the G262S substitution was explained as a “domino effect”: the presence of Ser262 supports His263, preserving the zinc ion coordination, and thus, stabilizes the enzyme-substrate intermediate complex, enhancing conversion of β-lactams with bulky or positively charged R2 groups like ceftazidime, penicillins, and imipenem [89, 90]. Interestingly, in a recent study Pegg et al. predicted that a Thr262 mutant that may be derived from IMP-6, would confer increased resistance toward ceftazidime, penicillins, and imipenem [91]. This result highlights the importance of position 262 in modulation of the substrate specificity in the IMP MBLs. On the other hand, position 235 is located on the ASL10, two residues away from Asn233, which is conserved in all but two IMP-type enzymes (IMP-14 and IMP-35) [88]. The Asn233 residue has been proposed to provide part of an “oxyanion hole” to stabilize a β-lactam reaction intermediate [64, 65, 77]. Substitutions at position 233 significantly alter the kinetic parameters, though most substituted enzymes still provide high levels of resistance [92]. Ser235 is proposed to form hydrogen bonds to Asn233 and alter the substrate-binding site, further explaining the kinetic characteristics of IMP-25. We note that IMP-25 is the only IMP-type enzyme with serine at position 235, but several other enzymes (IMP-11, IMP-12, IMP-16, IMP-21, IMP-22, and IMP-29) contain Asp at this position [88]. The extent to which mutations at this position change substrate specificity and the underlying molecular mechanism is not yet known.
Based on modeling and structures of inhibitor complexes, the side-chains of Val61 and Val67, located on the ASL3, and Phe87 are proposed to contribute to a hydrophobic surface found at the base of the ASL3 that putatively helps to bind substrates [77]. The addition of a longer bulky group in Phe compared to Val at position 67 may affect substrate binding resulting in the decreased inactivation of penicillins observed by IMP-10, a single variant of IMP-1 (V67F) [93]. Interestingly, IMP-43 possesses the same amino acid substitution (V67F) compared with IMP-7 (which is close to the IMP-1 cluster), and IMP-44 has two substitutions (V67F and F87S) compared with IMP-11 (which is related to IMP-2) [94]. The V67F substitution in IMP-43 significantly increases the catalytic efficiency for meropenem, whereas the double substitution V67F and F87S in IMP-44 also improves hydrolysis of imipenem and doripenem, compared to IMP-7 and IMP-11, respectively [94]. Position Phe87 forms a hydrophobic patch in the active site, near Zn2. The absence of a benzene ring due to the F87S substitution may affect stability and folding, thereby changing substrate specificity, similar to the effects demonstrated for Trp87 mutations in VIM-2 [95] (see below).
IMP-30 is another variant from the IMP-1 cluster with a mutation in the ASL3. An E59K substitution confers enhanced activity towards ceftazidime [96]. Residue 59 is located in a β strand that is part of the ASL3 and contributes to the binding pocket of the R1 side chains of β-lactams [77]. Pegg et al hypothesized that the E59K substitution in IMP-30 could stabilize the MBL-ceftazidime complex during the catalytic cycle by forming a salt bridge between Lys59 and the carboxylate in the R1 group of ceftazidime, resulting in enhanced catalytic efficiency [96].
IMP-12 and IMP-28 are two variants outside the IMP-1 cluster whose activities have been compared to that of IMP-1. IMP-12, which is very different (retaining 82 to 89% sequence identity) from all other known IMP variants, displays an overall lower catalytic efficiency towards penicillins, compared to IMP-1 and other IMP variants. A unique structural feature of IMP-12, an N62S substitution in the ASL3, which is highly conserved in all IMP variants, might contribute to the reduced activity toward those substrates observed for IMP-12 when compared to IMP-1 [97]. On the other hand, IMP-28, which differs by 15 substitutions from IMP-1, is a less efficient β-lactamase. Given the involvement of Gln306 in hydrogen bonds with residues at the ASL10 in IMP-1, it has been proposed that the Q306H mutation may be responsible for the low overall activity of IMP-28 [98].
Only two variants of the IMP-2 cluster (IMP-2 and IMP-19) are characterized to date. Despite a relatively low amino acid sequence identity (85%) and different phylogenetic origin, the substrate spectrum of IMP-2 is similar to that of IMP-1 (penicillins, cephalosporins, and carbapenems are processed, but not aztreonam) [24]. IMP-2, however, displays significantly lower catalytic efficiencies toward ampicillin and cephaloridine and significantly higher catalytic efficiencies toward carbenicillin and meropenem, as compared to IMP-1 [84]. IMP-19 differs from IMP-2 only by an R38A mutation, and has lower catalytic efficiencies toward imipenem and meropenem [99]. Given the position of residue 38 close to the N-terminus of the mature protein, which is located on top of the ASL3, this residue could contribute to substrate binding [24].
The IMP-13 MBL possesses 92% amino acid sequence identity with IMP-2 and 83% with IMP-1, being quite divergent from other variants. IMP-13 has higher hydrolysis efficiency towards cefuroxime and imipenem, compared to IMP-1 and IMP-2, respectively. IMP-13 owes its distinctive functional features to 11 unique substitutions, out of which G177E and V266K may have a substantial impact on the enzyme properties due to the non-conservative nature of the substitutions [100].
Finally, the divergent IMP-18 (91% sequence identity to IMP-14 and IMP-32) is characterized by overall lower turnover (kcat values) than those of other IMP-type variants, especially toward the carbapenem meropenem. One unique mutation (W242G) may account for this unusual selectivity. Since Trp242 is conserved in other subclass B1 enzymes, the low activity of IMP-18 might be due to this non-conservative substitution found in IMP-18 [101].
Site-directed saturation mutagenesis studies are critical to characterize key residues in protein stability, substrate binding, and catalysis. Palzkill and co-workers used a codon randomization and selection strategy with IMP-1 to determine the context-dependent sequence requirements for the hydrolysis of several substrates including ampicillin, imipenem, cephaloridine and cefotaxime. Results confirmed that all of the zinc-chelating positions, His116, His118, His196, Cys221, His263 and Asp120, as well as the ASL3 residue Gly65 are essential for IMP-1 function [102]. Gly63 was demonstrated to be essential for hydrolysis of all substrates except ampicillin, a result that supports its contribution, along with Gly65, to the formation of a β-hairpin turn at the end of the ASL3 [77, 102]. Likewise, the presence of a large, hydrophobic amino acid residue at position 64 at the apex of the ASL3, was found to be required for hydrolysis of all antibiotics tested [102]. This result supports the proposed role of Trp64 in the flexible ASL3 for binding structurally-diverse ligands by providing a hydrophobic binding surface that can adopt multiple conformations [103].
Crystal structures of IMP-1 display the conserved αββα protein fold described above, and display the characteristic features of B1 MBLs, such as the conserved primary zinc ligands and Lys224 [77]. This conserved Lys224, located in the ASL10, is proposed to form an electrostatic interaction with the negatively charged carboxylate group that is common to all β-lactam drugs, except the monobactams [77, 104]. In line with this proposed role, Materon et al. demonstrated that the presence of Lys224 is critical for effective hydrolysis of ampicillin, imipenem and cefotaxime [102].
The following structural studies provided evidence for the proposed roles of residues Asp120 and Cys221 in IMP-1. Yamaguchi et al., investigated the functional role of Asp120 using site-directed mutagenesis, reaction kinetics and high-resolution crystallography of D120A and D120E mutants. These studies revealed that Asp120 not only serves as a primary Zn2 ligand, but also orientates the bridging nucleophilic hydroxide for its attack at the carbonyl carbon of the β-lactam ring [105]. Using a similar strategy, Horton at al., demonstrated that Cys221 in IMP-1 is an important primary Zn2 ligand that maintains the stability and catalytic efficiency of the enzyme. Out of 19 substitutions made at this position, only the C221D and C221G mutants retained catalytic activity, which was highly dependent on the Zn2+ concentration and was associated with the ability of these residues to allow Zn2 binding -either through direct coordination (C221D) or by creating a space in which a water molecule is predicted to serve as a Zn2 ligand (C221G) [106]. These studies with IMP-1 can be compared with a previous study on BcII in which the corresponding C221A and C221S mutants retained decreased activity that was augmented by supplemental Zn2+ [107]. In comparison, the C221A mutation in IMP-1 resulted in an unstable protein that did not accumulate during expression, and the C221S mutation in IMP-1 did not retain enough activity to confer resistance to β-lactam antibiotics, and so was not investigated further.
VIM-type MBLs
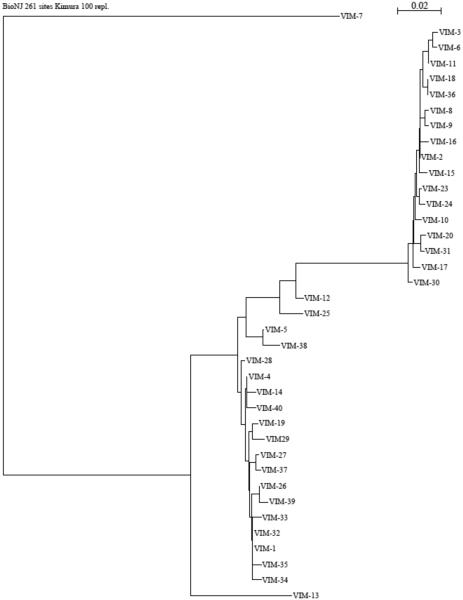
Full-length VIM-type enzymes usually consist of 266 residues (except VIM-7 and VIM-18, which contain 265 and 262 residues, respectively). In its mature form, VIM-2 contains 240 residues and has a molecular mass of about 25 kDa [78]. To date, 41 variants have been assigned (http://www.lahey.org/studies), of which 37 protein sequences have been published. Compared to IMP-type enzymes, the phylogenetic tree constructed based on amino acid similarity shows a more homogeneous distribution, with two well-defined major clusters (the VIM-1- and VIM-2-like enzymes), a minor cluster enclosing VIM-12, -5, -25 and -38, and the divergent VIM-7 and VIM-13 (Figure 8). A summary of the information of some of the characterized variants and key residues are shown in Table 2 and Figure 7D, E and F.
Figure 8.
Phylogenetic tree of currently known VIM enzymes, constructed as described for Figure 5.
Table 2.
Summary of the structure-function information available for the VIM-1 variants.
| Variant | Closest variant for reference |
Mutation(s) of interest |
Localization | Comments a |
|---|---|---|---|---|
| VIM-2 | VIM-1 | H224Y | ASL10 | Increased affinity for ceftazidime (KM value of 98 vs. 794 μM), compared to VIM-1 [40, 108]. |
| VIM-31 | VIM-2 | Y224H | ASL10 | Overall decreased activity (e.g. ampicillin: 0.9 vs. 1.4 μM−1 s−1; ceftazidime: 0.005 vs. 0.05 μM−1 s−1; meropenem: 0.3 vs. 2.5 μM−1 s−1), compared to VIM-2 [113]. |
| VIM-4 VIM-13 VIM-19 VIM-23 |
VIM-1 | S228R | ASL10 | Decreased activity for extended-spectrum cephalosporins, compared to VIM-1 (e.g. cefepime: 0.03 vs. 0.4 μM−1 s−1, for VIM-13 and VIM-1, respectively [117]). |
| VIM-11 | VIM-2 | N165S | Near ASL10 | Increased activity for ceftazidime (0.13 vs. 0.03 μM−1 s−1); cefepime (0.083 vs. 0.03 μM−1 s−1); and cefpirome (0.6 vs. 0.28 μM−1 s−1), compared to VIM-2 [119]. |
| S60K F61L |
Enlarged active-site cavity, likely due to increased flexibility of ASL3 and loss of H- bonding at ASL10. Overall decreased activity for most cephalosporins (e.g. cephalothin: 4 vs. 12 μM−1 s−1; ceftazidime: 0.012 vs. 0.05 μM−1 s−1; cefotaxime: 2.6 vs. 5.8 μM−1 s−1), compared to VIM-2 [171]. |
|||
| D62G | ASL3 | |||
| VIM-7 | VIM-2 | G63D | ||
| A64T | ||||
| S68P | ||||
| Y224H | ASL10 | |||
| Y218P |
Based on kcat/KM values, unless noted.
The initial biochemical characterization of VIM-1 by Franceschini et al., showed that VIM-1 exhibits a broad substrate specificity, which includes virtually all β-lactams except monobactams [108]. Unlike IMP-1, VIM-1 hydrolyses carbenicillin more efficiently than penicillin G and ampicillin, and displays high affinity for temocillin [109], indicating that the presence of an α-methoxy group at position C-6 does not prevent the interaction of penicillins with VIM-1 [108]. Cephalosporins were found to be good substrates for VIM-1, although lower catalytic efficiencies were observed for oxymino-cephalosporins (cefotaxime and ceftazidime) and cefoxitin, suggesting that the presence of an α-methoxy group at position C-7 decreases the catalytic efficiency of VIM-1 towards those substrates. This feature constitutes another difference between VIM-1 and IMP-1, which is not affected by the presence of an α-methoxy group at position C-7 [109]. Carbapenems were shown to be good substrates for VIM-1; efficient hydrolysis of imipenem is based on a very high affinity for the substrate (low KM) associated with relatively low turnover (low kcat values) [108].
Comparison of the kinetic parameters of VIM-1 and VIM-2 (93% amino acid identity) revealed similar kcat/KM values for several substrates, including carbapenems, piperacillin, ticarcillin, narrow-spectrum cephalosporins, and cefotaxime. However, VIM-1 displays higher kcat/KM values for cefuroxime, cefepime, and cefpirome, whereas VIM-2 has higher kcat/KM values for penicillin G and ceftazidime [40, 108].
In 2008, García-Saez et al. reported the crystal structure of VIM-2 [78], and revealed structural features conserved with IMP-1, such as the presence of Asp84 which adapts a strained position buried between two β-strands buried in the hydrophobic core of the protein. However, position 121, a second shell ligand that is a Ser in IMP-1, is instead an Arg in all the VIM variants [77, 78].
As mentioned before, in many subclass B1 MBLs, the conserved Lys224 in the ASL10 plays an important role in substrate binding [102, 110]. At this position, most of the variants at the VIM-1 cluster (except VIM-26 and VIM-28) have a His residue, whereas all variants at the VIM-2 cluster, but VIM-31, possess a Tyr. The aforementioned exceptions and the other VIM variants contain a Leu residue at this position. The side chains of His, Tyr and Leu residues are much shorter than that of a Lys residue, and may prevent the interaction of these two amino acids with the conserved carboxylate moiety (at C4 or C3) of the substrate [78, 111]. However, important kinetic differences have been attributed to changes at this position. For instance, Tyr224 in VIM-2 was previously suggested to enhance the binding affinity of certain substrates, such as ceftazidime [78, 112], for which VIM-2 exhibits a lower KM value than that for VIM-1 (His224). Similarly, compared to the parental VIM-2, VIM-31 contains two mutations (Y224H and H252R) in the C-terminal part of the protein and exhibits globally lower catalytic efficiencies. The catalytic features of this variant are thought to derive from the Y224H substitution, since the H252R substitution, situated on the α4 helix and distant from the active site of the VIM enzyme, renders unlikely any effect on substrate binding and/or catalysis [113].
For VIM enzymes, it is believed that the side chain of Arg228 may structurally replace that of Lys224 by interacting with the substrate [78, 111], despite coming from a different part of the primary structure. Crystal structures of VIM-2, VIM-4 and VIM-7 have shown that Arg228 located at the ASL10, defines a precise positively charged space for substrate binding by forming with its side chain a kind of “wall” that encloses one side of the active site [111, 114, 115]. This position may help define the substrate specificity in VIM MBLs. Indeed, the substitution of Ser228 at this position has been shown to result in a lesser-charged and more open active site, and has been associated with increased activity against substrates with bulky substituents at C3 such as ceftazidime and cefepime [111, 114, 115]. Moreover, most of the variants harboring Arg228, like VIM-4, VIM-7, VIM-13, VIM-19 and VIM-23, display less efficient hydrolysis of several expanded-spectrum cephalosporins, whereas the penicillinase and carbapenemase activities are comparable to the Ser228-containing variants [111, 114-118].
Mutations outside the active site also contribute to tuning the catalytic activity in VIM MBLs. For instance, VIM-11 differs from VIM-2 by one substitution at the outside of the ASL10 (N165S), which confers to VIM-11 a better catalytic efficiency against ceftazidime, cefepime and cefpirome, as compared to its ancestor (VIM-2) [119]. Likewise, VIM-12 has 97 and 94% sequence identity with VIM-1 and VIM-2, respectively, and displays narrow substrate specificity, limited to penicillins. Remarkably, the N-terminal and C-terminal portions of the VIM-12 sequence have high sequence identity to the corresponding residues in VIM-1 and VIM-2, respectively, and thus VIM-12 has been called a “hybrid” enzyme. It is hypothesized that the atypical kinetic behavior of this enzyme is due, in part, to disturbances in the hydrogen bonding network of second shell ligands [120].
Comparison of the crystal structures of VIM-2, VIM-4 (from the VIM-1 cluster) and VIM-7 (a more divergent enzyme) highlights interesting structural features of VIM MBLs. For example, VIM-7 has a number of non-conservative substitutions in the ASL3, including S60K, F61L, D62G, G63D, A64T, and P68S, as compared to VIM-2 and VIM-4 (where these two enzymes are identical). These changes lead to a farther localization of this loop away from the active site metal center in VIM-7, causing this enzyme to have a more open active site [114]. Furthermore, the P68S substitution may also increase the flexibility of the ASL3 and influence enzymatic activity, as the main-chain carbonyl at this position acts as a “second-shell” Zn2 ligand by making a hydrogen-bonding contact with His263 in all VIM-7, VIM-2, and VIM-4 structures, and in other B1 MBLs [86, 111, 114, 115, 121].
Relative to VIM-2, VIM-7 has two more substitutions near the active site (Y218P and Y224H). The former change interrupts three hydrogen bonds involving Asn70, Asp84 and Arg121, which position the ASL3, and thus confers more flexibility to that region in VIM-7. Of note, the F218Y substitution in IMP-1 has been associated with increased activity for cephalosporins with bulky substituents at C3 [122]. On the other hand, in the VIM-2 crystal structure, the Tyr224 phenol group is within hydrogen-bonding distance of the backbone NH of Gly232 in ASL10, and a water molecule that makes hydrogen bonds with the backbone carbonyl of Asn233 (also in ASL10) and with Nδ of His196, a direct Zn1 ligand [114]. Disruption of this hydrogen-bonding network due to the presence of His224 in VIM-7 may influence interactions with substrates and/or inhibitors because residues in the ASL10 are associated with substrate and inhibitor binding [77, 104, 123]. Furthermore, His224 slightly decreases the overall negatively charge of the binding pocket in VIM-7, which may diminish affinity for positively charged cephalosporins, possibly explaining the lower efficiency of this enzyme against cephalosporins containing a charged R2 group, as compared to VIM-2 [114].
Using saturation mutagenesis, Borgianni et al. tested the importance of residues Phe61, Ala64, Tyr67 (located in the ASL3), and Trp87, which form a hydrophobic patch neighboring the active site of VIM-2 [95]. Results showed that substitutions at positions 61, 64 and 67 do not significantly affect substrate hydrolysis or binding, which is a remarkable difference from IMP-1 [77, 103, 124]. The aromatic residue Trp87 was found to be critical for the stability and folding of VIM-2, although it is not a determinant for substrate binding and catalysis [95].
NDM-type MBLs
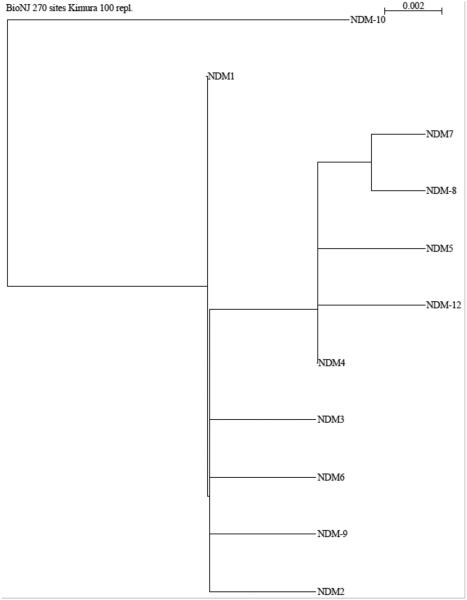
Full-length NDM-type enzymes usually consist of 270 residues. Mature NDM-1 consisting of 254, 252, 250 or 234 amino acid residues, with corresponding molecular mass ranging from 29 to 27 kDa have been reported [125]. This finding is consistent with several predicted cleavage sites in the signal peptide, and suggests that at least in Enterobacteriacea, NDM-1 is heterogeneously processed. NDM-1 shares very little sequence identity with other MBLs and is most closely related to VIM-1/ VIM-2, with which it shares 32% amino acid sequence identity. To date, 12 variants have been assigned (http://www.lahey.org/studies), of which 10 protein sequences have been published. The phylogenetic relationship among these enzymes is shown in Figure 9.
Figure 9.
Phylogenetic tree of currently known NDM enzymes, constructed as described for Figure 5.
The initial characterization of NDM-1 by Yong et al., pointed out some unique features of this MBL. For instance, NDM-1 has a sequence insertion (FAAN) between residues 160 and 164, an alanine between the two His residues in the defining HXHXD motif of MBLs, and a Tyr at position 242 rather than the conserved Trp [49]. Compared to VIM-2 and IMP-1, NDM-1 has lower KM and kcat values for most β-lactam substrates, with the exception of imipenem and meropenem [49].
At the time of writing, five variants of NDM-1 have been characterized by biochemical and/or microbiological methods. Their main features, compared to NDM-1, are summarized as follows: NDM-3 (D89N) has a slightly lower catalytic efficiency (kcat /KM) for most substrates (e.g. 1.1 vs. 4.3 μM−1s−1 for penicillin; 0.17 vs. 0.25 μM−1s−1 for ceftazidime; 0.17 vs. 0.48 μM−1s−1 for imipenem), due to a decreased kcat. One exception is doripenem, for which NDM-3 displays approximately the same efficiency (0.37 vs. 0.35 μM−1s−1) [126]. In contrast, NDM-4 (M151L) displays an improved hydrolytic activity against several substrates such as cefotaxime (1.2 vs. 0.6 μM−1s−1), and imipenem (0.46 vs. 0.20 μM−1s−1) due to an increased kcat; and ceftazidime (0.06 vs. 0.03 μM−1s−1) mainly due to a decreased KM. However, NDM-4 displays significantly lower catalytic efficiency towards cefepime (0.04 vs. 0.20 μM−1s−1) due to an increased KM value [127]. NDM-5 has two substitutions (V82L and M151L) and confers higher resistance against expanded-spectrum cephalosporins and carbapenems to E. coli transformants [127]. NDM-8 (D136G and M151L) displays similar hydrolytic activity, with slightly lower kcat /KM values for penicillin G (1.2 vs. 2.7 μM−1s−1) due to a higher KM, and ceftazidime (0.2 vs. 0.45 kcat /KM) due to a decreased kcat. [128]. Lastly, NDM-9 (E149K) shows higher kcat /KM ratios for meropenem (3.12 vs. 1.57 μM−1s−1) and imipenem (2.02 vs. 1.46 μM−1s−1) mainly due to an increased affinity; and for cefoxitin (0.48 vs. 0.19 μM−1s−1) and cefotaxime (0.30 vs. 0.18 μM−1s−1) due to higher turnover numbers [129]. Since none of these mutations are located at residues typically thought to be responsible for substrate specificity (such as those in ASL3 or ASL10), their influence on binding and catalysis may be indirect.
To assess the impact of these mutations on the structure, stability and catalytic activity of NDM-1, Makena et al. used site-directed mutagenesis to generate NDM variants [130]. Comparison of circular dichroism scans showed that the mutations do not substantially affect the overall secondary structure of the NDM variants, although significant differences in their thermal stability (>10 °C) were found. Interestingly, the more stable variants (NDM-5 and NDM-7) also conferred the highest resistance levels towards selected carbapenems (MIC values for imipenem >32 mg/L vs. 8 mg/L with the wild-type NDM-1). In contrast with previous studies, Makena et al. did not find significant differences in the catalytic efficiency of the variants towards any of the substrates tested. However, variants with the M154L substitution (NDM-4, -5, -7, and -8) were found to display an apparent substrate inhibition by nitrocefin (Ki values of 102, 139, 79 and 146 μM, respectively). Overall, the changes, or lack thereof, in catalytic rate constants among NDM variants, as reported by various laboratories, are small and could reflect minor differences in methodology.
The first crystal structure of NDM-1 was reported in 2011 by Guo et al. [131]. This structure displayed the conserved αβ/βα protein fold, with two catalytic zinc ions bound as a dinuclear center in the active site coordinated by His116, His118, Asp120, His196, Cys221 and His263, [131]. Site-directed mutagenesis on the unique HAHQD motif of NDM-1 suggested a possible evolutionary advantage for NDM-1, as the Q119D and A117F mutations decreased the imipenem hydrolysis activity, while the Q119D/A117F double mutant retained only 0.7% of the wild-type activity [131], although this result might also reflect the inability of NDM-1 to accommodate these non-conservative mutations. The unusual FAAN insertion at position 160 is located far away from the active site, and thus led to the proposal that it may not have any effect on the enzymatic activity of NDM-1 [131]. Based on their structural models, King et al. proposed that this insertion forms an extra eighth loop (L8) and contributes to key contacts required for dimerization of the full-length enzyme [79]. Their work also identified a Type I signal peptide and a lipidation signal peptide that is predicted to localize the processed and lipidated enzyme into the bacterial outer membrane. The full-length enzyme containing the lipidation site was shown to dimerize, but variants with shorter truncations of the N-terminus appear to be fully active and monomeric [79, 132]. To the best of our knowledge, only one other β-lactamase (a Ser β-lactamase) is also lipidated at the N-terminus [133]. The implications of these unique features and their impact on antibiotic resistance remain unknown.
As mentioned previously, NDM-1 possesses a Tyr at position 242 (as seen in B2 and B3 MBLs), rather the conserved tryptophan of B1 MBLs (Figure 5). This residue shows extended hydrophobic and hydrophilic interactions that appear to stabilize or orient the ASL10 loop, restricting the flexibility of the active-site residue Asn233, and thus possibly affect enzymatic activity [134]. However, a Y242W mutation did not show any effect on catalysis, so the precise role of that particular position in NDM-1 remains undetermined [131].
X-ray crystallography has identified multiple different conformations for the ASL3, found between Ser57 and Ala68 in NDM-1, which neighbors the active site [79, 131, 134, 135]. Phe64 is located at the apex of the loop, at the same position as Trp64 in IMP-1 [77], and likely contributes to substrate stabilization through hydrophobic interactions. However, this role for Phe64 has not been experimentally demonstrated (see below). In comparison with other B1 MBLs, NDM-1 has a somewhat deeper active site cavity with a hydrophobic wall comprised of Trp87, Gln119, Asp120 and His263 [79, 131, 134, 135]. A comparison of an unliganded NDM-1 structure to those obtained for VIM-2 and VIM-4 [78, 111] reveals three notable structural differences [79, 134-136] (Figure 7, panels D to I). First, ASL3 and ASL10 are positioned further away from the Zn active-site center, resulting in a more open active site for NDM-1. Second, VIM-2 has several residues such as Phe61, Asp62, Tyr67, and Arg228, which are projected into the active site, reducing the size of the VIM-2 binding cavity. In comparison, NDM-1 possesses less bulky and more hydrophobic residues at equivalent positions, including M61, P62, V67, and A228. Third, NDM-1 has more positive charges located near Zn1, which are proposed to help attract and position the anionic β-lactam substrate for more efficient catalysis [79].
Taking these attributes into account, NDM-1 may be an optimized enzyme that has achieved a balance between presenting an open active site, surrounded by loops flexible enough to accommodate substrates with different substituents, yet preserving the precise interactions needed to position substrates for productive catalysis. The optimal nature of these loops may be one explanation that accounts for the scarcity of naturally occurring mutations discovered in the ASL3, ASL10 and second-shell zinc ligands. This deficit is in stark contrast to the IMP and VIM MBLs, in which mutations implicated in modulating the substrate specificity, are either second-shell ligands (e.g. S262 in IMP), ASL3 mutations (e.g. Val67 in IMP), or ASL10 mutations (Gly235 in IMP, His/Tyr224, and Arg228 in VIM-like MBLs). However, the fewer number of currently identified loop and active-site mutations may also reflect the relatively shorter time that NDM has been recognized and studied.
Substrate binding, catalysis and inhibitor design
Generalizations about substrate binding among the B1 β-lactamases are somewhat difficult to make, due to the sequence variation within this subfamily and the structural variation of different β-lactam substrates. For a number of years, the complete lack of any MBL crystal structures that contained a bound substrate-derived molecule served as a considerable barrier toward understanding these interactions. The first structure of a MBL:reaction-intermediate complex was reported in 2004 for the B2 β-lactamase CphA and biapenem (a carbapenem) [137]. This complex contains a structurally rearranged species that molecular modeling studies suggest was produced through a kinetically-disfavored reaction pathway leading to accumulation of a thermodynamically-stable rearranged intermediate / product species trapped in the crystal [138]. This structural rearrangement may enable interactions with the protein that are not made by the parent substrate. The second MBL structure was reported in 2005 with the B3 β-lactamase L1 and the hydrolysis product of moxalactam (an oxycephem, in which the cephem sulfur is replaced by an oxygen) [139]. Unfortunately, these product-bound structures were not readily applicable to understanding substrate binding to the B1 β-lactamases. Fortunately, this deficiency has recently (2011 ff.) been addressed by the publication of a series of product complexes bound to the B1 β-lactamase NDM-1. In general, it should be noted that the observed conformations of bound product molecules may be different than those of the parent substrates due to chemical differences (e.g. introduction of a carboxylate) and due to increased flexibility subsequent to ring opening. However, these structures provide considerable insight into ligand recognition by NDM-1.
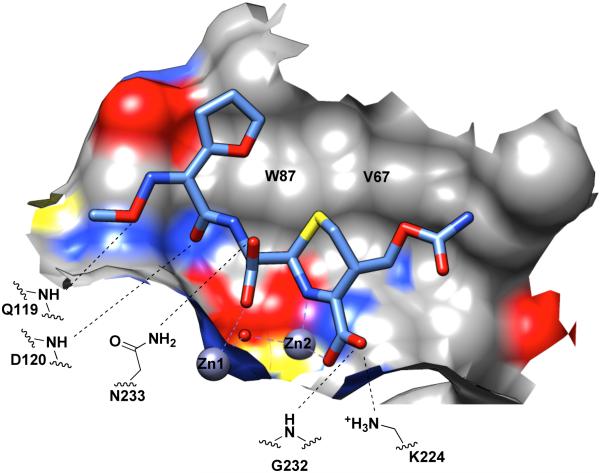
To date, structures have been reported for dizinc NDM-1 in complex with the hydrolyzed products of penams (benzylpenicillin [140], ampicillin [71, 134], methicillin [140] and oxacillin [140]), cephems (cefuroxime and cephalexin) [141], and a carbapenem (meropenem) (Figure 10) [140]. An artificial metalloform of NDM-1 with cadmium ions substituted for zinc was also used to characterize complexes with the hydrolysis products of a penem (faropenem) and of ampicillin [71]. Comparison of these structures reveals some binding interactions that are held in common for all product ligands, and other interactions that appear to vary depending on the product substituents. As an example, see Figure 11. Considering complexes with dizinc NDM-1, the common features observed in product-bound structures include interactions with the product’s newly formed carboxylate and the amine that were created upon cleavage of the β-lactam bond. In all cases, this carboxylate is coordinated to Zn1 and is in H-bonding distance with the side chain of Asn233. The newly formed amine is found coordinated (at 2.1 – 2.4 Å) to Zn2. Other common interactions are those with the conserved carboxylate found in all β-lactam drugs, and with the bicyclic β-lactam core structure. In all cases, this conserved carboxylate in the product complexes is found coordinated to Zn2 and is within H-bonding distance of both the side chain of Lys224 and the backbone nitrogen of Asn233. The bicyclic β-lactam core found in penam, penem and cephem parent compounds, presents one side of the rings (as their hydrolysis products) to the broad hydrophobic surface created by the side chains of Trp87 and Val67, the latter of which forms the base of the active-site β-hairpin loop ASL3.
Figure 10.
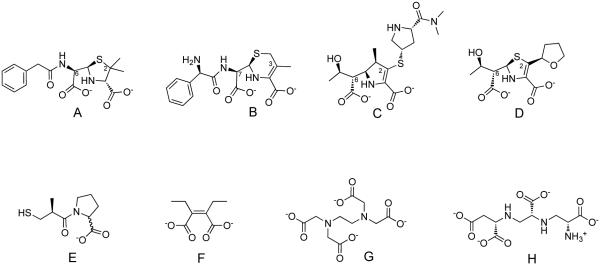
Selected structures of MBL hydrolysis products and inhibitors. A. Example of a hydrolyzed penam (benzylpenicillin); B. A hydrolyzed cephem (cephalexin); C. A hydrolyzed carbapenem (meropenem); D. A hydrolyzed penem (faropenem); Numbering is shown according to the parent substrates to identify substituents mentioned in the text. E. A thiol / carboxylate inhibitor (DL-captopril); F. A dicarboxylate inhibitor (ME1071 or (Z)-2,3-diethylbut-2-enediolate); note similarities to the dicarboxylates found in hydrolysis products; G. Example of a metal-stripping inhibitor (EDTA); H. A structurally-similar metal stripping inhibitor (aspergillomarasmine A).
Figure 11.
Substrate/Product Binding Site of B1 MBLs. The hydrolysis product of cefuroxime (shown as sticks) bound to dizinc NDM-1 [141] is shown as an example. A portion of the NDM-1 binding surface is shown. Coloring is by heteroatom (or the underlying heteroatom of the surface) (carbon in grey; oxygen in red; nitrogen in blue; sulfur in yellow). Coordination to the active-site zinc ions is shown with a purple dashed line. Distances between heteroatoms close enough to allow H-bonding are indicated by black dashed lines.
In contrast to the common binding interactions observed with the core β-lactam bicycle of penam, penem, carbapenem, and cephem products, NDM-1 appears to make many fewer interactions with substituents appended to these cores. The bound hydrolysis products of meropenem, faropenem and cefuroxime have substituents at positions 2, 2, or 3, respectively, that are placed in the wide substrate-binding groove extending away from Zn2 [71, 140, 141]. The substituent at this position is totally disordered in the faropenem complex, and does not make any H-bonds in the meropenem or cefuroxime complexes. Substituents extending away from the other side of the β-lactam bicycle (position 6 in penams, penems and carbapenems and position 7 in cephems) are placed in the wide substrate binding groove extending away from Zn1, and do reveal a few conserved H-bonding partners. Structures of the ampicillin product place the primary amine substituent within H-bonding distance of the side chain of Gln119 [71, 134]. Also, the substituents of cefuroxime, oxacillin, and methicillin H-bond with the backbone nitrogens of either Gln119, Asp120, or both [134, 140]. Other products (derived from meropenem, benzylpenicillin, and cephalexin) that lack suitable H-bonding groups at the analogous positions do not make any of these interactions with NDM-1 [134, 140].
As is true for all protein:ligand structures, it is important to note that the NDM-1:product structures may not reveal all of the structural conformations of the ligand or the protein that are relevant to substrate binding in solution. This may be a particular issue in the case of NDM-1: product complexes. Extensive characterization of other B1 β-lactamases by X-ray crystallography, NMR, and functional studies have indicated that the active-site β-hairpin loop (ASL3) closes down over inhibitors upon binding and over substrates during catalysis [81, 83, 142]. However, in all of the NDM-1:product complexes characterized by X-ray crystallography to date, ASL3 is found in an open and extended position. In a number of these structures, this loop appears to be artificially constrained. In some structures, an N-terminal extension of the NDM-1 sequence that is not required for maximum catalytic efficiency [132, 136] interacts with this β-hairpin to stabilize formation of an extended β-sheet [71, 134, 141]. In several others, the β-hairpin of one crystallographic monomer interacts with the active-site of a second monomer, making hydrophobic and H-bonding interactions directly with the product molecule of the opposing monomer [140, 141]. Since these observed conformations are likely artifacts of crystallization, the details of how ASL3 interacts with bound substrate/product will require further study for elucidation. Regardless, the observed interactions between β-lactam products and the B1 β-lactamase NDM-1 can be summarized as follows. NDM-1 makes its most extensive and specific interactions with the bicyclic β-lactam core and the conserved carboxylate of substrates/products. Many fewer binding interactions are made between the protein and substituents appended to the β-lactam core. Because of the structural similarities of the bicyclic core structures in penams, penems, carbapenems and cephems, these diverse substrates are all well recognized and processed despite the diverse structural variations found in their substituents, revealing the mechanism used for broad substrate recognition by the B1 MBLs to provide antibiotic resistance against a structurally diverse class of drugs.
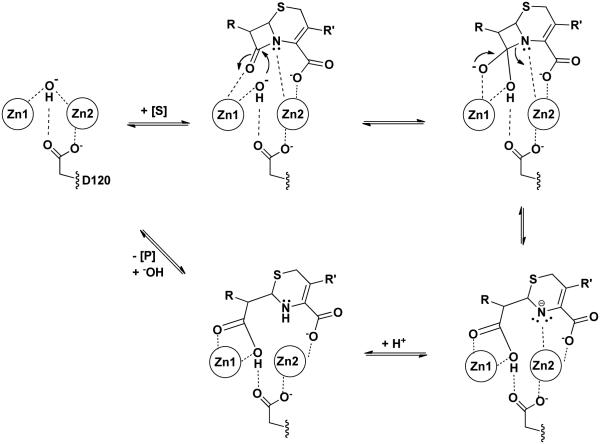
Similar to the limited number of important binding interactions noted above, subsequent β-lactam hydrolysis by B1 MBLs is also catalyzed by only a very focused set of interactions. We will consider the proposed mechanism for dinuclear B1 MBLs, with evidence for the key steps reviewed previously by Crowder and co-workers [16] and expanded by Dal Peraro et al [123] (Figure 12). Mechanisms for mononuclear B1 MBL catalysis are described elsewhere [16, 64, 123, 143, 144]. The resting unliganded B1 MBL contains a dizinc-bridging hydroxide ion within hydrogen bonding distance of the side chain of Asp120. Similar to the product-bound complexes described above, substrates are proposed to bind with their β-lactam carbonyl oxygen coordinating to Zn1, and the leaving group nitrogen to Zn2, filling open coordination sites on each zinc ion. In addition, the conserved substrate carboxylate also coordinates to Zn2 as well as a backbone amide nitrogen. A structurally conserved Lys/Arg residue also stabilizes the substrate carboxylate, but this residue can be varied while still maintaining significant catalysis (e.g. [145]), likely due to the remaining Zn2 interaction.
Figure 12.
Proposed Catalytic Mechanism for B1 MBLs. Only the essential catalytic zinc and D120 features are shown in a proposed hydrolysis mechanism of a cephem. Dashed lines indicate proposed interactions that facilitate binding and catalysis.
Upon the subsequent attack of the bridging hydroxide on the β-lactam carbonyl, a tetrahedral adduct is presumably formed and stabilized through Zn1 interactions, and collapse of this species leads to cleavage of the β-lactam C-N bond. The proposed tetrahedral adduct has not been well established and may not be long lived [16]. The Asn233 side chain that makes a conserved interaction with the carbonyl of the product that corresponds to the β-lactam bond carbonyl in the substrate, contributes to both kcat and KM kinetic parameters, but most variants at this position still provide sufficient activity to confer resistance when expressed in cell culture [92]. In serine β-lactamases, C-N bond cleavage is catalyzed, likely by a general acid that protonates the leaving group nitrogen [146]. However, in a defining mechanistic feature of the MBLs, instead of general acid catalysis by an amino acid side chain, the Zn2 ion acts instead as a Lewis acid to accept the lone electron pair developed on the leaving group nitrogen [147]. In substrates with electron withdrawing groups conjugated to this nitrogen (e.g. the cephems nitrocefin and chromacef), the nitrogen anion is strongly stabilized and protonation of this anionic intermediate, observed through kinetic, spectroscopic, and isotope effect techniques, is the rate limiting step in the overall reaction [70, 132, 147-149]. This protonation does not appear to be mediated by an amino acid side chain [105]. In substrates without conjugated electron withdrawing groups, the Zn2 ion is presumably still positioned to help polarize the β-lactam bond and to stabilize the developing charge on the leaving group. In some examples, an anionic intermediate is not observed [150, 151], so it is not clear if the Zn2 or the substrate is positioned differently in these cases and the anionic species is not formed, or if an anionic reaction intermediate just does not have a long enough lifetime to be detected. Subsequent dissociation of the product from the metal center and regeneration of the bridging hydroxide then returns the enzyme to its original state. The reactions summarized in Figure 12 are drawn to formally allow for microscopic reversibility of each step, but pre-steady state kinetic determinations typically assign the decay of the intermediate with the anionic nitrogen, and often the formation of the same intermediate, as irreversible. Some variations of this mechanism are proposed elsewhere [71, 149, 152]. The most striking aspect of the proposed mechanism is that all of the key catalytic steps are carried out only through direct interactions with the bound Zn ions and not through amino acid side chains, which presumably have roles more limited to binding and positioning the substrate and to providing a scaffold for the precise formation and “tuning” of the dinuclear zinc ion cluster.
Inhibitors of MBLs, including those for the B1 subclass, have been extensively and recently reviewed, and several common structural motifs noted [153-155]. These motifs include zinc-binding sulfur atoms, zinc-binding dicarboxylates, other zinc-binding moieties, natural products, fragment-sized inhibitors and other miscellaneous inhibitors, including those with covalent inactivation mechanisms as well as non- and un-competitive inhibitors (Figure 10). One often-expressed goal is the development of an inhibitor effective for all MBLs, and some progress has been made toward this end [156]. However, the disparity in zinc stoichiometry and coordination, the differences in substrate-binding loops, and the sequence divergence between B1, B2 and B3 subclasses of β-lactamases make this a difficult goal to reach. Such efforts may also have diminishing returns since the B1 subclass contains the most clinically relevant enzymes. A short term goal is to develop inhibitors with selectivity that focuses on the B1 subclass. Recent work has shown that inhibitors can be developed that maintain high potency for a broad spectrum of B1 MBLs [157]. The above consideration of the molecular epidemiology, identification of conserved substrate binding interactions, and conserved aspects of the catalytic mechanisms should be able to provide additional design constraints for improving inhibitors in terms of their potency, selectivity, and insensitivity to mutation within this subclass of β-lactamase.
Probably the most difficult challenge in designing B1 MBL inhibitors is the lack of amino acid sequence conservation within this subfamily. Because the major subgroups, IMP, VIM and NDM, share only approximately 30% sequence identity, the development of non-competitive or allosteric inhibitors (e.g. [158-160]) is likely to be too limiting since binding surfaces away from the active site are already quite divergent and can more easily tolerate a wider array of mutations than the active site. Even the substrate-binding groove presents a challenge because its shallowness leads to ligand binding interactions comprised of more distributive interactions than those seen in more typical deep pockets. As a result, the bound product molecules are observed to make only very few specific H-bonding interactions within this shallow groove. In addition to the structural features that impede a class B1 inhibitor design, the sequence divergence within each subgroup is also a potential problem. As an extreme example, Palzkill and co-workers constructed artificial libraries, which demonstrated that almost every residue in the IMP-1 active site, except for the primary zinc ligands, could be mutated and still provide resistance against at least a subset of β-lactam antibiotics [92, 102, 124]. Because a significant number of variants have already emerged for each subgroup, because these enzymes will continue to evolve, and because the identities of very few residues are tightly constrained by function, the appearance of resistance toward inhibitors that makes key contacts with variable residues is a significant concern.
Therefore, the design of B1 MBL inhibitors may best be achieved by mimicking the focused nature of interactions observed in both substrate recognition and the catalytic mechanism. This suggests that the key binding interactions, which provide the majority of inhibitor affinity, should be made at the conserved active site zinc ion cluster. Affinity could then be augmented by optimizing substituents to interact with sites on the protein that are more refractory to mutagenesis, such as backbone amides like those shown to H-bond to products (discussed above). Other active-site residue side chains shown to be evolutionarily constrained, in that they tolerate only a subset of mutations or that their mutation results in catalytic impairment, would also make good partners with which to design specific interactions with inhibitors, since there will be a selective pressure to retain the identity of these residues. Two examples are the structurally conserved Lys/Arg residue neighboring the Zn2 ion and the residues that provide the hydrophobic surface at the base of ASL3.
In light of these proposed design constraints for developing B1 MBL inhibitors, two particular types of inhibitors should also be given special attention: zinc-stripping chelators and covalent irreversible inhibitors. Zinc-binding inhibitors can work by two limiting mechanisms. In the first, the inhibitor binds zinc at the active site of the MBL resulting in a ternary complex that competitively blocks substrate from binding [161]. In the second, a strong zinc chelator binds and sequesters free zinc separate from the protein, essentially stripping the zinc out of the active site of β-lactamases. This process may proceed through a transient ternary complex with the enzyme [162], or might instead just sequester zinc as it dissociates during equilibrium, without making any true interactions with the enzyme. Such zinc-stripping chelators can result in very potent observed inhibition, but one significant drawback is that their affinity and selectivity may only be derived from interactions with the zinc ion, and not the protein. So, this type of inhibitors is much more likely to have nonspecific off-target effects by inhibiting other metalloproteins as well. One such example is the recent report of an NDM-1 inhibitor aspergillomarasmine A, which is a structural analog of the metal ion chelator EDTA (Figure 10) [163]. This compound inhibits NDM-1 by stripping and sequestering the active-site zinc ions. Although a KI value is reported, the magnitude of this value is on the same order of the enzyme concentration used, and likely represents a titration of the enzyme rather than any affinity for forming a true ternary complex with the protein.
The second special class to be considered is inhibitors that use a covalent irreversible mechanism. Although designing covalent inhibitors might seem to be a non-starter due to the non-covalent mechanism used by MBLs, several examples have been given in literature whereby time-dependent irreversible covalent modification of either a primary Zn2 ligand Cys or a conserved active-site Lys results in irreversible inhibition [145, 164-167]. Although these current compounds do not have the selectivity, stability, or potency required for clinical treatments, their mechanism of action might provide a roadmap for inhibitor development since changes in affinity due to mutation can be overcome by using longer treatment times with irreversible inhibitors [168], and because covalent inhibition by β-lactams has certainly had a long history of success [13].
Conclusions
This review of the molecular epidemiology, substrate binding interactions and catalytic mechanisms of subclass B1 MBLs highlights conserved features relating to the structure and function of the subclass of MBLs that contains the most clinically-relevant threats. The sequence diversity within this family and the continued evolution of β-lactamase allotypes, present “shifting sands” on which it is difficult to build inhibitor interactions that will survive the ensuing selective pressures. However, our analysis above highlights the focused nature of known binding interactions, and the focused catalytic site, both of which are conserved across the B1 MBL subclass and provide “solid ground” on which to stand in the continued efforts to develop more potent, selective, effective, and lasting inhibitors for this growing antibiotic resistant threat.
Acknowledgements
Work in WF’s laboratory is supported, in part, by a grant from the Robert. A. Welch Foundation (F1572). WF and RAB are supported, in part, by funds from the National Institutes of General Medical Sciences of the National Institutes of Health under award number GM111926. RAB is supported in part by funds from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award numbers R01AI063517 and R01AI100560. RAB is also supported by funds and/or facilities provided by the Cleveland Department of Veterans Affairs, the Veterans Affairs Merit Review Program Award 1I01BX001974 and the Geriatric Research Education and Clinical Center VISN 10. The content is solely the responsibility of the authors and does not represent the official views of the National Institutes of Health or the Department of Veterans Affairs.
REFERENCES
- [1].Fleming A. Classics in infectious diseases: on the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae by Alexander Fleming, Reprinted from the British Journal of Experimental Pathology 10:226-236, 1929. Rev Infect Dis. 1980;2:129–39. [PubMed] [Google Scholar]
- [2].Hare R. New light on the history of penicillin. Med Hist. 1982;26:1–24. doi: 10.1017/s0025727300040758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Waxman DJ, Strominger JL. Penicillin-binding proteins and the mechanism of action of beta-lactam antibiotics. Annu Rev Biochem. 1983;52:825–69. doi: 10.1146/annurev.bi.52.070183.004141. [DOI] [PubMed] [Google Scholar]
- [4].Page MI. The Mechanisms of Reactions of Beta-Lactam Antibiotics. Adv Phys Org Chem. 1987;23:165–270. [Google Scholar]
- [5].Frere JM, Joris B. Penicillin-sensitive enzymes in peptidoglycan biosynthesis. Crit Rev Microbiol. 1985;11:299–396. doi: 10.3109/10408418409105906. [DOI] [PubMed] [Google Scholar]
- [6].Tipper DJ, Strominger JL. Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-D-alanyl-D-alanine. Proc Natl Acad Sci U S A. 1965;54:1133–41. doi: 10.1073/pnas.54.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Tipper DJ. Mode of action of beta-lactam antibiotics. Pharmacol Ther. 1985;27:1–35. doi: 10.1016/0163-7258(85)90062-2. [DOI] [PubMed] [Google Scholar]
- [8].Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev. 2008;32:234–58. doi: 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- [9].Bayles KW. The bactericidal action of penicillin: new clues to an unsolved mystery. Trends Microbiol. 2000;8:274–8. doi: 10.1016/s0966-842x(00)01762-5. [DOI] [PubMed] [Google Scholar]
- [10].Cho H, Uehara T, Bernhardt TG. Beta-lactam antibiotics induce a lethal malfunctioning of the bacterial cell wall synthesis machinery. Cell. 2014;159:1300–11. doi: 10.1016/j.cell.2014.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Brown AG. beta-Lactam nomenclature. J Antimicrob Chemother. 1982;10:365–8. doi: 10.1093/jac/10.5.365. [DOI] [PubMed] [Google Scholar]
- [12].Ehmann DE, Jahic H, Ross PL, Gu RF, Hu J, Kern G, Walkup GK, Fisher SL. Avibactam is a covalent, reversible, non-beta-lactam beta-lactamase inhibitor. Proc Natl Acad Sci U S A. 2012;109:11663–8. doi: 10.1073/pnas.1205073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Drawz SM, Bonomo RA. Three decades of beta-lactamase inhibitors. Clin Microbiol Rev. 2010;23:160–201. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Llarrull LI, Testero SA, Fisher JF, Mobashery S. The future of the beta-lactams. Curr Opin Microbiol. 2010;13:551–7. doi: 10.1016/j.mib.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ambler RP, Coulson AF, Frere JM, Ghuysen JM, Joris B, Forsman M, Levesque RC, Tiraby G, Waley SG. A standard numbering scheme for the class A beta-lactamases. Biochem J. 1991;276:269–70. doi: 10.1042/bj2760269. Pt 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Crowder MW, Spencer J, Vila AJ. Metallo-beta-lactamases: novel weaponry for antibiotic resistance in bacteria. Acc Chem Res. 2006;39:721–8. doi: 10.1021/ar0400241. [DOI] [PubMed] [Google Scholar]
- [17].Palzkill T. Metallo-beta-lactamase structure and function. Ann N Y Acad Sci. 2013;1277:91–104. doi: 10.1111/j.1749-6632.2012.06796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Massidda O, Rossolini GM, Satta G. The Aeromonas hydrophila cphA gene: molecular heterogeneity among class B metallo-beta-lactamases. J Bacteriol. 1991;173:4611–7. doi: 10.1128/jb.173.15.4611-4617.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Walsh TR, Gamblin S, Emery DC, MacGowan AP, Bennett PM. Enzyme kinetics and biochemical analysis of ImiS, the metallo-beta-lactamase from Aeromonas sobria 163a. J Antimicrob Chemother. 1996;37:423–31. doi: 10.1093/jac/37.3.423. [DOI] [PubMed] [Google Scholar]
- [20].Saavedra MJ, Peixe L, Sousa JC, Henriques I, Alves A, Correia A. Sfh-I, a subclass B2 metallo-beta-lactamase from a Serratia fonticola environmental isolate. Antimicrob Agents Chemother. 2003;47:2330–3. doi: 10.1128/AAC.47.7.2330-2333.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Walsh TR, Hall L, Assinder SJ, Nichols WW, Cartwright SJ, MacGowan AP, Bennett PM. Sequence analysis of the L1 metallo-beta-lactamase from Xanthomonas maltophilia. Biochim Biophys Acta. 1994;1218:199–201. doi: 10.1016/0167-4781(94)90011-6. [DOI] [PubMed] [Google Scholar]
- [22].Bellais S, Aubert D, Naas T, Nordmann P. Molecular and biochemical heterogeneity of class B carbapenem-hydrolyzing beta-lactamases in Chryseobacterium meningosepticum. Antimicrob Agents Chemother. 2000;44:1878–86. doi: 10.1128/aac.44.7.1878-1886.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cornaglia G, Giamarellou H, Rossolini GM. Metallo-beta-lactamases: a last frontier for beta-lactams? Lancet Infect Dis. 2011;11:381–93. doi: 10.1016/S1473-3099(11)70056-1. [DOI] [PubMed] [Google Scholar]
- [24].Oelschlaeger P, Ai N, Duprez KT, Welsh WJ, Toney JH. Evolving carbapenemases: can medicinal chemists advance one step ahead of the coming storm? J Med Chem. 2010;53:3013–27. doi: 10.1021/jm9012938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Walsh TR, Weeks J, Livermore DM, Toleman MA. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis. 2011;11:355–62. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- [26].Abraham EP, Chain E. An Enzyme from Bacteria able to Destroy Penicillin. Nature. 1940;146:837. [PubMed] [Google Scholar]
- [27].Sabath LD, Abraham EP. Zinc as a cofactor for cephalosporinase from Bacillus cereus 569. Biochem J. 1966;98:11C–3C. doi: 10.1042/bj0980011c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Rasmussen BA, Gluzman Y, Tally FP. Cloning and sequencing of the class B beta-lactamase gene (ccrA) from Bacteroides fragilis TAL3636. Antimicrob Agents Chemother. 1990;34:1590–2. doi: 10.1128/aac.34.8.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Watanabe M, Iyobe S, Inoue M, Mitsuhashi S. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1991;35:147–51. doi: 10.1128/aac.35.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Osano E, Arakawa Y, Wacharotayankun R, Ohta M, Horii T, Ito H, Yoshimura F, Kato N. Molecular characterization of an enterobacterial metallo beta-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob Agents Chemother. 1994;38:71–8. doi: 10.1128/aac.38.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Senda K, Arakawa Y, Ichiyama S, Nakashima K, Ito H, Ohsuka S, Shimokata K, Kato N, Ohta M. PCR detection of metallo-beta-lactamase gene (blaIMP) in gram-negative rods resistant to broad-spectrum beta-lactams. J Clin Microbiol. 1996;34:2909–13. doi: 10.1128/jcm.34.12.2909-2913.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Cornaglia G, Riccio ML, Mazzariol A, Lauretti L, Fontana R, Rossolini GM. Appearance of IMP-1 metallo-beta-lactamase in Europe. Lancet. 1999;353:899–900. doi: 10.1016/s0140-6736(98)05954-6. [DOI] [PubMed] [Google Scholar]
- [33].Da Silva GJ, Correia M, Vital C, Ribeiro G, Sousa JC, Leitao R, Peixe L, Duarte A. Molecular characterization of bla(IMP-5), a new integron-borne metallo-beta-lactamase gene from an Acinetobacter baumannii nosocomial isolate in Portugal. FEMS Microbiol Lett. 2002;215:33–9. doi: 10.1111/j.1574-6968.2002.tb11366.x. [DOI] [PubMed] [Google Scholar]
- [34].Kouda S, Ohara M, Onodera M, Fujiue Y, Sasaki M, Kohara T, Kashiyama S, Hayashida S, Harino T, Tsuji T, Itaha H, Gotoh N, Matsubara A, Usui T, Sugai M. Increased prevalence and clonal dissemination of multidrug-resistant Pseudomonas aeruginosa with the blaIMP-1 gene cassette in Hiroshima. J Antimicrob Chemother. 2009;64:46–51. doi: 10.1093/jac/dkp142. [DOI] [PubMed] [Google Scholar]
- [35].Zhao W-H, Hu Z-Q. IMP-type metallo-β-lactamases in Gram-negative bacilli: distribution, phylogeny, and association with integrons. Crit Rev Microbiol. 2011;37:214–26. doi: 10.3109/1040841X.2011.559944. [DOI] [PubMed] [Google Scholar]
- [36].Bonomo RA, Tolmasky M. Enzyme-mediated resistance to antibiotics : mechanisms, dissemination, and prospects for inhibition. ASM Press; Washington, D.C.: 2007. [Google Scholar]
- [37].Carattoli A. Plasmids and the spread of resistance. Int J Med Microbiol. 2013;303:298–304. doi: 10.1016/j.ijmm.2013.02.001. [DOI] [PubMed] [Google Scholar]
- [38].Espedido BA, Partridge SR, Iredell JR. bla(IMP-4) in different genetic contexts in Enterobacteriaceae isolates from Australia. Antimicrob Agents Chemother. 2008;52:2984–7. doi: 10.1128/AAC.01634-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lauretti L, Riccio ML, Mazzariol A, Cornaglia G, Amicosante G, Fontana R, Rossolini GM. Cloning and characterization of bla VIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob Agents Chemother. 1999;43:1584–90. doi: 10.1128/aac.43.7.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Poirel L, Naas T, Nicolas D, Collet L, Bellais S, Cavallo J-D, Nordmann P. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid-and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob Agents Chemother. 2000;44:891–7. doi: 10.1128/aac.44.4.891-897.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Bush K, Jacoby G. Beta-Lactamse Classification and Amino Acid Sequences for TEM, SHV and OXA Extened-Spectrum and Inhibitor Resistant Enzymes 2014. [cited 2014 Nov. 7, 2014]. Available from: http://www.lahey.org/studies/ [Google Scholar]
- [42].Strateva T, Yordanov D. Pseudomonas aeruginosa - a phenomenon of bacterial resistance. J Med Microbiol. 2009;58:1133–48. doi: 10.1099/jmm.0.009142-0. [DOI] [PubMed] [Google Scholar]
- [43].Patel G, Bonomo RA. Status report on carbapenemases: challenges and prospects. Expert Rev Anti Infect Ther. 2011;9:555–70. doi: 10.1586/eri.11.28. [DOI] [PubMed] [Google Scholar]
- [44].Ktari S, Arlet G, Mnif B, Gautier V, Mahjoubi F, Ben Jmeaa M, Bouaziz M, Hammami A. Emergence of multidrug-resistant Klebsiella pneumoniae isolates producing VIM-4 metallo-beta-lactamase, CTX-M-15 extended-spectrum beta-lactamase, and CMY-4 AmpC beta-lactamase in a Tunisian university hospital. Antimicrob Agents Chemother. 2006;50:4198–201. doi: 10.1128/AAC.00663-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Pournaras S, Tsakris A, Maniati M, Tzouvelekis LS, Maniatis AN. Novel variant (bla(VIM-4)) of the metallo-beta-lactamase gene bla(VIM-1) in a clinical strain of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2002;46:4026–8. doi: 10.1128/AAC.46.12.4026-4028.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20:440–58. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Walsh TR. Clinically significant carbapenemases: an update. Curr Opin Infect Dis. 2008;21:367–71. doi: 10.1097/QCO.0b013e328303670b. [DOI] [PubMed] [Google Scholar]
- [48].Miro E, Segura C, Navarro F, Sorli L, Coll P, Horcajada JP, Alvarez-Lerma F, Salvado M. Spread of plasmids containing the bla(VIM-1) and bla(CTX-M) genes and the qnr determinant in Enterobacter cloacae, Klebsiella pneumoniae and Klebsiella oxytoca isolates. J Antimicrob Chemother. 2010;65:661–5. doi: 10.1093/jac/dkp504. [DOI] [PubMed] [Google Scholar]
- [49].Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53:5046–54. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Castanheira M, Deshpande LM, Mathai D, Bell JM, Jones RN, Mendes RE. Early dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY Antimicrobial Surveillance Program, 2006-2007. Antimicrob Agents Chemother. 2011;55:1274–8. doi: 10.1128/AAC.01497-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Johnson AP, Woodford N. Global spread of antibiotic resistance: the example of New Delhi metallo-beta-lactamase (NDM)-mediated carbapenem resistance. J Med Microbiol. 2013;62:499–513. doi: 10.1099/jmm.0.052555-0. [DOI] [PubMed] [Google Scholar]
- [52].Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, Krishnan P, Kumar AV, Maharjan S, Mushtaq S, Noorie T, Paterson DL, Pearson A, Perry C, Pike R, Rao B, Ray U, Sarma JB, Sharma M, Sheridan E, Thirunarayan MA, Turton J, Upadhyay S, Warner M, Welfare W, Livermore DM, Woodford N. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Nordmann P, Naas T, Poirel L. Global spread of Carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011;17:1791–8. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Patel G, Bonomo RA. "Stormy waters ahead": global emergence of carbapenemases. Front Microbiol. 2013;4:48. doi: 10.3389/fmicb.2013.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Darley E, Weeks J, Jones L, Daniels V, Wootton M, MacGowan A, Walsh T. NDM-1 polymicrobial infections including Vibrio cholerae. Lancet. 2012;380:1358. doi: 10.1016/S0140-6736(12)60911-8. [DOI] [PubMed] [Google Scholar]
- [56].Decousser JW, Jansen C, Nordmann P, Emirian A, Bonnin RA, Anais L, Merle JC, Poirel L. Outbreak of NDM-1-producing Acinetobacter baumannii in France, January to May 2013. Euro Surveill. 2013:18. doi: 10.2807/1560-7917.es2013.18.31.20547. [DOI] [PubMed] [Google Scholar]
- [57].Flateau C, Janvier F, Delacour H, Males S, Ficko C, Andriamanantena D, Jeannot K, Merens A, Rapp C. Recurrent pyelonephritis due to NDM-1 metallo-beta-lactamase producing Pseudomonas aeruginosa in a patient returning from Serbia, France, 2012. Euro Surveill. 2012:17. [PubMed] [Google Scholar]
- [58].Toleman MA, Spencer J, Jones L, Walsh TR. blaNDM-1 is a chimera likely constructed in Acinetobacter baumannii. Antimicrob Agents Chemother. 2012;56:2773–6. doi: 10.1128/AAC.06297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Liu Z, Li W, Wang J, Pan J, Sun S, Yu Y, Zhao B, Ma Y, Zhang T, Qi J, Liu G, Lu F. Identification and characterization of the first Escherichia coli strain carrying NDM-1 gene in China. PLoS One. 2013;8:e66666. doi: 10.1371/journal.pone.0066666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Daiyasu H, Osaka K, Ishino Y, Toh H. Expansion of the zinc metallo-hydrolase family of the beta-lactamase fold. FEBS Lett. 2001;503:1–6. doi: 10.1016/s0014-5793(01)02686-2. [DOI] [PubMed] [Google Scholar]
- [61].Baier F, Tokuriki N. Connectivity between catalytic landscapes of the metallo-beta-lactamase superfamily. J Mol Biol. 2014;426:2442–56. doi: 10.1016/j.jmb.2014.04.013. [DOI] [PubMed] [Google Scholar]
- [62].Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–40. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- [63].Massova I, Mobashery S. Kinship and diversification of bacterial penicillin-binding proteins and beta-lactamases. Antimicrob Agents Chemother. 1998;42:1–17. doi: 10.1128/aac.42.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Carfi A, Pares S, Duee E, Galleni M, Duez C, Frere JM, Dideberg O. The 3-D structure of a zinc metallo-beta-lactamase from Bacillus cereus reveals a new type of protein fold. EMBO J. 1995;14:4914–21. doi: 10.1002/j.1460-2075.1995.tb00174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Concha NO, Rasmussen BA, Bush K, Herzberg O. Crystal structure of the wide-spectrum binuclear zinc beta-lactamase from Bacteroides fragilis. Structure. 1996;4:823–36. doi: 10.1016/s0969-2126(96)00089-5. [DOI] [PubMed] [Google Scholar]
- [66].Kim Y, Tesar C, Mire J, Jedrzejczak R, Binkowski A, Babnigg G, Sacchettini J, Joachimiak A. Structure of apo- and monometalated forms of NDM-1--a highly potent carbapenem-hydrolyzing metallo-beta-lactamase. PLoS One. 2011;6:e24621. doi: 10.1371/journal.pone.0024621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].de Seny D, Heinz U, Wommer S, Kiefer M, Meyer-Klaucke W, Galleni M, Frere JM, Bauer R, Adolph HW. Metal ion binding and coordination geometry for wild type and mutants of metallo-beta-lactamase from Bacillus cereus 569/H/9 (BcII): a combined thermodynamic, kinetic, and spectroscopic approach. J Biol Chem. 2001;276:45065–78. doi: 10.1074/jbc.M106447200. [DOI] [PubMed] [Google Scholar]
- [68].Paul-Soto R, Bauer R, Frere JM, Galleni M, Meyer-Klaucke W, Nolting H, Rossolini GM, de Seny D, Hernandez-Valladares M, Zeppezauer M, Adolph HW. Mono- and binuclear Zn2+-beta-lactamase. Role of the conserved cysteine in the catalytic mechanism. J Biol Chem. 1999;274:13242–9. doi: 10.1074/jbc.274.19.13242. [DOI] [PubMed] [Google Scholar]
- [69].Rasia RM, Vila AJ. Exploring the role and the binding affinity of a second zinc equivalent in B. cereus metallo-beta-lactamase. Biochemistry. 2002;41:1853–60. doi: 10.1021/bi010933n. [DOI] [PubMed] [Google Scholar]
- [70].Aitha M, Marts AR, Bergstrom A, Moller AJ, Moritz L, Turner L, Nix JC, Bonomo RA, Page RC, Tierney DL, Crowder MW. Biochemical, mechanistic, and spectroscopic characterization of metallo-beta-lactamase VIM-2. Biochemistry. 2014;53:7321–31. doi: 10.1021/bi500916y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Kim Y, Cunningham MA, Mire J, Tesar C, Sacchettini J, Joachimiak A. NDM-1, the ultimate promiscuous enzyme: substrate recognition and catalytic mechanism. FASEB J. 2013;27:1917–27. doi: 10.1096/fj.12-224014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Gonzalez LJ, Moreno DM, Bonomo RA, Vila AJ. Host-specific enzyme-substrate interactions in SPM-1 metallo-beta-lactamase are modulated by second sphere residues. PLoS Pathog. 2014;10:e1003817. doi: 10.1371/journal.ppat.1003817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Meini MR, Llarrull LI, Vila AJ. Evolution of Metallo-beta-lactamases: Trends Revealed by Natural Diversity and Evolution. Antibiotics (Basel) 2014;3:285–316. doi: 10.3390/antibiotics3030285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Tomatis PE, Rasia RM, Segovia L, Vila AJ. Mimicking natural evolution in metallo-beta-lactamases through second-shell ligand mutations. Proc Natl Acad Sci U S A. 2005;102:13761–6. doi: 10.1073/pnas.0503495102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Garau G, García-Sáez I, Bebrone C, Anne C, Mercuri P, Galleni M, Frère J-M, Dideberg O. Update of the standard numbering scheme for class B β-lactamases. Antimicrob Agents Chemother. 2004;48:2347–9. doi: 10.1128/AAC.48.7.2347-2349.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Cadag E, Vitalis E, Lennox KP, Zhou CL, Zemla AT. Computational analysis of pathogen-borne metallo beta-lactamases reveals discriminating structural features between B1 types. BMC Res Notes. 2012;5:96. doi: 10.1186/1756-0500-5-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Concha NO, Janson CA, Rowling P, Pearson S, Cheever CA, Clarke BP, Lewis C, Galleni M, Frere JM, Payne DJ, Bateson JH, Abdel-Meguid SS. Crystal structure of the IMP-1 metallo beta-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: binding determinants of a potent, broad-spectrum inhibitor. Biochemistry. 2000;39:4288–98. doi: 10.1021/bi992569m. [DOI] [PubMed] [Google Scholar]
- [78].Garcia-Saez I, Docquier JD, Rossolini GM, Dideberg O. The three-dimensional structure of VIM-2, a Zn-beta-lactamase from Pseudomonas aeruginosa in its reduced and oxidised form. J Mol Biol. 2008;375:604–11. doi: 10.1016/j.jmb.2007.11.012. [DOI] [PubMed] [Google Scholar]
- [79].King D, Strynadka N. Crystal structure of New Delhi metallo-beta-lactamase reveals molecular basis for antibiotic resistance. Protein Sci. 2011;20:1484–91. doi: 10.1002/pro.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Huntley JJ, Scrofani SD, Osborne MJ, Wright PE, Dyson HJ. Dynamics of the metallo-beta-lactamase from Bacteroides fragilis in the presence and absence of a tight-binding inhibitor. Biochemistry. 2000;39:13356–64. doi: 10.1021/bi001210r. [DOI] [PubMed] [Google Scholar]
- [81].Scrofani SD, Chung J, Huntley JJ, Benkovic SJ, Wright PE, Dyson HJ. NMR characterization of the metallo-beta-lactamase from Bacteroides fragilis and its interaction with a tight-binding inhibitor: role of an active-site loop. Biochemistry. 1999;38:14507–14. doi: 10.1021/bi990986t. [DOI] [PubMed] [Google Scholar]
- [82].Huntley JJ, Fast W, Benkovic SJ, Wright PE, Dyson HJ. Role of a solvent-exposed tryptophan in the recognition and binding of antibiotic substrates for a metallo-beta-lactamase. Protein Sci. 2003;12:1368–75. doi: 10.1110/ps.0305303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Rydzik AM, Brem J, van Berkel SS, Pfeffer I, Makena A, Claridge TD, Schofield CJ. Monitoring conformational changes in the NDM-1 metallo-beta-lactamase by 19F NMR spectroscopy. Angew Chem Int Ed Engl. 2014;53:3129–33. doi: 10.1002/anie.201310866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Riccio ML, Franceschini N, Boschi L, Caravelli B, Cornaglia G, Fontana R, Amicosante G, Rossolini GM. Characterization of the metallo-β-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of bla IMP allelic variants carried by gene cassettes of different phylogeny. Antimicrob Agents Chemother. 2000;44:1229–35. doi: 10.1128/aac.44.5.1229-1235.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Iyobe S, Kusadokoro H, Ozaki J, Matsumura N, Minami S, Haruta S, Sawai T, O'Hara K. Amino acid substitutions in a variant of IMP-1 metallo-β-lactamase. Antimicrob Agents Chemother. 2000;44:2023–7. doi: 10.1128/aac.44.8.2023-2027.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Oelschlaeger P, Mayo SL, Pleiss J. Impact of remote mutations on metallo-β-lactamase substrate specificity: Implications for the evolution of antibiotic resistance. Protein Sci. 2005;14:765–74. doi: 10.1110/ps.041093405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Yano H, Kuga A, Okamoto R, Kitasato H, Kobayashi T, Inoue M. Plasmid-encoded metallo-β-lactamase (IMP-6) conferring resistance to carbapenems, especially meropenem. Antimicrob Agents Chemother. 2001;45:1343–8. doi: 10.1128/AAC.45.5.1343-1348.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Liu EM, Pegg KM, Oelschlaeger P. The sequence-activity relationship between metallo-β-lactamases IMP-1, IMP-6, and IMP-25 suggests an evolutionary adaptation to meropenem exposure. Antimicrob Agents Chemother. 2012;56:6403–6. doi: 10.1128/AAC.01440-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Oelschlaeger P, Pleiss J. Hydroxyl groups in the betabeta sandwich of metallo-beta-lactamases favor enzyme activity: Tyr218 and Ser262 pull down the lid. J Mol Biol. 2007;366:316–29. doi: 10.1016/j.jmb.2006.11.027. [DOI] [PubMed] [Google Scholar]
- [90].Oelschlaeger P, Schmid RD, Pleiss J. Modeling domino effects in enzymes: molecular basis of the substrate specificity of the bacterial metallo-beta-lactamases IMP-1 and IMP-6. Biochemistry. 2003;42:8945–56. doi: 10.1021/bi0300332. [DOI] [PubMed] [Google Scholar]
- [91].Pegg KM, Liu EM, George AC, LaCuran AE, Bethel CR, Bonomo RA, Oelschlaeger P. Understanding the determinants of substrate specificity in IMP family metallo-β-lactamases: The importance of S262. Protein Sci. 2014;23:1451–60. doi: 10.1002/pro.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Brown NG, Horton LB, Huang W, Vongpunsawad S, Palzkill T. Analysis of the functional contributions of Asn233 in metallo-beta-lactamase IMP-1. Antimicrob Agents Chemother. 2011;55:5696–702. doi: 10.1128/AAC.00340-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Iyobe S, Kusadokoro H, Takahashi A, Yomoda S, Okubo T, Nakamura A, O'Hara K. Detection of a variant metallo-β-lactamase, IMP-10, from two unrelated strains of Pseudomonas aeruginosa and an Alcaligenes xylosoxidans strain. Antimicrob Agents Chemother. 2002;46:2014–6. doi: 10.1128/AAC.46.6.2014-2016.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Tada T, Miyoshi-Akiyama T, Shimada K, Shimojima M, Kirikae T. IMP-43 and IMP-44 Metallo-β-Lactamases with Increased Carbapenemase Activities in Multidrug-Resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2013;57:4427–32. doi: 10.1128/AAC.00716-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Borgianni L, Vandenameele J, Matagne A, Bini L, Bonomo RA, Frère J-M, Rossolini GM, Docquier J-D. Mutational analysis of VIM-2 reveals an essential determinant for metallo-β-lactamase stability and folding. Antimicrob Agents Chemother. 2010;54:3197–204. doi: 10.1128/AAC.01336-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Pegg KM, Liu EM, LaCuran AE, Oelschlaeger P. Biochemical Characterization of IMP-30, a Metallo-β-lactamase with Enhanced Activity toward Ceftazidime. Antimicrob Agents Chemother. 2013;57:5122–6. doi: 10.1128/AAC.02341-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Docquier J-D, Riccio ML, Mugnaioli C, Luzzaro F, Endimiani A, Toniolo A, Amicosante G, Rossolini GM. IMP-12, a new plasmid-encoded metallo-β-lactamase from a Pseudomonas putida clinical isolate. Antimicrob Agents Chemother. 2003;47:1522–8. doi: 10.1128/AAC.47.5.1522-1528.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Pérez-Llarena FJ, Fernández A, Zamorano L, Kerff F, Beceiro A, Aracil B, Cercenado E, Miro E, Oliver A, Oteo J. Characterization of a novel IMP-28 metallo-β-lactamase from a Spanish Klebsiella oxytoca clinical isolate. Antimicrob Agents Chemother. 2012;56:4540–3. doi: 10.1128/AAC.00776-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Neuwirth C, Siebor E, Robin F, Bonnet R. First occurrence of an IMP metallo-β-lactamase in Aeromonas caviae: IMP-19 in an isolate from France. Antimicrob Agents Chemother. 2007;51:4486–8. doi: 10.1128/AAC.01462-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Santella G, Docquier J-D, Gutkind G, Rossolini GM, Radice M. Purification and biochemical characterization of IMP-13 metallo-β-lactamase. Antimicrob Agents Chemother. 2011;55:399–401. doi: 10.1128/AAC.00421-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Borgianni L, Prandi S, Salden L, Santella G, Hanson ND, Rossolini GM, Docquier J-D. Genetic context and biochemical characterization of the IMP-18 metallo-β-lactamase identified in a Pseudomonas aeruginosa isolate from the United States. Antimicrob Agents Chemother. 2011;55:140–5. doi: 10.1128/AAC.00858-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Materon IC, Beharry Z, Huang W, Perez C, Palzkill T. Analysis of the context dependent sequence requirements of active site residues in the metallo-beta-lactamase IMP-1. J Mol Biol. 2004;344:653–63. doi: 10.1016/j.jmb.2004.09.074. [DOI] [PubMed] [Google Scholar]
- [103].Moali C, Anne C, Lamotte-Brasseur J, Groslambert S, Devreese B, Van Beeumen J, Galleni M, Frère J-M. Analysis of the importance of the metallo-β-lactamase active site loop in substrate binding and catalysis. Chem Biol. 2003;10:319–29. doi: 10.1016/s1074-5521(03)00070-x. [DOI] [PubMed] [Google Scholar]
- [104].Fabiane SM, Sohi MK, Wan T, Payne DJ, Bateson JH, Mitchell T, Sutton BJ. Crystal structure of the zinc-dependent beta-lactamase from Bacillus cereus at 1.9 Å resolution: binuclear active site with features of a mononuclear enzyme. Biochemistry. 1998;37:12404–11. doi: 10.1021/bi980506i. [DOI] [PubMed] [Google Scholar]
- [105].Yamaguchi Y, Kuroki T, Yasuzawa H, Higashi T, Jin W, Kawanami A, Yamagata Y, Arakawa Y, Goto M, Kurosaki H. Probing the role of Asp-120(81) of metallo-beta-lactamase (IMP-1) by site-directed mutagenesis, kinetic studies, and X-ray crystallography. J Biol Chem. 2005;280:20824–32. doi: 10.1074/jbc.M414314200. [DOI] [PubMed] [Google Scholar]
- [106].Horton LB, Shanker S, Mikulski R, Brown NG, Phillips KJ, Lykissa E, Prasad BV, Palzkill T. Mutagenesis of zinc ligand residue Cys221 reveals plasticity in the IMP-1 metallo-β-lactamase active site. Antimicrob Agents Chemother. 2012;56:5667–77. doi: 10.1128/AAC.01276-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].de Seny D, Heinz U, Wommer S, Kiefer M, Meyer-Klaucke W, Galleni M, Frère J-M, Bauer R, Adolph H-W. Metal Ion Binding and Coordination Geometry for Wild Type and Mutants of Metallo-β-lactamase from Bacillus cereus 569/H/9 (BcII) J Biol Chem. 2001;276:45065–78. doi: 10.1074/jbc.M106447200. [DOI] [PubMed] [Google Scholar]
- [108].Franceschini N, Caravelli B, Docquier J-D, Galleni M, Frère J-M, Amicosante G, Rossolini GM. Purification and biochemical characterization of the VIM-1 metallo-β-lactamase. Antimicrob Agents Chemother. 2000;44:3003–7. doi: 10.1128/aac.44.11.3003-3007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Laraki N, Franceschini N, Rossolini GM, Santucci P, Meunier C, de Pauw E, Amicosante G, Frère JM, Galleni M. Biochemical Characterization of the Pseudomonas aeruginosa 101/1477 Metallo-β-Lactamase IMP-1 Produced by Escherichia coli. Antimicrob Agents Chemother. 1999;43:902–6. doi: 10.1128/aac.43.4.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Bebrone C, Anne C, Kerff F, Garau G, De Vriendt K, Lantin R, Devreese B, Van Beeumen J, Dideberg O, Frere J. Mutational analysis of the zinc-and substrate-binding sites in the CphA metallo-beta-lactamase from Aeromonas hydrophila. Biochem J. 2008;414:151–9. doi: 10.1042/BJ20080375. [DOI] [PubMed] [Google Scholar]
- [111].Lassaux P, Traoré DA, Loisel E, Favier A, Docquier J-D, Sohier JS, Laurent C, Bebrone C, Frère J-M, Ferrer J-L. Biochemical and structural characterization of the subclass B1 metallo-β-lactamase VIM-4. Antimicrob Agents Chemother. 2011;55:1248–55. doi: 10.1128/AAC.01486-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Docquier J-D, Lamotte-Brasseur J, Galleni M, Amicosante G, Frère J-M, Rossolini GM. On functional and structural heterogeneity of VIM-type metallo-β-lactamases. J Antimicrob Chemother. 2003;51:257–66. doi: 10.1093/jac/dkg067. [DOI] [PubMed] [Google Scholar]
- [113].Bogaerts P, Bebrone C, Huang T-D, Bouchahrouf W, DeGheldre Y, Deplano A, Hoffmann K, Glupczynski Y. Detection and Characterization of VIM-31, a New Variant of VIM-2 with Tyr224His and His252Arg Mutations, in a Clinical Isolate of Enterobacter cloacae. Antimicrob Agents Chemother. 2012;56:3283–7. doi: 10.1128/AAC.06249-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Borra PS, Leiros H-KS, Ahmad R, Spencer J, Leiros I, Walsh TR, Sundsfjord A, Samuelsen Ø . Structural and computational investigations of VIM-7: insights into the substrate specificity of VIM metallo-β-lactamases. J Mol Biol. 2011;411:174–89. doi: 10.1016/j.jmb.2011.05.035. [DOI] [PubMed] [Google Scholar]
- [115].Rodriguez-Martinez J-M, Nordmann P, Fortineau N, Poirel L. VIM-19, a metallo-β-lactamase with increased carbapenemase activity from Escherichia coli and Klebsiella pneumoniae. Antimicrob Agents Chemother. 2010;54:471–6. doi: 10.1128/AAC.00458-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Castanheira M, Deshpande LM, Mendes RE, Rodriguez-Noriega E, Jones RN, Morfin-Otero R. Comment on: Role of changes in the L3 loop of the active site in the evolution of enzymatic activity of VIM-type metallo-β-lactamases. J Antimicrob Chemother. 2011;66:684–5. doi: 10.1093/jac/dkq393. [DOI] [PubMed] [Google Scholar]
- [117].Juan C, Beceiro A, Gutiérrez O, Albertí S, Garau M, Pérez JL, Bou G, Oliver A. Characterization of the new metallo-β-lactamase VIM-13 and its integron-borne gene from a Pseudomonas aeruginosa clinical isolate in Spain. Antimicrob Agents Chemother. 2008;52:3589–96. doi: 10.1128/AAC.00465-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Merino M, Pérez-Llarena FJ, Kerff F, Poza M, Mallo S, Rumbo-Feal S, Beceiro A, Juan C, Oliver A, Bou G. Role of changes in the L3 loop of the active site in the evolution of enzymatic activity of VIM-type metallo-β-lactamases. J Antimicrob Chemother. 2010;65:1950–4. doi: 10.1093/jac/dkq259. [DOI] [PubMed] [Google Scholar]
- [119].Marchiaro P, Tomatis PE, Mussi MA, Pasteran F, Viale AM, Limansky AS, Vila AJ. Biochemical characterization of metallo-β-lactamase VIM-11 from a Pseudomonas aeruginosa clinical strain. Antimicrob Agents Chemother. 2008;52:2250–2. doi: 10.1128/AAC.01025-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Kontou M, Pournaras S, Kristo I, Ikonomidis A, Maniatis AN, Stathopoulos C. Molecular cloning and biochemical characterization of VIM-12, a novel hybrid VIM-1/VIM-2 metallo-β-lactamase from a Klebsiella pneumoniae clinical isolate, reveal atypical substrate specificity. Biochemistry. 2007;46:13170–8. doi: 10.1021/bi701258w. [DOI] [PubMed] [Google Scholar]
- [121].Murphy TA, Catto LE, Halford SE, Hadfield AT, Minor W, Walsh TR, Spencer J. Crystal Structure of Pseudomonas aeruginosa SPM-1 Provides Insights into Variable Zinc Affinity of Metallo-β-lactamases. J Mol Biol. 2006;357:890–903. doi: 10.1016/j.jmb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- [122].Oelschlaeger P, Mayo SL. Hydroxyl groups in the ββ sandwich of metallo-β-lactamases favor enzyme activity: a computational protein design study. J Mol Biol. 2005;350:395–401. doi: 10.1016/j.jmb.2005.04.044. [DOI] [PubMed] [Google Scholar]
- [123].Dal Peraro M, Vila AJ, Carloni P, Klein ML. Role of zinc content on the catalytic efficiency of B1 metallo beta-lactamases. J Am Chem Soc. 2007;129:2808–16. doi: 10.1021/ja0657556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Materon IC, Palzkill T. Identification of residues critical for metallo-beta-lactamase function by codon randomization and selection. Protein Sci. 2001;10:2556–65. doi: 10.1110/ps.40884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Thomas PW, Zheng M, Wu S, Guo H, Liu D, Xu D, Fast W. Characterization of purified New Delhi metallo-β-lactamase-1. Biochemistry. 2011;50:10102–13. doi: 10.1021/bi201449r. [DOI] [PubMed] [Google Scholar]
- [126].Tada T, Miyoshi-Akiyama T, Shimada K, Kirikae T. Biochemical Analysis of Metallo-β-Lactamase NDM-3 from a Multidrug-Resistant Escherichia coli Strain Isolated in Japan. Antimicrob Agents Chemother. 2014;58:3538–40. doi: 10.1128/AAC.02793-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Nordmann P, Boulanger AE, Poirel L. NDM-4 metallo-β-lactamase with increased carbapenemase activity from Escherichia coli. Antimicrob Agents Chemother. 2012;56:2184–6. doi: 10.1128/AAC.05961-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Tada T, Miyoshi-Akiyama T, Dahal RK, Sah MK, Ohara H, Kirikae T, Pokhrel BM. NDM-8 metallo-β-lactamase in a multidrug-resistant Escherichia coli strain isolated in Nepal. Antimicrob Agents Chemother. 2013;57:2394–6. doi: 10.1128/AAC.02553-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Wang X, Li H, Zhao C, Chen H, Liu J, Wang Z, Wang Q, Zhang Y, He W, Zhang F. Novel NDM-9 metallo-β-lactamase identified from a ST107 Klebsiella pneumoniae strain isolated in China. Int J Antimicrob Agents. 2014;44:90–1. doi: 10.1016/j.ijantimicag.2014.04.010. [DOI] [PubMed] [Google Scholar]
- [130].Makena A, Brem J, Pfeffer I, Geffen RE, Wilkins SE, Tarhonskaya H, Flashman E, Phee LM, Wareham DW, Schofield CJ. Biochemical characterization of New Delhi metallo-beta-lactamase variants reveals differences in protein stability. J Antimicrob Chemother. 2015;70:463–9. doi: 10.1093/jac/dku403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Guo Y, Wang J, Niu G, Shui W, Sun Y, Zhou H, Zhang Y, Yang C, Lou Z, Rao Z. A structural view of the antibiotic degradation enzyme NDM-1 from a superbug. Prot Cell. 2011;2:384–94. doi: 10.1007/s13238-011-1055-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Yang H, Aitha M, Hetrick AM, Richmond TK, Tierney DL, Crowder MW. Mechanistic and spectroscopic studies of metallo-beta-lactamase NDM-1. Biochemistry. 2012;51:3839–47. doi: 10.1021/bi300056y. [DOI] [PubMed] [Google Scholar]
- [133].Bootsma HJ, Aerts PC, Posthuma G, Harmsen T, Verhoef J, van Dijk H, Mooi FR. Moraxella (Branhamella) catarrhalis BRO beta-lactamase: a lipoprotein of gram-positive origin? J Bacteriol. 1999;181:5090–3. doi: 10.1128/jb.181.16.5090-5093.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Zhang H, Hao Q. Crystal structure of NDM-1 reveals a common beta-lactam hydrolysis mechanism. FASEB J. 2011;25:2574–82. doi: 10.1096/fj.11-184036. [DOI] [PubMed] [Google Scholar]
- [135].Green VL, Verma A, Owens RJ, Phillips SE, Carr SB. Structure of New Delhi metallo--lactamase 1 (NDM-1) Acta Crystallogr F: Struct Biol Crystal Commun. 2011;67:1160–4. doi: 10.1107/S1744309111029654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Thomas PW, Zheng M, Wu S, Guo H, Liu D, Xu D, Fast W. Characterization of purified New Delhi metallo-beta-lactamase-1. Biochemistry. 2011;50:10102–13. doi: 10.1021/bi201449r. [DOI] [PubMed] [Google Scholar]
- [137].Garau G, Bebrone C, Anne C, Galleni M, Frere JM, Dideberg O. A metallo-beta-lactamase enzyme in action: crystal structures of the monozinc carbapenemase CphA and its complex with biapenem. J Mol Biol. 2005;345:785–95. doi: 10.1016/j.jmb.2004.10.070. [DOI] [PubMed] [Google Scholar]
- [138].Wu S, Xu D, Guo H. QM/MM studies of monozinc beta-lactamase CphA suggest that the crystal structure of an enzyme-intermediate complex represents a minor pathway. J Am Chem Soc. 2010;132:17986–8. doi: 10.1021/ja104241g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Spencer J, Read J, Sessions RB, Howell S, Blackburn GM, Gamblin SJ. Antibiotic recognition by binuclear metallo-beta-lactamases revealed by X-ray crystallography. J Am Chem Soc. 2005;127:14439–44. doi: 10.1021/ja0536062. [DOI] [PubMed] [Google Scholar]
- [140].King DT, Worrall LJ, Gruninger R, Strynadka NC. New Delhi metallo-beta-lactamase: structural insights into beta-lactam recognition and inhibition. J Am Chem Soc. 2012;134:11362–5. doi: 10.1021/ja303579d. [DOI] [PubMed] [Google Scholar]
- [141].Feng H, Ding J, Zhu D, Liu X, Xu X, Zhang Y, Zang S, Wang DC, Liu W. Structural and Mechanistic Insights into NDM-1 Catalyzed Hydrolysis of Cephalosporins. J Am Chem Soc. 2014;136:14694–7. doi: 10.1021/ja508388e. [DOI] [PubMed] [Google Scholar]
- [142].Fitzgerald PM, Wu JK, Toney JH. Unanticipated inhibition of the metallo-beta-lactamase from Bacteroides fragilis by 4-morpholineethanesulfonic acid (MES): a crystallographic study at 1.85-Å resolution. Biochemistry. 1998;37:6791–800. doi: 10.1021/bi9730339. [DOI] [PubMed] [Google Scholar]
- [143].Bounaga S, Laws AP, Galleni M, Page MI. The mechanism of catalysis and the inhibition of the Bacillus cereus zinc-dependent beta-lactamase. Biochem J. 1998;331:703–11. doi: 10.1042/bj3310703. Pt 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Dal Peraro M, Llarrull LI, Rothlisberger U, Vila AJ, Carloni P. Water-assisted reaction mechanism of monozinc beta-lactamases. J Am Chem Soc. 2004;126:12661–8. doi: 10.1021/ja048071b. [DOI] [PubMed] [Google Scholar]
- [145].Thomas PW, Cammarata M, Brodbelt JS, Fast W. Covalent Inhibition of New Delhi Metallo-beta-Lactamase-1 (NDM-1) by Cefaclor. Chembiochem. 2014;15:2541–8. doi: 10.1002/cbic.201402268. [DOI] [PubMed] [Google Scholar]
- [146].Lobkovsky E, Billings EM, Moews PC, Rahil J, Pratt RF, Knox JR. Crystallographic structure of a phosphonate derivative of the Enterobacter cloacae P99 cephalosporinase: mechanistic interpretation of a beta-lactamase transition-state analog. Biochemistry. 1994;33:6762–72. doi: 10.1021/bi00188a004. [DOI] [PubMed] [Google Scholar]
- [147].Wang Z, Fast W, Benkovic SJ. Direct Observation of an Enzyme-Bound Intermediate in the Catalytic Cycle of the Metallo-beta-Lactamase from Bacteroides fragilis. J Am Chem Soc. 1998;120:10788–9. [Google Scholar]
- [148].Wang Z, Fast W, Benkovic SJ. On the mechanism of the metallo-beta-lactamase from Bacteroides fragilis. Biochemistry. 1999;38:10013–23. doi: 10.1021/bi990356r. [DOI] [PubMed] [Google Scholar]
- [149].Yang H, Young H, Yu S, Sutton L, Crowder MW. Targeting metallo-carbapenemases via modulation of electronic properties of cephalosporins. Biochem J. 2014;464:271–9. doi: 10.1042/BJ20140364. [DOI] [PubMed] [Google Scholar]
- [150].Griffin DH, Richmond TK, Sanchez C, Moller AJ, Breece RM, Tierney DL, Bennett B, Crowder MW. Structural and kinetic studies on metallo-beta-lactamase IMP-1. Biochemistry. 2011;50:9125–34. doi: 10.1021/bi200839h. [DOI] [PubMed] [Google Scholar]
- [151].Rasia RM, Vila AJ. Mechanistic study of the hydrolysis of nitrocefin mediated by B. cereus metallo-beta-lactamase. ARKIVOC. 2003;10:507–16. [Google Scholar]
- [152].Zheng M, Xu D. New Delhi metallo-beta-lactamase I: substrate binding and catalytic mechanism. J Phys Chem B. 2013;117:11596–607. doi: 10.1021/jp4065906. [DOI] [PubMed] [Google Scholar]
- [153].Fast W, Sutton LD. Metallo-beta-lactamase: inhibitors and reporter substrates. Biochim Biophys Acta. 2013;1834:1648–59. doi: 10.1016/j.bbapap.2013.04.024. [DOI] [PubMed] [Google Scholar]
- [154].King DT, Strynadka NC. Targeting metallo-beta-lactamase enzymes in antibiotic resistance. Future Med Chem. 2013;5:1243–63. doi: 10.4155/fmc.13.55. [DOI] [PubMed] [Google Scholar]
- [155].Drawz SM, Papp-Wallace KM, Bonomo RA. New beta-lactamase inhibitors: a therapeutic renaissance in an MDR world. Antimicrob Agents Chemother. 2014;58:1835–46. doi: 10.1128/AAC.00826-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Mollard C, Moali C, Papamicael C, Damblon C, Vessilier S, Amicosante G, Schofield CJ, Galleni M, Frere JM, Roberts GC. Thiomandelic acid, a broad spectrum inhibitor of zinc beta-lactamases: kinetic and spectroscopic studies. J Biol Chem. 2001;276:45015–23. doi: 10.1074/jbc.M107054200. [DOI] [PubMed] [Google Scholar]
- [157].Makena A, van Berkel SS, Lejeune C, Owens RJ, Verma A, Salimraj R, Spencer J, Brem J, Schofield CJ. Chromophore-linked substrate (CLS405): probing metallo-beta-lactamase activity and inhibition. ChemMedChem. 2013;8:1923–9. doi: 10.1002/cmdc.201300350. [DOI] [PubMed] [Google Scholar]
- [158].Sohier JS, Laurent C, Chevigne A, Pardon E, Srinivasan V, Wernery U, Lassaux P, Steyaert J, Galleni M. Allosteric inhibition of VIM metallo-beta-lactamases by a camelid nanobody. Biochem J. 2013;450:477–86. doi: 10.1042/BJ20121305. [DOI] [PubMed] [Google Scholar]
- [159].Spicer T, Minond D, Enogieru I, Saldanha SA, Allais C, Liu Q, Mercer BA, Roush WR, Hodder P. Probe Reports from the NIH Molecular Libraries Program. Bethesda (MD): 2010. ML302, a Novel Beta-lactamase (BLA) Inhibitor. PMID 23762954. [PubMed] [Google Scholar]
- [160].Kim SK, Sims CL, Wozniak SE, Drude SH, Whitson D, Shaw RW. Antibiotic resistance in bacteria: novel metalloenzyme inhibitors. Chem Biol Drug Des. 2009;74:343–8. doi: 10.1111/j.1747-0285.2009.00879.x. [DOI] [PubMed] [Google Scholar]
- [161].Toney JH, Hammond GG, Fitzgerald PM, Sharma N, Balkovec JM, Rouen GP, Olson SH, Hammond ML, Greenlee ML, Gao YD. Succinic acids as potent inhibitors of plasmid-borne IMP-1 metallo-beta-lactamase. J Biol Chem. 2001;276:31913–8. doi: 10.1074/jbc.M104742200. [DOI] [PubMed] [Google Scholar]
- [162].Siemann S, Brewer D, Clarke AJ, Dmitrienko GI, Lajoie G, Viswanatha T. IMP-1 metallo-beta-lactamase: effect of chelators and assessment of metal requirement by electrospray mass spectrometry. Biochim Biophys Acta. 2002;1571:190–200. doi: 10.1016/s0304-4165(02)00258-1. [DOI] [PubMed] [Google Scholar]
- [163].King AM, Reid-Yu SA, Wang W, King DT, De Pascale G, Strynadka NC, Walsh TR, Coombes BK, Wright GD. Aspergillomarasmine A overcomes metallo-beta-lactamase antibiotic resistance. Nature. 2014;510:503–6. doi: 10.1038/nature13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Bush K, Macalintal C, Rasmussen BA, Lee VJ, Yang Y. Kinetic interactions of tazobactam with beta-lactamases from all major structural classes. Antimicrob Agents Chemother. 1993;37:851–8. doi: 10.1128/aac.37.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Payne DJ, Bateson JH, Gasson BC, Khushi T, Proctor D, Pearson SC, Reid R. Inhibition of metallo-beta-lactamases by a series of thiol ester derivatives of mercaptophenylacetic acid. FEMS Microbiol Lett. 1997;157:171–5. doi: 10.1111/j.1574-6968.1997.tb12769.x. [DOI] [PubMed] [Google Scholar]
- [166].Payne DJ, Bateson JH, Gasson BC, Proctor D, Khushi T, Farmer TH, Tolson DA, Bell D, Skett PW, Marshall AC, Reid R, Ghosez L, Combret Y, Marchand-Brynaert J. Inhibition of metallo-beta-lactamases by a series of mercaptoacetic acid thiol ester derivatives. Antimicrob Agents Chemother. 1997;41:135–40. doi: 10.1128/aac.41.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Kurosaki H, Yamaguchi Y, Higashi T, Soga K, Matsueda S, Yumoto H, Misumi S, Yamagata Y, Arakawa Y, Goto M. Irreversible inhibition of metallo-beta-lactamase (IMP-1) by 3-(3-mercaptopropionylsulfanyl)propionic acid pentafluorophenyl ester. Angew Chem Int Ed Engl. 2005;44:3861–4. doi: 10.1002/anie.200500835. [DOI] [PubMed] [Google Scholar]
- [168].Singh J, Petter RC, Baillie TA, Whitty A. The resurgence of covalent drugs. Nat Rev Drug Discov. 2011;10:307–17. doi: 10.1038/nrd3410. [DOI] [PubMed] [Google Scholar]
- [169].Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–4. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- [170].Gascuel O. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol Biol Evol. 1997;14:685–95. doi: 10.1093/oxfordjournals.molbev.a025808. [DOI] [PubMed] [Google Scholar]
- [171].Samuelsen Ø , Castanheira M, Walsh TR, Spencer J. Kinetic characterization of VIM-7, a divergent member of the VIM metallo-β-lactamase family. Antimicrob Agents Chemother. 2008;52:2905–8. doi: 10.1128/AAC.00166-08. [DOI] [PMC free article] [PubMed] [Google Scholar]