Summary
The Ndc80 complex (Ndc80, Nuf2, Spc24, Spc25) is a highly conserved kinetochore protein essential for end-on anchorage to spindle microtubule plus-ends and for force generation coupled to plus-end polymerization and depolymerization. Spc24/Spc25 at one end of the Ndc80 complex binds the kinetochore. The N-terminal tail and CH domains of Ndc80 bind microtubules, and an internal domain binds microtubule-associated proteins (MAPs) such as the Dam1 complex. To determine how the microtubule and MAP binding domains of Ndc80 contribute to force production at the kinetochore in budding yeast, we have inserted a FRET tension sensor into the Ndc80 protein about halfway between its microtubule binding and internal loop domains. The data support a mechanical model of force generation at metaphase where the position of the kinetochore relative to the microtubule plus-end reflects the relative strengths of microtubule depolymerization, centromere stretch and microtubule binding interactions with Ndc80 and Dam1 complexes.
Introduction
The Ndc80 complex is a hetero-tetramer protein complex of Ndc80, Nuf2, Spc24, and Spc25 that plays an essential role in end-on attachment of spindle microtubule (MT) plus ends to the kinetochore1, 2. The Ndc80 complex transmits kMT-dependent force to the kinetochore at its inner Spc24/Spc25 end from at least two sources3. One is the well-characterized MT binding domains (MTBDs) at the N-terminus of Ndc80, which includes the N-terminal tail and the Calponin-homology (CH) domain (Fig. 1a)2, 4. The second sources are internal domains, including the helical hairpin or loop domains of Ndc80, which are proposed to bind to MT associated proteins (MAPs)5–7.
Figure 1. The Ndc80 FRET Biosensor detects tension at the N-terminus of Ndc80 in vivo.
(a) Cartoon of Ndc80 protein complex. We inserted FRET tension sensor at 410 aa in Ndc80 protein. This site is located between the CH and Loop domains. For a zero tension control, we fused the FRET sensor to the C-terminus of Nuf2 (Nuf2 FRET control). (b) The Ndc80 FRET biosensor exhibits higher FRET at lower tension and lower FRET at higher tension. (c) Representative FRET images (left) and Emission Ratios (right) for separated sister kinetochore clusters at metaphase for the Ndc80 FRET sensor (n = 117 kinetochore clusters) and Nuf2-FRET control (n = 100 kinetochore clusters). *** Unpaired Student t-test (two-tailed), p < 0.01. Error bars are SD from the means. The mean values were calculated using data pooled from 3 independent experiments. Scale bar is 5μm (c).
For force production, the best-characterized MAP is the budding yeast Dam1 complex. Dam1 is recruited to the plus-ends of kMTs by the Ndc80 complex6, 8. Dam1, which is a ten-protein complex oligomerizes into rings or spirals that surround a MT in vitro9–14. Purified Dam1 interacts with Ndc80 on MTs in vitro to increase the force needed for Ndc80 detachment from MT plus ends8, 15–17.
The plus-ends of kMTs switch between persistent phases of depolymerization and polymerization18. During depolymerization, kinetochores are moved poleward along their kMTs while during polymerization, kinetochores are pulled away from the pole by the force from centromere chromatin stretch. This kMT dynamic instability drives sister chromosome oscillations between the poles at metaphase. Loss of tension upon sister chromosome separation at anaphase contributes to persistent kMT depolymerization that results in anaphase A poleward movement of sisters18, 19. Tubulin protofilaments at the plus-ends of kMTs are seen in electron micrographs to curve inside-out with variable degrees of curvature20. In vitro, the curvature of tubulin protofilaments at polymerizing MT ends is low while the curvature at depolymerizing ends is high20. In a reconstituted system of a cargo bead tethered to Dam1 on a MT, previous work showed that 100 nm long tethers between the bead and Dam1 increased the force six-fold relative to a short tether21. The 57 nm Ndc80 complex serves as such a long tether. In addition, MT polymerases, like XMAP215 (Stu2 in budding yeast), selectively bind to GTP-tubulin at the tips of polymerizing ends and not to GDP-tubulin at depolymerizing ends22, 23. Analysis of the nm-scale protein architecture of yeast kinetochores place Stu2 near the Spc24/Spc25 end of the Ndc80 complex, while the Dam1 complex is inside but closer to the MTBDs of Ndc8024, 25.
To determine how the MT binding and MAP binding domains in Ndc80 contribute to force production at budding yeast kinetochores, we have inserted a FRET (Fluorescence/Förster Resonance Energy Transfer) biosensor for tension into the Ndc80 protein about halfway between the CH and loop domains (Fig. 1a). During the cell cycle, the FRET sensor reported lower tension from late anaphase through interphase and high tension at metaphase, when pericentromeric chromatin is maximally stretched between sister kinetochores. Deletions of the N-terminal tail of Ndc80 showed that tension detected by the FRET sensor is produced by the Ndc80 MTBD. Further, in vivo studies and in silico simulations indicate the position of the kinetochore relative to the microtubule plus-end at metaphase is dictated mainly by three conditions: the force and frequency of MT depolymerization, the affinity of Dam1 complex for the MT lattice and the force from centromere stretch during MT polymerization. Our FRET tension sensor data suggest a mechanical model for the Ndc80 force coupler at kinetochores.
Results
FRET Tension-Sensor in Ndc80 Near Its MTBDs
The FRET tension sensor was inserted into Ndc80 at amino acid position 410 (Figs. 1a). The FRET tension sensor contains an unstructured 12 aa (amino acid) linker between mYPet and mECFP26. Including the 5 and 11 unstructured aa segments at the N and C termini of the FPs (Fluorescence Proteins), there are a total of 28 unstructured amino acids between the β-barrels of the two FPs that form an entropic spring with a persistence length of 1 nm. The FRET efficiency of this construct in solution (no tension) was measured to be 0.50, giving an estimated 5.0 nm distance between the donor (mECFP) and acceptor (mYpet) fluorophores26. Tension stretches the entropic spring, moving the donor and acceptor apart and reducing the FRET signal (Fig. 1b). The hybrid FRET protein, driven by its endogenous promoter, supported normal mitosis and cell growth and performed all essential functions of the Ndc80 complex (Supplementary Figs. 1a–c).
The nm-scale mean position in metaphase of GFP inserted into Ndc80 at the same position as the FRET tension sensor, aa 410, was 21 nm inside (towards chromatin) of the N-terminus of Ndc80 and 19 nm outside (towards the pole) of the C-terminus of Ndc80 (Supplementary Fig. 1d)25. The integrated intensity of GFP at aa 410 of Ndc80 was nearly identical to the value for GFP fused to the C-terminus of Ndc80 (Supplementary Fig. 1e)27. Thus, the insertion of the FP at aa 410 of Ndc80 did not affect the length or stoichiometry of Ndc80 at kinetochores.
Changes in tension at the Ndc80 FRET biosensor were monitored in live cells by measurement of Emission Ratio (see Methods). We confirmed that the Emission Ratio is proportional to change in FRET efficiency (Supplementary Figs. 2a–c). The Emission Ratio decreases with increasing tension as the distance between the donor and acceptor FP increases (Fig. 1b). The mean Emission Ratio at metaphase for the Ndc80 FRET sensor was significantly lower than the value reported by the FRET sensor fused to the C-terminus of Nuf2 (Fig. 1c). The C-terminal Nuf2-FRET sensor serves as a control for zero tension, and reveals that the Ndc80 FRET sensor detects tension at the MTBD of Ndc80 in vivo.
Changes in FRET Emission Ratios at Different Stages of Mitosis
Measured values for the Emission Ratio from the Ndc80 FRET sensor were binned by cell cycle stage. The mean Emission Ratio decreased slightly from interphase to prometaphase, and then decreased significantly to its lowest value in metaphase (Figs. 2a–c). The Emission Ratio increased in early/middle anaphase, to a peak in late anaphase, and finally decreased in telophase to a value near the interphase level. The Emission Ratios in all cycle stages are significantly higher than the value in metaphase. Although the Emission Ratio at late anaphase for the FRET sensor fused to the C-terminus of Nuf2 was slightly higher than the value at metaphase, the values at the other stages of the cell cycle were not significantly different (Fig. 2c).
Figure 2. Tension during cell cycle monitored by Ndc80 FRET biosensor.
(a) Cartoon of the yeast cell cycle. K-K distance is Ndc80-Ndc80 (FRET sensor) distance between sister kinetochore clusters. (b) Representative Ndc80 FRET images throughout the cell cycle. (c) The Emission Ratio for each cell cycle stage for the Ndc80 FRET biosensor (left) or Nuf2-FRET control (right, note the different scale). Interphase: n = 118 (Ndc80), 97 (Nuf2), prometaphase: n = 17 (Ndc80), metaphase: n = 117 (Ndc80), 98 (Nuf2), early/middle anaphase: n = 276 (Ndc80), 102 (Nuf2), late anaphase: n = 105 (Ndc80), 96 (Nuf2), telophase: n = 101 (Ndc80), 81 (Nuf2). n values represent the number of kinetochore clusters. *** Unpaired Student t-test (two-tailed), p < 0.01. (d) Cartoon of diploid cells expressing Ndc80-mECFP (inserted at aa 410) and Ndc80-mYPet (inserted at aa 410) (left, top). Representative FRET images of diploid cells are shown left, bottom for different cell cycle stages. The bar graph on the right shows the Emission Ratio for each stage (n = 100 kinetochore clusters each). The Emission Ratio value for no FRET (red bars) was measured by bleed-through from mECFP and cross excitation from mYPet (see Supplementary Fig. 2d and Methods). Scale bars are 2.5 μm. Error bars are SD from the means. The mean values were calculated using data pooled from 2 independent experiments. The mean values of Emission Ratio, FRET efficiency, K-K distance are listed in Supplementary Table 1.
To distinguish intra- vs. inter-molecular FRET we examined cell lines that had only mECFP or mYPet at the aa 410 site in Ndc80, and diploid cells expressing mECFP or mYPet in separate copies of Ndc80 (Fig. 2d). The average Emission Ratio in diploid cells was comparable to the value of no FRET (Emission Ratio = 1.14, n = 384) (Fig. 2d and Supplementary Fig. 2d). Thus, inter-molecular FRET was negligible throughout cell cycle.
Changes in the values of the Emission Ratio for the Ndc80 FRET sensor are consistent with expected changes in increased tension in metaphase. Highest tension is present at late metaphase where pericentromeric chromatin is maximally stretched, and tension drops in late anaphase and interphase after sister chromatin separation and spindle disassembly. The increase in the Emission Ratio from metaphase to late anaphase is predicted by the kinetics of decrease in intra-kinetochore stretch that occurs between metaphase and late anaphase (Supplementary Fig. 2e), a shortening that supports decreased tension on the Ndc80 complex24, 25. Surprisingly, the Emission Ratio for the Ndc80 FRET sensor measured from late anaphase through interphase is significantly lower than that of the zero tension by the FRET sensor at the C-terminus of Nuf2. We conclude that yeast kinetochores remain under tension at interphase. It is known that kinetochores maintain Ndc80 dependent attachments to short (~50nm) kMTs during interphase28. This is consistent with force measurements from chromatin dynamics in G1/interphase29.
Tension Detected by Ndc80 Sensor Depends on the MTBD of Ndc80
The N-terminal tail of Ndc80 in both budding yeast and humans enhances the affinity of Ndc80 binding to MTs, unless phosphorylated by Aurora B kinase16, 30. In budding yeast, the N-terminal tail contains 112 unstructured amino acids and includes along its length 7 serine or threonine targets for the Aurora B kinase (Fig. 3a)31. At metaphase, phosphorylation of the N-terminal tail is low to promote stable kMT attachment32. To test how this MTBD of Ndc80 contributes to the tension detected by the FRET sensor in Ndc80, we made two N-terminal tail deletions: 70 aa (70 Del) and 112 aa (the whole tail, 112 Del) (Supplementary Figs. 3a–b). Unlike Ndc80 in humans, the N-terminal tail in budding yeast is nonessential33. We found that at metaphase the Emission Ratio in 70 Del increased to near the value for full length Ndc80 at late anaphase (Fig. 3b). When the entire tail was deleted, the value increased further, very near the no-tension level. Surprisingly, the 7D (dephospho-mimic) mutant of yeast Ndc80, produces only a 38% reduction of force in vitro16. Our data indicates that the N-terminal tail, either directly or through the CH domain, makes a major contribution to force at this location. Surprisingly, for both 70 and 112 deletions, centromere stretch at metaphase was normal, 840-890 nm (Fig. 3c).
Figure 3. The Tension Detected by the Ndc80 FRET Sensor Depends on the N-terminal tail of Ndc80.
(a) Diagram of the yeast Ndc80 protein. The unstructured N-terminal tail domain has 7 serine or threonine targets for the Aurora B kinase. The 70 Del deleted the first 70 aa and the 112 Del deleted the first 112 aa of Ndc80 protein. (b) Representative images at metaphase, of control, 70 Del, and 112 Del mutants (left). The average Ndc80 FRET Emission Ratio for each condition is shown in the bar graph (control: 2.12 ± 0.54, n = 117, 70 Del: 2.89 ± 0.64, n = 120, 112 Del: 3.68 ± 0.76, n = 149). n values represent number of kinetochores clusters. (c) The average K-K distance for each condition of (b) (n = 100 kinetochore pairs). (d) Representative time-lapse images (top) and the mean duration from prometaphase (PM) to anaphase onset and anaphase onset to late anaphase in each condition of (b) (bottom). n = 29, 27, 25, 29, 32, 27 cells (from left to right). (e) The percentage of cells in PM (prometaphase) or M (metaphase) in each condition of (b). n = 100 kinetochore pairs. Scale bars are 1μm (a, c–d). Error bars are SD from the means, Unpaired Student t-test (two-tailed): ***p < 0.01, **p < 0.05, * < 0.1. The mean values were calculated using data pooled from 3 independent experiments (b–d) or 2 independent experiments (e). All N-terminal tail mutant proteins were expressed by the endogenous Ndc80 promoter to maintain expression level (Supplementary Figs. 3a–c). The mean values of Emission Ratio, FRET efficiency, K-K distance are listed in Supplementary Table 1.
Although the centromere stretch was normal in tail deletion mutants, these cells were significantly delayed in prometaphase-metaphase, compared to control cells (Fig. 3d). This delay corresponded to significantly higher mitotic indices in tail deletion mutants (Fig. 3e). The tail deletion mutant cells were also slower completing anaphase, but this contribution for the total mitotic arrest was small. We did not observe any apoptotic cells during time-lapse imaging and there was only a very small percentage of apoptotic cells in the cell population analysis (Supplementary Fig. 3c).
Together, the above results show that the tension reported by the Ndc80 FRET sensor depends on the Ndc80 MTBD and the tension contributed by the Ndc80 MTBD is not needed for normal metaphase centromere stretch.
Emission Ratio Is Different Between Polymerization and Depolymerization
If force at the Ndc80 MTBDs is different between polymerization and depolymerization of kMT ends, then the Emission Ratio from clusters of sister kinetochores at metaphase may fluctuate as kMTs stochastically switch between polymerization and depolymerization19, 34. To test this idea, we measured the fluorescence of Stu2-GFP at metaphase kinetochores as a function of time. Stu2 may preferentially bind to the tips of polymerizing, but not depolymerizing MT ends22, 23. The ratio of integrated intensity of sister clusters (K1/K2) fluctuated randomly over time as predicted by the dynamic instability of kMTs (Fig. 4a)19, 35. Note that we calculated K1 intensity divided by K2 intensity for normalization of noise (see detail in Methods). Similar fluctuations were seen for Bim1-GFP, another microtubule plus-end binding protein (Supplementary Fig. 4a)36. Fluctuations were also observed for the Emission Ratio from the Ndc80 FRET sensor for opposite sister kinetochore clusters during metaphase (Fig. 4b and Supplementary Fig. 4b). The fluctuations between Stu2-tdTomato intensity and FRET emission were inversely correlated. When FRET emission decreased, Stu2 intensity increased (Fig. 4c and Supplementary Figs. 4c–d). Using Stu2 fluorescence as an indicator of microtubule growth state, mean FRET intensity was decreased, and tension increased, when there were more kMTs polymerizing than depolymerizing within a kinetochore cluster. Ndc80-GFP intensity exhibited little fluctuation during the course of metaphase, indicative of persistent kMT attachment for bi-oriented chromosomes (Supplementary Fig. 4e)37.
Figure 4. Stu2-GFP Intensity and Ndc80 FRET Emission Ratio fluctuate in metaphase.
(a) Two examples of fluctuations in Stu2-GFP intensities for separated sister kinetochore clusters (K1 and K2) in a control metaphase cell, a cell treated with low-dose benomyl, and a control cell at anaphase. K1 intensity is normalized by K2 intensity at each time point to control for fluctuations in illumination intensity (See Methods). Note, that the integrated fluorescence intensity of a sister kinetochore cluster decreases with depth into the cell beneath the coverslip surface. At metaphase both sister kinetochore clusters are at about the same depth, but in anaphase, interpolar spindle elongation often occurs at an angle to the coverslip surface, making the intensity of the sister kinetochore clusters unequal, and reducing the average value of the K1/K2 ratio from near 1 at metaphase, where K2 was the larger value. (b) Plots of normalized fluctuations in Ndc80 FRET Emission Ratio (K2/K1) from Supplementary Fig. 4b images in a control cell and a cell treated with low-dose benomyl (55 μM). (c) Representative cell showing Stu2-tdTomato and Ndc80 FRET images during metaphase and a plot of FRET emission (K1/K2) and Stu2 signal at K1 (the upper kinetochore) (See Methods and Supplementary Fig. 4c). Another example is shown in Supplementary Fig. 4d. We analyzed 12 cells from 3 independent experiments (a–c). (d) Representative images of Ndc80 FRET, mECFP, and mYpet control metaphase cells, and cells treated with benomyl (55 μM, 165 μM, or 551 μM). The bar graph shows the average Ndc80 FRET Emission Ratio and K-K distance (n = 117, 103, 105, 105 kinetochores from left to right). The mean values were calculated using data pooled from 3 independent experiments. ** Unpaired Student t-test (two-tailed), p < 0.05. Scale bars are 1 μm (a, c) and 2.5 μm (d). The mean values of Emission Ratio, FRET efficiency, K-K distance are listed in Supplementary Table 1.
We used low doses of the tubulin-binding drug, benomyl, a concentration that does not stop cell growth, to inhibit dynamicity of kMTs at metaphase. Previous studies have shown that low dose benomyl (55 μM) decreases growth and shortening velocities ~5-fold and increases ~5-fold the half-life of fluorescence recovery after photobleaching (FRAP) of GFP-tubulin in kMTs35. Low dose of benomyl significantly reduced the fluctuations in both Stu2- and Bim1-GFP fluorescence and the Emission Ratio (Figs. 4a–b). The above results indicate that fluctuations in tension at the Ndc80 MTBDs depend on kMT dynamic instability.
There is evidence in budding yeast mitosis that kMT detachment occurs frequently when pole to pole (P-P) distance is < 1.2 μm (corresponding K-K distance < ~400 nm) and very rarely when P-P distance is > 1.6 μm (K-K distance > ~800 nm)37. The mean value of our metaphase K-K distance was 800-900 nm, suggesting that detachment occurs rarely. To confirm this hypothesis, we performed FRAP analysis to quantify Ndc80 turnover at the kinetochore. Although the Ndc80-GFP integrated intensity recovered and fluctuated at prometaphase, it did not recover during metaphase and late anaphase (Supplementary Fig. 5). The great majority of fluctuations that occur in FRET Emission ratio and Stu2-GFP (as well as Bim1-GFP) intensities appear to be produced by kMT dynamic instability at attached kinetochores, with kMT detachment making a very small contribution.
Centromere Stretch Is Maintained when Ndc80 MTBD Tension Is Reduced
Low dose benomyl (55 μM) had little effect on K-K centromere stretch at metaphase, but caused the Emission Ratio to increase significantly (to 2.62 ± 0.84) compared to control metaphase (Fig. 4d). A level of 2.62 is also typical of cells treated with higher concentrations of benomyl (165 μM) that partially reduce K-K centromere stretch or cause the spindles to collapse and sister kinetochores to cluster into a single spot (551 μM) (Fig. 4d). Thus, elevated tension reported by the Ndc80 FRET biosensor requires kMT dynamics. In addition, normal tension at the Ndc80 MTBDs is not required to maintain metaphase centromere stretch by kMTs when dynamicity is suppressed ~5-fold. These results corroborate the centromere stretch that occurs for the Ndc80 tail-less mutant (112 Del) despite a lack of tension at the biosensor (Figs. 3b–c). The kinetochore is not a simple force transmitter, rather it is a more complex force coupler in which load is non-linearly distributed among its protein sub-complexes.
A Mechanical Model of the Force Coupler for Metaphase Kinetochores
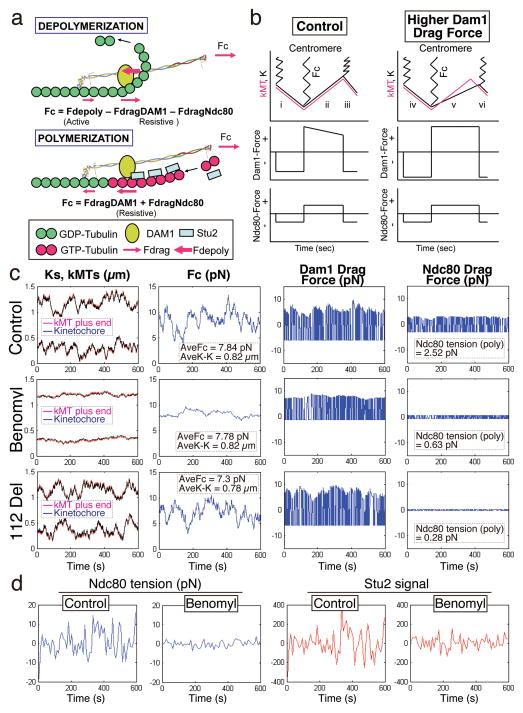
Our data suggest a mechanical model for the Ndc80 force coupler at kinetochores. Kinetochore movements relative to depolymerizing or polymerizing ends of kMTs are determined by two force balance equations: one for depolymerization and another for polymerization (Fig. 5a). Fig. 5b diagrams the kinetic movements at metaphase of a kinetochore and the end of its kMT relative to the pole during 1.5 cycles of kMT dynamic instability. Also included are the magnitude of FdragDAM1 and FdragNdc80 derived from the equations in Fig. 5a and the computer simulations described below for control cells and cells with elevated FdragDAM1.
Figure 5. Mechanical model and computer simulations for the Ndc80 force coupler at kinetochores of bi-oriented chromosomes in metaphase budding yeast.
(a) A cross-section diagram of the mechanical model along a single MT protofilament during depolymerization (top) and polymerization (bottom) (See Text and Methods for details). The Dam1 complex is anchored to Ndc80 inside of the tension sensor at a site like the Ndc80 loop domain. During depolymerization, pushing force on the Dam1 complex by curling protofilaments (Fdepoly), moves the Ndc80 complex poleward at the rate of depolymerization generating a pulling force on the centromere, Fc, and generates compressive drag forces on both the MTBDs of Ndc80 (FdragNdc80) and the DAM1 complex (FdragDam1). During polymerization, the force from centromere stretch, Fc, pulls the Ndc80 complex away from the pole generating tensile drag forces on the MTBDs of Ndc80 and the Dam1 complex. At the tips of polymerizing MTs, GTP-tubulin and Stu-2 proteins increase the force on the Dam1 complex to prevent detachment from the MT tip. (b) Kinetics predicted for the Ndc80 force coupler during kMT depolymerization and polymerization at constant velocity in control (left) and higher Dam1 drag force (right). (c) Computer simulations of mechanical model in A for all 16 sister kinetochore pairs at metaphase in budding yeast for wild type (top), for low dose benomyl to reduce dynamicity (middle), and for reduced Ndc80 drag force (112 Del) (bottom). (d) The difference in fluctuations in Ndc80 tension and Stu2 concentration in simulations of wild type and low dose benomyl treated cells (See Methods and Supplementary Table 2 for more details, parameter values, and simulation results.)
During depolymerization, the Ndc80 force coupler stretches the centromere poleward as Dam1 oligomers are pushed by the force (Fdepoly) of curling protofilaments (Fig. 5b, i, iii, iv and vi). The force coupler moves at the rate of depolymerization for both normal and elevated drag force since the force from curling protofilaments against a Dam1 ring can be several 10s of pN38. Fdepol equals the sum of the force from centromere stretch (Fc) at the Spc24/Spc25 end of the Ndc80 complex and the drag forces between tubulin-GDP within the MT lattice and the binding domains of Dam1 and Ndc80 (Fig. 5a, Depolymerization).
During polymerization, both drag forces change direction compared to their orientation during depolymerization and their sum equals the force from centromere stretch, Fc (Fig. 5a, Polymerization). In simulations, there is no directional dependence for the magnitude of drag force over the MT GDP-tubulin lattice. During polymerization, the force coupler can be pulled along the kMT by high levels of Fc force from centromere stretch at a rate faster than polymerization. When it comes within 20 nm of the polymerizing end, the force coupler is prevented from detaching because of additional Dam1 drag force, potentially from higher affinity of the Dam1 complex to tubulin-GTP39, resistance from the outward splaying of protofilaments, and resistance from Stu2 bound to the MT tips (Fig. 5b, ii). If the Fc force from centromere stretch becomes less than the sum of Dam1 and Ndc80 drag forces, the Ndc80 force coupler will lag behind the polymerizing end (Fig. 5b, v). When this kMT end switches to depolymerization, the Fc force from centromere stretch will continue to pull the kinetochore away from the pole along the kMT until the depolymerizing end reaches the kinetochore and then the kinetochore is moved poleward, re-stretching the centromere (Fig. 5b, v and vi).
Simulations of the model in Figs. 5a-c were performed based on reported parameters of kMT dynamic instability that best fit kMT dynamics and spindle MT morphology in normal and low benomyl treated cell (See Methods for more details)19, 35, 40, 41.
Simulations of control cells were compared to simulations of cells treated with low-dose benomyl, where the velocities of polymerization and depolymerization are reduced 5-fold 34 and to cells with tail-less Ndc80, where we reduced the Ndc80 drag coefficient 10-fold compared to normal (Fig. 3b). Figs. 5c–d show results for a simulation duration of 600 sec (see Supplementary Fig. 6a for a 100 sec duration). For each condition, the kinetics for a single pair of sister kinetochores and their kMTs are shown along with the corresponding fluctuations in Fc, FdragDAM1, and FdragNdc80 as well as the correlation of fluctuations in the mean levels of Ndc80 tension and Stu2 from all 16 kMTs within a half-spindle (Figs. 5c–d). The simulations all match the in vivo data. Fc from centromere stretch is maintained at control levels while Ndc80 tension is greatly reduced for both experimental conditions (112 Del and low-dose benomyl) and fluctuations in mean half-spindle Ndc80 tension and Stu2 signal all are substantially reduced by low-dose benomyl. Movies of the simulations presented in Fig. 5c vividly show the kinetics for all 16 sister kinetochore pairs and their centromere stretch for control, low dose benomyl, and entire tail deletion mutant (Videos 1–3 and Supplementary Fig. 6b).
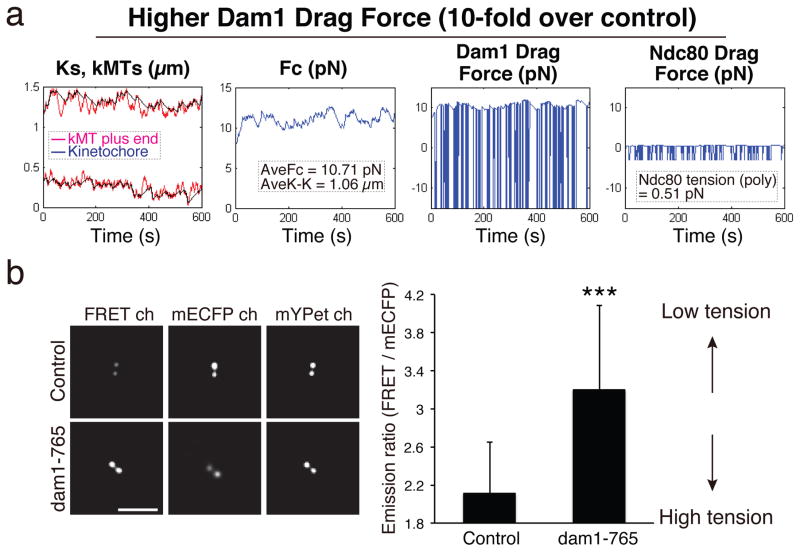
Tension at Ndc80 MTBD Is Very Low when Dam1 Drag Force is Increased
The above data suggest that the distribution of force within the kinetochore might be shifted depending on the relative affinities of MTBDs of individual sub-complexes. To test this hypothesis, we increased the Dam1 drag coefficient (10-fold over control) in the mechanical model without changing other parameters. Surprisingly, Ndc80 tension was significantly reduced and the mean length of kMTs became longer than the mean distance of kinetochores to their poles (Figs. 6a and 5b). To ascertain whether an in vivo condition recapitulates the simulation, we measured Ndc80 tension by the FRET biosensor in dam1-765 mutants. Dam1-765 was isolated based on its lethality with defects in binding of MT minus-ends to spindle poles42. These mutants achieved metaphase with kinetochores closer to their spindle poles compared to controls and then exhibited normal anaphase segregation, (Supplementary Figs. 6c–d)42. In addition, mean kinetochore tension at metaphase was higher than for controls because centromere-linked LacO/LacI-GFP markers were significantly more stretched apart (Supplementary Fig. 6e). In contrast, the mean Emission Ratio from the Ndc80 FRET sensor for the dam1-765 cells was significantly higher compared to controls (Fig. 6b). These in vivo results and in silico predictions indicate that the average kinetochore force at metaphase depends critically on the relative binding affinities of individual kinetochore complexes. In addition, kinetochores that remain bound to their kMTs can become uncoupled from polymerizing MT ends at metaphase (Supplementary Fig. 7a and Video 4).
Figure 6. The tension at Ndc80 MTBDs is dependent on Dam1 drag force.
(a) Computer simulations of mechanical model for metaphase budding yeast for the condition when Dam1 drag force was increased 10-fold over control. Ndc80 tension was significantly reduced and the mean length of kMTs became longer than the mean distance of kinetochores to their poles. (b) Representative Ndc80 FRET, mECFP, and mYpet images for a control cell and a dam1-765 cell (left). The average Emission Ratio at metaphase for control (2.12 ± 0.54, n = 117) and dam1-765 cells (3.20 ± 0.88, n = 80) (right). n values represent number of kinetochores clusters. *** Unpaired Student t-test (two-tailed), p < 0.01. Error bars are SD from the means. Scale bars are 5 μm. The mean values were calculated using data pooled from 2 independent experiments. The mean values of Emission Ratio, FRET efficiency, K-K distance are listed in Supplementary Table 1.
Discussion
The kinetochore must adjust to stochastic changes in the position of the microtubule plus-end, while maintaining attachment to the centromere on the surface of the chromosome. At metaphase, centromere stretch is coupled to kinetochore microtubule dynamics and has been considered as a surrogate for tension between sister kinetochores. The position of the force coupler relative to a kMT depends on a balance of forces that are different for the polymerization and depolymerization phases of dynamic instability. The Ndc80 force coupler is pulled toward polymerizing ends by centromere stretch at a velocity dependent on the Ndc80 and Dam1 drag forces along the MT lattice or at the plus end. In contrast, depolymerizing ends push the Dam1 complex and attached Ndc80 complex poleward, at the velocity of depolymerization (Supplementary Fig. 7b).
Our data and mechanical model predict that the Dam1 complex is attached to Ndc80 at a site (like the loop domain)6 interior (toward chromatin) to the position of the Ndc80 tension sensor. Experimental manipulations that caused loss of tension at the Ndc80 MTBD did not result in reduction of mean centromere stretch from control levels. In contrast, if the Dam1 complex was anchored to the Ndc80 complex at the hinge-loop domain proximal to the MT binding CH domain, the FRET sensor would be inside this site and detect metaphase tension from the nearly normal mean centromere stretch.
Although the yeast N-terminal tail of Ndc80 is considered not essential33, our results show that the N-terminal tail of Ndc80 is critical to the timely progression through mitosis and distribution of force like mammalian Ndc80/Hec1. Tension is borne within the unstructured tail itself43 or via interactions with the CH domain8. Our measurements do not distinguish whether the reduced tension with tail deletion is through the loss of tail-MT interactions or the inability of the CH domain to establish robust MT binding (Fig. 7).
Figure 7. Schematic of Force Coupler model in budding yeast metaphase.
Models for the Ndc80 force coupler during polymerization in wild-type, partial and whole N-terminal tail deletion mutants. The N-terminal tail has a critical role in Ndc80 tension. Normal mean K-K stretch was maintained despite reduction (partial tail deletion) or lack (whole N-terminal tail deletion) of tension in Ndc80 at the position of the FRET sensor. During depolymerization, forces from peeling protofilaments push the Dam1 and Ndc80 complexes along kMTs toward the pole to stretch the centromere; the MTBDs of both Dam1 and Ndc80 are under compression. During polymerization, force from centromere stretch pulls the Ndc80 force coupler along kMTs with the MTBDs of Dam1 and Ndc80 under tension.
Without the N-terminal tail on Ndc80, Dam1 becomes the primary load-bearing complex at metaphase as evidenced by the major loss of tension in the MTBD of Ndc80 together with nearly normal centromere stretch (Fig. 7). This finding is supported by in vitro studies showing that MT depolymerizing forces as high as 40 pN are borne by Dam1 rings coupled to 100 nm coiled-coil tethers like the Ndc80 complex21. MTBDs of Ndc80 themselves support much weaker detachment forces from MTs15, 44, 45. Mutations in Dam1 (dam1-765), as well as tail-less Ndc80 highlight the ability of forces to be differentially distributed within the Ndc80 coupler depending on the relative binding affinities of its sub-complexes.
What mechanism prevents centromere stretch from pulling the Ndc80 force coupler off the plus end of its kMT when the rate of polymerization is too slow? Our experimental results following loss of tension from the Ndc80 MTBDs show such a mechanism exists since mean centromere stretch is maintained. In our mechanical model (Figs. 5a–c), the drag coefficient for Dam1 was increased to a level that prevented detachment when the force coupler was within 20 nm of a polymerizing end. Detachment may be prevented because the Dam1 complex has a higher binding affinity to the GTP-tubulin at polymerizing ends compared to GDP-tubulin in the wall of MTs45, because of contributions from Stu2 or other MAPs like Bim1 that concentrate at growing tips, or because thermodynamic forces prevent detachment from ends46.
How well does our mechanical model for the Ndc80 coupler at budding yeast kinetochores represent the kMT attachment site in mammalian kinetochores? There is evidence that the outer mammalian kinetochore is less stretched during depolymerization in comparison to polymerization, a result proposed to be the consequence of poleward pushing forces from curling protofilaments at depolymerizing ends and only resistive forces during polymerization47. Importantly, the data from the force sensor substantially revises the role of the N-terminal tail of Ndc80 in budding yeast. Its loss has a major impact on the distribution of force in the kinetochore and illustrates the critical role the tail plays in the binding affinity of the Ndc80 MTBDs in vivo. This finding has been validated in vitro for budding yeast Ndc8016, as well as in vitro and in vivo for human Ndc8043, 48. The dominant load-bearing role played by the Dam1 complex likely masks this function in budding yeast. The load-bearing feature of the Ndc80 tail points to an evolutionarily conserved feature in force production, despite the presence or absence of oligomeric structures such as Dam1. A key discovery from the biosensor is the ability to dissect layers of protein functions and mechanisms that may be masked by evolutionarily acquired features.
Methods
Yeast strains and cell preparation
All strains are described in Supplementary Table 3. All yeast cells were grown to logarithmic phase at 24°C in rich YPD media. For benomyl treatment, we added 55 μM of benomyl for low concentration, 165 μM for intermediate, and 551 μM for high concentration in logarithmic phase cells and incubated for 1 hour at 24°C before imaging. Cells maintained bi-polar spindles in treatment of both low and intermediate benomyl. All target proteins fused with FPs (fluorescent proteins), including FRET biosensors, were inserted into the endogenous genome locus by homologous recombination. All Ndc80 FRET biosensors, including the N-terminus tail deletion mutants (70 Del and 112 Del) and Nuf2 C-terminus FRET protein, were expressed by the endogenous Ndc80 or Nuf2 promoter to maintain protein level and minimize the impact of free Ndc80 FRET or Nuf2 FRET protein (not incorporated at kinetochore) on the FRET analysis (Supplementary Table 3, Supplementary Figs. 1a–b, and 3a–b).
Imaging (FRET)
Imaging used a Nikon TE2000-E microscope equipped with a 100X/1.4NA (Planapo) DIC oil immersion objective. Images were recorded with MetaMorph 7.1 software (Molecular Devices) on an ANDOR iXON (DV897) EMCCD camera with total magnification giving a pixel size of 106.7 nm, an analog gain of 4, and no electron multiplication. FRET and mECFP fluorescent images were obtained from a Dual-View™ 2 imaging device (Photometrics) with an 89002 ET-ECFP/EYFP filter (Chroma Technology Corp). This allowed simultaneous imaging of both the FRET channel (CFP excitation, YFP emission) and the mECFP channel (CFP excitation, YFP emission). 10-frame 3D stacks at 200 nm steps along the z-axis were collected at 60 sec intervals (FRET imaging and Stu2-tdTomato).
Imaging (other)
Images were obtained sequentially at 200 nm steps along the z axis using MetaMorph 7.1 software (Molecular Devices), a Nikon Eclipse Ti microscope with image magnification yielding a 64 nm pixel size from a 100X/1.49NA (Apo TIRF) DIC oil immersion objective (Nikon) and an ANDOR Clara CCD camera (ANDOR). Time-lapse imaging collected 15-frame 3D stacks at 200 nm steps along the z-axis at 30 sec intervals.
The nm-scale measurements
The nm-scale analysis using the heat map method was described previously25. Images were aligned in MatLab by taking the brightest pixel for one spindle pole (labeled with Spc29-RFP) and rotating the spindle until the second spindle pole was horizontal to the first, then the GFP spots (Ndc80) were rotated to the same degree. The spindle poles were set at (0, 0 = x, y), and the distance to the GFP spots were taken relative to the spindle pole position as described25.
Fluorescence intensity measurement
The Z-axis position of best focus (maximum integrated intensity) was used for analysis of sister kinetochore clusters3, 27. A 7 x 7 pixel region was centered on a cluster of sister kinetochores to obtain integrated fluorescence, while a 9 x 9 pixel region centered on the 7 x 7 pixel region was used to obtain surrounding background intensity. Fi = integrated intensity for 7 x 7 region – (integrated counts for the 9 x 9 region – integrated counts for 7 x 7 region) x pixel area of the 7 x 7 region/(pixel area of the 9 x 9 region – pixel area of a 7 x 7 region). Measurements were made with MetaMorph 7.7 analysis software (Molecular Devices) using the region measurements tool.
For time-lapse image analysis for Stu2-GFP, Bim1-GFP, and Ndc80-GFP, we measured the ratio of integrated fluorescence intensity for pairs of sister kinetochore clusters (K1/K2) to minimize contributions from fluctuations in illumination intensity, photon noise, shifts in kinetochore cluster position from coverslip surface, and additional instrumental noise. We measured Fi for both sister kinetochore clusters (K1: kinetochore cluster and K2: sister kinetochore cluster) using the methods above and corrected for photobleaching, before K1 intensity was normalized by K2 intensity in each time point.
FRET Emission Ratio (FRET emission/mECFP emission)
Changes in tension at the Ndc80 FRET biosensor were monitored in live cells by measurement of Emission Ratio (defined as - FRET channel emission [CFP excitation, YFP emission]/mECFP channel emission [CFP excitation, CFP emission]). This is a standard method when the biosensor contains one donor and one acceptor FP. For most FRET measurements we used the FRET Emission Ratio for the following reasons. The FRET tension sensor is a single molecule FRET probe, where the number of donor and acceptor FPs are the same, eliminating variation in bleed through and cross excitation which depend on the number of FPs. In addition, we took both FRET and mECFP channel images at the same time using a dual view attachment, which prevented inaccuracy from specimen movement that occurs for sequential imaging needed to obtain mYpet emission. However, we also measured Cross Excitation and Bleed Through in our experimental condition. The FRET filter showed up to 12.6 ± 8.1 (n = 400) % cross excitation from mYpet, and up to 86.3 ± 24.0 (n = 54) % bleed through from mECFP (Supplementary Fig. 2d).
For experiments of Fig. 2d, the FRET Emission Ratio value of no FRET (1.14) was calculated using the value of both Bleed Through from ECFP (86.3%) and Cross Excitation from mYPet (12.6%) in Diploid cells (Supplementary Fig. 2d). For the experiment of Fig. 4c and Supplementary Fig. 4d (correlation between FRET emission and Stu2 signal), we used FRET emission (FRET channel intensity) instead of FRET Emission Ratio to eliminate the time interval for switching wavelength. We confirmed that FRET emission was directly proportional to the FRET Emission Ratio (Supplementary Fig. 4c).
FRET efficiency
To obtain a more quantitative estimate of tension induced extension of the FRET sensor, we used the more accurate measure of FRET Efficiency (EDD). This made use of cell lines where the mYpet was lost from the sensor, leaving Ndc80-mECFP(410), in which the sensor position had only mECFP. By mixing cell lines we could obtain in the same field cells containing the full sensor and cells with the mECFP only. From these we could calculate the FRET efficiency indicated by quenching of donor fluorescence.
| Eqn S1 |
where emCFP(FRET) is emission in the CFP channel of the full FRET construct, and emCFP(CFP) is emission in the CFP channel of the Ndc80-mECFP(410) cell.
Both cells with the Ndc80 FRET sensor and those isolated with Ndc80 mECFP were grown to logarithmic phase at 24°C in rich YPD media and mixed with the same number of cells just before imaging. We measured FRET efficiency separately for cells in metaphase, anaphase and metaphase treated with 55 uM benomyl. The FRET efficiency in these different conditions was directly proportional to the FRET Emission Ratio (Supplementary Fig. 2c), validating the use of FRET Emission Ratio in most experiments.
Computer simulations of metaphase kinetochore movements coupled to kMT plus ends by the Ndc80 Force Coupler
Computer simulations of the mechanical model as diagramed in Fig. 5 and described in Results were run in MatLab based on the Monte Carlo methods developed previously to account for the dynamic instability of kMT plus ends at budding yeast metaphase19, 35. As in the previous simulations, the model assumes that kMT plus-ends remain in one of two states, depolymerizing or polymerizing at all times. The previous simulations assumed the kinetochore perfectly tracked the plus-tips of kMTs. In our simulations, kinetochore movements relative to the ends of depolymerizing or polymerizing kMTs are determined by force balance equations. The force balance equation during depolymerization is:
| Eqn. S2 |
while during polymerization the force balance equation is
| Eqn S3 |
Fc is the force from centromere stretch applied to the Spc24/Spc25 end of the Ndc80 complex. During depolymerization, Fdepoly is the force on the Dam1 oligimers produced by the peeling towards the MT minus end of curved protofilaments typical of depolymerizing MT plus ends. FdragDam1 and FdragNdc80 are the collective drag forces from the MT binding domains of Dam1 and Ndc80 complexes respectively for tubulin-GDP in the MT lattice. The dynamic instability of kMT plus ends is described by four parameters: Vg, the velocity of growth, Vs, the velocity of shrinkage, kc, the probability of switching from polymerization to depolymerization (catastrophe transition) and kr, the probability of switching from depolymerization to polymerization (rescue transition). As in the previous simulations19, 35, Vg= Vs = 1.2 um/min constant velocity for wild type and the N-terminus tail deletion (112 Del) and the higher Dam1 drag force (proposed for dam1-765 mutant) and 0.2 um/min for the low dynamicity produced by low benomyl. To account for both the clustering of separated sisters on opposite sides of the spindle equator and to account for the spatial dependence of tubulin turnover within kMTs, kc and kr vary with position between the poles as described in previous studies19, 35. The probability of catastrophe is highest at the spindle equator and lower toward the poles19 and is given by:
| Eqn 3 |
where xkMT is the length of a kMT and bounded by the length of the spindle (PP). The probability of rescue when the Ndc80 force coupler is within 20 nm of the kMT tip is higher toward the poles and increases with centromere tension, Fc:
| Eqn 4 |
where kro is a basal level of rescue (12/min), Fo is a force where rescue rises sharply (~10 pN) and rescue always occurs if the xkMT is less than 50 nm since at that length the MT binding ends of the Ndc80 complex would be close to the pole. When the Ndc80 force coupler was greater than 20 nm from the tip, then the tip is no longer considered able to sense the tension from centromere stretch and the probability of rescue is given by:
| Eqn 5 |
The force, Fc, from the stretch of the centromere in between sister kinetochores is given by:
| Eqn 6 |
where kcen is the Hookean spring constant for chromatin, Lcen the separation between sister kinetochores and Lrest the rest length of the centromere, assumed to be 170 nm41.
Simulations of the model in Figs. 5 and 6 were performed based on reported parameters of kMT dynamic instability that best fit kMT dynamics and spindle MT morphology in normal and low dose benomyl treated cells19, 35. We used a mean force per kMT of 7.5 pN based on direct measurements in prometaphase insect spermatocytes and the 4–6 pN estimated from pericentromere chromatin stiffness in metaphase budding yeast40–41. Drag coefficients and the parameters of the centromere spring constant were adjusted to yield a mean value of 7.5 pN for Fc at the mean in vivo K-K centromere stretch of ~850 nm and spindle pole to pole length at metaphase of PP = 1500 nm19, 35, 40–41. The value of kcen = 12 pN/um in the simulation is very similar to a value of ~15.5 pN/um for centromere stiffness at metaphase in budding yeast based on thermal movements of centromeric fluorescent markers41
If the position of a plus end from its pole, xkMT, was greater than 20 nm from the position of the Ndc80 Force Coupler, xK, it was assumed that all the MT binding domains of Ndc80 and the Dam1 complex were bound to tubulin-GDP. Under this condition, the collective drag coefficients for the Ndc80 and DAM1 MT binding domains are 2.5 and 5 pN/um/min respectively for both wild-type and benomyl treated cells during both polymerization and depolymerization. If the position of the Ndc80 force coupler was pulled closer than 20 nm from the kMT end by centromere stretch, Fc, during polymerization, then the value for the Dam1 drag coefficient was increased to prevent detachment. For the simulation of the Dam1-765, the value of the Dam1 drag coefficient was increased 10-fold compared to normal.
The simulation was run for 1 sec intervals over a 10 min duration and the probability of a catastrophe or rescue was checked for each 1 sec interval for all 16 sister pairs (Left and Right) of kMTs. If a kMT end was in depolymerization then the Ndc80 Force Coupler moved poleward at the rate of depolymerization and the inner kinetochore remained 20 nm from the tip. If a kMT end was polymerizing, then the rate of away from the pole movement of the Ndc80 force coupler was determined by Eqn. 3 provided as described above, the coupler did not become closer than 20 nm to the end.
Both variable and fixed Parameter values and results from simulation in Fig. 5, 6a, and Supplementary Fig. 6 are shown in Supplementary Table 2.
The MatLab programs used for the simulations in Figs. 5–6 are available at the Bloom or Salmon Web sites.
Statistics and Reproducibility
All data were expressed as mean ± SD. Statistical significance was determined using Student’s t-test for comparison between two independent groups. ***P < 0.01 and **P < 0.05 were considered statistically significant and 0.05 < *P < 0.1 was considered marginally significant. Data were pooled across 2–3 independent experiments, as described in each figure legend.
Supplementary Material
Acknowledgments
We would like to thank Drs. Elaine Yeh, Karen Plevock, David Pellman, and Sue Biggins for critical reagents and valuable suggestions. We would also like to thank Dr Andrew McAnish for providing drawing tool for model. This work was supported by Uehara Memorial Foundation, Kazato research foundation, and Japan Society and Promotion of Science (A. Suzuki), Summer Undergraduate Research Fellowship from UNC (B.L. Badger), R37GM024364 and R01GM24364 (E.D. Salmon), R37GM32238 (K. Bloom) and R01GM66014 (H.P. Erickson) from the National Institutes of Health.
Footnotes
Author Contribution
A.S. performed entire experiments and analyzed the data. A.S. and B.L.B. performed experiments for Figs. 2–4 and Supplementary Figs. 4–5. J.H. performed the experiments for Supplementary Figs. 1d, 2e, and 6e experiments. T.O. and H.P.E. provided FRET probes and discussion of their use and interpretation. E.D.S. wrote the computer simulation of the mechanistic model. A.S., E.D.S. and K.B. designed all experiments and wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Wang HW, et al. Architecture and flexibility of the yeast Ndc80 kinetochore complex. J Mol Biol. 2008;383:894–903. doi: 10.1016/j.jmb.2008.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciferri C, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki A, Badger BL, Salmon ED. A quantitative description of Ndc80 complex linkage to human kinetochores. Nature communications. 2015;6:8161. doi: 10.1038/ncomms9161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nature structural & molecular biology. 2007;14:54–59. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- 5.Hsu KS, Toda T. Ndc80 internal loop interacts with Dis1/TOG to ensure proper kinetochore-spindle attachment in fission yeast. Current biology: CB. 2011;21:214–220. doi: 10.1016/j.cub.2010.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maure JF, et al. The Ndc80 loop region facilitates formation of kinetochore attachment to the dynamic microtubule plus end. Current biology: CB. 2011;21:207–213. doi: 10.1016/j.cub.2010.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang G, et al. The Ndc80 internal loop is required for recruitment of the Ska complex to establish end-on microtubule attachment to kinetochores. Journal of cell science. 2012;125:3243–3253. doi: 10.1242/jcs.104208. [DOI] [PubMed] [Google Scholar]
- 8.Lampert F, Mieck C, Alushin GM, Nogales E, Westermann S. Molecular requirements for the formation of a kinetochore-microtubule interface by Dam1 and Ndc80 complexes. The Journal of cell biology. 2013;200:21–30. doi: 10.1083/jcb.201210091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nogales E, Ramey VH. Structure-function insights into the yeast Dam1 kinetochore complex. Journal of cell science. 2009;122:3831–3836. doi: 10.1242/jcs.004689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheeseman IM, Enquist-Newman M, Muller-Reichert T, Drubin DG, Barnes G. Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex. The Journal of cell biology. 2001;152:197–212. doi: 10.1083/jcb.152.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enquist-Newman M, et al. Dad1p, third component of the Duo1p/Dam1p complex involved in kinetochore function and mitotic spindle integrity. Molecular biology of the cell. 2001;12:2601–2613. doi: 10.1091/mbc.12.9.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janke C, Ortiz J, Tanaka TU, Lechner J, Schiebel E. Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. The EMBO journal. 2002;21:181–193. doi: 10.1093/emboj/21.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hofmann C, et al. Saccharomyces cerevisiae Duo1p and Dam1p, novel proteins involved in mitotic spindle function. The Journal of cell biology. 1998;143:1029–1040. doi: 10.1083/jcb.143.4.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, et al. The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes & development. 2002;16:183–197. doi: 10.1101/gad.959402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tien JF, et al. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. The Journal of cell biology. 2010;189:713–723. doi: 10.1083/jcb.200910142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarangapani KK, Akiyoshi B, Duggan NM, Biggins S, Asbury CL. Phosphoregulation promotes release of kinetochores from dynamic microtubules via multiple mechanisms. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:7282–7287. doi: 10.1073/pnas.1220700110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franck AD, et al. Tension applied through the Dam1 complex promotes microtubule elongation providing a direct mechanism for length control in mitosis. Nature cell biology. 2007;9:832–837. doi: 10.1038/ncb1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inoue S, Salmon ED. Force generation by microtubule assembly/disassembly in mitosis and related movements. Molecular biology of the cell. 1995;6:1619–1640. doi: 10.1091/mbc.6.12.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gardner MK, et al. Tension-dependent regulation of microtubule dynamics at kinetochores can explain metaphase congression in yeast. Molecular biology of the cell. 2005;16:3764–3775. doi: 10.1091/mbc.E05-04-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McIntosh JR, et al. Conserved and divergent features of kinetochores and spindle microtubule ends from five species. The Journal of cell biology. 2013;200:459–474. doi: 10.1083/jcb.201209154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Volkov VA, et al. Long tethers provide high-force coupling of the Dam1 ring to shortening microtubules. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:7708–7713. doi: 10.1073/pnas.1305821110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slep KC, Vale RD. Structural basis of microtubule plus end tracking by XMAP215, CLIP-170, and EB1. Molecular cell. 2007;27:976–991. doi: 10.1016/j.molcel.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka K, et al. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 2005;434:987–994. doi: 10.1038/nature03483. [DOI] [PubMed] [Google Scholar]
- 24.Joglekar AP, Bloom K, Salmon ED. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Current biology: CB. 2009;19:694–699. doi: 10.1016/j.cub.2009.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haase J, et al. A 3D map of the yeast kinetochore reveals the presence of core and accessory centromere-specific histone. Current biology: CB. 2013;23:1939–1944. doi: 10.1016/j.cub.2013.07.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohashi T, Galiacy SD, Briscoe G, Erickson HP. An experimental study of GFP-based FRET, with application to intrinsically unstructured proteins. Protein science: a publication of the Protein Society. 2007;16:1429–1438. doi: 10.1110/ps.072845607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawrimore J, Bloom KS, Salmon ED. Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. The Journal of cell biology. 2011;195:573–582. doi: 10.1083/jcb.201106036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winey M, Bloom K. Mitotic spindle form and function. Genetics. 2012;190:1197–1224. doi: 10.1534/genetics.111.128710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verdaasdonk JS, et al. Centromere Tethering Confines Chromosome Domains. Molecular cell. 2013 doi: 10.1016/j.molcel.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeLuca KF, Lens SM, DeLuca JG. Temporal changes in Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis. Journal of cell science. 2011;124:622–634. doi: 10.1242/jcs.072629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akiyoshi B, Nelson CR, Ranish JA, Biggins S. Analysis of Ipl1-mediated phosphorylation of the Ndc80 kinetochore protein in Saccharomyces cerevisiae. Genetics. 2009;183:1591–1595. doi: 10.1534/genetics.109.109041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki A, Badger BL, Wan X, DeLuca JG, Salmon ED. The Architecture of CCAN Proteins Creates a Structural Integrity to Resist Spindle Forces and Achieve Proper Intrakinetochore Stretch. Developmental cell. 2014;30:717–730. doi: 10.1016/j.devcel.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Demirel PB, Keyes BE, Chaterjee M, Remington CE, Burke DJ. A redundant function for the N-terminal tail of Ndc80 in kinetochore-microtubule interaction in Saccharomyces cerevisiae. Genetics. 2012;192:753–756. doi: 10.1534/genetics.112.143818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pearson CG, Maddox PS, Zarzar TR, Salmon ED, Bloom K. Yeast kinetochores do not stabilize Stu2p-dependent spindle microtubule dynamics. Molecular biology of the cell. 2003;14:4181–4195. doi: 10.1091/mbc.E03-03-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pearson CG, et al. Measuring nanometer scale gradients in spindle microtubule dynamics using model convolution microscopy. Molecular biology of the cell. 2006;17:4069–4079. doi: 10.1091/mbc.E06-04-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maddox PS, Stemple JK, Satterwhite L, Salmon ED, Bloom K. The minus end-directed motor Kar3 is required for coupling dynamic microtubule plus ends to the cortical shmoo tip in budding yeast. Current biology: CB. 2003;13:1423–1428. doi: 10.1016/s0960-9822(03)00547-5. [DOI] [PubMed] [Google Scholar]
- 37.Marco E, et al. S. cerevisiae chromosomes biorient via gradual resolution of syntely between S phase and anaphase. Cell. 2013;154:1127–1139. doi: 10.1016/j.cell.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grishchuk EL, et al. The Dam1 ring binds microtubules strongly enough to be a processive as well as energy-efficient coupler for chromosome motion. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15423–15428. doi: 10.1073/pnas.0807859105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Westermann S, et al. Formation of a dynamic kinetochore- microtubule interface through assembly of the Dam1 ring complex. Molecular cell. 2005;17:277–290. doi: 10.1016/j.molcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 40.Nicklas RB, Lee GM, Rieder CL, Rupp G. Mechanically cut mitotic spindles: clean cuts and stable microtubules. Journal of cell science. 1989;94(Pt 3):415–423. doi: 10.1242/jcs.94.3.415. [DOI] [PubMed] [Google Scholar]
- 41.Chacon JM, Mukherjee S, Schuster BM, Clarke DJ, Gardner MK. Pericentromere tension is self-regulated by spindle structure in metaphase. The Journal of cell biology. 2014;205:313–324. doi: 10.1083/jcb.201312024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimogawa MM, et al. Mps1 phosphorylation of Dam1 couples kinetochores to microtubule plus ends at metaphase. Current biology: CB. 2006;16:1489–1501. doi: 10.1016/j.cub.2006.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zaytsev AV, et al. Multisite phosphorylation of the NDC80 complex gradually tunes its microtubule-binding affinity. Molecular biology of the cell. 2015;26:1829–1844. doi: 10.1091/mbc.E14-11-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Powers AF, et al. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell. 2009;136:865–875. doi: 10.1016/j.cell.2008.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lampert F, Hornung P, Westermann S. The Dam1 complex confers microtubule plus end-tracking activity to the Ndc80 kinetochore complex. The Journal of cell biology. 2010;189:641–649. doi: 10.1083/jcb.200912021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joglekar AP, Bloom KS, Salmon ED. Mechanisms of force generation by end-on kinetochore-microtubule attachments. Current opinion in cell biology. 2010;22:57–67. doi: 10.1016/j.ceb.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dumont S, Salmon ED, Mitchison TJ. Deformations within moving kinetochores reveal different sites of active and passive force generation. Science. 2012;337:355–358. doi: 10.1126/science.1221886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zaytsev AV, Sundin LJ, DeLuca KF, Grishchuk EL, DeLuca JG. Accurate phosphoregulation of kinetochore-microtubule affinity requires unconstrained molecular interactions. The Journal of cell biology. 2014;206:45–59. doi: 10.1083/jcb.201312107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.