Abstract
Huntington’s disease (HD) is an autosomal dominant neurodegenerative disorder, caused by an expansion of the CAG repeat in exon 1 of the huntingtin gene. The disease generally manifests in middle age with both physical and mental symptoms. There are no effective treatments or cures and death usually occurs 10–20 years after initial symptoms. Since the original identification of the Huntington disease associated gene, in 1993, a variety of models have been created and used to advance our understanding of HD. The most recent advances have utilized stem cell models derived from HD-patient induced pluripotent stem cells (iPSCs) offering a variety of screening and model options that were not previously available. The discovery and advancement of technology to make human iPSCs has allowed for a more thorough characterization of human HD on a cellular and developmental level. The interaction between the genome editing and the stem cell fields promises to further expand the variety of HD cellular models available for researchers. In this review, we will discuss the history of Huntington’s disease models, common screening assays, currently available models and future directions for modeling HD using iPSCs-derived from HD patients.
Keywords: Huntington’s disease, Stem cells, Induced pluripotent stem cells, Drug screening, Phenotypes
Introduction
Huntington’s disease (HD) is a devastating, dominantly inherited neurodegenerative disease caused by a polyglutamine expansion in the N-terminus of the huntingtin protein (HTT) (1, 2). Individuals with 36 CAG repeats or fewer are not affected, while those with 40 or more CAG repeats will eventually develop HD at some point in their lives. The HD mutation is fully penetrant with the time of onset correlating to repeat number. A higher number of CAG repeats results in an earlier age of onset and a more severe form of the disease. In HD patients, neurons in both the striatum and cortex are affected (Figure 1). Progressive atrophy of both the striatum and cortex results in movement incoordination (also known as chorea), a decline in cognitive function, and psychiatric disorders. Typically, most HD patients are diagnosed as middle age adults (40–45 years) and the disease slowly progresses over 10–20 years after initial symptoms (1). However, about 10% of HD cases occur in juveniles who typically have a more severe form of the disease.
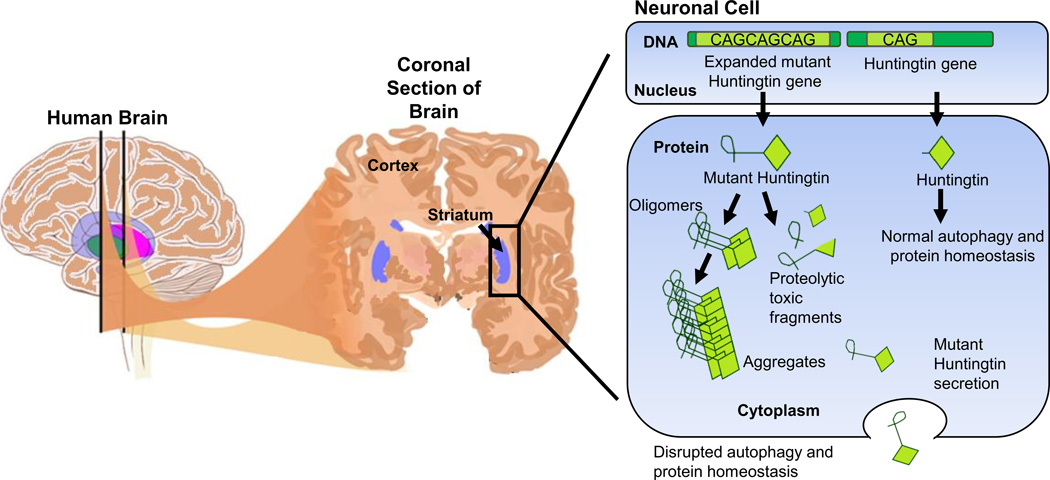
Figure 1. A schematic representation of the brain highlighting the striatum and the pathology of HD in a neuronal cell.
In affected neuronal cells, the mutant HTT gene with an expanded CAG trinucleotide tract is transcribed and translated to mutant HTT (mHTT) protein. mHTT protein does not fold correctly and forms aggregates and fragments which cannot be cleared properly from the cell and as a result is thought to cause certain disease phenotypes.
Unfortunately, there is no effective disease modifying therapy available for HD. One reason for these deficiencies may be the lack of accurate HD models that display all the phenotypes and pathology seen in human patients with HD. Post-mortem brain tissues from HD patients are useful to identify neuropathology of HD. However, post-mortem tissues generally provide late stage pathological changes for HD, which makes understanding early disease changes challenging. Phenotypes observed in post-mortem tissues may represent secondary phenotypes while primary causes might not be uncovered (3, 4). The use of post-mortem tissues as HD model is further impeded by limited availability. Patient derived cell types available are usually limited to blood and spinal fluid. Animal models that mimic some but not all HD phenotypes are used to help elucidate the mechanisms for HD pathogenesis. The HD mouse models most commonly used include the short HTT fragment transgenic models, full length HTT BAC/YAC transgenic models, and HTT knock-in models (5). However, no single HD mouse models is able to recapitulate all aspects of human HD disease progression most likely due to the fundamental genetic differences between rodents and humans (6). Additionally, and perhaps revealing why these models are inadequate, the HD rodent models require much longer CAG expansions to elicit phenotypes, and even then, disease phenotypes are still mild compared to human HD patients (7). Particularly striking is the fact that in human patients, there is a 90% loss of medium spiny neurons (MSNs) while in most rodent models the loss of MSNs is 10% (8).
The breakthrough in human embryonic stem cells (ESCs) technology (9) in the late 1990s when scientists were able to isolate and culture ESCs, provided new possibilities for modeling the various human neurological diseases in a dish, including HD (10–14). However, the use of human ESCs is hindered by ethical concerns regarding the destruction of human blastocysts in the process of ESC derivation and the fact that the disease relevant genotypes may not be represented. In 2006, the discovery of induced pluripotent stem cells (iPSCs) provided a more attractive and efficient way to model human diseases utilizing patient-derived stem cells without the previous ethical concerns. This technology can reprogram adult human somatic cells into ESC like iPSCs by ectopic expression of four transcription factors (15) without the use of embryonic tissue. The advancement of iPSC technology has truly revolutionized the disease-modeling field, making disease relevant human cell types and genotypes available for mechanistic studies and drug screening. Methods to improve and optimize the generation of iPSCs are ongoing (16–22).
The HD field has benefitted substantially from the recent growth and breakthroughs in the human stem cell field (23–31). The technologies in the stem cell field have allowed for the generation of more HD models, customization of specific HD models and the ability to study HD in many relevant cell types without the need of biopsies. This review will detail the current HD stem cell based models, considerations for screening technologies using these models, and the future directions for stem cells in HD modeling.
iPSC based human HD models
Many screens including high throughput screening (HTS) and high content screening (HCS) have already been conducted in HD invertebrate models, primary cells, or immortalized cell lines with chemical or genetic modifiers (32). However, lower organisms like yeast, worms, and fruit flies have limitations and may not accurately reflect the pathogenesis of HD in humans. Primary cells are physiologically relevant to HD, but their sources such as human biopsy samples or animal dissections are usually associated with poor availability, low quality, and inconsistency. Immortalized and easily available rodent and human cell lines such as HeLa, PC12 and HEK293 cells can be genetically modified to carry the HD mutation, but their relevance in the context of HD has not yet translated into a treatment for HD. Another consideration in terms of a cell lines suitability to model HD is the process of immortalization, which may introduce unpredictable variables (such as genetic mutations) to these cell lines (33). Likely due to nonphysiological expression of HTT and the prior mentioned drawbacks of immortalized cell line and invertebrate models, hits identified by previous screens has not yet translated into a treatment for HD (34). With these considerations, it is necessary for the field to have better models with which to understand the pathology of HD and screen for potential treatments.
Human pluripotent stem cells (PSCs), including ESCs and iPSCs, exhibit an unlimited potential for self-renewal and can differentiate into all cell types in the body including neurons. Therefore, they are an excellent source of cells to screen for HD therapies. One example is a screen in wild type human ESCs derived neural stem cells (NSCs) for chemical inhibitors of the transcriptional repressor REST (repressor element-1 silencing transcription factor) using a luciferase-based assay. One potent compound was identified, which could increase expression of brain-derived neurotrophic factor (BDNF) (35). This may be highly relevant to HD as BDNF is a key neurotrophic factor that is depleted during disease progression and restoring BDNF levels ameliorates disease phenotypes. Compared to ESCs, iPSCs have many advantages. As mentioned the generation of iPSCs does not require the destruction of human blastocysts, thus avoiding the major ethical issues associated with using human ESCs. More importantly, iPSCs have the same genetic background as the donor’s somatic cells. These autologous cell lines may be the foundation for the development of personalized diagnosis, screening and therapy.
Shortly after the discovery of human iPSCs, Park et al. generated a group of human iPSC lines from patients with various diseases including HD (31). Following this development, we demonstrated that HD-iPSCs when differentiated into NSCs had a number of features relevant to HD pathology, including susceptibility to cell death upon growth factor withdrawal as measured by caspase activity (30). In the years following, many iPSC-based human HD cell models have been generated and utilized (23–30, 36). These models together with human ESC based HD models are summarized in Table 1. Among these HD cell models, the ones generated by the HD iPSC Consortium are the best characterized. The HD Consortium generated fourteen iPSC lines derived from fibroblasts originating from seven individuals, healthy or affected by HD, representing cell models ranging from asymptomatic controls to HD models with varying CAG repeat numbers and disease severity. NSCs were differentiated from these iPSC donor lines with CAG repeat numbers 21, 33, 60, 109, and 180. These NSC lines were analyzed by microarray and clustered-based analysis of gene expression. Control NSCs (21 and 33 CAG repeats) were shown to be distinct from HD NSCs (60, 109 and 180 CAG repeats). It was found that HD NSCs and HD-derived neurons exhibited decreased cell adhesion and adenosine triphosphate (ATP) production, increased caspase-3 activation, increased cell death after prolonged culture or BDNF withdrawal, and increased vulnerability to stress/toxicity. Significantly, these disease-associated phenotypes correlated to CAG repeat number: more severe phenotypes were found in HD cells with longer CAG repeats (24). These iPSC lines are currently deposited at Coriell and represent a rich resource for the HD community including their use in potential drug screens.
Table 1.
Human stem cell models of Huntington’s disease and their phenotypes reported in the literature.
| Pluripotent Cell Type | CAG Repeat | Genetic Modification |
Differentiation Stage and Phenotypes | Reference |
|---|---|---|---|---|
| ESCs from HD embryos | 37 and 51 | no | Differentiated neurosphere cells showed expanded CAG repeat length instability |
11 |
| ESCs from HD embryos | 40, 45, 46 and 48 | no | No phenotype described | 12 |
| ESCs from HD embryos | 37 and 51 | no | CAG51 forebrain neurons had elevated glutamate-evoked responses such as intracellular calcium levels |
13 |
| H9 ESCs | 23, 73 and 145 |
HTT exonl under CAG promoter delivered by ePiggyBac system |
CAG73 and CAG145 neurons showed EM48 positive aggregates and increased cell death upon growth factor deprivation compared to CAG23 neurons |
14 |
| ESCs from HD embryos | 15/21, 18/21, 22/22, 12/40, 17/46, 17/48, 23/45, 19/41, 19/46,21/42 |
no | Proteomic analysis had mitochondria dysfunction in HD affected ESCs shown by reduction components of electron transport chain complex I, III and IV. Neurons from HD affected ESCs showed vulnerability to nonspecific kinase inhibitor STS. There were transcription dysregulation such as histone H1 family members and actin cytoskeletal signaling proteins in HD neurons |
10 |
| iPSCs from HD patient | 72 | no | No phenotype described | 31 |
| iPSCs from HD patient | 72 | no | Increased caspase activity upon growth factor deprivation in NSCs |
30 |
| iPSCs from HD patient | 50 and 109 | no | Increased cytoplasmic vacuolation seen in differentiated astrocytes |
29 |
| iPSCs from HD patient | 15/17, 15/18, 17/45,39/43,42/44 |
no | No difference in growth rate, differentiation, caspase activation in iPSCs. Higher lysosomal activity in HD-iPSCs and derived neurons shown by LysoTracker dye. Increased autophagesome formation in HD-iPSCs |
28 |
| iPSCs from HD patient | 72 | no | HD iPSCs form EM48+ aggregates upon treatment of proteasome inhibitor MG132. NPCs from HD iPSCs and developed EM48+ HD pathology characteristic at later stage of transplantation into rat quinolinic acid-induced HD model |
27 |
| iPSCs from HD patient | 72 | no | Dysregulated proteins involved in oxidative stress response, apoptosis in HD-iPSCs. Decreased neuronal differentiation and neurite outgrowth and increased apoptosis (TUNEL) in HD-iPSCs and neurons. Reduced cytoskeleton associated proteins in HD-neurons |
26 |
| iPSCs from HD patient | 72 | Expanded CAG was genetically corrected byHR |
At NSC stage, HD cells showed increased apoptosis (TUNEL), caspase activity, decreased BDNF expression, maximal oxygen consumption rate, altered TGF-β and cadherin signaling. Corrected NSCs reversed these DhenotvDes |
66 |
| iPSCs from HD patient | 17/21,21/28, 18/33, 18/60, 19/109, 18/180 |
no | Microarray analysis showed distinct gene expression pattern in differentiated NSCs. HD NSCs showed decrease in energy metabolism (lower intracellular ATP level and ATP/ADP ratio) and cell adhesion (smaller clump size). HD neurons had higher risk of cell death after prolonged culture (stained cleaved caspase-3), BDNF withdrawal (condensed nuclei and caspase activation) or stressed by H2O2 or 3-MA (condensed nuclei). Physiological or pathological glutamate treatment would increase percentage of HD neurons with calcium dyshomeostasis |
24 |
| iPSCs from HD patient | 72 | no | mHTT formed EM48 positive aggregates in differentiated neurons which could be reduced by microRNA miR-196a |
23 |
| iPSCs from HD patient (non-integrating method) |
18/21, 18/28,18/33, 18/60, 19/109, 18/180 |
no | More Nestin+ neural cells after differentiation of HD iPSCs. Withdrawal of BDNF caused more apoptosis (TUNEL) in HD neurons and reduced numbers of Nestin positive cells. |
36 |
At this time, most HD iPSC lines are studied without any genetic modification and require a direct comparison to age and sex-matched healthy control iPSC lines. Studies comparing disease and control iPSC lines from separate individuals have shown that genetic background differences between these individuals, even when controlled for by age, sex, and ethnicity, can skew results when trying to identify specific disease mechanisms. To circumvent this issue, isogenic iPSC lines, where disease-causing mutations are genetically corrected to produce a wild type allele, have been generated to produce genetically identical disease and control iPSC lines. Theoretically, any phenotypes identified in these isogenic iPSCs can be solely attributed to the disease-causing mutation. These cell lines are particularly suitable for screening of drugs or identifying mechanisms that target phenotypes caused by the disease mutation. With regards to HD, we generated two human HD isogenic iPSC lines using a homologous recombination based genetic correction method in which a 72 CAG repeat was replaced with a normal 21 CAG repeat in the gene HTT (25). It was demonstrated that at the NSC stage, genetic correction of the HD mutation could reverse disease-associated phenotypes such as elevated cell death and caspase-3/7 activity as well as lower BDNF levels and energy metabolism (25). As discussed further below, new genome editing tools such as CRISPR/Cas9 have made it easier to produce genetically modified cell models, which will allow for the efficient generation of iPSC allelic series of varying CAG repeat lengths and new models of HD (37).
An additional alternative to human HD iPSCs is HD iPSC lines derived from monkeys. These iPSCs were derived from skin cells of a transgenic monkey HD model with human HTT exon 1 with 84 CAG repeats (25). The iPSCs were able to differentiate into neurons and HD phenotypes were observed including aggregate formation of mutant HTT (mHTT) protein and increased cell death (38). These primate HD iPSC models provide another platform for drug screening with possibility of further exploring promising candidates in a non-human primate HD models. This is a potentially important drug-screening step as many candidate drugs are screened in non-human primates before entering Phase I human clinical trials.
HD phenotypes for screening
A pivotal consideration when utilizing HD stem cell models for drug screening is the selection of appropriate measurable phenotypes to use as endpoints. This may relate to the biology of the HTT protein or to well characterized phenotypes found in HD disease progression. Phenotypic endpoints may have benefits for drug discovery when compared to using known targets. Recent analysis of approved drugs suggests a decline in new drugs brought to market, which may be due to a switch in the pharmaceutical companies from using phenotypic assays to target-based assays for drug development (39, 40). After selecting endpoints, assays can be either designed or utilized for the most efficient screening methods. Previous studies have provided researchers with a robust selection of disease associated phenotypes as end points in primary cells and immortalized cell lines. The commonly used phenotypic endpoints for human stem cell derived HD models are listed in Table 1. Several of these commonly screened phenotypes first identified in HD post mortem or other models have also been validated in HD iPSC cell models.
HTT aggregation is arguably the most commonly used phenotype for HD screening. Similar to other neurodegenerative diseases, HD is characterized by abnormal protein (mHTT) folding, aggregation, and clearance (Figure 1). The aggregates formed by the N-terminal fragments of mHTT are found in cortical neurons and striatal medium spiny neurons in HD patients and various HD models (41). The clearance of mHTT aggregates correlates with disease reversal in HD mouse models (42). In some screens, aggregation of mHTT is monitored by fluorescence image based high content screening (43–45). Alternatively, sodium dodecyl sulfate (SDS) insoluble mHTT aggregates can be detected by a filter retardation assay coupled by dot immunoblotting (46–48). Research supports the concept that the mHTT aggregates are actually a coping mechanism for the cells and are less toxic than the smaller and more soluble HTT fragments (49, 50). When screening for potential therapeutic compounds, it would be advisable to screen for aggregation formation, mHTT, HTT post-translational modifications and toxic fragment levels. Since mHTT protein is the cause of HD pathology, screening for compounds capable of decreasing mHTT levels has been reported (32). Time-resolved fluorescence resonance energy transfer (FRET) assays that detect HTT levels were developed to screen small molecule libraries in the HN10 neuronal cell line in a high throughput format (51, 52). A western blot based assay aimed at reducing an N-terminal HTT fragment was utilized to screen a RNA interference (RNAi) library that targeted all human protease genes and it identified matrix metalloproteinases (MMPs) as modifiers of HTT proteolysis (53). These assays offer the researcher a variety of choices from which to select the best assay for their screening purposes.
Cell death and neuronal toxicity are other hallmark phenotypes of neurodegenerative diseases including HD. In HD, neuronal cell loss is observed in the cortex and striatum of patient brains. The decrease in striatal and cortical volume correlates with cell death and can be detected in patients many years before behavioral symptoms manifest (54). Early markers of cell death and upregulation of the apoptosis pathway including activation of caspases are typically associated with different kinds of HD cell models (41). Due to their obvious significance to HD pathology, cell death and neuronal toxicity are popular endpoints for screening. Using an HD PC12 cell model with an endpoint based on a fluorescent cell viability assay, five structurally distinct compounds were found to prevent apoptosis induced by mHTT. Their common target protein, disulfide isomerase, was further identified to be pro-apoptotic and is involved in the pathogenesis of a number of neurodegenerative diseases (44). An assay for caspase-3/7 activity, which are key execution caspases in apoptosis, was employed to screen a kinase inhibitor library in immortalized striatal cells from the Hdh111Q/111Q knock-in HD mouse model. The screen identified that DGK inhibitors decreased caspase-3/7 activity (55). The same caspase-3/7 activity assay was used on a larger scale to screen siRNA pools against over 7000 druggable human genes (56). Another chemical screen performed by the same group applied both a caspase-3/7 activity assay and an ATP assay, a measurement indicating mitochondrial function, in Hdh111Q/111Q striatal cells. This screen identified a number of structurally related compounds that may act through the serotonin receptor signaling pathway (57). All of these cell death and neuronal toxicity assays have been essential in understanding HD pathology. These assays can be utilized in further studies on HD iPSCs and corresponding disease derived cell types.
Autophagy is one of the major methods for cells to clear misfolded protein. In HD, the autophagy system is impaired by mHTT, which causes dysregulation in the clearance of misfolded and aggregated proteins including mHTT itself (58, 59). Inhibition of mammalian target of rapamycin (mTOR) can activate autophagy. The mTOR inhibitor rapamycin and its analog CCI-779 have been shown to reduce mHTT accumulation and toxicity in HD fly and mouse models (60). However rapamycin has many side effects due to its broad activity (61). It would be ideal to have a highly specific autophagy inducer for treating HD. Several screens in HD cell models have identified pharmacological (62, 63) or genetic (64) modifiers of autophagy. One group has also identified mTOR independent targets for autophagy induction in a screen of FDA-approved drugs (65).
Phenoytpes in human derived HD-iPSCs cells types
There are many phenotypes in human PSC derived HD models that can be measured by specific assays and these are listed in Table 1. The most commonly screened phenotypes in both human ESC and iPSCs were mentioned previously and are neuronal toxicity and HTT aggregation. Additionally, there are more phenotypes compatible with high throughput screening in iPSC based HD models. One of these phenotypes is BDNF expression, which is suppressed in HD NSCs compared to isogenic corrected NSCs with a normal CAG repeat length (30, 66). This effect is probably due to transcriptional dysregulation caused by mHTT (25) and has been confirmed in other human HD derived neurons from iPSCs (36). In HD NSCs, when growth factors in normal medium (full medium) are taken away, cells show phenotypes relevant to the disease. As shown by immunocytochemical staining in Figure 2, phenotypes such as increase in cleaved HTT, activated caspases and decrease in BDNF and polymer F-actin as measured by phalloidin conjugated fluorophore are suitable endpoints for screening in HD-NSCs (Figure 2). ATP levels were lower in NSCs bearing a longer CAG expansion (38), which indicates impaired mitochondrial function, as mitochondria are the major power source inside cells that produce ATP. Furthermore, neurite outgrowth was impaired in neurons differentiated from HD patient derived iPSCs (26), a phenomenon resembling defects in neuronal maturation and axon outgrowth in HD. These examples represent endpoints suitable for HD screens in a high throughput format, which is especially valuable when screening thousands of potential therapeutic candidates. Other phenotypes that have been identified in MSNs derived from human HD-iPSCs are increased expression of γH2AX and elevated oxidative stress (67). In this study the A2AR-selective agonists protected MSNs through the cAMP/PKA-dependent pathway (67). Also observed in human HD NSCs is a deficit is manganese-dependent activation of p53 (68).
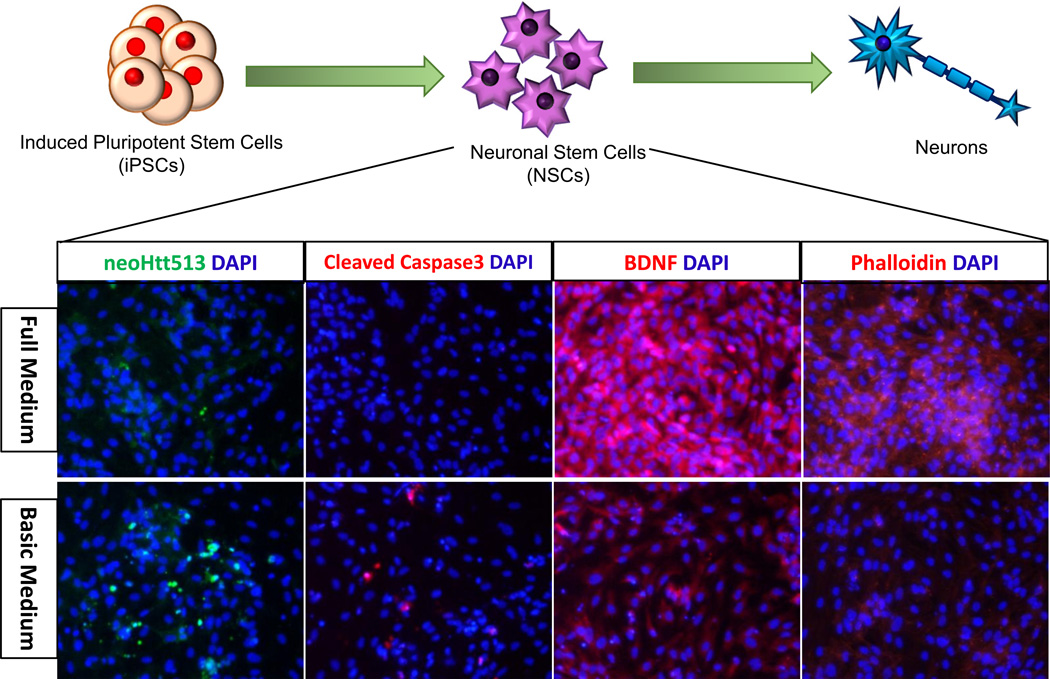
Figure 2. Diagram of iPSC differentiation and potential endpoints at the NSC stage.
Human HD iPSCs were differentiated into NSCs, then neurons and glia. At the NSC stage, we observed disease-associated phenotypes. When stressed (change from full medium to basic medium without growth factors) NSCs derived from HD iPSCs demonstrated upregulation of the N-terminal 513aa Htt fragment (recognized by neoHtt513 antibody) and cleaved caspase-3 and downregulation of BDNF and phalloidin labelled F-actin.
Recently, an unsuccessful screen funded by the California Institute for Regenerative Medicine (CIRM) and conducted by Numerate, Inc. demonstrated the importance of using more caution in screening plan design. This screen tried to identify novel small molecule modifiers of toxicity in neurons derived from HD patient iPSCs. In their counter screen, all previous hit compounds behaved the same as in their original screen, and thus these hits were identified as false positives (69). To reduce commonly seen false positive hits from primary screens, it is critical to plan the screening strategy carefully. It is advisable to have multiple end points for the primary screening. Alternatively, HD cell models from multiple sources can be screened simultaneously, to help account for cell line specific hits. Implementing these strategies should significantly lower the chance of generating false positive results. A counter screen should be included in the secondary screen to validate hits from the primary screen. Certainly expanding the number of end points and/or cell types will increase screening costs, but the cost will be even more if false positive candidates are carried over to later stages of the drug discovery process, where one starts to utilize model organisms and produce large quantities of the compounds.
Technological development of high content screening has made multiple end points in primary screen more feasible. For example, multiple disease biomarkers can be imaged simultaneously, and live imaging can be coupled with other conventional high throughput format screens. All these advances will make designing a successful screening plan more readily available and effective for potential iPSC based HD screens.
From iPSCs to differentiated cells
One important detail that should be considered when studying iPSC based HD models is the differentiation stage of the stem cells. In Table 1, the majority of HD disease phenotypes are not detected in pluripotent stem cells, but in either differentiated neurons and glia or lineage defined multipotent progenitor cells like NSCs. As one example, there is no significant difference in caspase-3/7 activity after growth factor deprivation between HD and corrected cells in iPSCs. The change in caspase-3/7 activation is only observed at the multipotent NSC stage (Ellerby lab unpublished data). This is in agreement with the fact that HD symptoms occur in the mature brain, affecting primarily the committed neural lineages. It should be noted that more subtle changes are detected in HD-iPSCs when compared to controls and this is noted in a number of signaling pathways such as TGF-β, β-cadherin, oxidative stress proteins and the p53 (66, 70). This information suggests that when using iPSC-based models for HD screening, it is essential to choose the appropriate differentiation stage starting from iPSCs and the method of differentiation.
Striatal neurons, specifically MSNs, are the main cell type in the brain affected by HD and MSN-like cells are likely the best candidates for the development of effective and predictive HD drug screens. MSNs of the striatum originate in the lateral ganglionic eminence (LGE) in the ventral part of telencephalon (71, 72). Sonic hedgehog (SHH) is one of the most important morphogens that determines dorsal-ventral patterning in developing neural tube through a concentration gradient with a high level of SHH on the ventral side and a low level of SHH on the dorsal side (73, 74). LGE formation is also under the control of the right concentration of SHH (75). In contrast to SHH, WNT signaling is critical in dorsal telencephalon development (76). Ectopic activation of WNT signaling in the ventral telencephalon can repress expression of ventral markers and induce expression of dorsal markers (77). Measurement of these proteins can be used to validate the correct development stage of iPSC-derived MSNs before using them to screen for therapeutic compounds.
Guided by knowledge of striatal development in the brain, multiple protocols are published that can derive striatal neurons from human ESCs or iPSCs. These ESC/iPSC-derived striatal neurons are positive for the MSN marker dopamine and cAMP-regulated neuronal phosphoprotein 32KD (DARPP-32) (Table 2) (64, 78–85). However, the efficiency of generating DARPP-32-positive striatal neurons in these protocols is still being optimized for conducting cell-based screening. The default in vitro differentiation of human ESCs to telencephalic progenitors is predominantly a dorsal fate because of endogenous WNT signaling (79). Therefore, almost all protocols aiming to generate striatal neurons from ESCs or iPSCs require supplementation of recombinant proteins or chemicals to suppress WNT signaling and activate SHH signaling at the appropriate concentration. Besides extrinsic signaling pathways, differentiation of striatal neurons is also regulated by intrinsic cues such as transcription factors. DLX1, DLX2, ASCL1, GSX2, FOXP1, and CTIP2 are all transcription factors that play key roles in the development of LGE progenitors and striatal neurons in the brain (86–93). Additionally, a recent study showed that Activin A is a critical factor for generating MSNs from human ESCs and iPSCs (83). One study involving direct conversion (or transdifferentiation) of human fibroblasts to striatal neurons involved expression of the brain enriched microRNA miR-9/9*-124 together with transcription factors DLX1, DLX2, CTIP2 and MYT1L (94). This strategy could also be used to optimize protocols for generating striatal neurons from iPSCs in combination with modulation of SHH and WNT signaling. Optimization of these pathways will allow for drug screening to take place in the most disease relevant cell type, increasing the likelihood of finding effective therapeutics.
Table 2.
Differentiation methods to produce medium spiny neurons, cortical neurons, astrocytes, and oligodendrocytes from ESC and iPSCs.
| Starting Cell Type |
Differentiated Cell Type |
Differentiation Method | Markers | Efficiency | Reference |
|---|---|---|---|---|---|
| ESC | MSN | d0-12: co-culture hESC with MS cells in DMEM-F12/KSR; d12–21: culture in DMEM-F12/N2; d21: isolate neural rosettes and plate on poly-ornithine/laminin surface; d21–46: culture in DMEM-F12/N2 with BDNF, SHH, DKK1; d46–62: differentiate into neuron in DMEM-F12/N2 with BDNF, db-cAMP, VPA |
DARPP32 GABA calretinin calbindin |
53% DARPP32+ in MAP2+ cells |
78 |
| ESC | MSN | d0-6: EB in suspension; d6: attach on plastic surface; d6–10: culture in DMEM-F12/N2; d10–30: induce ventral telencephalic progenitor in DMEM-F12/N2 with SHH, DKK1; d30–42: differentiate into neuron in Neurobasal/N2/B27/BDNF/GDNF/IGF1 |
DARPP32 GAD65/67 GABA |
not described |
79 |
| ESC | MSN | d0-12: primitive neuroepithelia induced in DMEM-F12/N2; d12–26: SHH or purmorphamine added in DMEM-F12/N2; d26–33 : Neurobasal/VPA; d33-: Neurobasal/BDNF/GDNF/IGF/cAMP |
DARPP32 GAD65/67 GABA Meis2 |
89.7% DARPP32+ in GABA neurons |
85 |
| ESC and iPSC | MSN | d0-3: PSCs expanded in MEF conditioned medium with Y27632; d3–15: KSR medium with dorsomorphin and SB431542; d8–29: SHH and DKK1 added; d29-d80 : terminal differentiation in N2/B27 medium with BDNF |
DARPP32 GAD65/67 GABA calbindin CTIP2 |
20% DARPP32+ |
80 |
|
Immortalized NPC line |
MSN | Differentiation in DMEM-F12/N2/B27/lnsulin; d0-d11: BDNF (30ng/ml)/SHH/DKK1; d0-d1: additional Y-27632 added; d11- 60: BDNF (50ng/ml) |
DARPP32 GABA Calbindin |
21% DARPP32+, 96% GABA+, 84% Calbindin+ in MAP2+ neurons |
82 |
| ESC and iPSC | MSN | d0-d5: 80% confluent PSCs induced in DMEM- F12/Neurobasal (2:1) supplemented with N2/B27/SB431542/LDN-193189; d5-d9: SB431542 removed; d9–28: LDN-193189 replaced by activin A; d28–36: BDNF/GDNF added |
DARPP32 CTIP2 Calbindin |
20–50% DARPP32+ |
83 |
| ESC and iPSC | Cortical Neuron | Differentiation in 3N medium (DMEM-F12/Neurobasal, 1;1, N2/B27/insulin/vitamin A). d0-d11: 95% confluent monolayer PSCs on Matrigel differentiated in 3N with retinoids, SB431542 and dorsomorphin or noggin; d11–13: replatedto poly-ornithine/laminin in 3N with bFGF; d13–100: maintained in3N |
Pax6 01×1/2 Emx1 Foxgl Tbr2 |
>95% Pax6+, Otx1/2+by d15 |
98 |
| ESC and iPSC | Cortical Neuron | d0-16: monolayer PSCs on Matrigel differentiated in DMEM- F12/N2/B27/BSA/Glutamax/noggin; d16–24: same medium with noggin removed; d24: replate cells to poly-lysine/laminin in same medium with noggin removed and Y27632 added; every 5–7d, half medium changed with Neurobasal/B27/glutamine |
Pax6 Otx1/2 Emx1/2 Foxgl Tbr1/2 vGlut1/2 |
Mostly Pax6+, Oxt1/2+ d10–19, 60% vGlut1 + d44 |
99 |
| NSC | Astrocyte | NSCs on poly-ornithine/laminin or Geltrex were induced for astrocyte differentiation with StemPro/activin A/Heregulin 1β/IGF1 |
GFAP S100β |
62% GFAP+ |
108 |
| ESC and iPSC | Astrocyte | d0-6: EB in suspension; d6: EB plated on Matrigel in Neurobasal/N2; d6–9: bFGF/EGF added; d9–12: bFGF/EGF/CNTF added; d12–15: bFGF/CNTF added; d15-: CNTF alone; |
GFAP S100β GLT-1 |
78% GFAP+ at 5weeks |
109 |
| iPSC | Astrocyte | iPSCs were induced with dorsomorphin/SB431542/NAC for 5–7d before enzymatic dissociation to form neurospheres; retinoic acid added to neurospheres for 7–12d; neurospheres mechanically chopped and replated in NSCR (AdDMEM- F12/N2/B27/NEAA/Glutamax) with EGF/LIF for 2–4w; propagated in NSCR with EGF/bFGF; dispase dissociation and CNTF induced differentiation |
GFAP sioop Vimentin NF1A |
>90% GFAP+, S100β+ |
110 |
| ESC and iPSC | Oligodendrocyte progenitor cells (OPCs), Oligodendrocytes (OL) |
d0-d8: neural induction using dual SMAD inhibition and 100 nM all-trans RA in adherent conditions; d8: added SAG to medium; d12: lift cells for sphere formation in medium with RA and SAG; d30: plate spheres on poly-L-ornithine/laminin dishes in PDGF medium for OPC formation; d75+: glial medium for oligodendrocyte maturation |
OPC: NKX2.2 SOX 10 OLIG2 OL: O4 MBP |
40–70% O4+ |
115 |
| ESC and iPSC | Oligodendrocyte progenitor cells (OPCs), Oligodendrocytes (OL) |
d0-d5: EB formation in ESC medium; d5-d19: neuroepithelial generation in NIM, FGF2; d19-d29: pre-OPC generation in neural induction medium (NIM), RA, B27, purmorphamine; d29-d40: pre-OPC expansion in NIM, bGFG, B27, purmorphamine; d40-d160: OPC generation in glial induction medium (GIM), T3, NT3, IGF, PDGF-AA, purmomorphamine; d160-d180: terminal differentiation in reduced GIM |
OPC: OLIG2 NKX2.2 SOX 10 PDGFRa 0L04 MBP |
OPC: >45% ESC derived >70% iPSC derived |
116 |
Cortical pyramidal projection neurons are also affected in HD. Specifically, the cortical layers III, V, and VI atrophy during HD disease progression and as much as 30% of cortical neurons in those layers die (95, 96). The cortex and striatum are linked by the corticostriatal pathway. The striatum receives excitatory glutamatergic inputs from the cortex via pyramidal projection neurons that innervate and excite MSNs. Dysfunction in striatal and cortical circuits or in striatal or cortical neurons can advance HD disease progression and neuropathology (97). Due to these findings, many studies of HD pathogenesis now incorporate toxicity screenings in cortical neurons as well as striatal neurons. Multiple protocols are published that can generate cortical neurons with high efficiency and purity from human ESCs and iPSCs (93, 98–100). By mimicking human cortical development, these protocols can generate cortical stem and progenitor cells that further differentiate into multiple classes of cortical projection neurons that express markers such as the glutamatergic marker vGlut1, deep-layer cortical neuron markers Tbr1 and CTIP2, and upper-layer neuron markers Cux1, Satb2, and Brn2. A commonly used method developed by Shi et al. was able to generate primary neuroepithelial cells and secondary cortical stem/progenitor cells using a method of dual SMAD inhibition to push cells towards the anterior neuroectodermal lineage (98). The primary cortical progenitor cells generated by this method express the transcription factors Foxg1, Pax6, Otx1/2, and Tbr2. The incorporation of cortical neurons along with striatal MSNs in HD drug screens will provide a more thorough model of pathology of HD, which may lead to more effective therapeutics.
In the process of making neurons, it is critical to monitor the maturity of these neurons. Terminally differentiated, mature neurons are usually mitotically inactive, have elaborated processes, form synapses with neighbouring neurons and are capable of firing action potentials. Mature neurons differ from immature neurons not only in function and marker expression, but also in their capacity to handle stress and apoptotic signals. Mature neurons are more resistant to cytotoxic stimuli and more likely to survive in stressful conditions (101). This is particularly important for HD models as immature and mature neurons may behave significantly different in screening assays. However, neuron aging is a distinct phenomenon, representing a decline rather than maturation of functions. In aging neurons, there is an increase in reactive oxygen species (ROS), DNA damage, misfolded proteins, mitochondrial instability and dysregulation of ion homeostasis. All of these factors contribute to neuron dysfunction and death (102). HD is a neurodegenerative disease associated with aging. Typically age of onset for HD is in middle age despite the presence of mHTT protein from birth. Elevated levels of ROS and senescence associated inflammatory cytokines are markers of aging and might also be targeted for accelerating aging of freshly differentiated neurons (103). Due to the association of age with onset of HD, it might be useful to age neurons in vitro cell models to more accurately recapitulate HD pathology.
While striatal and cortical neurons are the main cell types affected in HD, evidence of phenotypes in glia, specifically astrocytes, have also been reported. In human post-mortem HD brains, astrocytes express mHTT and gliosis is often found in late stage HD brains and in HD mouse models (104–106). Interestingly, HD mouse models that expressed mHTT in striatal astrocytes experienced a decrease in expression of glutamate transporters and a reduction in glutamate uptake. This resulted in increased striatal neuron dysfunction due to the inability of astrocytes to protect neurons from glutamate excitotoxicity (106, 107). In vitro, astrocytes derived from human HD iPSCs exhibit phenotypes such as a higher number of cytoplasmic vacuoles compared to wild type astrocytes (29). Multiple protocols that efficiently generate glial fibrillary acidic protein (GFAP) positive astrocytes from human ESCs and iPSCs have been published (Table 2) (108–110). Thus endpoints such as the presence of cytoplasmic vacuoles or reduced glutamate transporter expression could be used to screen HD astrocytes derived from patient iPSCs.
Investigating other glial cell types such as microglia and oligodendrocytes could prove worthwhile with regards to developing HD screens. Microglia are activated in presymptomatic HD patients (111) and microglial activation is correlated with HD diseases severity (112). In HD mice, mHTT impairs microglia migration in response to injury (113). The potential role that oligodendrocytes play in HD is more obscure. One study suggests that in preclinical stages of HD, myelin breakdown and increased iron levels occur and cause toxicity that could contribute to HD pathogenesis. During HD progression, neuronal loss is complemented by glial expansion of astrocytes and oligodendrocytes, which could further cause toxicity in HD patients, running endpoint assays in these cells will help elucidate if a therapeutic will be effective on all the cell types affected by HD (114). Protocols to generate oligodendrocytes are available (115, 116) and microglia from human ESCs or iPSCs are currently being developed.
Thus when considering how to model HD in vitro using ESCs or iPSCs, there are multiple different cell types to consider. As is the case with other neurodegenerative disorders, co-culture models where multiple cell types are cultured together could provide more sophisticated models of HD and more phenotypic endpoints for drug screening. Amyotrophic lateral sclerosis (ALS) is a prime example of the power of co-culture experiments for disease modeling. By culturing astrocytes from SOD1 mutant ALS mice with healthy motor neurons (the cell type affected in ALS patients), scientists discovered that superoxide dismutase 1 (SOD1) mutant astrocytes secrete toxic factors that cause the death of healthy motor neurons (117). Conducting a screen using this type of co-culture model could identify drug candidates that prevent astrocyte-mediated motor neuron death in ALS. In the case of HD, culturing glial cells with either striatal neurons, or cortical neurons, or both could be beneficial in a similar manner to ALS.
Another alternative to co-culture is the generation of cerebral organoids, which are 3D brain structures derived from human ESCs or iPSCs that exhibit similar developmental stages to endogenous neural development and generate multiple brain tissue types and regions (118, 119). Brain organoids can survive long periods in culture, thus providing “aged” neurodegenerative models with more mature, functional neurons. This system has already been utilized to model microcephaly, and organoids generated from microcephaly patient iPSCs exhibited premature neuronal differentiation (120). In a 3D organoid system using HD iPSCs, screens could be developed with neuron function or survival as endpoints among the others listed previously. Additionally, other cell types affected such as glial cells could be easily monitored at the same time. Due to the difficulty of obtaining human living brain tissue samples, cerebral organoids currently offers one of the most comprehensive model conditions to represent in-vivo HD pathogenesis. Further development of this technology and optimization for high throughput screening would be very beneficial not only for HD but for many other neurodegenerative diseases.
Genome Editing Tools for HD Models and Screening
When creating an ideal HD cell model for screening purposes, limiting the genetic background variance would be ideal while simultaneously having multiple CAG repeat lengths represented (37). Until very recently, creating a set of varying CAG repeat HD iPSC isogenic lines was a time consuming process and occurred with a low efficiency rate. However, the rapidly expanding genome engineering field has made the creation of multiple variations of a genetic disease model practical. The latest generation of genome engineering tools is referred to as “clustered regularly interspaced short palindromic repeats” (CRISPR) (121). Unlike the previous genome engineering tools, zinc finger nucleases (ZFNs) and TALENS (transcription activator-like effector nucleases), CRISPR is easier to utilize by the scientific community. The system targets any site in the genome with high efficiency, can be made to order, and has minimal off target effects (122). Cost and time have been drastically reduced, while accuracy and efficiency have been increased when utilizing this technology. CRISPR technology is a gene editing nuclease system, which uses a bacterial nuclease to cut at a specific site in the DNA. This specific site is dictated by a guide RNA (gRNA), which specifics 18–21bp in the DNA, making it possible to target a single unique site in the genome (123). Recent studies have shown that the off-target effects of CRISPR are minimal with as few as one off target site(s) that were cleaved (124, 125). Having a low off target cut rate is critically important when introducing a mutation for disease model creation, to ensure that only one mutation is introduced into the genome. Both ZFNs and TALENs are complicated to design, assemble and they exhibit significant off target binding and cleavage, making them less than ideal tools for disease model creation. The advances in genome sequencing would allow for each potential model to be fully sequenced ensuring that only one donor insertion event has occurred. CRISPR has been used successfully to make several disease relevant iPSC models and mammalian models including the generation of a mouse model with multiple specific mutations in a single generation (126–129). The high efficiency and accuracy of CRISPR makes it the ideal system to utilize for making a library of varying length CAG repeat models for HD (37) or engineering HD reporter cell lines for drug screening needs.
Increased availability of varying CAG repeat length models for HD and multiple isogenic cell lines will be valuable for understanding HD disease mechanisms and will improve the chances of discovering effective treatments for HD (37) (Figure 3). It is well documented that the CAG repeat length impacts the time of onset, progression and severity of HD, and thus it is possible that the repeat length may also affect drug response. Additionally, drug response may be different in individuals with modifier mutations (130). Many modifier genes have been identified for HD, meaning their expression or structure can affect when and how the disease manifests (131, 132). Models that reflect subsets of modifier genes may prove useful in drug screening as this subpopulation could respond differently to certain drugs. New genome editing tools such as CRISPR have made knock-in/knock-out of modifier genes in HD iPSC models more efficient and easier than before. CRISPR has also provided a powerful tool to screen genomes for HD genetic modifiers in an unbiased way. In fact CRISPR has been utilized in a genome wide knock-out screen to identified genes essential for viability to human PSCs (133). The same method can be readily adapted to screen for disease modifier genes in human iPSCs.
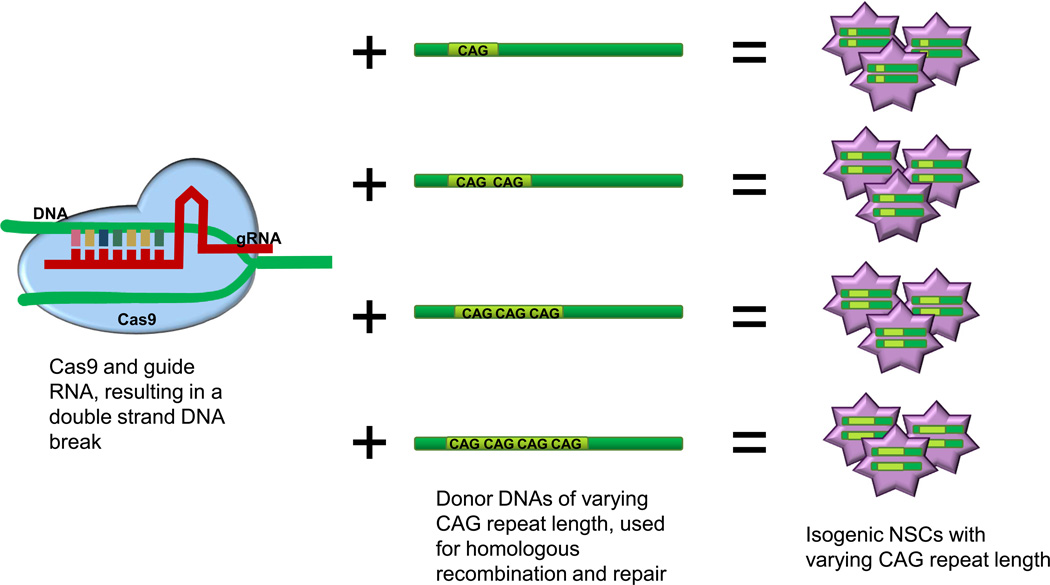
Figure 3. A representation of a potential genome editing method to create multiple isogenic HD iPSC lines.
CRISPR technology can target and modify specific genes by inducing DNA double strand breaks which triggers the cells DNA-repair machinery to correct the break via homologous recombination. With CRISPR and HTT specific gRNAs, introduction of a donor template DNA with varying lengths of CAG repeats allows generation of isogenic iPSC lines, which differ only in CAG repeat length in exon 1 the HTT gene.
Another application for genome editing tools in HD drug screening is generating reporter cell lines. The ease of genetic engineering would allow for the tagging of several potential therapeutic target proteins with fluorophores or other selection markers in HD. The flexibility of gRNA directed genomic modification with CRISPR makes it simple to insert selection markers either in endogenous loci or a safe harbour locus such as the AAVS1 site. Tagging certain proteins with a fluorophore, or even multiple proteins with different fluorophores in the same cell could allow for multiple fluorophore monitoring during screening of compounds (134). These are only a few of the potential benefits of the recent advances in the genome engineering field, which can significantly impact screening strategies and our basic understanding of HD.
Conclusion
The contributions that stem cells and specifically iPSCs have made to the HD field is substantial and has impacted our current understanding of HD and the therapeutic directions being pursued. As a tool available to researchers for under a decade, human iPSCs have been utilized in drug screens in neurodegenerative diseases including amyotrophic lateral sclerosis (ALS) (135) and familial dysautonomia (FD) (136). Given the advantages of iPSCs, including more relevant human phenotypic endpoints and the variety of differentiated cell types, iPSCs and their derivatives are a promising new direction for HD drug screening. The potential of iPSCs is just beginning to be thoroughly explored, both in phenotypic screens and in very early genetic manipulations to create even more representative disease models. iPSCs and their cell derivatives will undoubtedly help form the foundation on which new therapeutic investigations will occur and new insights to HD will be understood.
Acknowledgments
Funding
The funding for this research was provided by CHDI (L.M.E.) and NIH T32 AG000266 (L.M.E., N.Z., B.J.B. K.R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Warby SC, Graham RK, Hayden MR. Huntington Disease. 1993 [Google Scholar]
- 2.The Huntington’s Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72(6):971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 3.Marchetto MC, Brennand KJ, Boyer LF, Gage FH. Induced pluripotent stem cells (iPSCs) and neurological disease modeling: progress and promises. Hum Mol Genet. 20(R2):R109–R115. doi: 10.1093/hmg/ddr336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sterneckert JL, Reinhardt P, Scholer HR. Investigating human disease using stem cell models. Nat Rev Genet. 15(9):625–639. doi: 10.1038/nrg3764. [DOI] [PubMed] [Google Scholar]
- 5.Heng MY, Detloff PJ, Albin RL. Rodent genetic models of Huntington disease. Neurobiol Dis. 2008;32(1):1–9. doi: 10.1016/j.nbd.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 6.Kaye JA, Finkbeiner S. Modeling Huntington’s disease with induced pluripotent stem cells. Mol Cell Neurosci. 56:50–64. doi: 10.1016/j.mcn.2013.02.005. PMCID: 3791169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dodds L, Chen J, Berggren K, Fox J. Characterization of Striatal Neuronal Loss and Atrophy in the R6/2 Mouse Model of Huntington’s Disease. PLoS Curr. 6 doi: 10.1371/currents.hd.48727b68b39b82d5fe350f753984bcf9. PMCID: 3882322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vonsattel JP. Huntington disease models and human neuropathology: similarities and differences. Acta Neuropathol. 2008;115(1):55–69. doi: 10.1007/s00401-007-0306-6. PMCID: 2847401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 10.McQuade LR, Balachandran A, Scott HA, Khaira S, Baker MS, Schmidt U. Proteomics of Huntington’s Disease-Affected Human Embryonic Stem Cells Reveals an Evolving Pathology Involving Mitochondrial Dysfunction and Metabolic Disturbances. J Proteome Res. 13(12):5648–5659. doi: 10.1021/pr500649m. [DOI] [PubMed] [Google Scholar]
- 11.Niclis J, Trounson AO, Dottori M, Ellisdon A, Bottomley SP, Verlinsky Y, et al. Human embryonic stem cell models of Huntington disease. Reprod Biomed Online. 2009;19(1):106–113. doi: 10.1016/s1472-6483(10)60053-3. [DOI] [PubMed] [Google Scholar]
- 12.Bradley CK, Scott HA, Chami O, Peura TT, Dumevska B, Schmidt U, et al. Derivation of Huntington’s disease-affected human embryonic stem cell lines. Stem Cells Dev. 20(3):495–502. doi: 10.1089/scd.2010.0120. [DOI] [PubMed] [Google Scholar]
- 13.Niclis JC, Pinar A, Haynes JM, Alsanie W, Jenny R, Dottori M, et al. Characterization of forebrain neurons derived from late-onset Huntington’s disease human embryonic stem cell lines. Front Cell Neurosci. 7:37. doi: 10.3389/fncel.2013.00037. PMCID: 3617399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu B, Palacino J. A novel human embryonic stem cell-derived Huntington’s disease neuronal model exhibits mutant huntingtin (mHTT) aggregates and soluble mHTT-dependent neurodegeneration. FASEB J. 27(5):1820–1829. doi: 10.1096/fj.12-219220. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 16.Paull D, Sevilla A, Zhou H, Hahn AK, Kim H, Napolitano C, et al. Automated, high-throughput derivation, characterization and differentiation of induced pluripotent stem cells. Nature methods. 2015;12(9):885–892. doi: 10.1038/nmeth.3507. [DOI] [PubMed] [Google Scholar]
- 17.Kang X, Yu Q, Huang Y, Song B, Chen Y, Gao X, et al. Effects of Integrating and Non-Integrating Reprogramming Methods on Copy Number Variation and Genomic Stability of Human Induced Pluripotent Stem Cells. PLoS One. 2015;10(7):e0131128. doi: 10.1371/journal.pone.0131128. PMCID: 4488894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woltjen K, Kim SI, Nagy A. The piggyBac Transposon as a Platform Technology for Somatic Cell Reprogramming Studies in Mouse. Methods Mol Biol. 2015 doi: 10.1007/7651_2015_274. [DOI] [PubMed] [Google Scholar]
- 19.Drozd AM, Walczak MP, Piaskowski S, Stoczynska-Fidelus E, Rieske P, Grzela DP. Generation of human iPSCs from cells of fibroblastic and epithelial origin by means of the oriP/EBNA-1 episomal reprogramming system. Stem cell research & therapy. 2015;6:122. doi: 10.1186/s13287-015-0112-3. PMCID: 4515927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle MEPRI, et al. Highly efficient Cas9-mediated transcriptional programming. Nature methods. 2015;12(4):326–328. doi: 10.1038/nmeth.3312. PMCID: 4393883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caxaria S, Arthold S, Nathwani AC, Goh PA. Generation of Integration-Free Patient Specific iPS Cells Using Episomal Plasmids Under Feeder Free Conditions. Methods Mol Biol. 2015 doi: 10.1007/7651_2015_204. [DOI] [PubMed] [Google Scholar]
- 22.Buganim Y, Markoulaki S, van Wietmarschen N, Hoke H, Wu T, Ganz K, et al. The developmental potential of iPSCs is greatly influenced by reprogramming factor selection. Cell Stem Cell. 2014;15(3):295–309. doi: 10.1016/j.stem.2014.07.003. PMCID: 4170792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng PH, Li CL, Chang YF, Tsai SJ, Lai YY, Chan AW, et al. miR-196a ameliorates phenotypes of Huntington disease in cell, transgenic mouse, and induced pluripotent stem cell models. Am J Hum Genet. 93(2):306–312. doi: 10.1016/j.ajhg.2013.05.025. PMCID: 3738820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Induced pluripotent stem cells from patients with Huntington’s disease show CAG-repeat-expansion-associated phenotypes. Cell Stem Cell. 11(2):264–278. doi: 10.1016/j.stem.2012.04.027. PMCID: 3804072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan AW, Cheng PH, Neumann A, Yang JJ. Reprogramming Huntington monkey skin cells into pluripotent stem cells. Cell Reprogram. 12(5):509–517. doi: 10.1089/cell.2010.0019. PMCID: 2993046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chae JI, Kim DW, Lee N, Jeon YJ, Jeon I, Kwon J, et al. Quantitative proteomic analysis of induced pluripotent stem cells derived from a human Huntington’s disease patient. Biochem J. 446(3):359–371. doi: 10.1042/BJ20111495. [DOI] [PubMed] [Google Scholar]
- 27.Jeon I, Lee N, Li JY, Park IH, Park KS, Moon J, et al. Neuronal properties, in vivo effects, and pathology of a Huntington’s disease patient-derived induced pluripotent stem cells. Stem Cells. 30(9):2054–2062. doi: 10.1002/stem.1135. [DOI] [PubMed] [Google Scholar]
- 28.Camnasio S, Delli Carri A, Lombardo A, Grad I, Mariotti C, Castucci A, et al. The first reported generation of several induced pluripotent stem cell lines from homozygous and heterozygous Huntington’s disease patients demonstrates mutation related enhanced lysosomal activity. Neurobiol Dis. 46(1):41–51. doi: 10.1016/j.nbd.2011.12.042. [DOI] [PubMed] [Google Scholar]
- 29.Juopperi TA, Kim WR, Chiang CH, Yu H, Margolis RL, Ross CA, et al. Astrocytes generated from patient induced pluripotent stem cells recapitulate features of Huntington’s disease patient cells. Mol Brain. 5:17. doi: 10.1186/1756-6606-5-17. PMCID: 3506453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang N, An MC, Montoro D, Ellerby LM. Characterization of Human Huntington’s Disease Cell Model from Induced Pluripotent Stem Cells. PLoS Curr. 2:RRN1193. doi: 10.1371/currents.RRN1193. PMCID: 2966296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134(5):877–886. doi: 10.1016/j.cell.2008.07.041. PMCID: 2633781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calamini B, Lo DC, Kaltenbach LS. Experimental models for identifying modifiers of polyglutamine-induced aggregation and neurodegeneration. Neurotherapeutics. 10(3):400–415. doi: 10.1007/s13311-013-0195-4. PMCID: 3701774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.An WF, Tolliday NJ. Introduction: cell-based assays for high-throughput screening. Methods Mol Biol. 2009;486:1–12. doi: 10.1007/978-1-60327-545-3_1. [DOI] [PubMed] [Google Scholar]
- 34.Bard J, Wall MD, Lazari O, Arjomand J, Munoz-Sanjuan I. Advances in huntington disease drug discovery: novel approaches to model disease phenotypes. J Biomol Screen. 19(2):191–204. doi: 10.1177/1087057113510320. [DOI] [PubMed] [Google Scholar]
- 35.Charbord J, Poydenot P, Bonnefond C, Feyeux M, Casagrande F, Brinon B, et al. High throughput screening for inhibitors of REST in neural derivatives of human embryonic stem cells reveals a chemical compound that promotes expression of neuronal genes. Stem Cells. 31(9):1816–1828. doi: 10.1002/stem.1430. [DOI] [PubMed] [Google Scholar]
- 36.Mattis VB, Tom C, Akimov S, Saeedian J, Ostergaard ME, Southwell AL, et al. HD iPSC-derived neural progenitors accumulate in culture and are susceptible to BDNF withdrawal due to glutamate toxicity. Hum Mol Genet. 24(11):3257–3271. doi: 10.1093/hmg/ddv080. PMCID: 4424959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.An MC, O’Brien RN, Zhang N, Patra BN, De La Cruz M, Ray A, et al. Polyglutamine Disease Modeling: Epitope Based Screen for Homologous Recombination using CRISPR/Cas9 System. PLoS Curr. 6 doi: 10.1371/currents.hd.0242d2e7ad72225efa72f6964589369a. PMCID: 3994193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carter RL, Chen Y, Kunkanjanawan T, Xu Y, Moran SP, Putkhao K, et al. Reversal of cellular phenotypes in neural cells derived from Huntington’s disease monkey-induced pluripotent stem cells. Stem Cell Reports. 3(4):585–593. doi: 10.1016/j.stemcr.2014.07.011. PMCID: 4223707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Priest BT, Erdemli G. Phenotypic screening in the 21st century. Front Pharmacol. 5:264. doi: 10.3389/fphar.2014.00264. PMCID: 4249253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swinney DC. The value of translational biomarkers to phenotypic assays. Front Pharmacol. 5:171. doi: 10.3389/fphar.2014.00171. PMCID: 4097030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zuccato C, Valenza M, Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol Rev. 90(3):905–981. doi: 10.1152/physrev.00041.2009. [DOI] [PubMed] [Google Scholar]
- 42.Yamamoto A, Lucas JJ, Hen R. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington’s disease. Cell. 2000;101(1):57–66. doi: 10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]
- 43.Schulte J, Sepp KJ, Wu C, Hong P, Littleton JT. High-content chemical and RNAi screens for suppressors of neurotoxicity in a Huntington’s disease model. PLoS One. 6(8):e23841. doi: 10.1371/journal.pone.0023841. PMCID: 3166080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoffstrom BG, Kaplan A, Letso R, Schmid RS, Turmel GJ, Lo DC, et al. Inhibitors of protein disulfide isomerase suppress apoptosis induced by misfolded proteins. Nat Chem Biol. 6(12):900–906. doi: 10.1038/nchembio.467. PMCID: 3018711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X, Smith DL, Meriin AB, Engemann S, Russel DE, Roark M, et al. A potent small molecule inhibits polyglutamine aggregation in Huntington’s disease neurons and suppresses neurodegeneration in vivo. Proc Natl Acad Sci U S A. 2005;102(3):892–897. doi: 10.1073/pnas.0408936102. PMCID: 545525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heiser V, Engemann S, Brocker W, Dunkel I, Boeddrich A, Waelter S, et al. Identification of benzothiazoles as potential polyglutamine aggregation inhibitors of Huntington’s disease by using an automated filter retardation assay. Proc Natl Acad Sci U S A. 2002;(99 Suppl 4):16400–16406. doi: 10.1073/pnas.182426599. PMCID: 139900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J, Gines S, MacDonald ME, Gusella JF. Reversal of a full-length mutant huntingtin neuronal cell phenotype by chemical inhibitors of polyglutamine-mediated aggregation. BMC Neurosci. 2005;6:1. doi: 10.1186/1471-2202-6-1. PMCID: 548150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ehrnhoefer DE, Duennwald M, Markovic P, Wacker JL, Engemann S, Roark M, et al. Green tea (−)-epigallocatechin-gallate modulates early events in huntingtin misfolding and reduces toxicity in Huntington’s disease models. Hum Mol Genet. 2006;15(18):2743–2751. doi: 10.1093/hmg/ddl210. [DOI] [PubMed] [Google Scholar]
- 49.Saudou F, Finkbeiner S, Devys D, Greenberg ME. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95(1):55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 50.Kuemmerle S, Gutekunst CA, Klein AM, Li XJ, Li SH, Beal MF, et al. Huntington aggregates may not predict neuronal death in Huntington’s disease. Ann Neurol. 1999;46(6):842–849. [PubMed] [Google Scholar]
- 51.Baldo B, Weiss A, Parker CN, Bibel M, Paganetti P, Kaupmann K. A screen for enhancers of clearance identifies huntingtin as a heat shock protein 90 (Hsp90) client protein. J Biol Chem. 287(2):1406–1414. doi: 10.1074/jbc.M111.294801. PMCID: 3256905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paganetti P, Weiss A, Trapp M, Hammerl I, Bleckmann D, Bodner RA, et al. Development of a method for the high-throughput quantification of cellular proteins. Chembiochem. 2009;10(10):1678–1688. doi: 10.1002/cbic.200900131. [DOI] [PubMed] [Google Scholar]
- 53.Miller JP, Holcomb J, Al-Ramahi I, de Haro M, Gafni J, Zhang N, et al. Matrix metalloproteinases are modifiers of huntingtin proteolysis and toxicity in Huntington’s disease. Neuron. 67(2):199–212. doi: 10.1016/j.neuron.2010.06.021. PMCID: 3098887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang C, Feigin A. Monitoring Huntington’s disease progression through preclinical and early stages. Neurodegener Dis Manag. 2(4):421–435. doi: 10.2217/nmt.12.34. PMCID: 3519443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang N, Li B, Al-Ramahi I, Cong X, Held JM, Kim E, et al. Inhibition of lipid signaling enzyme diacylglycerol kinase epsilon attenuates mutant huntingtin toxicity. J Biol Chem. 287(25):21204–21213. doi: 10.1074/jbc.M111.321661. PMCID: 3375542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller JP, Yates BE, Al-Ramahi I, Berman AE, Sanhueza M, Kim E, et al. A genome-scale RNA-interference screen identifies RRAS signaling as a pathologic feature of Huntington’s disease. PLoS Genet. 8(11):e1003042. doi: 10.1371/journal.pgen.1003042. PMCID: 3510027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sarantos MR, Papanikolaou T, Ellerby LM, Hughes RE. Pizotifen Activates ERK and Provides Neuroprotection in vitro and in vivo in Models of Huntington’s Disease. J Huntingtons Dis. 1(2):195–210. doi: 10.3233/JHD-120033. PMCID: 3564659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martin DD, Ladha S, Ehrnhoefer DE, Hayden MR. Autophagy in Huntington disease and huntingtin in autophagy. Trends Neurosci. 38(1):26–35. doi: 10.1016/j.tins.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 59.Cortes CJ, La Spada AR. The many faces of autophagy dysfunction in Huntington’s disease: from mechanism to therapy. Drug Discov Today. 19(7):963–971. doi: 10.1016/j.drudis.2014.02.014. PMCID: 4096219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36(6):585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 61.Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005;17(6):596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 62.Sarkar S, Perlstein EO, Imarisio S, Pineau S, Cordenier A, Maglathlin RL, et al. Small molecules enhance autophagy and reduce toxicity in Huntington’s disease models. Nat Chem Biol. 2007;3(6):331–338. doi: 10.1038/nchembio883. PMCID: 2635561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang L, Yu J, Pan H, Hu P, Hao Y, Cai W, et al. Small molecule regulators of autophagy identified by an image-based high-throughput screen. Proc Natl Acad Sci U S A. 2007;104(48):19023–19028. doi: 10.1073/pnas.0709695104. PMCID: 2141901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yamamoto A, Cremona ML, Rothman JE. Autophagy-mediated clearance of huntingtin aggregates triggered by the insulin-signaling pathway. J Cell Biol. 2006;172(5):719–731. doi: 10.1083/jcb.200510065. PMCID: 2063704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Williams A, Sarkar S, Cuddon P, Ttofi EK, Saiki S, Siddiqi FH, et al. Novel targets for Huntington’s disease in an mTOR-independent autophagy pathway. Nat Chem Biol. 2008;4(5):295–305. doi: 10.1038/nchembio.79. PMCID: 2635566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.An MC, Zhang N, Scott G, Montoro D, Wittkop T, Mooney S, et al. Genetic correction of Huntington’s disease phenotypes in induced pluripotent stem cells. Cell Stem Cell. 11(2):253–263. doi: 10.1016/j.stem.2012.04.026. PMCID: 3608272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chiu FL, Lin JT, Chuang CY, Chien T, Chen CM, Chen KH, et al. Elucidating the role of the A2A adenosine receptor in neurodegeneration using neurons derived from Huntington’s disease iPSCs. Hum Mol Genet. 2015 doi: 10.1093/hmg/ddv318. [DOI] [PubMed] [Google Scholar]
- 68.Tidball AM, Bryan MR, Uhouse MA, Kumar KK, Aboud AA, Feist JE, et al. A novel manganese-dependent ATM-p53 signaling pathway is selectively impaired in patient-based neuroprogenitor and murine striatal models of Huntington’s disease. Hum Mol Genet. 2015;24(7):1929–1944. doi: 10.1093/hmg/ddu609. PMCID: 4355025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. [cited]; Available from: < http://www.cirm.ca.gov/our-progress/awards/use-human-ipsc-derived-neurons-huntington%E2%80%99s–disease-patients-develop-novel%3E.
- 70.Szlachcic WJ, Switonski PM, Krzyzosiak WJ, Figlerowicz M, Figiel M. Huntington disease iPSCs show early molecular changes in intracellular signaling, the expression of oxidative stress proteins and the p53 pathway. Disease models & mechanisms. 2015 doi: 10.1242/dmm.019406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Deacon TW, Pakzaban P, Isacson O. The lateral ganglionic eminence is the origin of cells committed to striatal phenotypes: neural transplantation and developmental evidence. Brain Res. 1994;668(1–2):211–219. doi: 10.1016/0006-8993(94)90526-6. [DOI] [PubMed] [Google Scholar]
- 72.Olsson M, Bjorklund A, Campbell K. Early specification of striatal projection neurons and interneuronal subtypes in the lateral and medial ganglionic eminence. Neuroscience. 1998;84(3):867–876. doi: 10.1016/s0306-4522(97)00532-0. [DOI] [PubMed] [Google Scholar]
- 73.Echelard Y, Epstein DJ, St-Jacques B, Shen L, Mohler J, McMahon JA, et al. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell. 1993;75(7):1417–1430. doi: 10.1016/0092-8674(93)90627-3. [DOI] [PubMed] [Google Scholar]
- 74.Ericson J, Muhr J, Placzek M, Lints T, Jessell TM, Edlund T. Sonic hedgehog induces the differentiation of ventral forebrain neurons: a common signal for ventral patterning within the neural tube. Cell. 1995;81(5):747–756. doi: 10.1016/0092-8674(95)90536-7. [DOI] [PubMed] [Google Scholar]
- 75.Kohtz JD, Baker DP, Corte G, Fishell G. Regionalization within the mammalian telencephalon is mediated by changes in responsiveness to Sonic Hedgehog. Development. 1998;125(24):5079–5089. doi: 10.1242/dev.125.24.5079. [DOI] [PubMed] [Google Scholar]
- 76.Gunhaga L, Marklund M, Sjodal M, Hsieh JC, Jessell TM, Edlund T. Specification of dorsal telencephalic character by sequential Wnt and FGF signaling. Nat Neurosci. 2003;6(7):701–707. doi: 10.1038/nn1068. [DOI] [PubMed] [Google Scholar]
- 77.Backman M, Machon O, Mygland L, van den Bout CJ, Zhong W, Taketo MM, et al. Effects of canonical Wnt signaling on dorso-ventral specification of the mouse telencephalon. Dev Biol. 2005;279(1):155–168. doi: 10.1016/j.ydbio.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 78.Aubry L, Bugi A, Lefort N, Rousseau F, Peschanski M, Perrier AL. Striatal progenitors derived from human ES cells mature into DARPP32 neurons in vitro and in quinolinic acid-lesioned rats. Proc Natl Acad Sci U S A. 2008;105(43):16707–16712. doi: 10.1073/pnas.0808488105. PMCID: 2575484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li XJ, Zhang X, Johnson MA, Wang ZB, Lavaute T, Zhang SC. Coordination of sonic hedgehog and Wnt signaling determines ventral and dorsal telencephalic neuron types from human embryonic stem cells. Development. 2009;136(23):4055–4063. doi: 10.1242/dev.036624. PMCID: 2778748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Delli Carri A, Onorati M, Lelos MJ, Castiglioni V, Faedo A, Menon R, et al. Developmentally coordinated extrinsic signals drive human pluripotent stem cell differentiation toward authentic DARPP-32+ medium-sized spiny neurons. Development. 140(2):301–312. doi: 10.1242/dev.084608. [DOI] [PubMed] [Google Scholar]
- 81.Delli Carri A, Onorati M, Castiglioni V, Faedo A, Camnasio S, Toselli M, et al. Human pluripotent stem cell differentiation into authentic striatal projection neurons. Stem Cell Rev. 9(4):461–474. doi: 10.1007/s12015-013-9441-8. [DOI] [PubMed] [Google Scholar]
- 82.Lin L, Yuan J, Sander B, Golas MM. In Vitro Differentiation of Human Neural Progenitor Cells Into Striatal GABAergic Neurons. Stem Cells Transl Med. doi: 10.5966/sctm.2014-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Arber C, Precious SV, Cambray S, Risner-Janiczek JR, Kelly C, Noakes Z, et al. Activin A directs striatal projection neuron differentiation of human pluripotent stem cells. Development. 142(7):1375–1386. doi: 10.1242/dev.117093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.El-Akabawy G, Medina LM, Jeffries A, Price J, Modo M. Purmorphamine increases DARPP-32 differentiation in human striatal neural stem cells through the Hedgehog pathway. Stem Cells Dev. 2011;20(11):1873–1887. doi: 10.1089/scd.2010.0282. [DOI] [PubMed] [Google Scholar]
- 85.Ma L, Hu B, Liu Y, Vermilyea SC, Liu H, Gao L, et al. Human embryonic stem cell-derived GABA neurons correct locomotion deficits in quinolinic acid-lesioned mice. Cell Stem Cell. 10(4):455–464. doi: 10.1016/j.stem.2012.01.021. PMCID: 3322292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Anderson SA, Qiu M, Bulfone A, Eisenstat DD, Meneses J, Pedersen R, et al. Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late born striatal neurons. Neuron. 1997;19(1):27–37. doi: 10.1016/s0896-6273(00)80345-1. [DOI] [PubMed] [Google Scholar]
- 87.Long JE, Swan C, Liang WS, Cobos I, Potter GB, Rubenstein JL. Dlx1&2 and Mash1 transcription factors control striatal patterning and differentiation through parallel and overlapping pathways. J Comp Neurol. 2009;512(4):556–572. doi: 10.1002/cne.21854. PMCID: 2761428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Casarosa S, Fode C, Guillemot F. Mash1 regulates neurogenesis in the ventral telencephalon. Development. 1999;126(3):525–534. doi: 10.1242/dev.126.3.525. [DOI] [PubMed] [Google Scholar]
- 89.Fode C, Ma Q, Casarosa S, Ang SL, Anderson DJ, Guillemot F. A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev. 2000;14(1):67–80. PMCID: 316337. [PMC free article] [PubMed] [Google Scholar]
- 90.Szucsik JC, Witte DP, Li H, Pixley SK, Small KM, Potter SS. Altered forebrain and hindbrain development in mice mutant for the Gsh-2 homeobox gene. Dev Biol. 1997;191(2):230–242. doi: 10.1006/dbio.1997.8733. [DOI] [PubMed] [Google Scholar]
- 91.Corbin JG, Gaiano N, Machold RP, Langston A, Fishell G. The Gsh2 homeodomain gene controls multiple aspects of telencephalic development. Development. 2000;127(23):5007–5020. doi: 10.1242/dev.127.23.5007. [DOI] [PubMed] [Google Scholar]
- 92.Arlotta P, Molyneaux BJ, Jabaudon D, Yoshida Y, Macklis JD. Ctip2 controls the differentiation of medium spiny neurons and the establishment of the cellular architecture of the striatum. J Neurosci. 2008;28(3):622–632. doi: 10.1523/JNEUROSCI.2986-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tamura S, Morikawa Y, Iwanishi H, Hisaoka T, Senba E. Foxp1 gene expression in projection neurons of the mouse striatum. Neuroscience. 2004;124(2):261–267. doi: 10.1016/j.neuroscience.2003.11.036. [DOI] [PubMed] [Google Scholar]
- 94.Victor MB, Richner M, Hermanstyne TO, Ransdell JL, Sobieski C, Deng PY, et al. Generation of human striatal neurons by microRNA-dependent direct conversion of fibroblasts. Neuron. 84(2):311–323. doi: 10.1016/j.neuron.2014.10.016. PMCID: 4223654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cudkowicz M, Kowall NW. Degeneration of pyramidal projection neurons in Huntington’s disease cortex. Ann Neurol. 1990;27(2):200–204. doi: 10.1002/ana.410270217. [DOI] [PubMed] [Google Scholar]
- 96.Hedreen JC, Peyser CE, Folstein SE, Ross CA. Neuronal loss in layers V and VI of cerebral cortex in Huntington’s disease. Neurosci Lett. 1991;133(2):257–261. doi: 10.1016/0304-3940(91)90583-f. [DOI] [PubMed] [Google Scholar]
- 97.Cepeda C, Wu N, Andre VM, Cummings DM, Levine MS. The corticostriatal pathway in Huntington’s disease. Prog Neurobiol. 2007;81(5–6):253–271. doi: 10.1016/j.pneurobio.2006.11.001. PMCID: 1913635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shi Y, Kirwan P, Livesey FJ. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat Protoc. 7(10):1836–1846. doi: 10.1038/nprot.2012.116. [DOI] [PubMed] [Google Scholar]
- 99.Espuny-Camacho I, Michelsen KA, Gall D, Linaro D, Hasche A, Bonnefont J, et al. Pyramidal neurons derived from human pluripotent stem cells integrate efficiently into mouse brain circuits in vivo. Neuron. 77(3):440–456. doi: 10.1016/j.neuron.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 100.Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 78(5):785–798. doi: 10.1016/j.neuron.2013.05.029. PMCID: 3751803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kole AJ, Annis RP, Deshmukh M. Mature neurons: equipped for survival. Cell Death Dis. 4:e689. doi: 10.1038/cddis.2013.220. PMCID: 3702294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mattson MP, Magnus T. Ageing and neuronal vulnerability. Nat Rev Neurosci. 2006;7(4):278–294. doi: 10.1038/nrn1886. PMCID: 3710114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Campos PB, Paulsen BS, Rehen SK. Accelerating neuronal aging in in vitro model brain disorders: a focus on reactive oxygen species. Front Aging Neurosci. 6:292. doi: 10.3389/fnagi.2014.00292. PMCID: 4209886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Singhrao SK, Thomas P, Wood JD, MacMillan JC, Neal JW, Harper PS, et al. Huntingtin protein colocalizes with lesions of neurodegenerative diseases: An investigation in Huntington’s, Alzheimer’s, and Pick’s diseases. Exp Neurol. 1998;150(2):213–222. doi: 10.1006/exnr.1998.6778. [DOI] [PubMed] [Google Scholar]
- 105.Faideau M, Kim J, Cormier K, Gilmore R, Welch M, Auregan G, et al. In vivo expression of polyglutamine-expanded huntingtin by mouse striatal astrocytes impairs glutamate transport: a correlation with Huntington’s disease subjects. Hum Mol Genet. 19(15):3053–3067. doi: 10.1093/hmg/ddq212. PMCID: 2901144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shin JY, Fang ZH, Yu ZX, Wang CE, Li SH, Li XJ. Expression of mutant huntingtin in glial cells contributes to neuronal excitotoxicity. J Cell Biol. 2005;171(6):1001–1012. doi: 10.1083/jcb.200508072. PMCID: 2171327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tong X, Ao Y, Faas GC, Nwaobi SE, Xu J, Haustein MD, et al. Astrocyte Kir4.1 ion channel deficits contribute to neuronal dysfunction in Huntington’s disease model mice. Nat Neurosci. 17(5):694–703. doi: 10.1038/nn.3691. PMCID: 4064471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shaltouki A, Peng J, Liu Q, Rao MS, Zeng X. Efficient generation of astrocytes from human pluripotent stem cells in defined conditions. Stem Cells. 31(5):941–952. doi: 10.1002/stem.1334. [DOI] [PubMed] [Google Scholar]
- 109.Emdad L, D’Souza SL, Kothari HP, Qadeer ZA, Germano IM. Efficient differentiation of human embryonic and induced pluripotent stem cells into functional astrocytes. Stem Cells Dev. 21(3):404–410. doi: 10.1089/scd.2010.0560. [DOI] [PubMed] [Google Scholar]
- 110.Serio A, Bilican B, Barmada SJ, Ando DM, Zhao C, Siller R, et al. Astrocyte pathology and the absence of non-cell autonomy in an induced pluripotent stem cell model of TDP-43 proteinopathy. Proc Natl Acad Sci U S A. 110(12):4697–4702. doi: 10.1073/pnas.1300398110. PMCID: 3607024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tai YF, Pavese N, Gerhard A, Tabrizi SJ, Barker RA, Brooks DJ, et al. Microglial activation in presymptomatic Huntington’s disease gene carriers. Brain. 2007;130(Pt 7):1759–1766. doi: 10.1093/brain/awm044. [DOI] [PubMed] [Google Scholar]
- 112.Pavese N, Gerhard A, Tai YF, Ho AK, Turkheimer F, Barker RA, et al. Microglial activation correlates with severity in Huntington disease: a clinical and PET study. Neurology. 2006;66(11):1638–1643. doi: 10.1212/01.wnl.0000222734.56412.17. [DOI] [PubMed] [Google Scholar]
- 113.Kwan W, Trager U, Davalos D, Chou A, Bouchard J, Andre R, et al. Mutant huntingtin impairs immune cell migration in Huntington disease. J Clin Invest. 122(12):4737–4747. doi: 10.1172/JCI64484. PMCID: 3533551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bartzokis G, Lu PH, Tishler TA, Fong SM, Oluwadara B, Finn JP, et al. Myelin breakdown and iron changes in Huntington’s disease: pathogenesis and treatment implications. Neurochem Res. 2007;32(10):1655–1664. doi: 10.1007/s11064-007-9352-7. [DOI] [PubMed] [Google Scholar]
- 115.Douvaras P, Wang J, Zimmer M, Hanchuk S, O’Bara MA, Sadiq S, et al. Efficient generation of myelinating oligodendrocytes from primary progressive multiple sclerosis patients by induced pluripotent stem cells. Stem Cell Reports. 2014;3(2):250–259. doi: 10.1016/j.stemcr.2014.06.012. PMCID: 4176529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang S, Bates J, Li X, Schanz S, Chandler-Militello D, Levine C, et al. Human iPSC-derived oligodendrocyte progenitor cells can myelinate and rescue a mouse model of congenital hypomyelination. Cell Stem Cell. 2013;12(2):252–264. doi: 10.1016/j.stem.2012.12.002. PMCID: 3700553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nagai M, Re DB, Nagata T, Chalazonitis A, Jessell TM, Wichterle H, et al. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat Neurosci. 2007;10(5):615–622. doi: 10.1038/nn1876. PMCID: 3799799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc. 9(10):2329–2340. doi: 10.1038/nprot.2014.158. PMCID: 4160653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kadoshima T, Sakaguchi H, Nakano T, Soen M, Ando S, Eiraku M, et al. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc Natl Acad Sci U S A. 110(50):20284–20289. doi: 10.1073/pnas.1315710110. PMCID: 3864329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, et al. Cerebral organoids model human brain development and microcephaly. Nature. 501(7467):373–379. doi: 10.1038/nature12517. PMCID: 3817409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Segal DJ. Bacteria herald a new era of gene editing. eLife. 2013;2:e00563. doi: 10.7554/eLife.00563. PMCID: 3557904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Segal DJ, Meckler JF. Genome engineering at the dawn of the golden age. Annual review of genomics and human genetics. 2013;14:135–158. doi: 10.1146/annurev-genom-091212-153435. [DOI] [PubMed] [Google Scholar]
- 123.Bassett AR, Tibbit C, Ponting CP, Liu JL. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 System. Cell Rep. 2013;4:220–228. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wu X, Scott DA, Kriz AJ, Chiu AC, Hsu PD, Dadon DB, et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nature biotechnology. 2014;32(7):670–676. doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nature biotechnology. 2014;32(7):677–683. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- 126.Wang H. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhou J, Shen B, Zhang W, Wang J, Yang J, Chen L, et al. One-step generation of different immunodeficient mice with multiple gene modifications by CRISPR/Cas9 mediated genome engineering. The international journal of biochemistry & cell biology. 2014;46:49–55. doi: 10.1016/j.biocel.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 128.Ye L, Wang J, Beyer AI, Teque F, Cradick TJ, Qi Z, et al. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Delta32 mutation confers resistance to HIV infection. Proc Natl Acad Sci U S A. 2014;111(26):9591–9596. doi: 10.1073/pnas.1407473111. PMCID: 4084478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. PMCID: 3712628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Metzger S, Rong J, Nguyen HP, Cape A, Tomiuk J, Soehn AS, et al. Huntingtin-associated protein-1 is a modifier of the age-at-onset of Huntington’s disease. Hum Mol Genet. 2008;17(8):1137–1146. doi: 10.1093/hmg/ddn003. [DOI] [PubMed] [Google Scholar]
- 131.Gusella JF, MacDonald ME. Huntington’s disease: the case for genetic modifiers. Genome Med. 2009;1(8):80. doi: 10.1186/gm80. PMCID: 2768966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Identification of Genetic Factors that Modify Clinical Onset of Huntington’s Disease. Cell. 2015;162(3):516–526. doi: 10.1016/j.cell.2015.07.003. PMCID: 4524551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 343(6166):84–87. doi: 10.1126/science.1247005. PMCID: 4089965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Huang HS, Allen JA, Mabb AM, King IF, Miriyala J, Taylor-Blake B, et al. Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature. 481(7380):185–189. doi: 10.1038/nature10726. PMCID: 3257422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Egawa N, Kitaoka S, Tsukita K, Naitoh M, Takahashi K, Yamamoto T, et al. Drug screening for ALS using patient-specific induced pluripotent stem cells. Sci Transl Med. 4(145):145ra04. doi: 10.1126/scitranslmed.3004052. [DOI] [PubMed] [Google Scholar]
- 136.Lee G, Ramirez CN, Kim H, Zeltner N, Liu B, Radu C, et al. Large-scale screening using familial dysautonomia induced pluripotent stem cells identifies compounds that rescue IKBKAP expression. Nature biotechnology. 30(12):1244–1248. doi: 10.1038/nbt.2435. PMCID: 3711177. [DOI] [PMC free article] [PubMed] [Google Scholar]