Abstract
Fungal plant pathogens are major threats to food security worldwide. Sclerotinia sclerotiorum and Botrytis cinerea are closely related Ascomycete plant pathogens causing mold diseases on hundreds of plant species. There is no genetic source of complete plant resistance to these broad host range pathogens known to date. Instead, natural plant populations show a continuum of resistance levels controlled by multiple genes, a phenotype designated as quantitative disease resistance. Little is known about the molecular mechanisms controlling the interaction between plants and S. sclerotiorum and B. cinerea but significant advances were made on this topic in the last years. This minireview highlights a selection of nine themes that emerged in recent research reports on the molecular bases of plant-S. sclerotiorum and plant-B. cinerea interactions. On the fungal side, this includes progress on understanding the role of oxalic acid, on the study of fungal small secreted proteins. Next, we discuss the exchanges of small RNA between organisms and the control of cell death in plant and fungi during pathogenic interactions. Finally on the plant side, we highlight defense priming by mechanical signals, the characterization of plant Receptor-like proteins and the hormone abscisic acid in the response to B. cinerea and S. sclerotiorum, the role of plant general transcription machinery and plant small bioactive peptides. These represent nine trends we selected as remarkable in our understanding of fungal molecules causing disease and plant mechanisms associated with disease resistance to two devastating broad host range fungi.
Keywords: fungal pathogen, necrotroph, broad host range, quantitative disease resistance, virulence
Introduction
A majority of studies on plant interactions with fungal pathogens over the last years have focused on specialized host–pathogen interactions. For instance the powdery mildew fungus Blumeria graminis, the cereal rust fungi of the Puccinia spp., and the corn smut fungus Ustilago maydis are among the most studied fungal pathogens and are obligate biotrophic pathogens restricted to a single host genus (Dean et al., 2012). Such interactions only represents a fraction of plant-fungal pathogen interactions encountered in nature and a number of broad host range fungal pathogens also are major threats for food security (Barrett et al., 2009; Dean et al., 2012). Understanding how broad host range pathogens successfully infect multiple plant lineages is a major challenge in plant pathology (Dong et al., 2015).
Among Leotiomycete, the gray mold fungus Botrytis cinerea and the white mold fungus Sclerotinia sclerotiorum stand out for having a remarkably broad host range, encompassing over 200 species. Each of these pathogens causes yearly several 100 millions of US dollars crop losses worldwide (Bolton et al., 2006; Dean et al., 2012). They are considered as typical necrotrophs, secreting an arsenal of cell wall-degrading enzymes, and toxins to kill host cells and derive energy. Host plants typically exhibit quantitative disease resistance (QDR) to B. cinerea and S. sclerotiorum, leading to a reduction rather than absence of disease (Roux et al., 2014). How these broad host range fungal pathogens cause disease and what are the genetic bases of plant QDR is still poorly understood.
In recent years, remarkable progress has been achieved in the characterization of fungal virulence factors and the dissection of plant response mechanisms. In this minireview, we chose to report on nine advances specifically related to pathosystems involving B. cinerea or S. sclerotiorum, concerning either the molecular bases of fungal virulence or plant QDR (summarized in Figure 1). The nine points presented hereafter are not meant to represent a complete overview of our current knowledge, and a number of significant discoveries could not be covered in this article. We selected nine trends based on convergent findings from multiple studies and as a source of inspiration for future studies on plant interactions with broad host range fungal pathogens.
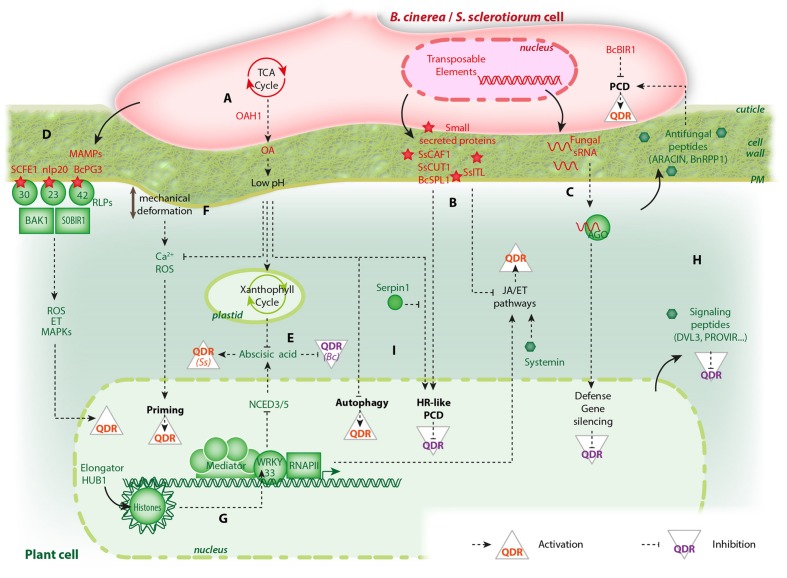
FIGURE 1.
An overview of the molecular players involved in Botrytis cinerea/Sclerotinia sclerotiorum interactions with plants and their effect on quantitative disease resistance (QDR). Only pathways discussed in this review are shown, some elements were omitted for clarity. Fungal molecules are shown in red, plant molecules in green. (A) Effects of oxalic acid (OA) biosynthesis and secretion by fungi. (B) Small proteins secreted by fungi can activate hypersensitive response (HR)-like programmed cell death (PCD) or suppress jasmonic acid (JA) and ethylene (ET) signaling pathways to suppress QDR. (C) Fungal small RNAs hijack plant argonaute (AGO) proteins to suppress QDR. (D) Perception of microbe associated molecular patterns (MAMPs) such as SCFE1, nlp20, and BcPG3 by receptor-like proteins (RLPs) activate QDR. (E) The plant hormone abscisic acid can either activate QDR against S. sclerotiorum (Ss) or suppress QDR against B. cinerea (Bc). (F) Mechanical deformation caused by appressorium formation and fungal colonization of plant tissues prime plant cells for QDR through calcium and reactive oxygen species (ROS). (G) The plant general transcription machinery involves multiprotein complexes such as Elongator and Mediator that recruit the RNA polymerase II (RNAPII) to modulate gene expression upon fungal challenge. Some histone post-translational modifications are epigenetic marks altered after inoculation that regulate the activity of plant general transcription factors and control QDR. (H) Plant small bioactive peptides (green hexagons) have contrasted effects on QDR. (I) PCD in plant cells can either have a positive or negative effect on QDR depending on the type of cell death program activated.
Putting the Role of Fungal Oxalate Secretion to the Acid Test
Oxalic acid (OA) is considered as a major virulence factor in species of the Sclerotiniaceae (Hegedus and Rimmer, 2005; Andrew et al., 2012). This central role of OA is further supported by the association of plant OA oxidase-related enzymes with disease resistance (Foster et al., 2012; Rietz et al., 2012). The roles of OA secretion in S. sclerotiorum virulence are still a matter of debate and likely include the suppression of plant defenses (Cessna et al., 2000; Williams et al., 2011), the induction of plant programmed cell death (PCD; Kim et al., 2008), the deregulation of guard cells function (Guimaraes and Stotz, 2004), and calcium detoxification (Heller and Witt-Geiges, 2013). Evidences for several of these conclusions were obtained using a UV-induced OA-deficient mutant of S. sclerotiorum (Godoy et al., 1990). Recently, Liang et al. (2015) generated disruptive mutants in the gene encoding oxaloacetate acetylhydrolase 1 (OAH1), an enzyme mediating OA biosynthesis (Joosten et al., 2008). This oah1 mutant was completely abolished in OA synthesis and impaired in pathogenicity. Several phenotypic differences were noted compared to the UV-induced OA-deficient mutants, prompting for a re-evaluation of OA function during host colonization (Liang et al., 2015). To this end, Xu et al. (2015) generated oah1 deletion mutants in another S. sclerotiorum strain and confirmed that their virulence varied with the pH of the host tissues. The virulence phenotype of oah1 mutants was restored by genetic complementation and partially restored by the exogenous acidification of host tissue (Liang et al., 2015; Xu et al., 2015), suggesting that low pH, rather than a specific acidic molecule, is required for S. sclerotiorum full virulence. Why acidification is mediated by OA and not another organic acid during S. sclerotiorum infection remains unclear. B. cinerea was proposed to rely primarily on the production of citrate and succinate during the colonization of sunflower. The production of different organic acids suggests that S. sclerotiorum and B. cinerea differ significantly in TCA cycle regulation (Billon-Grand et al., 2012). These findings call for investigations on metabolic organization in S. sclerotiorum and B. cinerea.
The Plenty and Hazy Fungal Small Secreted Proteins
Secondary metabolite toxins and cell wall-degrading enzymes are known to contribute to virulence of necrotrophic fungi (Choquer et al., 2007; Kabbage et al., 2015), but knowledge on the repertoires and mode of action of S. sclerotiorum and B. cinerea proteins secreted during host colonization remains limited. Mutant screens for pathogenicity defects in S. sclerotiorum have identified the secreted putative Ca2+-binding protein SsCAF1 and the secreted Cu/Zn superoxide dismutase SsSOD1 (Xu and Chen, 2013; Xiao et al., 2014). The analysis of genes down-regulated in a hypovirulent S. sclerotiorum strain identified the secreted integrin-like gene SsITL as required for full virulence (Zhu et al., 2013). Genomic and transcriptomic approaches have revealed S. sclerotiorum cutinase SsCUT1 (Zhang H. et al., 2014) and the secreted cyanovirin-N homolog SsCVNH (Lyu et al., 2015) to be associated with plant infection. Proteomic analyses of B. cinerea secretome and cell wall proteome led to the identification the cerato-platanin BcSPL1 required for full virulence (Espino et al., 2010; Frías et al., 2011, 2014). SsCAF1, SsCUT1, and BcSPL1 elicited cell death in plants whereas SsITL was proposed to suppress plant resistance pathways, illustrating the diverse activities of fungal small secreted proteins. Given that mutations in SsCAF1, SsITL, and SsCVNH caused morphological defects in vitro, it is unclear whether these genes are directly linked to host interaction. Since the release of genome sequences for S. sclerotiorum and B. cinerea (Amselem et al., 2011), bioinformatics analyses aimed at systematically identifying candidate proteins associated with virulence. Aguileta et al. (2012) identified 21 B. cinerea genes harboring signatures of positive selection, including 7 encoding predicted secreted proteins. Multicriterion analyses of S. sclerotiorum and B. cinerea secretomes highlighted over 400 secreted proteins including nearly 80 virulence factor candidates (Guyon et al., 2014; Heard et al., 2015). Specific patterns of amino acid usage and conformation may also serve as a filter to classify virulence factor candidates (Badet et al., 2015). Such bioinformatics approaches remain nevertheless challenging in fungi (Sperschneider et al., 2015). Functional analyses, improved genome annotation and extensive gene expression analyses should prove useful in understanding the role of small secreted proteins in B. cinerea and S. sclerotiorum virulence.
Host and Pathogen Cell Death Control
Pathogen recognition often triggers a hypersensitive response (HR), the rapid release of antimicrobial compounds and local PCD. The HR efficiently suppresses the growth of most biotrophic pathogens while facilitating plant colonization by B. cinerea and S. sclerotiorum (Govrin and Levine, 2000). The secretion of OA and cell death-eliciting proteins by B. cinerea and S. sclerotiorum were proposed to promote virulence by inducing host PCD (Kim et al., 2008; Frías et al., 2011). The molecular mechanisms by which B. cinerea and S. sclerotiorum manipulate host PCD remain, however, elusive. The protease inhibitor AtSerpin1 limits cell death induction by OA and plant colonization by B. cinerea and S. sclerotiorum by modulating the activity of the cysteine protease RD21 (Lampl et al., 2013). In addition, Arabidopsis mutants impaired in autophagy, a form of PCD involving the degradation of cytoplasmic components, were more susceptible to B. cinerea (Lai et al., 2011). Furthermore, S. sclerotiorum OA-deficient mutants trigger restricted autophagic cell death, suggesting that OA may suppress autophagy in host cells (Kabbage et al., 2013). In Arabidopsis acd5 mutant, ceramides were associated with autophagy and enhanced susceptibility to B. cinerea, but these two activities may be independent (Bi et al., 2014; Magnin-Robert et al., 2015). These findings indicate that certain mechanisms of host cell death may be favorable to necrotrophic fungi whereas others would be detrimental (Dickman and de Figueiredo, 2013). The control of PCD in fungal cells also plays a crucial role in pathogenicity. Massive fungal PCD was observed at the early stages of B. cinerea infection, and the anti-apoptotic protein BcBIR1 was found to positively contribute to B. cinerea virulence. These findings suggest that fungal PCD could be triggered by plant defense molecules, and that fungal anti-apoptotic machinery is required to prevent it (Shlezinger et al., 2011). This places the control of host and fungal cell death programs at the center of the arms race taking place during plant interactions with necrotrophic fungi.
Two-Ways Gene Silencing by Small RNAs
In eukaryotic cells, small RNAs (sRNAs) regulate a large number of biological processes, from development to immunity and pathogen virulence. These sRNAs trigger multiple RNA interference (RNAi) pathways ultimately leading to gene silencing. Using Arabidopsis and tomato plants infected by B. cinerea, Weiberg et al. (2013) showed that fungal sRNAs silence specific host immunity genes. Mutants of plant and fungal RNAi components showed reduced silencing of host immunity genes and reduced disease symptoms, respectively, (Weiberg et al., 2013). Together these results support the action of sRNAs across the species and kingdom barrier (cross-kingdom RNAi) in plant–fungi interactions, mediated by a yet unclear transfer mechanism (Sarkies and Miska, 2014; Weiberg et al., 2014, 2015). Several of these B. cinerea sRNA effectors originate from loci within Boty transposable elements (TEs), a family of mobile genetic elements associated with virulence in B. cinerea natural populations (Martinez et al., 2005; Weiberg et al., 2015). This suggests that TEs may contribute to the rapid evolution of sRNA effectors, similar to what has been observed for filamentous pathogen protein effectors (Raffaele et al., 2010; Raffaele and Kamoun, 2012). The genome of S. sclerotiorum experienced recent expansion of TEs (Amselem et al., 2011) and sRNA-producing loci have been experimentally identified in this fungus (Zhou et al., 2012). Nevertheless, whether S. sclerotiorum uses sRNAs as effectors and the extent to which they contribute to pathogenicity await analysis. Cross-kingdom RNAi has also been exploited to generate transgenic plants producing sRNAs that trigger fungal genes silencing (Nowara et al., 2010; Nunes and Dean, 2012; Koch et al., 2013). This strategy of host-induced gene silencing (HIGS) has been tested in model plants and crops to silence genes of various fungal and oomycete species, providing a promising approach to control diseases and study gene function in non-transformable pathogen species (Koch and Kogel, 2014; Yin et al., 2014). Whether HIGS occurs in natural plant-fungus interactions and whether it would provide an efficient way to control B. cinerea and S. sclerotiorum has not been reported yet.
Defense Priming by Mechanical Signals
Defense priming consists in establishing a physiological state in which plants are able to mount defense responses more rapidly or more efficiently (Conrath et al., 2006). Priming follows the perception of chemical and molecular signals linked to the presence of microbes interacting with plant cells, and also the perception of physical cues (Conrath, 2011). Plant cells can also perceive strains (mechanical deformation) caused by mechanical loads (Moulia et al., 2015). During their interaction with plants, and prior to plant tissue penetration or degradation, fungal pathogens develop important mechanical loads susceptible to emit mechanical signals (MS) and prime plant defense. Mechanical loads are due to the tremendous turgor pressure (up to 8 MPa) created by water in the vacuole of appressoria and fungal cell wall mechanical properties. This mechanical stress is generally sufficient to penetrate plant cells (Tariq and Jeffries, 1984; Bastmeyer et al., 2002; Sanati Nezhad and Geitmann, 2013). Mechanosensing occurs at the plant cell level and relies on the internal mechanical state of the cell (Coutand, 2010; Hamant, 2013; Monshausen and Haswell, 2013). Mechanosensing is involved in many plant core functions including seed development, morphogenesis, gravitropism, proprioception, and interaction with symbiotic microbes (Boudaoud, 2010; Hamant and Traas, 2010; Bastien et al., 2013; Jayaraman et al., 2014; Creff et al., 2015; Landrein et al., 2015). Recent studies demonstrated the link between mechanosensing and plant immune response to B. cinerea in Arabidopsis thaliana: plants submitted to MS exhibited higher resistance to fungal infection suggesting a priming effect operated by sterile mechanosensing (Chehab et al., 2012; Benikhlef et al., 2013). Relations between mechanosensing and immune response pathways are mediated by calcium (Chehab et al., 2011; Beneloujaephajri et al., 2013), reactive oxygen species (ROS; Chehab et al., 2012; Benikhlef et al., 2013) and are jasmonic acid (JA)-independent (Benikhlef et al., 2013). Future work should aim at addressing whether mechanosensing for fungal contact or penetration per se, in addition to PAMP perception, leads to enhanced plant immunity. Quantifying the MS intensity perceived by the plant will be required to this end. Quantitative biomechanical plant cell models (Barbacci et al., 2013; Ali et al., 2014) are required first milestones toward a better understanding of the molecular mechanisms underlying plant resistance priming by MS.
Perception of Fungi by Plant Receptor Like Proteins
Detection of microbe-associated molecular patterns (MAMPs) is an important part of the plant defense against pathogens (Boller and Felix, 2009). It relies on plasma membrane resident pattern recognition receptors (PRRs) able to perceive pathogen signatures in the apoplastic space and to activate a downstream signaling through their kinase domain (Monaghan and Zipfel, 2012). In plants, early detection of fungal pathogens occurs mainly through PRRs-mediated perception of chitin (Sanchez-Vallet et al., 2015). Peptide-mediated fungal perception is also becoming increasingly documented. S. sclerotiorum produces a protein elicitor (SCFE1) that triggers oxidative burst, ethylene production, mitogen-activated protein kinase activation and gene induction (Zhang et al., 2013). Natural variation among Arabidopsis accessions and mutants identified RLP30 as required for all SCFE1 responses. RLP30 encodes for a PRR devoid of a kinase domain, suggesting the involvement of co-regulators for intracellular signaling. In a similar approach Albert et al. (2015) screened for Arabidopsis mutants and natural accessions unresponsive to nlp20, a 20 amino acid conserved peptide derived from a class of necrosis inducing proteins found in bacteria, oomycetes and fungi (Albert et al., 2015). Upon nlp20 binding, the RLP23 receptor forms a ternary complex required for signaling with two other kinase-containing co-receptors, BAK1 and SOBIR1 (Albert et al., 2015). Finally, the Arabidopsis RLP42 gene is required for Arabidopsis responses to exogenous application of B. cinerea polygalacturonase 3 (Zhang L. et al., 2014). Given that RLP30, RLP42, and RLP23 are homologs, it is reasonable to hypothesize that all use BAK1 or SOBIR1 as co-receptors. Because SCFE1-responsive plants included Arabidopsis accessions with various levels of resistance, Zhang et al. (2013) proposed that redundant elicitor perception systems involve other PRRs in addition to RLP30. This is consistent with several perception and response mechanisms acting simultaneously to trigger QDR (Roux et al., 2014). The discoveries of RLP23, RLP42, and soybean RLPs identified through quantitative trait loci mapping strengthen this view (Zhang L. et al., 2014; Albert et al., 2015; Zhao et al., 2015). Pyramiding several PRRs into plants would likely yield increased and long-lived resistance to devastating pathogens.
Contrasted Impacts of Abscisic Acid on QDR
Plant hormones classically associated with resistance to necrotrophic pathogens are JA and ethylene (ET), whereas salicylic acid is associated with stimulation of resistance against biotrophic pathogens (Glazebrook, 2005). Abscisic acid (ABA) has roles in plant development and response to abiotic stress, and has also contrasted impact on plant diseases, depending notably on the pathogen infection strategy (Robert-Seilaniantz et al., 2011). ABA was shown to promote susceptibility to B. cinerea in tomato through alterations of the plant cuticle and C:N metabolism (Audenaert et al., 2002; Curvers et al., 2010; Seifi et al., 2013). In A. thaliana, Liu et al. (2015) revealed that loss of WRKY33 results in elevated ABA levels and high susceptibility to B. cinerea. WRKY33 limits ABA accumulation in B. cinerea-challenged plants by binding to the ABA biosynthesis genes NCED3 and NCED5 to suppress their expression. Consistently, the transmembrane receptor-like kinase AtLYK3 was proposed to act as a positive regulator of late responses to ABA and negative regulator of defenses to B. cinerea (Paparella et al., 2014). By contrast, inoculation of A. thaliana mutants revealed that ABA contributes to resistance to S. sclerotiorum, although it remains unclear whether ABA synthesis or perception is required for resistance (Guimaraes and Stotz, 2004; Perchepied et al., 2010). OA secreted by S. sclerotiorum was proposed to favor infection by inhibiting ABA-mediated stomatal closure (Guimaraes and Stotz, 2004). Zhou et al. (2015) showed that pH modulation by S. sclerotiorum correlates with increased synthesis of photoprotective compounds of the xanthophyll cycle that serve as precursors for ABA synthesis. Depletion in ABA precursors was suggested to account for reduced ABA levels in S. sclerotiorum-infected leaves and plant susceptibility (Zhou et al., 2015). Differences in the dynamics of OA secretion (Billon-Grand et al., 2012) may contribute to the contrasting impact of ABA on resistance toward S. sclerotiorum and B. cinerea. Furthermore, B. cinerea, but not S. sclerotiorum, synthesizes ABA (Siewers et al., 2004). Further studies will be required to fully understand how plants and fungi interfere with ABA pathways to modulate the outcome of infection.
Interactions with Fungi Shed Light on Host General Transcription Machinery
Host transcriptional reprogramming after pathogen challenge is paramount in the establishment of plant defense. The general transcription machinery relies on the mediator complex, a multiprotein co-activator scaffold acting as a bridge between RNA polymerase II (RNAPII) and transcription factors (Samanta and Thakur, 2015). Plants mutated in some mediator subunits show compromised resistance to both bacterial and fungal pathogens (Zhang et al., 2012). Further, the mediator complex is a target of the HaRL44 downy mildew effector (Caillaud et al., 2013), highlighting its relevance in plant defense. Mutations in MED25 or MED16 subunits abolished the induction of JA-responsive genes and reduced resistance to B. cinerea (Kidd et al., 2009; Zhang et al., 2012). MED16 physically associates with the plant defense regulator WRKY33 to recruit RNAPII and activate plant genes involved in JA/ET cross-talks (Wang et al., 2015b). By contrast, mutation in the CDK8 mediator subunit caused enhanced resistance to B. cinerea via the regulation of the biosynthesis of cuticular waxes and secondary metabolites (Zhu et al., 2014). The Elongator is another RNAP II-interacting complex required for the induction of JA/ET defense pathways and resistance to B. cinerea. The Elongator subunit ELP2 is required for histone acetylation and the induction of WRKY33 and defensin genes, suggesting that Elongator-mediated histone acetylation may be required for full activation of transcriptional responses to B. cinerea (Wang et al., 2015a,b). HUB1 encodes a RING E3 ligase that monoubiquitinates histone H2B, interacts with mediator subunit MED21, and positively controls resistance to B. cinerea in an ET-dependent manner (Dhawan et al., 2009). Consistently, silencing of HUB1 orthologs in tomato increased susceptibility to B. cinerea and downregulated JA/ET pathway genes (Zhang et al., 2015). Pathogen responses helped deciphering the function of general transcription complexes in plants but whether B. cinerea and S. sclerotiorum are able to manipulate these complexes or associated epigenetic processes is not known.
Plant Small Bioactive Peptides
Small bioactive peptides are defined as proteins of about 100 amino acids (aa) with roles in plant development, reproduction or interaction with the environment (Tavormina et al., 2015). They can exhibit a direct antifungal activity in the extracellular space, such as the Brassicaceae-specific ARACIN1 and 2. These ∼80 aa peptides display antifungal activity in vitro (Neukermans et al., 2015). Ectopic expression of ARACIN1 in Arabidopsis reduced infection by B. cinerea. Antifungal activity against S. sclerotiorum and B. cinerea has also been shown in vitro for the Brassica napus 35 aa proline-rich peptide BnPRP1. The BnPRP1 gene is induced upon S. sclerotiorum inoculation in susceptible but not in resistant plants (Cao et al., 2015). Small bioactive peptides can also serve as intra- or intercellular signals modulating plant defense signaling pathways. In tomato, systemin-mediated activation of the JA signaling pathways is required for resistance against B. cinerea (El Oirdi et al., 2011). Systemin is a Solanaceae-specific 18 aa peptide derived from the prosystemin precursor released into the vascular system at sites of cell damage (Pearce et al., 2001). Overexpression of prosystemin increased resistance to B. cinerea suggesting that systemin may serve as damage signal during fungal infection (Coppola et al., 2015). Dobon et al. (2015) identified four transcription factors that confer enhanced resistance to B. cinerea and found 77 genes up-regulated in the four corresponding mutants. Among these were several small signaling peptides (such as devil/rotundifolia peptide DVL3) and 13 small predicted secreted proteins of unknown function of 34–123 aa, named PROVIR1 to 13. Most of these small peptide genes were induced upon fungal infection and their over-expression caused enhanced susceptibility to necrotrophic fungi (Dobon et al., 2015).
Concluding Statement
Due their phylogenetic proximity and similarities in lifestyle, B. cinerea and S. sclerotiorum are often used interchangeably as models of broad host range necrotrophic fungi. Recent progress has revealed commonalities in their virulence strategies and in the corresponding plant responses, but also differences providing valuable insights into the diversity of the molecular bases of broad of host range pathogenicity and plant QDR.
Author Contributions
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to the many colleagues whose work could not be cited in this minireview.
Footnotes
Funding. This work is supported by the French Laboratory of Excellence project TULIP (ANR-10-LABX-41; ANR-11-IDEX-0002-02), a Starting Grant of the European Research Council (ERC-StG 336808 project VariWhim), a Marie Curie grant (MC-CIG 334036 project SEPAraTE) and a Labex TULIP New Frontiers grant (project ScleRNAi) to SR.
References
- Aguileta G., Lengelle J., Chiapello H., Giraud T., Viaud M., Fournier E., et al. (2012). Genes under positive selection in a model plant pathogenic fungus, Botrytis. Infect. Genet. Evol. 12 987–996. 10.1016/j.meegid.2012.02.012 [DOI] [PubMed] [Google Scholar]
- Albert I., Böhm H., Albert M., Feiler C. E., Imkampe J., Wallmeroth N., et al. (2015). An RLP23–SOBIR1–BAK1 complex mediates NLP-triggered immunity. Nat. Plants 1 15140 10.1038/nplants.2015.140 [DOI] [PubMed] [Google Scholar]
- Ali O., Mirabet V., Godin C., Traas J. (2014). Physical models of plant development. Annu. Rev. Cell Dev. Biol. 30 59–78. 10.1146/annurev-cellbio-101512-122410 [DOI] [PubMed] [Google Scholar]
- Amselem J., Cuomo C. A., van Kan J. A., Viaud M., Benito E. P., Couloux A., et al. (2011). Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 7:e1002230 10.1371/journal.pgen.1002230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew M., Barua R., Short S. M., Kohn L. M. (2012). Evidence for a common toolbox based on necrotrophy in a fungal lineage spanning necrotrophs, biotrophs, endophytes, host generalists and specialists. PLoS ONE 7:e29943 10.1371/journal.pone.0029943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audenaert K., De Meyer G. B., Hofte M. M. (2002). Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol. 128 491–501. 10.1104/pp.010605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badet T., Peyraud R., Raffaele S. (2015). Common protein sequence signatures associate with Sclerotinia borealis lifestyle and secretion in fungal pathogens of the Sclerotiniaceae. Front. Plant Sci. 6:776 10.3389/fpls.2015.00776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbacci A., Lahaye M., Magnenet V. (2013). Another brick in the cell wall: biosynthesis dependent growth model. PLoS ONE 8:e74400 10.1371/journal.pone.0074400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett L. G., Thrall P. H., Dodds P. N., van der Merwe M., Linde C. C., Lawrence G. J., et al. (2009). Diversity and evolution of effector loci in natural populations of the plant pathogen Melampsora lini. Mol. Biol. Evol. 26 2499–2513. 10.1093/molbev/msp166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastien R., Bohr T., Moulia B., Douady S. (2013). Unifying model of shoot gravitropism reveals proprioception as a central feature of posture control in plants. Proc. Natl. Acad. Sci. U.S.A. 110 755–760. 10.1073/pnas.1214301109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastmeyer M., Deising H. B., Bechinger C. (2002). Force exertion in fungal infection. Annu. Rev. Biophys. Biomol. Struct. 31 321–341. 10.1146/annurev.biophys.31.091701.170951 [DOI] [PubMed] [Google Scholar]
- Beneloujaephajri E., Costa A., L’Haridon F., Metraux J. P., Binda M. (2013). Production of reactive oxygen species and wound-induced resistance in Arabidopsis thaliana against Botrytis cinerea are preceded and depend on a burst of calcium. BMC Plant Biol. 13:160 10.1186/1471-2229-13-160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benikhlef L., L’Haridon F., Abou-Mansour E., Serrano M., Binda M., Costa A., et al. (2013). Perception of soft mechanical stress in Arabidopsis leaves activates disease resistance. BMC Plant Biol. 13:133 10.1186/1471-2229-13-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi F. C., Liu Z., Wu J. X., Liang H., Xi X. L., Fang C., et al. (2014). Loss of ceramide kinase in Arabidopsis impairs defenses and promotes ceramide accumulation and mitochondrial H2O2 bursts. Plant Cell 26 3449–3467. 10.1105/tpc.114.127050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billon-Grand G., Rascle C., Droux M., Rollins J. A., Poussereau N. (2012). pH modulation differs during sunflower cotyledon colonization by the two closely related necrotrophic fungi Botrytis cinerea and Sclerotinia sclerotiorum. Mol. Plant Pathol. 13 568–578. 10.1111/j.1364-3703.2011.00772.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60 379–406. 10.1146/annurev.arplant.57.032905.105346 [DOI] [PubMed] [Google Scholar]
- Bolton M. D., Thomma B. P. H. J., Nelson B. D. (2006). Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 7 1–16. 10.1111/j.1364-3703.2005.00316.x [DOI] [PubMed] [Google Scholar]
- Boudaoud A. (2010). An introduction to the mechanics of morphogenesis for plant biologists. Trends Plant Sci. 15 353–360. 10.1016/j.tplants.2010.04.002 [DOI] [PubMed] [Google Scholar]
- Caillaud M. C., Asai S., Rallapalli G., Piquerez S., Fabro G., Jones J. D. (2013). A downy mildew effector attenuates salicylic acid-triggered immunity in Arabidopsis by interacting with the host mediator complex. PLoS Biol. 11:e1001732 10.1371/journal.pbio.1001732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Ke T., Liu R., Yu J., Dong C., Cheng M., et al. (2015). Identification of a novel proline-rich antimicrobial peptide from Brassica napus. PLoS ONE 10:e0137414 10.1371/journal.pone.0137414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cessna S. G., Sears V. E., Dickman M. B., Low P. S. (2000). Oxalic acid, a pathogenicity factor for Sclerotinia sclerotiorum, suppresses the oxidative burst of the host plant. Plant Cell 12 2191–2200. 10.2307/3871114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chehab E. W., Wang Y., Braam J. (2011). “Mechanical force responses of plant cells and plants,” in Mechanical Integration of Plant Cells and Plants, ed. Wojtaszek P. (Heidelberg: Springer-Verlag Berlin; ), 173–194. [Google Scholar]
- Chehab E. W., Yao C., Henderson Z., Kim S., Braam J. (2012). Arabidopsis touch-induced morphogenesis is jasmonate mediated and protects against pests. Curr. Biol. 22 701–706. 10.1016/j.cub.2012.02.061 [DOI] [PubMed] [Google Scholar]
- Choquer M., Fournier E., Kunz C., Levis C., Pradier J. M., Simon A., et al. (2007). Botrytis cinerea virulence factors: new insights into a necrotrophic and polyphageous pathogen. FEMS Microbiol. Lett. 277 1–10. 10.1111/j.1574-6968.2007.00930.x [DOI] [PubMed] [Google Scholar]
- Conrath U. (2011). Molecular aspects of defence priming. Trends Plant Sci. 16 524–531. 10.1016/j.tplants.2011.06.004 [DOI] [PubMed] [Google Scholar]
- Conrath U., Beckers G. J., Flors V., Garcia-Agustin P., Jakab G., Mauch F., et al. (2006). Priming: getting ready for battle. Mol. Plant Microbe Interact. 19 1062–1071. 10.1094/MPMI-19-1062 [DOI] [PubMed] [Google Scholar]
- Coppola M., Corrado G., Coppola V., Cascone P., Martinelli R., Digilio M. C., et al. (2015). Prosystemin overexpression in tomato enhances resistance to different biotic stresses by activating genes of multiple signaling pathways. Plant Mol. Biol. Rep. 33 1270–1285. 10.1007/s11105-014-0834-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutand C. (2010). Mechanosensing and thigmomorphogenesis, a physiological and biomechanical point of view. Plant Sci. 179 168–182. 10.1016/j.plantsci.2010.05.001 [DOI] [Google Scholar]
- Creff A., Brocard L., Ingram G. (2015). A mechanically sensitive cell layer regulates the physical properties of the Arabidopsis seed coat. Nat. Commun. 6 6382 10.1038/ncomms7382 [DOI] [PubMed] [Google Scholar]
- Curvers K., Seifi H., Mouille G., de Rycke R., Asselbergh B., Van Hecke A., et al. (2010). Abscisic acid deficiency causes changes in cuticle permeability and pectin composition that influence tomato resistance to Botrytis cinerea. Plant Physiol. 154 847–860. 10.1104/pp.110.158972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean R., Van Kan J. A. L., Pretorius Z. A., Hammond-Kosack K. E., Di Pietro A., Spanu P. D., et al. (2012). The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13 414–430. 10.1111/j.1364-3703.2011.00783.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan R., Luo H., Foerster A. M., Abuqamar S., Du H. N., Briggs S. D., et al. (2009). HISTONE MONOUBIQUITINATION1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell 21 1000–1019. 10.1105/tpc.108.062364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickman M. B., de Figueiredo P. (2013). Death be not proud -cell death control in plant fungal interactions. PLoS Pathog. 9:e1003542 10.1371/journal.ppat.1003542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobon A., Canet J. V., Garcia-Andrade J., Angulo C., Neumetzler L., Persson S., et al. (2015). Novel disease susceptibility factors for fungal necrotrophic pathogens in Arabidopsis. PLoS Pathog. 11:e1004800 10.1371/journal.ppat.1004800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S., Raffaele S., Kamoun S. (2015). The two-speed genomes of filamentous pathogens: waltz with plants. Curr. Opin. Genet. Dev. 35 57–65. 10.1016/j.gde.2015.09.001 [DOI] [PubMed] [Google Scholar]
- El Oirdi M., El Rahman T. A., Rigano L., El Hadrami A., Rodriguez M. C., Daayf F., et al. (2011). Botrytis cinerea manipulates the antagonistic effects between immune pathways to promote disease development in tomato. Plant Cell 23 2405–2421. 10.1105/tpc.111.083394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espino J. J., Gutierrez-Sanchez G., Brito N., Shah P., Orlando R., Gonzalez C. (2010). The Botrytis cinerea early secretome. Proteomics 10 3020–3034. 10.1002/pmic.201000037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J., Kim H. U., Nakata P. A., Browse J. (2012). A previously unknown oxalyl-CoA synthetase is important for oxalate catabolism in Arabidopsis. Plant Cell 24 1217–1229. 10.1105/tpc.112.096032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frías M., Brito N., Gonzalez M., Gonzalez C. (2014). The phytotoxic activity of the cerato-platanin BcSpl1 resides in a two-peptide motif on the protein surface. Mol. Plant Pathol. 15 342–351. 10.1111/mpp.12097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frías M., González C., Brito N. (2011). BcSpl1, a cerato-platanin family protein, contributes to Botrytis cinerea virulence and elicits the hypersensitive response in the host. New Phytol. 192 483–495. 10.1111/j.1469-8137.2011.03802.x [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43 205–227. 10.1146/annurev.phyto.43.040204.135923 [DOI] [PubMed] [Google Scholar]
- Godoy G., Steadman J., Dickman M., Dam R. (1990). Use of mutants to demonstrate the role of oxalic acid in pathogenicity of Sclerotinia sclerotiorum on Phaseolus vulgaris. Physiol. Mol. Plant Pathol. 37 179–191. 10.1016/0885-5765(90)90010-U [DOI] [Google Scholar]
- Govrin E. M., Levine A. (2000). The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr. Biol. 10 751–757. 10.1016/S0960-9822(00)00560-1 [DOI] [PubMed] [Google Scholar]
- Guimaraes R. L., Stotz H. U. (2004). Oxalate production by Sclerotinia sclerotiorum deregulates guard cells during infection. Plant Physiol. 136 3703–3711. 10.1104/pp.104.049650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyon K., Balagué C., Roby D., Raffaele S. (2014). Secretome analysis reveals effector candidates associated with broad host range necrotrophy in the fungal plant pathogen Sclerotinia sclerotiorum. BMC Genomics 15:336 10.1186/1471-2164-15-336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamant O. (2013). Widespread mechanosensing controls the structure behind the architecture in plants. Curr. Opin. Plant Biol. 16 654–660. 10.1016/j.pbi.2013.06.006 [DOI] [PubMed] [Google Scholar]
- Hamant O., Traas J. (2010). The mechanics behind plant development. New Phytol. 185 369–385. 10.1111/j.1469-8137.2009.03100.x [DOI] [PubMed] [Google Scholar]
- Heard S., Brown N. A., Hammond-Kosack K. (2015). An interspecies comparative analysis of the predicted secretomes of the necrotrophic plant pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS ONE 10:e0130534 10.1371/journal.pone.0130534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegedus D. D., Rimmer S. R. (2005). Sclerotinia sclerotiorum: when “to be or not to be” a pathogen? FEMS Microbiol. Lett. 251 177–184. 10.1016/j.femsle.2005.07.040 [DOI] [PubMed] [Google Scholar]
- Heller A., Witt-Geiges T. (2013). Oxalic acid has an additional, detoxifying function in Sclerotinia sclerotiorum pathogenesis. PLoS ONE 8:e72292 10.1371/journal.pone.0072292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman D., Gilroy S., Ane J. M. (2014). Staying in touch: mechanical signals in plant-microbe interactions. Curr. Opin. Plant Biol. 20 104–109. 10.1016/j.pbi.2014.05.003 [DOI] [PubMed] [Google Scholar]
- Joosten H. J., Han Y., Niu W., Vervoort J., Dunaway-Mariano D., Schaap P. J. (2008). Identification of fungal oxaloacetate hydrolyase within the isocitrate lyase/PEP mutase enzyme superfamily using a sequence marker-based method. Proteins 70 157–166. 10.1002/prot.21622 [DOI] [PubMed] [Google Scholar]
- Kabbage M., Williams B., Dickman M. B. (2013). Cell death control: the interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum. PLoS Pathog. 9:e1003287 10.1371/journal.ppat.1003287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabbage M., Yarden O., Dickman M. B. (2015). Pathogenic attributes of Sclerotinia sclerotiorum: switching from a biotrophic to necrotrophic lifestyle. Plant Sci. 233 53–60. 10.1016/j.plantsci.2014.12.018 [DOI] [PubMed] [Google Scholar]
- Kidd B. N., Edgar C. I., Kumar K. K., Aitken E. A., Schenk P. M., Manners J. M., et al. (2009). The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21 2237–2252. 10.1105/tpc.109.066910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. S., Min J. Y., Dickman M. B. (2008). Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Mol. Plant Microbe Interact. 21 605–612. 10.1094/MPMI-21-5-0605 [DOI] [PubMed] [Google Scholar]
- Koch A., Kogel K. H. (2014). New wind in the sails: improving the agronomic value of crop plants through RNAi-mediated gene silencing. Plant Biotechnol. J. 12 821–831. 10.1111/pbi.12226 [DOI] [PubMed] [Google Scholar]
- Koch A., Kumar N., Weber L., Keller H., Imani J., Kogel K. H. (2013). Host-induced gene silencing of cytochrome P450 lanosterol C14alpha-demethylase-encoding genes confers strong resistance to Fusarium species. Proc. Natl. Acad. Sci. U.S.A. 110 19324–19329. 10.1073/pnas.1306373110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z., Wang F., Zheng Z., Fan B., Chen Z. (2011). A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. 66 953–968. 10.1111/j.1365-313X.2011.04553.x [DOI] [PubMed] [Google Scholar]
- Lampl N., Alkan N., Davydov O., Fluhr R. (2013). Set-point control of RD21 protease activity by AtSerpin1 controls cell death in Arabidopsis. Plant J. 74 498–510. 10.1111/tpj.12141 [DOI] [PubMed] [Google Scholar]
- Landrein B., Kiss A., Sassi M., Chauvet A., Das P., Cortizo M., et al. (2015). Mechanical stress contributes to the expression of the STM homeobox gene in Arabidopsis shoot meristems. Elife 4:e07811 10.7554/eLife.07811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Liberti D., Li M., Kim Y. T., Hutchens A., Wilson R., et al. (2015). Oxaloacetate acetylhydrolase gene mutants of Sclerotinia sclerotiorum do not accumulate oxalic acid, but do produce limited lesions on host plants. Mol. Plant Pathol. 16 559–571. 10.1111/mpp.12211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Kracher B., Ziegler J., Birkenbihl R. P., Somssich I. E. (2015). Negative regulation of ABA signaling by WRKY33 is critical for Arabidopsis immunity towards Botrytis cinerea 2100. Elife 4:e07295 10.7554/eLife.07295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu X., Shen C., Fu Y., Xie J., Jiang D., Li G., et al. (2015). Comparative genomic and transcriptional analyses of the carbohydrate-active enzymes and secretomes of phytopathogenic fungi reveal their significant roles during infection and development. Sci. Rep. 5 15565 10.1038/srep15565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnin-Robert M., Le Bourse D., Markham J., Dorey S., Clement C., Baillieul F., et al. (2015). Modifications of sphingolipid content affect tolerance to hemibiotrophic and necrotrophic pathogens by modulating plant defense responses in Arabidopsis. Plant Physiol. 169 2255–2274. 10.1104/pp.15.01126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez F., Dubos B., Fermaud M. (2005). The role of saprotrophy and virulence in the population dynamics of Botrytis cinerea in vineyards. Phytopathology 95 692–700. 10.1094/PHYTO-95-0692 [DOI] [PubMed] [Google Scholar]
- Monaghan J., Zipfel C. (2012). Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 15 349–357. 10.1016/j.pbi.2012.05.006 [DOI] [PubMed] [Google Scholar]
- Monshausen G. B., Haswell E. S. (2013). A force of nature: molecular mechanisms of mechanoperception in plants. J. Exp. Bot. 64 4663–4680. 10.1093/jxb/ert204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulia B., Coutand C., Julien J. L. (2015). Mechanosensitive control of plant growth: bearing the load, sensing, transducing, and responding. Front. Plant Sci. 6:52 10.3389/fpls.2015.00052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neukermans J., Inze A., Mathys J., De Coninck B., van de Cotte B., Cammue B. P., et al. (2015). ARACINs, Brassicaceae-specific peptides exhibiting antifungal activities against necrotrophic pathogens in Arabidopsis. Plant Physiol. 167 1017–1029. 10.1104/pp.114.255505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowara D., Gay A., Lacomme C., Shaw J., Ridout C., Douchkov D., et al. (2010). HIGS: host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 22 3130–3141. 10.1105/tpc.110.077040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes C. C., Dean R. A. (2012). Host-induced gene silencing: a tool for understanding fungal host interaction and for developing novel disease control strategies. Mol. Plant Pathol. 13 519–529. 10.1111/j.1364-3703.2011.00766.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paparella C., Savatin D. V., Marti L., De Lorenzo G., Ferrari S. (2014). The Arabidopsis LYSIN MOTIF-CONTAINING RECEPTOR-LIKE KINASE3 regulates the cross talk between immunity and abscisic acid responses. Plant Physiol. 165 262–276. 10.1104/pp.113.233759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce G., Moura D. S., Stratmann J., Ryan C. A. (2001). Production of multiple plant hormones from a single polyprotein precursor. Nature 411 817–820. 10.1038/35081107 [DOI] [PubMed] [Google Scholar]
- Perchepied L., Balagué C., Riou C., Claudel-Renard C., Rivière N., Grezes-Besset B., et al. (2010). Nitric oxide participates in the complex interplay of defense-related signaling pathways controlling disease resistance to Sclerotinia sclerotiorum in Arabidopsis thaliana. Mol. Plant Microbe Interact. 23 846–860. 10.1094/MPMI-23-7-0846 [DOI] [PubMed] [Google Scholar]
- Raffaele S., Farrer R. A., Cano L. M., Studholme D. J., MacLean D., Thines M., et al. (2010). Genome evolution following host jumps in the Irish potato famine pathogen lineage. Science 330 1540–1543. 10.1126/science.1193070 [DOI] [PubMed] [Google Scholar]
- Raffaele S., Kamoun S. (2012). Genome evolution in filamentous plant pathogens: why bigger can be better. Nat. Rev. Microbiol. 10 417–430. 10.1038/nrmicro2790 [DOI] [PubMed] [Google Scholar]
- Rietz S., Bernsdorff F. E., Cai D. (2012). Members of the germin-like protein family in Brassica napus are candidates for the initiation of an oxidative burst that impedes pathogenesis of Sclerotinia sclerotiorum. J. Exp. Bot. 63 5507–5519. 10.1093/jxb/ers203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A., Grant M., Jones J. D. (2011). Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 49 317–343. 10.1146/annurev-phyto-073009-114447 [DOI] [PubMed] [Google Scholar]
- Roux F., Voisin D., Badet T., Balagué C., Barlet X., Huard-Chauveau C., et al. (2014). Resistance to phytopathogens e tutti quanti: placing plant quantitative disease resistance on the map. Mol. Plant Pathol. 15 427–432. 10.1111/mpp.12138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta S., Thakur J. K. (2015). Importance of Mediator complex in the regulation and integration of diverse signaling pathways in plants. Front. Plant Sci. 6:757 10.3389/fpls.2015.00757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanati Nezhad A., Geitmann A. (2013). The cellular mechanics of an invasive lifestyle. J. Exp. Bot. 64 4709–4728. 10.1093/jxb/ert254 [DOI] [PubMed] [Google Scholar]
- Sanchez-Vallet A., Mesters J. R., Thomma B. P. (2015). The battle for chitin recognition in plant-microbe interactions. FEMS Microbiol. Rev. 39 171–183. 10.1093/femsre/fuu003 [DOI] [PubMed] [Google Scholar]
- Sarkies P., Miska E. A. (2014). Small RNAs break out: the molecular cell biology of mobile small RNAs. Nat. Rev. Mol. Cell Biol. 15 525–535. 10.1038/nrm3840 [DOI] [PubMed] [Google Scholar]
- Seifi H. S., Curvers K., De Vleesschauwer D., Delaere I., Aziz A., Hofte M. (2013). Concurrent overactivation of the cytosolic glutamine synthetase and the GABA shunt in the ABA-deficient sitiens mutant of tomato leads to resistance against Botrytis cinerea. New Phytol. 199 490–504. 10.1111/nph.12283 [DOI] [PubMed] [Google Scholar]
- Shlezinger N., Minz A., Gur Y., Hatam I., Dagdas Y. F., Talbot N. J., et al. (2011). Anti-apoptotic machinery protects the necrotrophic fungus Botrytis cinerea from host-induced apoptotic-like cell death during plant infection. PLoS Pathog. 7:e1002185 10.1371/journal.ppat.1002185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siewers V., Smedsgaard J., Tudzynski P. (2004). The P450 monooxygenase BcABA1 is essential for abscisic acid biosynthesis in Botrytis cinerea. Appl. Environ. Microbiol. 70 3868–3876. 10.1128/AEM.70.7.3868-3876.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperschneider J., Dodds P. N., Gardiner D. M., Manners J. M., Singh K. B., Taylor J. M. (2015). Advances and challenges in computational prediction of effectors from plant pathogenic fungi. PLoS Pathog. 11:e1004806 10.1371/journal.ppat.1004806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq V., Jeffries P. (1984). Appressorium formation by Sclerotinia sclerotiorum: scanning electron microscopy. Trans. Br. Mycol. Soc. 82 645–651. 10.1016/S0007-1536(84)80105-9 [DOI] [Google Scholar]
- Tavormina P., De Coninck B., Nikonorova N., De Smet I., Cammue B. P. (2015). The plant peptidome: an expanding repertoire of structural features and biological functions. Plant Cell 27 2095–2118. 10.1105/tpc.15.00440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Ding Y., Yao J., Zhang Y., Sun Y., Colee J., et al. (2015a). Arabidopsis Elongator subunit 2 positively contributes to resistance to the necrotrophic fungal pathogens Botrytis cinerea and Alternaria brassicicola. Plant J. 83 1019–1033. 10.1111/tpj.12946 [DOI] [PubMed] [Google Scholar]
- Wang C., Yao J., Du X., Zhang Y., Sun Y., Rollins J. A., et al. (2015b). The Arabidopsis mediator complex subunit16 is a key component of basal resistance against the necrotrophic fungal pathogen Sclerotinia sclerotiorum. Plant Physiol. 169 856–872. 10.1104/pp.15.00351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiberg A., Bellinger M., Jin H. (2015). Conversations between kingdoms: small RNAs. Curr. Opin. Biotechnol. 32 207–215. 10.1016/j.copbio.2014.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiberg A., Wang M., Bellinger M., Jin H. (2014). Small RNAs: a new paradigm in plant-microbe interactions. Annu. Rev. Phytopathol. 52 495–516. 10.1146/annurev-phyto-102313-045933 [DOI] [PubMed] [Google Scholar]
- Weiberg A., Wang M., Lin F.-M., Zhao H., Zhang Z., Kaloshian I., et al. (2013). Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342 118–123. 10.1126/science.1239705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B., Kabbage M., Kim H. J., Britt R., Dickman M. B. (2011). Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathog. 7:e1002107 10.1371/journal.ppat.1002107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Xie J., Cheng J., Li G., Yi X., Jiang D., et al. (2014). Novel secretory protein Ss-Caf1 of the plant-pathogenic fungus Sclerotinia sclerotiorum is required for host penetration and normal sclerotial development. Mol. Plant Microbe Interact. 27 40–55. 10.1094/MPMI-05-13-0145-R [DOI] [PubMed] [Google Scholar]
- Xu L., Chen W. (2013). Random T-DNA mutagenesis identifies a Cu/Zn superoxide dismutase gene as a virulence factor of Sclerotinia sclerotiorum. Mol. Plant Microbe Interact. 26 431–441. 10.1094/MPMI-07-12-0177-R [DOI] [PubMed] [Google Scholar]
- Xu L., Xiang M., White D., Chen W. (2015). pH dependency of sclerotial development and pathogenicity revealed by using genetically defined oxalate-minus mutants of Sclerotinia sclerotiorum. Environ. Microbiol. 17 2896–2909. 10.1111/1462-2920.12818 [DOI] [PubMed] [Google Scholar]
- Yin C., Park J. J., Gang D. R., Hulbert S. H. (2014). Characterization of a tryptophan 2-monooxygenase gene from Puccinia graminis f. sp. tritici involved in auxin biosynthesis and rust pathogenicity. Mol. Plant Microbe Interact. 27 227–235. 10.1094/MPMI-09-13-0289-FI [DOI] [PubMed] [Google Scholar]
- Zhang H., Wu Q., Cao S., Zhao T., Chen L., Zhuang P., et al. (2014). A novel protein elicitor (SsCut) from Sclerotinia sclerotiorum induces multiple defense responses in plants. Plant Mol. Biol. 86 495–511. 10.1007/s11103-014-0244-3 [DOI] [PubMed] [Google Scholar]
- Zhang L., Kars I., Essenstam B., Liebrand T. W., Wagemakers L., Elberse J., et al. (2014). Fungal endopolygalacturonases are recognized as microbe-associated molecular patterns by the Arabidopsis receptor-like protein RESPONSIVENESS TO BOTRYTIS POLYGALACTURONASES1. Plant Physiol. 164 352–364. 10.1104/pp.113.230698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Fraiture M., Kolb D., Loffelhardt B., Desaki Y., Boutrot F. F., et al. (2013). Arabidopsis receptor-like protein30 and receptor-like kinase suppressor of BIR1-1/EVERSHED mediate innate immunity to necrotrophic fungi. Plant Cell 25 4227–4241. 10.1105/tpc.113.117010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wang C., Zhang Y., Sun Y., Mou Z. (2012). The Arabidopsis mediator complex subunit16 positively regulates salicylate-mediated systemic acquired resistance and jasmonate/ethylene-induced defense pathways. Plant Cell 24 4294–4309. 10.1105/tpc.112.103317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Li D., Zhang H., Hong Y., Huang L., Liu S., et al. (2015). Tomato histone H2B monoubiquitination enzymes SlHUB1 and SlHUB2 contribute to disease resistance against Botrytis cinerea through modulating the balance between SA- and JA/ET-mediated signaling pathways. BMC Plant Biol. 15:252 10.1186/s12870-015-0614-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Han Y., Li Y., Liu D., Sun M., Zhao Y., et al. (2015). Loci and candidate gene identification for resistance to Sclerotinia sclerotiorum in soybean (Glycine max L. Merr.) via association and linkage maps. Plant J. 82 245–255. 10.1111/tpj.12810 [DOI] [PubMed] [Google Scholar]
- Zhou J., Fu Y., Xie J., Li B., Jiang D., Li G., et al. (2012). Identification of microRNA-like RNAs in a plant pathogenic fungus Sclerotinia sclerotiorum by high-throughput sequencing. Mol. Genet. Genomics 287 275–282. 10.1007/s00438-012-0678-8 [DOI] [PubMed] [Google Scholar]
- Zhou J., Zeng L., Liu J., Xing D. (2015). Manipulation of the xanthophyll cycle increases plant susceptibility to Sclerotinia sclerotiorum. PLoS Pathog. 11:e1004878 10.1371/journal.ppat.1004878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Wei W., Fu Y., Cheng J., Xie J., Li G., et al. (2013). A secretory protein of necrotrophic fungus Sclerotinia sclerotiorum that suppresses host resistance. PLoS ONE 8:e53901 10.1371/journal.pone.0053901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Schluttenhoffer C. M., Wang P., Fu F., Thimmapuram J., Zhu J. K., et al. (2014). CYCLIN-DEPENDENT KINASE8 differentially regulates plant immunity to fungal pathogens through kinase-dependent and -independent functions in Arabidopsis. Plant Cell 26 4149–4170. 10.1105/tpc.114.128611 [DOI] [PMC free article] [PubMed] [Google Scholar]