Abstract
Occult hepatitis B virus infection (OBI), characterized as the persistence of hepatitis B virus (HBV) surface antigen (HBsAg) seronegativity and low viral load in blood or liver, is a special form of HBV infection. OBI may be related mainly to mutations in the HBV genome, although the underlying mechanism of it remains to be clarified. Mutations especially within the immunodominant “α” determinant of S protein are “hot spots” that could contribute to the occurrence of OBI via affecting antigenicity and immunogenicity of HBsAg or replication and secretion of virion. Clinical reports account for a large proportion of previous studies on OBI, while functional analyses, especially those based on full-length HBV genome, are rare.
Keywords: Hepatitis B virus, Occult, Variation, Hepatitis B surface antigen
Core tip: With error-prone reverse transcription during replication, hepatitis B virus (HBV) displays a high rate of mutation, and a single mutation may affect the biological properties of HBV. Occult HBV infection (OBI) is a special form of HBV infection and a frequent phenomenon. Many previous publications have explored the association of OBI with the “hot spots” mutations that occur within the immunodominant “α” determinant of S proteins. However, there are no reviews available that elaborate on the relationship between OBI and mutations throughout the entire HBV genome. This review attempts to provide a comprehensive summary of HBV genetic variants that have been associated with OBI.
INTRODUCTION
Hepatitis B virus (HBV) is classified as a DNA virus, and it replicates via reverse transcription, which is an error-prone process, to generate an RNA intermediate. Thus, the mutation rate of HBV is in between those of RNA viruses and conventional DNA viruses[1]. The nucleotide mutation rate of HBV is estimated to be 10-5-10-6 per site per year, about 10 fold higher than that of other DNA viruses, and this rate might increase 100 fold after liver transplantation[2,3]. Mutations are commonly encountered, especially in the basal core promoter (BCP), the pre-core region (Pre-C), the polymerase (P) gene, and the “α” determinant of S protein, and single or multiple mutations may affect the biological properties of HBV[4]. Occult HBV infection (OBI), a special form of HBV infection and a frequent phenomenon, was first reported in 1978 and has been studied further since then[5]. In 2008, the Taormina expert meeting defined OBI as the presence of HBV DNA in the liver of individuals negative for HBV surface antigen (HBsAg) testing with the amount of HBV DNA in the serum usually lower than 200 IU/mL or undetectable[6]. OBI is characterized by the persistence of low level viremia and HBsAg seronegativity and can be grouped into two types: seropositive [anti-hepatitis B core antigen (HBc) and/or anti-hepatitis B surface antigen (HBs) positive] and seronegative (anti-HBc and anti-HBs negative)[7]. OBI harbors the potential risk of HBV transmission through blood transfusion, organ transplantation, and hemodialysis as well as from occult infected or HBsAg-positive mothers to newborns[8,9]. Although the clinical and biochemical symptoms of most OBIs are not severe, there is still a risk of developing serious liver diseases, such as liver cirrhosis (LC) and hepatocellular carcinoma (HCC)[10]. In fact, a meta-analysis and other findings have revealed that OBI retains the pro-oncogenic properties of HBV and serves as a risk factor for the development of HCC[11-13]. Suppression of viral replication is reversible in the case of OBI, and reactivation of OBI and/or development of fulminant hepatitis failure (FHF) has been reported in patients following chemotherapy or immunosuppressive therapy and after transplantation as well as in patients co-infected with human immunodeficiency virus (HIV) or hepatitis C virus (HCV)[14-16].
PREVALENCE OF OBI
OBI is a health problem worldwide, and its prevalence is affected by several factors, such as the prevalence of HBV infection, the study population, and the sensitivity of diagnostic assays[17,18]. OBI is significantly associated with the endemicity of HBV infection but not restricted to countries highly endemic for HBV[10,17,18]. The prevalence of OBI in blood donors from China, South Korea, and Japan was reported to be 0.11%-0.13%, 0.016%, and 1.01%, respectively[19-22]. The incidence of OBI in anti-HBc positive donors was reported to be from 0% to 15%[23,24], and the detection rate of OBI in HIV infection was reported to be from 0% to 89%[25,26]. In the context of chronic HCV infection, the percentage of OBI ranged from 0% to 52%[27,28]. Previous studies conducted in hemodialysis patients indicated a prevalence of OBI from 0% to 36%[29,30]. OBI was detected in 32% of patients with cryptogenic LC in Hong Kong, an area with a high prevalence of HBV infection[31]. The prevalence of OBI was reported to be 70.4% among HBsAg-negative HCC patients in China[32].
The identification of OBI largely depends on the tests for HBsAg and HBV DNA. There are several commercially available HBsAg assays, which mainly depend on the use of anti-HBs antibodies, with different sensitivities and specificities. Some diagnostic HBsAg assays are based on monoclonal antibodies and recognize limited types of HBsAg variants. In comparison, assays that utilize polyclonal antibodies show higher sensitivity and specificity for the detection of various types of HBsAg mutants[33,34]. Because the level of viremia is low in OBI, a small amount of HBsAg below the detection limit of tests may not be reliably recognized. Although highly sensitive real-time quantitative polymerase chain reaction (PCR) is strongly recommended for HBV nucleic acid testing (NAT) for its capability to uncover low level of HBV DNA in serum, this method harbors a risk of false-positive results[35,36]. Anti-HBc screening can reduce the risk of HBV transmission but may possibly provide false-positive results as well. It is worth noting that the nature of the specimen tested (i.e., blood sample or liver tissue), the amount of specimen, as well as contamination risks can also affect the detection of OBI.
Many studies have been conducted on OBI prevalence. However, cautions must be taken when results from different studies are compared, since differences in groups of people studied, areas selected, and diagnostic assays can significantly affect the results.
MECHANISM OF OBI
The mechanism of OBI is complicated and remains to be clarified. Both host and viral factors contribute to the occurrence of OBI. For example, the persistence of covalently closed circular DNA (cccDNA) in hepatocytes, suppression and detection failure of virion replication, and protein secretion are involved[37,38]. In addition, methylation of the HBV genome, microRNAs (miRNAs), and histone modification have been reported to affect the regulation of HBV replication and expression through epigenetic mechanisms, increasing the possibility that epigenetic regulation may also be associated with OBI[39].
Viral factors
Genetic mutations may contribute to OBI. Mutations have an impact on the expression, secretion, and antigenicity of HBsAg and cause failure in detecting HBsAg by commercial diagnostic tests. HBV is prone to mutations and exists as quasispecies to facilitate viral survival. Point mutations and insertion as well as deletion mutations are commonly encountered in OBI, and although mutations in all open reading frames (ORFs) of HBV could directly or indirectly affect detection of HBsAg, most studies have focused on mutations in the Pre-S/S gene. Besides, OBI mutations are associated with the suppression of viral replication typical of OBI, which in turn affects HBsAg expression and secretion[40]. Circulating immune complexes consisting of HBsAg and anti-HBs may be associated with Pre-S/S gene mutations and impair the binding efficiency of HBsAg capture antibodies to HBsAg in serum, leading to immune escape[41,42].
Low levels of HBV replication may be correlated with OBI. Compared to HBsAg-positive infection, OBI always has lower (< 200 IU/mL) or even undetectable HBV DNA in serum[18]. Although HBV DNA from the liver is more reliable than circulating HBV DNA for recognition of OBI, the levels of intra-hepatic HBV DNA and cccDNA in OBI were also found to be lower than those in HBsAg-positive infection[38,43]. Thus, the low level of HBV replication may be related to OBI.
HBV genome methylation is related to the development of OBI. Hypermethylation of HBV has been observed in isolates from OBI patients, and HBV CpG islands 1 and 2 were more frequently methylated in HBsAg positive infections and OBIs, respectively[44,45].
Host factors
The alternation of host immunologic responses is associated with OBI. In patients with spontaneous and therapy-induced HBsAg clearance, residual HBV may present in the serum, liver, and lymphatic system for a long time[46,47]. Similarly, using a woodchuck model, the closet natural model of HBV infection, Michalak[48] and Mulrooney-Cousins et al[49] described the low-level but life-long persistence of woodchuck hepatitis B virus (WHV) DNA and cccDNA in both the liver and lymphatic system in animals recovered from acute WHV invasion. Based on these data, they suggested that the hard-to-eliminate cccDNA made the complete clearance of WBV difficult to accomplish and might contribute to WBV reactivation and further speculated that HBV cccDNA may act similarly to WBV cccDNA. Therefore, latency of HBV in liver and in extra-hepatic tissues and cells provide a favorable condition for the occurrence of OBI[50]. Reactivation of overt HBV infection and OBI has been frequently described in HBV infected patients with suppressed immune systems, suggesting that HBV was inhibited rather than cleared in the setting of immunosuppression. Colson et al[51] investigated 16 anti-HBc positive patients who developed HBV reactivation following chemotherapy and detected an increased variability within HBsAg and HBV reverse transcriptase (RT) in these patients. OBI has also been reported in vaccinated children from Taiwan and Iran[52,53]. It is speculated that the patients’ immune response to HBV infection may be related to OBI by greatly controlling HBV activity, putting pressure on secretion of HBsAg and other HBV-secreted proteins, as well as promoting the screening of mutants.
In addition to altered host immunologic responses, host genetic factors are also linked to OBI. Related studies have focused on human leukocyte antigen (HLA), which plays an important role in the modulation of immune responses, and HLA polymorphisms are considered as vital factors in determining HBV infections outcome (persistence or clearance)[54]. Previous studies showed that HLA-A2 expression was correlated with protection against HBV infection, while significantly decreased intensity of HLA-A2 expression on peripheral blood mononuclear cells (PBMCs) was detected in some OBI patients compared to healthy controls, perhaps contributing to HBV resistance[55]. Serum level of interleukin (IL)-10 was markedly higher in some OBI patients than that in healthy controls, and polymorphisms of the IL-10 promoter were found with significant differences between OBIs and healthy controls, implying a relationship between IL-10 and OBI[56,57].
Host epigenetic modification may be related to OBI. miRNAs of host cells could regulate expression of some host genes and exert effects on HBV replication and expression[58]. A profile of HBV-specific serum miRNAs has been reported with a high accuracy to separate OBI cases from healthy controls (sensitivity: 99.9%, specificity: 99.8%), raising the possibility that they may serve as effective biomarkers in OBI detection[59]. Epigenetic modifications of cccDNA-bound histones may regulate HBV replication and transcription, with acetylation and methylation being the most common kinds of histone modification. Previous studies showed that HBV replication could be regulated by the acetylation status of cccDNA-bound H3 and H4 histones in chronic hepatitis B (CHB) patients, and this regulation has been confirmed by transfection experiments[60].
Other factors
Other factors may be connected with OBI, such as interference by other viruses. Concomitant presence of chronic HCV infection exerts a negative effect on HBV. HBV DNA levels are increased during interferon-ribavirin therapy of HCV but decreased after a viral breakthrough of HCV[61,62]. Moreover, in vitro studies have demonstrated the inhibitory effect exerted by HCV core protein on HBV replication and expression[63].
PRE-S/S MUTATIONS
Pre-S/S point mutation
Pre-S/S gene encodes three envelope proteins named large (L), middle (M), and small (S) protein, and S protein corresponds to HBsAg[64]. Pre-S/S has a relatively high mutation rate among all ORFs, and mutations of the Pre-S/S gene have been studied extensively. Point mutations that occur in the Pre-S/S gene may affect HBsAg in two different aspects, (1) mutations may affect antigenicity, immunogenicity, secretion, and/or expression of HBsAg, leading to detection failure of HBsAg[33,65]; (2) mutations may reduce or even abolish the replication and/or secretion of virion, exerting a negative effect on HBsAg[66,67]. Amino acid (aa) substitutions of HBsAg are mostly frequently clustered in the “α” determinant, which is located at aa124-147 of the S protein. The “α” determinant is rich in cysteine, and two loops are formed by disulfide bonds between C124 and C137 as well as C139 and C147 to maintain the conformational structure and antigenicity of HBsAg. The “α” determinant is a relatively conserved region within the major hydrophilic region (MHR) between aa100 to aa169, which serves as the most important antigenic determinant in all HBV strains and is essential to the detection of HBsAg and development of HBV vaccines[64,68]. Amino acids within the region between 120 to 123 were shown to be crucial for the antigenicity of HBsAg[69]. Therefore, single or multiple point mutations occurring within or adjacent to the “α” determinant may change the antigenicity and conformation of HBsAg, resulting in failure to detect HBsAg. Various point mutations of HBsAg have been observed in OBIs (Figure 1).
Figure 1.

Comparison of the amino acid sequences of the 226AA hepatitis B virus, hepatitis B virus surface antigen between occult hepatitis B virus infections, and controls. Amino acid substitutions are indicated with one-letter codes. Mutations identified in occult hepatitis B virus infections group and control group are in red and black, respectively. Mutations detected in both groups are in blue. Black dots denote amino acid identity with any of the reference strains of genotype A to H retrieved from GenBank (accession numbers: A, X02763; Ba, D00330; Bc, GQ205440; Bj, AB073858; C1, KM999990; C2, KM999991; D, X02496; E, X75657; F, X69798; G, AF160501; H, AY090454). Genotype-specific substitutions are listed. CON indicates control.
Amino acid mutations were detected mostly in the MHR in OBIs, and many previous reports have documented that the mutation rate in the MHR, especially in the “α” determinant, was higher in OBIs compared to that of HBsAg-positive patients[66,70]. A number of Pre-S/S mutations were observed in both OBIs and HBsAg-positive patients (Figure 1), while it has been speculated that they may not be associated with OBI. Clinically observed Pre-S/S mutations in OBIs summarized in Figure 1 indicate that: (1) Some mutations occurred in both OBIs and HBsAg-positive patients, while some mutations were only detected in OBIs; (2) Some mutations were found outside the “α” determinant and were located at the N-terminus or C-terminus of MHR or outside MHR; (3) Some mutations corresponded to amino acid substitutions in the P region; (4) Some amino acid substitutions were specific for genotypes, subgenotypes, or subtypes, while the clinical significance of them in OBIs remains to be determined; (5) Point mutations in the first loop of the “α” epitope (AA124-137) occurred more frequently in OBI isolates from unvaccinated patients[71], while point mutations in the second loop of the “α” epitope (AA138-147) were more frequently isolated from OBI patients after immune prophylaxis[72]; and (6) Apart from point substitutions, there were other types of mutations or combined mutations. However, further in vivo and in vitro studies are required to explore the effects of point mutations on the occurrence of OBI.
In order to elucidate the role of mutations in the occurrence of OBI, plasmids of Pre-S/S variants isolated from the sera and liver tissues of OBI patients were constructed by site-directed mutagenesis and transfected into cell lines or introduced into animal models. The antigenicity, immunogenicity, and secretion of HBsAg as well as the replication and secretion of virions were then analyzed. This strategy has been extensively applied to point mutations in the Pre-S/S gene (Table 1 and Figure 2). Similarly, these functional analyses also can be applied to research on other HBV-related mutations[73]. To date, most in vivo and in vitro studies have been limited to the Pre-S/S gene, and only a few functional studies have been performed with full-length, replication-competent HBV genomes. It is difficult to know whether some point mutations interfere with the replication of the virion and secretion of antigens or if they play a role in the occurrence of OBI[40,74].
Table 1.
Summary of hepatitis B virus surface antigen variants involved in functional studies
| Variants | Source | Model | Antigenicity | Immuno -genicity | HBsAg Intra- | HBsAg Extra- | Viral replication /secretion | References |
| W741 | CHB | Huh7 | L | L | L | [65] | ||
| M75T | Huh7 | ↑ | ↓ | ↓2 | [75] | |||
| Y100C | OBI | Huh7 | ↑ | [76] | ||||
| Y100S | OBI | Huh7 | L | [77] | ||||
| I110M | Huh7 | L3 | [78] | |||||
| T114R | Huh7 | ↓3 | [79] | |||||
| T115A | Huh7 | ↓3 | [79] | |||||
| T116N | OBI | Huh7 | ↓ | [77] | ||||
| G119E | Huh7 | ↓ | ↓3 | [78] | ||||
| G119R | OBI | Huh7/mice | ↓ | ↑ | ↓ | ↓3 | [66] | |
| P120T | OBI | Huh7/mice | ↓ | [66] | ||||
| P120T | Huh7 | ↓ | [79] | |||||
| C121S | HepG2/Hela | ↓ | ↓ | ↓ | [69] | |||
| C1211 | OBI | Pichia | ↓ or L | [80] | ||||
| C121A | Huh7 | ↓ | ↓ or L3 | [79] | ||||
| K122I | HepG2/Hela | ↓ | ↓ | ↓ | [69] | |||
| K122I | Huh7 | ↓ | ↓ | ↓ | ↓ | [81] | ||
| K122I | Huh7/Hela | ↓ or L | ↓ | ↓ | ↓ | [82] | ||
| R122P | OBI | Huh7 | ↓ | [77] | ||||
| T123N | HepG2/Hela | ↓ | ↓ | ↓ | [69] | |||
| T123N | Huh7/Hela | ↓ or L | ↓3 | [82] | ||||
| C1241 | OBI | Pichia | ↓ or L | [80] | ||||
| C124A | Huh7 | ↓ | ↓ or L3 | [79] | ||||
| C124R | OBI | Huh7/mice | ↓ | ↓3 | [66] | |||
| C124Y | OBI | Huh7/mice | ↓ | ↓3 | [66] | |||
| T125A | CHB | Huh7 | ↓ | ↓ | [65] | |||
| I126S | OBI | Huh7/mice | ↑ | ↓ | ↓3 | [66] | ||
| P127S | Huh7 | ↓3 | [79] | |||||
| Q129H | Huh7 | ↓3 | [79] | |||||
| Q129R | OBI | Huh7/mice | ↑ | ↓ | ↓3 | [66] | ||
| G130R | Huh7 | L3 | [79] | |||||
| T131I | Yeast | ↓ | [83] | |||||
| M133T4 | Huh7 | [78] | ||||||
| S136P | OBI | Huh7/mice | ↑ | ↓ | ↓3 | [66] | ||
| C1371 | OBI | Pichia | ↓ or L | [80] | ||||
| C138Y | Huh7 | ↓ | L3 | [79] | ||||
| C139R | CHB | HepG2 | ↓ | [84] | ||||
| C139R | OBI | Huh7/mice | ↓ | ↓3 | [66] | |||
| T140I | OBI | Huh7/mice | ↑ | ↓ | ↓3 | [66] | ||
| K141E | OBI | Huh7/mice | ↓ | ↑ | ↓ | ↓3 | [66] | |
| K141E | Huh7 | ↓ | ↓3 | [79] | ||||
| K141E | Yeast | ↓ | [83] | |||||
| P142S | Yeast | ↓ | [83] | |||||
| P142S | Huh7 | ↓ | [79] | |||||
| P142A | KT | Huh7 | ↓ | [85] | ||||
| D144V | KT | Huh7 | ↓ | [85] | ||||
| D144A | OBI | Huh7/mice | ↓ | ↑ | ↓ | ↓3 | [66] | |
| D144G | Huh7 | ↓ | L3 | [79] | ||||
| D144E | LC | HepG2 | ↓ | [84] | ||||
| G145R | Yeast | ↓ | [83] | |||||
| G145R | HepG2/Hela | ↓ | ↓ | ↓ | [69] | |||
| G145R | LT | Huh7 | ↓3 | [67] | ||||
| G145R | Huh7 | ↓ | ↓ | ↓ | [81] | |||
| G145R | Huh7 | ↓ | [79] | |||||
| G145R | OBI | Huh7/mice | ↓ | ↑ | ↓ | ↓3 | [66] | |
| G145A | OBI | Huh7/mice | ↓ | [66] | ||||
| G145A | OBI | Huh7/HepG2 | ↓ | ↑ | ↓ | [83] | ||
| G145A | Yeast | ↓ | [81] | |||||
| G145W | Huh7 | ↓ | ↓ | [81] | ||||
| G145I | Huh7 | ↓ | ↓ | [81] | ||||
| G145P | Huh7 | ↓ | [81] | |||||
| G145N | Huh7 | ↓ | [81] | |||||
| G145D | Huh7 | ↓ | [81] | |||||
| G145E | Huh7 | ↓ | [79] | |||||
| N146S | Huh7 | L3 | [79] | |||||
| C147A | Huh7 | ↓ or L3 | [79] | |||||
| C147R | Huh7 | ↓ | ↓3 | [79] | ||||
| C149A | Huh7 | ↓ | ↓ or L3 | [79] | ||||
| C149R | Huh7 | ↓ | L3 | [79] | ||||
| A159G | Huh7/Hela | ↓ | ↓ | ↓3 | [82] | |||
| K160N | Huh7/Hela | ↓ | ↑5 | ↓3 | [82] | |||
| R169P/G | Huh7 | ↓ | L3 | [78] | ||||
| Y100S + S143L | OBI | Huh7 | L | [77] | ||||
| M103I + G145A | OBI | Huh7/HepG2 | ↓ | ↓ | ↓ | [86] | ||
| M103I + R122K + G145A | OBI | Huh7/HepG2 | ↓ | ↓ | ↓ | [86] | ||
| T115I + T116N | CHB | HepG2 | ↓ or L | [87] | ||||
| T116N + S143L | OBI | Huh7 | ↓ | [77] | ||||
| R122P + Q101R | OBI | Huh7 | ↓ | [77] | ||||
| R122P + S167L | OBI | Huh7 | ↓ | [77] | ||||
| R122K + G145A | OBI | Huh7/HepG2 | ↓ | ↓ | ↓ | [86] | ||
| P142A + D144V | KT | Huh7 | ↓ | [85] | ||||
| A159G + K160N | Huh7/Hela | ↓ | ↑5 | ↓3 | [82] |
Stop codon mutation;
Viral replication;
Viral secretion;
Correct that I110M, G119E, G145R, N146Q/S, R169H mutations lead to the decrease of viral secretion;
Cell-mediated immunity. ↑: Increase; ↓: Decrease. Intra-HBsAg: Intracellular HBsAg; Extra-HBsAg: Extracellular HBsAg; L: Dramatic decrease; CHB: Chronic hepatitis B; OBI: Occult HBV infection; LC: Liver cirrhosis; KT: Kidney transplant; LT: Liver transplant; HBsAg: Hepatitis B virus surface antigen.
Figure 2.
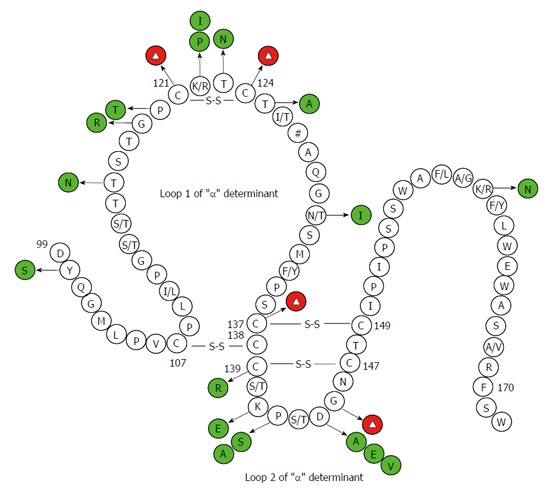
Secondary structure of the major hydrophilic region of the hepatitis B virus, hepatitis B virus surface antigen. Mutations have been confirmed by functional studies to exert negative effects on antigenicity of hepatitis B virus surface antigen (HBsAg) are shown in green and red circles. Red circles with the Δ symbol denote multiple kinds of amino acid substitutions. These have been substantiated to negatively correlate with HBsAg antigenicity at some sites. The # symbol indicates multiple genotype-specific substitutions. The numbers denote amino acid sites.
Many studies have documented that substitutions of wild-type cysteines and G145R in the “α” determinant cause conformational and phenotypic changes in HBsAg (Table 1 and Figure 2)[33,66,71,78]. El Chaar et al[80] restored the cysteines at positions 124 and 137 in recombinant surface protein (rS protein) from OBIs and expressed them in yeast and showed that antigenic reactivity of HBsAg was improved. However, restoration of cysteine at position 147 and G145R did not improve the impaired reactivity of HBsAg. Y100C was commonly found in OBIs, but transfection with Y100C plasmid resulted in higher extracellular HBsAg levels than the wild-type plasmid, implying that the mutation alone could not explain the decrease of HBsAg secretion and/or affinity to antibodies[76]. In vitro studies have found that some point mutations could strongly influence the secretion of HBsAg and that these mutations may interfere with the secretion of HBsAg in some cases, leading to the failure of HBsAg detection[86]. Some point mutations were found to affect HBV replication in hepatoma cell lines and/or in mice and the vesicular transport of infectious virions of HBV (i.e., Dane particles) from the cell[66].
Glycosylation, the most common kind being N-linked glycosylation, is required for crucial biological functions of many enveloped viruses, since it can impart various advantages to virus survival and virulence[88]. Point mutations can affect biogenesis, stability, and antigenicity of HBV through modification of the envelope protein glycosylation pattern[89,90]. The envelope protein of HBV possesses an N-glycosylation site at N146 of the “α” determinant, and the removal of this site decreased the production of virion without affecting the synthesis and stability of HBsAg (Table 1 and Figure 2)[78-80]. Several positions in the envelope proteins can substitute for position 146 as the potential glycosylation site[78]. Previous studies reported that T123N and K160N were each capable of creating new glycosylation sites and causing a decrease of HBsAg antigenicity. These two point substitutions were found to mainly affect virion assembly and secretion without interfering with virion replication. K122I and A159G appeared to affect biological properties of HBsAg and facilitated glycosylation of HBV. A159G was shown to impair greatly the assembly and secretion of virion, but K160N and A159G/K160N slightly reduced the virion production. Thus K160N could partially rescue the negative effect exerted by A159G on secretion of virion[82]. M133T mutation could create an additional glycosylation site 131NST133 and partially rescued the impaired secretion of virion induced by some point mutations, such as I110M, G119E, G145R, N146Q/S, and R169H. Preventing glycosylation could destroy the effect of the M133T mutation[78,79,82]. M133T alone was not able to generate a glycosylation site, unless there was an N at AA131 (as in the reference sequences of HBV genotype A) The T131N/M133T double mutations were frequently observed in OBIs infected with non-A genotypes and changing T131N could also abrogate the secretion-rescue effect exerted by M133T[78,79].
Among OBI related mutations, some point mutations cause HBsAg secretion deficiency by affecting the start codons of Pre-S/S gene or causing amino acid substitutions[91]. A purine at the -3 position (i.e., 3-nt upstream from the translational start site) of the AUG initiator codon is essential for protein expression, and substitution with a pyrimidine at this site makes translation more sensitive to changes at positions -1, -2, and +4 (i.e., 4-nt downstream from the translational start site)[92]. Point mutations around the initiator codons of HBV S gene (i.e., nt155-157), such as A152T and A152G/G158C double mutations, reduced HBsAg secretion by 70% and 30%, respectively, but maintained efficient virion secretion, while A152T combined with C154A or G158A completely abolished HBsAg secretion[91]. Some point mutations have an effect on RNA splicing. Hass et al[93] found that ntG458A inactivated 5’ splice site of the Pre-S2/S mRNA and reduced the level of Pre-S2/S mRNA as well as HBsAg expression and caused a low-replication phenotype. Among some genotype D OBIs, ntG173T was able to activate in cis a previously inactive splice acceptor site at neighboring nt202 and promoted RNA splicing from nt2986 to 202. The novel splicing could abolish the L, M, and S proteins without affecting the functions of polymerase[94].
Pre-S/S deletion
Deletion mutations in Pre-S/S have been frequently reported in OBI and deletions covering the Pre-S/S promoters may affect RNA splicing and exert an effect on expression of one or more envelope proteins (Figure 3)[50,74]. In cases of OBI, in-frame 183-bp deletions (nt3019-3201 or nt2983-3167) were commonly detected in the Pre-S1 region. Functional analyses found that variants harboring the 183-bp deletion mutation did not express HBsAg or secrete viral particles when transfected in cell lines, although they appeared to be replication-competent[50,95-97]. A CCAAT element (nt3143-3147) and binding sites for SP1 transcription factor are located in the Pre-S2/S promoter (nt2960-3180), which are completely and partially overlapped by the 183-bp deletion mutation, respectively[98,99]. The CCAAT element is where cellular transcription factor NF-Y binds to and plays a key role in activating the S promoter and regulating the ratio of Pre-S and S mRNA. Mutations in the CCAAT element downregulate the level of Pre-S1 mRNA while upregulate that of Pre-S2/S mRNA, resulting in a decreased amount of L protein but increased amounts of M and S proteins[100]. The SP1 sites also regulate S promoter activity and production of Pre-S and S mRNA but exert a weaker effect than the CCAAT element. Mutations that occur at SP1 sites affect the ratio of Pre-S/S mRNA as well as expression of envelope proteins[98,99]. The specific ratio of L protein and S protein is essential for assembly of the envelope particles, since an excessively high or low L/S proteins ratio could alter HBsAg assembly and secretion and reduce virion secretion[91,101,102]. Similarly, Xu and Yen[103] reported a 129-bp deletion within the Pre-S2/S promoter that affected Pre-S2/S mRNA production, leading to complete abolishment of HBsAg secretion. Chaudhuri et al[74] described several fragment deletions containing Pre-S2/S promoter in OBIs and demonstrated that full-length HBV genomes harboring these deletion variants could drastically decrease the expression of HBsAg by transfected HepG2 cells. Besides, the Pre-S1 region possesses a hepatocyte-binding site (AA21-47) that is vital to virion assembly and transport from the hepatocyte, and deletions covering this site may affect virion binding to and being secreted out of the hepatocyte[104].
Figure 3.
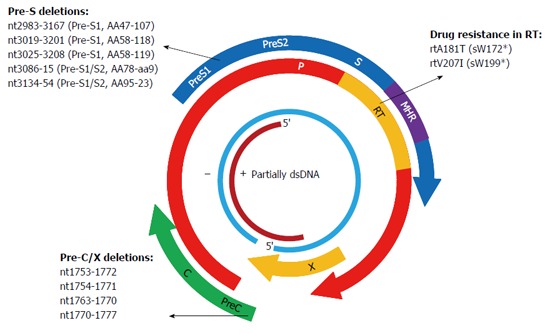
Mutations in X, P, and C genes of hepatitis B virus have been substantiated in association with the occurrence of occult hepatitis B virus infections by in vivo and in vitro studies. Plus and minus strands are denoted by + and -, respectively.
In OBIs, deletions in Pre-S2 region were frequently detected at aa8-23, which have also been associated with progression of liver diseases[74,105]. These deletions overlap with the binding site for cross-linked human serum albumin (AA17-28) in the Pre-S2 region, which is involved in the attachment of HBV to the human hepatocyte membrane, and mutations affecting this site may decrease the infectivity of HBV[106]. Deletions in Pre-S2 region may influence expression of M protein, which is dispensable for formation and secretion of virion in vivo and in vitro[91,107]. Deletions in the Pre-S2 region may be related to OBI by affecting the production of L protein. Deletions in the Pre-S2 region shorten the gap between the Pre-S2/S promoter and the start codon of S mRNA, in turn leading to an overproduction of L protein and an alternation of L/S ratio, which may decrease the secretion of HBsAg[108,109]. Deletions in the Pre-S2 region may be associated with antiviral-resistance, which is in turn linked to OBI. Mutations corresponding to lamivudine (LMV)-resistance were commonly detected in OBIs[110], and Pre-S2 deletion mutants were frequently found in patients with antiviral-resistance and shown to play a supportive role in the replication of LMV-resistant viruses by transfection analysis[111].
Pre-S/S insertion
Two to 8 amino acid insertions have been observed between codons 121 and 124 located upstream of the “α” determinant in OBI patients from the Far East, and they caused detection failure of HBsAg by conventional assays utilizing monoclonal and polyclonal antibodies[112]. Insertions occurring at these sites strongly influence the binding efficiency of HBsAg antibodies and may affect the conformation of the “α” determinant as well as the subtype determinant d/y at aa122. In addition, insertions detected in the Pre-S/S gene in OBI patients may lead to frameshift mutations or stop codon mutations and interfere with HBsAg detection.
Pre-S/S stop codon
Stop codon mutations in the Pre-S/S gene result in truncations of envelop proteins and may affect the expression and/or secretion of HBsAg. A stop codon mutation at aa216 in the C-terminus of HBsAg was observed in occult HBV/HIV co-infected patients and displayed undetectable or extremely low level of HBsAg in transfection experiments[73]. Other stop codon mutations at amino acid positions 61, 69, 181 etc. were also detected in OBI patients. These mutations may be correlated with failure in HBsAg detection, but in vivo and in vitro studies are required to confirm this association[113]. Furthermore, the Pre-S/S gene overlaps with the RT region of the P gene, and mutations corresponding to antiviral-resistance in the RT domain commonly result in stop codon mutations in HBsAg and changes in the antigenicity and secretion of HBsAg[114].
C GENE MUTATIONS
A1762T/G1764A double mutations in the BCP region and G1896A in the Pre-C region are the most frequently observed point substitutions in the C gene. These mutations prevent the production of the hepatitis B e-antigen (HBeAg) and are linked to HCC[115]. Genotypes C and D of HBV have been reported to have relatively high mutation rates in the BCP region[116]. However, A1762T/G1764A and G1896A were not common in genotype D-infected OBI patients from Turkey and India[117,118]. A study from Taiwan conducted on OBI related HCC and HBsAg-positive HCC patients found that mutations commonly found in HBsAg-positive HCC, such as A1762T/G1764A, G1896A, etc., were not frequently detected in OBI related HCC patients[119]. Therefore, point mutations in BCP/Pre-C in CHB may be not associated with OBI and are required to be explored by further functional studies. Several deletion mutations in the BCP region, such as nt1753-1772, nt1751-1770, nt1754-1771, etc., have been reported in OBIs, and variants containing deletions of nt1754-1771 were shown to reduce virion replication and expression levels of HBsAg and HBeAg (Figure 3)[97]. The BCP region recruits initiators for the transcription of Pre-C mRNA and pregenomic (pg) RNA and is rich in AT content[120,121]. Deletion mutations in the BCP region may prevent transcription of pgRNA, leading to a reduction in HBV DNA replication and HBsAg expression. Moreover, BCP region overlaps with the X gene, and deletions in the BCP region lead to truncated X protein, which also has an effect on virion replication and antigen expression[122].
X GENE MUTATIONS
X gene is a regulatory gene and generates the X protein, which is made up of 146 to 154 aa. X protein acts as a transcription factor and trans-activates gene expression of HBV and several other viruses and cells[123]. The X gene harbors enhancer elements and the C promoter, and deletion mutations covering the C promoter were frequently encountered in various HBV infections, such as OBI, HBeAg-negative infection, and so on[124]. The C-terminal end of X protein is essential for the trans-activation function of X protein[122]. Eight base pair (nt1763-1770) and 20-bp deletion (nt1753-1772) mutations at the C-terminal end of X protein were frequently observed in OBIs, and variants containing these two deletions led to truncation of X protein and a decrease in Pre-C promoter activity as well as secretion of HBsAg, HBeAg, and HBcAg in transfected cell lines (Figure 3)[125,126]. Eight nucleotide deletions (nt1640-1647 or nt1770-1777) were frequently detected in occult HBV/HCV co-infected patients, and they exerted a positive effect on HCV replication but a negative effect on HBV replication[127,128]. Besides, partial and large deletion mutations in the X gene have been reported in OBIs in renal dialysis and HBV-vaccinated thalassemia patients[129].
P GENE MUTATIONS
Since the P gene overlaps with the S gene, mutations that occur in one gene may affect the other gene. The RT region of the P gene completely spans the S region, thus mutations resistant to nucleos(t)ide analogues (NAs) in the RT region frequently lead to mutations of HBsAg (including stop codon mutations) and affect the antigenicity of HBsAg (Figure 3)[114,130]. Transient selection of a V542I mutant in the HBV polymerase has been described in a CHB patient with prolonged administration of famciclovir. The mutation corresponded to a stop codon mutation at aa199 in the S gene, and a transfection experiment with the full-length HBV genome harboring the mutation demonstrated a negative effect on replication capacity and failed to produce HBsAg[131]. The rtA181T variant was frequently found in patients with adefovir (ADV)-resistance, corresponding to a stop codon mutation at aaW172 in the S gene (sW172*) and leading to truncation of 55 aa at the C-terminal end of HBsAg. The variant not only resulted in secretion deficiency of the virion but also exerted a dominant negative effect on virion secretion despite co-transfection with wild-type isolates[132]. Other common antiviral-resistant mutations that lead to truncation of S proteins include entecavir (ETV)-resistant rtT184M corresponding to sL176*, rtM204I (YIDD) both resistant to LMV and telbivudine (LdT) corresponding to sW196*, and so on[114]. The rtM204V (YVDD), accompanied by rtV173L (corresponding to sI195M and sE164D) and/or rtL180M, were detected in both HBsAg-negative patients and HBsAg-positive patients untreated with LMV[133,134]. Variants harboring YIDD or YVDD caused a drastic reduction in virion replication capacity in transfection experiments[135]. Some studies found that LMV-selected HBsAg protein changes, including E164D, I195M, and E164D/I195M, significantly reduced the binding efficiency to anti-HBs, implying the potential to escape HBV vaccine like G145R[133]. Point mutations in the RNase H domain of P gene were observed in vaccinated children with OBIs[52]. In vitro studies demonstrated that the RNase H region is essential for both RNA packaging and DNA synthesis and that the P gene of HBV and duck hepatitis B virus (DHBV) have similar biological properties. Point mutations in the RNase H region of DHBV, such as L697Y, V719Y etc., led to deficiency of RNA packaging and significant reduction of DNA synthesis[136].
Deletion mutations in the P gene have been rarely reported. A 281-bp deletion (nt2068-2349) covering the start codon of the P gene was reported in genotype B infected OBIs, but the relationship remains to be confirmed by functional studies[137].
NO OBI-RELEVANT MUTATIONS
Mutations in the “α” determinant of the S protein are “hot spots” of OBI related mutations, but some clinical reports demonstrated that no mutation was detected in the “α” determinant or the mutation rate of the “α” determinant in OBI group was not significantly different from HBsAg-positive infections[31]. In some OBI patients, no mutation was detected in the Pre-S/S gene[117]. Moreover, no OBI-relevant mutation was found throughout the entire genome of HBV strains isolated from some OBI patients[138]. As no OBI-relevant mutation was encountered in some cases of OBI and some mutations occurred in both OBIs and HBsAg-positive infections, some reports concluded that there were no OBI-specific mutations[45]. Furthermore, in vivo and in vitro studies have demonstrated that some mutations identified in clinical reports did not interfere with HBV replication and expression or HBsAg secretion and were not associated with OBI. In a recent study, we investigated a number of OBI patients with a family history of HBV infection by cloning and sequencing the Pre-S/S region of HBV DNA isolated from serum samples. Sequence comparison between OBI patients and their family members with CHB failed to identify common genetic variations that were specific for OBI[139].
PROBLEMS
The mechanism of OBI is complex and remains to be clarified. Previous related studies have several limitations, as follows: (1) It is difficult to amplify the full-length HBV genome due to the extremely low viral load in OBI patients, and most clinical reports are limited to mutations in part of the viral genome (mainly the S gene coding for HBsAg). Studies based on full-length HBV genomes are rare; (2) Most previous studies of OBI were clinical reports with limited case numbers and no comparable controls, and conclusions about some mutations were not consistent across reports; (3) Most mutations were not studied in vivo or in vitro, and some mutations occurred both in OBIs and HBsAg-positive infections; (4) Functional studies of mutations were mostly focused on the secretion, antigenicity, or immunogenicity of HBsAg, while only a few of them addressed virion replication and secretion. Opposite conclusions may be drawn by studies on the same mutations, depending on whether a fragment or the entire genome was used (data not available); (5) Most studies focused on individual mutations, and it remained to be determined whether multiple mutations interact with each other or whether there is secondary function; (6) HBV genotype and subgenotype were ignored in most studies; (7) There is a lack of suitable reference sequences for genotype and subgenotype of HBV, and it is difficult to determine whether some amino acid substitutions were genotype-specific or OBI-related[140]; and (8) The levels of serum HBsAg in some OBI patients were fluctuant, and it remains to be elucidated whether HBV methylation, glycosylation, acetylation, or miRNA plays a role in this phenomenon.
The ability to screen for OBI is strongly affected by the assays used for detecting HBsAg and HBV DNA. For instance, the Abbott assay is highly sensitive for G145R detection. It is our opinion that multiple assays should be performed to verify the accuracy of the screening results and to reduce false positive or false negative results. NAT of HBV is highly sensitive and able to detect extremely low levels of HBV DNA but has a potential risk of false-positive results due to quality-control problems. HBV NAT is still important for recognition of OBI in HBsAg-negative blood donations worldwide especially in high-endemic countries. However, NAT has not been implemented in most of these countries due to its high cost[141]. In these countries, blood donations are screened by serological testing of HBsAg alone or combined with anti-HBc. In low-endemic regions of HBV or high-endemic regions of anti-HBc, anti-HBc testing may be not as effective as NAT[142]. Anti-HBc screening is not capable to detect pre-seroconversion window period infections and anti-HBc assays based on radioimmunoassay or enzyme immunoassay may suffer from false-positive results[143,144]. Although liver biopsy is the best way for detecting OBI, it is hard to obtain liver tissue and standardized assays for it are not yet available[145].
CONCLUSION
OBI is a special and complex form of HBV infection with worldwide distribution. Its reported prevalence significantly varies depending on study locations, detection assays used, and study population. The underlying mechanism of OBI is complex and unclear. Genetic variants may be relevant but future research is still need both in vivo and in vitro.
ACKNOWLEDGMENTS
We are deeply thankful to Dr. Meng-Ji Lu and Dr. Chun-Chen Wu for proof reading the manuscript.
Footnotes
Supported by the Specialized Research Fund for the Doctoral Program of Higher Education of China, No. 20093420120005; and the National Science Foundation of China, No. 81273142.
Conflict-of-interest statement: The authors have declared that no potential conflict of interest exists.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: October 30, 2015
First decision: November 27, 2015
Article in press: December 30, 2015
P- Reviewer: Bock CT, Georgopoulou U, Stalke P S- Editor: Ma YJ L- Editor: Filipodia E- Editor: Wang CH
References
- 1.Summers J, Mason WS. Replication of the genome of a hepatitis B--like virus by reverse transcription of an RNA intermediate. Cell. 1982;29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 2.Okamoto H, Imai M, Kametani M, Nakamura T, Mayumi M. Genomic heterogeneity of hepatitis B virus in a 54-year-old woman who contracted the infection through materno-fetal transmission. Jpn J Exp Med. 1987;57:231–236. [PubMed] [Google Scholar]
- 3.Sterneck M, Günther S, Gerlach J, Naoumov NV, Santantonio T, Fischer L, Rogiers X, Greten H, Williams R, Will H. Hepatitis B virus sequence changes evolving in liver transplant recipients with fulminant hepatitis. J Hepatol. 1997;26:754–764. doi: 10.1016/s0168-8278(97)80239-5. [DOI] [PubMed] [Google Scholar]
- 4.Kay A, Zoulim F. Hepatitis B virus genetic variability and evolution. Virus Res. 2007;127:164–176. doi: 10.1016/j.virusres.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 5.Hoofnagle JH, Seeff LB, Bales ZB, Zimmerman HJ. Type B hepatitis after transfusion with blood containing antibody to hepatitis B core antigen. N Engl J Med. 1978;298:1379–1383. doi: 10.1056/NEJM197806222982502. [DOI] [PubMed] [Google Scholar]
- 6.Raimondo G, Allain JP, Brunetto MR, Buendia MA, Chen DS, Colombo M, Craxì A, Donato F, Ferrari C, Gaeta GB, et al. Statements from the Taormina expert meeting on occult hepatitis B virus infection. J Hepatol. 2008;49:652–657. doi: 10.1016/j.jhep.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 7.Raimondo G, Navarra G, Mondello S, Costantino L, Colloredo G, Cucinotta E, Di Vita G, Scisca C, Squadrito G, Pollicino T. Occult hepatitis B virus in liver tissue of individuals without hepatic disease. J Hepatol. 2008;48:743–746. doi: 10.1016/j.jhep.2008.01.023. [DOI] [PubMed] [Google Scholar]
- 8.Walz A, Wirth S, Hucke J, Gerner P. Vertical transmission of hepatitis B virus (HBV) from mothers negative for HBV surface antigen and positive for antibody to HBV core antigen. J Infect Dis. 2009;200:1227–1231. doi: 10.1086/605698. [DOI] [PubMed] [Google Scholar]
- 9.Candotti D, Allain JP. Transfusion-transmitted hepatitis B virus infection. J Hepatol. 2009;51:798–809. doi: 10.1016/j.jhep.2009.05.020. [DOI] [PubMed] [Google Scholar]
- 10.Chemin I, Trépo C. Clinical impact of occult HBV infections. J Clin Virol. 2005;34 Suppl 1:S15–S21. doi: 10.1016/s1386-6532(05)80005-8. [DOI] [PubMed] [Google Scholar]
- 11.Wong DK, Huang FY, Lai CL, Poon RT, Seto WK, Fung J, Hung IF, Yuen MF. Occult hepatitis B infection and HBV replicative activity in patients with cryptogenic cause of hepatocellular carcinoma. Hepatology. 2011;54:829–836. doi: 10.1002/hep.24551. [DOI] [PubMed] [Google Scholar]
- 12.Chemin I, Zoulim F, Merle P, Arkhis A, Chevallier M, Kay A, Cova L, Chevallier P, Mandrand B, Trépo C. High incidence of hepatitis B infections among chronic hepatitis cases of unknown aetiology. J Hepatol. 2001;34:447–454. doi: 10.1016/s0168-8278(00)00100-8. [DOI] [PubMed] [Google Scholar]
- 13.Pollicino T, Squadrito G, Cerenzia G, Cacciola I, Raffa G, Craxi A, Farinati F, Missale G, Smedile A, Tiribelli C, et al. Hepatitis B virus maintains its pro-oncogenic properties in the case of occult HBV infection. Gastroenterology. 2004;126:102–110. doi: 10.1053/j.gastro.2003.10.048. [DOI] [PubMed] [Google Scholar]
- 14.Westhoff TH, Jochimsen F, Schmittel A, Stoffler-Meilicke M, Schafer JH, Zidek W, Gerlich WH, Thiel E. Fatal hepatitis B virus reactivation by an escape mutant following rituximab therapy. Blood. 2003;102:1930. doi: 10.1182/blood-2003-05-1403. [DOI] [PubMed] [Google Scholar]
- 15.Altfeld M, Rockstroh JK, Addo M, Kupfer B, Pult I, Will H, Spengler U. Reactivation of hepatitis B in a long-term anti-HBs-positive patient with AIDS following lamivudine withdrawal. J Hepatol. 1998;29:306–309. doi: 10.1016/s0168-8278(98)80017-2. [DOI] [PubMed] [Google Scholar]
- 16.Kidd-Ljunggren K, Simonsen O. Reappearance of hepatitis B 10 years after kidney transplantation. N Engl J Med. 1999;341:127–128. doi: 10.1056/NEJM199907083410213. [DOI] [PubMed] [Google Scholar]
- 17.Schmeltzer P, Sherman KE. Occult hepatitis B: clinical implications and treatment decisions. Dig Dis Sci. 2010;55:3328–3335. doi: 10.1007/s10620-010-1413-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu KQ. Occult hepatitis B virus infection and its clinical implications. J Viral Hepat. 2002;9:243–257. doi: 10.1046/j.1365-2893.2002.00344.x. [DOI] [PubMed] [Google Scholar]
- 19.Yuen MF, Lee CK, Wong DK, Fung J, Hung I, Hsu A, But DY, Cheung TK, Chan P, Yuen JC, et al. Prevalence of occult hepatitis B infection in a highly endemic area for chronic hepatitis B: a study of a large blood donor population. Gut. 2010;59:1389–1393. doi: 10.1136/gut.2010.209148. [DOI] [PubMed] [Google Scholar]
- 20.Zheng X, Ye X, Zhang L, Wang W, Shuai L, Wang A, Zeng J, Candotti D, Allain JP, Li C. Characterization of occult hepatitis B virus infection from blood donors in China. J Clin Microbiol. 2011;49:1730–1737. doi: 10.1128/JCM.00145-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seo DH, Whang DH, Song EY, Kim HS, Park Q. Prevalence of antibodies to hepatitis B core antigen and occult hepatitis B virus infections in Korean blood donors. Transfusion. 2011;51:1840–1846. doi: 10.1111/j.1537-2995.2010.03056.x. [DOI] [PubMed] [Google Scholar]
- 22.Satake M, Taira R, Yugi H, Hino S, Kanemitsu K, Ikeda H, Tadokoro K. Infectivity of blood components with low hepatitis B virus DNA levels identified in a lookback program. Transfusion. 2007;47:1197–1205. doi: 10.1111/j.1537-2995.2007.01276.x. [DOI] [PubMed] [Google Scholar]
- 23.O’Brien SF, Fearon MA, Yi QL, Fan W, Scalia V, Muntz IR, Vamvakas EC. Hepatitis B virus DNA-positive, hepatitis B surface antigen-negative blood donations intercepted by anti-hepatitis B core antigen testing: the Canadian Blood Services experience. Transfusion. 2007;47:1809–1815. doi: 10.1111/j.1537-2995.2007.01396.x. [DOI] [PubMed] [Google Scholar]
- 24.Hollinger FB. Hepatitis B virus infection and transfusion medicine: science and the occult. Transfusion. 2008;48:1001–1026. doi: 10.1111/j.1537-2995.2008.01701.x. [DOI] [PubMed] [Google Scholar]
- 25.Hofer M, Joller-Jemelka HI, Grob PJ, Lüthy R, Opravil M. Frequent chronic hepatitis B virus infection in HIV-infected patients positive for antibody to hepatitis B core antigen only. Swiss HIV Cohort Study. Eur J Clin Microbiol Infect Dis. 1998;17:6–13. doi: 10.1007/BF01584356. [DOI] [PubMed] [Google Scholar]
- 26.Núñez M, Ríos P, Pérez-Olmeda M, Soriano V. Lack of ‘occult’ hepatitis B virus infection in HIV-infected patients. AIDS. 2002;16:2099–2101. doi: 10.1097/00002030-200210180-00024. [DOI] [PubMed] [Google Scholar]
- 27.Cacciola I, Pollicino T, Squadrito G, Cerenzia G, Orlando ME, Raimondo G. Occult hepatitis B virus infection in patients with chronic hepatitis C liver disease. N Engl J Med. 1999;341:22–26. doi: 10.1056/NEJM199907013410104. [DOI] [PubMed] [Google Scholar]
- 28.De Maria N, Colantoni A, Friedlander L, Leandro G, Idilman R, Harig J, Van Thiel DH. The impact of previous HBV infection on the course of chronic hepatitis C. Am J Gastroenterol. 2000;95:3529–3536. doi: 10.1111/j.1572-0241.2000.03371.x. [DOI] [PubMed] [Google Scholar]
- 29.Gutiérrez-García ML, Fernandez-Rodriguez CM, Lledo-Navarro JL, Buhigas-Garcia I. Prevalence of occult hepatitis B virus infection. World J Gastroenterol. 2011;17:1538–1542. doi: 10.3748/wjg.v17.i12.1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minuk GY, Sun DF, Greenberg R, Zhang M, Hawkins K, Uhanova J, Gutkin A, Bernstein K, Giulivi A, Osiowy C. Occult hepatitis B virus infection in a North American adult hemodialysis patient population. Hepatology. 2004;40:1072–1077. doi: 10.1002/hep.20435. [DOI] [PubMed] [Google Scholar]
- 31.Chan HL, Tsang SW, Leung NW, Tse CH, Hui Y, Tam JS, Chan FK, Sung JJ. Occult HBV infection in cryptogenic liver cirrhosis in an area with high prevalence of HBV infection. Am J Gastroenterol. 2002;97:1211–1215. doi: 10.1111/j.1572-0241.2002.05706.x. [DOI] [PubMed] [Google Scholar]
- 32.Fang Y, Shang QL, Liu JY, Li D, Xu WZ, Teng X, Zhao HW, Fu LJ, Zhang FM, Gu HX. Prevalence of occult hepatitis B virus infection among hepatopathy patients and healthy people in China. J Infect. 2009;58:383–388. doi: 10.1016/j.jinf.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 33.Ireland JH, O’Donnell B, Basuni AA, Kean JD, Wallace LA, Lau GK, Carman WF. Reactivity of 13 in vitro expressed hepatitis B surface antigen variants in 7 commercial diagnostic assays. Hepatology. 2000;31:1176–1182. doi: 10.1053/he.2000.6407. [DOI] [PubMed] [Google Scholar]
- 34.Weber B. Diagnostic impact of the genetic variability of the hepatitis B virus surface antigen gene. J Med Virol. 2006;78 Suppl 1:S59–S65. doi: 10.1002/jmv.20610. [DOI] [PubMed] [Google Scholar]
- 35.Kaneko S, Miller RH, Feinstone SM, Unoura M, Kobayashi K, Hattori N, Purcell RH. Detection of serum hepatitis B virus DNA in patients with chronic hepatitis using the polymerase chain reaction assay. Proc Natl Acad Sci USA. 1989;86:312–316. doi: 10.1073/pnas.86.1.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roth WK, Weber M, Seifried E. Feasibility and efficacy of routine PCR screening of blood donations for hepatitis C virus, hepatitis B virus, and HIV-1 in a blood-bank setting. Lancet. 1999;353:359–363. doi: 10.1016/S0140-6736(98)06318-1. [DOI] [PubMed] [Google Scholar]
- 37.Zerbini A, Pilli M, Boni C, Fisicaro P, Penna A, Di Vincenzo P, Giuberti T, Orlandini A, Raffa G, Pollicino T, et al. The characteristics of the cell-mediated immune response identify different profiles of occult hepatitis B virus infection. Gastroenterology. 2008;134:1470–1481. doi: 10.1053/j.gastro.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 38.Levrero M, Pollicino T, Petersen J, Belloni L, Raimondo G, Dandri M. Control of cccDNA function in hepatitis B virus infection. J Hepatol. 2009;51:581–592. doi: 10.1016/j.jhep.2009.05.022. [DOI] [PubMed] [Google Scholar]
- 39.Zhang X, Hou J, Lu M. Regulation of hepatitis B virus replication by epigenetic mechanisms and microRNAs. Front Genet. 2013;4:202. doi: 10.3389/fgene.2013.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pollicino T, Raffa G, Costantino L, Lisa A, Campello C, Squadrito G, Levrero M, Raimondo G. Molecular and functional analysis of occult hepatitis B virus isolates from patients with hepatocellular carcinoma. Hepatology. 2007;45:277–285. doi: 10.1002/hep.21529. [DOI] [PubMed] [Google Scholar]
- 41.Lada O, Benhamou Y, Poynard T, Thibault V. Coexistence of hepatitis B surface antigen (HBs Ag) and anti-HBs antibodies in chronic hepatitis B virus carriers: influence of “a” determinant variants. J Virol. 2006;80:2968–2975. doi: 10.1128/JVI.80.6.2968-2975.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang JM, Xu Y, Wang XY, Yin YK, Wu XH, Weng XH, Lu M. Coexistence of hepatitis B surface antigen (HBsAg) and heterologous subtype-specific antibodies to HBsAg among patients with chronic hepatitis B virus infection. Clin Infect Dis. 2007;44:1161–1169. doi: 10.1086/513200. [DOI] [PubMed] [Google Scholar]
- 43.Zanella I, Rossini A, Domenighini D, Albertini A, Cariani E. Real-time quantitation of hepatitis B virus (HBV) DNA in tumorous and surrounding tissue from patients with hepatocellular carcinoma. J Med Virol. 2002;68:494–499. doi: 10.1002/jmv.10243. [DOI] [PubMed] [Google Scholar]
- 44.Kaur P, Paliwal A, Durantel D, Hainaut P, Scoazec JY, Zoulim F, Chemin I, Herceg Z. DNA methylation of hepatitis B virus (HBV) genome associated with the development of hepatocellular carcinoma and occult HBV infection. J Infect Dis. 2010;202:700–704. doi: 10.1086/655398. [DOI] [PubMed] [Google Scholar]
- 45.Vivekanandan P, Kannangai R, Ray SC, Thomas DL, Torbenson M. Comprehensive genetic and epigenetic analysis of occult hepatitis B from liver tissue samples. Clin Infect Dis. 2008;46:1227–1236. doi: 10.1086/529437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bläckberg J, Kidd-Ljunggren K. Occult hepatitis B virus after acute self-limited infection persisting for 30 years without sequence variation. J Hepatol. 2000;33:992–997. doi: 10.1016/s0168-8278(00)80134-8. [DOI] [PubMed] [Google Scholar]
- 47.Mason A, Yoffe B, Noonan C, Mearns M, Campbell C, Kelley A, Perrillo RP. Hepatitis B virus DNA in peripheral-blood mononuclear cells in chronic hepatitis B after HBsAg clearance. Hepatology. 1992;16:36–41. doi: 10.1002/hep.1840160108. [DOI] [PubMed] [Google Scholar]
- 48.Michalak TI. Occult persistence and lymphotropism of hepadnaviral infection: insights from the woodchuck viral hepatitis model. Immunol Rev. 2000;174:98–111. doi: 10.1034/j.1600-0528.2002.017406.x. [DOI] [PubMed] [Google Scholar]
- 49.Mulrooney-Cousins PM, Michalak TI. Persistent occult hepatitis B virus infection: experimental findings and clinical implications. World J Gastroenterol. 2007;13:5682–5686. doi: 10.3748/wjg.v13.i43.5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cabrerizo M, Bartolomé J, Caramelo C, Barril G, Carreno V. Molecular analysis of hepatitis B virus DNA in serum and peripheral blood mononuclear cells from hepatitis B surface antigen-negative cases. Hepatology. 2000;32:116–123. doi: 10.1053/jhep.2000.8541. [DOI] [PubMed] [Google Scholar]
- 51.Colson P, Borentain P, Coso D, Motte A, Aurran-Schleinitz T, Charbonnier A, Stoppa AM, Chabannon C, Serrero M, Bertrand J, et al. Hepatitis B virus reactivation in HBsAg-negative patients is associated with emergence of viral strains with mutated HBsAg and reverse transcriptase. Virology. 2015;484:354–363. doi: 10.1016/j.virol.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 52.Shahmoradi S, Yahyapour Y, Mahmoodi M, Alavian SM, Fazeli Z, Jazayeri SM. High prevalence of occult hepatitis B virus infection in children born to HBsAg-positive mothers despite prophylaxis with hepatitis B vaccination and HBIG. J Hepatol. 2012;57:515–521. doi: 10.1016/j.jhep.2012.04.021. [DOI] [PubMed] [Google Scholar]
- 53.Mu SC, Lin YM, Jow GM, Chen BF. Occult hepatitis B virus infection in hepatitis B vaccinated children in Taiwan. J Hepatol. 2009;50:264–272. doi: 10.1016/j.jhep.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 54.Ramezani A, Banifazl M, Mamishi S, Sofian M, Eslamifar A, Aghakhani A. The influence of human leukocyte antigen and IL-10 gene polymorphisms on hepatitis B virus outcome. Hepat Mon. 2012;12:320–325. doi: 10.5812/hepatmon.6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Askari A, Hassanshahi GH, Ghalebi SR, Jafarzadeh A, Mohit M, Hajghani M, Kazemi Arababadi M. Intensity of HLA-A2 Expression Significantly Decreased in Occult Hepatitis B Infection. Jundishapur J Microbiol. 2014;7:e10298. doi: 10.5812/jjm.10298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arababadi MK, Pourfathollah AA, Jafarzadeh A, Hassanshahi G. Serum Levels of IL-10 and IL-17A in Occult HBV-Infected South-East Iranian Patients. Hepat Mon. 2010;10:31–35. [PMC free article] [PubMed] [Google Scholar]
- 57.Ahmadabadi BN, Hassanshahi G, Arababadi MK, Leanza C, Kennedy D. The IL-10 promoter polymorphism at position -592 is correlated with susceptibility to occult HBV infection. Inflammation. 2012;35:818–821. doi: 10.1007/s10753-011-9381-x. [DOI] [PubMed] [Google Scholar]
- 58.Zhang X, Zhang E, Ma Z, Pei R, Jiang M, Schlaak JF, Roggendorf M, Lu M. Modulation of hepatitis B virus replication and hepatocyte differentiation by MicroRNA-1. Hepatology. 2011;53:1476–1485. doi: 10.1002/hep.24195. [DOI] [PubMed] [Google Scholar]
- 59.Chen Y, Li L, Zhou Z, Wang N, Zhang CY, Zen K. A pilot study of serum microRNA signatures as a novel biomarker for occult hepatitis B virus infection. Med Microbiol Immunol. 2012;201:389–395. doi: 10.1007/s00430-011-0223-0. [DOI] [PubMed] [Google Scholar]
- 60.Pollicino T, Belloni L, Raffa G, Pediconi N, Squadrito G, Raimondo G, Levrero M. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology. 2006;130:823–837. doi: 10.1053/j.gastro.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 61.Khattab E, Chemin I, Vuillermoz I, Vieux C, Mrani S, Guillaud O, Trepo C, Zoulim F. Analysis of HCV co-infection with occult hepatitis B virus in patients undergoing IFN therapy. J Clin Virol. 2005;33:150–157. doi: 10.1016/j.jcv.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 62.Sagnelli E, Coppola N, Scolastico C, Filippini P, Santantonio T, Stroffolini T, Piccinino F. Virologic and clinical expressions of reciprocal inhibitory effect of hepatitis B, C, and delta viruses in patients with chronic hepatitis. Hepatology. 2000;32:1106–1110. doi: 10.1053/jhep.2000.19288. [DOI] [PubMed] [Google Scholar]
- 63.Shih CM, Lo SJ, Miyamura T, Chen SY, Lee YH. Suppression of hepatitis B virus expression and replication by hepatitis C virus core protein in HuH-7 cells. J Virol. 1993;67:5823–5832. doi: 10.1128/jvi.67.10.5823-5832.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seeger C, Mason WS. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000;64:51–68. doi: 10.1128/mmbr.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hsu CW, Yeh CT. Emergence of hepatitis B virus S gene mutants in patients experiencing hepatitis B surface antigen seroconversion after peginterferon therapy. Hepatology. 2011;54:101–108. doi: 10.1002/hep.24363. [DOI] [PubMed] [Google Scholar]
- 66.Huang CH, Yuan Q, Chen PJ, Zhang YL, Chen CR, Zheng QB, Yeh SH, Yu H, Xue Y, Chen YX, et al. Influence of mutations in hepatitis B virus surface protein on viral antigenicity and phenotype in occult HBV strains from blood donors. J Hepatol. 2012;57:720–729. doi: 10.1016/j.jhep.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 67.Kalinina T, Iwanski A, Will H, Sterneck M. Deficiency in virion secretion and decreased stability of the hepatitis B virus immune escape mutant G145R. Hepatology. 2003;38:1274–1281. doi: 10.1053/jhep.2003.50484. [DOI] [PubMed] [Google Scholar]
- 68.Norder H, Couroucé AM, Magnius LO. Molecular basis of hepatitis B virus serotype variations within the four major subtypes. J Gen Virol. 1992;73(Pt 12):3141–3145. doi: 10.1099/0022-1317-73-12-3141. [DOI] [PubMed] [Google Scholar]
- 69.Tian Y, Xu Y, Zhang Z, Meng Z, Qin L, Lu M, Yang D. The amino Acid residues at positions 120 to 123 are crucial for the antigenicity of hepatitis B surface antigen. J Clin Microbiol. 2007;45:2971–2978. doi: 10.1128/JCM.00508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bes M, Vargas V, Piron M, Casamitjana N, Esteban JI, Vilanova N, Pinacho A, Quer J, Puig L, Guardia J, et al. T cell responses and viral variability in blood donation candidates with occult hepatitis B infection. J Hepatol. 2012;56:765–774. doi: 10.1016/j.jhep.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 71.Carman WF, Korula J, Wallace L, MacPhee R, Mimms L, Decker R. Fulminant reactivation of hepatitis B due to envelope protein mutant that escaped detection by monoclonal HBsAg ELISA. Lancet. 1995;345:1406–1407. doi: 10.1016/s0140-6736(95)92599-6. [DOI] [PubMed] [Google Scholar]
- 72.Carman WF, Zanetti AR, Karayiannis P, Waters J, Manzillo G, Tanzi E, Zuckerman AJ, Thomas HC. Vaccine-induced escape mutant of hepatitis B virus. Lancet. 1990;336:325–329. doi: 10.1016/0140-6736(90)91874-a. [DOI] [PubMed] [Google Scholar]
- 73.Märschenz S, Endres AS, Brinckmann A, Heise T, Kristiansen G, Nürnberg P, Krüger DH, Günther S, Meisel H. Functional analysis of complex hepatitis B virus variants associated with development of liver cirrhosis. Gastroenterology. 2006;131:765–780. doi: 10.1053/j.gastro.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 74.Chaudhuri V, Tayal R, Nayak B, Acharya SK, Panda SK. Occult hepatitis B virus infection in chronic liver disease: full-length genome and analysis of mutant surface promoter. Gastroenterology. 2004;127:1356–1371. doi: 10.1053/j.gastro.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 75.Qiu J, Qin B, Rayner S, Wu CC, Pei RJ, Xu S, Wang Y, Chen XW. Novel evidence suggests Hepatitis B virus surface proteins participate in regulation of HBV genome replication. Virol Sin. 2011;26:131–138. doi: 10.1007/s12250-011-3190-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mello FC, Martel N, Gomes SA, Araujo NM. Expression of Hepatitis B Virus Surface Antigen Containing Y100C Variant Frequently Detected in Occult HBV Infection. Hepat Res Treat. 2011;2011:695859. doi: 10.1155/2011/695859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Svicher V, Cento V, Bernassola M, Neumann-Fraune M, Van Hemert F, Chen M, Salpini R, Liu C, Longo R, Visca M, et al. Novel HBsAg markers tightly correlate with occult HBV infection and strongly affect HBsAg detection. Antiviral Res. 2012;93:86–93. doi: 10.1016/j.antiviral.2011.10.022. [DOI] [PubMed] [Google Scholar]
- 78.Ito K, Qin Y, Guarnieri M, Garcia T, Kwei K, Mizokami M, Zhang J, Li J, Wands JR, Tong S. Impairment of hepatitis B virus virion secretion by single-amino-acid substitutions in the small envelope protein and rescue by a novel glycosylation site. J Virol. 2010;84:12850–12861. doi: 10.1128/JVI.01499-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kwei K, Tang X, Lok AS, Sureau C, Garcia T, Li J, Wands J, Tong S. Impaired virion secretion by hepatitis B virus immune escape mutants and its rescue by wild-type envelope proteins or a second-site mutation. J Virol. 2013;87:2352–2357. doi: 10.1128/JVI.02701-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.El Chaar M, Candotti D, Crowther RA, Allain JP. Impact of hepatitis B virus surface protein mutations on the diagnosis of occult hepatitis B virus infection. Hepatology. 2010;52:1600–1610. doi: 10.1002/hep.23886. [DOI] [PubMed] [Google Scholar]
- 81.Wu C, Deng W, Deng L, Cao L, Qin B, Li S, Wang Y, Pei R, Yang D, Lu M, et al. Amino acid substitutions at positions 122 and 145 of hepatitis B virus surface antigen (HBsAg) determine the antigenicity and immunogenicity of HBsAg and influence in vivo HBsAg clearance. J Virol. 2012;86:4658–4669. doi: 10.1128/JVI.06353-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu C, Zhang X, Tian Y, Song J, Yang D, Roggendorf M, Lu M, Chen X. Biological significance of amino acid substitutions in hepatitis B surface antigen (HBsAg) for glycosylation, secretion, antigenicity and immunogenicity of HBsAg and hepatitis B virus replication. J Gen Virol. 2010;91:483–492. doi: 10.1099/vir.0.012740-0. [DOI] [PubMed] [Google Scholar]
- 83.Seddigh-Tonekaboni S, Waters JA, Jeffers S, Gehrke R, Ofenloch B, Horsch A, Hess G, Thomas HC, Karayiannis P. Effect of variation in the common “a” determinant on the antigenicity of hepatitis B surface antigen. J Med Virol. 2000;60:113–121. doi: 10.1002/(sici)1096-9071(200002)60:2<113::aid-jmv2>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 84.Kim KH, Chang HY, Park JY, Park ES, Park YK, Han KH, Ahn SH. Spontaneous HBsAg loss in Korean patients: relevance of viral genotypes, S gene mutations, and covalently closed circular DNA copy numbers. Clin Mol Hepatol. 2014;20:251–260. doi: 10.3350/cmh.2014.20.3.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schories M, Peters T, Rasenack J. Isolation, characterization and biological significance of hepatitis B virus mutants from serum of a patient with immunologically negative HBV infection. J Hepatol. 2000;33:799–811. doi: 10.1016/s0168-8278(00)80313-x. [DOI] [PubMed] [Google Scholar]
- 86.Martin CM, Welge JA, Rouster SD, Shata MT, Sherman KE, Blackard JT. Mutations associated with occult hepatitis B virus infection result in decreased surface antigen expression in vitro. J Viral Hepat. 2012;19:716–723. doi: 10.1111/j.1365-2893.2012.01595.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tang JH, Yeh CT, Chen TC, Hsieh SY, Chu CM, Liaw YF. Emergence of an S gene mutant during thymosin alpha1 therapy in a patient with chronic hepatitis B. J Infect Dis. 1998;178:866–869. doi: 10.1086/515345. [DOI] [PubMed] [Google Scholar]
- 88.Vigerust DJ, Shepherd VL. Virus glycosylation: role in virulence and immune interactions. Trends Microbiol. 2007;15:211–218. doi: 10.1016/j.tim.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Julithe R, Abou-Jaoudé G, Sureau C. Modification of the hepatitis B virus envelope protein glycosylation pattern interferes with secretion of viral particles, infectivity, and susceptibility to neutralizing antibodies. J Virol. 2014;88:9049–9059. doi: 10.1128/JVI.01161-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yu DM, Li XH, Mom V, Lu ZH, Liao XW, Han Y, Pichoud C, Gong QM, Zhang DH, Zhang Y, et al. N-glycosylation mutations within hepatitis B virus surface major hydrophilic region contribute mostly to immune escape. J Hepatol. 2014;60:515–522. doi: 10.1016/j.jhep.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 91.Garcia T, Li J, Sureau C, Ito K, Qin Y, Wands J, Tong S. Drastic reduction in the production of subviral particles does not impair hepatitis B virus virion secretion. J Virol. 2009;83:11152–11165. doi: 10.1128/JVI.00905-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 93.Hass M, Hannoun C, Kalinina T, Sommer G, Manegold C, Günther S. Functional analysis of hepatitis B virus reactivating in hepatitis B surface antigen-negative individuals. Hepatology. 2005;42:93–103. doi: 10.1002/hep.20748. [DOI] [PubMed] [Google Scholar]
- 94.van Hemert FJ, Zaaijer HL, Berkhout B, Lukashov VV. Occult hepatitis B infection: an evolutionary scenario. Virol J. 2008;5:146. doi: 10.1186/1743-422X-5-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Melegari M, Scaglioni PP, Wands JR. The small envelope protein is required for secretion of a naturally occurring hepatitis B virus mutant with pre-S1 deleted. J Virol. 1997;71:5449–5454. doi: 10.1128/jvi.71.7.5449-5454.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gerner PR, Friedt M, Oettinger R, Lausch E, Wirth S. The hepatitis B virus seroconversion to anti-HBe is frequently associated with HBV genotype changes and selection of preS2-defective particles in chronically infected children. Virology. 1998;245:163–172. doi: 10.1006/viro.1998.9126. [DOI] [PubMed] [Google Scholar]
- 97.Fang Y, Teng X, Xu WZ, Li D, Zhao HW, Fu LJ, Zhang FM, Gu HX. Molecular characterization and functional analysis of occult hepatitis B virus infection in Chinese patients infected with genotype C. J Med Virol. 2009;81:826–835. doi: 10.1002/jmv.21463. [DOI] [PubMed] [Google Scholar]
- 98.Raney AK, Le HB, McLachlan A. Regulation of transcription from the hepatitis B virus major surface antigen promoter by the Sp1 transcription factor. J Virol. 1992;66:6912–6921. doi: 10.1128/jvi.66.12.6912-6921.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lu CC, Yen TS. Activation of the hepatitis B virus S promoter by transcription factor NF-Y via a CCAAT element. Virology. 1996;225:387–394. doi: 10.1006/viro.1996.0613. [DOI] [PubMed] [Google Scholar]
- 100.Bock CT, Kubicka S, Manns MP, Trautwein C. Two control elements in the hepatitis B virus S-promoter are important for full promoter activity mediated by CCAAT-binding factor. Hepatology. 1999;29:1236–1247. doi: 10.1002/hep.510290426. [DOI] [PubMed] [Google Scholar]
- 101.Bock CT, Tillmann HL, Maschek HJ, Manns MP, Trautwein C. A preS mutation isolated from a patient with chronic hepatitis B infection leads to virus retention and misassembly. Gastroenterology. 1997;113:1976–1982. doi: 10.1016/s0016-5085(97)70018-0. [DOI] [PubMed] [Google Scholar]
- 102.Sengupta S, Rehman S, Durgapal H, Acharya SK, Panda SK. Role of surface promoter mutations in hepatitis B surface antigen production and secretion in occult hepatitis B virus infection. J Med Virol. 2007;79:220–228. doi: 10.1002/jmv.20790. [DOI] [PubMed] [Google Scholar]
- 103.Xu Z, Yen TS. Intracellular retention of surface protein by a hepatitis B virus mutant that releases virion particles. J Virol. 1996;70:133–140. doi: 10.1128/jvi.70.1.133-140.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ganem D. Assembly of hepadnaviral virions and subviral particles. Curr Top Microbiol Immunol. 1991;168:61–83. doi: 10.1007/978-3-642-76015-0_4. [DOI] [PubMed] [Google Scholar]
- 105.Bruss V. Revisiting the cytopathic effect of hepatitis B virus infection. Hepatology. 2002;36:1327–1329. doi: 10.1053/jhep.2002.37351. [DOI] [PubMed] [Google Scholar]
- 106.Itoh Y, Takai E, Ohnuma H, Kitajima K, Tsuda F, Machida A, Mishiro S, Nakamura T, Miyakawa Y, Mayumi M. A synthetic peptide vaccine involving the product of the pre-S(2) region of hepatitis B virus DNA: protective efficacy in chimpanzees. Proc Natl Acad Sci USA. 1986;83:9174–9178. doi: 10.1073/pnas.83.23.9174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bruss V, Ganem D. The role of envelope proteins in hepatitis B virus assembly. Proc Natl Acad Sci USA. 1991;88:1059–1063. doi: 10.1073/pnas.88.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pollicino T, Cacciola I, Saffioti F, Raimondo G. Hepatitis B virus PreS/S gene variants: pathobiology and clinical implications. J Hepatol. 2014;61:408–417. doi: 10.1016/j.jhep.2014.04.041. [DOI] [PubMed] [Google Scholar]
- 109.Kim H, Lee SA, Kim DW, Lee SH, Kim BJ. Naturally occurring mutations in large surface genes related to occult infection of hepatitis B virus genotype C. PLoS One. 2013;8:e54486. doi: 10.1371/journal.pone.0054486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Besisik F, Karaca C, Akyüz F, Horosanli S, Onel D, Badur S, Sever MS, Danalioglu A, Demir K, Kaymakoglu S, et al. Occult HBV infection and YMDD variants in hemodialysis patients with chronic HCV infection. J Hepatol. 2003;38:506–510. doi: 10.1016/s0168-8278(02)00457-9. [DOI] [PubMed] [Google Scholar]
- 111.Zhang D, Dong P, Zhang K, Deng L, Bach C, Chen W, Li F, Protzer U, Ding H, Zeng C. Whole genome HBV deletion profiles and the accumulation of preS deletion mutant during antiviral treatment. BMC Microbiol. 2012;12:307. doi: 10.1186/1471-2180-12-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hou J, Wang Z, Cheng J, Lin Y, Lau GK, Sun J, Zhou F, Waters J, Karayiannis P, Luo K. Prevalence of naturally occurring surface gene variants of hepatitis B virus in nonimmunized surface antigen-negative Chinese carriers. Hepatology. 2001;34:1027–1034. doi: 10.1053/jhep.2001.28708. [DOI] [PubMed] [Google Scholar]
- 113.Cassini R, De Mitri MS, Gibellini D, Urbinati L, Bagaglio S, Morsica G, Domenicali M, Verucchi G, Bernardi M. A novel stop codon mutation within the hepatitis B surface gene is detected in the liver but not in the peripheral blood mononuclear cells of HIV-infected individuals with occult HBV infection. J Viral Hepat. 2013;20:42–49. doi: 10.1111/j.1365-2893.2012.01623.x. [DOI] [PubMed] [Google Scholar]
- 114.Zoulim F, Locarnini S. Hepatitis B virus resistance to nucleos(t)ide analogues. Gastroenterology. 2009;137:1593–608.e1-2. doi: 10.1053/j.gastro.2009.08.063. [DOI] [PubMed] [Google Scholar]
- 115.Yang HI, Yeh SH, Chen PJ, Iloeje UH, Jen CL, Su J, Wang LY, Lu SN, You SL, Chen DS, et al. Associations between hepatitis B virus genotype and mutants and the risk of hepatocellular carcinoma. J Natl Cancer Inst. 2008;100:1134–1143. doi: 10.1093/jnci/djn243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu CJ, Kao JH. Global perspective on the natural history of chronic hepatitis B: role of hepatitis B virus genotypes A to J. Semin Liver Dis. 2013;33:97–102. doi: 10.1055/s-0033-1345716. [DOI] [PubMed] [Google Scholar]
- 117.Pinarbasi B, Onel D, Cosan F, Akyuz F, Dirlik N, Cakaloglu Y, Badur S, Besisik F, Demir K, Okten A, et al. Prevalence and virological features of occult hepatitis B virus infection in female sex workers who work uncontrolled in Turkey. Liver Int. 2009;29:227–230. doi: 10.1111/j.1478-3231.2008.01809.x. [DOI] [PubMed] [Google Scholar]
- 118.Chandra PK, Biswas A, Datta S, Banerjee A, Panigrahi R, Chakrabarti S, De BK, Chakravarty R. Subgenotypes of hepatitis B virus genotype D (D1, D2, D3 and D5) in India: differential pattern of mutations, liver injury and occult HBV infection. J Viral Hepat. 2009;16:749–756. doi: 10.1111/j.1365-2893.2009.01129.x. [DOI] [PubMed] [Google Scholar]
- 119.Chen CH, Changchien CS, Lee CM, Tung WC, Hung CH, Hu TH, Wang JH, Wang JC, Lu SN. A study on sequence variations in pre-S/surface, X and enhancer II/core promoter/precore regions of occult hepatitis B virus in non-B, non-C hepatocellular carcinoma patients in Taiwan. Int J Cancer. 2009;125:621–629. doi: 10.1002/ijc.24416. [DOI] [PubMed] [Google Scholar]
- 120.Okamoto H, Tsuda F, Akahane Y, Sugai Y, Yoshiba M, Moriyama K, Tanaka T, Miyakawa Y, Mayumi M. Hepatitis B virus with mutations in the core promoter for an e antigen-negative phenotype in carriers with antibody to e antigen. J Virol. 1994;68:8102–8110. doi: 10.1128/jvi.68.12.8102-8110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.López-Cabrera M, Letovsky J, Hu KQ, Siddiqui A. Multiple liver-specific factors bind to the hepatitis B virus core/pregenomic promoter: trans-activation and repression by CCAAT/enhancer binding protein. Proc Natl Acad Sci USA. 1990;87:5069–5073. doi: 10.1073/pnas.87.13.5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Arii M, Takada S, Koike K. Identification of three essential regions of hepatitis B virus X protein for trans-activation function. Oncogene. 1992;7:397–403. [PubMed] [Google Scholar]
- 123.Colgrove R, Simon G, Ganem D. Transcriptional activation of homologous and heterologous genes by the hepatitis B virus X gene product in cells permissive for viral replication. J Virol. 1989;63:4019–4026. doi: 10.1128/jvi.63.9.4019-4026.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang Y, Chen P, Wu X, Sun AL, Wang H, Zhu YA, Li ZP. A new enhancer element, ENII, identified in the X gene of hepatitis B virus. J Virol. 1990;64:3977–3981. doi: 10.1128/jvi.64.8.3977-3981.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Moriyama K. Reduced antigen production by hepatitis B virus harbouring nucleotide deletions in the overlapping X gene and precore-core promoter. J Gen Virol. 1997;78(Pt 6):1479–1486. doi: 10.1099/0022-1317-78-6-1479. [DOI] [PubMed] [Google Scholar]
- 126.Laskus T, Rakela J, Tong MJ, Nowicki MJ, Mosley JW, Persing DH. Naturally occurring hepatitis B virus mutants with deletions in the core promoter region. J Hepatol. 1994;20:837–841. doi: 10.1016/s0168-8278(05)80158-8. [DOI] [PubMed] [Google Scholar]
- 127.Fukuda R, Ishimura N, Niigaki M, Hamamoto S, Satoh S, Tanaka S, Kushiyama Y, Uchida Y, Ihihara S, Akagi S, et al. Serologically silent hepatitis B virus coinfection in patients with hepatitis C virus-associated chronic liver disease: clinical and virological significance. J Med Virol. 1999;58:201–207. doi: 10.1002/(sici)1096-9071(199907)58:3<201::aid-jmv3>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 128.Uchida T, Kaneita Y, Gotoh K, Kanagawa H, Kouyama H, Kawanishi T, Mima S. Hepatitis C virus is frequently coinfected with serum marker-negative hepatitis B virus: probable replication promotion of the former by the latter as demonstrated by in vitro cotransfection. J Med Virol. 1997;52:399–405. doi: 10.1002/(sici)1096-9071(199708)52:4<399::aid-jmv10>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 129.Feitelson M, Lega L, Guo J, Resti M, Rossi ME, Azzari C, Blumberg BS, Vierucci A. Pathogenesis of posttransfusion viral hepatitis in children with beta-thalassemia. Hepatology. 1994;19:558–568. doi: 10.1002/hep.1840190304. [DOI] [PubMed] [Google Scholar]
- 130.Zhang ZH, Deng WY, Lu MJ, Yang DL. Genetic variation and significance of hepatitis B surface antigen. J Clin Hepatol. 2013;29:810–815. [Google Scholar]
- 131.Pichoud C, Seignères B, Wang Z, Trépo C, Zoulim F. Transient selection of a hepatitis B virus polymerase gene mutant associated with a decreased replication capacity and famciclovir resistance. Hepatology. 1999;29:230–237. doi: 10.1002/hep.510290119. [DOI] [PubMed] [Google Scholar]
- 132.Warner N, Locarnini S. The antiviral drug selected hepatitis B virus rtA181T/sW172* mutant has a dominant negative secretion defect and alters the typical profile of viral rebound. Hepatology. 2008;48:88–98. doi: 10.1002/hep.22295. [DOI] [PubMed] [Google Scholar]
- 133.Torresi J, Earnest-Silveira L, Deliyannis G, Edgtton K, Zhuang H, Locarnini SA, Fyfe J, Sozzi T, Jackson DC. Reduced antigenicity of the hepatitis B virus HBsAg protein arising as a consequence of sequence changes in the overlapping polymerase gene that are selected by lamivudine therapy. Virology. 2002;293:305–313. doi: 10.1006/viro.2001.1246. [DOI] [PubMed] [Google Scholar]
- 134.Motta JS, Mello FC, Lago BV, Perez RM, Gomes SA, Figueiredo FF. Occult hepatitis B virus infection and lamivudine-resistant mutations in isolates from renal patients undergoing hemodialysis. J Gastroenterol Hepatol. 2010;25:101–106. doi: 10.1111/j.1440-1746.2009.05972.x. [DOI] [PubMed] [Google Scholar]
- 135.Melegari M, Scaglioni PP, Wands JR. Hepatitis B virus mutants associated with 3TC and famciclovir administration are replication defective. Hepatology. 1998;27:628–633. doi: 10.1002/hep.510270243. [DOI] [PubMed] [Google Scholar]
- 136.Chen Y, Robinson WS, Marion PL. Selected mutations of the duck hepatitis B virus P gene RNase H domain affect both RNA packaging and priming of minus-strand DNA synthesis. J Virol. 1994;68:5232–5238. doi: 10.1128/jvi.68.8.5232-5238.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chen SJ, Zhao YX, Fang Y, Xu WZ, Ma YX, Song ZW, Teng X, Gu HX. Viral deletions among healthy young Chinese adults with occult hepatitis B virus infection. Virus Res. 2012;163:197–201. doi: 10.1016/j.virusres.2011.09.029. [DOI] [PubMed] [Google Scholar]
- 138.Raimondo G, Caccamo G, Filomia R, Pollicino T. Occult HBV infection. Semin Immunopathol. 2013;35:39–52. doi: 10.1007/s00281-012-0327-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang Z, Zhang L, Dai Y, Jin L, Sun B, Su Q, Li X. Occult hepatitis B virus infection among people with a family history of chronic hepatitis B virus infection. J Med Virol. 2015;87:1890–1898. doi: 10.1002/jmv.24245. [DOI] [PubMed] [Google Scholar]
- 140.Zhang ZH, Zhang L, Lu MJ, Yang DL, Li X. [Establishment of reference sequences of hepatitis B virus genotype B and C in China] Zhonghua Gan Zang Bing Za Zhi. 2009;17:891–895. [PubMed] [Google Scholar]
- 141.Roth WK, Busch MP, Schuller A, Ismay S, Cheng A, Seed CR, Jungbauer C, Minsk PM, Sondag-Thull D, Wendel S, et al. International survey on NAT testing of blood donations: expanding implementation and yield from 1999 to 2009. Vox Sang. 2012;102:82–90. doi: 10.1111/j.1423-0410.2011.01506.x. [DOI] [PubMed] [Google Scholar]
- 142.Allain JP. Occult hepatitis B virus infection: implications in transfusion. Vox Sang. 2004;86:83–91. doi: 10.1111/j.0042-9007.2004.00406.x. [DOI] [PubMed] [Google Scholar]
- 143.Torbenson M, Thomas DL. Occult hepatitis B. Lancet Infect Dis. 2002;2:479–486. doi: 10.1016/s1473-3099(02)00345-6. [DOI] [PubMed] [Google Scholar]
- 144.Weber B. Recent developments in the diagnosis and monitoring of HBV infection and role of the genetic variability of the S gene. Expert Rev Mol Diagn. 2005;5:75–91. doi: 10.1586/14737159.5.1.75. [DOI] [PubMed] [Google Scholar]
- 145.Hollinger FB, Sood G. Occult hepatitis B virus infection: a covert operation. J Viral Hepat. 2010;17:1–15. doi: 10.1111/j.1365-2893.2009.01245.x. [DOI] [PubMed] [Google Scholar]


