Abstract
Better the drugs you know than the drugs you do not know. Drug repurposing is a promising, fast, and cost effective method that can overcome traditional de novo drug discovery and development challenges of targeting neuropsychiatric and other disorders. Drug discovery and development targeting neuropsychiatric disorders are complicated because of the limitations in understanding pathophysiological phenomena. In addition, traditional de novo drug discovery and development are risky, expensive, and time-consuming processes. One alternative approach, drug repurposing, has emerged taking advantage of off-target effects of the existing drugs. In order to identify new opportunities for the existing drugs, it is essential for us to understand the mechanisms of action of drugs, both biologically and pharmacologically. By doing this, drug repurposing would be a more effective method to develop drugs against neuropsychiatric and other disorders. Here, we review the difficulties in drug discovery and development in neuropsychiatric disorders and the extent and perspectives of drug repurposing.
1. Introduction
The principle of “polypharmacology” (i.e., one drug, multiple hits, or off-target effects) has been understood since the advent of drug discovery. Traditionally, the goal of drug discovery and development was to identify the potential therapeutic agents using a one drug for one target model, suggesting that high selectivity (and/or affinity) would maximize efficacy and minimize side effects. In a series of efforts to identify such specific compounds (like using high throughput screening), there was a problem that a vast majority of compounds mediated unexpected and often undesired effects. The concept of polypharmacology emerged from this observation (i.e., drug promiscuity); however, polypharmacology should be distinguished from drug promiscuity. In our definition, drug promiscuity represents either good or bad effects mediated by compounds binding to both therapeutic and nontherapeutic targets, whereas polypharmacology represents beneficial effects mediated by compounds binding to multiple therapeutic targets. Various drug classes such as selective serotonin reuptake inhibitors [1, 2], antipsychotic [3], cholinesterase inhibitors [4], and thrombolytic agents [5] show polypharmacological features. In addition, amantadine was initially developed for influenza; however, after redirection, it is useful for Parkinson's disease [6, 7]. Zidovudine was intended to cancer treatment, and now it is redirected to targeting HIV/AIDS [8–10]. Additional, but well-known example is Viagra (Sildenafil) that was intended to antianginal medication but redirected to penile erections [11]. This growing evidence is against the simplicity of Ehrlich's “magic bullet concept” and it redirects our attention from the one drug-multiple target model to multiple mechanisms of action. Since every drug is able to hit multiple targets with and without our sense and knowledge [12], pursuing multiple targets for drugs should be accompanied by addressing a fundamental question: whether promiscuous drugs are able to contribute to their clinical efficacy out of the original scopes. A direct application of polypharmacology is drug repurposing which is also referred to as drug repositioning, drug reprofiling, and therapeutic switching. Generally, drug repurposing refers to a reinvestigation of existing drugs for new therapeutic interventions [13–17]. However, drug repurposing does not have to narrow down to take advantage of the off-target effects of the existing drugs as discussed later. In order to expand our knowledge and drug potentials, drug repurposing is a very productive method in drug discovery and development. It is useful for identifying and classifying drugs based on their actions to multiple therapeutic targets (i.e., leading to better efficacy and/or safety) or their action to nontherapeutic targets (i.e., leading to adverse effects). Drug repurposing can reduce the cost and risk intrinsic to drug discovery and development. This is especially valid, regarding the targeting of neurological and psychiatric disorders due to the complexity in their etiology and pathology. In this review, we will discuss the difficulties of the drug discovery and the development process with respect to neuropsychiatric disorders and the extent of drug repurposing as an alternative approach in drug discovery and development.
2. Challenges in Clinical Development for Neuropsychiatric Disorders
Due to the great progress and development of modern technology, our understanding of biological, physiological, and metabolic processes has advanced tremendously. However, we still face many challenges in drug discovery and development targeting neuropsychiatric disorders. There are four primary reasons why it is difficult to develop therapeutic agents against neuropsychiatric disorders: (1) CNS disorders have a complex etiology (heterogeneity; gene to environment), (2) limitations of understanding pathophysiology in neuropsychiatric disorders, (3) lack of appropriate biomarkers and/or molecular targets, and (4) lack of appropriate animal models. The etiological complexity of CNS disorders (e.g., schizophrenia) stems from multiple genetic and environmental factors [18, 19], sequentially giving rise to the other three problem categories mentioned above. Since the etiological complexity restricts our understanding of pathophysiology of CNS disorders, it is difficult to identify and/or characterize appropriate biomarkers and/or molecular targets. As a result, it is difficult to evaluate the mechanism(s) of action of therapeutic agents [20, 21]. In addition, their pharmacological manipulations targeting appropriate markers become risky. The absence of biomarkers and/or molecular targets prevents us from producing appropriate animal models that are genetically manipulated to recapitulate human disease conditions [22]. The clinical ineffectiveness of therapeutic agents may also occur due to the potential disagreements with the biological basis in the animal models and the mechanism(s) of action of therapeutic agents [23].
3. Promiscuity of Drug-Target Interactions
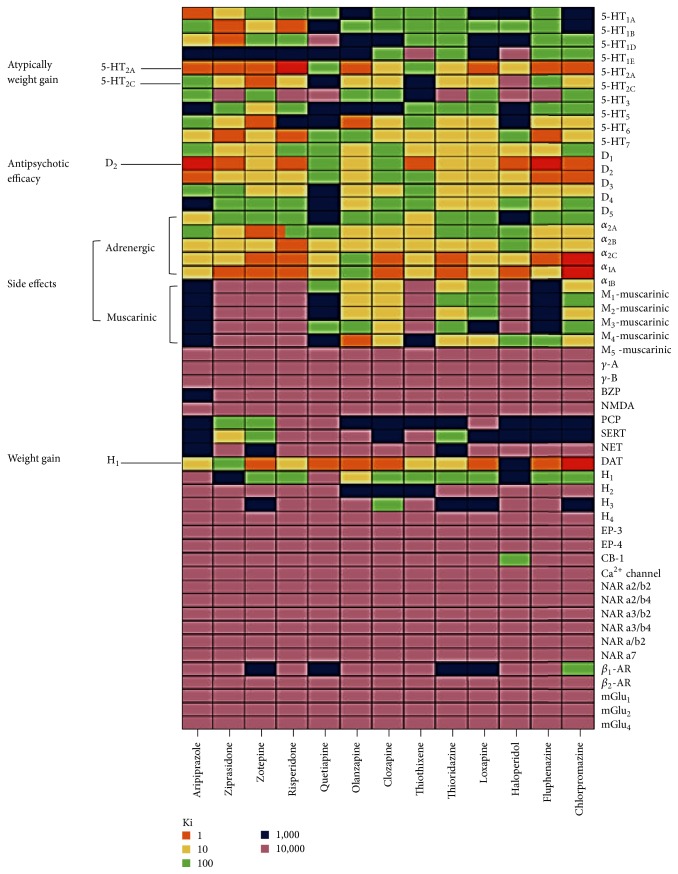
Due to the complex nature of neuropsychiatric disorders (e.g., schizophrenia, depression, anxiety disorders, insomnia, migraine headaches, chronic pain and seizure disorders, and/or other complex mental disorders), the goal of therapeutic intervention is daunting. Although our knowledge and understanding of their pathophysiological phenomena have been greatly advanced and a series of efforts in CNS drug discovery and development adapted cutting edge technology of preclinical research and development, the discovery of specific CNS agents was not very successful and even depended on serendipity. This may be partially due to promiscuity of drug interactions. Multiple aspects (e.g., drugs and their target selectivity and affinity versus drugs' molecular targets and their mediated signaling pathways) must be considered in clinical drug development for neuropsychiatric disorders. Since the first introduction of the serotonin reuptake inhibitors (SSRIs, zimelidine, discovered in 1971 [24, 25]) and atypical antipsychotic agents (clozapine, discovered in 1958 [26]), the five most frequently prescribed CNS agents (olanzapine, quetiapine, risperidone, sertraline, and venlafaxine) share common mechanisms of action with the first two drugs (zimelidine and clozapine). Sharing mechanisms of action implicates that at least CNS agents may have common molecular targets or alter target-mediated signaling pathway(s) in one way or another. Furthermore, they may have marginal therapeutic effects on most of the common neuropsychiatric disorders. Indeed, about 70 compounds with common targets (serotonin receptors and dopamine receptors) are under development or in current use for treating schizophrenia and about 30 drugs aim at multiple therapeutic targets in schizophrenia [27, 28]. This is because compounds that aimed at one target can interact with others, supported by the golden standard of clozapine that it has polypharmacological profile with high affinity for several receptors as shown in Figure 1. These multiple actions of clozapine to multiple targets lead to a well-defined pharmacological signature in schizophrenia and related disorders in agreement with possible molecular targets in schizophrenia highlighted in Table 1 [29–31]. However, for example, after several trials of discovering and developing novel therapeutics like clozapine, none of the new compounds showed unique effectiveness compared with clozapine [32]. The promiscuity of drug-target interactions sometimes stems from a similarity; therapeutic and nontherapeutic molecular targets share high homology. Many molecular targets display their sequence similarity (and thus presumably similar conformation) and drugs themselves show structural similarity. For example, therapeutic agents targeting aminergic G-protein coupled receptors (GPCRs) are more promiscuous than other drug classes in spite of a diversity of GPCRs [33–36]. The similar moiety among drugs stems from the designated advantages over the existing drugs that aim at known molecular targets and/or are already effective. These drug classes can obtain polypharmacological profiles. Defining their actions to multiple targets (therapeutic targets versus nontherapeutic targets) makes gaining new opportunities in CNS agent discovery in the current efforts of clinical development for neuropsychiatric disorders difficult. Thus, the increase in promiscuity of drug-target interactions makes the discovery of CNS agents more complex.
Figure 1.
Polypharmacological profiles of antipsychotic drugs. Receptoromic screening identified multiple molecular targets for several antipsychotic drugs. In particulr, clozapine has the high affinity (Ki) to 5-HT serotonin receptors (5-HT2A, 5-HT2C, 5-HT6, and 5-HT7), dopamine receptor (D4), muscarinic receptors, (M1, M2, M3, M4, and M5), adrenergic receptors (α-1 and α-2), and other aminergic receptors. Other antipsychotic drugs also interact with multiple targets. More information can be found at NIMH PDSP database (http://pdspdb.unc.edu/pdspWeb/?site=kidb) (reprinted with permission from Nature Publishing Group).
Table 1.
Potential molecular targets in schizophrenia.
| Molecular targets proposed for schizophrenia | |
|---|---|
| Target family | Drug actions |
|
| |
| Dopamine receptors | D1 agonists |
| D2 antagonists | |
| D4 antagonists | |
| D4 agonists | |
|
| |
| Serotonin receptors | 5-HT1A agonists |
| 5-HT1A antagonists | |
| 5-HT2A antagonists | |
| 5-HT4 agonists | |
| 5-HT6 agonists | |
| 5-HT7 agonists | |
|
| |
| Muscarinic receptors | M1 agonists |
| M4 agonists | |
| M5 antagonists | |
|
| |
| GABA receptors | GABA.a (α2) agonists |
| GABA.a (α5) antagonists | |
|
| |
| Adrenergic receptors | α2 adrenergic antagonists |
|
| |
| Glutamate receptors | Glycine transporter inhibitors |
| mGluR2/3 agonists | |
| mGluR5 agonists | |
| NMDA enhancers | |
|
| |
| Others | Nicotinic α7 agonists |
| Nicotinic α4β2 agonists | |
| Ampakines | |
| COMT inhibitors | |
4. Other Aspects in Drug-Target Interactions
We also should consider some other aspects that can provide us with the insights into the scope of CNS agent discovery. First two important aspects are functional selectivity and allosterism. Functional selectivity is defined as unique signaling pathways which are ligand-mediated. The ligand can be agonist or antagonist. They can modulate different signaling pathways through a single GPCR, resulting in different biological and/or physiological processes, depending on which pathway is activated [37–39]. Recently, Allen et al. studied the scaffold-based novel β-arrestin-biased ligand aripiprazole. A structure–functional–selectivity relationship (SFSR) of these novel compounds revealed distinct β-arrestin-bias toward the dopamine D2 receptor (D2R) [40]. Moreover, these unique D2R β-arrestin–biased agonists displayed atypical antipsychotic drug-like activities in vivo [41, 42]. Thus, it is often possible that drugs which may be thought to be nonselective to a particular target can actually interact with that target. However, due to the complexity of phenomena involved in biased signaling, the complete picture of the interactions between a receptor and a ligand can be difficult to access [38, 43, 44]. In addition, the results from inappropriate assays methodologically may provide precarious insights into their functional and/or molecular signatures, misdirecting researchers in the process of CNS drug discovery. However, the use of chimeric and/or promiscuous G proteins may partially overcome some of the potential difficulties of assessing drug-target interactions [45, 46] although considerable challenges remain. Allosteric modulation is the positively or negatively synergic effects on target function that are the result of a ligand binding to the binding site (allosteric) other than orthosteric binding site. Based on their binding effects, these ligand molecules are categorized as being positive of negative allosteric modulators. While the allosteric binding site does not accommodate endogenous ligands, allosteric modulators contribute to biological and physiological processes. Although the positive allosteric modulators do not activate receptors, they can enhance receptor-mediated activity at the presence of endogenous ligand. Since advantages over allosteric modulation are recently recognized in drug discovery and development, allosteric modulators appear to be more favorable therapeutic agents [47, 48]. The benzodiazepines are well known as allosteric modulators for GABA receptors which are in clinical use. However, pursuing allosteric modulators cannot be a panacea in CNS agent discovery because the presence of an orthosteric ligand is required for identifying allosteric modulators. This issue specifically applies to the targeting orphan GPCRs. There are additional problems in pursuing allosteric modulators. Allosteric binding sites are not evolutionarily conserved [48, 49], suggesting that they may be more diverse than orthosteric binding sites. Thus, it is possible that newly designed allosteric modulators can be more selective. However, this diversity and selectivity may cause other issues. For example, low evolutionary pressure of allosteric binding sites may result in many differences of receptor structure between species, which can make it difficult to employ in vivo animal models. The selectivity of allosteric modulators may not be verified because they bind to allosteric sites for their own targets and they may bind to orthosteric sites for nontargets. Then, allosteric modulator-dependent signaling pathways could lead to adverse side effects [50, 51]. The divergence of molecular targets also affects the efforts of CNS agent discovery. Many molecular targets have differential expression patterns and/or undergo posttranscriptional and/or posttranslational modifications. Such modifications and/or differential expression have significant effects on their confirmation and functions and eventually alter the properties of potential molecular targets [52–54]. It may be more difficult to identify and characterize drug-target interactions due to their divergence. For example, copy-number variations (CNVs) are recognized in risk for human diseases like schizophrenia [55–57], suggesting that some molecular targets among patients can be variable [58]; as a result, different or more promiscuous therapeutic agents are needed to aim at these variants or personalized medications can be an alternative solution. Receptor dimerization or oligomerization may contribute to the divergence of molecular targets, which is another challenge for CNS agent discovery. Even though the details of such mechanisms remain elusive, the receptor complex that mediates different signaling pathways may be important in the effectiveness of some CNS therapeutic agents [59, 60]. Designed multiple ligands (DMLs), which are the linked two therapeutic compounds, may be useful for manipulating receptor functions in targeting receptor complexes or two different receptors [60]. However, it may not be guaranteed whether DMLs have polypharmacological profile. Taken together, due to the complex nature of CNS disorders and challenges in basic and clinical researches, it is not surprising that we still face obstacle to the prospect of developing novel therapeutics with novel mechanisms. To accomplish major breakthrough, major challenges should be addressed to gain greater insight into the complexity of CNS disorders. In the next section, we will discuss an alternative approach (drug repurposing) in drug discovery and development in part to overcome the current drug discovery issues in neuropsychiatric disorders.
5. Drug Repurposing: Visit Your Drug Recycling Bins
As discussed above, many hurdles still exist to develop CNS therapeutic agents, resulting in the fact that drug pipelines have gradually run out. The current approaches and methodology (e.g., high throughput screening and structure-based drug design) may not be an ideal solution for developing CNS agents because they are not cost and time-effective [61–64]. In addition, despite the extensive efforts of drug development, several drugs were withdrawn from the market due to severe side effects. For example, thalidomide was thought to be very safe [65]; however, it was withdrawn due to teratogenic effects (although it was appreciated on the cancer therapy market later). Thus, alternative approaches are necessary for overcoming such challenges. In agreement with the concept of Sir James Black, “the best way to discovery a new drug is start with an existing drug” [66], one of the alternatives, but a very attractive approach is drug repurposing that is a direct application of polypharmacological features of drugs in drug discovery and development against neuropsychiatric disorders [67–69]. Due to complex etiology of neuropsychiatric disorders, multiple therapeutic approaches may be essential so that polypharmacological approaches are well suited. A compound hitting multiple targets may have improved efficacy and/or safety in the view of therapeutic intervention for neuropsychiatric disorders. Different levels of polypharmacology could lead to better efficacy and/or safety or adverse effects. These differences in effectiveness may be due to the interaction of a compound with different therapeutic and/or nontherapeutic targets and/or dose dependence [70]. As mentioned earlier, polypharmacology-based drug repurposing refers to the reinvestigation of existing drugs for novel therapeutic purpose [13–17]. The existing drugs can be currently approved drugs or even “failed” compounds that their original scopes have already been identified. Failed compounds indicate abandoned drugs due to the fact that there is no clinical effectiveness even though the original scopes (e.g., initial targets in symptom/disorder) might be appropriate. Since it is highly possible that every drug displays off-target effects, the concept of a magic bullet (one drug, one target) is too simple and therefore archaic. Indeed, one drug can interact with diverse targets (average 6–13 multiple targets as predicted, [36]) and lead to multiple side effects. It does not matter if these effects are good or bad, however, as those side effects allow us to see different angles of drug usages [71–73]. Thus, polypharmacological profiling of these drugs against any druggable targets is greatly beneficial to drug discovery and development in order to decrease cost and time and enhance therapeutic potentials. Repurposing can decrease the time line for drug discovery and development. From target discovery to market, it takes 10 to 17 years in traditional drug discovery and development, whereas the duration of drug discovery for repurposed drugs takes from 3 to 12 years [13]. In addition, known safety and proven bioavailability of the existing drugs allow for an acceleration of the developmental process, reducing failure risk [74], although traditionally drug repurposing depended on serendipity, indicating a lack of processes and information. Presently, there are a variety of innovative approaches (e.g., cheminformatics, bioinformatics, and drug-target network) which may be very useful for drug repurposing. There are several tools to recycle the existing drugs to identify new therapeutic intervention for them as Jin and Wong summarized [75]. In order to expand therapeutic indication of existing drugs successfully, we need to be able to integrate multidisciplinary information derived from diverse fields such as cheminformatics, bioinformatics, chemistry, and in vivo and in vitro pharmacological assays and clinical trials. Based on this profiling information of the drugs (e.g., their known targets, toxicity, safety, and bioavailability) and/or diseases of interest, we can begin to find success in implementing drug repurposing. Here, we will highlight several tools and methods for drug repurposing and discuss how they can contribute to CNS drug discovery.
6. NIHM PDSP Database and Receptoromics as Drug Repurposing Tool
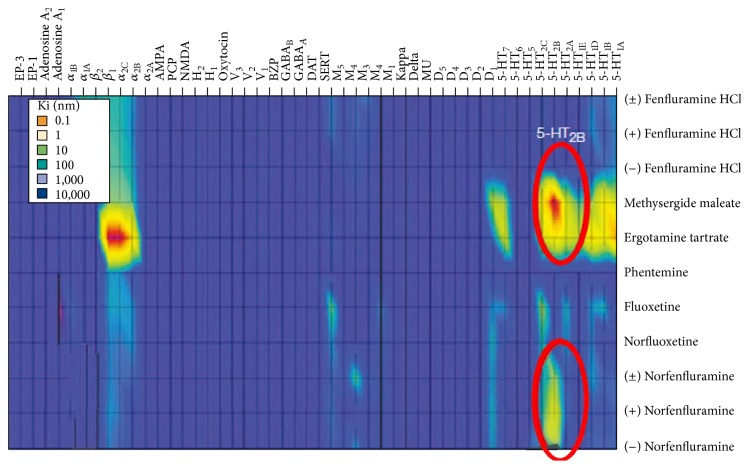
As various tools and methods have been developed and refined, corresponded databases have been constructed. Two-pioneered approach was dubbed receptoromics [76–81] and the NIMH PDSP database (National Institute of Mental Health Psychoactive Drug Screening Program) was created [77, 82, 83]. Since receptoromic approach encompasses computational and physical screening tools, it is an ideal tool for drug repurposing. In one way of a computational tool (or in silico approach), it has been referred as a receptor structure-function basis to be able to screen compounds virtually. At the beginning, several GPCR molecular models were based on homology modeling with bovine rhodopsin [84, 85]. However, as more validated molecular structures are solved [86–91], compound libraries can be screened in silico against a virtual receptorome in order to identify potential drug and target interactions. In line with in silico approach, the drug information databases are also extremely useful. Such databases may facilitate finding of target-specific drugs, identification of lead compounds, and characterization of ligand's structural features. The publically available database, NIMH PDSP Ki was constructed from data obtained through high or medium throughput receptoromic screening; this database is continually expanding. This information provides valuable insights into drug repurposing. Another way of a physical screening tool has been referred to as phenotypic/target-based screening that is used for receptoromic profiling. It can help in the identification of molecular targets for endogenous ligands. The data from physical screening also comes from ligand binding screening, functional screening, and validation of molecular targets. By using such computational and physical readouts, many targets can be screened in parallel. In addition, we can identify possible therapeutic or adverse effects of the existing compounds by blindly screening them against multiple targets in a nonbiased way. As a result, significant discoveries have already been reported. For example, 5-HT2B serotonin receptors may serve as a molecular target responsible for fenfluramine-induced valvular heart disease as receptoromic screening indicated that norfenfluramine, via activating 5-HT2B serotonin receptors, was a potential agent responsible for valvular heart disease and pulmonary hypertension (Figure 2; Fen-phen [92–95]). Additionally, H1 histamine receptors may serve as a molecular target responsible for weight gain in patients taking atypical antipsychotics [96]. Kappa-1 opioid receptors may serve as a molecular target on the hallucinogenic effects of salvinorin A [97]. Thus, receptoromic screening of drugs can provide insight for repurposing drugs because this nonbiased and parallel screening allows us to discover the molecular mechanisms responsible for and to predict drug-mediated side effects.
Figure 2.
The molecular targets for cardiopulmonary-associated drugs. Recetoromic screening revealed the molecular targets implicated in fenfluramine. 5-HT2B serotonin receptor was identified as a molecular target for the norfenfluramine (a metabolite of fenfluramine), methylergonovine (a metabolite of the valvular heart disease- and pulmonary hypertension-associated drug methysergide), and dihydroergotamine (potentially associated with valvular heart disease and pulmonary hypertension). The drugs associated with cardio diseases also show high affinity to α-2B adrenergic receptors, whereas fluoxetine and the metabolite norfluoxetine do not. In this heat map, the affinity of the drugs is mapped by color gradient, that is, blue (lower affinity, higher Ki), red (higher affinity, lower Ki), and intermediated color (reprinted with permission from Nature Publishing Group).
7. Other Databases and Potential Tools for Drug Repurposing
In addition to the NIMH PDSP database, other chemical databases are available. For example, Ligand Expo [98], ZINC [99], and KEGG DRUG [100] databases integrate diverse information such as molecular pathways, binding experiments, and drug targets. One of the large public databases, PubChem, containing the results from many screens and assays [101, 102], can also facilitate drug repurposing. The PubChem database includes 61 psychiatric drugs that were assessed in a number of different bioassays, resulting in that those drugs were identified as “active” in multiple bioassays. These data can be a fundamental resource of drug repurposing. The European College of Neuropsychopharmacology (ECNP) also provides valuable tools (e.g., ECNP Medicines chest) to help clinical researchers obtain access to pharmacological tools important for the pursuit of their studies (https://www.ecnp.eu/). Additionally, Pouliot and colleagues already used PubChem bioassay data for predicting adverse drug reactions (ADRs) [103]. Growing availability of such databases enables various tools to be applied to drug repurposing. For example, Keiser et al. reported Similarity Ensemble Approach (SEA) to categorize molecular drug targets through the similarity of their known molecular ligands [104–106]. This similarity based drug-target interaction database is further used for predicting novel drug-target interactions. Practically, out of 30 predictions, the authors confirmed 23 in experimental assays [105]. Drug-target binary association was created to build bipartite graphs, indicating the linkage between FDA approved drugs and target proteins [107]. Side effect similarity allowed inferring if two drugs share their targets [108]. Additionally, in order to predict the potential off-target effects for any given drug, ligand target interaction/molecular networks [109, 110] were reported. The comparative toxicogenomics database (CTD) was developed to provide insight into complex chemical, gene, and protein interaction [111]. Adverse reactions of drugs have been mapped by bioinformatic mining of approved drug information. Based on 7,684 approved drug labels, they mapped the adverse reactions corresponding to 988 unique drugs onto 174 side effects [112, 113]. Weighted network based inference methods enable us to predict chemical and protein interactions [114]. Recently, a self-organizing map based prediction of drug equivalence relationships (SPiDER) was developed to identify targets, both for known drugs and computer-generated molecular scaffolds [115]. Genome-based drug reposition is additional strength regarding the potential tools for drug repurposing [15, 116]. The integrative genome-wide metrics can provide insightful scalar theories of massive biological information. These drug repurposing tools and multiple databases can be applied to identify new therapeutic interventions for the existing drugs and can accelerate CNS drug discovery.
8. Conclusions
Despite the exponential growth of advanced technology and the resultant molecular database, we still do not fully understand the signaling pathways and molecular mechanisms behind many disease states, especially neuropsychiatric disorders. This lack of knowledge makes repurposing drugs difficult and that is a direct application of polypharmacology. Drug repurposing does not eliminate the risk of compound development; however, it reduces the risk of the lack of compound development. There are already several CNS therapeutic drugs which have been repurposed (summarized in Table 2). Moreover, another application of polypharmacology could be designed for multitarget ligands [67, 117]. If we designed ligands against profiles of multiple therapeutic targets, it would accelerate CNS drug discovery and enhance drug repurposing [60]. Indeed, Apsel and coworkers reported compounds that inhibit both tyrosine kinases and phosphatidylinositol-3-OH kinases [118]. This work is a successful example of rational design of multitarget ligands in the human kinome context. In addition, Besnard et al. successfully launched a new approach for the automated design of ligands with polypharmacological profiles [119]. Although it will be a challenge to rationally design multitarget ligands with therapeutic benefits, advancing the automated design of ligands and other areas in developing, optimizing, and validating target combinations will be the key in the future for rational design of drugs with polypharmacological profiles. The potential advantages in computer-aided drug repurposing enable us to prioritize CNS drug discovery. In addition, experimental approaches such as phenotypic screening are investigated for drug repurposing [52]. Therefore, the application of these combinatorial approaches to neuropsychiatric disorders will not only provide great opportunities in drug discovery and development but also lead to the identification of nontherapeutic targets that can cause adverse effects.
Table 2.
Some examples of repurposed drugs for neuropsychiatric disorders.
| Drugs (alphabetic order) | Actions/classes | First intervention | New intervention | References |
|---|---|---|---|---|
| Amantadine | Anticholinergic-like agent | Influenza | Parkinson's disease, ADHD | [6, 7] |
| Amphotericin B | NSAID∗ | Antifungal | Bipolar disorder | [120] |
| Arbaclofen | GABA agonist | Cerebral palsy | Fragile X syndrome | [116, 121–123] |
| Atomoxetine | NSRI∗∗ | Parkinson's diseases | ADHD | [124] |
| Dexmecamylamine | Nicotinic receptor modulator | Hypertension | Depression | [125, 126] |
| Galantamine | Acetylcholinesterase inhibitor | Polio, paralysis | Alzheimer's disease | [127] |
| Mecamylamine | Nicotinic receptor antagonist | Hypertension | ADHD Depression |
[128–131] |
| Mifepristone | Glucocorticoid receptor type II antagonist | Pregnancy termination | Psychotic major depression, Cushing's syndrome | [132–135] |
| Ropinirole | D2 agonist | Hypertension | Parkinson's disease, idiopathic restless leg syndrome | [136–138] |
| Tamoxifen | Estrogen receptor | Breast tumor | Bipolar disorder Mania |
[139] |
| Valsartan | Angiotensin receptor blocker | Hypertension | Alzheimer's disease | [140] |
∗NSAID is nonsteroidal anti-inflammatory drug.
∗∗NSRI is norepinephrine-selective reuptake inhibitor.
Acknowledgments
This work was supported by the Brain & Behavior Research Foundation (BBRF/NARSAD) Young Investigator Award (Grant no. 22587). The authors would like to thank to Noah Sciaky, Ph.D. at Department of Pharmacology, University of North Carolina, Chapel Hill, for proofreading.
Abbreviations
- 5-HT1A:
5-Hydroxytryptamine (serotonin) receptors
- D2:
Dopamine receptors
- α2AR:
α Adrenergic receptors
- β2-AR:
β-Adrenergic receptors
- M1:
Muscarinic receptors
- γ-A, B:
γ-Aminobutyric acid receptors
- BZP:
Benzodiazepines
- NMDAR:
N-Methyl-D-aspartate receptor
- AMPAR:
α-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor
- PCP:
Phencyclidine
- SERT:
Serotonin transporter
- NET:
Norepinephrine transporter
- DAT:
Dopamine transporter
- H1R:
Histamine receptors
- EP-3, 4:
Prostaglandin receptors
- CB-1:
Cannabinoid receptors
- NAR:
Nicotinic acetylcholine receptors
- mGluR:
Metabotropic glutamate receptors
- V1:
Vasopressin receptors
- κ, δ, μ:
Opioid receptors (kappa, delta, and mu).
Disclosure
As we focused on drug repurposing for CNS drug discovery, we may miss other valuable studies for drug repurposing.
Competing Interests
The authors confirm that this paper content has no competing interests.
References
- 1.Bianchi M. T. Non-serotonin anti-depressant actions: direct ion channel modulation by SSRIs and the concept of single agent poly-pharmacy. Medical Hypotheses. 2008;70(5):951–956. doi: 10.1016/j.mehy.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 2.Rammes G., Rupprecht R. Modulation of ligand-gated ion channels by antidepressants and antipsychotics. Molecular Neurobiology. 2007;35(2):160–174. doi: 10.1007/s12035-007-0006-1. [DOI] [PubMed] [Google Scholar]
- 3.Bianchi M. T. Promiscuous modulation of ion channels by anti-psychotic and anti-dementia medications. Medical Hypotheses. 2010;74(2):297–300. doi: 10.1016/j.mehy.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Zhang H. Y., Tang X. C. Neuroprotective effects of huperzine A: new therapeutic targets for neurodegenerative disease. Trends in Pharmacological Sciences. 2006;27(12):619–625. doi: 10.1016/j.tips.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Benarroch E. E. Tissue plasminogen activator: beyond thrombolysis. Neurology. 2007;69(8):799–802. doi: 10.1212/01.wnl.0000269668.08747.78. [DOI] [PubMed] [Google Scholar]
- 6.Crosby N., Deane K. H., Clarke C. E. Amantadine in Parkinson's disease. Cochrane Database of Systematic Reviews. 2003;(1) doi: 10.1002/14651858.CD003468.CD003468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwab R. S., England A. C., Jr., Poskanzer D. C., Young R. R. Amantadine in the treatment of Parkinson's disease. The Journal of the American Medical Association. 1969;208(7):1168–1170. doi: 10.1001/jama.1969.03160070046011. [DOI] [PubMed] [Google Scholar]
- 8.Mitsuya H., Weinhold K. J., Furman P. A., et al. 3′-Azido-3′-deoxythymidine (BW A509U): an antiviral agent that inhibits the infectivity and cytopathic effect of human T-lymphotropic virus type III/lymphadenopathy-associated virus in vitro. Proceedings of the National Academy of Sciences of the United States of America. 1985;82(20):7096–7100. doi: 10.1073/pnas.82.20.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun R., Eriksson S., Wang L. Identification and characterization of mitochondrial factors modulating thymidine kinase 2 activity. Nucleosides, Nucleotides & Nucleic Acids. 2010;29(4–6):382–385. doi: 10.1080/15257771003741018. [DOI] [PubMed] [Google Scholar]
- 10.Yarchoan R., Klecker R. W., Weinhold K. J., et al. Administration of 3′-azido-3′-deoxythymidine, an inhibitor of HTLV-III/LAV replication, to patients with AIDS or AIDS-related complex. The Lancet. 1986;1(8481):575–580. doi: 10.1016/s0140-6736(86)92808-4. [DOI] [PubMed] [Google Scholar]
- 11.DeBusk R. F., Pepine C. J., Glasser D. B., Shpilsky A., DeRiesthal H., Sweeney M. Efficacy and safety of sildenafil citrate in men with erectile dysfunction and stable coronary artery disease. The American Journal of Cardiology. 2004;93(2):147–153. doi: 10.1016/j.amjcard.2003.09.030. [DOI] [PubMed] [Google Scholar]
- 12.Kroeze W. K., Roth B. L. Polypharmacological drugs: “magic shotguns” for psychiatric diseases. In: Peters J.-U., editor. Polypharmacology in Drug Discovery. Somerset, NJ, USA: John Wiley & Sons; 2012. pp. 135–148. [Google Scholar]
- 13.Ashburn T. T., Thor K. B. Drug repositioning: identifying and developing new uses for existing drugs. Nature Reviews: Drug Discovery. 2004;3(8):673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 14.Hopkins A. L. Drug discovery: predicting promiscuity. Nature. 2009;462(7270):167–168. doi: 10.1038/462167a. [DOI] [PubMed] [Google Scholar]
- 15.Marusina K., Welsch D. J., Rose L., Brock D., Bahr N. The CTSA Pharmaceutical Assets Portal—a public-private partnership model for drug repositioning. Drug Discovery Today: Therapeutic Strategies. 2011;8(3-4):77–83. doi: 10.1016/j.ddstr.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Connor K. A., Roth B. L. Finding new tricks for old drugs: an efficient route for public-sector drug discovery. Nature Reviews Drug Discovery. 2005;4(12):1005–1014. doi: 10.1038/nrd1900. [DOI] [PubMed] [Google Scholar]
- 17.Oprea T. I., Bauman J. E., Bologa C. G., et al. Drug repurposing from an academic perspective. Drug Discovery Today. Therapeutic Strategies. 2011;8(3-4):61–69. doi: 10.1016/j.ddstr.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrison P. J., Owen M. J. Genes for schizophrenia? Recent findings and their pathophysiological implications. The Lancet. 2003;361(9355):417–419. doi: 10.1016/s0140-6736(03)12379-3. [DOI] [PubMed] [Google Scholar]
- 19.Lewis C. M., Levinson D. F., Wise L. H., et al. Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: schizophrenia. American Journal of Human Genetics. 2003;73(1):34–48. doi: 10.1086/376549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coyle J. T., Duman R. S. Finding the intracellular signaling pathways affected by mood disorder treatments. Neuron. 2003;38(2):157–160. doi: 10.1016/S0896-6273(03)00195-8. [DOI] [PubMed] [Google Scholar]
- 21.Roth B. L., Sheffler D., Potkin S. G. Atypical antipsychotic drug actions: unitary or multiple mechanisms for ‘atypicality’? Clinical Neuroscience Research. 2003;3(1-2):108–117. doi: 10.1016/s1566-2772(03)00021-5. [DOI] [Google Scholar]
- 22.Setola V., Roth B. L. Why mice are neither miniature humans nor small rats: a cautionary tale involving 5-hydroxytryptamine-6 serotonin receptor species variants. Molecular Pharmacology. 2003;64(6):1277–1278. doi: 10.1124/mol.64.6.1277. [DOI] [PubMed] [Google Scholar]
- 23.Shorter E. Looking backwards: a possible new path for drug discovery in psychopharmacology. Nature Reviews Drug Discovery. 2002;1(12):1003–1006. doi: 10.1038/nrd964. [DOI] [PubMed] [Google Scholar]
- 24.Carlsson A., Wong D. T. A note on the discovery of selective serotonin reuptake inhibitors. Life Sciences. 1997;61(12):p. 1203. doi: 10.1016/S0024-3205(97)00662-0. [DOI] [PubMed] [Google Scholar]
- 25.Wong D. T., Bymaster F. P., Engleman E. A. Prozac (fluoxetine, lilly 110140), the first selective serotonin uptake inhibitor and an antidepressant drug: twenty years since its first publication. Life Sciences. 1995;57(5):411–441. doi: 10.1016/0024-3205(95)00209-o. [DOI] [PubMed] [Google Scholar]
- 26.Hippius H. A historical perspective of clozapine. The Journal of Clinical Psychiatry. 1999;60(supplement 12):22–23. doi: 10.4088/jcp.v60n0105. [DOI] [PubMed] [Google Scholar]
- 27.Conn P. J., Roth B. L. Opportunities and challenges of psychiatric drug discovery: roles for scientists in academic, industry, and government settings. Neuropsychopharmacology. 2008;33(9):2048–2060. doi: 10.1038/sj.npp.1301638. [DOI] [PubMed] [Google Scholar]
- 28.Gray J. A., Roth B. L. The pipeline and future of drug development in schizophrenia. Molecular Psychiatry. 2007;12(10):904–922. doi: 10.1038/sj.mp.4002062. [DOI] [PubMed] [Google Scholar]
- 29.Gray J. A., Roth B. L. Molecular targets for treating cognitive dysfunction in schizophrenia. Schizophrenia Bulletin. 2007;33(5):1100–1119. doi: 10.1093/schbul/sbm074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kane J., Honigfeld G., Singer J., Meltzer H. Clozapine for the treatment-resistant schizophrenic. A double-blind comparison with chlorpromazine. Archives of General Psychiatry. 1988;45(9):789–796. doi: 10.1001/archpsyc.1988.01800330013001. [DOI] [PubMed] [Google Scholar]
- 31.Meltzer H. Y., Alphs L., Green A. I., et al. Clozapine treatment for suicidality in schizophrenia: International Suicide Prevention Trial (InterSePT) Archives of General Psychiatry. 2003;60(1):82–91. doi: 10.1001/archpsyc.60.1.82. [DOI] [PubMed] [Google Scholar]
- 32.Brea J., Castro M., Loza M. I., et al. QF2004B, a potential antipsychotic butyrophenone derivative with similar pharmacological properties to clozapine. Neuropharmacology. 2006;51(2):251–262. doi: 10.1016/j.neuropharm.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 33.Anighoro A., Rastelli G. Enrichment factor analyses on G-protein coupled receptors with known crystal structure. Journal of Chemical Information and Modeling. 2013;53(4):739–743. doi: 10.1021/ci4000745. [DOI] [PubMed] [Google Scholar]
- 34.Gregori-Puigjané E., Mestres J. A ligand-based approach to mining the chemogenomic space of drugs. Combinatorial Chemistry & High Throughput Screening. 2008;11(8):669–676. doi: 10.2174/138620708785739952. [DOI] [PubMed] [Google Scholar]
- 35.Selvam B., Porter S. L., Tikhonova I. G. Addressing selective polypharmacology of antipsychotic drugs targeting the bioaminergic receptors through receptor dynamic conformational ensembles. Journal of Chemical Information and Modeling. 2013;53(7):1761–1774. doi: 10.1021/ci400282q. [DOI] [PubMed] [Google Scholar]
- 36.Vogt I., Mestres J. Drug-target networks. Molecular Informatics. 2010;29(1-2):10–14. doi: 10.1002/minf.200900069. [DOI] [PubMed] [Google Scholar]
- 37.Roth B. L., Chuang D.-M. Multiple mechanisms of serotonergic signal transduction. Life Sciences. 1987;41(9):1051–1064. doi: 10.1016/0024-3205(87)90621-7. [DOI] [PubMed] [Google Scholar]
- 38.Urban J. D., Clarke W. P., von Zastrow M., et al. Functional selectivity and classical concepts of quantitative pharmacology. Journal of Pharmacology and Experimental Therapeutics. 2007;320(1):1–13. doi: 10.1124/jpet.106.104463. [DOI] [PubMed] [Google Scholar]
- 39.Urban J. D., Vargas G. A., von Zastrow M., Mailman R. B. Aripiprazole has functionally selective actions at dopamine D2 receptor-mediated signaling pathways. Neuropsychopharmacology. 2007;32(1):67–77. doi: 10.1038/sj.npp.1301071. [DOI] [PubMed] [Google Scholar]
- 40.Jin J., Roth B. L., Frye S. Novel Functionally Selective Ligands of Dopamine D2 Receptors. US Patent 20,130,137,679, 2013.
- 41.Allen J. A., Yost J. M., Setola V., et al. Discovery of β-arrestin-biased dopamine D2 ligands for probing signal transduction pathways essential for antipsychotic efficacy. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(45):18488–18493. doi: 10.1073/pnas.1104807108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen X., Sassano M. F., Zheng L., et al. Structure-functional selectivity relationship studies of β-arrestin-biased dopamine D2 receptor agonists. Journal of Medicinal Chemistry. 2012;55(16):7141–7153. doi: 10.1021/jm300603y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allen J. A., Roth B. L. Strategies to discover unexpected targets for drugs active at G protein-coupled receptors. Annual Review of Pharmacology and Toxicology. 2011;51:117–144. doi: 10.1146/annurev-pharmtox-010510-100553. [DOI] [PubMed] [Google Scholar]
- 44.Mailman R. B. GPCR functional selectivity has therapeutic impact. Trends in Pharmacological Sciences. 2007;28(8):390–396. doi: 10.1016/j.tips.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coward P., Chan S. D., Wada H. G., Humphries G. M., Conklin B. R. Chimeric G proteins allow a high-throughput signaling assay of Gi-coupled receptors. Analytical Biochemistry. 1999;270(2):242–248. doi: 10.1006/abio.1999.4061. [DOI] [PubMed] [Google Scholar]
- 46.Kostenis E., Waelbroeck M., Milligan G. Techniques: promiscuous Gα proteins in basic research and drug discovery. Trends in Pharmacological Sciences. 2005;26(11):595–602. doi: 10.1016/j.tips.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 47.Kenakin T. P. Ligand detection in the allosteric world. Journal of Biomolecular Screening. 2010;15(2):119–130. doi: 10.1177/1087057109357789. [DOI] [PubMed] [Google Scholar]
- 48.May L. T., Leach K., Sexton P. M., Christopoulos A. Allosteric modulation of G protein-coupled receptors. Annual Review of Pharmacology and Toxicology. 2007;47:1–51. doi: 10.1146/annurev.pharmtox.47.120505.105159. [DOI] [PubMed] [Google Scholar]
- 49.Lewis J. A., Lebois E. P., Lindsley C. W. Allosteric modulation of kinases and GPCRs: design principles and structural diversity. Current Opinion in Chemical Biology. 2008;12(3):269–280. doi: 10.1016/j.cbpa.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 50.Fang Z., Grütter C., Rauh D. Strategies for the selective regulation of kinases with allosteric modulators: exploiting exclusive structural features. ACS Chemical Biology. 2013;8(1):58–70. doi: 10.1021/cb300663j. [DOI] [PubMed] [Google Scholar]
- 51.Melancon B. J., Hopkins C. R., Wood M. R., et al. Allosteric modulation of seven transmembrane spanning receptors: theory, practice, and opportunities for central nervous system drug discovery. Journal of Medicinal Chemistry. 2012;55(4):1445–1464. doi: 10.1021/jm201139r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anighoro A., Bajorath J., Rastelli G. Polypharmacology: challenges and opportunities in drug discovery. Journal of Medicinal Chemistry. 2014;57(19):7874–7887. doi: 10.1021/jm5006463. [DOI] [PubMed] [Google Scholar]
- 53.Arya T., Kishor C., Saddanapu V., Reddi R., Addlagatta A. Discovery of a new genetic variant of methionine aminopeptidase from Streptococci with possible post-translational modifications: biochemical and structural characterization. PLoS ONE. 2013;8(10) doi: 10.1371/journal.pone.0075207.e75207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nisticò R., Ferraina C., Marconi V., et al. Age-related changes of protein SUMOylation balance in the AβPP Tg2576 mouse model of Alzheimer's disease. Frontiers in Pharmacology. 2014;5, article 63 doi: 10.3389/fphar.2014.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bassett A. S., Scherer S. W., Brzustowicz L. M. Copy number variations in schizophrenia: critical review and new perspectives on concepts of genetics and disease. The American Journal of Psychiatry. 2010;167(8):899–914. doi: 10.1176/appi.ajp.2009.09071016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grayton H. M., Fernandes C., Rujescu D., Collier D. A. Copy number variations in neurodevelopmental disorders. Progress in Neurobiology. 2012;99(1):81–91. doi: 10.1016/j.pneurobio.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 57.Henrichsen C. N., Chaignat E., Reymond A. Copy number variants, diseases and gene expression. Human Molecular Genetics. 2009;18(1):R1–R8. doi: 10.1093/hmg/ddp011. [DOI] [PubMed] [Google Scholar]
- 58.Cook E. H., Jr., Scherer S. W. Copy-number variations associated with neuropsychiatric conditions. Nature. 2008;455(7215):919–923. doi: 10.1038/nature07458. [DOI] [PubMed] [Google Scholar]
- 59.Liu Z., Zhang J., Zhang A. Design of multivalent ligand targeting G-protein-coupled receptors. Current Pharmaceutical Design. 2009;15(6):682–718. doi: 10.2174/138161209787315639. [DOI] [PubMed] [Google Scholar]
- 60.Morphy R., Rankovic Z. Designed multiple ligands. An emerging drug discovery paradigm. Journal of Medicinal Chemistry. 2005;48(21):6523–6543. doi: 10.1021/jm058225d. [DOI] [PubMed] [Google Scholar]
- 61.Adams C. P., Brantner V. V. Estimating the cost of new drug development: is it really 802 million dollars? Health Affairs. 2006;25(2):420–428. doi: 10.1377/hlthaff.25.2.420. [DOI] [PubMed] [Google Scholar]
- 62.Kapetanovic I. M. Computer-aided drug discovery and development (CADDD): in silico-chemico-biological approach. Chemico-Biological Interactions. 2008;171(2):165–176. doi: 10.1016/j.cbi.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pirmohamed M., James S., Meakin S., et al. Adverse drug reactions as cause of admission to hospital: prospective analysis of 18 820 patients. British Medical Journal. 2004;329(7456):15–19. doi: 10.1136/bmj.329.7456.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shankar R., Frapaise X., Brown B. LEAN drug development in R&D. Drug Discovery & Development. 2006;9(5):57–60. [Google Scholar]
- 65.Rowan A. Of Mice, Models and Men: A Critical Evaluation of Animal Research. Albany, NY, USA: State University of New York Press; 1984. [Google Scholar]
- 66.Raju T. N. The Nobel chronicles. 1988: James Whyte Black, (b 1924), Gertrude Elion (1918–99), and George H Hitchings (1905–98) The Lancet. 2000;355(9208):p. 1022. doi: 10.1016/s0140-6736(05)74775-9. [DOI] [PubMed] [Google Scholar]
- 67.Hopkins A. L., Mason J. S., Overington J. P. Can we rationally design promiscuous drugs? Current Opinion in Structural Biology. 2006;16(1):127–136. doi: 10.1016/j.sbi.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 68.Nobeli I., Favia A. D., Thornton J. M. Protein promiscuity and its implications for biotechnology. Nature Biotechnology. 2009;27(2):157–167. doi: 10.1038/nbt1519. [DOI] [PubMed] [Google Scholar]
- 69.Peters J.-U., Schnider P., Mattei P., Kansy M. Pharmacological promiscuity: dependence on compound properties and target specificity in a set of recent Roche compounds. ChemMedChem. 2009;4(4):680–686. doi: 10.1002/cmdc.200800411. [DOI] [PubMed] [Google Scholar]
- 70.Merino A., Bronowska A. K., Jackson D. B., Cahill D. J. Drug profiling: knowing where it hits. Drug Discovery Today. 2010;15(17-18):749–756. doi: 10.1016/j.drudis.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 71.Reddy A. S., Zhang S. Polypharmacology: drug discovery for the future. Expert Review of Clinical Pharmacology. 2013;6(1):41–47. doi: 10.1586/ecp.12.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Simon Z., Peragovics Á., Vigh-Smeller M., et al. Drug effect prediction by polypharmacology-based interaction profiling. Journal of Chemical Information and Modeling. 2012;52(1):134–145. doi: 10.1021/ci2002022. [DOI] [PubMed] [Google Scholar]
- 73.Yildirim M. A., Goh K.-I., Cusick M. E., Barabási A.-L., Vidal M. Drug-target network. Nature Biotechnology. 2007;25(10):1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 74.Padhy B. M., Gupta Y. K. Drug repositioning: re-investigating existing drugs for new therapeutic indications. Journal of Postgraduate Medicine. 2011;57(2):153–160. doi: 10.4103/0022-3859.81870. [DOI] [PubMed] [Google Scholar]
- 75.Jin G., Wong S. T. C. Toward better drug repositioning: prioritizing and integrating existing methods into efficient pipelines. Drug Discovery Today. 2014;19(5):637–644. doi: 10.1016/j.drudis.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Armbruster B. N., Roth B. L. Mining the receptorome. The Journal of Biological Chemistry. 2005;280(7):5129–5132. doi: 10.1074/jbc.r400030200. [DOI] [PubMed] [Google Scholar]
- 77.Jensen N. H., Roth B. L. Massively parallel screening of the receptorome. Combinatorial Chemistry & High Throughput Screening. 2008;11(6):420–426. doi: 10.2174/138620708784911483. [DOI] [PubMed] [Google Scholar]
- 78.Roth B. L. Receptor systems: will mining the receptorome yield novel targets for pharmacotherapy? Pharmacology & Therapeutics. 2005;108(1):59–64. doi: 10.1016/j.pharmthera.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 79.Roth B. L., Kroeze W. K. Screening the receptorome yields validated molecular targets for drug discovery. Current Pharmaceutical Design. 2006;12(14):1785–1795. doi: 10.2174/138161206776873680. [DOI] [PubMed] [Google Scholar]
- 80.Roth B. L., Sheffer D. J., Kroeze W. K. Magic shotguns versus magic bullets: selectively non-selective drugs for mood disorders and schizophrenia. Nature Reviews Drug Discovery. 2004;3(4):353–359. doi: 10.1038/nrd1346. [DOI] [PubMed] [Google Scholar]
- 81.Strachan R. T., Ferrara G., Roth B. L. Screening the receptorome: an efficient approach for drug discovery and target validation. Drug Discovery Today. 2006;11(15-16):708–716. doi: 10.1016/j.drudis.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 82.Roth B. L., Lopez E., Beischel S., Westkaemper R. B., Evans J. M. Screening the receptorome to discover the molecular targets for plant-derived psychoactive compounds: a novel approach for CNS drug discovery. Pharmacology & Therapeutics. 2004;102(2):99–110. doi: 10.1016/j.pharmthera.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 83.Roth B. L., Lopez E., Patel S., Kroeze W. K. The multiplicity of serotonin receptors: uselessly diverse molecules or an embarrassment of riches? Neuroscientist. 2000;6(4):252–262. doi: 10.1177/107385840000600408. [DOI] [Google Scholar]
- 84.Ballesteros J. A., Shi L., Javitch J. A. Structural mimicry in G protein-coupled receptors: Implications of the high-resolution structure of rhodopsin for structure-function analysis of rhodopsin-like receptors. Molecular Pharmacology. 2001;60(1):1–19. [PubMed] [Google Scholar]
- 85.Shapiro D. A., Kristiansen K., Weiner D. M., Kroeze W. K., Roth B. L. Evidence for a model of agonist-induced activation of 5-hydroxytryptamine 2A serotonin receptors that involves the disruption of a strong ionic interaction between helices 3 and 6. Journal of Biological Chemistry. 2002;277(13):11441–11449. doi: 10.1074/jbc.M111675200. [DOI] [PubMed] [Google Scholar]
- 86.Fenalti G., Giguere P. M., Katritch V., et al. Molecular control of δ-opioid receptor signalling. Nature. 2014;506(7487):191–196. doi: 10.1038/nature12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vardy E., Mosier P. D., Frankowski K. J., et al. Chemotype-selective modes of action of κ-opioid receptor agonists. The Journal of Biological Chemistry. 2013;288(48):34470–34483. doi: 10.1074/jbc.m113.515668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang C., Jiang Y., Ma J., et al. Structural basis for molecular recognition at serotonin receptors. Science. 2013;340(6132):610–614. doi: 10.1126/science.1232807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang C., Wu H., Evron T., et al. Structural basis for Smoothened receptor modulation and chemoresistance to anticancer drugs. Nature Communications. 2014;5, article 4355 doi: 10.1038/ncomms5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang C., Wu H., Katritch V., et al. Structure of the human smoothened receptor bound to an antitumour agent. Nature. 2013;497(7449):338–343. doi: 10.1038/nature12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu H., Wacker D., Mileni M., et al. Structure of the human κ-opioid receptor in complex with JDTic. Nature. 2012;485(7398):327–332. doi: 10.1038/nature10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hutcheson J. D., Setola V., Roth B. L., Merryman W. D. Serotonin receptors and heart valve disease—it was meant 2B. Pharmacology & Therapeutics. 2011;132(2):146–157. doi: 10.1016/j.pharmthera.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rothman R. B., Baumann M. H., Savage J. E., et al. Evidence for possible involvement of 5-HT(2B) receptors in the cardiac valvulopathy associated with fenfluramine and other serotonergic medications. Circulation. 2000;102(23):2836–2841. doi: 10.1161/01.CIR.102.23.2836. [DOI] [PubMed] [Google Scholar]
- 94.Setola V., Hufeisen S. J., Grande-Allen K. J., et al. 3,4-Methylenedioxymethamphetamine (MDMA, ‘Ecstasy’) induces fenfluramine-like proliferative actions on human cardiac valvular interstitial cells in vitro. Molecular Pharmacology. 2003;63(6):1223–1229. doi: 10.1124/mol.63.6.1223. [DOI] [PubMed] [Google Scholar]
- 95.Setola V., Roth B. L. Screening the receptorome reveals molecular targets responsible for drug-induced side effects: focus on ‘fen-phen’. Expert Opinion on Drug Metabolism & Toxicology. 2005;1(3):377–387. doi: 10.1517/17425255.1.3.377. [DOI] [PubMed] [Google Scholar]
- 96.Kroeze W. K., Hufeisen S. J., Popadak B. A., et al. H1-histamine receptor affinity predicts short-term weight gain for typical and atypical antipsychotic drugs. Neuropsychopharmacology. 2003;28(3):519–526. doi: 10.1038/sj.npp.1300027. [DOI] [PubMed] [Google Scholar]
- 97.Roth B. L., Baner K., Westkaemper R., et al. Salvinorin A: a potent naturally occurring nonnitrogenous κ opioid selective agonist. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(18):11934–11939. doi: 10.1073/pnas.182234399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Feng Z., Chen L., Maddula H., et al. Ligand Depot: a data warehouse for ligands bound to macromolecules. Bioinformatics. 2004;20(13):2153–2155. doi: 10.1093/bioinformatics/bth214. [DOI] [PubMed] [Google Scholar]
- 99.Irwin J. J., Shoichet B. K. ZINC—a free database of commercially available compounds for virtual screening. Journal of Chemical Information and Modeling. 2005;45(1):177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Research. 2012;40(1):D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang Y., Xiao J., Suzek T. O., et al. PubChem's BioAssay database. Nucleic Acids Research. 2012;40(1):D400–D412. doi: 10.1093/nar/gkr1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wheeler D. L., Barrett T., Benson D. A., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Research. 2008;36(1):D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pouliot Y., Chiang A. P., Butte A. J. Predicting adverse drug reactions using publicly available PubChem BioAssay data. Clinical Pharmacology and Therapeutics. 2011;90(1):90–99. doi: 10.1038/clpt.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Keiser M. J., Roth B. L., Armbruster B. N., Ernsberger P., Irwin J. J., Shoichet B. K. Relating protein pharmacology by ligand chemistry. Nature Biotechnology. 2007;25(2):197–206. doi: 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- 105.Keiser M. J., Setola V., Irwin J. J., et al. Predicting new molecular targets for known drugs. Nature. 2009;462(7270):175–181. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lounkine E., Keiser M. J., Whitebread S., et al. Large-scale prediction and testing of drug activity on side-effect targets. Nature. 2012;486(7403):361–367. doi: 10.1038/nature11159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Barabási A.-L., Oltvai Z. N. Network biology: understanding the cell's functional organization. Nature Reviews Genetics. 2004;5(2):101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 108.Campillos M., Kuhn M., Gavin A.-C., Jensen L. J., Bork P. Drug target identification using side-effect similarity. Science. 2008;321(5886):263–266. doi: 10.1126/science.1158140. [DOI] [PubMed] [Google Scholar]
- 109.Morrow J. K., Tian L., Zhang S. Molecular networks in drug discovery. Critical Reviews in Biomedical Engineering. 2010;38(2):143–156. doi: 10.1615/critrevbiomedeng.v38.i2.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tian L., Zhang S. Mapping drug-target interaction networks. Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC '09); September 2009; Minneapolis, Minn, USA. IEEE; pp. 2336–2339. [DOI] [PubMed] [Google Scholar]
- 111.Davis A. P., King B. L., Mockus S., et al. The comparative toxicogenomics database: update 2011. Nucleic Acids Research. 2011;39(1):D1067–D1072. doi: 10.1093/nar/gkq813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Oprea T. I., Mestres J. Drug repurposing: far beyond new targets for old drugs. The AAPS Journal. 2012;14(4):759–763. doi: 10.1208/s12248-012-9390-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Oprea T. I., Kim Nielsen S., Ursu O., et al. Associating drugs, targets and clinical outcomes into an integrated network affords a new platform for computer-aided drug repurposing. Molecular Informatics. 2011;30(2-3):100–111. doi: 10.1002/minf.201100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cheng F., Zhou Y., Li W., Liu G., Tang Y. Prediction of chemical-protein interactions network with weighted network-based inference method. PLoS ONE. 2012;7(7) doi: 10.1371/journal.pone.0041064.e41064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Reker D., Rodrigues T., Schneider P., Schneider G. Identifying the macromolecular targets of de novo-designed chemical entities through self-organizing map consensus. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(11):4067–4072. doi: 10.1073/pnas.1320001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Erickson C. A., Veenstra-Vanderweele J. M., Melmed R. D., et al. STX209 (arbaclofen) for autism spectrum disorders: an 8-week open-label study. Journal of Autism and Developmental Disorders. 2014;44(4):958–964. doi: 10.1007/s10803-013-1963-z. [DOI] [PubMed] [Google Scholar]
- 117.Morphy J. R., Harris C. J. Designing Multi-Target Drugs. London, UK: Royal Society of Chemistry; 2012. [Google Scholar]
- 118.Apsel B., Blair J. A., Gonzalez B., et al. Targeted polypharmacology: discovery of dual inhibitors of tyrosine and phosphoinositide kinases. Nature Chemical Biology. 2008;4(11):691–699. doi: 10.1038/nchembio.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Besnard J., Ruda G. F., Setola V., et al. Automated design of ligands to polypharmacological profiles. Nature. 2012;492(7428):215–220. doi: 10.1038/nature11691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.McLaren K. D., Marangell L. B. Special considerations in the treatment of patients with bipolar disorder and medical co-morbidities. Annals of General Hospital Psychiatry. 2004;3(1, article 7) doi: 10.1186/1475-2832-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Berry-Kravis E. M., Hessl D., Rathmell B., et al. Effects of STX209 (arbaclofen) on neurobehavioral function in children and adults with fragile X syndrome: a randomized, controlled, phase 2 trial. Science Translational Medicine. 2012;4(152) doi: 10.1126/scitranslmed.3004214.152ra127 [DOI] [PubMed] [Google Scholar]
- 122.Healy A., Rush R., Ocain T. Fragile X syndrome: an update on developing treatment modalities. ACS Chemical Neuroscience. 2011;2(8):402–410. doi: 10.1021/cn200019z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lozano R., Hare E. B., Hagerman R. J. Modulation of the GABAergic pathway for the treatment of fragile X syndrome. Neuropsychiatric Disease and Treatment. 2014;10:1769–1779. doi: 10.2147/NDT.S42919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mohammadi M. R., Kashani L., Akhondzadeh S., Izadian E. S., Ohadinia S. Efficacy of theophylline compared to methylphenidate for the treatment of attention-deficit hyperactivity disorder in children and adolescents: a pilot double-blind randomized trial. Journal of Clinical Pharmacy and Therapeutics. 2004;29(2):139–144. doi: 10.1111/j.1365-2710.2004.00545.x. [DOI] [PubMed] [Google Scholar]
- 125.Moller H. J., Demyttenaere K., Olausson B., et al. Two phase III randomised double-blind studies of fixed-dose TC-5214 (dexmecamylamine) adjunct to ongoing antidepressant therapy in patients with major depressive disorder and an inadequate response to prior antidepressant therapy. The World Journal of Biological Psychiatry. 2015;16(7):483–501. doi: 10.3109/15622975.2014.989261. [DOI] [PubMed] [Google Scholar]
- 126.Vieta E., Thase M. E., Naber D., et al. Efficacy and tolerability of flexibly-dosed adjunct TC-5214 (dexmecamylamine) in patients with major depressive disorder and inadequate response to prior antidepressant. European Neuropsychopharmacology. 2014;24(4):564–574. doi: 10.1016/j.euroneuro.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 127.Corbett A., Smith J., Ballard C. New and emerging treatments for Alzheimers disease. Expert Review of Neurotherapeutics. 2012;12(5):535–543. doi: 10.1586/ern.12.43. [DOI] [PubMed] [Google Scholar]
- 128.George T. P., Sacco K. A., Vessicchio J. C., Weinberger A. H., Shytle R. D. Nicotinic antagonist augmentation of selective serotonin reuptake inhibitor-refractory major depressive disorder: a preliminary study. Journal of Clinical Psychopharmacology. 2008;28(3):340–344. doi: 10.1097/jcp.0b013e318172b49e. [DOI] [PubMed] [Google Scholar]
- 129.Levin E. D., Simon B. B. Nicotinic acetylcholine involvement in cognitive function in animals. Psychopharmacology. 1998;138(3-4):217–230. doi: 10.1007/s002130050667. [DOI] [PubMed] [Google Scholar]
- 130.Singh A., Das D. K., Kelley M. E. Mecamylamine: Targacept. IDrugs. 2006;9(3):205–217. [PubMed] [Google Scholar]
- 131.Ueno K.-I., Togashi H., Matsumoto M., Ohashi S., Saito H., Yoshioka M. α4β2 Nicotinic acetylcholine receptor activation ameliorates impairment of spontaneous alternation behavior in stroke-prone spontaneously hypertensive rats, an animal model of attention deficit hyperactivity disorder. The Journal of Pharmacology and Experimental Therapeutics. 2002;302(1):95–100. doi: 10.1124/jpet.302.1.95. [DOI] [PubMed] [Google Scholar]
- 132.Belanoff J. K., Flores B. H., Kalezhan M., Sund B., Schatzberg A. F. Rapid reversal of psychotic depression using mifepristone. Journal of Clinical Psychopharmacology. 2001;21(5):516–521. doi: 10.1097/00004714-200110000-00009. [DOI] [PubMed] [Google Scholar]
- 133.Blasey C. M., Block T. S., Belanoff J. K., Roe R. L. Efficacy and safety of mifepristone for the treatment of psychotic depression. Journal of Clinical Psychopharmacology. 2011;31(4):436–440. doi: 10.1097/JCP.0b013e3182239191. [DOI] [PubMed] [Google Scholar]
- 134.Carroll B. J., Rubin R. T. Mifepristone in psychotic depression? Biological Psychiatry. 2008;63(1):e1–e3. doi: 10.1016/j.biopsych.2007.03.032. [DOI] [PubMed] [Google Scholar]
- 135.Howland R. H. Mifepristone as a therapeutic agent in psychiatry. Journal of Psychosocial Nursing and Mental Health Services. 2013;51(6):11–14. doi: 10.3928/02793695-20130513-01. [DOI] [PubMed] [Google Scholar]
- 136.Clarke C. E., Deane K. H. Ropinirole for levodopa-induced complications in Parkinson's disease. The Cochrane Database of Systematic Reviews. 2000;3 doi: 10.1002/14651858.CD001516.CD001516 [DOI] [PubMed] [Google Scholar]
- 137.Clarke C. E., Deane K. H. Ropinirole for levodopa-induced complications in Parkinson's disease. The Cochrane Database of Systematic Reviews. 2001;(1) doi: 10.1002/14651858.CD001518.CD001516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tomasiuk R., Szlufik S., Friedman A., Koziorowski D. Ropinirole treatment in Parkinson's disease associated with higher serum level of inflammatory biomarker NT-proCNP. Neuroscience Letters. 2014;566:147–150. doi: 10.1016/j.neulet.2014.02.053. [DOI] [PubMed] [Google Scholar]
- 139.Yildiz A., Guleryuz S., Ankerst D. P., Öngür D., Renshaw P. F. Protein kinase C inhibition in the treatment of mania: a double-blind, placebo-controlled trial of tamoxifen. Archives of General Psychiatry. 2008;65(3):255–263. doi: 10.1001/archgenpsychiatry.2007.43. [DOI] [PubMed] [Google Scholar]
- 140.Corbett A., Williams G., Ballard C. Drug repositioning: an opportunity to develop novel treatments for Alzheimer's disease. Pharmaceuticals. 2013;6(10):1304–1321. doi: 10.3390/ph6101304. [DOI] [PMC free article] [PubMed] [Google Scholar]