Abstract
Salmonella spp. is one of the most important food-borne pathogens causing digestive tract and invasive infections in both humans and animals. Extended-spectrum β-lactamases (ESBLs) especially the CTX-M-type ESBLs are increasingly being reported worldwide and in China. These studies seldom focused on Salmonella isolates from food-producing animals. The aim of this study was to characterize the antimicrobial resistance profiles, serotypes and ESBLs and in particular, CTX-M producing Salmonella isolates from chickens and pigs in China. Salmonella isolates were identified by API20E system and polymerase chain reaction (PCR) assay; serotypes were determined using slide agglutination with hyperimmune sera; antimicrobial susceptibility was tested using the ager dilution method; the prevalence of ESBLs and PMQR genes were screened by PCR; CTX-M-producing isolates were further characterized by conjugation along with genetic relatedness and plasmid replicon type. In total, 159 Salmonella strains were identified, among which 95 strains were Salmonella enterica serovar Typhimurium, 63 strains were S. enterica serovar Indiana, and 1 strain was S. enterica serovar Enteritidis. All of these isolates presented multi-drug resistant phenotypes. Forty-five isolates carried blaCTX-M genes, the most common subtype was CTX-M-27(34), followed by CTX-M-65(7) and CTX-M-14(4). Most blaCTX-M genes were transmitted by non-typeable or IncN/IncFIB/IncP/IncA/C/IncHI2 plasmids with sizes ranging from 80 to 280 kb. In particular, all the 14 non-typeable plasmids were carrying blaCTX-M-27 gene and had a similar size. PFGE profiles indicated that CTX-M-positive isolates were clonally related among the same serotype, whilst the isolates of different serotypes were genetically divergent. This suggested that both clonal spread of resistant strains and horizontal transmission of the resistance plasmids contributed to the dissemination of blaCTX-M-9G-positive Salmonella isolates. The presence and spread of CTX-M, especially the CTX-M-27 in S. enterica serovars Typhimurium and Indiana from food-producing animals poses a potential threat for public health. Control strategies to limit the dissemination of these strains through the food chain are necessary.
Keywords: Salmonella, food-producing animals, serotype, antimicrobial resistance, CTX-M
Introduction
The emergence of antibiotic-resistant bacteria has become a serious challenge in human and veterinary medicine globally and poses a serious public health threat. Non-typhoidal Salmonella (NTS) are a leading cause of bacterial diarrhea worldwide and one of the most important foodborne pathogens. The majorities of human infections by NTS are associated with food product consumption of meat, eggs, milk, seafood, and other fresh products derived from animals (Foley and Lynne, 2008). In the animal husbandry industry, antibiotics are used both for therapy and as growth promoters. Additionally, animals are always a large repository of resistant bacteria. Domestic farm animals, especially poultry and pigs have been shown to be major environmental reservoirs of food-borne NTS (Vo et al., 2006).
Serotyping is a useful classification scheme that allows for trends in Salmonella surveillance data to be followed over time. These patterns are also related to the ability to cause disease and with the associated antibiotic resistance profile. Over 2,600 serotypes of Salmonella have been identified based on the reactivity of antisera to O and H antigens (Stevens et al., 2009), and there are numerous overlaps between animal and human Salmonella serotypes (Alcaine et al., 2006). This suggests that Salmonella transmission from animals to humans occurs via the food chain. Salmonella enterica serovars Enteritidis and Typhimurium are the most common serotypes in Europe (European Centre for Disease Prevention and Control, 2013). However, serotype distribution is varied in different provinces of China and in total the most prevalent serovars were also Typhimurium and Enteritidis with a rising rate of reports of S. enterica serovar Indiana in isolates obtained from animals (Xia et al., 2009; Yang et al., 2010; Lu et al., 2014; Zhang et al., 2014).
It is estimated that that between 1994 and 2005 roughly 22% of foodborne diseases in China were caused by Salmonella though there was no official surveillance data (Wang et al., 2007). In 2013, unpublished data from the China CDC surveillance system showed that the carriage rate of human salmonellosis was 549 per 100,000 people. This is more than 33 times higher than human infections in the USA in 2012 (16.4 per 100,000; Centers for Disease Control and Prevention, 2013; Sun et al., 2014). There is also an increasing risk of Salmonella spread from animals to humans via the food chain due to worldwide distribution of animal food products from China. Extended spectrum cephalosporins (ESCs) and fluoroquinolones are the drugs of choice for treatment of invasive Salmonella infections in food animals and for people at risk of such infections (Harrois et al., 2014). This is the focus of current concerns on the emergence of resistance to these drugs. ESCs- and fluoroquinolone-resistant Salmonella populations have increased dramatically worldwide (Arlet et al., 2006; Hendriksen et al., 2009; Jiang et al., 2014; Lu et al., 2014). Resistance to cephalosporins is mainly due to the acquisition of extended-spectrum β-lactamases (ESBLs) genes especially the CTX-M type that is primarily carried by transferable plasmids and transposons (Liebana et al., 2013). These plasmids and transposons, in many cases, also carry resistance genes for other antimicrobial classes such as the fluoroquinolones which sometimes limits treatment options (Harrois et al., 2014). Plasmid-mediated quinolone resistance (PMQR) has recently been categorized as containing three distinct resistance mechanisms: (1) Qnr proteins that mediate target protection, (2) A variant of an aminoglycoside acetyltransferase designated AAC(6’)-Ib-cr that acetylates ciprofloxacin and norfloxacin and (3) Drug efflux mediated by OqxAB and QepA (Strahilevitz et al., 2009). Although these PMQR determinants confer only low-level fluoroquinolone resistance, their presence can provide a selective advantage for bacteria to develop to high-level quinolone resistance. This occurs by chromosomal mutations in the quinolone resistance-determining regions (QRDRs) of genes encoding target enzymes (Strahilevitz et al., 2009). The situation is especially troubling when ESBL genes and PMQR genes are co-transmitted through transferable plasmids.
Indiscriminate use of antibiotics in both human and veterinary medicine make the ESBLs especially the CTX-M-producing bacteria including Salmonella increase rapidly accelerating the spread of antibiotic resistance. There have been several reports regarding characteristics of CTX-M-producing Salmonella isolated from food-producing animals in China. However, data on distribution of serotypes and antimicrobial susceptibility of Salmonella recovered from animals are available in most of these reports (Yan et al., 2010; Yang et al., 2010; Li R. et al., 2013; Li et al., 2014; Lu et al., 2014; Kuang et al., 2015). The aim of the present study was to investigate the prevalence of antibiotic resistance, serotypes and ESBLs among Salmonella isolates from chickens and pigs in China. Plasmids that carried blaCTX-M genes were also further studied to characterize the mechanism of transfer and dissemination of β-lactamases.
Materials and Methods
Salmonella Isolation, Identification, and Serotyping
In 2014, a total of 3850 non-repetitive fecal swabs were collected from healthy chickens and pigs in Guangdong, Shandong, Henan, Hubei provinces of China. Of these samples, 2090 samples were from chickens and 1760 samples were from pigs.
Cotton swabs of feces were inoculated into sterile selenite cystine broth for 24 h at 37°C, and then streaked onto chromogenic medium selective for Salmonella (CHROMagar Microbiology, France) and incubated for another 24 h at 37°C. One purple colony was selected from each plate and then confirmed using the API20E system (bioMérieux, Marcy l’Étoile, France) and a PCR assay targeting the invA gene (Rahn et al., 1992). Salmonella isolates were serotyped using slide agglutination with hyperimmune sera (S&A Company, Bangkok, Thailand) and the results were interpreted according to the Kauffmann–White scheme. All identified isolates were stored at –80°C in Luria–Bertani (LB) broth containing 30% glycerol.
Antimicrobial Susceptibility Testing
The minimum inhibitory concentrations (MICs) of Salmonella isolates were determined using the agar dilution method following Clinical and Laboratory Standards Institution guidelines. Sixteen antimicrobials were tested: ampicillin (AMP), cefotaxime (CTX), cefoxitin (CXT), ceftiofur (CTF), ceftazidime (CAZ), ceftriaxone (CTR), nalidixic acid (NAL), ciprofloxacin (CIP), enrofloxacin (ENR), kanamycin (KAN), gentamycin (GEN), amikacin (AMK), tetracycline (TET), chloramphenicol (CHL), florfenicol (FFC) and olaquindox (OLA). The results were interpreted according to the standards described by CLSI (M100-S25) (Clinical and Laboratory Standards Institute [CLSI], 2015) (ampicillin, cefotaxime, ceftiofur, ceftriaxone, cefoxitin, ceftazidime, nalidixic acid, ciprofloxacin, kanamycin, amikacin) and VET01-A4/VET01-S2 (Clinical and Laboratory Standards Institute [CLSI], 2013) (gentamicin, enrofloxacin, tetracycline, chloramphenicol, florfenicol) and the DANMAP 98 (olaquindox). Escherichia coli ATCC 25922 was used as quality control.
Detection of β-Lactamase Genes and PMQR Genes
The Salmonella isolates harvested from Mueller Hinton (MH) agar plates supplied with 2 mg/L cefotaxime were subjected to screening for ESBL-genes using PCR as described previously (Jiang et al., 2012). The DNA sequences and deduced amino acid sequences were compared with the reported sequences from GenBank1. The presence of PMQR determinants [qnrA, qnrB, qnrS, qnrD, qnrC, aac(6’)-Ib-cr,qepA, and oqxAB] was also analyzed using PCR primers and conditions described previously (Jiang et al., 2012). Mutations in QRDR of target genes (gyrA and parC) were further analyzed in both PMQR and CTX-M-positive isolates as described previously (O’Regan et al., 2009).
Molecular Typing
Genetic relatedness of all blaCTX-M -positive isolates was analyzed by pulsed-field gel electrophoresis (PFGE) after Xbal-digested genomic DNA using a CHEF-MAPPER System (Bio-Rad Laboratories, Hercules, CA, USA) as described previously (Jiang et al., 2014). The resulting PFGE patterns were compared using the Dice similarity coefficient of BioNumerics software (Applied Maths, Sint-Martens-Latem, Belgium).
Conjugation and Transformation Experiments
Conjugation experiments were conducted in blaCTX-M-positive strains by liquid mating-out assay in LB-medium using a sodium azide-resistant E. coli J53 as the recipient. Transconjugants were selected on MacConkey agar containing cefotaxime (2 mg/L) and sodium azide (300 mg/L). For transformation experiments, plasmid DNA from the blaCTX-M-positive strains were extracted using Qiagen Plasmid Midi Kit according to the manufacturer’s instructions (Qiagen, Germany). Purified plasmids were transformed into E. coli DH5a (TaKaRa Biotechnology, Dalian, China). Selection of transformants was performed on MacConkey agar containing 2 mg/L cefotaxime. MICs and the presence of blaCTX-M gene of transconjugants and transformants were determined as described above.
Plasmid Characterization
PCR-based replicon typing (PBRT) was performed on transconjugants and transformants using primers as described previously (Carattoli et al., 2005). Then PFGE with S1 nuclease (Takara Biotechnology, Dalian, China) digestion of whole genomic DNA was carried out as described previously (Barton et al., 1995). The resulting gels were analyzed by Southern transfer and probing with a DIG-labeled blaCTX-M gene fragment according to the manufacturer’s instructions (DIG High Prime DNA Labeling and Detection Starter Kit I, Roche Applied Science, Mannheim, Germany).
Results
Salmonella Isolation, Identification, and Serotyping
A total of 159 Salmonella strains were obtained among which 90 (56.6%) were isolated from pigs and 69 (43.4%) were recovered from chickens. In all, three serotypes were identified in the 159 Salmonella isolates and this accounted for 95 strains of S. enterica serovar Typhimurium, 63 strains of S. enterica serovar Indiana, and 1 strain of S. enterica serovar Enteritidis.
The S. enterica serovar Typhimurium isolates were distributed as follows: 73.7% from pigs and 26.3% from chickens while the Indiana isolates were almost evenly distributed (50.8% from chickens and 49.2% from pigs, relatively).
Antimicrobial Susceptibility Phenotypes
Among the159 Salmonella isolates, resistance was most frequently observed to tetracycline (89.9%), ampicillin (84.9%), and gentamicin (81.1%). Resistance rates of the quinolone antibiotics were all above 50% including resistance to nalidixic acid (77.3%), ciprofloxacin (64.8%), and enrofloxacin (62.9%). Cephalosporin resistance rates ranged from 17.6 to 76.7% with high resistance rates to ceftriaxone (76.7%), ceftiofur (52.8%), and cefotaxime (47.2%). Resistance rates to ceftazidime and cefoxitin were 30.8 and 17.6%, respectively. The presence of resistance to olaquindox, a growth promoter used extensively in pigs and recently prohibited for poultry use, was detected in 65.4% of all isolates (Table 1).
Table 1.
Antimicrobial resistance of Salmonella enterica isolates from food-producing animals in China.
| Antimicrobial agents | Number(%) of resistant isolates (n = 159) |
|---|---|
| β-Lactams | |
| Ampicillin | 135 (84.9) |
| Cefotaxime | 75 (47.2) |
| Cefoxitin | 28 (17.6) |
| Ceftiofur | 84 (52.8) |
| Ceftazidime | 49 (30.8) |
| Ceftriaxone | 122 (76.7) |
| Quinolones | |
| Nalidixic acid | 123 (77.4) |
| Ciprofloxacin | 103 (64.8) |
| Enrofloxacin | 100 (62.9) |
| Aminoglycosides | |
| Kanamycin | 124 (78.0) |
| Gentamicin | 129 (81.1) |
| Amikacin | 9 (5.7) |
| Other Antibiotics | |
| Tetracycline | 143 (89.9) |
| Chloramphenicol | 126 (79.2) |
| Florfenicol | 124 (78.0) |
| Olaquindox | 104 (65.4) |
All the 159 Salmonella isolates were multi-drug resistant displaying resistance to three or more classes of antimicrobials. Higher resistance frequencies were found among S. enterica serovar Indiana isolates compared with S. enterica serovar Typhimurium isolates. For example, cefotaxime resistance in Indiana and Typhimurium isolates were 71.4 and 30.5%, respectively, while the ciprofloxacin resistance rates were 73.0 and 43.2%, respectively (Table 2).
Table 2.
Antibiotic resistance rate of Salmonella enterica serotype Typhimurium and Indiana isolates (%).
| Serotype Antibiotics | AMP | CTF | CTX | CXT | CTZ | CTR | GEN | KAN | AMI | CIP | ENR | NAL | TET | CHL | FFC | OQX |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Indiana (63) | 93.7 | 76.2 | 71.4 | 19.0 | 33.3 | 82.5 | 88.9 | 84.1 | 6.3 | 73.0 | 77.8 | 87.3 | 96.8 | 90.5 | 84.1 | 68.3 |
| Typhimurium (95) | 89.5 | 37.9 | 30.5 | 11.6 | 29.5 | 73.7 | 76.8 | 74.7 | 5.3 | 60.0 | 52.6 | 70.5 | 86.3 | 72.6 | 74.7 | 64.2 |
ESBL Characterization
Seventy-five isolates displayed non-wild type MICs for cefotaxime and 45 of these carried blaCTX-M genes, three harbored blaCMY-2 one of which coexisted with the CTX-M-encoding gene. For other β-lactamase genes, narrow-spectrum β-lactamase blaTEM-1 was present in 86 of the 159 isolates, blaSHV-1 and blaOXA-1 genes were found in 62 and 55 of these isolates, respectively. No other type of ESBLs gene was found.
For the PMQR genes, three classes of resistance mechanisms were represented: oqxAB (78.0%, n = 124); aac(6’)-Ib-cr(69.2%, n = 110) and qnr(17.6%, n = 28). For 28 qnr genes, 12 were qnrD, 7 were qnrB, 5 were qnrS, and the remaining 4 were qnrA. All qnr-positive strains also carried oqxAB and/or aac(6’)-Ib-cr. At least one PMQR gene was found in each CTX-M-producing strains, especially the aac(6’)-Ib-cr +oqxAB and qnr+aac(6’)-Ib-cr +oqxAB were found in 17 and 4 of all 45 CTX-M-producing strains, respectively. Mutations in the QRDRs of gyrA and parC were analyzed among the 45 PMQR and CTX-M-positive isolates. A combination of mutations in gyrA83 (S83F) and 87 (D87G) and in parC 80(S80R) were found in 35 (77.8%) isolates, a signal mutation in gyrA 87 (D87N) without mutation in parC was detected in the remaining 10 isolates (Table 3).
Table 3.
Characteristics of CTX-M-producing Salmonella enterica isolates from food-producing animals in China.
| Strain | Source | Serotype | ESBL gene | PMQR | Amino acid substitutions |
Co-transferred resistant gene | Plasmid replicon type | Plasmid approx. size(kb) | |
|---|---|---|---|---|---|---|---|---|---|
| gyrA | parC | ||||||||
| SP5 | Pig | Typhimurium | blaCTX-M-14 | aac-(6’)-Ib-cr,oqxAB | D87N | – | blaSHV -1, aac-(6’)-Ib-cr | FIB | 100 |
| SP85 | Pig | Indiana | blaCTX-M-27 | aac-(6’)-Ib-cr oqxAB | S83F,D87G | S80R | blaOXA-1,aac-(6’)-Ib-cr | NT | 100 |
| SP96 | Pig | Indiana | blaCTX-M-27 | oqxAB | S83F,D87G | S80R | blaSHV -1, oqxAB | P | 150 |
| SP99 | Pig | Typhimurium | blaCTX-M-27 | qnrB, aac-(6’)-Ib-cr,oqxAB | S83F,D87G | S80R | aac-(6’)-Ib-cr oqxAB | NT | 100 |
| SP101 | Pig | Typhimurium | blaCTX-M-65 | oqxAB | D87N | – | oqxAB | FIB | 150 |
| SP108 | Pig | Indiana | blaCTX-M-27 | aac-(6’)-Ib-cr,oqxAB | S83F,D87G | S80R | blaSHV -1, aac-(6’)-Ib-cr | A/C | 150 |
| SP110 | Pig | Indiana | blaCTX-M-65 | oqxAB | S83F,D87G | S80R | blaSHV -1 | N | 100 |
| SP115 | Pig | Indiana | blaCTX-M-27 | aac-(6’)-Ib-cr,oqxAB | S83F,D87G | S80R | aac-(6’)-Ib-cr | NT | 100 |
| SP123 | Pig | Indiana | blaCTX-M-27 | oqxAB | D87N | – | – | FIB | 150 |
| SP125 | Pig | Indiana | blaCTX-M-27 | oqxAB | D87N | – | – | FIB | 150 |
| SP129 | Pig | Typhimurium | blaCTX-M-27 | aac-(6’)-Ib-cr,oqxAB | S83F,D87G | S80R | blaSHV -1,aac-(6’)-Ib-cr | P | 80 |
| SP131 | Pig | Indiana | blaCTX-M-65 | aac-(6’)-Ib-cr,oqxAB | S83F,D87G | S80R | blaSHV -1, aac-(6’)-Ib-cr | N | 150 |
| SP132 | Pig | Indiana | blaCTX-M-27 | oqxAB | S83F,D87G | S80R | blaSHV -1, oqxAB | N | 150 |
| SP2019 | Pig | Typhimurium | blaCTX-M-14 | aac-(6’)-Ib-cr,oqxAB | D87N | – | blaOXA-1, oqxAB | HI2 | 280 |
| CL108 | Chicken | Indiana | blaCTX-M-27 | oqxAB | S83F,D87G | – | – | NT | 100 |
| CL129 | Chicken | Indiana | blaCTX-M-27 | aac-(6’)-Ib-cr | S83F,D87G | S80R | blaSHV -1,aac-(6’)-Ib-cr | FIB | 150 |
| CL135 | Chicken | Indiana | blaCTX-M-27 | oqxAB | S83F,D87G | S80R | – | NT | 100 |
| CL140 | Chicken | Indiana | blaCTX-M-27 | aac-(6’)-Ib-cr | S83F,D87G | S80R | – | NT | 100 |
| CL146 | Chicken | Indiana | blaCTX-M-27 | qnrD,aac-(6’)-Ib-cr,oqxAB | S83F,D87G | S80R | blaSHV -1,aac-(6’)-Ib-cr | NT | 100 |
| CL189 | Chicken | Indiana | blaCTX-M-27 | oqxAB | S83F,D87G | S80R | oqxAB | HI2 | 200 |
| XC48 | Chicken | Typhimurium | blaCTX-M-27 | aac-(6’)-Ib-cr, oqxAB | S83F,D87G | S80R | aac-(6’)-Ib-cr | N | 150 |
| XC87 | Chicken | Typhimurium | blaCTX-M-65 | aac-(6’)-Ib-cr,oqxAB | S83F,D87G | S80R | aac-(6’)-Ib-cr oqxAB | N, | 150 |
| XC134 | Chicken | Typhimurium | blaCTX-M-14 | aac-(6’)-Ib-cr,oqxAB | S83F,D87G | S80R | oqxAB | N,A/C | 150 |
| XC164 | Chicken | Typhimurium | blaCTX-M-27 | oqxAB | S83F,D87G | S80R | blaTEM-1b,oqxAB | HI2 | 194 |
| HB73 | Chicken | Indiana | blaCTX-M-27 | oqxAB | S83F,D87G | S80R | – | A/C | 130 |
| HB137 | Chicken | Indiana | blaCTX-M-27 | aac-(6’)-Ib-cr,oqxAB | S83F,D87G | S80R | oqxAB | NT | 100 |
| HB154 | Chicken | Indiana | blaCTX-M-65 | aac-(6’)-Ib-cr,oqxAB | S83F,D87G | S80R | aac-(6’)-Ib-cr oqxAB | N, | 130 |
| SG36 | Chicken | Indiana | blaCTX-M-27 | oqxAB | S83F,D87G | S80R | blaSHV -1 | NT | 100 |
| SG119 | Chicken | Indiana | blaCTX-M-27 | aac-(6’)-Ib-cr | S83F,D87G | S80R | – | NT | 100 |
| MM62 | Chicken | Enteritidis | blaCTX-M-27 | aac-(6’)-Ib-cr | D87N | – | aac-(6’)-Ib-cr | P | 80 |
| PY12 | Chicken | Indiana | blaCTX-M-14 | aac-(6’)-Ib-cr,oqxAB | D87N | – | blaSHV -1 | N,A/C | 130 |
| K13 | Chicken | Indiana | blaCTX-M-27 | aac-(6’)-Ib-cr,oqxAB | S83F,D87G | S80R | – | NT | 100 |
| K14 | Chicken | Indiana | blaCTX-M-27 | oqxAB | S83F,D87G | S80R | – | NT | 100 |
| K17 | Chicken | Typhimurium | blaCTX-M-27 | aac-(6’)-Ib-cr,oqxAB | S83F,D87G | S80R | aac-(6’)-Ib-cr | NT | 100 |
| K19 | Chicken | Indiana | blaCTX-M-65 | aac-(6’)-Ib-cr,oqxAB | S83F,D87G | S80R | aac-(6’)-Ib-cr,oqxAB | N | 150 |
| K21 | Chicken | Indiana | blaCTX-M-27 | aac-(6’)-Ib-cr | D87N | – | aac-(6’)-Ib-cr | FIB | 150 |
| K46 | Chicken | Indiana | blaCTX-M-27 | aac-(6’)-Ib-cr,oqxAB | S83F,D87G | S80R | aac-(6’)-Ib-cr,oqxAB | P | 100 |
| K47 | Chicken | Indiana | blaCTX-M-27 | aac-(6’)-Ib-cr | S83F,D87G | S80R | aac-(6’)-Ib-cr | NT | 100 |
NT, non-typeable incompatibility (Inc) group plasmid. The underline isolates were transconjugants, the others were transformants.
A total of three different CTX-M ESBLs were detected in these isolates and they all belonged to the CTX-M-9 group with the CTX-M-encoding genes distributed as 34 blaCTX-M-27, 7 blaCTX-M-65, and 4 blaCTX-M-14. The distribution of these three CTX-M subtypes showed no specificity between serotypes and animals and were found in both Indiana and Typhimurium isolates from pigs and chickens. Antimicrobial susceptibility testing demonstrated that CTX-M-27-positive isolates had significantly higher resistance levels than the CTX-M-14 or CTX-M-65 -positive isolates.
Plasmid Characterization
Six transconjugants and 32 transformants were successfully obtained from 45 blaCTX-M-positive isolates by conjugation/transformation experiments with frequencies of 10-8 to 10-3 per donor cell. PMQR genes were found to be co-transferred with blaCTX-M genes in 26 (68.4%) transconjugants/transformants and the most common co-transferring pattern was blaCTX-M +aac(6’)-Ib-cr accounting for 13 of 26 transconjugants/transformants, blaCTX-M +oqxAB and blaCTX-M +aac(6’)-Ib-cr +oqxAB accounting for eight and five transconjugants/transformants, respectively. For narrow-spectrum β-lactamases, blaSHV -1, blaOXA-1, and blaTem-1b were co-transferred with blaCTX-M in 11, 2, and 1 isolates, respectively.
Plasmid replicon typing of transconjugants/transformants revealed that IncN, IncFIB, IncP, IncA/C, and IncHI2 were detected in 9, 6, 4, 4, and 3 transconjugants/transformants, respectively. Two plasmid replicons (IncN in combination with IncA/C) were simultaneously present in 2 CTX-M-14-producing transformants. Surprisingly, the replicon types could not be determined by PBRT in 14 transconjugants/transformants all of which carried the blaCTX-M-27 gene (Table 3).
Southern blot hybridization suggested that the plasmid sizes of blaCTX-M genes varied between 80 and 280 kb among which the 100 kb (n = 17) and 150 kb (n = 13) were the most common sizes. Interestingly, the size of all the 14 non-typeable blaCTX-M-27 plasmids was ∼100 kb.
Pulsed-Field Gel Electrophoresis (PFGE)
A total of 34 different PFGE profiles were obtained from the 45 blaCTX-M-positive isolates (Figure 1). These could be grouped into 15 PFGE clusters designated A–O having 85% genetic similarity. Clusters G, D, J, and A accounted for 28.9, 15.6, 11.1, and 8.9% of the isolates, respectively. In addition, all isolates in each cluster had the same serotype and were also isolated from the same province (same or different cities). The exceptions were clusters C and M both of which were comprised of two isolates each from different provinces (Hubei, Henan, or Shan dong provinces).
FIGURE 1.
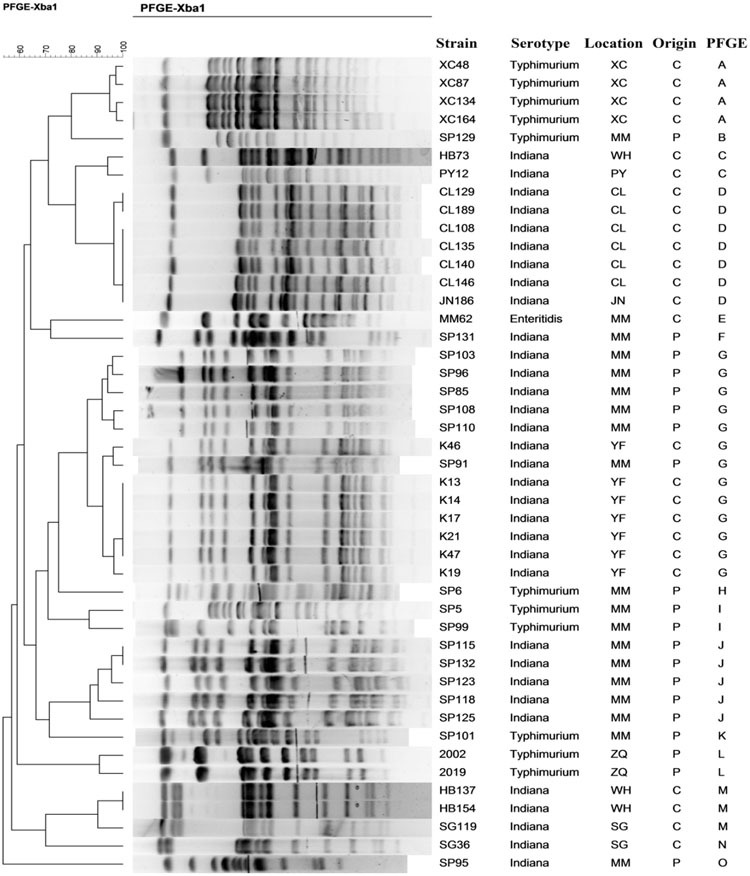
Pulsed-field gel electrophoresis fingerprinting patterns of Xba I-digested total DNA preparations from Salmonella isolates harboring CTX-M-encoding genes.
In the most predominant cluster G, a high similarity of PFGE profiles of the isolates could be found and the isolates were recovered from pigs or chickens in two cities of Guangdong province. Seven isolates in this cluster were recovered from chickens and six were from pigs. In the remaining 14 PFGE clusters, isolates were all recovered from the same origin; either chickens or pigs.
Discussion
In this study, we examined Salmonella isolates recovered from food-producing animals to determine their serotypes, antimicrobial susceptibility phenotypes, and genotypes. The results showed that S. enterica serovars Typhimurium and Indiana were the prevailing serotypes of Salmonella in pigs and chickens. Both serovars were resistant to multiple antimicrobials including ESCs and fluoroquinolones which are often antimicrobials of choice for treating salmonellosis. Resistance to cefotaxime could be explained by the presence of non-typeable and IncN/IncFIB/IncP/IncA/C/IncHI2 plasmids carrying blaCTX-M-9G genes. The high detection rate of PMQR determinants especially aac(6’)-Ib-cr and oqxAB and mutations in QRDR of target genes made an important contribution to the non-wild type MICs of the fluoroquinolones.
Salmonella has various serotypes, and some serotypes may also relate to multidrug-resistant phenotypes (Meunier et al., 2002). The predominant serotypes change over time and differ from one geographical area to another. Additionally, a particular serovar can prevail and emerge within a region for a certain period while being completely absent in other regions. S. enterica serovar Indiana is rarely reported worldwide but there have been large-scale food poisoning outbreaks caused by this serotype in both the USA and Europe (Price and Carter, 1967; Campbell and Eckman, 1975). In China, little information about S. enterica serovar Indiana was available until this serotype was obtained from a food animal (Yan et al., 2010) and showed resistance to multiple antimicrobial agents. Following this, isolation of S. enterica serovar Indiana increased rapidly and it became more and more prevalent in China especially in isolates from veterinary clinics and animal foods. The characteristics of multidrug-resistant Indiana in China were also reported (Xia et al., 2009; Yang et al., 2010; Lu et al., 2014). In the present study, all of the 159 Salmonella isolates displayed resistance to three or more classes of antimicrobials, but serovar Indiana isolates showed higher resistance levels than serovar Typhimurium isolates. Our results indicate that the multi-resistant serotype Indiana along with Typhimurium isolates are of high prevalence among food-producing animals in China.
CTX-M type ESBLs have been widely reported globally for many years and there is a strong linkage between their emergence and increasing quinolone resistance in the Enterobacteriaceae (Lavilla et al., 2008). The co-existence or co-transfer of PMQR genes in ESBLs and especially in CTX-M-producing isolates has been also previously described (Liu et al., 2013; Jiang et al., 2014; Li et al., 2014; Yang et al., 2015). These may promote the development of multidrug-resistant isolates under selective pressure of the quinolone and/or cephalosporin (Liu et al., 2013). We found all CTX-M-encoding strains contained at least one PMQR genes especially the aac(6’)-Ib-cr and oqxAB genes. These two PMQR genes have been shown to accelerate the development of fluoroquinolone resistance in S. typhimurium (Wong et al., 2014a) and were frequently co-transferred with blaCTX-M genes. Previous reports showed that aac(6’)-Ib-cr and oqxAB were also prevalent PMQR genes in E. coli isolates from humans, animals and the environment (Chen et al., 2012). In general, detection rates of PMQR genes in Salmonella were less than those observed in E. coli. However, these rates in Salmonella increased quickly in China especially aac(6’)-Ib-cr and oqxAB. In the present study, 78.0 and 69.2% harbored oqxAB and aac(6’)-Ib-cr, which is higher than levels previously reported in 2009–2010 (32.3%, 25.8%), 2007–2011(30.8%, 30.8%) and in 2012–2013 (31.7%, 36.5%) in China (Li et al., 2013; Jiang et al., 2014; Li et al., 2014). Our results may indicate that ESBL genes along with PMQR genes are increasing among Salmonella strains in China.
The CTX-M type β-lactamases are the most prevalent ESBLs among the Enterobacteriaceae recovered from both animals and humans worldwide (D’Andrea et al., 2013). The spread and characteristics of CTX-M type ESBLs producing pathogenic bacteria had been widely studied previously in China, but works seldom focused on food-borne Salmonella. In the present study, 60% of cefotaxime-resistant isolates produced CTX-M type ESBLs and three different CTX-M-9 group variants were detected among which the CTX-M-27 enzyme was the most dominant. Consistent with the results in this study, the CTX-M-27 enzyme was also the main type of ESBL in our previous study with Salmonella (Jiang et al., 2014) and we also previously found the CTX-M-27 was the only CTX-M-type ESBLs detected in Salmonella isolates of retail pork (unpublished data). However, only two reports were found about CTX-M-27-producing human clinical Salmonella strains. One study showed that CTX-M-27-producing isolates of S. enterica serotype Livingstone were the cause of a nosocomial outbreak in the neonatal ward, the other study showed blaCTX-M-27 along with oqxAB on the IncHI2 plasmid which was similar to that of the strain CL189 in our study (Bouallegue-Godet et al., 2005; Wong et al., 2014b). Many reports suggested that CTX-M-14 and CTX-M-55 enzymes were the two most dominant types in E. coli obtained from food animals in China (Zheng et al., 2012; Rao et al., 2014). CTX-M-27 in E. coli was only sporadically detected in several reports (Liu et al., 2007; Lu et al., 2010; Ho et al., 2011; Ma et al., 2012; Zheng et al., 2012; Rao et al., 2014) among which two studies found its high presence in E. coli isolated from duck and environmental samples and aquatic sediment samples (Lu et al., 2010; Ma et al., 2012). Study on isolates from human beings and environmental samples suggested that most CTX-M-27-positive E. coli from different hospitals and from rivers and lakes belonged to the O25b-ST131-B2 or B2:ST131 which was strongly associated with potentially severe infections in both humans and animals (Matsumura et al., 2012; Zurfluh et al., 2013; Micenkova et al., 2014). The high prevalence of blaCTX-M-27 in Salmonella obtained from food animals and in E. coli isolates from different sources (human beings, environment, and animals) should be a cause for concern which may be a potential threat to public health.
CTX-M-27 which was first found in a clinical isolate from a French hospital differed from its ancestor-CTX-M-14 only by the substitution D240G, but had significant higher hydrolytic activity against ceftazidime (Bonnet et al., 2003). Previous studies (Rao et al., 2014) suggested that CTX-M-1G-positive isolates had significantly higher resistance rates to cefquinome, ceftazidime, amikacin and fosfomycin when compared to the CTX-M-9G-positive isolates. These authors also compared the activity of different CTX-M variants on cephalosporin resistance and proved that CTX-M-1 group enzymes, especially CTX-M-55, have higher hydrolytic activities against cefquinome than CTX-M-9 group enzymes (except CTX-M-27). All the blaCTX-M genes detected in this study belonged to CTX-M-9 group. When we analyzed the MICs of cephalosporin among different CTX-M-9G variants, we concluded that blaCTX-M-27-positive isolates had a higher MIC for cephalosporin in particular to ceftazidime than that in CTX-M-14 and CTX-M-65-producing isolates. There were few differences in the resistance phenotypes of different variants of the CTX-M-9G family of β-lactamases isolates. This may also explain the prevalence of different variants.
In this study, 38 blaCTX-M-positive transconjugants/transformants were obtained, among which 68.4% co-transferred blaCTX-M with aac(6’)-Ib-cr and/or oqxAB, indicating that these fluoroquinolone resistant determinants can be transferred horizontally simultaneous with the transfer of CTX-M-encoding genes. Characterization of blaCTX-M plasmids in the transconjugants/transformants revealed that blaCTX-M genes were located on plasmids of un-typeable and IncN/IncFIB/IncP/IncA/C/IncHI2 replicon types with different plasmid sizes. In particular, we observed that plasmids carrying blaCTX-M-27 from epidemiologically unrelated strains (from different geographic regions and sources) had surprisingly similar properties (replicon types and sizes) indicating the presence of epidemic blaCTX-M-27 plasmids in China. It’s worth noting that the replicon types were untypeable using the current PBRT scheme in half (14/28) of the transconjugants/transformants carrying blaCTX-M-27. Previous studies on blaCTX-M-27 also found some plasmids could not be determined by PBRT (Ma et al., 2012). Therefore, there may be a new type of plasmid-carrying blaCTX-M-27. We have sequenced two blaCTX-M-27 plasmids from different sources to further study their characteristics (unpublished data).
Among the five detected plasmid replicons in present study, the IncP type plasmid carrying blaCTX-M in Salmonella isolates was reported for the first time. In general, most blaCTX-M genes can be horizontally transferred. Untypeable plasmids and IncN/IncFIB/IncP/IncA/C/IncHI2 plasmids can drive such increased dissemination in Salmonella isolates from food animals. To the best of our knowledge, the current study is also the first report about the spread of non-typeable or IncN/IncFIB/IncP/IncA/C/IncHI2 plasmids carrying blaCTX-M genes in S. enterica serovars Typhimurium and Indiana isolated from pigs and chickens in China.
PFGE is a gold-standard for Salmonella subtyping and has been widely used to determine the relatedness of pathogens to confirm the outbreaks and trace the source of the isolates (White et al., 2007). Fifteen different PFGE clusters (85% genetic similarity) were obtained in this study and these represented a wide variety of genotypes. However, isolates recovered from the same or even different provinces in each identified serovars were highly genetically similar (cluster A, D, G, J and cluster C, M). These may have derived from a specific clone, so the horizontal transmission of the resistance plasmids along with clonal spread of resistant strains were both responsible for dissemination of blaCTX-M-9G-positive strains.
All of the 45 blaCTX-M-9G-positive Salmonella strains were isolated from food animals from different provinces. Therefore, humans can be infected through the consumption of food of animal origin (Thorns, 2000), accelerating the transmission of such Salmonella strains. Along with this, food animals trading may be an important source of Salmonella colonization as has been previously assumed (Wegener et al., 2003). Though studies proving definitive evidence of ESC-R bacterial transmission from food animals to humans are limited; differences can always be shown when comparing Salmonella isolates from humans and the animals. Importantly, previous studies have shown that the same IncN plasmids harboring blaCTX-M are simultaneously present in isolates from pigs and farm workers (Moodley and Guardabassi, 2009). Therefore, further study monitoring and comparing antimicrobial susceptibility, serotypes and ESBL-producing Salmonella isolates from both veterinary settings (pets, wild animals, and livestock) and humans (communities and hospitals) in the same geographic region could lead to the discovery of dissemination routes.
Conclusion
Multi-drug resistant S. enterica serotype Typhimurium and Indiana and especially CTX-M-producing isolates were predominant among food-producing animals in China. Untypeable and IncN/IncFIB/IncP/IncA/C/IncHI2 resistant plasmids harboring blaCTX-M genes and clonal spread of strains were both responsible for dissemination of resistant Salmonella isolates. The presence and spread of blaCTX-M, especially the blaCTX-M-27 on non-typeable plasmids in Salmonella isolates from food-producing animals may pose a potential threat for public health. Proper molecular approaches and hygienic practices are urgently needed to control the dissemination of resistant Salmonella strains through food chain. As ceftiofur and cefquinome have been widely used as therapeutic antibiotic in veterinary medicine, the third- and fourth-generation cephalosporins should be used more prudently in animal husbandry in China.
Author Contributions
Conceived and designed the experiments: Z-LZ, H-XJ, and Y-HL. Performed the experiments: W-HZ, X-YL, LX, and X-XG. Analyzed the data: W-HZ and X-YL. Contributed reagents/materials/analysis tools: LY, WL, and S-QR. Wrote the paper: W-HZ and H-XJ.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported in-part by the National Natural Science Foundation of China (31272602) and 973 Program (2013CB127203).
Footnotes
References
- Alcaine S. D., Soyer Y., Warnick L. D., Su W. L., Sukhnanand S., Richards J., et al. (2006). Multilocus sequence typing supports the hypothesis that cow- and human-associated Salmonella isolates represent distinct and overlapping populations. Appl. Environ. Microbiol. 72 7575–7585. 10.1128/AEM.01174-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlet G., Barrett T. J., Butaye P., Cloeckaert A., Mulvey M. R., White D. G. (2006). Salmonella resistant to extended-spectrum cephalosporins: prevalence and epidemiology. Microbes Infect. 8 1945–1954. 10.1016/j.micinf.2005.12.029 [DOI] [PubMed] [Google Scholar]
- Barton B. M., Harding G. P., Zuccarelli A. J. (1995). A general method for detecting and sizing large plasmids. Anal. Biochem. 226 235–240. 10.1006/abio.1995.1220 [DOI] [PubMed] [Google Scholar]
- Bonnet R., Recule C., Baraduc R., Chanal C., Sirot D., De Champs C., et al. (2003). Effect of D240G substitution in a novel ESBL CTX-M-27. J. Antimicrob. Chemother. 52 29–35. 10.1093/jac/dkg256 [DOI] [PubMed] [Google Scholar]
- Bouallegue-Godet O., Ben S. Y., Fabre L., Demartin M., Grimont P. A., Mzoughi R., et al. (2005). Nosocomial outbreak caused by Salmonella enterica serotype Livingstone producing CTX-M-27 extended-spectrum beta-lactamase in a neonatal unit in Sousse, Tunisia. J. Clin. Microbiol. 43 1037–1044. 10.1128/JCM.43.3.1037-1044.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell C. W., Eckman M. R. (1975). Acute acalculous cholecystitis caused by Salmonella Indiana. JAMA 233 815 10.1001/jama.1975.03260070073030 [DOI] [PubMed] [Google Scholar]
- Carattoli A., Bertini A., Villa L., Falbo V., Hopkins K. L., Threlfall E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63 219–228. 10.1016/j.mimet.2005.03.018 [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (2013). Trends in Foodborne Illness in the United States 2012. Available at: http://www.cdc.gov/Features/dsfoodnet2012/reportcard.html [Google Scholar]
- Chen X., Zhang W., Pan W., Yin J., Pan Z., Gao S., et al. (2012). Prevalence of qnr, aac(6’)-Ib-cr, qepA, and oqxAB in Escherichia coli isolates from humans, animals, and the environment. Antimicrob. Agents Chemother. 56 3423–3427. 10.1128/AAC.06191-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute [CLSI] (2013). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals; Approved Standard 4th Edn and supplement Documents VET 01-A4 and VET01-S2 (Wayne, PA: Clinical, and Laboratory Standards Institute; ). [Google Scholar]
- Clinical and Laboratory Standards Institute [CLSI] (2015). Performance Standards for Antimicrobial SusceptibilityTesting Twenty-fifth Informational Supplement. CLSI document M100-S25 Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- D’Andrea M. M., Arena F., Pallecchi L., Rossolini G. M. (2013). CTX-M-type beta-lactamases: a successful story of antibiotic resistance. Int. J. Med. Microbiol. 303 305–317. 10.1016/j.ijmm.2013.02.008 [DOI] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control (2013). Annual epidemiological report Reporting on 2011 Surveillance Data and 2012 Epidemic Intelligence Data. Stockholm: European Centre for Disease Prevention and Control; 103–108. Available at: http://ecdc.europa.eu/en/publications/_layouts/forms/Publication_DispForm.aspx?List=4f55ad51-4aed-4d32-b960-af70113dbb90&ID=989 [Google Scholar]
- Foley S. L., Lynne A. M. (2008). Food animal-associated Salmonella challenges: pathogenicity and antimicrobial resistance. J. Anim. Sci. 86 E173–E187. 10.2527/jas.2007-0447 [DOI] [PubMed] [Google Scholar]
- Harrois D., Breurec S., Seck A., Delaune A., Le Hello S., Pardos D. L. G. M., et al. (2014). Prevalence and characterization of extended-spectrum beta-lactamase-producing clinical Salmonella enterica isolates in Dakar. Senegal, from 1999 to 2009. Clin. Microbiol. Infect. 20 O109–O116. 10.1111/1469-0691.12339 [DOI] [PubMed] [Google Scholar]
- Hendriksen R. S., Mikoleit M., Kornschober C., Rickert R. L., Duyne S. V., Kjelso C., et al. (2009). Emergence of multidrug-resistant salmonella concord infections in Europe and the United States in children adopted from Ethiopia, 2003–007. Pediatr. Infect. Dis. J. 28 814–818. 10.1097/INF.0b013e3181a3aeac [DOI] [PubMed] [Google Scholar]
- Ho P. L., Chow K. H., Lai E. L., Lo W. U., Yeung M. K., Chan J., et al. (2011). Extensive dissemination of CTX-M-producing Escherichia coli with multidrug resistance to ’critically important’ antibiotics among food animals in Hong Kong, 2008–2010. J. Antimicrob. Chemother. 66 765–768. 10.1093/jac/dkq539 [DOI] [PubMed] [Google Scholar]
- Jiang H. X., Song L., Liu J., Zhang X. H., Ren Y. N., Zhang W. H., et al. (2014). Multiple transmissible genes encoding fluoroquinolone and third-generation cephalosporin resistance co-located in non-typhoidal Salmonella isolated from food-producing animals in China. Int. J. Antimicrob. Agents 43 242–247. 10.1016/j.ijantimicag.2013.12.005 [DOI] [PubMed] [Google Scholar]
- Jiang H. X., Tang D., Liu Y. H., Zhang X. H., Zeng Z. L., Xu L., et al. (2012). Prevalence and characteristics of beta-lactamase and plasmid-mediated quinolone resistance genes in Escherichia coli isolated from farmed fish in China. J. Antimicrob. Chemother. 67 2350–2353. 10.1093/jac/dks250 [DOI] [PubMed] [Google Scholar]
- Kuang X., Hao H., Dai M., Wang Y., Ahmad I., Liu Z., et al. (2015). Serotypes and antimicrobial susceptibility of Salmonella spp. isolated from farm animals in China. Front. Microbiol. 6:602 10.3389/fmicb.2015.00602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavilla S., Gonzalez-Lopez J. J., Sabate M., Garcia-Fernandez A., Larrosa M. N., Bartolome R. M., et al. (2008). Prevalence of qnr genes among extended-spectrum beta-lactamase-producing enterobacterial isolates in Barcelona. Spain. J. Antimicrob. Chemother. 61 291–295. 10.1093/jac/dkm448 [DOI] [PubMed] [Google Scholar]
- Li L., Liao X., Yang Y., Sun J., Li L., Liu B., et al. (2013). Spread of oqxAB in Salmonella enterica serotype Typhimurium predominantly by IncHI2 plasmids. J. Antimicrob. Chemother. 68 2263–2268. 10.1093/jac/dkt209 [DOI] [PubMed] [Google Scholar]
- Li L., Liao X. P., Liu Z. Z., Huang T., Li X., Sun J., et al. (2014). Co-spread of oqxAB and blaCTX-M-9G in non-Typhi Salmonella enterica isolates mediated by ST2-IncHI2 plasmids. Int. J. Antimicrob. Agents 44 263–268. 10.1016/j.ijantimicag.2014.05.014 [DOI] [PubMed] [Google Scholar]
- Li R., Lai J., Wang Y., Liu S., Li Y., Liu K., et al. (2013). Prevalence and characterization of Salmonella species isolated from pigs, ducks and chickens in Sichuan Province. China. Int. J. Food Microbiol. 163 14–18. 10.1016/j.ijfoodmicro.2013.01.020 [DOI] [PubMed] [Google Scholar]
- Liebana E., Carattoli A., Coque T. M., Hasman H., Magiorakos A. P., Mevius D., et al. (2013). Public health risks of enterobacterial isolates producing extended-spectrum beta-lactamases or AmpC beta-lactamases in food and food-producing animals: an EU perspective of epidemiology, analytical methods, risk factors, and control options. Clin. Infect. Dis. 56 1030–1037. 10.1093/cid/cis1043 [DOI] [PubMed] [Google Scholar]
- Liu B. T., Yang Q. E., Li L., Sun J., Liao X. P., Fang L. X., et al. (2013). Dissemination and characterization of plasmids carrying oqxAB-bla CTX-M genes in Escherichia coli isolates from food-producing animals. PLoS ONE 8:e73947 10.1371/journal.pone.0073947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. H., Wei S. Y., Ma J. Y., Zeng Z. L., Lu D. H., Yang G. X., et al. (2007). Detection and characterisation of CTX-M and CMY-2 beta-lactamases among Escherichia coli isolates from farm animals in Guangdong Province of China. Int. J. Antimicrob. Agents 29 576–581. 10.1016/j.ijantimicag.2006.12.015 [DOI] [PubMed] [Google Scholar]
- Lu S. Y., Zhang Y. L., Geng S. N., Li T. Y., Ye Z. M., Zhang D. S., et al. (2010). High diversity of extended-spectrum beta-lactamase-producing bacteria in an urban river sediment habitat. Appl. Environ. Microbiol. 76 5972–5976. 10.1128/AEM.00711-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Zhao H., Sun J., Liu Y., Zhou X., Beier R. C., et al. (2014). Characterization of multidrug-resistant Salmonella enterica serovars Indiana and Enteritidis from chickens in Eastern China. PLoS ONE 9:e96050 10.1371/journal.pone.0096050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Liu J. H., Lv L., Zong Z., Sun Y., Zheng H., et al. (2012). Characterization of extended-spectrum beta-lactamase genes found among Escherichia coli isolates from duck and environmental samples obtained on a duck farm. Appl. Environ. Microbiol. 78 3668–3673. 10.1128/AEM.07507-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura Y., Yamamoto M., Higuchi T., Komori T., Tsuboi F., Hayashi A., et al. (2012). Prevalence of plasmid-mediated AmpC beta-lactamase-producing Escherichia coli and spread of the ST131 clone among extended-spectrum beta-lactamase-producing E. coli in Japan. Int. J. Antimicrob. Agents 40 158–162. 10.1016/j.ijantimicag.2012.04.013 [DOI] [PubMed] [Google Scholar]
- Meunier D., Boyd D., Mulvey M. R., Baucheron S., Mammina C., Nastasi A., et al. (2002). Salmonella enterica serotype Typhimurium DT 104 antibiotic resistance genomic island I in serotype paratyphi B. Emerg. Infect. Dis. 8 430–433. 10.3201/eid0804.010375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micenkova L., Siskova P., Bosak J., Jamborova I., Cernohorska L., Smajs D. (2014). Characterization of human uropathogenic ESBL-producing Escherichia coli in the Czech Republic: spread of CTX-M-27-producing strains in a university hospital. Microb. Drug Resist. 20 610–617. 10.1089/mdr.2014.0013 [DOI] [PubMed] [Google Scholar]
- Moodley A., Guardabassi L. (2009). Transmission of IncN plasmids carrying blaCTX-M-1 between commensal Escherichia coli in pigs and farm workers. Antimicrob. Agents Chemother. 53 1709–1711. 10.1128/AAC.01014-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Regan E., Quinn T., Pages J. M., McCusker M., Piddock L., Fanning S. (2009). Multiple regulatory pathways associated with high-level ciprofloxacin and multidrug resistance in Salmonella enterica serovar enteritidis: involvement of RamA and other global regulators. Antimicrob. Agents Chemother. 53 1080–1087. 10.1128/AAC.01005-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J. J., Carter H. J. (1967). An outbreak of gastroenteritis caused by Salmonella indiana. Public Health Rep. 82 551–554. 10.2307/4593068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahn K., De Grandis S. A., Clarke R. C., McEwen S. A., Galan J. E., Ginocchio C., et al. (1992). Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol. Cell. Probes 6 271–279. 10.1016/0890-8508(92)90002-F [DOI] [PubMed] [Google Scholar]
- Rao L., Lv L., Zeng Z., Chen S., He D., Chen X., et al. (2014). Increasing prevalence of extended-spectrum cephalosporin-resistant Escherichia coli in food animals and the diversity of CTX-M genotypes during 2003-2012. Vet. Microbiol. 172 534–541. 10.1016/j.vetmic.2014.06.013 [DOI] [PubMed] [Google Scholar]
- Stevens M. P., Humphrey T. J., Maskell D. J. (2009). Molecular insights into farm animal and zoonotic Salmonella infections. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364 2709–2723. 10.1098/rstb.2009.0094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahilevitz J., Jacoby G. A., Hooper D. C., Robicsek A. (2009). Plasmid-mediated quinolone resistance: a multifaceted threat. Clin. Microbiol. Rev. 22 664–689. 10.1128/CMR.00016-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Ke B., Huang Y., He D., Li X., Liang Z., et al. (2014). The molecular epidemiological characteristics and genetic diversity of Salmonella typhimurium in Guangdong, China, 2007-2011. PLoS ONE 9:e113145 10.1371/journal.pone.0113145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorns C. J. (2000). Bacterial food-borne zoonoses. Rev. Sci. Tech. 19 226–239. [DOI] [PubMed] [Google Scholar]
- Vo A. T., van Duijkeren E., Fluit A. C., Heck M. E., Verbruggen A., Maas H. M., et al. (2006). Distribution of Salmonella enterica serovars from humans, livestock and meat in Vietnam and the dominance of Salmonella Typhimurium phage type 90. Vet. Microbiol. 113 153–158. 10.1016/j.vetmic.2005.10.034 [DOI] [PubMed] [Google Scholar]
- Wang S., Duan H., Zhang W., Li J. W. (2007). Analysis of bacterial foodborne disease outbreaks in China between 1994 and 2005. FEMS Immunol. Med. Microbiol. 51 8–13. 10.1111/j.1574-695X.2007.00305.x [DOI] [PubMed] [Google Scholar]
- Wegener H. C., Hald T., Lo F. W. D., Madsen M., Korsgaard H., Bager F., et al. (2003). Salmonella control programs in Denmark. Emerg. Infect. Dis. 9 774–780. 10.3201/eid0907.030024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White P. L., Naugle A. L., Jackson C. R., Fedorka-Cray P. J., Rose B. E., Pritchard K. M., et al. (2007). Salmonella Enteritidis in meat, poultry, and pasteurized egg products regulated by the U.S. Food Safety and Inspection Service, 1998 through 2003. J. Food Prot. 70 582–591. [DOI] [PubMed] [Google Scholar]
- Wong M. H., Chan E. W., Liu L. Z., Chen S. (2014a). PMQR genes oqxAB and aac(6’)Ib-cr accelerate the development of fluoroquinolone resistance in Salmonella typhimurium. Front. Microbiol. 5:521 10.3389/fmicb.2014.00521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M. H., Yan M., Chan E. W., Biao K., Chen S. (2014b). Emergence of clinical Salmonella enterica serovar Typhimurium isolates with concurrent resistance to ciprofloxacin, ceftriaxone, and azithromycin. Antimicrob. Agents Chemother. 58 3752–3756. 10.1128/AAC.02770-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Hendriksen R. S., Xie Z., Huang L., Zhang J., Guo W., et al. (2009). Molecular characterization and antimicrobial susceptibility of Salmonella isolates from infections in humans in Henan Province, China. J. Clin. Microbiol 47 401–409. 10.1128/JCM.01099-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Li L., Alam M. J., Shinoda S., Miyoshi S., Shi L. (2010). Prevalence and antimicrobial resistance of Salmonella in retail foods in northern China. Int. J. Food Microbiol. 143 230–234. 10.1016/j.ijfoodmicro.2010.07.034 [DOI] [PubMed] [Google Scholar]
- Yang B., Qu D., Zhang X., Shen J., Cui S., Shi Y., et al. (2010). Prevalence and characterization of Salmonella serovars in retail meats of marketplace in Shaanxi. China. Int. J. Food Microbiol. 141 63–72. 10.1016/j.ijfoodmicro.2010.04.015 [DOI] [PubMed] [Google Scholar]
- Yang L., Yang L., Lu D. H., Zhang W. H., Ren S. Q., Liu Y. H., et al. (2015). Co-prevalance of PMQR and 16S rRNA methylase genes in clinical Escherichia coli isolates with high diversity of CTX-M from diseased farmed pigeons. Vet. Microbiol. 178 238–245. 10.1016/j.vetmic.2015.05.009 [DOI] [PubMed] [Google Scholar]
- Zhang J., Jin H., Hu J., Yuan Z., Shi W., Ran L., et al. (2014). Serovars and antimicrobial resistance of non-typhoidal Salmonella from human patients in Shanghai, China, 2006-2010. Epidemiol. Infect. 142 826–832. 10.1017/S0950268813001659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Zeng Z., Chen S., Liu Y., Yao Q., Deng Y., et al. (2012). Prevalence and characterisation of CTX-M beta-lactamases amongst Escherichia coli isolates from healthy food animals in China. Int. J. Antimicrob. Agents 39 305–310. 10.1016/j.ijantimicag.2011.12.001 [DOI] [PubMed] [Google Scholar]
- Zurfluh K., Hachler H., Nuesch-Inderbinen M., Stephan R. (2013). Characteristics of extended-spectrum beta-lactamase- and carbapenemase-producing Enterobacteriaceae Isolates from rivers and lakes in Switzerland. Appl. Environ. Microbiol. 79 3021–3026. 10.1128/AEM.00054-13 [DOI] [PMC free article] [PubMed] [Google Scholar]


