Abstract
Hydrogen sulfide (H2S) impairs mitochondrial respiration by potently inhibiting the heme-copper cytochrome c oxidase. Since many prokaryotes, including Escherichia (E.) coli, generate H2S and encounter high H2S levels particularly in the human gut, herein we tested whether bacteria can sustain sulfide-resistant O2-dependent respiration. E. coli has three respiratory oxidases, the cyanide-sensitive heme-copper bo3 enzyme and two bd oxidases much less sensitive to cyanide. Working on the isolated enzymes, we found that, whereas the bo3 oxidase is inhibited by sulfide with half-maximal inhibitory concentration IC50 = 1.1 ± 0.1 μM, under identical experimental conditions both bd oxidases are insensitive to sulfide up to 58 μM. In E. coli respiratory mutants, both O2-consumption and aerobic growth proved to be severely impaired by sulfide when respiration was sustained by the bo3 oxidase alone, but unaffected by ≤200 μM sulfide when either bd enzyme acted as the only terminal oxidase. Accordingly, wild-type E. coli showed sulfide-insensitive respiration and growth under conditions favouring the expression of bd oxidases. In all tested conditions, cyanide mimicked the functional effect of sulfide on bacterial respiration. We conclude that bd oxidases promote sulfide-resistant O2-consumption and growth in E. coli and possibly other bacteria. The impact of this discovery is discussed.
Along with nitric oxide (NO) and carbon monoxide (CO), hydrogen sulfide (H2S) has been recognized as an important gaseous signalling molecule, playing a major role in human (patho)physiology1. Like NO and CO, H2S is a key regulator of many physiological processes in the cardiovascular, nervous, respiratory and gastrointestinal systems, among others. While exerting beneficial physiological effects at lower levels, at higher concentrations H2S can cause detrimental effects. In eukaryotes, depending on its concentration, H2S can have opposite effects on respiration (reviewed in2): at nanomolar concentrations it can sustain energy metabolism both as a substrate for the mitochondrial respiratory chain and as a vasodilator favouring O2 supply, whereas at higher levels it impairs cellular respiration via direct binding to and inhibition of mitochondrial cytochrome c oxidase (mtCcOX) (see3 and references therein). Sulfide inhibition of mtCcOX is very effective (Ki = 0.2–0.45 μM at pH = 7.43,4), leading to dissipation of the mitochondrial membrane potential, consequent arrest of aerobic ATP production and eventually cell death2.
In mammalian tissues, H2S is enzymatically produced by cystathionine β-synthase (CBS), cystathionine γ-lyase (CSE) and via the combined action of 3-mercaptopyruvate sulfurtransferase (3-MST) and cysteine aminotransferase (reviewed in5). At variance from other compartments in the human body, in the intestinal lumen H2S is also generated by the gut microbiota through bacterial amino acid metabolism and via dissimilatory sulfate reduction by ‘sulfate-reducing bacteria’ (SRB)6. H2S levels in the gut are therefore high. Whereas the total sulfide pool content in the colon is around one millimolar7, the concentration of free H2S in the intestinal lumen was reported to be ca. 40–60 μM, as estimated by direct measurement of the gas in the rat cecum8,9 and analysis of human faecal samples10.
E. coli is a ubiquitous member of the human gut microbiota, with more than one strain commonly colonizing the large intestine at the same time. Since E. coli, like the other microorganisms inhabiting the gut, lives in a particularly H2S-enriched microaerobic niche, the question arises as to whether this microorganism can accomplish O2-dependent respiration without being inhibited by H2S. The E. coli respiratory chain possesses three terminal oxygen reductases, utilizing quinols as reducing substrates: the cyanide-sensitive cytochrome bo3 enzyme and the bd-I and bd-II oxidases, much less sensitive to cyanide11,12. Cytochrome bo3 belongs to the superfamily of heme–copper oxygen reductases that includes mtCcOX. The enzyme contains three redox-active metal centres: the low-spin heme b involved in quinol oxidation and a binuclear site composed of heme o3 and CuB, where O2 reduction to water takes place. On the contrary, bd-I and bd-II are cytochrome bd-type O2-reductases phylogenetically unrelated to heme–copper oxidases12. They have no copper, but contain three hemes: the low-spin heme b558 (the primary electron acceptor from the quinol), and the two high-spin hemes b595 and d (possibly forming a di-heme site for O2 reduction, see12 and references therein). Cytochrome bo3 predominates in E. coli under high aeration, whereas O2-limiting conditions such as those found in the human gut stimulate the expression of the cytochromes bd-I and bd-II13,14,15. The three E. coli terminal oxidases all generate a proton motive force, but cytochrome bo3 is the only one able to pump protons, thus being twice as effective as bd-type cytochromes in terms of energy transduction16. Besides its role in bacterial energy metabolism, cytochrome bd-I was suggested to serve other physiological functions, being implicated in the bacterial response to oxidative and nitrosative stress17,18,19,20.
In this work, we examined the effect of sulfide on the O2 reductase activity of the three terminal oxidases of E. coli and tested the ability of these enzymes to sustain bacterial growth and O2 consumption in the presence of sulfide.
Results
Effect of NaHS on isolated E. coli terminal oxidases
The effect of sulfide on the O2 reductase activity of the E. coli respiratory oxidases, cytochromes bo3, bd-I and bd-II, was initially investigated testing the ability of each purified oxidase to consume O2 before and after addition of the sulfide donor NaHS. In these assays, O2 consumption was measured in the presence of dithiothreitol (DTT) and 2,3-dimethoxy-5-methyl-6-(3-methyl-2-butenyl)-1,4-benzoquinone (Q1) as the reducing system. As shown in Fig. 1A, NaHS (~7 μM) rapidly and effectively inhibits the O2 reductase activity of the isolated cytochrome bo3. The enzyme is inhibited with an apparent half-maximal inhibitory concentration IC50 = 1.1 ± 0.1 μM (Fig. 2). The inhibition of cytochrome bo3 is fully reversible. A rapid and complete recovery of the O2 reductase activity of the isolated enzyme was observed, when sulfide was quickly removed from solution by addition of an excess of O-acetyl-L-serine (OAS) and catalytic amounts of the sulfide-consuming O-acetylserine sulfhydrylase enzyme from Entamoeba histolytica (EhOASS, Fig. 1A). Sulfide consumption by EhOASS in the presence of OAS was assessed independently using a H2S-selective electrode (Figure S1). Notably, while being an effective inhibitor of E. coli cytochrome bo3, NaHS proved to be unable to inhibit the two E. coli bd-type oxidases. Addition of NaHS, even at high concentration (58 μM), did not alter the O2 consumption catalyzed by the bd-I or bd-II enzyme in the presence of DTT and Q1 (Fig. 1A). No O2 consumption stimulation by the OAS/EhOASS sulfide-scavenging system was observed in control oxygraphic experiments carried out in the absence or presence of the isolated oxidases (not shown).
Figure 1. Effect of NaHS on E. coli terminal oxidases.
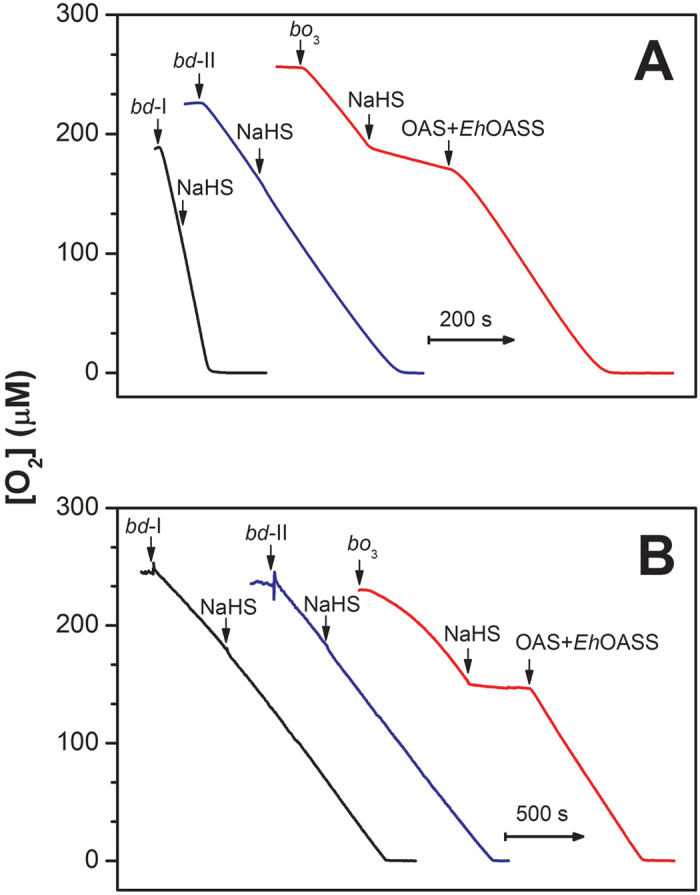
(A) O2 reductase activity of the isolated cytochromes bd-I (20 nM), bd-II (2.5 nM) and bo3 (6 nM) as measured at 25 °C in the presence of DTT (10 mM) and Q1 (0.25 mM). O2 consumption rates measured prior to NaHS addition (mean ± standard deviation, n = 3): 1.62 ± 0.07 μM O2/s (bd-I); 0.54 ± 0.02 μM O2/s (bd-II) and 0.46 ± 0.02 μM O2/s (bo3). O2 consumption by cytochrome bo3 is rapidly inhibited (~85%) by 7.2 μM NaHS, to be quickly and completely restored upon removal of sulfide from solution following the addition of 200 μM OAS and 216 nM EhOASS. On the contrary, NaHS (58 μM) does not affect the oxidase activity of either cytochrome bd-I or cytochrome bd-II. (B) O2 consumption by cell suspensions of the mutant strains expressing cytochrome bd-I (400 μl cells with OD600 = 1.85), cytochrome bd-II (600 μl cells with OD600 = 1.17) or cytochrome bo3 (200 μl cells with OD600 = 2.45), as the only terminal oxidase. O2 consumption rates measured prior to NaHS addition (mean ± standard deviation, n = 3): 0.20 ± 0.07 μM O2/s (bd-I); 0.18 ± 0.02 μM O2/s (bd-II) and 0.19 ± 0.02 μM O2/s (bo3). When sustained solely by cytochrome bo3, cell respiration is rapidly inhibited by 50 μM NaHS, to be quickly and completely restored following sulfide removal on addition of OAS (200 μM) and EhOASS (216 nM). No inhibition is observed following the addition of NaHS (50 μM), when respiration is sustained by the only cytochrome bd-I or bd-II.
Figure 2. NaHS inhibition of isolated cytochrome bo3.
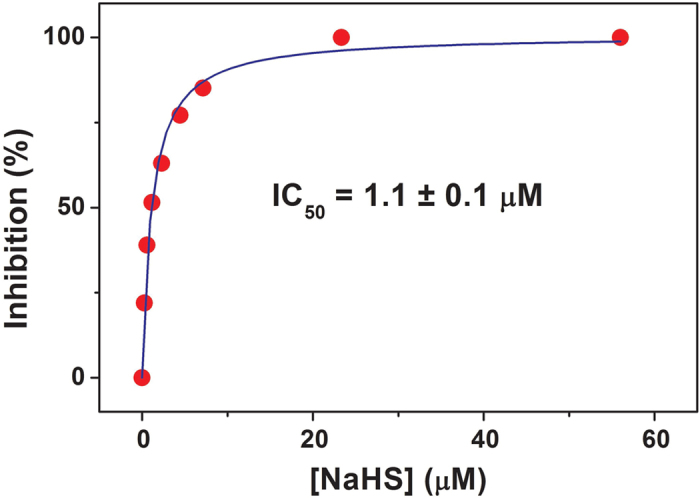
Percentage inhibition of the O2 reductase activity of isolated cytochrome bo3 (6 nM) measured at increasing concentration of NaHS, in the presence of the 10 mM DTT and 0.25 mM Q1.
Effect of NaHS on E. coli respiration
The striking results obtained with the isolated enzymes prompted us to explore the effect of sulfide on E. coli cell respiration. To this end, we investigated aerobic cultures of E. coli (see Methods for details) and tested the effect of NaHS on cell respiration along cell growth, i.e., at increasing cell density. We initially assayed three mutant strains each expressing a single terminal oxidase (bo3, bd-I or bd-II). The results were remarkably similar to those obtained with the isolated enzymes. O2 consumption by E. coli cells expressing solely cytochrome bo3 was quickly and fully inhibited upon addition of 50 μM NaHS (Fig. 1B). As observed with the isolated bo3 enzyme, the inhibition was promptly and fully restored upon sulfide depletion by the EhOASS/OAS system (Fig. 1B). In contrast, no inhibition was observed following the addition of 50 μM NaHS to E. coli cells expressing either bd-I or bd-II as the only terminal oxidase (Fig. 1B). The results on the three mutant strains proved to be independent of the density at which cells were collected and assayed (Fig. 3, top panel). Similarly to NaHS, cyanide (50 μM) almost completely abolished O2-consumption in E. coli cells expressing only the bo3 oxidase, whereas it was essentially ineffective when respiration was sustained by either bd oxidase (Fig. 3, bottom panel).
Figure 3. Effect of NaHS and cyanide on respiration of E. coli cells.
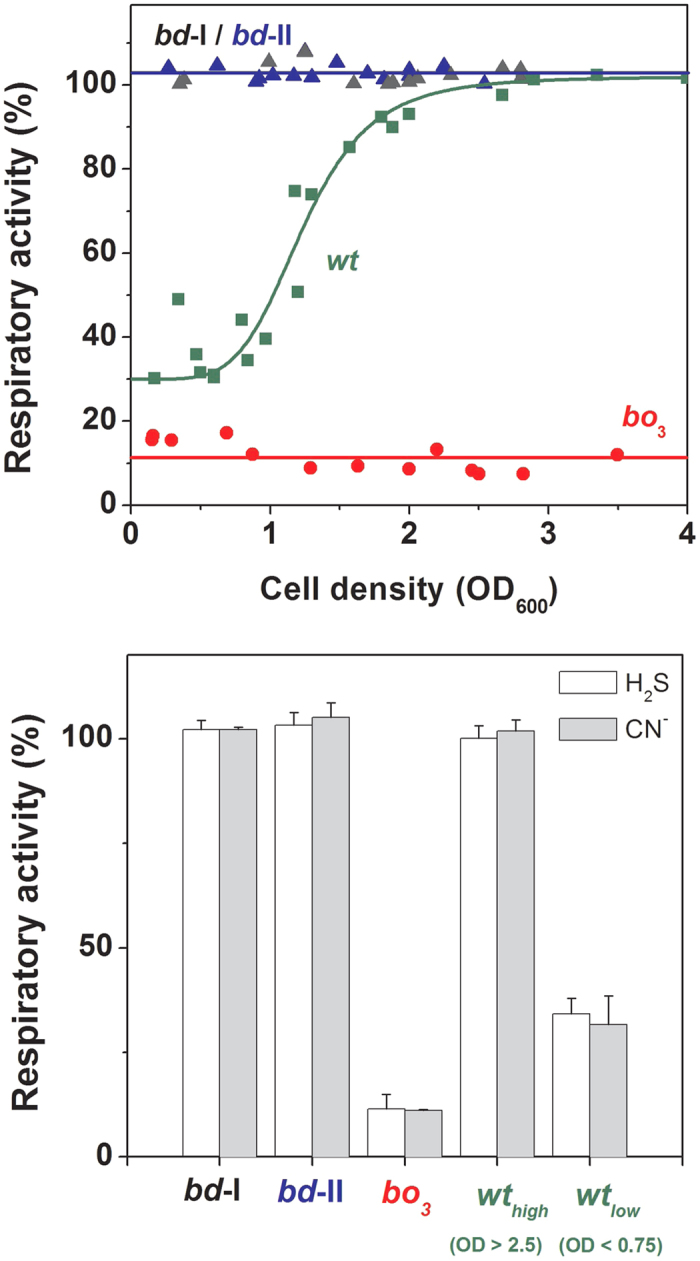
(Top) Residual respiratory activity measured after the addition of 50 μM NaHS to E. coli cells collected at the reported cell density. (Bottom) Comparison of the effect of cyanide and sulfide on cell respiration: respiratory activity measured after the addition of 50 μM NaHS or 50 μM NaCN to wild-type and mutant E. coli cells. Data (mean ± standard deviation) refer to the control activity measured before the addition of inhibitors (taken as 100%).
The effect of NaHS on respiration of the wild-type strain was assessed in the same way. Namely, we investigated aerobic cultures in which a change in oxidase expression from cytochrome bo3 to the cytochromes of the bd-type is expected to take place along cell growth, following a progressive reduction in O2 availability in the medium21,22. Accordingly, when cells were assayed in an early phase of the culture (OD600 < 0.7), most of respiration (65–70%) proved to be sensitive to NaHS or cyanide (both at 50 μM, Fig. 3). In contrast, with cell growth bacterial O2-consumption became progressively less sensitive to sulfide inhibition and, in a late phase of the culture (OD600 > 2.5), NaHS or cyanide caused only marginal effects on respiration (Fig. 3).
Altogether these results show that, unlike the heme-copper bo3 oxidase, E. coli bd oxidases enable O2-dependent respiration in the presence of sulfide.
Effect of NaHS on E. coli cell growth
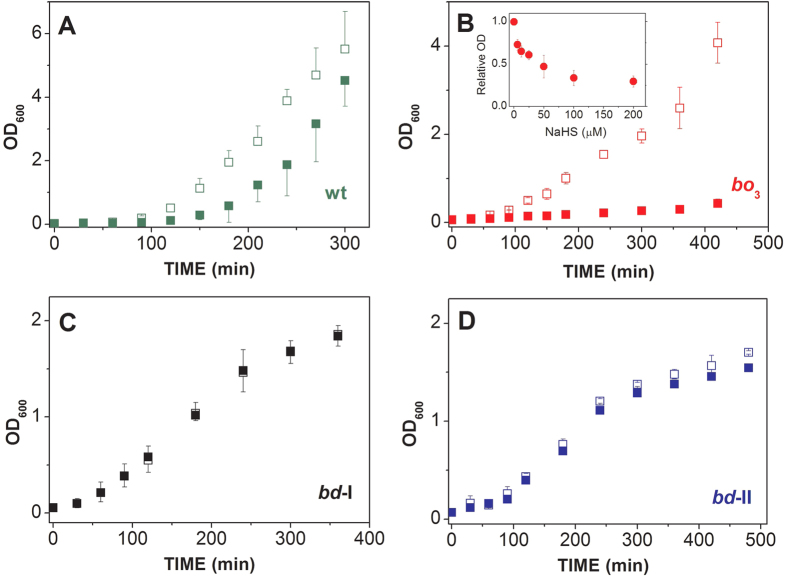
The lack of sulfide inhibition of cytochromes bd-I and bd-II, as opposed to the high sensitivity displayed by the bo3 oxidase, prompted us to test whether the bd-type oxidases, besides enabling respiration, promote E. coli cell growth in the presence of sulfide. We investigated the effect of sulfide on the growth of both the wild-type and the three respiratory mutant strains. Following the addition of 200 μM NaHS, the wild-type strain showed a delayed growth (Fig. 4A), while the growth of the bo3-expressing strain was severely impaired (Fig. 4B). Lacking bd oxidases, the latter strain proved to be highly sensitive to sulfide, with 6 μM NaHS causing ~25% reduced cell growth, as evaluated at 2 hours after NaHS addition (inset Fig. 4B). In contrast, no or very little effect on cell growth was observed over the same time window after addition of 200 μM NaHS to the strains expressing either bd-I or bd-II as the only terminal oxidase (Fig. 4C,D). Altogether, these data show that, unlike the bo3 oxidase, the cytochromes bd-I and bd-II sustain E. coli growth in the presence of sulfide.
Figure 4. Effect of NaHS on E. coli cell growth.
Cell growth of E. coli wild-type (A) and mutant strains with bo3 (B), bd-I (C) or bd-II (D) as the only terminal oxidase, assayed in the presence (‘closed symbols’) or absence (‘open symbols’) of 200 μM NaHS. Inset to panel B: Effect of NaHS on the growth of the bo3-only expressing mutant, as evaluated at 2 hours after addition of NaHS used at the indicated concentrations. ‘Relative OD’ indicates the ratio between the optical density measured at 600 nm in the presence of NaHS and the one recorded after the same period of time (2 hours) in the absence of NaHS. Data expressed as mean ± standard deviation.
Discussion
Together with NO and CO, H2S is presently considered a highly relevant signalling molecule in human (patho)physiology. It has long been recognized that many prokaryotes, including the model organism E. coli and numerous other members of the human gut microbiota, generate H2S (see6 and references therein). Bacteria can accomplish H2S production by several pathways, including cysteine degradation by L-cysteine desulfhydrase, and dissimilatory sulfate reduction by SRB (see6 and references therein). In a recent study, it was reported that orthologs of the mammalian H2S-synthesizyng enzymes CBS, CSE and 3-MST are widespread in the bacterial world and contribute to H2S generation, as demonstrated for several bacteria by genetic manipulation23. As an example, E. coli was shown to harbour an ortholog of 3-MST significantly contributing to bacterial H2S synthesis. Notably, in the same study H2S production was shown to enhance antibiotic resistance in all tested bacteria, thereby providing an adaptive advantage.
The presence of numerous H2S-producing bacteria in the human gut makes this compartment particularly enriched in H2S compared to other tissues, with the free gas reaching in the intestinal lumen concentrations as high as 40–60 μM8,9,10. Relevant to human (patho)physiology, bacteria-derived H2S is emerging as a key regulator of several physiological functions not only in the gastrointestinal system, but also throughout the human body1. Moreover, it has been recently suggested that the differential susceptibility of mutualistic microbes to sulfide toxicity may contribute to shape the human gut microbiota6, a recognized factor contributing to human health and disease. In turn, the host H2S systemic bioavailability and metabolism have been found to be profoundly affected by the microbiota in studies on germ-free mice24. Altogether these observations provide evidence for interplay between H2S and the human microbiota, with important consequences on human health.
Though currently considered a key signalling molecule, H2S has long been known as a mere poison. Toxicity has been related to the ability of H2S to bind heme proteins and inhibit cellular respiration targeting mtCcOX (see3 and references therein). Indeed, H2S is a potent (Ki = 0.2–0.45 μM3,4), non-competitive inhibitor of this respiratory enzyme, the inhibition being reversible, independent of oxygen concentration25, but dependent on pH26. Sulfide inhibition of isolated mtCcOX in turnover with ascorbate and cytochrome c is relatively fast, occurring at an initial rate constant of 2.2 × 104 M−1 s−1, as measured at pH 7.4 3. The inhibited enzyme exhibits sulfide bound to ferric heme a327,28, with CuB in the cuprous state possibly bound to a second H2S molecule, as revealed by electron paramagnetic resonance (EPR) spectroscopy29. The mechanism of inhibition of mtCcOX is only partly understood, yet the reaction was suggested to involve the binding of H2S to the enzyme in turnover at cupric or cuprous CuB, followed by intramolecular transfer of H2S to ferric heme a3, eventually blocking the reaction with O23.
The well-known toxicity of H2S on mitochondrial respiration prompted us to address whether bacterial O2-dependent respiration can be accomplished in a H2S-enriched environment such as the human gut, thereby providing an adaptive advantage in terms of bacterial growth. This issue was addressed in the present study working on the model organism E. coli, a ubiquitous member of the human gut microbiota. Namely, we investigated the effect of sulfide on the O2 reductase activity of each of the three terminal respiratory oxidases of this bacterium (cytochromes bo3, bd-I and bd-II), and tested the ability of these enzymes to sustain O2 consumption and bacterial cell growth in the presence of sulfide. Using NaHS as a H2S donor, we carried out experiments on the isolated enzymes, as well as on the wild-type and three respiratory mutant E. coli strains each expressing only a single terminal oxidase. NaHS is commonly used as a donor of the cell permeant H2S, because in aqueous solution HS− equilibrates with H2S and S2−, according to the pKa1 ~7.0 (H2S/HS−) and pKa2 ~19 (HS−/S2−) measured at 25 °C. At pH = 7.0–7.4, ~30–50% of HS− is thus expected to be protonated to H2S, with S2− being present in negligible amounts.
As a new finding we report that, whereas the heme-copper bo3 oxidase is highly sensitive to sulfide inhibition (IC50 =1.1 ± 0.1 μM, Figs 1 and 2), the two bd oxidases (bd-I and bd-II) are remarkably insensitive to sulfide (Fig. 1), as confirmed by measuring the effect of NaHS on O2 consumption by the purified terminal oxidases (Fig. 1A) or by whole cells (Figs 1B and 3). In agreement with these finding, cell growth proved to be severely impaired by sulfide in an E. coli mutant strain expressing only the bo3 oxidase (Fig. 4B), but unaffected in mutant strains expressing either bd-I or bd-II as the only terminal oxidase (Fig. 4, panel C,D). Consistently, in the wild-type strain, H2S affected cell growth and respiration only in the early phase of the culture, when O2 availability is expected to be still sufficiently high to favour the expression of the bo3 oxidase, but it caused no effect in a late phase of the culture, when O2 limitation is expected to stimulate the expression of bd oxidases (Fig. 4A).
Altogether, these observations led us to conclude that, at variance with the heme-copper bo3 oxidase that is potently and reversibly inhibited by sulfide, both E. coli bd oxidases are sulfide-insensitive and thus able to sustain cell respiration and growth in the presence of considerably high levels of sulfide. Although the molecular basis for the remarkable sulfide insensitivity of the E. coli bd oxidases remains to be elucidated, it may originate from the lack of CuB, which was indeed suggested to be implicated in sulfide inhibition of mtCcOX3. In this regard, still possibly due to the lack of CuB, it is noteworthy that bd oxidases are not only more resistant to NO inhibition than heme-copper oxidases30,31,32, but also poorly sensitive to other commonly used oxidase inhibitors, such as cyanide12 and azide33. On this basis, cyanide and sulfide are expected to exert similar inhibitory effects on E. coli respiration, as observed in the present study (Fig. 3).
As shown here for E. coli, it is likely that bd oxidases confer sulfide resistance also to other microorganisms. The bd oxidases are indeed widespread in the prokaryotic world and have been identified in numerous enterobacteria34, where expression of these oxidases is likely stimulated in the microaerobic conditions found in the human colon. In view of the novel results presented here, it will be important to test whether bd oxidases, by conferring sulfide resistance, play a role in shaping the human gut microbiota, thereby impacting human (patho)physiology. Furthermore, based on these data, bd oxidases may represent very attractive targets for the development of next-generation antimicrobials against pathogenic enterobacteria18,20,35. Finally, the finding that bd oxidases enhance bacterial resistance to sulfide, if representing a hallmark of this protein family, may pave the way to biotechnological applications aimed at increasing bacterial sulfide resistance.
Methods
Materials, bacterial strains and growth conditions
All chemicals were purchased from Sigma unless otherwise indicated. NaHS stock solutions were prepared by dissolving NaHS in degassed water or phosphate buffer saline, and the overall concentration of sulfide species (H2S/HS−/S2−) in solution was determined spectrophotometrically according to36. All E. coli strains used were K-12 derivatives; MG1655 (RKP5416) was the wild type37 from which the respiratory mutants, TBE025 (MG1655 ΔcydB nuoB appB::kan), TBE026 (MG1655 ΔcydB nuoB cyoB::kan) and TBE037 (MG1655 ΔappB nuoB cyoB::kan) were derived, respectively expressing cytochrome bo3, bd-II and bd-I as the only terminal oxidase (mutants kindly given by Alex Ter Beek and Joost Teixeira de Mattos, University of Amsterdam). These strains carry the same mutant alleles as described by Bekker et al.38. E. coli cells were grown in 50 mL-Falcon tubes, in 5 mL Luria Bertani (LB) medium supplemented with 30 μg/mL kanamycin, at 37 °C and 200 rpm. For growth studies, cells were grown as described above in the absence or presence of NaHS (6–200 μM) added to cells at an OD600 of about 0.05.
Purification of terminal oxidases from E. coli
The cytochromes bd-I, bd-II and bo3 were isolated from the E. coli strains GO105/pTK1, MB37 and GO105/pJRhisA, respectively, as previously described39,40,41. The concentration of the cytochromes bd-I and bd-II was determined from the difference absorption spectrum using Δε628-607 = 10.8 mM−1 cm−1 for the dithionite-reduced minus ‘as prepared’ proteins. Cytochrome bo3 concentration was estimated from the Soret absorption band of the oxidized enzyme using ε407 = 183 mM−1 cm−1. UV-visible absorption spectra were acquired in an Agilent Cary 60 spectrophotometer.
Purification and H2S consumption by recombinant O-acetylserine sulfhydrylase from Entamoeba histolytica
The O-acetylserine sulfhydrylase-encoding gene (EhOASS, Genbank XM_643199.1) was PCR-amplified from Entamoeba histolytica HM-1:IMSS genomic DNA using the forward primer 5′-CATATGATGGAACAAATAAGTATTAGC and the reverse primer 5′-AACGTTTTATTCATTCAATAATGAATCAAG, containing the NdeI and HindIII restriction sites respectively. The PCR product was cloned into the Topo TA pCR2.1 vector, digested with the NdeI and HindIII restriction enzymes, and gel purified. The DNA insert was subcloned into the NdeI and HindIII restriction sites of the pET28b expression vector, yielding the pET-EhOASS construct encoding N-terminally 6xHis-tagged EhOASS. pET-EhOASS was used to transform E. coli BL21 (DE3). Cells were grown at 37 °C in LB broth supplemented with 25 mg/L kanamycin (Nzytech) until OD600 reached 0.4–0.5. EhOASS expression was induced with 0.1 mM isopropyl-β-D-thiogalactoside addition and the cultures moved to 30 °C, 130 rpm for 4 h. Cells were harvested and the pellet resuspended in 10 mL/L culture of buffer A (50 mM potassium phosphate, 300 mM KCl, pH 7.5, 10% glycerol) containing 1 mg/mL lysozyme, 1 mM phenylmethylsulfonyl fluoride and deoxyribonuclease I. After 30-min incubation on ice, cells were disrupted by sonication, centrifuged at 8200 g (5 min, 4 °C) and imidazole was added to the supernatant to a final concentration of 10 mM. Protein purification steps were performed in an Åkta Prime (GE Healthcare) chromatography system. Affinity purification of the His-tagged protein was performed using a HisTrap FF crude 1-mL column previously equilibrated with buffer A containing 10 mM imidazole (buffer B). The cleared supernatant was loaded onto the column at 1 mL/min and the column was washed with 25 column volumes of buffer B followed by a linear gradient of 15 column volumes up to 500 mM imidazole. Pooled protein fractions were loaded onto a PD10 (GE Healthcare) desalting column for imidazole removal, equilibrated and washed with buffer A. EhOASS-containing fractions were concentrated with Amicon Ultra-15 centrifugal filter units (30 kDa cut-off) and loaded onto a size-exclusion 120-ml Superdex S-200 (GE Healthcare) column, equilibrated and eluted with buffer A at 0.7 mL/min. EhOASS fractions were pooled; protein purity was assessed by SDS-PAGE and protein concentration was determined by the Bradford assay. As previously reported42, pure EhOASS eluted as a dimer of ~38 kDa monomers (Figure S1).
H2S consumption by EhOASS was measured at 20 °C in 100 mM HEPES, 260 U/mL catalase, 100 μM EDTA pH 7.0, using an ISO-H2S-2 hydrogen sulfide sensor coupled to an Apollo 4000 Free Radical Analyzer (World Precision Instruments). In these assays the concentration of H2S in solution was obtained from the nominal concentration of the NaHS added, assuming 1:1 partition between HS- and H2S at pH 7.0, according to the pKa of H2S.
O2 consumption measurements
Oxygraphic measurements were carried out at 25 °C in 100 mM Na/phosphate pH 7.4, using a high-resolution respirometer (Oxygraph-2k, Oroboros Instruments) with a 1.5 mL chamber. The buffer was supplemented with 0.1 mM EDTA and either 0.05% N-lauroyl-sarcosine (cytochrome bd-I) or 0.02% dodecyl-β-D-maltoside (cytochrome bd-II and cytochrome bo3) in the assays on isolated oxidases. The apparent IC50 of NaHS for the O2-reductase activity of the isolated bo3 oxidase was obtained by plotting the percentage inhibition of the enzyme as a function of NaHS concentration and fitting the data to the Hill equation43, assuming a Hill coefficient n = 1.
Additional Information
How to cite this article: Forte, E. et al. The Terminal Oxidase Cytochrome bd Promotes Sulfide-resistant Bacterial Respiration and Growth. Sci. Rep. 6, 23788; doi: 10.1038/srep23788 (2016).
Supplementary Material
Acknowledgments
This work was partly supported by: Ministero dell’Istruzione, dell’Università e della Ricerca of Italy (PNR-CNR Aging Program 2012–2014 to A.G. and PRIN 20107Z8XBW_005 to P.S.); Regione Lazio of Italy (FILAS-RU-2014 – 1020); Russian Foundation for Basic Research (research projects № 14-04-00153 and 15-04-06266 to V.B.B.); Fundação para a Ciência e Tecnologia (FCT) of Portugal (Grant PTDC/SAU-MIC/111447/2009 to J.B.V.); a bilateral grant award by Consiglio Nazionale delle Ricerche of Italy (CNR) of Italy and FCT of Portugal (to A.G. and J.B.V.). iNOVA4Health - UID/Multi/04462/2013, a program financially supported by FCT/Ministério da Educação e Ciência, through national funds and co-funded by FEDER under the PT2020 Partnership Agreement is acknowledged. V.B.B. was the recipient of a short-term fellowship by CNR of Italy. We thank Prof. R.B. Gennis (Urbana, USA) for the E. coli strain GO105/pTK1, Dr. M. Bekker (Amsterdam, Netherlands) for the E. coli strain MB37 and the E. coli oxidase mutants, Dr. M. Verkhovskaya (Helsinki, Finland) for the purified E. coli bo3 oxidase, and Dr. Upinder Singh (Stanford University, CA, USA) for the Entamoeba histolytica HM-1:IMSS genomic DNA.
Footnotes
Author Contributions E.F., V.B.B., P.S. and A.G. conceived the study and designed the experimental plan. E.F., V.B.B. and M.F. performed and analyzed the experiments with isolated oxidases and E. coli strains. V.B.B. produced the recombinant bd oxidases. H.G.C. and J.B.V. produced the recombinant O-acetylserine sulfhydrylase from Entamoeba histolytica and performed and analyzed the experiments with this enzyme. M.T.T. and R.K.P. contributed to the implementation of the experiments with the E. coli strains and their interpretation. A.G., V.B.B., E.F. and J.B.V. wrote the paper. All authors reviewed the results, contributed to data interpretation and critical revision of the manuscript, and approved the final version of the manuscript.
References
- Wallace J. L. & Wang R. Hydrogen sulfide-based therapeutics: exploiting a unique but ubiquitous gasotransmitter. Nat. Rev. Drug. Discov. 14, 329–345 (2015). [DOI] [PubMed] [Google Scholar]
- Szabo C. et al. Regulation of mitochondrial bioenergetic function by hydrogen sulfide. Part I. Biochemical and physiological mechanisms. Br. J. Pharmacol. 171, 2099–2122 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls P., Marshall D. C., Cooper C. E. & Wilson M. T. Sulfide inhibition of and metabolism by cytochrome c oxidase. Biochem. Soc. Trans. 41, 1312–1316 (2013). [DOI] [PubMed] [Google Scholar]
- Cooper C. E. & Brown G. C. The inhibition of mitochondrial cytochrome oxidase by the gases carbon monoxide, nitric oxide, hydrogen cyanide and hydrogen sulfide: chemical mechanism and physiological significance. J. Bioenerg. Biomembr. 40, 533–539 (2008). [DOI] [PubMed] [Google Scholar]
- Kabil O. & Banerjee R. Enzymology of H2S biogenesis, decay and signaling. Antioxid. Redox Signal. 20, 770–782 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonero F., Benefiel A. C., Alizadeh-Ghamsari A. H. & Gaskins H. R. Microbial pathways in colonic sulfur metabolism and links with health and disease. Front. Physiol. 3, 448 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane G. T., Gibson G. R. & Cummings J. H. Comparison of fermentation reactions in different regions of the human colon. J. Appl. Bacteriol. 72, 57–64 (1992). [DOI] [PubMed] [Google Scholar]
- Suarez F., Furne J., Springfield J. & Levitt M. Production and elimination of sulfur-containing gases in the rat colon. Am. J. Physiol. 274, G727–733 (1998). [DOI] [PubMed] [Google Scholar]
- Levitt M. D., Springfield J., Furne J., Koenig T. & Suarez F. L. Physiology of sulfide in the rat colon: use of bismuth to assess colonic sulfide production. J. Appl. Physiol. 92, 1655–1660 (2002). [DOI] [PubMed] [Google Scholar]
- Jorgensen J. & Mortensen P. B. Hydrogen sulfide and colonic epithelial metabolism: implications for ulcerative colitis. Dig. Dis. Sci. 46, 1722–1732 (2001). [DOI] [PubMed] [Google Scholar]
- Poole R. K. & Cook G. M. Redundancy of aerobic respiratory chains in bacteria? Routes, reasons and regulation. Adv. Microb. Physiol. 43, 165–224 (2000). [DOI] [PubMed] [Google Scholar]
- Borisov V. B., Gennis R. B., Hemp J. & Verkhovsky M. I. The cytochrome bd respiratory oxygen reductases. Biochim. Biophys. Acta 1807, 1398–1413 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfe M. D. et al. Transcript profiling and inference of Escherichia coli K-12 ArcA activity across the range of physiologically relevant oxygen concentrations. J. Biol. Chem. 286, 10147–10154 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ederer M. et al. A mathematical model of metabolism and regulation provides a systems-level view of how Escherichia coli responds to oxygen. Front. Microbiol. 5, 124 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettenbrock K. et al. Towards a systems level understanding of the oxygen response of Escherichia coli. Adv. Microb. Physiol. 64, 65–114 (2014). [DOI] [PubMed] [Google Scholar]
- Puustinen A., Finel M., Haltia T., Gennis R. B. & Wikström M. Properties of the two terminal oxidases of Escherichia coli. Biochemistry 30, 3936–3942 (1991). [DOI] [PubMed] [Google Scholar]
- Lindqvist A., Membrillo-Hernandez J., Poole R. K. & Cook G. M. Roles of respiratory oxidases in protecting Escherichia coli K12 from oxidative stress. Antonie Van Leeuwenhoek 78, 23–31 (2000). [DOI] [PubMed] [Google Scholar]
- Giuffrè A., Borisov V. B., Arese M., Sarti P. & Forte E. Cytochrome bd oxidase and bacterial tolerance to oxidative and nitrosative stress. Biochim. Biophys. Acta 1837, 1178–1187 (2014). [DOI] [PubMed] [Google Scholar]
- Borisov V. B., Forte E., Siletsky S. A., Sarti P. & Giuffrè A. Cytochrome bd from Escherichia coli catalyzes peroxynitrite decomposition. Biochim. Biophys. Acta 1847, 182–188 (2015). [DOI] [PubMed] [Google Scholar]
- Borisov V. B. et al. Cytochrome bd protects bacteria against oxidative and nitrosative stress: a potential target for next-generation antimicrobial agents. Biochemistry-Moscow 80, 565–575 (2015). [DOI] [PubMed] [Google Scholar]
- Alexeeva S., Hellingwerf K. & Teixeira de Mattos M. J. Quantitative assessment of oxygen availability: Perceived aerobiosis and its effect on flux distribution in the respiratory chain of Escherichia coli. J. Bacteriol. 184, 1402–1406 (2002). [DOI] [PMC free article] [PubMed]
- Cotter P. A., Chepuri V., Gennis R. B. & Gunsalus R. P. Cytochrome o (cyoABCDE) and d (cydAB) oxidase gene expression in Escherichia coli is regulated by oxygen, pH, and the fnr gene product. J. Bacteriol. 172, 6333–6338 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatalin K., Shatalina E., Mironov A. & Nudler E. H2S: a universal defense against antibiotics in bacteria. Science 334, 986–990 (2011). [DOI] [PubMed] [Google Scholar]
- Shen X. et al. Microbial regulation of host hydrogen sulfide bioavailability and metabolism. Free Radic. Biol. Med. 60, 195–200 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen L. C. The effect of inhibitors on the oxygen kinetics of cytochrome c oxidase. Biochim. Biophys. Acta 460, 299–307 (1977). [DOI] [PubMed] [Google Scholar]
- Nicholls P. & Kim J. K. Sulphide as an inhibitor and electron donor for the cytochrome c oxidase system. Can. J. Biochem. 60, 613–623 (1982). [DOI] [PubMed] [Google Scholar]
- Nicholls P. The effect of sulphide on cytochrome aa3. Isosteric and allosteric shifts of the reduced a-peak. Biochim. Biophys. Acta 396, 24–35 (1975). [DOI] [PubMed] [Google Scholar]
- Nicholls P., Petersen L. C., Miller M. & Hansen F. B. Ligand-induced spectral changes in cytochrome c oxidase and their possible significance. Biochim. Biophys. Acta 449, 188–196 (1976). [DOI] [PubMed] [Google Scholar]
- Hill B. C. et al. Interactions of sulphide and other ligands with cytochrome c oxidase. An electron-paramagnetic-resonance study. Biochem. J. 224, 591–600 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisov V. B. et al. Redox control of fast ligand dissociation from Escherichia coli cytochrome bd. Biochem. Biophys. Res. Commun. 355, 97–102 (2007). [DOI] [PubMed] [Google Scholar]
- Mason M. G. et al. Cytochrome bd confers nitric oxide resistance to Escherichia coli. Nat. Chem. Biol. 5, 94–96 (2009). [DOI] [PubMed] [Google Scholar]
- Giuffrè A., Borisov V. B., Mastronicola D., Sarti P. & Forte E. Cytochrome bd oxidase and nitric oxide: From reaction mechanisms to bacterial physiology. FEBS Lett. 586, 622–629 (2012). [DOI] [PubMed] [Google Scholar]
- Poole R. K., Williams H. D., Downie J. A. & Gibson F. Mutations affecting the cytochrome d-containing oxidase complex of Escherichia coli K12: Identification and mapping of a fourth locus, cydD. J. Gen. Microbiol. 135, 1865–1874 (1989). [DOI] [PubMed] [Google Scholar]
- Degli Esposti M. et al. Molecular evolution of cytochrome bd oxidases across proteobacterial genomes. Genome Biol. Evol. 7, 801–820 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook G. M., Greening C., Hards K. & Berney M. Energetics of pathogenic bacteria and opportunities for drug development. Adv. Microb. Physiol. 65, 1–62 (2014). [DOI] [PubMed] [Google Scholar]
- Nashef A. S., Osuga D. T. & Feeney R. E. Determination of hydrogen sulfide with 5,5′-dithiobis-(2-nitrobenzoic acid), N-ethylmaleimide, and parachloromercuribenzoate. Anal. Biochem. 79, 394–405 (1977). [DOI] [PubMed] [Google Scholar]
- Blattner F. R. et al. The complete genome sequence of Escherichia coli K-12. Science 277, 1453–1462 (1997). [DOI] [PubMed] [Google Scholar]
- Bekker M., de Vries S., Ter Beek A., Hellingwerf K. J. & de Mattos M. J. Respiration of Escherichia coli can be fully uncoupled via the nonelectrogenic terminal cytochrome bd-II oxidase. J. Bacteriol. 191, 5510–5517 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puustinen A., Verkhovsky M. I., Morgan J. E., Belevich N. P. & Wikström M. Reaction of the Escherichia coli quinol oxidase cytochrome bo3 with dioxygen: The role of a bound ubiquinone molecule. Proc. Natl. Acad. Sci. USA 93, 1545–1548 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisov V. B. Interaction of bd-type quinol oxidase from Escherichia coli and carbon monoxide: Heme d binds CO with high affinity. Biochemistry-Moscow 73, 14–22 (2008). [DOI] [PubMed] [Google Scholar]
- Borisov V. B. et al. Aerobic respiratory chain of Escherichia coli is not allowed to work in fully uncoupled mode. Proc. Natl. Acad. Sci. USA 108, 17320–17324 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna C. et al. Crystallization and preliminary crystallographic analysis of cysteine synthase from Entamoeba histolytica. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 63, 512–515 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goutelle S. et al. The Hill equation: a review of its capabilities in pharmacological modelling. Fundam. Clin. Pharmacol. 22, 633–648 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.