Abstract
The clinical challenge posed by p53 abnormalities in hematological malignancies requires therapeutic strategies other than standard genotoxic chemotherapies. ONC201 is a first-in-class small molecule that activates p53-independent apoptosis, has a benign safety profile, and is in early clinical trials. We found that ONC201 caused p53-independent apoptosis and cell cycle arrest in cell lines and in mantle cell lymphoma (MCL) and acute myeloid leukemia (AML) samples from patients; these included samples from patients with genetic abnormalities associated with poor prognosis or cells that had developed resistance to the nongenotoxic agents ibrutinib and bortezomib. Moreover, ONC201 caused apoptosis in stem and progenitor AML cells and abrogated the engraftment of leukemic stem cells in mice while sparing normal bone marrow cells. ONC201 caused changes in gene expression similar to those caused by the unfolded protein response (UPR) and integrated stress responses (ISRs), which increase the translation of the transcription factor ATF4 through an increase in the phosphorylation of the translation initiation factor eIF2α. However, unlike the UPR and ISR, the increase in ATF4 abundance in ONC201-treated hematopoietic cells promoted apoptosis and did not depend on increased phosphorylation of eIF2α. ONC201 also inhibited mammalian target of rapamycin complex 1 (mTORC1) signaling, likely through ATF4-mediated induction of the mTORC1 inhibitor DDIT4. Overexpression of BCL-2 protected against ONC201-induced apoptosis, and the combination of ONC201 and the BCL-2 antagonist ABT-199 synergistically increased apoptosis. Thus, our results suggest that by inducing an atypical ISR and p53-independent apoptosis, ONC201 has clinical potential in hematological malignancies.
INTRODUCTION
p53 is a critical effector molecule for inducing apoptosis in tumors. Unfortunately, TP53 is mutated with consequent loss of function in about 50% of solid tumors, 14% of leukemias [5 to 10% of acute myeloid leukemias (AMLs) (1–3), ~5% of acute lymphoblastic leukemias (2), and 10% of chronic lymphocytic leukemias (CLLs) (4)], and 12.5% of non-Hodgkin’s lymphomas (5). Although the frequency of TP53 mutations in hematological malignancies appears to be relatively low, it plays a major role in resistant and poor prognosis cases. For example, AML patients whose tumor cells have a complex karyotype, and who have a much shorter survival than patients with noncomplex karyotypes (6), reportedly have a TP53 mutation incidence of >70% (3). Indeed, AML cases with TP53 mutations or deletions had the shortest survival among the entire AML spectrum in a large-scale sequencing project (3, 7). Mantle cell lymphoma (MCL), a disease incurable by standard chemotherapies with a median survival of 3 to 5 years, is also characterized by a high incidence (>30%) of TP53 mutations or deletions (8) that are associated with the clinically aggressive blastoid variant (9) and shorter overall survival (8, 10). Correlations between TP53 mutations (or deletions) and poor prognosis have also been reported for diffuse large B cell lymphomas and CLL (5). Hence, there is an urgent need to develop agents that are active independently of TP53 status.
ONC201 (previously referred to as TIC10) is a first-in-class small molecule that was identified in a high-throughput small-molecule library screen as potent inducer of p53-independent apoptosis in tumor cells, with a remarkable safety profile (11, 12). In solid tumors, ONC201 caused late-stage induction of tumor necrosis factor–related apoptosis-inducing ligand (TRAIL) death receptor 5 (DR5) and promoted the transcription of the TRAIL gene, the latter through activation of the transcription factor FOXO3a caused by late-stage inactivation of signaling by the kinases AKT and MAPK (mitogen-activated protein kinase) (12). ONC201 has substantial antitumor activity in preclinical models in various advanced solid tumors with infrequent oral dosing and without toxicity in normal cells in culture and in vivo (12). Preclinical studies demonstrated broad synergism of ONC201 with established anticancer therapies, including the depletion of colorectal cancer stem cells (13, 14).
Here, we examined the efficacy and tumoricidal mechanism of ONC201 in leukemias and lymphomas, in both cultured cell lines and primary cells bearing either wild-type or mutant p53. ONC201 exerted antileukemia and antilymphoma activity regardless of p53 status and selectively killed AML stem cells [namely, cells that can engraft and reconstitute AML in nonobese diabetic/severe combined immunodeficient γ (NSG) mice] and progenitor cells (enriched in CD34+CD38− cells) while sparing normal bone marrow (BM) cells. However, mechanisms previously identified in solid tumors (induction of TRAIL and DR5) were not operational in leukemia and lymphomas.
RESULTS
ONC201 exerts p53-independent apoptotic and antiproliferative effects in lymphoma and leukemia
Four MCL and AML cell lines were each treated with ONC201 in vitro. Measures of apoptosis or viable cell number indicated that ONC201 exerted both cytotoxic and antiproliferative effects (Fig. 1, A and B). TP53 mutant AML lines were slower to undergo apoptosis, but the onset of viable cell reduction was similar for AML and MCL, regardless of TP53 status (Fig. 1, A and B). Among MCL cell lines, the TP53 mutant lines JeKo-1 and MINO were actually more prone to ONC201-induced apoptosis than were the TP53 wild-type lines Z-138 and JVM-2 (Fig. 1A). Overall, except for JVM-2 cells, the half-maximal inhibitory concentration doses on the basis of cell viability were less than 2.5 μM. Stable TP53 knockdown in Z-138 and JVM-2 cells (Fig. 1C) did not affect their sensitivity to ONC201, thus confirming that ONC201-induced apoptosis was p53-independent (Fig. 1D).
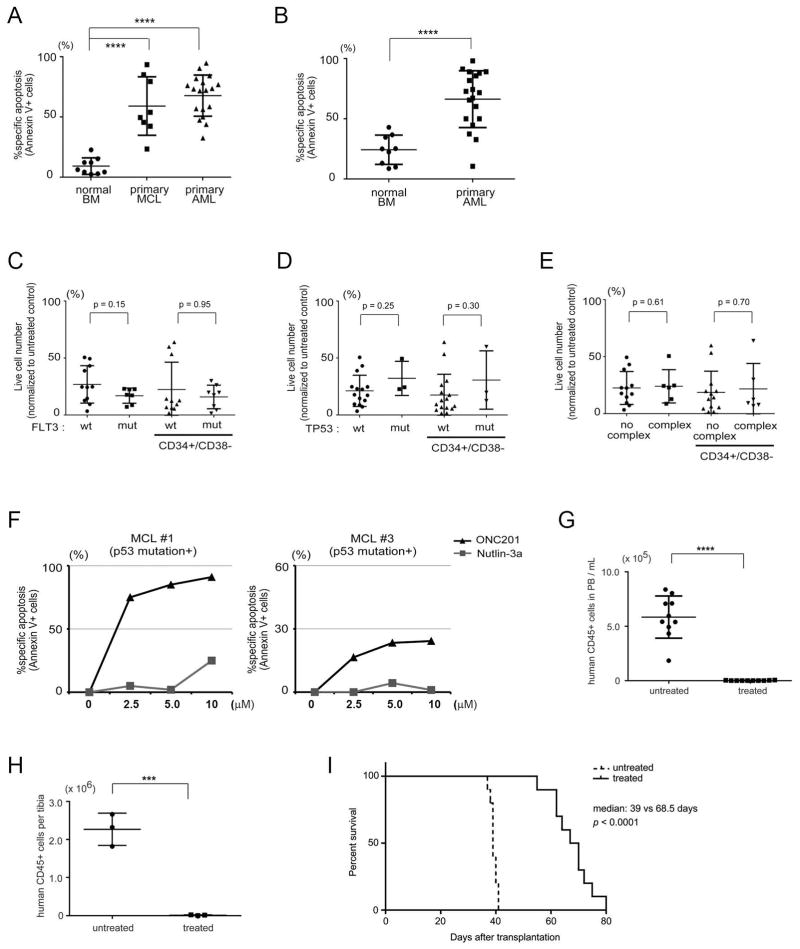
Fig. 1. ONC201 induces apoptosis in MCL and AML cells independent of TP53 mutation status.
(A) Annexin V–positive cells were counted by flow cytometry after a 72-hour treatment with ONC201. HL-60 and THP-1 cells were treated with the same concentrations for 72 and 120 hours (n = 3 experiments for each cell line). (B) Annexin V/propidium iodide (PI) double-negative cells were counted as live cells and normalized by the live cell count in untreated control samples (n = 3 experiments for each cell line). (C) Immunoblot of p53 in Z-138 and JVM-2 cells transduced with lentivirus expressing control (C) short hairpin RNA (shRNA) or shRNA specific to TP53. (D) Specific apoptosis (see Materials and Methods) induced by ONC201 treatment for 72 hours in TP53 knockdown or intact Z-138 and JVM-2 cells (n = 3 experiments for each isogenic line).
ONC201 inhibits DNA synthesis but does not induce DNA double-strand breaks
To test how ONC201 affects the cell cycle, we measured the incorporation of 5-ethynyl-2′-deoxyuridine (EdU) and DNA content in MCL cell lines. In Z-138 and JeKo-1 cells, ONC201 decreased the incorporation of EdU in S phase and increased cell numbers in G1 phase while reducing cell numbers in S phase (fig. S1, A to C), specifically in early S phase (fig. S1D), together indicating a slowing of S-phase progression. To assess whether ONC201 induced DNA damage, we analyzed γH2A.X positivity in ONC201-treated cells. Nutlin-3a (an inhibitor of the E3 ubiquitin ligase MDM2 that can increase p53 abundance independently of DNA damage) and doxorubicin (a topoisomerase II inhibitor that induces DNA strand breaks) were used as negative and positive controls, respectively. ONC201 minimally increased γH2A.X positivity, to an extent similar to that induced by Nutlin-3a in both Z-138 and JeKo-1 cells (fig. S1E).
ONC201 is toxic to primary MCL and AML stem/progenitor cells
To investigate the effects of ONC201 on primary tumor cells, we treated 18 primary AML and 8 MCL samples with ONC201 for 72 hours. Adverse prognostic factors in the AML samples included five cases with complex karyotype, seven cases with FLT3 mutations, and three cases with TP53 mutations detected by next-generation sequencing (table S1). All MCL samples were sequenced for TP53 mutations from exon 5 to 8, in which hotspot mutations occur, and mutations were found in two cases. To assess the toxicity of ONC201 against normal hematopoietic cells, we treated BM mononuclear cells (BMMCs) from eight healthy donors. Cells from both AML and MCL were significantly more sensitive to ONC201 than normal BMMCs (Fig. 2A and fig. S2A).
Fig. 2. Effects of ONC201 on primary MCL and AML cells and normal BM cells.
(A and B) Specific apoptosis induced by a 72-hour in vitro treatment with ONC201 (5 μM) in normal BMMCs (n = 9 samples), primary MCL cells (n = 8 samples), and AML cells (n = 18 samples) (A) or in stem/progenitor (CD45+CD34+CD38−) populations in normal BM samples and primary AML cells (B). (C to E) Comparison of effects of ONC201 treatment (5 μM for 72 hours) on live cell numbers of primary AML cells according to FLT3 mutation (mut) status (C), TP53 mutation status (D), or complex/noncomplex karyotype (E). (F) Specific apoptosis by ONC201 or Nutlin-3a treatment (72 hours) in two primary MCL samples with TP53 mutation. Representative of two technical replicates. (G and H) Absolute number of human CD45-positive cells in peripheral blood (PB) (per milliliter, n = 10 mice for each group) (G) and in BM (per tibia, n = 3 for each group) (H) at 4 weeks after transplantation into NSG mice of viable AML cells after treatment with (5 μM for 48 hours, then washed) or without ONC201 in vitro into NSG mice. (I) Survival curves of mice (n = 10 mice for each group) after transplantation of AML cells as in (G) and (H). **P < 0.01, ***P < 0.001, ****P < 0.0001.
Hematologic toxicity and anti-AML efficacy of ONC201 may be mediated by effects on normal and malignant stem cells, respectively. Therefore, we tested the in vitro toxicity of ONC201 to stem/progenitor cell–containing subpopulations (CD45+CD34+CD38−) from AML and normal BM, and found that AML cells were significantly more sensitive (Fig. 2B and fig. S2B). No significant difference in cell sensitivity was detected in AML samples according to mutations in FLT3 or TP53, or to complex karyotype (Fig. 2, C to E), suggesting that the effects of ONC201 were independent of these resistance factors. Both TP53 mutant MCL samples were sensitive to ONC201, whereas they were resistant to Nutlin-3a, as expected (Fig. 2F).
To confirm that ONC201 was toxic to leukemic stem cells (LSCs), defined as primary AML cells that can engraft in immunodeficient mice, we recovered unfractionated LSC-containing populations of AML cells [t(9;11)(p22; q23), CEBPA and ATM mutant] from secondarily engrafted mice and cultured them in vitro for 48 hours, with or without ONC201. For both groups, the same number of washed, trypan blue–negative (viable) cells was then retransplanted. After 4 weeks, both the absolute number (Fig. 2G) and the percentage (fig. S2C) of human CD45+ cells were significantly lower in both the peripheral blood (Fig. 2G and fig. S2C) and tibial BM samples (Fig. 2H and fig. S2D) from the ONC201-treated group. Spleens collected from untreated mice were significantly larger because of infiltration by AML cells (fig. S2, E and F). Moreover, mice injected with ONC201-treated cells survived significantly longer (Fig. 2I). These results indicated that ONC201 was effective against LSC, whether by direct toxicity or attenuation of self-renewal.
Furthermore, because LSCs have increased expression of the gene encoding the drug transporter MDR1 [multidrug resistance protein 1; also known as P-glycoprotein or permeability protein 1 (Pgp)] (15), which could mediate resistance to ONC201, we tested the efficacy of ONC201 in MDR1-overexpressing myeloma cells (RPMI8226/DOX6 cells) (16). ONC201 was toxic to RPMI8226/DOX6 cells, which, in contrast, were resistant to doxorubicin (fig. S2G). In addition, the MDR1 inhibitor PSC-833 had no effect on ONC201 activity, whereas PSC-833 sensitized RPMI8226/DOX6 cells to doxorubicin (fig. S2G), demonstrating independence of ONC201 from MDR1-mediated drug resistance.
ONC201 is active in cells resistant to other targeted inhibitors
Proteasome inhibitors like bortezomib are a mainstay of therapy for multiple myeloma (MM), in which they induce a terminal (apoptotic) unfolded protein response (UPR) (17) but ultimately are limited by the frequent development of resistance. The MM cell line ANBL-6, made bortezomib-resistant by chronic exposure, was only slightly less sensitive than drug-naïve ANBL-6 cells to ONC201 (fig. S3A), suggesting that ONC201 may be an effective option for bortezomib-resistant MM patients.
Likewise, the Bruton’s tyrosine kinase inhibitor ibrutinib, which acts in part by interrupting B cell receptor signaling, shows high clinical efficacy in CLL but is also limited by the frequent development of resistance, which is associated with very short survival (18). A similar scenario is frequently observed in MCL. We treated four primary samples of MCL with ONC201, ibrutinib, or in combination. The combination was more effective than either drug alone in all samples (fig. S3B), two of which were resistant to ibrutinib alone, suggesting that ONC201 could be effective in ibrutinib-resistant patients.
ONC201-induced apoptosis in hematological malignancies is independent of caspase-8 activation and FOXO3a
In solid tumors, ONC201 induces the transcription of the genes encoding TRAIL and its death receptor DR5, which induce an extrinsic pathway of programmed cell death (12). However, ONC201 induced expression of DR5 in only one of four hematologic cell lines, although ONC201 caused apoptosis in all, and none exhibited transcriptional induction of TRAIL (Fig. 3A).
Fig. 3. ONC201-induced apoptosis is independent of the extrinsic pathway of apoptosis.
(A) Real-time quantitative polymerase chain reaction (PCR) for DR5 and TRAIL mRNA after exposure to ONC201 (5 μM) of Z-138, MINO, JeKo-1, and OCI-AML3 cells. Relative quantity (RQ) values of mRNA expression are calculated as the fold change relative to mRNA expression at time 0 (normalized to GAPDH) (n = 3 experiments). (B) Specific apoptosis of MCL cell lines treated for 72 hours with ONC201 (5 μM), with or without Z-IETD-FMK (25 μM). JeKo-1 cells were also treated with TRAIL (10 nM), with or without Z-IETD-FMK, for 72 hours (n = 3 experiments). (C) Specific apoptosis and number of live cells (PI and annexin V double-negative cells) in parental Jurkat and caspase-8–deficient Jurkat I9.2 cells after treatment with ONC201 (5 μM for 72 hours) (n = 3 experiments). (D) Specific apoptosis induced by ONC201 with or without the pan-caspase inhibitor Z-VAD-FMK (50 μM) in JeKo-1 and Jurkat I9.2 cells (n = 3 experiments). ***P < 0.001, ****P < 0.0001.
As further evidence that ONC201-induced cell death did not depend on the extrinsic pathway of apoptosis, treatment with the caspase-8 inhibitor Z-IETD-FMK did not reduce apoptosis induction by ONC201 in three (Z-138, JVM-2, and MINO) of four MCL lines (Fig. 3B). Z-IETD-FMK partially reduced ONC201-induced apoptosis in JeKo-1 cells, but Z-IETD-FMK can inhibit caspases other than caspase-8 at the concentration used (19). Addition of soluble TRAIL induced apoptosis in JeKo-1 and Jurkat cells, indicating that the extrinsic pathway is intact in JeKo-1 and Jurkat cells and was blocked almost completely by Z-IETD-FMK (Fig. 3B), suggesting that the partial effects of Z-IETD-FMK on ONC201-induced apoptosis was not through inhibition of the extrinsic pathway. Caspase-8–deficient Jurkat I9.2 cells, which were resistant to TRAIL (fig. S4A), were as sensitive as wild-type Jurkat cells to apoptosis induction by ONC201 (Fig. 3C), and the pan-caspase inhibitor Z-VAD-FMK significantly inhibited ONC201-induced apoptosis in both cell lines (Fig. 3D). The proteins BAX (BCL-2–associated X protein) and BAK (BCL-2 homologous antagonist/killer) promote the intrinsic pathway of apoptosis by permeabilizing mitochondria. We found that Bax and Bak double-knockout mouse embryonic fibroblasts (MEFs) were resistant to ONC201, in contrast to wild-type MEFs (fig. S4, B and C), as assessed by flow cytometric detection of sub-G1 populations. Together, these results indicate that ONC201 engages the intrinsic rather than extrinsic pathway of apoptosis in the malignant hematopoietic cells tested.
A previous report on ONC201’s mechanism of action showed that dephosphorylation of the kinases ERK (extracellular signal–regulated kinase) and AKT activates FOXO3a by inducing its nuclear translocation, resulting in the transcriptional induction of TRAIL (12). In cell lines derived from hematologic malignancies, the absence of TRAIL induction by ONC201 was accompanied by an absence of upstream changes in the AKT/ERK/FOXO3a axis (fig. S5, A and B). Stable FOXO3a knockdown in OCI-AML3 and Z-138 cells (fig. S5C) did not change their sensitivity to ONC201 (fig. S5D), further showing that FOXO3a did not participate in the effects of ONC201. These results indicate that the mechanism of action of ONC201 in hematological malignancies appears to be different from that previously observed in solid tumors, although there may be a common ONC201 target in all malignancies.
The gene expression profile of the response to ONC201 resembles the response to ER stress or amino acid deprivation and mTOR inhibition
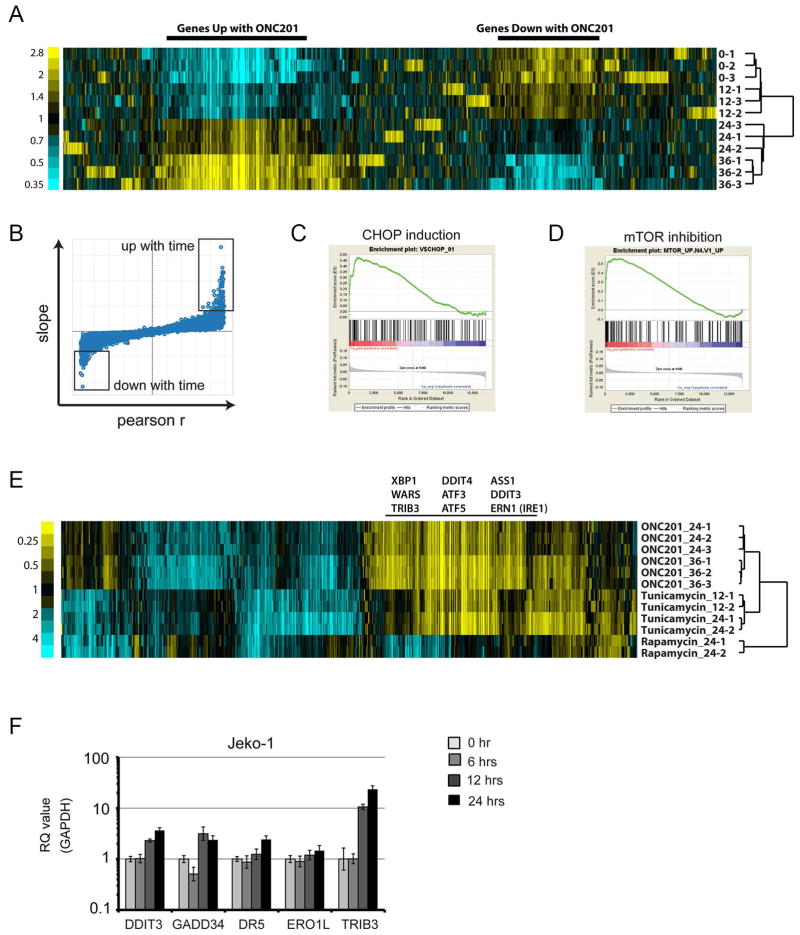
We used gene expression profiling to investigate in more detail the effects of ONC201 in hematological malignancies, using JeKo-1 (TP53−/−) and Z-138 (TP53 wild type) MCL lines. In a time course experiment using JeKo-1 cells treated with ONC201, a mean-centered heat map of variant genes showed two large areas of genes with similarly progressive time-dependent changes (up or down) in expression (Fig. 4A). To identify genes and pathways affected by ONC201, we used data from all four time points for Pearson’s correlation of expression compared to time and chose genes at the extremes of slope and r values (boxes in Fig. 4B and data files S1 and S2) for analysis by the hypergeometric distribution test (20) of overlap with gene sets of known associations. Highly significant results were obtained for genes up-regulated by amino acid depletion in the AML cell line HL-60 or by treatment with the aminopeptidase inhibitor tosedostat (CHR-2797), which induces endoplasmic reticulum (ER) stress and inhibits phosphorylation of mammalian target of rapamycin (mTOR) substrates (21). Gene Set Enrichment Analysis (GSEA) using gene ranks based on slope or r values indicated that ONC201 up-regulated genes were enriched for those related to ER stress, such as those encoding ER component proteins and targets of the ER stress–induced transcription factor CHOP (DDIT3; Fig. 4C). GSEA also suggested mTOR inhibition, by results such as enrichment of the Molecular Signatures Database (MSigDB) gene set “mTOR_UP.N4.V1_UP” (Fig. 4D), which consists of genes up-regulated in a T cell line by the mTOR inhibitor everolimus. This gene set was also enriched by the hypergeometric distribution test of up-regulated genes in Fig. 4B. Similar results resembling an ER stress response were generated by ONC201 treatment of Z-138 cells, and many genes up-regulated by ER stress were up-regulated by ONC201 exposure in both JeKo-1 and Z-138 cells, but not in a population of Z-138 cells made resistant to ONC201 by chronic exposure (data file S3).
Fig. 4. Gene expression profiling of JeKo-1 cells treated with ONC201.
(A) Mean-centered heat map of variant genes from a time course treatment of JeKo-1 cells with ONC201 (5 μM for 12, 24, and 36 hours) (n = 3 for each time point). (B) Distribution of slope and Pearson’s r values of correlation between gene expression and time. Boxes indicate probes with extreme values of slope and r (data files S1 and S2). (C) Enrichment plot of target genes of CHOP. False discovery rate (FDR) q = 0.016. (D) Enrichment plot of genes that positively correlate with mTOR inhibition. FDR q = 0.000. (E) Subtracted heat map of expression changes (absolute log2 mean-centered value >1 in at least 1 of 12 replicates) in genes of JeKo-1 cells treated with ONC201 (5 μM), rapamycin (10 nM), or tunicamycin (1 μM) for the times indicated (12, 24, or 36 hours) (n ≥ 2 for each condition). (F) Real-time PCR analysis of DDIT3, GADD34, DR5, and ERO1L mRNA expression in JeKo-1 cells during treatment with ONC201 relative to that of GAPDH (n = 3 experiments).
We investigated these findings further by comparing the gene expression profile in JeKo-1 cells after exposure to ONC201 to that after exposure to the ER stress inducer tunicamycin or the allosteric mTOR inhibitor rapamycin. ONC201 and tunicamycin induced similar changes (both up and down) in gene expression, and common up-regulated genes included those associated with ER stress (Fig. 4E). In contrast, there was little overlap between changes induced by ONC201 and those of rapamycin. These results suggested that the principal effect of ONC201 resembled an ER stress response and implied that inhibition of the mTOR pathway by ONC201 might involve effects different from those induced by rapamycin, such as those resulting from mTOR active-site inhibitors (22).
To verify the ER stress–related gene expression profiling data, we used real-time PCR to confirm that a 12- to 24-hour exposure to ONC201 in JeKo-1 cells increased the expression of CHOP (DDIT3) and its target genes GADD34, DR5, and TRIB3 (Fig. 4F), as well as the protein abundance of CHOP and DR5 (fig. S6A). We also found that the abundance of the mRNA splice variant of XBP-1 (XBP-1s) and the protein abundance of IRE1α, other key players of the UPR, were increased as early as 6 or 12 hours, respectively, after exposure to ONC201 (fig. S6, A and B). Together, these data indicated that ER stress and the UPR, or similar stress pathways, were induced by ONC201.
ONC201 induces ATF4 and inhibits mTORC1
In the classical UPR to ER stress pathway, accumulation of unfolded proteins in the ER causes the chaperone protein BiP to detach from the eIF2α kinase PERK, which is then activated by autophosphorylation and in turn phosphorylates eIF2α, resulting in selective enhancement of activating transcription factor 4 (ATF4) translation while general protein translation slows down (23–25). Other stresses such as nutrient deprivation and viral infections activate integrated stress responses (ISRs) using different eIF2α kinases such as GCN2 and PKR (24, 26, 27), but generally have similar effects on eIF2α, ATF4, and CHOP. Accumulating evidence suggests that the UPR and ISR affect the mTOR complex 1 (mTORC1) kinase complex, but the mechanism is complex; whether the UPR and ISR promote or inhibit mTORC1 seems to vary depending on tissue context (27–32), and which of the eIF2α kinases (PERK, PKR, or GCN2) is (or are) involved in affecting mTORC1 also varies (33–38).
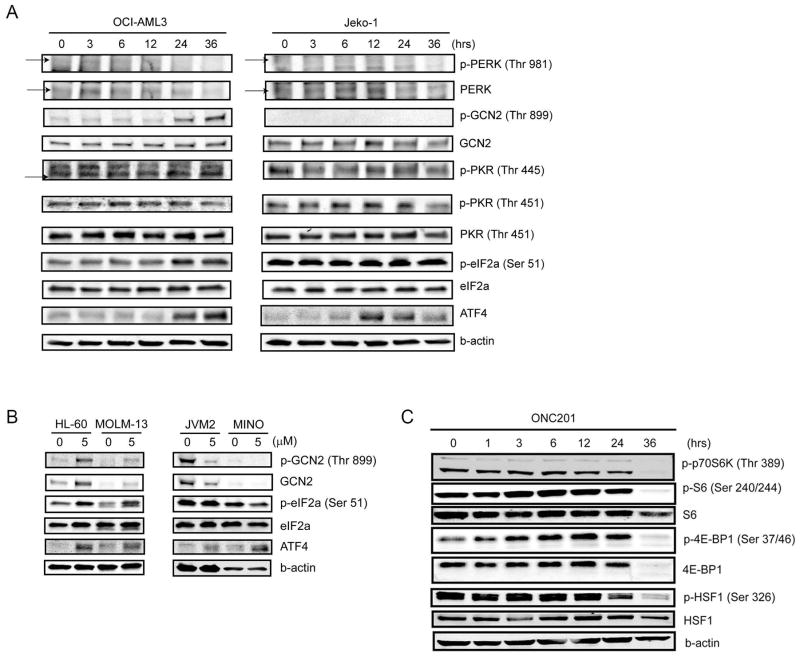
To investigate how ONC201 affects these proteins, we performed immunoblot analysis in OCI-AML3 and JeKo-1 cells treated with ONC201. In OCI-AML3 cells treated with ONC201, eIF2α phosphorylation and ATF4 abundance were increased in parallel at 24 and 36 hours (Fig. 5A and fig. S7A). The phosphorylation of GCN2, but not that of PERK and PKR, was increased, suggesting that GCN2 may be the kinase involved in ONC201-induced phosphorylation of eIF2α in OCI-AML3. In JeKo-1 cells, however, the phosphorylation of eIF2α did not increase, even though ATF4 protein abundance increased between 12 and 36 hours (Fig. 5A and fig. S7A), suggesting that the ATF4 induction may be independent of eIF2α phosphorylation. Consistent with this hypothesis, ONC201 did not increase the phosphorylation of PERK, GCN2, or PKR in JeKo-1 cells (Fig. 5A and fig. S7A). We confirmed these differences in the response to ONC201 between the AML and MCL cell type in additional cell lines: in the AML lines HL-60 and MOLM-13, ONC201 similarly increased the phosphorylation of GCN2 and eIF2α, with ATF4 induction at 24 hours, whereas in the MCL lines JVM-2 and MINO, no phosphorylation of GCN2 or eIF2α was detected, but ATF4 was induced at 24 hours (Fig. 5B and fig. S7B).
Fig. 5. Effects of ONC201 on eIF2α kinases, eIF2α, ATF4, and mTORC1.
(A) Representative immunoblot analysis (from three experiments) of the abundance and phosphorylation (p) of eIF2α kinases, eIF2α, and ATF4 in OCI-AML3 and JeKo-1 cells from a time course of treatment with ONC201 (5 μM for 36 hours). (B) Representative immunoblot analysis (from three experiments) of GCN2, eIF2α, and ATF4 in HL-60, MOLM-13, JVM-2, and MINO cells treated with control or ONC201 (5 μM) for 24 hours. (C) Representative immunoblot analysis (from three experiments) of mTORC1 target proteins in JeKo-1 cells from a time course of treatment with ONC201 (5 μM for 36 hours).
Notably, treatment of JeKo-1 and OCI-AML3 cells with ONC201 reduced phosphorylation of mTORC1 targets p70 S6 kinase (p70S6K), S6, and 4E-BP1, indicating inhibition of mTORC1 (Fig. 5C), as was also implicated by our gene expression profiling results. mTOR inhibition also results in the dephosphorylation and inactivation of heat shock factor 1 (HSF1) (39, 40); we found that HSF1 was indeed dephosphorylated after 24 hours (Fig. 5C).
The effects of classical UPR and ISR inducers differ from those of ONC201
Deprivation of amino acids or glucose activates the GCN2/eIF2α/ATF4 pathway. However, in some circumstances, GCN2 plays a role in a UPR (41, 42), in collaboration with PERK. Therefore, we performed immunoblot analyses of OCI-AML3 and JeKo-1 cells after amino acid or glucose deprivation or treatment with the ER stress inducers tunicamycin and thapsigargin. As expected, but unlike ONC201, thapsigargin and tunicamycin induced phosphorylation of eIF2α with coincident induction of ATF4 in JeKo-1 cells by 24 hours (fig. S8, A and B), confirming that eIF2α was not maximally phosphorylated in untreated JeKo-1 cells and that this antibody could detect the increase of eIF2α phosphorylation by ER stress. We also found that tunicamycin and thapsigargin reduced the phosphorylation of mTORC1 targets p70S6K, S6, and 4E-BP1 in JeKo-1 cells (fig. S8C).
When various AML and MCL cell lines were treated with tunicamycin for 24 hours, phosphorylation of eIF2α but not that of GCN2 was increased (fig. S8, D and E). Because GCN2 phosphorylation is most often reported in nutrient-related ISRs, we cultured OCI-AML3 and JeKo-1 cells in medium lacking glucose or a few essential amino acids (L-arginine, L-glutamine, and L-lysine). Unexpectedly, amino acid deprivation did not cause phosphorylation of GCN2 or eIF2α in either OCI-AML3 or JeKo-1 cells, although ATF4 was significantly induced by 24 hours (fig. S9, A and B). Especially in JeKo-1 cells, the phosphorylation of eIF2α was even reduced in a time-dependent manner, opposite to that of ATF4 induction and in contrast to the response to ONC201 treatment. However, amino acid deprivation in these lines reduced the phosphorylation of p70S6K as expected (fig. S9, A and B) (21, 43). In OCI-AML3 cells, glucose deprivation increased the phosphorylation of GCN2 and eIF2α within 8 hours, concurrent with ATF4 induction (fig. S9, C and D) but transiently, unlike the sustained effects of ONC201. In contrast, in JeKo-1 cells, the phosphorylation of GCN2 or eIF2α was not increased by 24 hours after exposure, whereas ATF4 was transiently induced (fig. S9, C and D). Rather than a decrease as observed in amino acid–deprived cells, the phosphorylation of p70S6K was increased in response to glucose starvation in JeKo-1 cells (fig. S9, A and B). These results indicated that the response to ONC201 in AML or MCL cells was only partially similar to changes induced by the UPR to classical ER stress inducers or by the ISRs to amino acid or glucose deprivation (table S2).
ATF4 up-regulation by ONC201 is independent of an increase in eIF2α phosphorylation but uses the same posttranscriptional mechanism as UPR
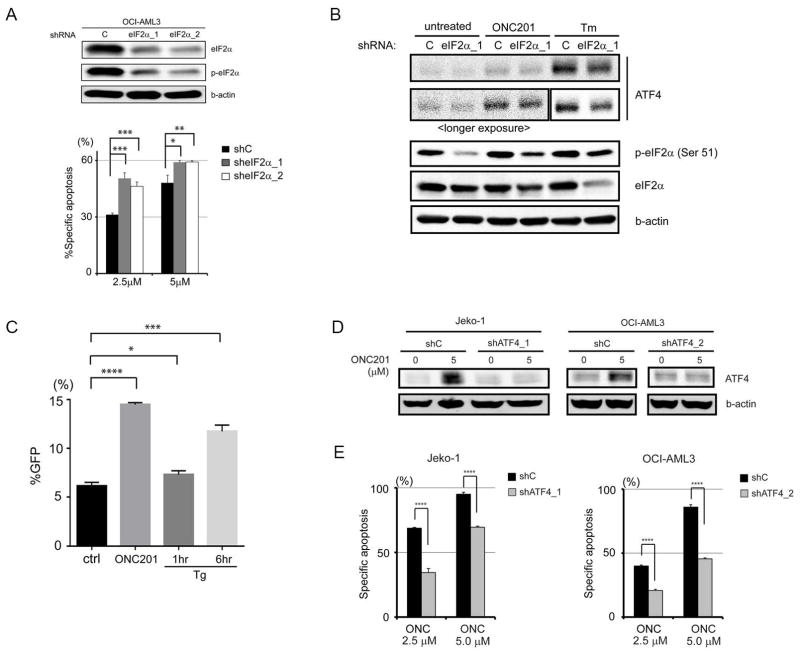
Phosphorylation of eIF2α is thought to be an adaptive response helping cells to survive during cellular stresses, by promoting inhibition of protein translation in general. However, eIF2α phosphorylation during ER stress (or other stresses) is typically short-lived; induction of the phosphatase regulator GADD34 by CHOP promotes eIF2α dephosphorylation, and the resulting recovery of protein translation leads to apoptosis through increased abundance of proapoptotic proteins (24, 25, 44–46). eIF2α phosphorylation remained increased in ONC201-treated OCI-AML3 cells at 36 hours (Fig. 5A), and knockdown of eIF2α in OCI-AML3 cells increased ONC201-induced apoptosis (Fig. 6A), indicating that increased phosphorylation of eIF2α may mediate an adaptive mechanism reducing ONC201-induced apoptosis, as in classical ER stress and ISRs. However, eIF2α knockdown did not reduce the ONC201-induced increase in ATF4 abundance, whereas it did reduce tunicamycin-induced ATF4 up-regulation (Fig. 6B and fig. S10). We interpreted these results as evidence that even in OCI-AML3 cells, in which eIF2α phosphorylation (by GCN2) occurred in response to ONC201, induction of ATF4 by ONC201 is independent of an increase in eIF2α phosphorylation. Although ATF4 induction without increased phosphorylation of eIF2α is atypical for the UPR or most ISRs (table S2), there are other examples of ISRs that can induce ATF4 or CHOP without increased phosphorylation of eIF2α in hematopoietic cells (macrophages), fibroblasts, and solid tumor cells (47–49).
Fig. 6. ATF4 induction by ONC201 and its role in apoptosis.
(A) Immunoblot analysis of total and phosphorylated eIF2α in OCI-AML3 cells transfected with control shRNA and shRNAs against eIF2α (eIF2α_1 and eIF2α_2) and effects of eIF2α knockdown on apoptosis of cells treated with ONC201 (5 μM) for 72 hours. (B) Representative immunoblots (from three experiments) of ATF4 and eIF2α in OCI-AML3 cells transfected with control or eIF2α-targeting shRNA and treated with ONC201 (5 μM for 24 hours) or tunicamycin (Tm; 1 μM for 6 hours). (C) Green fluorescent protein (GFP) reporter assay of ATF4 translation. 4′,6-Diamidino-2-phenylindole (DAPI)–negative and GFP-positive cells were counted by flow cytometry after treatment with dimethyl sulfoxide (ctrl), ONC201 (5 μM for 27 hours), or thapsigargin (Tg; 1 μM for 1 or 6 hours) (n = 3 experiments). (D) Immunoblot verification of ATF4 knockdown in JeKo-1 (shATF4_1) and OCI-AML3 (shATF4_2) cells. (E) Specific apoptosis induced by ONC201 in control and ATF4 knockdown JeKo-1 (shATF_1) and OCI-AML3 (shATF_2) cells (n = 3 experiments). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Transcriptional up-regulation might explain the eIF2α phosphorylation–independent induction of ATF4 because ONC201 increased ATF4 mRNA expression in JeKo-1 and OCI-AML3 cells in a time-dependent manner (fig. S11). However, especially in JeKo-1 cells, the increase in ATF4 mRNA was minimal after 12 hours, whereas protein abundance had begun to increase by 6 hours (Fig. 5A), suggesting that increased transcription was not the main mechanism of ATF4 induction by ONC201.
eIF2α phosphorylation–dependent induction of ATF4 in UPR or ISR occurs through a translational mechanism that uses specific open reading frames (ORFs) called “μORF” found upstream of the start codon of ATF4 (50–52). In the absence of stress, one of the μORFs works as an inhibitory motif that traps ribosomes from moving forward to the start codon; however, in stressed conditions that result in phosphorylation of eIF2α and global translation inhibition, phosphorylation of eIF2α enables ribosomes to bypass the inhibitory μORF and selectively accelerate translation of ATF4 (51, 52), which then induces transcription of its target genes, including CHOP. To address whether translational up-regulation is involved in ATF4 induction by ONC201, despite the absence of eIF2α phosphorylation, we used a reporter plasmid (5′ATF4-GFP) (52) in which the murine μORF-containing upstream ATF4 sequence is fused in frame to GFP. Transcription of this reporter is constitutively driven by a cytomegalovirus promoter, and 3′ mRNA processing is specified by SV40 viral signals, but translation is induced only when the bypass to the second μORF occurs. After transfection of the reporter into MEFs, treatment with ONC201 (for 27 hours) or thapsigargin (for 6 hours) both significantly increased GFP expression compared to the untreated control (Fig. 6C), indicating that ATF4 induction by ONC201 also depends on this posttranscriptional mechanism.
ONC201 causes apoptosis through ATF4
Because CHOP induction by ER stress leads to GADD34 transcription and eIF2α dephosphorylation, enabling the translation of proapoptotic proteins (25, 44–46), we speculated that CHOP knockdown would inhibit ONC201-induced apoptosis. However, the sensitivity of OCI-AML3 and JeKo-1 cells to ONC201 (and to tunicamycin) was actually increased by CHOP knockdown (fig. S12, A and B). In a similar paradoxical fashion, although ATF4 typically promotes adaptation to cellular stress, both JeKo-1 and OCI-AML3 cells were significantly less sensitive to ONC201-induced apoptosis after stable knockdown of ATF4 (Fig. 6, D and E, and fig. S12, C and D). ATF4 induction by ONC201 was higher in cells with CHOP knockdown (fig. S12, E and F), perhaps explaining the greater sensitivity of these cells to ONC201. We also examined the role of IRE1α, another ER stress–related protein, which has both proapoptotic and prosurvival functions in UPR-induced apoptosis, especially in B cells (24, 46, 53, 54). IRE1α abundance was increased in response to ONC201 in JeKo-1 cells but not in OCI-AML3 cells (figs. S6A and S13A). Stable IRE1α knockdown rendered JeKo-1 cells significantly less sensitive to ONC201-induced apoptosis (fig. S13, B and C).
mTORC1 inhibition by ONC201 in MCL and AML cells is partially dependent on ATF4 induction
mTORC1 target proteins are dephosphorylated within an hour in cells exposed to the direct mTORC1 inhibitor rapamycin (55). Because phosphorylation of these proteins was decreased by thapsigargin exposure starting at 3 hours and by tunicamycin and ONC201 at a later time point (fig. S8C and Fig. 5C), these results suggested that ONC201 inhibited mTORC1 through an indirect mechanism, likely by using a pathway shared with the UPR. ER stress can transcriptionally induce DDIT4 (also known as REDD1) (56, 57), an inhibitor of mTORC1, through ATF4 induction (58), and we found that DDIT4 mRNA was induced in both JeKo-1 and OCI-AML3 cells after exposure to ONC201 in a time-dependent manner (fig. S14A), suggesting that DDIT4 induction may explain the mechanism of mTORC1 inhibition by ONC201. We confirmed that the reduction in the phosphorylation of p70S6K and S6 in response to ONC201 in OCI-AML3 and JeKo-1 cells was substantially blocked by ATF4 knockdown (fig. S14, B and C), indicating that ONC201 inhibited mTORC1 at least partially through ATF4. However, ONC201 did not inhibit mTORC1 in ONC201-resistant JeKo-1 cells (fig. S14D).
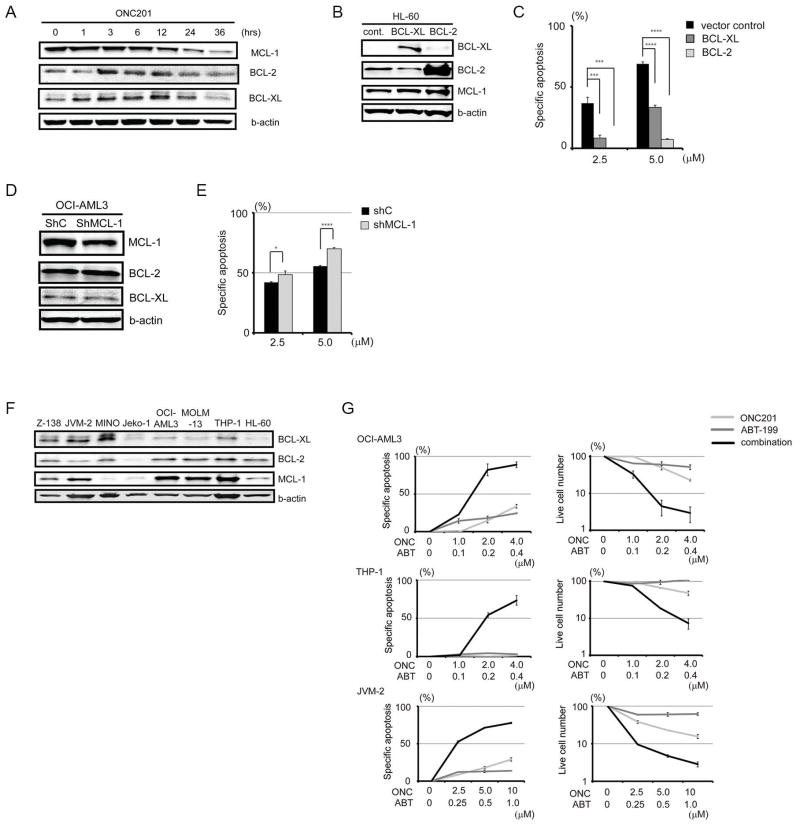
Dual targeting of MCL-1 and BCL-2, by ONC201 and ABT-199, is synergistic
Because ONC201 triggered apoptosis, we assessed the protein abundance of antiapoptotic BCL-2 family members (BCL-2, BCL-XL, and MCL-1) in JeKo-1 cells after ONC201 treatment. MCL-1 was reduced most notably after 12 hours (Fig. 7A and fig. S15). We then tested the effect of ONC201 on cells with overexpression or knockdown of these BCL-2 family proteins. ONC201 efficacy was reduced in BCL-2–overexpressing HL-60 cells (Fig. 7, B and C), more so than in BCL-XL–overexpressing HL-60 cells, supporting the notion that BCL-2 is a protective factor for cells under ONC201-induced stress. MCL-1 knockdown in OCI-AML3 cells increased their sensitivity to ONC201 only slightly (Fig. 7, D and E). Therefore, we investigated whether ONC201 sensitivity could be increased by ABT-199, a small-molecule BH3 mimetic that specifically inhibits BCL-2 and that is ineffective in cell lines with MCL-1 overexpression (59). The combination of ONC201 and ABT-199 in AML lines OCI-AML3 and THP-1, and in the MCL line JVM-2 [which has the highest MCL-1 abundance among the MCL lines examined (Fig. 7F) and is resistant to ABT-199], was substantially more toxic than was either drug alone (Fig. 7G). This synergy was reflected in the low combination indices (table S3).
Fig. 7. Effects of antiapoptotic BCL-2 family proteins on ONC201-induced apoptosis.
(A) Representative immunoblots (from three experiments) of BCL-XL, BCL-2, and MCL-1 in JeKo-1 cells during treatment with ONC201 (5 μM). (B) Immunoblots of BCL-XL, BCL-2, and MCL-1 in BCL-XL– or BCL-2–overexpressing HL-60 cells. HL-60 cells transfected with vector control were used as control. (C) Specific apoptosis of BCL-2– or BCL-XL–overexpressing HL-60 cells after a 72-hour ONC201 treatment (2.5 and 5 μM) (n = 3 experiments). (D) Immunoblots of BCL-XL, BCL-2, and MCL-1 after MCL-1 knockdown in OCI-AML3 cells. (E) Specific apoptosis after MCL-1 knockdown in OCI-AML3 cells after a 72-hour ONC201 treatment (2.5 and 5 μM) (n = 3 experiments). (F) Representative immunoblots (from three experiments) of BCL-XL, BCL-2, and MCL-1 in MCL or AML cell lines. (G) Synergistic effects on specific apoptosis of OCI-AML3, THP-1, and JVM-2 cells by combination of ABT-199 and ONC201 (n = 3 experiments). *P < 0.05, ***P < 0.001, ****P < 0.0001.
DISCUSSION
ONC201 was initially reported to transcriptionally induce the abundance of TRAIL and its receptor DR5 in a FOXO3a-dependent manner in solid tumors (12). However, in hematological malignancies, we found that ONC201 did not induce TRAIL, that DR5 mRNA was only modestly induced, and that apoptosis induction did not depend on either caspase-8 (part of the extrinsic pathway of apoptosis) or FOXO3a. Instead, our investigations implicated an atypical ISR and secondary mTOR pathway inhibition as prominent mechanisms induced by ONC201. These mechanisms likely explain our preclinical findings that ONC201 was effective against MCL and AML cells despite several abnormalities that commonly confer resistance and/or poor outcomes to standard chemotherapy in these cancers: p53 mutation, complex karyotype, the small-molecule efflux protein Pgp, and resistance to other agents including ibrutinib. Along with the ability of ONC201 to deplete AML stem and progenitor cells while relatively sparing normal hematopoietic precursors and the absence of genotoxicity, these findings support the potential of ONC201 to serve as a robust antitumor agent in hematological malignancies, particularly in aggressive and resistant disease that cannot be successfully treated with conventional therapy.
The term “integrated stress response” was introduced by Harding et al. (26) to describe the response to nutrient deprivation and oxidative stress. Because these stresses, like protein misfolding, which causes the UPR, can produce cell death if prolonged, it is generally believed that the UPR and ISR are adaptive, compensatory mechanisms that help cells to survive. ATF4 up-regulation is a common aspect of the UPR and ISR and can protect cells by promoting transcription of genes that encode proteins involved in protein folding or amino acid metabolism (for example, glutamine transporters, SLC7A5, SLC7A11, and asparaginyl-tRNA synthetase) and by inhibition of oxidative stress (through interaction with the oxidative stress–activated transcription factor NRF-2, which activates glutathione metabolism) (23, 24, 26, 60).
The response to ONC201 differs from the classical UPR and ISR in several molecular features (table S2), the most important of which is that the ATF4-dependent stress response induced by ONC201 appeared to be causal of its toxicity rather than a protective response. Knockdown of ATF4 reduced ONC201-induced toxicity and up-regulation of DDIT4, a transcriptional target of ATF4 (58) that is induced in response to hypoxia, oxidative stress, or ER stress. DDIT4 is likely the direct cause of mTORC1 inhibition by ONC201, by inducing the expression of the gene encoding tuberous sclerosis complex 2 (57). ONC201 cytotoxicity also appeared to be mediated by ATF4 because CHOP knockdown increased both sensitivity to ONC201 and ATF4 induction by ONC201 in OCI-AML3 and JeKo-1 cells. ATF4 can cause cell death in certain conditions, especially when stress exposure is prolonged, usually because of transcriptional induction of CHOP (24, 60).
Another feature of the response to ONC201 that was discordant with classical UPR or ISR was that ATF4 induction appeared to be independent of an increase in eIF2α phosphorylation. ATF4 can be regulated through transcriptional, posttranscriptional (translational), and posttranslational means, and in our study, ONC201 induced a time-dependent increase in ATF4 mRNA. However, increased ATF4 protein abundance preceded increased expression of ATF4 mRNA in response to ONC201, indicating a posttranscriptional mechanism. Similar to mechanisms triggered by the UPR and ISR, ONC201 enhanced ATF4 translation through a mechanism involving μORFs of the ATF4 gene (50–52), but differs in that an increase in eIF2α phosphorylation appeared to not be involved. Evidence for this conclusion comes largely from MCL lines, in which ONC201 increased ATF4 abundance in the absence of increased eIF2α phosphorylation, although both were increased by UPR inducers. However, in AML lines in which eIF2α phosphorylation was increased by ONC201, eIF2α knockdown increased ONC201-induced apoptosis without affecting ATF4 induction, which we interpret as further evidence that an increase in eIF2α phosphorylation was not involved in ATF4 induction by ONC201. Knockdown (or knockout) of eIF2α with reconstitution of wild-type and S51A eIF2α would clarify the role of phosphorylation of eIF2α in ATF4 induction by ONC201.
How ONC201 induces ATF4 independently of eIF2α phosphorylation is not clear, but its mechanism of ATF4 induction might be more compatible with those of the UPR and ISR if eIF2α phosphorylation is not the direct cause of ATF4 induction. Such a possibility is provided by a work suggesting that ATF4 can also be induced without an increase in eIF2α phosphorylation through the inhibition of eIF2B, a guanine nucleotide exchange factor (GEF) (Fig. 8) (47–49). In the UPR and typical ISR, phosphorylation of eIF2α causes it to bind to eIF2B, inhibiting the GEF activity of eIF2B and consequently protein translation in general while up-regulating ATF4 (42, 61). We speculate that ONC201 may cause eIF2α phosphorylation–independent inhibition of eIF2B. In support of this possibility, the ISR to amino acid deprivation does not always result in phosphorylation of eIF2α but usually inhibits eIF2B (47). Another possibility is that translational ATF4 induction after ONC201 may not be completely independent of eIF2α phosphorylation, but simply does not require its increase. The phosphorylation of eIF2α was increased in AML cells after ONC201 and was still present after eIF2α knockdown, which did not affect ATF4 induction; also, phosphorylated eIF2α was abundant in MCL cells before and after ONC201 treatment. Therefore, we also speculate that ONC201 treatment may result in inactivation of an unknown factor that “uncouples” eIF2α phosphorylation and ATF4 induction, at least in MCL cells, allowing ATF4 induction to proceed. In addition, because ATF4 can be posttranslationally regulated by the F-box protein βTrCP, the receptor component of the SCF E3 ubiquitin ligase (62), ONC201 may act to inhibit ubiquitin-proteasome pathway–mediated degradation of ATF4. Finally, ATF4 can be posttranslationally stabilized by anoxia (63).
Fig. 8. Schematic of atypical ISR induced by ONC201 leading to ATF4-mediated apoptosis, and comparison to other causes of ATF4 induction.

ONC201 appears to up-regulate ATF4 translation through a mechanism involving μORFs of the ATF4 gene that partially resembles that used by the classical ISR and UPR. However, ONC201-induced ATF4 up-regulation does not require an increase in the phosphorylation of eIF2a, indicating a mechanism of action of ONC201 that indirectly activates this pathway. ONC201-induced ATF4 serves as an apoptosis-promoting factor that also inhibits mTORC1 activity. ONC201 also reduces MCL-1 protein abundance, causing synergistic effects with the BCL-2 inhibitor ABT-199. AA, amino acids; dsRNA, double-stranded RNA.
Because Kline et al. (64) also found that ATF4 activation and mTORC1 inhibition were involved in ONC201 effects on solid tumors, ATF4 may be a common driver of ONC201-induced apoptosis and might serve as a pharmacodynamic marker of response to ONC201 in solid and hematopoietic tumors. The mechanism of ATF4 induction by ONC201 found by Kline et al. (64) differed from our results in that inhibition of eIF2α phosphorylation reduced ATF4 induction by ONC201. We acknowledge that this difference might be due to the different experimental strategies used (eIF2α knockdown compared to eIF2α S51A overexpression). However, the effects of ONC201 differed between solid tumors and hematopoietic malignancies with respect to whether TRAIL and DR5 were induced: therefore, it is not surprising that they differed in how eIF2α phosphorylation was involved in ATF4 induction in response to ONC201.
Of clinical importance, we found that BCL-2 overexpression inhibited ONC201-induced apoptosis, consistent with reports that BCL-2 is a major resistance factor to ER stress–induced apoptosis (45, 65, 66). The combination of ONC201 with ABT-199 exerted synergistic effects; whereas ABT-199 is a specific BCL-2 inhibitor, ONC201 reduces protein abundance of MCL-1, against which ABT-199 is ineffective. ABT-199 is a clinically promising drug especially in CLL (67) and AML (68), but resistance to ABT-199 has been observed, in part related to MCL-1 in both preclinical (59) and clinical studies (68). The combination of ABT-199 and ONC201 could potentially achieve greater initial tumor reduction and prevent or delay the development of resistance.
Proteasome inhibitors cause ER stress, and the present study suggests the potential clinical activity of ONC201 and other inducers of the UPR or ISR in hematological malignancies. The induction of an ISR reported here is also operational in solid tumors [see Kline et al. (64)]. Ongoing phase 1/2 clinical trials of ONC201 (NCT02038699) are enrolling patients with advanced solid tumors, and phase 1/2 studies in AML and lymphomas have opened at MD Anderson Cancer Center (NCT02392572), providing the opportunity to corroborate the mechanism of action and other effects identified here in translational studies.
MATERIALS AND METHODS
Reagents
Pharmaceutical-grade ONC201 was manufactured and provided by Oncoceutics Inc. The BCL-2 inhibitor ABT-199 was purchased from Selleckchem. Nutlin-3a was from Cayman Chemical Company, PSC-833 was from Santa Cruz Biotechnology, Z-IETD-FMK was from EMD Millipore, Z-VAD-FMK was from Enzo Life Science, recombinant human TRAIL was from Life Technologies, and doxorubicin was purchased from Bedford Laboratories.
Cell culture and treatment
Cell lines were purchased from Leibniz-Institut Deutsche Sammlung von Mikroorganismen und Zellkulturen or the American Type Culture Collection. The authenticity of the cell lines was confirmed by DNA fingerprinting with the short tandem repeat method, using a PowerPlex 16 HS System (Promega) within 6 months before the experiments. The cells were cultured in RPMI 1640 medium containing 20% fetal bovine serum for MCL lines (Z-138, JVM-2, MINO, and JeKo-1) and primary MCL samples, or 10% fetal bovine serum for AML lines (OCI-AML3, MOLM-13, HL-60, and THP-1), Jurkat cells (parental and I9.2), and primary AML samples. Glucose-free RPMI (11879-020, purchased from Thermo Fisher Scientific) or RPMI lacking three essential amino acids (L-arginine, L-glutamine, L-lysine; purchased from Thermo Fisher Scientific) was used for glucose/amino acid deprivation assay. Heparinized peripheral blood and pleural effusion samples were obtained from MCL and AML patients after informed consent, according to the University of Texas MD Anderson Cancer Center guidelines in accordance with the Declaration of Helsinki. Mononuclear cells were purified by density gradient centrifugation. All of the experiments were performed at least in triplicate. For 72-hour drug treatment experiments, cell lines were harvested at log-phase growth, plated at a density of 1.5 × 105/ml for Z-138, MINO, JeKo-1, OCI-AML3, and MOLM-13, 2.0 × 105/ml for JVM-2, HL-60, and THP-1, or 1.0 × 106/ml for primary AML and MCL cells, and treated with the indicated compounds. shRNA-mediated stable knockdown of target genes was performed by lentiviral infection as previously reported (69). shRNAs specifically targeting TP53, FoxO3a, eIF2α, DDIT3, IRE1α, ATF4, and MCL-1 were obtained from The RNAi Consortium (TRC) arrayed human genome-wide shRNA collection (purchased from Sigma-Aldrich; see also table S4). Transformed cells were selected by puromycin treatment, and knockdown was confirmed by immunoblot analysis of target proteins. HL-60 cells with stable overexpression of either BCL-XL or BCL-2, or with an empty vector, were provided by K. N. Bhalla (The University of Texas MD Anderson Cancer Center). Bortezomib-naïve and bortezomib-resistant ANBL-6 cells were provided by H. C. Lee and R. Orlowski.
Reporter assay with an ATF4 μORF construct
The “ATF4 5: 5′ATF4.GFP” plasmid (#21852) was purchased from Addgene (52). MEFs were transfected with the ATF4-GFP–expressing plasmid using jetPRIME transfection reagents (Polyplus-transfection) and, 24 hours later, were pretreated with 5 μM ONC201 for 24 hours and 1 μM thapsigargin for 1 and 6 hours. DAPI-negative and GFP-positive cells were measured by flow cytometry.
Flow cytometric analysis
For apoptosis analysis, annexin V and PI (purchased from Sigma-Aldrich) binding assays were performed to assess apoptosis as described previously (70). Annexin V– and PI-negative cells were counted as live cells. Apoptosis (A) in a sample was quantified as the proportion of cells that were annexin V–positive cells, and specific apoptosis was calculated by the following formula: % specific apoptosis = (Atest - Acontrol) × 100/(100 - Acontrol). Thus, by subtracting spontaneous apoptosis (= apoptosis observed in untreated control samples), “specific apoptosis” in the present study indicates specifically induced apoptosis by ONC201. For gating stem/progenitor population (CD45+CD34+CD38− cells) in primary AML samples, phycoerythrin (PE)–conjugated anti-human CD45, PE-Cyanine7 (PE-Cy7)–conjugated anti-human CD34, and fluorescein isothiocyanate–conjugated CD38 antibodies were used. Allophycocyanin-conjugated annexin V (purchased from BD Biosciences) and DAPI were used in this setting to measure apoptosis. For cell cycle analysis, DNA content analysis by staining with PI was performed as reported previously (71). EdU incorporation was measured by using Click-iT EdU Alexa Fluor 647 Flow Cytometry Assay Kit (Life Technologies). γH2A.X staining was performed by following the manufacturer’s instruction. Briefly, cells were fixed with 2% paraformaldehyde at room temperature for 5 min and permeabilized by adding ice-cold 90% methanol while gently vortexing. After 30 min of incubation on ice, cells were stained with the Alexa Fluor 488–conjugated rabbit monoclonal anti–phospho–histone H2A.X (Ser139) (20E3) antibody (1:50 dilution; purchased from Cell Signaling) for 30 min.
In vivo engraftment experiment
Primary AML cells were serially transplanted in female NSG mice (6 weeks old; Jackson Laboratory) to confirm self-renewal ability, and leukemia cells were harvested from secondarily transplanted mice. Leukemic cells were cultured for 48 hours, with or without 5 μM ONC201, then washed and assessed for viability by trypan blue exclusion; 0.7 million trypan blue–negative cells were then injected via tail vein into each of 13 6-week-old female NSG mice per group. At 4 weeks after transplantation, human CD45-positive and DAPI-negative cells in peripheral blood were counted by flow cytometry. Three mice in each group were sacrificed to collect BM, spleens, and livers at 4 weeks after transplantation, and then human CD45-positive and DAPI-negative cells in the BM were counted by flow cytometry. The in vivo mouse study was performed following the guidelines approved by the Institutional Animal Care and Use Committees at MD Anderson Cancer Center.
Immunoblot analysis
Immunoblot analysis was performed as reported previously (70) and quantitated using the Odyssey Imaging System (LI-COR Biotechnology). Densitometry data with mean error were provided by quantifying blots from three independent immunoblot experiments. The signals for phosphorylated form of proteins were normalized to those for the total proteins; otherwise, they were normalized to those of β-actin. Protease/phosphatase inhibitor cocktail was used (purchased from Cell Signaling Technology). The antibodies used for the studies are described in table S5.
Gene sequencing and expression
Mutation analysis of TP53 was performed as described previously (69). mRNA expression was quantified as reported previously (72), using TaqMan gene expression assays (listed in table S6) as previously reported (73).
Gene expression profiling
Total RNA was extracted from MCL lines by TRIzol (Invitrogen) at various times after ONC201 treatment. Subsequent RNA cleanup, quality assessment, generation of biotinylated aRNA for hybridization on Illumina HT-12 BeadArrays (Illumina Inc.), and data processing were as previously described (74). Data from microarray studies are submitted to Gene Expression Omnibus as GSE68091. After log2 transformation, genes expressed above background in at least 25% of samples were used for heat mapping and correlation analysis. From the time course of JeKo-1 cells treated with ONC201, all values for each gene probe were correlated with time of exposure, yielding for each gene a slope of the regression line and a Pearson’s r value. Up and down genes were identified with high absolute values from the extremes of the plot of slope versus r. GSEA (75) was performed on the ranked list of probe-averaged gene values for slope or r, using gene sets in the C2, C3, C5, and C6 categories in the MSigDB web service release version 3.84.
Statistical analyses
Statistical analyses were performed using the two-tailed Student’s t test or Mann-Whitney test by the Prism (version 6.0; GraphPad Software) statistical software programs. The Kaplan-Meier method was used to generate survival curves, and log-rank test was used for comparison of the two groups. P values less than 0.05 were considered statistically significant (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). Unless otherwise indicated, values are expressed as the mean ± SD calculated by performing three independent experiments.
Supplementary Material
Acknowledgments
We thank N. Hail for editorial assistance, K. N. Bhalla for providing HL-60 cells stably expressing BCL-XL or BCL-2, and H. C. Lee and R. Orlowski for providing the bortezomib-naïve and bortezomib-resistant ANBL-6 cells.
Funding: This work was supported in part by grants from the Japan Society for the Promotion of Science Postdoctoral Fellowship for Research Abroad Award (to J.I.); the Ministry of Education, Culture, Sports, Science and Technology in Japan (26461425), the Princess Takamatsu Cancer Research Fund (14-24610), and the Osaka Cancer Research Foundation (to K.K.); the Cancer Prevention Research Institute of Texas (RP130397 to P.L.L.); and the NIH (CA49639, CA100632, CA136411, and CA16672) and the Paul and Mary Haas Chair in Genetics (to M.A.).
Footnotes
Competing interests: J.E.A. is an employee and stockholder of Oncoceutics, the manufacturer of ONC201; M.A. is a stockholder of Oncoceutics and serves on its board and scientific advisory board. The other authors declare that they have no competing interests.
Data and materials availability: ONC201 requires a materials transfer agreement from Oncoceutics. Data from microarray studies have been submitted to Gene Expression Omnibus (GSE68091).
www.sciencesignaling.org/cgi/content/full/9/415/ra17/dc1
Fig. S1. Cell cycle changes and DNA damage induced by ONC201.
Fig. S2. Effect of ONC201 on primary MCL and AML cells and normal BM cells.
Fig. S3. Efficacy of ONC201 in bortezomib-resistant MM cells and combination therapy with ibrutinib in primary MCL cells.
Fig. S4. Apoptosis induced by ONC201 in caspase-8– or Bax/Bak-deficient cells.
Fig. S5. Changes in the AKT/ERK/FOXO3a pathway with ONC201 treatment.
Fig. S6. Immunoblot and PCR analysis of ER stress–related molecules.
Fig. S7. Quantification of the immunoblots in Fig. 5 (A and B).
Fig. S8. Effects of ER stress inducers on GCN2, eIF2α, ATF4, and mTORC1.
Fig. S9. Immunoblot analysis of ISR-related molecules after amino acid or glucose deprivation in OCI-AML3 and JeKo-1 cells.
Fig. S10. ATF4 induction by ONC201 or tunicamycin in OCI-AML3 cells with eIF2α knockdown.
Fig. S11. ATF4 expression over time in response to ONC201 in OCI-AML3 and JeKo-1 cells.
Fig. S12. Effects of CHOP knockdown on ONC201-induced apoptosis.
Fig. S13. Effects of ONC201 on IRE1α.
Fig. S14. The role of ATF4 in mTORC1 inhibition by ONC201.
Fig. S15. Quantification of the immunoblots in Fig. 7A.
Table S1. Clinical information for the primary AML samples.
Table S2. Summary of changes in the GCN2/eIF2α/ATF4 axis and mTORC1 targets by tunicamycin, amino acid or glucose deprivation, and ONC201 in AML and MCL cells.
Table S3. Combination index of each cell line treated with ONC201 and ABT-199.
Table S4. shRNA sequences used.
Table S5. Antibodies for immunoblot analysis.
Table S6. Primers used for PCR.
Data file S1. Genes with most positive slope and r in Fig. 4B.
Data file S2. Genes with most negative slope and r in Fig. 4B.
Data file S3. Common up-regulated genes in ONC201-sensitive MCL cell lines.
Author contributions: J.I., K.K., R.E.D., and M.A. conceived and designed the study. J.I., D.C., R.E.D., and L.H. acquired the data. J.I., K.K., Z.Z., Z.W., W.M., P.L.L., X.H., R.E.D., J.E.A., M.K., and M.A. analyzed and interpreted the data. J.I., K.K., J.E.A., R.E.D., and M.A. wrote, reviewed, and/or revised the manuscript. P.R., V.R., R.O.J., G.B., H.M., Z.Z., Y.T., D.D.S., S.S.N., T.M., R.N.M., M.W., H.K., and R.O. provided administrative, technical, or material support.
REFERENCES AND NOTES
- 1.The Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krug U, Ganser A, Koeffler HP. Tumor suppressor genes in normal and malignant hematopoiesis. Oncogene. 2002;21:3475–3495. doi: 10.1038/sj.onc.1205322. [DOI] [PubMed] [Google Scholar]
- 3.Rücker FG, Schlenk RF, Bullinger L, Kayser S, Teleanu V, Kett H, Habdank M, Kugler C-M, Holzmann K, Gaidzik VI, Paschka P, Held G, von Lilienfeld-Toal M, Lübbert M, Fröhling S, Zenz T, Krauter J, Schlegelberger B, Ganser A, Lichter P, Döhner K, Döhner H. TP53 alterations in acute myeloid leukemia with complex karyotype correlate with specific copy number alterations, monosomal karyotype, and dismal outcome. Blood. 2012;119:2114–2121. doi: 10.1182/blood-2011-08-375758. [DOI] [PubMed] [Google Scholar]
- 4.Rossi D, Khiabanian H, Spina V, Ciardullo C, Bruscaggin A, Famà R, Rasi S, Monti S, Deambrogi C, De Paoli L, Wang J, Gattei V, Guarini A, Foà R, Rabadan R, Gaidano G. Clinical impact of small TP53 mutated subclones in chronic lymphocytic leukemia. Blood. 2014;123:2139–2147. doi: 10.1182/blood-2013-11-539726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheung KJ, Horsman DE, Gascoyne RD. The significance of TP53 in lymphoid malignancies: Mutation prevalence, regulation, prognostic impact and potential as a therapeutic target. Br J Haematol. 2009;146:257–269. doi: 10.1111/j.1365-2141.2009.07739.x. [DOI] [PubMed] [Google Scholar]
- 6.Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, Wheatley K, Harrison CJ, Burnett AK National Cancer Research Institute Adult Leukaemia Working Group. Refinement of cytogenetic classification in acute myeloid leukemia: Determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116:354–365. doi: 10.1182/blood-2009-11-254441. [DOI] [PubMed] [Google Scholar]
- 7.Grossmann V, Schnittger S, Kohlmann A, Eder C, Roller A, Dicker F, Schmid C, Wendtner CM, Staib P, Serve H, Kreuzer KA, Kern W, Haferlach T, Haferlach C. A novel hierarchical prognostic model of AML solely based on molecular mutations. Blood. 2012;120:2963–2972. doi: 10.1182/blood-2012-03-419622. [DOI] [PubMed] [Google Scholar]
- 8.Halldórsdóttir AM, Lundin A, Murray F, Mansouri L, Knuutila S, Sundström C, Laurell A, Ehrencrona H, Sander B, Rosenquist R. Impact of TP53 mutation and 17p deletion in mantle cell lymphoma. Leukemia. 2011;25:1904–1908. doi: 10.1038/leu.2011.162. [DOI] [PubMed] [Google Scholar]
- 9.Hernandez L, Fest T, Cazorla M, Teruya-Feldstein J, Bosch F, Peinado MA, Piris MA, Montserrat E, Cardesa A, Jaffe ES, Campo E, Raffeld M. p53 gene mutations and protein overexpression are associated with aggressive variants of mantle cell lymphomas. Blood. 1996;87:3351–3359. [PubMed] [Google Scholar]
- 10.Rubio-Moscardo F, Climent J, Siebert R, Piris MA, Martín-Subero JI, Nieländer I, Garcia-Conde J, Dyer MJS, Terol MJ, Pinkel D, Martinez-Climent JA. Mantle-cell lymphoma genotypes identified with CGH to BAC microarrays define a leukemic subgroup of disease and predict patient outcome. Blood. 2005;105:4445–4454. doi: 10.1182/blood-2004-10-3907. [DOI] [PubMed] [Google Scholar]
- 11.Allen JE, Krigsfeld G, Patel L, Mayes PA, Dicker DT, Wu GS, El-Deiry WS. Identification of TRAIL-inducing compounds highlights small molecule ONC201/TIC10 as a unique anti-cancer agent that activates the TRAIL pathway. Mol Cancer. 2015;14:99. doi: 10.1186/s12943-015-0346-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allen JE, Krigsfeld G, Mayes PA, Patel L, Dicker DT, Patel AS, Dolloff NG, Messaris E, Scata KA, Wang W, Zhou J-Y, Wu GS, El-Deiry WS. Dual inactivation of Akt and ERK by TIC10 signals Foxo3a nuclear translocation, TRAIL gene induction, and potent antitumor effects. Sci Transl Med. 2013;5:171ra117. doi: 10.1126/scitranslmed.3004828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allen JE, Prabhu VV, Talekar M, van den Heuvel APJ, Lim B, Dicker DT, Fritz JL, Beck A, El-Deiry WS. Genetic and pharmacological screens converge in identifying FLIP, BCL2 and IAP proteins as key regulators of sensitivity to the TRAIL-inducing anti-cancer agent ONC201/TIC10. Cancer Res. 2015;75:1668–1674. doi: 10.1158/0008-5472.CAN-14-2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prabhu VV, Allen JE, Dicker DT, El-Deiry WS. Small-molecule ONC201/TIC10 targets chemotherapy-resistant colorectal cancer stem–like cells in an Akt/Foxo3a/TRAIL–dependent manner. Cancer Res. 2015;75:1423–1432. doi: 10.1158/0008-5472.CAN-13-3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho MM, Hogge DE, Ling V. MDR1 and BCRP1 expression in leukemic progenitors correlates with chemotherapy response in acute myeloid leukemia. Exp Hematol. 2008;36:433–442. doi: 10.1016/j.exphem.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 16.Drach D, Zhao S, Drach J, Mahadevia R, Gattringer C, Huber H, Andreeff M. Subpopulations of normal peripheral blood and bone marrow cells express a functional multidrug resistant phenotype. Blood. 1992;80:2729–2734. [PubMed] [Google Scholar]
- 17.Obeng EA, Carlson LM, Gutman DM, Harrington WJ, Jr, Lee KP, Boise LH. Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells. Blood. 2006;107:4907–4916. doi: 10.1182/blood-2005-08-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jain P, Keating M, Wierda W, Estrov Z, Ferrajoli A, Jain N, George B, James D, Kantarjian H, Burger J, O’Brien S. Outcomes of patients with chronic lymphocytic leukemia after discontinuing ibrutinib. Blood. 2015;125:2062–2067. doi: 10.1182/blood-2014-09-603670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pereira NA, Song Z. Some commonly used caspase substrates and inhibitors lack the specificity required to monitor individual caspase activity. Biochem Biophys Res Commun. 2008;377:873–877. doi: 10.1016/j.bbrc.2008.10.101. [DOI] [PubMed] [Google Scholar]
- 20.Ma W, Yang D, Gu Y, Guo X, Zhao W, Guo Z. Finding disease-specific coordinated functions by multi-function genes: Insight into the coordination mechanisms in diseases. Genomics. 2009;94:94–100. doi: 10.1016/j.ygeno.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 21.Krige D, Needham LA, Bawden LJ, Flores N, Farmer H, Miles LEC, Stone E, Callaghan J, Chandler S, Clark VL, Kirwin-Jones P, Legris V, Owen J, Patel T, Wood S, Box G, Laber D, Odedra R, Wright A, Wood LM, Eccles SA, Bone EA, Ayscough A, Drummond AH. CHR-2797: An antiproliferative aminopeptidase inhibitor that leads to amino acid deprivation in human leukemic cells. Cancer Res. 2008;68:6669–6679. doi: 10.1158/0008-5472.CAN-07-6627. [DOI] [PubMed] [Google Scholar]
- 22.Janes MR, Limon JJ, So L, Chen J, Lim RJ, Chavez MA, Vu C, Lilly MB, Mallya S, Ong ST, Konopleva M, Martin MB, Ren P, Liu Y, Rommel C, Fruman DA. Effective and selective targeting of leukemia cells using a TORC1/2 kinase inhibitor. Nat Med. 2010;16:205–213. doi: 10.1038/nm.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chevet E, Hetz C, Samali A. Endoplasmic reticulum stress–activated cell reprogramming in oncogenesis. Cancer Discov. 2015;5:586–597. doi: 10.1158/2159-8290.CD-14-1490. [DOI] [PubMed] [Google Scholar]
- 24.Clarke HJ, Chambers JE, Liniker E, Marciniak SJ. Endoplasmic reticulum stress in malignancy. Cancer Cell. 2014;25:563–573. doi: 10.1016/j.ccr.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 25.Wang WA, Groenendyk J, Michalak M. Endoplasmic reticulum stress associated responses in cancer. Biochim Biophys Acta. 2014;1843:2143–2149. doi: 10.1016/j.bbamcr.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 26.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, Ron D. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 27.Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Appenzeller-Herzog C, Hall MN. Bidirectional crosstalk between endoplasmic reticulum stress and mTOR signaling. Trends Cell Biol. 2012;22:274–282. doi: 10.1016/j.tcb.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 29.Goldfinger M, Shmuel M, Benhamron S, Tirosh B. Protein synthesis in plasma cells is regulated by crosstalk between endoplasmic reticulum stress and mTOR signaling. Eur J Immunol. 2011;41:491–502. doi: 10.1002/eji.201040677. [DOI] [PubMed] [Google Scholar]
- 30.Kato H, Nakajima S, Saito Y, Takahashi S, Katoh R, Kitamura M. mTORC1 serves ER stress-triggered apoptosis via selective activation of the IRE1-JNK pathway. Cell Death Differ. 2012;19:310–320. doi: 10.1038/cdd.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Høyer-Hansen M, Jäättelä M. Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium. Cell Death Differ. 2007;14:1576–1582. doi: 10.1038/sj.cdd.4402200. [DOI] [PubMed] [Google Scholar]
- 32.Jeon YJ, Khelifa S, Ratnikov B, Scott DA, Feng Y, Parisi F, Ruller C, Lau E, Kim H, Brill LM, Jiang T, Rimm DL, Cardiff RD, Mills GB, Smith JW, Osterman AL, Kluger Y, Ronai ZA. Regulation of glutamine carrier proteins by RNF5 determines breast cancer response to ER stress-inducing chemotherapies. Cancer Cell. 2015;27:354–369. doi: 10.1016/j.ccell.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bobrovnikova-Marjon E, Pytel D, Riese MJ, Vaites LP, Singh N, Koretzky GA, Witze ES, Diehl JA. PERK utilizes intrinsic lipid kinase activity to generate phosphatidic acid, mediate Akt activation, and promote adipocyte differentiation. Mol Cell Biol. 2012;32:2268–2278. doi: 10.1128/MCB.00063-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kazemi S, Mounir Z, Baltzis D, Raven JF, Wang S, Krishnamoorthy JL, Pluquet O, Pelletier J, Koromilas AE. A novel function of eIF2α kinases as inducers of the phosphoinositide-3 kinase signaling pathway. Mol Biol Cell. 2007;18:3635–3644. doi: 10.1091/mbc.E07-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koromilas AE, Mounir Z. Control of oncogenesis by eIF2α phosphorylation: Implications in PTEN and PI3K-Akt signaling and tumor treatment. Future Oncol. 2013;9:1005–1015. doi: 10.2217/fon.13.49. [DOI] [PubMed] [Google Scholar]
- 36.Mounir Z, Krishnamoorthy JL, Wang S, Papadopoulou B, Campbell S, Muller WJ, Hatzoglou M, Koromilas AE. Akt determines cell fate through inhibition of the PERK-eIF2α phosphorylation pathway. Sci Signal. 2011;4:ra62. doi: 10.1126/scisignal.2001630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rajesh K, Krishnamoorthy J, Kazimierczak U, Tenkerian C, Papadakis AI, Wang S, Huang S, Koromilas AE. Phosphorylation of the translation initiation factor eIF2α at serine 51 determines the cell fate decisions of Akt in response to oxidative stress. Cell Death Dis. 2015;6:e1591. doi: 10.1038/cddis.2014.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao F, Huang Z, Li H, Yu J, Wang C, Chen S, Meng Q, Cheng Y, Gao X, Li J, Liu Y, Guo F. Leucine deprivation increases hepatic insulin sensitivity via GCN2/mTOR/S6K1 and AMPK pathways. Diabetes. 2011;60:746–756. doi: 10.2337/db10-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chou S-D, Prince T, Gong J, Calderwood SK. mTOR is essential for the proteotoxic stress response, HSF1 activation and heat shock protein synthesis. PLOS One. 2012;7:e39679. doi: 10.1371/journal.pone.0039679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santagata S, Mendillo ML, Tang Y-c, Subramanian A, Perley CC, Roche SP, Wong B, Narayan R, Kwon H, Koeva M, Amon A, Golub TR, Porco JA, Jr, Whitesell L, Lindquist S. Tight coordination of protein translation and HSF1 activation supports the anabolic malignant state. Science. 2013;341:1238303. doi: 10.1126/science.1238303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamanaka RB, Bennett BS, Cullinan SB, Diehl JA. PERK and GCN2 contribute to eIF2α phosphorylation and cell cycle arrest after activation of the unfolded protein response pathway. Mol Biol Cell. 2005;16:5493–5501. doi: 10.1091/mbc.E05-03-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Donnelly N, Gorman AM, Gupta S, Samali A. The eIF2α kinases: Their structures and functions. Cell Mol Life Sci. 2013;70:3493–3511. doi: 10.1007/s00018-012-1252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R, Nagata K, Harding HP, Ron D. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004;18:3066–3077. doi: 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sano R, Reed JC. ER stress-induced cell death mechanisms. Biochim Biophys Acta. 2013;1833:3460–3470. doi: 10.1016/j.bbamcr.2013.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tabas I, Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol. 2011;13:184–190. doi: 10.1038/ncb0311-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X, Proud CG. A novel mechanism for the control of translation initiation by amino acids, mediated by phosphorylation of eukaryotic initiation factor 2B. Mol Cell Biol. 2008;28:1429–1442. doi: 10.1128/MCB.01512-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woo CW, Kutzler L, Kimball SR, Tabas I. Toll-like receptor activation suppresses ER stress factor CHOP and translation inhibition through activation of eIF2B. Nat Cell Biol. 2012;14:192–200. doi: 10.1038/ncb2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lehman SL, Ryeom S, Koumenis C. Signaling through alternative Integrated Stress Response pathways compensates for GCN2 loss in a mouse model of soft tissue sarcoma. Sci Rep. 2015;5:11781. doi: 10.1038/srep11781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 51.Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci USA. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu PD, Harding HP, Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol. 2004;167:27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han D, Lerner AG, Vande Walle L, Upton JP, Xu W, Hagen A, Backes BJ, Oakes SA, Papa FR. IRE1α kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138:562–575. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leung-Hagesteijn C, Erdmann N, Cheung G, Keats JJ, Stewart AK, Reece DE, Chung KC, Tiedemann RE. Xbp1s-negative tumor B cells and pre-plasmablasts mediate therapeutic proteasome inhibitor resistance in multiple myeloma. Cancer Cell. 2013;24:289–304. doi: 10.1016/j.ccr.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc Natl Acad Sci USA. 2008;105:17414–17419. doi: 10.1073/pnas.0809136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG., Jr Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18:2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DeYoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW. Hypoxia regulates TSC1/2–mTOR signaling and tumor suppression through REDD1-mediated 14–3–3 shuttling. Genes Dev. 2008;22:239–251. doi: 10.1101/gad.1617608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whitney ML, Jefferson LS, Kimball SR. ATF4 is necessary and sufficient for ER stress-induced upregulation of REDD1 expression. Biochem Biophys Res Commun. 2009;379:451–455. doi: 10.1016/j.bbrc.2008.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pan R, Hogdal LJ, Benito JM, Bucci D, Han L, Borthakur G, Cortes J, DeAngelo DJ, Debose L, Mu H, Döhner H, Gaidzik VI, Galinsky I, Golfman LS, Haferlach T, Harutyunyan KG, Hu J, Leverson JD, Marcucci G, Müschen M, Newman R, Park E, Ruvolo PP, Ruvolo V, Ryan J, Schindela S, Zweidler-McKay P, Stone RM, Kantarjian H, Andreeff M, Konopleva M, Letai AG. Selective BCL-2 inhibition by ABT-199 causes on-target cell death in acute myeloid leukemia. Cancer Discov. 2014;4:362–375. doi: 10.1158/2159-8290.CD-13-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ye J, Koumenis C. ATF4, an ER stress and hypoxia-inducible transcription factor and its potential role in hypoxia tolerance and tumorigenesis. Curr Mol Med. 2009;9:411–416. doi: 10.2174/156652409788167096. [DOI] [PubMed] [Google Scholar]
- 61.Kilberg MS, Shan J, Su N. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol Metab. 2009;20:436–443. doi: 10.1016/j.tem.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lassot I, Ségéral E, Berlioz-Torrent C, Durand H, Groussin L, Hai T, Benarous R, Margottin-Goguet F. ATF4 degradation relies on a phosphorylation-dependent interaction with the SCFβTrCP ubiquitin ligase. Mol Cell Biol. 2001;21:2192–2202. doi: 10.1128/MCB.21.6.2192-2202.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ameri K, Lewis CE, Raida M, Sowter H, Hai T, Harris AL. Anoxic induction of ATF-4 through HIF-1-independent pathways of protein stabilization in human cancer cells. Blood. 2004;103:1876–1882. doi: 10.1182/blood-2003-06-1859. [DOI] [PubMed] [Google Scholar]
- 64.Kline CLB, van den Heuvel APJ, Allen JE, Prabhu VV, Dicker DT, El-Deiry WS. ONC201 kills solid tumor cells by triggering an integrated stress response dependent on ATF4 activation by specific eIF2α kinases. Sci Signal. 2016;9:ra18. doi: 10.1126/scisignal.aac4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.EH-Y, Cheng A, Wei MC, Weiler S, Flavell RA, Mak TW, Lindsten T, Korsmeyer SJ. BCL-2, BCL-XL sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol Cell. 2001;8:705–711. doi: 10.1016/s1097-2765(01)00320-3. [DOI] [PubMed] [Google Scholar]
- 66.Akl H, Vervloessem T, Kiviluoto S, Bittremieux M, Parys JB, De Smedt H, Bultynck G. A dual role for the anti-apoptotic Bcl-2 protein in cancer: Mitochondria versus endoplasmic reticulum. Biochim Biophys Acta. 2014;1843:2240–2252. doi: 10.1016/j.bbamcr.2014.04.017. [DOI] [PubMed] [Google Scholar]
- 67.Seymour JF, Davids MS, Pagel JM, Kahl BS, Wierda WG, Puvvada S, Gerecitano JF, Kipps TJ, Anderson MA, Huang DCS, Rudersdorf N, Gressick LA, Montalvo NP, Yang J, Zhu M, Dunbar M, Cerri E, Enschede SH, Humerickhouse R, Roberts AW. ABT-199 (GDC-0199) in relapsed/refractory (R/R) chronic lymphocytic leukemia (CLL) and small lymphocytic lymphoma (SLL): High complete response rate and durable disease control. J Clin Oncol. 2014;32:7015. [Google Scholar]
- 68.Konopleva M, Pollyea DA, Potluri J, Chyla BJ, Busman T, McKeegan E, Salem A, Zhu M, Ricker JL, Blum W, DiNardo CD, Dunbar M, Kirby R, Falotico N, Leverson JD, Humerickhouse RA, Mabry M, Stone RM, Kantarjian HM, Letai AG. A phase 2 study of ABT-199 (GDC-0199) in patients with acute myelogenous leukemia (AML) Blood. 2014;124:118. [Google Scholar]
- 69.Kojima K, Kornblau SM, Ruvolo V, Dilip A, Duvvuri S, Davis RE, Zhang M, Wang Z, Coombes KR, Zhang N, Qiu YH, Burks JK, Kantarjian H, Shacham S, Kauffman M, Andreeff M. Prognostic impact and targeting of CRM1 in acute myeloid leukemia. Blood. 2013;121:4166–4174. doi: 10.1182/blood-2012-08-447581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kojima K, Konopleva M, Samudio IJ, Shikami M, Cabreira-Hansen M, McQueen T, Ruvolo V, Tsao T, Zeng Z, Vassilev LT, Andreeff M. MDM2 antagonists induce p53-dependent apoptosis in AML: Implications for leukemia therapy. Blood. 2005;106:3150–3159. doi: 10.1182/blood-2005-02-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ishizawa J, Kuninaka S, Sugihara E, Naoe H, Kobayashi Y, Chiyoda T, Ueki A, Araki K, Yamamura K-i, Matsuzaki Y, Nakajima H, Ikeda Y, Okamoto S, Saya H. The cell cycle regulator Cdh1 controls the pool sizes of hematopoietic stem cells and mature lineage progenitors by protecting from genotoxic stress. Cancer Sci. 2011;102:967–974. doi: 10.1111/j.1349-7006.2011.01884.x. [DOI] [PubMed] [Google Scholar]
- 72.Yoshimura M, Ishizawa J, Ruvolo V, Dilip A, Quintás-Cardama A, McDonnell TJ, Neelapu SS, Kwak LW, Shacham S, Kauffman M, Tabe Y, Yokoo M, Kimura S, Andreeff M, Kojima K. Induction of p53-mediated transcription and apoptosis by exportin-1 (XPO1) inhibition in mantle cell lymphoma. Cancer Sci. 2014;105:795–801. doi: 10.1111/cas.12430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lai WL, Wong NS. The PERK/eIF2α signaling pathway of unfolded protein response is essential for N-(4-hydroxyphenyl)retinamide (4HPR)-induced cytotoxicity in cancer cells. Exp Cell Res. 2008;314:1667–1682. doi: 10.1016/j.yexcr.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 74.Ma W, Wang M, Wang ZQ, Sun L, Graber D, Matthews J, Champlin R, Yi Q, Orlowski RZ, Kwak LW, Weber DM, Thomas SK, Shah J, Kornblau S, Davis RE. Effect of long-term storage in TRIzol on microarray-based gene expression profiling. Cancer Epidemiol Biomarkers Prev. 2010;19:2445–2452. doi: 10.1158/1055-9965.EPI-10-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.