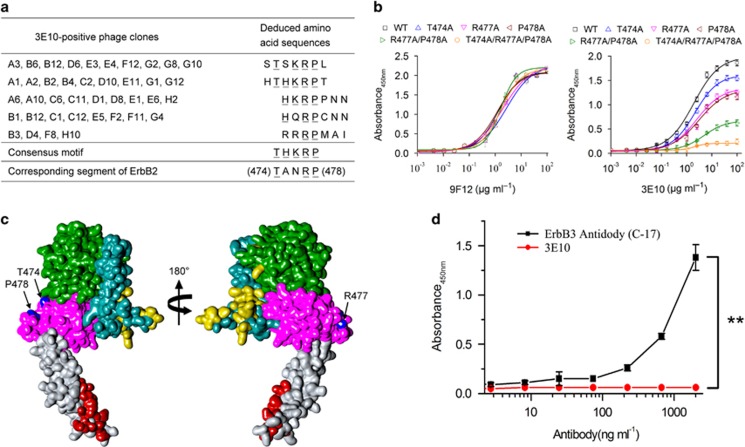
Figure 2.
Mapping of 3E10-specific epitope on the extracellular domain of ErbB2. (a) Amino-acid sequences of the insert from 3E10-positive phage clones. Sequences were aligned for the consensus motif, which is indicated by underlined letters. (b) Effect of alanine substitutions on 3E10 binding to ErbB2. Data are expressed as means±s.d. (c) Surface representation of the extracellular domain of ErbB2 (PDB accession code 1N8Z). Domains I, II, III and IV are green, dark cyan, magenta and gray, respectively. The ErbB2 residues within 5 Å of trastuzumab (PDB accession code 1N8Z) and pertuzumab (PDB accession code 1S78) are colored red and yellow, respectively. The 3E10 epitope residues, T474, R477 and P478, are colored blue. (d) 3E10 did not cross-react with ErbB3. Different concentrations of the 3E10 antibody were added to 96-well plates precoated with 3 μg/ml of recombinant human ErbB3/Her3 Fc chimera Protein (R&D Systems), followed by incubation at 37 °C for 1 h. After washing, horseradish peroxidase-labeled goat anti-mouse IgG H&L (Abcan) was added and the plates were further incubated for 1 h at 37 °C. Finally, 3,3′,5,5′-tetramethylbenzidine (TMB) was added as a substrate and the absorbance was read at 450 nm. As a positive control, different concentrations of horseradish peroxidase-labeled ErbB-3 antibody (C-17) (sc-285; Santa Cruz Biotechnology) were added to 96-well plates precoated with 3 μg/ml of recombinant human ErbB3/Her3 Fc chimera Protein, followed by incubation at 37 °C for 1 h. After washing, TMB was added as a substrate and the absorbance was read at 450 nm.