Phosphorylation of WRKYs by MAPK regulates an NADPH oxidase, demonstrating that the MAPK-WRKY pathway is required for the ROS burst in effector-triggered immunity via upregulation of RBOHB.
Abstract
Pathogen attack sequentially confers pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) after sensing of pathogen patterns and effectors by plant immune receptors, respectively. Reactive oxygen species (ROS) play pivotal roles in PTI and ETI as signaling molecules. Nicotiana benthamiana RBOHB, an NADPH oxidase, is responsible for both the transient PTI ROS burst and the robust ETI ROS burst. Here, we show that RBOHB transactivation mediated by MAPK contributes to R3a/AVR3a-triggered ETI (AVR3a-ETI) ROS burst. RBOHB is markedly induced during the ETI and INF1-triggered PTI (INF1-PTI), but not flg22-tiggered PTI (flg22-PTI). We found that the RBOHB promoter contains a functional W-box in the R3a/AVR3a and INF1 signal-responsive cis-element. Ectopic expression of four phospho-mimicking mutants of WRKY transcription factors, which are MAPK substrates, induced RBOHB, and yeast one-hybrid analysis indicated that these mutants bind to the cis-element. Chromatin immunoprecipitation assays indicated direct binding of the WRKY to the cis-element in plants. Silencing of multiple WRKY genes compromised the upregulation of RBOHB, resulting in impairment of AVR3a-ETI and INF1-PTI ROS bursts, but not the flg22-PTI ROS burst. These results suggest that the MAPK-WRKY pathway is required for AVR3a-ETI and INF1-PTI ROS bursts by activation of RBOHB.
INTRODUCTION
Plants have specific defense mechanisms that are activated after sensing pathogens, initially on the cell surface. This perception depends on pattern recognition receptors (PRRs) that recognize pathogen-associated molecular patterns (PAMPs) derived from pathogens and activate first level immunity referred to as pattern-triggered immunity (PTI) (Jones and Dangl, 2006). Plants have developed resistance (R) proteins that recognize pathogen effector molecules as weapons to compromise PTI, dealing with infection strategies evolved by pathogens, and activate second level immunity referred to as effector-triggered immunity (ETI) (Jones and Dangl, 2006). Both plant immunities share common signaling components, such as Ca2+, reactive oxygen species (ROS), and mitogen-activated protein kinase (MAPK) cascades, and lead to robust transcriptional reprogramming and production of antimicrobial metabolites (Tsuda and Katagiri, 2010). In many cases, ETI induces hypersensitive response (HR) accompanied by a localized cell death at the infection sites (Coll et al., 2011).
Respiratory burst oxidase homolog (RBOH), which is a plant NADPH oxidase located on the plasma membrane (Kobayashi et al., 2006), has a pivotal role in ROS production during biotic and abiotic stresses (Suzuki et al., 2011). RBOH-dependent ROS function as key signaling factors for local and systemic resistances against plant pathogens (Park et al., 1998; Miller et al., 2009; Mersmann et al., 2010). In Arabidopsis thaliana, RBOHD and RBOHF are capable of generating ROS in response to pathogen attacks (Torres et al., 2002) and PAMPs (Zhang et al., 2007). In Nicotiana benthamiana, RBOHA and RBOHB, the orthologs of Arabidopsis RBOHF and RBOHD, respectively, are essential for ROS production against oomycete pathogen Phytophthora infestans (Yoshioka et al., 2003). ROS bursts are invoked during both PTI and ETI, often referred as first and second bursts, respectively (Torres, 2010). While the first burst is rapidly and transiently induced within a few minutes after PAMP recognition, the massive second burst sustainably occurs hours after pathogen attacks and plays an important role in regulating HR cell death (Mur et al., 2008).
MAPK cascades play pivotal roles in signaling pathways of plant defense (Pedley and Martin, 2005). In tobacco (Nicotiana tabacum), wound-induced protein kinase (WIPK) and salicylic acid-induced protein kinase (SIPK) are pathogen-responsive MAPKs (Seo et al., 1995; Zhang and Klessig, 1997). MEK2, a tobacco MAPK kinase, functions upstream of SIPK and WIPK (Ren et al., 2006). The MEK2-SIPK/WIPK cascade (MEK2 cascade) is highly conserved in diverse plant species and participates in defense responses (Tanaka et al., 2009). Some WRKY transcription factors appear to be regulated by MAPKs at the transcriptional and posttranscriptional levels in defense-related signaling pathways (Pandey and Somssich, 2009; Ishihama and Yoshioka, 2012) and positively or negatively regulate defense responses (Eulgem and Somssich, 2007; Shen et al., 2007). WRKY8, a group I WRKY transcriptional factor in N. benthamiana, is a substrate of pathogen-responsive MAPKs and is specifically phosphorylated by SIPK and WIPK in plants (Ishihama et al., 2011). Proline-directed serine (SP cluster), in the N-terminal region of WRKY8, is a target of MAPKs (Ishihama et al., 2011). Phosphorylation of WRKY8 increases W-box binding and transactivation activities, and the phospho-mimicking mutant of WRKY8 significantly induces expression of target genes 3-hydroxy-3-methylglutaryl CoA reductase2 (HMGR2) and NADP-malic enzyme (NADP-ME) (Ishihama et al., 2011). WRKY33, the closest Arabidopsis WRKY to WRKY8, also contains the SP cluster, and phosphorylation of the SP cluster by MPK3/MPK6 is required to induce PHYTOALEXIN-DEFICIENT3 in camalexin production (Mao et al., 2011) and 1-AMINO-CYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE2 (ACS2) in ethylene synthesis (Li et al., 2012). These studies suggest that the MAPK cascade regulates transcriptional reprogramming via the WRKY transcription factor in plant immunity.
RBOH is regulated at the transcriptional and posttranslational levels to induce pathogen-responsive ROS bursts (Yoshioka et al., 2011; Adachi and Yoshioka, 2015). Biphasic ROS bursts occur in the interaction between potato and P. infestans, and only the second ROS burst requires new protein synthesis (Chai and Doke, 1987). P. infestans INF1 elicitin induces HR cell death in N. benthamiana (Kamoun et al., 1998), and the expression level of RBOHB is strongly upregulated later after INF1 treatment (Yoshioka et al., 2003). The expression of MEK2DD, a constitutively active mutant of MEK2, induces HR-like cell death, transactivation of RBOHB, and ROS burst (Yoshioka et al., 2003). Not only the MEK2 cascade, but also the MEK1-NTF6 cascade, activates transcription of RBOHB; INF1-triggered RBOHB expression and ROS burst are compromised in SIPK/NTF6-silenced leaves (Asai et al., 2008). These reports indicated that MAPK cascades are involved in transcriptional activation of RBOHB, which may be responsible for the second burst. However, the regulatory mechanism of the RBOHB downstream of MAPK and its role in the first and second bursts are unclear. In this study, we found that four WRKYs, including WRKY8 phosphorylated by MAPKs, bind to the W-box in the RBOHB promoter in plants and positively regulate the RBOHB. Gain- and loss-of-function analyses indicated that these WRKYs redundantly participate in the potato (Solanum tuberosum) R protein/oomycete effector pair, R3a/AVR3a- and INF1-triggered RBOHB transactivation, and ROS bursts, but not in bacterial PAMP, flg22-triggered events. We propose that transactivation of RBOHB via the MAPK-WRKY pathway is required for the long-lasting and robust second burst, but not the rapid first burst.
RESULTS
Expression of the RBOHB Gene Induced by INF1 and R3a/AVR3a Depends on MAPK Cascades
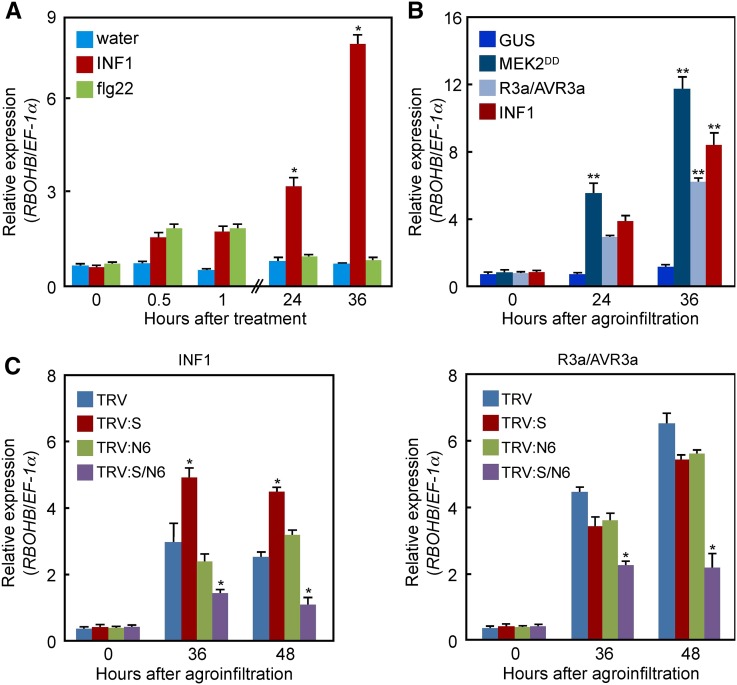
Previously we indicated that the expression of RBOHB, which is a main player for ROS burst in N. benthamiana leaves, is induced by INF1 and MEK2DD (Yoshioka et al., 2003). To investigate if the expression of RBOHB is induced during PTI or ETI, leaves were treated with flg22 peptide or INF1 protein as PAMPs to generate PTI signaling (Kamoun et al., 1998; Zipfel et al., 2004), and R3a and AVR3a were expressed as the R protein/effector pair to activate ETI (Bos et al., 2006; Yaeno et al., 2011). The transcript level of RBOHB increased by 0.5 to 1 h after flg22 and INF1 treatments (Figure 1A). Then, INF1 significantly upregulated RBOHB 24 and 36 h later. R3a/AVR3a and INF1 were expressed using Agrobacterium tumefaciens infiltration (agroinfiltration) in leaves. MEK2DD and GUS were also expressed as positive and negative controls, respectively. Expression of MEK2DD, R3a/AVR3a, and INF1 resulted in significant RBOHB activation compared with the GUS control (Figure 1B). These results suggest that cell death-inducing signals, such as INF1 and R3a/AVR3a, strongly upregulate RBOHB expression.
Figure 1.
Induction of RBOHB by INF1 and R3a/AVR3a.
(A) Expression of RBOHB in response to flg22 and INF1 proteins. INF1 (100 nM) and 100 nM flg22 were infiltrated into leaves by a needleless syringe, and total RNAs were used for RT-qPCR.
(B) Expression of RBOHB in response to MEK2DD, R3a/AVR3a, and INF1. Total RNAs were extracted from leaves at the indicated hours after agroinfiltration and were used for RT-qPCR.
(C) Effects of single or multiple MAPK gene silencing on RBOHB expression induced by INF1 and R3a/AVR3a. Total RNAs were extracted from SIPK (S)- or NTF6 (N6)-silenced leaves 0, 36, and 48 h after agroinfiltration and were used for RT-qPCR.
Asterisks indicate statistically significant differences compared with water (A), GUS (B), or TRV (C) (t test, *P < 0.05 and **P < 0.01). Data are means ± sd from at least three experiments.
Previously we showed that the MEK2-SIPK and MEK1-NTF6 cascades regulate INF1-triggered RBOHB expression (Asai et al., 2008). In addition to INF1, we confirmed whether the expression of RBOHB induced by R3a/AVR3a depends on both MAPK cascades. SIPK-, NTF6-, and SIPK/NTF6-silenced leaves were inoculated with Agrobacterium strains carrying R3a/AVR3a or INF1. Compared with the Tobacco rattle virus (TRV) control, the expression of RBOHB was statistically significantly compromised in SIPK/NTF6-silenced leaves, but not in the single MAPK gene-silenced leaves (Figure 1C). This result suggests that INF1- and R3a/AVR3a-induced RBOHB expression is mediated by the two different MAPK cascades.
Identification of MEK2DD-Responsive cis-Element in the RBOHB Promoter
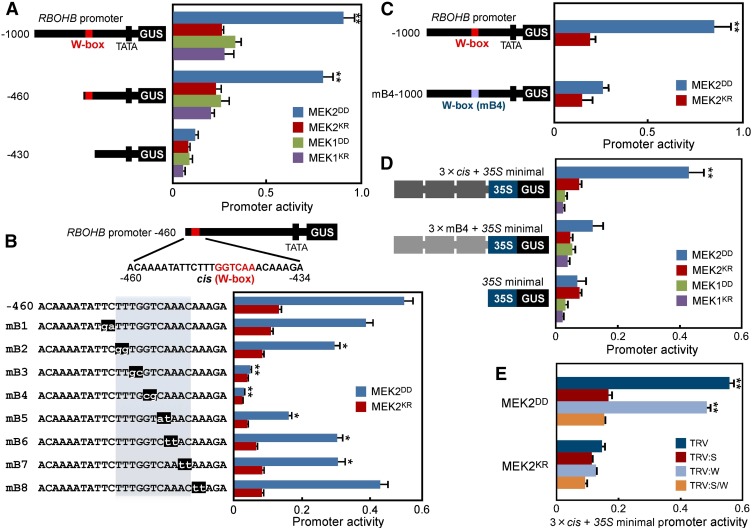
To investigate if the RBOHB promoter responds to constitutively active MAPKKs MEK2DD or MEK1DD, we prepared a 1000-bp 5′-flanking fragment of RBOHB fused to GUSint, which contains an intron (int) to avoid expression in Agrobacterium, as a reporter. Agrobacterium strains containing the −1000 RBOHB promoter-GUSint (reporter), pER8:MEK2DD, or MEK1DD (effector) and CaMV 35S promoter-luciferase (LUC) int (reference) were coinfiltrated into leaves and then the promoter activity was estimated by GUS activity normalized to LUC activity. Kinase-inactive mutants MEK2KR and MEK1KR were expressed as effector controls. To synchronize the expression of effector genes, we used an estradiol-inducible system and made a time lag of the expression between the reporter and effector genes. The −1000 RBOHB promoter activity was significantly induced by MEK2DD, but not by MEK1DD (Figure 2A), suggesting that there is MEK2DD-responsive cis-element in the −1000 RBOHB promoter. We found a W-box sequence (TTGACC/T), which is bound by WRKY transcription factors (Rushton et al., 2010), in the −1000 RBOHB promoter. These findings made us examine the possibility that WRKY transcription factors are involved in the activation of the −1000 RBOHB promoter. We generated 5′ deletions of the promoter with or without the W-box (−460 or −430). A deletion to −430 (lacking the W-box) resulted in a clear decrease in the effector-responsive GUS activity (Figure 2A). To determine the regulatory element, 2-bp substitutions were introduced into the 15 bp (mB1 to mB8). The −460 RBOHB promoter activity induced by MEK2DD significantly decreased for constructs carrying the mB2 to mB7 mutations (Figure 2B), suggesting that a MEK2DD-responsive 11-bp estimated cis-element (TTTGGTCAAAC) containing a W-box is in the RBOHB promoter. We introduced an mB4 mutation (in the W-box) into the −1000 RBOHB promoter (mB4-1000). The mB4-1000 promoter lacks responsiveness to MEK2DD (Figure 2C), indicating involvement of the W-box in activation of the RBOHB promoter.
Figure 2.
Promoter Activity of RBOHB Induced by MEK2DD via cis-Element.
(A) Deletion analysis of RBOHB promoter activity. The number indicates the distance from the RBOHB translation start site. A mixture of Agrobacterium cultures containing RBOHB promoter-GUSint (reporter), pER8:MEK2DD (effector), and CaMV 35S promoter-LUCint (reference) was coinfiltrated into leaves and 24 h later 20 μM estradiol was injected into the leaves. MEK2KR, MEK1DD, and MEK1KR were also expressed as the effector. GUS activities and LUC activities were determined 24 h after estradiol treatments, and RBOHB promoter activities were the values of GUS activities divided by LUC activities.
(B) Analysis of RBOHB promoter and mutated promoters containing two-base substitutions. Mutant bases are shown on a black background.
(C) Analysis of RBOHB −1000 and mB4-1000 promoter in response to MEK2DD.
(D) Analysis of three-tandem repeats of the cis-element and mB4 with 35S minimal promoter in response to MEK2DD and MEK1DD.
(E) Effects of SIPK and WIPK silencing on the three-tandem promoter activity. SIPK (S)- and WIPK (W)-silenced leaves were inoculated with Agrobacterium carrying promoter assay constructs.
Asterisks indicate statistically significant differences compared with MEK2KR and MEK1KR ([A] and [D]), −460 (B), mB4-1000 (C), and TRV (E) (t test, *P < 0.05 and **P < 0.01). Data are means ± sd from at least three experiments.
To check whether the estimated cis-element is sufficient for responsiveness to MEK2DD, we prepared a three-tandem (3 × cis) promoter of 19-bp sequences (TATTCTTTGGTCAAACAAA) containing the estimated cis-element and generated a 3 × cis-fused −46 minimal CaMV 35S promoter (3 × cis + 35S minimal promoter) -GUSint reporter. Consistent with Figure 2A, the expression of MEK2DD strongly induced activation of the 3 × cis promoter, while MEK1DD did not activate (Figure 2D). The mB4 mutation in the 3 × cis promoter (3 × mB4) suppressed MEK2DD-responsive activation, indicating that the cis-element containing the W-box is responsible for MEK2DD-triggered RBOHB promoter activity. To test whether the identified cis-element depends on SIPK or WIPK, we measured MEK2DD-induced 3 × cis promoter activity in SIPK-, WIPK-, or SIPK/WIPK-silenced leaves. The 3 × cis promoter activity induced by MEK2DD decreased in SIPK- and SIPK/WIPK-silenced leaves (Figure 2E). By contrast, WIPK silencing did not affect MEK2DD-induced 3 × cis promoter activity. These results suggest that the cis-element responds downstream of the MEK2-SIPK cascade.
St-RBOHC, a potato ortholog of Nb-RBOHB, is markedly expressed in response to P. infestans in potato leaves (Yamamizo et al., 2006). Similar to the Nb-RBOHB promoter, the expression of MEK2DD significantly increased St-RBOHC promoter activity; the same 11-bp sequences also exist in the St-RBOHC promoter (Supplemental Figure 1A). Deletion and mutation analyses showed that the 11-bp sequences are essential for St-RBOHC promoter activity induced by MEK2DD (Supplemental Figures 1A and 1B). These findings indicate that the same transcriptional regulatory mechanism through common cis-element is conserved in at least solanaceous plants.
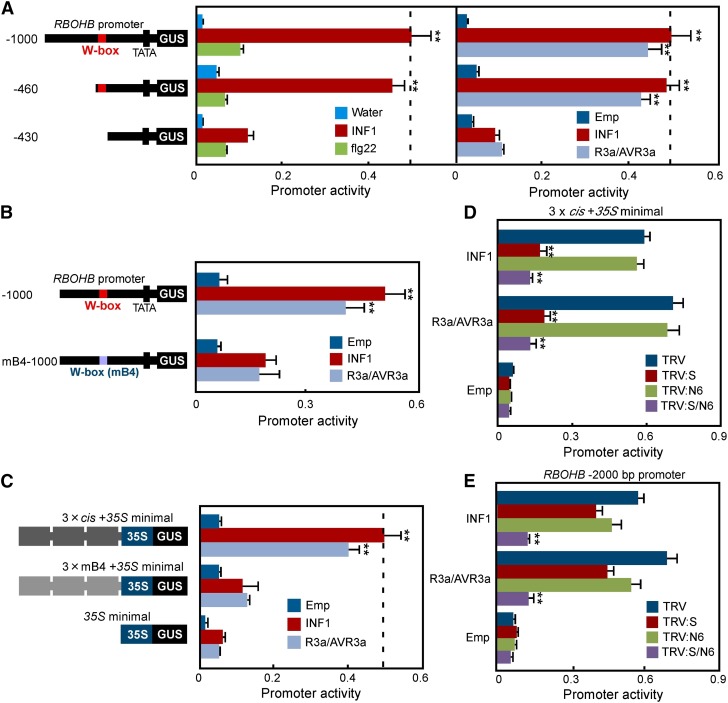
Involvement of the MEK2DD-Responsive cis-Element in RBOHB Promoter Activity during PTI and ETI
To examine whether the identified cis-element participates in PTI- or ETI-dependent activation of the RBOHB promoter, we evaluated the effects of 5′ deletion on the RBOHB promoter in response to flg22, INF1, or R3a/AVR3a. As in the case of Figure 2, the reporter and reference constructs were expressed in N. benthamiana leaves for the promoter assay. At 24 h after agroinfiltration, leaves were treated with flg22 peptide or INF1 protein and then GUS and LUC activities were measured 24 h after injection. The INF1 treatment strongly induced activation of the −1000 RBOHB promoter compared with the water control, while flg22 treatment did not significantly induce the activity (Figure 3A). The INF1-triggered promoter activity significantly decreased in the −430 deletion construct, but not in the −460 construct. Similarly, RBOHB promoter activity induced by Agrobacterium-mediated INF1 and R3a/AVR3a expressions decreased in the −430 deletion construct (Figure 3A). To confirm whether the decrease depends on the W-box between −460 and −430, we used the mB4-1000 promoter (Figure 3B). As a control for effector genes, we inoculated an Agrobacterium strain containing an empty binary vector. Activation of the −1000 RBOHB promoter induced by INF1 or R3a/AVR3a was compromised by mB4 mutation, while significant inductions of the −1000 RBOHB and mB4 promoters were not observed in the empty vector control. These results suggest that the W-box in the identified cis-element is essential for responsiveness to INF1 or R3a/AVR3a. To confirm the responsiveness of the cis-element to INF1 or R3a/AVR3a, we measured three-tandem promoter activities (Figure 3C). The expressions of INF1 and R3a/AVR3a increased the 3 × cis promoter activity, but not the 3 × mB4 promoter activity, indicating that the identified cis-element is required for activation of the RBOHB promoter in response to INF1 and R3a/AVR3a.
Figure 3.
Promoter Activity of RBOHB Induced by INF1 and R3a/AVR3a via the cis-Element.
(A) Deletion analysis of the RBOHB promoter in response to flg22, INF1, and R3a/AVR3a. Leaves were treated with 100 nM INF1 and 100 nM flg22 or were inoculated with Agrobacterium carrying INF1 and R3a/AVR3a. Promoter activities were analyzed as described in Figure 2A.
(B) Analysis of the RBOHB −1000 and mB4-1000 promoter in response to INF1 and R3a/AVR3a.
(C) Activation of three tandem repeats of the cis-element by INF1 and R3a/AVR3a. Promoter activity was represented as a relative value, using the comparative 3 × cis of INF1.
(D) Analysis of INF1- or R3a/AVR3a-induced three-tandem promoter activities in single or multiple MAPK gene-silenced leaves.
(E) Analysis of INF1- or R3a/AVR3a-induced RBOHB −2000 promoter activities in single or multiple MAPK gene-silenced leaves.
Asterisks indicate statistically significant differences compared with water and empty (Emp) (A), mB4-1000 (B), 35S minimal (C), or TRV ([D] and [E]) (t test, *P < 0.05 and **P < 0.01). Data are means ± sd from at least three experiments.
The INF1- and R3a/AVR3a-dependent activation of the RBOHB gene was mediated by SIPK and NTF6 (Figure 1C). To study the involvement of SIPK and NTF6 in an increase in three-tandem promoter activity induced by INF1 and R3a/AVR3a, we examined the effects of SIPK, NTF6, or SIPK/NTF6 silencing on the promoter activity (Figure 3D). The 3 × cis promoter activities induced by INF1 and R3a/AVR3a were significantly compromised by silencing SIPK or SIPK/NTF6, but not by silencing NTF6. The 3 × cis promoter activities between SIPK- and SIPK/NTF6-silenced plants showed no statistical difference, suggesting that INF1- and R3a/AVR3a-induced 3 × cis promoter activities depend on only SIPK. Furthermore, the −1000 RBOHB promoter responded to MEK2DD, but not MEK1DD (Figure 2A), and the −1000 RBOHB promoter activities induced by INF1 and R3a/AVR3a were compromised in SIPK-silenced leaves (Supplemental Figure 2). These results do not agree with Figure 1C, showing NTF6 is also involved in the expression of RBOHB induced by INF1 and R3a/AVR3a, suggesting that the other cis-element in response to NTF6 may exist and be in the <−1000 region of the RBOHB promoter. Therefore, we isolated the −2000 RBOHB promoter and examined the effects of SIPK, NTF6, or SIPK/NTF6 silencing on the promoter activities induced by INF1 and R3a/AVR3a. Predictably, the activation of the −2000 RBOHB promoter was significantly suppressed in SIPK/NTF6-silenced plants, and SIPK- and NTF6-silenced plants showed a slight reduction (Figure 3E). These results suggest that two MAPK cascades participate in regulating RBOHB promoter activity via different cis-elements, and the WRKY transcription factor plays pivotal roles in transactivation of RBOHB downstream of the MEK2-SIPK cascade.
Isolation of WRKYs Phosphorylated by MAPKs
WRKY8 is specifically phosphorylated by SIPK and WIPK and has a phosphorylation motif, the SP cluster (Ishihama et al., 2011). The SP cluster is highly conserved in some group I WRKYs, such as N. tabacum WRKY1 and Arabidopsis WRKY33, which are substrates of MAPK (Ishihama and Yoshioka, 2012). These findings indicated the possibility that WRKYs containing the SP cluster may be substrates of MAPKs. In this study, we first explored group I WRKY genes containing the SP cluster by BLAST searches in the EST database of solanaceous plants (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Based on obtained DNA sequence information, seven N. benthamiana WRKY genes, WRKY9-15, were isolated (Supplemental Figure 3A). Similar to WRKY8, the SP cluster is conserved in the N-terminal region of each WRKY protein. Ser-79 and Ser-86 in the SP cluster of WRKY8 (Supplemental Figure 3B) are phosphorylated by pathogen-responsive MAPKs in plants (Ishihama et al., 2011). We constructed a phylogenetic tree of the group I WRKY family containing the SP cluster (Supplemental Figure 4). WRKY9 is closest to WRKY7 (95% amino acid identity), and amino acid sequences of both genes are closely similar to the amino acid of N. tabacum WRKY1.
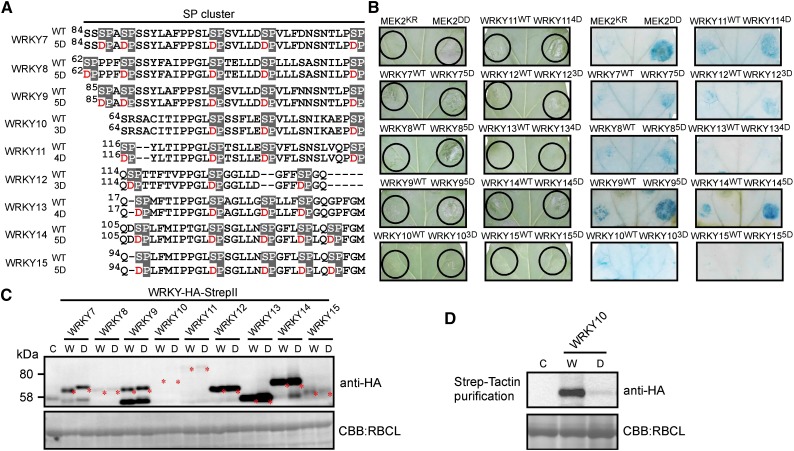
HR-like cell death is induced downstream of the MEK2-SIPK/WIPK cascade (Yang et al., 2001; Yoshioka et al., 2003), and N. tabacum WRKY1, a substrate of SIPK, is involved in inducing HR-like cell death (Menke et al., 2005). These studies suggest that MAPK substrates play pivotal roles in cell death induction downstream of the MAPK cascade. The transactivation of WRKY8 is enhanced by phospho-mimicking mutations in the SP cluster (Ishihama et al., 2011). To examine whether WRKY7-15 have cell death-inducible activity, phospho-mimicking mutants (WRKYnD) of the nine WRKYs were generated (Figure 4A). Then, we introduced these WRKY7-15 variants in leaves by agroinfiltration. Four days after agroinfiltration, cell death was seen in regions expressing phospho-mimicking mutants of WRKY7, 8, 9, 11, 12, and 14 and was visualized by trypan blue staining (Figure 4B). Table 1 shows the percentage of leaves developing cell death by expression of each WRKYnD gene, suggesting that these WRKYs are involved in inducing cell death as MAPK substrates. Immunoblot analysis showed that WRKYWT and WRKYnD showed similar accumulation of WRKY proteins (Figure 4C). These results indicated that phospho-mimicking mutations of WRKYs result in strong induction of cell death. Although the protein level of WRKY103D was low (Figure 4D), cell death was observed in 31% of the leaves, meaning that WRKY10 might have the potential to induce cell death (Table 1). We examined the expression profile of each WRKY gene. The expression levels of some cell death-inducing WRKYs, especially WRKY7, 8, and 9, significantly increased during PTI and ETI (Supplemental Figure 5), indicating that these WRKYs may play a role in plant immune responses. On the other hand, basal level of WRKY10 expression drastically decreased during ETI.
Figure 4.
Promotion of Cell Death by Phospho-Mimicking Mutations on the SP Cluster of WRKY Transcription Factors.
(A) Phospho-mimicking mutations in a SP cluster. Putative phosphorylated Ser residues in SP cluster were substituted with Asp (nD).
(B) Induction of cell death by overexpression of phospho-mimicking mutants. Leaves were infiltrated with Agrobacterium carrying the indicated gene expression constructs. Photographs were taken 4 d after agroinfiltration (left). After taking the photographs, WRKY-induced cell death was detected by trypan blue staining (right).
(C) Detection of WRKY-HA-StrepII proteins by anti-HA antibody. Total proteins were prepared 48 h after agroinfiltration. Protein loads were monitored by Coomassie blue (CBB) staining of the bands corresponding to the ribulose-1,5-bisphosphate carboxylase large subunit (RBCL). Red asterisks indicate WRKY-HA-StrepII proteins. C, control plant; W, wild-type WRKYs; D, phospho-mimicking mutants.
(D) Detection of WRKY10-HA-StrepII protein. Total proteins were purified by Strep-Tactin and then immunoblot analysis was done.
Table 1. The Percentage of N. benthamiana Leaves Developing Cell Death by Transient Expression of WRKYnD Genes.
| Overexpressing Construct | Total Leaves Infiltrated | Leaves Developing Cell Death | % of Leaves Developing Cell Death |
|---|---|---|---|
| WRKY7 | 13 | 9 | 69 |
| WRKY8 | 16 | 13 | 81 |
| WRKY9 | 15 | 11 | 73 |
| WRKY10 | 13 | 4 | 31 |
| WRKY11 | 11 | 11 | 100 |
| WRKY12 | 15 | 12 | 80 |
| WRKY13 | 13 | 0 | 0 |
| WRKY14 | 16 | 15 | 93 |
| WRKY15 | 11 | 0 | 0 |
SIPK and WIPK phosphorylate WRKY8 in vitro and in vivo (Ishihama et al., 2011). To confirm whether WRKY7, 9, 11, 12, and 14 are phosphorylated by pathogen-responsive MAPKs, we prepared a thioredoxin-fused N-terminal half of recombinant WRKYs (N-WRKYs) containing the SP cluster (WRKY71-200, WRKY91-199, WRKY111-203, WRKY121-200, and WRKY141-200). For pathogen-responsive MAPKs, we generated active SIPK, WIPK, and NTF6 that had been phosphorylated by upstream constitutively active MAPKK in vitro. All active MAPKs had the kinase activity to phosphorylate myelin basic protein in vitro (Supplemental Figure 6). All N-WRKYs were phosphorylated by each active MAPK (Supplemental Figure 7), but the phosphorylation level of each N-WRKY differed among the type of MAPKs (Supplemental Table 1). These results suggest that not only WRKY8, but also WRKY7, 9, 11, 12, and 14 are substrates of pathogen-responsive MAPKs. In particular, the phospho-Ser of N-WRKY11 was detected using anti-WRKY8 phospho-Ser-98 (pSer98) antibody in vitro (Supplemental Figure 8A). To determine if WRKY11 is phosphorylated by MAPKs in vivo, we prepared total proteins from N. benthamiana leaves coexpressing WRKY11-HA-StrepII with FLAG-MEK2DD or FLAG-MEK2KR. After purification of StrepII-tagged proteins with Strep-Tactin chromatography, we performed immunoblot analyses using anti-pSer98 antibody. Phosphorylation of WRKY11 was only detected in leaves expressing WRKY11-HA-StrepII with FLAG-MEK2DD, but not with FLAG-MEK2KR (Supplemental Figure 8B), suggesting that WRKY11 is phosphorylated by downstream MAPKs of MEK2 in vivo.
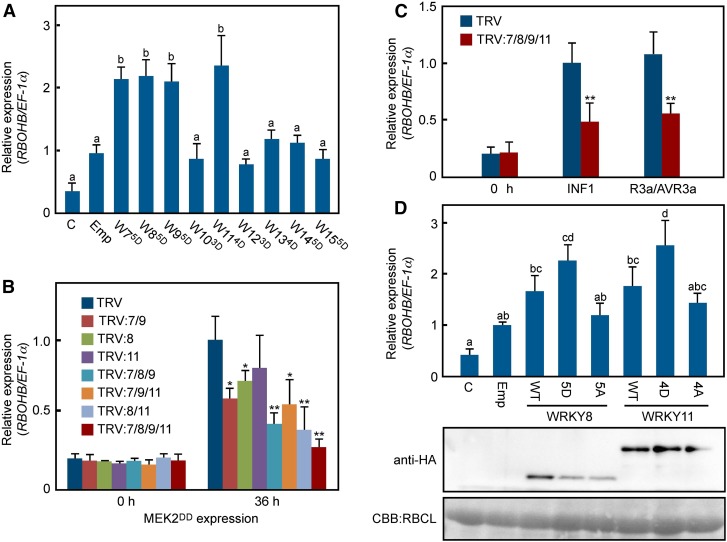
Silencing of Multiple WRKY Genes Compromises the Expression of RBOHB Induced by INF1 and R3a/AVR3a
To assess whether WRKYs containing the SP cluster participate in RBOHB expression, phospho-mimicking mutants of WRKYs were expressed in N. benthamiana leaves (Figure 5A). The RBOHB gene was significantly induced by WRKY75D, 85D, 95D, and 114D, but not by WRKY103D, 123D, 134D 145D, and 155D. Multiple WRKYs function redundantly downstream of MAPK cascades (Popescu et al., 2009). Therefore, we generated plants in which WRKY8 and closely related WRKY genes are simultaneously silenced. The expression levels of targeted WRKY genes were specifically reduced in single or multiple WRKY gene-silenced plants compared with the TRV control (Supplemental Figure 9). The expression of RBOHB induced by MEK2DD was slightly compromised in single WRKY gene-silenced plants (Figure 5B). However, the expression was suppressed in multiple WRKY gene-silenced plants, especially in WRKY7/8/9/11-silenced plants (Figure 5B). We investigated whether WRKY7, 8, 9, and 11 participate in INF1- and R3a/AVR3a-induced RBOHB expression because INF1 and R3a/AVR3a induced RBOHB via activation of its promoter (Figures 1 and 3). The expression of RBOHB was compromised in WRKY7/8/9/11-silenced plants, suggesting that multiple WRKYs are involved in the induction of RBOHB expression by INF1 and R3a/AVR3a (Figure 5C). We also confirmed that MEK2DD-triggered expression of NADP-ME and ACS2 was compromised in WRKY7/8/9/11-silenced plants (Supplemental Figure 10). These results suggest that multiple WRKYs positively regulate expression of diverse genes downstream of the MEK2 cascade.
Figure 5.
Regulation of RBOHB Expression by Multiple WRKY Transcription Factors.
(A) Induction of the RBOHB gene by phospho-mimicking mutants of SP cluster-carrying WRKYs. Total RNAs were extracted from leaves 24 h after agroinfiltration and were used for RT-qPCR. nD indicates phospho-mimicking mutants of WRKYs.
(B) Involvement of multiple WRKY genes in MEK2DD-induced RBOHB expression. Total RNAs were extracted from the WRKY-silenced leaves 0 and 36 h after agroinfiltration and were used for RT-qPCR.
(C) Suppression of INF1- and R3a/AVR3a-induced RBOHB expression by silencing of multiple WRKY genes. Total RNAs were extracted from leaves 0 or 36 h after agroinfiltration and were used for RT-qPCR.
(D) Expression of RBOHB in response to variants of WRKY8 and WRKY11. Total RNAs were extracted from leaves 24 h after agroinfiltration and were used for RT-qPCR. Anti-HA antibody was used to detect accumulations of WRKY-HA variants. nD and nA indicate that WRKY mutants mimic the phosphorylated form and nonphosphorylated form, respectively.
Letters represent each significance group, determined through Tukey's multiple range test. Asterisks indicate statistically significant differences compared with TRV ([B] and [C]) (t test, *P < 0.05 and **P < 0.01). Data are means ± sd from at least three independent experiments.
To examine the effect of phosphorylation of WRKYs on RBOHB expression, wild-type WRKYs (WRKY8WT and WRKY11WT), phospho-mimicking mutants (WRKY85D and WRKY114D), and alanine-substituted mutants (WRKY85A and WRKY114A) were transiently expressed in leaves (Figure 5D). WRKY85D and WRKY114D induced the expression of RBOHB compared with each alanine-substituted mutant, suggesting that phosphorylated WRKY8 and WRKY11 accelerate the expression of RBOHB.
Multiple WRKY Genes Are Required for INF1-PTI and AVR3a-ETI ROS Bursts
WRKY7, 8, 9, and 11 are required for MEK2DD-induced RBOHB expression (Figure 5B), suggesting that these WRKYs are involved in the ROS burst via the MEK2 cascade. We examined the effects of silencing single or multiple WRKY genes on the MEK2DD-induced ROS burst (Supplemental Figure 11). The ROS burst was significantly suppressed in multiple WRKY gene-silenced plants compared with TRV control plants. By contrast, single WRKY gene-silenced plants showed no decrease, indicating that WRKY7, 8, 9, and 11 are required for the ROS burst downstream of the MEK2 cascade.
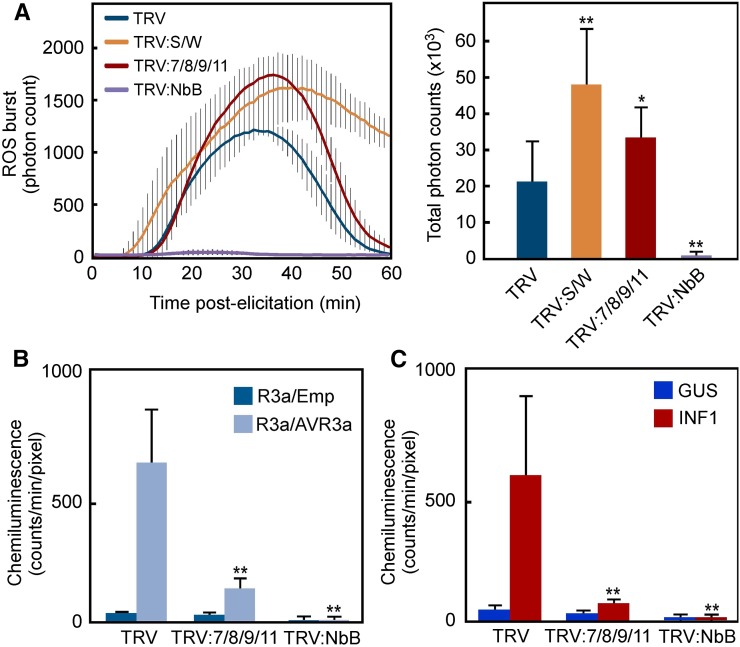
To assess if the MAPK-WRKY pathway is required for the PTI or ETI ROS burst, we investigated the effects of silencing multiple WRKY genes on flg22-triggered PTI (flg22-PTI) or R3a/AVR3a-triggered ETI (AVR3a-ETI) ROS burst (Figure 6A). In TRV control leaves, ROS burst was induced between 10 and 60 min after flg22 treatment and peaked at 30 min after elicitation. In agreement with a previous study (Segonzac et al., 2011), SIPK/WIPK silencing amplified the flg22-PTI ROS burst. A similar trend was observed in WRKY7/8/9/11-silenced leaves. To estimate AVR3a-ETI ROS burst, R3a and AVR3a were driven by the CaMV 35S promoter and estradiol-inducible promoter, respectively. Then, RBOH-mediated ROS were measured using the chemiluminescence probe L-012, which detects apoplastic ROS (Supplemental Figure 12). SIPK and WIPK were activated 12 h after estradiol treatment and increased from 12 to 24 h. AVR3a-ETI ROS burst was initiated with elevation of MAPK activation. Then, we examined the AVR3a-ETI ROS burst in multiple WRKY-silenced leaves 24 h after estradiol treatment. WRKY7/8/9/11 silencing significantly compromised AVR3a-ETI ROS burst compared with the TRV control (Figure 6B), suggesting that the MAPK-WRKY pathway is involved in AVR3a-ETI ROS burst, but not in the flg22-PTI ROS burst. However, both bursts depended on RBOHB (Figures 6A and 6B).
Figure 6.
Involvement of Multiple WRKY Transcription Factors in INF1-PTI and AVR3a-ETI ROS Bursts.
(A) Enhancement of flg22-triggered ROS burst in MAPK- or WRKY-silenced plants. Flg22-triggered ROS bursts were measured for 60 min in leaves silencing SIPK/WIPK (S/W), WRKY7, 8, 9, and 11 (7/8/9/11), or RBOHB (NbB). Total photon counts of each treatment during 60 min were graphed in the right panel.
(B) Effects of multiple WRKY gene silencing on effector-triggered ROS burst. Silenced leaves were coinoculated with Agrobacterium carrying pER8:AVR3a-HA and pBinPlus:R3a and then were injected with 20 μM estradiol 24 h later. To detect ROS generation, inoculation sites were infiltrated with 0.5 mM L-012 solution 24 h after estradiol injection and were monitored using a CCD camera. Chemiluminescence intensities were quantified by a program equipped with a photon image processor.
(C) Effects of multiple WRKY gene silencing on INF1-triggered ROS burst. INF1-ROS burst was detected in silenced leaves 24 h after agroinfiltration.
Asterisks indicate statistically significant differences compared with TRV (t test, *P < 0.05 and **P < 0.01). Data are means ± se from at least five experiments.
INF1-triggered PTI (INF1-PTI) ROS burst is observed in an RBOHB-dependent manner, and two MAPK cascades, MEK2-SIPK and MEK1-NTF6, are involved in the ROS burst (Asai et al., 2008). Therefore, we hypothesized that the INF1-PTI ROS burst requires WRKYs, which were essential for the MEK2DD-induced ROS burst. As expected, the INF1-PTI ROS burst was significantly compromised in the WRKY7/8/9/11-silenced leaves (Figure 6C), indicating that although INF1 is categorized as a PAMP-like flg22, the INF1-PTI ROS burst requires the MAPK-WRKY pathway. To confirm the difference between flg22-PTI and INF1-PTI ROS bursts, N. benthamiana leaves were treated with flg22 and INF1 proteins side by side, and ROS were monitored in the early phase and late phase after the treatments with or without α-amanitin, which is an inhibitor of RNA synthesis. INF1 induced a ROS burst during the first 1 and 12 h after the treatment, while flg22 also induced a ROS burst in the early phase but not the late phase (Supplemental Figure 13). The presence of α-amanitin inhibited only the late-phase ROS burst induced by INF1. These results support the idea that the INF1-PTI ROS burst is different from the flg22-PTI ROS burst, and the long-lasting ROS production induced by INF1 may depend on de novo protein synthesis including resupply of RBOHB via activation of RBOHB.
Multiple WRKYs Bind to and Activate the RBOHB Promoter
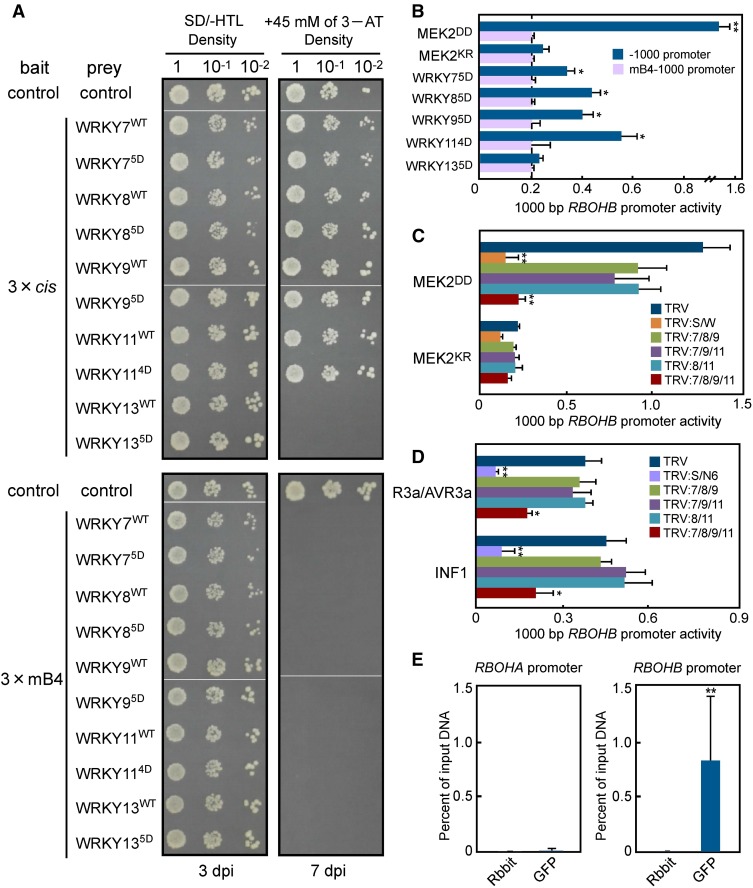
We hypothesized that WRKY7, 8, 9, and 11 might bind to the W-box in the cis-element and directly regulate RBOHB promoter activity because MEK2DD-, INF1-, and R3a/AVR3a-induced RBOHB expression and ROS burst were compromised by silencing of WRKY7/8/9/11 (Figures 5 and 6). First, to examine whether these WRKYs bind to the cis-element, we performed yeast one-hybrid analysis. The 3 × cis promoter was used as the bait, and wild-type or phospho-mimicking mutants of WRKY7, 8, 9, 11, and 13 were used as the prey. WRKY13 was used as a negative control because overexpression of WRKY13 did not induce cell death (Figure 4B, Table 1) and RBOHB expression in N. benthamiana leaves (Figure 5A). Each variant of WRKY7, 8, 9, and 11 interacted with the 3 × cis promoter (Figure 7A). The mB4 mutation in the cis-element inhibited the interactions, suggesting that WRKY7, 8, 9, and 11 bind to the cis-element of the RBOHB promoter. The −1000 RBOHB promoter activity significantly increased in response to WRKY75D, WRKY85D, WRKY95D, and WRKY114D in a cis-element-dependent manner, while WRKY134D did not induce the −1000 RBOHB promoter (Figure 7B). We further investigated RBOHB promoter activation by wild-type, phospho-mimicking, and alanine-substituted WRKYs. These WRKYs were individually expressed in N. benthamiana leaves, and activity of the −1000 RBOHB promoter was measured. The −1000 RBOHB promoter activities increased in response to phospho-mimicking WRKY7, 8, 9, and 11 compared with the wild-type and alanine-substituted WRKYs (Supplemental Figure 14). These results suggest that phosphorylation of the WRKYs promotes the activity of the RBOHB promoter. To test if WRKY7, 8, 9, and 11 are required for activation of the −1000 RBOHB promoter, we investigated the effects of multiple WRKY gene silencing on −1000 RBOHB promoter activities induced by MEK2DD, INF1, and R3a/AVR3a (Figures 7C and 7D). The MEK2DD-induced −1000 RBOHB promoter activity was modestly compromised in WRKY7/8/9-, WRKY7/9/11-, and WRKY8/11-silenced leaves, whereas drastic reduction was observed in WRKY7/8/9/11-silenced leaves to the same degree as SIPK/WIPK-silenced leaves (Figure 7C). The INF1- and R3a/AVR3a-induced activation of the promoter was suppressed by WRKY7/8/9/11 silencing, but not by WRKY7/8/9, WRKY7/9/11, and WRKY8/11 silencing (Figure 7D). These results indicate that WRKY7, 8, 9, and 11 redundantly function in regulating RBOHB promoter activity.
Figure 7.
Involvement of WRKY Transcription Factors in MEK2DD-, INF1-, or R3a/AVR3a-Dependent Activation of the RBOHB Promoter.
(A) Yeast one-hybrid analysis using a three-tandem promoter as bait and WRKY7, 8, 9, 11, or 13 as prey. The representative growth status of yeast cells is shown on SD/-HTL agar media with or without 3-amino-1,2,4-triazole from triplicate independent trials. Numbers on the top of each photograph indicate relative densities of the cells. dpi, days postinoculation.
(B) Analysis of RBOHB −1000 and mB4-1000 promoter in response to WRKY transcription factors. N. benthamiana leaves were inoculated with Agrobacterium carrying promoter assay constructs. WRKY genes were expressed as the effector. Promoter activities were analyzed as described in Figure 2A.
(C) Involvement of multiple WRKY genes in MEK2DD-induced RBOHB promoter activity. Silenced leaves were inoculated with Agrobacterium carrying promoter assay constructs.
(D) Effects of multiple WRKY gene silencing on RBOHB promoter activity induced by INF1 and R3a/AVR3a.
(E) ChIP-qPCR analysis in GFP-WRKY85D-expressed plants. Leaves were inoculated with Agrobacterium carrying pER8:GFP-WRKY85D and then were injected with 20 μM estradiol 24 h later. Input chromatin was isolated from the leaves 12 h after estradiol treatment. GFP-tagged WRKY8-chromatin complex was immunoprecipitated with an anti-GFP antibody. A control reaction was processed at the same time using rabbit IgG. ChIP- and input-DNA samples were quantified by qPCR using primers specific to the promoters of RBOHA and RBOHB genes. ChIP results are shown as percentages of input DNA.
Asterisks indicate statistically significant differences compared with mB4-1000 promoter (B), TRV ([C] and [D]), and rabbit (E) (t test, *P < 0.05 and **P < 0.01). Data are means ± sd from three experiments.
To further investigate the interaction between the WRKYs and the RBOHB promoter in plants, we performed chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) analysis. For this experiment, we constructed pER8:GFP-WRKY85D, in which the expression of WRKY85D fused with GFP at the N terminus was driven by an estradiol-inducible promoter. We first tested the effects of the fusion of the GFP tag on WRKY8 functions. WRKY85D-induced ROS burst and RBOHB expression were not affected by the GFP tag fusion (Supplemental Figure 15). In addition, GFP-WRKY85D localized in nuclei in leaves (Supplemental Figure 16). These results suggest that GFP-tagged WRKY85D can function to the same degree as WRKY85D. We immunoprecipitated the WRKY85D-DNA complex using anti-GFP antibody 12 h after estradiol treatment and then the DNA was purified. Previously we showed that RBOHA is constitutively expressed at a low level and does not respond to MEK2DD (Yoshioka et al., 2003). Therefore, the RBOHA promoter was used as a control for qPCR. The RBOHB promoter region was specifically immunoprecipitated by anti-GFP antibody, but not by control anti-rabbit antibody (Figure 7E). In contrast, although the RBOHA promoter contains two W-boxes, the promoter region was not collected by immunoprecipitation using anti-GFP antibody. These results indicate that WRKY8 directly binds to the RBOHB promoter in plants and positively regulates promoter activity.
WRKY Activates the RBOHB-Dependent ROS Burst
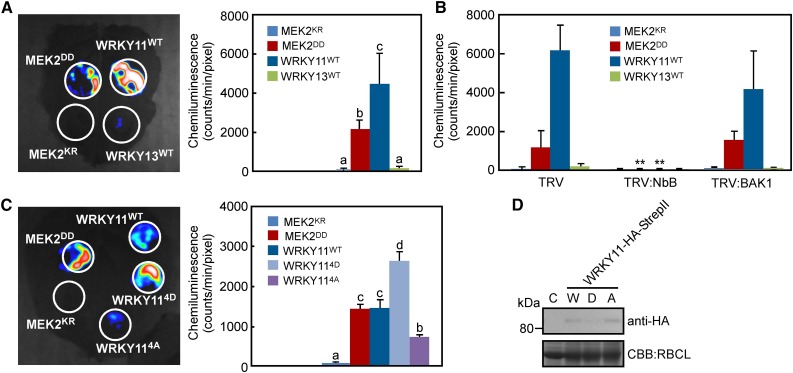
Ectopic expression of WRKY7, 8, 9, and 11 caused cell death in N. benthamiana leaves (Figure 4). We tested if these WRKYs possess ROS burst-inducing activity. WRKY11 induced the most severe cell death in leaves, while WRKY13 did not (Table 1). Therefore, WRKY11 and WRKY13 were transiently expressed by agroinfiltration, and ROS production was detected. MEK2DD and MEK2KR were used as positive and negative controls, respectively. Significant ROS production was observed after expression of MEK2DD and WRKY11, but not MEK2KR and WRKY13 (Figure 8A). To confirm involvement of RBOHB in the WRKY11-induced ROS burst, the WRKY genes were expressed in RBOHB-silenced leaves. Silencing RBOHB completely inhibited ROS production induced by WRKY11 (Figure 8B), suggesting that WRKY11 not only activates expression of RBOHB, but that it also induces an RBOHB-dependent ROS burst. To eliminate PTI signals against Agrobacterium infection, we silenced the BAK1 gene, which encodes a coreceptor of PRRs and is essential to induce PTI (Heese et al., 2007). BAK1 silencing rarely affected the WRKY11-dependent ROS burst (Figure 8B), suggesting that the WRKY-triggered ROS burst does not result from PTI after agroinfiltration.
Figure 8.
Involvement of Multiple WRKY Transcription Factors in RBOHB-Dependent ROS Burst.
(A) ROS generation by overexpression of WRKY transcription factors. Leaves were inoculated with Agrobacterium carrying the indicated gene expression constructs. ROS generation was detected 24 h after agroinfiltration as described in Figure 6B.
(B) WRKY-induced ROS generation via RBOHB. TRV control, RBOHB (NbB)-, and BAK1-silenced leaves were inoculated with Agrobacterium, and ROS were measured 24 h after agroinfiltration.
(C) ROS generation by overexpression of phospho-mimicking mutants.
(D) Detection of phospho-mimicking mutants by anti-HA antibody. C, control plant; W, wild-type WRKY11; D, phospho-mimicking mutant; A, nonphosphorylated mutant.
Letters represent each significance group, determined through Tukey's multiple range test. Asterisks indicate statistically significant differences compared with TRV ([B] and [C]) (t test, **P < 0.01). Data are means ± se from at least three experiments.
Phospho-mimicking mutants of WRKYs strongly induced cell death and RBOHB expression (Figures 4 and 5). To examine the effect of phosphorylation in the SP cluster on the WRKY11-induced ROS burst, we used WRKY114D and WRKY114A. Compared with WRKY11WT, the WRKY11-triggered ROS burst was enhanced by WRKY114D but was compromised by WRKY114A (Figure 8C). Immunoblot analysis indicated that proteins of the variants accumulated at similar levels (Figure 8D). These results suggest that WRKY11 is a positive regulator of ROS burst and that phosphorylation in the SP cluster facilitates ROS production.
Relevance of Redundant Function of WRKYs and of Cell Death-Mediated Immunity to P. infestans
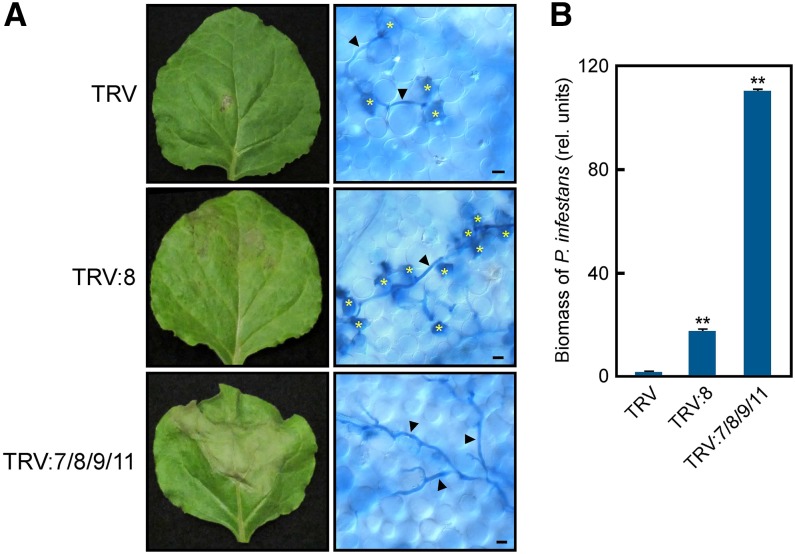
Next, we tested the redundant roles of WRKY7, 8, 9, and 11 in the basal defense against P. infestans, a potent pathogen of N. benthamiana (Kamoun et al., 1998). The oomycete P. infestans is a near-obligate pathogen (i.e., a potential biotroph), and INF1 derived from the pathogen induces an RBOHB-dependent ROS burst and HR cell death in N. benthamiana leaves (Figure 6C; Kamoun et al., 1998; Asai et al., 2008), suggesting that ROS burst and HR cell death occur during P. infestans and N. benthamiana interactions. We previously reported that WRKY8 silencing shows a decrease in resistance to P. infestans (Ishihama et al., 2011). Here, zoospores of the virulent isolate of P. infestans were inoculated on the surface of WRKY8- and WRKY7/8/9/11-silenced leaves. Severe disease symptoms were observed in WRKY7/8/9/11-silenced leaves compared with TRV control leaves (Figure 9A). Trypan blue staining to visualize dead cells and P. infestans hyphae showed that HR cell death is induced in mesophyll cells around the invaded hyphae in TRV control and WRKY8-silenced leaves, but not in WRKY7/8/9/11-silenced leaves (Figure 9A). Unlike TRV control leaves, the invaded hyphae significantly extended in WRKY8-silenced leaves in spite of the induction of cell death. To analyze P. infestans biomass, we conducted qPCR using primers specific to highly repetitive sequences in the P. infestans genome. In agreement with a previous study (Ishihama et al., 2011), the growth rate of P. infestans increased in WRKY8-silenced leaves compared with TRV control leaves (Figure 9B). WRKY7/8/9/11-silenced leaves showed a higher growth rate in similar patterns of severity of the disease symptoms (Figure 9B). These results suggest that WRKY7, 8, 9, and 11 play redundant roles in cell death-mediated immunity to P. infestans.
Figure 9.
Increased Disease Susceptibility to a Virulent Strain of P. infestans by Silencing of Multiple WRKY Genes.
(A) Susceptibility to P. infestans in the silenced plants. Inoculated leaves were photographed 6 d after the inoculation (left). N. benthamiana leaves were stained with lactophenol-trypan blue 6 d after inoculation (right). Arrowheads indicate hyphae of P. infestans. Asterisks indicate HR cell death of mesophyll cells. Bars = 20 μm.
(B) Effects of single or multiple WRKY gene silencing on P. infestans infection. Biomasses were determined by qPCR 6 d after inoculation. Asterisks indicate statistically significant differences compared with TRV (t test, **P < 0.01). Data are means ± sd from three independent experiments.
DISCUSSION
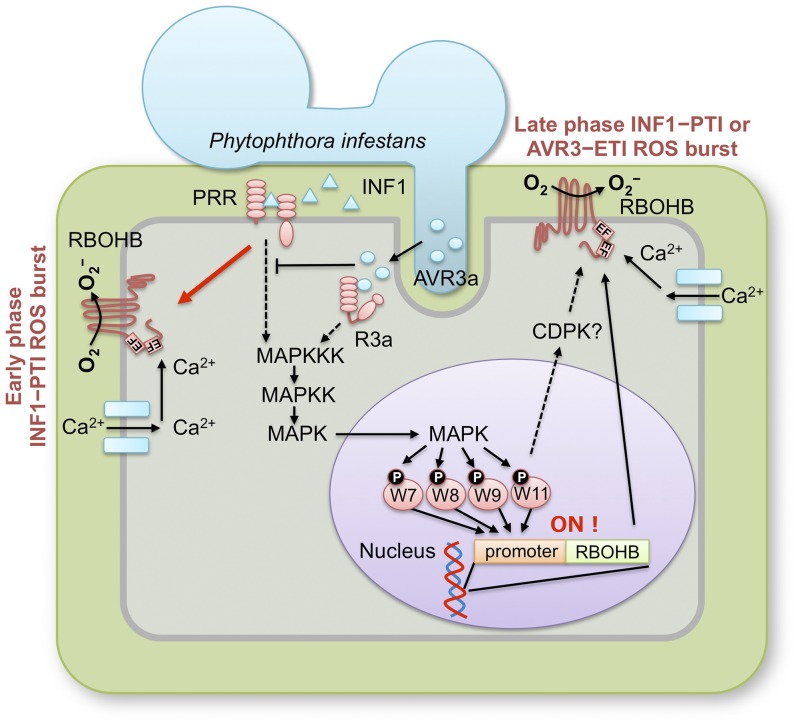
Transcription of the plant immune NADPH oxidase gene RBOHB is activated after INF1 perception and contributes to ROS burst and resistance to P. infestans in N. benthamiana leaves (Yoshioka et al., 2003; Asai et al., 2008). Although previously we reported information on MAPK-mediated RBOHB upregulation, the detailed mechanisms and roles of the RBOH activation in PTI and ETI ROS bursts have not been addressed. This work provided evidence that phosphorylation of WRKYs by MAPK accelerates WRKY-dependent RBOHB expression via binding to the cognate W-box in the promoter, resulting in ROS burst (Figure 10). Our findings show that this activation mechanism via the MAPK-WRKY pathway is important for the late phase INF1-PTI and AVR3a-ETI ROS bursts.
Figure 10.
Model of the Regulatory Mechanism of INF1-PTI and AVR3a-ETI ROS Bursts.
Sensing INF1 by the PRR triggers the early phase ROS burst, which is not required for de novo RNA synthesis, and the late phase ROS burst. Perception of INF1 and AVR3a by receptors leads to activation of the MAPK cascade. Activated MAPK phosphorylates and activates WRKY7, 8, 9, and 11. These WRKYs bind to the W-box in the RBOHB promoter, resulting in upregulation of the RBOHB gene. Supply of newly synthesized RBOHB to the plasma membrane may contribute to the late phase INF1-PTI and AVR3a-ETI ROS bursts.
Resupply of RBOHB Is Required for AVR3a-ETI and INF1-PTI ROS Bursts
Previous reports showed that ROS bursts occur biphasically. The first burst is transient and does not require protein synthesis, while the second burst is massive and depends on de novo protein synthesis (Chai and Doke, 1987; Yoshioka et al., 2001). PTI ROS bursts induced by flg22 and chitin are negatively regulated by, or do not depend on, the MAPK cascade (Segonzac et al., 2011). In agreement, our loss-of-function analysis also showed that the MAPK-WRKY pathway is not required for flg22-PTI ROS burst (Figure 6A). These results indicate that transactivation of RBOHB followed by protein synthesis is not involved in the rapid flg22-PTI ROS burst. Recent studies demonstrated that plasma membrane-associated kinase BIK1, which is a substrate of the PRR complex, directly phosphorylates and activates Arabidopsis RBOHD to induce PTI ROS burst (Kadota et al., 2014; Li et al., 2014). Confocal images of At-RBOHD promoter:GFP-At-RBOHD transgenic seedlings showed that GFP-At-RBOHD proteins accumulated in untreated Arabidopsis leaves (Hao et al., 2014), indicating that some amount of RBOHD exists on the plasma membrane for rapid reaction mediated by BIK1 upon the PAMP sensing. ROS bursts induced by INF1 treatment (Supplemental Figure 13; Chaparro-Garcia et al., 2011) and R3a/AVR3a coexpression (Figures 6B and 6C) are sustained and require the transcriptional activation of RBOHB via the MAPK-WRKY pathway (Figure 6C). INF1 is a PAMP similar to flg22 because its signaling depends on PRR and coreceptor BAK1/SERK3 (Chinchilla et al., 2007; Chaparro-Garcia et al., 2011; Du et al., 2015). What is the difference between flg22 signaling and INF1 signaling? Kamoun et al. (1998) isolated INF1 from P. infestans as an extracellular protein, which induces HR cell death and other biochemical changes related to defense responses in N. benthamiana. Several reports revealed that INF1-induced HR cell death is suppressed in SGT1- and HSP90-silenced N. benthamiana leaves (Peart et al., 2002; Kanzaki et al., 2003; Kanneganti et al., 2006). An HSP90 forms a chaperone complex with SGT1 and RAR1 to stabilize client proteins, including immune sensing R proteins, and the chaperone complex components contribute to ETI (Takahashi et al., 2003; Kadota and Shirasu, 2012). Taking these results together, we speculate that INF1 generates not only PTI signaling, but also massive ETI-like signaling, as in the case of R protein-mediated responses. In WRKY7/8/9/11-silenced plants, HR cell death was not induced in the mesophyll cells around P. infestans invaded hyphae, and the pathogen growth increased (Figure 9). A previous report showed that INF1-deficient strains of P. infestans developed disease symptoms in inoculated N. benthamiana leaves (Kamoun et al., 1998), suggesting that INF1 functions as an avirulence factor in the interaction between N. benthamiana and P. infestans. Thus, WRKY7/8/9/11-dependent immune responses induced by P. infestans inoculation might be derived from INF1.
The flg22-PTI ROS burst peaked 30 to 40 min after treatment and returned to nearly basal levels within 60 min (Figure 6A), in that activated RBOHs on the plasma membrane might have been rapidly inactivated or degraded. The dynamics of RBOHs after elicitation were observed recently in living Arabidopsis cells: Heterogeneously distributed RBOHD on the plasma membrane appeared to internalize into the cytoplasm, leading to endocytosis after ROS generation (Hao et al., 2014). In tobacco cultured cells, RBOHD-GFP seems to exist on the plasma membrane and at the periphery of Golgi cisternae, and elicitation with oomycete protein cryptogein activates transcriptional upregulation of RBOHD and distribution of RBOHD-GFP to the plasma membrane from the Golgi pool; a protein synthesis inhibitor accelerates the decrease in RBOHD-GFP fluorescence in intracellular compartments (Noirot et al., 2014). These lines of evidence suggest that activated RBOHs on the plasma membrane are rapidly turned over by endocytosis, and preexisting RBOHs are replenished by delivery from the Golgi pool, and even by de novo RBOH synthesis. Consistent with this, FLS2, which is associated with Arabidopsis RBOHD in vivo (Kadota et al., 2014), is rapidly degraded after flg22 treatment via ubiquitination by E3 ligases or is internalized by endocytosis, followed by degradation of FLS2 (Robatzek et al., 2006; Lu et al., 2011), suggesting that the activated FLS2-RBOHD complex is recruited into a turnover system after ligand perception. Consistent with this hypothesis, after the initial fast and transient flg22-PTI ROS burst, reelicitation of the same tissue with flg22 at 60 min did not result in significant ROS production (Smith et al., 2014). Although the detailed mechanism of RBOH turnover after PTI ROS burst is largely unknown, a resupply system of newly synthesized RBOHB to the plasma membrane may be required for the subsequent ETI ROS burst. We propose that transcriptional regulation of RBOHB via the MAPK-WRKY pathway could explain the biphasic ROS burst and that the increase in the amount of RBOHB on the plasma membrane permits a sustained and massive ROS burst, such as INF1-PTI and AVR3a-ETI ROS bursts.
WRKYs Redundantly Regulate Downstream Target Genes and the ROS Burst
Most WRKY transcription factors are regulated at the transcriptional level during PTI and ETI (Chen and Chen, 2000; Asai et al., 2002; Gao et al., 2013). This study also showed that WRKY7, 8, 9, and 11 genes are upregulated during both immunities (Supplemental Figure 5). Group I WRKY transcription factors containing the SP cluster, such as WRKY8, At-WRKY33, and Nt-WRKY1, are activated by MAPK-mediated phosphorylation (Andreasson et al., 2005; Menke et al., 2005; Ishihama et al., 2011; Mao et al., 2011). SIPK-mediated phosphorylation of Nt-WRKY1 and WRKY8 increases DNA binding activity (Menke et al., 2005; Ishihama et al., 2011), and phospho-mimicking mutant WRKY85D increases in transcription of downstream target genes NADP-ME and HMGR2 (Ishihama et al., 2011). In this study, phospho-mimicking mutation enhanced WRKY-dependent RBOHB expression and promoter activity, ROS burst, and cell death (Figures 4B, 5D, and 8C; Supplemental Figure 14). Our results strongly support the idea that phosphorylation of group I WRKYs by MAPK alters their DNA binding and transcriptional activities and results in induction of downstream phenotypes. On the other hand, loss-of-function analyses using single or multiple WRKY gene-silenced plants indicated that multiple WRKYs redundantly or additively functions in transactivation of RBOHB and ROS burst (Figure 5B; Supplemental Figure 11). Arabidopsis WRKY11 and WRKY17 function redundantly (Journot-Catalino et al., 2006), and the redundancy of transcription factors carrying similar DNA binding domains is indicated (Eulgem and Somssich, 2007). Recent research showed that the Ralstonia solanacearum effector PopP2 selectively acetylates Lys within the WRKY domain (WRKYGQK) of target WRKYs, including WRKY8, and impairs their DNA binding activities (Le Roux et al., 2015). We hypothesize that diversification of WRKYs binding to the RBOHB promoter may be a consequence of the struggle against pathogens to escape from the Lys acetylation strategy.
RBOHB promoter analyses indicated that INF1-induced −1000 RBOHB promoter activity depends on only SIPK (Supplemental Figure 2), while the −2000 RBOHB promoter showed dependency on not only SIPK, but NTF6 (Figure 3E), suggesting the other cis-element in response to NTF6 may exist in the −2000 to −1000 region of the promoter. Unlike −2000 RBOHB promoter activity (Figure 3E), SIPK silencing enhanced INF1-induced RBOHB expression and that was compromised by SIPK/NTF6 silencing (Figure 1C). Previous work also showed that INF1-induced RBOHB expression is modestly elevated in SIPK-silenced plants (Asai et al., 2008). These results suggest that upstream region of the −2000 RBOHB promoter might contain an enhancer sequence for an NTF6-dependent transacting factor triggered by SIPK silencing. Unexpectedly, the INF1-PTI ROS burst was partially compromised by SIPK silencing in disagreement with RBOHB expression profile (Asai et al., 2008). Taking these results together, we hypothesize that SIPK silencing could cause complex signaling crosstalk leading to transcriptional and posttranscriptional regulation of RBOHB under INF1 stimulation.
In addition to transactivation of the RBOH gene, posttranslational regulation of RBOH is required for its activation and ROS production. Ca2+, phosphatidic acid, and direct interactors Rac1 and RACK1 (Receptor for Activated C-Kinase 1) have been reported to be positive regulators of RBOHs (Sagi and Fluhr, 2001; Wong et al., 2007; Nakashima et al., 2008; Zhang et al., 2009; Nakano et al., 2013; de la Torre et al., 2013). Our results indicated that transient expression of phospho-mimicking WRKY11 significantly induced ROS burst than MEK2DD-mediated ROS burst, suggesting that the WRKY activates RBOHB not only at the transcriptional level, but also at the posttranslational level (Figure 8). We reported that N termini of RBOHs (St-RBOHA, B, C, and D and Nb-RBOHA and B) are directly phosphorylated by calcium-dependent protein kinase (CDPK or CPK), resulting in the activation of these RBOHs (Kobayashi et al., 2007, 2012; Asai et al., 2013). In this study, the CDPK4 gene, which encodes an ortholog of the St-RBOH-activating potato St-CDPK4 (Kobayashi et al., 2007), was transactivated downstream of the MAPK-WRKY pathway in N. benthamiana leaves (Supplemental Figure 10). Activation of CDPK depends on Ca2+, and Ca2+ influx is rapidly induced after recognition of PAMPs and pathogens by PRRs (Blume et al., 2000; Grant et al., 2000). Agrobacterium also has common PAMPs, such as flagellin, and Agrobacterium strain GV3101 likely primes the defense responses, such as ROS production in N. tabacum leaves (Sheikh et al., 2014). Therefore, we cannot rule out the possibility that WRKY-triggered ROS burst is derived from a Ca2+ influx induced by agroinfiltration. Loss-of-function analysis of BAK1, a coreceptor of PRRs, was performed to eliminate PTI signals by agroinfiltration and did not result in suppression of WRKY-triggered ROS burst (Figure 8B). In addition, we previously reported that overexpression of RBOHs (St-RBOHA, B, C, and D) by agroinfiltration does not induce a ROS burst (Kobayashi et al., 2012; Asai et al., 2013). We speculate that certain posttranslational regulatory mechanisms, such as CDPK-mediated phosphorylation, might exist downstream of MAPK-WRKY pathway, coordinately or synergistically enhancing RBOH activity with various positive regulators during immune responses.
Differential Regulation of the MAPK-WRKY Pathway in PTI and ETI
The flg22 treatment upregulated WRKY7, 8, 9, and 11 genes in N. benthamiana leaves (Supplemental Figure 5), suggesting that these WRKYs might function downstream of MAPK during PTI. Actually, flg22 induces NADP-ME, HMGR2, and ACS2 genes in N. benthamiana in a WRKY8-dependent manner (Le Roux et al., 2015). However, unlike ETI signaling, flg22 did not fully activate RBOHB, the same as its promoter activity (Figures 1A and 3A), indicating that PTI-dependent upregulation of these WRKYs does not seem to participate in robust RBOHB transactivation. Currently, we do not know how these WRKYs play different roles in PTI and ETI.
PTI and ETI share extensively common signaling mechanisms, but the kinetics and intensity of the responses appear to be different. Generally, ETI differs from PTI in accompanying HR cell death. One of the different features between PTI and ETI is duration of MAPK activity (Tsuda and Katagiri, 2010). Although MPK3 and MPK6 are activated in both immunities, duration of the activation is more prolonged during ETI than during PTI (Tsuda et al., 2013). In animals and yeast, switching common downstream signaling by differential duration of MAPK activity has been reported. In one case, long-lasting ERK MAPK activity leads to phosphorylation and stabilization of downstream c-Fos transcription factor (Murphy et al., 2002, 2004). In another case, the sustained MAPK activity causes nuclear translocation of MAPK, facilitating phosphorylation of the target in the nucleus (Traverse et al., 1992; Sabbagh et al., 2001; Glotin et al., 2006). Thus, MAPK substrates can function as a molecular sensor for signaling duration or are distinct between transient and sustained activation, which could lead to different downstream signaling. Comparative analyses of the phosphorylation-dependent activation mechanisms of WRKYs during PTI and ETI will provide new insight into differentiating downstream responses between PTI and ETI.
METHODS
Plant Growth Condition
Nicotiana benthamiana plants were grown at 23°C under a 16-h photoperiod and an 8-h dark period in environmentally controlled growth cabinets.
RT-qPCR
Total RNA from N. benthamiana leaves was prepared using TRIzol reagent (Invitrogen) according to the manufacturer’s procedure. Reverse transcription was done using the ReverTra Ace qPCR RT kit (Toyobo). RT-qPCR analysis was done using the StepOnePlus Real-Time PCR system (Applied Biosystems) with Power SYBR Green PCR Master Mix (Applied Biosystems). The expression of WRKYs, RBOHB, NADP-ME, ACS2, and CDPK4 was normalized to the expression of EF-1α. Supplemental Table 2 lists the gene-specific primers used for each sequence.
Virus-Induced Gene Silencing
Virus-induced gene silencing was done as described by Ratcliff et al. (2001) and Ishihama et al. (2011). Supplemental Table 3 lists the primers used to amplify cDNA fragments from the N. benthamiana cDNA library (Yoshioka et al., 2003). Restriction sites were added to the 5′-ends of the forward and reverse primers for cloning into TRV vector pTV00. The pTV00 vectors previously reported were used to silence SIPK, WIPK, and SIPK/WIPK (Tanaka et al., 2009), NTF6, SIPK/NTF6, and RBOHB (Asai et al., 2008), and WRKY8 (Ishihama et al., 2011).
Reporter Constructs and Measurement of Promoter Activity
Promoter activity was assayed as described by Kobayashi et al. (2010). For reporter plasmids, the RBOHB promoter or three-tandem repeats of the cis-element (TATTCTTTGGTCAAACAAA) and their variants with the first 14 bp of the RBOHB coding region were ligated into the EcoRI/ClaI sites of pGreen and were fused to an intron-containing GUS reporter gene (Supplemental Figure 17). For effector plasmids, cDNA fragments of WRKY variants, MEK1DD, MEK1KR, and AVR3aKI, were cloned into pER8 vector (Zuo et al., 2000) behind the estradiol-inducible promoter. The pER8 containing Nt-MEK2DD, Nt-MEK2KR, WRKY8, or its variants was described by Ishihama et al. (2011). As a reference plasmid, pGreen carrying an intron-containing LUC gene was used under the control of the CaMV 35S promoter (Ishihama et al., 2011).
Transient assays were done as described below. N. benthamiana leaves were infiltrated with Agrobacterium tumefaciens mixture, in which each Agrobacterium (OD600 = 0.5) containing reporter, effector (with pBINplus:R3a), and reference was mixed in a 50:1(2):10 ratio. At 24 h after agroinfiltration, leaves were injected with 20 µM β-estradiol and were incubated for 24 h. Total proteins were isolated from frozen tissues with extraction buffer (100 mM potassium phosphate, pH 7.8, 1 mM EDTA, 10% glycerol, 0.5% Triton X-100, and 1 mM DTT). Supernatant after centrifugation at 20,000g for 10 min at 4°C was used to measure GUS and LUC activities. GUS activity was measured by fluorometric quantitation of 4-methylumbelliferone produced from the glucuronide precursor using a multiplate reader (355-nm excitation filter and 460-nm emission filter; TriStar LB941; Berthold Technologies). LUC activity was measured using a Pica Gene Luminescence Kit (Toyo Ink) according to the manufacturer’s protocol with a multiplate reader. The promoter activity was evaluated by normalization of the level of GUS activity with LUC activity.
cDNA Cloning of WRKY Genes and Site-Directed Mutagenesis
The cDNAs of WRKYs were amplified by PCR from an N. benthamiana cDNA library as a template (Yoshioka et al., 2003) using the primers listed in Supplemental Table 4. The PCR products were cloned into pGEM-T Easy (Promega) and were sequenced. The WRKY variants were generated using GeneArt Seamless Cloning and Assembly Kit (Invitrogen).
Agrobacterium-Mediated Transient Expression in N. benthamiana Leaves
The cDNA fragments of WRKYs and their variants were cloned into pEL2-MCS with an HA-StrepII tag (Ishihama et al., 2011). pEL2 containing Nt-MEK2DD, Nt-MEK2KR, WRKY8, or its variants and pGreen containing Nt-MEK2DD were described by Ishihama et al. (2011). pGreen containing GUS or INF1 was described by Asai et al. (2008). Transformation of Agrobacterium GV3101 by electroporation and infiltration of Agrobacterium suspensions were done as described by Asai et al. (2008). Immunodetection of WRKYs-HA-StrepII was done by coinfiltration of Agrobacterium expressing p19, the suppressor of posttranscriptional gene silencing of Tomato bushy stunt virus (Voinnet et al., 2003).
Trypan Blue Staining
Trypan blue staining was done as described by Yoshioka et al. (2003), but was modified as follows. Leaves were transferred to a trypan blue solution (10 mL of lactic acid, 10 mL of glycerol, 10 g of phenol, 10 mL of water, and 10 mg of trypan blue) diluted in ethanol 1:1 and were boiled for 1 h. The leaves were then destained for 24 to 48 h in chloral hydrate.
Preparation of Proteins and Immunoblotting
Protein samples were prepared from two leaf discs (8 mm in diameter) of N. benthamiana leaves and were finely crushed with 62 μL of 2× SDS-PAGE sample buffer. Supernatant after centrifugation at 12,000g for 10 min was used for SDS-PAGE. Strep-Tactin affinity chromatography was done as described by Ishihama et al. (2014) using MagStrep type 2 beads (IBA). Immunoblotting was done as reported by Ishihama et al. (2014) with antibodies against WRKYs-HA (Clone H 9658; Sigma-Aldrich), FLAG-MEK2 (clone M2; Sigma- Aldrich), and phosphorylated MAPK (Phospho-p44/42 MAPK Erk 1/2 Thr202/Tyr204 XP Rabbit mAb #4370; Cell Signaling Technology). The pSer-98 antibody for WRKY8 was purified by affinity chromatography using pSer-98 peptide (ILPS[PO3H2]PTTGTFPAQAFNWK) as described by Ishihama et al. (2014).
Measurement of ROS
ROS production after 1 μM flg22 peptide treatment was monitored by a luminol-based assay as previously reported (Segonzac et al., 2011). Chemiluminescence using leaf discs was measured by a multiplate reader (TriStar LB941; Berthold Technologies). ROS production in detached leaves was monitored using a CCD camera as described by Asai et al. (2008).
Yeast One-Hybrid Assays
The cDNA fragments of WRKY variants were cloned into the pGADT7-Rec2 vector in frame with the GAL4 activation domain. Linkers of 3 × cis and 3 × mB4 were introduced into the pHIS2 vector upstream of the HIS3 reporter gene. Supplemental Table 5 lists the sequences of the linkers. These vectors were cotransformed into yeast Y187 (Clontech) using Matchmaker One-Hybrid Library Construction and Screening Kit (Clontech). Cotransformants were selected on SD-HLW (synthetic dropout medium lacking His, Leu, and Trp). The interactions were tested on SD-HLW by adding 45 mM 3-amino-1, 2,4-triazole, according to the manufacturer’s instructions.
ChIP-qPCR
N. benthamiana leaves infiltrated with Agrobacterium strain containing pER8:GFP-WRKY85D were used for ChIP assay. Leaf tissues treated with 20 µM β-estradiol for 12 h were processed as described by Kaufmann et al. (2010). Briefly, chromatin complex was isolated from 0.8 g fixed and frozen tissue and was sonicated for 60 min (30-s on and 30-s off cycles) with a Bioruptor UCW-310 (Cosmo Bio). Immunoprecipitation of chromatin complex was done using Anti-GFP mAb-Magnetic beads (MBL) or Goat Anti-Rabbit IgG magnetic beads (BioLabs) as a negative control. The immunoprecipitated DNA was purified using the Wizard SV Gel and a PCR Clean-Up system (Promega), and the purified DNA was used for qPCR analysis. Supplemental Table 6 lists the primers used for qPCR.
Pathogen Inoculation
Phytophthora infestans zoospore inoculation was done as described by Yoshioka et al. (2003). P. infestans race 1.2.3.4 zoospore suspension (1 × 104 zoospores/mL) was applied to the upper side of the attached leaves under high humidity at 20°C. Determination of P. infestans biomass was done as described by Asai et al. (2008).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL/DDBJ data libraries under accession numbers AB711130 (Nb-WRKY9), AB711131 (Nb-WRKY10), AB711132 (Nb-WRKY11), AB711133 (Nb-WRKY12), AB711134 (Nb-WRKY13), AB711135 (Nb-WRKY14), and AB711136 (Nb-WRKY15).
Supplemental Data
Supplemental Figure 1. Promoter Activity of St-RBOHC Induced by MEK2DD via the cis-Element.
Supplemental Figure 2. Analysis of INF1- or R3a/AVR3a-Induced RBOHB -1000 Promoter Activities in SIPK-Silenced Leaves.
Supplemental Figure 3. Identification of WRKY Transcriptional Factors.
Supplemental Figure 4. Phylogenetic Tree of Group I WRKYs Containing the SP Cluster.
Supplemental Figure 5. Induction of WRKY Genes in Response to Flg22, INF1, and R3a/AVR3a.
Supplemental Figure 6. In Vitro Phosphorylation of MBP by Recombinant MAPKs.
Supplemental Figure 7. Phosphorylation of WRKYs Inducing Cell Death by Pathogen-Responsive MAPKs in Vitro.
Supplemental Figure 8. MAPK-Dependent Phosphorylation of WRKY11 in Vitro and in Vivo.
Supplemental Figure 9. Specific Gene Silencing of Single WRKY Gene or Multiple WRKY Genes in TRV:WRKY-Infected Plants.
Supplemental Figure 10. Regulation of Defense-Related Gene Expression by Multiple WRKYs.
Supplemental Figure 11. Effects of Single or Multiple WRKY Gene Silencing on MEK2DD-Dependent ROS Burst.
Supplemental Figure 12. Activation of SIPK and WIPK Induced by R3a/AVR3a.
Supplemental Figure 13. Early and Late Phase ROS Bursts in Response to Flg22 and INF1.
Supplemental Figure 14. Analysis of RBOHB −1000 Promoter in Response to WRKY Variants.
Supplemental Figure 15. Induction of ROS by GFP-Tagged WRKY85D or Nontagged WRKY85D.
Supplemental Figure 16. Subcellular Localization of GFP-WRKY85D.
Supplemental Figure 17. Schematic Representation of the Construct of RBOHB Promoter:GUSint Reporter Gene.
Supplemental Table 1. Phosphorylation Levels of WRKYs by Pathogen-Responsive MAPKs.
Supplemental Table 2. Primers Used for RT-qPCR.
Supplemental Table 3. Primers Used for PCR Targeting Silencing Inserts.
Supplemental Table 4. Primers Used for PCR Targeting WRKY cDNA.
Supplemental Table 5. Sequences of Linkers for Y1H Assay.
Supplemental Table 6. Primers Used for ChIP-qPCR.
Supplemental Table 7. Accession Numbers of the Amino Acid Sequences Included in the Phylogenetic Tree.
Supplemental Data Set 1. Text File of Alignment Used to Generate Supplemental Figure 4.
Supplementary Material
Acknowledgments
We thank Phil Mullineaux and Roger Hellens for pGreen vector, Sophien Kamoun for INF1 and AVR3a/R3a constructs, Andrew O. Jackson for pGD binary vector, David C. Baulcombe for TRV vector and p19 construct, and the Leaf Tobacco Research Center, Japan, for N. benthamiana seeds. We also thank Kenta Hatae and members of the Radioisotope Research Center, Nagoya University, for technical assistance. This work was supported by a Grant-in-Aid for Scientific Research on Innovative Areas “Oxygen Biology: a new criterion for integrated understanding of life” (15H01398 to H.Y.) and Scientific Research (20425630 to K.S.) from MEXT of Japan and by a Grant-in-Aid for Scientific Research (26292023 to H.Y. and 264206 to H.A.) from the Japan Society of the Promotion of Science, by Council for Science, Technology, and Innovation, Cross-ministerial Strategic Innovation Promotion Program of Japan (to H.Y.).
AUTHOR CONTRIBUTIONS
H.A., T.N., N.M., T.Y., K.S., and H.Y. designed the research. H.A., T.N., N.M., T.Y., Y.K., M.Y., and N.I. performed the research. H.A., T.N., N.M., T.Y., and H.Y. analyzed the data. H.A. and H.Y. wrote the article.
Glossary
- PRR
pattern recognition receptor
- PAMP
pathogen-associated molecular pattern
- PTI
pattern-triggered immunity
- ETI
effector-triggered immunity
- ROS
reactive oxygen species
- MAPK
mitogen-activated protein kinase
- HR
hypersensitive response
- WIPK
wound-induced protein kinase
- SIPK
salicylic acid-induced protein kinase
- TRV
Tobacco rattle virus
- ChIP-qPCR
chromatin immunoprecipitation-quantitative PCR
Footnotes
Articles can be viewed online without a subscription.
References
- Adachi H., Yoshioka H. (2015). Kinase-mediated orchestration of NADPH oxidase in plant immunity. Brief. Funct. Genomics 14: 253–259. [DOI] [PubMed] [Google Scholar]
- Andreasson E., et al. (2005). The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J. 24: 2579–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai S., Ichikawa T., Nomura H., Kobayashi M., Kamiyoshihara Y., Mori H., Kadota Y., Zipfel C., Jones J.D.G., Yoshioka H. (2013). The variable domain of a plant calcium-dependent protein kinase (CDPK) confers subcellular localization and substrate recognition for NADPH oxidase. J. Biol. Chem. 288: 14332–14340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai S., Ohta K., Yoshioka H. (2008). MAPK signaling regulates nitric oxide and NADPH oxidase-dependent oxidative bursts in Nicotiana benthamiana. Plant Cell 20: 1390–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983. [DOI] [PubMed] [Google Scholar]
- Blume B., Nürnberger T., Nass N., Scheel D. (2000). Receptor-mediated increase in cytoplasmic free calcium required for activation of pathogen defense in parsley. Plant Cell 12: 1425–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos J.I., Kanneganti T.D., Young C., Cakir C., Huitema E., Win J., Armstrong M.R., Birch P.R., Kamoun S. (2006). The C-terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a-mediated hypersensitivity and suppress INF1-induced cell death in Nicotiana benthamiana. Plant J. 48: 165–176. [DOI] [PubMed] [Google Scholar]
- Chai H.B., Doke N. (1987). Activation of the potential of potato leaf tissue to react hypersensitively to Phytophthora infestans by cytospore germination fluid and the enhancement of this potential by calcium ions. Physiol. Mol. Plant Pathol. 30: 27–37. [Google Scholar]
- Chaparro-Garcia A., Wilkinson R.C., Gimenez-Ibanez S., Findlay K., Coffey M.D., Zipfel C., Rathjen J.P., Kamoun S., Schornack S. (2011). The receptor-like kinase SERK3/BAK1 is required for basal resistance against the late blight pathogen phytophthora infestans in Nicotiana benthamiana. PLoS One 6: e16608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Chen Z. (2000). Isolation and characterization of two pathogen- and salicylic acid-induced genes encoding WRKY DNA-binding proteins from tobacco. Plant Mol. Biol. 42: 387–396. [DOI] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J.D.G., Felix G., Boller T. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500. [DOI] [PubMed] [Google Scholar]
- Coll N.S., Epple P., Dangl J.L. (2011). Programmed cell death in the plant immune system. Cell Death Differ. 18: 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre F., Gutiérrez-Beltrán E., Pareja-Jaime Y., Chakravarthy S., Martin G.B., del Pozo O. (2013). The tomato calcium sensor Cbl10 and its interacting protein kinase Cipk6 define a signaling pathway in plant immunity. Plant Cell 25: 2748–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., et al. (2015). Elicitin recognition confers enhanced resistance to Phytophthora infestans in potato. Nat. Plants 1: 15034. [DOI] [PubMed] [Google Scholar]
- Eulgem T., Somssich I.E. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10: 366–371. [DOI] [PubMed] [Google Scholar]
- Gao X., Chen X., Lin W., Chen S., Lu D., Niu Y., Li L., Cheng C., McCormack M., Sheen J., Shan L., He P. (2013). Bifurcation of Arabidopsis NLR immune signaling via Ca²⁺-dependent protein kinases. PLoS Pathog. 9: e1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotin A.L., Calipel A., Brossas J.Y., Faussat A.M., Tréton J., Mascarelli F. (2006). Sustained versus transient ERK1/2 signaling underlies the anti- and proapoptotic effects of oxidative stress in human RPE cells. Invest. Ophthalmol. Vis. Sci. 47: 4614–4623. [DOI] [PubMed] [Google Scholar]
- Grant M., Brown I., Adams S., Knight M., Ainslie A., Mansfield J. (2000). The RPM1 plant disease resistance gene facilitates a rapid and sustained increase in cytosolic calcium that is necessary for the oxidative burst and hypersensitive cell death. Plant J. 23: 441–450. [DOI] [PubMed] [Google Scholar]
- Hao H., Fan L., Chen T., Li R., Li X., He Q., Botella M.A., Lin J. (2014). Clathrin and membrane microdomains cooperatively regulate RbohD dynamics and activity in Arabidopsis. Plant Cell 26: 1729–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese A., Hann D.R., Gimenez-Ibanez S., Jones A.M., He K., Li J., Schroeder J.I., Peck S.C., Rathjen J.P. (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA 104: 12217–12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama N., Adachi H., Yoshioka M., Yoshioka H. (2014). In vivo phosphorylation of WRKY transcription factor by MAPK. Methods Mol. Biol. 1171: 171–181. [DOI] [PubMed] [Google Scholar]
- Ishihama N., Yamada R., Yoshioka M., Katou S., Yoshioka H. (2011). Phosphorylation of the Nicotiana benthamiana WRKY8 transcription factor by MAPK functions in the defense response. Plant Cell 23: 1153–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama N., Yoshioka H. (2012). Post-translational regulation of WRKY transcription factors in plant immunity. Curr. Opin. Plant Biol. 15: 431–437. [DOI] [PubMed] [Google Scholar]
- Jones J.D.G., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Journot-Catalino N., Somssich I.E., Roby D., Kroj T. (2006). The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 18: 3289–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota Y., Shirasu K. (2012). The HSP90 complex of plants. Biochim. Biophys. Acta 1823: 689–697. [DOI] [PubMed] [Google Scholar]
- Kadota Y., Sklenar J., Derbyshire P., Stransfeld L., Asai S., Ntoukakis V., Jones J.D.G., Shirasu K., Menke F., Jones A., Zipfel C. (2014). Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell 54: 43–55. [DOI] [PubMed] [Google Scholar]
- Kamoun S., van West P., Vleeshouwers V.G., de Groot, K.E., Govers F. (1998). Resistance of Nicotiana benthamiana to Phytophthora infestans is mediated by the recognition of the elicitor protein INF1. Plant Cell 10: 1413–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanneganti T.D., Huitema E., Cakir C., Kamoun S. (2006). Synergistic interactions of the plant cell death pathways induced by Phytophthora infestans Nepl-like protein PiNPP1.1 and INF1 elicitin. Mol. Plant Microbe Interact. 19: 854–863. [DOI] [PubMed] [Google Scholar]
- Kanzaki H., Saitoh H., Ito A., Fujisawa S., Kamoun S., Katou S., Yoshioka H., Terauchi R. (2003). Cytosolic HSP90 and HSP70 are essential components of INF1-mediated hypersensitive response and non-host resistance to Pseudomonas cichorii in Nicotiana benthamiana. Mol. Plant Pathol. 4: 383–391. [DOI] [PubMed] [Google Scholar]
- Kaufmann K., Muiño J.M., Østerås M., Farinelli L., Krajewski P., Angenent G.C. (2010). Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat. Protoc. 5: 457–472. [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Ishihama N., Yoshioka H., Takabatake R., Tsuda S., Seo S., Ohashi Y., Mitsuhara I. (2010). Analyses of the cis-regulatory regions responsible for the transcriptional activation of the N resistance gene by Tobacco mosaic virus. J. Phytopathol. 158: 826–828. [Google Scholar]
- Kobayashi M., Kawakita K., Maeshima M., Doke N., Yoshioka H. (2006). Subcellular localization of Strboh proteins and NADPH-dependent O2(-)-generating activity in potato tuber tissues. J. Exp. Bot. 57: 1373–1379. [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Ohura I., Kawakita K., Yokota N., Fujiwara M., Shimamoto K., Doke N., Yoshioka H. (2007). Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell 19: 1065–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M., Yoshioka M., Asai S., Nomura H., Kuchimura K., Mori H., Doke N., Yoshioka H. (2012). StCDPK5 confers resistance to late blight pathogen but increases susceptibility to early blight pathogen in potato via reactive oxygen species burst. New Phytol. 196: 223–237. [DOI] [PubMed] [Google Scholar]
- Le Roux C., et al. (2015). A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell 161: 1074–1088. [DOI] [PubMed] [Google Scholar]
- Li G., Meng X., Wang R., Mao G., Han L., Liu Y., Zhang S. (2012). Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genet. 8: e1002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Li M., Yu L., Zhou Z., Liang X., Liu Z., Cai G., Gao L., Zhang X., Wang Y., Chen S., Zhou J.M. (2014). The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15: 329–338. [DOI] [PubMed] [Google Scholar]
- Lu D., Lin W., Gao X., Wu S., Cheng C., Avila J., Heese A., Devarenne T.P., He P., Shan L. (2011). Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao G., Meng X., Liu Y., Zheng Z., Chen Z., Zhang S. (2011). Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23: 1639–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke F.L., Kang H.G., Chen Z., Park J.M., Kumar D., Klessig D.F. (2005). Tobacco transcription factor WRKY1 is phosphorylated by the MAP kinase SIPK and mediates HR-like cell death in tobacco. Mol. Plant Microbe Interact. 18: 1027–1034. [DOI] [PubMed] [Google Scholar]
- Mersmann S., Bourdais G., Rietz S., Robatzek S. (2010). Ethylene signaling regulates accumulation of the FLS2 receptor and is required for the oxidative burst contributing to plant immunity. Plant Physiol. 154: 391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G., Schlauch K., Tam R., Cortes D., Torres M.A., Shulaev V., Dangl J.L., Mittler R. (2009). The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci. Signal. 2: ra45. [DOI] [PubMed] [Google Scholar]
- Mur L.A., Kenton P., Lloyd A.J., Ougham H., Prats E. (2008). The hypersensitive response; the centenary is upon us but how much do we know? J. Exp. Bot. 59: 501–520. [DOI] [PubMed] [Google Scholar]
- Murphy L.O., MacKeigan J.P., Blenis J. (2004). A network of immediate early gene products propagates subtle differences in mitogen-activated protein kinase signal amplitude and duration. Mol. Cell. Biol. 24: 144–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy L.O., Smith S., Chen R.H., Fingar D.C., Blenis J. (2002). Molecular interpretation of ERK signal duration by immediate early gene products. Nat. Cell Biol. 4: 556–564. [DOI] [PubMed] [Google Scholar]
- Nakano M., Nishihara M., Yoshioka H., Takahashi H., Sawasaki T., Ohnishi K., Hikichi Y., Kiba A. (2013). Suppression of DS1 phosphatidic acid phosphatase confirms resistance to Ralstonia solanacearum in Nicotiana benthamiana. PLoS One 8: e75124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima A., Chen L., Thao N.P., Fujiwara M., Wong H.L., Kuwano M., Umemura K., Shirasu K., Kawasaki T., Shimamoto K. (2008). RACK1 functions in rice innate immunity by interacting with the Rac1 immune complex. Plant Cell 20: 2265–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noirot E., Der C., Lherminier J., Robert F., Moricova P., Kiêu K., Leborgne-Castel N., Simon-Plas F., Bouhidel K. (2014). Dynamic changes in the subcellular distribution of the tobacco ROS-producing enzyme RBOHD in response to the oomycete elicitor cryptogein. J. Exp. Bot. 65: 5011–5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S.P., Somssich I.E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150: 1648–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.J., Miura Y., Kawakita K., Yoshioka H., Doke N. (1998). Physiological mechanisms of a sub-systemic oxidative burst triggered by elicitor-induced local oxidative burst in potato tuber slices. Plant Cell Physiol. 39: 1218–1225. [Google Scholar]
- Peart J.R., et al. (2002). Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc. Natl. Acad. Sci. USA 99: 10865–10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedley K.F., Martin G.B. (2005). Role of mitogen-activated protein kinases in plant immunity. Curr. Opin. Plant Biol. 8: 541–547. [DOI] [PubMed] [Google Scholar]
- Popescu S.C., Popescu G.V., Bachan S., Zhang Z., Gerstein M., Snyder M., Dinesh-Kumar S.P. (2009). MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 23: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff F., Martin-Hernandez A.M., Baulcombe D.C. (2001). Technical Advance. Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 25: 237–245. [DOI] [PubMed] [Google Scholar]
- Ren D., Yang K.Y., Li G.J., Liu Y., Zhang S. (2006). Activation of Ntf4, a tobacco mitogen-activated protein kinase, during plant defense response and its involvement in hypersensitive response-like cell death. Plant Physiol. 141: 1482–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S., Chinchilla D., Boller T. (2006). Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20: 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton P.J., Somssich I.E., Ringler P., Shen Q.J. (2010). WRKY transcription factors. Trends Plant Sci. 15: 247–258. [DOI] [PubMed] [Google Scholar]
- Sabbagh W. Jr., Flatauer L.J., Bardwell A.J., Bardwell L. (2001). Specificity of MAP kinase signaling in yeast differentiation involves transient versus sustained MAPK activation. Mol. Cell 8: 683–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagi M., Fluhr R. (2001). Superoxide production by plant homologues of the gp91(phox) NADPH oxidase. Modulation of activity by calcium and by tobacco mosaic virus infection. Plant Physiol. 126: 1281–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segonzac C., Feike D., Gimenez-Ibanez S., Hann D.R., Zipfel C., Rathjen J.P. (2011). Hierarchy and roles of pathogen-associated molecular pattern-induced responses in Nicotiana benthamiana. Plant Physiol. 156: 687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S., Okamoto M., Seto H., Ishizuka K., Sano H., Ohashi Y. (1995). Tobacco MAP kinase: a possible mediator in wound signal transduction pathways. Science 270: 1988–1992. [DOI] [PubMed] [Google Scholar]
- Sheikh A.H., Raghuram B., Eschen-Lippold L., Scheel D., Lee J., Sinha A.K. (2014). Agroinfiltration by cytokinin-producing Agrobacterium sp. strain GV3101 primes defense responses in Nicotiana tabacum. Mol. Plant Microbe Interact. 27: 1175–1185. [DOI] [PubMed] [Google Scholar]
- Shen Q.H., Saijo Y., Mauch S., Biskup C., Bieri S., Keller B., Seki H., Ulker B., Somssich I.E., Schulze-Lefert P. (2007). Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315: 1098–1103. [DOI] [PubMed] [Google Scholar]
- Smith J.M., Salamango D.J., Leslie M.E., Collins C.A., Heese A. (2014). Sensitivity to Flg22 is modulated by ligand-induced degradation and de novo synthesis of the endogenous flagellin-receptor FLAGELLIN-SENSING2. Plant Physiol. 164: 440–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Miller G., Morales J., Shulaev V., Torres M.A., Mittler R. (2011). Respiratory burst oxidases: the engines of ROS signaling. Curr. Opin. Plant Biol. 14: 691–699. [DOI] [PubMed] [Google Scholar]
- Takahashi A., Casais C., Ichimura K., Shirasu K. (2003). HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc. Natl. Acad. Sci. USA 100: 11777–11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Ishihama N., Yoshioka H., Huser A., O’Connell R., Tsuji G., Tsuge S., Kubo Y. (2009). The Colletotrichum orbiculare SSD1 mutant enhances Nicotiana benthamiana basal resistance by activating a mitogen-activated protein kinase pathway. Plant Cell 21: 2517–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M.A. (2010). ROS in biotic interactions. Physiol. Plant. 138: 414–429. [DOI] [PubMed] [Google Scholar]
- Torres M.A., Dangl J.L., Jones J.D.G. (2002). Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 99: 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traverse S., Gomez N., Paterson H., Marshall C., Cohen P. (1992). Sustained activation of the mitogen-activated protein (MAP) kinase cascade may be required for differentiation of PC12 cells. Comparison of the effects of nerve growth factor and epidermal growth factor. Biochem. J. 288: 351–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K., Katagiri F. (2010). Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 13: 459–465. [DOI] [PubMed] [Google Scholar]
- Tsuda K., Mine A., Bethke G., Igarashi D., Botanga C.J., Tsuda Y., Glazebrook J., Sato M., Katagiri F. (2013). Dual regulation of gene expression mediated by extended MAPK activation and salicylic acid contributes to robust innate immunity in Arabidopsis thaliana. PLoS Genet. 9: e1004015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956. [DOI] [PubMed] [Google Scholar]
- Wong H.L., Pinontoan R., Hayashi K., Tabata R., Yaeno T., Hasegawa K., Kojima C., Yoshioka H., Iba K., Kawasaki T., Shimamoto K. (2007). Regulation of rice NADPH oxidase by binding of Rac GTPase to its N-terminal extension. Plant Cell 19: 4022–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaeno T., Li H., Chaparro-Garcia A., Schornack S., Koshiba S., Watanabe S., Kigawa T., Kamoun S., Shirasu K. (2011). Phosphatidylinositol monophosphate-binding interface in the oomycete RXLR effector AVR3a is required for its stability in host cells to modulate plant immunity. Proc. Natl. Acad. Sci. USA 108: 14682–14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamizo C., Kuchimura K., Kobayashi A., Katou S., Kawakita K., Jones J.D.G., Doke N., Yoshioka H. (2006). Rewiring mitogen-activated protein kinase cascade by positive feedback confers potato blight resistance. Plant Physiol. 140: 681–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K.Y., Liu Y., Zhang S. (2001). Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc. Natl. Acad. Sci. USA 98: 741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka H., Mase K., Yoshioka M., Kobayashi M., Asai S. (2011). Regulatory mechanisms of nitric oxide and reactive oxygen species generation and their role in plant immunity. Nitric Oxide 25: 216–221. [DOI] [PubMed] [Google Scholar]
- Yoshioka H., Numata N., Nakajima K., Katou S., Kawakita K., Rowland O., Jones J.D.G., Doke N. (2003). Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell 15: 706–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka H., Sugie K., Park H.J., Maeda H., Tsuda N., Kawakita K., Doke N. (2001). Induction of plant gp91 phox homolog by fungal cell wall, arachidonic acid, and salicylic acid in potato. Mol. Plant Microbe Interact. 14: 725–736. [DOI] [PubMed] [Google Scholar]
- Zhang J., et al. (2007). A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1: 175–185. [DOI] [PubMed] [Google Scholar]
- Zhang S., Klessig D.F. (1997). Salicylic acid activates a 48-kD MAP kinase in tobacco. Plant Cell 9: 809–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhu H., Zhang Q., Li M., Yan M., Wang R., Wang L., Welti R., Zhang W., Wang X. (2009). Phospholipase dalpha1 and phosphatidic acid regulate NADPH oxidase activity and production of reactive oxygen species in ABA-mediated stomatal closure in Arabidopsis. Plant Cell 21: 2357–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C., Robatzek S., Navarro L., Oakeley E.J., Jones J.D.G., Felix G., Boller T. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767. [DOI] [PubMed] [Google Scholar]
- Zuo J., Niu Q.W., Chua N.H. (2000). Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24: 265–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.