The plastid HCF145 protein, composed of two ligand binding domains followed by a newly defined transcript binding motif repeat (TMR) domain, is essential for plant growth, binds to the 5′ UTR of the plastid tricistronic psaA-psaB-rps14 mRNA, and protects it from degradation.
Abstract
The seedling-lethal Arabidopsis thaliana high chlorophyll fluorescence145 (hcf145) mutation leads to reduced stability of the plastid tricistronic psaA-psaB-rps14 mRNA and photosystem I (PSI) deficiency. Here, we genetically mapped the HCF145 gene, which encodes a plant-specific, chloroplast-localized, modular protein containing two homologous domains related to the polyketide cyclase family comprising 37 annotated Arabidopsis proteins of unknown function. Two further highly conserved and previously uncharacterized tandem repeat motifs at the C terminus, herein designated the transcript binding motif repeat (TMR) domains, confer sequence-specific RNA binding capability to HCF145. Homologous TMR motifs are often found as multiple repeats in quite diverse proteins of green and red algae and in the cyanobacterium Microcoleus sp PCC 7113 with unknown function. HCF145 represents the only TMR protein found in vascular plants. Detailed analysis of hcf145 mutants in Arabidopsis and Physcomitrella patens as well as in vivo and in vitro RNA binding assays indicate that HCF145 has been recruited in embryophyta for the stabilization of the psaA-psaB-rps14 mRNA via specific binding to its 5′ untranslated region. The polyketide cyclase-related motifs support association of the TMRs to the psaA RNA, presumably pointing to a regulatory role in adjusting PSI levels according to the requirements of the plant cell.
INTRODUCTION
The chloroplast is the product of an endosymbiotic event by which a cyanobacterium was ingested by a eukaryotic cell. Chloroplast biogenesis and function requires adaption of transcription rates and posttranscriptional events in the development- and environment-dependent control of gene expression (Stern et al., 2010; Barkan, 2011; Yagi and Shiina, 2014; Börner et al., 2015). Chloroplasts have retained some parts of the general eubacterial RNA degradation system, like marking of RNAs by polyadenylation and the ribonucleases involved in RNA processing and degradation (Schuster and Stern, 2009; Stoppel and Meurer, 2012).
Unlike in cyanobacteria, nearly, if not all, polycistronic precursor transcripts are processed by endo- and exonucleases, splicing activities, and editing events (Stern et al., 2010; Stoppel and Meurer, 2012, 2013; Germain et al., 2013). In several cases, processing within intergenic regions is required for subsequent translation of the 5′ processed product (Germain et al., 2013) and regulation of stability of individual mRNAs at both ends (Barkan, 2011; Zhelyazkova et al., 2012). The 3′ processing has been hypothesized to be a prerequisite also for efficient translation of plastid transcripts (Stoppel et al., 2011). Degradation of plastid RNAs is often initiated by endonucleolytic processing and by the subsequent action of unspecific 5′ → 3′ and 3′ → 5′ exonucleases (Germain et al., 2013). The resulting processing products have obtained phylogenetically new 5′ and 3′ untranslated regions (UTRs) that have to be stabilized either by complex secondary stem loop structures and/or by the recruitment of factors that have gained sequence-specific RNA binding capabilities (Stern et al., 2010; Barkan, 2011; Germain et al., 2013). Accordingly, many UTR binding proteins also evolved de novo. This is supported by the presence of only few RNases with little sequence specificity in chloroplasts (Stoppel and Meurer, 2012) and the action of nuclear-encoded proteins in a gene-specific manner (Stern et al., 2010; Barkan, 2011; Germain et al., 2013).
Although the chloroplast still retains its own genome as well as a transcription and translation apparatus of cyanobacterial origin, most nuclear-encoded proteins involved in posttranscriptional RNA processing within this organelle evolved during endosymbiotic evolution and homologs are rarely found in cyanobacteria or even algae. Examples are PALE CRESS, ACCUMULATION OF PHOTOSYSTEM ONE1 (APO1) to APO4, PEPTIDE CHAIN RELEASE FACTOR B3 (PrfB3), RHO-N domain protein (RHON1), PLANT ORGANELLE RNA RECOGNITION domains, and many chloroplast PENTATRICOPEPTIDEREPEAT (PPR) proteins (Meurer et al., 1998; Kroeger et al., 2009; Stoppel et al., 2011, 2012; Watkins et al., 2011; Shikanai and Fujii, 2013; Barkan and Small, 2014). Recruitment of novel factors for the management and regulation of the plastid and mitochondrial RNA metabolism was presumably driven by the acquisition of introns, massive endonucleolytic cleavage of precursor transcripts, numerous editing sites, and the fast divergence of noncoding sequences in the intergenic as well as the 5′ and 3′ UTRs in the genome of the endosymbiont (Barkan, 2011; Manavski et al., 2012a). Homologs of only few factors for chloroplast RNA metabolism are also found in cyanobacteria. However, they significantly diversified by recruiting novel domains and/or extensions and changed their functions, as was found for CHLOROPLAST RNA SPLICING2 (Jenkins and Barkan, 2001), HCF109 (Meurer et al., 2002), CHLOROPLAST RNA SPLICING AND RIBOSOME MATURATION (Barkan et al., 2007), PrfB3 (Stoppel et al., 2011), and RNase E (Mudd et al., 2008; Schein et al., 2008; Stoppel et al., 2012).
Plastome-genome coevolution is an ongoing process that diverges in different lineages and retains species-specific interactions on the level of the RNA metabolism since it represents a fast-evolving process (Manavski et al., 2012a). Little is known about factors that authentically regulate plastid mRNA stability and even less about how metabolic processes as well as endogenous and external stimuli are involved in this regulation. This regulation is, for instance, essential to cope with photosystem II (PSII) excitation pressure and plastidic redox stress induced by changes in temperature and light intensity (Stoppel et al., 2011; Kupsch et al., 2012). In summary, divergence of plastid UTRs and appearance of phylogenetically novel RNA processing sites provide new platforms at the RNA extremities for RNA binding factors to implement molecular processes in response to environmental stimuli, emphasizing the importance of RNA metabolism and translation in adjusting chloroplast homeostasis (Stern et al., 2010; Stoppel and Meurer, 2013).
The previously described nuclear hcf145-1 mutant in Arabidopsis thaliana shows severely decreased stability of the tricistronic psaA-psaB-rps14 transcript encoding the two major reaction center proteins, PsaA and PsaB, of photosystem I (PSI) and the ribosomal subunit Rps14 (Lezhneva and Meurer, 2004). The rps14 transcript is efficiently cleaved from the precursor transcript and accumulates at almost normal levels, allowing wild-type-comparable translation efficiency in the mutant. The resulting dicistronic psaA-psaB transcript accumulates neither in the wild type nor in the mutant, indicating fast degradation of this processing product (Lezhneva and Meurer, 2004). The decreased RNA stability of the tricistronic precursor RNA causes a specific PSI deficiency and seedling lethality, demonstrating an essential role of HCF145 in plastid gene expression. Here, to analyze the function of the protein, the HCF145 gene was identified by high-resolution mapping. HCF145 encodes a plant-specific chloroplast protein of unknown function. HCF145 was found to be a modular protein containing two types of tandem repeat domains of unknown function: one displayed significant structural similarity to the ubiquitous SRPBCC superfamily encoding a deep hydrophobic ligand binding pocket and is related to the polyketide cyclase subfamily. The other tandemly repeated motif, located at the C terminus of HCF145, confers specific binding capability to the psaA 5′ UTR. Based on our phylogenetic and molecular analysis, we suggest that HCF145 is composed of modules of cyanobacterial origin and evolved to stabilize the psaA-psaB-rps14 mRNA.
RESULTS
Molecular Mapping of HCF145
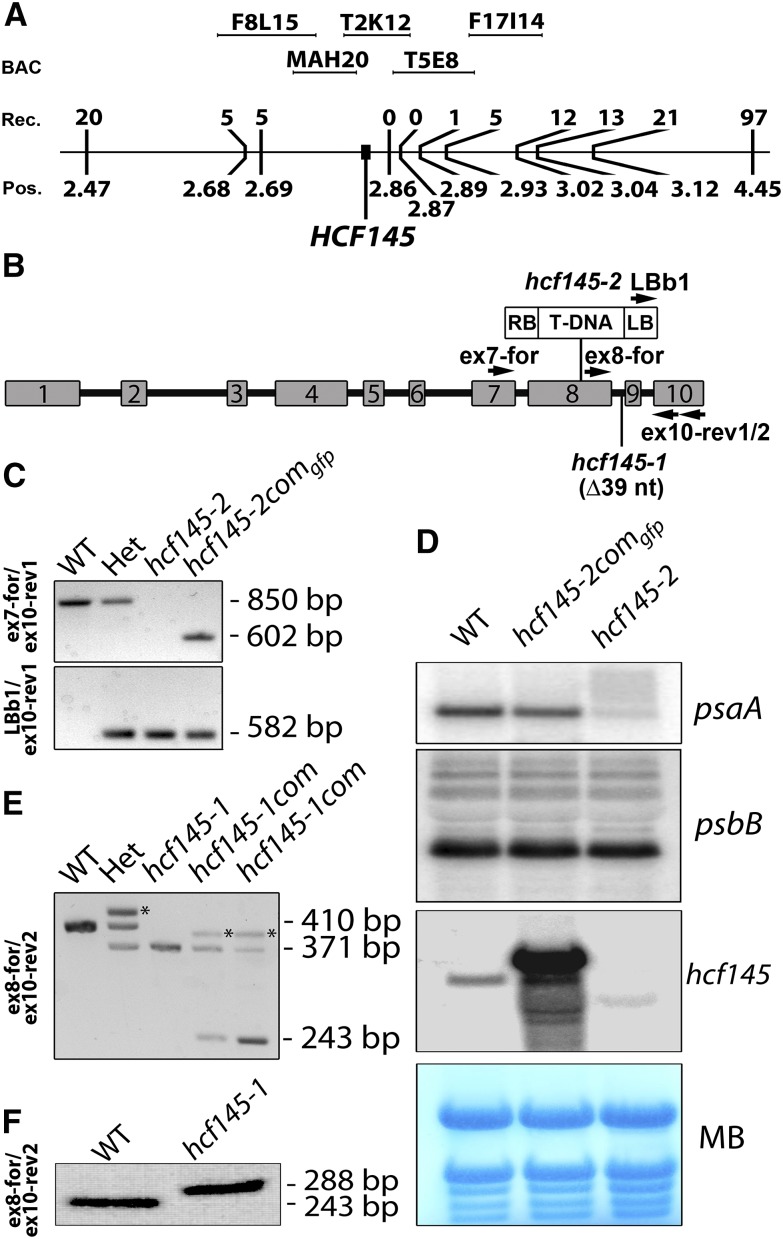
The previously described hcf145-1 mutant accumulates <10% of the tricistronic psaA-psaB-rps14 mRNA compared with the wild type (Lezhneva and Meurer, 2004). This precursor RNA is produced by the plastid-encoded polymerase (PEP). Run-on analysis revealed that the nucleus-encoded factor HCF145 is involved primarily in psaA-psaB-rps14 mRNA stabilization rather than in transcription of this tricistronic RNA in Arabidopsis (Lezhneva and Meurer, 2004). The tetracistronic ycf3-psaA-psaB-rps14 precursor, which is suggested to be generated by the nuclear-encoded phage-type RNA polymerase(s) (NEP) in vascular plants (Summer et al., 2000; Legen et al., 2002; Cho et al., 2009), and processing products accumulate at slightly higher rates. Both precursors have the same 3′ end, indicating that the determinant for RNA stabilization is located at the 5′ end of the tricistronic transcript (Lezhneva and Meurer, 2004). The reduction of the psaA-psaB-rps14 mRNA levels leads to a corresponding reduction of PSI amounts. To identify the nuclear HCF145 gene, the hcf145-1 mutant was previously mapped roughly to the upper arm of chromosome 5 between the molecular simple sequence length polymorphic markers nga158 and nga151 (http://www.arabidopsis.org) located at positions 1.69 and 4.67 Mb, respectively (Lezhneva and Meurer, 2004). Here, further mapping using 1281 F2 mutant plants derived from backcrosses to the Arabidopsis accession Landsberg erecta localized the mutation at the map position of ∼2.8 Mb, with zero recombinants in the region covered by the two bacterial artificial chromosomes T2K12 and T5E8 (Figure 1A).
Figure 1.
Mapping, PCR, and RNA Gel Blot Analysis of the Arabidopsis hcf145 Mutants.
(A) Genetic mapping of the HCF145 mutation localized at map position (Pos.) 2.83 Mb on chromosome 5. Backcrosses to the Arabidopsis accession Landsberg erecta using 1281 F2 mutants obtained 0 recombinants (Rec.) in the region covered by the BACs T2K12 and T5E8.
(B) Schematic representation of the HCF145 gene, indicating the 39-nucleotide deletion (∆39 nt) in hcf145-1, the T-DNA insertion in hcf145-2, and the location of the oligonucleotides used for PCR analysis.
(C) Genotyping of wild-type, heterozygous, hcf145-2, and complemented hcf145-2comgfp lines by PCR analysis. Genomic and T-DNA primers used are shown on the left and in (B). The data demonstrate homozygosity of hcf145-2, successful complementation (upper panel), and the presence of the T-DNA in hcf145-2 (lower panel).
(D) Expression of psaA and hcf145 in wild-type, mutant, and complemented lines. RNA gel blot analysis was performed using 10 µg total leaf RNA with strand-specific 80-mer oligonucleotides (psaA) and a full-length HCF145 cDNA (hcf145) as hybridization probes. MB, methylene blue.
(E) Selection of the homozygous and complemented hcf145-1 mutants by PCR analysis using the primers ex8-for and ex10-rev2. A 410-bp fragment was amplified in the wild type and plants heterozygous for hcf145-1 (Het). A smaller fragment of 371 bp was amplified in hcf145-1, hcf145-1com, and Het lines. A 243-bp fragment, corresponding to the cDNA, was amplified in the hcf145-1com lines. The asterisks mark mismatching products of the amplified fragments in the heterozygous and hcf145-1com lines, migrating at higher molecular weight.
(F) RT-PCR analysis of the wild type and hcf145-1. Exon-specific primers were used to amplify the cDNA of mutant and wild-type plants. The sizes of the products are indicated.
Identification of a Second HCF145 Allele
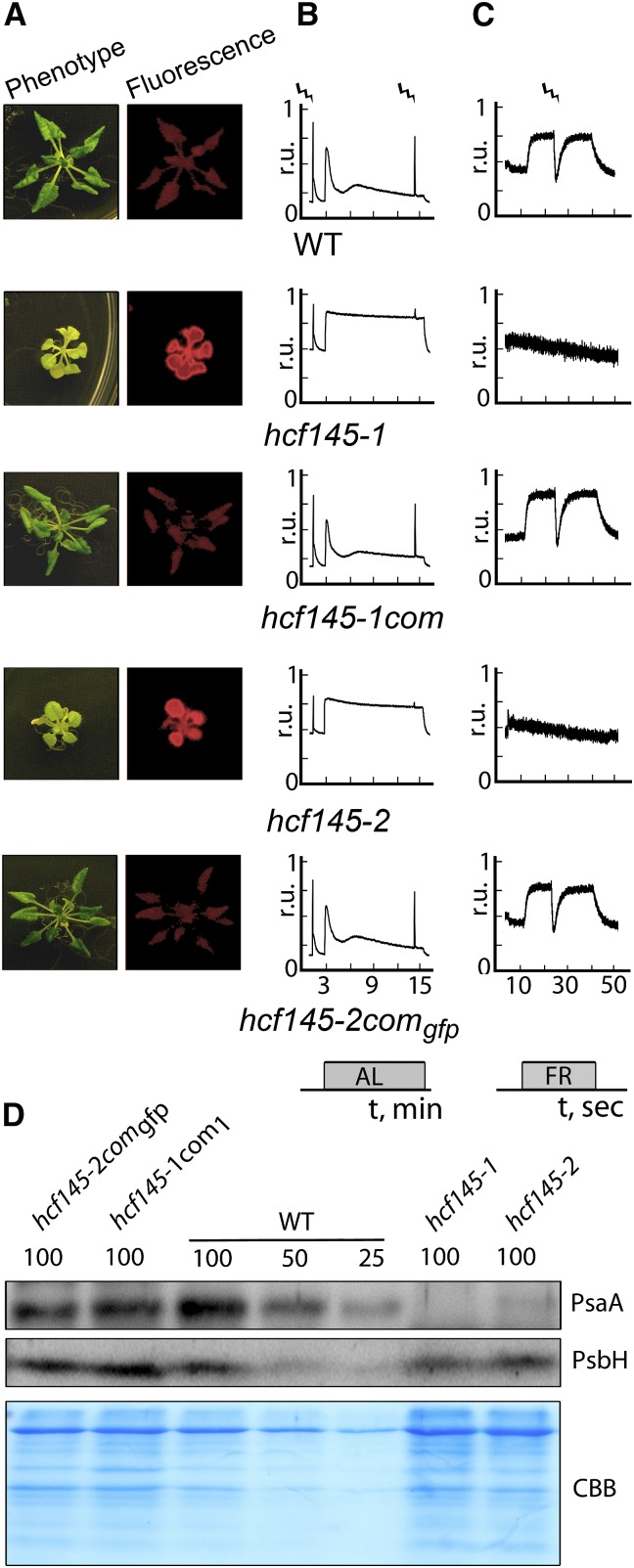
A dozen T-DNA insertion lines around this region were analyzed preliminarily using chlorophyll fluorescence imaging. The Salk_01411 line of the AT5G08720 locus (hcf145-2) appeared pale, grew only on medium supplemented with sucrose, and showed high chlorophyll fluorescence, thus resembling the hcf145-1 phenotype (Meurer et al., 1996a; Lezhneva and Meurer, 2004). We confirmed the T-DNA insertion in exon eight and the homozygosity of the locus in the segregating hcf145-2 mutants by sequencing PCR products (Figures 1B and 1C). The mature transcripts of hcf145 were below the detection limit but some truncated transcripts appeared, presumably because of the gene dissection by the T-DNA in hcf145-2 (Figure 1D). RNA gel blot analysis confirmed the severe reduction of the psaA-psaB-rps14 transcript levels in hcf145-2, similar to what had been shown to be the primary cause of the hcf145-1 mutation (Lezhneva and Meurer, 2004) (Figure 1D). Levels of the psbB and ribosomal transcripts were not affected, further suggesting that hcf145-2 is allelic to hcf145-1 (Figure 1D). Sequencing of the AT5G08720 locus in hcf145-1 identified a deletion of 39 nucleotides including the guanine residue of the 3′ junction of intron eight (Figures 1B and 1E). Sequencing of RT-PCR products revealed that hcf145-1 is unable to splice this intron and therefore produces a premature stop codon (Figure 1F). The hcf145-1 and hcf145-2 mutant plants displayed very similar growth phenotypes (Figure 2A). They were both able to survive only on medium supplemented with an alternate carbon source. The photosynthetic performances of the mutants were also similar as revealed by chlorophyll a fluorescence measurements and P700 absorbance kinetics (Figures 2B and 2C). The singlet-excited states of chlorophyll were hardly quenched upon application of actinic light in both mutants, resulting in a strong hcf phenotype (Figure 2B). Similar to hcf145-1 (Lezhneva and Meurer 2004), the maximum and the effective quantum yield of PSII in hcf145-2 were reduced to 0.54 ± 0.05 and 0.11 ± 0.04 compared with 0.81 ± 0.01 and 0.76 ± 0.03 in the wild type, respectively (Figure 2B). The P700 absorbance changes were below the limit of detection in both mutants indicating a specific deficiency of PSI (Amann et al., 2004; Lezhneva and Meurer, 2004) (Figure 2C). In accordance with the reduced PSI activity and psaA-psaB-rps14 transcript levels, amounts of the PsaA protein were also reduced to much below 25% in both mutants, whereas the PSII protein PsbH (Figure 2D) and many other chloroplast proteins (Lezhneva and Meurer, 2004) appeared at levels comparable to the wild type, confirming the specific loss of PSI in both mutant lines.
Figure 2.
Spectroscopic and Immunological Analyses of the Arabidopsis hcf145 Mutants.
(A) Photographs and chlorophyll a fluorescence images of 3-week-old plants grown on sucrose-supplemented medium at 10 µmol photons m−2 s−1.
(B) Chlorophyll a fluorescence measurements. Strokes indicate saturating light pulses, and bars indicate the duration of actinic light (AL) application. r.u., relative units.
(C) P700 redox kinetics. Strokes indicate saturating light pulses, and bars indicate the duration of far-red light (FR) application. r.u., relative units.
(D) Immunoblot analyses using specific antisera for the large PSI core subunit PsaA and the PSII subunit PsbH. Protein loading corresponds to 4, 2, and 1 µg chlorophyll (100, 50, and 25, respectively). CBB, Coomassie blue.
Molecular Complementation of hcf145 Mutants
To complement the mutant and to characterize the subcellular localization of HCF145 in the plant cell, we stably expressed a full-length HCF145 cDNA in homozygous hcf145-1 lines (hcf145-1com) and a full-length HCF145 protein C-terminally fused to GFP in homozygous hcf145-2 lines (hcf145-2comgfp). Both transgenes were expressed under the control of a constitutive 35S promoter. The homozygous mutants were selected by genotyping and RNA gel blot analysis (Figures 1C to 1E). The constitutive expression of the corresponding cDNAs fully restored photoautotrophic growth, green leaf color and the wild-type-comparable levels of chlorophyll fluorescence in both complemented mutants (Figure 2A). The hcf145 transcripts were greatly upregulated and of larger size due to the GFP fusion in the complemented hcf145-2comgfp lines compared with the wild type (Figure 1D). Transcripts of psaA reappeared in hcf145-2comgfp at wild-type-comparable levels (Figure 1D). The complemented hcf145-1com and hcf145-2comgfp lines recovered the maximum quantum yield of PSII (Fv/Fm = 0.81 ± 0.02 and 0.80 ± 0.02) and quenching of singlet-excited states of chlorophyll (Figures 2A and 2B). P700 absorbance changes and PsaA protein levels were also comparable to the wild type in both complemented lines, demonstrating that PSI activity and levels were also recovered (Figures 2C and 2D).
HCF145 Is a Chloroplast Protein and Has a Modular Composition
Publicly available programs such as PSORT (http://psort.hgc.jp/) and Predotar (https://urgi.versailles.inra.fr/predotar/predotar.html) did not provide conclusive information about the subcellular localization of HCF145. ChloroP (http://www.cbs.dtu.dk/services/ChloroP) predicted a chloroplast transit peptide with a length of 57 amino acid residues. However, based on a previous proteomic study in Arabidopsis (Heazlewood et al., 2004), HCF145 was annotated as a mitochondrial protein in the TAIR and UniProt databases (http://www.arabidopsis.org/index.jsp and http://www.uniprot.org). In a large-scale proteome approach, peptides of HCF145 were found in triton-insoluble fractions, representing the nucleoid fraction of the chloroplast (Phinney and Thelen, 2005).
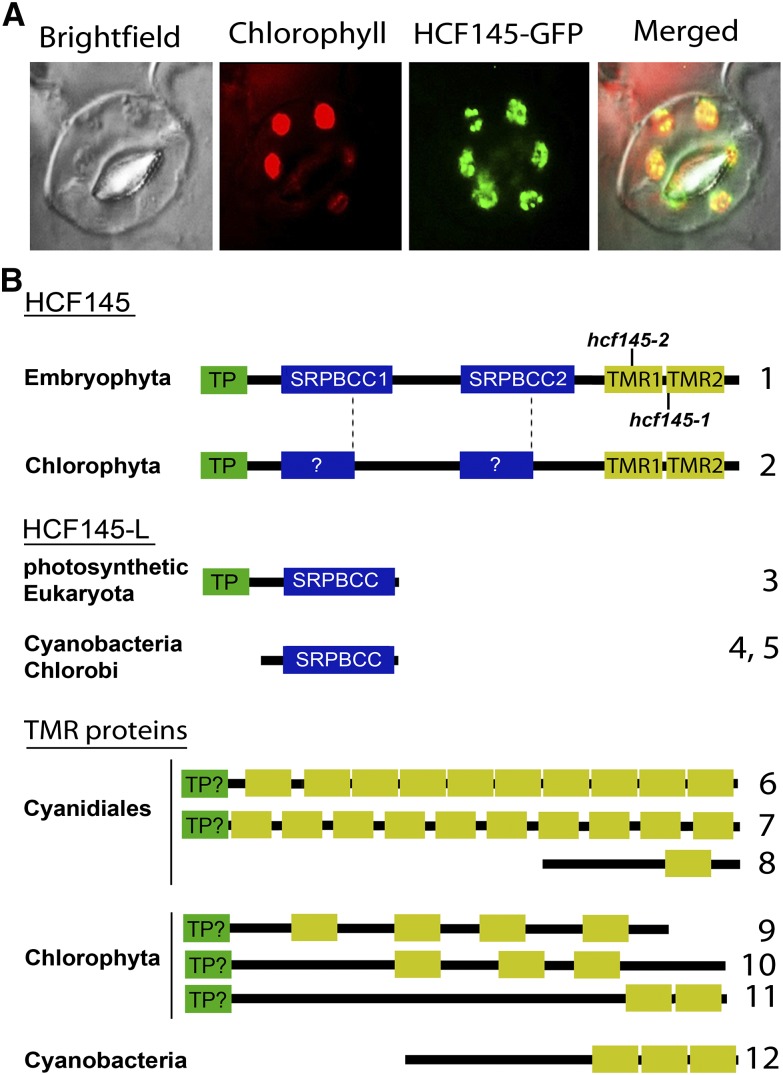
To clarify these discrepancies, we performed confocal microscopy analyses on the homozygous progeny of the stable hcf145-2comgfp transformants. Very bright GFP fluorescence perfectly overlapped with the autofluorescence of the chlorophyll (Figure 3A), demonstrating the chloroplast localization of HCF145. In agreement with the previous data suggesting nucleoid localization of HCF145 (Phinney and Thelen, 2005), we also observed spots of GFP signal within the chloroplast. Even when the fluorescence microscope settings were set on the highest sensitivity, not even traces of GFP fluorescence were detected anywhere else within the cell, demonstrating that HCF145 is very likely exclusively localized in the chloroplast (Figure 3A).
Figure 3.
Subcellular Localization of HCF145 and Its Motifs in HCF145, HCF145-L, and TMR Proteins.
(A) Fluorescence micrographs demonstrating presence of the HCF145-GFP fusion in the chloroplasts of the hcf145-2comgfp guard cells.
(B) Schematic representation and motif organization of HCF145, HCF145-L, and representative TMR proteins in photosynthetic organisms. N-terminal chloroplast transit peptide (TP; green boxes), SRPBCC motifs in HCF145 and HCF145-L proteins (blue boxes), and repeated TMR domains (yellow boxes) are shown. The positions of the mutations found in the hcf145-1 and hcf145-2 lines are indicated. (For multiple sequence alignments of the SRPBCC motifs in HCF145 and HCF145-L, and the TMR proteins, see Supplemental Figures 4, 5, and 7). The accession numbers correspond to the proteins numbered from 1 to 12: 1, Arabidopsis HCF145 (At5g08720); 2, C. subellipsoidea (XM_005645014); 3. Arabidopsis (At4g01650); 4, C. tepidum (AE006470); 5, Synechococcus sp CC9902 (CP000097); 6, C. merolae (XM_005538687); 7, G. sulphuraria (XM_005705295); 8, C. merolae (XM_005538035); 9, C. variabilis (XM_005846292); 10, C. subellipsoidea (C-169 XP_005645828); 11, Ostreococcus lucimarinus (XM_001415510); 12, Microcoleus sp PCC 7113 (AFZ22266).
The mature plant-specific HCF145 protein has a modular composition. It harbors two repeated motifs of unknown function that are related to polyketide cyclases; each comprises ∼160 amino acids, and they are herein named SRPBCC (START/RHO_alpha_C/PITP/Bet_v1/CoxG/CalC) domains, SRPBCC1 and SRPBCC2 (Figure 3B). A visual inspection and sequence analysis of HCF145 using RADAR (http://www.ebi.ac.uk/Tools/pfa/radar), BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi), and HMMER (http://hmmer.janelia.org) identified two additional repeated motifs at the C terminus, which have a length of ∼70 amino acids. These and homologous motifs have not been described nor has a related domain of unknown function been defined (http://pfam.xfam.org). We designated these newly defined motifs as TMR1 and TMR2 (see below; Figure 3B). Multiple TMR repeats that were not associated with SRPBCC motifs were found in more than two dozen proteins in algae and cyanobacteria (http://blast.ncbi.nlm.nih.gov/Blast.cgi; http://hmmer.janelia.org) (Figure 3B). In addition, HCF145 homologs were present in land plants and as truncated forms in green algae. Homologs of both domains could be found in proteins of all photosynthetic lineages (Figure 3B).
The Function of HCF145 Is Conserved in Embryophyta
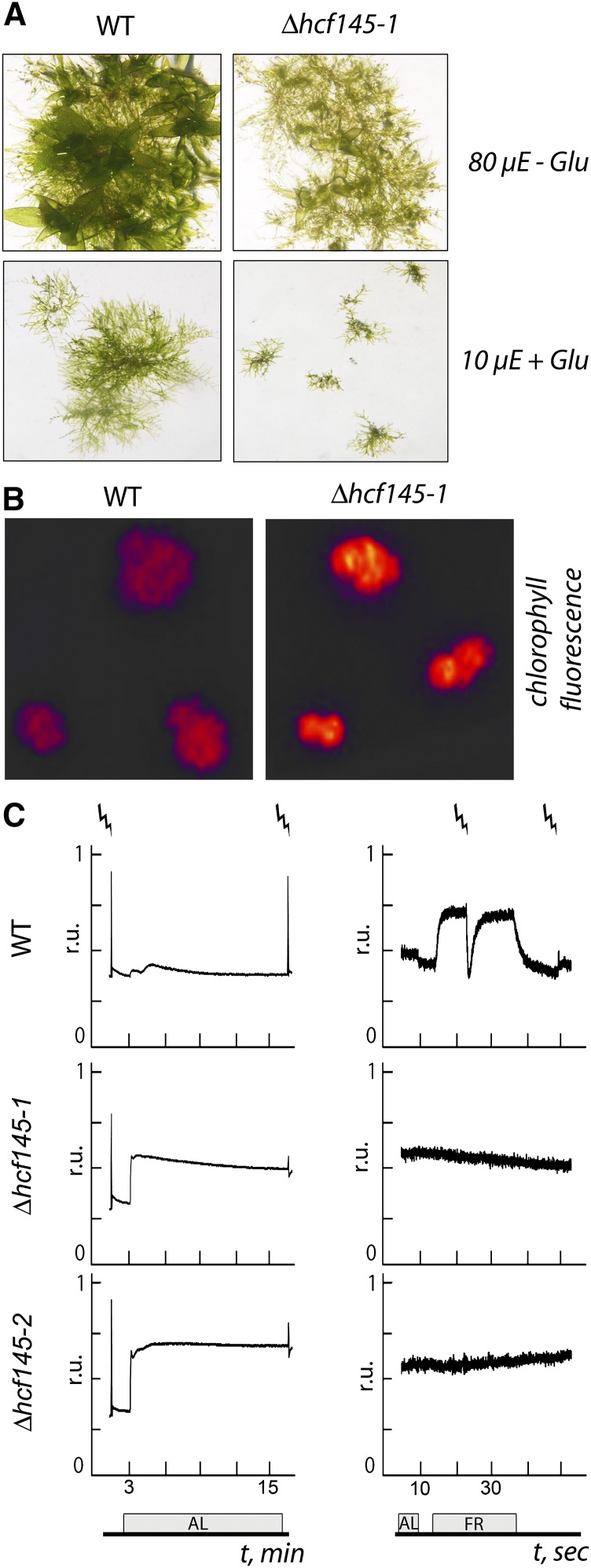
Only one HCF145 homolog exists in the moss Physcomitrella patens. To study the conservation of the HCF145 function, we generated knockout mutants in this moss taking advantage of homologous recombination. Three targeted knockout lines showing precise 5′ and 3′ integration of the knockout construct and lack of hcf145 mRNA were selected (Supplemental Figures 1A to 1C). All three independent mutants, Δhcf145-1 to Δhcf145-3, showed the same growth retardation, pale appearance, and hcf phenotype (Figures 4A and 4B). In strong contrast to the Arabidopsis hcf145 and other PSI mutants, the maximum quantum yield of PSII was unchanged in Δhcf145-1 and Δhcf145-2 (0.70 ± 0.00 and 0.69 ± 0.01, respectively) compared with the wild type (0.69 ± 0.02) (Figure 4C). Thus, even under high growth light conditions P. patens mutants retained normal PSII activity and did not show photoinhibition, which is always observed as a secondary effect in the PSI mutants of vascular plants (Meurer et al., 1996a). The PSII yield and the photochemical quenching were 0.62 ± 0.02 and 0.95 ± 0.02, respectively, in the wild type and reduced by about half in the mutants (sd < 0.05), indicating disturbances in the photosynthetic electron transport rate (Figure 4C). PSI activity was estimated by measuring light-induced changes in P700 absorbance (Figure 4C). The detected signal in the wild type was comparable to that obtained in Arabidopsis (Figure 2C). Under growth light conditions, PSI in the wild type was ∼18% oxidized, and short saturating light pulses of 600 ms were sufficient to induce a complete reduction of PSI in the far-red background light. In the mutants, the signal was below the limit of detection even when the highest sensitivity for measurements was used, demonstrating a severe loss of the PSI activity (Figure 4C).
Figure 4.
Phenotype of the Moss Δhcf145 Mutants.
(A) Wild type and Δhcf145-1 grown for 3 weeks in the absence (-Glu) and presence of glucose (+ Glu) at 80 and 10 μmol photons m−2 s−1, respectively. The phenotype of Δhcf145-1 is in all respects representative for all three knockout lines.
(B) Chlorophyll fluorescence imaging of the wild type and Δhcf145-1, which is representative for all three knockout lines.
(C) Chlorophyll a fluorescence (left) and P700 redox kinetics (right) in the wild type, Δhcf145-1, and Δhcf145-2. Strokes indicate saturating light pulses, and bars the duration of actinic light (AL) or far-red light (FR) application.
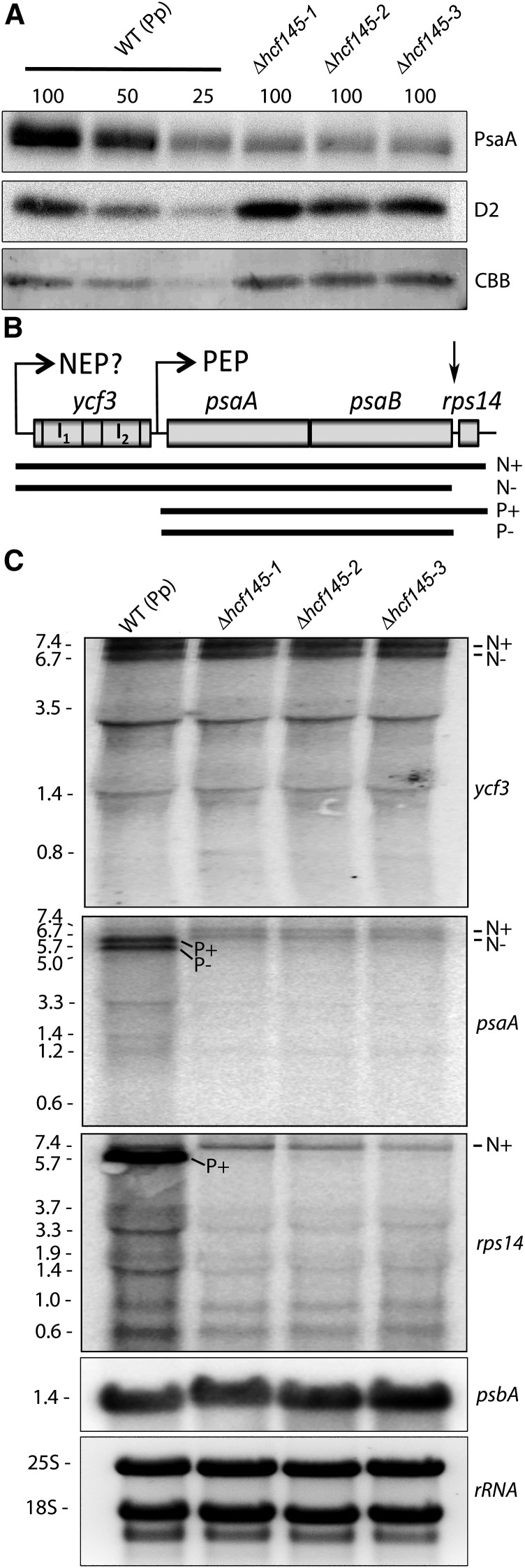
Immunological analysis confirmed that levels of PsaA were reduced to ∼25% in all three mutant lines, whereas those of the PSII protein D2 (PsbD) remained unchanged compared with the wild type (Figure 5A), indicating the specific deficiency of PSI in all three mutants. The organization of the plastid ycf3-psaA-psaB-rps14 gene cluster, the localization and sizes of the two ycf3 introns, and the psaA 5′ UTR are conserved in land plants including mosses (Figure 5B; Supplemental Figure 2A). A ycf3-specific probe labeled two abundant bands of 7.4 and 6.7 kb and several more or less abundant smaller processed and/or spliced forms ranging from 0.8 to 3.5 kb in the wild type (Figure 5C). No marked differences could be detected in the mutants, indicating normal expression of ycf3. Hybridization with a psaA probe yielded two lower (7.4 and 6.7 kb) and two higher abundance (5.7 and 5.0 kb) transcripts, respectively, and some smaller forms between 1.2 and 3.3 kb in the wild type (Figure 5C). According to their sizes, the 7.4- and 5.7-kb transcripts represent the primary messages generated by polymerases using the ycf3 and the psaA promoters, respectively (Figures 5B and 5C). Presumably, as in vascular plants, these transcripts are generated by the actions of the NEP and the PEP, respectively, and so here we term them NEP- and PEP-generated transcripts (Figures 5B and 5C). A marked difference to vascular plants was the presence of two instead of one abundant primary PEP and NEP transcript (Lezhneva and Meurer, 2004; Cho et al., 2009). Therefore, it seemed likely that endonucleolytic processing in the psaB-rps14 intergenic region resulted in the accumulation of additional tricistronic ycf3-psaA-psaB and dicistronic psaA-psaB transcripts in P. patens (Figure 5B). To test this, we also probed with rps14, which resulted, as expected, in only one less abundant primary tetracistronic NEP and one more abundant primary tricistronic PEP transcript (Figure 5C). Thus, processing of rps14 from both precursor transcripts produces tricistronic ycf3-psaA-psaB and dicistronic psaA-psaB transcripts. These transcripts are assumed to be unstable in vascular plants. Several smaller and less abundant transcripts between 0.6 and 3.7 kb were also detectable when using the rps14 probe. According to the size, the 0.6-kb band should represent the monocistronic rps14 transcript. The three Δhcf145 moss mutants were unable to accumulate the two abundant PEP-generated psaA-psaB-rps14 and psaA-psaB transcripts, and levels of processed transcripts were reduced, as revealed by the use of the psaA and rps14 probes. Only the two larger, low-abundance NEP transcripts appeared at normal levels (Figure 5C). Amounts of processed rps14-containing transcripts were somehow reduced. However, these amounts and the unchanged presence of the primary rps14-containing NEP transcripts seem to be sufficient to allow translation of rps14 required for normal ribosome function in the mutant (Figure 5A). Similar to the situation observed in the Arabidopsis hcf145 mutant (Lezhneva and Meurer, 2004), precursors generated by NEP were stable and only PEP transcripts starting with the psaA 5′ UTR were unstable in the moss Δhcf145 mutants.
Figure 5.
Immunological and RNA Gel Blot Analysis of Δhcf145 Mutants.
(A) Immunoblot analysis with PsaA- and D2-specific antibodies. Protein loading corresponds to 4, 2, and 1 µg chlorophyll (100, 50, and 25, respectively). CBB, Coomassie blue.
(B) Organization of the ycf3-psaA-psaB-rps14 gene cluster in P. patens. I1 and I2, intron 1 and intron 2, respectively. Primary transcripts generated by the NEP and the PEP and proposed processing products are shown below. N+ and N-, proposed NEP transcripts with and without rps14, respectively; P+ and P-, PEP transcripts with and without rps14, respectively. The ycf3, psaA, psbA, and rps14 probes were generated by PCR using primer pairs ppYcf3_f and -r, ppPsaA_f and -r, ppPsba_f and -r, and ppRps14_f and -r. For the amplification of an intron-less ycf3 probe, we used a cDNA as template.
(C) RNA gel blot analysis using 10 µg total RNA from the wild-type and Δhcf145 lines and hybridization probes specific for ycf3, psaA, rps14, and psbB. The band sizes are given in kilobases on the left.
In conclusion, we hypothesize from these observations that both At-HCF145 and Pp-HCF145 are primarily required for the protection of the psaA 5′ UTR from 5′ → 3′ exonucleolytic degradation. Sizes and levels of the psbA transcripts were unchanged in the Δhcf145 mutants, emphasizing the specificity of the HCF145 function in plants (Figure 5C). The phenotype of the moss Δhcf145 mutants illustrates that the function of HCF145 is conserved among embryophyta and that the target of HCF145 is very likely located in the psaA 5′ UTR. Inspection of this region demonstrates a high conservation downstream of the psaA transcription start site in embryophyta but not in algae (Supplemental Figures 2A and 2B).
PsaA/B Transcripts Are Truncated in the 5′ UTR in hcf145-2
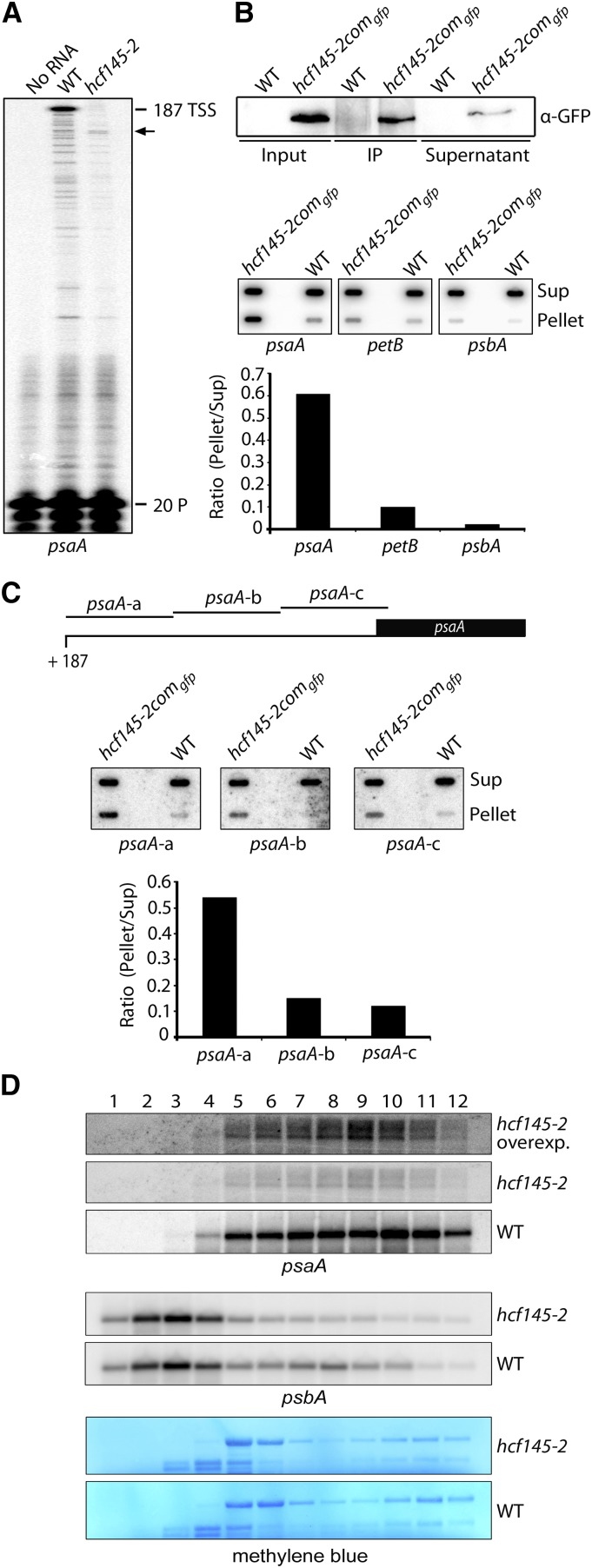
To confirm whether HCF145 is required for conferring stability to the psaA 5′ UTR, we performed primer extension analysis in Arabidopsis (Figure 6A; for sequences of oligonucleotides used in this work, see Supplemental Table 1). As expected, we detected the transcription start site 187 nucleotides upstream of the psaA start codon in the wild type. It was also the major transcript besides several shorter forms of much lower abundance, which normally represent premature termination products of the primer extension reaction. By contrast, the hcf145 mutant, which accumulates <10% of the psaA-containing RNAs (Lezhneva and Meurer, 2004), also failed to accumulate full-length transcripts (Figure 6A). Instead, at least one truncated transcript could be detected, which was not found in the wild type and, therefore, very likely represented a metastable degradation product. This independent finding confirms an important role of HCF145 in the stabilization of the tricistronic psaA-psaB-rps14 transcript most likely in the psaA 5′ UTR (Lezhneva and Meurer, 2004) (Figure 6A). Although only transcripts truncated in their 5′ extremities could be detected in the hcf145-2 mutant, there were still significant amounts of PsaA proteins detectable. This suggests that translation can also occur from the 5′ truncated mRNA.
Figure 6.
Determination of the psaA 5′ Termini, RNA Immunoprecipitation, and psaA Polysome Loading of hcf145-2.
(A) Primer extension analysis with radiolabeled primer (20 P) indicates that the 5′ UTR of psaA is truncated in the Arabidopsis hcf145-2 mutant as indicated by arrows. TSS, transcription start site.
(B) Detection of RNA coimmunoprecipitating with HCF145. Upper panel: Immunoprecipitation of the HCF145-GFP fusions from chloroplast extracts of hcf145-2comgfp using GFP antibodies. HCF145-GFP expression was driven by the 35S promoter. IP, immunoprecipitate. Lower panel: Coprecipitated RNAs of the supernatant (Sup) and the pellet were applied to slot blots. Filters were hybridized with the psaA 5′ UTR, petB ORF-, and psbA 5′-region-specific probes. The signals were quantified using ImageQuant software and the ratio of bound versus unbound RNA is indicated in the bar graph.
(C) Fine-mapping of the psaA 5′ UTR RNAs that coimmunoprecipitate with HCF145. Slot-blot hybridization (as in [B]) with three probes covering the entire psaA 5′ UTR. The ratio of bound versus unbound RNA is indicated in the bar graph.
(D) RNA gel blot analysis of polysome fractions 1 to 12 taken from the sucrose gradient of the wild-type and hcf145-2 plants. The probes used are indicated. rRNA was stained with methylene blue. Due to the low expression of psaA in the hcf145-2 mutant, the filter was overexposed (overexp.) for better comparison.
HCF145 Is Associated with the psaA 5′ UTR in Vivo
To strengthen our interpretation of the HCF145 function and to provide further experimental evidence for the association of HCF145 with the 5′ UTR of the psaA transcripts in vivo, RNA-immunoprecipitation studies were performed. For this, we first immunoprecipitated HCF145-GFP fusions from isolated chloroplasts of the complemented Arabidopsis hcf145-2comgfp lines and wild-type control plants using GFP antibodies. The HCF145-GFP fusion was greatly enriched in the pellet of the transgenic plants (Figure 6B), while no enrichment was obtained in the pellet of the wild-type sample. Coprecipitated RNAs were isolated from the pellets and the supernatants and spotted onto nylon membranes. Hybridization of the filters with radiolabeled probes revealed that the HCF145-GFP fusions preferentially precipitated the psaA 5′ UTR and much less efficiently the petB open reading frame and the 5′ region of the psbA mRNA, indicating that HCF145 itself or in association with other factors binds to the psaA 5′ UTR with a higher affinity than to the petB and psbA RNA (Figure 6B). We then asked which region of the psaA 5′ UTR is recognized by HCF145 or its interaction partners in vivo. For this purpose, we hybridized filters from RNA-immunoprecipitation with three nonoverlapping probes (70-mer psaA-a to -c) of comparable size covering the entire UTR of 187 nucleotides (Figure 6C). It appeared that HCF145-GFP coprecipitated with significantly higher amounts of transcripts containing sequences of the very left part of the psaA 5′ UTR (psaA-a) compared with the middle (psaA-b) and the right part (psaA-c) (Figure 6C, upper panel). Quantification of bound versus unbound RNA revealed a 3.6- and 4.5-fold enrichment of probe psaA-a compared with psaA-b and psaA-c, respectively (Figure 6C, lower panel). Therefore, we hypothesize that HCF145-GFP protects the psaA-psaB-rps14 RNA by binding to the 5′ region of the psaA 5′ UTR.
Role of HCF145 in Translation of the Tricistronic psaA-psaB-rps14 Transcript
Association of HCF145 to the psaA 5′ UTR could affect not only stability but also translation of the psaA message. Therefore, we investigated the association of the tricistronic psaA-psaB-rps14 RNA with polysomes via sedimentation in sucrose gradients. In the wild type, the psbA RNA encoding the PSII protein D1 was mainly found in monosomes and only a small portion was present in the polysomal fraction, whereas the psaA RNA was predominantly associated with the polysomes (Figure 6D). By contrast, the hcf145-2 mutant in Arabidopsis showed a slightly decreased association of the psbA mRNA to the polysomes but apparently no shift of the psaA polysomes to lower sucrose concentrations (Figure 6D). The fact that the psaA RNA in hcf145-2 was found in polysomes and its pattern of distribution along the fractions was similar to that of the wild type reveals that translation of psaA generally does occur in the mutants. This is also congruent with a concomitant reduction of the psaA-psaB-rps14 transcripts and PsaA proteins (Figures 1D and 2D) (Lezhneva and Meurer, 2004). Since translation in chloroplasts is under redox control (Marín-Navarro et al., 2007), it is likely that the slight differences of the psbA polysomal loading between the mutant and wild type result from a general downregulation of the psbA translation when photosynthesis is severely affected. We conclude that HCF145 plays either no or only a minor role in translation of PsaA.
Recombinant HCF145 Binds Directly to the psaA 5′ UTR
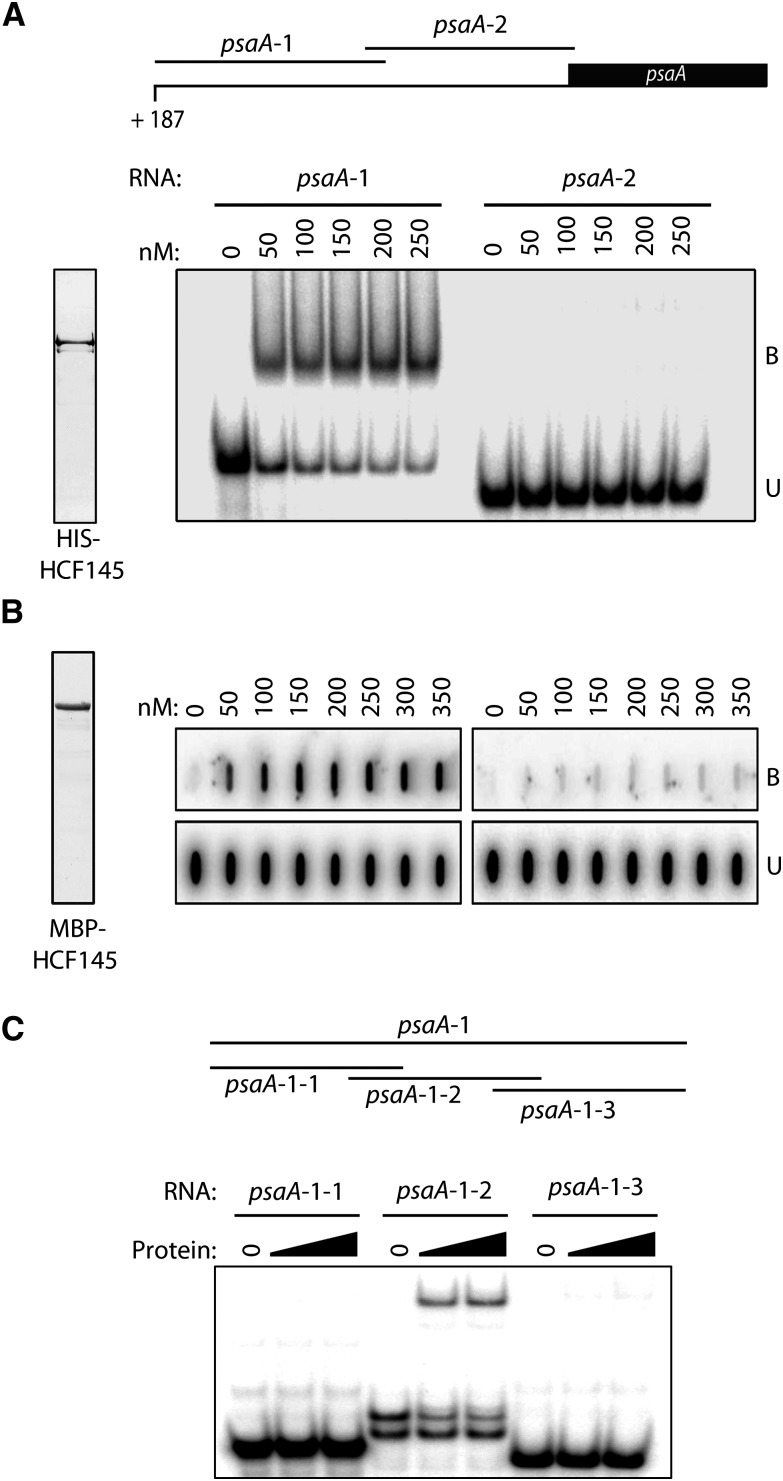
To confirm that HCF145 itself or in association with other proteins binds to the psaA 5′ RNA target, we performed in vitro RNA binding studies with purified recombinant HCF145. The mature HCF145 protein was fused to a His-tag and overexpressed in Escherichia coli. Then, the purified protein was used for electrophoretic mobility shift assays (EMSAs) with RNA probes of the psaA 5′ UTR divided into two overlapping parts of comparable lengths (Figure 7A). HCF145 strongly bound to the 5′ region of the psaA 5′ UTR with high specificity, whereas no binding could be detected for the 3′ region of the psaA 5′ UTR (Figure 7A). Thus, it appears that HCF145 represents a plant-specific RNA binding protein that recognizes and directly binds specific targets in chloroplasts. We hypothesized that HCF145 stabilizes the psaA-psaB-rps14 mRNA via direct binding to the psaA 5′ UTR. To test this using a different fusion, we performed filter binding assays using highly purified maltose binding protein (MBP)-HCF145. Indeed, this fusion efficiently bound to the 5′ part of the psaA 5′ UTR, whereas virtually no binding was observed to the 3′ part of the psaA 5′ UTR (Figure 7B), thus confirming the high specificity by which HCF145 directly binds to the 5′ end of the psaA 5′ UTR. To further narrow down the binding site, we subdivided the psaA-1 probe (Figure 7A) covering the first 124 nucleotides downstream of the transcription start site into three overlapping probes (psaA-1-1 to psaA-1-3), which are shown in Supplemental Figure 2. EMSA experiments showed specific binding to the middle probe, psaA-1-2, but not to the adjacent regions (psaA-1-1 and psaA-1-3), indicating that the binding site is located within the region +29 to +88 nucleotides downstream of the transcription start site (Figure 7C).
Figure 7.
Recombinant HCF145 Binds with High Specificity to the 5′ End of the psaA 5′ UTR.
(A) Generation of two overlapping transcripts covering the entire psaA 5′ UTR for binding assays (upper panel). EMSA using the two psaA 5′ UTR probes (lower panel). The binding reaction contained the indicated concentrations of recombinant HCF145 (rHCF145). Purified protein was stained with Coomassie blue (left panel).
(B) Filter binding assay performed with the purified MBP-HCF145 fusion. Purified protein was visualized with Coomassie blue (left panel). psaA 5′ UTR probes and amounts of rHCF145 are indicated. B, bound RNA; U, unbound RNA.
(C) EMSA using rHCF145 (see [B]) and three overlapping probes covering the psaA-1 probe in the 5′ region of the 5′ UTR (see [A]). A schematic representation of the probes is shown in the upper panel and the sequences of the probes are indicated in Supplemental Figure 2.
The C-Terminal Tandem Repeated Motifs of HCF145 Confer RNA Binding Capability
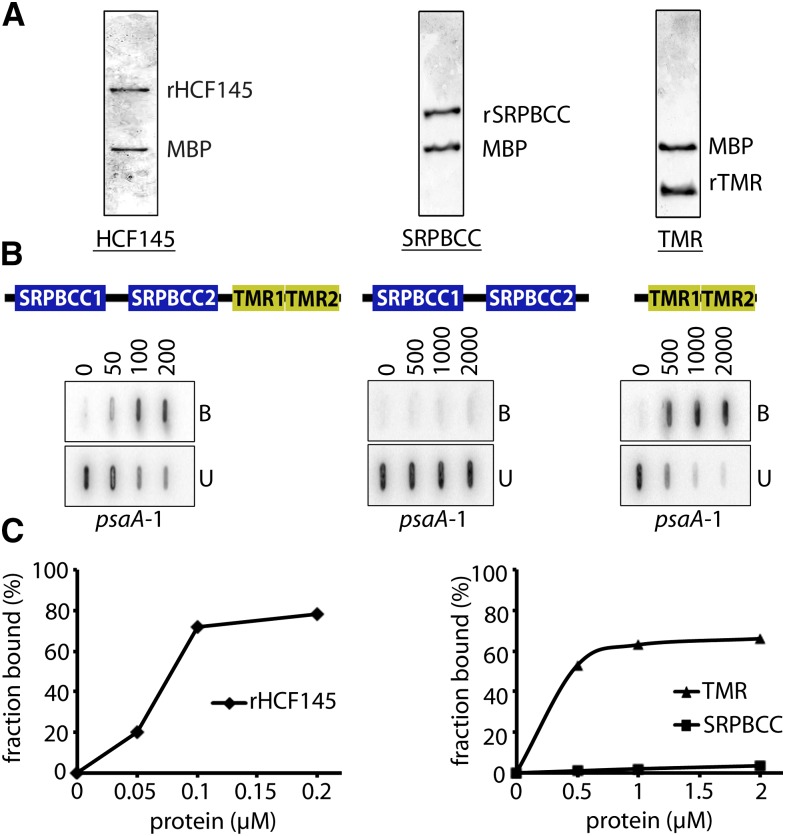
We next examined which of the two tandem repeated regions of HCF145 is required for RNA binding, the SPRBCC domains, or the two newly defined short C-terminal TMR domains. For this, we aimed to compare the filter binding efficiency of the mature protein with that of the two SRPBCC and the two TMR motifs. All three recombinant forms were expressed in E. coli as MBP fusions, purified and released from the tag before performing filter binding assays (Figure 8A). For a better comparison and to exclude that the spacer between the two tandem repeated SPRBCC and TMR domains itself binds to the RNA, both truncated recombinant forms contained the entire spacer between the motifs SRPBCC2 and TMR1 (Figure 8B). The mature HCF145 bound with high affinity to its target. At protein concentrations of 100 nM, more than half of the RNA was already bound. Virtually no binding occurred with the tandem repeated SRPBCC domains even at higher protein concentrations. By contrast, the recombinant C-terminal tandem repeated domains alone were sufficient to confer RNA binding capability (Figure 8B). To estimate the binding affinity, we calculated the equilibrium binding constant (Kd) of HCF145 and its C terminus to the 5′ region of the psaA 5′ UTR (psaA-1; Figure 7A) using increasing concentrations of the recombinant proteins in RNA binding assays (Figure 8B). The calculated Kd value of 107 nM for the mature HCF145 protein was in the range of known RNA binding proteins (Ostersetzer et al., 2005; Hammani et al., 2012) and reflected its high affinity to the psaA 5′ UTR (Figure 8C). Interestingly, the apparent Kd value of 493 nM for the recombinant TMR domains was ∼5 times higher compared with that of the entire protein, indicating an important role of the SRPBCC domains for binding efficiency (Figure 8C). Alternatively, structural misfolding of the isolated recombinant form could be responsible for less efficient binding by the TMR domains.
Figure 8.
Purification and Filter Binding Assays of Defined Motifs of rHCF145 Protein.
(A) Release of the entire recombinant HCF145 protein, a fragment containing the SRPBCC domains, as well as the tandem repeated TMR motifs from the MBP fusion.
(B) Filter binding assays of purified proteins shown in (A) using increasing concentrations of protein up to 200 nM for the full-length protein and up to 2000 nM for the truncated versions. B, bound RNA; U, unbound RNA.
(C) Quantitative determination of the RNA fraction bound to HCF145 to evaluate the dissociation constant (Kd) value.
Levels of HCF145 Decrease upon Light Induction
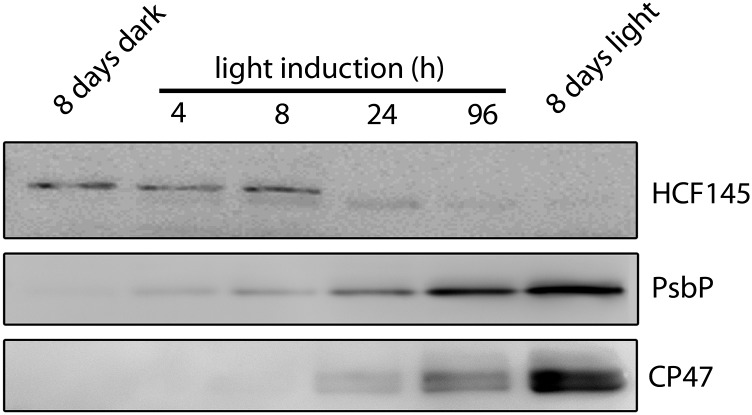
Antibodies were raised against an epitope of the C terminus of HCF145 partially covering the second TMR domain. Antisera for CSP41, an abundant factor for stabilizing plastid RNAs (Qi et al., 2012), and of the thylakoid-associated PSII assembly factor PsbN (Torabi et al., 2014) were used as a control. The limit of detection of HCF145 is in the range of ∼20 fmol, as revealed by titration analysis, indicating excellent sensitivity of the antibodies. Immunoblot analysis of total chloroplast fractions of dark-grown Arabidopsis seedlings illuminated for 8 h resulted in a signal of the expected size of 76 kD (Supplemental Figure 3A). When equal amounts of chloroplast proteins were separated into membrane and soluble fractions, no signal could be detected in the membrane fraction. The signal intensity in the soluble fraction was comparable to that of the total fraction, indicating that the HCF145 protein is exclusively found in the stroma of dark-grown seedlings (Supplemental Figure 3A). Wild-type and mutant seedlings successfully complemented with a GFP-, FLAG-, and Strep/HA-tagged HCF145 protein were also subjected to immunoblot analysis. All three recombinant forms were expressed under the control of the 35S promoter and their corresponding sizes could be confirmed, whereas the mature wild-type protein of 76 kD was lacking in all three complemented mutant lines demonstrating the specificity of the antiserum (Supplemental Figure 3B). However, when grown under continuous light, almost no signal was obtained in leaves of the wild type, indicating that levels of HCF145 are of quite low abundance in light-exposed plants. Unlike PsbP, CP47, and many other photosynthetic proteins, which usually accumulate only after several hours of light induction of dark-grown seedlings, the mature form of HCF145 was found already in the dark and almost completely disappeared upon light induction of 24 h (Figure 9). Instead, a slightly smaller form of HCF145, which presumably represents a specific degradation product, appeared after 4 h but was hardly detectable after 8 d of continuous illumination (Figure 9). Therefore, it seems likely that the function of HCF145 is already important in etiolated seedlings before and/or during the onset of light-induced chloroplast biogenesis and only trace amounts are sufficient to fulfill its function in the light.
Figure 9.
HCF145 Is Predominantly Expressed in Dark-Grown Seedlings.
Immunological analysis of HCF145, PsbP, and CP47 accumulation in Arabidopsis seedlings etiolated for 8 d and subsequently illuminated for 4, 8, 24, and 96 h at 50 μmol photons m−2 s−1. Seedlings grown for 8 d in the light were used for comparison.
DISCUSSION
Phylogenetic Origin, Conservation, and Functional Analysis of the HCF145 Modules
In our previous study, we showed that the nuclear HCF145 locus encodes a trans-acting factor specifically required for protecting the psaA-psaB-rps14 mRNA from degradation (Lezhneva and Meurer, 2004). Our genetic and molecular studies using two allelic lines hcf145-1 and hcf145-2 in Arabidopsis demonstrate that HCF145 is encoded by the locus AT5G08720. HCF145 is specific for the green lineage and has a modular composition of cyanobacterial origin. It harbors an N-terminal transit peptide for plastid import followed by two homologous motifs that are related to the huge and ubiquitously distributed SRPBCC ligand binding domain superfamily consisting of 11 Pfam families, including START domain, phosphatidylinositol transfer protein, Bet_v_1, CalC related, CoxG, and polyketide cyclase-related families (http://pfam.janelia.org) (Iyer et al., 2001; Radauer et al., 2008) (Figure 3B; Supplemental Figure 4A). The structural similarity of the domains to already crystalized members of known function and ligands suggests that they bind hydrophobic ligands like lipids, phytohormones, steroids, coenzyme Q, alkaloids, and polyketides, and/or produce complex polyaromatic substances of the secondary metabolism, such as aromatic hydrocarbon hydroxylating enzymes (Radauer et al., 2008). This is exemplified by experimentally verified binding of structurally unrelated phytohormones and secondary metabolites by members even within the Bet_v_1 family, which together with the polyketide cyclase family represents the most ancient family (Radauer et al., 2008). The SRPBCC domains of HCF145 are around 130 amino acids long and belong to the largest plant SRPBCC domain family of aromatic polyketide cyclase-like proteins found in 37 structurally related Arabidopsis proteins, all of unknown function (http://www.arabidopsis.org/index.jsp) (Supplemental Figure 4A). The SRPBCC1 and SRPBCC2 motifs in Arabidopsis share 41% sequence identity and 53% similarity. Interestingly, the conservation of the individual domains between plants is much higher, ranging from ∼86% sequence identity and more than 90% similarity. The relatively low sequence similarity between the two SRPBCC domains within the HCF145 proteins compared with the high conservation of the individual domains between different plant species could reflect a specific adaptation of the domains to the requirement of the plants and/or binding of different but related ligands. Overall, this domain belongs to the families whose members show the highest degree of sequence similarity between species of all superkingdoms (16% median identity and 28% median similarity) (Radauer et al., 2008).
A closer inspection of the HCF145 protein at the C terminus detected additional tandemly repeated motifs, which have not been described previously (Figure 3B). Based on the RNA binding capability shown here, we named the repeated motifs transcript binding motif repeat (TMR) domains and proteins containing related repeats TMR proteins. Protein homologs with a modular organization similar to that of HCF145 were found exclusively in Viridiplantae (green lineage) but not in the red algae lineage or in cyanobacteria (Figure 3B; Supplemental Figures 4A to 4C). Notably, HCF145 homologs could be found in all land plant genomes analyzed, including the moss P. patens, but only in some green algae, such as Ostreococcus tauri (Ot01g03250), Coccomyxa subellipsoidea (COCSUDRAFT_18573), and Chlorella variabilis (CHLNCDRAFT_57236). However, the C-terminal part of the two SRPBCC domains appeared to be truncated in green algae (Figure 3B; Supplemental Figure 4A). The absence of the HCF145 gene in the sequenced genomes of some green algae, such as Chlamydomonas reinhardtii, suggests that HCF145 was lost secondarily in some lineages presumably followed by diversification of the psaA 5′ UTR (Supplemental Figure 2B) and the acquisition of other factors required for regulation of the RNA stability at the 5′ extremity. Inspection of large-scale genomic and transcriptomic data of the holoparasitic and nonphotosynthetic Orobanche Phelipanche aegyptiaca revealed that HCF145 has been lost from this genome, whereas hemiparasites of this family still retain this gene. This indicates a close correlation of the HCF145 function with photosynthesis.
The closest relatives of the SRPBCC domains of HCF145 were found in all photosynthetic lineages, including cyanobacteria and Chlorobi, here designated HCF145-like (HCF145-L; At4G01650) (Figure 3B; Supplemental Figures 5A to 5C and Supplemental Data Set 1). With the exception of an N-terminal extension in eukaryotes, which encodes a chloroplast transit peptide in Arabidopsis as revealed by transient expression of a GFP fusion (Supplemental Figure 6A), proteins mainly consist of a unique SRPBCC domain with unknown function and target (Figure 3B). At-HCF145-L shows ∼60% similarity to both SRPBCC domains of HCF145 (Figure 3B; Supplemental Figures 5A to 5C). Knockout plants of HCF145-L in Arabidopsis showed no obvious deficiencies, growth retardation, or photosynthetic alteration when compared with the wild type, indicating that its function is either not related to or not essential for photosynthesis (Supplemental Figure 6B). The next closest relatives are found in Chlorobium tepidum and several cyanobacteria such as Anabaena sp PCC 7120, Thermosynechococcus elongatus sp, Synechococcus elongatus sp, and Prochlorococcus marinus sp (Supplemental Figures 5A to 5C). As homologs of HCF145-L could be found in some cyanobacteria and based on the high level of conservation, we hypothesize that they very likely represent the direct ancestor of the SRPBCC modules of HCF145 in plants. The finding that not all cyanobacteria possess a HCF145-L homolog indicates that the function is dispensable in some species. The bacterial form is also mainly composed of the SRPBCC domain and shares ∼50 to 58% similarity with the domains of HCF145 and HCF145-L in Arabidopsis (Figure 3B; Supplemental Figure 5A).
The two newly identified tandem repeated TMR motifs at the very C terminus of HCF145 are separated by only one amino acid (Figure 3B; Supplemental Figure 4B). No proteins other than HCF145 in vascular plants were found to contain similar domains. Remarkably, homologous motifs with up to 10 repeats are found in more than two dozen and quite diverse proteins exclusively in cyanidiales, chlorophyta, and cyanobacteria and therefore form a family of TMR proteins of unknown function in photosynthetic organisms (Figure 3B; Supplemental Figure 7). In one protein of the red algae Cyanidioschyzon merolae (XM_005538035), only one TMR motif was found. Existing algorithms for prediction of transit peptides are oriented toward land plants and tend to mispredict the localization of nuclear-encoded proteins, especially in red algae. Recently, the PredAlgo algorithm was developed based on proteomics data to predict transit peptides relatively reliably in green algae (https://giavap-genomes.ibpc.fr). Using this server, most of the TMR proteins of green algae are predicted to encode plastid proteins, which in turn could be true for TMR proteins also in red algae.
We propose to number the corresponding domains within the TMR proteins according to their arrangement in HCF145, namely, TMR1 and TMR2. They show around 56% sequence similarity within HCF145 proteins but again a higher conservation of the individual modules when compared with other plant species (around 90% sequence similarity) (Figure 3B; Supplemental Figure 4B). As defined by Seq2Logo (http://www.cbs.dtu.dk/biotools/Seq2Logo), the internal consensus sequence of TMR motifs is MP-x(4)-L-x(3)-GR-x-DL-x(2)-AI-x(2-4)-HGG-x(3)-VA-x(2)-LGL-x(5-27)-GYW (Supplemental Figure 7).
With the exception of the HCF145 homologs in some green algae (e.g., C. variabilis and C. subellipsoidea), none of the TMR proteins present in algae and cyanobacteria contain sequences with similarities to the SRPBCC domain. Only rarely are they composed of additional conserved motifs, like a domain in the TMR protein of the cyanobacterium Microcoleus sp PCC 7113 (WP_015211581.1) with homology to prokaryotic DNA binding proteins belonging to the xenobiotic response element family of transcriptional regulators. They rarely contain only one C-terminal TMR motif, such as in C. merolae (XM_005538035), but rather consist mainly of multiple TMR motifs with up to 10 copies, such as in the red algae C. merolae (XM_005538687) and Galdieria sulphuraria (Figure 3B; Supplemental Figure 7). Interestingly, the repeats of different proteins are separated by less or nonconserved spacers of quite diverse lengths from 0 to ∼80 amino acids, but the length of the spacers is more or less constant within individual proteins (Figure 3B).
In contrast to PPR proteins, which were found only in eukaryotic organisms, TMR proteins are of cyanobacterial origin and seem to be restricted to organisms performing oxygenic photosynthesis. The TMR motifs in HCF145 most likely originated from cyanobacteria, as three C-terminal TMR motif repeats can be found in one protein of Microcoleus sp PCC 7113 (Figure 3B; Supplemental Figure 7). The high diversity of the chloroplast genome organization found among green algae (Letsch and Lewis, 2012) can explain why HCF145 became dispensable in individual members. It appears that HCF145 has coevolved with its mRNA target in land plants. This is supported by several lines of evidence: (1) the psaA 5′ UTRs are conserved, (2) high sequence identity and similarity among homologous HCF145 proteins in land plants, and (3) the comparable phenotypes in the hcf145 knockouts in Arabidopsis and moss. Most likely, a high evolutionary pressure during endosymbiosis caused a bias for the recruitment of plant-specific nucleus-encoded factors, like many PPR proteins, and other factors for stabilization and processing of organellar RNAs (Germain et al., 2013; Shikanai and Fujii, 2013).
In summary, we suggest that the modular HCF145 protein most likely originated from the polyketide cyclase and TMR modules of cyanobacterial origin and evolved relatively late during endosymbiosis in the green linage leading to green algae and embryophyta.
HCF145 Binds to and Protects the psaA-psaB-rps14 Transcript
The coimmunoprecipitation analysis with HCF145, deficiency of the psaA-psaB-rps14 RNA in the hcf145 mutants, and RNA binding studies, such as EMSA and filter binding assays, all documented that the TMR motifs of HCF145 bind to the psaA 5′ UTR, presumably to protect the RNA from degradation. Primer extension experiments have shown that the psaA transcription start site is missing almost completely in the mutant (Figure 6A). Apparently, the transcription start site is protected in the presence of HCF45, although it binds farther downstream (Figures 6C and 7C). One possible explanation is that binding changes the folding of the RNA and results in masking the 5′ end that is otherwise vulnerable to exonucleases. However, the precise mechanism remains elusive so far.
Taking all this together, our data suggest that the TMR motifs represent a new class of repeated RNA binding domains specific for photosynthetic organisms, including cyanobacteria. However, the function and the RNA binding capability and specificity of the TMR proteins in algae and cyanobacteria still have to be investigated. HCF145 is unique among the TMR proteins in combining the TMR motifs with the SRPBCC domains. In vitro association studies indicate that the presence of the SRPBCC1 and SRPBCC2 motifs is important for the RNA binding efficiency, although alone they do not show binding (Figure 8). Several scenarios could explain the significance of the SRPBCC1 and SRPBC2 motifs for the HCF145 function. (1) Perhaps they structurally support RNA binding by motifs TMR1 and TMR2; consequently, they might play a stabilizing role and not necessarily bind metabolic ligands. (2) The SRPBCC domains might bind RNA only in conjunction with the TMR domains, e.g., sequentially, because they need a TMR-RNA interaction so that certain RNA segments get liberated for SRPBCC access. (3) RNA binding capability of TMR1 and TMR2 could be regulated by ligand binding to the SRPBCC motifs. For example, ligand binding by the structurally related START domain in modular proteins has been suggested to regulate the function of other domains, such as the DNA binding homeodomain, the thioesterase domain, and the RhoGAP domain (Iyer et al., 2001).
Similar to TMR proteins, multiple degenerated repeats are also found in the RNA binding PPR, OPR, and PUF domain families (Filipovska and Rackham, 2012; Barkan and Small, 2014; Hammani et al., 2014). Interestingly, one TMR motif spans about twice the length of a PUF or a PPR repeat and they most probably form four α-helices as predicted by the I-Tasser algorithm (Supplemental Figures 4 and 8).
HCF145 Is Predominantly Expressed in Dark-Grown Seedlings
Large-scale expression data provide only poor or no information about the At5g08720 locus, indicating a low expression and abundance of hcf145 mRNA. In contrast to most chloroplast proteins with photosynthesis-related functions, HCF145 accumulated already in dark-grown seedlings but disappeared during the first 24 h of illumination (Figure 9). Only trace amounts of a slightly smaller truncated form was detectable in the light. Therefore, it seems very likely that the function of HCF145 is already important in etiolated seedlings, during germination and/or upon chloroplast development. Notably, the psaA mRNA also accumulates in etiolated barley (Hordeum vulgare) seedlings and gradually disappears after 16 h of continuous light application due to decreased stability (Klein and Mullet, 1987). This suggests that stabilization of the psaA mRNA is important in darkened seedlings before the onset of the chloroplast development and that HCF145 fulfills its function already in the proplastid. Interestingly, the half-life of the psaA mRNA appeared to be much shorter than that of many other chloroplast mRNAs after illumination of etiolated barley seedlings, suggesting a coregulation of transcription activity and stability of the psaA mRNA (Klein and Mullet, 1987; Mullet and Klein, 1987). In this respect, it will be interesting to investigate a possible regulatory function of HCF145 in adjusting psaA mRNA levels in future experiments.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana hcf145-1 line, accession Wassilewskija, originated from a T-DNA collection (Errampalli et al., 1991) as previously described (Lezhneva and Meurer, 2004). The T-DNA insertion line hcf145-2 (SALK_011411), accession Columbia, was obtained from the SALK collection (http://signal.salk.edu). The Arabidopsis hcf145-like line (TTT370), accession Wassilewskija, was obtained from The Versailles Arabidopsis Stock Center. Unless stated otherwise, selection, propagation, and growth of the Arabidopsis wild-type and mutant plants were performed as previously described (Lezhneva and Meurer, 2004). For coimmunoprecipitation analyses, the wild-type and complemented lines were grown on soil in a 16-h-light (20°)/8-h-dark (18°) cycle for 1 week and another 3 to 4 weeks in a 8-h-light/16-h-dark cycle with a PFD of 100 µmol photons m−2 s−1. Physcomitrella patens cultivation, protoplast isolation, polyethylene glycol-mediated transformation, and regeneration of stably transformed plants were performed according to standard procedures (Frank et al., 2005).
Map-Based Cloning of HCF145
Mapping populations were generated by crossing the wild-type plants (accession Landsberg erecta) with plants heterozygous for the hcf145-1 mutation as pollen donor and subjected to PCR-based analysis with polymorphic markers.
Complementation of hcf145-1 and hcf145-2
The cDNA of At5g08720 was obtained from the RIKEN BioResource Center (Seki et al., 2002). For complementation of hcf145-1, the cDNA was amplified with Pfu polymerase (Fermentas) using the primers 720-ATG-f-P and 720-3UTR-r-P and blunt end ligated with the SmaI-linearized pSEX001-VS (Meurer et al., 1998). For complementation of hcf145-2, the cDNA was amplified with the Phusion High-Fidelity DNA Polymerase (Finnzymes) using the primers Cacc-145-for and 145-rev. The resulting PCR product was cloned into pENTR/D-TOPO (Invitrogen) followed by insertion into the binary Gateway vector pB7FWG2,0 (Plant System Biology) in frame with the GFP coding region using LR Clonase II (Invitrogen) according to the manufacturer’s instructions. Both vectors were introduced into Agrobacterium tumefaciens GV3101 (pMP90RK) and subsequently transformed into the corresponding heterozygous hcf145 mutants using the floral dip method (Clough and Bent, 1998). Transformed seedlings were selected with sulfadiazine or BASTA and the complemented homozygous mutants were identified by PCR analyses. All primers are listed in Supplemental Table 1.
RNA Isolation and Gel Blot Analyses
Total leaf RNA was isolated with TriPure Isolation Reagent (Roche) as recommended by the manufacturer. For gel blot analyses, glyoxylated RNA was electrophoresed as described (Meurer et al., 1996b) and transferred onto a nylon membrane (Biodyne A, 0.45µM; PALL) with a capillary blot setup using 20× SSC transfer buffer. After the transfer, the membrane was washed in 2× SSC and UV-cross-linked (Stratagene UV 1800). Hybridization probes were generated by T4 polynucleotide kinase (New England Biolabs)-mediated end labeling of oligonucleotides with [γ-32P]ATP (Hartmann) or by random labeling of PCR products using Klenow fragment polymerase with [α-32P]dCTP (Hartmann). Hybridization was performed with ExpressHyb hybridization solution (BD Biosciences) according to the user’s manual.
Spectroscopic Analysis
Chlorophyll a fluorescence induction kinetics were measured using a pulse-modulated fluorometer (Imaging-PAM, Dual-PAM-100, and PAM101; Walz). For the measurement of P700 absorbance changes, PAM101 (Walz) was used. All spectroscopic analyses were performed as described (Meurer et al., 1996a).
Subcellular Localization of HCF145 and HCF145-L
The stably expressed HCF145-GFP fusion in the hcf145-2comgfp plants was visualized using a fluorescence microscope (Axio Imager; Zeiss). The sequence encoding the N-terminal part of HCF145-L consisting of 462 nucleotides was amplified from genomic DNA by PCR using primers at4-Kpn1-f and at4-Kpn1-f (Supplemental Table 1). The resulting product was digested with KpnI and cloned in frame into the KpnI site of the GFP expression vector pOL-LT (Mollier et al., 2002). Transient expression of the HCF145-L GFP fusions was performed in polyethylene glycol-treated Nicotiana tabacum protoplasts, and fluorescence was visualized 18 h after transformation as described (Gross et al., 2006).
Protein and Immunological Analysis
Thylakoid membrane proteins and soluble chloroplast proteins were isolated as described (Stoppel et al., 2011). For total protein isolation, fresh or frozen plant material was homogenized in isolation buffer (10 mM EDTA, 2 mM EGTA, 50 mM Tris-HCl, pH 8.0, 10 mM DTT, and proteinase inhibitor cocktail [Roche]). Soluble and membrane proteins were separated in 10% and 15% SDS-PAGE (Laemmli, 1970) and transferred to a PVDF membrane (Immobilon, 0.45µm; Millipore). Immunoblotting and antisera used were described previously (Torabi et al., 2014). HCF145 antibodies were raised against the epitope RQLNSRKDNGNTILRTC in rabbit and immunoaffinity purified as described by the manufacturer (Pineda Antikörper-Service). Titration analysis was performed via quantitative dot blot analysis (Porat et al., 1995).
Generation of Δpphcf145 Mutant Lines in P. patens
The Pp-HCF145 knockout construct was generated using a Gibson assembly cloning kit (NEB) that allows joining of DNA fragments with overlapping DNA ends. The knockout construct was designed to harbor an nptII selection marker cassette that is flanked by Pp-HCF145 genomic fragments at both sides. Initially, three DNA fragments (nptII cassette, a 538-bp Pp-HCF145 5′ fragment, and a 569-bp Pp-HCF145 3′-fragment) were amplified by PCR with primers that harbor overlapping ends. Primers npt5′ and npt3′ were used to amplify the nptII selection marker cassette from the vector pBSNNNEV (Egener et al., 2002). The 5′ and 3′ flanking fragments derived from Pp-HCF145 were amplified from genomic DNA with primers pp5′f and pp5′r, and pp3′f and pp3′r, respectively. The resulting PCR products were assembled according to the manufacturer’s protocol and cloned into the CloneJET plasmid (Thermo Scientific). The Pp-HCF145 gene knockout construct was released from the vector backbone by SalI restriction and transfected into P. patens protoplasts. Protoplasts were regenerated and selected on G418-containing medium (12.5 mg/L). Targeted insertion of the knockout construct via homologous recombination caused the replacement of a 1402-bp genomic Pp-HCF145 region by the nptII cassette. PCR was performed with DNA from regenerated antibiotic resistant lines with the primers ppF1 and ppR1 to confirm precise 5′ integration of the Pp-HCF145 knockout construct. Correct 3′ integration was analyzed by PCR using the primers ppF2 and ppR2. Subsequently, loss of the Pp-HCF145 mRNA was analyzed by RT-PCR using the primers ppF3 and ppR3. The primers EF1α-f and EF1α-r were used to monitor cDNA synthesis.
Preparation of Recombinant Proteins
To produce recombinant MBP-HCF145-Strep proteins, the coding sequence of At-HCF145 lacking the signal sequence (amino acids 1 to 56) was PCR amplified using primers Fw-BclI-hcf145 and Rev-SalI-hcf145-strep, digested with SalI and BclI, and inserted into the SalI/BamHI sites of pMAL-TEV (kindly provided by Alice Barkan, University of Oregon). Protein expression and purification via affinity chromatography were performed as described (Chi et al., 2014). HCF145-Strep was subsequently cleaved from the MBP moiety using AcTEV Protease (LifeTechnologies). Cloning using the primers Fw-BclI-hcf145, Rev-SalI-hcf145-strepA2, Fw-BclI-hcf145-B1, and Rev-SalI-hcf145-strep and expression and purification of MBP-SRPBCC-Strep and MBP-TMR-Strep were performed as described above.
His-HCF145 was expressed from pDEST17 in BL21(DE3)pLysS cells at 37°C. Cells were lysed by sonication in lysis buffer (50 mM NaH2PO4, pH 8, 300 mM NaCl, 10 mM imidazole, and Complete Protease Inhibitor Cocktail Tablets [Roche]). The cleared lysate was incubated with Ni-NTA agarose (Qiagen). Washing and elution steps were performed with the lysis buffer supplemented with 20 and 250 mM imidazole, respectively. All buffers were supplemented with 10 mM β-mercaptoethanol. Recombinant proteins were further purified via size exclusion chromatography using Superose 6, 10/300 GL (GE Healthcare Life Sciences). Proteins were stored at −20°C in elution buffers indicated above supplemented with 50% glycerol.
Primer Extension
For the primer extension assay, a psaA-specific primer (psaA ATG rev) was 5′ radio-end-labeled and annealed to the RNA for 2.5 h at 55°C in 150 mM KCl, 10 mM Tris, pH 8.3, and 1 mM EDTA. Primer extension was performed with the Transcriptor reverse transcriptase (Roche), and products were resolved on denaturing polyacrylamide gel. To accurately determine the size of the primer extension products, the same procedure was performed with the 6-Fam-labeled primers. Samples were analyzed on an ABI 3730 48 capillary sequencer using a mixture of length standards.
RNA Coimmunoprecipitation and Slot-Blot Hybridization
Chloroplasts were isolated from the wild-type and hcf145-2comgfp plants as described previously (Stoppel et al., 2012). The chloroplast pellet was lysed by incubation for 15 min on ice in coimmunoprecipitation buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.5% [v/v] Nonidet P‑40, and Complete Protease Inhibitor Cocktail Tablets [Roche]). Membranes were then pelleted by centrifugation at 36,000g for 30 min. One milligram of protein was incubated with 25 μL coimmunoprecipitation buffer-washed GFP-Trap-M beads (Chromotek) for 1 h at 4°C with rotation. The beads were washed three times with 0.5 mL coimmunoprecipitation buffer and were subsequently used for the SDS-PAGE and/or for the RNA extraction.
For the slot-blot hybridization, RNA from the pellet und supernatant was extracted by phenol-chloroform treatment and subsequent ethanol precipitation. For analysis, one-twentieth of the supernatant and half of the pellet were spotted onto nylon membranes using a slot-blot device (Bio-Rad) and hybridized as described above with the probes indicated. The psaA probe was generated with the primers T7_5UTRpsaA 3 and psaA 5′ 1, the petB probe with the primers B3-T7-petB-for and petB-ex-rev, and the psbA probe (psbA 80-mer) was 5′ end labeled. For fine mapping of the HCF145 binding site, chloroplast extracts were treated with 2 units of RNaseT1 (Ambion).
EMSA and Filter Binding Assay
EMSA experiments were performed as described previously (Manavski et al., 2012b). Briefly, 32P-RNA probes were generated by in vitro transcription using PCR products generated with primers T7_5UTRpsaA 3 and psaA5UTRrev, T7_5UTR-psaA 2 and psaA ATG rev, T7_5UTRpsaA 3 and rev psaA3_1, T7_5UTRpsaA 3_1 and rev psaA3_2, and T7_5UTRpsaA 3_2 and psaA5UTRrev. Trace amounts of labeled RNA were incubated with increasing protein concentrations as indicated in a buffer containing 40 mM Tris, pH 7.5, 150 mM NaCl, 0.1 mg/mL BSA, 4 mM DTT, and 0.5 mg/mL heparin at 25°C for 15 min. Samples were then either separated on nondenaturing 5% polyacrylamide gels containing 0.5× TBE buffer or filtered through nitrocellulose and nylon membrane using a slot-blot device. Filter binding assays were performed under the same condition but with 30 µg/mL heparin-containing binding buffer. The Kd values were estimated as the protein concentration at which half of the RNA was bound.
Polysome Analysis
Polysome extraction from leaf tissue was performed as described previously (Barkan, 1993). Aliquots (0.4 mL) of polysomes were loaded onto 3.6 mL 15 to 55% sucrose gradients. After centrifugation 12 fractions of 0.3 mL were collected, and the extracted RNA was used for the RNA gel blot analysis with end-labeled 80-mer oligonucleotides (psaA80-mer and psbA80-mer).
Phylogenetic Analysis and Sequence Alignments
Alignments were generated using Clustal Omega (Sievers and Higgins, 2014). The evolutionary history was inferred using the maximum likelihood method based on the JTT matrix-based model (Jones et al., 1992). Evolutionary analyses were conducted in MEGA6 (Tamura et al., 2013).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: HCF145 in Arabidopsis thaliana (AT5G08720), Brachypodium distachyon (XM_003559134), Chlorella variabilis (XM_005849478), Coccomyxa subellipsoidea (XM_005645014), Glycine max (XM_003528024), Oryza sativa (Os03g0837900), Physcomitrella patens (XM_001783694), Populus trichocarpa (XM_002307027), Selaginella moellendorffii (XM_002987569), and Sorghum bicolor (XM_002466088), HCF145-L in Arabidopsis (At4G01650), B. distachyon (XM_003567477), Calothrix sp PCC7507 (CP003943), Chlamydomonas reinhardtii (XM_001699680), Chlorobium tepidum (AE006470), C. subellipsoidea (XM_005645755), Cylindrospermum stagnale PCC7417 (CP003642), Dactylococcopsis salina PCC8305 (CP003944), Galdieria sulphuraria (XM_005707359), Nostoc punctiforme PCC73102 (CP001037), O. sativa (AK059198), P. patens (XM_001775661), P. trichocarpa (XM_002320890), Prochlorococcus marinus sp AS9601 (CP000551), Prosthecochloris aestuarii (CP001108), S. moellendorffii (XM_002987229), Setaria italica (XM_004971363), Sorghum bicolor (XM_002459131), Synechococcus sp CC9902 (CP000097), Vitis vinifera (XM_002280685), TMR proteins in C. variabilis (XM_005846292), C. subellipsoidea (C-169 XP_005645828), Cyanidioschyzon merolae (XM_005538049, XM_005538687, and XM_005538035), G. sulphuraria (XM_005705295), Microcoleus sp PCC 7113 (AFZ22266), and Ostreococcus lucimarinus (XM_001415510), and the psaA 5′ UTR in Arabidopsis (NC_000932), Chara vulgaris (NC_008097), Chlorella vulgaris (NC_001865), C. subellipsoidea C-169 (NC_015084), Euglena gracilis (NC_001603), Marchantia polymorpha (NC_001319), O. sativa (NC_001776), P. patens (NC_005087), P. trichocarpa (NC_009143), Psilotum nudum (NC_003386), S. moellendorffii (NC_013086), S. bicolor (NC_008602), and Thalassiosira pseudonana (NC_008589).
Supplemental Data
Supplemental Figure 1. Generation of the P. patens Δhcf145 Knockout Lines.
Supplemental Figure 2. Conservation of the psaA 5′ UTR in Embryophyta.
Supplemental Figure 3. Immunological Analysis of HCF145 in Wild-Type and Recombinant Forms in Complemented Lines.
Supplemental Figure 4. Conservation of the Tandem Repeated SRPBCC and TMR Motifs in HCF145.
Supplemental Figure 5. Conservation of the SRPBCC Motif in HCF145-L Proteins of Photosynthetic Organisms.
Supplemental Figure 6. Localization of HCF145-L and Genotyping of hcf145-like Knockouts in Arabidopsis.
Supplemental Figure 7. Multiple Alignment of Transcript Binding Motif Repeats Present in Representative TMR Proteins of Photosynthetic Organisms in Eukaryotes and Cyanobacteria.
Supplemental Figure 8. Proposed Three-Dimensional Structures of Arabidopsis HCF145 Motifs SRPBCC1, SRPBCC2, and TMR1.
Supplemental Table 1. Oligonucleotides Used for PCR, RT-PCR, Primer Extension, Probe
Generation, Genotyping, and Other Applications.
Supplemental Data Set 1. Text File of the Alignment Used for the Phylogenetic Analysis in Supplemental Figure 5C.
Supplementary Material
Acknowledgments
We thank Elisabeth Gerick and Jaroslav Mráček for help with initial mapping analysis. We also thank Dario Leister for providing laboratory space and CSP41 sera. This research was supported by the German Science Foundation (Deutsche Forschungsgemeinschaft; ME1794/6-1) to J.M.
AUTHOR CONTRIBUTIONS
N.M., S.T., L.L., M.A.A., and J.M. performed the research. N.M., S.T., L.L., M.A.A., and J.M. analyzed the data. N.M., S.T., W.F., and J.M. designed the work. N.M., S.T., and J.M. wrote the article.
Glossary
- UTR
untranslated region
- PSII
photosystem II
- PSI
photosystem I
- PEP
plastid-encoded polymerase
- NEP
nuclear-encoded phage-type RNA polymerase
- EMSA
electrophoretic mobility shift assay
References
- Amann K., Lezhneva L., Wanner G., Herrmann R.G., Meurer J. (2004). ACCUMULATION OF PHOTOSYSTEM ONE1, a member of a novel gene family, is required for accumulation of [4Fe-4S] cluster-containing chloroplast complexes and antenna proteins. Plant Cell 16: 3084–3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. (1993). Nuclear mutants of maize with defects in chloroplast polysome assembly have altered chloroplast RNA metabolism. Plant Cell 5: 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. (2011). Studying the structure and processing of chloroplast transcripts. Methods Mol. Biol. 774: 183–197. [DOI] [PubMed] [Google Scholar]
- Barkan A., Klipcan L., Ostersetzer O., Kawamura T., Asakura Y., Watkins K.P. (2007). The CRM domain: an RNA binding module derived from an ancient ribosome-associated protein. RNA 13: 55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A., Small I. (2014). Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 65: 415–442. [DOI] [PubMed] [Google Scholar]
- Börner T., Aleynikova A.Y., Zubo Y.O., Kusnetsov V.V. (2015). Chloroplast RNA polymerases: Role in chloroplast biogenesis. Biochim. Biophys. Acta 1847: 761–769. [DOI] [PubMed] [Google Scholar]
- Chi W., He B., Manavski N., Mao J., Ji D., Lu C., Rochaix J.D., Meurer J., Zhang L. (2014). RHON1 mediates a Rho-like activity for transcription termination in plastids of Arabidopsis thaliana. Plant Cell 26: 4918–4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W.K., Geimer S., Meurer J. (2009). Cluster analysis and comparison of various chloroplast transcriptomes and genes in Arabidopsis thaliana. DNA Res. 16: 31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Egener T., et al. (2002). High frequency of phenotypic deviations in Physcomitrella patens plants transformed with a gene-disruption library. BMC Plant Biol. 2: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errampalli D., Patton D., Castle L., Mickelson L., Hansen K., Schnall J., Feldmann K., Meinke D. (1991). Embryonic lethals and T-DNA insertional mutagenesis in Arabidopsis. Plant Cell 3: 149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipovska A., Rackham O. (2012). Modular recognition of nucleic acids by PUF, TALE and PPR proteins. Mol. Biosyst. 8: 699–708. [DOI] [PubMed] [Google Scholar]
- Frank W., Decker E.L., Reski R. (2005). Molecular tools to study Physcomitrella patens. Plant Biol (Stuttg) 7: 220–227. [DOI] [PubMed] [Google Scholar]
- Germain A., Hotto A.M., Barkan A., Stern D.B. (2013). RNA processing and decay in plastids. Wiley Interdiscip. Rev. RNA 4: 295–316. [DOI] [PubMed] [Google Scholar]
- Gross J., Cho W.K., Lezhneva L., Falk J., Krupinska K., Shinozaki K., Seki M., Herrmann R.G., Meurer J. (2006). A plant locus essential for phylloquinone (vitamin K1) biosynthesis originated from a fusion of four eubacterial genes. J. Biol. Chem. 281: 17189–17196. [DOI] [PubMed] [Google Scholar]
- Hammani K., Cook W.B., Barkan A. (2012). RNA binding and RNA remodeling activities of the half-a-tetratricopeptide (HAT) protein HCF107 underlie its effects on gene expression. Proc. Natl. Acad. Sci. USA 109: 5651–5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammani K., Bonnard G., Bouchoucha A., Gobert A., Pinker F., Salinas T., Giegé P. (2014). Helical repeats modular proteins are major players for organelle gene expression. Biochimie 100: 141–150. [DOI] [PubMed] [Google Scholar]
- Heazlewood J.L., Tonti-Filippini J.S., Gout A.M., Day D.A., Whelan J., Millar A.H. (2004). Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 16: 241–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer L.M., Koonin E.V., Aravind L. (2001). Adaptations of the helix-grip fold for ligand binding and catalysis in the START domain superfamily. Proteins 43: 134–144. [DOI] [PubMed] [Google Scholar]
- Jenkins B.D., Barkan A. (2001). Recruitment of a peptidyl-tRNA hydrolase as a facilitator of group II intron splicing in chloroplasts. EMBO J. 20: 872–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.T., Taylor W.R., Thornton J.M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8: 275–282. [DOI] [PubMed] [Google Scholar]
- Klein R.R., Mullet J.E. (1987). Control of gene expression during higher plant chloroplast biogenesis. Protein synthesis and transcript levels of psbA, psaA-psaB, and rbcL in dark-grown and illuminated barley seedlings. J. Biol. Chem. 262: 4341–4348. [PubMed] [Google Scholar]
- Kroeger T.S., Watkins K.P., Friso G., van Wijk K.J., Barkan A. (2009). A plant-specific RNA-binding domain revealed through analysis of chloroplast group II intron splicing. Proc. Natl. Acad. Sci. USA 106: 4537–4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupsch C., Ruwe H., Gusewski S., Tillich M., Small I., Schmitz-Linneweber C. (2012). Arabidopsis chloroplast RNA binding proteins CP31A and CP29A associate with large transcript pools and confer cold stress tolerance by influencing multiple chloroplast RNA processing steps. Plant Cell 24: 4266–4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. [DOI] [PubMed] [Google Scholar]
- Legen J., Kemp S., Krause K., Profanter B., Herrmann R.G., Maier R.M. (2002). Comparative analysis of plastid transcription profiles of entire plastid chromosomes from tobacco attributed to wild-type and PEP-deficient transcription machineries. Plant J. 31: 171–188. [DOI] [PubMed] [Google Scholar]
- Letsch M.R., Lewis L.A. (2012). Chloroplast gene arrangement variation within a closely related group of green algae (Trebouxiophyceae, Chlorophyta). Mol. Phylogenet. Evol. 64: 524–532. [DOI] [PubMed] [Google Scholar]
- Lezhneva L., Meurer J. (2004). The nuclear factor HCF145 affects chloroplast psaA-psaB-rps14 transcript abundance in Arabidopsis thaliana. Plant J. 38: 740–753. [DOI] [PubMed] [Google Scholar]
- Manavski N., Torabi S., Stoppel R., Meurer J. (2012a). Phylogenetic and ontogenetic integration of organelles into the compartmentalized genome of the eukaryotic cell. Endocytobiosis Cell Res. 23: 25–31. [Google Scholar]
- Manavski N., Guyon V., Meurer J., Wienand U., Brettschneider R. (2012b). An essential pentatricopeptide repeat protein facilitates 5′ maturation and translation initiation of rps3 mRNA in maize mitochondria. Plant Cell 24: 3087–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín-Navarro J., Manuell A.L., Wu J., P Mayfield S. (2007). Chloroplast translation regulation. Photosynth. Res. 94: 359–374. [DOI] [PubMed] [Google Scholar]
- Meurer J., Berger A., Westhoff P. (1996b). A nuclear mutant of Arabidopsis with impaired stability on distinct transcripts of the plastid psbB, psbD/C, ndhH, and ndhC operons. Plant Cell 8: 1193–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurer J., Grevelding C., Westhoff P., Reiss B. (1998). The PAC protein affects the maturation of specific chloroplast mRNAs in Arabidopsis thaliana. Mol. Gen. Genet. 258: 342–351. [DOI] [PubMed] [Google Scholar]
- Meurer J., Lezhneva L., Amann K., Gödel M., Bezhani S., Sherameti I., Oelmüller R. (2002). A peptide chain release factor 2 affects the stability of UGA-containing transcripts in Arabidopsis chloroplasts. Plant Cell 14: 3255–3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurer J., Meierhoff K., Westhoff P. (1996a). Isolation of high-chlorophyll-fluorescence mutants of Arabidopsis thaliana and their characterisation by spectroscopy, immunoblotting and northern hybridisation. Planta 198: 385–396. [DOI] [PubMed] [Google Scholar]
- Mollier P., Hoffmann B., Debast C., Small I. (2002). The gene encoding Arabidopsis thaliana mitochondrial ribosomal protein S13 is a recent duplication of the gene encoding plastid S13. Curr. Genet. 40: 405–409. [DOI] [PubMed] [Google Scholar]
- Mudd E.A., Sullivan S., Gisby M.F., Mironov A., Kwon C.S., Chung W.I., Day A. (2008). A 125 kDa RNase E/G-like protein is present in plastids and is essential for chloroplast development and autotrophic growth in Arabidopsis. J. Exp. Bot. 59: 2597–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullet J.E., Klein R.R. (1987). Transcription and RNA stability are important determinants of higher plant chloroplast RNA levels. EMBO J. 6: 1571–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostersetzer O., Cooke A.M., Watkins K.P., Barkan A. (2005). CRS1, a chloroplast group II intron splicing factor, promotes intron folding through specific interactions with two intron domains. Plant Cell 17: 241–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phinney B.S., Thelen J.J. (2005). Proteomic characterization of a triton-insoluble fraction from chloroplasts defines a novel group of proteins associated with macromolecular structures. J. Proteome Res. 4: 497–506. [DOI] [PubMed] [Google Scholar]
- Porat Y.B., Zan-Bar I., Ravid A. (1995). Quantitative dot-blot assay for low titer anti-lipopolysaccharide antibodies in human plasma. J. Immunol. Methods 180: 213–218. [DOI] [PubMed] [Google Scholar]
- Qi Y., Armbruster U., Schmitz-Linneweber C., Delannoy E., de Longevialle A.F., Rühle T., Small I., Jahns P., Leister D. (2012). Arabidopsis CSP41 proteins form multimeric complexes that bind and stabilize distinct plastid transcripts. J. Exp. Bot. 63: 1251–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radauer C., Lackner P., Breiteneder H. (2008). The Bet v 1 fold: an ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evol. Biol. 8: 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schein A., Sheffy-Levin S., Glaser F., Schuster G. (2008). The RNase E/G-type endoribonuclease of higher plants is located in the chloroplast and cleaves RNA similarly to the E. coli enzyme. RNA 14: 1057–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster G., Stern D. (2009). RNA polyadenylation and decay in mitochondria and chloroplasts. Prog. Mol. Biol. Transl. Sci. 85: 393–422. [DOI] [PubMed] [Google Scholar]
- Seki M., et al. (2002). Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141–145. [DOI] [PubMed] [Google Scholar]
- Shikanai T., Fujii S. (2013). Function of PPR proteins in plastid gene expression. RNA Biol. 10: 1446–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Higgins D.G. (2014). Clustal omega. Curr. Protoc. Bioinformatics 48: 3.1–3.13, 16. [DOI] [PubMed] [Google Scholar]
- Stern D.B., Goldschmidt-Clermont M., Hanson M.R. (2010). Chloroplast RNA metabolism. Annu. Rev. Plant Biol. 61: 125–155. [DOI] [PubMed] [Google Scholar]
- Stoppel R., Meurer J. (2012). The cutting crew - ribonucleases are key players in the control of plastid gene expression. J. Exp. Bot. 63: 1663–1673. [DOI] [PubMed] [Google Scholar]
- Stoppel R., Meurer J. (2013). Complex RNA metabolism in the chloroplast: an update on the psbB operon. Planta 237: 441–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppel R., Lezhneva L., Schwenkert S., Torabi S., Felder S., Meierhoff K., Westhoff P., Meurer J. (2011). Recruitment of a ribosomal release factor for light- and stress-dependent regulation of petB transcript stability in Arabidopsis chloroplasts. Plant Cell 23: 2680–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppel R., Manavski N., Schein A., Schuster G., Teubner M., Schmitz-Linneweber C., Meurer J. (2012). RHON1 is a novel ribonucleic acid-binding protein that supports RNase E function in the Arabidopsis chloroplast. Nucleic Acids Res. 40: 8593–8606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summer H., Pfannschmidt T., Link G. (2000). Transcripts and sequence elements suggest differential promoter usage within the ycf3-psaAB gene cluster on mustard (Sinapis alba L.) chloroplast DNA. Curr. Genet. 37: 45–52. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torabi S., Umate P., Manavski N., Plöchinger M., Kleinknecht L., Bogireddi H., Herrmann R.G., Wanner G., Schröder W.P., Meurer J. (2014). PsbN is required for assembly of the photosystem II reaction center in Nicotiana tabacum. Plant Cell 26: 1183–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins K.P., Rojas M., Friso G., van Wijk K.J., Meurer J., Barkan A. (2011). APO1 promotes the splicing of chloroplast group II introns and harbors a plant-specific zinc-dependent RNA binding domain. Plant Cell 23: 1082–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi Y., Shiina T. (2014). Recent advances in the study of chloroplast gene expression and its evolution. Front. Plant Sci. 5: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhelyazkova P., Hammani K., Rojas M., Voelker R., Vargas-Suárez M., Börner T., Barkan A. (2012). Protein-mediated protection as the predominant mechanism for defining processed mRNA termini in land plant chloroplasts. Nucleic Acids Res. 40: 3092–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.