The historical DNA changes that created the black rice trait and its local spread into rice subspecies involved rearrangement in the promoter region of Kala4 and subsequent introgression events.
Abstract
The origin and spread of novel agronomic traits during crop domestication are complex events in plant evolution. Wild rice (Oryza rufipogon) has red grains due to the accumulation of proanthocyanidins, whereas most cultivated rice (Oryza sativa) varieties have white grains induced by a defective allele in the Rc basic helix-loop-helix (bHLH) gene. Although the events surrounding the origin and spread of black rice traits remain unknown, varieties with black grains due to anthocyanin accumulation are distributed in various locations throughout Asia. Here, we show that the black grain trait originated from ectopic expression of the Kala4 bHLH gene due to rearrangement in the promoter region. Both the Rc and Kala4 genes activate upstream flavonol biosynthesis genes, such as chalcone synthase and dihydroflavonol-4-reductase, and downstream genes, such as leucoanthocyanidin reductase and leucoanthocyanidin dioxygenase, to produce the respective specific pigments. Genome analysis of 21 black rice varieties as well as red- and white-grained landraces demonstrated that black rice arose in tropical japonica and its subsequent spread to the indica subspecies can be attributed to the causal alleles of Kala4. The relatively small size of genomic fragments of tropical japonica origin in some indica varieties indicates that refined introgression must have occurred by natural crossbreeding in the course of evolution of the black trait in rice.
INTRODUCTION
The grain color of cereals is determined by the pigmentation of certain phytochemicals. In rice (Oryza sativa), most varieties have white grains, but some have brown, red, or black grains. The red grain color is due to the deposition and oxidative polymerization of proanthocyanidins in the pericarp, whereas the black grain color is caused by deposition of anthocyanins (Supplemental Figure 1A) (Reddy et al., 1995; Rahman et al., 2013; Goufo and Trindade, 2014). The anthocyanin biosynthesis pathway has been elucidated in various plant species (Supplemental Figure 1B and Supplemental Table 1) (Dooner et al., 1991; Shih et al., 2008). Transcriptional regulation related to anthocyanin biosynthesis has also been extensively studied, particularly for the maize (Zea mays) R/B and C1/Pl gene families, and orthologs in other plant species, which encode basic helix-loop-helix (bHLH)-type and R2R3-Myb-type transcription factors, respectively (Styles et al., 1973; Paz-Ares et al., 1987; Chandler et al., 1989; Cone et al., 1993). These regulatory proteins act in a ternary complex, as MBW (MYB-bHLH-WD40) complex transcription factors (Hichri et al., 2011; Petroni and Tonelli, 2011). In rice, R/B homolog genes have been reported to be involved in the regulation of anthocyanin biosynthesis (Hu et al., 1996, 2000). Moreover, analysis of three alleles of Purple leaf (Pl) in rice, namely, Plj, Pli, and Plw, which differentially confer purple-colored leaves, has revealed two candidate R genes, OSB1 and OSB2 (Sakamoto et al., 2001). Although the OSB1 and OSB2 genes activate the biosynthesis of anthocyanins when the maize C1 is cobombarded into the rice aleurone layer cells (Sakamoto et al., 2001), the causal functional nucleotide polymorphisms (FNPs) and genetic complementation have not been reported yet.
Three loci of the maize anthocyanin biosynthetic regulator gene, namely, b1, r1, and pl1, are involved in the epigenetic behaviors of the trait (Supplemental Table 1) (Brink, 1956; Coe, 1959; Hollick and Chandler, 2001). These epialleles have strong genetic linkages to changes in grain color and the DNA methylation status in the regulatory sequences of the genes (Walker, 1998; Stam et al., 2002). These results suggest that alteration of the DNA methylation status in these genes can change the activities of the anthocyanin biosynthetic pathway. Furthermore, genome-wide methylome analysis in maize and rice has suggested that the DNA methylation in the promoter region often represses gene expression, whereas moderate gene-body methylation has a positive effect on the gene expression (Li et al., 2012; Regulski et al., 2013). Therefore, DNA methylation status may affect gene expression.
The rice domestication process is reflected in the early history of artificial selection of key agronomically important traits, such as seed shattering, plant architecture, yield, and grain color (Izawa et al., 2009). Several genes related to rice domestication, including sh4, qSH1, PLOG1, qSW5, GIF1, and Rc, have been cloned and the corresponding causal FNPs have also been identified (Konishi et al., 2006; Li et al., 2006; Sweeney et al., 2007; Jin et al., 2008; Shomura et al., 2008; Tan et al., 2008; Wang et al., 2008). Among them, a 14-bp deletion within the Rc gene, which induced a premature stop codon, resulted in the change of pericarp color from red to white. The red grain color is ubiquitous in wild rice (Oryza rufipogon) varieties, which share common ancestors with cultivated rice, and the defect in Rc was spread broadly into most rice varieties in the process of domestication (Sweeney et al., 2007). To date, only two loci for the red rice trait have been reported; Rc, which encodes a bHLH transcription factor, and Rd, which encodes dihydroflavonol-4-reductase (DFR) (Sweeney et al., 2006; Furukawa et al., 2007). The black (or purple) grain color has not been observed in any accessions of O. rufipogon (see Oryzabase, http://www.gramene.org/). Thus, the black rice trait is most likely newly acquired, incorporated either during or after rice domestication. Compared with the broad distribution of white rice cultivars, black rice cultivars are sporadically distributed throughout Asia. Black rice was used in ancient China before Chinese dynastic times (Newman, 2004) and is sometimes called “emperor’s rice” (or “forbidden rice”) since it was used as a tribute food and prized for its rarity in ancient China. It is completely unknown how this aesthetic trait arose and has been maintained for such a long time.
Previously, the ‘Hong Xie Nuo’ (a black rice cultivar) alleles of three loci, namely, Kala1, Kala3, and Kala4, were introgressed into an elite temperate japonica cultivar ‘Koshihikari’ (Maeda et al., 2014). The resulting ‘Black rice NIL’ showed complete conversion of the white pericarp into black pericarp. It has been speculated that the Kala1 and Kala3 genes encode a DFR and an R2R3-Myb transcriptional factor, respectively (Maeda et al., 2014). The ‘Hong Xie Nuo’ alleles of Kala1 and Kala3 loci play subsidiary roles in the black rice trait as shown by the brown grain color of the ‘AAB-type NIL,’ which possesses the ‘Koshihikari’ alleles of Kala1 and Kala3 and the ‘Hong Xie Nuo’ allele of Kala4. Meanwhile, Kala4 acts as a main contributor of the black trait in the ‘Black rice NIL’ since the grain color of the ‘BBA-type NIL’ with the ‘Hong Xie Nuo’ alleles of Kala1 and Kala3 and ‘Koshihikari’ allele of Kala4 is completely white, whereas the grain color of the ‘BBB-type NIL or (Black rice NIL)’ with the ‘Hong Xie Nuo’ alleles of Kala1, Kala3, and Kala4 loci is black (Maeda et al., 2014). However, identification of Kala4 has been unexplored, and the underlying molecular mechanism of the origin of black rice has not yet been fully elucidated.
Here, we demonstrated genetically that the Kala4 gene involved in the origin of black rice corresponds to Os04g0557500. Kala4 encodes a bHLH transcription factor, which is a rice homolog of the maize R/B gene, previously reported as the OSB2 gene (Sakamoto et al., 2001). We further demonstrated that a structural change in the Kala4 promoter induced ectopic expression of the bHLH protein and the subsequent activation of the anthocyanin biosynthesis pathway. With this structural change in Kala4, rice acquired the brand new black grain color trait, resulting in the birth of black rice. We also demonstrated that a relatively small genomic segment around the Kala4 gene that originated from tropical japonica was transferred into the indica subspecies, probably as a result of refined introgression through multiple natural crossbreeding processes. The results described here reveal the process that led to the neofunctionalization of Kala4 and subsequent spread of the black rice allele of Kala4 to other subspecies of rice.
RESULTS
The Gene Responsible for the Origin of Black Rice Is Kala4
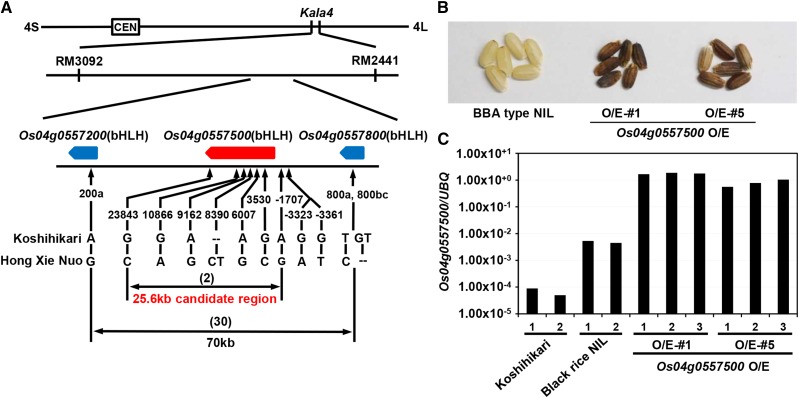
The Kala4 gene was previously mapped between two simple sequence repeat (SSR) markers, RM3092 and RM2441 on chromosome 4, and the ‘Hong Xie Nuo’ allele of Kala4 is known to be inherited in a semidominant manner (Maeda et al., 2014). Using the Os-Nipponbare-Reference-build5 genome assembly (http://rapdb.dna.affrc.go.jp/), we found three homologs of the maize R/B genes encoding bHLH transcription factors, between the two SSR markers (Figure 1A). We then performed fine mapping of the Kala4 gene (Figure 1A; Supplemental Figure 2). Approximately 3000 seedlings in the F2 generation, produced by crossing the ‘BBB-type NIL (or Black rice NIL)’ and the ‘BBA-type NIL’ as the parental plants, were genotyped for two PCR marker loci (200a/800a and 800bc) to genetically dissect the genomic region of the bHLH genes (Figure 1A). Thirty recombinant plants were selected and genotyped further using seven markers in the Os04g0557500 gene region (Figure 1A). We examined the grain color of the F3 seeds from the F2 recombinant plants (Supplemental Figure 2A) with the homozygous ‘Koshihikari’ genotype at either of these two loci and the grain color of the F4 seeds (Supplemental Figure 2B) from the F2 recombinant plants with the homozygous ‘Hong Xie Nuo’ genotype. As a result, we were able to narrow down the candidate region for the causal FNP to an ∼25.6-kb genomic region containing the Os04g0557500 gene (Figure 1A).
Figure 1.
Identification of Kala4.
(A) Fine mapping. The Kala4 locus was previously localized to chromosome 4 between two SSR markers, RM3092 and RM2441. This region contains three bHLH protein-encoding genes. Fine mapping narrowed the Kala4 locus to an ∼25.6-kb region between two SNP markers (+23843 and −1707). Six SNPs and one in/del between ‘Koshihikari’ and ‘Hong Xie Nuo’ in this region were mapped onto the reference ‘Nipponbare’ genome. The number in parentheses indicates the number of recombinant plants.
(B) Grain color of transgenic plants overexpressing Os04g0557500. The transgenic lines (O/E#1 and O/E#5) show black seed color in contrast to the white seeds of the parental ‘BBA-type NIL.’
(C) Quantification of mRNAs by RT-qPCR analysis. Total RNAs were extracted from leaves of three independent hygromycin-resistant plants of the T1 generation for O/E. The y axis is shown in a logarithmic scale.
For microarray analysis, we used total RNAs extracted from the white, black, and red pericarps of the recurrent cultivar ‘Koshihikari’ (BBA-type NIL), the ‘Black rice NIL’ (BBB-type NIL) with three ‘Hong Xie Nuo’ genomic loci, and the ‘Red rice NIL’ with two ‘Kasalath’ genomic loci, carrying functional alleles of Rd and Rc (Maeda et al., 2014; Yamaguchi et al., 2015), respectively (Supplemental Figure 3). Some of the upstream anthocyanin biosynthesis genes such as chalcone synthase, flavanone 3-hydroxylase, flavanone 3′-hydroxylase, and DFR were upregulated in the red, black, and the red/black pericarps derived from the F1 plants compared with the recurrent white grain rice lines (Supplemental Figures 3A to 3D). In particular, leucoanthocyanidin reductase (LAR) was specifically upregulated in the F1 lines with red and red/black pericarps, whereas leucoanthocyanidin dioxygenase (LDOX) and UDP-glucosyl transferase were specifically upregulated in the F1 lines with black pericarp (Supplemental Figures 3E to 3G). These results indicate that the proanthocyanidin and anthocyanin biosynthesis pathways are activated in the red and black rice pericarps, respectively. Among the three bHLH genes located in the Kala4 locus, only Os04g0557500 expression was upregulated in the pericarps of the black and F1 rice lines (Supplemental Figure 3H). Therefore, it is likely that the upregulated expression of Os04g0557500 may have induced the activation of the anthocyanin biosynthesis pathway.
To further confirm that Os04g0557500 is Kala4, we generated transgenic plants overexpressing the Os04g0557500 coding sequence in the ‘BBA-type NIL’ (Figures 1B and 1C). Nine of the 11 independent T0 transgenic rice plants exhibited black grains (Figure 1B) with the expected upregulation of Os04g0557500 (Figure 1C).
The Relationship between DNA Methylation and the Black Rice Trait
The Os04g0557500 gene has eight exons and seven introns, with seven polymorphisms between ‘Koshihikari’ and ‘Hong Xie Nuo’ in the ∼25.6-kb candidate region (Supplemental Figure 4). We analyzed the sequence of these polymorphic regions in 21 black rice landraces obtained from the National Institute of Agrobiological Sciences Gene Bank (http://www.gene.affrc.go.jp/). Based on five marker positions (+23843, +9162, +6007, +3530, and −1707), we found that all the black rice landraces and seven heritage white and red rice landraces of tropical japonica had the same genotypes (Konishi et al., 2008) (Supplemental Table 2). The other two markers, +10866 and +8390, were specific for some but not all of the black rice landraces (Supplemental Table 2). Therefore, none of the seven polymorphisms in the 25.6-kb candidate region is the causal FNP for the black rice trait.
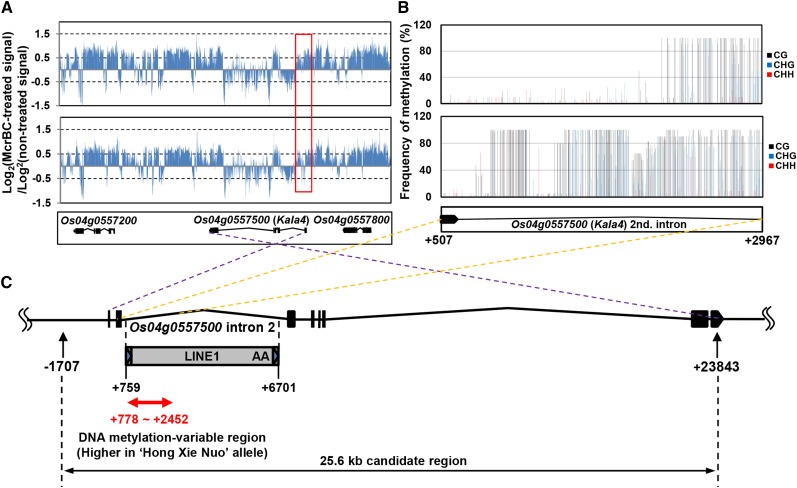
This result implies that an epigenetic change may be involved in the expression of the black pericarp in rice. Accordingly, we investigated the DNA methylation status for the candidate region by designing a tilling array covering an ∼85-kb region containing the three bHLH genes. We performed McrBC-comparative genomic hybridization (McrBC-CGH) using ‘Koshihikari’ and ‘Hong Xie Nuo’ genomic DNAs (Figure 2A) to detect the methylated CpG islands after digestion of the DNA containing methyl-cytosine (Sutherland et al., 1992). The signal pattern of ‘Hong Xie Nuo’ was very similar to that of ‘Koshihikari’ in the 85-kb region except in the peripheral region of the Os04g0557500 intron 2 (Figure 2A). Bisulfite sequencing analysis further revealed an increase in the DNA methylation level around this intron in ‘Hong Xie Nuo’ (Figure 2B). The increased DNA methylation was observed in the 1.7-kb region of the 5′-end of the intron 2 (Figure 2C). Homology searches of this region showed an insertion of the LINE1-type retrotransposon in intron 2 of Os04g0557500 (Figure 2C; Supplemental Figure 5).
Figure 2.
Analysis of DNA Methylation in Kala4.
(A) McrBC-CGH analysis. The DNA methylation patterns in pericarps of ‘Koshihikari’ (upper panel) and ‘Hong Xie Nuo’ (lower panel) differ around the 85-kb region at the Kala4 locus. The relative positions of the three bHLH genes are shown below the lower panel. Red box indicates a region where difference of DNA methylation pattern between ‘Koshihikari’ and ‘Hong Xie Nuo’ was observed.
(B) Analysis of the intron 2 region of Os04g0557500. The DNA methylation patterns upstream of intron 2 in ‘Koshihikari’ (upper panel) and ‘Hong Xie Nuo’ (lower panel) were analyzed by bisulfite sequencing of the region between markers +507 and +2967.
(C) Diagram of the DNA methylation-variable region in the 25.6-kb candidate region. Black boxes and lines indicate exons and introns of Os04g0557500. The LINE1 retrotransposon insertion is located in intron 2 of Os04g0557500 and the DNA methylation-variable region is located in 5′-upstream of intron 2 (shown as red bidirectional arrow). The numbers indicate the position from the start site of Kala4 transcription.
We then investigated the DNA methylation status in the 21 black rice landraces using McrBC-DNA gel blot analysis (Supplemental Figure 6). All 21 black landraces similarly exhibited a higher DNA methylation frequency in intron 2, indicating a strong association between the DNA methylation status and the black rice trait. However, three of the seven red or white heritage landraces exhibited higher DNA methylation similar to that of the black rice landraces (Supplemental Figure 6). Furthermore, the Os04g0557500 transcript levels were low in both leaves and pericarps of these three heritage landraces (Supplemental Figure 7). These results suggest that although the higher DNA methylation status of intron 2 in ‘Hong Xie Nuo’ is closely linked to the black rice trait, it may not be the causal genetic variation for this specific trait.
Reevaluation of the Structure of the ‘Hong Xie Nuo’ Allele of Os04g0557500
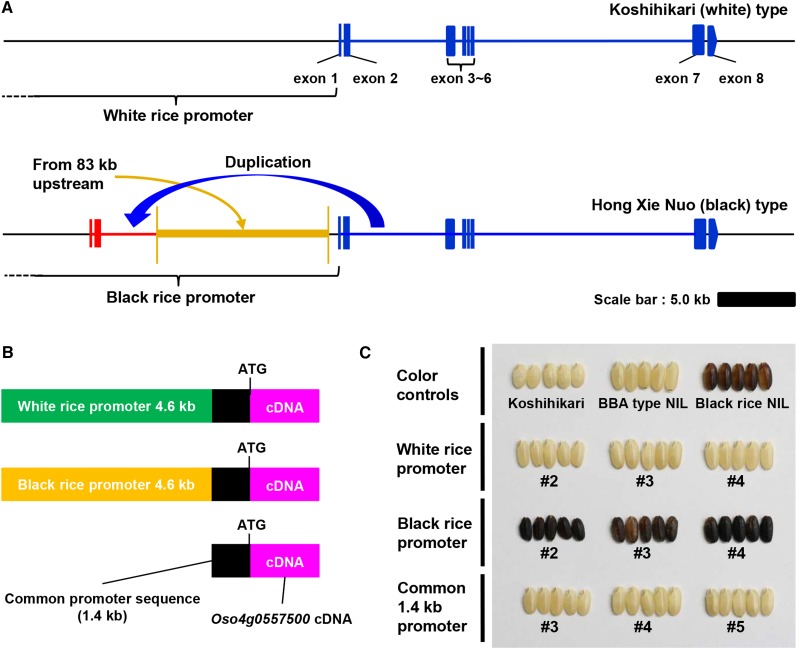
When the McrBC-DNA gel blot analysis was performed as described above, we noticed an unexpected band detected in both the ‘Black rice NIL’ and ‘Hong Xie Nuo’ genomes (Supplemental Figure 8). This result raised the possibility of duplication of the genomic region related to the Os04g0557500 gene in the ‘Hong Xie Nuo’ genome. We thus attempted to identify the possible duplicated sequence. Using a combination of the fusion primer and nested integrated (FPNI)-PCR method (Wang et al., 2011) and DNA gel blot analysis, we were able to identify a duplication from the promoter region 4.9 kb upstream of exon 1 to within intron 2 of Os04g0557500 in ‘Hong Xie Nuo’ (Figure 3A). In addition, an 11.0-kb genomic fragment originated from ∼83 kb upstream of Os04g0557500 was duplicated and then inserted at the 954 bp upstream of the transcription start point of Os04g0557500 (Figure 3A). To determine the promoter structure in ‘Hong Xie Nuo’ in detail, DNA gel blot analysis was also performed using six kinds of probes covering the entire Os04g0557500 gene and the promoter region (Supplemental Figures 9 and 10). We were able to confirm the results using five distinct restriction enzymes (Supplemental Figures 10A to 10D and 10F and Supplemental Table 3) and determine the correct structure of the Os04g0557500 promoter in ‘Hong Xie Nuo’ (Figure 3A).
Figure 3.
Relationship of the Os04g0557500 Promoter Structure and Black Rice Trait.
(A) Structures of the Os04g0557500 promoter regions in ‘Koshihikari’ and ‘Hong Xie Nuo.’ Upper bar indicates genomic structure of Os04g0557500 gene in ‘Koshihikari’. Lower bar indicates that in ‘Hong Xie Nuo.’ Blue lines and boxes indicate the ORF region. The promoter region in ‘Hong Xie Nuo’ possesses a duplicated region of Os04g0557500 (4679 bp) including exon 1, intron 1, exon 2, and the part of intron 2 (red boxes and lines). In addition to this duplication, the Os04g0557500 promoter region in ‘Hong Xie Nuo’ has an ∼11.02-kb insertion of a genome fragment originating from ∼83 kb upstream of Os04g0557500 (orange bar). Bar = 5.0 kb.
(B) Promoter sequence constructs of Os04g0557500. Schematic diagram of the three types of promoter:Os04g0557500 cDNA constructs with the white rice-type promoter sequence from ‘Koshihikari’ (green box), black rice-type promoter sequence from ‘Black rice NIL’ (orange box), and the 1.4-kb common promoter sequence shared between ‘Koshihikari’ and ‘Hong Xie Nuo’ (black box).
(C) Grain color of transgenic plants harboring the three promoter:Os04g0557500 constructs. Grains of three representative lines for each promoter:Os04g0557500 construct (T1) are shown. ‘Koshihikari,’ ‘BBA-type NIL,’ and ‘Black rice NIL’ are shown as white and black color controls.
When we first sequenced the corresponding genome region in Os04g0557500 to examine the FNPs (Figure 1), we used direct sequencing of PCR-amplified genome fragments (Supplemental Figure 11). Thus, the 11.0-kb insertion observed in ‘Hong Xie Nuo’ allele was not amplified by PCR due to the partial duplication of Os04g0557500 (Supplemental Figure 11). Taken together with this new data and fine mapping, this complex rearrangement could be the causal FNP for the black rice trait.
We next introduced three types of promoter (‘Koshihikari,’ ‘Hong Xie Nuo,’ and a 1.4-kb common promoter) fused to the Os04g0557500 cDNA (Figure 3B) into the ‘BBA-type NIL.’ All transgenic lines with black rice ‘Hong Xie Nuo’ promoter had black rice grains, whereas all transgenic lines with white rice ‘Koshihikari’ or the 1.4-kb common promoter had completely white rice grains (Figure 3C), indicating that the rearranged promoter of the Os04g0557500 in ‘Hong Xie Nuo’ could confer the black rice trait and that the transformed region from the 11.0-kb insertion found in ‘Hong Xie Nuo’ was essential for the black rice trait. Thus, it is likely that the 11.0-kb insertion has a positive role in the expression of Kala4.
The Structural Change in the Os04g0557500 Promoter, but Not the Increased DNA Methylation, Is Critical for the Black Rice Trait
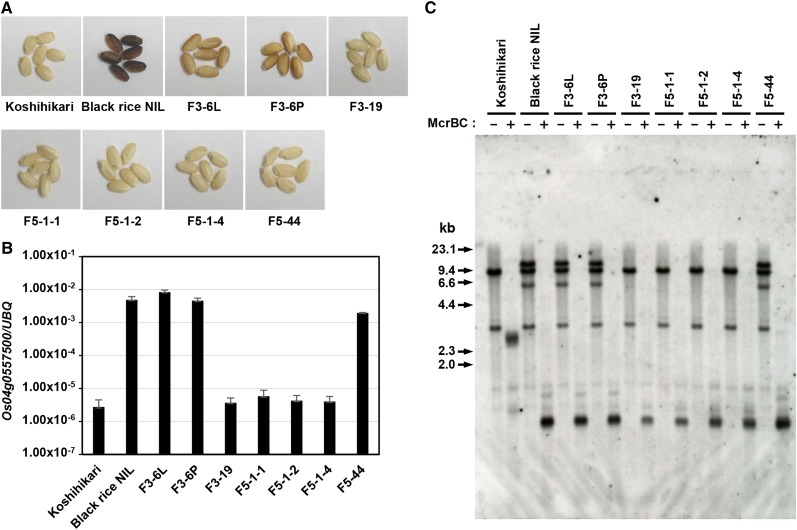
To obtain genetic resources related to the black rice trait, we screened thousands of M2 progeny of ‘Black rice NIL’ mutated by γ-ray irradiation and identified seven suppressor mutants that resulted in white grains (Figure 4A). Analysis of these suppressor mutants provided direct evidence that the black grain was caused by the structural change in Os04g0557500 but not by the increased DNA methylation in intron 2. In four of these seven suppressor mutants, namely, F3-19, F5-1-1, F5-1-2, and F5-1-4, the expression of Os04g0557500 was decreased compared with the ‘Black rice NIL’ (Figure 4B). To examine the genomic structure of Os04g0557500 in the suppressor mutants, several primer sets were designed (Supplemental Figure 12A) for PCR amplifications. The results suggest that the 11.0-kb insertion was possibly deleted from the Os04g0557500 promoter region in the four suppressor mutants (Supplemental Figures 12B and 12C), although the open reading frame (ORF) region was intact (Supplemental Figures 12D to 12G). To further elucidate the structure of Os04g0557500 in the suppressor mutants, DNA gel blot hybridization was performed using three kinds of probes (Supplemental Figure 13A). After NdeI digestion and hybridization with the exon1N or D9K probes, the duplicated fragments could not be detected in the four suppressor mutants (Supplemental Figures 13B and 13C). Moreover, upon digestion using EcoRI and hybridization with the “Kala4pro+” probe, a ‘Koshihikari’-type fragment was identified in the four suppressor lines (Supplemental Figure 13D). To exclude the possibility that this reversion (or deletion) was due to possible outcrossing between ‘Black rice NIL’ and other white cultivars grown in the same paddy field, we examined the three single nucleotide polymorphism (SNP) markers and two PCR markers (Supplemental Figure 14). We found that the seven suppressor mutants possess the ‘Black rice NIL’-type polymorphisms, indicating that the deletion in the four suppressor mutants resulted from genomic recombination in the ‘Black rice NIL’ by γ-irradiation and not from outcrossing.
Figure 4.
Characterization of Black Rice Suppressor Mutants.
(A) Grain color of the suppressor mutants. The seven suppressor mutants showed variation in grain color from the white grain of ‘Koshihikari’ and the black grain of ‘Black rice NIL.’
(B) Expression of Os04g0557500 mRNA in the suppressor mutants. Total RNAs were extracted from three to five 10-d-old seedlings. Three independent total RNAs extractions and RT-qPCRs were performed. The y axis is shown in a logarithmic scale. Means + se of three independent experiments are shown.
(C) McrBC-DNA gel blot hybridization. Genomic DNAs were extracted from 10 to 15 12-d-old seedlings. The extracted DNAs were digested with NdeI with (+) or without (−) subsequent McrBC digestion. Hybridization was performed with the exon1N probe. In the case of digestion with McrBC, the distinct band sizes were detected according to the DNA methylation status of intron 2 in Os04g0557500
We then performed McrBC-DNA gel blot analysis using the suppressor mutant lines (Figure 4C). Since DNA methylation of intron 2 was maintained in the four suppressor lines, we could exclude the possibility that the DNA methylation in intron 2 was the cause of the increased transcripts and the black rice trait. Taken together, we concluded that the black rice trait in ‘Black rice NIL’ is due to the structural change in the Os04g0557500 promoter and not from the higher DNA methylation status in intron 2. With this identification of the FNP, we were able to conclude that Kala4 corresponds to Os04g0557500.
The Relationship between the Structural Change of Kala4 and the Origin of Black Rice
To investigate the relationship between the structural change in the Kala4 gene and the origin of black rice, DNA gel blot analysis was performed using exon1N probe for the 21 black rice landraces (Supplemental Figure 15). In addition to the expected band from the ‘Nipponbare’ genomic sequence, several bands not detected in the white and red rice lines were observed in the 21 black rice landraces, indicating at least a partial duplication of the exon 2 region of the Kala4 gene (Supplemental Figure 15).
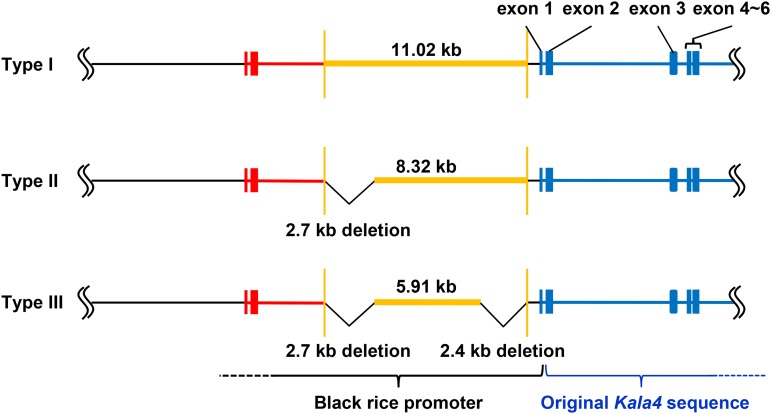
Using specific PCR amplification with junction 1 and junction 2 primer pairs (Supplemental Figures 12A and 16A), we were able to divide the 21 black rice landraces into three groups based on the types of ends of insertion, namely, type I (‘Hong Xie Nuo’ type), type II, and type III (Supplemental Figure 16B and Supplemental Table 4). We selected ‘Mitak’ as a representative variety for type II and determined the Kala4 promoter structure in this black rice using DNA gel blot analysis followed by FPNI-PCR. As a representative of type III, the promoter structure of ‘KH. Kam’ variety was also determined. As a result, three types of promoter structures in Kala4 were identified with variations of insertion length in the duplications and insertions detected in ‘Hong Xie Nuo’ (Figure 5). These results strongly suggest that the structural changes in the promoter of Kala4 induced the black rice trait in rice. Furthermore, the type I insertion (i.e., the longest one) was likely to be the original genetic change that occurred to give rise to black rice.
Figure 5.
Structural Variations of the Kala4 Promoters in 21 Black Rice Varieties.
The Kala4 promoters of 21 black rice varieties were classified into three types based on the length of inserted fragments. Type I promoters have the longest insertion (11.02 kb) with the same size as ‘Hong Xie Nuo.’ Type II promoters have a deletion of ∼2.7 kb at the 5′ junction. Type III promoters have the 2.7-kb deletion at the 5′ junction and a 2.4-kb deletion at the 3′ junction. The original Kala4 ORF sequence (blue region), insertion from ∼83 kb upstream of Kala4 (orange), and the duplicated segment of Kala4 (red) for each type are shown.
The Origin and Spread of the Black Rice Gene
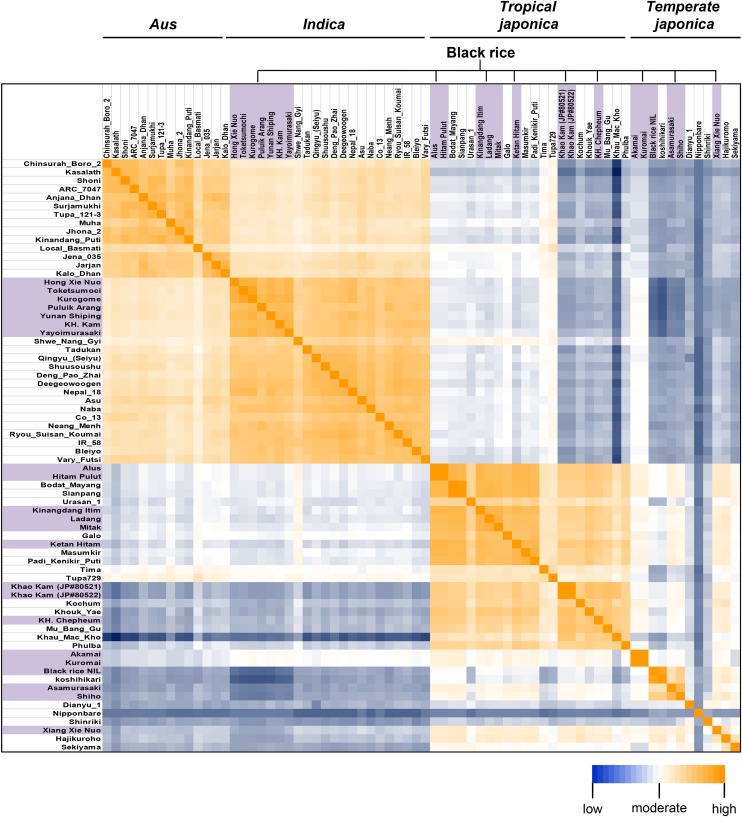
Since closely related structural changes of the Kala4 promoter were observed in the 21 black rice landraces, it is likely that black rice possessed the red trait in the course of its evolution. Subsequently, several structural changes must have occurred and spread to other varieties with some DNA variations. To address this possibility, the 21 black rice landraces were examined by genome-wide genotyping using the Golden Gate system. We used 642 SNPs from ‘Nipponbare’ (the reference temperate japonica) and ‘Kasalath’ (an aus subspecies of rice) and 468 SNPs from ‘Nipponbare’ and ‘Khau Mac Kho’ (tropical japonica) to reduce possible bias of genetic distance that may result from the original pairs used for SNP information. The 21 black rice lines were then subjected to genotyping together with other varieties including 14 aus, 15 indica, 12 tropical japonica, and 5 temperate japonica cultivars (Figure 6; Supplemental Table 5). The aus, indica, tropical japonica, and temperate japonica cultivars were grouped into four distinct clusters, indicating that the probe sets used were suitable to address the origin of black rice. The results also indicated that the 21 black rice landraces could be distinctly classified into subspecies indica, tropical japonica, and temperate japonica (Figure 6). Therefore, the black rice trait must have spread to these rice subspecies, probably by introgression after natural outcrossing. Since the breeding of rice using artificial crossing methods started around 150 years ago, it is noteworthy that the ancient landraces may not have undergone selection after artificial crossing. We found that the peripheral regions of Kala4 in the 21 black rice landraces had quite similar SNP patterns on chromosome 4 to representative tropical japonica cultivars (Supplemental Figure 17), implying multiple introgressions of the Kala4 region into the other subspecies. We next sequenced seven indica black rice landraces between 28.0 and 29.0 Mb on chromosome 4 at 0.2-Mb intervals and found that relatively small genomic fragments of the japonica haplotype group (minimum length of ∼200 kb) containing the Kala4 gene were introgressed into the indica genomic background between 28.0 and 29.0 Mb (Figure 7A). Therefore, the Kala4 allele conferring the black pericarp trait must have been transferred from subspecies japonica to indica by multiple introgressions.
Figure 6.
Clustering Analysis of White, Red, and Black Rice Landraces.
Golden Gate assay results revealed clustering of the white, red, and black rice landraces into four subspecies. The heat map shows genome similarity between varieties based on Pearson’s correlation coefficient of genetic distance of the genome-wide Golden Gate genotyping data. The 21 black rice landraces (Supplemental Table 5) were classified into subspecies indica, tropical japonica, and temperate japonica. Yellow and blue colors indicate higher and lower correlation coefficients, respectively. The black rice varieties are highlighted in gray.
Figure 7.
Haplotype Analysis of Black Rice Varieties.
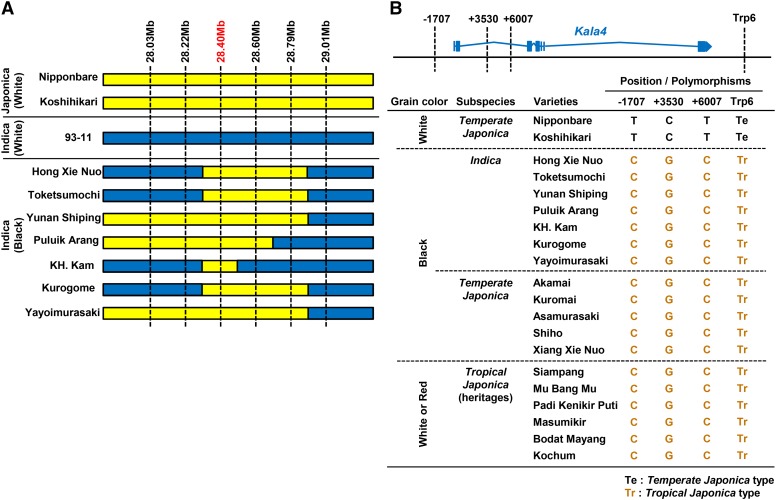
(A) Introgression of the japonica allele of Kala4 into indica subspecies. Six sequence markers were constructed at ∼0.2-Mb intervals in the 28.0 to 29.0 Mb region of chromosome 4. Seven indica black rice lines were sequenced at these markers with ‘Nipponbare’ (japonica), ‘Koshihikari’ (japonica), and ‘93-11’ (indica) as reference sequences. Yellow and blue bars indicate japonica and indica type genotypes, respectively. The marker of 28.40 Mb was the closest to Kala4.
(B) DNA polymorphisms between temperate japonica and tropical japonica in the proximal region of Kala4. Four SNP markers were examined in the two white temperate japonica, seven black indica, five black temperate japonica, and six white (or red) tropical japonica varieties.
To clarify these events further, four distinct DNA polymorphisms between the tropical and temperate japonica alleles were further examined at the proximal region of the Kala4 gene by direct sequencing (Figure 7B). Although black rice landraces belong to both the indica and temperate japonica backgrounds, the polymorphism among the DNA markers showed the tropical japonica genotype (Figure 7B). Taken together, these results strongly suggest that the original structural changes resulting in the black rice trait must have occurred in a rice line belonging to tropical japonica. Furthermore, the causal structural changes must have spread to subspecies indica and subsequently spread to temperate japonica from continuous natural crossing and artificial selection of the black rice trait by the ancient people.
DISCUSSION
Activation of the Anthocyanin Biosynthesis Pathway in Pericarps of Black Rice
Black rice varieties accumulate anthocyanins in their pericarps (Reddy et al., 1995; Rahman et al., 2013; Goufo and Trindade, 2014). In ‘Black rice NIL,’ several anthocyanin biosynthesis genes are upregulated in the pericarp (Supplemental Figures 1 and 3), indicating that the black rice trait is conferred by activation of the anthocyanin biosynthesis pathway. Phenotypic comparison of the ‘BAB-type NIL’ (with the ‘Hong Xie Nuo’ alleles of Kala1 and Kala4) and the ‘BBA-type NIL’ (with the ‘Hong Xie Nuo’ alleles of Kala1 and Kala3) revealed a more critical role for Kala4 in black rice (Maeda et al., 2014). The Kala4-specific activation of LDOX and other common enzymes in the anthocyanin biosynthesis pathway (Supplemental Figure 1B) further supports its importance in the origin of black rice. It is noteworthy that both Kala4 and Rc (Sweeney et al., 2006), which confers the red rice trait, have conserved bHLH-Myc N-terminal and bHLH domains with 53 and 46% identity, respectively (Supplemental Figure 18). Both genes can regulate several enzymes in the anthocyanin and proanthocyanidin biosynthesis pathways and can also specifically regulate the key enzyme genes LDOX and LAR to produce anthocyanins or proanthocyanidins, respectively (Supplemental Figure 3). The fact that the functional changes in both Rc and Kala4 have occurred in the common biosynthesis pathway within a relatively evolutionary short period implies that such transcriptional regulators that share gene regulation in common biological pathways, but produce distinct products (or traits), could be effective targets of selection during plant evolution to produce diversity within plant species.
DNA Methylation of Kala4
Epialleles have been reported in several plant species, including maize (Brink, 1956; Banks et al., 1988; Das and Messing, 1994; Hollick and Chandler, 2001) and rice (Miura et al., 2009, 2010; Wang et al., 2010; Zhang et al., 2012; Li et al., 2014). The phenotypes of these epialleles are affected by DNA methylation status and/or histone modification resulting in the alteration of gene expression. Here, we found that the DNA methylation status in the 25.6-kb candidate region of the Kala4 locus differed between the ‘Hong Xie Nuo’ and ‘Koshihikari’ alleles (Figure 2). This DNA methylation status is closely linked to the black pericarp trait among the black rice landraces examined (Supplemental Figure 6). However, we also confirmed that the higher DNA methylation status was not the causal FNP for the black rice trait, suggesting that such apparent epigenetic linkage may not be enough to conclude a causal relationship between a trait and epigenetic polymorphism.
The Structural Change in the Promoter of Kala4 Confers the Black Pericarp in Rice
We conclude that the origin of black rice can be attributed to the structural change in the Kala4 promoter region based on the following: (1) the structural changes in the Kala4 promoter were observed in all 21 black rice landraces (Figure 5); (2) the Kala4 promoter reverted to the white rice Koshihikari-type without apparent changes in the DNA methylation status in the four suppressor mutant lines with white grain phenotype (Figure 4; Supplemental Figures 12 to 14); (3) based on natural variations in seven loci among all black rice varieties, the seven polymorphisms observed in the 25.6-kb candidate region cannot be the FNP for the black pericarp trait in ‘Black rice NIL’ (Supplemental Table 2); and (4) upon transformation, only the black rice type promoter fused to the Kala4 cDNA conferred the black trait in rice grains (Figures 3B and 3C).
Alterations in promoter structure leading to modulation of gene expression can underlie evolutionary changes in diverse aspects of phenotypes (Wray et al., 2003). Kala4 genes both with and without the LINE1 insertion were observed in rice landraces (Supplemental Table 6). Thus, the LINE insertion itself is not a causal FNP for the black rice trait. However, the LINE1 retroelement can generate genomic instability in many ways (reviewed in Cordaux and Batzer, 2009). Thus, the partial duplication of Kala4 is probably a consequence of an “ectopic recombination” event related to LINE1. Indeed, parts of the LINE element are duplicated in this recombination event. Furthermore, the 11.0-kb insertion, which probably occurred after the ectopic recombination event, may have led to the creation of the functional black rice promoter of Kala4 (Figure 3). Several functional polymorphisms that cause regulatory alterations have been reported to produce domestication traits (Konishi et al., 2006; Studer et al., 2011; Meyer and Purugganan, 2013; Chakrabarti et al., 2013; Xu et al., 2015). In the case of teosinte branched 1 in maize, an insertion of a transposon called Hopscotch acts as an enhancer of gene expression and contributed to the change in apical dominance of maize upon domestication (Studer et al., 2011). Thus, it could be often the case that transposons have acted as modulators of gene expression during domestication of crop species.
The Origin of Black Rice
Considering the results described so far, we have proposed a model for the evolutionary process that lead to the creation of black rice and its local and sporadic spread into rice landraces in Asia (Supplemental Figure 19). Black rice landraces exist in at least three subspecies of rice, i.e., indica, tropical japonica, and temperate japonica (Figure 6). All 21 black rice landraces, but not the white and red rice landraces, have related structural changes accompanying the duplication and the insertion of genomic fragments of tropical japonica in Kala4 (Figure 5; Supplemental Table 4). Considering the causal relationship among related genetic events, the structural changes in Kala4 must have started from the insertion of LINE1 at intron 2 of Kala4 in a single ancestral line of tropical japonica rice. However, this insertion was broadly observed in varieties including the black, red, and white varieties (Supplemental Table 6), suggesting that this insertion has spread into subspecies independent of the black rice traits. In contrast to tropical japonica and temperate japonica, there are relatively few instances of the LINE1 insertion in Kala4 of indica and aus subspecies (Supplemental Table 6), supporting the above idea that the LINE1-inserted allele of Kala4 originated from an ancient japonica (or tropical japonica) rice and transferred to the indica and aus subspecies. The LINE1 insertion in Kala4 was followed by partial duplication of the LINE1 element. Subsequently (or simultaneously), the insertion of the 11.0-kb fragment that originated ∼83 kb upstream of the Kala4 gene occurred; thus, the black rice was created. It is very likely that the 11.0-kb insertion event occurred specifically in black rice because no related insertions have been observed in any of the red and white varieties investigated so far (Supplemental Figure 20).
We demonstrated that the Kala4 allele of black rice is the origin of the black trait in tropical japonica (Supplemental Figure 17; Figure 7B). Moreover, relatively small japonica genomic segments including the Kala4 gene were observed in the indica black rice (Figure 7A). Therefore, after the creation of the black rice Kala4 gene, this tropical japonica Kala4 allele may have spread into indica subspecies via refined introgressions, probably through multiple natural crosses and artificial selection. With the five black temperate japonica varieties tested, we were not able to obtain any strong evidence to show the introgression of Kala4 through indica from tropical japonica. Thus, based on current knowledge, black rice alleles of Kala4 in the temperate japonica subspecies could have been transferred from either indica or tropical japonica by natural and artificial crossing (Supplemental Figure 19).
It has been reported that many introgression events from ancient japonica to proto-indica occurred during domestication (Huang et al., 2012). An example of agronomic spreading of a single mutation into other subspecies is the domestication process related to white rice pericarp trait at the rc locus (Sweeney et al., 2007). Less than 1 Mb of the japonica DNA segment hitchhiked with the rc allele into most indica varieties. In this work, we revealed that extensive introgression events have also occurred for the local trait that led to the black rice cultivars, especially from tropical japonica to indica (Figure 7). It is likely that a number of selection events for other agronomic traits along with many natural outcrossings must be involved in the global prevalence of major agronomic traits such as white grain color due to the defective mutation in Rc. Thus, the introgression of relatively tiny genomic fragments for such global traits would be a natural consequence (Sweeney et al., 2007). However, it is a surprise that an agronomic trait like the black rice trait that produced local varieties such as “Forbidden rice,” which has been preferred in tributes, festivals, and religious ceremonies (Newman, 2004), arose with extensive introgression, similar to global agronomic traits during rice domestication and subsequent early breeding. It is of note that cultivated rice tends to self-pollinate, unlike maize. Furthermore, since Kala4 was inherited in a semidominant manner, it may have caused distinct selection pressure on introgression as compared with the recessive rc mutation. Therefore, elucidating the origin of black rice provides new insights not only into rice domestication but also into genome evolution of plants.
METHODS
Plant Materials
The near-isogenic lines (NIL) of rice (Oryza sativa), namely, ‘Black rice NIL,’ ‘BBA-type NIL,’ ‘BAB-type NIL,’ and ‘ABB-type NIL,’ were developed by introgression of the ‘Hong Xie Nuo’ alleles in three loci on chromosomes 1, 3, and 4 (Kala1, Kala3, and Kala4) as described previously (Maeda et al. 2014). The NIL identifiers correspond to the ‘Hong Xie Nuo’ (B) and ‘Koshihikari’ (A) alleles of Kala1, Kala2, and Kala3 in respective order. The R71 (cv Akamusubi) and R1284 (cv Tokei-1284) lines used for reciprocal crossing (Supplemental Figure 3) with ‘Black rice NIL’ were provided by Yamaguchi et al. (2015). The 21 black rice landraces were obtained from the Gene Bank of the National Institute of Agrobiological Sciences. The seven tropical japonica landraces have been previously reported to retain the ancestral functional alleles for three domestication-related genes (qSW5, Wx, and qSH1) after domestication (Konishi et al., 2008).
Transcriptome Analysis
Total RNAs were extracted from rice grains from which developing endosperm cells were squeezed out (thus, mainly pericarp cells) with the RNeasy Mini Kit (Qiagen). Total RNAs (400 ng) were used for synthesis of Cy5- or Cy3-labeled cRNA. Hybridization to the rice 4x44K microarray platform (Agilent Technologies) was conducted following the manufacturer’s protocol (Agilent Technologies). After washing, the hybridized slides were scanned in an Agilent G2505C DNA microarray scanner and analyzed using the Agilent Feature Extraction Software. The oligonucleotides used as probes are listed in Supplemental Table 7.
Vector Construction and Rice Transformation
The vector for overexpression of the Os04g0557500 (Kala4) cDNA was constructed by inserting ∼1.4 kb of the cDNA downstream of a maize Ubiquitin promoter in the pEASY-PUbiTNos binary vector using LR clonase (Invitrogen). For promoter analysis, ∼6.0-kb fragments of the promoters from white and black rice were amplified from genomic DNA of ‘Koshihikari’ and ‘Black rice NIL,’ respectively. The common 1.4-kb promoter was amplified from the white rice promoter fragment. The amplified promoter fragments were connected to the Kala4 cDNA by PCR-based mutagenesis and cloned into the pCR8 entry vector (Invitrogen). The resulting promoter:Kala4 cDNA fragments were then introduced into the pGWB501 binary vector (Nakagawa et al., 2007) using LR clonase (Invitrogen). The primers used for construction of promoters are listed in Supplemental Data Set 1.
Rice transformation was performed as described by Hiei et al. (1994). The transgenic calli were selected on medium containing hygromycin and transferred to regeneration medium. The regenerated plants were grown in a growth chamber.
Fine Mapping of Kala4
The ‘Black rice NIL’ was crossed with ‘BBA-type NIL’ and the F1 plants were self-pollinated. Using allele-specific PCR primers, ∼3000 plants in the F2 generation were genotyped for marker ‘200a’ and either marker ‘800a’ or ‘800bc.’ The primers were designed by SNPs-ici (Rizo Co.) and could amplify the specific PCR products from each allele of ‘Koshihikari’ and ‘Hong Xie Nuo.’ Thirty recombinant F2 plants were grown in a paddy field. Among them, 15 recombinant lines homozygous for the ‘Koshihikari’ allele in either marker were checked for grain color in the F3 generation. The recombinant plants homozygous for the ‘Hong Xie Nuo’ allele were further confirmed for segregation of grain color in the next generation. The PCR primers used for fine mapping are listed in Supplemental Data Set 1.
Quantitative RT-PCR
The total RNAs were extracted using the phenol-SDS method. Samples (4 µg) of the total RNAs were treated with RQ1 DNase I (Promega) to synthesize the first-strand cDNA using SuperScript II reverse transcriptase (Invitrogen) and Oligo(dT)12-18 primer (Invitrogen). RT-qPCR was performed using Power SYBR Green Master Mix (Life Technologies) and an ABI PRISM 7900 sequence detection system (Applied Biosystems). The gene specific primers used in this assay are listed in Supplemental Data Set 1.
McrBC-CGH
Genomic DNAs from pericarps of ‘Koshihikari’ and ‘Hong Xie Nuo’ were extracted using DNeasy Plant Mini Kit (Qiagen). Genomic DNAs (4 µg) were digested with RsaI restriction enzyme (Promega). After phenol-chloroform extraction and ethanol precipitation, digested DNAs were dissolved in 40 μL water. The DNA solution was subjected to McrBC digestion and used in two-color CGH microarray analysis. The RsaI/McrBC double-digested and RsaI single-digested DNAs were labeled by Cy5 and Cy3, respectively, using a BioPrime array CGH genome labeling system (Invitrogen). A custom tiling array with 2 × 8500 probes was designed from the 85-kb region of the Kala4 locus containing the three bHLH genes. The Cy5- and Cy3-labeled DNAs were hybridized to the tiling array at 65°C for 40 h and scanned using the Agilent Microarray Scanner.
Bisulfite Sequencing
Genomic DNAs were extracted from leaf samples using cetyltrimethylammonium bromide (Wako). Genomic DNA (2 µg) was subjected to bisulfite conversion using the EpiTect Bisulfite Kit (Qiagen) and amplified using the EpiTect Whole Bisulfitome Kit (Qiagen). The PCR-amplified fragments were cloned into pCR-Blunt II-TOPO cloning vector (Invitrogen) and sequenced using the PCR primers listed in Supplemental Data Set 1.
FPNI-PCR
FPNI-PCR was performed as described by Wang et al. (2011). The PCR products were cloned into pCR-Blunt II TOPO cloning vector and were sequenced using the PCR primers listed in Supplemental Data Set 1.
DNA Gel Blot Analysis
Approximately 10 μg genomic DNAs were digested with the appropriate enzymes, electrophoresed on 0.8% agarose gels (Nacalai), and transferred to Hybond-N+ positively charged nylon membrane (GE Healthcare). Hybridization and detection were performed using Alkphos Direct Labeling and Detection System (GE Healthcare). The primers used for preparation of the DNA probes are listed in Supplemental Data Set 1.
Isolation of Suppressor Mutants
Approximately 3000 seeds of ‘Black rice NIL’ were mutagenized by γ-ray irradiation. The M1 seeds were grown in a paddy field. Rice plants with white seed color were selected from among around 10,000 plants in the M2 generation. Seven suppressor plants from at least four independent mutational events were selected, although F5-1-1, F5-1-2, and F5-1-4 might have been screened from the same mutational event.
Golden Gate Assay
The ‘Black rice NIL,’ 21 black rice lines, 14 aus lines, 15 indica lines, 12 tropical japonica lines, and 51 temperate japonica lines were examined by the Golden Gate assay (Supplemental Table 5). Genomic DNAs were prepared using DNeasy Plant mini Kit (Qiagen) and 15 μL of the 50 ng/μL DNA solution was subjected to prehybridization and hybridization using the Golden Gate Genotyping Universal-32 and custom Bead Chip according to the manufacturer’s protocol (Illumina).
Sequencing Analysis
All the sequence data were obtained using an ABI PRISM 3130xl genetic analyzer (Life Technologies) and analyzed using GENETYX software.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession number: NC_008397 for Kala4 (Os04g0557500) locus in Nipponbare. All microarray data generated in this study have been submitted to the NCBI Gene Expression Omnibus under accession number GSE67987 (http://www.ncbi.nlm.nih.gov/geo/).
Supplemental Data
Supplemental Figure 1. Black rice cultivars and anthocyanin biosynthesis.
Supplemental Figure 2. Genotypes and phenotypes of thirty recombinant plants obtained by fine mapping.
Supplemental Figure 3. Expression of genes related to anthocyanin and proanthocyanidin biosynthesis in the pericarps of white, black, and red rice lines.
Supplemental Figure 4. Exon-intron structure of Os04g0557500.
Supplemental Figure 5. Structure of the LINE1 element inserted into intron 2 of Os04g0557500.
Supplemental Figure 6. DNA methylation status of the 21 black rice varieties and seven heritage landraces based on McrBC-DNA gel blot analysis.
Supplemental Figure 7. Expression of Os04g0557500 in seven heritage landraces.
Supplemental Figure 8. Discovery of duplicated fragment related to Os04g0557500 ORF by McrBC-DNA gel blot analysis.
Supplemental Figure 9. Restriction maps of the ‘Koshihikari’ and the ‘Hong Xie Nuo’ types of the Os04g0557500 gene.
Supplemental Figure 10. Confirmation of the Os04g0557500 structure in ‘Black rice NIL’ by DNA gel blot analysis.
Supplemental Figure 11. Binding sites of the primers used for sequencing of Os04g0557500 and its promoter.
Supplemental Figure 12. Confirmation of the structure of the Os04g0557500 promoter and ORF in the seven suppressor mutants by PCR.
Supplemental Figure 13. Confirmation of the Os04g0557500 promoter structure in the suppressor mutants by DNA gel blot analysis.
Supplemental Figure 14. Genotypes of Os04g0557500 and the two PCR markers in the seven suppressor mutants.
Supplemental Figure 15. DNA gel blot analysis of the Kala4 promoter in the 21 black rice varieties.
Supplemental Figure 16. Classification of 21 black rice varieties based on PCR analysis.
Supplemental Figure 17. Local genetic distances of the black rice varieties on chromosome 4.
Supplemental Figure 18. Sequence alignment of the two bHLH proteins.
Supplemental Figure 19. Schematic diagram of a model showing the processes in the origin of black rice and the spread of the Kala4 black allele.
Supplemental Figure 20. PCR analysis of the promoter in white and red rice varieties with LINE1 element insertion in intron 2 of the Kala4 gene.
Supplemental Table 1. Anthocyanin biosynthesis-related genes in rice and maize.
Supplemental Table 2. Polymorphisms in the 25.6-kb candidate region of the Kala4 locus.
Supplemental Table 3. Deduced band sizes for DNA gel blot analysis.
Supplemental Table 4. Classification of the 21 black rice varieties according to three types of insertions.
Supplemental Table 5. White and red rice cultivars used in Golden Gate assay.
Supplemental Table 6. Analysis of LINE1 insertion at intron 2 of the Kala4 gene in rice varieties.
Supplemental Table 7. Information on probes in the 4 × 44K microarray platform related to anthocyanin and proanthocyanidin biosynthesis genes.
Supplemental Data Set 1. Primers used in this work.
Supplementary Material
Acknowledgments
We thank M. Yamazaki for kindly providing the binary vector and T. Shibukawa for supporting the experiments. We also thank J. Matsuzaki, H. Itoh, and Y. Nemoto for valuable comments and suggestions. We especially thank Bal Antonio for his English editing of our article. This work was supported by a Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science, and Technology “Genome Adaptation.”
AUTHOR CONTRIBUTIONS
T.E. and T.I. conceived the original plan of this work. H.M. and T.Y. produced the NILs used in this work. H.M. also performed rough mapping of Kala4. T. Oguchi performed microarray analysis, preliminary fine mapping of Kala4, and CGH analysis. T.I. and N.T. screened the suppressor mutants. K.E. and M.Y. performed the Golden Gate analysis of the rice varieties. T. Oikawa performed the latter part in fine mapping of Kala4, identified the structural changes of Kala4 promoter, performed extensive DNA gel blot analysis, developed the transgenic plants, and analyzed the suppressor mutants. T.I. organized all the work.
Glossary
- bHLH
basic helix-loop-helix
- FNP
functional nucleotide polymorphism
- SSR
simple sequence repeat
- McrBC-CGH
McrBC-comparative genomic hybridization
- FPNI
fusion primer and nested integrated
- SNP
single nucleotide polymorphism
- ORF
open reading frame
References
- Banks J.A., Masson P., Fedoroff N. (1988). Molecular mechanisms in the developmental regulation of the maize Suppressor-mutator transposable element. Genes Dev. 2: 1364–1380. [DOI] [PubMed] [Google Scholar]
- Brink R.A. (1956). A genetic change associated with the R locus in maize which is directed and potentially reversible. Genetics 41: 872–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti M., Zhang N., Sauvage C., Muños S., Blanca J., Cañizares J., Diez M.J., Schneider R., Mazourek M., McClead J., Causse M., van der Knaap E. (2013). A cytochrome P450 regulates a domestication trait in cultivated tomato. Proc. Natl. Acad. Sci. USA 110: 17125–17130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler V.L., Radicella J.P., Robbins T.P., Chen J., Turks D. (1989). Two regulatory genes of the maize anthocyanin pathway are homologous: isolation of B utilizing R genomic sequences. Plant Cell 1: 1175–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe E.H. (1959). A regular and continuing conversion-type phenomenon at the B locus in maize. Proc. Natl. Acad. Sci. USA 45: 828–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone K.C., Cocciolone S.M., Burr F.A., Burr B. (1993). Maize anthocyanin regulatory gene pl is a duplicate of c1 that functions in the plant. Plant Cell 5: 1795–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R., Batzer M.A. (2009). The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 10: 691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das O.P., Messing J. (1994). Variegated phenotype and developmental methylation changes of a maize allele originating from epimutation. Genetics 136: 1121–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner H.K., Robbins T.P., Jorgensen R.A. (1991). Genetic and developmental control of anthocyanin biosynthesis. Annu. Rev. Genet. 25: 173–199. [DOI] [PubMed] [Google Scholar]
- Furukawa T., Maekawa M., Oki T., Suda I., Iida S., Shimada H., Takamure I., Kadowaki K. (2007). The Rc and Rd genes are involved in proanthocyanidin synthesis in rice pericarp. Plant J. 49: 91–102. [DOI] [PubMed] [Google Scholar]
- Goufo P., Trindade H. (2014). Rice antioxidants: phenolic acids, flavonoids, anthocyanins, proanthocyanidins, tocopherols, tocotrienols, γ-oryzanol, and phytic acid. Food Sci. Nutr. 2: 75–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hichri I., Barrieu F., Bogs J., Kappel C., Delrot S., Lauvergeat V. (2011). Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 62: 2465–2483. [DOI] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kumashiro T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6: 271–282. [DOI] [PubMed] [Google Scholar]
- Hollick J.B., Chandler V.L. (2001). Genetic factors required to maintain repression of a paramutagenic maize pl1 allele. Genetics 157: 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Anderson B., Wessler S.R. (1996). Isolation and characterization of rice R genes: evidence for distinct evolutionary paths in rice and maize. Genetics 142: 1021–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Reddy V.S., Wessler S.R. (2000). The rice R gene family: two distinct subfamilies containing several miniature inverted-repeat transposable elements. Plant Mol. Biol. 42: 667–678. [DOI] [PubMed] [Google Scholar]
- Huang X., et al. (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490: 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T., Konishi S., Shomura A., Yano M. (2009). DNA changes tell us about rice domestication. Curr. Opin. Plant Biol. 12: 185–192. [DOI] [PubMed] [Google Scholar]
- Jin J., Huang W., Gao J.-P., Yang J., Shi M., Zhu M.-Z., Luo D., Lin H.-X. (2008). Genetic control of rice plant architecture under domestication. Nat. Genet. 40: 1365–1369. [DOI] [PubMed] [Google Scholar]
- Konishi S., Ebana K., Izawa T. (2008). Inference of the japonica rice domestication process from the distribution of six functional nucleotide polymorphisms of domestication-related genes in various landraces and modern cultivars. Plant Cell Physiol. 49: 1283–1293. [DOI] [PubMed] [Google Scholar]
- Konishi S., Izawa T., Lin S.Y., Ebana K., Fukuta Y., Sasaki T., Yano M. (2006). An SNP caused loss of seed shattering during rice domestication. Science 312: 1392–1396. [DOI] [PubMed] [Google Scholar]
- Li C., Zhou A., Sang T. (2006). Rice domestication by reducing shattering. Science 311: 1936–1939. [DOI] [PubMed] [Google Scholar]
- Li R.-Q., Huang J.-Z., Zhao H.-J., Fu H.-W., Li Y.-F., Liu G.-Z., Shu Q.Y. (2014). A down-regulated epi-allele of the genomes uncoupled 4 gene generates a xantha marker trait in rice. Theor. Appl. Genet. 127: 2491–2501. [DOI] [PubMed] [Google Scholar]
- Li X., et al. (2012). Single-base resolution maps of cultivated and wild rice methylomes and regulatory roles of DNA methylation in plant gene expression. BMC Genomics 13: 300–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H., et al. (2014). Genetic dissection of black grain rice by the development of a near isogenic line. Breed. Sci. 64: 134–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R.S., Purugganan M.D. (2013). Evolution of crop species: genetics of domestication and diversification. Nat. Rev. Genet. 14: 840–852. [DOI] [PubMed] [Google Scholar]
- Miura K., Agetsuma M., Kitano H., Yoshimura A., Matsuoka M., Jacobsen S.E., Ashikari M. (2009). A metastable DWARF1 epigenetic mutant affecting plant stature in rice. Proc. Natl. Acad. Sci. USA 106: 11218–11223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Ikeda M., Matsubara A., Song X.J., Ito M., Asano K., Matsuoka M., Kitano H., Ashikari M. (2010). OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42: 545–549. [DOI] [PubMed] [Google Scholar]
- Nakagawa T., et al. (2007). Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71: 2095–2100. [DOI] [PubMed] [Google Scholar]
- Newman J.M. (2004). Black rice. Flavor & Fortune 11: 5–9. [Google Scholar]
- Paz-Ares J., Ghosal D., Wienand U., Peterson P.A., Saedler H. (1987). The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 6: 3553–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroni K., Tonelli C. (2011). Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 181: 219–229. [DOI] [PubMed] [Google Scholar]
- Rahman M.M., Lee K.E., Lee E.S., Martin M.N., Lee D.S., Yun J.S., Kim J.B., Kang S.G. (2013). The genetic constitutions of complementary genes Pp and Pb determine the purple color variation in pericarps with cyanidin-3-O-glucoside depositions in rice. J. Plant Biol. 56: 24–31. [Google Scholar]
- Reddy V.S., Dash S., Reddy A.R. (1995). Anthocyanin pathway in rice (Oryza sativa L): identification of a mutant showing dominant inhibition of anthocyanins in leaf and accumulation of proanthocyanidins in pericarp. Theor. Appl. Genet. 91: 301–312. [DOI] [PubMed] [Google Scholar]
- Regulski M., et al. (2013). The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res. 23: 1651–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W., Ohmori T., Kageyama K., Miyazaki C., Saito A., Murata M., Noda K., Maekawa M. (2001). The Purple leaf (Pl) locus of rice: the Plw allele has a complex organization and includes two genes encoding basic helix-loop-helix proteins involved in anthocyanin biosynthesis. Plant Cell Physiol. 42: 982–991. [DOI] [PubMed] [Google Scholar]
- Shih C.H., Chu H., Tang L.K., Sakamoto W., Maekawa M., Chu I.K., Wang M., Lo C. (2008). Functional characterization of key structural genes in rice flavonoid biosynthesis. Planta 228: 1043–1054. [DOI] [PubMed] [Google Scholar]
- Shomura A., Izawa T., Ebana K., Ebitani T., Kanegae H., Konishi S., Yano M. (2008). Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 40: 1023–1028. [DOI] [PubMed] [Google Scholar]
- Stam M., Belele C., Dorweiler J.E., Chandler V.L. (2002). Differential chromatin structure within a tandem array 100 kb upstream of the maize b1 locus is associated with paramutation. Genes Dev. 16: 1906–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer A., Zhao Q., Ross-Ibarra J., Doebley J. (2011). Identification of a functional transposon insertion in the maize domestication gene tb1. Nat. Genet. 43: 1160–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styles E.D., Ceska O., Seah K.T. (1973). Developmental difference in action of R and B alleles in maize. Can. J. Genet. Cytol. 15: 59–72. [Google Scholar]
- Sutherland E., Coe L., Raleigh E.A. (1992). McrBC: a multisubunit GTP-dependent restriction endonuclease. J. Mol. Biol. 225: 327–348. [DOI] [PubMed] [Google Scholar]
- Sweeney M.T., Thomson M.J., Pfeil B.E., McCouch S. (2006). Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell 18: 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney M.T., Thomson M.J., Cho Y.G., Park Y.J., Williamson S.H., Bustamante C.D., McCouch S.R. (2007). Global dissemination of a single mutation conferring white pericarp in rice. PLoS Genet. 3: e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Li X., Liu F., Sun X., Li C., Zhu Z., Fu Y., Cai H., Wang X., Xie D., Sun C. (2008). Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 40: 1360–1364. [DOI] [PubMed] [Google Scholar]
- Walker E.L. (1998). Paramutation of the r1 locus of maize is associated with increased cytosine methylation. Genetics 148: 1973–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E., et al. (2008). Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat. Genet. 40: 1370–1374. [DOI] [PubMed] [Google Scholar]
- Wang K., Tang D., Hong L., Xu W., Huang J., Li M., Gu M., Xue Y., Cheng Z. (2010). DEP and AFO regulate reproductive habit in rice. PLoS Genet. 6: e1000818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Ye S., Li J., Zheng B., Bao M., Ning G. (2011). Fusion primer and nested integrated PCR (FPNI-PCR): a new high-efficiency strategy for rapid chromosome walking or flanking sequence cloning. BMC Biotechnol. 11: 109–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray G.A., Hahn M.W., Abouheif E., Balhoff J.P., Pizer M., Rockman M.V., Romano L.A. (2003). The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 20: 1377–1419. [DOI] [PubMed] [Google Scholar]
- Xu C., et al. (2015). A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 47: 784–792. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T., Omoteno M., Maeda H., Morikawa M., Kidani Y., Ozaki H., Murata K., Iyama Y., Kojima Y., Takarada T., Mukaino N., Ebitani T. (2015). Development of a new rice cultivar “AKA-MUSUBI”. Bull. Agric. Res. Inst. Toyama Pref. Agr. For. Fish. Res. Ctr. 6: in press. [Google Scholar]
- Zhang L., et al. (2012). Identification and characterization of an epi-allele of FIE1 reveals a regulatory linkage between two epigenetic marks in rice. Plant Cell 24: 4407–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.